Abstract
The recent discovery that the Treponema pallidum genome encodes 12 orthologs of the Treponema denticola major sheath protein (Msp) prompted us to reexamine the cellular location and topology of the T. denticola polypeptide. Experiments initially were conducted to ascertain whether Msp forms an array on or within the T. denticola outer membrane. Transmission electron microscopy (EM) of negatively stained and ultrathin-sectioned organisms failed to identify a typical surface layer, whereas freeze-fracture EM revealed that the T. denticola outer membrane contains heterogeneous transmembrane proteins but no array. In contrast, a lattice-like structure was observed in vesicles released from mildly sonicated treponemes; combined EM and biochemical analyses demonstrated that this structure was the peptidoglycan sacculus. Immunoelectron microscopy (IEM) subsequently was performed to localize Msp in T. denticola. Examination of negatively stained whole mounts identified substantial amounts of Msp in sonicated organisms. IEM of ultrathin-sectioned, intact treponemes also demonstrated that the preponderance of antigen was unassociated with the outer membrane. Lastly, immunofluorescence analysis of treponemes embedded in agarose gel microdroplets revealed that only minor portions of Msp are surface exposed. Taken as a whole, our findings challenge the widely held belief that Msp forms an array within the T. denticola outer membrane and demonstrate, instead, that it is predominantly periplasmic with only limited surface exposure. These findings also have implications for our evolving understanding of the contribution(s) of Msp/Tpr orthologs to treponemal physiology and disease pathogenesis.
During the past decade, the search for rare outer membrane proteins of Treponema pallidum as potential virulence determinants and vaccinogens has become a major focus of syphilis research (38). Although diverse experimental strategies have been developed to achieve this important objective (5, 6, 14, 24, 44, 54), none has resulted in the identification of T. pallidum proteins which are unequivocally surface exposed (1, 14, 51). The recently completed T. pallidum genomic sequence (21) has provided an alternative means of searching for these low-abundance, poorly immunogenic polypeptides (21). In this regard, a potential major advance has been the finding that the T. pallidum chromosome encodes 12 polypeptides with sequence relatedness to the major sheath protein (Msp) of the cultivatable oral treponeme Treponema denticola (12, 21), a bacterium implicated in the progression of periodontal disease. This abundant 55-kDa polypeptide has been reported to form a hexagonal array within the T. denticola outer membrane (or outer sheath), to exhibit pore-forming activity in artificial and HeLa cell membranes, to bind extracellular-matrix components, to function as a cytadhesin, and to induce cytopathic effects in cultured epithelial cells (16, 17, 23, 32). Not surprisingly, there has been speculation that one or more of the T. pallidum Msp orthologs (designated T. pallidum repeat [Tpr] proteins) perform analogous physiological and/or virulence-related functions (21).
A number of considerations prompted us to undertake a detailed ultrastructural investigation of T. denticola Msp. Given the protein’s abundance and the cultivability of T. denticola, we reasoned that it could serve as a prototype for the ostensibly more complex Tpr system of the noncultivatable syphilis spirochete and that the resulting information could provide a conceptual backdrop for studies of the T. pallidum orthologs. A systematic reexamination from a morphological perspective appeared to be warranted further by the fact that an analogous lattice has never been observed on or within the T. pallidum outer membrane (26, 38). A review of previously published data also led us to question whether this protein had been correctly localized in T. denticola. It is well known that spirochetal outer membranes are easily disrupted during experimental manipulations and that subsurface antigens may be inadvertently localized to the spirochetal surface when precautions to ensure the integrity of the outer membrane are not taken (2, 14, 38). It should be noted, therefore, that micrographs purporting to localize Msp to the T. denticola outer membrane show immunogold labeling of treponemes disrupted by sonication or released material whose putative outer membrane origin was not rigorously determined (16, 17, 23). Also problematic is the notion that Msp forms an electron-dense polygonal array with porin-like properties. Proteinaceous two-dimensional arrays typically are extrinsic to bacterial outer membranes (hence their designation as surface- or S-layers) rather than embedded within them (46). Porin trimers, on the other hand, are tightly packed within gram-negative bacterial outer membranes (50, 56) but do not normally form true polygonal arrays (34, 49). Lastly, the recent demonstration by Fenno et. al. (17) that Msp lacks amphiphilic character by Triton X-114 phase partitioning suggests that it is not a conventional outer membrane protein.
In the present study, we provide evidence that challenges the contention that Msp forms a lattice or array within the T. denticola outer membrane. Our findings demonstrate instead that this protein is predominantly periplasmic and has only limited surface exposure. In addition to resolving apparent inconsistencies involving this protein, these results have important implications for our evolving appreciation of the contribution(s) of Msp/Tpr orthologs to treponemal physiology and disease pathogenesis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
T. denticola ATCC 35405 was grown in new oral spirochete (NOS) broth (23) supplemented with 10% heat-inactivated normal rabbit serum (Pel-Freeze Biologicals, Rogers, Ark.) by using the GasPak Plus anaerobic system (Becton Dickinson, Cockeysville, Md.). Wild-type Campylobacter rectus ATCC 33238 and an S-layer-deficient spontaneous mutant (35) grown on mycoplasma-formate-fumarate agar were the generous gifts of Stanley C. Holt (University of Texas Health Sciences Center at San Antonio). Escherichia coli DH10B (Gibco/BRL, Gaithersburg, Md.) was used for transformation experiments. Except where otherwise stated, transformants were grown on yeast-tryptone agar or broth supplemented with the appropriate antibiotic. Plasmid pProEx (Gibco/BRL) was used to generate a His6-tagged Msp fusion protein. The plasmid pCR2.1-TOPO (Invitrogen, San Diego, Calif.) was used for the cloning of PCR products.
Construction of a His6-tagged Msp fusion protein.
A His6-tagged Msp fusion protein was constructed by cloning a DNA fragment encoding amino acids 21 (the residue following the proposed signal peptidase I cleavage site) to 543 of T. denticola 35405 Msp (18) into the BamHI and NotI sites of pProEx. The DNA fragment was PCR amplified from T. denticola DNA with the following primers: 5′-GGATCCCCCCAACTTACACCTCAAGTTACAGC-3′ (BamHI site is underlined, plus nucleotides 61 to 83) and 5′-GCGGCCGCTTATTAGTAGATAACTTTAACACCGATTAC-3′ (NotI site is underlined, plus nucleotides complementary to bases 1607 to 1632) (18). The nucleotide sequence of the PCR product matched exactly that reported for the msp gene of the ATCC 35405 isolate (18). The resulting fusion protein was purified by using a nickel-NTA agarose affinity matrix (Qiagen, Santa Clarita, Calif.) according to the manufacturer’s instructions.
Immunologic reagents.
Rat antiserum against whole T. denticola was generated by priming Sprague Dawley rats with 1011 organisms in a 1:1 mixture of Freund complete adjuvant (Difco Laboratories, Detroit, Mich.) and phosphate-buffered saline (PBS; pH 7.4) (total volume, 0.5 ml) administered intraperitoneally. After 3 and 6 weeks, the animals were boosted intraperitoneally with the same number of organisms in a 1:1 mixture of Freund incomplete adjuvant (Difco Laboratories). To generate anti-Msp antiserum, rats were primed by an intraperitoneal injection of 100 μg of Msp fusion protein in a 1:1 mixture of Freund complete adjuvant and PBS (total volume, 0.5 ml). After 3 and 6 weeks, the animals were boosted intraperitoneally with the same amount of protein in a 1:1 mixture of Freund incomplete adjuvant. Immunoblot analysis revealed that the resulting antiserum reacted with a single 55-kDa polypeptide in T. denticola whole-cell lysates. The generation of rabbit antiserum directed against T. pallidum flagellin was as described previously (42).
