Abstract

Two zinc (Zn) complexes, [Zn2(DAT)2Cl4] (I) and [Zn2(DAT)2(NO3)4] (II), were prepared by grinding 3,5-diamino-1,2,4-triazole (C2H5N5, DAT) with Zn precursors such as ZnCl2 and Zn(NO3)2, respectively. This solid-state reaction gives the corresponding Zn complex as the sole product in over 99% yield. This mechanochemical method promotes the selective formation of Zn complexes different from those obtained using the conventional solution-based route. The crystal structures of the two complexes were analyzed by single-crystal X-ray diffraction. Complex (I) crystallizes in the monoclinic space group P21/c, whereas complex (II) crystallizes in the triclinic space group P1̅. Each complex is characterized by the presence of a characteristic DAT-bridged dimer with one DAT ligand per Zn atom, and the DAT ligand provides a bridge between the two Zn metals. All Zn centers of (I) and (II) adopted a slightly distorted tetrahedral geometry. Both complexes contain a hexanuclear Zn2N4 ring, but their ring structures are different. Complex (I) possesses a boat geometry, while complex (II) has a nearly planar structure. The Zn-bound chlorides of complex (I) form intermolecular N–H···Cl hydrogen bonds that link neighboring molecules. In complex (II), the O atoms in the nitrate groups are hydrogen-bonded to the DAT ligand via O···H–N linkages. Both complexes exhibit blue emissions in the solid state at ambient temperature. They were evaluated as anticancer agents in HeLa, NCCIT, and MCF-7 cancer cell lines, exhibiting promising anticancer activities.
Introduction
Triazoles are five-membered heterocyclic compounds composed of two carbon atoms and three nitrogen atoms. Many nitrogen-rich triazole derivatives can act as weak Lewis bases and are found to be biologically active species.1−5 They are considered pharmacophores capable of interacting with specific biological targets, which are widely used as antitumor,6−8 antibacterial,9 antifungal,10 and prebiotic agents.11 Several commercial drugs with triazole cores have been demonstrated.
Metal complexes bearing triazole ligands are also considered one of the most biologically active materials. These complexes can be utilized in various agricultural and pharmaceutical applications because their activity can be easily tuned by varying metals, ancillary ligands, and coordination geometries.12−14 Particularly, zinc (Zn) complexes have been extensively investigated due to their chemical stability and physiological merits.15−20 Zinc ions exist in many chemical and biological systems as Zn2+ species with a d10 configuration, making them difficult to either oxidize or reduce. Since Zn2+ ions can act as Lewis acids, they have a strong tendency to form stable adducts with triazole ligands. In addition, zinc is essential for almost all biological functions and has beneficial therapeutic and preventive effects on a variety of diseases.
Although many Zn triazole complexes have been reported,21,22 the reactivity of 3,5-diamino-1,2,4-triazole (DAT, C2H5N5) with Zn has been less well explored. DAT adopts a planar structure containing two amino (−NH2) groups attached to carbon positions (Scheme 1), acts as a weak Lewis base, and exhibits antitumor activities similar to those found in other triazole derivatives.23 The DAT ligand might produce structurally tunable coordination complexes with many metals, leading to the discovery of noble materials with superior chemical and biological properties. Thus, it is desirable to design and synthesize new Zn–DAT complexes that possess promising antitumor properties with minimal side effects. Here, we report two novel Zn–DAT complexes containing bidentate DAT ligands, which were prepared through simple grinding of zinc precursor and DAT (Scheme 1). The grinding plays an essential role in increasing the contact surface between the two reactants, followed by growing the Zn–DAT phase through the interfacial dissolution. The biological properties of the Zn–DAT complexes were evaluated with three different cancer cells and exhibit promising antitumor activities. In addition, they show intense blue emissions in the range of 375–390 nm.
Scheme 1. Formation of Complexes (I) and (II) by Solid-State Reaction of DAT with Zinc Chloride and Zinc Nitrate, Respectively.
Results and Discussion
Synthesis
The synthesis of [Zn2(DAT)2Cl4] (I) was achieved through neat grinding of zinc chloride with DAT in an argon-filled glovebox using a mortar and pestle. The resulting powder with a 1:1 molar ratio of zinc chloride and DAT was stored in a vial without any mechanical agitation. The formation of a new complex was carefully monitored by powder X-ray diffraction (XRD). Complete conversion of the reactants to complex (I) was accomplished via the interfacial reaction within 3 h at room temperature. Neither byproducts nor reactants were detected in the XRD data. The solid-state reaction affords complex (I) as the sole product with greater than 99% yield. It is worth mentioning that the Zn complex can be prepared within 20 min by heating the ground powder at 80 °C. To prove that the product obtained by heating at 80 °C is identical to the one obtained at room temperature, the powder XRD data of both samples were overlaid (Figure S1).
To determine whether the solution-based reaction could also produce the same complex (I), the reaction between zinc chloride and DAT was carefully monitored in an aqueous solution. In contrast to the solid-state route, the solution-based reaction gave unidentified products (Figure S2) rather than complex (I). This is because ZnCl2 in aqueous solution readily dissociates into Zn2+ and Cl– species, and the dissociated ions directly react with DAT. As a result, the solution-based reaction could lead to the formation of products different from those obtained via solid-state grinding.
Similarly, we were able to synthesize [Zn2(DAT)2(NO3)4] (II) by grinding DAT with zinc nitrate in an argon-filled glovebox, followed by storing the mixed powder. Complete conversion to complex (II) was achieved at room temperature within 3 h, without using additives. Like complex (I), the solid-state reaction produces selectively complex (II) with a yield of 99% or more, and heating the powder at 80 °C yields complex (II) within 20 min. This mechanochemical method demonstrates a straightforward and solvent-free method for the synthesis of Zn–DAT complexes. In addition, this synthetic route based on solid-state grinding is easily scalable and environmentally benign,24,25 which might provide a noble approach for preparing various hybrid metal-DAT materials in the absence of solvents. It is worth mentioning that the high yield for the complexes obtained via mechanochemical synthesis might be due to liquid-assisted grinding. Perhaps a small amount of water in the reactants acts as a promoter and plays an important role in accelerating the solid-state reaction.
