Abstract
DNA end-joining is the major repair pathway for double-strand breaks (DSBs) in higher eukaryotes. To understand how DSB structure affects the end-joining process in human cells, we have examined the in vivo repair of linearized plasmids containing complementary as well as several different configurations of non-complementary DNA ends. Our results demonstrate that, while complementary and blunt termini display comparable levels of error-free rejoining, end-joining fidelity is decreased to varying extents among mismatched non-complementary ends. End structure also influences the kinetics of repair, accurately recircularized substrates for blunt and complementary termini being detected significantly earlier than for mismatched non-complementary ends. These results suggest that the end-joining process is composed of an early component, capable of efficiently repairing substrates requiring a single ligation event, and a late component, involved in the rejoining of complex substrates requiring multiple processing steps. Finally, these two types of repair events may have different genetic requirements as suggested by the finding that exposure of cells to wortmannin, a potent inhibitor of phosphatidylinositol 3-related kinases (PI 3-related kinases), blocks the repair of complex substrates while having little or no effect on those requiring a simple ligation event.
INTRODUCTION
DNA double-strand breaks (DSBs), one of the most severe types of DNA damage, result from the disruption of the phosphodiester backbone on both strands of the double helix. To cope with this type of DNA injury, cells have developed two major repair pathways, homologous recombination and DNA end-joining (for review see 1). The contribution of each of these repair mechanisms differs not only from organism to organism but also from cell type to cell type. For example, in prokaryotes and lower eukaryotes, DSBs are repaired mainly by homologous recombination. This repair process also plays an important role in mammalian embryonic and germ cells, while in mammalian somatic cells, DNA end-joining is the major DSB repair mechanism (2,3). Two important characteristics of DSB repair are efficiency and fidelity. Repair efficiency, which reflects the capacity of cells to eliminate the lesions, is directly correlated to the survival of cells after DNA damage, while repair fidelity represents the capability of cells to precisely restore the damaged DNA and thus is an important determinant for the faithful transmission of genetic information.
Several important components of the end-joining machinery have so far been identified, including XRCC4, DNA ligase IV and DNA-dependent protein kinase (DNA-PKcs/Ku70/Ku80) (4). XRCC4, a DNA-binding protein, has been shown to interact tightly with DNA ligase IV and stimulate its ligase activity (5,6). DNA-PK is a heterotrimeric enzyme composed of a large catalytic subunit (DNA-PKcs), encoding a serine/threonine protein kinase, and a dimeric regulatory component consisting of the Ku70 and Ku80 proteins (for review see 7). It has been proposed that the Ku70/80 heterodimer, which binds to DNA ends with high affinity, protects DNA ends from degradation and/or promotes DNA ligation through the alignment of DNA ends, while DNA-PKcs has been suggested to participate in regulating the accessibility and/or processing of DNA ends through its kinase activity.
DSBs can be generated by extrinsic agents such as ionizing radiation or radiomimetic drugs, but the majority result as a consequence of normal cellular metabolism, for example, as intermediates during the repair of DNA lesions (DNA cross-links), as a result of errors in DNA replication as well as during programmed processes such as meiosis or V(D)J recombination. Since most of these DSBs are enzymatically induced, the DNA ends with which the cells are confronted will be of varying configurations [blunt, 5′ protruding single-strand (PSS), 3′PSS]. In the process of DNA end-joining, the structure of DSB termini can play a particularly important role; it can determine the nature of the resulting products as well as the enzymatic activities [nucleases, DNA polymerase(s) and accessory factor(s)] that participate in the repair.
To elucidate the molecular events involved in the processing of different DNA ends via DNA end-joining, we have examined, in human cells, the in vivo repair of linearized plasmids containing complementary as well as several different configurations of non-complementary DNA ends. Our results demonstrate that while the efficiency of repair is similar for all DNA ends, the fidelity of rejoining is conditioned by the structure of the input DSB termini. We also show that complementary and blunt substrates, which can be recircularized by a single ligation event, are repaired at significantly earlier time points than those containing various configurations of mismatched complementary ends, which require multiple processing steps. Finally, the idea that these two types of repair events may have different requirements is suggested by the finding that exposure of cells to wortmannin, a potent inhibitor of phosphatidylinositol 3 related kinases (PI 3-related protein kinases) such as DNA-PK, blocks the repair of complex substrates while having little or no effect on those requiring a simple ligation event.
MATERIALS AND METHODS
Cell culture
AHH-1 is a human lymphoblastoid cell line derived from a healthy donor (8). Lymphoblasts were routinely grown in suspension in RPMI 1640 medium (Gibco BRL) supplemented with glutamine (2 mM), gentamycin (40 µg/ml) and 10% (v/v) fetal calf serum. They were maintained in exponential growth at 37°C in a humidified 7% CO2 atmosphere.
Drug treatment and assessment of radiation sensitivity
Wortmannin (Sigma) was dissolved in dimethyl sulfoxide (DMSO) at a stock concentration of 20 mM and added to cell cultures at a final concentration of 20 µM, with the addition of a similar concentration of DMSO (0.1%) to control cultures. For the analysis of DNA end-joining, cells were pretreated for 2 h with either wortmannin or DMSO (0.1%) before transfection and subsequently maintained in medium containing wortmannin or DMSO.
To evaluate radiation sensitivity, cells were resuspended at 5 × 105 cells/ml, pre-incubated with either wortmannin or DMSO for 2 h at 37°C and then exposed to the appropriate doses of ionizing radiation using a 137Cs source (dose rate 1.83 Gy/min) or left unirradiated (0 Gy). Cell survival was determined immediately after treatment by plating cells in 96-well plates at the appropriate cell densities (2–10 cells per well) in the absence of wortmannin. Cells were then incubated for 18–20 days and colonies were scored. Assuming a Poisson distribution of the clonable cells, the plating efficiency (PE) was calculated by the formula PE = –ln Po/n, where Po is the ratio of the number of negative wells over total wells and n is the number of cells plated per well. Survival is the ratio of the PE of irradiated cells and the PE of the unirradiated cells.
