Abstract
Allele-specific amplification (ASA) is a generally applicable technique for the detection of known single nucleotide polymorphisms (SNPs), deletions, insertions and other sequence variations. Conventionally, two reactions are required to determine the zygosity of DNA in a two-allele system, along with significant upstream optimisation to define the specific test conditions. Here, we combine single tube bi-directional ASA with a ‘matrix-based’ optimisation strategy, speeding up the whole process in a reduced reaction set. We use sickle cell anaemia as our model SNP system, a genetic disease that is currently screened using ASA methods. Discriminatory conditions were rapidly optimised enabling the unambiguous identification of DNA from homozygous sickle cell patients (HbS/S), heterozygous carriers (HbA/S) or normal DNA in a single tube. Simple downstream mathematical analyses based on product yield across the optimisation set allow an insight into the important aspects of priming competition and component interactions in this competitive PCR. This strategy can be applied to any polymorphism, defining specific conditions using a multifactorial approach. The inherent simplicity and low cost of this PCR-based method validates bi-directional ASA as an effective tool in future clinical screening and pharmacogenomic research where more expensive fluorescence-based approaches may not be desirable.
INTRODUCTION
The successful implementation of single nucleotide polymorphism (SNP) screening for clinical research and the administration of medicine tailored to an individual’s genotype, so-called ‘personalised medicine’, lies in the ability to deliver highly reliable, accurate and inexpensive assays that can discriminate between key DNA polymorphisms. Biotechnology and pharmaceutical companies have addressed this challenge with the development of methodologies including single-strand conformation polymorphism analysis, heteroduplex analysis and denaturing gel electrophoresis. Each of these techniques relies on conformation-induced gel mobility changes (1). Simple homogeneous assays based on fluorescence resonance energy transfer, mass spectrometry or direct sequencing of PCR products have also been shown to be effective tools for SNP detection (2). Implementation of these methods can require specific reagent synthesis and downstream purification, making them complicated and expensive.
However, assays based on PCR for SNP diagnosis have broad potential in clinical diagnostics because of their inherent simplicity and potential low cost. Established PCR-based methodologies include certain ligation assays (3,4), genetic bit analysis (5), arrayed primer extension (6) and restriction fragment length polymorphisms (7), and use of sequence-specific fluorescent probes (8).
PCR has also been adapted for the detection of well-characterised SNPs using allele-specific amplification (ASA); oligonucleotides complementary to a given DNA sequence except for a mismatch at their 3′-hydroxyl residue will not function as primers in the PCR under appropriate conditions. A typical ASA test consists of two complementary reactions, each containing a common primer, an allele-specific primer and Taq DNA polymerase lacking 3′→5′ proofreading activity. The first reaction contains a primer specific for the normal (or wild-type) DNA sequence and refractory to amplification from mutant DNA at a given locus. Similarly, the second reaction contains a mutant-specific primer unable to amplify wild-type DNA. Molecular conformation is achieved by analysis of the resulting PCR amplicon profiles. A normal individual will generate product in the first reaction only; a heterozygote amplifies products in both reactions; and a homozygous mutant individual does so only in the mutant-specific reaction. Internal control amplification is necessary, providing a positive control for the PCR test (9).
More recently, single tube adaptations have been introduced for known SNP detection in a two-allele system including competitive oligonucleotide priming(10), multi-coloured fluorescent oligo labelling systems (2), tetra-primer PCR (11) and overlapping PCR strategies (12,13). The latter both use bi-directional primer arrangements in which common ‘outer’ primers define the size of each allele-specific fragment allowing simple identification using electrophoretic methods.
Maintaining the integrity of all ASA techniques, particularly for diagnostic application, requires the optimisation of many experimental parameters. The literature suggests that a magnesium and oligonucleotide titration is sufficient to obtain discriminatory conditions. Generally, further mismatches are incorporated at the 3′-end of the allele-specific primer to weaken hydrogen bonding between the primer and template, increasing the likelihood of discrimination (14). However, these conventional approaches are simplistic considering the complex component interactions in a PCR reaction and prove time consuming and expensive. This has limited the applicability of PCR-based SNP diagnosis for routine clinical application. In particular, bi-directional systems require significant optimisation due the effects of primer competition caused by multiple primer sets in the PCR amplification.
