Abstract
The sequence logo for DNA binding sites of the bacteriophage P1 replication protein RepA shows unusually high sequence conservation (∼2 bits) at a minor groove that faces RepA. However, B-form DNA can support only 1 bit of sequence conservation via contacts into the minor groove. The high conservation in RepA sites therefore implies a distorted DNA helix with direct or indirect contacts to the protein. Here I show that a high minor groove conservation signature also appears in sequence logos of sites for other replication origin binding proteins (Rts1, DnaA, P4 α, EBNA1, ORC) and promoter binding proteins (σ70, σD factors). This finding implies that DNA binding proteins generally use non-B-form DNA distortion such as base flipping to initiate replication and transcription.
INTRODUCTION
DNA replication is thought to begin by the sequence-specific binding of proteins to an origin of replication, followed by untwisting, which opens the nearby DNA (1,2). After further unwinding by a helicase, RNA primers are synthesized and then extended with deoxyribonucleotides to switch to DNA synthesis.
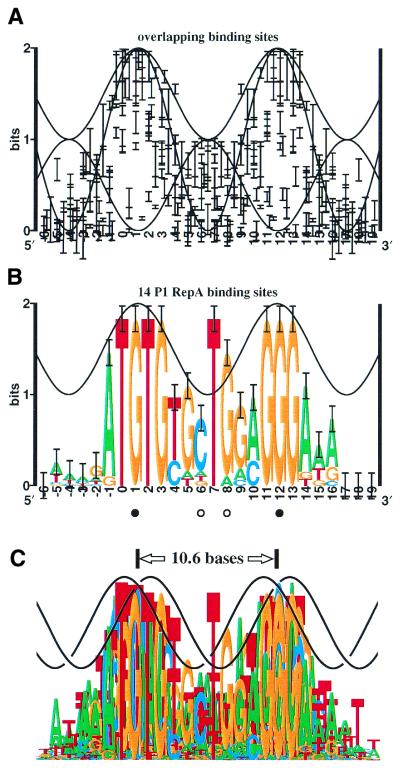
Bacteriophage P1 replicates as a plasmid by using the RepA protein (3–6). As with a number of other DNA binding proteins that bind on one face of DNA (Fig. 1A), the RepA protein binds to sites that show high sequence conservation at two positions spaced 10 bp apart. These regions correspond to the protein binding into two successive major grooves (7,8) (Fig. 1B). In addition, RepA binding sites display an unusually high sequence conservation (∼2 bits) of a T in the intervening minor groove, at position +7. In B-form DNA more than 1 bit of conservation is unlikely for contacts entering the minor groove because such contacts allow ambiguity in the orientation of the base pair (7,9,10). That is, AT resembles TA and CG resembles GC as viewed from the minor groove. Although the high conservation might be explained by a second protein binding to the back side of the DNA (11–13), this cannot be the entire explanation because substituting the T with an A leads to faster RepA dissociation kinetics (7,14). Furthermore, when RepA was used to select random DNA sequences, T was favored over A at +7 (7), suggesting that this position affects binding despite the near inaccessibility of the major groove on the opposite face of the DNA. Together, these results imply that the RepA binding site DNA is distorted. The nature of this distortion, which could lead to DNA strand opening in conjunction with DnaA (15), is the subject of this and the companion paper (16).
Figure 1.
Sequence conservation at transcriptional activator and repressor DNA binding sites versus sequence logo for P1 RepA sites. (A) The sequence conservation of 12 protein binding sites was computed. The resulting 12 information curves were aligned to match the experimentally determined DNA binding faces and the sequence conservation was plotted as error bars (11) using the makelogo program (version 9.22) with the letters made invisible (37). The proteins used were from previously computed sequence logos: λcI and cro, 434 repressor, CRP, FNR, λ O, ArgR, TrpR, LexA (7); Fis (25); OxyR (10); Lrp (112); and SoxS (113). Only the central two domains of OxyR were used, and asymmetrical sites (Lrp, SoxS) were represented only once in the orientation previously published. A set of three sine waves represent minor groove accessibility (0 to 1 bit, of the form y = cosx + in bits, where x = 2π(b – 1)/10.6 and b is base position), major groove accessibility (0 to 2 bits, y = –cosx + 1) and total accessibility (1 to 2 bits, y = –cosx + 1.5). (B) The sequence logo for the P1 RepA binding sites (4,7) demonstrates how sequence conservation is derived mainly from contacts penetrating the major groove (positions –1 to +3 and +11 to +13 match the wave peaks), while contacts at +7 and +8 would face the minor groove. (C) Overlay of sequence logos for transcriptional factors [from (A)] and RepA [from (B)], with two sine waves artistically representing B-form DNA, demonstrates the exceptional nature of the T at position +7.
High minor groove conservation has been observed in binding sites for IHF (17), which causes extreme DNA bending (18); for TATA binding protein, which widens the groove (19,20); for purR, which intercalates into the DNA (21); and for proteins that open DNA (22). RepA has been shown to bend DNA by 40° (23), which one might suppose could provide enough distortion to account for the unusual conservation. However, Fis deflects DNA up to 90° (24) and yet there is no minor groove conservation spike in the Fis sequence logo (25). DNA distortion, therefore, does not invariably lead to strong minor groove sequence conservation. Nevertheless, a main starting point of this paper is the converse, that the presence of excess information, where the minor groove faces a single protein, implies non-B-form DNA distortion.
The main kind of DNA distortion observed in DNA–protein complexes occurs as a roll at TG or TA base steps (26). Because the highly conserved T at RepA sites is adjacent to a G or A (Fig. 1B), roll may be important for RepA bending. An alternative kind of DNA distortion for a replication protein to make is one in which the helix remains in place but individual bases break their hydrogen contacts with the complementary base and swivel outwards, a phenomenon seen in the X-ray co-crystal structure of DNA-bound HhaI methyltransferase and several other DNA modification proteins (27–30), 30S ribosomal subunits (31) and during DNA replication (32). This base flipping could constitute the second step of DNA replication, concurrent with or following protein binding, as suggested by Roberts (33,34). Base flipping could also explain the unusual 1.27 ± 0.08-fold excess information content found at RepA sites (7,14): the initial binding would use the predicted amount (1.00), and binding to the flipped out base(s) might account for the excess (0.27 ± 0.08).
Experimental data for RepA already suggest that the T at position +7, but not the complementary A, may flip out. Missing-base experiments show that removal of T+7 reduces binding, whereas removal of the A on the opposite strand has no effect or slightly increases binding (8,16). These data suggest that the T is flipped out and binds into a pocket of RepA, so that if the T is lost binding energy is decreased. If the A on the opposite strand is removed instead, its base pairing with the T would be gone, allowing the T to swivel out more easily. In contrast, HhaI protein binds more strongly when there are weakened base pairs (35), and binding is strongest when the flipped base is removed. We investigate the contact energetics between RepA and the +7 T/A base pair in the companion paper (16).
In light of the flipping hypothesis, the finding of high sequence conservation at the minor groove of the RepA binding site prompted me to analyze sequence logos from DNA replication and transcription proteins to see if they might also have unusually strong sequence conservation in regions where minor grooves face the protein. This paper presents such sequence logos and correlates them with experimental data on helix orientation, base contacts and base pair opening. The general picture that emerges is that the sites are indeed distorted and that, in many cases, base flipping may account for the data.
MATERIALS AND METHODS
In this section I review previously published quantitative measures of sequence conservation and discuss how they often reflect a simple DNA binding mechanism. A new wrinkle on the previous interpretation (10) is presented which shows that proteins rarely contact a base from both sides simultaneously but that when they do, the effects can be detectable. We then consider how the sequence logos from RepA and the related replication protein Rts1 reveal that B-form DNA binding is not sufficient to explain their binding mechanisms and that some form of DNA distortion is involved. Finally, we consider the case of IHF, which reveals that DNA bending does not necessarily correlate with sequence conservation.
