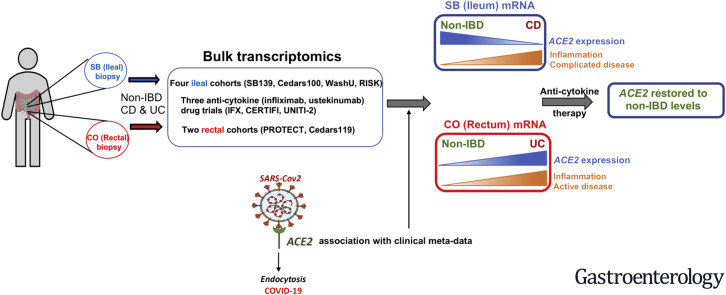
Abstract
Background And Aims
The host receptor for severe acute respiratory syndrome coronavirus 2, angiotensin-converting enzyme 2 (ACE2), is highly expressed in small bowel (SB). Our aim was to identify factors influencing intestinal ACE2 expression in Crohn’s disease (CD), ulcerative colitis (UC), and non–inflammatory bowel disease (IBD) controls.
Methods
Using bulk RNA sequencing or microarray transcriptomics from tissue samples (4 SB and 2 colonic cohorts; n = 495; n = 387 UC; n = 94 non-IBD), we analyzed the relationship between ACE2 with demographics and disease activity and prognosis. We examined the outcome of anti–tumor necrosis factor and anti–interleukin-12/interleukin-23 treatment on SB and colonic ACE2 expression in 3 clinical trials. Univariate and multivariate regression models were fitted.
Results
ACE2 levels were consistently reduced in SB CD and elevated in colonic UC compared with non-IBD controls. Elevated SB ACE2 was also associated with demographic features (age and elevated body mass index) associated with poor coronavirus disease 2019 outcomes. Within CD, SB ACE2 was reduced in patients subsequently developing complicated disease. Within UC, colonic ACE2 was elevated in active disease and in patients subsequently requiring anti–tumor necrosis factor rescue therapy. SB and colonic ACE2 expression in active CD and UC were restored by anti-cytokine therapy, most notably in responders.
Conclusions
Reduced SB but elevated colonic ACE2 levels in IBD are associated with inflammation and severe disease, but normalized after anti-cytokine therapy, suggesting compartmentalization of ACE2-related biology in SB and colonic inflammation. The restoration of ACE2 expression with anti-cytokine therapy might be important in the context of severe acute respiratory syndrome coronavirus 2 infection and potentially explain reports of reduced morbidity from coronavirus disease 2019 in IBD patients treated with anti-cytokines.
Keywords: Crohn’s Disease, Ulcerative Colitis, Infliximab, Ustekinumab
Abbreviations used in this paper: ACE, angiotensin-converting enzyme; BMI, body mass index; CD, Crohn’s disease; cCD, Crohn’s disease with no small bowel or ileal disease; COVID-19, coronavirus disease 2019; CSMC, Cedars-Sinai Medical Center; FC, fold-change; GI, gastrointestinal; IBD, inflammatory bowel disease; iCD, Crohn’s disease with ileum involvement; IL, interleukin; ISG, interferon-stimulated gene; mRNA, messenger RNA; SARS-CoV2, severe acute respiratory syndrome coronavirus 2; RAAS, renin–angiotensin–aldosterone system; RNA-seq, RNA sequencing; SB, small bowel; TNF, tumor necrosis factor; UC, ulcerative colitis; WashU, Washington University
Graphical abstract
What You Need to Know.
Background and Context:
ACE2 is highly expressed in small bowel (SB); however, demographic and clinical characteristics associated with altered intestinal ACE2 expression in Crohn’s disease (CD) and ulcerative colitis (UC) are unknown.
New Findings:
Analysis of bulk transcriptomics from biopsies revealed that within SB CD, reduced ACE2 but within colonic UC, elevated ACE2, correlated with worse prognosis. ACE2 was ‘restored’ via anti-cytokine therapy. In SB, elevated ACE2 correlated with age and BMI – predictors of poor COVID-19 outcome. Biologically, we uncovered profound differences in homeostatic mucosal ACE2 levels and their response to inflammation.