Here, we modify current systems at a number of levels with the application of a multifactorial optimisation procedure to single tube bi-directional ASA. Our strategy enables the rapid identification of component ratios favouring ASA in a greatly reduced experiment set using unmodified oligonucleotides. This low cost optimisation strategy combined with a single tube genotyping system introduces improvements to PCR-based SNP screening.
SNP model
We demonstrate our system by detecting the single base pair mutation that causes the autosomal recessive disease sickle cell anaemia (GenBank accession no. M34058). A single A→T transversion in the sequence encoding codon 6 of the human β-globin gene causes the substitution of amino acid glutamine by valine, forming a mutant globin chain termed HbS. Haemoglobin S is freely soluble when fully oxygenated, but under conditions of low oxygen tension the erythrocytes assume an irregular ‘sickle’ shape, leading to aggregation and haemolysis. The sickled erythrocytes become trapped in the microcirculation, depriving organs of essential oxygen, causing pain and chronic anaemia (15). Homozygous HbS is a serious haemoglobinopathy found almost exclusively in the Black population. About 8% of American Blacks are carriers and ∼0.2% are affected. In the UK sickle cell disease affects ∼10 000 people (actionresearch.co.uk/camp/sick.html). Heterozygotes (sickle cell carriers) are clinically normal, although their red cells will sickle when subjected to very low oxygen pressure in vitro (genelink.com/technical/sickle.html). PCR-based techniques for prenatal diagnosis of sickle cell disease were introduced in 1989 following the invention of the amplification refractory mutation system (9) and are still the method of choice in clinical research laboratories (16). A single tube adaptation of conventional ASA would add significant benefits to routine testing for sickle cell disease.
MATERIALS AND METHODS
Reagents
PCR reagents (Taq DNA polymerase, HS TaQUANT-OFF, 10× Mg-free Taq buffer, 25 mM MgCl2) were obtained from Q-Biogene (CA), except for dNTPs (2 mM each dATP, dCTP, dGTP and dTTP), which were from Promega (UK). Primer selection was accomplished using the PrimerCalc© software package (Q-Biogene). For standard gel electrophoresis, molecular biology grade agarose was used (Q-Biogene) in 0.5× TBE buffer (45 mM Tris, 45 mM borate, 1 mM Na2EDTA, pH 8.3). Double deionised water (18 MΩ) was used for all buffers and solutions for electrophoresis and PCR amplifications. Homozygous HbS human genomic DNA and heterozygous HbA/S DNA were a generous gift from the National Haemoglobinopathies Reference Service (Oxford, UK).
Bi-directional allele-specific PCR amplifications
WT-AS (5′-ATG GTG CAC CTG ACT CCT GA-3′) and WT-CP517 (5′-CCC CTT CCT ATG ACA TGA ACT-3′) were designed for amplification of a 517 bp fragment from the normal β-globin gene (wild-type primer set). MUT-AS (5′-CAG TAA CGG CAG ACT TCT CCA-3′) and MUT-CP267 (5′-GGG TTT GAA GTC CAA CTC CTA-3′) were designed for amplification of a 267 bp fragment from homozygous mutant DNA (HbS/S) conferring sickle cell disease (mutant primer set). All reactions were performed in a volume of 25 µl containing 10 ng total human genomic DNA template, 10× PCR reaction buffer, 1 U µl–1 Taq DNA polymerase and water. The wild-type primer set, mutant primer set, MgCl2 and dNTP concentrations are shown in Table 1 and were aliquoted according to an orthogonal array (Table 2) (17). Amplification was performed using an MJ Thermal Cycler (GRI, UK) through an initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 30 s, primer annealing at 60°C for 30 s and extension at 72°C for 35 s. A small blind trial was performed in which 14 human genomic DNA samples of mixed zygosity were used as substrate DNA under discriminatory conditions that were considered optimal (reaction 9, Tables 1 and 2). The exact template concentration of each sample was not specified, varying between 5 and 25 ng µl–1.