Programs
All programs are available at http://www.lecb.ncifcrf.gov/~toms/ and sequences were obtained from GenBank or as indicated. Delila instructions (36) that allow exact replication of each aligned sequence set and logo are available at http://www.lecb.ncifcrf.gov/~toms/paper/baseflip. Sequence logos were created as described previously (10,37).
Sequence logos
As shown in Figure 1B, and Figures 2–6, sequence logos quantitatively and reproducibly summarize the sequence conservation in a set of binding sites by depicting stacks of letters (37). The height of each letter within a stack is proportional to the base frequency at that position, and the letters are sorted by size, with the tallest (i.e. most frequent) on top. The height of the stack is the sequence conservation measured in bits of information; 1 bit measures the choice between two equally likely possibilities (11,38–40). Because bits are additive when positions are independent, as they are in many binding sites (41), it is legitimate to compare the relative heights of the stacks. Other measures of sequence conservation, such as counting the number of matches to a consensus (42), frequently give inconsistent or incorrect results (43).
Figure 2.
Sequence logo of GAL4 binding sites. The sites are those given by Bram et al. (114). Two sine waves (solid and dashed curves) represent total accessibility on two faces of the DNA. The GAL4 protein wraps around the DNA (49), accounting for the high sequence conservation by contacts on opposite faces.
Figure 6.
Sequence logos for RNA transcription binding sites. Details are given in Results.
Analysis of binding site sequence conservation: transcription activation and repression sites
Proteins that bind to DNA to activate or repress RNA transcription do not necessarily need to distort the DNA structure. Repression can be accomplished by occluding the RNA polymerase, while activation may require only that a protein contact is placed into the proper region of space relative to the promoter. When proteins use such simple contact mechanisms, the sequence conservation of their DNA binding sites has three distinctive features.
First, a protein facing a major groove can distinguish all four possible bases by their chemical moieties (9). Using information theory, sequence conservation can be precisely measured in bits of information. To select one of the four bases is a log24 = 2 bit choice (11,38,39). In contrast, when a minor groove faces the protein, the conservation is lower because the orientation of base pairs is nearly indistinguishable: AT resembles TA while GC resembles CG (9). A protein facing a minor groove can select only 2 of the 4 possible base pairs, which is equivalent to a 1 in 2 choice, or 1 bit of information. When the frequencies of bases are not 100, 50 or 0%, information theory provides a consistent and continuous method for computing the average sequence conservation.
Proteins tend to approach and bind to DNA from one face of the helix (44) so they encounter alternating major and minor grooves. The maximum sequence conservation should therefore vary smoothly between 1 and 2 bits, with a periodicity of 10.6 bp (45,46). This is demonstrated in Figure 1A, which shows error ranges for the sequence conservation of 12 protein–DNA binding sites aligned so that all proteins would be on one face of the DNA. Positions 0 to 3 and 11 to 14, both at the major groove, show high conservation, near 2 bits, while the intervening minor region dips down to 1 bit, as expected from the structure of DNA.
These quantitative measurements account for the varying frequencies of the four bases across binding sites, and they unveil the second distinctive feature of simple binding sites: the maximum sequence conservation is sinusoidal, and will often run parallel to a sine wave between 1 and 2 bits (10). (The effect is clearer on the individual sequence logos for sites used to construct the figure rather than on Figure 1A itself.) This effect occurs because DNA is approximately a cylinder, and ‘accessibility’ (the ease with which a protein can form a contact) depends on the angle between the base pair and the approaching protein binding surface. B-form DNA can be modeled using two sinusoidal accessibility curves. The minor groove accessibility ranges from 0 to 1 bit, while the major groove accessibility ranges from 0 to 2 bits but 180° out of phase. These two curves and their sum (which is also a sine wave) are shown in Figure 1A. Note that information, measured in bits, is the only scale for sequence conservation that allows the independent accessibility curves to be added (38). Figure 1A shows that, for the most part, the sequence conservation remains below the upper sine wave.
The three accessibility curves reveal the third feature of sequence conservation at simple DNA binding sites. Just adjacent to the central two bases—that is, in positions 4, 5, 8 and 9—the sequence conservation tends to leave a gap or clear zone in a roughly triangular area above both the major and minor groove accessibility sine waves and below the sine wave that represents their sum. These gaps are easily understood if proteins usually contact the DNA in either the minor groove or the major groove but not both simultaneously. Instead of strictly following the total sum sine wave, therefore, the sequence conservation is below the maximum of the two curves. From crystal studies, however, several cases are known in which a protein arm wraps around to the back face. The λ repressor wraps in the major groove (47), accounting for a small (0.8 bit) G predominated conservation at ±1 (7). The Epstein–Barr virus (EBV) nuclear antigen 1 (EBNA1) protein contacts the major groove, and has an arm that runs through the minor groove (48). In a third case, the dimeric GAL4 protein contacts two surfaces 14 bp apart (49). Although the λ repressor logo does not have high information, since the minor groove accessibility at ±1 is 0.9 bits, the situation is noticeable because a G preference is unusual for a minor groove contact (10). In contrast, EBNA1 positions –2 (and symmetrical position +3) and –1 (+2) significantly exceed the major groove accessibility sine wave (by 8 and 23 standard deviations of the sequence conservation, respectively; see Fig. 5C) and they are much closer to the sum (deviating by –5 and 4 standard deviations, respectively), consistent with the idea that the protein contacts the DNA from both faces. Finally, when one patch of the GAL4 sites is aligned with the major groove, sequence conservation of three bases on the other end exceeds the major groove accessibility (Fig. 2). However, within each GAL4 contact patch the conservation follows the major groove accessibility curve, making the case easy to spot. In Figure 1, only ArgR conservation at positions 4 and 9 (7) is high above the major groove accessibility sine wave (12 standard deviations) but is not significantly above the sum (2 standard deviations), suggesting that the DNA is non-B-form or that ArgR makes a pincer contact to both the major and minor grooves.
Figure 5.
Sequence logos for DNA replication binding sites. Details are given in Results.
Binding sites generally contain just enough information for proteins to locate them in the genome (11). Moreover, difficult contacts never form or tend to be lost during evolution, while more accessible contacts are gained or retained (10,50). These considerations show that the quantitative measure of sequence conservation by the information theory method gives results that can be understood geometrically. That is, Figure 1A shows that during evolution most protein contacts relax and mold into the available major and minor groove geometrical structure of helical B-form DNA. As a result, exceptional cases can be detected.
Plasmid P1 RepA binding sites are highly exceptional
Figure 1B shows the sequence logo for the RepA binding sites of bacteriophage P1 (7). As in Figure 1A, the standard deviation of each stack height is indicated by I-beams, and again the sine wave represents the informational accessibility of B-form DNA, which varies between 1 and 2 bits and has a periodicity of 10.6 bases (10). The crests represent the major groove facing the RepA protein. The positions of these crests were predicted by matching the accessibility curve to the logo, and they were then confirmed experimentally (7,8). Dimethyl sulfate (DMS) methylates the N–7 position of guanines in the major groove. Filled circles indicate bases that interfere with RepA binding when methylated by DMS or that are protected from DMS methylation by RepA binding, so the major groove of these bases faces RepA. Methylation of bases indicated by open circles does not have these effects, so the minor groove of these bases faces RepA. Hydroxyl radical and ethylation interference footprinting also indicate that RepA binds to the proposed DNA face and does not wrap around the DNA (7,8).