Preparation of linearized DNA substrates
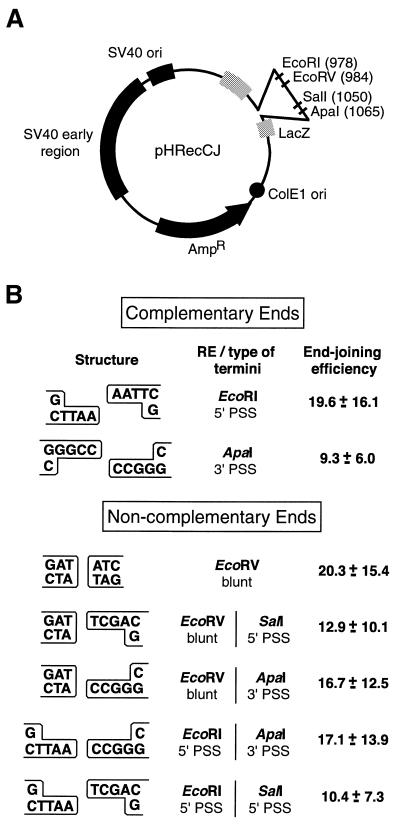
The recombination substrate used in this study, pHRecCJ (Fig. 1A), has been described previously (9,10). Linearized plasmid DNA was prepared by digesting with restriction enzymes (New England Biolabs) that recognize a unique recognition site within the substrate. In the case of complementary termini, the plasmid was linearized by digestion with a single appropriate restriction enzyme while substrates with non-complementary ends required digestion with either EcoRV or two different restriction enzymes. For double digests, complete digestion with the first restriction enzyme was verified, the sample dialyzed and then redigested with the second enzyme. All linearized substrates were analyzed by agarose gel electrophoresis and by transformation into Escherichia coli, which indicated that the completeness of digestion was >99%.
Figure 1.
(A) The substrate pHRecCJ. This plasmid contains the prokaryotic ColEI origin, the β-lactamase gene (AmpR), the SV40 origin of replication and large T-antigen coding sequence. The target LacZ gene is interrupted by an intervening sequence that contains the unique restriction sites EcoRV, EcoRI, SalI and ApaI. (B) End structures of the linearized substrates used to examine end-joining in human cells and their associated repair efficiencies. Ends are categorized as complementary when there is homology along the entire length of the PSS and non-complementary when the ends display little or no terminal homology. The restriction enzyme(s) (RE) used to create the depicted end structures are indicated as well as the mean ± SD of the observed end-joining efficiencies 48 h after transfection.
Preparation of dideoxy-capped DNA substrates
The DNA substrate, pHRecCJ, was digested with ApaI and then dialyzed to eliminate the digestion buffer. Subsequently, the 3′PSS were capped by adding dideoxynucleotide C (ddCTP) at a molar ratio of 103 ddCTP/substrate 3′OH, terminal deoxynucleotidyl transferase (TdT; Boehringer Mannheim) at a concentration of 3 U/µg DNA and TdT reaction buffer (Boehringer Mannheim). After incubation at 37°C for 2 h the reaction was stopped by the addition of EDTA to a final concentration of 40 mM. The sample was then extracted with phenol and chloroform, precipitated with ethanol and treated with T4 DNA ligase (New England Biolabs) to ligate molecules that had not been capped. Capped linear monomers were subsequently purified from an agarose gel and digested with the restriction enzyme EcoRV to generate the final blunt/3′PSS capped substrate. The addition of ddCTP results in the regeneration of the ApaI recognition site (GGGCC↓C) and thus allows recovered recombinant plasmids to be screened for the presence of an intact 3′PSS by digestion with ApaI. All plasmids containing this site were then sequenced to verify that error-free rejoining had occurred.
Transient transfection and plasmid recovery
Cells were transfected by electroporation at 300 V and 950 µF using 1.25 µg DNA and 1 × 107 cells per point as described previously (9). Unless otherwise noted, cells were harvested 48 h after transfection, lysed and plasmid DNA extracted by a rapid alkali lysis procedure (11). Since linearized plasmids have to be recircularized in order to replicate in human cells, recovered DNA samples were digested with the restriction enzyme DpnI, which recognizes and cuts only bacterially methylated GATC sequences (dam+). Thus, the DpnI digestion ensures that only plasmids repaired and replicated in human cells will be recovered. Following dialysis, plasmids were electroporated into XL1-blue competent bacteria (Stratagene) and plated on LB plates containing ampicillin (100 µg/ml). The end-joining efficiency was determined by dividing the number of bacterial colonies derived from linear plasmids by the number of colonies obtained with supercoiled plasmid. Average values are calculated from two to eight independent experiments.
Time course and wortmannin experiments were carried out in a similar manner except that cells were harvested at 0, 6, 12 ,24, 36 or 48 h after transfection. Additionally, due to the absence of replication at the earliest time points, extracted DNA samples were not digested with DpnI. Instead, any remaining linearized plasmid was rendered unligatable by incubation for 1 h with 5 U calf intestinal phosphatase (CIP) (New England Biolabs) at 37°C. Following inactivation of the enzyme at 75°C for 10 min, the samples were dialyzed and introduced into bacteria as described above. Based on results obtained at 48 h, the type of recombinants recovered are similar to those observed with DpnI treatment (data not shown). However, repair efficiencies following CIP treatment are lower than those obtained following DpnI digestion due to the recovery of replicated and non-replicated plasmids, which leads to an increased number of bacterial colonies for the supercoiled circular plasmid.
Analysis of recombinant plasmids
To distinguish error-free and error-prone repair events, small-scale DNA preparations were made from bacterial colonies (12) and analyzed at a molecular level. In the case of plasmid substrates generated by digestion with a single enzyme, the level of error-free rejoining was determined by screening for the restitution of the original restriction site. In contrast, for non-complementary substrates generated by double digestion, the level of error-free rejoining was determined by sequencing plasmids that exhibited small deletions upon restriction analysis. Error-prone repair events for all substrates were characterized by random sequencing of plasmids. Statistical analyses of data were performed using the χ2 test with Yates’ correction or Fisher’s exact test if needed. The P values for the test are indicated when appropriate.
Recombinant substrates were sequenced using the Big Dye Terminator Cycle Sequencing Kit and an ABI 373A Stretch automated sequencer (Applied Biosystems). The sequencing reactions were performed with one of five sense or anti-sense primers, which span the region between the SV40 and ColE1 origin using a Perkin-Elmer thermocycler as described (9). Sequences of each rearranged substrate were compared with the wild-type plasmid sequences using ABI version 1.0.1 Sequence Navigator software (Applied Biosystems).
RESULTS
Human cells are equally efficient in repairing complementary and non-complementary ends
To determine whether the overall capacity of human lymphoblasts to repair DSB by DNA end-joining is influenced by the structure of the termini, we have used a plasmid-based host cell end-joining assay. The plasmid substrate used, pHRecCJ (Fig. 1A), is capable of replicating in both human and bacterial cells, and contains several unique restriction sites, thus allowing the generation of DSBs with complementary termini of different polarities [3′ and 5′PSS] as well as various configurations of non-complementary ends (blunt, blunt associated with 5′ or 3′PSS, 5′PSS with 3′PSS or non-homologous 5′PSS) (Fig. 1B).
