Abstract

The development of L-lactate biosensors has been hampered in recent years by the lack of availability and knowledge about a wider range and diversity of L-lactate-oxidizing enzymes that can be used as bioelements in these sensors. For decades, L-lactate oxidase of Aerococcus viridans (AvLOx) has been used almost exclusively in the field of L-lactate biosensor development and has achieved somewhat like a monopoly status as a biocatalyst for these applications. Studies on other L-lactate-oxidizing enzymes are sparse and are often missing biochemical data. In this work, we made use of the vast amount of sequence information that is currently available on protein databases to investigate the naturally occurring diversity of L-lactate-utilizing enzymes of the flavin mononucleotide (FMN)-dependent α-hydroxy acid oxidoreductase (HAOx) family. We identified the HAOx sequence space specific for L-lactate oxidation and additionally discovered a not-yet described class of soluble and FMN-dependent L-lactate dehydrogenases, which are promising for the construction of second-generation biosensors or other biotechnological applications. Our work paves the way for new studies on α-hydroxy acid biosensors and proves that there is more to the HAOx family than AvLOx.
Introduction
L-Lactate is a central metabolite in the anaerobic metabolism of many organisms and as such is an important biomarker to monitor in various fields, including medicine.1−3 In clinical diagnostics and intensive care, lactate levels are used to detect lactic acidosis, which can be caused by tissue hypoxia, sepsis, or kidney and liver diseases.4,5 Additionally, monitoring lactate levels can draw a picture of the patients’ condition or response to treatment and can help to assess the risk of a shock or mortality.6,7 In sports medicine, athletes’ blood lactate levels are used as a measure for fitness and endurance, and in the food industry, lactate concentrations in fermented food products are an indication of freshness and product quality.1,3,8,9 During production of recombinant proteins in mammalian cell cultures, lactate is one of the critical parameters because of its toxic and growth-inhibiting effects on the cells.10−12 Currently, the preferred method to monitor L-lactate levels is via biosensors, in which a suitable enzyme as the biocatalyst is connected to an electrode to produce electric currents proportional to the lactate concentration in the analyte. The currently preferred enzyme used for the construction of L-lactate biosensors is L-lactate oxidase (LOx).
LOx belongs to the family of flavin mononucleotide (FMN)-dependent α-hydroxy acid oxidoreductases (HAOx; EC 1.1.3.1513), which oxidize various α-hydroxy acids to their corresponding α-keto acids via a ping-pong reaction mechanism.13 L-lactate, the preferred substrate of LOx, is oxidized to pyruvate, while FMN is reduced to FMNH2 in the first (reductive) half-reaction. In the second, ensuing (oxidative) half-reaction of LOx, oxygen is used as an electron acceptor to re-oxidize FMNH2, while it is concurrently reduced to hydrogen peroxide (H2O2). Furthermore, LOx was shown to react with several alternative electron acceptors, such as various benzoquinones or dichlorophenol-indophenol (DCIP) in its oxidative half-reaction.14 Commonly, the protein monomer of LOx has a mass of 39–44 kDa and shows a β8α8 TIM-barrel fold, which is typical for the HAOx family. LOx is active as a homotetramer with the FMN cofactor noncovalently bound to each subunit.15,16 The active site is formed by the isoalloxazine ring of FMN together with several well conserved substrate-binding residues at the edge of the β-barrel, and it is covered by the so-called “active-site lid”.15 This lid has been shown to be highly dynamic and to regulate substrate entry and product exit to and from the active site.17,18 Even though the loop forming the lid can be found in all members of the HAOx family, it is poorly conserved across the family, and due to its flexibility, it is usually disordered in HAOx crystal structures.
Apart from LOx, the HAOx family contains glycolate oxidase (GLO19), long- or medium-chain 2-hydroxy acid oxidase (HAO20,21), flavocytochrome b2 (FCB222,23), L-lactate monooxygenase (LMO24), mandelate dehydrogenase (MDH25), 4-hydroxymandelate oxidase (HMO26), glycolate dehydrogenase (GLDH27), and membrane-bound L-lactate dehydrogenase (mLDH28,29). Despite most of these different HAOx members being known since at least the 1990s, most of the individual groups are still poorly characterized with only 1–3 sequences studied experimentally. The only exceptions are GLO and LOx, both with >10 experimentally characterized sequences, yet most of these sequences are only briefly described in the literature and/or were never biochemically characterized in a purified form. The vast majority of all studies regarding the engineering of LOx or its application in biosensors has focused only on a single sequence, namely, LOx from Aerococcus viridans (AvLOx). One of the reasons for this narrow focus is probably that AvLOx is currently the only commercially available LOx on the market. This lack of availability and knowledge about other L-lactate-sensing enzymes has recently been noted as a limiting factor in the development of novel L-lactate sensors.30 This is because the current application of AvLOx in biosensors comes with several disadvantages. First of all, its kinetic stability is not as high as those of other enzymes typically used in biosensors, and hence, when developing multiplexed sensors, where multiple enzymes are combined, AvLOx is likely the limiting factor for the maximum time of continuous sensor operation, as was shown for a glucose-lactate sensor using glucose dehydrogenase and LOx31 in tandem on one sensor. Second, the low Km values for L-lactate of ∼1 mM together with moderate substrate and pronounced product inhibition of LOx13,31,32 are far from optimal for the determination of L-lactate concentrations in physiologically relevant ranges, usually between 1 and 25 mM for L-lactate in blood33 or even >100 mM in sweat after exhaustive exercise.34 Third, AvLOx suffers from low expression yields in E. coli (see, for example, Table S3), making the screening of high numbers of AvLOx variants laborious and limiting the manageable size of mutant libraries that can be screened for engineering purposes. Lastly, LOx, as an oxidase, is sensitive to oxygen being present in the analyte. This can become problematic when constructing second-generation biosensors, where electron mediators other than O2/H2O2 are applied to transfer electrons from the active site FMN to the electrode. In such systems, oxygen is usually present as well and competes with the alternative electron acceptor at the active site of LOx, where electrons “leak” from the detectable electron flow, resulting in a reduced sensor signal. Such an oxygen interference can be avoided by applying strict dehydrogenases, showing negligible reactivity with oxygen, for second-generation sensors. Unfortunately, the only L-lactate dehydrogenases (LDHs) described to date are either membrane-bound or NAD+-dependent, both of which are unsuitable for several applications, since production, purification, and application of membrane-bound proteins come with various challenges and NAD+ would have to be resupplied during measurement as it is continuously lost due to diffusion or degradation.2,35 Because of this need for better suited enzymes for second-generation lactate biosensors, recent efforts aimed at engineering AvLOx to reduce oxygen reactivity while improving reactivity with alternative electron acceptors, thus basically creating an FMN-dependent lactate dehydrogenase.30,36 It is interesting to note that up to now, shortcomings of L-lactate biosensors were mainly addressed by altering sensor architecture, like applying selective membranes, various immobilization techniques or utilizing different electron mediator systems, or by the engineering of AvLOx, while alternative L-lactate-oxidizing enzymes have hardly been studied as novel biocatalysts for these biosensor applications.
The aim of this study was to gain more knowledge on the variety of naturally occurring L-lactate-oxidizing HAOx enzymes by studying novel fungal and bacterial HAOx members of hitherto unexplored sequence space. We present a substrate specificity screening of 42 selected members, expanding the toolbox of these enzymes for various biocatalytic applications. Furthermore, we show a detailed characterization for five enzymes from a newly identified class of soluble L-lactate-specific, FMN-dependent α-hydroxy acid dehydrogenases.
Results
Sequence Similarity Network of the HAOx Family
The HAOx family is well conserved in sequence as well as structure; nevertheless, it contains members of different and variable substrate selectivities. In order to select HAOx sequences that are likely to utilize L-lactate and to discriminate them from HAOx enzymes of different functionalities, we first established an overview of the families’ sequence space together with an analysis of the distribution of different HAOx members within this sequence space. For this, a total of 34 previously described HAOx sequences, which had their functionalities confirmed experimentally (Table S1), were collected, and of these, 13 representative sequences were used as queries for database searches on NCBI and UniProt (search in July 2020). The resulting sequences (60,454) were used to calculate a sequence similarity network (SSN) of the HAOx family (Figure 1). To avoid duplicate nodes resulting from the use of two databases, the SSN was displayed using representative nodes with a sequence similarity cutoff of 95%. Different alignment score cutoffs (1 × 10–85–1 × 10–125) were used to visualize different levels of sequence relationships. At an alignment score cutoff of 1 × 10–105, the majority of the 34 characterized sequences within the SSN were clearly separated into distinct clusters according to their different functionalities. These distinct clusters were then functionally annotated by the characterized sequences they contained.
Figure 1.
Sequence similarity network of the α-hydroxy acid oxidoreductase family at an E-value cutoff of 1 × 10–105 showing the clustering into functionally distinct groups. Clusters are annotated according to their characterized sequences. LOx, lactate oxidases; mLDH, membrane-bound lactate dehydrogenases; GLDH, glycolate dehydrogenases; GLO, glycolate oxidases; HAO, (long/medium-chain) hydroxy acid oxidases; FCB2, flavocytochrome b2; LMO, lactate monooxygenase; MDH, mandelate dehydrogenases; HMO, hydroxymandelate oxidases.