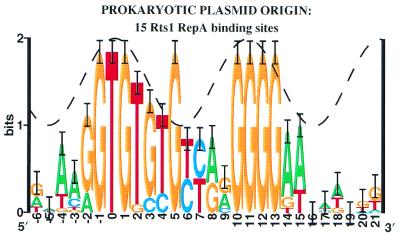
In contrast to the 12 site sets collected in Figure 1A, the RepA binding sites show sequence conservation at position +7 that significantly exceeds both the minor groove accessibility (6 standard deviations, p = 6.7 × 10–10) and the total accessibility. The major groove there is the most difficult position for a protein to reach if the DNA is in B-form, and contacts into the minor groove cannot account for the strict conservation at this position. RepA does not have the leisurely space that GAL4 uses to wrap around to the back face (Fig. 2). Thus base +7 (and perhaps +8) is highly unusual (Fig. 1C). The related plasmid Rts1 RepA sites (Fig. 3) and perhaps other iterons in plasmid replicons, also show anomalous bases (6). Interestingly, almost the entire left side of the Rts1 sites consists of TG pairs with sequence conservation that follows the sine wave closely but ends with a strikingly conserved guanine at +5 where the minor groove probably faces the protein. If the TG pairs allow flexibility in DNA structure, why are there so many, and why should position +5 be exceptional? One explanation is that both flexibility and base flipping are involved.
Figure 3.
Sequence logo for plasmid Rts1 RepA sites binding sites. The sequences for the RepA of E.coli plasmid Rts1 were obtained from GenBank K00053 (coordinates, followed by orientation: 28–, 63–, 135–, 163–, 200–, 233–, 1201–, 1223–, 1244–, 1265–, 1286–) and M60191 (79+, 177+, 271–, 355–) using the set of repeats originally noticed (115,116) and adding one more (1201) that is next to the others and is strong by the individual information method (117). The sequence logo resembles the P1 RepA logo in Figure 1B (6). The sine wave was placed by analogy with P1 RepA, and moved 1 bp left to match the majority of the conservation, which follows the sine wave smoothly on the left side (10). Because this orientation of the helix relative to the protein has not been proven experimentally, the sine wave is dashed. Given this placement, a spike of unusual sequence conservation appears in the middle of the site at position +5.
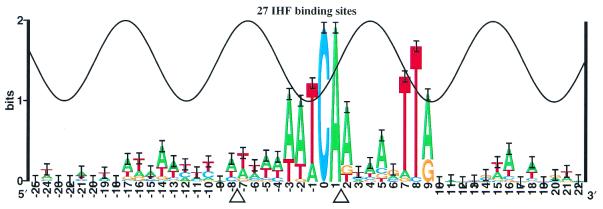
Example of a distorted DNA: IHF binding sites
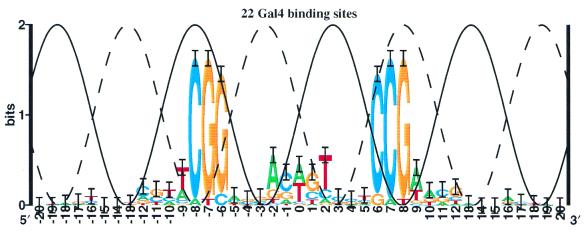
As a general nucleoprotein in Escherichia coli, the IHF protein binds to DNA and bends it severely (18). Information analysis of the binding sites shows two major peaks of sequence conservation (17) that are out of phase with the major groove (Fig. 4). The crystal structure reveals that bending is associated with the insertion of prolines into the helix (18). However, the location of the bends does not correlate to the positions of the strong mononucleotide sequence conservation. Significant correlations between bases are not detectable in the IHF binding sequences (41), so if dinucleotide sequence conservation exists it is probably minimal and cannot account for the absence of strong sequence conservation around the bend at –7. The conservation from –3 to +2 has been qualitatively accounted for by structural effects, including a TG/CA bend (18), while the conservation in positions +7 to +8 is thought to be accounted for first by the beta group of arginine 46, which interferes with G-C or C-G in the minor groove, and secondly by a large twist at the YR base step (18).
Figure 4.
Sequence logo for IHF binding sites. The 27 IHF sites collected by Goodrich et al. (17) are presented as a sequence logo. Points where IHF inserts a proline into the DNA to induce a bend are indicated by triangles.
The case of IHF demonstrates that when the conservation exceeds the sine wave, sequence logos indicate that simple B-form contacts on one face of the DNA are not being used. Mechanisms that can explain such excess conservation include wrapping arms, secondary contact by another protein (11,12), indirect readout of distorted DNA, non-B-form DNA contacts and base flipping.
RESULTS
To test the hypothesis that DNA opening is detectable in sequence conservation, this section presents new sequence logos for several replication and transcription initiation factors and correlates these to the available chemical footprinting and other data on DNA opening.
DNA replication factor binding sites
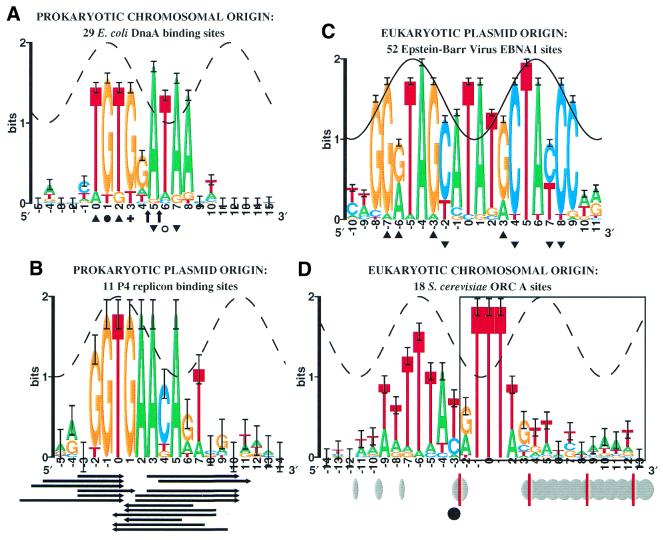
Escherichia coli DnaA sites. DnaA is responsible for replication of the E.coli origin, oriC (51). The orientation of the DnaA logo (Fig. 5A) was chosen to show its resemblance to the P1, Rts1 and related RepA-like sites (6). One major groove of each site contains high conservation of a TG motif, suggesting that the DNA may be bendable in this region (26). The base at +3 (marked by a +) preferentially reacts with DMS in the presence of DnaA; a base at +1 (marked with a filled circle) is protected from DMS methylation by DnaA binding (15,52). The data suggest that base +1 contacts the protein in the major groove. Placing the sine wave peak at +1 causes it to fit the conservation heights in 0 to +3 (10). This placement reveals that bases +5 to +8 are unusually conserved and likely to be distorted, suggesting that the DNA melting initiated by DnaA in adjacent regions (1) begins at the DnaA binding site.
In contrast, UDG footprints, which identify methyl group contacts of thymines (53,54), show that DnaA does not bind the thymine at +6 (open circle in Fig. 5A), but does bind thymines at 0, +2 (top strand, triangles), +5 and +7 (bottom strand, inverted triangles). It is therefore possible that positions +5 and +7 represent a major groove facing the protein while positions 0 to +3 represent a distortion, although this would not be consistent with the patterns at P1 RepA and Rts1 RepA. To indicate this alternative possibility, the sine wave on the sequence logo is dashed. In either case, the results suggest distortion or base flipping.
Enhanced DNase I cleavage is observed with DnaA protein at positions 4.5 and 5.5 in the oriC R2 site (arrows in Fig. 5A) (55). This suggests that the structure is open in the middle of the site. The excess information revealed by the logo at positions +5 to +8 shows that the sites of this replication protein are indeed distorted, and possibly flipped, as predicted by Roberts’ hypothesis (33,34).
Plasmid P4 α protein binding sites. The phage-plasmid P4 replicates in E.coli by the binding of the α protein to 11 iterons in ori1 and crr sites (56,57). The left half (positions –2 to +1) of the sequence logo (Fig. 5B) resembles those of P1 RepA (Fig. 1B) and Rts1 RepA (Fig. 3), but these and other plasmid replicon replication binding sites have two parts that correspond to two successive major grooves (6), whereas for the α protein the right side of the site is missing. Presumably the two parts of RepA-like sites correspond to two distinct protein components, A and B, since there are examples of inversion by 180° in various sequence logos (6) and, as predicted from the sequence logos, the RepE crystal structure contains two distinct domains that bind two successive major grooves (58). In the case of the P4 α protein, the B portion of the protein would be lacking or altered.