Purification and biochemical analysis of T. denticola peptidoglycan.
Peptidoglycan was purified from T. denticola as described by Joseph et al. (27), except that sodium dodecyl sulfate-extracted cell bodies were treated overnight at 55°C with proteinase K (1 mg/ml) instead of pronase. The peptidoglycan sacculi were collected by centrifugation at 108,000 × g for 1 h and washed extensively with MilliQ water; the final pellet was resuspended in 100 μl of water. For biochemical analysis, a 10-μl portion was mixed with 4 μl of a 250-pmol/μl concentration of α-aminobutyric acid as an internal standard. The sample was then hydrolyzed for 22 h at 110°C in vacuo by using vapor-phase HCl. Derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (supplied as the AccQ-Fluor reagent kit; Waters, Milford, Mass.) was performed in a volume of 100 μl as described by van Wandelen and Cohen (58). Three separate 5-μl samples were subjected to amino acid analysis on a Waters Alliance system consisting of a 2690XE Separations Module and a 474 scanning fluorescence detector. Separations were carried out on a Sentry guard column (20 by 3.9 mm; Nova-Pak C18 bonded silica) connected to an AccQ-Tag column (150 by 3.9 mm; both from Waters) with a step gradient comprised of buffers A (19% sodium acetate, 6.9% phosphoric acid, 17% triethylamine, 0.1% sodium azide, 72.3% water) and B (60% acetonitrile, 40% water). Eluted compounds were detected by fluorescence with excitation at 250 nm and emission at 395 nm. The Waters Millennium 2010 Chromatography Manager was used for systems operation and data management.
Localization of Msp by indirect immunofluorescence analysis of T. denticola encapsulated in gel microdroplets.
Mid-logarithmic-phase T. denticola was encapsulated in microdroplets of low-melting-point agarose as previously described (13, 14). Encapsulated organisms were probed in a two-step process. In the first step, rat anti-Msp, rat anti-T. denticola, or rabbit anti-T. pallidum flagellin antiserum diluted 1:200, 1:1,000, or 1:100, respectively, was added to the bead suspensions (0.2 to 0.3 ml) in the presence or absence of 0.05% (vol/vol) Triton X-100. In two experiments, encapsulated organisms were fixed with 0.5% formalin (3) and extensively washed with PBS–1 mM MgCl2 and NOS broth prior to the addition of primary antibody. Samples were incubated with gentle mixing in a 34°C water bath for 2 h. The beads were then washed three times by low-speed centrifugation (100 × g) and resuspensed in NOS broth followed by incubation for 1 h at 34°C with 1 μg of Alexa 488 conjugates of goat anti-rat or goat anti-rabbit immunoglobulin G (IgG; both from Molecular Probes, Eugene, Oreg.) per ml. The beads were then washed three times with NOS broth and observed with a Nikon Optiphot-2 fluorescence microscope (Nikon, Inc., Melville, N.Y.) equipped with ×15 oculars, a darkfield condenser, and a fluorescein filter block. Samples were enumerated at ×600 total magnification; photomicrographs were taken with a ×100 oil immersion objective at ×1,500 total magnification. For each sample, three slides were prepared, and approximately 100 organisms were scored for labeling.
Routine transmission electron microscopy (EM). (i) Intact treponemes.
First, 5-ml portions of a mid-logarithmic-phase culture were fixed by the addition of equal volumes of 4% glutaraldehyde–4% paraformaldehyde (vol/vol [final concentration, 2% glutaraldehyde–2% paraformaldehyde]). For the examination of whole mounts, 5-μl droplets were applied directly to formvar-carbon-coated 200-mesh copper grids (Energy Beam Sciences, Agawam, Mass.) which had been glow discharged immediately prior to use. The grids were floated on the suspension for 5 min at room temperature and then washed three times with filtered, distilled water before being stained with 1% uranyl acetate (Electron Microscopy Sciences, Ft. Washington, Pa.). For ultrathin sections, the fixed cells were pelleted in a microfuge; the pellets were gently dislodged and embedded in 2% low-melting-temperature agarose. The samples were cut into blocks of approximately 2 mm3 and placed into shell vials containing 0.2 M sodium cacodylate buffer (pH 7.3). The blocks were covered with 2% osmium tetroxide in 0.1 M sodium cacodylate and allowed to react overnight at 4°C; the following day, they were washed three times with Veronal acetate buffer (pH 5.0). After a final 2-h incubation with rocking in fresh buffer at 4°C, the blocks were placed in 2% uranyl acetate-Veronal acetate and allowed to rock for an additional 1 h at 4°C. The samples were then taken through a dehydration series of ethanol, followed by propylene oxide, and then embedded in Epon-Araldite. Ultrathin sections were then stained by submersion for 10 min in 50% ethanol-water saturated with uranyl acetate, rinsed for 15 s with deionized water, and counter-stained by submersion in Reynold’s lead citrate.
(ii) Sonicated treponemes.
Approximately 5 × 108 cells in mid-logarithmic phase were centrifuged at 20,000 × g for 20 min at 4°C. The pelleted bacteria were gently washed once with PBS and resuspended in 1 ml of PBS. The cells were then sonicated for 10 s by using a Model 550 Sonic Dismembrator (Fisher Scientific, Houston, Tex.) at a setting of 5 and with a 15% output. After sonication, cells were either mounted on copper grids for negative staining or embedded in Epon for ultrathin sectioning as described above.
(iii) Wild-type and S-layer-deficient C. rectus.
Isolated colonies were gently scraped from the agar surface and resuspended in 0.5 ml of PBS. Then, 5-μl portions of the suspensions were applied to grids for negative staining with 1% uranyl acetate. Alternatively, cells were fixed in suspension by the addition of an equal volume of 4% glutaraldehyde–4% paraformaldehyde and processed for ultrathin sectioning as described above.
(iv) Peptidoglycan.
A 5-μl portion of the washed peptidoglycan suspension was applied to glow-discharged carbon-formvar-coated grids and negatively stained with 1% uranyl acetate.
All specimens were examined with a JEOL JEM-100CX transmission electron microscope (JEOL USA, Inc., Peabody, Mass.) at an accelerating voltage of 80 kV.
IEM. (i) Sonicated whole mounts.
Immunoelectron microscopy (IEM) of whole mounts was performed by using a modification of the method of Cox et al. (15); mid-logarithmic-phase cells were sonicated and applied to copper grids as described above. After two washes with PBS, grids were floated for 5 min on droplets of 1× CMRL (Gibco/BRL) containing 10% fetal calf serum (CMRL-FCS). The grids were then floated for 10 min on droplets containing primary antibody diluted 1:100 in CMRL-FCS. After two washes with CMRL, the grids were then floated for 10 min on droplets containing colloidal-gold-conjugated anti-rat or anti-rabbit IgG (EY Laboratories, San Mateo, Calif.) diluted 1:5 in CMRL-FCS. After a final wash with CMRL and distilled water, the grids were negatively stained with 1% uranyl acetate.
(ii) Intact treponemes.
Five milliliters of a mid-logarithmic-phase culture was fixed for 2 h after the addition of an equal volume of 8% paraformaldehyde–0.4% glutaraldehyde (vol/vol). Fixed cells were washed once in PBS and then embedded in low-melting-temperature agarose. The agarose pellets were then cut into blocks of approximately 2 mm3. The blocks were suspended in 2% uranyl acetate for 2 h at 4°C with gentle rocking. The pellets were dehydrated in ethanol, followed by infiltration with LR Gold (London Resin Company, Basingstoke, Hampshire, United Kingdom) according to the manufacturer’s instructions. The LR Gold was polymerized under long UV light (366 nm) for 24 h at −20°C. After polymerization, the blocks were trimmed and thin sectioned at ca. 50 nm on a Reichert-Jung Ultracut E microtome (Reichert-Jung Optische Werke AG, Wien, Austria). Specimens were collected on formvar-carbon-coated grids and immunostained with 1:100 dilutions of rat or rabbit anti-Msp antiserum. The sections were then floated on 2% aqueous glutaraldehyde for 5 min, washed for 30 s with deionized water, floated for 15 min on 2% osmium tetroxide, washed once more with water for 30 s, and then stained for 3 min with Reynold’s lead citrate (4).