Table 1. Concentration levels of components used in a multifactorial array for the identification of discriminatory reaction conditions for bi-directional ASA.
| Level | A | B | C |
|---|---|---|---|
| WT-AS/WT-CP517 (µM) | 0.2 | 0.4 | 0.8 |
| MUT-AS/MUT-CP267 (µM) | 0.2 | 0.4 | 0.8 |
| MgCl2 (mM) | 0.5 | 1 | 2 |
| dNTPs (mM) | 80 | 120 | 200 |
Table 2. An orthogonal array for testing four reaction parameters, each at three different levels.
| Reaction | WT-AS/WT-CP517 | MUT-AS/MUT-CP267 | MgCl2 | dNTPs |
|---|---|---|---|---|
| 1 | A | A | A | A |
| 2 | A | B | B | B |
| 3 | A | C | C | C |
| 4 | B | A | B | C |
| 5 | B | B | C | A |
| 6 | B | C | A | B |
| 7 | C | A | C | B |
| 8 | C | B | A | C |
| 9 | C | C | B | A |
Analysis of PCR products
An aliquot (10 µl) of each PCR product was loaded with 6× loading buffer (Promega) onto a 2% agarose gel and run in 0.5× TBE buffer at 30 mA constant current. Amplification products were visualised by UV transillumination (305 nM) after staining with SYBR Gold (Molecular Probes, UK) for 30 min. The agarose gels were photographed and analysed by gel imaging software to obtain maximum band optical density measurements as a representation or ‘score’ of specific product yield.
RESULTS
Experimental design
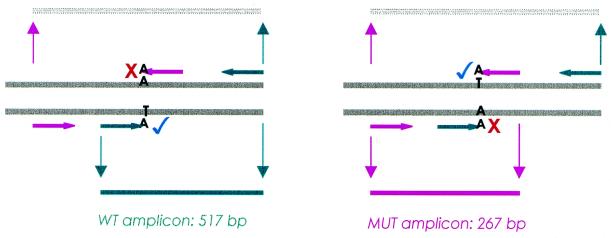
A schematic of bi-directional ASA is shown in Figure 1. A wild-type allele-specific primer is designed as complementary to the non-coding strand of the wild-type allele terminating at the known polymorphic site. This primer will be extended in the presence of wild-type DNA only under appropriate PCR conditions. A further allele-specific primer is included in the reaction, designed as complementary to the coding strand of the known mutant allele, again terminating at the SNP site. This primer will be extended in the presence of the mutant allele only, as it is refractory to extension of wild-type DNA due to a single base pair mismatch at the 3′-end. Two ‘outer primers’ are placed on the opposite strand at a pre-defined number of bases from their allele-specific pair in order that molecular confirmation can be achieved by analysis of the resulting amplicon profiles using gel-based methods. A difference of at least 100 bp is preferable to increase electrophoretic separation.
Figure 1.
Schematic representation of bi-directional allele-specific PCR. The wild-type primer set is shown in green and the mutant primer set in pink. Three possible fragments can be formed depending upon substrate DNA zygosity, as well as enzyme system and cycling parameters. The green amplicon defines the allele-specific product formed when only the wild-type primers are extended and the pink amplicon defines the allele-specific product formed when only the mutant primers are extended. A heterozygote is identified by the presence of both PCR products. The target region amplified by non-allele-specific outer primers is shown in pale grey. Our selected enzyme system was inefficient at amplifying longer sequences and in the presence of competition from a smaller fragment insufficient target region was amplified for visualisation on a stained gel.
Taq DNA polymerase will extend both outer primers in all reactions as they are non-allele-specific, but the yield of the large amplicon spanning the whole target region is subject to variation according to the enzyme system and cycling parameters employed. The HotStart enzyme system used in this study was less efficient at amplifying larger fragments and in the presence of competition from a smaller fragment did not amplify sufficient DNA for the larger band spanning the whole target region to be visualised on an agarose gel.
We designed two 20/21mer allele-specific primers, WT-AS and MUT-AS, complementary to the normal β-globin or sickle cell genes, respectively, arranged in a bi-directional orientation such that both primers terminate at the mutation site. Primer WT-AS has a single A nucleotide at the 3′-end, matching the T nucleotide on the non-coding strand of the normal β-globin gene. Primer MUT-AS has a single A nucleotide at the 3′-end, matching the T base on the coding strand of the sickle cell gene. The wild-type outer primer (WT-CP517) was positioned 517 bp downstream on the opposite strand of WT-AS. The mutant outer primer (MUT-CP267) was placed 267 bp upstream of MUT-AS. Unlike normal ASA, this assay has an inherent PCR control, as at least one allele-specific fragment should be yielded per reaction under optimised conditions. This negates the need for a distant pair of primers to direct the amplification of an internal control fragment.
