Abstract
The Threshold of Toxicological Concern (TTC) is a pragmatic approach used to establish safe thresholds below which there can be no appreciable risk to human health. Here, a large inventory of ~45,000 substances (referred to as the LRI dataset) was profiled through the Kroes TTC decision module within Toxtree v3.1 to assign substances into their respective TTC categories. Four thousand and two substances were found to be not applicable for the TTC approach. However, closer examination of these substances uncovered several implementation issues: substances represented in their salt forms were automatically assigned as not appropriate for TTC when many of these contained essential metals as counter ions which would render them TTC applicable. High Potency Carcinogens and dioxin-like substances were not fully captured based on the rules currently implemented in the software. Phosphorus containing substances were considered exclusions when many of them would be appropriate for TTC. Refinements were proposed to address the limitations in the current software implementation. A second component of the study explored a set of substances representative of those released from medical devices and compared them to the LRI dataset as well as other toxicity datasets to investigate their structural similarity. A third component of the study sought to extend the exclusion rules to address application to substances released from medical devices that lack toxicity data. The refined rules were then applied to this dataset and the TTC assignments were compared. This case study demonstrated the importance of evaluating the software implementation of an established TTC workflow, identified certain limitations and explored potential refinements when applying these concepts to medical devices.
Keywords: Threshold of Toxicological Concern (TTC), Toxtree, Exclusions, Extractables, Medical devices
1. Introduction
The Threshold of Toxicological Concern (TTC) is an approach intended to establish safe thresholds below which there can be no appreciable risk to human health [1,2]. There are 2 types of TTC – a DNA-reactive TTC and those for non-cancer effects. The DNA-reactive TTC devised by Rulis (1986) [3] was derived by low-dose extrapolation of carcinogenicity data from animal studies to define human exposures associated with an upper-bound estimate of one in a million cancer risk. The carcinogenicity data relied on the carcinogenicity potency database (CPDB) developed by Lois Gold and colleagues [4]. This TTC, termed the Threshold of Regulation (TOR) was set at 0.5 ppb. The workflow for the assessment of TTC proposed by Kroes et al. [2] has 0.0025 μg/kg-day as the lowest TTC tier for substances that raise a concern of genotoxicity determined on the basis of structural alerts for DNA reactivity. For substances without structural alerts for DNA reactivity, a series of non-cancer TTC tiers, are used which are predominantly based on the Cramer et al. decision tree [5]. Derivation of these non-cancer TTC values arises from work by Munro et al. (1996) who compiled a database of No Observed Effect Levels (NOELs) for 613 substances that had been tested in repeat-dose oral toxicity studies including subchronic, chronic, reproductive and developmental toxicity [6]. In cases where there were multiple NOELs for a given substance, Munro et al. [6] selected the lowest one (there were a total of 2941 NOELs for the 613 substances). The substances were then assigned to the appropriate Cramer structural class, and cumulative distributions of the logarithms of NOELs were plotted separately for each structural class. Adjustments were made to extrapolate subchronic NOELs to chronic, and Lowest Observed Effect Levels (LOELs) to NOELs as appropriate. The 5th percentile NOEL was estimated for each structural class, which then was converted into its respective TTC value by applying a safety factor of 100 (10X to account for extrapolation of animals to humans and 10X for human variability). The TTC values established by Munro et al. [6] were 30 μg/kg-day for Cramer Class I, 9 μg/kg-day for Cramer Class II, and 1.5 μg/kg-day for Cramer Class III substances. Certain chemicals are excluded from the TTC approach because they were not represented in the original toxicity databases supporting TTC (e.g., metals or metal containing compounds, organosilicons, proteins) or because standard risk assessment approaches are more appropriate (e.g., 2,3,7,8-dibenzop-dioxin (TCDD) and its analogues, high potency carcinogens such as N-nitroso compounds).
Although the origins of the TTC lie in food ingredients, food additives and food contact materials, the approach is increasingly applied in many other sectors such as cosmetics [7], fragrances [8], pesticides [9–12], and pharmaceuticals [13]. For example, the TTC approach is used to assess genotoxic impurities in pharmaceutical active ingredients [14]. More recently, an approach for prioritising large numbers of chemicals using a TTC approach has been proposed [15]. The TTC approach also formed a component of the EPA Proof-of-Concept case study integrating publicly available information to screen candidates for chemical prioritisation under TSCA [16]. The workflow in Patlewicz et al. [15] relied on that proposed by Kroes et al. [2] and highlighted a couple of key issues: 1) how to benchmark the approach against a large inventory of chemicals whose TTC assignment were already established and, 2) challenges in adapting the module within Toxtree to accommodate batch processing of substances for which the exposure was not defined a priori. The study also identified a number of issues around the way in which organophoshates and carbamates were identified, and refinements were made in Nelms et al. [17] to address some of these specific short comings.
A question raised in both Patlewicz et al. [15] and Nelms et al. [17] regarded the implementation of specific questions in the Kroes TTC decision tree module as implemented in Toxtree 3.1 (Ideaconsult Ltd – https://sourceforge.net/projects/toxtree/files/). This is challenging in that much of the TTC literature published has proposed refinements or evaluated TTC approaches focused solely on the Cramer structural classes without a clear articulation of how a determination was made for the remaining aspects of the Kroes workflow. Indeed, processing the enriched dataset used in Yang et al. [7] finds instances where substances might be excluded from the TTC approach or assigned as DNA-reactive based on the presence of structural alerts (unpublished). Another shortcoming in the Kroes module in Toxtree is that an exclusion exists to address substances that are dioxin-like; presumably due to their bio-accumulation potential and persistence; yet there are no explicit structure-based or property-based rules to capture these exclusions. The dataset used in Nelms et al. [17] contained Per- and polyfluoroalkyl substances (PFAS)-like substances but the Kroes decision tree module within Toxtree did not identify these substances as out of scope of the TTC approach. On a case-by-case basis, a substance likely to be bio-accumulative would be unlikely to be a candidate for assessment through the TTC. However, if the scenario is to use the TTC approach to prioritise large numbers of chemicals, then a more robust means of systematically addressing these types of considerations is needed. Reconsidering what constitutes an exclusion in the TTC approach and whether refinements to the software implementation within Toxtree are potentially needed particularly in the use case of profiling large numbers of chemicals.
One such example is the ability to identify whether a substance is a candidate for TTC in the context of a toxicological risk assessment of leachables released from an FDA-regulated medical device. The International Organization for Standardization (ISO) technical specification ISO/TS21726:2019 describes a set of TTC exclusions for assessing medical device constituents that have been compared against the more generic guiding principles described in Kroes et al. [2]. Kroes et al. [2] describes how polymers, proteins, radioactive constituents, high potency carcinogens, and dioxin-like substances are excluded. This is replicated in the European Food Safety Authority and World Health Organisation (EFSA/WHO) 2016 guidance [18].
This case study had three main aims to: 1) perform a closer evaluation of the exclusion rules as implemented in the Kroes module within the Toxtree software tool and propose refinements if necessary, 2) assess the chemistry landscape of substances released by medical devices relative to other inventories where TTC is used or can be applied and, 3) apply the revised set of exclusion rules to the Extractables and Leachables Safety Information Exchange (ELSIE) dataset (described in more detail in Masuda-Herrera et al. [19]), a dataset of extractables representative of the medical device chemical space.
The intent of the first aim was to evaluate the exclusions implemented in the Kroes Toxtree module and consider whether these needed to be refined in any manner. A large inventory of over 45,000 substances, first compiled by Scitovation as part of an ACC LRI funded project (since published in Nicolas et al. [20]), had already been profiled in Nelms et al. (2019) and served as a comprehensive inventory of chemicals representing multiple industrial sectors from pesticides, cosmetics and industrial chemicals. This inventory, referred to herein as the ‘LRI dataset’, would be a suitable one for use in this evaluation. Aim 2 was to explore whether the landscape of medical device substances being considered for TTC assessment is similar to other sectors based on their chemistry. This would help determine whether the chemistry is unique and, furthermore, to scrutinise whether there are specific types of chemical functional groups that are over or underrepresented in the TTC training set. Aim 3 would apply the learnings from the previous steps to the ELSIE dataset.
2. Materials and Methods
2.1. Datasets
The assessment of the chemistry landscape focused on several specific datasets: a dataset of representatives from the medical device chemical space, the ELSIE dataset, the LRI and Munro TTC datasets that had been profiled in Nelms et al. [17] and the EPA Toxicity Values Database (ToxValDB). An earlier version of ToxValDB (version 7) had been profiled in Nelms et al. [17]. Here a later version of the dataset (version 9.1) was used.
