SUMMARY
Vertebrate brain development depends on a complex program of cell proliferation and migration. Post-mitotic neuronal migration in the developing cerebral cortex involves Nesprin-2, which recruits cytoplasmic dynein, kinesin, and actin to the nuclear envelope (NE) in other cell types. However, the relative importance of these interactions in neurons has remained poorly understood. To address these issues we performed in utero electroporation into the developing rat brain to interfere with Nesprin-2 function. We find that a ~100 kDa “mini” form of the ~800 kDa Nesprin-2 protein, which binds dynein and kinesin, is sufficient, remarkably, to support neuronal migration. In contrast to dynein’s role in forward nuclear migration in these cells, we find that kinesin-1 inhibition accelerates neuronal migration, suggesting a novel role for the opposite-directed motor proteins in regulating migration velocity. In contrast to studies in fibroblasts, the actin-binding domain of Nesprin-2 was dispensable for neuronal migration. We find further that, surprisingly, the motor proteins interact with Nesprin-2 through the dynein/kinesin “adaptor” BicD2, both in neurons and in non-mitotic fibroblasts. Furthermore, mutation of the Nesprin-2 LEWD sequence, implicated in nuclear envelope kinesin recruitment in other systems, interferes with BicD2 binding. Although disruption of the Nesprin-2/BicD2 interaction severely inhibited nuclear movement, centrosome advance proceeded unimpeded, supporting an independent mechanism for centrosome advance. Our data together implicate Nesprin-2 as a novel and fundamentally important form of BicD2 cargo and help explain BicD2’s role in neuronal migration and human disease.
eTOC Blurb
The mechanisms by which microtubule motor proteins contribute to neuronal migration remain incompletely understood. Here, Gonçalves et al. find that the nucleus-cytoskeleton linker protein Nesprin-2 recruits dynein via its adaptor protein BicD2 for neuronal nuclear migration. Kinesin-1 counteracts dynein forces allowing fine tuning of migration.
Graphical Abstract
INTRODUCTION
Cell migration is critical to the formation and function of diverse tissues and organs. In the Ventricular Zone (VZ) of the developing brain, neurons are generated from Radial Glial Progenitor cells (RGPs) [1], which divide and proliferate at the Ventricular Surface (VS). Between divisions, the RGP nucleus undergoes cell cycle-dependent nuclear oscillations, referred to as Interkinetic Nuclear Migration (INM). Whereas symmetric divisions expand the progenitor pool, asymmetric divisions produce intermediate progenitors, and, in turn, neurons [2]. The latter undergo long-distance directed migration toward the cortical plate (CP), where they establish the laminar neuronal arrangement characteristic of the cerebral cortex. In the intermediate zone (IZ), the neurons extend a trailing axon and a thicker leading migratory process, which is directed toward the CP. As the neurons migrate, the centrosome advances into the leading process followed by the nucleus, in a coordinated saltatory fashion [3–6].
Work over the past several years has revealed important roles for cytoskeletal motor proteins in these behaviors and other aspects of brain development [7]. Importantly, human mutations in motor protein genes can lead to severe neurodevelopmental pathologies, such as lissencephaly and microcephaly [8–12]. Our laboratory found that cytoplasmic dynein-1 (hereafter “dynein”) is recruited to the G2 nuclear envelope (NE) by successive RanBP2-BicD2- and Nup133-CENP-F-Nde1-mediated mechanisms. Each is activated by Cdk1 [13,14] and together they are responsible for the apical migration of the RGP nucleus back to the ventricular surface for the next mitotic division.
Dynein also plays important roles in subsequent post-mitotic neuronal migration. In these cells, dynein is required for the multipolar-to-bipolar morphological transition of the neurons which takes place in the IZ, and for subsequent directed neuronal migration to the CP [12,15,16]. Nuclear transport at this stage requires both dynein and actomyosin [4–6,17,18]. Kinesin-1 has been reported to contribute to rotational nuclear behavior in migrating cerebellar neurons in vitro [19], though the purpose of this activity is not known. The role of kinesin-1 in cortical neuronal migration has remained elusive.
Neuronal migration at this stage also involves the LINC complex [20], comprised of the SUN (Sad1 and UNC-84) proteins at the inner nuclear membrane, and the associated KASH (Klarsicht, ANC-1, Syne Homology) proteins, such as the Nesprins at the outer nuclear membrane. The latter interact with microtubule motors and actin filaments for proper nuclear positioning [21–23]. Nesprin 2 has also been found to play an important role in the more complex, but less well-understood cellular behavior involved in brain development. This protein was implicated on the basis of lamination and migration defects observed in the brains of Syne2-null mice, which lack the gene encoding Nesprin-2 [24]. Our lab subsequently found that expression of the inhibitory Nesprin-2 KASH domain in post-mitotic rat cortical neurons severely inhibited post-mitotic neuronal migration [14]. However, the mechanisms for Nesprin control of neuronal migration have remained poorly understood.
We report here that Nesprin-2 contributes to nuclear displacement during neuronal migration by recruiting microtubule motors to the NE, through the well-known adaptor BicD2. Finally, we find kinesin-1 to retard nuclear migration, identifying a novel form of motor protein cooperativity with implications for tuning neuronal migration.
RESULTS
Roles of Nesprin-2 functional domains in neuronal migration.
To test the distribution of Nesprin-2 in migrating rat brain neurons, we performed anti-Nesprin-2 immunostaining [23]. We observed clear perinuclear Nesprin-2 decoration in cells throughout the neocortex (Figure 1 A; Figure S1 A), similar to results obtained in mouse [24].
Figure 1. Roles of Nesprin-2 functional domains in neuronal migration.
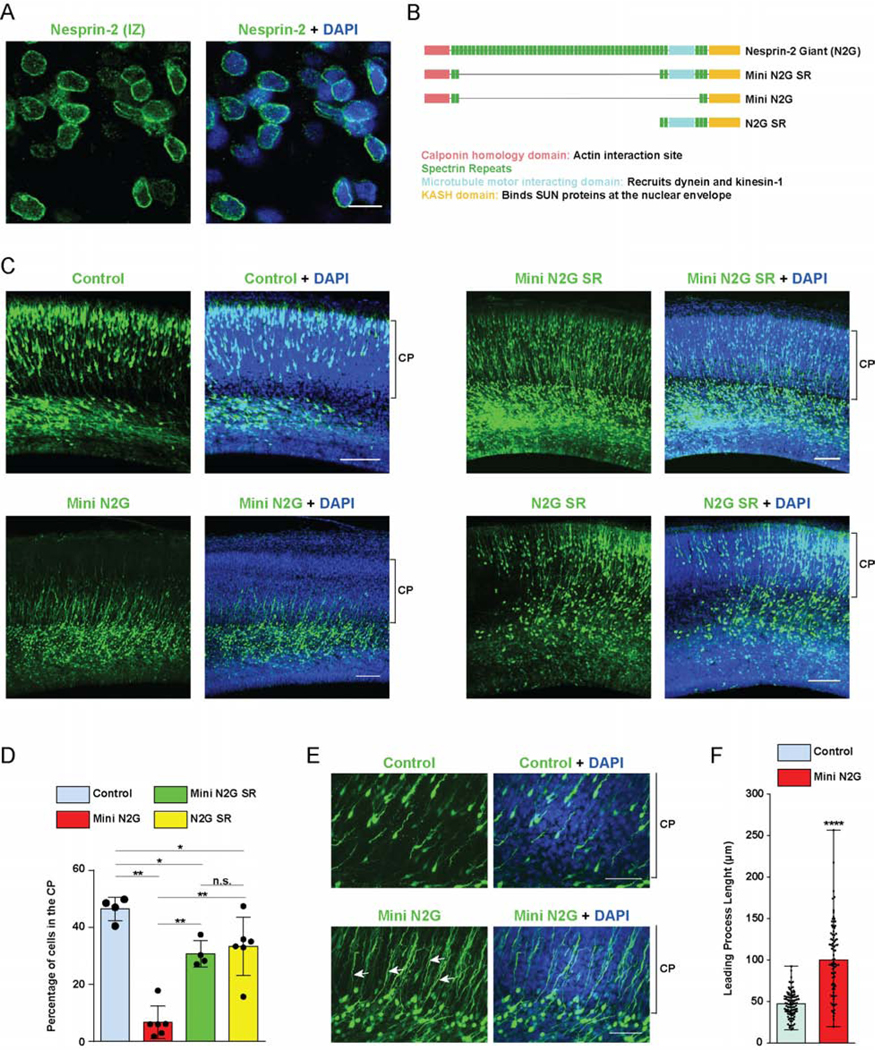
(A) E20 rat brain slices were immunostained for endogenous Nesprin-2 and with DAPI (blue). A representative image from the Intermediate Zone (IZ) where post-mitotic neurons start to migrate is shown. Clear Nesprin-2 decoration of the nuclear envelope (NE) is observed. (B) Diagrammatic representation of the Nesprin-2 Giant (N2G) isoform, and the engineered fragments designed for actin- (Mini N2G) or, microtubule motor- (N2G SR)-binding, or both (Mini N2G SR) [22]. (C-F) E16 rat brain was electroporated with IRES vectors expressing GFP alone or with the Mini N2G SR, Mini N2G or N2G SR constructs. Slices from fixed brains were imaged 4 days following electroporation at E20. (C) Representative images of the neocortex with electroporated cells in green. Brackets show Cortical Plate (CP) margins. (D) Proportion of cells reaching the CP. The Mini N2G expression substantially impaired neuronal migration, whereas Mini N2G SR and N2G SR caused only moderate inhibition. (E) Magnified images of the upper IZ and lower CP of control- and Mini N2G-electroporated brains. Mini N2G-expressing cells have markedly elongated leading processes (arrows). (F) Quantification of leading process length in electroporated neurons. Data presented as scatter dot plot with bar representing mean±s.d in D, and with bar representing median with range in F. Mann Whitney test for non-parametric distributions was used in D and unpaired T-Test with Welch’s correction in F (*P<0.05; **P<0.01; ****P<0.0001; n.s. non-significant). Data in D include at least of 967 electroporated cells from at least 4 embryos per condition. Data in F include at least 83 neurons from at least 3 embryos, per condition. A scale bar, 10μm. C scale bar, 100μm. E scale bar, 50μm. Related to Figure S1.
Because of the large size of the “giant” Nesprin-2 isoform (N2G ~800kDa), recent efforts to define its functions have involved the expression of shortened Nesprin functional domains, which retained physiological activity in cultured nonneuronal cells [22,23,25]. To address the roles of the Nesprin-2 cytoskeleton-interacting domains in neuronal migration, we used three chimeric cDNA constructs which allow for functional domain mapping (Figure 1 B). This approach was preferable in the developing brain because of the slow turnover of the large Nesprin-2 polypeptide precludes proper use of Nesprin-2 RNAi. The “Mini N2G SR” construct includes sequences encoding the actin-binding Calponin Homology (CH) domain and the microtubule motor-binding portion, spectrin repeats (SR) 52 to 56. “Mini N2G” encodes the CH domain alone, whereas “N2G SR”, the microtubule motor-binding region alone. All three constructs contain the KASH domain, which binds SUN proteins in the NE and displaces endogenous Nesprins (Figure S1 B) [14,23]. We used in utero electroporation to deliver our constructs, which are HA-tagged and co-express soluble GFP, into embryonic day 16 (E16) rat brain. Successful expression and NE targeting by the chimeric Nesprin constructs were confirmed in electroporated brain slices (Figure S1 C).