Optimisation strategy for discrimination between the normal and sickle cell alleles
Inclusion of multiple allele-specific primer sets in a single reaction tube requires the consideration of many experimental factors. For typical multiplex reactions, the key parameter for optimisation is primer concentration. At equimolar concentrations, amplification by the two primer sets may be weak or strong at a given locus, an event dependent upon primer stabilities, binding efficiencies, priming competition, product size and concentration of other reaction components (18). Normally, primer ratios are individually optimised for each reaction, however, an adjustment in primer concentration has associated implications considering the complex interactions between other components in a PCR. Unique to this assay, each primer is complementary to a different part of the DNA sequence, introducing additional variability to the reaction.
MgCl2 and dNTPs have been shown to affect the efficiency of priming and extension by altering the kinetics of association and dissociation of primer–template duplexes at annealing and extension temperatures. These components also alter the efficiency with which the polymerase recognises and extends such duplexes. Of particular relevance to allele-specific PCR methods, the concentration of MgCl2 and dNTPs required for optimal amplification depends largely on the target and primer sequences. The presence of excess magnesium in a reaction may result in the accumulation of non-specific amplification products and insufficient concentrations reduce product yield. It has also been demonstrated that dNTPs quantitatively bind Mg2+, so that any modification of dNTP concentration will require a compensatory adjustment of MgCl2 (17). Additionally, Taq polymerase requires free Mg2+ as a cofactor so any excess of dNTPS can have a detrimental effect on product yield by chelating cofactor ions (19). Standard approaches to multiplex or allele-specific PCR optimisation are unable to account for these multiple interactions, as they are largely based on trial-and-error strategies.
We applied a multifactorial optimisation strategy in which optimal component concentrations for allele discrimination were identified in a single trial with few reactions. The wild-type primer set, mutant primer set, MgCl2 and dNTPs were selected as four critical parameters, the interactions of which are most likely to affect the outcome of bi-directional PCR. The concentrations of these components were varied according to an orthogonal array and differences in product yield across the reaction set were used for two purposes, qualitatively in the identification of diagnostic reaction conditions and quantitatively to study the individual effect of each parameter on the assay.
Optimal conditions for zygosity determination
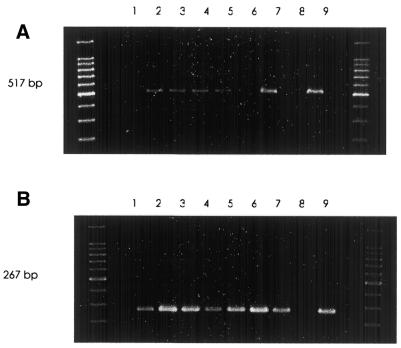
Figure 2 shows gel image data from reaction sets A and B. Both sets are identical in component ratios and cycling conditions, containing normal and HbS/S DNA templates, respectively. Although product yield varied considerably across the reaction sets, only the appropriate allele-specific amplicon was generated according to template zygosity. Set A amplified the 517 bp amplicon and set B generated the 267 bp fragment only. The larger fragment spanning the whole target region was not amplified in all cases using our enzyme system and cycling parameters.
Figure 2.
Gel image data of PCR amplifications. Outer lanes contain 100 bp ladders. Reactions 1–9 according to the matrix (Table 2) are labelled accordingly. Set A represents reactions in which wild-type template DNA was the substrate. Set B represents reactions in which HbS/S template DNA was included. The 517 and 267 bp allele-specific fragments were generated in each set, respectively. Product yield varied across each reaction set. Table 3 represents maximal optical density (OD) of the amplicon bands, used as a representation of product yield/reaction score.
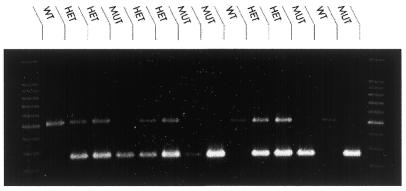
Diagnostic conditions were selected by qualitative analysis of PCR yields from set A (WT DNA) and set B (HbS/S DNA). Optimal conditions for zygosity determination are those that amplify each allele-specific amplicon at similar efficiencies, as measured by band intensity from the gel. In this study, the conditions and interactions of reaction 9 (Table 2) satisfied these criteria, producing 517 and 267 bp bands with maximum OD values of 160.67 and 165.93 units, respectively. The optimal nature of these conditions was confirmed unambiguously in a small blind trial of 14 DNA samples of different zygosity (Fig. 3). Reactions generating the 517 bp fragment identified a normal genotype. Amplification of the 267 bp fragment confirmed a homozygous mutation and sickle cell carriers (HbA/S) were identified by the amplification of both the 517 and 267 bp bands, diagnostic of heterozygous inheritance. The robustness of the optimised reaction conditions was demonstrated by the ability to amplify from a range of unknown template concentrations without compromising the allele specificity of the system.