Only the characterized enzymes GLO and HAO share a common cluster (Figure 1). This GLO/HAO cluster contains all higher eukaryotic HAOx sequences from metazoa and viridiplantae, which seem to be much closer related than any other HAOx members. Although mLDH and GLDH appear in the same cluster, GLDH does not represent a separate group of enzymes since it is the only sequence of microalgae (Phaeodactylum tricornutum) within a cluster that otherwise consists of proteobacterial and actinobacterial sequences and was therefore considered a unique occurrence. All other clusters, namely, LOx, FCB2, LMO, MDH, and HMO, contain only sequences of one distinct functional annotation. The taxonomic distribution of the annotated clusters is in good agreement with previous reports in the literature. While LOx, HMO, MDH, and mLDH occur predominantly in a single group of bacteria, GLO and HAO are mainly found in metazoa and viridiplantae, and FCB2 is almost exclusively present in fungi, with more than half of the sequences from saccharomyceta. To date, two LOx enzymes have been described that do not originate from firmicutes but from the green algae Chlamydomonas reinhardtii and the cyanobacterium Nostoc sp. PCC 7120.37 The only enzyme sequence which shows a higher variation of species in its cluster was LMO. This confirms a taxonomic distribution of LMO beyond bacteria, as was already suggested by Kean and Karplus in 2019.38 In contrast to this earlier publication, which found LMO to occur in various bacteria but only one fungal genus and a single archaeon, our analysis predicts LMOs in 161 bacterial genera, 83 fungal genera (mainly ascomycota), and 26 archaeal genera.
Substrate Preference of Novel HAOx Members
To select novel HAOx sequences from the SSN that are likely to exhibit L-lactate-oxidizing activity, we used the established annotation of functional clusters and concentrated on clusters that either directly showed LOx activity or are connected to a cluster doing so (see Figure 1). Since some GLOs and HAOs were also shown to exhibit activity with L-lactate in addition to their preferred substrates,39−41 the GLO/HAO cluster was considered as well. The sequences constituting selected clusters were extracted from the SSN and had their approximate-maximum-likelihood dendrograms calculated respectively to visualize the sequence variations within them. Finally, a total of 42 HAOx sequences were selected that are well distributed throughout the investigated sequence space (Table S2).
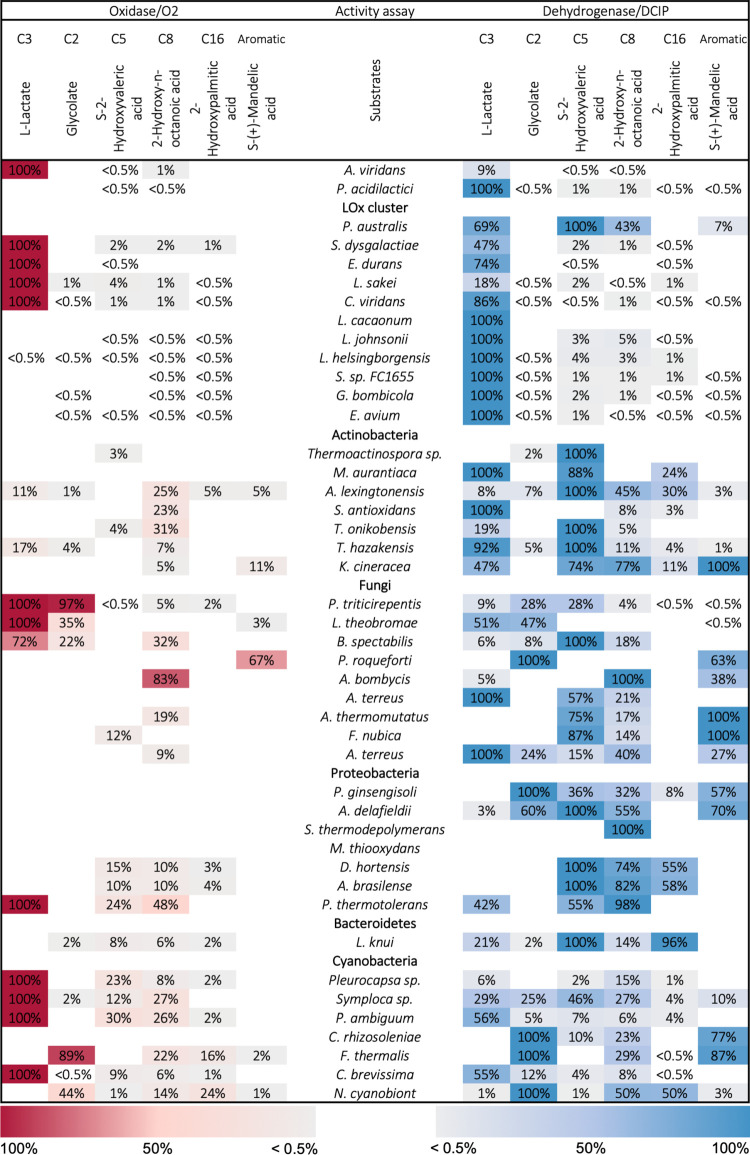
Genes of the so selected enzyme sequences were purchased from a commercial supplier and expressed in E. coli shaken flask cultures. Crude cell extracts were then used to screen for enzymatic activities using the α-hydroxy acids L-lactic acid, glycolic acid, S-2-hydroxyvaleric acid (2-hydroxypentanoic acid), 2-hydroxyoctanoic acid, 2-hydroxypalmitic acid (2-hydroxyhexadecanoic acid), and S-mandelic acid (structural comparison see Figure S1). These compounds were respectively used as electron-donating substrates in spectrophotometric dehydrogenase assays, using DCIP as an electron acceptor, as well as oxidase assays, using oxygen as an electron acceptor and a coupled reaction with Amplex Red to measure hydrogen peroxide formation (Figure 2). In addition to the 42 selected HAOx members, we also included two previously described enzymes in this screening, the extensively studied AvLOx and the putatively annotated LOx from Pediococcus acidilactici (PaLCTO), which recently had its crystal structure solved (PDB: 6RHT) but did not show activity for L-lactate or glycolate in an oxidase assay.16
Figure 2.
Substrate specificity screening of 42 novel HAOx enzymes. Activities are given relative to the maximum volumetric activity for each enzyme. Color intensities for oxidase (red) and dehydrogenase (blue) activities increase with higher activity. Enzymatic assays were measured in quadruplicates using crude enzymes and a 10 mM concentration of the respective electron donor substrate (1.6 mM concentration of 2-hydroxypalmitic acid) at 30 °C and pH 6.5. Background signal was subtracted for each electron donor individually using an empty vector expression as blank. The limit of quantification was defined as 10× the standard deviation of the blank.
In general, the effective expression of putative HAOx genes proved highly successful, with 41 enzymes out of the 42 novel sequences being active with at least one of the substrates. Only the enzyme from Methylophaga thiooxydans showed no activity using the current experimental setup. Interestingly, only 12 out of the total 44 screened enzymes showed true LOx activity, i.e., oxidizing L-lactate as the preferred α-hydroxy acid and accepting oxygen as an electron acceptor. This included the reference enzyme AvLOx, four additional enzymes from the LOx cluster, and two fungal, one proteobacterial, and four cyanobacterial enzymes. It should be noted that most of these oxidases also showed good activity with DCIP, as had been reported for AvLOx before.14 In contrast, a higher number of enzymes exhibited strict dehydrogenase activity with only low or negligible reactivity with oxygen (Figure 2). One of them was PaLCTO, which was previously reported to be inactive with L-lactate and oxygen. Our screening showed PaLCTO to be a highly L-lactate-specific dehydrogenase though, and we propose that it should be classified as an LDH. In addition to PaLCTO, six other enzymes from the LOx cluster displayed a negligible activity with oxygen but highly specific LDH activity using DCIP. Given that PaLCTO and AvLOx have highly similar crystal structures (C-alpha atom rmsd of 0.70 Å16) and that both appear in the LOx cluster of the HAOx family SSN, the LOx cluster must comprise an enzyme class of soluble, FMN-dependent LDHs, which is very closely related to LOx (see also Figure 3) but had not yet been described. Out of 41 active enzymes in the screening, we identified 33 oxidizing L-lactate at least to some extent and 23 oxidizing L-lactate as their preferred substrate. Among them, sequences from the LOx cluster showed the highest selectivity toward L-lactate and hardly accepted other α-hydroxy acids in the activity screening. The only exception is LDH from Photorhabdus australis (PauLDH), which is also a member of the LOx cluster but showed a more diverse substrate selectivity and preferred S-2-hydroxyvaleric acid over L-lactate. Screened sequences from actinobacteria gave a high preference for either L-lactate or S-2-hydroxyvaleric acid or showed activity with both substrates. Fungal sequences displayed the highest variability in substrate utilization, with varying substrate preference patterns for every member. Proteobacterial sequences typically showed highest activities with S-2-hydroxyvaleric acid and 2-hydroxyoctanoic acid (thus being medium- or long-chain α-hydroxy acid oxidoreductases), and cyanobacterial sequences mainly prefer L-lactate or glycolate. The only Bacteroidetes sequence that was tested also preferred medium- to long-chain α-hydroxy acids.