To analyze the end-joining efficiency, equal quantities of either linearized or supercoiled plasmid are transiently transfected into normal human lymphoblasts. After 48 h, the plasmid DNA is recovered and digested with the restriction enzyme DpnI. Since DpnI cleaves bacterially methylated DNA, only those substrates that have been repaired and replicated in human cells are resistant to DpnI cleavage and will give rise to ampicillin-resistant colonies following transformation of E.coli (see Materials and Methods). The repair efficiency can then be determined by dividing the number of colonies obtained with linear versus supercoiled plasmid. The data show that for all end structures analyzed (Fig. 1B), the ability of the cell to repair the DSB was not significantly different, averaging 15.2 ± 11.6%. Thus, in human cells, the overall efficiency of end-joining is independent of the structure of the DNA ends.
The fidelity of the end-joining reaction is dependent on the structure of the termini
Our results demonstrate that, quantitatively, human cells are capable of rejoining all DNA ends with the same efficiency. To determine whether the quality of the end-joining events is influenced by the structure of the termini, individual plasmids were recovered and examined by restriction and sequence analysis (see Materials and Methods). This analysis permits the differentiation of substrates that are precisely rejoined (i.e. without loss or gain of nucleotides), classified as error-free events and those associated with deletions and/or insertions, classified as error-prone events.
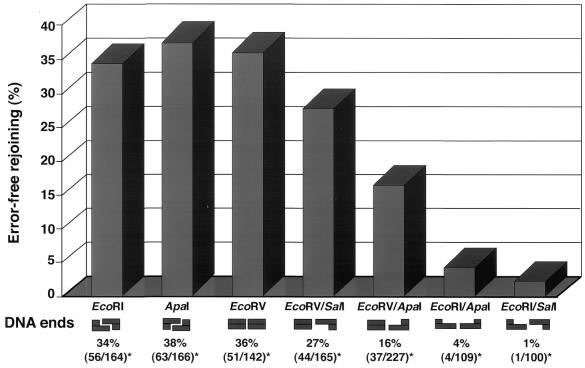
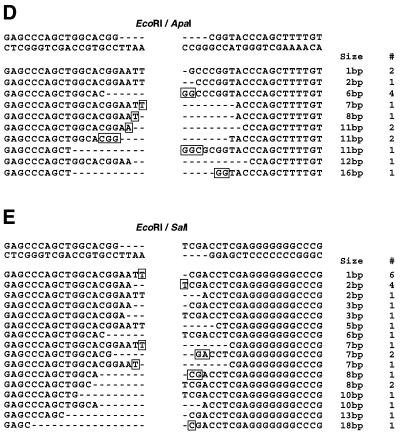
The molecular analysis of over 1000 plasmids allowed us to determine the proportion of recombinants that had been accurately recircularized for different DNA end structures. As depicted in Figure 2, similar levels of faithful repair were observed for substrates containing complementary termini with either 5′PSS (EcoRI) or 3′PSS (ApaI), 34 and 38%, respectively. Interestingly, among the substrates containing non-complementary termini, both blunt ends (EcoRV) and blunt/5′PSS (EcoRV/SalI) display a level of error-free rejoining (36 and 27%, respectively) that is not statistically different from that observed for complementary ends (P = 0.8). In contrast, only 16% of blunt/3′PSS (EcoRV/ApaI) plasmids are accurately repaired, a significantly lower level than for blunt or blunt/5′PSS substrates (P < 0.01). Finally, the lowest levels of error-free rejoining were detected with plasmids containing 3′PSS/5′PSS (EcoRI/ApaI) and non-complementary 5′PSS (EcoRI/SalI) ends, 4 and 1%, respectively. Together, these results indicate that the structure of DSB termini has a significant influence on the fidelity of the end-joining reaction.
Figure 2.
Percentage of error-free joining for different complementary and non-complementary substrates. The substrates examined are noted along with a physical representation of the end structures. In addition, the ratio of accurate error-free events observed among the total number of plasmids analyzed is indicated (*).
Evidence for the involvement of an alignment protein in the accurate repair of 3′PSS in human cells
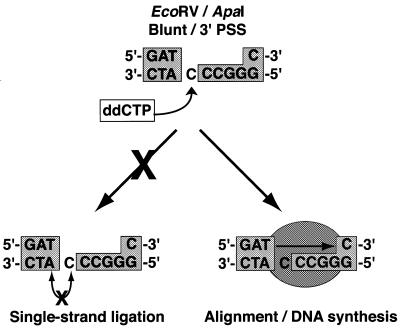
Error-free rejoining of complementary or blunt termini can occur by direct ligation, while accurate repair of 5′PSS is most easily explained by filling-in of the 3′ recessed end by a DNA polymerase to create a ligatable blunt end. More difficult to explain is the accurate repair of 3′PSS, for which the recessed end cannot be filled-in by normal 5′→3′-directed DNA synthesis (13). Ligation of a 3′PSS to a partner terminus would provide an appropriate primer for fill-in DNA synthesis. However, this type of repair mechanism would require a single-strand-specific ligase, an activity that is not known to be associated with the currently identified mammalian DNA ligases (14). Alternatively, it has been proposed that an alignment protein may stably and precisely associate the DNA ends and thus allow fill-in synthesis to be primed from the 3′ terminus of the associated DNA end (15).
To explore the mechanism involved in the accurate repair of 3′PSS in human cells, ddCTP was added to the 3′PSS end of a blunt/3′PSS (EcoRV/ApaI) substrate (Fig. 3). Thus, if error-free repair of the substrate requires single-strand ligation before filling-in of the gapped DNA, then the presence of a dideoxynucleotide at the 3′PSS would prevent accurate repair of the plasmid. However, if fill-in synthesis occurs before DNA ligation, a nicked plasmid would be formed and upon DNA replication would generate an intact error-free junction. Indeed, analysis of 89 recircularized plasmids indicated that 7% had been accurately repaired, maintaining both the blunt and 3′PSS sequence as well as the dideoxynucleotide position. Thus, this result provides the first in vivo evidence in favor of the involvement of an alignment protein in the end-joining process.
Figure 3.
