Abstract
In this paper we focus on the detection of differentially expressed genes according to changes in hybridization signals using statistical tests. These tests were applied to 14 208 zebrafish cDNA clones that were immobilized on a nylon support and hybridized with radioactively labeled target mRNA from wild-type and lithium-treated zebrafish embryos. The methods were evaluated with respect to 16 control clones that correspond to eight different genes which are known to be involved in dorso-ventral axis specification. Moreover, 4608 Arabidopsis thaliana clones on the same array were used to judge statistical significance of expression changes and to control the false positive rates of the test decisions. Utilizing this special array design we show that differential expression of a high proportion of cDNA clones (15/16) and the respective genes (7/8) were identified, with a false positive error of <5% using the constant control data. Furthermore, we investigated the influence of the number of repetitions of experiments on the accuracy of the procedures with experimental and simulated data. Our results suggest that the detection of differential expression with repeated hybridization experiments is an accurate and sensitive way of identifying even small expression changes (1:1.5) of a large number of genes in parallel.
INTRODUCTION
An important application of cDNA clone arrays is the identification of genes that show significant changes when their hybridization signals are compared using target mRNA from different tissues or different physiological states. Parallel detection of differential expression of the cDNA clones on the array offers the chance to identify a large number of possible markers, for example for a certain disease, and allows inferences as to biological pathways and gene function that are fundamental to scientific and industrial research. Hybridization-based studies of differential expression are manifold and vary with respect to the array support and labeling of the target mRNA pool: a widely used system is immobilization of PCR products on glass slides and hybridization with two-color fluorescently labeled mRNA (1–3). Other protocols utilize immobilization of a large number of short oligonucleotides on glass slides hybridized with fluorescently labeled mRNA (4,5). Nylon filter membranes and glass slides combined with radioactively labeled mRNA have been applied (6,7), as well as nylon microarrays combined with colorimetric detection (8). It has been shown that the sensitivity of all these methods, i.e. the amount of sample necessary to detect a given mRNA, is fairly similar (9). Furthermore, a number of publications treat the methodological aspects of these techniques (10–16).
The use of statistical tests is common in studies of differential expression via tag sampling experiments incorporating Fisher’s exact test and alternatives (17). In contrast, in hybridization-based experiments statistical tests are rarely used because of the lack of repetitions; in most studies changes of expression are judged via thresholding and experiments are repeated at most two or three times. However, hybridization experiments are typically noisy and therefore their proper evaluation is essential. An important aspect is the number of times hybridization should be repeated in order to allow sufficient reproducibility of the signals and thus distinction between true expression changes and errors in hybridization and data capture. Repetitions allow statistical modeling of the experiment within a well-prepared framework: the two-sample location test problem (18). Studies that incorporate tests use a regularized version of Student’s t-test and adjustment of the corresponding P values in order to determine true expression changes (19,20).
In this paper we present a comparison of four different tests (see Materials and Methods) with respect to their performance in detecting significant changes in hybridization levels.
Procedures were applied to PCR products of 14 208 zebrafish cDNA clones selected from a representative cDNA library from zebrafish gastrula stage embryos (5–6 h post-fertilization); this library was pre-screened using the oligonucleotide fingerprinting technique (21,22). The cDNA clones were immobilized on a nylon filter membrane and hybridized with target mRNA from wild-type and lithium-treated gastrula stage embryos. Lithium is a well-known hyperdorsalizing agent (23), which exerts its dorsalizing effect by mimicking the endogenous maternal dorsalizing signal. Sixteen well-defined cDNA clones corresponding to eight different genes were previously identified as being influenced by lithium treatment and served as control clones for testing the performance of the methods. We show that up to 15 of these 16 clones (seven of eight genes) were detected, with false positive rates of <5% as judged by the constant control data.
Furthermore, we show how the number of repetitions of experiments influences the accuracy of the methods with experimental data and with a simulation based on the Welch test. We show that repetition of hybridizations compensates for measurement errors to a high degree even when expression ratios are small (1:1.5). Our results suggest that repeated hybridizations followed by statistical testing are an accurate and sensitive method to detect large numbers of reliable expression changes in parallel.
MATERIALS AND METHODS
Biological background
The zebrafish has emerged in recent years as a major model organism for vertebrate development and human disease. In particular, the combination of externally developing, optically clear embryos with the possibility of forward genetics makes the zebrafish a good model for studying vertebrate gastrulation.