Figure 3.
Maximum likelihood phylogenetic tree of the LOx cluster from Figure 1 and relative activities of the screened enzymes using L-lactate and two electron acceptors, oxygen and DCIP. Positions of HAOx sequences studied in this work are indicated by black triangles, and selected literature-known sequences are indicated by black circles. The tree shows a clear separation of oxidases and dehydrogenases into two clades.
Phylogeny and Functional Distribution of the LOx Cluster
To get a more detailed view on the LOx cluster and its sequence/function distribution, a phylogenetic tree of the LOx cluster was inferred and sequences with their relative L-lactate activities from the initial screening were annotated in the tree (Figure 3). Based on this depiction, a functional split of the tree into two main clades is evident, one clade showing oxygen reactivity (LOx; 150 sequences) and the other showing strict dehydrogenase activity (LDH; 291 sequences). The difference in activity that was observed in the screening, separating LOx and LDH sequences, is therefore also supported by sequence information. One tested sequence does not belong to either of the two main clades of the tree though but instead appeared closer to the outgroup of glycolate oxidases. This sequence is PauLDH, which was also the only sequence in the LOx cluster that was not L-lactate specific. This indicates that the high L-lactate specificity of the LOx cluster observed in the activity screening may be limited to the two main clades of the tree, LOx and LDH.
Sequence Analysis of the LOx Cluster
Subsequently, we compared the aligned amino acid sequences of different clades in the tree of the LOx cluster by visualizing them in an alignment overview (Figure S2). Overall, LOx cluster sequences showed a uniform alignment, with ambiguity only appearing at the N- and C-termini as well as one region close to the center of the aligned sequences. Three clades in the tree showed additional amino acids at their N terminus. Two of these clades contained PauLDH and the LDH of Shigella sp. FC1655 (SsLDH), respectively, and analysis of these two sequences using SignalP 5.042 indicated that both N-termini have a high likelihood for a gram-negative bacterial signal peptide, a TAT signal peptide for PauLDH (likelihood: 0.972) and a Sec signal peptide for SsLDH (likelihood: 0.997). Removing the predicted signal peptides from PauLDH and SsLDH and repeating the expression experiment in E. coli and the substrate specificity screening did not change their pattern of substrate selectivity but in both cases decreased the volumetric activities obtained because of reduced expression levels as estimated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (data not shown). Sequences of two other clades showed an extension at their C-terminus when compared to the majority of sequences in the LOx cluster. These clades contain the LOx of Streptococcus dysgalactiae (SdLOx) and the LDHs from Lactobacillus johnsonii (LjLDH) and Lactobacillus helsingborgensis (LhLDH), respectively (dotted line in Figure S2). The third ambiguously aligned region, near the center of the aligned sequences, roughly corresponds to residues 190–220 in AvLOx (black arrow in Figure S2), which form the active-site lid in AvLOx that was shown to affect substrate binding and product release.15,17,18 Our analysis indicates that this lid is the most variable region of LOx cluster sequences, not considering terminal regions. In contrast to that, we found no substantial differences when assessing the conservation of substrate-binding residues. The five residues responsible for substrate binding in AvLOx, Tyr40, Tyr146, Asp174, His265, and Arg26815,16,43 are well conserved throughout the LOx cluster, with 97% conservation for Tyr40 and 100% conservation for the other residues (Figure S3).
Expression and Purification of LDHs
Five sequences covering a wide sequence space in the LDH clade were selected for a more detailed biochemical characterization. Genes of the enzymes of L. johnsonii (LjLDH), L. helsingborgensis (LhLDH), Shigella sp. FC1655 (SsLDH), Gilliamella bombicola (GbLDH), and Enterococcus avium (EaLDH) as well as for AvLOx were modified with an N-terminal purification tag consisting of 6xHis and a recognition site for the 3C protease of the human rhinovirus (HRV 3C). These six genes were then expressed in E. coli BL21 (DE3) shaken flask cultures and purified using immobilized-metal affinity chromatography (IMAC). Purified PaLCTO, as described previously,16 was thankfully received from the group of Lari Lehtiö from the University of Oulu in Finland. Expression of LjLDH, LhLDH, SsLDH, GbLDH, and EaLDH yielded significantly more recombinant protein than that of AvLOx (up to 119 times higher) when comparing production yields normalized with the amount of harvested cell pellet (mg of purified enzyme/g of wet cell pellet) (Table S3). UV–vis absorption spectra of the purified enzymes showed the typical peaks of flavoproteins with one maximum around 278 nm and two maxima around 374 and 458 nm, which are characteristic for FMN and disappeared upon the addition of 10 mM L-lactate due to the reduction of the flavin cofactor (Figure S4).
Oligomerization States of LDHs
Oligomerization states of the five LDHs in solution were determined by size exclusion chromatography coupled with static light scattering (SEC-SLS) (Figure S5). The results showed a single major SEC elution peak for all samples and masses that are 3.5 and 4.0 times the theoretical mass of the monomers for SsLDH and EaLDH, respectively, indicating tetrameric structures for these two enzymes. A tetrameric oligomerization, determined by SEC, has also been reported for PaLCTO.16 Elution peaks of LjLDH and LhLDH displayed 7.5 and 7.7 times the theoretical masses of their monomers, respectively, implying that they form octameric structures instead of tetramers. Intriguingly, the sequences of LjLDH and LhLDH both contain additional amino acids at the C-terminus (Figure S2), and hence, octamerization could be the result of an additional inter-domain interaction at the C-terminus. SEC-SLS measurement of GbLDH gave inconclusive results. The SEC elution peak was tailing and showed 5.8 times the theoretical mass of the monomer at its maximum; however, the measured mass decreased quickly to the mass of a dimer toward the end of the peak. This uneven distribution of molecular masses within a single peak indicates a mixed population of oligomers. Why such multiple oligomeric states were not separated during SEC is unclear but could be explained either by dynamic interactions between GbLDH subunits, resulting in a continuous rearrangement of the enzymes’ oligomerization states during SEC, or by unspecific interactions with the column matrix. It is also unclear whether the determined molecular mass at the peak maximum is the result of a hexameric structure or the measured mean value of octameric and tetrameric structures that eluted simultaneously from SEC. The native oligomerization state of GbLDH is therefore still unknown and needs further investigation.
Reactivity with Different Electron Acceptor and Donor Substrates
Specific activities were determined for the purified enzymes and the electron acceptors O2 (air), DCIP, 1,4-benzoquinone (1,4-BQ), and ferrocenium hexafluorophosphate (FcPF6) using spectrophotometric assays (Table 1). The highest specific dehydrogenase activities were measured for LjLDH, LhLDH, SsLDH, and EaLDH with DCIP, for GbLDH with FcPF6, and for PaLCTO with 1,4-BQ. Oxygen reactivity was the highest for AvLOx. LjLDH, LhLDH, SsLDH, GbLDH, EaLDH, and PaLCTO showed oxygen reactivities ranging from 0.07 to 0.71 U/mg, accounting for 0.37, 0.47, 0.07, 0.03, 0.05, and 0.37% of their maximal dehydrogenase activity, respectively.
Table 1. Specific Activities for Four Different Electron Acceptorsa.
| specific
activity (U/mg) |
||||
|---|---|---|---|---|
| FcPF6 | 1,4-BQ | DCIP | O2 | |
| LjLDH | 24 ± 2 | 19.7 ± 0.2 | 72 ± 3 | 0.26 ± 0.01 |
| LhLDH | 31 ± 9 | 27 ± 2 | 152 ± 5 | 0.71 ± 0.04 |
| SsLDH | 41 ± 1 | 125 ± 30 | 166 ± 2 | 0.12 ± 0.02 |
| GbLDH | 207 ± 11 | 109 ± 11 | 93 ± 23 | 0.07 ± 0.01 |
| EaLDH | 65 ± 1 | 45 ± 6 | 146 ± 14 | 0.07 ± 0.01 |
| PaLCTO | 11 ± 2 | 25 ± 4 | 6.9 ± 0.3 | 0.09 ± 0.01 |
| AvLOx | nd | nd | 14 ± 3 | 18.4 ± 1.8 |
Activities were measured with 10 mM L-lactate at 30 °C using 120 μM DCIP, 160 μM FcPF6, 500 μM 1,4-BQ, or ambient concentrations of oxygen. nd, not determined.
We further determined the LDHs’ L-lactate specificity by comparing their relative reactivities with various electron donor substrates, including glycolate, L-lactate, S-2-hydroxybutyric acid, R-2-hydroxybutyric acid, S-2-hydroxyvaleric acid, 2-hydroxyoctanoic acid, 2-hydroxypalmitic acid, and S-mandelic acid (Table 2). GbLDH, EaLDH, and PaLCTO showed the highest specificity for L-lactate, with activities with alternative α-hydroxy acids of no more than 3% relative activity compared to L-lactate. LhLDH showed remarkably high reactivity with S-2-hydroxybutyric acid, while LjLDH and SsLDH displayed moderate reactivities with S-2-hydroxybutyric acid and S-2-hydroxyvaleric acid, respectively. The only tested R-enantiomer, R-2-hydroxybutyric acid, was not accepted as the substrate by any of the enzymes. Note that the obtained results for electron donor substrate preferences are not fully in accordance with what was measured in the initial activity screening (Figure 2). This difference is most likely due to crude E. coli cell extracts being used in the initial screening, while purified enzymes and higher concentrations thereof were applied for the substrate reactivity measurements shown here.