Repair pathways for the accurate rejoining of substrates containing a 3′PSS. The error-free repair of 3′PSS substrates can occur either by single-strand ligation followed by fill-in synthesis or alternatively fill-in synthesis can be primed from the 3′ terminus of an associated DNA end followed by ligation. To differentiate these two repair pathways, ddCTP was added to the 3′PSS of the substrate EcoRV/ApaI (blunt/3′PSS). Since the DNA strand containing the ddCTP cannot be ligated, faithful rejoining of these ends is not possible by single-strand ligation. However, if a DNA binding protein (light gray oval) can align the termini, fill-in synthesis followed by ligation of a single strand will generate a nicked plasmid that upon DNA replication will result in an intact error-free recombinant.
Analysis of error-prone repair events
The sequence analysis of error-prone events obtained with linearized plasmids containing complementary and non-complementary ends is presented in Table 1. A small percentage of plasmids contain insertions (5–17%), the majority of which are 1–4 nucleotides. Among the remaining deletion events, ∼68% are associated with microhomology from 1 to 6 bp. The average deletion size is similar, ranging from 154 to 326 bp. Thus, in general, similar types of error-prone rejoining events are obtained with substrates containing complementary and non-complementary termini.
Table 1. Rejoining characteristics for error-prone recombinants.
| Enzyme(s) | DNA ends | Plasmids sequenceda | No. associated with insertions | Average deletion size | DRc |
|---|---|---|---|---|---|
| PSS, Protruding single strands. DR, direct repeats. | |||||
| EcoRI | 5′PSS/5′PSS | 48 (4)b | 6% (3/48) | 326 bp | 71% (32/45) |
| ApaI | 3′PSS/3′PSS | 55 (4)b | 5% (3/55) | 251 bp | 75% (39/52) |
| EcoRV | blunt/blunt | 45 (3)b | 7% (3/45) | 284 bp | 59% (25/42) |
| EcoRV/SalI | blunt/5′PSS | 101 (6)b | 17% (17/101) | 260 bp | 65% (55/84) |
| EcoRV/ApaI | blunt/3′PSS | 71 (4)b | 6% (4/71) | 240 bp | 66% (44/67) |
| EcoRI/ApaI | 5′PSS/3′PSS | 49 (3)b | 8% (4/49) | 298 bp | 71% (32/45) |
| EcoRI/SalI | 5′PSS/5′PSS | 79 (4)b | 11% (9/79) | 154 bp | 66% (46/70) |
aNumber of mis-rejoined plasmids sequenced. 11–17 plasmids randomly chosen per experiment.
bThe number of independent experiments performed are indicated in parentheses.
cPercentage of deletion events containing direct repeats.
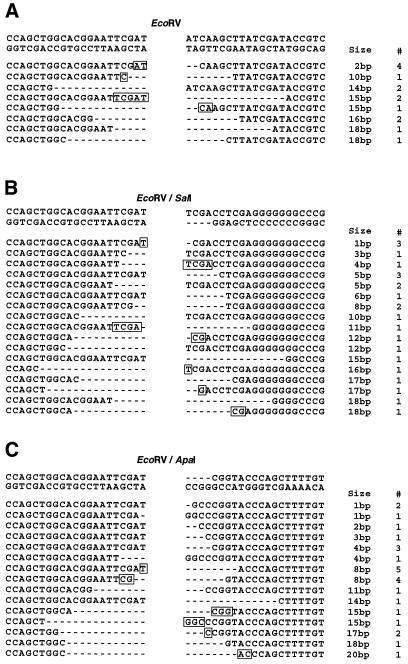
To understand better the molecular steps implicated in end-joining, in particular for non-complementary substrates, a detailed analysis of the smallest deletions (<20 bp) was undertaken (Fig. 4). Homology appears to play an important role in these repair events since more than half of the junctions involve sequences with at least 1 bp of homology. Additionally, for several of the substrates examined, homology at the DSB termini appears to play a particularly important role. This is especially evident in the case of the EcoRI/SalI substrate, for which 38% (10/26) of the error-prone events detected involve terminal homology of only a single base pair (Fig. 4E, lines 1 and 2). Another factor analyzed was the stability of the DNA ends among the linearized substrates. Interestingly, if we consider the stability of the ends with regard to their structure, we find significant differences; 5′PSS (SalI and EcoRI) and blunt ends (EcoRV) display similar levels of stability 42% (37/89) and 43% (33/77), respectively, while for 3′PSS (ApaI) only 19% (8/42) of the ends remain intact. Thus, among non-complementary substrates, 3′PSS are significantly less stable than other DNA end structures.
Figure 4.
Sequences of recombinant junctions for deletions of ≤20 bp. (A) EcoRV. (B) EcoRV/SalI. (C) EcoRV/ApaI. (D) EcoRI/ApaI. (E) EcoRI/SalI. The first line represents the sequence of junctions created by error-free rejoining, without loss of nucleotides from either end. To the right of the figure is noted the deletion size and the number of times each independent recombinant was detected (#). Dashes represent nucleotides lost. Sequence homologies at the junctions are noted with boxes and in all cases have been localized to the less deleted DNA end.
DNA end structure determines the kinetics of end-joining
To determine whether DNA end structure has an influence on the relative kinetics of end-joining, we examined the repair of EcoRI-, EcoRV-, EcoRV/SalI- and EcoRV/ApaI-linearized substrates 0, 6, 12, 24, 36 and 48 h after introduction into human cells (Materials and Methods). To monitor the kinetics of end-joining, two different parameters were examined: the percentage relative repair, which is defined as the number of bacterial colonies recovered at a given time point compared with the number of colonies obtained at 48 h, and the percentage error-free repair, which is the proportion of total recombinants at a given time point that have been accurately rejoined.
