Abstract
BACKGROUND:
Endomyocardial biopsy (EMB), the reference surveillance test for acute rejection (AR) in heart transplant (HTx) recipients, is invasive, costly, and shows significant interobserver variability. Recent studies indicate that donor-derived cell-free DNA (dd-cfDNA), obtained non-invasively from blood, is associated with AR and could reduce the frequency of EMB surveillance. The aim of this study was to examine the performance characteristics of a novel test for detecting AR in adult HTx recipients.
METHODS:
Plasma samples with contemporaneous EMBs were obtained from HTx recipients. A clinically available SNP-based massively multiplexed-PCR dd-cfDNA assay was used to measure dd-cfDNA fraction. dd-cfDNA fractions were compared with EMB-defined rejection status and test performance was assessed by constructing ROC curves and calculating accuracy measures.
RESULTS:
A total of 811 samples from 223 patients with dd-cfDNA testing and contemporaneous EMB were eligible for the study. dd-cfDNA fraction was significantly higher in AR (median 0.58%, IQR, 0.13%-1.68%) compared to non-AR (median 0.04%, IQR, 0.01%-0.11%, pc < 0.001). ROC analysis produced an area under the curve (AUC-ROC) of 0.86 (95% CI, 0.77–0.96). Defining samples with dd-cfDNA fraction ≥0.15% as AR yielded 78.5% sensitivity (95% CI, 60.7%–96.3%) and 76.9% specificity (95% CI, 71.1%–82.7%). Positive and negative predictive values were 25.1% (95% CI, 18.8%–31.5%) and 97.3% (95% CI, 95.1%–99.5%) respectively, calculated using the cohort AR prevalence of 9.0% (95% CI, 5.3%–12.8%) with adjustment for repeat samples.
CONCLUSIONS:
This novel dd-cfDNA test detects AR in HTx recipients with good accuracy and holds promise as a noninvasive test for AR in HTx recipients.
Keywords: acute rejection, heart transplant, cell free DNA, endomyocardial biopsy, acute cellular rejection, antibody mediated rejection, graft dysfunction
Heart transplantation (HTx) is the definitive treatment option for patients with advanced heartfailure.1,2 Despite continued advances in post-transplant outcomes, allograft rejection and allograft injury remain impediments to post-transplant survival. Endomyocardial biopsy (EMB) and histopathology, pioneered several decades ago by Caves and Billingham, are still the principal surveillance tools for rejection following heart transplant.3 EMB is regarded as the “gold standard” for the diagnosis of acute rejection (AR), but is invasive and costly.4–8 Further, histopathology reads are prone to interobserver variability.9,10 Although there are internationally accepted grading systems for acute cellular rejection (ACR) and antibody mediated rejection (AMR), recent studies using molecular diagnostic tools have demonstrated a discordance between rejection grades and clinical outcomes; furthermore, EMB does not detect early rejection.11,12 There remains an unmet need for a less invasive and more accurate approach for diagnosing or ruling out allograft rejection after HTx.
Donor-derived cell-free DNA (dd-cfDNA) has been evaluated in solid organ transplantation as a potential marker of donor organ injury and shows promise in HTx.13–17 Cell-free DNA is extracellular DNA found in blood originating from cellular apoptosis and necrosis, and is associated with tissue injury.13,18 Genetic differences between the donor and recipient allow quantification of cell-free DNA originating from the heart allograft, allowing dd-cfDNA to act as a biomarker of allograft injury.13–17 A recent study from the Genomic Research Alliance for Transplantation (GRAfT) found dd-cfDNA was strongly associated with AR with a high area under the receiver operating characteristic curve (AUC-ROC) and a high negative predictive value (NPV). In addition, a rise in dd-cfDNA was often seen before AR was confirmed histopathologically.19 These findings suggest that dd-cfDNA could reduce the need for frequent surveillance EMBs after HTx.
We report on a novel, clinically available test for measuring dd-cfDNA fraction in heart transplant recipients without the need for prior genotyping of the donor or recipient. The platform is run in a CLIA-certified, College of American Pathologists-accredited laboratory, has been analytically validated, and clinically validated in kidney transplant recipients.20,21 The test measures allele sequence read counts at over 13,000 single nucleotide polymorphisms (SNPs) selected to maximize the number of informative SNPs across ethnicities, with a limit of quantification of 0.05% and very low coefficient of variation (CV),20 which is critical as dd-cfDNA fractions in HTx recipients are the lowest of all solid organ transplants.
The purpose of the study was to determine the most appropriate cutoff threshold for AR, and measure performance characteristics of the test for detecting AR after HTx.
Materials and methods
Study design
This was an observational study of HTx patients at the University of Utah Medical Center and University of California, San Diego Health who had EMB and a contemporaneous blood draw for dd-cfDNA testing. Eligible patients were HTx recipients 18 years of age or older with no other solid organ transplant. dd-cfDNA samples were included if the blood draw was performed at least 28 days post-HTx. Retrospective samples from eligible patients collected from July 2017 to August 2020 with matched EMBs were obtained from the University of Utah Medical Center Biobank. Prospective samples were collected from October 2020 to December 2021 at University of Utah Medical Center and from March 2020 to January 2022 at UC San Diego Health. A dd-cfDNA test was considered to be contemporaneous with EMB if the blood sample was collected within the 4 days prior to EMB. Both for cause EMBs (where dd-cfDNA was not used in the decision-making process) and surveillance EMBs were included in the study cohort. Demographic and clinical data were collected for each patient.
All patients provided informed consent. Approval for these studies was provided by the University of Utah Medical Center and the UC San Diego Health Institutional Review Boards (IRB numbers 00094302 and 201821, respectively). This study adheres to the principles of the Declaration of Helsinki formulated by the World Medical Association, the Declaration of Istanbul, and the International Society for Heart and Lung Transplantation statement on Transplant Ethics.