Structure
The solid-state structures of the two Zn–DAT complexes were analyzed by single-crystal X-ray diffraction studies. The as-synthesized sample (I) is moderately soluble in ethanol, which was recrystallized from an ethanol solution. Slow evaporation of ethanol afforded transparent flower-like crystals suitable for X-ray diffraction analysis. Similarly, single crystals of complex (II) were grown from an ethanol solution. The acquisition of single-crystal diffraction data for each complex gave clear evidence for the formation of the Zn–DAT complex. The X-ray crystallographic data for the two complexes are described in Table S1.
Complex (I) crystallizes in the monoclinic space group P21/c with four formula units per unit cell and one molecule in the asymmetric unit. The molecular structure of complex (I) with the atom numbering scheme is illustrated in Figure 1, showing a dimeric structure bridged by two DAT ligands. Two Zn atoms are linked together via two DAT ligands to form a six-membered ring, which can be described as Zn2N4. From the viewpoint of the cyclohexane conformation, the cyclic ring might be described as a boat shape. Each Zn center shows a tetrahedral geometry coordinated by two Cl anions and two N atoms. The charge neutralization of complex (I) is attained by four Cl– anions and two neutral DAT ligands, which counterbalances the two Zn2+ cations in the complex. Selected bond lengths and bond angles of complex (I) are listed in Table S2. The Zn–Cl bond lengths are within the expected range for tetrahedral ZnCl2L2 (L = N-bonded ligand) complexes, i.e., 2.20–2.30 Å.26,27 The Zn atoms are doubly bridged by two DAT ligands, which gives rise to a significantly long Zn1···Zn2 distance of 3.568 Å.
Figure 1.

Molecular structure of the complex (I) with the atom-labeling scheme. Anisotropic displacement parameters are plotted as the 50% probability ellipsoids for non-H atoms and spheres of arbitrary size for H atoms.
The DAT ligand can bind to transition metal in various ways, such as monodentate, bidentate, and tridentate modes. As seen in Figure 1, two DAT molecules link two Zn ions through N1,N2-type exo-bidentate modes.28 Four Zn–N–N bond angles at the DAT bridging region range from 121 to 123°, which is close to the ideal bond angle for trigonal planar (120°). Two N–Zn–N bond angles at the Zn1 and Zn2 sites are 104.81 and 102.04°, respectively. Five other angles of the Zn-centered tetrahedral are slightly deviated from the ideal value (109.5°) but are similar to the reported value for typical tetrahedral ZnCl2L2 complexes.26,27 Two N–N bond lengths of the five-membered ring are 1.412 and 1.411 Å, which are slightly longer than that of the free DAT molecule (1.398 Å),29 but are within the single N–N bond range of other triazole complexes. The cyclic DAT rings in (I) consist of four longer C–N (C1–N3, C1–N4, C2–N3, C2–N5) and two shorter (C1–N1, C2–N2) bonds, which are similar to the typical single (C–N) and double (C=N) bond distances, respectively. These bonding properties are consistent with those commonly observed in similar 1,2,4-triazole-bridged complexes in an N1,N2-bidentate mode.
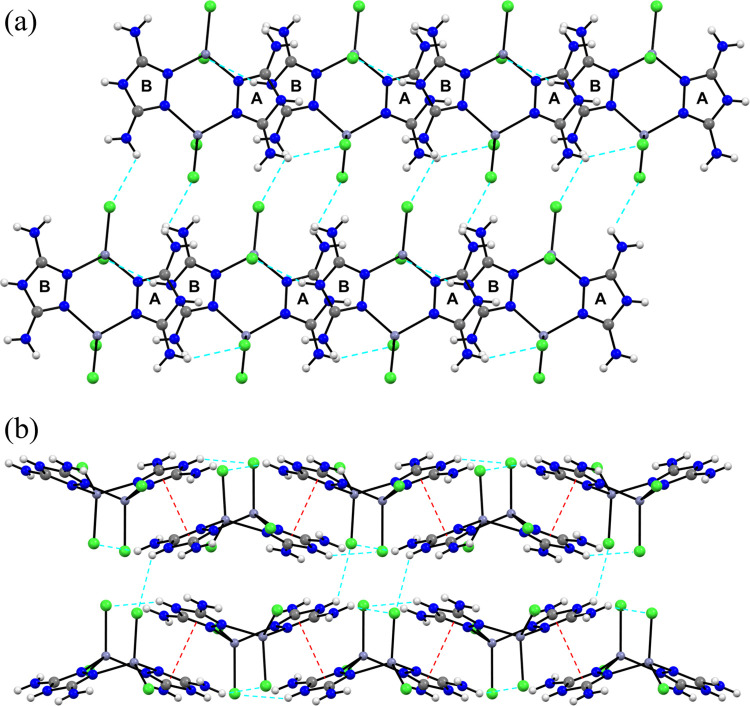
Figure 2a presents an enlarged view of several hydrogen-bonding (H-bonding) interactions between the hydrogen atom of the DAT ligand and the Zn-bound chlorine atom. The hydrogen-bonding network in complex (I) is connected via directional N–H···Cl–Zn bond linkages. Table S3 shows hydrogen-bond geometry for complex (I), where only N···Cl distances shorter than the sum of van der Waals radii for the N and Cl atoms (3.30 Å) are included. The N···Cl distances indicate that the Cl atoms are clearly involved in intermolecular H-bonding with adjacent DAT ligands. Each Cl atom forms an N–H···Cl–Zn hydrogen bond to bridge the adjacent molecule. Three of the four Cl atoms interact with hydrogen atoms of three respective NH2 groups. The remaining Cl atom participates in H-bond with the N–H hydrogen atom of the triazole ring. Weak intramolecular H-bonding is also feasible in complex (I), but the N···Cl distances are longer than the van der Waals sum.