Freeze-fracture EM.
A 50-ml culture of mid-logarithmic-phase cells was divided in half. One of the aliquots was transferred to an ice bath, while the other was maintained at 34°C. Thirty minutes later, both aliquots were pelleted by centrifugation at 10,000 × g in centrifuges which had been temperature adjusted to 0 or 34°C. The pellets were then resuspended in temperature-adjusted 30% aqueous glycerol solutions. After 30 min, the cells were harvested in a microcentrifuge, and the pellets transferred to gold Balzer support pins (Baltek, Middlebury, Conn.). The cells were snap frozen by plunging the pins into liquid ethane and then stored in liquid nitrogen prior to fracturing. Frozen samples were placed on a liquid nitrogen-cooled specimen support table and inserted into the chamber of a Balzer 400 freeze-fracture device which had been cooled to −170°C. The specimens were warmed to −105°C, and the chamber evacuated to 5.32 × 10−7 torrs prior to cleavage. Replicas were produced by stationary platinum shadowing at 45° and rotary carbon shadowing at 90°. Replicas were floated in undiluted bleach (Clorox) for approximately 12 h and washed three times in filtered, double-distilled water. They were then transferred to formvar-coated 200-mesh copper grids and examined by transmission EM as described above.
RESULTS
T. denticola lacks a typical surface layer.
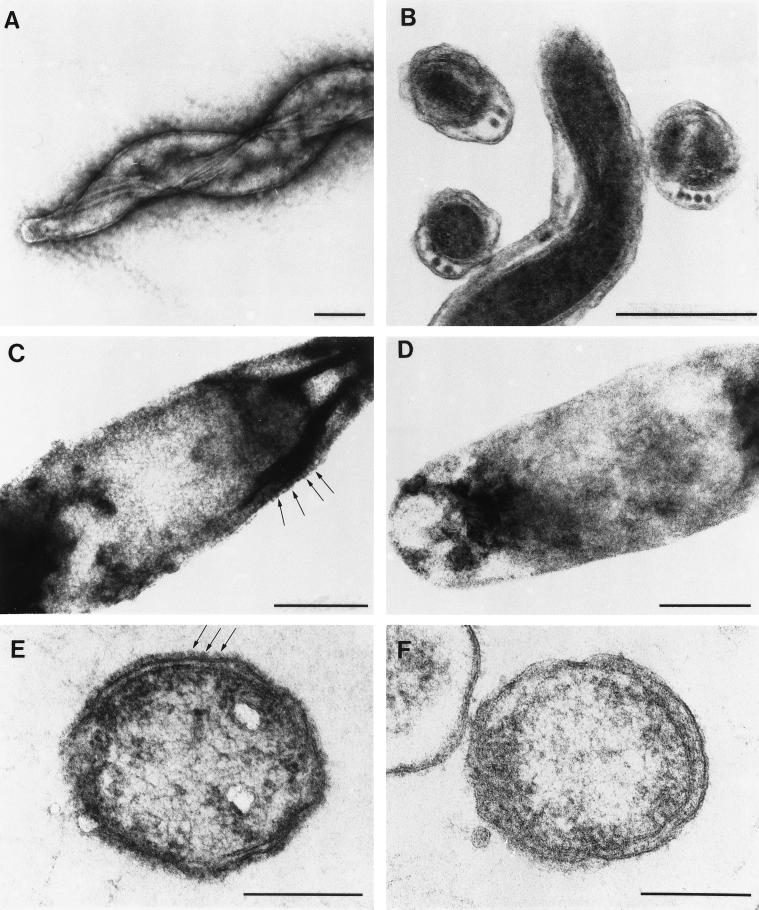
Msp oligomers reportedly form hexagonal arrays within the T. denticola outer membrane (16, 17). However, two-dimensional proteinaceous arrays in eubacteria typically are extrinsic to the outer membrane (46). In light of this apparent discrepancy, we began our investigation by determining whether the previously observed lattice is actually external to the T. denticola outer membrane rather than embedded within it. To prevent the possible loss of surface-associated components in these experiments, motile treponemes were fixed in suspension and subjected to minimal manipulations prior to EM analysis. The intactness of the negatively stained whole mounts was evident by the fact that flagella were tightly wound around the protoplasmic cylinder and an outer membrane surrounding the protoplasmic cylinder could be identified at multiple points along the length of the cell (Fig. 1A). Careful examination of these organisms failed to reveal a periodic structure consistent with an S-layer (53). The high degree of visible internal structure also argued against the presence of an external electron-dense layer. To confirm these findings, we also examined T. denticola in ultrathin sections. As shown in Fig. 1B, the outer membranes were smooth and lacked the characteristic external “beading” of S-layers (53). As a control for these experiments, we analyzed C. rectus, an anaerobic oral gram-negative bacterium which possesses an S-layer composed of hexagonally arrayed subunits, as well as an S-layer-deficient C. rectus spontaneous mutant (35). A hexagonal lattice was discernible on the surface of the negatively stained wild-type isolate (Fig. 1C) but was absent from the mutant (Fig. 1D); identification of subunits on the cell periphery (Fig. 1C) confirmed that this structure was external rather than internal. Subunits of the S-layer also were visualized in ultrathin sections containing wild-type C. rectus but were not observed in sections containing the mutant (Fig. 1E and F, respectively).
FIG. 1.
T. denticola lacks a typical S-layer. T. denticola (A and B), wild-type C. rectus (C and E), and an S-layer-deficient C. rectus mutant (D and F) were prepared as negatively stained whole mounts (A, C, and D) or as ultrathin sections (B, E, and F). Arrows in panels C and E indicate the S-layer subunits on the surface of wild-type C. rectus. Bars, 0.25 μm.
The T. denticola outer membrane contains transmembrane proteins but no array.
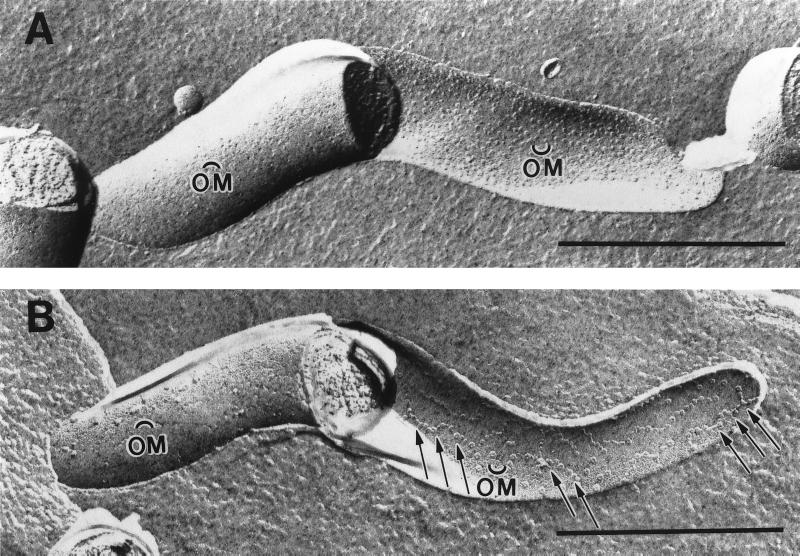
Having ruled out the existence of an S-layer, we next sought to confirm reports of an ordered structure within the T. denticola outer membrane (16). Freeze-fracture electron microscopy is an appropriate technique to address this issue because it visualizes proteins within the hydrophobic interior of a lipid bilayer (20). In fact, this methodology has been used to identify complex proteinaceous structures within biological membranes, including one (designated “linear bodies”) which we described in an earlier freeze-fracture study of the Borrelia burgdorferi outer envelope (39, 57). As shown in Fig. 2A, freeze-fractured T. denticola outer membranes contained numerous intramembranous particles representing proteins with membrane-spanning domains (20). The large majority of the outer membrane particles segregated with the concave or external leaflet, a phenomenon previously observed with other cultivatable spirochetes (7, 39, 60). The heterogeneous appearance and relatively high density of these particles in the concave outer membrane leaflet (2,142 particles/μ2) were distinctly different from the relatively uniform, low-density particles in T. pallidum outer membranes (43). It should be noted that the ostensibly random distribution of the T. denticola outer membrane particles was inconsistent with the presence of an array. To confirm that these particles did not form an ordered structure, we also performed freeze-fracture EM analysis of treponemes after immersion in an ice bath. Slow cooling in this manner induces lipid-phase separations; membrane proteins, when mobile, are excluded from enlarging crystalline domains and form aggregates separated by protein-free patches (30, 33). On the other hand, aggregation will not occur if the mobility of the membrane proteins is constrained by their organization into a rigid structure such as an array. As shown in Fig. 2B, aggregates of membrane proteins were plainly visible in the respective concave and convex outer membrane leaflets.