Table 2. Relative Substrate Reactivities of Purified LDHsa.
Activities were measured with 120 μM DCIP at 30 °C using 10 mM glycolate, L-lactate, S-2-hydroxybutyric acid, R-2-hydroxybutyric acid, S-2-hydroxyvaleric acid, 2-hydroxy-n-octanoic acid, S-(+)-mandelic acid, and 0.5 mM 2-hydroxypalmitic acid. Activities are given relative to L-lactate for each enzyme.
Michaelis–Menten Kinetics
Apparent steady-state reaction kinetics for L-lactate were measured using DCIP as the electron acceptor (Table 3). LhLDH showed the highest activity for L-lactate as judged from the catalytic efficiency, mainly because of its low Km value. The highest Km value was found for PaLCTO, which is considerably higher than that of the other LDHs studied. Identification of enzymes with Km values covering a wide concentration span could help overcome an issue of AvLOx, which was reported to have an unsuitably low Km (of about 1 mM) for its application in L-lactate sensors.31 Turnover numbers of the studied LDHs for L-lactate varied less drastically, with the highest kcat value measured for GbLDH.
Table 3. Apparent Steady-State Kinetic Constants for the Oxidation of L-Lactate by Various LDHsa.
| Km [mM] | kcat [s–1] | kcat/Km [mM–1 s–1] | |
|---|---|---|---|
| LjLDH | 1.35 ± 0.18 | 47.2 ± 2.0 | 35.0 |
| LhLDH | 0.52 ± 0.09 | 102 ± 8 | 196 |
| SsLDH | 5.67 ± 1.24 | 107 ± 10 | 18.9 |
| GbLDH | 16.9 ± 2.5 | 207 ± 29 | 12.3 |
| EaLDH | 21.6 ± 2.2 | 94.0 ± 7.8 | 4.35 |
| PaLCTO | 235 ± 45 | 61.8 ± 8.5 | 0.263 |
Reactions were measured in 11 mM PBS pH 7.4 at 30 °C and DCIP as the electron acceptor used at a constant concentration of 120 μM. L-Lactate concentrations were varied from 0.125 to 64 mM or from 4 to 500 mM for PaLCTO.
pH Dependence of LDH and AvLOx Activity
Determination of pH optima of the lactate-oxidizing activity was done in Britton–Robinson buffer (BRB) from pH 4.5 to 10.0 using DCIP and O2 as electron acceptors (Figure S6). The pH range where LDHs show 80–100% activity with DCIP is 4.5–6.0 for LjLDH, 4.5–7.0 for LhLDH, 7.5–8.0 for SsLDH, 5.5–7.0 for GbLDH, 6.0–7.0 for EaLDH, and 4.5–5.5 for PaLCTO. AvLOx showed 80–100% activity with O2 in a pH range of 7.0–8.5. Interestingly, measuring LDH activities with O2 and AvLOx activity with DCIP (i.e., switching their preferred electron acceptors) resulted in different pH profiles, showing that optimal pH is not only dependent on the respective enzyme but also on the applied electron acceptor. Additionally, the influence of different buffer species on the enzymes’ activity was investigated. To this end, activities in phosphate-buffered saline (PBS) at pH 7.4 and activities in BRB at pH 7.5 were compared and showed that LjLDH, LhLDH, and GbLDH activities in PBS were 7.8-, 2.0-, and 2.1-fold higher than in BRB pH 7.5. Other enzymes were hardly affected in their activity when comparing the two buffers (Figure S7).
Effect of Temperature on LDH Activity
The thermostability of LDHs was measured by incubating the purified enzymes at different temperatures for 30 min and determination of the residual activities. The temperature at half inactivation (T50), i.e., the temperature at which enzyme activity is reduced to 50% after the 30 min heat treatment, was obtained from an iterative sigmoidal fit of the observed data (Table 4 and Figure S8). The two highest T50 values were observed for SsLDH and EaLDH, while LhLDH and GbLDH showed substantially lower T50 values. The residual activities of PaLCTO could not be fitted to a sigmoidal curve; a T50 value could therefore not be determined.
Table 4. T50 Values of LDHsa.
| LjLDH | LhLDH | SsLDH | GbLDH | EaLDH | PaLCTO | |
|---|---|---|---|---|---|---|
| T50 [°C] | 47.2 | 36.6 | 61.0 | 39.7 | 59.4 |
Residual activities were determined after incubation at various temperatures for 30 min using 10 mM L-lactate and 120 μM DCIP.
Effect of N-Terminal Purification Tag on Activity
Finally, the effect of the N-terminally added purification tag on the enzyme activity was investigated, as it was shown for AvLOx that modifications at the N-terminus may influence its activity.32 The purification tag contained a 6xHis-tag as well as a cleavage site for the HRV 3C protease, which leaves an additional N-terminal Gly and Pro after digestion while completely cleaving off the 6xHis-tag. Purified enzymes were incubated overnight with and without HRV 3C, and their specific activities with L-lactate and DCIP were determined (Figure 4). The obtained results varied considerably. While AvLOx and LhLDH showed an approximately 2-fold increase in activity after the digest, compared to the undigested sample, and GbLDH and EaLDH showed 18- and 4-fold increases, respectively, and LjLDH and SsLDH showed slight decreases in activity. We could thus confirm that the nature of the N-terminus may affect the activity of an LDH, yet the extent of this seems to be individual for each enzyme.
Figure 4.

Effect of an overnight digest of the enzymes’ purification tags on their specific activities. The digest was performed for 22 h at 20 °C using the protease HRV 3C. Activities were measured using 10 mM l-lactate and 120 μM DCIP.
Discussion
We show that, by employing SSNs, members of the HAOx family can be separated into functionally distinct clusters and established a first concise overall picture of this family as well as the relationship between its members. Expression of hitherto uncharacterized HAOx sequences showed that 41 out of 42 selected genes could be actively expressed in E. coli, indicating a good general expressability of HAOx family members. This is not surprising, considering that also HAOx sequences of higher eukaryotes were already successfully expressed in E. coli,20,21,37,44,45 yet it shows that even when investigating unexplored sequence space of the HAOx family, expression of active enzymes in E. coli is not a limiting factor.
An activity screening of these newly expressed HAOx members, using α-hydroxy acids of various chain lengths and structures, showed that L-lactate-specific enzymes are only found in one cluster, which was termed the “LOx cluster”. Most of the enzymes investigated from other clusters showed a broader reactivity with different α-hydroxy acids, and many did not utilize L-lactate as their preferred substrate but other medium- or long-chain α-hydroxy acids. This selection of sequences displaying a variety of substrate specificities can assist future projects that aim at identifying HAOx enzymes with a substrate preference that is different or broader than that of LOx.
The most prominent finding of our activity screening was that a large fraction of the HAOx enzymes that were studied is in fact dehydrogenases and not oxidases (thus showing negligible activity with oxygen), and many of these have been misannotated in databases. It is also noteworthy that even within the LOx cluster—a cluster of sequences that exclusively contains oxidases as previously characterized members—oxygen reactivity is limited to only one specific clade in its phylogenetic tree. We predict that the majority of sequences in the LOx cluster are in fact lactate dehydrogenases, as was also experimentally proven for several representative sequences in this work. Consequently, it is more appropriate to henceforth term the LOx cluster the lactate oxidoreductase cluster. One of the reasons why LDHs have been overlooked up until now is probably that most sequences of the LOx cluster were functionally annotated as oxidases because of their high sequence similarity to a small number of characterized LOx sequences, resulting in an overestimation of oxidases in the LOx sequence space and to numerous misannotations of LDHs as oxidases. Such large-scale misannotations are a common problem in biological databases,46−50 and automated functional enzyme annotations can often be misleading without experimental validation. Recently, Rembeza and Engqvist51 described functional misannotations in the HAOx family (EC 1.1.3.15; S-2-hydroxy acid oxidases) regarding the annotation of the utilized electron donor substrates, with an estimated 78% of the family being misannotated. It seems that in cases such as the HAOx family, with high structural and sequence conservation, but a diverse set of functions, automated computational annotations, as found on databases, are not sufficient for a precise functional prediction.
According to BRENDA (https://www.brenda-enzymes.org/), LDHs are classified as EC 1.1.1.27, NAD-dependent LDH, EC 1.1.2.3, LDH (cytochrome), or EC 1.1.5.B5, PQQ-dependent lactate dehydrogenase or quinone-dependent LDH. These enzymes employ different coenzymes/cofactors—NAD+, FMN plus heme b2 or PQQ—as their primary electron acceptors. LDH (cytochrome) or flavocytochrome b2 is a flavohemoprotein composed of two individual domains, one containing a heme group and one containing an FMN in a separate domain. No L-lactate-oxidizing dehydrogenase containing only FMN is described in BRENDA, and thus, soluble, FMN-containing LDHs are a novel class of enzymes that have not been attributed an EC number yet even though they seem to be widely distributed.