Sample collection
University of Utah Medical Center Biobank plasma samples preserved with EDTA were stored at −80°C. The prospective plasma samples collected at either University of Utah Medical Center or UC San Diego Health, were centrifuged, isolated and frozen or collected in Streck cfDNA BCT tubes and stored at room temperature. Samples were shipped to Natera’s CLIA-certified and College of American Pathologists accredited laboratory (San Carlos, California, USA) for analysis.
dd-cfDNA analysis
Laboratory testing involved cfDNA extraction and library preparation as described previously for retrospective21 and prospective22 samples using the Prospera™ test (Natera Inc., Austin, Texas). This was followed by cfDNA amplification using massively multiplexed-PCR, targeting over 13,000 single nucleotide polymorphisms designed to maximize the number of informative SNPs across ethnicities and next-generation sequencing of the resultant amplicons,21 with sequencing performed on the Illumina Next-Seq500 on rapid run with an average of 14 to 15 million reads per sample. Laboratory technicians involved in processing samples were blinded to biopsy results. For all samples, the donor-derived cfDNA (dd-cfDNA) fraction (analyzed as the percentage of total cfDNA) was measured. A previous study has demonstrated that this test has high reproducibility (coefficient of variation <5%, upper confidence limit <6% for various DNA inputs), and linearity (donor vs. targeted mixture fractions: slope = 1.0813 95% CI, 0.9721 to 1.1906, R2= 0.9995 [95% CI, 0.9994–0.9996]).20
Biopsy defined rejection
The ISHLT revised classification scheme for ACR23 and pathologic diagnosis of AMR24 were used to grade biopsies. AR was defined as AMR ISHLT grade pAMR1 (H+), pAMR1 (I+), pAMR2 and pAMR3, ACR ISHLT grade 2R and grade 3R, or a combination thereof. Non-rejection (non-AR) was defined as AMR ISHLT grade pAMR 0 and ACR ISHLT grade 0 or 1R on biopsy histology.
Statistical analysis
Box plots, showing the interquartile range (IQR) and minimum and maximum whiskers (1st quartile − 1.5 × IQR, 3rd quartile + 1.5 × IQR), were constructed to compare dd-cfDNA fractions in samples grouped by matched EMB rejection status (AR, AMR and ACR subtypes, and no rejection) and the Mann-Whitney U test used to assess the statistical significance between groups. To account for correlations arising from resampling from the same patients, we also assessed the statistical significance amongst the different rejection categories by implementing bootstrapping for each patient.25 For these particular analyses bootstrapping did not substantially alter results, and we report p values from analyses that did not use bootstrapping. We did implement a bootstrapping approach to assess performance of dd-cfDNA to detect AR, and AMR and ACR subtypes. For performance evaluation we performed 10,000 iterations for bootstrapping and each iteration had only one sample from each patient. With 223 unique patients in our dataset, each distribution had 223 samples. ROC curves were constructed, and sensitivity and specificity were calculated for various thresholds. Positive predictive value (PPV), and negative predictive value (NPV) were calculated based on the observed cohort prevalence, after calculating the prevalence using bootstrapping to adjust for repeat sampling.26 Comparison of dd-cfDNA fraction and AR status across study sub-groups was performed using the χ2 test. A sensitivity analysis was used to investigate the impact of for cause biopsies and surveillance biopsies on the sensitivity of the test to detect AR. We also performed a post-hoc analysis using absolute quantification of dd-cfDNA for detection of AR, as opposed to dd-cfDNA fraction, and report on the performance of this test. Statistical analyses were performed using Python 3.8.2. We used the Benjamini-Hochberg procedure for correction whenever multiple comparisons were performed while implementing a particular statistical hypothesis test.27 The corrected p values are designated as pc. For single hypothesis tests we report the p value. p or pc < 0.05 are considered significant.
Results
Characteristics of study population
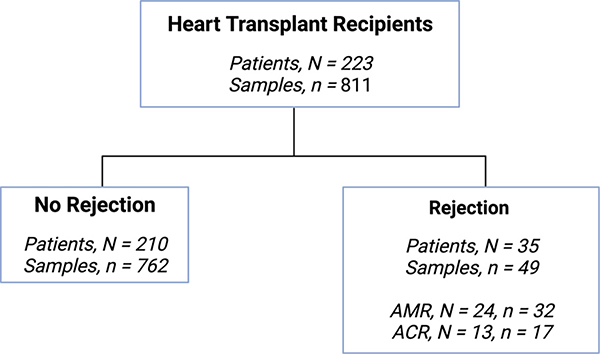
A total of 811 samples from 223 patients with dd-cfDNA testing and contemporaneous EMB were eligible for the study (Figure 1). The median time from transplant to first biopsy was 9.6 weeks (IQR, 5.5–29.1 weeks). Patients were predominantly male (73.1%) and white (54.3%), with a median age of 54.0 years (IQR, 41.0–63.0) and median body mass index of 27.3 kg/m2 (IQR, 23.6–31.7 kg/m2; Table 1). The most commonly reported indications for transplant were non-ischemic cardiomyopathy (65.4%) and ischemic cardiomyopathy (23.3%). Thirty-five percent of patients had durable ventricular assist devices at the time of transplant (Table 1).
Figure 1.
Study flow chart.
Table 1.
Demographic and Clinical. Characteristics of Patients
| Variable | Value |
|---|---|
|
| |
| Number of patients | 223 |
| Gender | |
| Male | 163 (73.1%) |
| Female | 60 (26.9%) |
| Age (y) | 54.0 (IQR,41.0–63.0) |
| BMI (kg/m2) | 27.3 (IQR, 23.6–31.7) |
| Race/ethnicity | |
| Asian | 14 (6.2%) |
| African American | 26 (11.7%) |
| Hispanic | 46 (20.6%) |
| White | 121 (54.3%) |
| Other | 11 (4.9%) |
| Indication for transplant | |
| Non-ischemic cardiomyopathy | 146 (65.4%) |
| Ischemic cardiomyopathy | 52 (23.3%) |
| Congenital | 10 (4.5%) |
| Retransplant | 4 (1.8%) |
| Other/missing | 11 (4.9%) |
| Panel-reactive Antibody (PRA) | |
| Sensitized patients (PRA≥10%) | 25 (11.2%) |
| Unsensitized patients (PRA<10%) | 194 (87.0%) |
| Unknown | 4 (1.8%) |
| VAD use | |
| Yes | 78 (35.0%) |
| No | 145 (65.0%) |
Association of dd-cfDNA with rejection
AR was observed in 49 biopsy matched samples from 35 patients while 762 samples from 210 patients did not show AR. Median dd-cfDNA fraction was significantly higher in samples with a matched biopsy showing AR (median 0.58%, IQR, 0.13%–1.68%) compared to samples where matched biopsies did not show AR (median 0.04%, IQR, 0.01–0.11%, pc < 0.001; Figure 2A).
Figure 2.