During gastrulation, induction and patterning, together with extensive cell movement, results in an embryo with the typical vertebrate body plan, where the three main body axes and the three germ layers are specified and patterned. To identify genes involved in the specification of the dorso-ventral axis we have compared gene expression data for lithium-treated gastrula stage embryos with that for normal embryos. Lithium is a well-known hyperdorsalizing agent (23), which exerts its dorsalizing effect by interfering with two pathways: it mimics an early dorsalizing Wnt signal by inhibiting glycogen synthase kinase 3β (24,25) and it inhibits a ventralizing phosphoinositide signaling pathway (26–28). Dorso-ventral axis specification is relatively well characterized and several of the genes and pathways involved have been described. As controls we have chosen 16 cDNA clones that correspond to eight different genes known to be expressed either in the dorsal organizer at the shield stage or known to be involved in the Wnt signaling pathway at this stage.
cDNA clone array design
PCR products of 14 208 zebrafish cDNA clones and 4608 copies of a cDNA clone from Arabidopsis thaliana were immobilized on 22 × 22 cm2 nylon filter membranes. All clones were spotted twice on the filter to improve reproducibility. 17 664 spot positions were kept empty and were used for the purpose of data normalization. The zebrafish clones were derived from a representative cDNA library from gastrula stage (5–6 h post-fertilization) embryos comprising an initial 55 000 cDNA clones. This library was normalized by the oligonucleotide fingerprinting technique in order to produce a low redundancy cDNA set that represents most genes activated in the tissue (29). Clones were spotted on a rectangular grid of blocks on the filter, each block containing 25 spots; clones were spotted in duplicates so that in each single block 12 different spots were present comprising six or seven zebrafish clones, two Arabidopsis clones, three or four empty spot positions on average and one empty position in the middle of each block. Raw data are accessible via the web site (http://www.molgen.mpg.de/~lh_bioinf/projects/statistics/zebrafish/zebra fish.html).
mRNA labeling and hybridization
For the lithium-treated and normal embryos 3 × 1 µg poly(A)+ RNA was labeled with [α-33P]dCTP as described previously (7). Aliquots of 60 ng A.thaliana DNA template were likewise labeled in a random priming labeling reaction (7) and 1/6 of the reaction added to each of the labeled zebrafish target mRNAs. The target mRNAs were denatured by addition of NaOH to a final concentration of 1.25 M and diluted in 10 ml of hybridization solution (modified Church buffer, 0.25 M Na2HPO4, pH 7.2, 5% SDS, 1 mM EDTA). Membranes were pre-hybridized in modified Church buffer for a minimum of 30 min at 65°C and then hybridized at 65°C for 16 h. The membranes were then washed twice in 40 mM Na2HPO4, pH 7.2, with 0.1% SDS at room temperature and twice in the same buffer at 65°C.
In situ hybridization
RNA in situ hybridization using wild-type and lithium-treated embryos was performed as described previously (30) in order to verify significantly differentially expressed clones.
Data capture and normalization
Six independent hybridizations were carried out with both the lithium-treated and wild-type target mRNAs. After hybridization the filter membranes were exposed to phosphor storage screens for 14–16 h at room temperature. The screens were scanned at a resolution of 100 µm using a Fuji BAS 5000 phosphor scanner and stored in Fuji BAS format. Image analysis was performed using VisualGrid image analysis software (GPC Biotech, Munich, Germany). The range of the resulting spot intensities was 0–400 (arbitrary units). Normalization of raw intensity values was done within each filter to eliminate the influence of factors not due to the probe–target interaction, such as labeling efficiency, exposure time to the phosphor storage screens, the scanning process, array quality, etc. Normalization was performed by subtraction of the mean of all intensity values derived from empty spot positions within the same block (local background) and by division by a filter-specific spot intensity (median type).
Calculating P values
Four different statistical tests were compared: Student’s t-test, the Welch test, Wilcoxon’s rank sum test and a version of Pitman’s permutation test. Student’s t-test assumes that the control and treatment signal series are normally distributed with the same variance, which restricts its application to a very special case. If these assumptions are fulfilled, however, this test procedure has the highest power of all the tests available. If the assumption of equal variances is not valid, the t-test can be approximated by the Welch test (31,32). It is still assumed that both series are normally distributed. P values for Student’s t-test and the Welch test were calculated according to the ‘Numerical Recipes in C’ functions ttest and tutest (33), respectively.
A well-known distribution-free alternative to the t-test is the unpaired Wilcoxon rank sum test. Here, the test statistic was calculated according to the ranks of the signals derived from the treatment sample within the combined sample of treatment and control signals. For calculation of the exact values of this test statistic we implemented a recursive function (18). The Wilcoxon test is more conservative than the t-test, i.e. it will detect a lower number of significant regulations, but if the assumption of a normal distribution is not applicable, it has higher efficiency. A less well-known distribution-free test procedure is the permutation test originally introduced by Pitman (34). In this approach the absolute difference of the means of the treatment and the control sample is used as the test statistic. Under the hypothesis of no regulation, any random division of the combined sample into a ‘treatment sample’ and a ‘control sample’ of respective sizes will lead to small values of the test statistic, whereas high values indicate differences in the series. P values for the permutation test were calculated by counting all combinations of such a random division that lead to an equal or higher value of the test statistic than the value observed divided by the number of all possible combinations.