When studying six selected LDHs in their purified form in more detail, we could confirm that they are indeed true LDHs, showing oxygen reactivities well below 1% of their maximal dehydrogenase activity. Additionally, we found that, even within this small selection of sequences, other enzymatic properties are varying strongly compared to to-date characterized LOx sequences. Apparent Km values for L-lactate, for example, were ranging over almost two orders of magnitude, from 0.52 to 235 mM, and determination of oligomerization states, thermostabilities, and pH optima also revealed a surprisingly high variation between these enzymes. Note that here the specific activities of LDHs were determined for physiological conditions, relevant for biosensor applications (pH 7.4 and 10 mM L-lactate), and do not necessarily represent the maximum activities for some of the enzymes. Furthermore, we used the His-tagged enzyme preparations for characterization, and as we showed, this tag can affect kinetic properties considerably in certain instances. Furthermore, kinetic and biophysical data of GbLDH presented in this work should be considered with care. The enzyme showed repeatedly high standard deviations in kinetic measurements, an unusual SEC elution peak that could not be explained, and an unexpected high loss of activity after overnight incubation at 20 °C. To date, we hypothesize that GbLDH undergoes unspecific surface interactions, leading to reduced soluble enzyme upon surface contact and unusual chromatographic behavior. If or to what extent such unspecific surface interactions are influenced by the N-terminal purification tag still needs to be determined.
The discovery of this novel class of soluble FMN-dependent LDHs will have important implications for the development of L-lactate biosensors and second-generation L-lactate biosensors in particular. Their application can solve the inherent disadvantages of oxygen interference and NAD+ dependence in biosensors that are encountered when LOxs or NAD+-dependent LDHs are used. Recently, AvLOx was engineered to display a more favorable dehydrogenase activity, thereby creating a L-lactate-oxidizing enzyme which is insensitive to oxygen.30,36 The here described naturally occurring LDHs eliminate the need of such time-consuming engineering in the future and show that a number of sequences with widely different properties, which can be useful for biosensor applications, already exist. In addition, LDH can be a useful enzyme for other biocatalytic applications, such as the conversion of L-lactate to pyruvate. Pyruvate is of interest for various medical applications and as a food supplement52 and an enzyme-based biocatalytic approach to produce this compound is claimed to be advantageous of the currently employed chemical or fermentative routes toward pyruvate.53−55
Our approach highlights the value of utilizing sequence information of an enzyme family to identify new enzyme classes and functionalities. Furthermore, we showed that SSNs can provide helpful guidance in an often-confusing sequence space of enzymatic functions and annotations.
Methods
Sequence Similarity Networks
In July 2020, 13 experimentally characterized HAOx sequences (LOx from Aerococcus viridans Q44467, Lactococcus lactis A0A4Y3JPV3, Nostoc sp. PCC 7120 Q8Z0C8, and Chlamydomonas reinhardtii F8WQN2; LMO from Mycolicibacterium smegmatis P21795; GLO from Homo sapiens Q9UJM8, Spinacia oleracea P05414, and Cyanidioschyzon merolae M1VAT1; FCB2 from Saccharomyces cerevisiae P00175; MDH from Pseudomonas putida P20932; HAO from Rattus norvegicus Q07523; HMO from Amycolatopsis orientalis O52792; and mLDH from Pseudomonas stutzeri ADL63037) were used as queries for individual database searches on NCBI and UniProt using blastp and phmmer56 with maximum E-values of 1 × 10–79, respectively. Search results were combined, and duplicate sequence IDs were removed. This selection was further restricted to sequences of 250 to 650 amino acids in length, and all sequences containing the nonproteinogenic letter “X” were deleted. An SSN of the selection was calculated using the Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST) (https://efi.igb.illinois.edu/efi-est/).57 It was visualized and graphically edited in Cytoscape58 using an alignment score weighted prefuse force-directed-Layout. Taxonomic information was automatically retrieved from UniProt.
Calculation of Phylogenetic Trees
In order to guide the novel HAOx sequence selection process, approximate phylogenetic trees were calculated from sequence selections extracted from the SSN. The extraction was done at three different alignment score cutoffs to ascertain that the extracted clusters contain their complete set of sequences but are also fully separated from sequences of other clusters. The LOx cluster was extracted at an alignment score cutoff of 1 × 10–90, two clusters of actinobacteria, one cluster of fungi and one cluster of proteobacteria at 1 × 10–110, and a cluster of cyanobacteria at 1 × 10–125. Extracted sequence selections were aligned by MAFFT v7.40259 using the FFT-NS-2 method, manually sorted for sequences showing large indels, had sequence redundancy of 99% removed in Jalview v.2.11.1.4,60 and were realigned by MAFFT using the G-INS-i algorithm. The final alignments were trimmed for positions with ≥90% gaps by trimAl 1.2,61 and approximately maximum-likelihood dendrogram trees were calculated with FastTree 2.1.1062 using the Whelan and Goldman (WAG) substitution model63 and standard settings for increased accuracy (−spr 4 −mlacc 2 −slownni). The sequence selection of the LOx cluster was additionally sorted for sequences not showing a start-methionine, was expanded by 11 functionally annotated LOx and 8 GLO sequences, and was trimmed for positions with ≥99% gaps by trimAl. The resulting trees were either rooted on the GLO outgroup, in the case of the LOx cluster, or on midpoint, for all other selections. A more elaborate bootstrapped maximum likelihood tree for the LOx cluster was calculated from the same trimmed alignment described above, including the GLO outgroup, using RAxML-NG v.0.9.0.64 The preceding selection of the best-fit amino acid substitution model according to AIC was done with ModelTest-NG (downloaded Oct 30, 2019).65 The ML-tree was inferred using default settings from RAxML-NG and the best-fit model LG + I + G4 + F. Bootstrap trees were calculated until the average weighted Robinson–Foulds distance (avg WRF) dropped below a default cutoff of 3% for >990/1000 permutations (560 bootstraps).66
Sequence Analysis
Sequence selections and corresponding phylogenetic trees were visualized in MEGA 767 or Jalview. Signal peptides were predicted by the online-tool SignalP – 5.0,42 and sequence logos were created with WebLogo v.2.8.2.68
Plasmids and Genes
Genes coding for the selected novel HAOx enzymes and AvLOx were ordered codon-optimized for E. coli expression in a pET-21(+) vector from Twist Bioscience (South San Francisco, USA). The N-terminal purification tag (-GSS-HHHHHH-G-LEVLFQGP-) was added between the start-Met and the second amino acid according to the protocol of Gibson Assembly (New England Biolabs) using 146 and 110 bp overlaps at the 5′ and 3′ end, respectively. The native form of PaLCTO was cloned in a pNIC-CH vector as described previously.16 Predicted bacterial signal peptides of PauLDH and SsLDH were removed by amplifying their plasmids without the signal-peptide-coding region by PCR and re-ligating the linearized plasmid using the KLD Enzyme Mix (New England Biolabs). Plasmids were transformed into chemically competent E. coli BL21 (DE3) cells by heat-shock transformation at 42 °C. All plasmid modifications and transformations were verified by Sanger sequencing (Microsynth, Austria).
Buffers, Media, and Chemicals
PBS 11 mM, pH 7.4 with 137 mM NaCl and 3 mM KCl was used as the standard buffer for all experiments unless stated otherwise. Britton–Robinson universal buffer (BRB) contains 40 mM phosphoric, boric, and acetic acid. Cultivation of bacteria was routinely done in the Luria-Bertani (LB) medium (10 g/L peptone from casein, 5 g/L yeast extract, and 10 g/L NaCl) with 100 mg/L ampicillin. In the case of cultivating bacteria carrying the pNIC-CH plasmid for PaLCTO, ampicillin was replaced with 50 mg/L kanamycin. General medium components were purchased from Carl Roth; sodium L-lactate, ferrocenium hexafluorophosphate (FcPF6), isopropyl β-D-1-thiogalactopytanoside (IPTG), 2,6-dichlorophenol-indophenol sodium salt hydrate (DCIP), horseradish peroxidase, sodium glycolate, 1,4-BQ, R-2-hydroxybutyric acid, and S-2-hydroxybutyric acid from Sigma-Aldrich (Germany); S-2-hydroxyvaleric acid from BLD Pharmatech Ltd. (Shanghai); 2-hydroxypalmitic acid and 2-hydroxy-n-octanoic acid from TCI (Japan); (S)-2-hydroxybutyric acid and S-(+)-mandelic acid from Fluorochem Ltd. (United Kingdom); and 10-acetyl-3,7-dihydroxyphenoxazine (AmplexRed) from Chemodex (Switzerland).
haox Gene Expression
Expression of recombinant genes in E. coli BL21 (DE3) was done in baffled shake flasks at a scale of 40 or 250 mL expression medium. The LB-amp (LB-kan for PaLCTO) medium was inoculated with bacterial culture to an optical density at 600 nm (OD600) of 0.05 and was incubated at 37 °C and 180 rpm until cells reached an OD600 of 0.45–0.5, where expression was induced by 100 μM IPTG (250 μM in the case of 250 mL expressions). Induced cultures were incubated overnight at 20 °C and 180 rpm for 19 h. Cells were harvested by centrifugation at 4000 rpm for 20 min at 4 °C and washed once with 50 mM potassium phosphate buffer (PPB) pH 6.5. Washed cell pellets were stored at −20 °C prior to cell disruption.