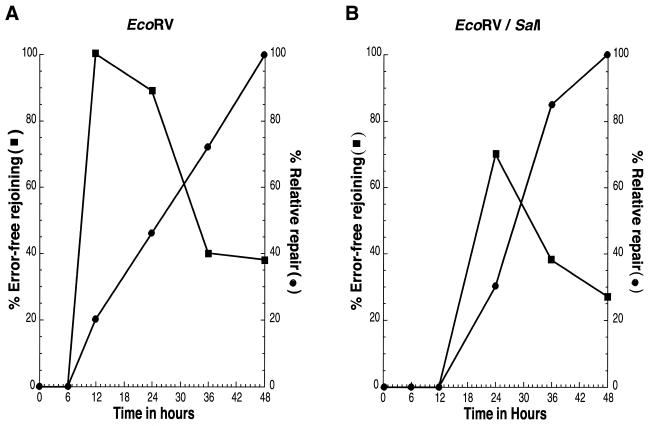
The repair kinetics observed for EcoRV- and EcoRV/SalI-linearized plasmids are represented in Figure 5. The data obtained for the percentage relative repair indicate that, while recombinants for EcoRV-linearized plasmids are first detected 12 h after transfection of human cells, the double-digested plasmid, EcoRV/SalI, was not repaired until the 24 h time point. Molecular analysis of rejoined plasmids at these early time points indicates that the large majority of recovered recombinants for both substrates are error-free repair events (100% for EcoRV, 70% for EcoRV/SalI). The EcoRI substrate displayed repair kinetics similar to EcoRV while that of EcoRV/ApaI closely resembled EcoRV/SalI (data not shown). This finding strongly suggests that substrates containing DNA ends that are directly ligatable (EcoRI and EcoRV) can be repaired more rapidly than those that require additional processing steps (EcoRV/SalI and EcoRV/ApaI). At subsequent time points, the total number of repaired plasmids increases, for both types of substrates, while the percentage of error-free events decreases. This finding indicates that error-prone repair events occur at later time points and with their accumulation, diminish the relative proportion of error-free events.
Figure 5.
Kinetics of DNA end-joining. (A) EcoRV. (B) EcoRV/SalI. Plasmid DNA was recovered 0, 6, 12, 24, 36 and 48 h after transfection of the indicated linearized substrate into human cells. Following introduction into bacteria, the kinetics of repair were monitored by determining percentage relative repair (square), the number of colonies recovered at a given time point compared with the number of colonies obtained at 48 h and percentage error-free joining (circle), the proportion of total recombinants at a given time point that have been accurately repaired.
Wortmannin treatment impedes rejoining of complex substrates
To dissect further the DNA end-joining process, we examined the repair of EcoRI-, EcoRV-, EcoRV/SalI- and EcoRV/ApaI-linearized substrates in cells that had been treated with the fungal metabolite wortmannin, a potent inhibitor of DNA-PK as well as other PI 3-related protein kinases. It has previously been shown that exposure of cells to wortmannin inhibits the repair of DSBs, resulting in a significant increase in radiation sensitivity (16–18). Indeed, pre-treatment of the human lymphablastoid cell line used in these studies, AHH-1, with 20 µΜ wortmannin results in a 2.7-fold decrease in cell survival following exposure to 1 Gy of ionizing radiation (9.7 ± 3.3) compared with control cells (26.2 ± 3.2) that have been exposed to a similar concentration of the solvent (0.1% DMSO).
Analysis of the efficiency of DNA end-joining in similarly treated cells indicates that exposure to wortmannin has significant effects on global repair efficiency (Table 2). At the 24 h time point, while there is no significant difference between wortmannin- and DMSO-treated cells for the EcoRV substrate, the EcoRI substrate displays a 9-fold reduction. An even greater decrease in the repair efficiency is observed for the substrates containing mismatched non-complementary ends; the EcoRV/SalI substrate exhibits a 12-fold reduction while a >20-fold decrease is observed for the EcoRV/ApaI substrate. At the 48 h time point, the observed end-joining defect following wortmannin treatment is significantly increased, EcoRV-linearized plasmids now display an ∼10-fold lower rejoining efficiency while EcoRI substrates display a 40-fold reduction. Double-digested plasmids exhibit an almost complete absence of recombinants resulting in a >150-fold reduction in the efficiency of repair.
Table 2. End-joining efficiency in human cells treated with wortmannin.
| Treatment | Timea | Circularb | EcoRIc | EcoRVc | EcoRV/ApaIc | EcoRV/SalIc |
|---|---|---|---|---|---|---|
| 0.1% DMSO | 24 h | 90 000 | 1.500 (1350) | 0.290 (265) | 0.400 (325) | 0.110 (103) |
| 48 650 | 0.640 (310) | 1.300 (614) | ND | ND | ||
| 170 000 | 0.400 (640) | 0.500 (800) | 0.100 (180) | 0.120 (210) | ||
| 0.850d | 0.700d | 0.250d | 0.120d | |||
| 48 h | 80 000 | 2.320 (1850) | 2.950 (2360) | 0.650 (522) | 0.860 (686) | |
| 140 000 | 2.70 (3780) | 2.450 (3430) | 0.821 (1150) | 0.721 (1010) | ||
| 2.510d | 2.700d | 0.736d | 0.791d | |||
| 20 µM wortmannin | 24 h | 144 000 | 0.080 (120) | 1.900 (2750) | 0.004 (6) | 0.009 (13) |
| 71 250 | 0.120 (88) | 0.510 (360) | 0.024 (18) | 0.014 (10) | ||
| 172 000 | 0.070 (114) | 0.140 (245) | 0.010 (19) | 0.008 (14) | ||
| 0.090d | 0.850d | 0.012d | 0.010d | |||
| 48 h | 27 500 | 0.040 (12) | 0.290 (80) | 0.005 (2) | 0.003 (1) | |
| 68 000 | 0.080 (56) | 0.310 (212) | 0.004 (3) | <0.001 (0) | ||
| 0.060d | 0.300d | 0.005d | 0.002d |
aNumber of hours after which plasmid DNA was recovered from human cells.
bTotal number of bacterial colonies recovered with a circular substrate.
cRepair efficiency as derived from the formula: [(Number of bacterial colonies obtained with linear substrate/Number of colonies with circular substrate) × 100]. The total number of colonies obtained with the indicated linearized substrate are indicated within parentheses.
dAverage repair efficiency.
The molecular analysis of EcoRI and EcoRV substrates indicate that, similar to controls, >90% of the plasmids recovered from wortmannin-treated cells at the 24 h time point were accurately repaired. However, while the level of error-free end-joining for control cells decreased to 33% at the 48 h time point, the level of accurate repair remains stationary (85%) for wortmannin-treated cells. In contrast, among substrates containing mismatched non-complementary ends, sequence analysis of the few recovered recombinants indicates that none had been accurately rejoined. Together these results suggest that early end-joining events, which only involve DNA ligation, display little (EcoRI) or no (EcoRV) inhibition by wortmannin, while late occurring events, which involve multiple processing steps (EcoRV/SalI and EcoRV/ApaI), are completely blocked in the presence of this DNA-PK inhibitor.