RESULTS
Simulations of the influence of sample size and fold change
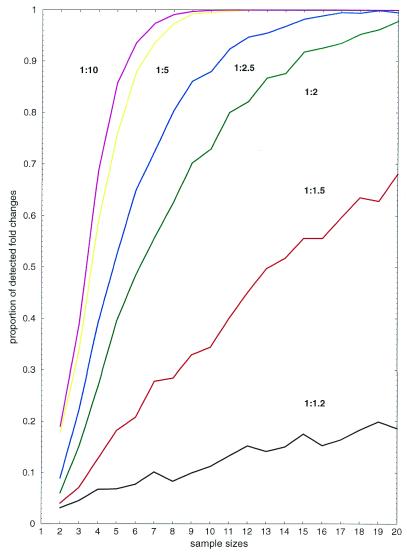
We simulated the influence of sample size and fold change on the performance of the Welch test and investigated how measurement error can be compensated for by an increase in the number of repetitions of the hybridization experiment (Fig. 1). Normal samples were derived by the Box–Muller method (35) with mean values corresponding to the intended level of expression change. For example, if a 1:1.5 expression change was simulated, then the control series was sampled from a normal distribution with mean value µ = 1 and the treatment series was sampled from a normal distribution with mean µ = 1.5. The standard deviation of both series was set to σ = 0.5 µ, since experimental observations indicate that this is a realistic value for measurement error. We repeated sampling and testing 1000 times for each pair of parameters, and counted the number of times an expression change was detected by the test with P < 0.05. For example, in 15.1% of the cases the test detected a significant variation factor of 1:2 (green line) if sample sizes were 3. This number was three times higher if sample sizes were increased to 6 (48.4%). If sample sizes were 15 then 91.8% of the expression changes were detected. The results give an upper boundary for the power of the Welch test, since the theoretical assumptions were completely satisfied and we did not take into account any false positive error. Figure 1 thus suggests that only a small number of relevant regulations (<50% up to the 10-fold level) will be detected when repeating the experiment only two or three times and that at least five or six replicated experiments should be done.
Figure 1.
Simulation results. True positive rates of the Welch test (y-axis) and dependence on sample sizes (x-axis). Curves show different levels of simulated fold changes: 1:1.2 (black), 1:1.5 (red), 1:2 (green), 1:2.5 (blue), 1:5 (yellow) and 1:10 (magenta).
Quality assessment of hybridization signals
Repetitions of hybridization experiments enabled us to analyze the reproducibility of the individual target–probe interactions before and after data normalization and thus to give an overall view of the performance of data normalization. For each clone we calculated the coefficient of variation (CV) as a reproducibility factor, i.e. the ratio of the standard deviation and the mean derived from the six replicated hybridization experiments. If reproducibility is perfect the CV is 0, if it is poor the CV tends to higher values. If the CV is close to 1, i.e. measurement deviation is on average of the order of the observed signal, no meaningful analysis is possible.
Figure 2 shows the global effect of normalization. The upper histograms show the CV values derived from hybridization signals with the target mRNA from wild-type embryos using raw (Fig. 2A) and normalized data (Fig. 2B). It is observable that there is a shift of the mode of the histograms to the left, from 0.28 to 0.21. In 10 741 of 14 208 cases (75.60%) the CV was decreased by normalization, whereas in 3467 cases (24.40%) it was increased.
Figure 2.
Global effects of data normalization. CV of 14 208 zebrafish clones according to six replicated experiments with raw data (A) and normalized data (B). (C) A scatter plot of two independent wild-type experiments with raw data. The line shows the regression line. (D) A scatter plot for the same experiments when normalized data were used. (E) Plot of the mean duplicate differences of the six replications for the wild-type hybridizations before (solid line) and after normalization (histogram). (F) Selection criterion for judging cDNA clone absence within one experiment. The left histogram shows the distribution of 17 664 log signals measured on empty spots, the right histogram shows the distribution of 14 208 zebrafish log signals. The line indicates the 95% upper quantile of the left histogram.
Another way of assessing the quality of normalization is a scatter plot of the intensity signals of different independent experiments before and after normalization. Figure 2 shows scatter plots of two replicated hybridization experiments with the wild-type target mRNA before (Fig. 2C) and after normalization (Fig. 2D). Normalization improved the linear dependency of the hybridization signals and thus the robustness between replications of the experiment. The lines show the regression lines of the respective linear fits. For the raw data the slope (a) and the intercept (b) of the regression lines were a = 0.8969 and b = 0.6254 and for the normalized data a = 0.9906 and b = 0.0473 (note that ideally we have a = 1 and b = 0). Additionally, the histogram (Fig. 2E) shows the mean difference of the duplicate pairs over all six experiments for the 14 208 clones when using normalized data. The line shows the shape of the histogram when using raw data. It is observable that normalization decreased duplicate differences by a significant amount. Furthermore, a shift effect for raw data (probably due to differing material transfer) is observable, which tends to assign one duplicate a higher intensity on average than the other, so that the duplicate differences were not symmetrical around zero but around a slightly negative value. This effect is also eliminated by data normalization.