Figure 3.
Gel image data from a small blind trial. Fourteen human DNA samples of varying zygosity were selected and amplified under conditions specified by reaction 9 (Table 2). Diagnosed template zygosity is shown on the gel image. Three normal samples were identified by amplification of the 517 bp product only. Six heterozygous samples were identified by amplification of both the 267 and 517 bp fragments. For heterozygotes, the 267 bp band was slightly more intense than the 517 bp band, likely due to staining disparity between optimisation sets A and B. Five homozygous sickle cell samples were identified by amplification of the 267 bp amplicon only.
Effect of each parameter on specificity
A quantitative analysis was performed in which the actual product yield for each reaction was used to estimate the effects of individual components on amplification in bi-directional PCR. Signal-to-noise (SNL) ratios were calculated using the maximal optical density (max OD) of the allele-specific PCR products (Table 3) to represent product yield (y) in the quadratic loss function,
Table 3. Maximal optical density (max OD) measurements of band intensities from amplification sets A and B in Figure 2.
| Reaction | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| All values are in arbitrary units. | |||||||||
| |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
| Set A, max OD wild-type | 5.13 | 90.83 | 80.30 | 63.40 | 63.70 | 1 | 132.27 | 1 | 160.67 |
| Set B, max OD mutant | 113.6 | 172.53 | 169.20 | 108.73 | 161.13 | 183.67 | 144.80 | 1 | 165.93 |
SNL = –10 log(1/n∑ni = 11/yi2)
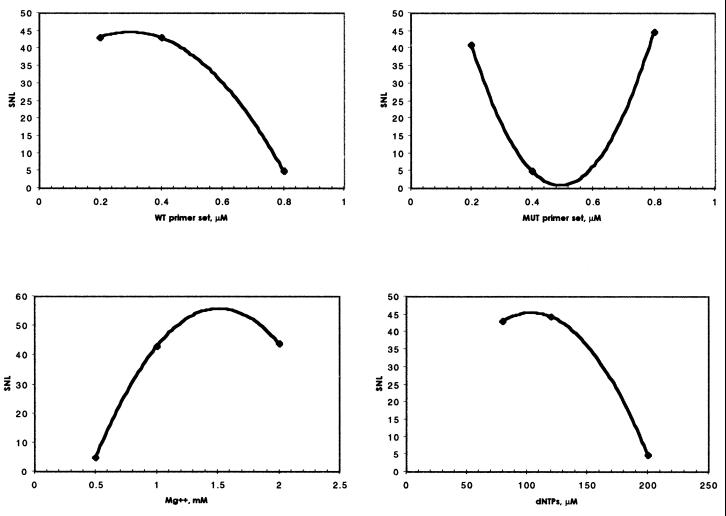
assuming that variable y denotes the performance of the system measured for a particular combination of factor settings in a given experiment (total of n repeated measurements per experiment) (17). Reactions giving non-allele-specific fragments or no amplification at all were given a score of 1. The effects of reaction components on the SNL ratio were determined by regression analysis for both normal and HbS/S DNA templates (Figs 4 and 5, respectively), using calculated SNL values (Table 4).
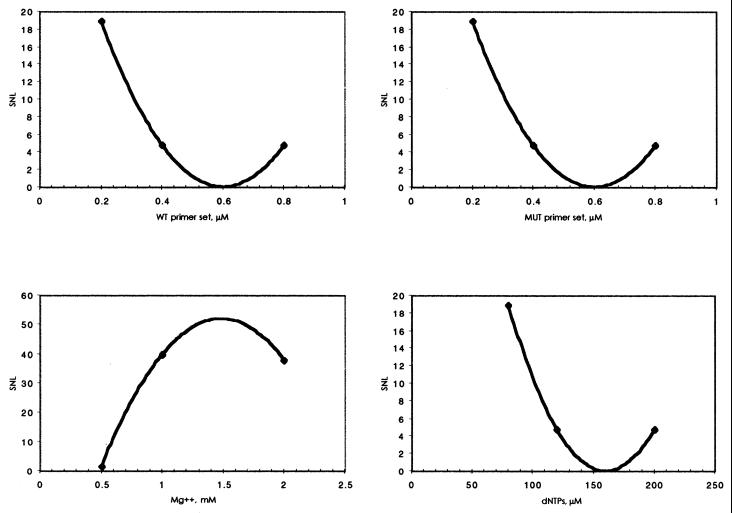
Figure 4.