Expression of Mini N2G SR resulted in near-normal migration to the CP, supporting the great utility of the miniature chimeric cDNA constructs in Nesprin functional analysis (Figure 1 C, D). Instead, expression of the Mini N2G construct, which is only capable of actin recruitment, caused a striking decrease in the number of neurons reaching the CP (Figure 1 C, D). These cells accumulated in the upper IZ extending abnormally elongated processes into the CP (Figure 1 E, F). Importantly, the successful migration of N2G SR-expressing cells strongly suggests that the Nesprin-2 dependent recruitment of microtubule motors is pivotal for neuronal migration (Figure 1 C, D). In contrast, actin-binding activity, which is critical in fibroblast nuclear displacement [22,23], makes a little-to-no contribution in the developing brain. Together, our data reveal that Nesprin-2-associated microtubule motors are required for nuclear transport within migrating neurons.
Roles for Nesprin-2 functional domains in nucleus vs. centrosome behavior.
During migration, neurons extend a leading process that contains one or more transiently-formed “dilations” (or “swellings”), toward which the centrosome advances, followed by the nucleus [4–6]. Analysis of Syne-2 KO mice suggested a role for Nesprin-2 in neuronal migration as a Nucleus-Centrosome (N-C) connector [24]. The physical nature of such a link and the mechanism responsible for Nesprin-mediated neuronal migration have remained incompletely understood. To address these basic issues we evaluated the role of Nesprin-2 and its functional domains in centrosome as well as nuclear behavior in embryonic rat brain. For this purpose, we co-expressed our constructs in E16 rat brain, along with DsRed-tagged PACT to label centrosomes [26–28], and analyzed N-C distance by fixed imaging at E20. Mini N2G expression dramatically increased the median N-C distance by more than 7-fold when compared to Mini N2G SR (Figure 2 A, B). In contrast, the absence of the actin-binding calponin homology domain had no apparent effect on N-C distance, as judged by the similar effects of the N2G SR and Mini N2G SR constructs (Figure 2 A, B). These results indicate that the Nesprin-2 microtubule motor-binding domain is necessary and sufficient for nuclear advance, and that centrosome and nuclear transport are largely independent.
Figure 2. Roles of Nesprin-2 functional domains in Nucleus-Centrosome coupling.
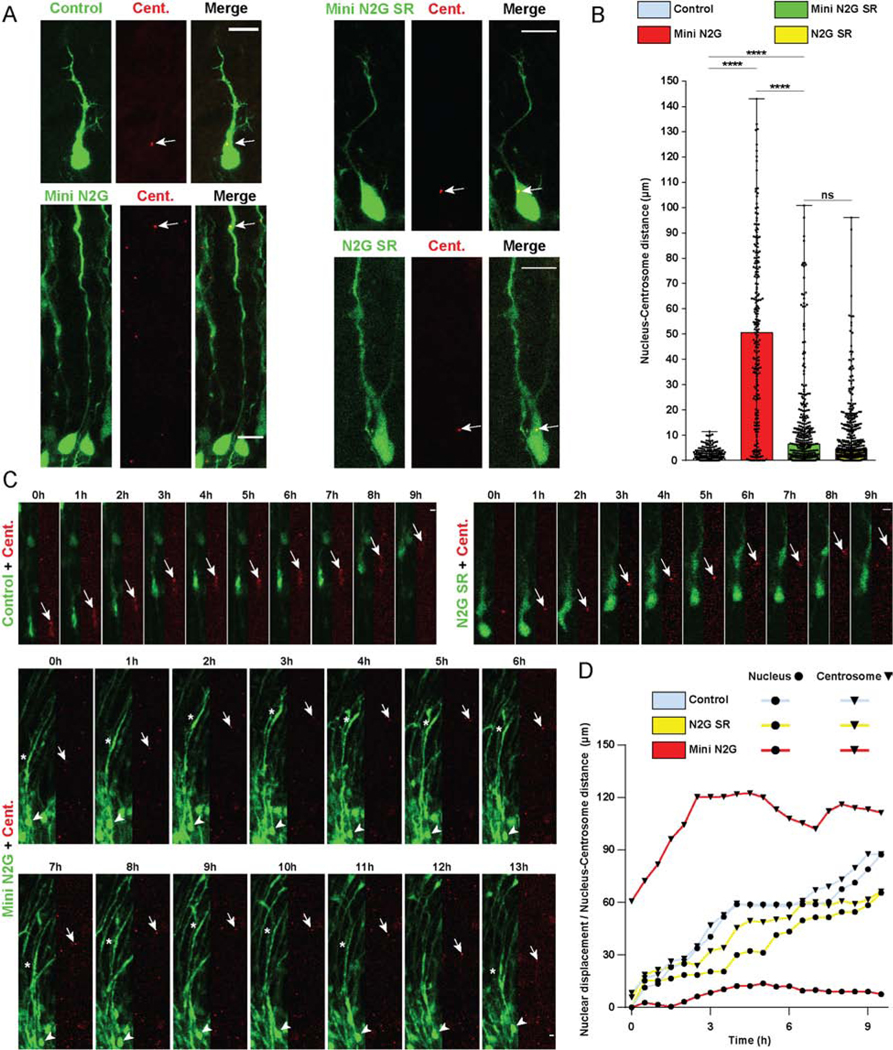
(A-D) E16 rat brain was electroporated with control vector or Nesprin-2 functional domains, in each case co-expressing soluble GPF to indicate position of soma, together with PACT-DsRed construct to mark centrosomes. Brain slices were imaged fixed or live at E20. (A) Representative images of electroporated neurons in the CP showing centrosome (Cent.) (arrow) and soma position within the migrating neuron. (B) Quantification of Nucleus-Centrosome (N-C) distance for each condition. Expression of the Mini N2G construct, lacking the microtubule motor-binding domain, increased N-C distance by more than 50-fold compared to empty vector, and more than 7-fold compared to the Mini N2G SR. (C) Time-lapse with dual-channel images for the soma and centrosome (red) are shown at 1h intervals (Videos S1–3). Arrows, arrowheads, and asterisks indicate, respectively, centrosomes, cell bodies, and leading process dilations. (D) Graphical representation of relative position of the nucleus and centrosome over time. For each condition, nuclear displacement (●) was measured over time, relative to the original position (t=0). For each time point, N-C distance was quantified and summed to the nuclear position (▼). Data presented as scatter dot plot with bar representing median with range in B. Mann Whitney test for non-parametric distributions was used in B (****P<0.0001; n.s. non-significant). Data in B include at least 217 electroporated neurons from at least 3 embryos, per condition. A scale bar, 10μm. C scale bar, 5μm. Related to Videos S1–3.
To test the relative roles of Nesprin-2 functional domains in nucleus vs. centrosome behavior more directly, we performed live imaging analysis of embryonic rat brain slices expressing control GFP, Mini N2G, or N2G SR, each plus PACT-DsRed. As expected in the control cells, the nucleus and centrosome each advanced in their normal saltatory fashion. However, in Mini N2G-expressing neurons the nucleus remained mostly immobile (Figure 2 C, D; Video S1-3) and separated by large distances from the centrosome and dilations. The latter exhibited marked oscillatory movements in the axis of the leading process (Figure 2 C, D; Video S3), and individual centrosomes advanced to reach the moving dilations. These data strongly suggest that the motive forces for nuclear and centrosome transport are at least partially independent. In contrast to these results, N2G SR expression had little effect on nuclear movement, and we only noticed a slight change in N-C distance (Figure 2 C, D; Video S2), consistent with our observations in fixed brain slices (Figure 2 A, B). Together our data demonstrate a fundamental role for the microtubule motor-binding domain of Nesprin-2 in nuclear, but not centrosome, migration in neurons.
Nesprin-2 recruits cytoplasmic dynein via the motor protein adaptor BicD2
The C. elegans KASH protein UNC-83 is responsible for recruiting dynein and kinesin to the hypodermal cell NE and for nuclear migration in these cells [29,30]. Motor protein recruitment was through BICD-1 and NUD-2, homologues of the mammalian dynein regulators BicD2 and Nde1/Ndel1, respectively [31]. We, therefore, tested whether either of these proteins might interact with mammalian Nesprin-2 and help mediate motor protein recruitment to the NE. To address this question, we first used HeLa cells, which have proven valuable in detecting dynein and its cofactors at the G2 NE [13,14,32–34] (Figure S2 A). The HeLa cells were transfected with GFP-tagged Mini N2G or -Mini N2G SR cDNAs and stained for endogenous BicD2. In Mini N2G-expressing cells, BicD2 was readily detected at the G2 NE (Figure S2 B, C). Interestingly, Mini N2G SR helped recruit endogenous BicD2 to the HeLa NE independent of cell cycle stage (Figure S2 D, E). We also observed comparable NE localization for the BicD2 paralogue, BicD1, which has little or no role in early cortical development [14] (Figure S2 F, G), but no NE recruitment of the dynein recruitment factors Nde1 and Ndel1 (Figure S2 H and I). These results indicate that the microtubule motor-binding domain of Nesprin-2 recruits BicD adaptor proteins to the HeLa NE.
To test further for a cell cycle independent component of Nesprin-2-mediated BicD2 recruitment, we transfected cultured NIH 3T3 fibroblasts with the Mini N2G and N2G SR constructs. We then arrested the cells in G0 by serum starvation [22], fixed and stained them for endogenous dynein and BicD2. As previously reported [22], cells expressing N2G SR, but not Mini N2G, showed clear dynein decoration at the NE (Figure 3 A, B). With regard to the current study, the N2G SR-transfected cells notably exhibited a strong BicD2 signal at the NE, which we did not observe in cells expressing Mini N2G (Figure 3 C, D). These results strongly suggest that dynein is recruited to the microtubule motor-binding domain of Nesprin-2 by BicD2.
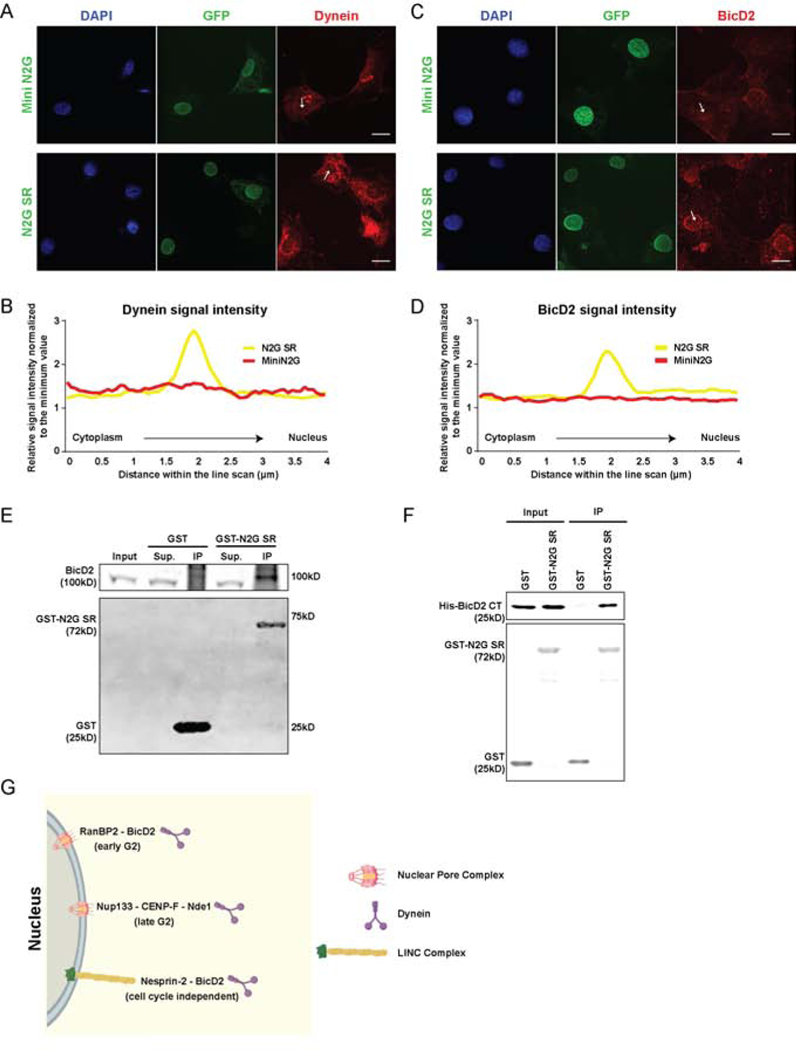
Figure 3. BicD2 interaction with Nesprin-2.
