ABSTRACT
Large-scale genomic changes, including copy number variations (CNVs), are frequently observed in long-term evolution experiments (LTEEs). We have previously reported the detection of recurrent CNVs in Saccharomyces cerevisiae populations adapting to glutamine-limited conditions over hundreds of generations. Here, we present the whole-genome sequencing (WGS) assemblies of 7 LTEE strains and their ancestor.
ANNOUNCEMENT
Large-scale genomic rearrangements frequently occur in single-celled organisms adapting to nutrient-limited conditions (1, 2). We previously adapted populations of Saccharomyces cerevisiae to nutrient-limited growth medium over hundreds of generations (3). We present the whole-genome sequencing (WGS) genome assemblies of 7 evolved strains of Saccharomyces cerevisiae and their ancestor.
The ancestor strain (DGY1657) is derived from the S288C haploid strain FY4 (4, 5), with a constitutively expressed fluorescent reporter (ACT1pr::mCitrine::ADH1term) and a drug resistance gene (TEFpr::KanMX::TEFterm) integrated into the upstream intergenic region of GAP1 (chromosome XI [ChrXI], position 513945). We inoculated DGY1657 into 20-mL chemostat vessels (6) containing glutamine-limited (Gln−) medium (400 μM glutamine, 1 g/L CaCl2·2H2O, 1 g/L NaCl, 5 g/L MgSO4·7H2O, 10 g/L KH2PO4; 2% glucose, metals, vitamins [7]). Chemostats were maintained at 30°C in aerobic conditions and diluted at a rate of 0.12 h−1 (approximate doubling time, 5.8 h). Steady-state populations of 