2.12. ELSIE dataset
The dataset of representatives from the medical device chemical space was taken from the EPA CompTox Chemicals Dashboard [21] (https://comptox.epa.gov/dashboard/chemical-lists/ELSIE). This list named ELSIE, the Extractables and Leachables Safety Information Exchange, comprised 457 substances (last updated 16th November 2019). The list contained preferred names, DTXSIDs (DSSTox Substance Identifiers), CAS Registry numbers and structural 1D representations as Simplified Molecular-Input Line Entry System (SMILES). The set of identifiers were queried against the Dashboard to retrieve QSAR_READY_SMILES. The final ELSIE list with SMILES comprised 414 substances. Substances for which SMILES could not be readily determined were mainly inorganics, reaction products and polymers. Examples included sulfur, fluoride, alumina, hydrocarbon waxes, Polymethylsilsesquioxane (silsesquioxanes, Me), epoxidised soybean, and talc. The ELSIE list from the Dashboard was used since a master list of extractables from devices does not exist. Whilst this set is representative of extractables that may be released from devices, the list primarily represents substances released from pharmaceutical packaging and drug delivery systems and as such may not cover the full chemical space of device-related extractables.
2.13. LRI dataset
A publicly available inventory of chemicals (~45,000 substances) along with their TTC category assignments that were reported by Scitovation as part of ACC LRI supported research (first accessed 28th June 2019) formed the benchmark comparison inventory. This inventory had been profiled into its respective TTC categories in Nelms et al. [17]. The dataset comprised DTXSIDs, SMILES and TTC assignments. The TTC assignments relied upon in this study were those reported in Nelms et al. [17]. There were 45,038 substances in the LRI dataset, of which TTC assignments were available for 45,017 substances.
2.14. Munro TTC
The dataset underpinning the TTC values published by Munro et al. [6] comprising 613 substances was used as one toxicity dataset comparator. SMILES had been assigned in Nelms et al. [17] hence the supporting data files associated with this publication were used directly (hosted at https://gaftp.epa.gov/Comptox/CCTE_Publication_Data/CCED_Publication_Data/PatlewiczGrace/CompTox-TTC_evaluation_refinements-regtoxpharm/).
2.15. EPA Toxicity Values Database (ToxValDB)
The EPA Toxicity Values Database as compiled by RS Judson and hosted at https://gaftp.epa.gov/Comptox/Staff/rjudson/datasets/ToxValDB/ was used as a second comparator toxicity dataset. ToxValDB consists of a collection of summary level in vivo test data from a variety of study types typically used in risk assessments. It comprises point of departure (POD) values such as the no observed (adverse) effect level NO(A)EL and lowest observed (adverse) effect level (LO(A)EL) data. These data have been aggregated from over 40 publicly available sources including US Federal and State agencies (e.g., US EPA, US Food and Drug Administration (FDA), and California EPA), alongside international organisations (e.g., World Health Organisation (WHO)), as well as data submitted under regulatory frameworks, such as the European Union’s REACH regulation (e.g., non-confidential registration data submitted to the European Chemicals Agency (ECHA) by industry registrants). This was filtered to retrieve repeated systemic dose toxicity studies conducted in rats, rabbits, mice or guinea pigs where the route of administration was via the oral route and where a NOAEL or LOAEL had been reported. Studies that were specifically developmental, reproductive or neurotoxicity in nature were excluded. There were 60,614 records for 8331 individual substances. SMILES identifiers were queried for the 8331 substances using the EPA CompTox Chemicals Dashboard which garnered 4344 substances with associated SMILES.
3. Chemical features
3.1. ToxPrints
A binary molecular fingerprint was generated for each chemical in the four datasets utilising the publicly available ToxPrint chemotype feature set (https://toxprint.org) and the ChemoTyper software (https://chemotyper.org/). These fingerprints were intended to facilitate the comparison of the chemistry landscape between the four datasets in a number of different ways – from profiling the datasets using fingerprints and projecting them in 2D for visualisation purposes or performing an enrichment analysis to compare the overrepresentation of specific fingerprints between datasets.
In practice, given the large numbers of substances being profiled, the commercial command line version of CORINA Symphony application licensed from Molecular Networks was used. ToxPrint chemotypes consist of a predefined library of 729 sub-structural features designed to encapsulate a broad range of chemical atoms and scaffolds, which were developed by Altamira and Molecular Networks under contract by the US Food and Drug Administration [22].
The full 729-bit ToxPrint fingerprint was condensed to a length of 70-bits (Level 2) for profiling purposes. To do this, the ToxPrints were condensed based upon the root of the ToxPrint name. For example, the ToxPrints “bond:C#N_cyano_cyanamide”, “bond:C#N_nitro_isonitrile”, and “bond:C#N_nitrile_generic” were concatenated to form the more generalised name “bond:C#N” that is common amongst these ToxPrints. The frequency of these condensed Level 2 fingerprints in each dataset were calculated and plotted.
3.2. International Chemical Identifier (InChI) Keys
International Chemical Identifier (InChI) Keys were generated for each chemical in the four datasets utilising the open-source python package, RDKit [23]. This was performed to enable a comparison of substances based on their structural representations between the four datasets since DTXSIDs were not available for the Munro dataset.
3.3. 2D visualisations
Visualisation of the four datasets was undertaken using the ToxPrint fingerprints and projecting them in a 2D scatter plot. Two approaches were undertaken – t-SNE and UMAP. T-SNE or t-Distributed Stochastic Neighbor Embedding is a machine learning algorithm for visualisation developed by van der Maaten and Hinton [24]. It is a nonlinear dimensionality reduction technique that is well suited to embedding high dimensional data for visualisation in a low dimensional space of 2 or 3 dimensions. It models each high dimensional object in a manner that similar objects are close together whereas dissimilar objects are far away in 2D space. Since t-SNE is very memory intensive, rather than reducing the entire dataset of ~50,000 substances across 729 dimensions, 10 % of the data was extracted at random whilst conserving the ratio between the inventories. For the ToxPrint dataset, a Variance Threshold was first used to exclude ToxPrints for which there was no variance. This removed 129 ToxPrints as containing zero variance. For the 600 ToxPrints remaining, t-SNE was performed to extract 2 features on the basis of a learning rate of 200 using scikit learning’s TSNE module [25].
Since t-SNE was only applied to 10 % of the dataset, a different dimensionality technique, UMAP, Uniform Manifold Approximation and Projection [26] was applied to the full dataset. This is a similar approach to t-SNE although UMAP aims to keep both globally and locally similar objects together in contrast to t-SNE which keeps locally similar objects together.
3.4. Chemotype enrichment analysis
Briefly, chemotype enrichment analysis identifies sub-structural features that are over-represented with respect to a given endpoint. Typically, this endpoint may show activity in a particular (suite of) assay (s); however, in this study the “endpoint” was the presence/absence of the chemical in the respective comparator datasets relative to the ELSIE dataset. Dummy variables indicating presence or absence of a substance in the datasets was denoted by a 1 for presence and 0 for absence. To conduct this analysis, the full 729-bit ToxPrint fingerprints generated above were annotated with an additional binary column representing which dataset the substance was present in. A Fisher exact test was performed to compute the odds ratio statistic and its associated p-value. The number of true positives (TPs, member of the inventory of interest) and containing a ToxPrint were computed from the corresponding confusion matrix for each ToxPrint. Enriched ToxPrints were identified by filtering for an odds ratio equal or greater than 3, a p-value equal to or less than 0.05 and the number of TPs greater than or equal to 3.
3.5. Assessment of the exclusion rules encoded in Kroes TTC decision tree within Toxtree
The assignments of the LRI dataset as reported by Nelms et al. [17] were used as a starting point. Examination of the assignments and the rationale reported were investigated by exploring the software implementation itself to determine how specific structural rules had been captured. Additional filters and rules were created making use of RDKit’s functionality to capture metals and inorganics and referencing both the original Kroes et al. [2] publication as well as the EFSA guidance [1].
Structural alerts for genotoxicity represented by SMILES arbitrary target specification (SMARTS) were taken from the ISS profiler implemented as another module within Toxtree, from publications such as Enoch and Cronin [27] or the ToxAlerts website embedded in OCHEM [28]. Dioxin-like substance alerts were compiled based on existing SMARTS in Toxtree or created de novo using https://smarts.plus/ [29] as a resource for creating and testing SMARTS. New exclusion rules were created using SMARTS already established for existing structural alerts encoding genotoxicity or skin sensitisation such as those available in OCHEM.
4. Software and code
Toxtree v3.1.0 was used to profile substances into their non-cancer TTC categories, using the Cramer rules [30] and Kroes TTC modules as well as custom modules developed by Patlewicz et al. [15] and Nelms et al. [19] to identify carbamates, organophosphates and steroids.
Data processing was conducted using the Anaconda distribution of Python 3.9 (anaconda.com) and associated libraries – RDKit [23], scikit-learn [25], pandas [31], numpy [32], visualisation tools matplotlib [33] and seaborn [34] and the statistical library scipy [35] within a Jupyter lab environment [36]. The code repository supporting this analysis is available at https://github.com/g-patlewicz/ttc_exclusions. All data files (including supplementary data files) are available at https://doi.org/10.23645/epacomptox.21215354.
5. Results and discussion
5.1. Evaluation of the exclusion rules
As noted in the Methods, the LRI dataset was used to probe the substances that were flagged as ‘exclusions’ since this dataset comprised over ~45,000 substances covering a diverse breadth of chemistry that was representative of many sectors of interest from industrial chemicals to pesticides. The assignment of TTC classes were taken from the evaluation in Nelms et al. [17] (Table 1, Supplementary Fig. S1, code Notebook 03).