(A-D) Cultured NIH 3T3 fibroblasts were transfected with GFP-tagged Mini N2G or N2G SR cDNAs and immunostained for endogenous dynein or BicD2. Intensity scans were performed along 4μm lines through the NE (arrows). (A, C) Representative fixed images and (B, D) line scan quantifications of dynein and BicD2 signal. Cells expressing N2G SR show NE dynein and BicD2 decoration, indicating that this Nesprin fragment is sufficient to recruit both factors. (E) GST-N2G SR pull-down from embryonic rat brain lysates showing BicD2 coprecipitation. (F) Purified bacterially-expressed GST-N2G SR pulls down bacterially-expressed BicD2 CT. (G) Schematic diagram depicting multiple dynein NE recruitment mechanisms in G2 vs. Non-G2 phases of the cell cycle. During G2, dynein is recruited by two consecutive Nuclear Pore Complex-dependent pathways [13,14]. An “early pathway”, involving the nucleoporin RanBP2 and BicD2, and a “late pathway” involving the nucleoporin Nup133 and CENP-F/Nde1. In the current study we find that BicD2 and dynein are also recruited by the microtubule motor-binding domain of Nesprin-2, independent of cell cycle stage. Data are presented as superimposed symbols at mean with a connecting line in B and D. Data in B and D include line scan analysis from at least 6 and 12 cells, respectively. A and C scale bar, 10μm. Related to Figure S2.
To test whether Nesprin-2 and BicD2 can interact physically, we performed pull-downs from embryonic rat brain cytosolic lysates using bacterially expressed GST-N2G SR, and immunoblotted for BicD2. Our data revealed a strong and specific signal for BicD2 (Figure 3 E), supporting its interaction with Nesprin-2 through the previously identified microtubule motor-binding region.
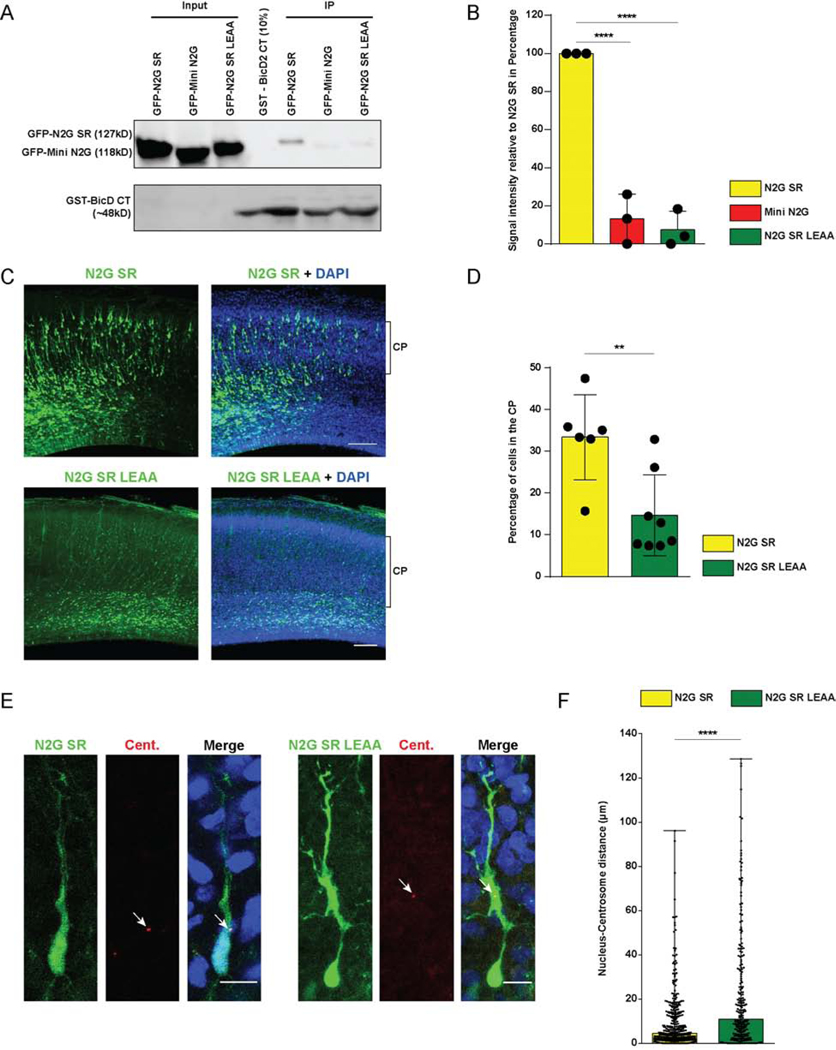
BicD2 consists of two N-terminal coiled-coil regions that recruit the dynein-dynactin supercomplex [35–37] and kinesin [38], respectively. The C-terminal BicD2 coiled-coil region, in contrast, binds motor protein cargoes, including Rab6 and RanBP2 [32,39]. To test whether Nesprin-2 might also interact with the BicD2 C-terminal region, we performed pull-downs with bacterially-expressed GST-BicD2 C-terminus (BicD2 CT) from HeLa cell lysates expressing GFP-tagged Mini N2G or N2G SR. We obtained clear evidence that GST-BicD2 CT is, indeed, able to pull-down the motor protein-binding N2G SR, but not Mini N2G, which lacks this capability (Figure 6 A, B). To determine whether the BicD2-Nesprin-2 interaction is direct or involves additional intermediary proteins, we incubated bacterially purified His-BicD2 CT with GST or GST-N2G SR and pulled down potential protein complexes. Using a BicD2 C-terminal domain-specific antibody we observed a clear BicD2 CT interaction with GST-N2G SR, but not GST alone (Figure 3 F). Together these data support a direct interaction between the C-terminal region of BicD2 and its novel form of cargo, Nesprin-2, which serves to recruit dynein to the NE independent of cell cycle stage (Figure 3 G).
Figure 6. Effect of Nesprin-2 LEAA mutation on interaction with BicD2.
(A, B) GST-BicD2 CT pull-downs from HeLa cell lysates expressing GFP-tagged N2G SR, Mini N2G or N2G SR LEAA were evaluated for GFP co-immunoprecipitation. (B) Quantification of the GFP signal intensity relative to N2G SR. The LEAA mutation severely disrupted the Nesprin-2-BicD2 interaction. (C, D) E16 rat brain was electroporated with N2G SR or N2G SR LEAA, and subsequently imaged in fixed brain slices at E20. (C) Representative images of the neocortex with electroporated cells in green (GFP) and stained with DAPI. Bracket shows the margins of the CP. (D) Quantification of the proportion of cells reaching the CP. Electroporation of the N2G SR LEAA construct causes a decrease in the number of cells reaching this region. (E, F) E16 rat brain was electroporated with N2G SR or N2G SR LEAA together with PACT-DsRed, and imaged in fixed brain slices at E20. (E) Representative images of electroporated neurons in the CP showing centrosome (Cent.) position (arrow) relative to that of the soma. (F) Quantification of the N-C distance. Electroporation of the N2G SR LEAA causes an increase in N-C spacing. Data presented as scatter dot plot with bar representing mean±s.d. in B and D, and as scatter dot plot with bar representing median with range in F. One-way ANOVA was used in B, and Mann Whitney test for non-parametric distributions was used in D and F (**P<0.01; ****P<0.0001). Data in B arises from n=3 independent experiments. The GFP signal for each condition was normalized to GST BicD2 CT signal. For each experiment, GFP signal of the different conditions was normalized to N2G SR. Data in D include at least 1666 electroporated cells from at least 6 embryos, per condition. Data in F include at least 239 electroporated cells from at least 4 embryos, per condition. C scale bar, 100μm. E scale bar, 10μm.
BicD2 participates in post-mitotic neuronal migration in a cell-autonomous fashion
Data from BicD2 knockout mice have supported a BicD2 contribution to the organization of the cerebral cortex and cerebellum [40]. However, the observed impairment in cerebellar granule cell migration was proposed to involve a non-cell autonomous defect in tenascin secretion by Bergmann glial cells. Our finding of a direct Nesprin-2-BicD2 interaction suggested, instead, a cell-autonomous role for the latter protein in post-mitotic neuronal migration. To test this possibility more directly, we electroporated E16 rat brain with a cDNA construct that conditionally expresses the BicD2 CT domain under control of the NeuroD1 promoter, resulting in specific expression in post-mitotic neurons [41,42]. Notably, BicD2 inhibition in neurons caused a marked reduction in their number within the CP (Figure 4 A, B), consistent with a novel cell-autonomous BicD2 role in post-mitotic neuronal migration.
Figure 4. Effects of BicD2 inhibition on neuronal migration.