Limitations:
mRNA does not always reflect protein levels hence further functional work is required.
Impact:
This study supports the anti-inflammatory/fibrotic role of ACE2 in SB (tissue with highest mRNA level) CD. If ACE2 has a similar role in COVID-19, then anti-cytokine therapy may be one strategy to ‘restore’ ACE2 following SARS-CoV-2 infection to mitigate the secondary cytokine storm observed in severe COVID-19.
As of October 12, 2020, more than 37 million people worldwide have confirmed coronavirus disease 2019 (COVID-19) infection with current (and likely conservative) estimates implicating the virus in more than 1 million deaths. COVID-19, caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), most commonly presents with respiratory symptoms. However, recent reports have suggested that patients can frequently have both respiratory and gastrointestinal (GI) symptoms (predominantly diarrhea and nausea) and in a proportion of patients GI symptoms may be the only symptoms.1, 2, 3 There has also been concern that detection of the virus in stool might implicate the fecal–oral route as an important mode of transmission.
There is very significant variation in outcomes from COVID-19, with the majority having mild symptoms, a minority with respiratory complications, and a small percentage dying as a consequence of secondary cytokine storm or superimposed infection. Increasing age, male sex, smoking, comorbidities, and an elevated body mass index (BMI) have all been implicated in increased morbidity and mortality, but it is likely that other factors also contribute to the variability in disease course.4, 5, 6, 7 There is understandable interest and concern in the role that immunosuppressive medications commonly used in immune-mediated diseases might have on the susceptibility and natural history of COVID-19.
Angiotensin-converting enzyme 2 (ACE2) is a putative receptor for SARS-CoV-2 entry into human cells. Other molecules interacting with ACE2 and plausible candidates in COVID-19 biology include the transmembrane serine proteases (TMPRSS2 and TMPRSS4) that help prime SARS-CoV-2 spike protein for host cell entry8 , 9; the ACE2 paralog in the renin–angiotensin–aldosterone system (RAAS), angiotensin converting enzyme I (ACE); and the solute carrier family 6 member 19 (SLC6A19), expression of which is dependent on ACE2.10 The expression of ACE2 is altered in fibrotic pulmonary disease and in the lung tissue of smokers.5 , 11 ACE2 is abundantly expressed in small bowel (SB) compared with other tissues, including whole blood.12 Previous work has suggested a critical role for the RAAS pathway and ACE2 in GI tract homeostasis and development of GI inflammation.13 , 14 Our aim was to determine factors, including inflammation and drug treatment, influencing intestinal ACE2 expression in SB and colon in Crohn’s disease (CD) and colon of patients with ulcerative colitis (UC), as well as non–inflammatory bowel disease (IBD) controls, and to investigate shared disease biology between IBD and COVID-19.
Methods
Tissue Samples and Study Subjects
We investigated the association of ACE2 messenger RNA (mRNA) with age at collection, sex, smoking, BMI, diagnosis, and disease sub-phenotypes in 6 independent transcriptomic data sets from either SB or colon contingent on cohort-specific meta-data availability (Table 1 ).
Table 1.
Small Bowel and Colon Transcriptomic Cohorts With Available Demographics and Disease Status
| Subjects, n | Meta-data availability, n | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name (tissue) | Platform (expression) | GEO/ArrayExpress | Non-IBD | CD | UC | Age at collection | Sex | BMI at collection | Smoking | Disease status | Sub-phenotypes |
| SB139 (SB) | Microarray (log2) | GSE120782 | — | 139 | — | Yes (139) | Yes (139) | No | Yes (127) | Yes (139) | Yes∗ |
| WashU (SB) | RNA-seq (FPKM) | E-MTAB-5783 | 32 | 38 | — | Yes (70) | Yes (55) | Yes (66) | Yes (34) | Yes (70) | No |
| Cedars100 (SB) | RNA-seq (RPKM) | Unpublished | — | 100 | — | Yes (99) | Yes (100) | No | Yes (78) | Yes (100) | Yes∗ |
| RISK (SB) | RNA-seq (RPKM) | GSE57945 | 42 | 218 | 62 | Yes (322) | Yes (322) | No | NA | Yes (322) | Yes∗ |
| PROTECT (CO) | RNA-seq (TPM) | GSE109142 | 20 | — | 206 | Yes (206) | Yes (206) | No | NA | Yes (226) | Yes∗ |
| Cedars119 (CO) | RNA-seq (FPKM) | Unpublished | — | — | 119 | Yes (105) | Yes (99) | No | Yes (119) | Yes (119) | Yes∗ |
CO, colon; FPKM, fragments per kilobase million; GEO, Gene Expression Omnibus; NA, not available; RPKM, reads per kilobase million; TPM, transcripts per kilobase million.