Table 1.
Initial assignment of TTC categories for the LRI dataset.
| TTC category | Number of substances |
|---|---|
| Cramer Class I | 7370 |
| Cramer Class II | 952 |
| Cramer Class III | 21,691 |
| Organophosphates/carbamates | 1580 |
| Substances presenting an alert for DNA reactivity | 9422 |
| Not appropriate for TTC | 4002 |
| Could not be processed by Toxtree | 21 |
| Total | 45,038 |
There were 4002 substances identified as ‘not appropriate for TTC’ i. e., an exclusion of some kind. These substances were examined by probing the decision point triggered within the Kroes TTC decision tree logic within Toxtree. The majority of substances (3759 out of 4002 substances) flagged as ‘not appropriate for TTC’ were due to the first question in the workflow (see Supplementary Table S1, code Notebook 03).
Question 1 in the Kroes TTC decision tree within Toxtree considers whether a substance is a non-essential metal or metal containing, or whether it is a polyhalogenated dibenzodioxin, -dibenzofuran or -biphenyl. Polyhalogenated substances are excluded due to their potential for bioaccumulation. This description is consistent with what is described in Kroes et al. [2]. Within the Toxtree module, a list of essential metals and non-essential metals are also provided. The EFSA guidance [1] provides additional context for the Q1 exclusion, specifically substances such as inorganics, metals in elemental, ionic or organic form, proteins, organosilicons, nanomaterials, radioactive substances are either outside of the domain of applicability of TTC or are simply substances that were not represented in the underlying TTC dataset. In the case of organic salts, where the counter ion is an essential metal such as sodium, the TTC approach is applicable to the organic ion. The Kroes module in Toxtree specifies 3 substructures by name ‘dibenzodioxin’, ‘dibenzofuran’ and ‘biphenyl’ to identify the polyhalogenated substances.
According to Zoroddu et al. [37], essential metals are as follows: Na, K, Mg, Ca, Fe, Mn, Co, Cu, Zn and Mo. The Toxtree Kroes module names these as essential metals but it also includes P as an essential metal even though phosphorus itself is not a metal. Further, Toxtree does not include Si in its listing of non-essential metals and includes B which is not a metal but recognised as a metalloid.
There were 241 substances tagged as high potency carcinogens (HPC). HPC are specifically named as N-nitroso, azoxy, aflatoxin-like and benzidines in the Kroes workflow. The Toxtree Kroes module has SMARTS patterns to identify N-nitroso, aflatoxin and azoxy containing substances following an initial profiling against 33 specific alerts for genotoxic carcinogenicity. Two substances were tagged as ‘Steroids’. This was captured by one of the alerts for genotoxic carcinogens (SA_gen_and_nogen).
The entire set of 4002 substances were re-evaluated in light of the manner in which the Q1 was implemented in Toxtree. A revised set of rules were proposed to a) identify the presence of a metal atom and distinguish it from an essential metal that was present as a salt form and b) identify inorganic substances such phosphoric acid. If the excluded substances were reconsidered, then of the 4002 substances, only 639 were true inorganics whereas 2220 substances were actually metal salts where the metal was an essential metal and could therefore be applicable for TTC purposes. The remaining 1143 substances were still not applicable for TTC on account of dioxin like features, alerts for steroids or high potency carcinogens (Table 2 - see code Notebook 03).
Table 2.
Counts following re-profiling through the revised inorganic rules.
| Toxtree exclusions | Number of substances |
|---|---|
| Inorganics | 639 |
| Metal salts | 2220 – should have been considered applicable for TTC |
| Not applicable to TTC | 1143 |
| Yes to Q1 | 900 |
| High Potency Carcinogens (HPC) | 241 |
| Steroids | 2 |
From the Kroes TTC decision tree profiling, 4002 substances were tagged as not appropriate for TTC due to 3 main reasons. Closer examination of these substances found that substances that contained metals as salts were still being flagged as inorganic and excluded from TTC consideration. Substances with P were being erroneously excluded from consideration; thus organophosphates would be assigned as not appropriate for TTC when they might be otherwise filtered into the OPs/carbamates category. Silicon containing substances would not be excluded from consideration despite being noted as not appropriate for TTC based on the EFSA guidance though a recent study has proposed that silicon containing compounds should be included [38].
An important consideration when using the Kroes TTC module within Toxtree therefore is for structural representations to be desalted and neutralised to avoid potential misclassification. QSAR_READY_SMILES as provided in the EPA CompTox Chemicals Dashboard [21] or generated using the OPERA software (https://github.com/kmansouri/OPERA) are two freely available resources that can be utilised to identify SMILES representations more suited to processing through the Kroes module.
Within the Kroes TTC module, a count of all the genotoxicity structural alerts was made to determine whether any of the alerts correspond to a HPC. To better understand how this was implemented in the workflow, the 4002 excluded substances were profiled through the ISS module (specifically the Carcinogenicity (genotox and nongenotox) and mutagenicity rulebase by Istituto Superiore di Sanita (ISS)) within Toxtree to identify what structural alerts were triggered and which of these were associated with alerts characterising HPCs. The output from Toxtree presents the alert id but to link this back to the alert name – a dictionary was created that mapped alert ids to alert names. The output of all alerts fired was cross referenced to this dictionary so that for each substance, a list of which DNA-reactive structural alerts were triggered could be identified.
For the 241 substances assigned as HPCs, 204 substances contained a N-nitroso alert and 8 contained azoxy alerts. Of the remaining substances, one substance did not flag any structural alerts whereas 28 substances fired other DNA reactive structural alerts including aromatic nitro, aliphatic nitro, hydrazine, aromatic diazo, aliphatic halogens, N-methylol derivatives. The module in Toxtree performs a count of the alerts before moving onto the next question and asking whether the chemical is an aflatoxin-like, N-Nitroso or azoxy using generic SMARTS patterns. A closer examination was made to identify how substances were being identified as HPCs. Within the Kroes TTC decision tree, there was no alert for benzidine, aflatoxin-like substances only identified aflatoxin itself and azoxy and N-nitroso flags were represented by generic functional group flags rather than specific structural alerts. A new set of queries was created to better capture the HPC alerts using a custom set of SMARTS patterns – either created manually using the SMARTS.plus resource or extracted from the existing ISS module. Seven SMARTS patterns were created or extracted from existing alerts for ‘aflatoxin-like’ substances, 5 SMARTS patterns were created for Azoxy substances, 2 SMARTS for Nitroso- substances and 1 pattern for Benzi-dine. The SMARTS patterns and the corresponding HPC alert name are available as supplementary information.
The set of 4002 substances tagged as excluded was re-profiled through this revised set of alerts for HPCs. HPC alerts were not flagged for 3708 substances, but alerts were flagged for 284 substances – i. e., more than the initial 241 substances due to their more limited coverage (Table 3, Fig. S2, code Notebook 03).
Table 3.
Reprofiled 4002 excluded substances based on revised HPC alerts.
| HPC Alert Name | Number of substances |
|---|---|
| No alert | 3708 |
| Azoxy or Nitroso | 207 |
| Benzidine | 44 |
| Azoxy | 36 |
| Aflatoxin-like | 6 |
| Nitroso | 1 |
Q1Y also addresses substances that are dioxin-like (DLC) including polyhalogenated – dibenzodioxins, dibenzofurans and biphenyls. The Kroes TTC decision tree module identified these substances on the basis of a structure data file (sdf) comprising dibenzodioxin, dibenzofuran and biphenyl themselves. New SMARTS patterns were constructed to capture these substances in a more generalised manner (Supplementary Table 2) of which 334 substances were found to fire an alert.
The 4002 substances originally tagged as being considered not appropriate for TTC were then re-profiled using the revised HPC alerts, the inorganics filters, and DLC flags. Table 4 summarises the outcomes.
Table 4.
Re-profiled substances that were assigned as not applicable for TTC.
| TTC category | Number of substances |
|---|---|
| Inorganic | 639 |
| HPC | 293 |
| DLC | 334 |
| Steroids | 0 |
| Substances firing 1 or more DNA-reactive structural alerts | 487 |
| Substances that should progress to OP/carbamates and/or Cramer class assignments | 2249 |
5.2. Cohort of Concern based on ISO/TS21726:2019
The ISO/TS21726:2019 describes an extended list of substances that would be tagged as ‘cohort of concern’ (CoC) and hence not appropriate for TTC. In addition to the inorganic substances, steroids, DLCs and HPCs, a number of additional alerts have been proposed.
Examples of proposed CoCs include azo compounds, strained heteronuclear rings, alpha-nitro furyl compounds, hydrazines, triazenes, azides, organophosphorus compounds and polycyclic amines.
A list of 102 CoC were compiled based on DNA alerts proposed in Enoch and Cronin (2010), alerts published on the OCHEM Toxalerts website as well as the revised organophosphate alerts from Nelms et al. (2019) (see supplementary information).