Normal DNA. Effects of reaction components on the SNL ratios for the amplification of a wild-type 517 bp amplicon from normal DNA. Increased SNL value represents a larger effect on product yield than a low SNL.
Figure 5.
HbS/S DNA. Effects of reaction components on the SNL ratios for the amplification of a mutant 267 bp amplicon from HbS/S DNA. An increased SNL value represents a larger effect on product yield than a low SNL.
Table 4. Signal-to-noise ratios (SNL) for reaction components used for the amplification of (A) the 517 bp wild-type allele-specific fragment and (B) the 267 bp mutant allele-specific fragment, calculated using product yields from Figure 2.
| Level | |||
|---|---|---|---|
| SNL |
A |
B |
C |
| (A) Normal DNA template | |||
| WT-AS/WT-CP517 | 18.94 | 4.77 | 4.77 |
| MUT-AS/MUT-CP267 | 18.95 | 4.77 | 4.77 |
| Mg2+ | 1.68 | 39.95 | 38.12 |
| dNTP | 18.94 | 4.77 | 4.77 |
| (B) HbS/S DNA template | |||
| WT-AS/WT-CP517 | 43.12 | 42.93 | 4.77 |
| MUT-AS/MUT-CP267 | 41.01 | 4.77 | 44.73 |
| Mg2+ | 4.77 | 42.88 | 43.94 |
| dNTP | 42.94 | 44.32 | 4.77 |
We were able to demonstrate the significance of priming competition and component interaction on the outcome of bi-directional ASA reactions. The SNL analysis revealed that the optimal magnesium requirement for the generation of maximal product yield without compromising specificity was 1.5 mM. This value did not vary according to template zygosity, which is in accordance with the multiple roles of magnesium in PCR. Likewise, the optimal concentration of dNTPs in sets A and B were similar, with lower levels being favoured. The quantitative binding of magnesium ions by dNTPs may account for this observation, as excess dNTPs may chelate the magnesium cofactor ions of Taq, decreasing processivity or simply reducing the yield and specificity.
The regression profiles of primer sets generated by normal and mutant DNA samples were quite different, demonstrating differences in primer stabilities, efficiencies and competition. For normal DNA the aim is to direct amplification of the 517 bp fragment only. The lower concentration of the wild-type primer set tested gave the largest SNL value relating to product yield. Intermediate concentrations decreased the SNL and the upper concentration showed an incremental increase in SNL. An identical profile was observed with the mutant primer set, optimally redundant in the presence of wild-type DNA. It is possible that at lower concentrations of both primer sets, i.e. conditions of minimal competition, the wild-type pair is able to direct allele-specific amplification of the 517 bp fragment.
When the mutant allele is present, the aim is to direct amplification of the 267 bp amplicon only. The optimal concentration of the mutant primer set was predictably at the upper level. Likewise, the inclusion of lower concentrations of the wild-type primer allowed maximal optimal amplification of the mutant fragment, as the wild-type set thereby exhibited least competition with the mutant primer set. We have observed similar trends using other SNP models, confirming that primer interactions have a significant role in determining the outcome of bi-directional amplifications.
DISCUSSION
We have successfully demonstrated the use of bi-directional ASA for single tube genotyping of the SNP responsible for sickle cell anaemia. Our simple matrix optimisation strategy offers advantages over conventional methods, allowing the determination of specific test conditions in a greatly reduced reaction set. Combined with single tube bi-directional allele-specific PCR, rapid diagnosis of normal, homozygous and heterozygous template DNA can be achieved simply and applied to the screening of any known SNP. Bi-directional ASA is based on the recent adaptation of ASA methods, using a single tube for the diagnosis of a two-allele system, differing from other protocols described in which modified oligonucleotides or cycling parameters are generally required to maintain specificity.