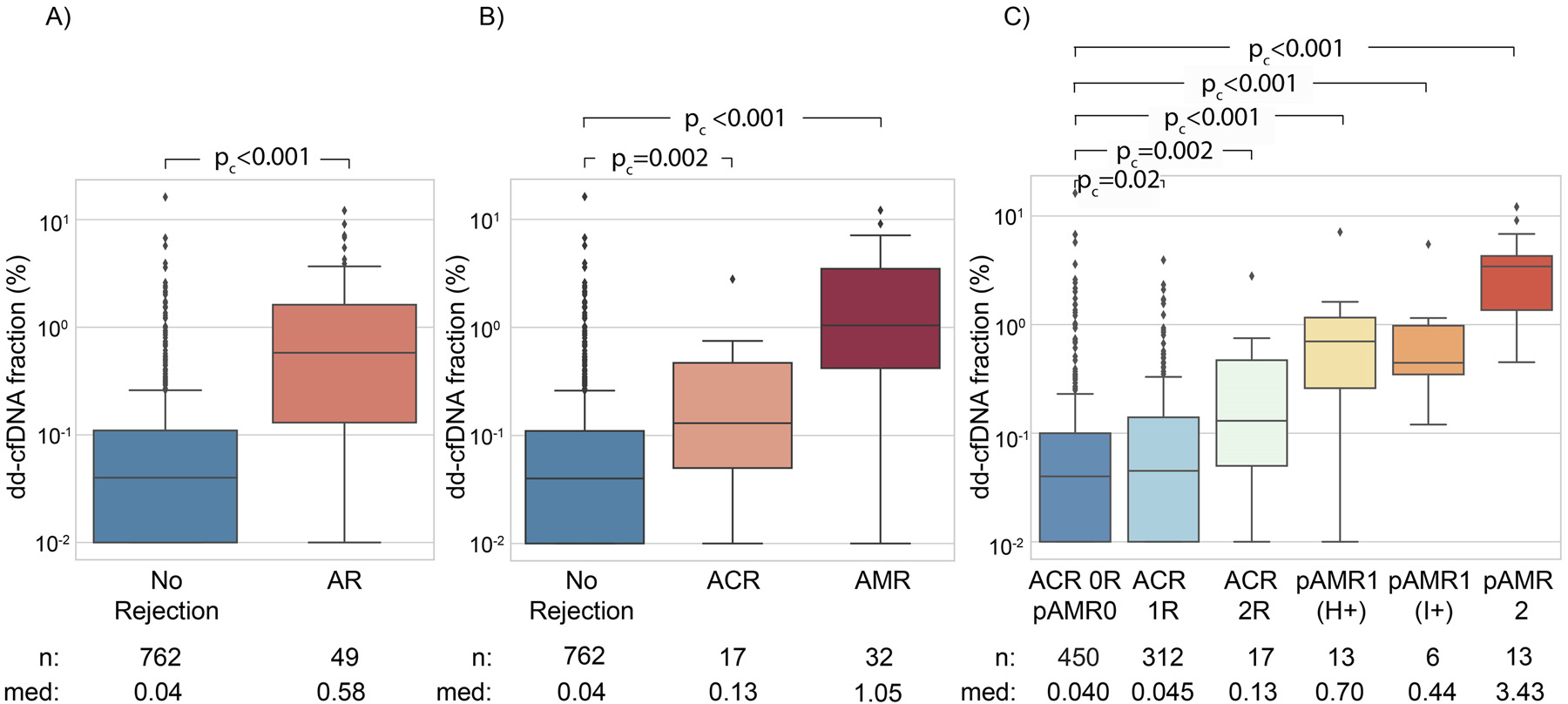
(A) dd-cfDNA fraction (%) stratified by rejection status (AR vs no rejection). (B) dd-cfDNA fraction (%) stratified by ACR and AMR vs. no rejection. (C) dd-cfDNA fraction (%) stratified by ACR and AMR grades vs ACR=0R / pAMR = 0. Boxes represent interquartile ranges, horizontal lines within boxes show medians, whiskers represent minimum and maximum as defined in methods and dots are outliers. Med refers to median dd-cfDNA fraction (%).
We examined whether dd-cfDNA fraction or prevalence of AR varied by sample source (retrospective vs. prospective samples) or biopsy type (surveillance vs. for cause). The median dd-cfDNA fraction in retrospective samples was 0.06% compared to 0.04% in prospective samples (pc = 0.01). The AR proportion in retrospective samples was 12.0% compared to 5.1% in prospective samples (pc = 0.03; Table S1). Median dd-cfDNA fraction and AR proportion were higher in for cause biopsies than in surveillance biopsies (dd-cfDNA: for cause 0.22%, surveillance 0.04%, pc < 0.001; AR proportion: for cause 28.4%, surveillance 3.8%, pc < 0.001; Table S2).
We also stratified false negative (n = 13) and false positive (n = 142) test results by biopsy type. No discernible trend was observed (Table S3). Among false negative results 4 were pAMR1, and 9 were ACR - 2R (Table S3). Of the 49 biopsy-proven ARs, 32 (65.3%) were AMR and 17 (34.7%) were ACR, including 4 biopsies with evidence of mild AMR (8.2%) in addition to the moderate ACR (Figure 1). Consistent with our findings for AR, median dd-cfDNA fraction was significantly higher for samples where matched biopsy showed AMR or ACR relative to samples that did not show rejection (AMR: pc < 0.001; ACR: pc = 0.002; Figure 2B). A more detailed breakdown of rejection subtypes, with comparison to ACR = 0R / pAMR = 0 is provided in Table 2 and Figure 2C. Of the 762 samples considered to be non-AR, 312 had ACR grade 1R histological determination. The median dd-cfDNA fraction in samples from patients with ACR grade 1R was 0.045% (IQR, 0.01%–0.14%), compared to the median dd-cfDNA of those with ACR = 0R / pAMR = 0 of 0.040% (IQR, 0.01%–0.10%, p = .02).
Table 2.