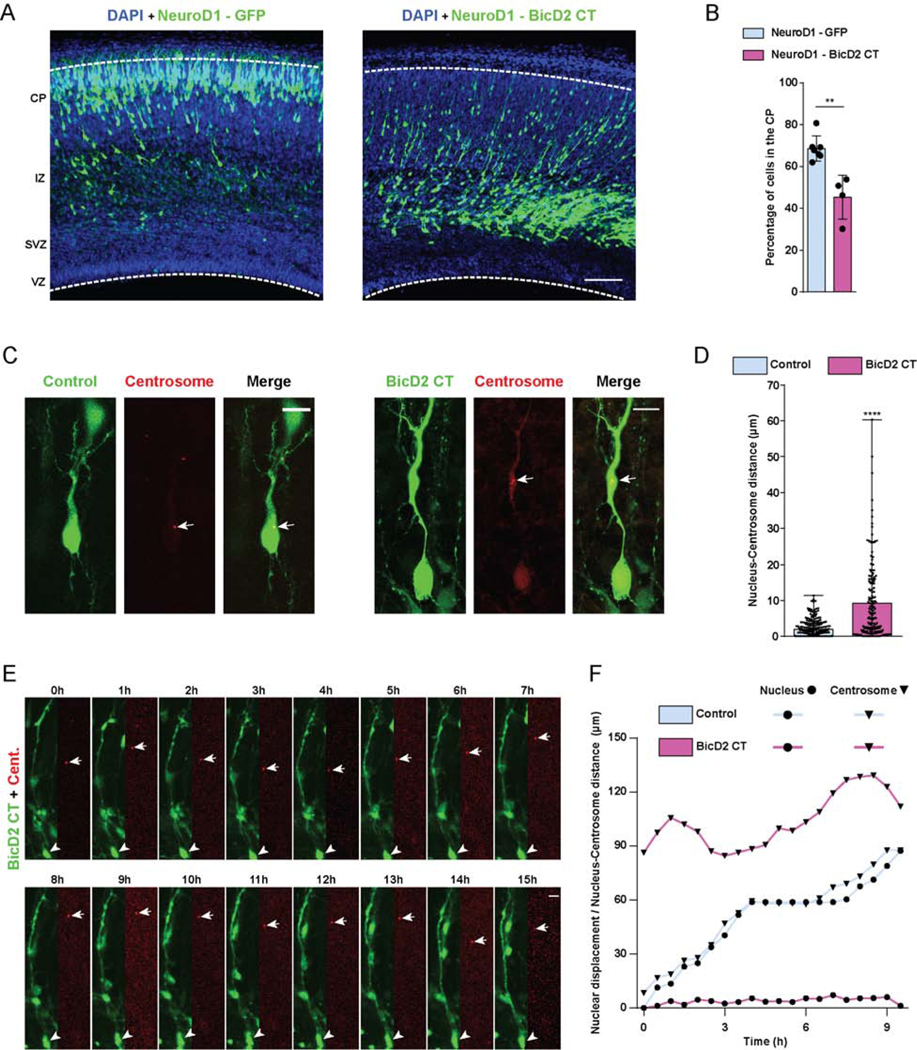
(A, B) E16 rat brain was electroporated with a cDNA encoding the BicD2 C-terminal domain under NeuroD1 promoter regulation for specific expression in post-mitotic neurons. Fixed brain slices were imaged at E20. (A) Representative images of the neocortex showing electroporated cells (green), co-stained with DAPI (blue). (B) Fraction of cells reaching the CP revealing that neuronal expression of BicD2-CT causes dramatic inhibition of neuronal migration. (C-F) E16 rat brain was electroporated with control GFP or the BicD2 CT fragment, along with PACT-DsRed, and imaged fixed at E20. (C) Representative images of electroporated neurons in the CP showing dramatically increased nucleus-centrosome spacing in BicD2-CT-expressing cells. (D) Quantification of N-C distance. (E) Time-lapse with dual-channel images for the soma and centrosome (red) are shown at 1h intervals (Video S4). Arrows and arrowheads indicate, respectively, centrosomes and cell bodies. (F) Graphical representation of relative position of the nucleus and centrosome over time. For each condition, nuclear displacement (●) was measured over time, relative to the original position (t=0). For each time point, N-C distance was quantified and summed to the nuclear position (▼). Data presented as mean±s.d. in B, and as scatter dot plot with bar representing median with range in D. Mann Whitney test for non-parametric distributions was used in B and D (**P<0.01; ****P<0.0001). Data in B include at least 3235 electroporated cells from at least 4 embryos, per condition. Data in D includes at least 169 electroporated cells from at least 4 embryos, per condition. A scale bar, 100μm. C and E scale bars, 10μm. Related to Figure S3 and Video S4.
We reasoned that the BicD2 CT domain might displace endogenous full-length BicD2 from its NE Nesprin-2 binding site, thereby interfering specifically with nuclear transport. To test this possibility, we electroporated E16 brains with the BicD2 CT-expressing cDNA and analyzed nuclear and centrosomal behavior by fixed and live imaging. Consistent with a specific role in nuclear migration alone, BicD2-CT caused a striking increase in N-C distance (Figure 4 C, D). Live monitoring of BicD2 CT-expressing neurons also showed clear inhibition of nuclear, but not centrosome movement, resulting in extreme N-C separation (Figure 4 E, F). Centrosomes and dilations still showed oscillatory behavior within the neuronal leading process, comparable to what we observed after expression of Mini N2G (Video S3, 4). These data further confirm the requirement for BicD2 in nuclear, but not centrosome, transport during neuronal migration.
As noted, BicD2 recruits dynein to the RGP NE via RanBP2 [13]. Although this behavior is G2-specific, we tested whether a comparable mechanism might be involved in the post-mitotic migrating neurons as well. For this purpose, we performed BicD2 and RanBP2 RNAi. Then we analyzed N-C separation in migrating post-mitotic neurons in each case. BicD2 RNAi caused a significant increase in N-C spacing, yet RanBP2 RNAi had no effect (Figure S3 A, B), arguing further for distinct BicD2 roles and NE interacting partners in INM vs. post-mitotic neuronal migration.
Effects of kinesin-1 inhibition on neuronal migration
KASH proteins, including Nesprin-2, have been reported to recruit kinesin-1 as well as dynein to the NE [19,30,43,44]. Nesprin-2-mediated kinesin-1 recruitment was found to be required for nuclear transport and positioning in myoblasts [44,45] and for nuclear rotation in migrating cerebellar granule cell neurons [19]. In view of these observations, we tested for a possible kinesin-1 contribution to Nesprin-mediated nuclear transport in cortical neurons.
The kinesin-1 sub-family consists of three heavy chain (KIF5A, B and C) and 4 light chain (KLC1–4) genes, each of which class has been implicated in cargo binding. Nesprin-2 was reported to interact with the KLCs via their TPR domains, which suggested a NE recruitment mechanism for kinesin-1 in myoblasts [43,45]. Kinesin-1 is also recruited to the G2 NE in other cell types via BicD2 [32,38]. Given these data, we tested whether kinesin-1 might contribute to neuronal migration in rat cerebral cortex by expression of the Kif5B tail, or the KLC1 TPR domains [19,45]. Expression of either substantially increased the number of electroporated cells reaching the CP (Figure 5 A, B). No change in N-C distance was detected (Figure S4 A, B). To gain insight into the mechanism responsible for these remarkable results, we performed live imaging of rat brain slices in utero electroporated with the KLC TPR construct. We observed a striking increase in migration velocity (Control 0.16±0.05 μm/min vs. KLC TPR 0.23±0.07 μm/min, mean ± SD) (Figure 5 C, D), consistent with our analysis of neuronal number in the CP of fixed brain tissue. These results suggest that kinesin-1 normally acts to oppose dynein and, thereby, constrain neuronal migration.
Figure 5. Role of Kinesin-1 in neuronal migration.
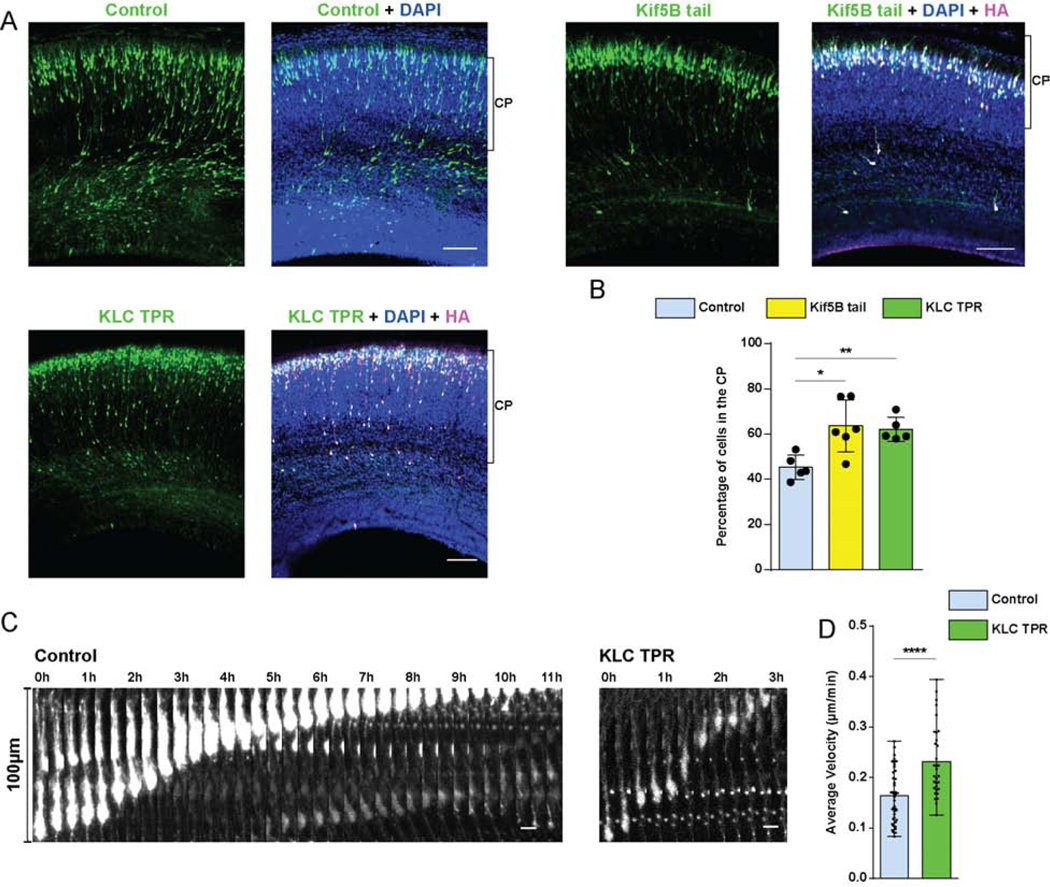
(A, B) E16 rat brain was electroporated with IRES vectors encoding GFP and HA-tagged Kif5B tail or HA-tagged KLC TPR cDNA constructs and empty vector control. Fixed images of brain slices were imaged at E20. (A) Representative images of the neocortex with electroporated cells in green, stained with DAPI (blue) and immunostained for HA, in magenta. Brackets show boundaries of CP. (B) Quantification of cells reaching CP. Expression of the Kif5B tail or KLC TPR domains caused a considerable increase of neurons within the CP. (C, D) E16 rat brain was electroporated with the KLC TPR construct and empty vector, and brain slices imaged live at E20. (C) Time-lapse images for control and KLC TPR-expressing neurons (Videos S5-6) over a time period sufficient in each case to complete 100μm of nuclear displacement. Images are shown at 20 min intervals. (D) Quantification of neuronal migration velocity, from initiation of nuclear migration in each cell until its termination, in control vs. KLC TPR expressing neurons. Expression of KLC-TPR resulted in significantly higher migration velocity vs that for the control. Data presented as scatter dot plot with bar representing mean±s.d. in B, and as scatter dot plot with bar representing mean with range in D. Mann Whitney test for non-parametric distributions was used in B (*P<0.05; **P<0.01). Unpaired T-Test with Welch’s correction was used in D (****P<0.0001). Data in B include at least 967 electroporated cells from at least 4 embryos, per condition. Data in D include 42 and 32 migrating neurons for control and KLC TPR, respectively. A scale bar, 100μm. C scale bar, 10μm. Related to Figure S4 and Videos S5-6.
The KLC TPR domain was reported to interact with Nesprin-2 through a conserved LEWD (Leu-Glu-Trp-Asp) motif in the microtubule motor-binding domain [45]. Therefore, as an additional means to address the contribution of Nesprin-associated motor proteins to neuronal migration, we mutated the LEWD motif in the N2G SR construct to LEAA (Leu-Glu-Ala-Ala). This change was reported to inhibit binding of kinesin-1 [45], but dynactin, used as a proxy for dynein, was unaffected [22]. To test the effect of the Nesprin LEAA mutation on BicD2 binding, we performed pull-downs with GST-BicD2 CT from HeLa cell lysates expressing GFP-tagged Mini N2G, N2G SR wild-type, or N2G SR LEAA mutant proteins. GST-BicD2 CT exhibited clear binding to N2G SR, an effect strongly reduced for N2G SR LEAA (Figure 6 A, B). These results indicate that the LEAA mutation also interferes with BicD2 binding.