FIG. 2.
Absence of an array within the T. denticola outer membrane. Micrographs showing mid-logarithmic-phase T. denticola incubated at 34°C (A) or 0°C (B) prior to processing for freeze-fracture TEM. OM and OM designate the respective convex and concave outer membrane leaflets. The arrows in panel B indicate particle aggregates in the concave outer membrane leaflet. Bars, 0.5 μm.
Identification of the peptidoglycan sacculus as the lattice-like structure in T. denticola.
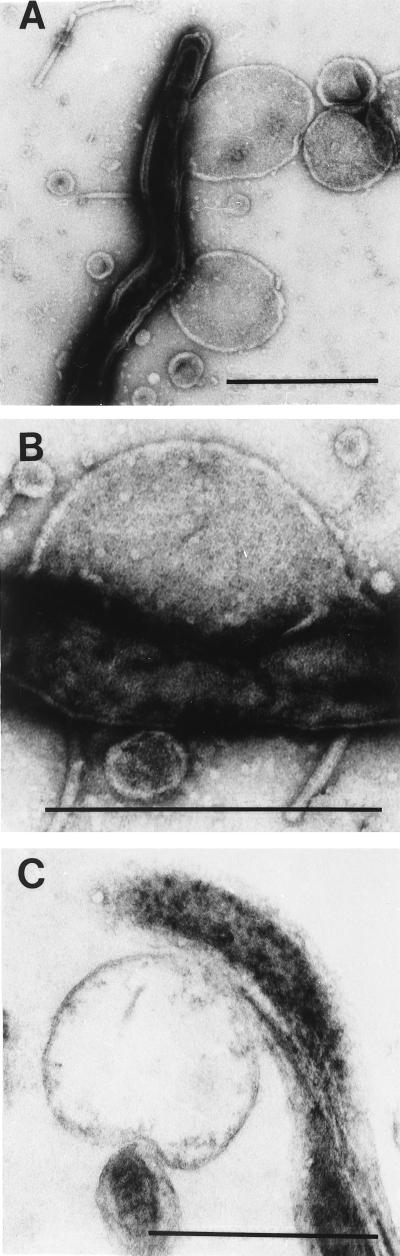
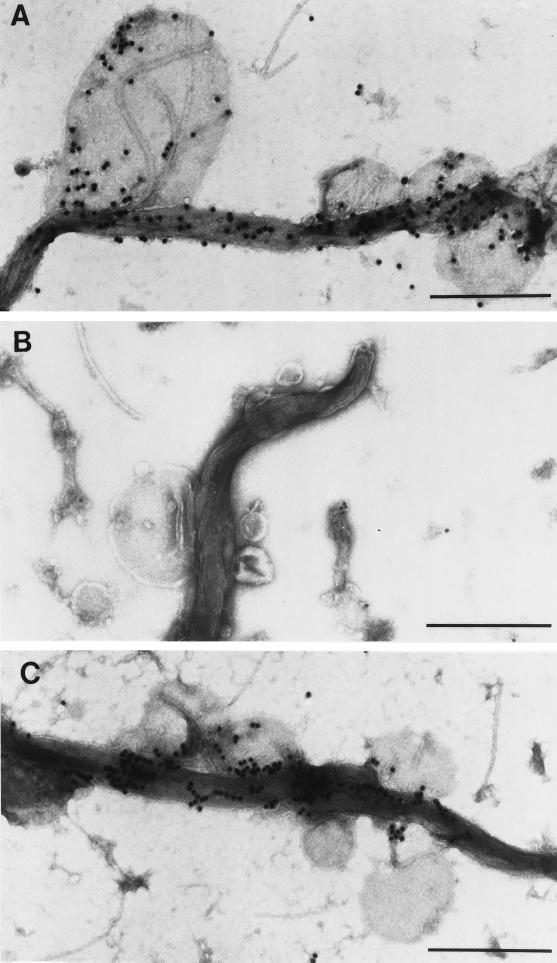
Because we were unable to identify a lattice on or within the T. denticola outer membrane, we next sought evidence that it was located intracellularly. As in prior studies (16, 17, 19), mild sonication was used to expose the presumptive Msp lattice for subsequent EM analysis. Negatively stained whole mounts contained cell cylinders with attached or closely associated membranous blebs of various sizes; identical free-standing vesicles also were visualized in all fields (Fig. 3A). An underlying lattice-like structure was discernible when attached and isolated vesicles were examined at high power, although a definite hexagonal array could not be visualized (Fig. 3B). In addition to confirming that the sonicated cells lacked outer membranes, it was evident from ultrathin sections that the blebs were extrusions from the cytoplasmic membrane (Fig. 3C).
FIG. 3.
T. denticola after mild sonication. (A) Negatively stained whole mount showing a cell cylinder, as well as attached and free-standing vesicles. (B) Negatively stained cell cylinder and attached vesicle at a higher magnification. (C) Ultrathin section of a cell cylinder and attached vesicle. Bars, 0.5 μm.