DISCUSSION
The host cell end-joining assay provides a unique tool to understand the mechanistic steps involved in the repair of DSBs with defined DNA end structures. This type of plasmid-based system has been used previously to analyze V(D)J recombination, a specialized form of DNA end-joining, where numerous studies have validated its relevance to chromosomal DSB repair (19–21). In this study, we have used the host cell end-joining assay in human cells to examine the fate of DSB with complementary termini of different polarities and lengths as well as various configurations of non-complementary termini. In particular, we have been able to determine, through the analysis of error-free repair events, the effect of different processing events on several parameters such as the timing, efficiency and accuracy of repair. Together these results have allowed us to define several end-joining mechanisms involved in the repair of DSB in human cells and provide an important basis for understanding the role of known DNA repair proteins in the process.
In this study, we demonstrate that in human cells, as in other mammalian systems (22), the end-joining efficiency 48 h after transfection is not influenced by DNA end structure. This parameter reflects the global ability of a cell to repair a DSB and includes both error-free and error-prone events. Error-prone repair events involve a loss or gain of nucleotides and thus involve a modification of DNA termini during the repair process. Thus, to better understand the influence of a precise end structure on the process of DNA end-joining we focused our initial analysis on error-free repair events.
Interestingly, our analysis of the fidelity of repair indicates that human cells are equally capable of accurately ligating both blunt and complementary termini. This finding is rather surprising since in vitro studies have shown that mammalian DNA ligases are much more proficient in ligating complementary than blunt ends (14,23). Thus, our results suggest that, in vivo, the involvement of additional cofactors significantly enhances the efficiency of blunt-end ligation. Indeed, biochemical analysis has shown that the addition of the heterodimeric DNA binding protein Ku to in vitro ligation reactions significantly stimulates blunt-end ligation (24).
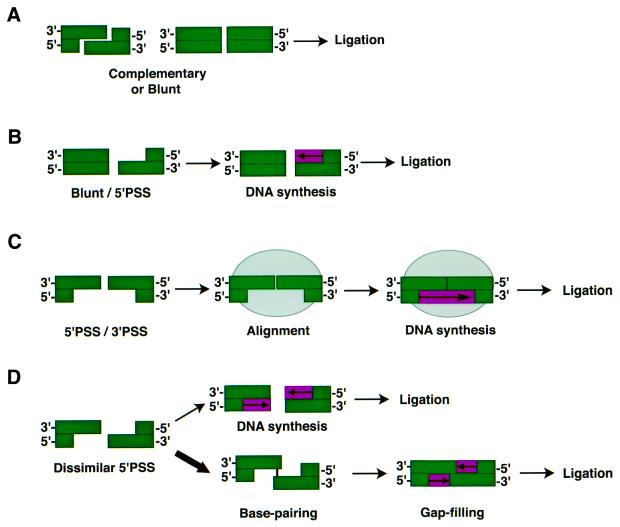
In contrast, among substrates containing dissimilar ends we find that the structure of the termini plays an important role in determining the fidelity of repair (Fig. 2). One possible explanation for this difference is that these substrates require additional processing steps as well as protein components in order to be accurately rejoined (Fig. 6). For example, the substrate containing one blunt end and one 5′PSS (EcoRV/SalI) requires an additional step of fill-in synthesis in order to be accurately repaired (Fig. 6B). In the case of substrates containing a 3′PSS (EcoRV/ApaI, EcoRI/ApaI), we have shown that error-free repair is consistent with the involvement of an alignment protein that stably and tightly aligns the DNA ends, allowing the priming of DNA synthesis from the 3′PSS (Fig. 6C). Additionally, it should be noted that this type of alignment activity may also be involved in the rejoining of other types of DNA ends including complementary, blunt and other complex DNA ends. At present, it is not clear what protein(s) may provide such an alignment activity; however, it seems likely that the implicated factors(s) will have DNA end-binding activity and thus one obvious candidate is the Ku heterodimer mentioned previously. In favor of this hypothesis are results showing that the rejoining of 5′PSS/3′PSS DNA ends was significantly inhibited in Xenopus laevis extracts depleted for the Ku protein (25) as well as in Ku80-deficient hamster cell extracts (26). Another possible candidate is the DNA ligase IV–XRCC4 complex, which has been recently shown to form a tetramer and thus is capable of serving as a bridging factor by simultaneously binding to two different DNA ends (27,28).
Figure 6.
Processing steps involved in the repair of different configurations of DNA ends. (A) Complementary or blunt ends. The faithful repair of these ends can occur by a single step of DNA ligation. (B) Blunt/5′PSS ends. Fill-in synthesis of the recessed strand of the 5′PSS end creates a blunt end that is now a suitable substrate for DNA ligation. (C) 5′PSS/3′PSS ends. In the first step, the termini are aligned by a DNA binding protein (light gray oval), which stabilizes their interaction and permits the second step of fill-in DNA synthesis, which is primed from the recessed strand of the 5′PSS end. The resulting intermediate can then be ligated. Although not depicted, a similar mechanism can explain the accurate repair of blunt, blunt/5′PSS and 3′PSS/5′PSS ends. (D) Dissimilar 5′PSS/5′PSS ends. A minor repair pathway (1 out of 100 events) involves fill-in synthesis of the 5′PSS ends to create two blunt termini that can be directly ligated. The major repair pathway for this substrate (10/100 events) appears to involve pairing of the terminal nucleotide from each single-stranded extension to form a base-paired intermediate containing two gaps. In the second step, these gaps are filled-in to create a ligatable intermediate.
The substrate showing the lowest level of accurate repair contains two 5′PSS (EcoRI/SalI). Only one single error-free event was recovered out of 100 recombinants examined. This finding was unexpected since fill-in synthesis of 5′PSS followed by blunt-end ligation should be a relatively efficient process. Instead, sequence analysis of error-prone recombinants indicates that the preferred repair pathway for EcoRI/SalI-generated DSB involves pairing of the ends via a single base pair homology at the termini. Approximately 10% (10/100) of the total recombinants are consistent with this type of end-joining event. In other words, the use of homology, even a single base pair, is preferred over a mechanism that involves more complex processing of the DNA ends (Fig. 6D).
An important role for terminal homology was also observed among the error-prone recombinants for both blunt (EcoRV) and blunt/5′PSS (EcoRV/SalI) substrates (Fig. 4A and B). Interestingly, however, similar end-joining events were not detected among the recombinants obtained with the blunt/3′PSS (EcoRV/ApaI) substrate (Fig. 4C). One explanation for this difference may be that, unlike the other substrates, the most terminal homology present between the EcoRV and ApaI ends involves a G nucleotide that is 2 bp removed from the DNA end. Alternatively, the reduced stability of the 3′PSS may prevent the effective use of this homology. In favor of this hypothesis are the results we have obtained concerning the number of recombinants that maintain an intact DNA end. Blunt (EcoRV) and 5′PSS (SalI and EcoRI) display similar levels of stability 42% (37/89) and 43% (33/77), respectively, while a significant reduction of intact ends, 19% (8/42), was detected for 3′PSS (ApaI). This decreased stability of 3′PSS compared with other termini may be due to the decreased protection of 3′PSS and/or the increased activity of a 3′ exonuclease(s) in human cells. In either case, this ensemble of results provides strong evidence that the structure of DSB termini plays an important role in determining the mechanism of repair.