To test the importance of the Nesprin-2 LEWD sequence physiologically, we determined the effect of the LEAA mutation in neuronal migration. For this purpose, we electroporated N2G SR or N2G SR LEAA cDNAs into E16 rat brain and analyzed cortical neuron redistribution and N-C spacing. Expression of the LEAA construct substantially reduced the numbers of neurons reaching the CP when compared to the LEWD control (Figure 6 B, C). We also observed a substantial increase in N-C spacing in these cells (Figure 6 D, E). Both results are consistent with a predominant role for the Nesprin-2 LEWD motif in the recruitment of dynein to the NE, with kinesin recruitment playing a subsidiary role in neuronal migration.
DISCUSSION
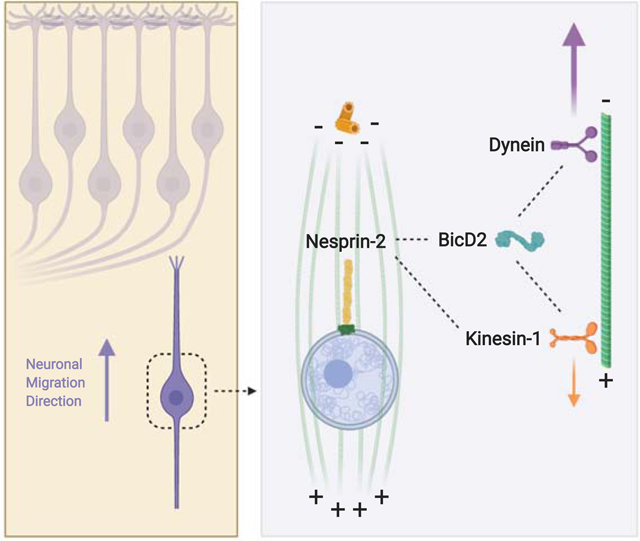
Our results demonstrate that, of the ~800 kDa Nesprin-2 polypeptide, the much smaller microtubule motor-binding domain (N2G SR ~ 100kDa) is necessary and sufficient, to support neuronal migration. We find no detectable role for Nesprin-2 actin-binding activity, in contrast to its importance in nuclear migration in non-neuronal cells [23]. We find further that Nesprin-2 binds the dynein adaptor BicD2 (Figure 7A), which has been implicated in diverse aspects of microtubule-mediated intracellular motility and human neurological disease [46]. We identified a novel role for kinesin-1 in constraining neuronal migration, which indicates that dynein and kinesin cooperate in neurons in a manner distinct from other systems. Overall, our data provide a new and potentially unifying model for the basic, but, until now, incompletely understood process of post-mitotic neuronal migration (Figure 7 B, B’).
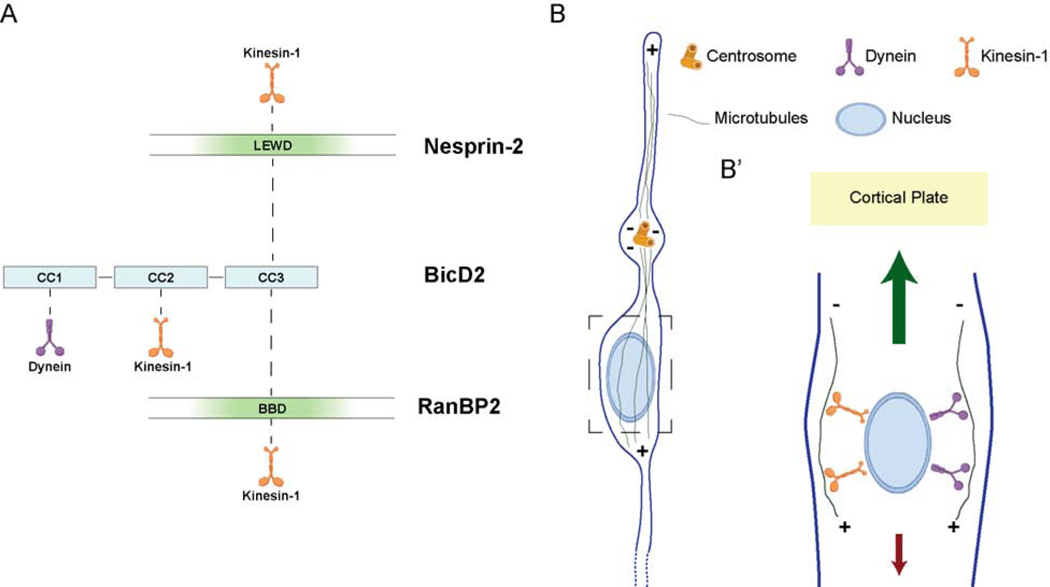
Figure 7. Model for microtubule motor recruitment during neuronal migration.
(A) Schematic depiction of the interactions between BicD2, Nesprin-2, RanBP2, and microtubule motors. The microtubule motor-binding domain (in green) within Nesprin-2, and the BicD2 Binding Domain (BBD, in green) of RanBP2 [13,32] are represented. BicD2 is shown as an elongated structure with three major coiled-coil (CC) sections implicated, respectively, in dynein, kinesin-1, or cargo binding. The LEWD-containing microtubule motor-binding region of Nesprin-2 is shown binding to BicD2 CC3 (this study). Kinesin-1 and BicD2 have been reported bind to the nucleoporin RanBP2 in a cooperative mechanism [32,54]. We speculate that Nesprin-2 might obtain a similar arrangement to explain the importance of the LEWD sequence in both myoblast [44,45] and neuronal (this study) nuclear migration. Finally, it is unclear how the less well characterized kinesin-1 binding CC2 domain of BicD2 contributes to nuclear migration in these systems. (B) Diagrammatic representation of a migrating neuron showing microtubules emanating from the centrosome, which is located ahead of the nucleus. Box (dashed line) represents the area enlarged in migrating neuron in B’. (B’) Microtubules are shown with minus ends directed upward in the direction of migration and interacting tangentially with the NE via dynein and kinesin-1. In this arrangement dynein would propel the nucleus toward the centrosome and Cortical Plate. Kinesin-1 should pull the nucleus downward, toward the microtubule plus ends, countering the dynein forces. Larger (green) and smaller (red) arrows represent the greater and lesser microtubule motor-mediated forces suggested by our data to act on the migrating cell nucleus.
Specific Role for Nesprin-2 recruitment of Microtubule Motors vs. Actomyosin in Neuronal Migration.
LINC complexes have been implicated in nuclear migration in diverse cell types and organisms [47]. The Nesprins serve to recruit dynein and/or kinesin-1 to the NE [19,22,30,44,45]. Our data in the developing brain support this important basic role for Nesprin-2 as well, though significant aspects of Nesprin function appear to differ in migrating neurons. Expression of the ~100 kDa N2G-SR construct, capable of microtubule motor binding, preserved near-normal neuronal migration. This suggests that much of the Nesprin-2 polypeptide may be unessential for cellular redistribution in the developing cortex. Our data also reveal an important new pathway for postmitotic neuronal migration and identify a novel kinesin-1 contribution to this process as well. Surprisingly, kinesin-1 inhibition enhanced neuronal migration to the cortical plate. We found this behavior to involve accelerated cellular migration as judged by monitoring live somal behavior, and, therefore, consistent with a kinesin role at the NE. These results imply that, under normal physiological conditions, kinesin-1 acts opposite to dynein in the migrating neuron, serving to restrain the advancing neuronal nucleus.
Actin and myosin have been found to contribute to the migration of isolated neuronal precursors cultured in vitro [4–6,17,48–50]. Activity of the non-muscle myosin IIb was reported to be necessary for nuclear advance in migrating neurons and was suggested to push the nucleus forward via contractile forces generated behind the nucleus [4–6,48]. Actin filaments were also observed to be enriched in the proximal portion of the leading process [17,49,50] and were suggested to pull the centrosome and the nucleus forward [17], perhaps by interacting with the microtubule cytoskeleton [51]. In the current study, expression of the Mini-N2G fragment, which supports rearward actin-mediated nuclear movement in non-neuronal cells [23], had no effect on neuronal migration in vivo in our hands. These results indicate that Nesprin-2 is dispensable for actin-mediated nuclear migration in neurons, and imply some other, as yet incompletely understood, actin role in these cells.
Nucleus-centrosome Coupling
Nesprin-2 was previously proposed to mediate the N-C connection in migrating neurons [24], but the purpose and mechanisms underlying this link have remained unclear. In the current study, removal of the Nesprin-2 microtubule motor-binding domain caused severe uncoupling of nuclear from centrosomal movement. These results can be simply explained by a role for Nesprin-2 in nuclear, but not centrosome behavior. Despite arrest of nuclear advance by Nesprin inhibition, extension of the leading migratory process in these cells persisted. Strikingly, the centrosome continued to advance on its own to distances up to 100μm ahead of the nucleus These findings suggest that the centrosome is driven by its own transport machinery, likely involving dynein pulling on microtubules from sites within the swellings and other parts of the migratory process [5].
Role of BicD2 in Nesprin Function.
BicD proteins have been implicated in the recruitment of motor proteins to a number of subcellular cargoes [52]. Here, we identify a clear, direct interaction between the Nesprin-2 microtubule motor-binding domain and the BicD2 C-terminal cargo binding region (Figure 7A). We also find that LEWD sequence mutation to LEAA disrupts BicD2 binding to Nesprin-2, and impairs the function of the latter in migrating neurons. Notably, the LEWD motif was implicated in kinesin-1 NE recruitment by Nesprin-2 [45]. Thus, our results raise the question whether kinesin-1 binds Nesprin-2 directly, via BicD2 [32,38] or both (Figure 7 A). Considering the role of BicD2 we now find for Nesprin, we suspect that the former may prove to be an important contributor to myoblast nuclear behavior [44,45].
Comparable to Nesprin-2, kinesin-1 and BicD2 are also known to be recruited to the G2 NE by RanBP2 [13,32]. This nucleoporin was reported to bind kinesin-1 directly through its KLCs [53], and also via BicD2 [32]. Recent in vitro evidence [54] has shown that LEAA mutation in RanBP2 does not affect BicD2 binding, but alters its stoichiometry. The physiological importance of these unusual arrangements remains to be fully elucidated. Nonetheless, they seem to be consistent with BicD2 roles in INM [13] and neuronal migration.
Models for the Role of Dynein and Kinesin in Nuclear Transport
Potential roles for kinesin-1 in brain development have been incompletely understood. Both kinesin-1 heavy and light chain genes have been implicated in cerebellar granule cell behavior [19]. We expressed the same truncated inhibitory regions of the Kif5b KHC as well as the KLC1 TPR in vivo. Each construct caused an increase in the number of neurons reaching the CP, associated with a faster rate of neuronal migration. Whether our study indicates a real difference in kinesin-1 roles in cortical vs. cerebellar neurons is uncertain, but we note that nuclei in cerebellar neurons exhibited distinctive behavior in switching position with their accompanying centrosomes, and in reversing migration direction [55]. We find that in cortical neurons the centrosome remains in physical proximity to the nucleus upon kinesin-1 inhibition, and, more strikingly, the velocity of nuclear transport and neuronal migration were each increased. Whether the different features of migration evaluated in the two studies reflect common or related forms of behavior must await further investigation to resolve.