If the bases around position zero of the P4 α binding site were to face the protein through the major groove (corresponding to P1 RepA and DnaA) then, as with the other replication protein binding sites, there is unusually strong sequence conservation at base +5 where a minor groove would face the protein. Protection of regions by the protein from DNase I (top strand, rightward arrows; bottom strand, leftward arrows) indicates that the middle of the site (positions +1 to +3) is exposed (59), suggesting a discontinuity in the nucleoprotein structure. A similar exposure occurs in the middle of the DnaA R2 site (arrows in Fig. 5A). In addition, the P4 α and DnaA logos resemble one another (compare Fig. 5A with 5B), suggesting that P4 may use a replication mechanism similar to DnaA. However, P4 plasmid can replicate without host DnaA (57), implying that the replication proteins may have a common mechanism and/or ancestry. The structure of both P4 and DnaA sequence logos and the shared presence of central regions that are exposed to nucleases suggest that, instead of flipping a single base, both of these molecules may open a ‘flap’ of four or more bases simultaneously (DnaA bases +5 to +8 and P4 bases +2 to +7).
Although the logo in conjunction with experimental data supports a hypothesis of base flipping in this case, positions +4 and +5 are often the complement of TG or TA and so may alternatively represent DNA bending or other forms of distortion.
Epstein–Barr virus EBNA1 sites. EBV plays a role in infectious mononucleosis, Burkitt’s lymphoma, nasopharyngeal carcinoma and other diseases (60). Unlike many mammalian and avian tumor viruses, EBV replicates as a covalently closed, circular double-stranded DNA plasmid during latent infection, much like bacteriophage P1 (61). DNase I footprinting experiments show that the dimeric EBNA1 protein, which is responsible for replication from oriP (62,63), binds to 26 sites in the EBV genome. The sites and their complements are shown in the logo (Fig. 5C). Hydroxyl radical footprinting shows binding on one face of the DNA (64). The orientation of EBNA1 binding to DNA was also determined by the X-ray crystal structure (48) and methylation protection (top strand, triangles; bottom strand, inverted triangles) (64,65). The co-crystal structure shows contacts to –8(+9) and –7(+8) but not to the region between, where the DNA faces the protein (48,66). From this and other experiments, it was proposed that the protein binds in the middle of the site and then distorts the DNA (67). Most of the sequence logo tracks the sine wave, but the center of the site (0 and +1 in Fig. 5C), where the protein faces the minor groove, shows exceptional sequence conservation. A 9-base peptide wraps around the DNA in the minor groove, and, although it does not reach to the center of the palindrome, it provides a residue (Arg-469) that stabilizes a water molecule that, in turn, contacts carbonyl 2 of thymine in the central bases (48). In addition, the central base pairs show a high helical twist. It is not known whether these effects are sufficient to conserve the bases to the observed 1.7 bits. Although the crystal structure does not show base flipping, in some systems DNA opening requires special conditions, such as superhelical DNA and binding of other proteins (68).
Is EBNA1 involved in DNA opening? Although EBNA1 binds in the origin region and helps DNA synthesis, it is not essential for replication (69). Current information for EBNA1 suggests that it has several other functions. EBNA1 is required for DNA segregating the episome (70), alternative replication origins exist, and EBNA1 slows replication forks (71). In addition, the origin sequence itself is easily unwound (72) and the specific replication function could be supplied by the human origin recognition complex (hsORC), which binds to the EBV origin (73,74) and is required for origin function (75). Since EBNA1 has other functions and DNA replication functions can be assigned to other molecules, it might not be directly involved in DNA replication. Still, EBNA1 does bind at the origin and the data do not preclude an additional DNA opening function by EBNA1, as suggested by the sequence logo. Multiple DNA opening components occur in many DNA origins (6) and they may be a general property of origins (16), suggesting that EBNA1 may be one such element.
Saccharomyces cerevisiae ORC sites. The origin replication complex (ORC) binds to specific sequences (ARS, autonomously replicating sequence) to initiate DNA replication in the yeast S.cerevisiae (2,76). The A elements of ORC sites were shown by mutational analysis to be essential for replication (77), and these were used to generate the sequence logo (Fig. 5D). A second binding site, B1, is associated with the A region, but no patterns have been identified in the region (78) and there are not enough footprint data (79,80) to create a reliable model. As at DnaA sites, ORC A sites contain two blocks of highly conserved bases that are separated by 5–6 bp, so there is unusual conservation (above the sine wave) no matter where the sine wave is placed. Position –6 was chosen for the wave crest because it is the highest point of the left conserved region and because the surrounding conservation follows the sine wave, as expected for B-form DNA (10). The approximate locations and sensitivity of mung bean nuclease hypersensitive bases in the absence of ORC are indicated in Figure 5D by ellipses for the rDNA ARS (42) and by red lines for the Two Micron ARS (81). The majority of the nuclease sensitive region (boxed region at right) extends 100 bases 3′ of the binding site. These changes to unprotected supercoiled DNA only show that the general region is exposed, but do not indicate what happens in the presence of ORC. An enhanced hydrazine signal appears at position –3 (indicated by the filled circle) when ORC binds to the ARS1 site in vitro (79). More recent in vivo experiments with KMnO4 also show that DNA distortions or ssDNA first appear at position –3, triggered by Cdc7p at the CDC7 execution point of the cell cycle (82). The sequence logo shows that these enhancements split the site at the open/closed boundary, which lies exactly at the central break in the sequence conservation. Consequently, ORC sites appear to be in the ‘flapper’ class defined above for DnaA and P4 α. Indeed, ORC binding proteins and DnaA are in the same protein superfamily (83). Interestingly, the CDC7 to CDC8 cell cycle transition is associated with a small DNA topology change (82).
Transcription initiation factor binding sites
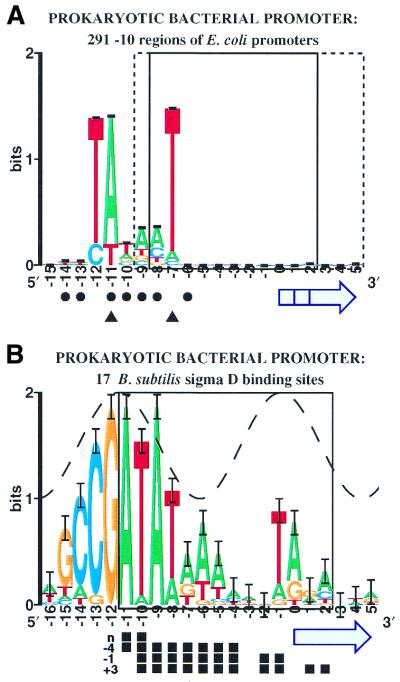
Escherichia coli Pribnow Box (σ70). Transcription by E.coli RNA polymerase requires the σ70 factor to bind to a region 10 bases upstream of the first transcribed base. σ70 –10 promoter regions from the Lisser–Margalit database (84) were aligned (Fig. 6A) to maximize their information content (85,86). Eight promoters that each have two nearby –10 regions became aligned. To avoid duplication artifacts, all of these were eliminated. The resulting information curve is comparable to one reported earlier (87). As determined by ethylated phosphates that interfere with polymerase binding and by the location of purines protected from DMS methylation or bromouracil substituted thymines protected by the polymerase (filled circles), the polymerase follows the major groove from –16 to –6. This was interpreted to mean that the polymerase was opening the DNA in the downstream region rather than wrapping (44). Whether or not that is the case, the location of a sine wave would be ambiguous and is not shown. Transcription initiates near base 0. The region unwound by the polymerase in the T7 A3 promoter is boxed (44); dashes indicate that the open T7 A1 promoter region extends to +5.5 (88) while the lac UV5 promoter extends back to –9.5 (44).