3 × 108 cells were maintained and sampled at generations 150 (~870 h; DGY1728, DGY1740, DGY1747) and 250 (~1,450 h; DGY1734, DGY1736, DGY1744, DGY1751) (Fig. 1A). Clones from these populations were isolated by plating the cultures onto rich medium (yeast extract-peptone-dextrose [YPD]; 30°C, 24 h) and picking individual colonies, which were used to inoculate batch cultures containing Gln− medium and incubated for 24 h (30°C). Amplification of the reporter gene was verified using flow cytometry (3).
FIG 1.
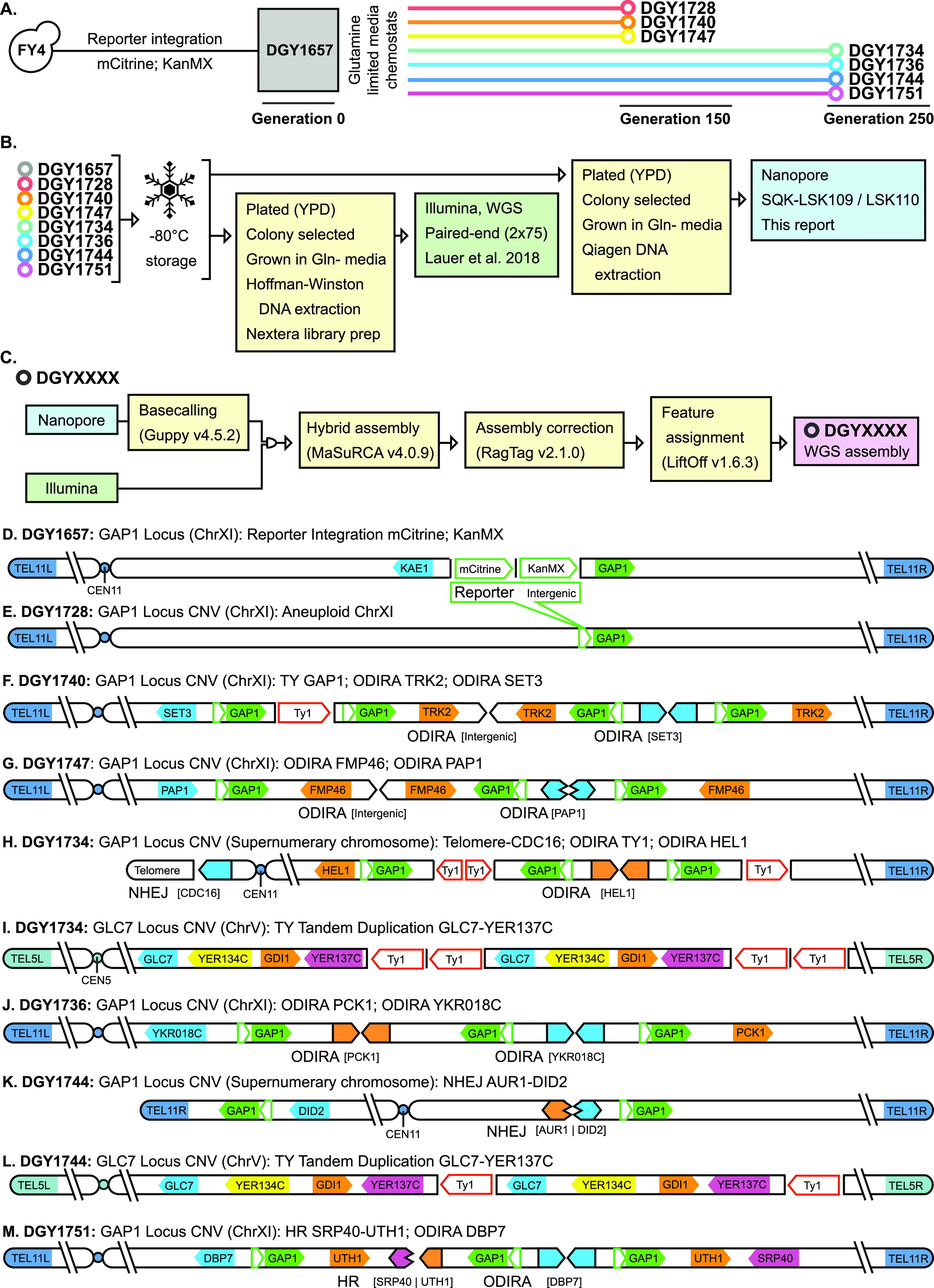
Schematic showing S. cerevisiae strain provenance. (A) Ancestral strain DGY1657 has a copy number variation (CNV) reporter construct in a FY4 background. DGY1657 was inoculated into separate chemostats and grown in glutamine-limited medium. The strains were isolated and sequenced at generations 150 (DGY1728, DGY1740, DGY1747) and 250 (DGY1734, DGY1736, DGY1744, DGY1751). (B) Flowchart showing the strain and independent library preparation paths for Illumina (green box) and Nanopore (blue box). (C) Flowchart for hybrid WGS assembly pipeline for a given strain. (D to M) Topology diagrams for evolved strains indicating CNV breakpoints, orientations, and the occurrence of transposon events. Green boxes represent the reporter; red arrows indicate transposon yeast. CNV breakpoints are annotated with their most likely mechanism. ODIRA, origin-dependent inverted-repeat amplification; NHEJ, nonhomologous end-joining; HR, homologous recombination.