Purification and Protein Concentration Measurements
Frozen cell pellets were thawed, resuspended in 50 mM PPB, 500 mM NaCl, 50 mM imidazole pH 6.5, and disrupted by 4–5 passages in a French press. Cell debris was removed by centrifugation (3000 rcf at 4 °C for 30 min), and the resulting supernatant was filtered with a 0.22 μm membrane filter and loaded onto 2 × 5 mL IMAC HisTrap FF columns (Cytiva, USA) using an Äkta FPLC system (GE Healthcare, USA). His-tagged proteins were eluted by a linear imidazole gradient (50–750 mM) in 50 mM PPB, 500 mM NaCl, pH 6.5, and fractions were pooled according to activity and elution peaks measured at 280 and 450 nm. Pooled fractions were concentrated in Amicon centrifugal filters (MWCO 10 kDa), rebuffered to 11 mM PBS, pH 7.4, and stored at 4 °C. Homogeneity of the enzymes was assessed by SDS-PAGE, and protein concentrations were calculated from their absorbance at 280 nm assuming theoretical extinction coefficients as determined by the ExPASy tool ProtParam69 from the amino acid sequence (48,360, 41,830, 24,870, 29,910, 28,420, 25,900, and 51,340 M–1 cm–1 for LjLDH, LhLDH, SsLDH, GbLDH, EaLDH, PaLCTO, and AvLOx, respectively). Purified PaLCTO was produced as described previously16 using E. coli BL21 (DE3) expression, His-tag purification followed by cleavage of the tag by TEV protease digestion and size exclusion chromatography.
Enzymatic Activity Measurements
For the activity screening of novel HAOx members, frozen cell pellets were thawed and resuspended in PPB pH 6.5 to 0.16 g wet cell pellet/mL and cells were disrupted by sonication (Sonopuls HD60; Bandelin, Berlin) on ice using 6 rounds at 50% power and 30% cycles for 30 s. Cell debris was removed by ultracentrifugation (25,000 × g at 4 °C for 20 min), and the resulting supernatants were directly utilized as crude enzymes. All other activity measurements were conducted using purified enzymes. Spectrophotometric enzyme activity assays were recorded at least in triplicate in 96-well microtiter plates at 30 °C using an EnSpire multimode plate reader (PerkinElmer) or Infinite M Quant plate reader (Tecan). Volumetric activities were calculated from linear correlations of changes in absorption over time (ΔAbs/Δt) at the monitored wavelength. In the activity screening of novel HAOx members, a measurement was considered active and quantifiable if linear regression showed an R2 coefficient ≥ 0.98 and a ΔAbs/Δt value 10 times higher than the standard deviation of the respective blank (using empty vector expression). All measured values were corrected for unspecific background reactions determined from blinds for each substrate.
Activity assays contained the electron donor substrates L-lactate, glycolate, S-2-hydroxybutyric acid, R-2-hydroxybutyric acid, S-2-hydroxyvaleric acid, 2-hydroxyoctanoic acid, and S-mandelic acid at 10 mM concentration dissolved in buffer and 2-hydroxypalmitic acid dissolved in 96% ethanol at 0.5 mM concentration unless stated otherwise. Oxidase activity was monitored using a peroxidase-coupled reaction containing 7.1 U/mL horseradish peroxidase (181 U/mg; Sigma) and 0.05 mM AmplexRed (resorufin: ε560 nm = 54.0 mM–1 cm–1). Oxygen was present at ambient concentrations of ∼250 μM. Dehydrogenase activities were measured by direct dye-mediated assays containing 120 μM (300 μM for screening of substrates) DCIP (ε520 nm = 6.8 mM–1 cm–1 or ε600 nm = 8.98 mM–1 cm–1), 500 μM 1,4-BQ (ε290 nm = 2.24 mM–1 cm–1), or 160 μM FcPF6 (ε300 nm = 4.3 mM–1 cm–1). Measurements of the enzyme activity screening, relative substrate specificities, and specific activities for FcPF6 and 1,4-BQ were conducted in 50 mM PPB pH 6.5. One unit of enzyme activity was defined as the amount of enzyme that catalyzes the oxidation of 1 μmol α-hydroxy acid per minute at 30 °C.
Apparent steady-state kinetic constants were determined by using 11 different concentrations of L-lactate from 0.125 to 64 mM (4–500 mM for PaLCTO) with DCIP at a constant concentration of 120 μM and fitting the Michaelis–Menten model (v = (vmax*[S]/(Km + [S])) to the observed data using iterative least-square regression fitting with the Microsoft Excel Solver plugin. Turnover rates are calculated based on the monomeric masses of the respective enzymes. Assessing the influence of pH on the L-lactate oxidizing activity with O2 and DCIP was done in 40 mM BRB by varying the pH from 4.5 to 10 in steps of 0.5.
Size-Exclusion Chromatography-Static Light Scattering (SEC-SLS) Analysis
SEC-SLS analysis was conducted using an OMNISEC multi-detector GPC/SEC (Malvern Panalytical, Worcestershire, UK) equipped with a refractive index, right angle light scattering, and UV/vis diode array detector. Proteins were separated on a Superdex S200 increase 10/300 GL column (Cytiva) at 25 °C using PBS at a flowrate of 0.5 mL/min. The instrument was calibrated using commercial BSA (2 mg/mL) (Thermo Scientific Pierce), and samples were applied at 2 mg/mL with sample injection volumes between 20 and 40 μL. Sample solutions were filtered through 0.45 μm pore size, hydrophilic PVDF centrifugal filters (Millipore) prior to analysis. The protein concentration was determined by using a refractive index detector and a dn/dc of 0.185. Data processing and calculation of molecular masses were done using the OMNISEC v.11.31 software.
Thermostability Measurements
Enzyme samples were diluted to a concentration of 1 mg/mL and incubated in duplicates for 30 min in a temperature range from 30 to 60 °C (43 to 73 °C in the case of SsLDH and EaLDH). As a reference, one additional duplicate was incubated at 4 °C. All samples were cooled on ice for 15 min and centrifuged before residual activities with L-lactate and DCIP were measured in duplicates. Estimation of the thermal inactivation temperature (T50) was done by an iterative least-square regression sigmoidal curve fit (min + (max – min)/(1 + 10(n*(log10(°C) – log10(EC50))))) using the Microsoft Excel Solver plugin.
HRV 3C Protease Digest
Purification tag cleavage was conducted at a small scale using 30 μg of enzyme and 1 μg of HRV 3C protease (in-house production) at 20 °C for 22 h. Determination of activity was conducted in quadruplicates using L-lactate and DCIP. Blinds were incubated with buffer instead of HRV 3C.
Acknowledgments
This work was supported by the Austrian Research Promotion Agency (FFG) through the BRIDGE Young Scientists project ROxanE. We want to thank Georg Mlynek from the BOKU Core Facility of Biomolecular & Cellular Analysis for the repeated help he provided regarding SEC-SLS measurements, even if it meant working until late at night. We also thank Erik Breslmayr for setting up our local phylogeny-server and for all his energy to make it more user-friendly. Your efforts will never be forgotten. A special thanks also goes to Lari Lehtiö and Mirko Maksimainen from the University of Oulu in Finland for providing us with purified PaLCTO and its expression plasmid.
Glossary
Abbreviations
- 1,4-BQ
1,4-benzoquinone
- AvLOx
lactate oxidase from Aerococcus viridans
- BRB
Britton–Robinson buffer
- DCIP
dichlorophenol-indophenol
- EaLDH
lactate dehydrogenase from Enterococcus avium
- FCB2
flavocytochrome b2
- FcPF6
ferrocenium hexafluorophosphate
- GbLDH
lactate dehydrogenase from Gilliamella bombicola
- GDH
glucose dehydrogenase
- GLDH
glycolate dehydrogenase
- GLO
glycolate oxidase
- HAO
long- or medium-chain 2-hydroxy acid oxidase
- HAOx
α-hydroxy acid oxidoreductase
- HMO
4-hydroxymandelate oxidase
- LDH
lactate dehydrogenase
- LhLDH
lactate dehydrogenase from Lactobacillus helsingborgensis
- LjLDH
lactate dehydrogenase from Lactobacillus johnsonii
- LMO
L-lactate monooxygenase
- LOx
lactate oxidase
- MDH
mandelate dehydrogenase
- mLDH
membrane bound L-lactate dehydrogenase
- PaLCTO
lactate dehydrogenase from Pediococcus acidilactici
- PBS
phosphate-buffered saline
- PPB
potassium phosphate buffer
- SEC-SLS
size exclusion chromatography static light scattering
- SsLDH
lactate dehydrogenase from Shigella sp. FC1655
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c05257.
Protein sequence IDs, substrate structures, alignment overview, sequence logo of active site residues, purification table, UV–vis spectra, SEC-SLS elution spectra, pH profiles, buffer comparisons, and thermal inactivation curves (PDF)
The authors declare the following competing financial interest(s): As a BRIDGE Young Scientists project, the project was conducted in cooperation with a company (DirectSens Biosensors GmbH). PH and BE are employees of DirectSens Biosensors GmbH, which deals in the development of enzyme based biosensors.