We found that the A:A or T:T mismatches designated by the sickle cell polymorphism were completely refractory to amplification under the optimised conditions. This is discordant with a report by Kwok et al. (20) in which only A:G, G:A and C:C mismatches reduced overall PCR product yield significantly. Other combinations required the incorporation of additional mismatches near the 3′-end to increase discrimination. This difference may be due to improvements in the process of oligonucleotide synthesis and the use of recombinant Taq DNA polymerases. In particular, the use of hot start systems was seen to improve the specificity of our allele-specific amplifications. The optimisation system used and the increased priming competition in our bi-directional assay will have positively contributed to the absolute discrimination.
The implementation of an orthogonal array in our optimisation system generated product yield data from two small reaction sets that served a dual purpose in our experiments. Firstly, we were able to use product yield data qualitatively to identify conditions under which both allele-specific fragments were amplified to similarly maximal efficiencies according to template zygosity. Secondly, reaction ‘scores’ were used quantitatively in a simple regression analysis, allowing the effects of ‘control factors’ influencing reaction performance to be assessed simultaneously. This provided an insight into the component interactions and priming competition that dominate bi-directional allele-specific amplifications.
The conditions selected for the blind trial were 0.8 µM for each primer from the wild-type and mutant sets, 1 mM magnesium ions and 80 µM dNTPs. This is unlikely to be in accordance with optimal component levels calculated by SNL values because of the different objectives of each enquiry. The theoretical target of the SNL analysis is to define conditions producing the maximum yield of each product. This differs from the practical target of specifying diagnostic conditions under which each allele must be amplified specifically and at equal efficiencies enabling a heterozygous mutation to be identified without a gross over-amplification of one allele over another. Thus, to achieve this objective, sub-optimal conditions for the amplification of one allele may be combined with optimal conditions for the amplification of another by a less efficient primer set, as in this study.
Two further variables that have an effect on reaction outcome are the enzyme system and instrumentation used for amplification. The HotStart Taq DNA polymerase used in this study was extremely accurate, amplifying allele-specific products with only minimal spurious products. Additionally, the HotStart system was inefficient at amplifying larger fragments (>600 bp) when in competition with a smaller fragment, thus the non-allele-specific fragment spanning the whole target region was not amplified in this case. In similar studies, this larger fragment has been designated an internal control amplification, avoiding the incorporation of a distant primer pair conventionally required for diagnostic tests (12). However, any successful bi-directional ASA reaction will yield a fragment serving as an internal positive control for the PCR. Thus we favour our system where the processivity of Taq is expended generating allele-specific products only. We tested a conventional enzyme system in similar experiments and found that the larger fragment was generally produced at the expense of the allele-specific fragment, with decreased specificity (data not shown). Application to other thermal cyclers may require further optimisation, since different reaction vessels and heating/cooling methods can dramatically change the dynamics of a PCR.
Here, genotyping was simply based on size discrimination of the resulting allele-specific PCR products, visualised by agarose gel electrophoresis. With the emergence of qPCR technologies, offering rapid thermal cycling and on-line product detection, our system can be adapted to detect PCR products based on their specific melting temperature using non-specific DNA-binding dyes. This would address high throughput issues whilst maintaining the advantages of low cost and the use of simple chemistries associated with the gel-based assay.
The technique described could be modified to detect more than one SNP simultaneously with the combination of our optimisation strategy and accurate primer design. It has recently been suggested that the grouping and interaction of several SNPs in haplotypes may be more important than single SNPs in the identification of drug response and disease susceptibility of individuals (21). The commercial applications of this are widespread, as the development of reliable, cost-effective tests will be considerably more complex if haplotypes need to be determined rather than single SNPs.
In conclusion, we have described various factors that have a combined influence on the outcome of bi-directional PCR amplifications. We have demonstrated the technique by identifying accurate conditions for determining the zygosity of sickle cell patients and carriers, a technique that can be simply applied to the detection of any known genetic mutation.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Drs John Old and Chris Fisher at the National Haemoglobinopathy Reference Service, Oxford, UK, for the kind gift of DNA from affected sickle cell individuals and sickle cell carriers. This work was supported by grants from the University of Bath and Molecular Sensing plc.