Figure 2.
(a) Expanded unit cell for complex (I) with a view of the (010) plane, highlighting the N–H···Cl interactions. Blue dashed lines indicate intermolecular hydrogen bonds. Two triazole rings, labeled A and B, are close to the same plane. Purple, gray, blue, and green spheres represent Zn, C, N, and Cl atoms, respectively. Small white spheres denote H atoms. Only N···Cl distances less than van der Waals sum (3.30 Å) are included. (b) Packing arrangement of complex (I) showing π···π interactions between adjacent triazole rings, which are indicated by red dashed lines.
Figure 2b shows the centroid-to-centroid interaction between adjacent triazole rings with a distance of 3.33 Å. The interlayer distance is virtually identical to that of graphite (3.34 Å) and is slightly shorter than other triazole compounds.22 This indicates that strong π···π interactions are present in complex (I). The π···π interaction and the intermolecular H-bonding interaction create a three-dimensional packing of the molecules, resulting in the supramolecular features of complex (I).
The molecular structure of complex (II), as illustrated in Figure 3a, shows a centrosymmetric DAT-bridged Zn dimer. Complex (II) crystallizes in the triclinic space group P1̅, where the asymmetric unit is one half of the molecule with the other half related by an inversion center. The coordination sphere around each Zn ion adopts a distorted tetrahedral geometry with ligation by two O atoms of two nitrate ions (NO3–) and two N atoms of two DAT ligands. The bond angles of N1–Zn1–N2 and O1B–Zn1–O1A are 114.59 and 104.78°, respectively. The average Zn–O bond length involving the monodentate nitrate ligands is 1.99 Å, which is slightly shorter than that reported for zinc complexes with nitrate ligands.30,31 Bond lengths associated with the DAT ligand of complex (II) are virtually identical to those of complex (I). Unlike complex (I), however, the Zn2N4 ring in complex (II) adopts a nearly planar geometry, which provides two terminal nitrate ligands to each Zn center (Figure 3b). This is evidenced by a sum of internal angles of the hexanuclear Zn2N4 ring (716.78°), close to the ideal hexagon value (720°). Selected bond lengths and bond angles of complex (II) are listed in Table S4.
Figure 3.
(a) Molecular structure of complex (II) with the atom-labeling scheme. Anisotropic displacement parameters are drawn at the 50% probability ellipsoids for non-H atoms and spheres of arbitrary size for H atoms. The unlabeled atoms are related by a center of inversion. (b) Crystal structure of complex (II), viewed nearly parallel to the Zn2N4 plane. Only Zn2N4 ring and four oxygen atoms of nitrate ligands are included for clarity.
Complex (II) contains the four Zn-bound nitrate groups that can be donor groups to hydrogen bonds. The nitrate group engages in both intermolecular and intramolecular H-bonding interactions with the DAT ligand through the N–O···H–N linkage. Oxygen atoms of the nitrate form two intramolecular hydrogen bonds to two hydrogen atoms of two respective NH2 groups (red dashed lines in Figure 4), which stabilizes the dinuclear structure. In addition, the oxygen atoms undergo strong intermolecular H-bonding interactions, indicated by blue dashed lines in Figure 4, with hydrogen atoms of adjacent molecules. These intermolecular interactions create an extensive three-dimensional network, demonstrating the supramolecular features of complex (II). Hydrogen-bond geometry for complex (II) is given in Table S3. Only N···O distances shorter than the sum of van der Waals radii for the N and O atoms (3.07 Å) are included in the H-bonding network.
Figure 4.
Expanded unit cell for complex (II) with a view of (a) (010) and (b) (100) planes, showing three molecules constructed from hydrogen-bonded networks and highlighting the N–H···O interactions. Blue and red dashed lines indicate intermolecular and intramolecular hydrogen bonds, respectively. Purple, gray, blue, and red spheres represent Zn, C, N, and O atoms, respectively. The small white sphere denotes H atom.
Thermal Stability
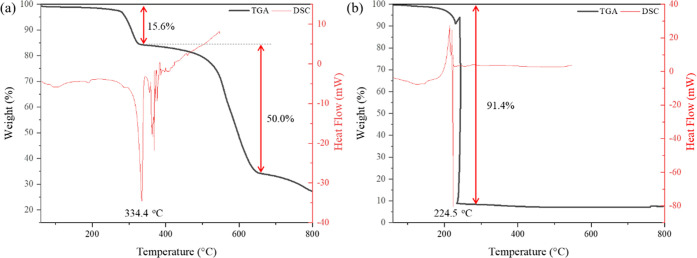
The thermal stabilities of complexes (I) and (II) were evaluated through thermogravimetric analysis (TGA) and differential scanning calorimetry (DSC). The TGA and DSC curves for complexes (I) and (II) are shown in Figure 5a,b, respectively. The TGA curve of (I) shows that the complex is stable up to 250 °C in nitrogen and then a gradual weight loss occurs in two steps. The sample first loses 15.6% of its weight from 270 to 330 °C, which is attributed to the desorption of two Zn-bound chlorides as hydrogen chloride. A further weight loss of 50.0% occurs between 350 and 650 °C. This may indicate the decomposition of remaining Cl and DAT ligands, followed by the oxidation of Zn species to ZnO. As a result of analyzing the decomposition product by powder X-ray diffraction, it was confirmed that ZnO is the only product. In contrast, the TGA curve of complex (II) shows a sharp transition at about 230 °C, analogous to those of typical energetic materials.32 During complete decomposition, 91.4% of the original mass was lost, which is higher than the theoretically expected value of the mass change for ZnO formation (71.8%). This suggests that complex (II) decomposes explosively upon heating. This is not surprising, as complex (II) contains four nitrate groups and two nitrogen-rich DAT ligands that can explode at high temperatures. The corresponding DSC curves of each TGA curve are generally similar to those observed in the thermal decomposition profiles. The DSC curve of complex (I) shows an endothermic peak at 334 °C, indicating an endothermic reaction caused by thermal dissociation of Zn–Cl bonds. During DSC measurements, the pan and lid are separated due to the swelling of the sample. As a result, data after the peak temperature are unreliable. The exothermic peak of complex (II) observed at 224 °C is indicative of the exothermic oxidative decomposition normally observed in energetic materials.32
Figure 5.