The kinetic analysis of DNA end-joining indicates the structure of DNA termini also has an important influence on the timing of the repair event. Precisely recircularized recombinants for EcoRI and EcoRV substrates can be detected significantly earlier than for the EcoRV/SalI and EcoRV/ApaI substrates. This result indicates that substrates that can be recircularized by a single ligation event (complementary and blunt) are repaired at significantly earlier time points than those requiring multiple processing steps (mismatched complementary ends). The additional time required for the repair of complex DSBs is possibly due to the necessity to recruit additional repair factors and/or to carry out extra processing steps in order to generate ligatable DNA ends. The idea that these two types of repair, simple and complex, may not have the same requirements is supported by the finding that exposure of cells to wortmannin blocks the repair of complex substrates (SalI/EcoRV and ApaI/EcoRV) while having little or no effect on those requiring a simple ligation event (EcoRI and EcoRV).
Wortmannin, a fungal metabolite, is a potent and selective inhibitor of mammalian PI 3-related protein kinases. Previous studies have demonstrated that exposure to wortmannin results in increased radiosensitivity (16–18). However, in this study we provide the first evidence that wortmannin treatment leads to a direct defect in the DNA end-joining pathway. In the case of radiosensitivity, this effect has been linked to the inactivation of the PI 3-related protein kinase family members DNA-PKcs and/or ATM (mutated in ataxia telangiectasia) (29–31). Of these two kinases, only DNA-PKcs has been shown to play a central role in DNA end-joining. Cells defective for DNA-PKcs have previously been shown to display faulty V(D)J recombination (7), which is involved in the rearrangement of immunoglobulin and T-cell receptor genes. Furthermore, analysis of DNA-PK-specific kinase activity in our cells following exposure to wortmannin indicates that the kinase activity of DNA-PK was almost completely abolished (data not shown).
The fact that DNA-PK-dependent kinase activity is not completely eliminated by treatment with wortmannin raises the possibility that residual levels of kinase activity may be adequate for simple ligation events but insufficient for complex repair events. Alternatively, there may be two genetically diverse end-joining pathways: a DNA-PK-independent pathway capable of supporting rejoining events that require a single ligation step in the absence of DNA-PK-dependent kinase activity, and a DNA-PK-dependent pathway that requires the kinase activity to recruit additional protein factors necessary for these complex repair events. We favor the second possibility, in part due to the fact that the end-joining defects we observe following exposure to wortmannin closely parallel the V(D)J recombination phenotype of DNA-PKcs-defective cell lines. During V(D)J recombination, RAG-mediated cleavage results in blunt double-stranded signal ends and covalently sealed hairpin coding ends. Subsequently, the signal ends are precisely ligated to each other while hairpins are opened and coding ends are joined in a reaction that frequently results in the loss or addition of nucleotides. Cells lacking DNA-PK activity are specifically defective in coding joint formation, displaying little or no effect on the formation of signal joints (32,33). Thus, similar to our end-joining results, DNA-PKcs-defective cells display a defect in the repair of more complex DNA ends while repair involving a single ligation event is affected much less.
Wortmannin has been shown previously to inhibit DNA-PKcs by covalently and irreversibly binding a lysine residue within the kinase domain that is critical for the phosphate transfer reaction (34). Thus, our results suggest that protein phosphorylation mediated by DNA-PK is crucial for efficient and accurate repair of complex DSB by DNA end-joining. The precise role of phosphorylation by DNA-PKcs in DNA end-joining is not known; however, several possibilities have been suggested based on in vitro biochemical analysis. For example, DNA-PKcs has been proposed to regulate accessibility of DNA ends to processing, by promoting its own dissociation following autophosphorylayion and/or by allowing translocation of Ku away from the DSB (35,36). Alternatively, phosphorylation of XRCC4 by DNA-PKcs may remove or temporarily relocate the ligase IV–XRCC4 complex from Ku-bound DNA ends, allowing necessary processing steps to occur (37).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Serge Gangloff for critical reading of the manuscript. This research was funded by the Centre National pour la Recherche Scientifique (CNRS) and grants from the Association pour la Recherche sur le Cancer, Ministère de l’Enseignement Superieur et de la Recherche and the European Community. J.S. was supported by a fellowship from the Fondation pour la Recherche Medicale and the Ligue Nationale Française Contre le Cancer. C.B. was a recipient of a fellowship from the Ministère de l’Enseignement Superieur et de la Recherche.