Although the –10 consensus sequence model TATAAT is widespread (89,90), the sequence logo of the σ70 binding site reveals that the sequence conservation is distinctly segregated into two parts, {–12, –11} and {–7}, with a region of low sequence conservation in between, as noted diagrammatically by McClure (91), so the site does not look like TATAAT. In addition, more mutations are observed in the high conservation regions than the low region (91). Since the downstream sequence conservation falls into the region opened by the polymerase, the strongly conserved T at –7 on the non-template strand could be distorted or flipped out. This interpretation is consistent with the fact that the non-template strand preferentially interacts with the polymerase (92–94). It is also consistent with the degree of DNA opening by RNA polymerase as measured by difference absorption spectroscopy: at 10°C core polymerase opened ∼2 bp whereas holoenzyme (core + σ70) opened ∼3.5 bp (95), implying that σ70 is responsible for an ∼1.5 bp opening at an early step in initiation. Also consistent with a base flipping model is the finding that nucleation of the open complex begins in the –10 region (94). Furthermore, non-template mismatches at positions –11 and –7 (triangles) increase the rate of open complex formation (96). Position T–7, which corresponds to T–11 in Zaychikov et al. (88) as assigned by using a sequence walker (43), has a lower melting temperature than bases downstream (88), suggesting that it is most liable to open first.
After DNA binding by RNA polymerase, several intermediate steps take place before an open complex is formed (91,97–100). A recent scheme (101) involves three intermediate complexes:
![]() 1
1
Binding of RNA polymerase enzyme (E) to a promoter (P) occurs at the first step b to form the closed complex ![]() . In step s a ‘structural alteration in the protein’ is thought to occur to form a second closed complex, EPc. Step f is proposed to consist of a ‘conformational change in the protein and nucleation of strand separation’, forming a third closed complex
. In step s a ‘structural alteration in the protein’ is thought to occur to form a second closed complex, EPc. Step f is proposed to consist of a ‘conformational change in the protein and nucleation of strand separation’, forming a third closed complex ![]() . This is followed by step o, which is strand separation and formation of the open complex, EPo. From the studies mentioned above, the f nucleation step may occur at the strongly conserved position –7 and so could represent an initially flipping base.
. This is followed by step o, which is strand separation and formation of the open complex, EPo. From the studies mentioned above, the f nucleation step may occur at the strongly conserved position –7 and so could represent an initially flipping base.
Thus a detailed analysis of a broad array of experimental data conjoined with a highly significant sequence logo (note the small error bars in Fig. 6A) strongly supports the hypothesis that opening at a single base initiates transcription (33,34).
Bacillus subtilis σD promoter. Seventeen sites for the σD promoter of B.subtilis have been identified (102). Four successive stages of permanganate reactivity have been proposed for the Phag promoter (103). On the sequence logo for σD promoters (Fig. 6B), DNA melting begins at position –11 and then expands downstream (filled squares; n: EδσD at 0°C; –4: EσD at 0°C; –1: EσD at 20°C; +3: EσD at 40°C). The peak of a sine wave was placed at –11.5 so that the wave matches the majority of positions in the logo. Given this assignment, position –9 appears to be mildly exceptional (ptotal = 2.4 × 10–3; pmajor = 4.1 × 10–12). As at σ70, the sequence conservation at –9 may reflect transcription bubble nucleation.
DISCUSSION
The sequence conservation for RepA binding sites at the origin of bacteriophage P1 (Fig. 1B) is strikingly and significantly different from those of the activators and repressors (Fig. 1A): all 14 sites have completely conserved a T exactly in the middle of a region where the minor groove has been demonstrated to face the protein (Fig. 1B). One explanation for this anomaly could be that a second protein—or RepA itself—binds into the major groove on the back face of the DNA, but experimental work appears to have ruled out this possibility (7). A second possible explanation is that the TG or TA at positions +7 and +8 allows the DNA to bend (26) in both P1 and Rts1. Alternatively, an elegant suggestion by Roberts is that RepA flips out the T after binding to the DNA (33).
It is also formally possible that the protein opens the DNA and inserts contact elements into the helix. However, the flip hypothesis is simpler, because DNA breathing opens single bases more frequently than many bases simultaneously (104,105). Once a single base pair has been disrupted and held open by specific contacts to the protein, other bases could follow more easily, to open the helix. I therefore investigated the sequence logos of other DNA replication and transcription factor binding sites to see if they also have unusual sequence conservation signatures.
Binding sites containing regions of high sequence conservation separated by ∼5 bases—that is, where high sequence conservation is present in both the major and minor grooves on the same DNA face—appear not only at RepA binding sites (Fig. 1B), but also at Rts1 binding sites (Fig. 3), the P4 replicon (Fig. 5B), other related prokaryotic plasmid replication systems (6) and DnaA protein binding sites (Fig. 5A), all of which are required for replication from origins in E.coli. The unusual sequence conservation at minor groove positions is not confined to prokaryotes, as it also appears in the human EBV origin (Fig. 5C) and the eukaryotic yeast S.cerevisiae ARSs (76), which are bound by the ORC (2,106–108) (Fig. 5D). Unusual sequence conservation appears to fall into two general classes: single base flipping (RepA, Rts1, EBNA1) and multiple base ‘flapping’ (DnaA, P4 α, ORC). Flapping could be nucleated by a single base flip, but this would not necessarily be visible on a logo.
EBNA1 is not only involved in binding to oriP as part of the replication process, but it is also involved in DNA segregation (70). This pleiotropy also occurs with the P1 RepA protein, which is involved not only in replication but also auto-transcriptional repression and replication control (6). This raises the question as to why there is sequence conservation of unusual bases [+7 in RepA, 0(+1) in EBNA1] over many sites if their function is primarily for replication at a few of the sites. First, these positions are part of the overall sequence conservation, so removal of the contacts would disrupt binding to some degree. Secondly, the protein structure is unlikely to be flexible and intelligent enough to distinguish between different binding sites. If the organism requires the function in one site, then that function or action may be performed at the other sites as well by default. In the case of RepA, all natural sites have a T at +7, but the logo shows that there is some variation in EBNA1 position 0(+1). One would expect the variation to be in sites away from the origin, and this is indeed the case, although clearly this is not a strong test.
To determine the generality of the anomalous base observation, the sequence logo analysis was extended to RNA transcription bubbles. The –10 region bound by E.coli σ70 (Fig. 6A) was found to have a high sequence conservation signature. A conserved spike of sequence conservation appears near the 3′ edge of the region opened by the polymerase, and transcription begins near the 5′ edge. Extensive experimental evidence points to a T at –7 being opened first during transcription initiation. This possibility was not fully recognized previously because creating the Pribnow ‘box’ consensus ‘TATAAT’ suppresses the dip that is clearly visible in the sequence logo (Fig. 6A). An analysis of B.subtilis σD promoters (Fig. 6B) also shows a high information spike, but it is close to the major groove and may not be significant because the exact placement of the sine wave is not known. Still, its position correlates well with the base pairs that are opened in succession since it is adjacent to the first two bases that are opened.
Polyamides have been synthesized that are capable of distinguishing all four bases in the minor groove of DNA (109,110) but comparable non-distorting interactions by natural proteins that produce 2 bit sequence conservation from the minor groove of B-form DNA have not been observed. In contrast, complete sequence conservation can be accomplished by proteins binding to the minor groove of strongly distorted DNA structures (22,111). It is possible that such contacts account for some of the high minor groove sequence conservation reported here. However, as shown in Figure 1A, the absence or rarity of ‘anomalous’ bases in the binding sites of many transcriptional activators and repressors (7,10,112,113), in contrast to their ubiquity in DNA sites bound by protein factors known to open DNA (Figs 5 and 6), suggests that the ‘anomalous’ bases may play a critical role in DNA opening.
These observations indicate that, after the initial protein binding step, the second step in both DNA replication and RNA transcription could be caused by structural perturbations at specific bases that can be revealed by sequence logos. One likely possibility is that in some cases the perturbation is one or more bases flipping out of the DNA helix with little other distortion (33,92). In the accompanying paper we report tests of this hypothesis for the P1 RepA system (16).