Combined roles for NE-associated kinesin-1 and dynein have been examined in C. elegans embryonic hypodermal cells [30]. Kinesin-1 played the predominant role in nucleus translocation while dynein controlled nuclear rotation, apparently helping the nucleus navigate past cytoplasmic impediments. In our system, microtubules emanate from the centrosome and surround the nucleus as a parallel, nearly unidirectional array [5](Fig 7 B, B’). This arrangement should allow the two motor proteins to exert force on the NE (Figure 7B), but in opposite directions. This arrangement is related to that in G2 HeLa cells, in which acute dynein inhibition caused clear centrosome displacement from the nucleus [32], behavior determined to be mediated by the underlying activity of kinesin-1. In this case the two motors appeared to act in controlling relative centrosome-nucleus position in preparation for mitosis. In our work, we find that kinesin-1 acts in opposition to dynein, perhaps with the purpose of fine-tuning radial neuronal migration velocity. The importance of this novel mechanism in brain development is not yet evident. Conceivably, it serves in a fail-safe mechanism to ensure that migrating neurons reach their destinations in a timely manner. Whatever its physiological role, our new mechanism might ultimately be of help in developing strategies to correct shortcomings in neuronal migration resulting from diverse forms of developmental insult or disease.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Richard Bert Vallee (rv2025@columbia.edu).
Materials availability
All unique reagents generated in this study are available from the Lead Contact.
Data and Code Availability
The authors declare that all relevant data supporting the findings of this study are provided in the Article and Supplementary files.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Timed Sprague Dawley pregnant rats used for in utero electroporation were obtained from Taconic, and were housed in Columbia University animal facilities. All the experiments conducted were done in accordance with the animal welfare guidelines and the guidance of the Institutional Animal Care and Use Committee (IACUC) at Columbia University.
Cell Culture
NIH 3T3 fibroblasts (gift from Dr. Gregg Gundersen, Columbia University) and HeLa cells (a gift from Dr. Viki Allan, University of Manchester) were cultured in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin and maintained at 37°C with 5% CO2.
METHOD DETAILS
In utero electroporation
Plasmids encoding for shRNAs or cDNA were electroporated into the developing brain at embryonic day 16 (E16) and electroporated as described [56]. In more detail, timed pregnant Sprague Dawley E16 rats were anesthetized with a ketamine xylazine cocktail administered intraperitoneally, and toe pinch was performed to ensure deep anesthesia. To avoid excessive heating loss during the surgical procedure, an external heating source was provided. For pain management buprenorphine and bupivacaine were administered subcutaneously, before the surgery. The abdominal cavity was opened and uterine horns exposed and trans-illuminated for clear identification of the embryonic brain ventricles. For easy visualization of the DNA in the brain ventricular space, a non-toxic dye (Sigma, F7252) was added to the DNA before surgery and electroporated with a sharpened glass needle. After injection, embryos were subjected to five electric impulses (50V, 50ms each, separated by 1s intervals) delivered by an electroporator (Harvard Apparatus ECM 830), to target the DNA to RGPs in the lateral neocortex. The embryos were returned to the abdominal cavity and the wound was closed. Rats were monitored every day post-surgery and buprenorphine was administered every 12h for the first 48h, for post-operative pain control.
Immunohistochemistry and live imaging
For embryonic brain harvesting, pregnant rats were re-anesthetized and the surgical wound was reopened 4 days post-injection, at E20, to expose the uterus.
For fixed imaging, embryonic (E20) rat brains were harvested and immersed in PBS with 4% PFA overnight. They were then embedded in 4% of agarose (Sigma, A9539) and sliced in a vibratome (Leica, VT 1200S) in 100μm slices. After blocking in 5% normal donkey serum (Sigma, D9663) in PBS-Triton X-100 0.5% for 1 hour, slices were incubated with primary antibodies diluted in the blocking solution, overnight on a shaker, at 4°C. Secondary antibodies (1:500) and DAPI (4’,6-diamidino-2-phenylindole, Thermo Scientific, 62248, 1:10.000 dilution) were diluted in PBS and incubated for 2 h at room temperature. Slices were mounted with Aqua-Poly mounting media (Polysciences, 18606).
For live imaging, the dissected embryonic rat brains were embedded in 4% low melting agarose (IBI Scientific, IB70057) diluted in artificial cerebrospinal fluid [56] and sliced into 300μm coronal sections. The slices were placed on porous filters (EMB Millipore, PICMORG50) in cortical culture medium containing 25% Hanks balanced salt solution (Life Technologies, 24020–117), 47% basal MEM (Life Technologies, 21010–046), 25% normal horse serum (Life Technologies, 26050–088), 1% penicillin/streptomycin/glutamine (Life Technologies, 10378–016), and 2% of 30% glucose (Sigma, G5767) in a 50mm glass-bottom dish (MatTek Corporation, P50G-0–14-F) and imaged on an IX80 laser scanning confocal microscope (Olympus FV1000 Spectral Confocal System) at intervals of 10 min for up to 24 h.
RNAi and cDNA expressing constructs
For the in utero electroporation experiments in the embryonic brain, the shRNAs for BicD2, RanBP2, and LIC1 as well as the KASH-RFP were previously described [13–15,23] and EGFP-C1 was used as control. The Mini N2G, N2G Spectrin Repeats (SR) 52–56, Mini N2G SR constructs were cloned into the pCAGIG vector (Addgene plasmid #11159)[57] by PCR amplification using KOD Hot Start DNA Polymerase (Millipore, 71086). Previously described EGFP-C1-Mini N2G and pMSCV-N2G SR 52–56 constructs were used as templates [22] and an HA-tag coding sequence was introduced. Point mutagenesis was used to introduce LEAA mutation within the pCAGIG N2G SR construct. BicD2 CT (630–820aa) was PCR amplified from full-length mouse BicD2 in pIRES2 DsRed-Express2 vector [14] and inserted into pCAGIG and pCALNL-GFP (Addgene plasmid #13770)[58] vectors. For the conditional expression of BicD2 CT, pNeuroD1-Cre (gift from Dr. Carlos Cardoso, Institut de Neurobiologie de la Méditerranée) was co-injected with pCALNL-GFP and pCALNL-BicD2 CT. C-terminal HA-tagged Kif5B tail (809–963 aa of rat Kif5B) and KLC-TPR (211–497 aa of rat KLC1) nucleotide sequences were gene synthesized and cloned into pCAGIG vector (Synbio Technologies, New Jersey, USA). To tag centrosomes, PACT domain (pericentrin-AKAP450 centrosomal targeting) sequence from rat pericentrin (2729–2933) was amplified and cloned into a pCAG-GFP vector (Addgene plasmid #11150)[57], in which the GFP sequence was removed and it was C-terminal tagged with DsRed.
The constructs for bacterial expression GST-BicD2-CT (gift from Dr. Anna Akhmanova, Utrecht University)[32], His-BicD2-CT [13] GST-N2G (gift from Dr. Gregg Gundersen, Columbia University) [22] were described previously. For experiments in NIH 3T3 fibroblasts, pMSCV-puro GFP-Mini N2G and pMSCV-puro GFP-N2G SR 52–56 constructs were used. For experiments in HeLa cells, pEGFP-C1 GFP-Mini N2G, pEGFP-C1 GFP-Mini N2G SR 51–56, pMSCV N2G SR 52–56 and pMSCV N2G SR 52–56 LEAA constructs were used. These constructs were a kind gift from Dr. Gregg Gundersen and they were described previously [22].
Cell Transfection and Immunostaining
HeLa cells transfection was performed with Effectene (Qiagen) as described by the manufacturer. NIH 3T3 fibroblasts were transfected with adenovirus and then cells were serum-starved for 2 days. Just before fixation, cells were treated with Nocodazole (5μM) for 1h for better nuclear envelope staining visualization. Then, cells were washed in PBS and fixed in −20°C methanol for 10 min. Permeabilization was done with PBS-TritonX-100 (0.1% for 5min), and stained in PBS-Tween (0.05%) supplemented with donkey serum.
Protein purification protocol
GST-tagged proteins were produced using IPTG-inducible BL-21 bacteria. Cells were grown in LB media, pelleted and lysed by sonication in solution (pH=7.5, Tris HCl 50mM, NaCl 100mM, DTT 1mM, EDTA-free protease Inhibitors, Lysozyme 100 ug/ml, DNase 5 ug/mL). Cleared lysates were added to Glutathione agarose beads (Affymetrix, 78820) for purification. Protein elution was performed by adding 20 mM reduced glutathione. Protein was dialyzed against a solution (pH=7.5, Tris HCl 50mM, NaCl 100mM, DTT 1mM) twice, 4 hrs each. His-BicD2-CT was obtained by cleavage of GST-His-BicD2-CT using HRV 3C Protease (PierceTM).
Western Blot and Co-Immunoprecipitation
For co-immunoprecipitation experiments in HeLa cells, these were lysed on ice with RIPA buffer (pH=7.4, 50mM Tris-HCL, 125mM NaCl, 1mM EGTA, 0.5% NP-40) containing 1mM DTT and a protease inhibitor cocktail (Sigma, P8340). GST or GST-BicD2 CT were incubated with Glutathione magnetic beads (Thermo Scientific, 78601) for 30min at 4°C to allow binding of the protein to beads. Subsequently, beads were incubated with the lysate expressing the desired constructs for 3 hrs at 4°C. For the pull-down with brain samples, rat brains were homogenized on ice using brain buffer (pH=7.4, HEPES 50mM, PIPES 50mM, MgCl 2mM, EDTA 1mM, NP-40 0.5%) containing 1mM DTT and a protease inhibitor cocktail (Sigma, P8340). Lysis proceeded for 30 min on ice, and then lysates were clarified removing the debris by centrifugation. Brain lysates were mixed with purified GST or GST-N2G proteins and incubated for 1 hr, at 4°C. After that, washed Glutathione magnetic beads were added and incubated for 2h, at 4°C. For recombinant protein interaction studies, purified proteins were incubated for 1 hr, at 4°C, in a buffer solution (pH=7.5, 50mM Tris-HCL, 100mM NaCl, 1 mM DTT). Washed Glutathione magnetic Beads were added and incubated for 2h, at 4°C. Beads were pelleted and washed 4 times with brain buffer before elution. Eluate was obtained boiling the beads with Laemmli sample buffer.
Purified lysates were loaded on a polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. The membrane was blocked in PBS with 5% milk, incubated with primary antibodies diluted in either PBS with 0.5% Tween or PBS with 1% milk, washed and incubated with secondary LI-COR antibodies in PBS. Imaging of the blots was carried out using an Odyssey system (LI-COR).
Antibodies
Antibodies used for immunofluorescence in brain slices include anti-Nesprin-2 (1:1000, gift from Dr. Gundersen, [23]), anti-HA (Sigma-Aldrich, H6908, 1:2000) and donkey fluorophore-conjugated secondary antibodies (Jackson Labs, 1:500).