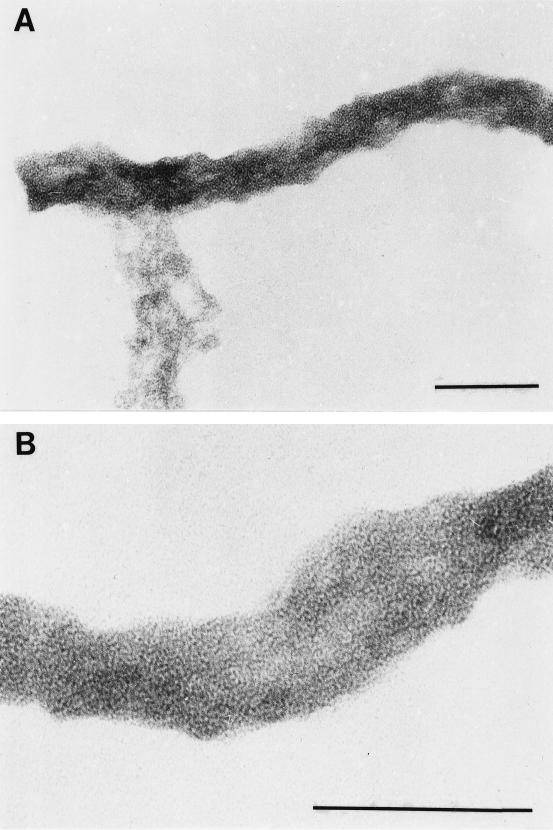
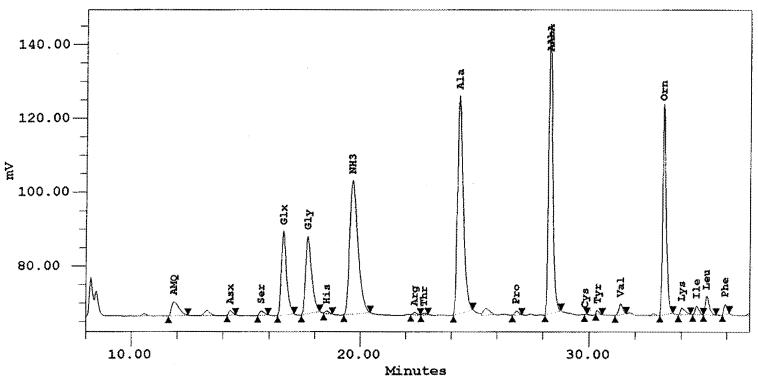
Reasoning that the murein sacculus was likely to be the lattice associated with cytoplasmic membrane blebs, we next isolated peptidoglycan for combined EM and biochemical analysis. The micrographs in Fig. 4 show that the material remaining after extensive detergent extraction and proteolysis of treponemes consisted of a reticular structure which retained the general shape of a spirochetal cell; indeed, it closely resembled the peptidoglycan sacculus previously isolated from T. pallidum (41). Ornithine, glutamine or glutamic acid (which both produce the same posthydrolysis derivative), glycine, and alanine were the principal amino acids in a hydrolysate of this material (Fig. 5). The presence of these four amino acids, as well as their molar ratios (1.0, 1.18, 1.25, and 2.29, respectively), are characteristic of spirochetal peptidoglycans (27, 41, 48).
FIG. 4.
Mid (A)- and high (B)-power micrographs of the structure isolated from T. denticola by extensive extraction with SDS and proteinase K digestion. Bars, 0.1 μm.
FIG. 5.
Amino acid profile obtained after hydrolysis of the material shown in Fig. 4. The glutamine-glutamic acid peak is designated Glx to indicate that the posthydrolysis derivatives of the two amino acids are indistinguishable. The peak labeled AAbA (second from the right) is the aminobutyric acid standard. The peak labeled AMQ (6-aminoquinoline) is an internal hydrolysis control. NH3 is an environmental contaminant.
Msp is predominantly a subsurface antigen.
In the next series of experiments, IEM was performed to localize Msp. Having previously shown that mild sonication causes significant disruption of the bacterium, including the removal of outer membranes, we first attempted to reproduce prior studies showing that sonicated treponemes react well with Msp-specific antibodies (17, 28). As shown in Fig. 6A, cell cylinders incubated with anti-Msp antibodies were strongly decorated with gold particles, whereas labeling was not observed with the preimmune serum (Fig. 6B). Interestingly, contrary to previous reports stating that Msp is abundant in the vesicles released from sonicated treponemes (17, 32), we frequently observed unlabeled vesicles alongside labeled cylinders (Fig. 6C), indicating that Msp was not an intrinsic vesicular component.
FIG. 6.
Subsurface localization of Msp. Negatively stained whole mounts of mildly sonicated treponemes incubated with rat anti-Msp antiserum (A and C) or preimmune serum (B). Bars, 0.5 μm.
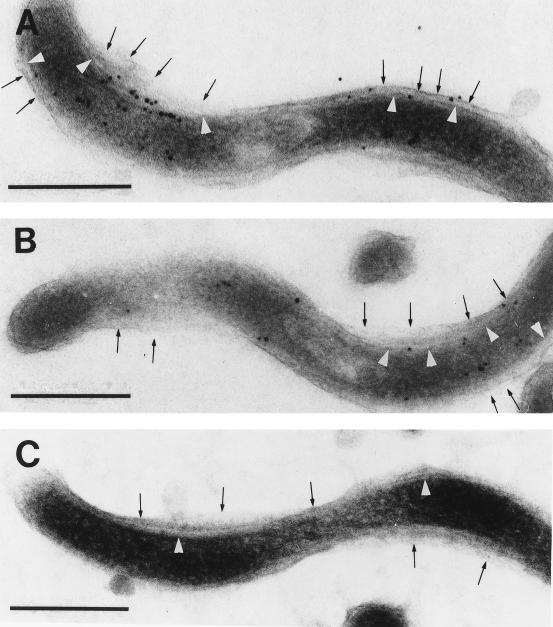
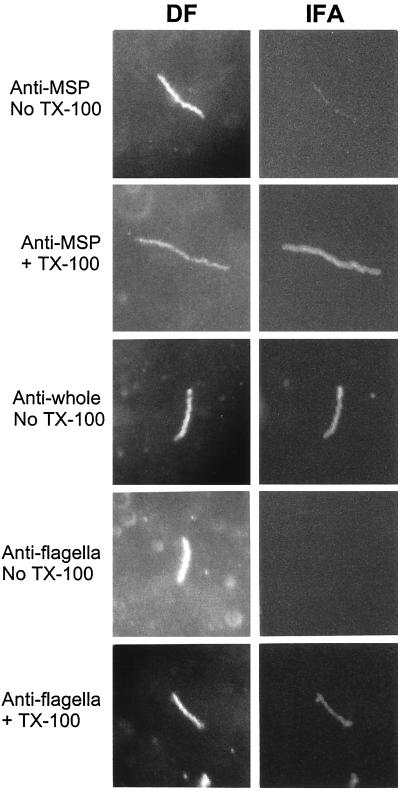
Although these results demonstrated that a substantial amount of Msp was located below the outer membrane, they did not preclude the possibility that some Msp was outer membrane associated and/or surface exposed. Two approaches were employed to examine this issue. The first involved immunogold labeling of ultrathin-sectioned, intact treponemes embedded in the hydrophilic resin LR Gold. Consistent with the IEM studies of whole mounts, gold particles were associated mainly with the cell cylinder, cytoplasmic membrane, and periplasmic space; occasional particles were, however, located on or in close proximity to the outer membrane (Fig. 7A and B). To determine whether any Msp was surface exposed, we performed immunofluorescence analysis of treponemes encapsulated in agarose beads (gel microdroplets) (13, 14). This method is advantageous because it not only preserves the integrity of fragile treponemal outer membranes during surface immunolabeling studies but it also enables the detection of periplasmic antigens when organisms are incubated with antibodies in the presence of a concentration of Triton X-100 sufficient to permeabilize the outer membrane (≥0.03%). Micrographs representative of three independent experiments are shown in Fig. 8. In the absence of detergent, all of the organisms were labeled by rat antisera directed against Msp or whole T. denticola. However, the labeling patterns produced by the two antisera differed markedly. In addition to being much weaker, the labeling produced by the anti-Msp antiserum was punctate rather than continuous. In two separate experiments, we quantitated the degree of labeling by the anti-Msp antibodies. A mean of 30% of the organisms had five or fewer fluorescent spots, whereas only 15% had more than 10 spots. Another 2% of the organisms were labeled uniformly by the anti-Msp antiserum; these were probably disrupted organisms, given that the same percentage of organisms fluoresced after incubation with antiflagellar antibodies in the absence of detergent. We considered the possibility that this unusual labeling pattern was due to antibody-mediated aggregation of antigens which are normally evenly distributed on the treponemal surface, a phenomenon well recognized with B. burgdorferi outer surface lipoproteins (3, 13). However, labeling was not affected when encapsulated organisms were fixed with 0.5% formalin prior to incubation with antibody (data not shown). Consistent with the IEM studies demonstrating large amounts of Msp distributed along the length of the cell cylinder, treponemes coincubated with anti-Msp antiserum and Triton X-100 fluoresced brightly and uniformly. This labeling pattern was indistinguishable from that produced when detergent-treated organisms were incubated with antiflagellar antibodies. Neither intact nor detergent-treated organisms reacted with preimmune sera (not shown).