Supplementary Material
References
- Rattu G.; Khansili N.; Maurya V. K.; Krishna P. M. Lactate detection sensors for food, clinical and biological applications: a review. Environ. Chem. Lett. 2021, 19, 1135–1152. 10.1007/s10311-020-01106-6. [DOI] [Google Scholar]
- Kucherenko I. S.; Topolnikova Y. V.; Soldatkin O. O. Advances in the biosensors for lactate and pyruvate detection for medical applications: A review. TrAC, Trends Anal. Chem. 2019, 110, 160–172. 10.1016/j.trac.2018.11.004. [DOI] [Google Scholar]
- Nikolaus N.; Strehlitz B. Amperometric lactate biosensors and their application in (sports) medicine, for life quality and wellbeing. Microchim. Acta 2008, 160, 15–55. 10.1007/s00604-007-0834-8. [DOI] [Google Scholar]
- Mizock B. A.; Falk J. L. Lactic acidosis. West J. Med. 1992, 20, 80–93. [DOI] [PubMed] [Google Scholar]
- Gómez H.; Mizock B. A.. Critical Care Nephrology, 3rd ed.; Elsevier Inc., 2019, [Google Scholar]
- Kruse O.; Grunnet N.; Barfod C. Blood lactate as a predictor for in-hospital mortality in patients admitted acutely to hospital: A systematic review. Scand. J. Trauma, Resusc. Emerg. Med. 2011, 19, 74. 10.1186/1757-7241-19-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Xu X. Lactate clearance is a useful biomarker for the prediction of all-cause mortality in critically Ill patients: A systematic review and meta-analysis. Crit. Care Med. 2014, 42, 2118–2125. 10.1097/CCM.0000000000000405. [DOI] [PubMed] [Google Scholar]
- Rathee K.; Dhull V.; Dhull R.; Singh S. Biosensors based on electrochemical lactate detection: A comprehensive review. Biochem. Biophys. Rep. 2016, 5, 35–54. 10.1016/j.bbrep.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löllgen H.; Leyk D. Ergometrische Belastungsuntersuchungen in der Sportmedizin. Dtsch. Arztebl. Int. 2018, 115, 409–416. 10.3238/arztebl.2018.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S.; Kildegaard H. F.; Andersen M. R. Impact of CHO Metabolism on Cell Growth and Protein Production: An Overview of Toxic and Inhibiting Metabolites and Nutrients. Biotechnol. J. 2018, 13, e1700499 10.1002/biot.201700499. [DOI] [PubMed] [Google Scholar]
- Ritacco F. V.; Wu Y.; Khetan A. Cell culture media for recombinant protein expression in Chinese hamster ovary (CHO) cells: History, key components, and optimization strategies. Biotechnol. Prog. 2018, 34, 1407–1426. 10.1002/btpr.2706. [DOI] [PubMed] [Google Scholar]
- Torres M.; Altamirano C.; Dickson A. J. Process and metabolic engineering perspectives of lactate production in mammalian cell cultures. Curr. Opin. Chem. Eng. 2018, 22, 184–190. 10.1016/j.coche.2018.10.004. [DOI] [Google Scholar]
- Maeda-Yorita K.; Aki K.; Sagai H.; Misaki H.; Massey V. L-lactate oxidase and L-lactate monooxygenase: Mechanistic variations on a common structural theme. Biochimie 1995, 77, 631–642. 10.1016/0300-9084(96)88178-8. [DOI] [PubMed] [Google Scholar]
- Stoisser T.; Rainer D.; Leitgeb S.; Wilson D. K.; Nidetzky B. The Ala 95-to-Gly substitution in Aerococcus viridans l -lactate oxidase revisited - Structural consequences at the catalytic site and effect on reactivity with O2 and other electron acceptors. FEBS J. 2015, 282, 562–578. 10.1111/febs.13162. [DOI] [PubMed] [Google Scholar]
- Leiros I.; Wang E.; Rasmussen T.; Oksanen E.; Repo H.; Petersen B. S.; Heikinheimo P.; Hough E. The 2.1 Å structure of Aerococcus viridans L-lactate oxidase (LOX). Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2006, 62, 1185–1190. 10.1107/S1744309106044678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok Y.; Maksimainen M. M.; Kallio T.; Kilpeläinen P.; Lehtiö L. FMN-dependent oligomerization of putative lactate oxidase from Pediococcus acidilactici. PLoS One 2020, 15, e0223870 10.1371/journal.pone.0223870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoisser T.; Klimacek M.; Wilson D. K.; Nidetzky B. Speeding up the product release: A second-sphere contribution from Tyr 191 to the reactivity of l- lactate oxidase revealed in crystallographic and kinetic studies of site-directed variants. FEBS J. 2015, 282, 4130–4140. 10.1111/febs.13409. [DOI] [PubMed] [Google Scholar]
- Stoisser T.; Brunsteiner M.; Wilson D. K.; Nidetzky B. Conformational flexibility related to enzyme activity: Evidence for a dynamic active-site gatekeeper function of Tyr 215 in Aerococcus viridans lactate oxidase. Sci. Rep. 2016, 6, 27892. 10.1038/srep27892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist Y.; Brändén C. I. The active site of spinach glycolate oxidase. J. Biol. Chem. 1989, 264, 3624–3628. 10.1016/s0021-9258(18)94112-8. [DOI] [PubMed] [Google Scholar]
- Belmouden A.; Lê K. H. D.; Lederer F.; Garchon H. Molecular cloning and nucleotide sequence of cDNA encoding rat kidney long-chain l-2-hydroxy acid oxidase: Expression of the catalytically active recombinant protein as a chimaera. Eur. J. Biochem. 1993, 214, 17–25. 10.1111/j.1432-1033.1993.tb17891.x. [DOI] [PubMed] [Google Scholar]
- Jones J. M.; Morrell J. C.; Gould S. J. Identification and characterization of HAOX1, HAOX2, and HAOX3, three human peroxisomal 2-hydroxy acid oxidases. J. Biol. Chem. 2000, 275, 12590–12597. 10.1074/jbc.275.17.12590. [DOI] [PubMed] [Google Scholar]
- Appleby C. A.; Morton R. K. Crystalline cytochrome b2 and lactic dehydrogenase of yeast. Nature 1954, 173, 749–752. 10.1038/173749a0. [DOI] [PubMed] [Google Scholar]
- Jacq C.; Lederer F. Cytochrome b2 from Bakers’ Yeast (L-Lactate Dehydrogenase): A Double-Headed Enzyme. Eur. J. Biochem. 1974, 41, 311–320. 10.1111/j.1432-1033.1974.tb03271.x. [DOI] [PubMed] [Google Scholar]
- Sullivan P. A. Crystallization and properties of L-lactate oxidase from Mycobacterium smegmatis. Biochem. J. 1968, 110, 363–371. 10.1042/bj1100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou A. Y.; Ransom S. C.; Gerlt J. A.; Buechter D. D.; Babbitt P. C.; Kenyon G. L. Mandelate Pathway of Pseudomonas putida: Sequence Relationships Involving Mandelate Racemase, (S)-Mandelate Dehydrogenase, and Benzoylformate Decarboxylase and Expression of Benzoylformate Decarboxylase in Escherichia coli. Biochemistry 1990, 29, 9856–9862. 10.1021/bi00494a015. [DOI] [PubMed] [Google Scholar]
- Yeh H. W.; Lin K. H.; Lyu S. Y.; Li Y. S.; Huang C. M.; Wang Y. L.; Shih H. W.; Hsu N. S.; Wu C. J.; Li T. L. Biochemical and structural explorations of a-hydroxyacid oxidases reveal a four-electron oxidative decarboxylation reaction. Acta Crystallogr., Sect. D: Struct. Biol. 2019, 75, 733–742. 10.1107/S2059798319009574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J.; Srikanth N. V.; Hüdig M.; Poschmann G.; Lercher M. J.; Maurino V. G. The ancestors of diatoms evolved a unique mitochondrial dehydrogenase to oxidize photorespiratory glycolate. Photosynth. Res. 2017, 132, 183–196. 10.1007/s11120-017-0355-1. [DOI] [PubMed] [Google Scholar]
- Dong J. M.; Taylor J. S.; Latour D. J.; Iuchi S.; Lin E. C. C. Three overlapping lct genes involved in L-lactate utilization by Escherichia coli. J. Bacteriol. 1993, 175, 6671–6678. 10.1128/jb.175.20.6671-6678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C.; Jiang T.; Dou P.; Ma C.; Li L.; Kong J.; Xu P. NAD-independent l-lactate dehydrogenase is required for l-lactate utilization in pseudomonas stutzeri SDM. PLoS One 2012, 7, e36519 10.1371/journal.pone.0036519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraka K.; Kojima K.; Tsugawa W.; Asano R.; Ikebukuro K.; Sode K. Rational engineering of Aerococcus viridans L-lactate oxidase for the mediator modification to achieve quasi-direct electron transfer type lactate sensor. Biosens. Bioelectron. 2020, 111974 10.1016/j.bios.2019.111974. [DOI] [PubMed] [Google Scholar]
- Hiraka K.; Tsugawa W.; Asano R.; Yokus A. M.; Ikebukuro K.; Daniele A. M.; Sode K. Rational design of direct electron transfer type L-lactate dehydrogenase for the development of multiplexed biosensor. Biosens. Bioelectron. 2021, 176, 112933 10.1016/j.bios.2020.112933. [DOI] [PubMed] [Google Scholar]