Breakdown of Cases by Rejection Subtype
| Rejection subtype | dd-cfDNA fraction (%) median (IQR) | pc value (comparison with No rejection) |
|---|---|---|
|
| ||
| AMR - pAMR1 (H+) (n = 13) | 0.70 (0.26–1.16) | <0.001 |
| AMR - pAMR1 (i+) (n = 6) | 0.44 (0.34–0.98) | <0.001 |
| AMR - pAMR 2 (n = 13) | 3.43 (1.36–4.28) | <0.001 |
| ACR - 1R (n = 312) | 0.045 (0.01–0.14) | 0.02 |
| ACR - 2R (n = 17) | 0.13 (0.05–0.47) | 0.002 |
| pAMR0 and ACR - 0 (n = 450) | 0.040 (0.01%–0.10%) | NA |
Performance of dd-cfDNA fraction to detect AR
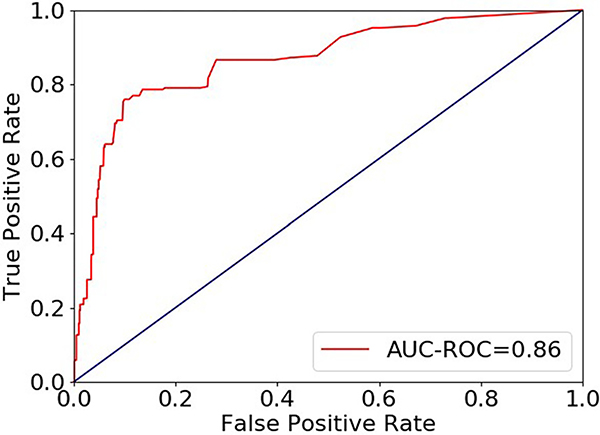
ROC analysis results for discrimination of AR from non-AR by dd-cfDNA testing based on bootstrapped performance estimates are shown in Figure 3—the AUC-ROC was 0.86 (95% CI, 0.77–0.96). Table 3 shows performance characteristics of this dd-cfDNA test to detect AR at different dd-cfDNA fraction thresholds (0.12%, 0.15%, 0.20%).
Figure 3.
ROC curve for discrimination of AR from non-AR by dd-cfDNA fraction (%) for 811 biopsy matched samples.
Table 3.
Performance of dd-cfDNA Fraction in Discriminating AR From Nonrejection for the Study Cohort
| Measure | dd-cfDNA fraction (%) cutoff |
||
|---|---|---|---|
| 0.12 | 0.15 | 0.20 | |
|
| |||
| Sensitivity | 86.59% (71.98%–100%) | 78.52% (60.73%–96.30%) | 78.58% (60.80%–96.35%) |
| Specificity | 72.03% (65.86%–78.20%) | 76.88% (71.08%–82.67%) | 82.12% (76.86%–87.39%) |
| PPV | 23.42% (18.44%– 28.40%) | 25.12% (18.76%–31.47%) | 30.27% (22.43%–38.11%) |
| NPV | 98.19% (96.26%–100%) | 97.31% (95.14%–99.49%) | 97.49% (95.45%–99.53%) |
| AUC-ROC | 0.86 (0.77–0.96) | ||
Brackets show 95% confidence limits.
For example, using a donor fraction threshold of ≥0.15% to indicate AR, our dd-cfDNA test discriminated AR from nonrejection with 78.5% sensitivity (95% CI, 60.7%–96.3%) and 76.9% specificity (95% CI, 71.1%–82.7%). The PPV and NPV were projected to be 25.1% (95% CI, 18.8%–31.5%) and 97.3% (95% CI, 95.1%–99.5%), respectively, using the cohort prevalence of 9.0% (95% CI, 5.3%–12.8%; Table 3). Performance was similar in prospectively (AUC=0.87 (95% CI:0.78–0.96)) and retrospectively (AUC=0.83 (95% CI:0.58–1.00)) collected samples. Furthermore, our sensitivity analysis indicated that biopsy type (for cause vs surveillance) did not substantially impact performance (sensitivity to detect AR: for cause 78.9%, surveillance 76.6%, p = .58).
dd-cfDNA fraction in patients without AR
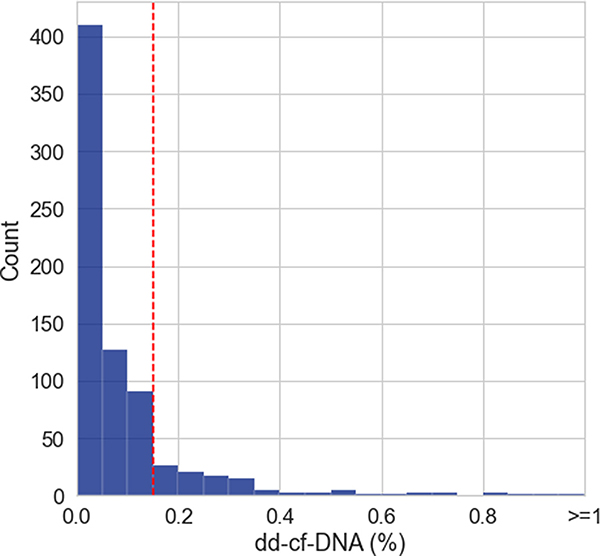
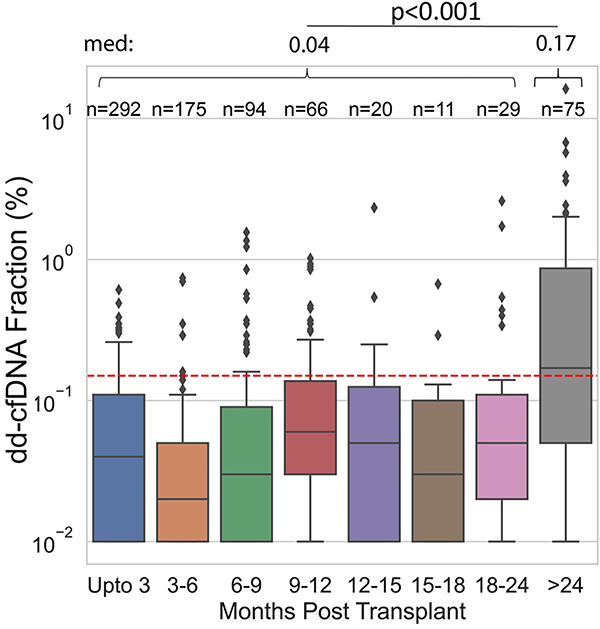
Figure 4A shows the distribution of dd-cfDNA fractions where matched biopsies did not show rejection. The distribution for dd-cfDNA fraction was strongly skewed with dd-cfDNA fraction well below the 0.15% threshold for most samples (median 0.04%, IQR, 0.01%–0.11%) and 18.6% (n = 142/762) of samples falling above the 0.15% threshold. In biopsy-matched non-rejection samples the dd-cfDNA fraction was stable up to 24 months post-transplant, and increased after 24 months. The difference in dd-cfDNA fraction between samples in these two time periods was found to be statistically significant (≤24 months: median 0.04%, IQR, 0.01%–0.10%; >24 months: median 0.17%, IQR, 0.05%–0.87%, p < .001; Figure 4B). We examined whether cardiac allograft vasculopathy (CAV) prevalence among cases ≥24 months post-transplant could in part explain elevated dd-cfDNA in the absence of rejection. We found that, overall, prevalence of CAV was associated with higher dd-cfDNA fraction (p < .001) in the non-rejection group (Figure S1) and was higher at 24 months post-transplant (28.8% (17/59) with CAV) relative to the <24 month time period (5.1% (8/157) with CAV, OR=7.5, p < .001).