FIG. 7.
Localization of Msp in ultrathin sections of intact treponemes. Cells shown in panels A and B were incubated with rat anti-Msp antiserum, while that in panel C was incubated with preimmune serum. Arrows and arrowheads indicate the outer and cytoplasmic membranes, respectively. Bars, 0.25 μm.
FIG. 8.
Immunofluorescence analysis of T. denticola encapsulated in gel microdroplets. Each pair of micrographs shows the same organism visualized by darkfield microscopy (DF) or indirect immunofluorescence (IFA).
DISCUSSION
The discovery that T. pallidum contains a family of polypeptides related to the T. denticola Msp (21) represents a potential breakthrough in the long and arduous search for the syphilis spirochete’s rare outer membrane proteins (21, 38). Stimulated by this finding, we reviewed the T. denticola Msp literature with the expectation that it would help to lay the theoretical groundwork for an analysis of the Tpr system. To our surprise, this analysis generated a number of concerns, noted in the introduction above, which led us to conclude that an ultrastructural reexamination of the T. denticola protein was the most appropriate starting point for our syphilis-related investigations. This report describes the two-pronged experimental strategy we implemented to address these perceived technical and conceptual issues. The first phase was an ultrastructural study intended to identify and localize the previously described array or lattice. This was followed by an extensive use of immunolabeling techniques to localize Msp on or within T. denticola.
Two-dimensional arrays of protein or glycoprotein subunits are commonly observed on prokaryotic cell surfaces (46). Moreover, S-layers have been detected on some spirochetes, including cultivatable treponemes (25). Thus, at the outset we considered it necessary to formally exclude the possibility that the reported T. denticola array was actually an S-layer which somehow had been overlooked by prior investigators. No external regular structure was visualized, however, by EM techniques which readily detected a hexagonal S-layer on a control bacterium. We next used freeze-fracture EM to look for a proteinaceous array within the T. denticola outer membrane. Although this powerful technique for examining membrane interiors has been available for several decades (20), it has never, to the best of our knowledge, been used to evaluate the widely held belief that the T. denticola outer membrane possesses a polygonal array. Here we show that the outer membrane architecture of T. denticola is similar to that of other cultivatable spirochetes (7, 39, 59) and consists of a moderately dense, heterogeneous mixture of randomly dispersed proteins. That these proteins could be redistributed by temperature-induced lipid-phase separations was further strong evidence against the existence of an intramembranous array composed of Msp or other T. denticola proteins.
If there is no array in the T. denticola outer membrane, how can one explain reports of outer-membrane-associated arrays as well as labeling of this structure by Msp-specific antibodies? We propose that published micrographs actually show cytoplasmic membranes and that peptidoglycan was mistakenly identified as the outer-membrane-associated array. Three findings presented here support this contention: (i) the peptidoglycan sacculus was the only lattice-like structure which we were able to identify in T. denticola; (ii) vesicles released from sonicated organisms originated from the cytoplasmic membrane; and (iii) allowing for differences in negative staining techniques, these vesicles bore a strong resemblance to the putative outer membranes isolated by others (31). Inspection of the published data also enables us to reconcile our results with the reported labeling of outer membrane arrays by anti-Msp antibodies; the immunolabeled amorphous material shown in some micrographs (16, 17) is not the same as the arrays shown by others (16, 17, 19). The notion that the T. denticola outer membrane possesses a polygonal array ultrastructure, now well entrenched in the literature, stemmed from a 1982 study by Masuda and Kawata (31) in which 40 freeze-thaw cycles were used to disrupt treponemes prior to sucrose gradient centrifugation. Viewed in the context of contemporary knowledge of spirochetal ultrastructure, our own extensive experience with spirochetal membrane fractionation (40, 44), and the EM work presented here, it seems unlikely that this would be a useful protocol for isolating outer membranes. Indeed, in the Masuda and Kawata study (31), as well as subsequent studies which used sonication to disrupt organisms, little or no evidence was presented to support claims that outer membranes had been isolated or identified.
We also used a battery of immunolabeling techniques to localize Msp. Overall, these methodologies yielded consistent results which meshed nicely with the ultrastructural studies discussed above. Taken together, these studies showed that Msp is predominantly periplasmic and more or less evenly distributed along the cell cylinder and that small amounts of antigen “poke through” the outer membrane to the treponemal surface. The notion that Msp has limited surface exposure is clearly at odds with reports of heavy surface labeling by anti-Msp antibodies (23). However, this dilemma can be resolved by pointing out that earlier IEM studies were conducted with less appreciation for the fragility of treponemal outer membranes and without the controls needed to assess outer membrane integrity. Although in the past we utilized T. pallidum and B. burgdorferi to illustrate this important point (8, 15), the findings here, as well as those from other preliminary studies (11), indicate that the T. denticola outer membrane is similarly susceptible to disruption by physical forces and chemical agents (e.g., detergents).
Fenno et al. (18) showed by DNA sequence analysis that Msp possesses a typical prokaryotic signal sequence with putative signal peptidase I cleavage site and that the mature (i.e., processed) polypeptide lacks the long hydrophobic stretches typical of cytoplasmic membrane proteins. From this sequence information, they predicted that Msp adopts the amphipathic beta-barrel secondary structure required for porin-like function (18). As porins have multiple surface-exposed loops (29), this membrane topology is also consistent with evidence that Msp performs virulence-related functions which require surface exposure. However, periplasmic and outer membrane proteins are equally hydrophilic at the sequence level and, for this reason, cannot be distinguished easily by existing computer algorithms (22, 47). Thus, there is no inherent contradiction between the sequence data and our assertion that Msp is mainly periplasmic. Taking this notion further, outer membrane proteins are exported as periplasmic intermediates that adopt their final conformations by folding into the outer membrane (37, 55). In order for our localization data to be consistent with this scenario, one need only postulate that the majority of Msp is constrained from inserting into the outer membrane (perhaps as a result of interactions with other peptidoglycan-cytoplasmic membrane constituents) and that some proportion adopts the beta-barrel outer membrane configuration. One can also envision a topology in which only limited stretches loop through the outer membrane. The B. burgdorferi outer membrane protein p66 may be a precedent for this novel form of outer membrane protein; evidence from two laboratories indicates that this protein contains a single surface-exposed, hypervariable domain (9, 10, 36). Ongoing studies to identify membrane-spanning and surface-exposed domains will enable us to distinguish between these alternatives and will set the stage for more comprehensive structure-function analyses potentially relevant to both treponemal physiology and periodontal disease pathogenesis.
Even at this early juncture in the characterization of the Tpr system, two points can be extrapolated from this work to T. pallidum. First, it resolves the dilemma created by the known lack of an array in T. pallidum. Second, not all Tpr proteins need be surface exposed. Indeed, this is already the case given that only a subset of Tpr proteins possess N-terminal export signals (21, 45). Based on the findings reported here, it is possible that Tpr proteins which do possess export signals also might be entirely or partly periplasmic. An interesting feature of the Tpr proteins is that their N and C termini are often highly conserved, whereas the central domains are highly variable (12, 21, 52). Sequence differences within the central domains might determine whether a specific Tpr does or does not become a rare outer membrane protein. Along these lines, it is interesting to note that Centurion-Lara and coworkers (12) recently presented evidence that TprK is surface exposed and capable of inducing protective immunity against syphilitic infection in the rabbit model. The T. denticola Msp protein could serve as a convenient model system for examining the relationship between particular domains and the membrane topologies of the T. pallidum paralogs.