TGA (black line) and DSC (red line) curves of (a) complex (I) and (b) complex (II) measured under flowing N2 gas with a heating rate of 10 °C min–1.
Spectroscopy
The UV–visible absorption and photoluminescence (PL) spectra of complexes (I) and (II) in the solid state are displayed in Figure 6a,b, respectively. The measured reflectance spectra were transformed into the corresponding absorption spectra. Two broad peaks are displayed in the absorption spectrum of complex (I). One peak starts in the UV region and ends at about 270 nm, and the other shows a broad weak band with maxima at around 330–350 nm. The horizontal intercept of the linear fit of the Kubelka–Munk function yields the estimated bandgap energy of complex (I), which is 4.86 eV (255 nm, Figure S3). This bandgap energy is close to that (4.50 eV) obtained from the projected density of state (PDOS) for the complex (I) using the structural data (Figure S4). The main contributors to the valence band of complex (I) are the DAT and Cl ligands, while the conduction band consists solely of the C and N orbitals of the DAT ring. Unlike complex (I), complex (II) does not exhibit a steep transition in the UV region. Instead, it shows a broad band with maxima at around 280 nm. The bandgap energy of complex (II) was also determined by extrapolating the plot to the horizontal axis. The optical bandgap energy is estimated to be 4.36 eV (284 nm, Figure S3), which is roughly equivalent to the magnitude (4.41 eV) calculated from the PDOS calculation (Figure S4). Both valence and conduction bands of complex (II) are strongly affected by the p orbital of O in the nitrate (NO3–) group. The bandgap is almost unaffected by the Zn ion. This is because the Zn2+ ion has a filled d orbital (d10). PL spectra of complexes (I) and (II) were recorded with excitation wavelengths of 310 and 325 nm, respectively. Both complexes have emission bands in the visible region with maxima at 373 and 390 nm for complexes (I) and (II), respectively.
Figure 6.
UV–visible absorption and photoluminescence (PL) spectra of (a) complex (I) and (b) complex (II) in the solid state at room temperature. Black and blue curves represent UV–visible absorption and PL emission spectra, respectively.
Biological Evaluation
To evaluate the cytotoxic properties of the Zn–DAT complexes, cell viability studies were conducted with the MTT assay. The in vitro cytotoxicity of the two complexes in HeLa cells was first tested because they are the most commonly used human cancer cell lines for the initial investigation of chemotherapy. In addition, NCCIT and MCF-7 cancer cells were assessed. The concentration required for 50% inhibition of the cell population (IC50 value) was determined for the two complexes and the DAT compound (Table 1 and Figures S5–S7). Each IC50 value was obtained from at least three independent experiments. All IC50 values of complexes (I) and (II) range from 118.5 to 367.2 μM, implying that both Zn–DAT complexes exhibit anticancer effects against all cancer cell lines tested. Complex (I) showed slightly higher activity than complex (II) in the same cell lines. The relative activities of the two complexes appear to be related to the difference between the chloride and nitrate ligands. To investigate the role of the DAT ligand, we tested the activities of free DAT compound against the three cell lines. As can be seen in Table 1, DAT alone is inactive in all investigated cell lines.
Table 1. IC50 Values of Complex (I), Complex (II), and DAT in NCCIT, MCF-7, and HeLa Cell Lines.
| IC50 (μM) |
|||
|---|---|---|---|
| compound | HeLaa | NCCITa | MCF-7a |
| complex (I)b | 128.8 ± 1.9 | 245.3 ± 4.0 | 118.5 ± 2.2 |
| complex (II)b | 217.1 ± 1.4 | 367.2 ± 6.5 | 210.8 ± 6.3 |
| DAT | N.D.c | N.D.c | N.D.c |
| [NiL2(H2O)4](NO3)433,d | 26.5 ± 1.2 | ||
| [Pd(AMTT)2]Cl2·2H2O34,e | 925 | ||
All of the MTT assays were performed after 72 h of incubation.
IC50 values are expressed as concentrations of zinc.
No cytotoxic activity was detected.
L = 4-amino-3,5-bis(2-pyridyl)-1,2,4-triazole. The MTT assay was performed for 24 h.
The MTT assay was performed for 48 h.
Conclusions
Two thermally stable and biologically active Zn–DAT complexes, [Zn2(DAT)2Cl4] (I) and [Zn2(DAT)2(NO3)4] (II), were synthesized through the solid-state reaction of DAT with zinc chloride and zinc nitrate, respectively. They were isolated directly from the corresponding ground mixture as the sole products in greater than 99% yield. Two DAT ligands are linked to two Zn metals in both molecular structures, forming a hexanuclear Zn2N4 ring. Depending on the Zn-bound ligand, the Zn2N4 structure adopts a boat shape for complex (I) and a planar geometry for complex (II). Both complexes have extensive hydrogen bonding between the Zn-bound ligand and the hydrogen atoms of the DAT ligand, leading to the construction of supramolecular architectures. They show intense photoluminescence properties in the solid state. The Zn–DAT complexes exhibit promising anticancer activity in three human cancer cells, whereas DAT does not possess any noticeable cytotoxic activity toward the same cell lines. Notably, the “mechanochemical” pathway can be used to accelerate the synthesis of a wide range of metal-DAT complexes, providing ample opportunities to develop highly efficient anticancer drugs and tunable luminescent materials with desired properties.