- Taurino I.; Reiss R.; Richter M.; Fairhead M.; Thöny-Meyer L.; De Micheli G.; Carrara S. Comparative study of three lactate oxidases from Aerococcus viridans for biosensing applications. Electrochim. Acta 2013, 93, 72–79. 10.1016/j.electacta.2013.01.080. [DOI] [Google Scholar]
- Goodwin M. L.; Harris J. E.; Hernández A.; Gladden L. B. Blood lactate measurements and analysis during exercise: A guide for clinicians. J. Diabetes Sci. Technol. 2007, 1, 558–569. 10.1177/193229680700100414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsubayashi K.; Suzuki M.; Tamiya E.; Karube I. Analysis of metabolites in sweat as a measure of physical condition The increasing desire to naturally enhance the performance of sportsmen and athletes through. Anal. Chiica Acta 1994, 289, 27–34. 10.1016/0003-2670(94)80004-9. [DOI] [Google Scholar]
- Cardosi M.; Liu Z.. Amperometric Glucose Sensors for Whole Blood Measurement Based on Dehydrogenase Enzymes. In Dehydrogenases; InTech, 2012. [Google Scholar]
- Hiraka K.; Kojima K.; Lin C. E.; Tsugawa W.; Asano R.; La Belle T. J.; Sode K. Minimizing the effects of oxygen interference on L-lactate sensors by a single amino acid mutation in Aerococcus viridans L-lactate oxidase. Biosens. Bioelectron. 2018, 103, 163–170. 10.1016/j.bios.2017.12.018. [DOI] [PubMed] [Google Scholar]
- Hackenberg C.; Kern R.; Hüge J.; Stal J. L.; Tsuji Y.; Kopka J.; Shiraiwa Y.; Bauwe H.; Hagemann M. Cyanobacterial lactate oxidases serve as essential partners in N2 fixation and evolved into photorespiratory glycolate oxidases in plants. Plant Cell 2011, 23, 2978–2990. 10.1105/tpc.111.088070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean K. M.; Karplus P. A. Structure and role for active site lid of lactate monooxygenase from Mycobacterium smegmatis. Protein Sci. 2019, 28, 135–149. 10.1002/pro.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P.; Chirat I.; Lederer F. Rat Kidney L-2-hydroxyacid Oxidase: Structural and Mechanistic Comparison With Flavocytochrome B2from Baker’S Yeast. Biochemistry 1988, 27, 7365–7371. 10.1021/bi00419a029. [DOI] [PubMed] [Google Scholar]
- Esser C.; Kuhn A.; Groth G.; Lercher M. J.; Maurino V. G. Plant and animal glycolate oxidases have a common eukaryotic ancestor and convergently duplicated to evolve long-chain 2-hydroxy acid oxidases. Mol. Biol. Evol. 2014, 31, 1089–1101. 10.1093/molbev/msu041. [DOI] [PubMed] [Google Scholar]
- Cao Y.; Han S.; Yu L.; Qian H.; Chen J. Z. MD and QM/MM studies on long-chain l -α-hydroxy acid oxidase: Substrate binding features and oxidation mechanism. J. Phys. Chem. B 2014, 118, 5406–5417. 10.1021/jp5022399. [DOI] [PubMed] [Google Scholar]
- Almagro Armenteros J. J.; Tsirigos K. D.; Sønderby C. K.; Nordahl Petersen T.; Winther O.; Brunak S.; von Heijne G.; Nielsen H. Signal P 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- Umena Y.; Yorita K.; Matsuoka T.; Kita A.; Fukui K.; Morimoto Y. The crystal structure of l-lactate oxidase from Aerococcus viridans at 2.1 Å resolution reveals the mechanism of strict substrate recognition. Biochem. Biophys. Res. Commun. 2006, 350, 249–256. 10.1016/j.bbrc.2006.09.025. [DOI] [PubMed] [Google Scholar]
- Kohler S. A.; Menotti E.; Kühn L. C. Molecular cloning of mouse glycolate oxidase: High evolutionary conservation and presence or an iron-responsive element-like sequence in the mRNA. J. Biol. Chem. 1999, 274, 2401–2407. 10.1074/jbc.274.4.2401. [DOI] [PubMed] [Google Scholar]
- Engqvist M. K. M.; Schmitz J.; Gertzmann A.; Florian A.; Jaspert N.; Arif M.; Balazadeh S.; Mueller-Roeber B.; Fernie A. R.; Maurino V. G. GLYCOLATE OXIDASE3, a glycolate oxidase homolog of yeast L-lactate cytochrome c oxidoreductase, supports L-lactate oxidation in roots of arabidopsis. Plant Physiol. 2015, 169, 1042–1061. 10.1104/pp.15.01003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoes A. M.; Brown S. D.; Dodevski I.; Babbitt P. C. Annotation error in public databases: Misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol. 2009, 5, e1000605 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P.; Clark W. T.; Oron T. R.; Schnoes A. M.; Wittkop T.; Sokolov A.; Graim K.; Funk C.; Verspoor K.; Ben-Hur A.; et al. A large-scale evaluation of computational protein function prediction. Nat. Methods 2013, 10, 221–227. 10.1038/nmeth.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M. J.; Lord P. On patterns and re-use in bioinformatics databases. Bioinformatics 2017, 33, 2731–2736. 10.1093/bioinformatics/btx310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R. Protein Function Prediction: Problems and Pitfalls. Curr. Protoc. Bioinf. 2015, 51, 4.12.1–4.12.8. 10.1002/0471250953.bi0412s51. [DOI] [PubMed] [Google Scholar]
- Furnham N.; Garavelli J. S.; Apweiler R.; Thornton J. M. Missing in action: Enzyme functional annotations in biological databases. Nat. Chem. Biol. 2009, 5, 521–525. 10.1038/nchembio0809-521. [DOI] [PubMed] [Google Scholar]
- Rembeza E.; Engqvist M. K. M. Experimental and computational investigation of enzyme functional annotations uncovers misannotation in the EC 1.1.3.15 enzyme class. PLoS Comput. Biol. 2021, 17, e1009446 10.1371/journal.pcbi.1009446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo F.; Liu J.; Chen J.; Li X.; Solem C.; Jensen P. R. Efficient Production of Pyruvate Using Metabolically Engineered Lactococcus lactis. Front. Bioeng. Biotechnol. 2021, 8, 611701 10.3389/fbioe.2020.611701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P.; Qiu J.; Gao C.; Ma C. Biotechnological routes to pyruvate production. J. Biosci. Bioeng. 2008, 105, 169–175. 10.1263/jbb.105.169. [DOI] [PubMed] [Google Scholar]
- Ma C. Q.; Xu P.; Qiu J. H.; Zhang Z. J.; Wang K. W.; Wang M.; Zhang Y. N. An enzymatic route to produce pyruvate from lactate. Appl. Microbiol. Biotechnol. 2004, 66, 34–39. 10.1007/s00253-004-1646-y. [DOI] [PubMed] [Google Scholar]
- Ogawa J.; Soong C. L.; Ito M.; Shimizu S. Enzymatic production of pyruvate from fumarate - An application of microbial cyclic-imide-transforming pathway. J. Mol. Catal. B: Enzym. 2001, 11, 355–359. 10.1016/S1381-1177(00)00024-2. [DOI] [Google Scholar]
- Potter S. C.; Luciani A.; Eddy S. R.; Park Y.; Lopez R.; Finn R. D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallot R.; Oberg N.; Gerlt J. A. The EFI Web Resource for Genomic Enzymology Tools: Leveraging Protein, Genome, and Metagenome Databases to Discover Novel Enzymes and Metabolic Pathways. Biochemistry 2019, 58, 4169–4182. 10.1021/acs.biochem.9b00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K.; Standley D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A. M.; Procter J. B.; Martin D. M. A. A.; Clamp M.; Barton G. J. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S.; Silla-Martínez J. M.; Gabaldón T. trim Al: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N.; Dehal P. S.; Arkin A. P. Fast Tree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S.; Goldman N. A General Empirical Model of Protein Evolution Derived from Multiple Protein Families Using a Maximum-Likelihood Approach. Mol. Biol. Evol. 2001, 18, 691–699. 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Kozlov A. M.; Darriba D.; Flouri T.; Morel B.; Stamatakis A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di D.; Posada D.; Kozlov A. M.; Stamatakis A.; Morel B.; Flouri T. Model Test-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattengale N. D.; Alipour M.; Bininda-Emonds O. R. P.; Moret B. M. E.; Stamatakis A. How many bootstrap replicates are necessary?. J. Comput. Biol. 2010, 17, 337–354. 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Stecher G.; Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G. E.; Hon G.; Chandonia J. M.; Brenner S. E. Web Logo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E.; Hoogland C.; Gattiker A.; Duvaud S.; Wilkins M. R.; Appel R. D.; Bairoch A.. Protein Identification and Analysis Tools on the ExPASy Server. Proteomics Protoc. Handb., 2005; pp 571–608.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.