Figure 4.
(A) Histogram of dd-cfDNA fraction (%) in nonrejection biopsy matched samples (n = 762). (B) dd-cfDNA fraction (%) in non-AR samples over time. dd-cfDNA fraction >24 months post-transplant was significantly higher than dd-cfDNA fraction for all other time periods combined. Med refers to median dd-cfDNA fraction (%). For both 4A and 4B, the red dashed lines represent the 0.15% dd-cfDNA cutoff.
dd-cfDNA fraction and graft dysfunction
Patients with left ventricular ejection fraction (LVEF) <40% had significantly higher median dd-cfDNA fraction compared to those with LVEF ≥40% (0.46% vs 0.04%, pc < 0.001) (Figure 5A). This difference was not significant among patients with concurrent AR on biopsy, as dd-cfDNA was elevated in AR patients regardless of LVEF (0.94% vs 0.45%, pc = 0.06; Figure 5B). Yet, dd-cfDNA was higher among patients with LVEF <40% compared to those with LVEF ≥40% in the absence of concurrent AR (0.26% vs 0.04%, pc = .003; Figure 5C).
Figure 5.
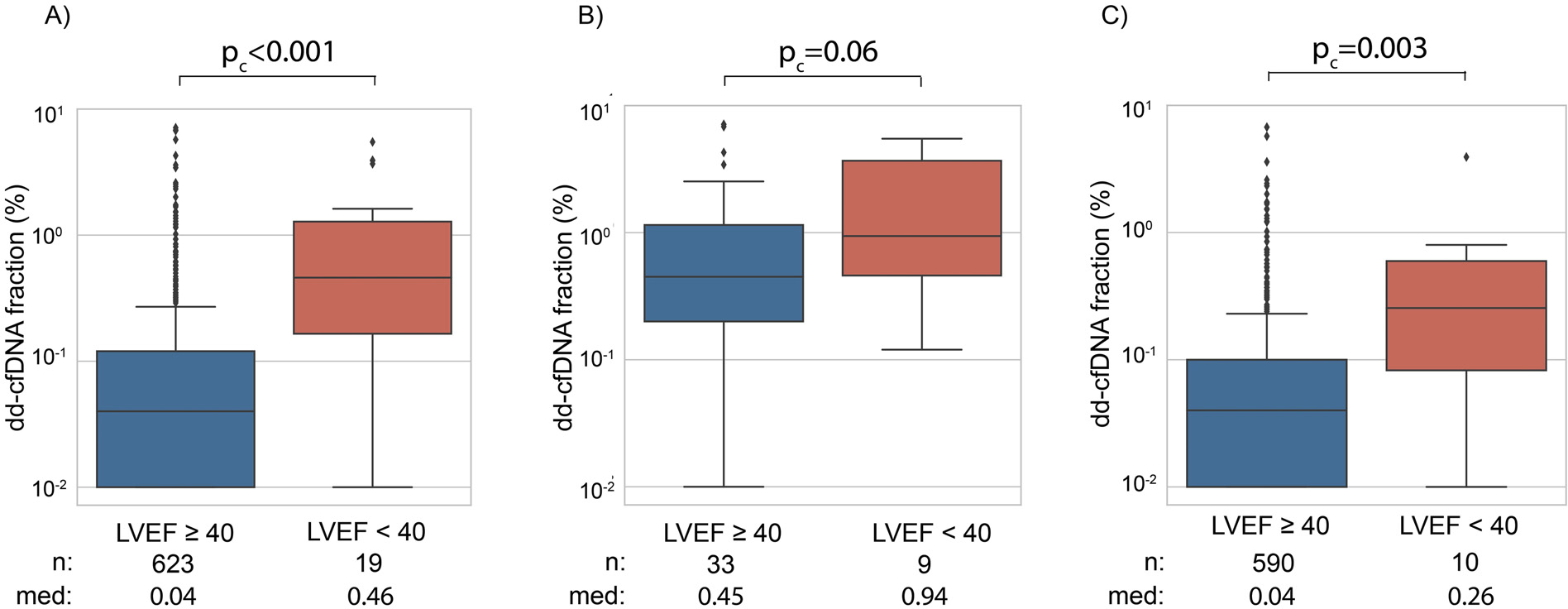
Relationship of dd-cfDNA fraction (%) with LVEF for (A) all samples, (B) AR and (C) non-AR samples. Med refers to median dd-cfDNA fraction (%).
Low dd-cfDNA fraction and stable LVEF in ACR 2R cases
Some ACR 2R cases in patients with stable LVEF (≥60%) had very low dd-cfDNA across serial measurements. For example, Figure S2 shows results for a patient with stable LVEF where dd-cfDNA fraction remains at or below 0.02 for nearly 5 months. During this time period the first and fourth biopsies indicated AR.
Post-hoc analysis of absolute quantity of dd-cfDNA and detection of AR
Since recipient cfDNA concentration can fluctuate, it has been suggested that absolute quantity of dd-cfDNA might be considered as an alternative measure. Using absolute quantity of dd-cfDNA to detect AR resulted in improved performance (AUC-ROC=0.88 95% CI, 0.79–0.97; Table S4, Figure S3) relative to using dd-cfDNA fraction. This included higher sensitivity, specificity, PPV and NPV than dd-cfDNA fraction using an absolute dd-cfDNA quantity cutoffof ≥13 cp/mL, determined by a performance analysis where 13 cp/mL outperformed several other thresholds (Table S4).