For sequencing, cells were recovered from −80°C glycerol stocks (Fig. 1B) by plating the stocks and incubating them (YPD, 30°C, 24 h). For Illumina sequencing, strains were inoculated into rich medium (YPD) and grown overnight to a concentration of >1 × 108 cells/mL (3). DNA was extracted using the Hoffman-Winston method (8), followed by a modified Nextera library preparation (9); the library was sequenced on a NextSeq 500 instrument in paired-end format (2 × 75 bp) (3). Adapters were trimmed using Cutadapt v1.12 with default settings (10). For Nanopore sequencing, strains were grown to a concentration of >1 × 107 cells/mL in Gln− medium (11). DNA from each strain was extracted from ~1.5 × 109 cells using the Qiagen Genomic-tip 20/G kit, following the manufacturer’s protocol. Nanopore was used for long-read sequencing following the manufacturer’s protocol (DGY1657, NBE_9065_v109_revB_23May2018; DGY1728 and DGY1744, NBE_9065_V109_revP_14Aug2019; all others, GDE_9108_v110_revJ_10Nov2020), with the following exceptions for DGY1657, DGY1728, and DGY1744: (i) incubation times for the enzymatic repair step were increased to 15 min, (ii) Agencourt AMPure XP beads were incubated for 30 min at 37°C before elution, and (iii) the adapter ligation time was increased to 10 min. All libraries were loaded onto FLO-MIN106D (R9.4) flow cells and sequenced using a MinION instrument (MIN-101B). Default parameters were used for all software unless otherwise specified. Demultiplexing of DGY1657, DGY1728, and DGY1744 was performed using Epi2Me (epi2me.nanoporetech.com). Base-calling was performed using Guppy v4.5.2, gpu (12). Long and short reads were used for hybrid assembly with MaSuRCA v4.0.9 (13). The assemblies were corrected with Ragtag v2.1.0 using the “remove-small” option to remove unique fragments shorter than 1,000 bp (14). LiftOff v1.6.3 was used for chromosome assignment, using the “copies” option to allow extra copies of genes (15). Proposed final assemblies of these genomes are shown in Fig. 1D to M.
Data availability.
This whole-genome shotgun project has been deposited at GenBank under the accession numbers JAMQBG000000000 (DGY1657), JAMQBF000000000 (DGY1728), JAMQBE000000000 (DGY1734), JAMQBD000000000 (DGY1736), JAMQBC000000000 (DGY1740), JAMQBB000000000 (DGY1744), JAMQBA000000000 (DGY1747), and JAMQAZ000000000 (DGY1751). The versions described in this paper are the first versions, JAMQBG010000000 (DGY1657), JAMQBF010000000 (DGY1728), JAMQBE010000000 (DGY1734), JAMQBD010000000 (DGY1736), JAMQBC010000000 (DGY1740), JAMQBB010000000 (DGY1744), JAMQBA010000000 (DGY1747), and JAMQAZ010000000 (DGY1751). The raw Illumina reads are available under the SRA accession numbers SRR7057840 (DGY1657), SRR7057779 (DGY1728), SRR7057829 (DGY1734), SRR7057832 (DGY1736), SRR7057777 (DGY1740), SRR7057833 (DGY1744), SRR7057780 (DGY1747), and SRR7057828 (DGY1751). The ONT reads can be found under the SRA accession numbers SRR10525523 (DGY1657), SRR19440503 (DGY1728), SRR19440502 (DGY1734), SRR19440501 (DGY1736), SRR19440500 (DGY1740), SRR10525517 (DGY1744), SRR19440499 (DGY1747), and SRR19440498 (DGY1751).The sequencing data and accession numbers are provided in Table 1. The computational pipeline is available on GitHub (https://github.com/pspealman/WGS_assemble_pipeline [16]).
TABLE 1.