Experimental Section
Chemicals and Materials
All reagents were used as received without further purification. Zinc chloride (>98%, ZnCl2) and zinc nitrate (>99%, Zn(NO3)2(H2O)6) were purchased from Merck KGaA (Darmstadt, Germany). DAT (>98%, C2H5N5) was purchased from Acros Organics (Geel, Belgium), and anhydrous ethanol (99.9%) was purchased from Dae Jung Chemicals & Metals (Seoul, Korea). HeLa, NCCIT, and MCF-7 cell lines were purchased from the American Type Culture Collection (Manassas, Virginia).
Synthesis of [Zn2(DAT)2Cl4] (I)
A 1:1 mixture of ZnCl2 (0.136 g, 1.0 mmol) and DAT (0.099 g, 1.0 mmol) was ground at room temperature using a mortar and pestle. The ground mixture was placed in a vial and allowed to react at room temperature for 3 h. The same product was also obtained by heating the ground mixture at 80 °C for 20 min in an oven. Both yield and selectivity are typically over 99%. The resulting product was initially characterized by powder XRD. Single crystals of (I) were obtained by the recrystallization of the solid product from an ethanol solution. Colorless flower-like crystals suitable for single-crystal X-ray diffraction studies were grown by slow evaporation. Elemental analysis: C4H10Cl4N10Zn2 Calcd C 10.20, H 2.12, N 29.74. Found C 10.19, H 2.22, N 29.75. DSC (10 °C min–1) peak temperature: 334.4 °C (dec.)
Synthesis of [Zn2(DAT)2(NO3)4] (II)
A 1:1 mixture of Zn(NO3)2·6H2O (0.297 g, 1.0 mmol) and DAT (0.099 g, 1.0 mmol) was ground at room temperature using a mortar and pestle. The ground mixture was placed in a vial and allowed to react at room temperature for 3 h. The same product was also obtained by heating the ground mixture at 80 °C for 20 min in an oven. Both yield and selectivity are typically over 99%. The resulting product was initially characterized by powder XRD. Single crystals of (II) were obtained by the recrystallization of the solid product from an ethanol solution. Colorless block-shaped crystals suitable for single-crystal X-ray diffraction studies were grown by slow evaporation. Elemental analysis: C4H10N14O12Zn2 Calcd C 8.32, H 1.73, N 33.97. Found C 8.56, H 1.90, N 33.26. DSC (10 °C min–1) peak temperature: 224.5 °C (dec.)
Cell Culture and Cell Viability Assay
HeLa, NCCIT, and MCF-7 cells (1 × 104) were plated in 96-well plates and cultured in the presence of indicated concentrations of complex (I) or complex (II). After 3 days (for HeLa cells) or 4 days (for NCCIT and MCF-7 cells), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays were performed according to the manufacturer’s instructions using Cell Proliferation Kit I (MTT, Roche). Briefly, 15 μL of MTT labeling reagent (5 mg/mL MTT in phosphate-buffered saline) was added to each well of the 96-well plate to measure cell viability. For metabolization of soluble MTT compound (yellow) to an insoluble formazan salt (blue) by viable HeLa, NCCIT, or MCF-7 cells, the plate was incubated at 37 °C for 4 h in a cell culture incubator (Thermo Fisher Scientific). The reduced blue formazan crystals were solubilized overnight with 150 μL of solubilization buffer (10% sodium dodecyl sulfate (SDS) in 0.01 M HCl) in each well. The intensity of solubilized formazan product in the culture medium proportionally represented the number of living cells and was quantified using a microplate reader (Molecular Devices) at 570 nm wavelength with background subtraction at 660 nm.
Characterization
Elemental analyses (C, H, and N) were performed on Thermo Flash EA 1112/2000 element analyzer at the Organic Chemistry Research Center at Sogang University. Fourier transform infrared (FT-IR) spectra were recorded with Nicolet Summit FT-IR Spectrophotometer with an attenuated total reflectance (ATR) accessory in the region on 650–4000 cm–1. 1H and 13C NMR spectra were recorded on a Varian 400 MHz Gemini operating at 400 MHz for 1H and 100 MHz for 13C, respectively. Electrospray ionization mass spectrometry (ESI-MS) was performed using an ion trap mass spectrometer, where the accurate mass was measured using a Varian 500-MS with a heated electrospray ionization source. Thermogravimetric analyses (TGA) were carried out on TGA Q50 thermal analyzer. The powder sample was placed on a platinum pan and analyzed under nitrogen flow from 60 to 800 °C at a heating rate of 10 °C min–1. Differential scanning calorimeter (DSC) experiments were performed with DSC Q2000 under nitrogen flow from 20 to 550 °C at a heating rate of 10 °C min–1. UV–Vis spectra were measured using a Jasco V-660 spectrophotometer by the diffuse reflectance method in the region on 200–700 nm with 1 nm intervals. The bandgap energy was converted from the reflection spectra by the Kubelka–Munk functions.35 Luminescent spectra were recorded on a Hitachi F-7000 spectrophotometer. Powder X-ray diffraction spectra were recorded with a Rigaku Miniflex 600 diffractometer with Cu Kα radiation (λ = 1.5406 Å) operated at 40 kV and 15 mA. Samples were scanned in the 2θ range of 5–70° at a scan speed of 10° min–1 and a scan width of 0.02°.