Discussion
This study describes the performance of the Prospera dd-cfDNA test in detecting AR in adult heart transplant recipients. Analysis of 811 plasma samples from 223 patients ≥28 days post-transplant showed a significant elevation in dd-cfDNA fraction in patients with AR, defined as either ACR grade >1R (ISHLT 200423) or pAMR grade >0 (ISHLT 201324), compared to patients with no rejection, defined as ACR ≤1R and pAMR 0. In this study population, the AUC-ROC of the dd-cfDNA test to distinguish AR from non-AR was 0.86. Using a dd-cfDNA fraction threshold of 0.15%, dd-cfDNA detected histologically-proven rejection with a sensitivity of 78.5% (95% CI, 60.7%–96.3%) and a specificity of 76.9% (95% CI, 71.1%–82.7%). A PPV of 25.1% (95% CI, 18.8%–31.5%) and NPV of 97.3% (95% CI, 95.1%–99.5%) were calculated for this cohort. These findings corroborate previous studies of dd-cfDNA in heart transplantation, which used a variety of methodologies to quantify donor fraction, affirming the robustness of this biomarker to detect biopsy-proven rejection.14–17,19 This assay has also been shown to accurately detect AR in kidney transplant.21
Median dd-cfDNA was elevated for each rejection type –AMR and ACR– relative to the non-AR group (pAMR1 (H+) = 0.70%, pAMR1 (I+) = 0.44%, pAMR2 = 3.43%, ACR - 2R = 0.13%, non-AR = 0.04%). Consistent with findings by others,19 we find that dd-cfDNA tends to be elevated more in AMR than in ACR, and that levels are higher for pAMR2 compared with pAMR1. Interestingly, dd-cfDNA fraction in pAMR1 (H+) was higher than in pAMR (I+), a finding that is in line with the report by Loupy et al. that rejection-associated intragraft gene expression alteration in some pAMR1 (H+) cases is similar to changes seen in pAMR2/3 cases.28 We also show that median dd-cfDNA fraction in ACR 1R was 0.045% (IQR, 0.01%–0.14%), compared to 0.040% (IQR, 0.01%–0.10%, pc = 0.02) for ACR = 0R / pAMR = 0. While statistically significant, the absolute difference is small. Whether this marginal increase in dd-cfDNA fraction translates to different outcomes when ACR 1R is not treated needs further study.
We also observed a statistically significant difference in median dd-cfDNA between patients with LVEF <40% and ≥40%, (0.46% vs 0.04%, pc < 0.001; Figure 5), indicating that graft injury is detected through dd-cfDNA in patients with allograft dysfunction, even in the absence of biopsy-proven AR. The implications of elevated dd-cfDNA in patients with left ventricular dysfunction in the absence of AR requires further study.
Additionally, we noted that some ACR 2R cases in patients with stable LVEF (≥60%) had very low dd-cfDNA across serial measurements (Figure S2). Whether ACR 2R histology in the setting of low dd-cfDNA may be self-limited even without treatment needs further study. Indeed, anecdotal evidence from the CTOT-05 study demonstrated favorable outcomes in patients with stable graft function in whom ACR 2R diagnosis was assigned by the research core pathologist, but where no treatment for AR was given by the clinical team.29
We also present performance characteristics for this cohort including 3 cutoffs for dd-cfDNA: 0.12%, 0.15%, and 0.20%. As with other diagnostic tools, clinicians will likely adopt a Bayesian approach to dd-cfDNA measurement interpretation and use a cutoff value that best suits the full clinical picture.
Several dd-cfDNA platforms have been tested in heart transplantation and other solid organ transplantation, and results correlated with biopsy-based diagnosis of rejection.13–17 In this context it is important to define the information we expect dd-cfDNA to provide the clinician. While dd-cfDNA does not always correlate with AR, there are possible explanations for seemingly discrepant results. For example, as reported by Agbor-Enoh et al.,13 some patients with a positive dd-cfDNA result and a negative EMB finding do develop AR subsequently, indicating that dd-cfDNA may be an early marker of AR, that is, allograft tissue injury resulting in dd-cfDNA elevation starts before histopathological changes are apparent on EMB. Also, cases where dd-cfDNA is within the normal range while the EMB read indicates AR, could be explained by variability in the histopathology read, or histologic changes suggestive of AR not resulting in clinically significant tissue injury. An example could be seen in our cohort where a patient had 2 episodes of ACR 2R in the setting of stable graft function and very low dd-cfDNA (Figure S2). This hypothesis is supported by gene expression profiling studies, which have also demonstrated discrepancies between histologic grading and molecular expression in the graft, for example, finding that some cases of pAMR1 (I+) were quiescent on the molecular level, or that some pAMR1 (H+) cases equaled pAMR2/3 in gene expression.28
Similar to others,15 we also observed an increase in dd-cfDNA >2 years after transplant, which suggests events other than rejection may influence dd-cfDNA after this time point. dd-cfDNA was previously observed to be associated with CAV30 and we found CAV was associated with higher dd-cfDNA fraction and more frequently observed greater than 2 years after transplant. Increased white blood cell fragility due to long-term tacrolimus use could also be a potential factor.31 Since surveillance biopsies are mostly performed in the first 2 years after transplant, this dd-cfDNA elevation may be of lesser practical importance. Still, assessment of dd-cfDNA kinetics, as performed here, is needed to better understand changes in dd-cfDNA fraction over time, with the potential benefit of optimizing the test for patients whose time since transplant is more than 2 years.
With the development of modern immunosuppression protocols and standardized care which have resulted in relatively low rates of AR and favorable survival, our study adds to the growing body of literature that suggests routine screening by EMB after the first month following transplant should be re-evaluated. dd-cfDNA testing offers the promise of an alternative to screening EMB by non-invasively assessing allograft health and reserving EMB for patients with elevation of dd-cfDNA, allograft dysfunction or other presentations that indicate allograft injury. This complementary approach could compensate for some of the drawbacks of EMB-based surveillance which include its invasiveness, potential for complications, interobserver variability in interpretation, and cost.4–9
While different dd-cfDNA analysis methodologies may be calibrated differently, recent studies that have evaluated dd-cfDNA performance in heart transplantation all raise the same question: could dd-cfDNA testing replace surveillance EMB?13–17 We believe that this question should be tested in a prospective, randomized fashion. The upcoming DETECT trial (NCT05081739) is a multicenter, randomized controlled clinical trial whose aim is to determine whether dd-cfDNA based surveillance starting as early as 4 weeks after heart transplant is noninferior to EMB-based screening for rejection. The follow-up question to be answered is perhaps even more consequential: could treatment decisions be made on the basis of dd-cfDNA? For example, does intensification of immunosuppression in the setting of elevated dd-cfDNA and absence of histologic rejection mitigate future episodes of biopsy-proven rejection, graft injury and/or graft dysfunction, with consequently lower lifetime immunosuppression burden? Or, alternatively, could low dd-cfDNA be used to guide safe weaning of immunosuppression?
Our study has limitations, which include the fact that the study cohort is restricted to two heart transplant programs in the United States. Our prevalence of AR, particularly AMR, was higher than in some other cohorts. Our assessment for AR included immunofluorescence staining for C4d in patients perceived to be at higher risk of AMR– those with DSA, history of AMR or a clinical suspicion of AMR. In one center (University of Utah) immunofluorescence staining for C4d was routinely performed up to 2 months post-transplant. Further, as enrollment in the study was based on a scheduled biopsy, there may have been a selection bias for patients with previous history or suspicion of AMR, who would be more frequently biopsied at our centers. The higher prevalence of AR in this cohort compared with the overall transplant population might have implications for generalizing the findings of this study to patient cohorts with a different risk of rejection. There was also a limited number of sequential samples. We anticipate that the results of the Quantitative Detection of Circulating Donor-Specific DNA in Organ Transplant Recipients (NCT02109575), an NIH-supported observational study that includes approximately 1,000 samples, will provide corresponding complementary information to address these limitations.