Acknowledgments
ACKNOWLEDGEMENTS
I thank Rich Roberts for suggesting that RepA might flip a base; Mark Shaner for collecting the GAL4 sequences; Hanah Margalit for sending the promoter database; John Helmann for the list of σD promoters prior to publication; Becky Chasan for many editorial comments; Boris Steipe for help with the DNA structure of the cover graphic; and Lakshmanan Iyer, Ilya Lyakhov, Denise Rubens, Dhruba Chattoraj, Paul N. Hengen and Brandon Cunningham for comments on the manuscript.
REFERENCES
- 1.Bramhill D. and Kornberg,A. (1988) Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell, 52, 743–755. [DOI] [PubMed] [Google Scholar]
- 2.Bell S.P. and Stillman,B. (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- 3.Abeles A.L., Snyder,K.M. and Chattoraj,D.K. (1984) P1 plasmid replication: Replicon structure. J. Mol. Biol., 173, 307–324. [DOI] [PubMed] [Google Scholar]
- 4.Abeles A.L. (1986) P1 plasmid replication. Purification and DNA-binding activity of the replication protein RepA. J. Biol. Chem., 261, 3548–3555. [PubMed] [Google Scholar]
- 5.Abeles A.L., Reaves,L.D. and Austin,S.J. (1989) Protein–DNA interactions in regulation of P1 plasmid replication. J. Bacteriol., 171, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chattoraj D.K. and Schneider,T.D. (1997) Replication control of plasmid P1 and its host chromosome: the common ground. Prog. Nucleic Acid Res. Mol. Biol., 57, 145–186. [DOI] [PubMed] [Google Scholar]
- 7.Papp P.P., Chattoraj,D.K. and Schneider,T.D. (1993) Information analysis of sequences that bind the replication initiator RepA. J. Mol. Biol., 233, 219–230. [DOI] [PubMed] [Google Scholar]
- 8.Papp P.P. and Chattoraj,D.K. (1994) Missing-base and ethylation interference footprinting of P1 plasmid replication initiator. Nucleic Acids Res., 22, 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seeman N.C., Rosenberg,J.M. and Rich,A. (1976) Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl Acad. Sci. USA, 73, 804–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider T.D. (1996) Reading of DNA sequence logos: Prediction of major groove binding by information theory. Methods Enzymol., 274, 445–455. [DOI] [PubMed] [Google Scholar]
- 11.Schneider T.D., Stormo,G.D., Gold,L. and Ehrenfeucht,A. (1986) Information content of binding sites on nucleotide sequences. J. Mol. Biol., 188, 415–431. [DOI] [PubMed] [Google Scholar]
- 12.Schneider T.D. and Stormo,G.D. (1989) Excess information at bacteriophage T7 genomic promoters detected by a random cloning technique. Nucleic Acids Res., 17, 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman N.D. and Schneider,T.D. (1992) High information conservation implies that at least three proteins bind independently to F plasmid incD repeats. J. Bacteriol., 174, 3558–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papp P.P., Mukhopadhyay,G. and Chattoraj,D.K. (1994) Negative control of plasmid DNA replication by iterons. Correlation with initiator binding affinity. J. Biol. Chem., 269, 23563–23568. [PubMed] [Google Scholar]
- 15.Mukhopadhyay G., Carr,K.M., Kaguni,J.M. and Chattoraj,D.K. (1993) Open-complex formation by the host initiator, DnaA at the origin of P1 plasmid replication. EMBO J., 12, 4547–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyakhov I.G., Hengen,P.N., Rubens,D. and Schneider,T.D. (2001) The P1 phage replication protein RepA contacts an otherwise inaccessible thymine N3 proton by DNA distortion or base flipping. Nucleic Acids Res., 29, 4892––4900.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodrich J.A., Schwartz,M.L. and McClure,W.R. (1990) Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res., 18, 4993–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice P.A., Yang,S., Mizuuchi,K. and Nash,H.A. (1996) Crystal structure of an IHF–DNA complex: a protein-induced DNA U-turn. Cell, 87, 1295–1306. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y., Geiger,J.H., Hahn,S. and Sigler,P.B. (1993) Crystal structure of a yeast TBP/TATA-box complex. Nature, 365, 512–520. [DOI] [PubMed] [Google Scholar]
- 20.Kim J.L., Nikolov,D.B. and Burley,S.K. (1993) Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature, 365, 520–527. [DOI] [PubMed] [Google Scholar]
- 21.Schumacher M.A., Choi,K.Y., Zalkin,H. and Brennan,R.G. (1994) Crystal structure of LacI member, PurR, bound to DNA: Minor groove binding by α helices. Science, 266, 763–770. [DOI] [PubMed] [Google Scholar]
- 22.Bewley C.A., Gronenborn,A.M. and Clore,G.M. (1998) Minor groove-binding architectural proteins: structure, function and DNA recognition. Annu. Rev. Biophys. Biomol. Struct., 27, 105–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhopadhyay G. and Chattoraj,D.K. (1993) Conformation of the origin of P1 plasmid replication. Initiator protein induced wrapping and intrinsic unstacking. J. Mol. Biol., 231, 19–28. [DOI] [PubMed] [Google Scholar]
- 24.Thompson J.F. and Landy,A. (1988) Empirical estimation of protein-induced DNA bending angles: applications to λ site-specific recombination complexes. Nucleic Acids Res., 16, 9687–9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hengen P.N., Bartram,S.L., Stewart,L.E. and Schneider,T.D. (1997) Information analysis of Fis binding sites. Nucleic Acids Res., 25, 4994–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickerson R.E. (1998) DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res., 26, 1906–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng X., Kumar,S., Posfai,J., Pflugrath,J.W. and Roberts,R.J. (1993) Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell, 74, 299–307. [DOI] [PubMed] [Google Scholar]
- 28.Klimaauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 29.Verdine G.L. (1994) The flip side of DNA methylation. Cell, 76, 197–200. [DOI] [PubMed] [Google Scholar]
- 30.Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) The crystal structure of HaeIII methyltransferase covalently complexed to DNA: An extrahelical cytosine and rearranged base pairing. Cell, 82, 143–153. [DOI] [PubMed] [Google Scholar]
- 31.Carter A.P., Clemons,W.M.,Jr, Brondersen,D.E., Morgan-Warren,R.J., Wimberly,B.T. and Ramakrishnan,V. (2000) Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature, 407, 340–348. [DOI] [PubMed] [Google Scholar]
- 32.Patel P.H., Suzuki,M., Adman,E., Shinkai,A. and Loeb,L.A. (2001) Prokaryotic DNA polymerase I: evolution, structure and “base flipping” mechanism for nucleotide selection. J. Mol. Biol., 308, 823–837. [DOI] [PubMed] [Google Scholar]
- 33.Roberts R.J. (1995) On base flipping. Cell, 82, 9–12. [DOI] [PubMed] [Google Scholar]
- 34.Roberts R.J. and Cheng,X. (1998) Base flipping. Annu. Rev. Biochem., 67, 181–198. [DOI] [PubMed] [Google Scholar]
- 35.Klimaauskas S. and Roberts,R.J. (1995) Disruption of the target G-C base-pair by the HhaI methyltransferase. Gene, 157, 163–164. [DOI] [PubMed] [Google Scholar]
- 36.Schneider T.D., Stormo,G.D., Haemer,J.S. and Gold,L. (1982) A design for computer nucleic-acid sequence storage, retrieval and manipulation. Nucleic Acids Res., 10, 3013–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider T.D. and Stephens,R.M. (1990) Sequence logos: A new way to display consensus sequences. Nucleic Acids Res., 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shannon C.E. (1948) A mathematical theory of communication. Bell System Tech. J., 27, 379–423, 623–656. [Google Scholar]
- 39.Pierce J.R. (1980) An Introduction to Information Theory: Symbols, Signals and Noise, 2nd edn. Dover Publications, Inc., New York, NY.
- 40.Sloane N.J.A. and Wyner,A.D. (1993) Claude Elwood Shannon: Collected Papers. IEEE Press, Piscataway, NJ.