Sensitivity of the clone array
An important consideration in array experiments is obtaining a sufficient number of positive signals. In our study we used a tissue-specific array that represented the majority of the genes in the target mRNA under analysis (see Materials and Methods). The numerical criterion for judging cDNA clone presence was based on the empty positions. Within each experiment we determined the 95% quantile of the sample of empty spots (total 17 664 signals) and marked each zebrafish clone signal as ‘positive’ for that experiment if it was above this threshold and ‘negative’ if it was below (Fig. 2F). If more than half of the replicated signals of the clone were marked positive we tagged this clone as ‘present’, otherwise we tagged it as ‘absent’. Exploring all replications we found that 13 061 of 14 208 (91.93%) of the clones on the array were present within the wild-type and the lithium-treated target mRNA pools, clearly an effect of the preselection of cDNAs.
Error analysis and adjustment of P values using control data
An important issue in testing is the false positive rate. A fixed error level would only be valid if the distributional assumptions were valid (see Materials and Methods). In practice, however, these assumptions are hardly ever given, so that any calculated P value must be adjusted. Here, we used 4608 copies of an A.thaliana cDNA clone that were spotted on each filter membrane to control the false positive rates of the various tests and also to judge the significance of the calculated P values. Since the amount of Arabidopsis target was spiked at a constant level in the target mRNA, values obtained from these data represent the test statistics given the null hypothesis of no differential expression. To judge whether a given P value indicated a significant change or not we applied the following procedure:
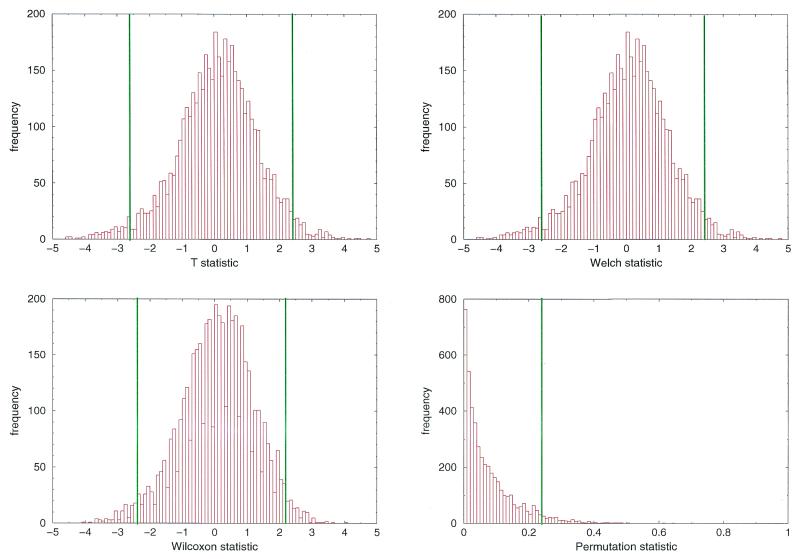
(i) For each test statistic we calculated the values for the 4608 Arabidopsis data (Fig. 3).
Figure 3.
Baseline distributions of the test statistics under the null hypothesis. Histograms of the t-statistic (upper left), the Welch statistic (upper right), the Wilcoxon statistic (lower left) and the permutation statistic (lower right) when using 4608 Arabidopsis samples that served as control data (null hypothesis). The lines indicate the lower and upper 2.5% proportion of the sample, the area between the lines thus indicates the 95% area of the samples. In the case of the permutation statistic the line indicates the upper 5% area of the sample.
(ii) Let α1,…, αN be the corresponding P values for the inner 95% range of the Arabidopsis data. The adjusted P value αad = min{α1,…, αN} was defined as the minimal P value of the inner 95% range of these Arabidopsis data.
(iii) Each zebrafish cDNA clone with a P value < αad was marked ‘significantly differentially expressed’.
With this procedure we forced the false positive rate of our decision process to be <5% for the experimental data, which is far more accurate than simply setting the significance value of the tests to 0.05. Adjusted P values at that level decreased considerably: they were 0.0156 for the t-test statistic, 0.0161 for the Welch test statistic, 0.0173 for the Wilcoxon test statistic and 0.0121 for the permutation test statistic.
Comparison of tests and influence of sample size
Table 1 shows the results for the procedure with dependency on sample size for the different tests. In each table entry we give the number of significantly differentially expressed control clones (maximum 16) and the total number of differentially expressed zebrafish clones (maximum 14 208). Since the control clones should be differentially expressed, the proportion of detected significant fold changes indicates the power of the tests. Results show that there is a considerable increase in detected fold changes when sample size was increased from 6 to 12 (in the cases of sample sizes >6 we incorporated the duplicate signals for each clone on the same array). The true positive rate of the 16 control clones improved from 8 (sample size 6) to 15 (sample size 12) when, for example, using Student’s t-test. The distribution-free tests seemed to perform better, especially when sample sizes were small; however, this difference disappeared when the number of repetitions increased.