Crystallographic Refinement and Structure Solution (X-ray Crystallography)
Prepared colorless crystals were mounted on glass fiber and used for data collection on a Bruker D8 QUEST diffractometer with a graphite monochromated Mo Kα (λ = 0.71703 Å) radiation source and a PHOTON-II CPAD detector at the Advanced Bio-Interface Core Research Facility, Sogang University. The collected data were integrated using the SAINT program,36 and an absorption correction was applied using the SADABS program.37 The crystal structures were solved using the SHELXS-201338 and refined using the SHELXL-201539 implemented in the WinGX-201340 program.
Density Functional Theory (DFT) Calculation
The density of state investigation was performed by the Quantum Espresso package,41 using the data collected by single-crystal X-ray diffraction for structural optimization. The electronic structures of the complexes were calculated by the ultrasoft pseudopotential42 and Perdew–Burke–Ernzerhof (PBE)43 exchange–correlation functional. The kinetic energy and charge density cutoff were set to 45.656 and 410.902 Ry for complex (I), and 46.659 and 419.846 Ry for complex (II), respectively. The self-consistent function (SCF) convergence thresholds were set to 10–6 Ry for both complexes, and calculations were implemented in 3 × 3 × 3 k-point grids of the Brillouin zone for complex (I) and 4 × 4 × 3 for complex (II).
Acknowledgments
This work (N.H.H.) was supported by the NRF funded by the Korean Government (MSIT) (grant no. NRF-2019R1A2C1003666) and the COMPA funded by the Korea government (MSIT) (grant no. 2021-JDH-1-SB-1). H.S.P. also acknowledges the NRF funded by the Korean Government (MSIT) (NRF-2020R1A6A3A13070878).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c03715.
Additional experimental method and characterization information; crystallographic data; collected and calculated powder XRD pattern; Kubelka–Munk function; diffuse reflectance spectrum; total and projected density of state; effects of complexes on the viability of HeLa, NCCIT, and MCF-7 cells; FT-IR spectra; 1H and 13C NMR spectra; and ESI-MS data (PDF)
Accession Codes
CCDC 2176496 and CCDC 2176497 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif, by emailing data_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: +44 1223 336033.
The authors declare no competing financial interest.
Supplementary Material
References
- Moulin A.; Bibian M.; Blayo A.-L.; Habnouni S. E.; Martinez J.; Fehrentz J.-A. Synthesis of 3,4,5-Trisubstituted-1,2,4-triazoles. Chem. Rev. 2010, 110, 1809–1827. 10.1021/cr900107r. [DOI] [PubMed] [Google Scholar]
- Gautier R.; Clérac R. Tuning the Crystal Structure Dimensionality of Cobalt(II)/1,2,4-Triazole Complexes. Cryst. Growth Des. 2017, 17, 864–869. 10.1021/acs.cgd.6b01723. [DOI] [Google Scholar]
- Kaur J.; Saxena M.; Rishi N. An Overview of Recent Advances in Biomedical Applications of Click Chemistry. Bioconjugate Chem. 2021, 32, 1455–1471. 10.1021/acs.bioconjchem.1c00247. [DOI] [PubMed] [Google Scholar]
- Haasnoot J. G. Mononuclear, Oligonuclear and Polynuclear Metal Coordination Compounds with 1, 2, 4-Triazole Derivatives as Ligands. Coord. Chem. Rev. 2000, 200-202, 131–185. 10.1016/S0010-8545(00)00266-6. [DOI] [Google Scholar]
- Padmaja R. D.; Chanda K. A Short Review on Synthetic Advances toward the Synthesis of Rufinamide, an Antiepileptic Drug. Org. Process Res. Dev. 2018, 22, 457–466. 10.1021/acs.oprd.7b00373. [DOI] [Google Scholar]
- Grob N. M.; Schmid S.; Schibli R.; Behe M.; Mindt T. L. Design of Radiolabeled Analogs of Minigastrin by Multiple Amide-to-Triazole Substitutions. J. Med. Chem. 2020, 63, 4496–4505. 10.1021/acs.jmedchem.9b01937. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Keenan S. M.; Peng Y.; Nair A. C.; Yu S. J.; Howells R. D.; Welsh W. J. Discovery of Novel Triazole-Based Opioid Receptor Antagonists. J. Med. Chem. 2006, 49, 4044–4047. 10.1021/jm0601250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne Y.; Sharpless K. B.; Taylor P.; Marchot P. Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase. J. Am. Chem. Soc. 2016, 138, 1611–1621. 10.1021/jacs.5b11384. [DOI] [PubMed] [Google Scholar]
- Guo H.; Dong Y.; Zhu S.; Que H.; Lu X.; Zhu X.; Cheng K.; Gu X. Synthesis, Compound, Emulsification, and Antibacterial Activity of Modified 1,2,4-Trizaole Derivatives. ACS Omega 2019, 4, 9680–9685. 10.1021/acsomega.9b00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo M.-M.; Faria-Ramos I.; Cruz L. C.; Pina-Vaz C.; Rodrigues A. G. Genesis of Azole Antifungal Resistance from Agriculture to Clinical Settings. J. Agric. Food Chem. 2015, 63, 7463–7468. 10.1021/acs.jafc.5b02728. [DOI] [PubMed] [Google Scholar]