- 41.Stephens R.M. and Schneider,T.D. (1992) Features of spliceosome evolution and function inferred from an analysis of the information at human splice sites. J. Mol. Biol., 228, 1124–1136. [DOI] [PubMed] [Google Scholar]
- 42.Miller C.A. and Kowalski,D. (1993) cis-acting components in the replication origin from ribosomal DNA of Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 5360–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider T.D. (1997) Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences [published erratum appears in Nucleic Acids Res., 1998, 26, 1135]. Nucleic Acids Res., 25, 4408–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siebenlist U., Simpson,R.B. and Gilbert,W. (1980) E. coli RNA polymerase interacts homologously with two different promoters. Cell, 20, 269–281. [DOI] [PubMed] [Google Scholar]
- 45.Peck L.J. and Wang,J.C. (1981) Sequence dependence of the helical repeat of DNA in solution. Nature, 292, 375–378. [DOI] [PubMed] [Google Scholar]
- 46.Rhodes D. and Klug,A. (1981) Sequence-dependent helical periodicity of DNA. Nature, 292, 378–380. [DOI] [PubMed] [Google Scholar]
- 47.Pabo C.O., Krovatin,W., Jeffrey,A. and Sauer,R.T. (1982) The N-terminal arms of l repressor wrap around the operator DNA. Nature, 298, 441–443. [DOI] [PubMed] [Google Scholar]
- 48.Bochkarev A., Barwell,J.A., Pfuetzner,R.A., Bochkareva,E., Frappier,L. and Edwards,A.M. (1996) Crystal structure of the DNA binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell, 84, 791–800. [DOI] [PubMed] [Google Scholar]
- 49.Marmorstein R., Carey,M., Ptashne,M. and Harrison,S.C. (1992) DNA recognition by GAL4: structure of a protein–DNA complex. Nature, 356, 408–414. [DOI] [PubMed] [Google Scholar]
- 50.Schneider T.D. (2000) Evolution of biological information. Nucleic Acids Res., 28, 2794–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Messer W. and Weigel,C. (1996) Initiation of chromosome replication. In Neidhardt,F.C., Curtiss,R.,III, Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. American Society for Microbiology Press, Washington, DC, Vol. 2, pp. 1579–1601.
- 52.Samitt C.E., Hansen,F.G., Miller,J.F. and Schaechter,M. (1989) In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J., 8, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Speck C., Weigel,C. and Messer,W. (1997) From footprint to toeprint: a close-up of the DnaA box, the binding site for the bacterial initiator protein DnaA. Nucleic Acids Res., 25, 3242–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savva R., McAuley-Hecht,K., Brown,T. and Pearl,L. (1995) The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature, 373, 487–493. [DOI] [PubMed] [Google Scholar]
- 55.Fuller R.S., Funnell,B.E. and Kornberg,A. (1984) The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell, 38, 889–900. [DOI] [PubMed] [Google Scholar]
- 56.Ziegelin G. and Lanka,E. (1995) Bacteriophage P4 DNA replication. FEMS Microbiol. Rev., 17, 99–107. [DOI] [PubMed] [Google Scholar]
- 57.Tocchetti A., Galimberti,G., Deho,G. and Ghisotti,D. (1999) Characterization of the oriI and oriII origins of replication in phage-plasmid P4. J. Virol., 73, 7308–7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komori H., Matsunaga,F., Higuchi,Y., Ishiai,M., Wada,C. and Miki,K. (1999) Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6 Å resolution. EMBO J., 18, 4597–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziegelin G., Scherzinger,E., Lurz,R. and Lanka,E. (1993) Phage P4 α protein is multifunctional with origin recognition, helicase and primase activities. EMBO J., 12, 3703–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawa K. (2000) Epstein-Barr virus-associated diseases in humans. Int. J. Hematol., 71, 108–117. [PubMed] [Google Scholar]
- 61.Lindahl T., Adams,A., Bjursell,G., Bornkamm,G.W., Kaschka-Dierich,C. and Jehn,U. (1976) Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J. Mol. Biol., 102, 511–530. [DOI] [PubMed] [Google Scholar]
- 62.Rawlins D.R., Milman,G., Hayward,S.D. and Hayward,G.S. (1985) Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell, 42, 859–868. [DOI] [PubMed] [Google Scholar]
- 63.Hsieh D.J., Camiolo,S.M. and Yates,J.L. (1993) Constitutive binding of EBNA1 protein to the Epstein-Barr virus replication origin, oriP, with distortion of DNA structure during latent infection. EMBO J., 12, 4933–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kimball A.S., Milman,G. and Tullius,T.D. (1989) High-resolution footprints of the DNA-binding domain of Epstein-Barr virus nuclear antigen 1. Mol. Cell. Biol., 9, 2738–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frappier L. and O‘Donnell,M. (1992) EBNA1 distorts oriPi, the Epstein-Barr virus latent replication origin. J. Virol., 66, 1786–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dean F.B. and O‘Donnell,M. (1996) Two steps to binding replication origins? DNA–protein interactions. Curr. Biol., 6, 931–934. [DOI] [PubMed] [Google Scholar]
- 67.Cruickshank J., Shire,K., Davidson,A.R., Edwards,A.M. and Frappier,L. (2000) Two domains of the Epstein-Barr virus origin DNA-binding protein, EBNA1, orchestrate sequence-specific DNA binding. J. Biol. Chem., 275, 22273–22277. [DOI] [PubMed] [Google Scholar]
- 68.Hwang D.S. and Kornberg,A. (1992) Opening of the replication origin of Escherichia coli by DnaA protein with HU or IHF. J. Biol. Chem., 267, 23083–23086. [PubMed] [Google Scholar]
- 69.Aiyar A., Tyree,C. and Sugden,B. (1998) The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J., 17, 6394–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu H., Ceccarelli,D.F. and Frappier,L. (2000) The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep., 1, 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Little R.D. and Schildkraut,C.L. (1995) Initiation of latent DNA replication in the Epstein-Barr virus genome can occur at sites other than the genetically defined origin. Mol. Cell. Biol., 15, 2893–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams D.L. and Kowalski,D. (1993) Easily unwound DNA sequences and hairpin structures in the Epstein-Barr virus origin of plasmid replication. J. Virol., 67, 2707–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schepers A., Ritzi,M., Bousset,K., Kremmer,E., Yates,J.L., Harwood,J., Diffley,J.F. and Hammerschmidt,W. (2001) Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J., 20, 4588–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chaudhuri B., Xu,H., Todorov,I., Dutta,A. and Yates,J.L. (2001) Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl Acad. Sci. USA, 98, 10085–10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dhar S.K., Yoshida,K., Machida,Y., Khaira,P., Chaudhuri,B., Wohlschlegel,J.A., Leffak,M., Yates,J. and Dutta,A. (2001) Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell, 106, 287–296. [DOI] [PubMed] [Google Scholar]
- 76.Stinchcomb D.T., Struhl,K. and Davis,R.W. (1979) Isolation and characterisation of a yeast chromosomal replicator. Nature, 282, 39–43. [DOI] [PubMed] [Google Scholar]
- 77.Newlon C.S. and Theis,J.F. (1993) The structure and function of yeast ARS elements. Curr. Opin. Genet. Dev., 3, 752–758. [DOI] [PubMed] [Google Scholar]
- 78.Kelly T.J. and Brown,G.W. (2000) Regulation of chromosome replication. Annu. Rev. Biochem., 69, 829–880. [DOI] [PubMed] [Google Scholar]
- 79.Lee D.G. and Bell,S.P. (1997) Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol., 17, 7159–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rao H., Marahrens,Y. and Stillman,B. (1994) Functional conservation of multiple elements in yeast chromosomal replicators. Mol. Cell. Biol., 14, 7643–7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Umek R.M. and Kowalski,D. (1987) Yeast regulatory sequences preferentially adopt a non-B conformation in supercoiled DNA. Nucleic Acids Res., 15, 4467–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Geraghty D.S., Ding,M., Heintz,N.H. and Pederson,D.S. (2000) Premature structural changes at replication origins in a yeast minichromosome maintenance (MCM) mutant. J. Biol. Chem., 275, 18011–18021. [DOI] [PubMed] [Google Scholar]
- 83.Neuwald A.F., Aravind,L., Spouge,J.L. and Koonin,E.V. (1999) AAA+: A class of chaperone-like ATPases associated with assembly, operation and disassembly of protein complexes. Genome Res., 9, 27–43. [PubMed] [Google Scholar]
- 84.Lisser S. and Margalit,H. (1993) Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res., 21, 1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Toledano M.B., Kullik,I., Trinh,F., Baird,P.T., Schneider,T.D. and Storz,G. (1994) Redox-dependent shift of OxyR-DNA contacts along an extended DNA binding site: A mechanism for differential promoter selection. Cell, 78, 897–909. [DOI] [PubMed] [Google Scholar]
- 86.Schneider T.D. and Mastronarde,D. (1996) Fast multiple alignment of ungapped DNA sequences using information theory and a relaxation method. Discrete Appl. Math., 71, 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hertz G.Z. and Stormo,G.D. (1996) Escherichia coli promoter sequences: Analysis and prediction. Methods Enzymol., 273, 30–42. [DOI] [PubMed] [Google Scholar]
- 88.Zaychikov E., Denissova,L., Meier,T., Gotte,M. and Heumann,H. (1997) Influence of Mg2+ and temperature on formation of the transcription bubble. J. Biol. Chem., 272, 2259–2267. [DOI] [PubMed] [Google Scholar]
- 89.Lewin B. (1997) Genes VI. Oxford University Press, Oxford, UK.