Sequencing metrics for the S. cerevisiae strains in this study
| Characteristic | Data for strain: |
|||||||
|---|---|---|---|---|---|---|---|---|
| DGY1657 | DGY1728 | DGY1734 | DGY1736 | DGY1740 | DGY1744 | DGY1747 | DGY1751 | |
| Illumina data | ||||||||
| SRA accession no. | SRR7057840 | SRR7057779 | SRR7057829 | SRR7057832 | SRR7057777 | SRR7057833 | SRR7057780 | SRR7057828 |
| Total no. of reads | 3,653,321 | 6,229,610 | 1,760,584 | 5,401,867 | 4,531,223 | 5,447,512 | 4,709,699 | 3,272,815 |
| Nanopore data | ||||||||
| SRA accession no. | SRR10525523 | SRR19440503 | SRR19440502 | SRR19440501 | SRR19440500 | SRR10525517 | SRR19440499 | SRR19440498 |
| Total no. of reads | 47,436 | 194,350 | 44,825 | 40,245 | 88,706 | 140,456 | 30,289 | 343,924 |
| N50 (bp) | 8,324 | 625 | 1,929 | 1,954 | 1,198 | 414 | 2,449 | 2,074 |
| Estimated coverage depth (×) | 56.6 | 30.1 | 15.4 | 15.5 | 26.9 | 48.5 | 17.2 | 137.3 |
| Assembly N50 (bp) | 740,333 | 818,015 | 266,864 | 751,856 | 813,575 | 810,678 | 666,227 | 781,298 |
| BUSCO (%) | 96.10 | 99.30 | 90.20 | 98.60 | 99.40 | 99.40 | 98.90 | 97.50 |
| GC (%) | 38.20 | 38.10 | 38.30 | 38.20 | 38.30 | 38.30 | 38.30 | 38.20 |
| No. of contigs | 20 | 32 | 68 | 30 | 22 | 20 | 30 | 36 |
| GenBank accession no. | JAMQBG000000000 | JAMQBF000000000 | JAMQBE000000000 | JAMQBD000000000 | JAMQBC000000000 | JAMQBB000000000 | JAMQBA000000000 | JAMQAZ000000000 |
ACKNOWLEDGMENTS
This work was made possible by grants from the NIH and NSF to P.S. (F32-GM131573), G.A. (DGE1342536), and D.G. (R01-GM134066, R01-GM107466, MCB1818234).
Contributor Information
David Gresham, Email: dgresham@nyu.edu.
David Rasko, University of Maryland School of Medicine.
REFERENCES
- 1.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. 2009. Mechanisms of change in gene copy number. Nat Rev Genet 10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lauer S, Gresham D. 2019. An evolving view of copy number variants. Curr Genet 65:1287–1295. doi: 10.1007/s00294-019-00980-0. [DOI] [PubMed] [Google Scholar]
- 3.Lauer S, Avecilla G, Spealman P, Sethia G, Brandt N, Levy SF, Gresham D. 2018. Single-cell copy number variant detection reveals the dynamics, and diversity of adaptation. PLoS Biol 16:e3000069. doi: 10.1371/journal.pbio.3000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winston F, Dollard C, Ricupero-Hovasse SL. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 5.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132. doi:. [DOI] [PubMed] [Google Scholar]
- 6.Miller AW, Befort C, Kerr EO, Dunham MJ. 2013. Design and use of multiplexed chemostat arrays. J Vis Exp:e50262. doi: 10.3791/50262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong J, Gresham D. 2014. Molecular specificity, convergence and constraint shape adaptive evolution in nutrient-poor environments. PLoS Genet 10:e1004041. doi: 10.1371/journal.pgen.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman CS, Winston F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 9.Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. 2015. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10:e0128036. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 11.Spealman P, Burrell J, Gresham D. 2020. Inverted duplicate DNA sequences increase translocation rates through sequencing nanopores resulting in reduced base calling accuracy. Nucleic Acids Res 48:4940–4945. doi: 10.1093/nar/gkaa206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wick RR, Judd LM, Holt KE. 2019. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol 20:129. doi: 10.1186/s13059-019-1727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimin AV, Puiu D, Luo M-C, Zhu T, Koren S, Marçais G, Yorke JA, Dvořák J, Salzberg SL. 2017. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res 27:787–792. doi: 10.1101/gr.213405.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, Lippman ZB, Schatz MC. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol 20:224. doi: 10.1186/s13059-019-1829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shumate A, Salzberg SL. 2021. Liftoff: accurate mapping of gene annotations. Bioinformatics 37:1639–1643. doi: 10.1093/bioinformatics/btaa1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spealman P. 2022. pspealman/WGS_assemble_pipeline: release for MRA. Zenodo doi: 10.5281/ZENODO.6613405. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This whole-genome shotgun project has been deposited at GenBank under the accession numbers JAMQBG000000000 (DGY1657), JAMQBF000000000 (DGY1728), JAMQBE000000000 (DGY1734), JAMQBD000000000 (DGY1736), JAMQBC000000000 (DGY1740), JAMQBB000000000 (DGY1744), JAMQBA000000000 (DGY1747), and JAMQAZ000000000 (DGY1751). The versions described in this paper are the first versions, JAMQBG010000000 (DGY1657), JAMQBF010000000 (DGY1728), JAMQBE010000000 (DGY1734), JAMQBD010000000 (DGY1736), JAMQBC010000000 (DGY1740), JAMQBB010000000 (DGY1744), JAMQBA010000000 (DGY1747), and JAMQAZ010000000 (DGY1751). The raw Illumina reads are available under the SRA accession numbers SRR7057840 (DGY1657), SRR7057779 (DGY1728), SRR7057829 (DGY1734), SRR7057832 (DGY1736), SRR7057777 (DGY1740), SRR7057833 (DGY1744), SRR7057780 (DGY1747), and SRR7057828 (DGY1751). The ONT reads can be found under the SRA accession numbers SRR10525523 (DGY1657), SRR19440503 (DGY1728), SRR19440502 (DGY1734), SRR19440501 (DGY1736), SRR19440500 (DGY1740), SRR10525517 (DGY1744), SRR19440499 (DGY1747), and SRR19440498 (DGY1751).The sequencing data and accession numbers are provided in Table 1. The computational pipeline is available on GitHub (https://github.com/pspealman/WGS_assemble_pipeline [16]).
TABLE 1.
Sequencing metrics for the S. cerevisiae strains in this study
| Characteristic | Data for strain: |
|||||||
|---|---|---|---|---|---|---|---|---|
| DGY1657 | DGY1728 | DGY1734 | DGY1736 | DGY1740 | DGY1744 | DGY1747 | DGY1751 | |
| Illumina data | ||||||||
| SRA accession no. | SRR7057840 | SRR7057779 | SRR7057829 | SRR7057832 | SRR7057777 | SRR7057833 | SRR7057780 | SRR7057828 |
| Total no. of reads | 3,653,321 | 6,229,610 | 1,760,584 | 5,401,867 | 4,531,223 | 5,447,512 | 4,709,699 | 3,272,815 |
| Nanopore data | ||||||||
| SRA accession no. | SRR10525523 | SRR19440503 | SRR19440502 | SRR19440501 | SRR19440500 | SRR10525517 | SRR19440499 | SRR19440498 |
| Total no. of reads | 47,436 | 194,350 | 44,825 | 40,245 | 88,706 | 140,456 | 30,289 | 343,924 |
| N50 (bp) | 8,324 | 625 | 1,929 | 1,954 | 1,198 | 414 | 2,449 | 2,074 |
| Estimated coverage depth (×) | 56.6 | 30.1 | 15.4 | 15.5 | 26.9 | 48.5 | 17.2 | 137.3 |
| Assembly N50 (bp) | 740,333 | 818,015 | 266,864 | 751,856 | 813,575 | 810,678 | 666,227 | 781,298 |
| BUSCO (%) | 96.10 | 99.30 | 90.20 | 98.60 | 99.40 | 99.40 | 98.90 | 97.50 |
| GC (%) | 38.20 | 38.10 | 38.30 | 38.20 | 38.30 | 38.30 | 38.30 | 38.20 |
| No. of contigs | 20 | 32 | 68 | 30 | 22 | 20 | 30 | 36 |
| GenBank accession no. | JAMQBG000000000 | JAMQBF000000000 | JAMQBE000000000 | JAMQBD000000000 | JAMQBC000000000 | JAMQBB000000000 | JAMQBA000000000 | JAMQAZ000000000 |