Another potential limitation when measuring dd-cfDNA fraction is the effect of recipient-derived cfDNA, which can be influenced by factors such as infection and BMI.32–34 Some have suggested that dd-cfDNA quantity (cp/mL) may be a better marker than dd-cfDNA fraction, as it is independent of changes in background cfDNA.35 A recent study in kidney transplantation incorporated recipient cfDNA levels for detecting rejection, which increased sensitivity, albeit in a small cohort. A “two-threshold” algorithm was employed, which combines a cutoff for dd-cfDNA fraction with a cutoff for absolute quantity of dd-cfDNA.36 In the present study, a post-hoc analysis using dd-cfDNA quantity indicated that incorporation of this measure could increase the sensitivity of the assay. Finally, this study, and indeed most studies assessing novel tests for rejection, is limited by the use of EMB as the comparator. While histology is a commonly accepted gold standard, its variability may lead to an underestimation of performance of the test under investigation. One option would be to follow patients for several months after blood draw and use clinical outcomes data to confirm or refute the accuracy of individual biopsy reads.14 Alternatively, tissue gene expression profiling may also be a more appropriate comparator, as Kobashigawa et al. showed, in a small series of adult heart transplant patients, that concordance between dd-cfDNA and intragraft mRNA transcripts was better than between dd-cfDNA and histology.37
In conclusion, this study affirms an association between elevated levels of dd-cfDNA and histologic evidence of rejection after heart transplant, and extends previous findings showing that dd-cfDNA is a valuable biomarker of allograft health.14–17,19 Prospective controlled studies are needed to confirm the clinical utility of dd-cfDNA assessment in the management of heart transplant recipients.
Supplementary Material
Abbreviations:
- ACR
acute cellular rejection
- AR
acute rejection
- AMR
value
- PPV
positive predictive value
- ROC
Receiver operating characteristic
- SNP
single nucleotide polymorphism
- pc
corrected p-value antibody mediated rejection
- AUC
area under the curve
- AUC-ROC
area under the receiver operating characteristic curve
- EMB
Endomyocardial biopsy
- CLIA
Clinical Laboratory Improvement Act
- CV
coefficient of variation
- dd-cfDNA
donor-derived cell-free DNA
- IQR
Interquartile range
- ISHLT
The International Society for Heart and Lung Transplantation
- LVEF
left ventricular ejection fraction
- NPV
negative predictive
Footnotes
Disclosure Statement
MO, NL, RS, JSternberg, NK, EAhmed, YC, GF, ZPD, and PRB are full time employees at Natera Inc. with stocks or options to own stocks in the company. ZPD and EAhmed report they may have 1 or more patents assigned to Natera, Inc. which may be broadly relevant to this work. AS reports receiving or working at an institution that received payment from Natera Inc and receiving travel expenses from Pfizer Inc.
JStehlik and PJK reports working at an institution that received research payments from Natera Inc. JStehlik reports consulting fees from Natera, Medtronic and Sanofi-Aventis. PJK reports research funding from CareDx and serving on the CareDx Scientific Advisory Board.
This study was funded by Natera Inc and also partially supported by the American Heart Association Grant #16SFRN31890003/Josef Stehlik, American Heart Association Grant #18CDA34110250/Paul J. Kim, MD/2018 and by the Altman Clinical & Translational Research Institute (ACTRI) at UC San Diego Health (PJK). The ACTRI is funded from awards issued by the National Center for Advancing Translational Sciences, NIH KL2TR001444.
The authors acknowledge Dr. Carol Battikha and Nicholas Rodgers from UC San Diego Health and Habeeb Mohammed, Christian Gallagher and Monica Verma from the University of Utah Health for their significant contributions in patient sample and data collection.
Supplementary materials
Supplementary material associated with this article can be found in the online version at https://doi.org/10.1016/j.healun.2022.04.002.
References
- 1.Crespo-Leiro MG, Metra M, Lund LH, et al. Advanced heart failure: a position statement of the Heart Failure Association of the European Society of Cardiology. Eur J Heart Fail 2018;20:1505–35. 10.1002/ejhf.1236. [DOI] [PubMed] [Google Scholar]
- 2.Mehra MR, Canter CE, Hannan MM, et al. The 2016 International Society for Heart Lung Transplantation listing criteria for heart transplantation: A 10-year update. J Heart Lung Transplant 2016;35:1–23. 10.1016/j.healun.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Kittleson MM, Garg S, Solid gold, or liquid gold?: towards a new diagnostic standard for heart transplant rejection. Circulation 2021;143:1198–201. 10.1161/CIRCULATIONAHA.120.052925. [DOI] [PubMed] [Google Scholar]
- 4.Lampert BC, Teuteberg JJ, Shullo MA, Holtz J, Smith KJ. Cost- effectiveness of routine surveillance endomyocardial biopsy after 12 months post-heart transplantation. Circ Heart Fail 2014;7:807–13. 10.1161/CIRCHEARTFAILURE.114.001199. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed T, Goyal A. Endomyocardial biopsy. StatPearls 2021. Available at: https://www.statpearls.com/ArticleLibrary/viewarticle/93193. [PubMed]
- 6.Bennett MK, Gilotra NA, Harrington C, et al. Evaluation of the role of endomyocardial biopsy in 851 patients with unexplained heart failure from 2000–2009. Circ Heart Fail 2013;6:676–84. 10.1161/CIRCHEARTFAILURE.112.000087. [DOI] [PubMed] [Google Scholar]
- 7.Chimenti C, Frustaci A. Contribution and risks of left ventricular endomyocardial biopsy in patients with cardiomyopathies: a retrospective study over a 28-year period. Circulation 2013;128:1531–41. 10.1161/CIRCULATIONAHA.13.001414. [DOI] [PubMed] [Google Scholar]
- 8.Yilmaz A, Kindermann I, Kindermann M, et al. Comparative evaluation of left and right ventricular endomyocardial biopsy: differences in complication rate and diagnostic performance. Circulation 2010;122:900–9. 10.1161/CIRCULATIONAHA.109.924167. [DOI] [PubMed] [Google Scholar]