5.3. Extending and revising the genotoxicity structural alerts
Genotoxicity alerts were extracted from Enoch and Cronin (2010), and OCHEM and grouped manually into specific alert classes. There were 207 unique SMARTS patterns associated with 109 alert classes. 34 of the SMARTS could not be resolved by RDKit and were excluded from further consideration. The full set of genetox structural alerts collected are provided in the supplementary information.
5.4. Reprofiling the LRI dataset with the revised workflow
The 45,038 substances were re-profiled using the revised set of alerts and schemes. One substance could not be resolved by RDKit hence was dropped from further consideration. Firstly, the inorganic filters developed were applied to the 45,037 substances to identify whether substances were inorganic and should be excluded from inclusion by the TTC approach.
From this initial profiling, 1012 substances were found to be inorganic and should be excluded from further consideration of the TTC, 2388 were found to be metal salts. These together with the remaining 41,638 substances could be processed further through the remaining TTC workflow. Substances were then processed through the extended CoC filter. The CoC filter identified 5120 substances as possessing 1 or more alerts whereas the remaining 39,917 substances flagged no CoC alerts. Substances were next profiled with the extended collection of structural alerts for genotoxicity which identified 26,600 substances triggering an alert. The outcomes from the various filters and schemes were combined together to assign a final assignment for each substance (see Table 5, code Notebook 04). If a substance was identified as an inorganic substance – the final assignment was tagged as ‘inorganic’. If a substance was not inorganic but flagged a CoC alert, it was tagged as a CoC. If a substance was not inorganic, flagged no CoC alerts but did trigger structural alerts for genotoxicity (including those for OPs and carbamates), it was tagged as presenting an alert. Substances that were not inorganic and did not trigger any structural alerts were assigned as applicable for Cramer structural class consideration. Those substances found to be applicable for Cramer class assignment were then profiled using the original Cramer module within Toxtree.
Table 5.
Re-profiled substances from the LRI dataset.
| TTC category | Number of substances |
|---|---|
| Inorganic | 1012 |
| CoC | 4993 |
| Substances presenting a genotoxicity alert (including carbamates/OPs) | 22,093 |
| Cramer class applicable* | 16,940 |
| Cramer Class III | 11,152 |
| Cramer Class II | 738 |
| Cramer Class I | 5049 |
1 substance not processed by Toxtree’s Cramer class module.
Since the CoC is substantially broader to address the medical device need for additional exclusions, the number of substances not applicable for TTC in this application is far higher than before 5995 vs 4002. With the revised set of genotoxicity structural alerts, the number of substances presenting an alert is also much higher 22,093 vs 9422. As such the number of substances that would proceed to a Cramer class designation is far lower (16,940). The study highlights a need for further evaluation of the genotoxicity structural alerts used within a TTC workflow, harmonising which alerts are used and how specific they need to be i.e., generic DNA-reactive based on chemistry considerations vs supported by empirical genotoxicity data.
5.5. Profiling the ELSIE dataset with the revised workflow
The 416 ELSIE substances for which SMILES could be computed were profiled with the revised workflow developed herein (code Notebook 08). Table 6 lists the counts for the dataset. Over half of the substances in this representative data are applicable to Cramer class assignment.. For the 45 % of substances that are not eligible for assignment of Cramer class values, other approaches such as read-across and QSAR models could be considered to assess these substances.
Table 6.
Reprofiled substances from the ELSIE dataset.
| TTC category | Number of substances |
|---|---|
| Inorganic | 27 |
| CoC | 24 |
| Substances presenting a genotoxicity alert (including carbamates/OPs) | 139 |
| Cramer class applicable | 226 |
| Cramer Class III | 81 |
| Cramer Class II | 11 |
| Cramer Class I | 134 |
The 226 substances that were applicable to Cramer class assignment were merged with the toxicity data already extracted from ToxValDB (see code Notebook 11). As a worst case scenario and to maximise the number of returned toxicity records, all 226 substances were assumed to belong to Cramer Class III. Repeated dose toxicity with reported NOAELs was available for 143 of the 226 substances. Subacute (risk assessment class in ToxValDB was denoted as either subacute, short-term or repeat dose) and subchronic NOAEL values were extrapolated to chronic values using 6 and 2 as assessment factors respectively (per ECHA REACH technical guidance for extrapolating between subacute, subchronic and chronic values). The minimum NOAEL value was then computed on a per chemical basis and this was converted to the log10(NOAEL) equivalent. The 5th percentile of the distribution of log10(NOAEL) values was then calculated and compared with the 5th percentile values reported in Munro et al. (1996). The 5th percentile of the distribution was found to be 0.1697 mg/kg-bw-day (TTC 102 ug/person/day; 1.7 ug/kg-bw-day) which was most comparable to the Cramer Class III 5th percentile (0.15 mg/kg-bw-day) value (TTC 90 ug/person/day; 1.5 ug/kg-bw/day) reported by Munro et al. (1996). The 5th percentile value from Munro et al. (1996) was compared with the 5th percentile derived from the distribution of the log10(NOAEL) values using a bootstrap hypothesis test. In this case the distribution of log10(NOAEL) values was shifted with the 5th percentile from the Munro et al. (1996) study and 1000 bootstrapped replicates were drawn from the difference between the 5th percentiles. The p-value was taken as the sum of the bootstrapped replicates that exceeded the actual observed difference in percentiles. Since the p-value was calculated to be 0.438, it is possible that there is no difference between the 5th percentiles derived from the ToxValDB and Munro datasets and hence their associated TTC values. Bootstrapping the log10(NOAEL), 1000 times resulted in a median TTC percentile of 1.697 ug/kg-bw-day but with a very wide 95th confidence interval [0.25 ug/kg-bw-day, 16.67 ug/kg-bw-day] highlighting the limitations of such a small dataset. There were eight substances found to exceed the TTC threshold proposed based on the ELSIE dataset. These were namely acrylic acid [DTXSID0039229], methacrylic acid [DTXSID3025542], cyclohexanol [DTXSID4021894], dimethyl sulfide [DTXSID9026398], DEHP [DTXSID5020607], styrene [DTXSID2021284], acrylonitrile [DTXSID5020029] and Bisphenol A [DTXSID7020182]. Although these were not CoC substances per se, they might be exempt from the TTC approach. Closer examination of the underlying toxicity data beyond taking the minimum NOAEL as a conservative value would be warranted before excluding them from the TTC approach. Note: A similar investigation was undertaken using the LRI substances for which min log10 (NOAEL) values were extracted for Cramer class applicable substance, 47 of these were found to exceed the TTC ELSIE threshold. The set of 47 substances can be found in the supplementary information.
5.6. Profiling the ELSIE and other datasets using chemical fingerprints: ToxPrints
The four datasets were first profiled using the condensed Level 2 ToxPrints. A countplot of their profile is shown in Fig. 1 (see code Notebook 01). Based on the profiling outcomes, the distribution of the features across the datasets appears quite different. Moreover, there are a number of features present in the other datasets that are not necessarily present in the ELSIE dataset. This is more readily observed in the heatmap in Fig. 2 where the counts (represented as a log10 value) are depicted for each of the datasets. Grey colours in the heatmap are indicative of no data, whereas light to dark colours signify high to low count numbers. For instance, the highest counts are for hetero ring structures, chain:alkaneLinear, bond:CH_amine and bond:COH_alcohol whereas the lowest counts are for substances containing bond:N=[N+]=[N-]_azide and bond:P=C_phosphorane_generic features. Phosphate and sulphate containing substances are absent in the ELSIE dataset as are many Nitrogen containing substances such as triazenes, nitriles, nitro, azide and azo compounds whereas these functional groups are present at lower count levels in the LRI dataset and to a lesser extent in the ToxValDB and Munro datasets. An enrichment analysis using the full set of ToxPrints was conducted to provide greater resolution in terms of which features were more or less represented between the respective datasets.
Fig. 1.