REFERENCES
- 1.Brookes A.J. (1999) The essence of SNPs. Gene, 234, 177–186. [DOI] [PubMed] [Google Scholar]
- 2.Myakishev M.V., Khripin,Y., Hu,S. and Hamer,D.H. (2001) High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer labelled primers. Genome Res., 11, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee H.H. (1996) Ligase chain reaction. Biologicals, 24, 197–199. [DOI] [PubMed] [Google Scholar]
- 4.Bi W. and Stambrock,P.J. (1997) CCR: a rapid and simple approach for mutation detection. Nucleic Acids Res., 25, 2949–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikiforov T.T., Rendle,R.B., Goelet,P., Rogers,Y.H., Kotewicz,M.L., Anderson,S., Trainor,G.L. and Knapp,M.R. (1994) Genetic Bit Analysis: a solid phase method for typing single nucleotide polymorphisms. Nucleic Acids Res., 22, 4167–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tonisson N., Kurg,A., Kaasik,K., Lohmussar,E. and Metspalu,A. (2000) Unravelling genetic data by arrayed primer extension. Clin. Chem. Lab. Med., 38, 165–170. [DOI] [PubMed] [Google Scholar]
- 7.Gu W., Aguirre,G.D. and Ray,K. (1998) Detection of single-nucleotide polymorphisms. Biotechniques , 24, 836–837. [DOI] [PubMed] [Google Scholar]
- 8.Kuklin A., Munson,K., Gjerde,D., Haefele,R. and Taylor,P. (1997/98) Detection of single-nucleotide polymorphisms with the WAVE™ DNA fragment analysis system. Genet. Test., 1, 201–205. [DOI] [PubMed] [Google Scholar]
- 9.Newton C.R., Graham,A., Heptinstall,L.E., Powell,S.J., Summers,C., Kalsheker,N., Smith,J.C. and Markham,A.F. (1989) Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res., 17, 2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibbs R.A., Nguyen,P. and Caskey,C.T. (1989) Detection of single DNA base differences by competitive oligonucleotide priming. Nucleic Acids Res., 17, 2437–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye S., Humphries,S., and Green,F. (1992) Allele specific amplification by tetra-primer PCR. Nucleic Acids Res., 20, 1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Q., Thorland,E.C., Heit,J.A. and Sommer,S.S. (1997) Overlapping PCR for bidirectional PCR amplification of specific alleles: a rapid one-tube method for simultaneously differentiating homozygotes and heterozygotes. Genome Res., 7, 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasvari-Szekely M., Gerstner,A., Ronai,Z., Staub,M. and Guttman,A. (2000) Rapid genotyping of factor V Leiden mutation using single-tube bi-directional allele-specific amplification and automated ultrathin-layer agarose gel electrophoresis. Electrophoresis, 21, 816–821. [DOI] [PubMed] [Google Scholar]
- 14.Sommer S.S., Groszbach,A.R. and Bottema,C.D.K. (1992) PCR amplification of specific alleles (PASA) is a general method for rapidly detecting known single-base changes. Biotechniques, 12, 1152. [PubMed] [Google Scholar]
- 15.Wu Y., Ugozzoli,L., Pal,B.K. and Wallace,R.B. (1989) Allele-specific enzymatic amplification of β-globin genomic DNA for diagnosis of sickle cell anemia. Proc. Natl Acad. Sci. USA, 86, 2757–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Old J., Petrou,M., Varnavides,L., Layton,M. and Modell,B. (2000) Accuracy of prenatal diagnosis for haemoglobin disorders in the UK: 25 years’ experience. Prenat. Diagn. 20, 986–991. [PubMed] [Google Scholar]
- 17.Cobb B.D. and Clarkson,J.M. (1994) A simple procedure for optimising the polymerase chain reaction (PCR) using modified Taguchi methods. Nucleic Acids Res., 22, 3801–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henegariu O., Heerema,N.A., Dlouhy,S.R., Vance,G.H. and Vogt,P.H. (1997) Multiplex PCR: critical parameters and step-by-step protocol. Biotechniques, 23, 504–511. [DOI] [PubMed] [Google Scholar]
- 19.Linz U., Delling,U. and Rubsamen-Waigmann,H. (1990) Systematic studies on parameters influencing the performance of the polymerase chain reaction. J. Clin. Chem. Clin. Biochem., 28, 5–13. [PubMed] [Google Scholar]
- 20.Kwok S., Kellogg,D.E., McKinney,N., Spasic,D., Goda,L., Levenson,C. and Sninsky,J.J. (1990) Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res., 18, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson S. (2000) Research suggests importance of haplotypes over SNPs. Nature Biotechnol., 18, 1134–1135. [DOI] [PubMed] [Google Scholar]