Table 1. Dependency of detection rates on sample size for 16 control cDNA clones and the total proportion of differentially expressed zebrafish clones.
| Sample size | Student’s t-test | Welch test | Wilcoxon test | Permutation test | Intersection of tests |
|---|---|---|---|---|---|
| Significance was judged by adjusted P values. | |||||
| 12 | 1026 (7.22%) | 1002 (7.05%) | 962 (6.77%) | 1115 (7.85%) | 878 (6.16%) |
| 15 | 15 | 14 | 15 | 14 | |
| 10 | 789 (5.55%) | 751 (5.29%) | 776 (5.46%) | 841 (5.92%) | 656 (4.62%) |
| 13 | 13 | 13 | 13 | 13 | |
| 8 | 670 (4.72%) | 641 (4.51%) | 696 (4.90%) | 745 (5.24%) | 563 (3.96%) |
| 11 | 11 | 11 | 11 | 11 | |
| 6 | 341 (2.40%) | 283 (1.99%) | 324 (2.28%) | 324 (2.28%) | 227 (1.60%) |
| 8 | 7 | 9 | 9 | 7 |
It should be pointed out that sample signals cannot in general be considered as independent signals when derived from replicate clones on the same array (see Discussion), so that the upper three cases in Table 2 (sample sizes 8, 10 and 12) cannot be straightforwardly compared to the case where only the six independent signals from replicate experiments were used. However, using all signal samples increased the performance by a considerable amount.
Table 2. Results for the control clones: expression ratios, reproducibility factors and detection rates.
| Gene (GenBank identifier) | Ratio (treated versus controls) | Coefficient of variation | No. of cDNA clones | Identified by t-test | |
|---|---|---|---|---|---|
| |
|
Treatment |
Control |
|
|
| chordin (AF034606) | 2.22 | 0.33 | 0.35 | 6 | 6 |
| forkhead-4 (AF052247) | 2.09 | 0.39 | 0.35 | 2 | 2 |
| frizzled-8b (AF060696) | 2.08 | 0.34 | 0.40 | 1 | 1 |
| forkhead-2 (AF052245) | 2.01 | 0.38 | 0.49 | 2 | 2 |
| lim-1 (L37802) | 1.92 | 0.40 | 0.41 | 1 | 1 |
| one-eyed pinhead (AF041440) | 1.47 | 0.21 | 0.32 | 2 | 2 |
| otx-3 (D26174) | 0.69 (–1.45) | 0.22 | 0.34 | 1 | 1 |
| floating head (L48017) | 1.16 | 0.28 | 0.66 | 1 | 0 |
Of the clones, 878 (6.18%) had a significant fold change based on all tests using all available data. These were the candidates for further characterization by in situ hybridization.
Comparison of P values and expression ratios
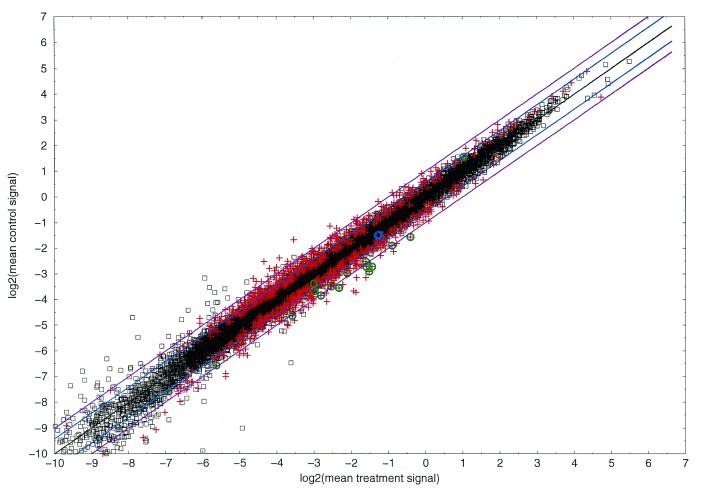
Figure 4 shows a plot of the mean wild-type signals versus the mean lithium signals (log–log scale) for the normalized values; significant (plus signs) and non-significant clones (squares) as judged by P value are marked. The results show that most significant clones had a ratio within the 2–1.5-fold range and indicate that simple detection of fold changes for these clones would have failed in this study (see Discussion). On the other hand, checks on control data by in situ hybridization indicated significant regulation, although the ratio did not exceed the 2-fold up or down threshold, so that these detected regulations are truly given (see below). Thus, Figure 4 implies that P values are far more sensitive and reliable than expression ratios.
Figure 4.