- Fialho D. M.; Roche T. P.; Hud N. V. Prebiotic Syntheses of Noncanonical Nucleosides and Nucleotides. Chem. Rev. 2020, 120, 4806–4830. 10.1021/acs.chemrev.0c00069. [DOI] [PubMed] [Google Scholar]
- McDonald K. A.; Seth S.; Matzger A. J. Coordination Polymers with High Energy Density: An Emerging Class of Explosives. Cryst. Growth Des. 2015, 15, 5963–5972. 10.1021/acs.cgd.5b01436. [DOI] [Google Scholar]
- Orselli E.; Kottas G. S.; Konradsson A. E.; Coppo P.; Fröhlich R.; Cola L. D.; Dijken A.; Büchel M.; Börner H. Blue-Emitting Iridium Complexes with Substituted 1,2,4-Triazole Ligands: Synthesis, Photophysics, and Devices. Inorg. Chem. 2007, 46, 11082–11093. 10.1021/ic701110p. [DOI] [PubMed] [Google Scholar]
- Kim S.; Joarder B.; Hurd J. A.; Zhang J.; Dawson K. W.; Gelfand B. S.; Wong N. E.; Shimizu G. K. H. Achieving Superprotonic Conduction in Metal–Organic Frameworks through Iterative Design Advances. J. Am. Chem. Soc. 2018, 140, 1077–1082. 10.1021/jacs.7b11364. [DOI] [PubMed] [Google Scholar]
- Fernández B.; Fernández I.; Cepeda J.; Medina-O’Donnell M.; Rufino-Palomares E. E.; Raya-Barón Á.; Gómez-Ruiz S.; Pérez-Jiménez A.; Lupiáñez J. A.; Reyes-Zurita F. J.; Rodríguez-Diéguez A. Modulating Anticancer Potential by Modifying the Structural Properties of a Family of Zinc Metal–Organic Chains Based on 4-Nitro-1H-pyrazole. Cryst. Growth Des. 2018, 18, 969–978. 10.1021/acs.cgd.7b01443. [DOI] [Google Scholar]
- Zhang R.-B.; Li Z.-J.; Qin Y.-Y.; Cheng J.-K.; Zhang J.; Yao Y.-G. Synthesis, Structure, and Physical Properties of a New Anions-Controlled Cd(II)-Guanazole (3,5-Diamino-1,2,4-triazole) Hybrid Family. Inorg. Chem. 2008, 47, 4861–4876. 10.1021/ic8001042. [DOI] [PubMed] [Google Scholar]
- Hernández-Gil J.; Ovèjak N.; Ferrer S.; Lloret F.; Castiñeiras A. Novel Hexanuclear Copper(II) Complex Built from a Simple Tetrachelating Triazole Ligand: Synthesis, Structure, and Magnetism. Inorg. Chem. 2013, 52, 2289–2291. 10.1021/ic3027946. [DOI] [PubMed] [Google Scholar]
- Aznar E.; Ferrer S.; Borrás J.; Lloret F.; Liu-González M.; Rodríguez-Prieto H.; García-Granda S. Coordinative Versatility of Guanazole [3, 5-Diamino-1, 2, 4-triazole]: Synthesis, Crystal Structure, EPR, and Magnetic Properties of a Dinuclear and a Linear Trinuclear Copper (II) Complex Containing Small Bridges and Triazole Ligands. Eur. J. Inorg. Chem. 2006, 2006, 5115–5125. 10.1002/ejic.200600711. [DOI] [Google Scholar]
- Ray A.; Mitra S.; Rosair G. M. A Novel Hydroxo-Bridged Cyclic Tetranuclear Copper (II) Complex Using the Guanazole Ligand: Synthesis, Crystal Structure and Thermal Analysis. Inorg. Chem. Commun. 2008, 11, 1256–1259. 10.1016/j.inoche.2008.07.010. [DOI] [Google Scholar]
- Klapötke T. M.; Schmid P. C.; Stierstorfer J.; Szimhardt N. Synthesis and Characterization of Tetrahedral Zinc (II) Complexes with 3, 6, 7-Triamino-7H-[1, 2, 4]triazolo[4, 3-b][1, 2, 4]triazole as Nitrogen-Rich Ligand. Z. Anorg. Allg. Chem. 2016, 642, 383–389. 10.1002/zaac.201600006. [DOI] [Google Scholar]
- Zhao H.; Dong Y.; Liu H. Two New Luminescent Zn(II) Compounds Constructed from Guanazole and Aromatic Polycarboxylate Ligands. J. Mol. Struct. 2016, 1105, 112–117. 10.1016/j.molstruc.2015.10.046. [DOI] [Google Scholar]
- Gusev A.; Braga E.; Baluda Y.; Kiskin M.; Kryukova M.; Karaush-Karmazin N.; Baryshnikov G.; Kuklin A.; Minaev B.; Ågren H.; Linert W. Structure and Tuneable Luminescence in Polymeric Zinc Compounds Based on 3-(3-Pyridyl)-5-(4-pyridyl)-1,2,4-triazole. Polyhedron 2020, 191, 114768 10.1016/j.poly.2020.114768. [DOI] [Google Scholar]
- Li H.; Gao J.; Tang H.; Li J.; Zhang T.; Yang X. Investigating the Synthetic Mechanism of 3,5-Diamino-1,2,4-triazole by Using Fibre Optic ATR-IR Spectroscopy Combined with Kernel Independent Component Analysis. Anal. Methods 2015, 7, 4152–4158. 10.1039/C4AY02956A. [DOI] [Google Scholar]
- Martí-Rujas J.; Guo F. Dehydrohalogenation Reactions in Second-Sphere Coordination Complexes. Dalton Trans. 2021, 50, 11665–11680. 10.1039/D1DT02099D. [DOI] [PubMed] [Google Scholar]
- Loya J. D.; Li S. J.; Unruh D. K.; Hutchins K. M. Mechanochemistry as a Tool for Crystallizing Inaccessible Solids from Viscous Liquid Components. Cryst. Growth Des. 2022, 22, 285–292. 10.1021/acs.cgd.1c00929. [DOI] [Google Scholar]