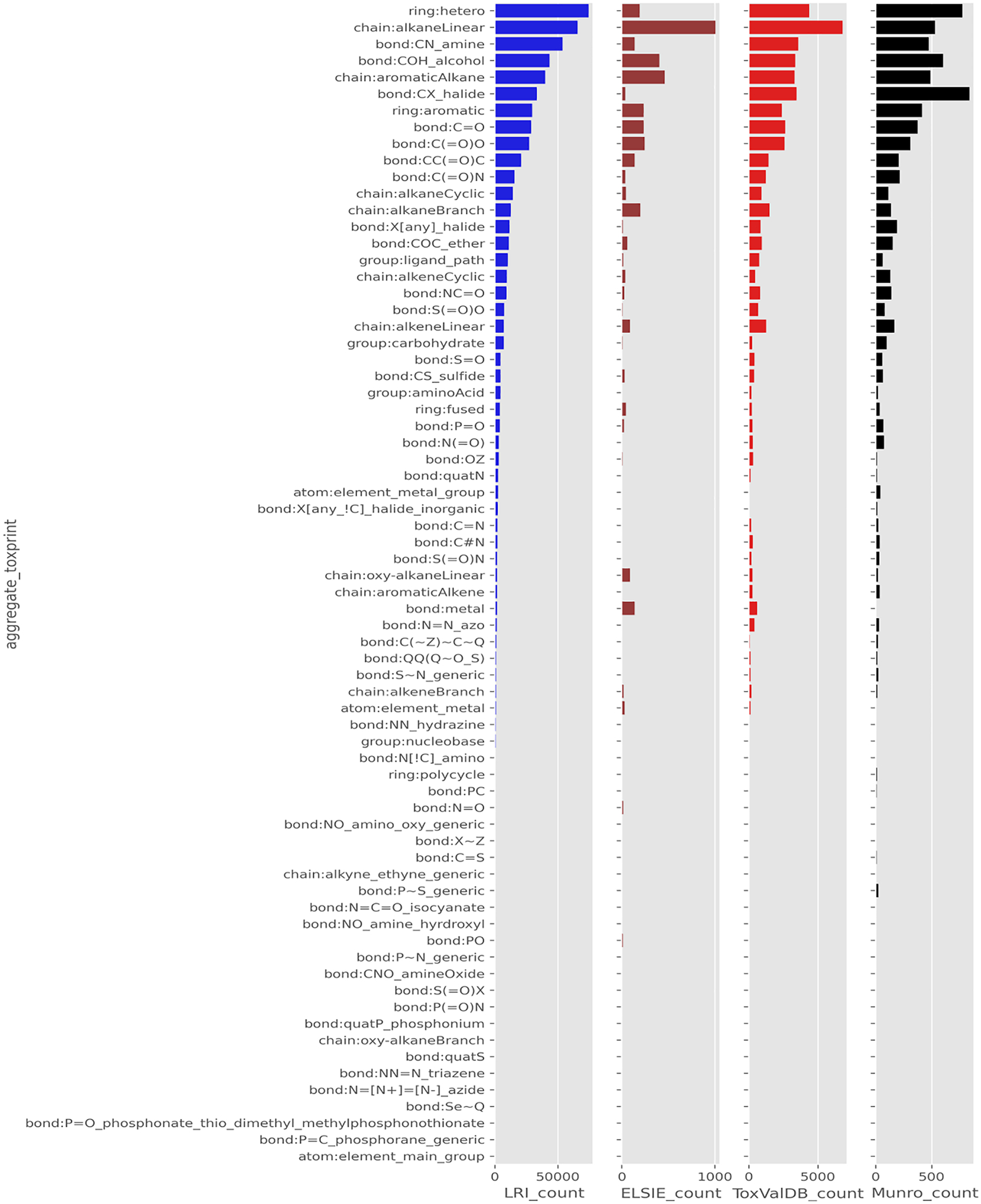
Countplot of Level 2 ToxPrints for the four datasets.
Fig. 2.
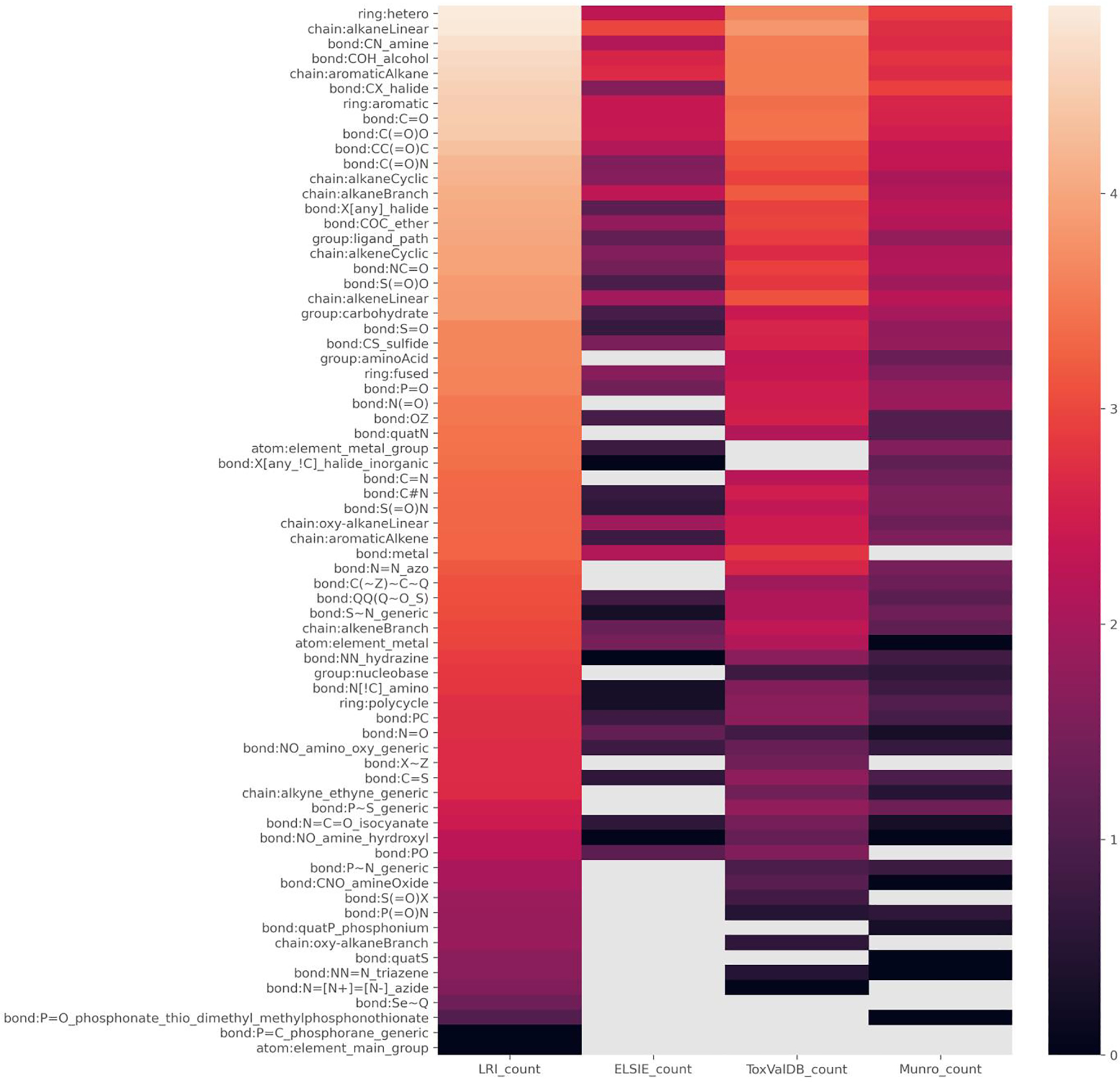
Heatmap of Level 2 ToxPrint counts for the four datasets.
Grey colours are indicative of zero counts whereas dark colours are indicative of low counts.
5.7. Visualisation of landscapes of the datasets through non-linear dimension reduction techniques
Visualisation of the datasets was undertaken using the ToxPrints and projected on 2D scatterplots was conducted using 2 techniques. For the t-SNE approach, a sample of the dataset was used for visualisation purposes where the ratio between membership in one or other dataset was maintained.
The coverage of ELSIE vs LRI datasets using ToxPrints can be seen in Fig. 3 (see code Notebook 01).
Fig. 3.
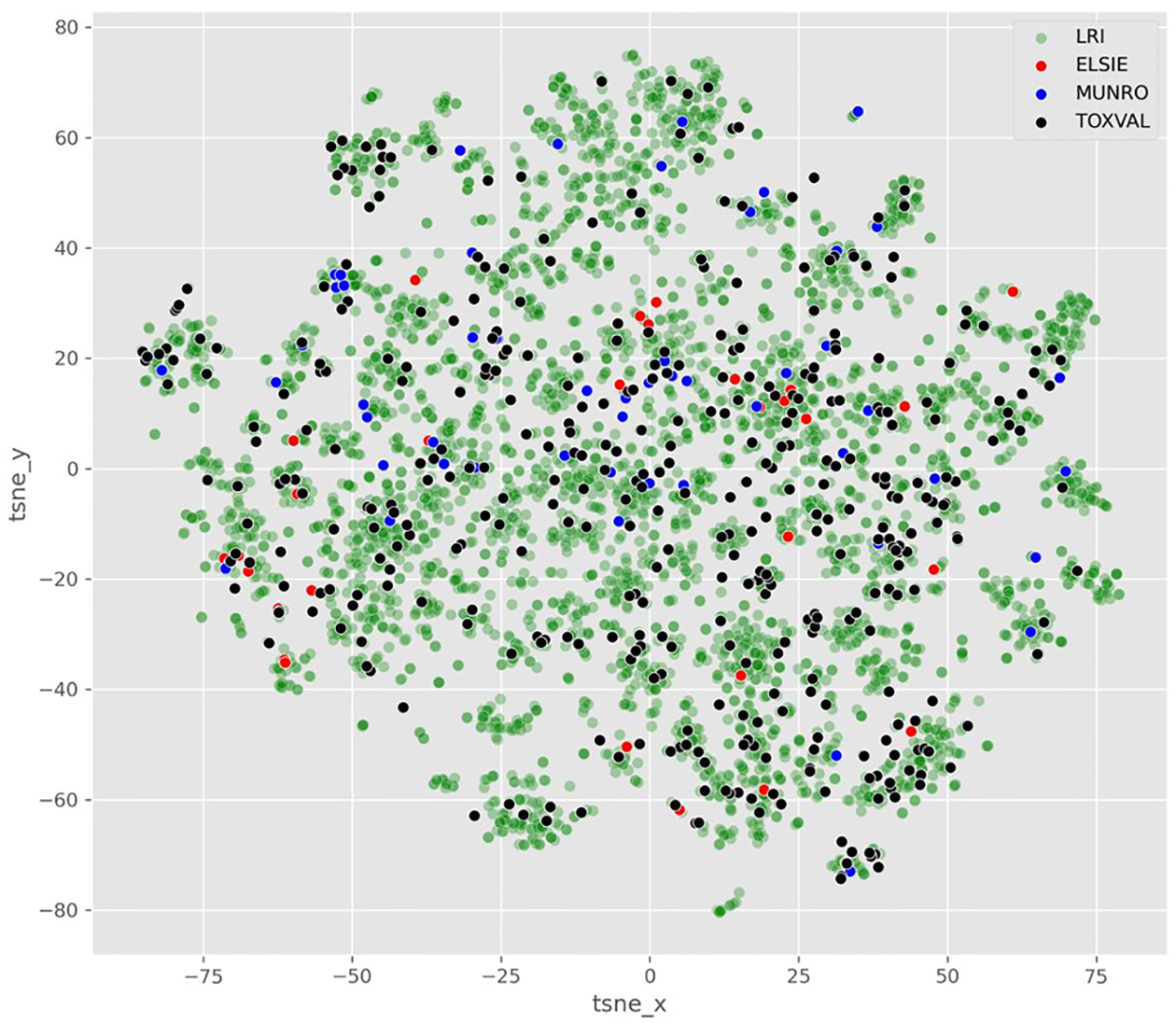
t-SNE 2D scatterplot based on ToxPrint fingerprints colour coded by dataset.