P values versus expression ratios. Log–log plot (base 2) of the mean normalized signal values of the treatment (x-axis) and the control samples (y-axis). Each square denotes a data point that showed no significant regulation judged by the adapted P value of Student’s t-test; each plus denotes a data point that showed significant regulation. Zebrafish control data are marked with a green circle when they were truly detected as significant (total 15); one data point was not detected and is marked by a blue circle. The violet and blue lines indicate the 2- and 1.5-fold up- and down-regulated thresholds, the black line denotes the identity.
Verifying test results by in situ hybridization
Lithium treatment at the early blastula stage causes hyperdorsalization of the embryo, converting ventro-lateral cells to a dorsal fate, with a concomitant expansion of expression domains of dorsal marker genes. Whole mount in situ hybridization can thus be used to verify the test results for genes that have a localized expression pattern. Figure 5 shows in situ hybridization images for three control clones. It is observable that the expression of all three genes was clearly amplified when comparing the lithium-treated (right) with the wild-type (left) embryos. Table 2 shows a summary. The results for the redundant control clones were consistent. For example, chordin was spotted six times on the array while forkhead-2, forkhead-4 and one-eyed pinhead were spotted twice. All these clones showed significant changes in expression according to all four tests. The ratio of the mean signals indicated that even small expression changes, such as a 1.47-fold up-regulation in the case of the one-eyed pinhead clones and a 1.45-fold down-regulation in the case of the otx-3 clone, were verified. Furthermore, of 15 clones (apart from the control clones) that have been tested as being differentially expressed all showed clear regulation when judged by in situ hybridization. One false negative clone (floating head) showed no significant regulation. This might be due to poor data reproducibility since the wild-type signals had a CV of 0.66, i.e. the standard deviation was 66% of the observed mean signal across the six replications.
Figure 5.
In situ images of zebrafish control clones. forkhead-4 (top), forkhead-2 (middle) and chordin (bottom) showed significant variation of expression when comparing the wild-type pictures (left) and the lithium treatment pictures (right).
Dependency of reliability on signal range
An important issue is the reliability of the observed signal with respect to the signal range. For example, the independence of the variation from the absolute signal value is a theoretical prerequisite for applying statistics based on ‘normality’ assumptions. In Table 3 we summarize the results of our experimental observations. For example, 5213 (36.69%) of the zebrafish clones had mean control signals between 10- and 100-fold above background level, the average standard deviation across the six replicated experiments with the wild-type target mRNA was 1.076139 and this average standard deviation was reduced to 0.096985 when normalized data were used. Additionally, we counted the number of significantly differentially expressed clones for the respective signal range (total 878, cf. Table 1). In the 10- to 100-fold bin 387 significant clones (44.08%) were detected. It is observable that: (i) signal variation increases with signal range; (ii) signal variation is almost one order of magnitude lower for normalized data; (iii) significant clones are spread proportionally across the bins so that the selection criteria for choosing statistical significance of differential expression is robust across the signal range.
Table 3. Dependency of signal standard deviation on signal range.
| Signal range | Total signals | σcr | σcn | σtr | σtn | Significant clones |
|---|---|---|---|---|---|---|
| The signal range describes the ratio of the average control signal and the average background signal across six replications. Data is shown for treatment and control samples as well as for raw and normalized data (σcr, average standard deviation of control sample with raw data; σcn, control sample, normalized data; σtr, treatment sample, raw data; σtn, treatment sample, normalized data). | ||||||
| ≤1 | 136 | 0.025448 | 0.001828 | 0.016401 | 0.001239 | 0 |
| 1–10 | 7768 | 0.170669 | 0.013216 | 0.148963 | 0.013897 | 436 |
| 10–100 | 5213 | 1.076139 | 0.096985 | 1.064368 | 0.104894 | 387 |
| 100–1000 | 1070 | 8.840333 | 0.868110 | 9.5821168 | 0.979571 | 51 |
| >1000 | 21 | 48.037467 | 5.421007 | 50.066056 | 5.078543 | 4 |
DISCUSSION AND CONCLUSION
We have shown that standard statistical test procedures can be used efficiently in order to detect a large number of differentially expressed genes on cDNA clone arrays. The calculation of exact P values is far more accurate than simple binary classification via thresholding of expression ratios for treated and control probes, which seems somewhat heuristic (for a discussion see 11). It should be pointed out that the actual expression ratio is only a rough indicator of the strength of the expression changes. Especially in cases where high biological variation is present, as in our case, since we worked with target mRNA derived from a pool of embryos, the expression ratio will not distinguish significant from non-significant clones as sharply as necessary.
P values for both distribution-free test procedures (i.e. the Wilcoxon and permutation tests) cannot be calculated exactly when sample sizes are large because the number of possible combinations for the test statistic increases rapidly with increasing sample size (e.g. 924 for a sample size of 6, 2 704 156 for a sample size of 12, and 155 117 520 for a sample size of 15); thus, P values should be approximated by asymptotic distributions of the test statistics. The approximation for the Wilcoxon rank sum test statistic using the standard normal distribution is, for example, known to be accurate if the sizes of the individual samples are greater than four and if the combined sample size is more than 20. However, in most gene expression studies the sample sizes are smaller and it would be necessary to perform exact calculations.