- 9.Crespo-Leiro MG, Zuckermann A, Bara C, et al. Concordance among pathologists in the second Cardiac Allograft Rejection Gene Expression Observational Study (CARGO II). Transplantation 2012;94:1172–7. 10.1097/TP.0b013e31826el9e2. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham KS, Veinot JP, Butany J. An approach to endomyocardial biopsy interpretation. J Clin Pathol Feb 2006;59(2):121–9. 10.1136/jcp.2005.026443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crespo-Leiro MG, Stypmann J, Schulz U, et al. Clinical usefulness of gene-expression profile to rule out acute rejection after heart transplantation: CARGO II. Eur Heart J 2016;37:2591–601. 10.1093/eurheartj/ehv682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marboe CC, Billingham M, Eisen H, et al. Nodular endocardial infiltrates (Quilty lesions) cause significant variability in diagnosis of ISHLT Grade 2 and 3A rejection in cardiac allograft recipients. J Heart Lung Transplant Jul 2005;24(7 suppl):S219–26. 10.1016/j.healun.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Agbor-Enoh S, Chan JL, Singh A, et al. Circulating cell-free DNA as a biomarker of tissue injury: assessment in a cardiac xenotransplantation model. J Heart Lung Transplant 2018;37:967–75. 10.1016/j.healun.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Vlaminck I, Valantine HA, Snyder TM, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med 2014;6:241–77. 10.1126/scitranslmed.3007803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khush KK, Patel J, Pinney S, et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: A prospective multicenter study. Am J Transplant 2019;19:2889–99. 10.1111/ajt.15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.North PE, Ziegler E, Mahnke DK, et al. Cell-free DNA donor fraction analysis in pediatric and adult heart transplant patients by multiplexed allele- specific quantitative PCR: Validation of a rapid and highly sensitive clinical test for stratification of rejection probability. PLoS One 2020;15:e0227385. 10.1371/journal.pone.0227385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richmond ME, Zangwill SD, Kindel SJ, et al. Donor fraction cell-free DNA and rejection in adult and pediatric heart transplantation. J Heart Lung Transplant 2020;39:454–63. 10.1016/j.healun.2019.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Ungerer V, Bronkhorst AJ, Van den Ackerveken P, Herzog M, Holdenrieder S. Serial profiling of cell-free DNA and nucleosome histone modifications in cell cultures. Sci Rep 2021;11:9460. 10.1038/s41598-021-88866-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agbor-Enoh S, Shah P, Tunc I, et al. Cell-free DNA to detect heart allograft acute rejection. Circulation 2021;143:1184–97. 10.1161/CIRCULATIONAHA.120.049098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altug Y, Liang N, Ram R, et al. Analytical validation of a single-nucleotide polymorphism-based donor-derived cell-free DNA assay for detecting rejection in kidney transplant patients. Transplantation 2019;103:2657–65. 10.1097/TP.0000000000002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sigdel TK, Archila FA, Constantin T, et al. Optimizing detection of kidney transplant injury by assessment of donor-derived cell-free DNA via massively multiplex PCR. J Clin Med 2018;8. 10.3390/jcm8010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017:545:446–51. 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart S, Winters GL, Fishbein MC, et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant 2005;24:1710–20. 10.1016/j.healun.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 24.Berry GJ, Burke MM, Andersen C, et al. The 2013 International Society for Heart and Lung Transplantation Working Formulation for the standardization of nomenclature in the pathologic diagnosis of antibody-mediated rejection in heart transplantation. J Heart Lung Transplant 2013;32:1147–62. 10.1016/j.healun.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Efron B, Tibshirani R. Bootsrap methods for standard errors confidence intervals and other measures of statistical accuracy. Statistical Sci 1986;1:54–77. [Google Scholar]
- 26.Khush KK, Cherikh WS, Chambers DC, et al. The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: Thirty-sixth adult heart transplantation report - 2019; focus theme: Donor and recipient size match. J Heart Lung Transplant 2019;38:1056–66. 10.1016/j.healun.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling for the false discovery rate: a practical and powerful approach to mulitple testing. J R Stat S Series B 1995;57:289–300. [Google Scholar]
- 28.Loupy A, Duong Van Huyen JP, Hidalgo L, et al. Gene expression profiling for the identification and classification of antibody-mediated heart rejection. Circulation 2017;135:917–35. 10.1161/CIRCULATIONAHA.116.022907. [DOI] [PubMed] [Google Scholar]
- 29.Starling RC, Stehlik J, Baran DA, et al. Multicenter analysis of immune biomarkers and heart transplant outcomes: results of the Clinical Trials in Organ Transplantation-05 Study. Am J Transplant 2016;16:121–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holzhauser L, Clerkin KJ, Fujino T, et al. Donor-derived cell-free DNA is associated with cardiac allograft vasculopathy. Clin Transplant 2021;35:e14206. 10.1111/ctr.14206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schutz E, Asendorf T, Beck J, et al. Time-dependent apparent increase in dd-cfDNA percentage in clinically stable patients between one and five years following kidney transplantation. Clin Chem 2020;66:1290–9. 10.1093/clinchem/hvaa175. [DOI] [PubMed] [Google Scholar]
- 32.Vora NL, Johnson KL, Basu S, Catalano PM, Hauguel-De Mouzon S, Bianchi DW. A multifactorial relationship exists between total circulating cell-free DNA levels and maternal BMI. Prenat Diagn 2012;32:912–4. 10.1002/pd.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi J, Zhang Y, Zhang Y, et al. Increased plasma cell-free DNA level during HTNV infection: correlation with disease severity and virus load. Viruses 2014;6:2723–34. 10.3390/v6072723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang N, Zhu L, Zhang Y, et al. Circulating rather than alveolar extracellular deoxyribonucleic acid levels predict outcomes in influenza. J Infect Dis 2020;222:1145–54. 10.1093/infdis/jiaa241. [DOI] [PubMed] [Google Scholar]
- 35.Oellerich M, Shipkova M, Asendorf T, et al. Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. Am J Transplant 2019;19:3087–99. 10.1111/ajt.15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bunnapradist S, Homkrailas P, Ahmed E, Fehringer G, Billings P, Tabriziani H. Using both the fraction and quantity of donor-derived cell-free DNA to detect kidney allograft rejection. J Am Soc Nephrol 2021. 10.1681/ASN.2021050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobashigawa JA, Patel J, Kittleson M, et al. Donor-derived cell free DNA correlates more closely with intragraft mRNA transcripts rather than pathology read biopsies. J Heart Lung Transplant 2018;37:S198. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.