ACKNOWLEDGMENTS
This research was supported in part by U.S. Public Health Service grant AI-26756 to J.D.R. M.J.C. was supported by Molecular Microbiology Training grant AI-07520 (NIAID).
We are indebted to Clive Slaughter for assistance with the peptidoglycan analysis. We also express our gratitude to Michael Norgard and Deborah Bouis for their critical reading of the manuscript.
REFERENCES
- 1.Akins D R, Robinson E, Shevchenko D V, Elkins C, Cox D L, Radolf J D. Tromp1, a putative rare outer membrane protein, is anchored by an uncleaved leader peptide to the Treponema pallidum cytoplasmic membrane. J Bacteriol. 1997;179:576–586. doi: 10.1128/jb.179.16.5076-5086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour A G, Hayes S F. Biology of Borrelia species. Microbiol Rev. 1986;50:381–400. doi: 10.1128/mr.50.4.381-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour A G, Tessier S L, Todd W J. Lyme disease spirochetes and ixodid tick spirochetes share a common surface antigenic determinant defined by a monoclonal antibody. Infect Immun. 1983;41:795–804. doi: 10.1128/iai.41.2.795-804.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berryman M A, Rodewald R D. An enhanced method for post-embedding immunocytochemical staining which preserves cell membranes. J Histochem Cytochem. 1990;38:159–170. doi: 10.1177/38.2.1688894. [DOI] [PubMed] [Google Scholar]
- 5.Blanco D R, Giladi M, Champion C I, Haake D A, Chikami G K, Miller J N, Lovett M A. Identification of Treponema pallidum subspecies pallidum genes encoding signal peptides and membrane-spanning sequences using a novel alkaline phosphatase expression vector. Mol Microbiol. 1991;5:2405–2415. doi: 10.1111/j.1365-2958.1991.tb02086.x. [DOI] [PubMed] [Google Scholar]
- 6.Blanco D R, Reimann K, Skare J, Champion C I, Foley D, Exner M M, Hancock R E W, Miller J N, Lovett J N. Isolation of the outer membranes from Treponema pallidum and Treponema vincentii. J Bacteriol. 1994;176:6088–6099. doi: 10.1128/jb.176.19.6088-6099.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourell K W, Schulz W, Norgard M V, Radolf J D. Treponema pallidum rare outer membrane proteins: analysis of mobility by freeze-fracture electron microscopy. J Bacteriol. 1994;176:1598–1608. doi: 10.1128/jb.176.6.1598-1608.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brusca J S, McDowall A W, Norgard M V, Radolf J D. Localization of outer surface proteins A and B in both the outer membrane and intracellular compartments of Borrelia burgdorferi. J Bacteriol. 1991;173:8004–8008. doi: 10.1128/jb.173.24.8004-8008.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bunikis J, Luke C J, Bunikiene E, Bergstrom S, Barbour A G. A surface-exposed region of a novel outer membrane protein (P66) of Borrelia spp. is variable in size and sequence. J Bacteriol. 1998;180:1618–1623. doi: 10.1128/jb.180.7.1618-1623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunikis J, Noppa L, Ostberg Y, Barbour A G, Bergstrom S. Surface exposure and species specificity of an immunoreactive domain of a 66-kilodalton outer membrane protein (P66) of the Borrelia species that cause Lyme disease. Infect Immun. 1996;64:5111–5116. doi: 10.1128/iai.64.12.5111-5116.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caimano, M. J., K. W. Bourell, and J. D. Radolf. Unpublished observations.
- 12.Centurion-Lara A, Castro C, Barrett L, Cameron C, Mostowfi M, Van Voorhis W C, Lukehart S A. Treponema pallidum major sheath protein homologues Tpr K is a target of opsonic antibody and the protective immune response. J Exp Med. 1999;189:647–656. doi: 10.1084/jem.189.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox D L, Akins D R, Bourell K W, Lahdenne P, Norgard M V, Radolf J D. Limited surface exposure of Borrelia burgdorferi outer surface lipoproteins. Proc Natl Acad Sci USA. 1996;93:7973–7978. doi: 10.1073/pnas.93.15.7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox D L, Akins D R, Porcella S F, Norgard M V, Radolf J D. Treponema pallidum in gel microdroplets: a novel strategy for investigation of treponemal molecular architecture. Mol Microbiol. 1995;15:1151–1164. doi: 10.1111/j.1365-2958.1995.tb02288.x. [DOI] [PubMed] [Google Scholar]
- 15.Cox D L, Chang P, McDowall A, Radolf J D. The outer membrane, not a coat of host proteins, limits the antigenicity of virulent Treponema pallidum. Infect Immun. 1992;60:1076–1083. doi: 10.1128/iai.60.3.1076-1083.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egli C, Leung W K, Muller K H, Hancock R E, McBride B C. Pore-forming properties of the major 53-kilodalton surface antigen from the outer sheath of Treponema denticola. Infect Immun. 1993;61:1694–1699. doi: 10.1128/iai.61.5.1694-1699.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenno J C, Hannam P M, Leung W K, Tamura M, Uitto V-J, McBride B C. Cytopathic effects of the major surface protein and the chymotrypsinlike protease of Treponema denticola. Infect Immun. 1998;66:1869–1877. doi: 10.1128/iai.66.5.1869-1877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fenno J C, Muller K H, McBride B C. Sequence analysis, expression, and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J Bacteriol. 1996;178:2489–2497. doi: 10.1128/jb.178.9.2489-2497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenno J C, Wong G W, Hannam P M, Muller K H, Leung W K, McBride B C. Conservation of msp, the gene encoding the major outer membrane protein of oral Treponema spp. J Bacteriol. 1997;179:1082–1089. doi: 10.1128/jb.179.4.1082-1089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher K A. Membrane-splitting analyses of membrane spanning proteins. In: Hui S W, editor. Freeze-fracture studies of membranes. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 11–39. [Google Scholar]
- 21.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G C, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith H O, Venter J C. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 22.Gromiha M, Ponnuswamy P K. Prediction of transmembrane β-strands from hydrophobic characteristics of proteins. Int J Peptide Protein Res. 1993;42:420–431. doi: 10.1111/j.1399-3011.1993.tb00149.x. [DOI] [PubMed] [Google Scholar]
- 23.Haapasalo M, Muller K H, Uitto V J, Leung W K, McBride B C. Characterization, cloning, and binding properties of the major 53-kilodalton Treponema denticola surface antigen. Infect Immun. 1992;60:2058–2065. doi: 10.1128/iai.60.5.2058-2065.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardham J M, Stamm L V. Identification and characterization of the Treponema pallidum tpn50 gene, an ompA homolog. Infect Immun. 1994;62:1015–1025. doi: 10.1128/iai.62.3.1015-1025.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holt S C. Anatomy and chemistry of spirochetes. Microbiol Rev. 1978;42:114–160. doi: 10.1128/mr.42.1.114-160.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hovind-Hougen K. Morphology. In: Schell R F, Musher D M, editors. Pathogenesis and immunology of treponemal infection. New York, N.Y: Marcel Dekker, Inc.; 1983. pp. 3–28. [Google Scholar]