In Fig. 3, it can be seen that the ELSIE substances are clustered within the boundaries of the broader LRI dataset and typically close in space to substances from the other 2 comparator toxicity datasets.
Since t-SNE was only applied to 10 % of the dataset, UMAP, Uniform Manifold Approximation and Projection was used to create a 2D plots with ToxPrints containing the full datasets (Fig. 4) (see code Notebook 02).
Fig. 4.
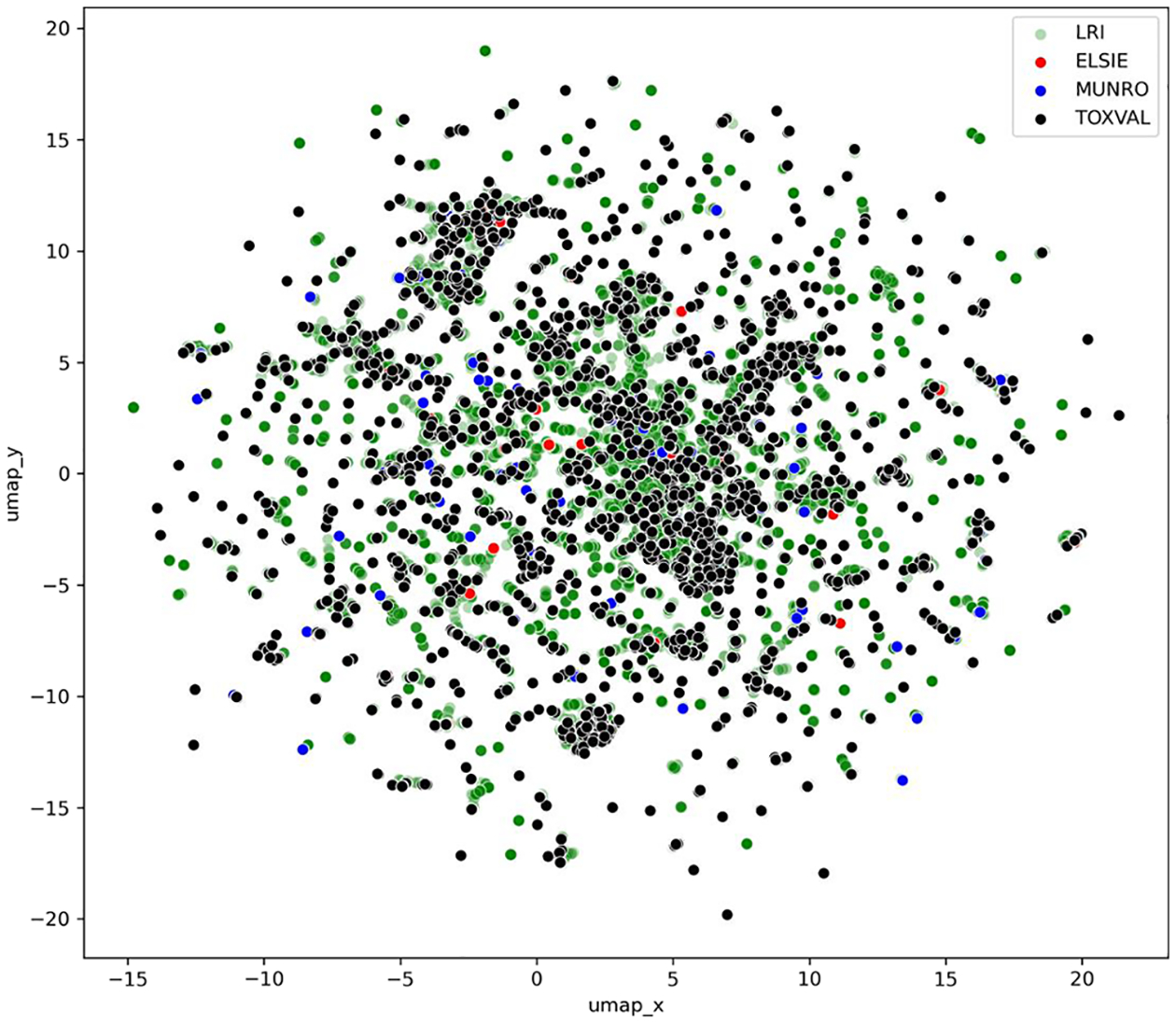
UMAP 2D scatterplot based on ToxPrint fingerprints colour coded by dataset.
The ToxPrints representation shows the ELSIE dataset to be more evenly distributed throughout the LRI landscape.
Overall, based on these representations, the ELSIE substances are not distinct relative to the other datasets. Had there been differences then the ELSIE would be expected to form their own distinct cluster within the overall projection. This is not such a surprise given that there is significant overlap in the substances themselves in this set and the other datasets. After computing InChI Keys and dropping duplicate keys from each dataset, the concatenated dataset comprised 50,382 records for 46,197 unique InChI Keys. There were 47 substances that overlapped across all 4 datasets, 491 that overlapped across 3 datasets (185 were matches between the ELSIE, TOXVAL and LRI datasets and 3 were matches between the ELSIE, MUNRO and LRI datasets) and 3062 that overlapped across 2 datasets (108 substances included matches with ELSIE). Overall, only 71 substances were distinct to the ELSIE dataset. Exploring the predicted LogKow values and Molecular weights across the ELSIE set relative to these 71 (LogKow could only be predicted for 63 substances) also showed no apparent differences (Fig. 5).
Fig. 5.