The origin of the cDNA clones on the array is important. The advantage of using tissue-specific arrays lies in the fact that they ensure a large number of true positive signals and thus allow extraction of meaningful information.
The method of normalization of experimental data was optimized to our special application, but in general our results indicate that gene array data should be normalized ‘locally’ rather than ‘globally’, since variations in the array area are locally influenced (pin effects, local contamination, differences in the spot environments, masking by neighboring bright or large spots).
Our results reveal how an increasing number of repetitions improves the performance of the procedure. Practically, repetitions can be achieved either by repeating the hybridization experiment or by spotting clones multiple times on the array. In theory, test procedures require independent repetitions of experiments so that the number of hybridizations should be increased rather than the redundancy on the array, since spots on the same array correlate substantially. It was shown by simulations as well as with experimental data that six signal values should be available for both the treated and the control samples. In practice, however, it is far more efficient to increase redundancy on the array rather than to repeat hybridization experiments a large number of times. For example, it is conceivable that in the near future there will exist clone arrays with only a few (<500) disease- or pathway-specific genes that can then be spotted at high redundancy, at say 20 or more times. This would enhance the sensitivity of the procedure to a high degree and would allow the identification of very small fold changes, for example in the early stages of a disease, which would be extremely useful in medical research.
Acknowledgments
ACKNOWLEDGEMENTS
The authors want to thank E. Brundke for technical assistance. Discussions with Arif Malik on the topic of evaluation of gene expression data related to this work were a great help. Mario Drungowski and Wasco Wruck helped with computational assistance. Patrizia Ruiz and Wilfried Nietfeld made valuable remarks on the manuscript. Marcus Albrecht helped set up the web site. The work was financed by the Max-Planck Society.
REFERENCES
- 1.Schena M., Shalon,D., Heller,R., Chai,A., Brown,P. and Davis,R. (1996) Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl Acad. Sci. USA, 93, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeRisi J., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 3.Ross D.T., Scherf,U., Eisen,M.B., Perou,C.M., Rees,M., Spellman,P., Iyer,V., Jeffrey,S., Van de Rijn,M., Waltham,M. et al. (2000) Systematic variation in gene expression patterns in human cancer cell lines. Nature Genet., 24, 227–235. [DOI] [PubMed] [Google Scholar]
- 4.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,A., Horton,H. and Brown,E.L. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nature Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 5.Lipshutz R.J., Fodor,S.P., Gingeras,T.R. and Lockhart,D.J. (1999) High density synthetic oligonucleotide arrays. Nature Genet., 21, 20–24. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen C., Rocha,D., Granjeaud,S., Baldit,M., Bernard,K., Naquet,P. and Jordan,B.R. (1996) Differential gene expression in the murine thymus assayed by quantitative hybridization of arrayed cDNA clones. Genomics, 29, 207–216. [DOI] [PubMed] [Google Scholar]
- 7.Eickhoff H., Schuchardt,J., Ivanov,I., Meier-Ewert,S., O’Brien,J., Malik,A.,Tandon,N., Wolski,E., Rohlfs,E., Nyarsik,L. et al. (2000) Tissue gene expression analysis using arrayed normalized cDNA libraries. Genome Res., 10, 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J.J., Wu,R., Yang,P., Huang,J., Sher,Y., Han,M., Kao,W., Lee,P., Chiu,T.F., Chang,F. et al. (1998) Profiling expression patterns and isolating differentially expressed genes by cDNA microarray system with colorimetry detection. Genomics, 51, 313–324. [DOI] [PubMed] [Google Scholar]
- 9.Bertucci F., Bernard,K., Loriod,B., Chang,Y., Granjeaud,S., Birnbaum,D., Nguyen,C., Pech,K. and Jordan,B. (1999) Sensitivity issues in DNA array-based expression measurements and performance of nylon microarrays for small samples. Hum. Mol. Genet., 8, 1715–1722. [DOI] [PubMed] [Google Scholar]
- 10.Greller L.D. and Tobin,F.L. (1999) Detecting selective expression of genes and proteins. Genome Res., 9, 282–296. [PMC free article] [PubMed] [Google Scholar]
- 11.Claverie J.M. (1999) Computational methods for the identification of differential and coordinated gene expression. Hum. Mol. Genet., 8, 1821–1832. [DOI] [PubMed] [Google Scholar]
- 12.Vingron M. and Hoheisel,J. (1999) Computational aspects of expression data. J. Mol. Med., 77, 3–7. [DOI] [PubMed] [Google Scholar]
- 13.Schuchardt J., Beule,D., Malik,A., Wolski,E., Eickhoff,H., Lehrach,H. and Herzel,H. (2000) Normalization strategies for cDNA microarrays. Nucleic Acids Res., 28, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manduchi E., Grant,G.R., McKenzie,S.E., Overton,G.C., Surrey,S. and Stoeckert,C. (2000) Generation of patterns from gene expression data by assigning confidence to differentially expressed genes. Bioinformatics, 16, 685–698. [DOI] [PubMed] [Google Scholar]
- 15.Yue H., Eastman,P.S., Wang,B.B., Minor,J., Doctolero,M.H., Nuttall,R.L., Stack,R., Becker,J.W., Montgomery,J.R., Vainer,M. and Johnston,R. (2001) An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res., 29, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton M.A., Kendziorski,C.M., Richmond,C.S., Blattner,F.R. and Tsui,K.W. (2001) On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J. Comp. Biol., 8, 37–52. [DOI] [PubMed] [Google Scholar]
- 17.Audic S. and Claverie,J.M. (1997) The significance of digital gene expression profiles. Genome Res., 7, 986–995. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann E.L. (1975) Nonparametrics: Statistical Methods Based on Ranks. Holden-Day, San Francisco, CA.