- Zick P. L.; Geiger D. K. Structural Characterization of Two Benzene-1,2-diamine Complexes of Zinc Chloride: A Molecular Compound and a Co-Crystal Salt. Acta Crystallogr., Sect. E: Crystallogr. Commun. 2016, 72, 1037–1042. 10.1107/S2056989016010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivšić T.; Bi D. W.; Magrez A. New Refinement of the Crystal Structure of Zn(NH3)2Cl2 at 100K. Acta Crystallogr., Sect. E: Crystallogr. Commun. 2019, 75, 1386–1388. 10.1107/S2056989019011757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela J.; Vaddadi S.; Kingsley S.; Flaschenriem C. J.; Lachicotte R. J.; Cundari T. R.; Holland P. L. Bidentate Coordination of Pyrazolate in Low-Coordinate Iron(II) and Nickel(II) Complexes. Angew. Chem., Int. Ed. 2006, 45, 1607–1611. 10.1002/anie.200503535. [DOI] [PubMed] [Google Scholar]
- Starova G. L.; Frank-Kamenetskaya O. V.; Shibanova E. F.; Lopyrev V. A.; Voronkov M. G.; Makarskii V. V. X-ray Diffraction Examination of the Molecular Structure of Guanazole (3, 5-Diamino-1H-1, 2, 4-triazole). Chem. Heterocycl. Compd. 1979, 15, 1149–1150. 10.1007/BF00471922. [DOI] [Google Scholar]
- Zhou L. Crystal Structure of Bis(2,2′-bipyridine-N,N′)-(nitrato-O)zinc(II) Nitrate Monohydrate, [Zn(C10H8N2)(NO3)][NO3] · H2O. Z. Kristallogr. - New Cryst. Struct. 2011, 226, 27–28. 10.1524/ncrs.2011.0014. [DOI] [Google Scholar]
- Mousavi S. A.; Montazerozohori M.; Masoudiasl A.; Mahmoudi G.; White J. M. Sonication-Assisted Synthesis of a New Cationic Zinc Nitrate Complex with a Tetradentate Schiff Base Ligand: Crystal Structure, Hirshfeld Surface Analysis and Investigation of Different Parameters Influence on Morphological Properties. Ultrason. Sonochem. 2018, 46, 26–35. 10.1016/j.ultsonch.2018.02.050. [DOI] [PubMed] [Google Scholar]
- Tang Y.; Yin Z.; Chinnam A. K.; Staples R. J.; Shreeve J. M. A Duo and a Trio of Triazoles as Very Thermostable and Insensitive Energetic Materials. Inorg. Chem. 2020, 59, 17766–17774. 10.1021/acs.inorgchem.0c03014. [DOI] [PubMed] [Google Scholar]
- Poornima S.; Anbu S.; Ravishankaran R.; Sundaramoorthy S.; Vennila K. N.; Karande A. A.; Velmurugan D.; Kandaswamy M. DNA and Protein Targeting 1,2,4-Triazole Based Water Soluble Dinickel(II) Complexes Enhances Antiproliferation and Lactate Dehydrogenase Inhibition. Polyhedron 2013, 62, 26–36. 10.1016/j.poly.2013.06.017. [DOI] [Google Scholar]
- Sajadiyeh E.; Tabatabaee M.; Seifati S. M.; Derikvand Z. Cytotoxic Effect of Palladium(II) Complex with 4-Amino-5-methyl-2H-1,2,4-triazole-3(4H)-thione Ligand on MCF-7 Cell Line. Pharm. Chem. J. 2020, 54, 145–147. 10.1007/s11094-020-02171-5. [DOI] [Google Scholar]
- Yoo J. B.; Yang H. J.; Hur N. H. Controlled Synthesis of Luminescent Mn-Doped Zn2SiO4 Microspheres by Thermal Hydrolysis of Urea. J. Lumin. 2022, 243, 118608. 10.1016/j.jlumin.2021.118608. [DOI] [Google Scholar]
- SAINT v 8.40B; Area-Detector Control and Integration Software; Bruker Analytical X-ray Instruments Inc.: Madison, WI, 2018.
- Krause L.; Herbst-Irmer R.; Sheldrick G. M.; Stalke D. Comparison of Silver and Molybdenum Microfocus X-ray Sources for Single-Crystal Structure Determination. J. Appl. Crystallogr. 2015, 48, 3–10. 10.1107/S1600576714022985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M. SHELXS-2013/1, Program for the Solution of Crystal Structures; University of Göttingen: Germany, 2013.
- Sheldrick G. M. Crystal Structure Refinement with SHELXL. Acta Crystallogr., Sect. C: Struct. Chem. 2015, C71, 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrugia L. J. WinGX and ORTEP for Windows: an Update. J. Appl. Crystallogr. 2012, 45, 849–854. 10.1107/S0021889812029111. [DOI] [Google Scholar]
- Giannozzi P.; Baroni S.; Bonini N.; Calandra M.; Car R.; Cavazzoni C.; Ceresoli D.; Chiarotti G. L.; Cococcioni M.; Dabo I.; Dal Corso A.; de Gironcoli S.; Fabris S.; Fratesi G.; Gebauer R.; Gerstmann U.; Gougoussis C.; Kokalj A.; Lazzeri M.; Martin-Samos L.; Marzari N.; Mauri F.; Mazzarello R.; Paolini S.; Pasquarello A.; Paulatto L.; Sbraccia C.; Scandolo S.; Sclauzero G.; Seitsonen A. P.; Smogunov A.; Umari P.; Wentzcovitch R. M. QUANTUM ESPRESSO: A Modular and Open-Source Software Project for Quantum Simulations of Materials. J. Phys.: Condens. Matter 2009, 21, 395502 10.1088/0953-8984/21/39/395502. [DOI] [PubMed] [Google Scholar]
- Vanderbilt D. Soft Self-Consistent Pseudopotentials in a Generalized Eigenvalue Formalism. Phys. Rev. B 1990, 41, 7892–7895. 10.1103/PhysRevB.41.7892. [DOI] [PubMed] [Google Scholar]
- Perdew J. P.; Burke K.; Ernzerhof M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 77, 3865–3868. 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.