- 90.Doree S.M. and Mulks,M.H. (2001) Identification of an Actinobacillus pleuropneumoniae consensus promoter structure. J. Bacteriol., 183, 1983–1989. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.McClure W.R. (1985) Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem., 54, 171–204. [DOI] [PubMed] [Google Scholar]
- 92.deHaseth P.L. and Helmann,J.D. (1995) Open complex formation by Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA. Mol. Microbiol., 16, 817–824. [DOI] [PubMed] [Google Scholar]
- 93.Marr M.T. and Roberts,J.W. (1997) Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science, 276, 1258–1260. [DOI] [PubMed] [Google Scholar]
- 94.Helmann J.D. and deHaseth,P.L. (1999) Protein–nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry, 38, 5959–5967. [DOI] [PubMed] [Google Scholar]
- 95.Wheeler A.R., Woody,A.-Y.M. and Woody,R.W. (1987) Salt-dependent binding of Escherichia coli RNA polymerase to DNA and specific transcription by the core enzyme and holoenzyme. Biochemistry, 26, 3322–3330. [DOI] [PubMed] [Google Scholar]
- 96.Roberts C.W. and Roberts,J.W. (1996) Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell, 86, 495–501. [DOI] [PubMed] [Google Scholar]
- 97.Buc H. and McClure,W.R. (1985) Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry, 24, 2712–2723. [DOI] [PubMed] [Google Scholar]
- 98.Spassky A., Kirkegaard,K. and Buc,H. (1985) Changes in the DNA structure of the lac UV5 promoter during formation of an open complex with Escherichia coli RNA polymerase. Biochemistry, 24, 2723–2731. [DOI] [PubMed] [Google Scholar]
- 99.Buckle M., Pemberton,I.K., Jacquet,M.A. and Buc,H. (1999) The kinetics of sigma subunit directed promoter recognition by E. coli RNA polymerase. J. Mol. Biol., 285, 955–964. [DOI] [PubMed] [Google Scholar]
- 100.Craig M.L., Tsodikov,O.V., McQuade,K.L., Schlax,P.E.,Jr, Capp,M.W., Saecker,R.M. and Record,M.T.,Jr (1998) DNA footprints of the two kinetically significant intermediates in formation of an RNA polymerase-promoter open complex: evidence that interactions with start site and downstream DNA induce sequential conformational changes in polymerase and DNA. J. Mol. Biol., 283, 741–756. [DOI] [PubMed] [Google Scholar]
- 101.Johnson R.S. and Chester,R.E. (1998) Stopped-flow kinetic analysis of the interaction of Escherichia coli RNA polymerase with the bacteriophage T7 A1 promoter. J. Mol. Biol., 283, 353–370. [DOI] [PubMed] [Google Scholar]
- 102.Helmann J.D. and Moran,C.P.,Jr (2001) Bacillus subtilis and its closest relatives: from genes to cells. In Sonenshein,A.L., Hoch,J.A and Losick,R. (eds), RNA Polymerase and Sigma Factors. ASM Press, Washington, DC, pp. 289–312.
- 103.Chen Y.F. and Helmann,J.D. (1997) DNA-melting at the Bacillus subtilis flagellin promoter nucleates near –10 and expands unidirectionally. J. Mol. Biol., 267, 47–59. [DOI] [PubMed] [Google Scholar]
- 104.Guéron M., Kochoyan,M. and Leroy,J.L. (1987) A single mode of DNA base-pair opening drives imino proton exchange. Nature, 328, 89–92. [DOI] [PubMed] [Google Scholar]
- 105.Leroy J.L., Kochoyan,M., Huynh-Dinh,T. and Guéron,M. (1988) Characterization of base-pair opening in deoxynucleotide duplexes using catalyzed exchange of the imino proton. J. Mol. Biol., 200, 223–238. [DOI] [PubMed] [Google Scholar]
- 106.Diffley J.F.X. and Cocker,J.H. (1992) Protein–DNA interactions at a yeast replication origin. Nature, 357, 169–172. [DOI] [PubMed] [Google Scholar]
- 107.Newlon C.S. (1993) Two jobs for the origin replication complex. Science, 262, 1830–1831. [DOI] [PubMed] [Google Scholar]
- 108.Rowley A., Crocker,J.H., Harwood,J. and Diffley,J.F.X. (1995) Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J., 14, 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kielkopf C.L., White,S., Szewczyk,J.W., Turner,J.M., Baird,E.E., Dervan,P.B. and Rees,D.C. (1998) A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA. Science, 282, 111–115. [DOI] [PubMed] [Google Scholar]
- 110.White S., Szewczyk,J.W., Turner,J.M., Baird,E.E. and Dervan,P.B. (1998) Recognition of the four Watson–Crick base pairs in the DNA minor groove by synthetic ligands. Nature, 391, 468–471. [DOI] [PubMed] [Google Scholar]
- 111.Travers A. (2000) Recognition of distorted DNA structures by HMG domains. Curr. Opin. Struct. Biol., 10, 102–109. [DOI] [PubMed] [Google Scholar]
- 112.Shultzaberger R.K. and Schneider,T.D. (1999) Using sequence logos and information analysis of Lrp DNA binding sites to investigate discrepancies between natural selection and SELEX. Nucleic Acids Res., 27, 882–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wood T.I., Griffith,K.L., Fawcett,W.P., Jair,K.-W., Schneider,T.D. and Wolf,R.E. (1999) Interdependence of the position and orientation of SoxS binding sites in the transcriptional activation of the class I subset of Escherichia coli superoxide-inducible promoters. Mol. Microbiol., 34, 414–430. [DOI] [PubMed] [Google Scholar]
- 114.Bram R.J., Lue,N.F. and Kornberg,R.D. (1986) A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J., 5, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kamio Y., Tabuchi,A., Itoh,Y., Katagiri,H. and Terawaki,Y. (1984) Complete nucleotide sequence of mini-Rts1 and its copy mutant. J. Bacteriol., 158, 307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nozue H., Tsuchiya,K. and Kamio,Y. (1988) Nucleotide sequence and copy control function of the extension of the incI region (incI-b) of Rts1. Plasmid, 19, 46–56. [DOI] [PubMed] [Google Scholar]
- 117.Schneider T.D. (1997) Information content of individual genetic sequences. J. Theor. Biol., 189, 427–441. [DOI] [PubMed] [Google Scholar]