- 27.Joseph R, Holt S C, Canale-Parola E. Peptidoglycan of free-living anaerobic spirochetes. J Bacteriol. 1973;115:426–435. doi: 10.1128/jb.115.1.426-435.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennell W L, Egli C, Hancock R E, Holt S C. Pore-forming ability of major outer membrane proteins from Wolinella recta ATCC 33238. Infect Immun. 1992;60:380–384. doi: 10.1128/iai.60.2.380-384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klebba P E, Benson S A, Bala S, Abdullah T, Reid J, Singh S P, Nikaido H. Determinants of OmpF porin antigenicity and structure. J Biol Chem. 1990;265:6800–6810. [PubMed] [Google Scholar]
- 30.Kleemann W, Grant C W M, McConnell H M. Lipid phase separations and protein distribution in membranes. J Supramol Struct. 1974;2:609–616. doi: 10.1002/jss.400020508. [DOI] [PubMed] [Google Scholar]
- 31.Masuda K, Kawata T. Isolation, properties, and reassembly of outer sheath carrying a polygonal array from an oral treponeme. J Bacteriol. 1982;150:1405–1413. doi: 10.1128/jb.150.3.1405-1413.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathers D A, Leung W K, Fenno J C, Hong Y, McBride B C. The major surface protein complex of Treponema denticola depolarizes and induces ion channels in HeLa cell membranes. Infect Immun. 1996;64:2904–2910. doi: 10.1128/iai.64.8.2904-2910.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maul G G. Temperature-dependent changes in intramembrane particle distribution. In: Rash J E, Hudson C S, editors. Freeze fracture: methods, artifacts, and interpretations. New York, N.Y: Raven Press; 1979. pp. 37–42. [Google Scholar]
- 34.Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. J Biol Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 35.Nitta H, Holt S C, Ebersole J L. Purification and characterization of Campylobacter rectus surface layer proteins. Infect Immun. 1998;65:478–483. doi: 10.1128/iai.65.2.478-483.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Probert W S, Allsup K M, LeFebvre R B. Identification and characterization of a surface-exposed 66-kilodalton protein from Borrelia burgdorferi. Infect Immun. 1995;63:1933–1939. doi: 10.1128/iai.63.5.1933-1939.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radolf J D. Treponema pallidum and the quest for outer membrane proteins. Mol Microbiol. 1995;16:1067–1073. doi: 10.1111/j.1365-2958.1995.tb02332.x. [DOI] [PubMed] [Google Scholar]
- 39.Radolf J D, Bourell K W, Akins D R, Brusca J S, Norgard M V. Analysis of Borrelia burgdorferi membrane architecture by freeze-fracture electron microscopy. J Bacteriol. 1994;176:21–31. doi: 10.1128/jb.176.1.21-31.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radolf J D, Goldberg M S, Bourell K W, Baker S I, Jones J D, Norgard M V. Characterization of outer membranes isolated from Borrelia burgdorferi, the Lyme disease spirochete. Infect Immun. 1995;63:2154–2163. doi: 10.1128/iai.63.6.2154-2163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radolf J D, Moomaw C, Slaughter C A, Norgard M V. Penicillin-binding proteins and peptidoglycan of Treponema pallidum subsp. pallidum. Infect Immun. 1989;57:1248–1254. doi: 10.1128/iai.57.4.1248-1254.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radolf J D, Norgard M V, Brandt M E, Isaacs R D, Thompson P A, Beutler B. Lipoproteins of Borrelia burgdorferi and Treponema pallidum activate cachectin/tumor necrosis factor synthesis: analysis using a CAT reporter construct. J Immunol. 1991;147:1968–1974. [PubMed] [Google Scholar]
- 43.Radolf J D, Norgard M V, Schulz W W. Outer membrane ultrastructure explains the limited antigenicity of virulent Treponema pallidum. Proc Natl Acad Sci USA. 1989;86:2051–2055. doi: 10.1073/pnas.86.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radolf J D, Robinson E J, Bourell K W, Akins D R, Porcella S F, Weigel L M, Jones J D, Norgard M V. Characterization of outer membranes isolated from Treponema pallidum, the syphilis spirochete. Infect Immun. 1995;63:4244–4252. doi: 10.1128/iai.63.11.4244-4252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radolf J D, Steiner B, Shevchenko D V. Treponema pallidum: doing a remarkable job with what it’s got. Trends Microbiol. 1999;7:7–9. doi: 10.1016/s0966-842x(98)01422-x. [DOI] [PubMed] [Google Scholar]
- 46.Sara M, Sleytr U B. Crystalline bacterial cell surface layers (S-layers): from cell structure to biomimetics. Prog Biophys Mol Biol. 1996;65:83–111. doi: 10.1016/s0079-6107(96)00007-7. [DOI] [PubMed] [Google Scholar]
- 47.Schirmer T, Cowan S W. Prediction of membrane-spanning β-strands and its application to maltoporin. Protein Sci. 1993;2:1361–1363. doi: 10.1002/pro.5560020820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schulz G E. Bacterial porins: structure and function. Curr Opin Cell Biol. 1993;5:701–707. doi: 10.1016/0955-0674(93)90143-e. [DOI] [PubMed] [Google Scholar]
- 50.Schweizer M, Schwarz H, Sonntag I, Henning U. Mutational change of membrane architecture. Mutants of Escherichia coli K12 missing major proteins of the outer cell envelope membrane. Biochim Biophys Acta. 1976;448:474–491. doi: 10.1016/0005-2736(76)90301-1. [DOI] [PubMed] [Google Scholar]
- 51.Shevchenko D V, Akins D R, Robinson E J, Li M, Shevchenko O V, Radolf J D. Identification of homologs for thioredoxin, peptidyl prolyl cis-trans isomerase, and glycerophosphodiester phosphodiesterase in outer membrane fractions from Treponema pallidum, the syphilis spirochete. Infect Immun. 1997;67:4179–4189. doi: 10.1128/iai.65.10.4179-4189.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shevchenko, D. V., M. J. Caimano, and J. D. Radolf. 1999. Unpublished results.
- 53.Sleytr U B, Messner P, Pum D, Sara M. Crystalline bacterial cell surface layers. Mol Microbiol. 1993;10:911–916. doi: 10.1111/j.1365-2958.1993.tb00962.x. [DOI] [PubMed] [Google Scholar]
- 54.Stebeck C E, Shaffer J M, Arroll T W, Lukehart S A, Van Voorhis W C. Identification of the Treponema pallidum subsp. pallidum glycerophosphodiester phosphodiesterase homologue. FEMS Microbiol Lett. 1997;154:303–310. doi: 10.1111/j.1574-6968.1997.tb12660.x. [DOI] [PubMed] [Google Scholar]
- 55.Surrey T, Schmid A, Jahnig F. Folding and membrane insertion of the trimeric beta-barrel protein OmpF. Biochemistry. 1996;35:2283–2288. doi: 10.1021/bi951216u. [DOI] [PubMed] [Google Scholar]
- 56.van Alphen L, Van Alphen W, Verkleij A, Lugtenberg B. Architecture of the outer membrane of Escherichi coli K12 IV. Relationship between outer membrane particles and aqueous pores. Biochim Biophys Acta. 1979;556:233–243. doi: 10.1016/0005-2736(79)90045-2. [DOI] [PubMed] [Google Scholar]
- 57.Van Deurs B, Koehler J K. Tight junctions in the choroid plexus epithelium. J Cell Biol. 1979;80:662–673. doi: 10.1083/jcb.80.3.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Wandelen C, Cohen S A. Using quaternary high-performance liquid chromatography eluent systems for separating 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate-derivatized amino acid mixtures. J Chromatogr A. 1997;763:11–22. [Google Scholar]
- 59.Walker E M, Borenstein L A, Blanco D R, Miller J N, Lovett M A. Analysis of outer membrane ultrastructures of pathogenic Treponema and Borrelia species by freeze-fracture electron microscopy. J Bacteriol. 1991;173:5585–5588. doi: 10.1128/jb.173.17.5585-5588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walker E M, Zampighi G A, Blanco D R, Miller J N, Lovett M A. Demonstration of rare protein in the outer membrane of Treponema pallidum subsp. pallidum by freeze-fracture analysis. J Bacteriol. 1989;171:5005–5011. doi: 10.1128/jb.171.9.5005-5011.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]