Antibodies used for immunofluorescence in cells include anti-dynein intermediate chain clone 74.1 (University of Virginia, 1:250), anti-BicD1 (1:250, Abcam, ab170878), anti-BicD2 (1:250, Abcam, ab117818), anti-Nde1/Ndel1 (1:250, [59]), anti-cyclin B1 (1:100, BD Biosciences, 554177), chicken polyclonal against GFP (1:150, Millipore, 16901), and Donkey fluorophore-conjugated secondary antibodies (Jackson Labs, 1:250 dilution).
Antibodies used for western blotting include anti GST (1:3000, Santa Cruz, sc-53909), anti GFP (1:3000, Invitrogen, A11122), anti-BicD2 (1:2000, Abcam, ab117818), anti BicD2 CT (1:2000, GeneTex, GTX120683). To develop in a LI-COR system, fluorescent secondary antibodies (1:10,000) were acquired from Invitrogen and Rockland.
Microscope image acquisition
The majority of the images were collected with an IX80 laser scanning confocal microscope (Olympus FV100 Spectral Confocal System). Brain sections were imaged using a 60× 1.42 N.A. oil objective or a 10× 0.40 N.A. air objective. Fixed cell images for Figure 3 and Figure S2 were collected using an IX83 Andor Revolution XD Spinning Disk Confocal System with a 1.49 N.A. 100x oil objective and a 2x magnifier coupled to an iXon Ultra 888 EMCCD Camera.
All images were analyzed using ImageJ software (NIH, Bethesda, MD, USA).
The Graphical abstract and the diagrams in Figures 3 G and 7 A, B and B’ were created with BioRender.com.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analysis was performed using Prism (GraphPad Software, La Jolla, CA, USA). Values from the populations under analysis were subjected to the D’Agostino-Pearson omnibus normality test, to determine whether they followed a normal distribution. If the values respected a Gaussian distribution, the Two-tailed parametric unpaired T-test with Welch’s correction was used. But, when the normality test failed, the non-parametric Mann-Whitney test or one-way ANOVA were used. Significance was accepted at the level of P < 0.05.
Somal distribution control raw data was used in Figure 1 D and Figure 5 B. Somal distribution N2G SR raw data was used in Figure 1 D and Figure 6 D. N-C distance control raw data was used in Figure 2 B, Figure 4 D and Figure S4 B. Data for GFP N-C position in Figure 2 D was also depicted in Figure 4 F for better comparison. N-C distance N2G SR raw data was used in Figure 2 B and Figure 6 F.
Supplementary Material
Transfected neurons were continuously imaged in the CP for 9hr50min at 10 min intervals. The arrow indicates the centrosome. Electroporated neurons migrate in a saltatory fashion, in which the centrosome advances followed by the nucleus. Scale bar 10μm.
Transfected neurons were continuously imaged in the CP for 18hr at 10 min intervals. The arrow indicates the centrosome. Although slightly slower, electroporated neurons migrate in a saltatory fashion similar to control cells (Video S1). Scale bar 10μm.
Transfected neurons were continuously imaged in the CP for 12hr40min at 10 min intervals. Arrow and arrowhead indicate, respectively centrosome and cell body position. In contrast to the control (Video S1) and N2G SR-expressing cells (Video S2), the centrosome migrates into the leading process, while the nucleus remains immotile during the live imaging period. At a very high distance from the nucleus, the centrosome and the dilation can be seen making back and forth excursions within the leading process, without net forward movement. Scale bar 10μm.
Transfected neurons were continuously imaged in the CP for 15hr50min at 10 min intervals. Arrow and arrowhead indicate, respectively centrosome and cell body position. The centrosome and the centrosome associated dilation migrate into the leading process, but the nucleus remains immotile during the live imaging period. Similar to the Mini N2G-expressing neurons (Video S3), at a substantial distance from the nucleus, the centrosome can be seen oscillating within the leading process, without net forward movement. Scale bar 10μm.
Transfected neurons were continuously imaged in the CP for 13hr50min at 10 min intervals. Electroporated neurons migrate toward the CP. Scale bar 10μm.
Transfected neurons were continuously imaged in the CP for 12hr40min at 10 min intervals. Electroporated neurons migrate toward the CP, but at a significantly higher velocity (Figure 5 C, D). Scale bar 10μm.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Nesprin-2 | Gift of Dr. Gundersen [23] | N.A. |
| Rabbit polyclonal anti-HA | Sigma-Aldrich | Cat#6908 |
| Mouse monoclonal anti-DIC 74.1 | Univ. of Virginia | N.A. |
| Rabbit monoclonal anti-BicD1 | Abcam | Cat#170878 |
| Rabbit polyclonal anti-BicD2 | Abcam | Cat#117818 |
| Rabbit polyclonal anti-BicD2 CT | GeneTex | Cat#120683 |
| Rabbit polyclonal anti-Nde1/Ndel1 | [59] | N.A. |
| Mouse monoclonal anti-Cyclin B1 | Biosciences | Cat#554177 |
| Chicken polyclonal anti-GFP | Millipore | Cat#16901 |
| Rabbit polyclonal anti-GFP | Invitrogen | Cat#11122 |
| Mouse monoclonal anti-GST | Santa Cruz | Cat#53909 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Aqua-Poly mounting media | Polysciences | Cat#18606 |
| Low melting agarose | IBI Scientific | Cat#70057 |
| Penicillin/streptomycin/glutamine | Life Technologies | Cat#10378-016 |
| Agarose | Sigma | Cat#9539 |
| Dye | Sigma | Cat#7252 |
| Normal Donkey Serum | Sigma | Cat#9663 |
| Hanks balanced salt solution | Life Technologies | Cat#24020-117 |
| Basal MEM | Life Technologies | Cat#21010-046 |
| Horse serum | Life Technologies | Cat#26050-088 |
| Glucose | Sigma | Cat#5767 |
| Critical Commercial Assays | ||
| KOD Hot Start DNA Polymerase | Millipore | Cat#71086 |
| Effectene | Qiagen | Cat#301425 |
| Glutathione agarose beads | Affymetrix | Cat#78820 |
| HRV 3C Protease | PierceTM | Cat#88946 |
| Glutathione magnetic beads | Thermo Scientific | Cat#78601 |
| Experimental Models: Cell Lines | ||
| NIH 3T3 | Gift of Dr. Gundersen, Columbia Univ. | ATCC CRL-1658 |
| HeLa | Gift of Dr. Allan, Univ of Manchester | HELAM |
| Experimental Models: Organisms/Strains | ||
| Sprague Dawley Pregnant Rats | Taconic | N.A. |
| Recombinant DNA | ||
| pCAGIG vector | Addgene [57] | Cat#11159 |
| pCAGIG-BicD2CT | This paper | N.A. |
| pCAGIG Mini N2G SR 52–56 | This paper | N.A. |
| pCAGIG N2G SR 52–56 | This paper | N.A. |
| pCAGIG N2G SR 52–56 LEAA | This paper | N.A. |
| pCAGIG Mini N2G | This paper | N.A. |
| pCAGIG Kif5B | This Paper | Synthesized by Synbio Technologies |
| pCAGIG KLC-TPR | This Paper | Synthesized by Synbio Technologies |
| pCALNL-GFP | Addgene [58] | Cat#13770 |
| pCALNL-BicD2CT | This paper | N.A. |
| pNeuroD1-Cre | Gift of Dr. Cardoso, Institut de Neurobiologie de la Méditerranée | N.A. |
| pRNAT-U6.1/Neo BicD2 | [14] | N.A |
| pGFP-V-RS RanBP2 | [13] | N.A. |
| pRETRO LIC1 | [15] | N.A. |
| mRFP-KASH | [23] | N.A. |
| GST-BicD2-CT | Gift from Dr. Anna Akhmanova, Utrecht University [32] | N.A. |
| His-BicD2-CT | [13] | N.A. |
| GST-N2G | gift from Dr. Gundersen, Columbia University [22] | N.A. |
| pCAG DsRed-PACT | This Paper | N.A. |
| pMSCV-puro GFP-Mini N2G | Gift of Dr. Gundersen, Columbia University [22] | N.A. |
| pMSCV-puro GFP-N2G SR 52–56 | Gift of Dr. Gundersen, Columbia University [22] | N.A. |
| pEGFP-C1 GFP-Mini N2G | Gift of Dr. Gundersen, Columbia University [22] | N.A. |
| pEGFP-C1 GFP-Mini N2G SR 51–26 | Gift of Dr. Gundersen, Columbia University [22] | N.A. |
| pMSCV N2G SR 52–56 | Gift of Dr. Gundersen, Columbia University [22] | N.A. |
| pMSCV N2G SR 52–56 LEAA | Gift of Dr. Gundersen, Columbia University [22] | N.A. |
| Software and Algorithms | ||
| Fiji | Image J | https://fiji.sc |
Highlights.
Nesprin-2 recruits microtubule motors, but not actin, for neuronal migration.
BicD2 mediates dynein and kinesin-1 binding to Nesprin-2.
Kinesin-1 opposes the forward forces exerted by dynein.
Nuclear and centrosome transport are largely independent in neurons.
ACKNOWLEDGMENTS
We thank the members of the Vallee laboratory for the technical expertise and productive discussions. We thank Dr. Aditi Falnikar and Paige Helmer for critical reading of the manuscript. We thank Dr. Tiago Dantas for the PACT-DsRed construct. We thank Dr. Gregg Gundersen for the very useful Nesprin-2 reagents and the extensive critical input. We thank Dr. Ruijun Zhu for the NIH 3T3 cells infected with Nesprin constructs and helpful technical expertise. We thank Dr. Tiago Gil Oliveira for the critical input.
This project was supported by National Institutes of Health grants HD40182 and GM105536 to R.B. Vallee; Fundação para Ciência e a Tecnologia MDPhD Scholarship PD/BD/113766/2015 to J.C. Gonçalves; Fulbright CONICYT Becas Chile 72170520 scholarship to S.Q. This work was also supported by the Columbia University Medical Center MD-PhD program.