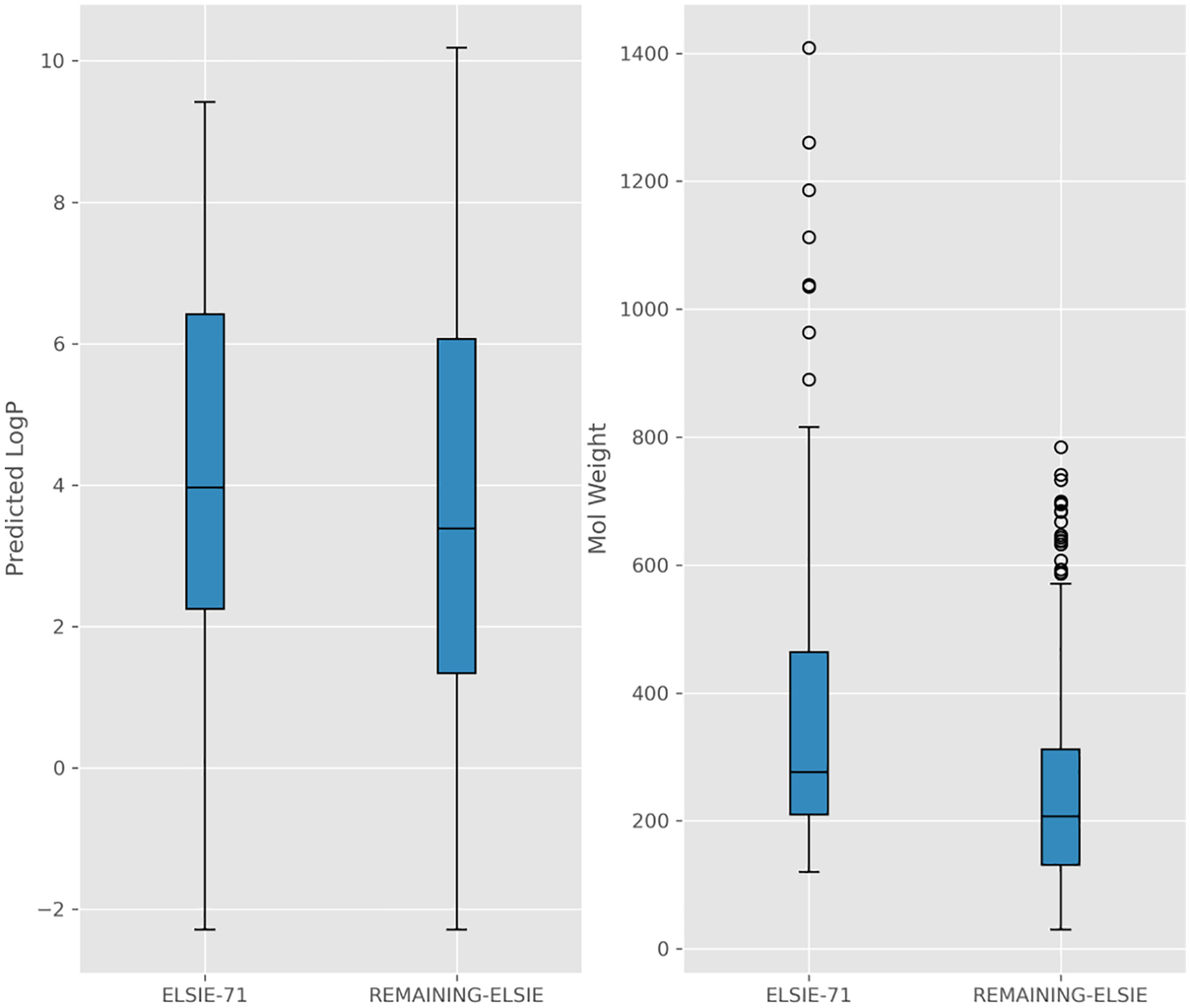
Boxplots for the ELSIE datasets showing the distribution of parameters for the unique substances relative to the remaining set.
5.8. ToxPrint enrichment analysis
The four datasets were concatenated together as one single data-frame comprising 50,291 substances characterised by 729 ToxPrints. A final column provided a tag of membership in one or other dataset. The tag column was converted into a dummy variable, such that presence in the ELSIE dataset was denoted with a ‘1’ and presence in the other datasets was denoted by ‘0’. An enrichment analysis was performed to identify the most enriched ToxPrints in the ELSIE dataset relative to the LRI/Munro/ToxValDB dataset. A Fisher exact test was performed to compute the odds ratio statistic and its associated p-value. There were 31 enriched ToxPrints for the ELSIE dataset.
The enriched ELSIE ToxPrints provide more resolution to explain the differences between the inventories namely that there were a number of silicon containing substances (cyclic siloxanes such as hexamethylcy-clotrisiloxane DTXSID6027185), long chain fatty carboxylates (e.g. oleic acid DTXSID1025809) as well as fused aromatic structures such as benzo (a)pyrene (DTXSID2020139) in this set that were not significantly present in the remaining datasets. The top 3 enriched ToxPrints were ring: fused_PAH_acenaphthylene, ring:fused_PAH_pyrene and bond:metal_metalloid_alkylSiloxane. The full set of 31 enriched ToxPrints are provided in the supplementary files (see code Notebook 02).
6. Conclusion
This study was focused on three main objectives. The first was to evaluate the exclusions implemented in the Kroes Toxtree module. A large inventory of ~45,000 substances (termed the LRI dataset) was profiled through the Kroes TTC decision module within Toxtree to assign substances into their respective non-cancer TTC categories. Closer examination of the 4002 substances found to be not applicable for the TTC approach uncovered several issues. Substances represented as their salt forms were automatically assigned as not appropriate for TTC when many of these contained essential metals as counter ions and would be considered applicable for TTC. Structural representations should be presented as their desalted equivalents to avoid this error when using the Kroes module within Toxtree. High Potency Carcinogens and dioxin-like substances were not entirely captured based on the rules implemented in the software. Refinements were made to the exclusion rules to address these issues.
The second objective of the study sought understand the structural similarity between chemical inventories and medical device constituents, prior to extending the refined exclusion rules to address these constituents in the final objective. Our comparison of the coverage and relevance of the LRI dataset coupled with 2 toxicity datasets against the ELSIE dataset showed an overall reasonable overlap in terms of the actual substances, as well as more generally when comparing their ToxPrint fingerprints in a 2D UMAP projection. However, profiling on the basis of Level 2 ToxPrint showed that there were some differences in the structural features between the datasets which were further explored by performing an enrichment analysis. The main difference in the datasets were that siloxanes, fused polyaromatic substances and long chain fatty acid carboxylates were significantly enriched in the ELSIE dataset relative to the remaining datasets.
A third and final objective of the study was profiling the ELSIE dataset through the refined workflow which resulted in 226 substances denoted as Cramer class applicable for TTC. Merging these substances with repeated dose toxicity studies extracted from the ToxValDB identified relevant studies for 143 substances from which a TTC value was derived which was comparable to the Cramer class III TTC value reported by Munro et al. (1996). Considering the chemical similarity of the ELSIE substances to those in the broader dataset in concert with deriving a very similar TTC value to the Cramer Class III threshold provides greater confidence in the use of TTC for these medical device substances.
In summary, this study demonstrated the importance of evaluating the software implementation of established workflows, identified and addressed certain limitations in the TTC workflow, as well as extended the approach for medical device constituents.
Supplementary Material
Acknowledgments
This project was supported in part by an appointment to the Research Participation Program at the Center for Computational Toxicology and Exposure, US Environmental Protection Agency, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and EPA.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Disclaimer
The findings and conclusions in this article have not been formally disseminated by the US Food & Drug Administration and the U.S. Environmental Protection Agency and they should not be construed to represent any Agency determination or policy. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services, US Food & Drug Administration or the U.S. Environmental Protection Agency.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.comtox.2022.100246 and https://doi.org/10.23645/epacomptox.21215354.
References
- [1].EFSA ES; More SJ; Bampidis V; Benford D; Bragard C; Halldorsson TI; Hernández-Jerez AF; Hougaard Bennekou S; Koutsoumanis KP; Machera K; Naegeli H; Nielsen SS; Schlatter JR; Schrenk D; Silano V; Turck D; Younes M; Gundert-Remy U; Kass GEN; Kleiner J; Rossi AM; Serafimova R; Reilly L; Wallace HM Guidance on the use of the threshold of toxicological concern approach in food safety assessment. EFSA J 2019, 17 (6), e05708. 10.2903/j.efsa.2019.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kroes R, Renwick AG, Cheeseman M, Kleiner J, Mangelsdorf I, Piersma A, Schilter B, Schlatter J, van Schothorst F, Vos JG, Würtzen G, European branch of the international life sciences institute. Structure-based thresholds of toxicological concern (TTC): guidance for application to substances present at low levels in the diet, Food Chem. Toxicol 42 (1) (2004) 65–83, 10.1016/j.fct.2003.08.006. [DOI] [PubMed] [Google Scholar]
- [3].Rulis AM, De Minimis and the threshold of regulation. In Food Protection Technology; Michigan, 1986, pp. 29–37. [Google Scholar]
- [4].Gold LS, Sawyer CB, Magaw R, Backman GM, De Veciana M, Levinson R, Hooper NK, Havender WR, Bernstein L, Peto R, Pike MC, Ames BN,A carcinogenic potency database of the standardized results of animal bioassays, Environ. Health Perspect 58 (1984) 9–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cramer GM, Ford RA, Hall RL, Estimation of toxic hazard–a decision tree approach, Food Cosmet. Toxicol 16 (3) (1978) 255–276, 10.1016/s0015-6264(76)80522-6. [DOI] [PubMed] [Google Scholar]
- [6].Munro IC, Ford RA, Kennepohl E, Sprenger JG, Correlation of structural class with no-observed-effect levels: a proposal for establishing a threshold of concern, Food Chem. Toxicol 34 (9) (1996) 829–867, 10.1016/s0278-6915(96)00049-x. [DOI] [PubMed] [Google Scholar]
- [7].Yang C, Barlow SM, Muldoon Jacobs KL, Vitcheva V, Boobis AR, Felter SP, Arvidson KB, Keller D, Cronin MTD, Enoch S, Worth A, Hollnagel HM, Thresholds of toxicological concern for cosmetics-related substances: new database, thresholds, and enrichment of chemical space, Food Chem. Toxicol 109 (Pt 1) (2017) 170–193, 10.1016/j.fct.2017.08.043. [DOI] [PubMed] [Google Scholar]
- [8].Patel A, Joshi K, Rose J, Laufersweiler M, Felter SP, Api AM, Bolstering the existing database supporting the non-cancer threshold of toxicological concern values with toxicity data on fragrance-related materials, Regul. Toxicol. Pharm 116 (2020), 104718, 10.1016/j.yrtph.2020.104718. [DOI] [PubMed] [Google Scholar]