REFERENCES
- 1.Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Essers J., van Steeg,H., de Wit,J., Swagemakers,S.M., Vermeij,M., Hoeijmakers,J.H. and Kanaar,R. (2000) Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J., 19, 1703–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pluth J.M., Fried,L.M. and Kirchgessner,C.U. (2001) Severe combined immunodeficient cells expressing mutant hRAD54 exhibit a marked DNA double-strand break repair and error-prone chromosome repair defect. Cancer Res., 15, 2649–2655. [PubMed] [Google Scholar]
- 4.Jeggo P.A. (1998) Identification of genes involved in repair of DNA double-strand breaks in mammalian cells. Radiat. Res., 150, S80–91. [PubMed] [Google Scholar]
- 5.Grawunder U., Wilm,M., Wu,X., Kulesza,P., Wilson,T.E., Mann,M. and Lieber,M.R. (1997) Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature, 388, 492–495. [DOI] [PubMed] [Google Scholar]
- 6.Critchlow S.E., Bowater,R.P. and Jackson,S.P. (1997) Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol., 7, 588–598. [DOI] [PubMed] [Google Scholar]
- 7.Smith G.C. and Jackson,S.P. (1999) The DNA-dependent protein kinase. Genes Dev., 13, 916–934. [DOI] [PubMed] [Google Scholar]
- 8.Crespi C.L. and Thilly,W.G. (1984) Assay for gene mutation in a human lymphoblast line, AHH-1, competent for xenobiotic metabolism. Mutat. Res., 128, 221–230. [DOI] [PubMed] [Google Scholar]
- 9.Escarceller M., Buchwald,M., Singleton,B.K., Jeggo,P.A., Jackson,S.P., Moustacchi,E. and Papadopoulo,D. (1998) Fanconi anemia C gene product plays a role in the fidelity of blunt DNA end-joining. J. Mol. Biol., 279, 375–385. [DOI] [PubMed] [Google Scholar]
- 10.Smith J., Andrau,J.C., Kallenbach,S., Laquerbe,A., Doyen,N. and Papadopoulo,D. (1998) Abnormal rearrangements associated with V(D)J recombination in Fanconi anemia. J. Mol. Biol., 281, 815–825. [DOI] [PubMed] [Google Scholar]
- 11.Birnboim H.C. and Doly,J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res., 7, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes D.S. and Quigley,M. (1981) A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem., 114, 193–197. [DOI] [PubMed] [Google Scholar]
- 13.Wang T.S. (1991) Eukaryotic DNA polymerases. Annu. Rev. Biochem., 60, 513–552. [DOI] [PubMed] [Google Scholar]
- 14.Tomkinson A.E. and Mackey,Z.B. (1998) Structure and function of mammalian DNA ligases. Mutat. Res., 407, 1–9. [DOI] [PubMed] [Google Scholar]
- 15.Thode S., Schafer,A., Pfeiffer,P. and Vielmetter,W. (1990) A novel pathway of DNA end-to-end joining. Cell, 60, 921–928. [DOI] [PubMed] [Google Scholar]
- 16.Powis G., Bonjouklian,R., Berggren,M.M., Gallegos,A., Abraham,R., Ashendel,C., Zalkow,L., Matter,W.F., Dodge,J., Grindey,G. et al. (1994) Wortmannin, a potent and selective inhibitor of phosphatidylinositol-3-kinase. Cancer Res., 54, 2419–2423. [PubMed] [Google Scholar]
- 17.Price B.D. and Youmell,M.B. (1996) The phosphatidylinositol 3-kinase inhibitor wortmannin sensitizes murine fibroblasts and human tumor cells to radiation and blocks induction of p53 following DNA damage. Cancer Res., 56, 246–250. [PubMed] [Google Scholar]
- 18.Boulton S., Kyle,S., Yalcintepe,L. and Durkacz,B.W. (1996) Wortmannin is a potent inhibitor of DNA double strand break but not single strand break repair in Chinese hamster ovary cells. Carcinogenesis, 17, 2285–2290. [DOI] [PubMed] [Google Scholar]
- 19.Bogue M.A., Wang,C., Zhu,C. and Roth,D.B. (1997) V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal and hybrid joint formation. Immunity, 7, 37–47. [DOI] [PubMed] [Google Scholar]
- 20.Gu Y., Seidl,K.J., Rathbun,G.A., Zhu,C., Manis,J.P., van der Stoep,N., Davidson,L., Cheng,H.L., Sekiguchi,J.M., Frank,K., Stanhope-Baker,P., Schlissel,M.S., Roth,D.B. and Alt,F.W. (1997) Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity, 7, 653–665. [DOI] [PubMed] [Google Scholar]
- 21.Gao Y., Chaudhuri,J., Zhu,C., Davidson,L., Weaver,D.T. and Alt,F.W. (1998) A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity, 9, 367–376. [DOI] [PubMed] [Google Scholar]
- 22.Roth D.B. and Wilson,J.H. (1986) Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell. Biol., 6, 4295–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindahl T. and Barnes,D.E. (1992) Mammalian DNA ligases. Ann. Rev. Biochem., 61, 251–281. [DOI] [PubMed] [Google Scholar]
- 24.Ramsden D.A. and Gellert,M. (1998) Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks EMBO J., 17, 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labhart P. (1999) Ku-dependent nonhomologous DNA end joining in Xenopus egg extracts. Mol. Cell. Biol., 19, 2585–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldmann E., Schmiemann,V., Goedecke,W., Reichenberger,S. and Pfeiffer,P. (2000) DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res., 28, 2585–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L., Trujillo,K., Sung,P. and Tomkinson,A.E. (2000) Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J. Biol. Chem., 275, 26196–26205. [DOI] [PubMed] [Google Scholar]
- 28.Lee K.J., Huang,J., Takeda,Y. and Dynan,W.S. (2000) DNA ligase IV and XRCC4 form a stable mixed tetramer that functions synergistically with other repair factors in a cell-free end-joining system. J. Biol. Chem., 275, 34787–34796. [DOI] [PubMed] [Google Scholar]
- 29.Sarkaria J.N., Tibbetts,R.S., Busby,E.C., Kennedy,A.P., Hill,D.E. and Abraham,R.T. (1998) Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res., 58, 4375–4382. [PubMed] [Google Scholar]
- 30.Chernikova S.B., Wells,R.L. and Elkind,M.M. (1999) Wortmannin sensitizes mammalian cells to radiation by inhibiting the DNA-dependent protein kinase-mediated rejoining of double-strand breaks. Radiat. Res., 151, 159–166. [PubMed] [Google Scholar]
- 31.DiBiase S.J., Zeng,Z.C., Chen,R., Hyslop,T., Curran,W.J. and Iliakis,G. (2000) DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer Res., 60, 1245–1253. [PubMed] [Google Scholar]
- 32.Lieber M.R., Hesse,J.E., Lewis,S., Bosma,G.C., Rosenberg,N., Mizuuchi,K., Bosma,M.J. and Gellert,M. (1988) The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell, 55, 7–16. [DOI] [PubMed] [Google Scholar]
- 33.Poltoratsky V.P., Shi,X., York,J.D., Lieber,M.R. and Carter,T.H. (1995) Human DNA-activated protein kinase (DNA-PK) is homologous to phosphatidylinositol kinases. J. Immunol., 155, 4529–4533. [PubMed] [Google Scholar]
- 34.Wymann M.P., Bulgarelli-Leva,G., Zvelebil,M.J., Pirola,L., Vanhaesebroeck,B., Waterfield,M.D. and Panayotou,G. (1996) Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol. Cell. Biol., 16, 1722–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan D.W. and Lees-Miller,S.P. (1996) The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem., 271, 8936–8941. [DOI] [PubMed] [Google Scholar]
- 36.Calsou P., Frit,P., Humbert,O., Muller,C., Chen,D.J. and Salles,B. (1999) The DNA-dependent protein kinase catalytic activity regulates DNA end processing by means of Ku entry into DNA. J. Biol. Chem., 274, 7848–7856. [DOI] [PubMed] [Google Scholar]
- 37. Nick McElhinny S.A., Snowden,C.M., McCarville,J. and Ramsden,D.A. (2000) Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol., 20, 2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]