Footnotes
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Noctor SC, Martínez-Cerdeño V, Ivic L, and Kriegstein AR (2004). Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci 7, 136–144. [DOI] [PubMed] [Google Scholar]
- 2.Kriegstein A, and Alvarez-buylla A. (2009). The Glial Nature of Embryonic and Adult Neural Stem Cells. Annu. Rev. Neurosci, 149–184. [DOI] [PMC free article] [PubMed]
- 3.Solecki DJ, Model L, Gaetz J, Kapoor TM, and Hatten ME (2004). Par6α signaling controls glial-guided neuronal migration. Nat. Neurosci 7, 1195–1203. [DOI] [PubMed] [Google Scholar]
- 4.Schaar BT, and McConnell SK (2005). Cytoskeletal coordination during neuronal migration. Proc. Natl. Acad. Sci 102, 13652–13657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai J-W, Bremner KH, and Vallee RB (2007). Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci 10, 970–979. [DOI] [PubMed] [Google Scholar]
- 6.Bellion A, Baudoin J-P, Alvarez C, Bornens M, and Métin C. (2005). Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J. Neurosci 25, 5691–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertipaglia C, Gonçalves JC, and Vallee RB (2018). Nuclear migration in mammalian brain development. Semin. Cell Dev. Biol 82, 57–66. [DOI] [PubMed] [Google Scholar]
- 8.Poirier K, Lebrun N, Broix L, Tian G, Saillour Y, Boscheron C, Parrini E, Valence S, Pierre B. Saint Oger M., et al. (2013). Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet 45, 639–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jamuar SS, Lam A-TN, Kircher M, D’Gama AM, Wang J, Barry BJ, Zhang X, Hill RS, Partlow JN, Rozzo A, et al. (2014). Somatic Mutations in Cerebral Cortical Malformations. N. Engl. J. Med 371, 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, and Ledbetter DH (1993). Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature 364, 717–721. [DOI] [PubMed] [Google Scholar]
- 11.Doobin DJ, Kemal S, Dantas TJ, and Vallee RB (2016). Severe NDE1-mediated microcephaly results from neural progenitor cell cycle arrests at multiple specific stages. Nat. Commun 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai JW, Chen Y, Kriegstein AR, and Vallee RB (2005). LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J. Cell Biol 170, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baffet AD, Hu DJ, and Vallee RB (2015). Cdk1 Activates Pre-mitotic Nuclear Envelope Dynein Recruitment and Apical Nuclear Migration in Neural Stem Cells. Dev. Cell 33, 703–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu DJ-K, Baffet ADD, Nayak T, Akhmanova A, Vallee RBB, Doye V, and Vallee RBB (2013). Dynein Recruitment to Nuclear Pores Activates Apical Nuclear Migration and Mitotic Entry in Brain Progenitor Cells. Cell 154, 1300–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonçalves JC, Dantas TJ, and Vallee RB (2019). Distinct roles for dynein light intermediate chains in neurogenesis, migration, and terminal somal translocation. J. Cell Biol 218, 808–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu T, Ayala R, Nguyen MD, Xie Z, Gleeson JG, and Tsai LH (2004). Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44, 263–277. [DOI] [PubMed] [Google Scholar]
- 17.Solecki DJ, Trivedi N, Govek E-E, Kerekes RA, Gleason SS, and Hatten ME (2009). Myosin II Motors and F-Actin Dynamics Drive the Coordinated Movement of the Centrosome and Soma during CNS Glial-Guided Neuronal Migration. Neuron 63, 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, and Gleeson JG (2004). Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J. Cell Biol 165, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu YK, Umeshima H, Kurisu J, and Kengaku M. (2018). Nesprins and opposing microtubule motors generate a point force that drives directional nuclear motion in migrating neurons. Development 145, dev158782. [DOI] [PubMed] [Google Scholar]
- 20.Chang W, Worman HJ, and Gundersen GG (2015). Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol 208, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomes ER, Jani S, and Gundersen GG (2005). Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell 121, 451–463. [DOI] [PubMed] [Google Scholar]
- 22.Zhu R, Antoku S, and Gundersen GG (2017). Centrifugal Displacement of Nuclei Reveals Multiple LINC Complex Mechanisms for Homeostatic Nuclear Positioning. Curr. Biol, 1–14. [DOI] [PMC free article] [PubMed]
- 23.Luxton GWG, Gomes ER, Folker ES, Vintinner E, and Gundersen GG (2010). Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 329, 956–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, and Han M. (2009). SUN1/2 and Syne/Nesprin-1/2 Complexes Connect Centrosome to the Nucleus during Neurogenesis and Neuronal Migration in Mice. Neuron 64, 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kutscheidt S, Zhu R, Antoku S, Luxton GWG, Stagljar I, Fackler OT, and Gundersen GG (2014). FHOD1 interaction with nesprin-2G mediates TAN line formation and nuclear movement. Nat. Cell Biol 16, 708–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baudoin JP, Viou L, Launay PS, Luccardini C, Espeso Gil S, Kiyasova V, Irinopoulou T, Alvarez C, Rio JP, Boudier T, et al. (2012). Tangentially Migrating Neurons Assemble a Primary Cilium that Promotes Their Reorientation to the Cortical Plate. Neuron 76, 1108–1122. [DOI] [PubMed] [Google Scholar]
- 27.Gillingham AK, and Munro S. (2000). The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1, 524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, and Matsuzaki F. (2008). Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat. Cell Biol 10, 93–101. [DOI] [PubMed] [Google Scholar]
- 29.Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, and Starr DA (2009). UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development 136, 2725–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fridolfsson HN, and Starr DA (2010). Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J. Cell Biol 191, 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fridolfsson HN, Ly N, Meyerzon M, and Starr DA (2010). UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev. Biol 338, 237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu K. Lou Grigoriev I., Engelsma D, Haasdijk ED, Keijzer N, et al. (2010). Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, and Doye V. (2011). A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J. Cell Biol 192, 855–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wynne CL, and Vallee RB (2018). Cdk1 phosphorylation of the dynein adapter Nde1 controls cargo binding from G2 to anaphase. J. Cell Biol, 1–11. [DOI] [PMC free article] [PubMed]
- 35.Splinter D, Razafsky DS, Schlager MA, Serra-Marques A, Grigoriev I, Demmers J, Keijzer N, Jiang K, Poser I, Hyman AA, et al. (2012). BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol. Biol. Cell 23, 4226–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, and Vale RD (2014). Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, and Carter AP (2015). The structure of the dynactin complex and its interaction with dynein. Science (80-. ). 347, 1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grigoriev I, Splinter D, Keijzer N, Wulf PS, Demmers J, Ohtsuka T, Modesti M, Maly IV, Grosveld F, Hoogenraad CC, et al. (2007). Rab6 Regulates Transport and Targeting of Exocytotic Carriers. Dev. Cell 13, 305–314. [DOI] [PubMed] [Google Scholar]
- 39.Matanis T, Akhmanova A, Wulf P, Del Nery E, Weide T, Stepanova T, Galjart N, Grosveld F, Goud B, De Zeeuw CI, et al. (2002). Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol 4, 986–992. [DOI] [PubMed] [Google Scholar]
- 40.Jaarsma D, van den Berg R, Wulf PS, van Erp S, Keijzer N, Schlager M. a, de Graaff E, De Zeeuw CI, Pasterkamp RJ, Akhmanova A, et al. (2014). A role for Bicaudal-D2 in radial cerebellar granule cell migration. Nat. Commun 5, 3411. [DOI] [PubMed] [Google Scholar]
- 41.Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, and Polleux F. (2009). The F-BAR Domain of srGAP2 Induces Membrane Protrusions Required for Neuronal Migration and Morphogenesis. Cell 138, 990–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carabalona A, Hu DJ-K, and Vallee RB (2016). KIF1A inhibition immortalizes brain stem cells but blocks BDNF-mediated neuronal migration. Nat. Neurosci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider M, Lu W, Neumann S, Brachner A, Gotzmann J, Noegel AA, and Karakesisoglou I. (2011). Molecular mechanisms of centrosome and cytoskeleton anchorage at the nuclear envelope. Cell. Mol. Life Sci 68, 1593–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson MH, and Holzbaur ELF (2012). Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J. Cell Sci 125, 4158–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson MH, and Holzbaur ELF (2015). Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development 142, 218–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lipka J, Kuijpers M, Jaworski J, and Hoogenraad CC (2013). Mutations in cytoplasmic dynein and its regulators cause malformations of cortical development and neurodegenerative diseases. Biochem. Soc. Trans 41, 1605–12. [DOI] [PubMed] [Google Scholar]
- 47.Bone CR, and Starr DA (2016). Nuclear migration events throughout development. J. Cell Sci 129, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martini FJ, and Valdeolmillos M. (2010). Actomyosin Contraction at the Cell Rear Drives Nuclear Translocation in Migrating Cortical Interneurons. J. Neurosci 30, 8660–8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He M, Zhang Z. -h, Guan C. -b., Xia D, and Yuan X. -b. (2010). Leading Tip Drives Soma Translocation via Forward F-Actin Flow during Neuronal Migration. J. Neurosci 30, 10885–10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang J, Zhang Z. hong Yuan, bin X, and Poo M. ming(2015). Spatiotemporal dynamics of traction forces show three contraction centers in migratory neurons. J. Cell Biol 209, 759–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trivedi N, Stabley DR, Cain B, Howell D, Laumonnerie C, Ramahi JS, Temirov J, Kerekes RA, Gordon-Weeks PR, and Solecki DJ (2017). Drebrin-mediated microtubule-actomyosin coupling steers cerebellar granule neuron nucleokinesis and migration pathway selection. Nat. Commun 8, 14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoogenraad CC, and Akhmanova A. (2016). Bicaudal D Family of Motor Adaptors: Linking Dynein Motility to Cargo Binding. Trends Cell Biol. 26, 327–340. [DOI] [PubMed] [Google Scholar]
- 53.Cai Y, Singh BB, Aslanukov A, Zhao H, and Ferreira PA (2001). The Docking of Kinesins, KIF5B and KIF5C, to Ran-binding Protein 2 (RanBP2) Is Mediated via a Novel RanBP2 Domain. J. Biol. Chem 276, 41594–41602. [DOI] [PubMed] [Google Scholar]
- 54.Umeshima H, Hirano T, and Kengaku M. (2007). Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc. Natl. Acad. Sci 104, 16182–16187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui H, Noell CR, Behler RP, Zahn JB, Terry LR, Russ BB, and Solmaz SR (2019). Adapter Proteins for Opposing Motors Interact Simultaneously with Nuclear Pore Protein Nup358. Biochemistry 58, 5085–5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baffet AD, Carabalona A, Dantas TJ, Doobin DD, Hu DJ, and Vallee RB (2016). Cellular and subcellular imaging of motor protein-based behavior in embryonic rat brain. Methods Cell Biol. 131, 349–363. [DOI] [PubMed] [Google Scholar]
- 57.Matsuda T, and Cepko CL (2004). Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci 101, 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsuda T, and Cepko CL (2007). Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci 104, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stehman S, Chen Y, McKenney RJ, and Vallee RB (2007). NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J. Cell Biol 178, 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transfected neurons were continuously imaged in the CP for 9hr50min at 10 min intervals. The arrow indicates the centrosome. Electroporated neurons migrate in a saltatory fashion, in which the centrosome advances followed by the nucleus. Scale bar 10μm.
Transfected neurons were continuously imaged in the CP for 18hr at 10 min intervals. The arrow indicates the centrosome. Although slightly slower, electroporated neurons migrate in a saltatory fashion similar to control cells (Video S1). Scale bar 10μm.
Transfected neurons were continuously imaged in the CP for 12hr40min at 10 min intervals. Arrow and arrowhead indicate, respectively centrosome and cell body position. In contrast to the control (Video S1) and N2G SR-expressing cells (Video S2), the centrosome migrates into the leading process, while the nucleus remains immotile during the live imaging period. At a very high distance from the nucleus, the centrosome and the dilation can be seen making back and forth excursions within the leading process, without net forward movement. Scale bar 10μm.
Transfected neurons were continuously imaged in the CP for 15hr50min at 10 min intervals. Arrow and arrowhead indicate, respectively centrosome and cell body position. The centrosome and the centrosome associated dilation migrate into the leading process, but the nucleus remains immotile during the live imaging period. Similar to the Mini N2G-expressing neurons (Video S3), at a substantial distance from the nucleus, the centrosome can be seen oscillating within the leading process, without net forward movement. Scale bar 10μm.
Transfected neurons were continuously imaged in the CP for 13hr50min at 10 min intervals. Electroporated neurons migrate toward the CP. Scale bar 10μm.
Transfected neurons were continuously imaged in the CP for 12hr40min at 10 min intervals. Electroporated neurons migrate toward the CP, but at a significantly higher velocity (Figure 5 C, D). Scale bar 10μm.
Data Availability Statement
The authors declare that all relevant data supporting the findings of this study are provided in the Article and Supplementary files.