- [9].Czaja K, Struciński P, Korcz W, Minorczyk M, Hernik A, Wiadrowska B, Alternative toxicological methods for establishing residue definitions applied for dietary risk assessment of pesticides in the European Union, Food Chem. Toxicol 137 (2020), 111120, 10.1016/j.fct.2020.111120. [DOI] [PubMed] [Google Scholar]
- [10].Feigenbaum A, Pinalli R, Giannetto M, Barlow S, Reliability of the TTC approach: learning from inclusion of pesticide active substances in the supporting database, Food Chem. Toxicol 75 (2015) 24–38, 10.1016/j.fct.2014.10.016. [DOI] [PubMed] [Google Scholar]
- [11].Melching-Kollmuss S, Dekant W, Kalberlah F, Application of the “threshold of toxicological concern” to derive tolerable concentrations of “non-relevant metabolites” formed from plant protection products in ground and drinking water, Regul. Toxicol. Pharm 56 (2) (2010) 126–134, 10.1016/j.yrtph.2009.09.011. [DOI] [PubMed] [Google Scholar]
- [12].Yang C, Cheeseman M, Rathman J, Mostrag A, Skoulis N, Vitcheva V, Goldberg S, A new paradigm in threshold of toxicological concern based on chemoinformatics analysis of a highly curated database enriched with antimicrobials, Food Chem. Toxicol 143 (2020), 111561, 10.1016/j.fct.2020.111561. [DOI] [PubMed] [Google Scholar]
- [13].Delaney EJ, An impact analysis of the application of the threshold of toxicological concern concept to pharmaceuticals, Regul. Toxicol. Pharm 49 (2) (2007) 107–124, 10.1016/j.yrtph.2007.06.008. [DOI] [PubMed] [Google Scholar]
- [14].Müller L, Mauthe RJ, Riley CM, Andino MM, Antonis DD, Beels C, DeGeorge J, De Knaep AGM, Ellison D, Fagerland JA, Frank R, Fritschel B, Galloway S, Harpur E, Humfrey CDN, Jacks AS, Jagota N, Mackinnon J, Mohan G, Ness DK, O’Donovan MR, Smith MD, Vudathala G, Yotti L, A rationale for determining, testing, and controlling specific impurities in pharmaceuticals that possess potential for genotoxicity, Regul. Toxicol. Pharm 44 (3) (2006) 198–211, 10.1016/j.yrtph.2005.12.001. [DOI] [PubMed] [Google Scholar]
- [15].Patlewicz G, Wambaugh JF, Felter SP, Simon TW, Becker RA, Utilizing Threshold of toxicological concern (TTC) with high throughput exposure predictions (HTE) as a risk-based prioritization approach for thousands of chemicals, Comput. Toxicol 7 (2018) 58–67, 10.1016/j.comtox.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].US EPA. A Proof-of-Concept Case Study Integrating Publicly Available Information to Screen Candidates for Chemical Prioritization under TSCA EPA/600/R-21–106 2021. 10.23645/epacomptox.14878125. [DOI] [PubMed] [Google Scholar]
- [17].Nelms MD, Pradeep P, Patlewicz G, Evaluating potential refinements to existing threshold of toxicological concern (TTC) values for environmentally-relevant compounds, Regul. Toxicol. Pharm 109 (2019), 104505, 10.1016/j.yrtph.2019.104505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].EFSA/WHO report makes recommendations on Threshold of Toxicological Concern approach | EFSA https://www.efsa.europa.eu/en/press/news/160310-0 (accessed 2021-10-14).
- [19].Masuda-Herrera MJ; Bercu JP; Broschard TH; Burild A; Hasselgren C; Parris P; Ford LC; Graham J; Stanard B; Comerford M; Lettiere D; Erler S; Callis CM; Morinello E; Muster W; Martin EA; Griffin TR; Nagao L; Cruz M Development of Duration-Based Non-Mutagenic Thresholds of Toxicological Concern (TTC) Relevant to Parenteral Extractables and Leachables (E&Ls). PDA J Pharm Sci Technol 2022, pdajpst.2021.012693. 10.5731/pdajpst.2021.012693. [DOI] [PubMed] [Google Scholar]
- [20].Nicolas CI, Bronson K, Pendse SN, Efremenko A, Fitzpatrick JM, Minto MS, Mansouri K, Yoon M, Phillips MB, Clewell RA, Andersen ME, Clewell HJ, McMullen PD, The TTC data mart: an interactive browser for threshold of toxicological concern calculations, Comput. Toxicol 15 (2020), 100128, 10.1016/j.comtox.2020.100128. [DOI] [Google Scholar]
- [21].Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, Patlewicz G, Shah I, Wambaugh JF, Judson RS, Richard AM, The CompTox chemistry dashboard: a community data resource for environmental chemistry, J Cheminform 9 (1) (2017) 61, 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yang C, Tarkhov A, Marusczyk J, Bienfait B, Gasteiger J, Kleinoeder T, Magdziarz T, Sacher O, Schwab CH, Schwoebel J, Terfloth L, Arvidson K, Richard A, Worth A, Rathman J, New publicly available chemical query language, CSRML, to support chemotype representations for application to data mining and modeling, J. Chem. Inf. Model 55 (3) (2015) 510–528, 10.1021/ci500667v. [DOI] [PubMed] [Google Scholar]
- [23].Landrum G, RDKit: Open-Source Cheminformatics; http://Www.Rdkit.Org.
- [24].van er Maaten L, Hinton G, Visualizing data using T-SNE. J. Mach. Learn. Res 2018, 8, 2579–2605. [Google Scholar]
- [25].Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res https://dl.acm.org/doi/10.5555/1953048.2078195 (accessed 2022-06-14). [Google Scholar]
- [26].McInnes L; Healy J; Melville J UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv:1802.03426 [cs, stat] 2020.
- [27].Enoch SJ, Cronin MTD, A review of the electrophilic reaction chemistry involved in covalent DNA binding, Crit. Rev. Toxicol 40 (8) (2010) 728–748, 10.3109/10408444.2010.494175. [DOI] [PubMed] [Google Scholar]
- [28].Sushko I, Salmina E, Potemkin VA, Poda G, Tetko IV, ToxAlerts: a web server of structural alerts for toxic chemicals and compounds with potential adverse reactions, J. Chem. Inf Model 52 (8) (2012) 2310–2316, 10.1021/ci300245q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Schomburg K, Ehrlich H-C, Stierand K, Rarey M, From structure diagrams to visual chemical patterns, J. Chem. Inf. Model 50 (9) (2010) 1529–1535, 10.1021/ci100209a. [DOI] [PubMed] [Google Scholar]
- [30].Patlewicz G, Jeliazkova N, Safford RJ, Worth AP, Aleksiev B, An evaluation of the implementation of the Cramer classification scheme in the Toxtree software, SAR QSAR Environ. Res 19 (5–6) (2008) 495–524, 10.1080/10629360802083871. [DOI] [PubMed] [Google Scholar]
- [31].McKinney W, Data Structures for Statistical Computing in Python. Proceedings of the 9th Python in Science Conference 2010, 56–61. 10.25080/Majora-92bf1922-00a. [DOI] [Google Scholar]
- [32].Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, Smith NJ, Kern R, Picus M, Hoyer S, van Kerkwijk MH, Brett M, Haldane A, del Río JF, Wiebe M, Peterson P, Gérard-Marchant P, Sheppard K, Reddy T, Weckesser W, Abbasi H, Gohlke C, Oliphant TE, Array programming with NumPy, Nature 585 (7825) (2020) 357–362, 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hunter JD, Matplotlib: a 2D graphics environment, Comput. Sci. Eng 9 (3) (2007) 90–95, 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- [34].Waskom ML, Seaborn: Statistical Data Visualization. Journal of Open Source Software 2021, 6 (60), 3021. 10.21105/joss.03021. [DOI] [Google Scholar]
- [35].Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, Brett M, Wilson J, Millman KJ, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat İ, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, van Mulbregt P, SciPy 1.0: Fundamental algorithms for scientific computing in Python, Nat. Methods 17 (3) (2020) 261–272, 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kluyver T; Ragan-Kelley B; Pérez F; Granger B; Bussonnier M; Frederic J; Kelley K; Hamrick J; Grout J; Corlay S; Ivanov P; Avila D; Abdalla S; Willing C; Jupyter development team. Jupyter Notebooks – a Publishing Format for Reproducible Computational Workflows; Loizides F, Scmidt B, Eds.; IOS Press, 2016; pp 87–90. 10.3233/978-1-61499-649-1-87. [DOI] [Google Scholar]
- [37].Zoroddu MA, Aaseth J, Crisponi G, Medici S, Peana M, Nurchi VM, The essential metals for humans: a brief overview, J. Inorg. Biochem 195 (2019) 120–129, 10.1016/j.jinorgbio.2019.03.013. [DOI] [PubMed] [Google Scholar]
- [38].Schmitt BG, Jensen E, Laufersweiler MC, Rose JL, Threshold of toxicological concern: extending the chemical space by inclusion of a highly curated dataset for organosilicon compounds, Regul. Toxicol. Pharm 127 (2021), 105074, 10.1016/j.yrtph.2021.105074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