- 19.Callow M.J., Dudoit S., Gong,E.L., Speed,T.P. and Rubin,E.M. (2000) Microarray expression profiling identifies genes with altered expression in HDL-deficient mice. Genome Res., 10, 2022–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldi P. and Long,A.D. (2001) A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics, 17, 509–519. [DOI] [PubMed] [Google Scholar]
- 21.Lennon G. and Lehrach,H. (1991) Hybridization analyses of arrayed cDNA libraries. Trends Genet., 7, 314–317. [DOI] [PubMed] [Google Scholar]
- 22.Meier-Ewert S., Maier,E., Ahmadi,A., Curtis,J. and Lehrach,H. (1993) An automated approach to generating expressed sequence catalogues. Nature, 361, 375–376. [DOI] [PubMed] [Google Scholar]
- 23.Stachel S.E., Grunwald,D.J. and Myers,P.Z. (1993) Lithium perturbation and goosecoid expression identify a dorsal specification pathway in the pregastrula zebrafish. Development, 117, 1261–1274. [DOI] [PubMed] [Google Scholar]
- 24.Klein P.S. and Melton,D.A. (1996) A molecular mechanism for the effect of lithium on development. Proc. Natl Acad. Sci. USA, 93, 8455–8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stambolic V., Ruel,L. and Woodgett,J.R. (1996) Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr. Biol., 6, 1664–1668. [DOI] [PubMed] [Google Scholar]
- 26.Maslanski J.A., Leshko,L. and Busa,W.B. (1992) Lithium-sensitive production of inositol phosphate during amphibian embryonic mesoderm induction. Science, 256, 243–245. [DOI] [PubMed] [Google Scholar]
- 27.Ault K.T., Durmowicz,G., Galione,A., Harger,P.L. and Busa,W.B. (1996) Modulation of Xenopus embryo mesoderm-specific gene expression and dorsoanterior patterning by receptors that activate the phosphatidylinositol cycle signal transduction pathway. Development, 122, 2033–2041. [DOI] [PubMed] [Google Scholar]
- 28.Kume S., Muto,A., Inoue,T., Suga,K., Okano,S. and Mikoshiba,K. (1997) Role of inositol 1,4,5-trisphosphate receptor in ventral signaling in Xenopus embryos. Science, 278, 1940–1943. [DOI] [PubMed] [Google Scholar]
- 29.Clark M.D., Hennig,S., Herwig,R., Clifton,S.W., Marra,M.A., Lehrach,H., Johnson,S.L. and the WU-GSC EST group (2001) An oligonucleotide fingerprint normalized and EST characterized zebrafish cDNA library. Genome Res., 11, 1594–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aanstad P. and Whitaker,M. (1999) Predictability of dorso-ventral asymmetry in the cleavage stage zebrafish embryo: an analysis using lithium sensitivity as a dorso-ventral marker. Mech. Dev., 88, 33–41. [DOI] [PubMed] [Google Scholar]
- 31.Welch B.L. (1947) The generalization of Student’s problem when several different population variances are involved. Biometrika, 34, 28–35. [DOI] [PubMed] [Google Scholar]
- 32.Best D.I. and Rayner,C.W. (1987) Welch’s approximate solution for the Behrens–Fisher problem. Technometrics, 29, 205–220. [Google Scholar]
- 33.Press W.H., Teukolsky,S.A., Vetterling,W.T. and Flannery,B.P. (1992) Numerical Recipes in C. Cambridge University Press, New York, NY.
- 34.Pitman E.J.G. (1937) Significance tests which may be applied to samples from any population. J. R. Statist. Soc., B4, 119–130, 225–237. [Google Scholar]
- 35.Fishman G.S. (1996) Monte Carlo: Concepts, Algorithms and Applications. Springer, New York, NY.