See Methods for details.
The SB13915 data set was generated using whole Human Genome 4x44k Microarrays (Agilent) from formalin-fixed paraffin-embedded tissue taken from the unaffected margin of SB tissue resected during ileo-cecal or SB resection for complicated CD. Median age at time of surgery (performed at Cedars-Sinai Medical Center [CSMC], Los Angeles, CA) was 32 years. The Washington University (WashU) data set16 was generated by RNA sequencing (RNA-seq) from formalin-fixed paraffin-embedded tissue from the unaffected proximal margin of resected CD tissues and control (non-IBD) subjects. These subjects had a median age of 51 years at time of surgery (performed at WashU, St Louis, MO). The SB139 and WashU samples were all reviewed by a single pathologist (T.S.S.)16 excluding any samples with microscopic evidence of inflammation. The RISK17 , 18 data set was generated by RNA-seq from ileal biopsies taken from pediatric subjects in an inception cohort from multiple centers across North America (median age of 12 years at the time of biopsy). Being an inception cohort, age of diagnosis is same as age at specimen collection. The CD subjects in RISK cohort were divided into 2 groups: those with no SB/ileal disease (cCD) and those where the ileum was involved (iCD). The Cedars100 data set has not been published previously but similarly used formalin-fixed paraffin-embedded from un-involved proximal resection margins from complicated CD procedures (performed at CSMC) and transcriptomics were generated by RNA-seq after review by T.S.S., as described earlier (unpublished data, MIRIAD Biobank, Cedars-Sinai Medical Center). All study subjects in SB139 and Cedars100 had CD; the WashU cohort consisted of CD and non-IBD controls and the RISK cohort is a mix of CD, UC, and non-IBD controls (Table 1). In 3 of the 4 SB cohorts, specimens were taken from macroscopically normal-appearing tissue. The RISK cohort had samples from both inflamed (iCD) as well macroscopically normal-appearing tissue (cCD).17
The PROTECT19 , 20 cohort consists of pediatric subjects with varying degrees of disease severity in a UC inception cohort from multiple centers across North America (median age at time of biopsy was 13 years). We used transcriptomics from a sub-cohort of 206 UC subjects with baseline rectal biopsies before instigation of any IBD therapy, along with 20 non-IBD controls. The Cedars119 cohort has not been published previously and consists of 119 UC subjects with varying disease severity (median age of 42 years, Mayo endoscopy subscore range of 0–3) treated at CSMC. Transcriptomics for Cedars119 cohort was generated from rectal biopsies using RNA-seq.
We looked at the effect of drug exposure on SB and colonic ACE2 by analyzing transcriptomics from 3 clinical trials investigating biologic therapies used in IBD. The biologic used along with ClinicalTrials.gov number and Gene Expression Omnibus accession for associated transcriptomics are as follows: infliximab (referred to as IFX trial here), NCT00639821,21 GSE1687921; and ustekinumab (CERTIFI22 trial), NCT00771667, GSE100833,23 and ustekinumab (UNITI-216 , 24 , 25 induction and maintenance) NCT01369342, GSE112366.16 For the UNITI-2 trial, ileal histologic activity was quantified based on modified Global Histology Activity Score and endoscopic activity was quantified by Simple Endoscopic Score for Crohn’s Disease.16 , 24 The details of the drug trials can be found in the Supplementary Material.
Study Approval
For SB139, Cedars100 and Cedars119 cohorts, tissue samples and genetic data were obtained by the MIRIAD (Material and Information Resources for Inflammatory and Digestive Diseases) IBD Biobank after the patient’s informed consent and approval by the Institutional Review Board of the CSMC (protocol #3358). The other data sets were all published previously, and details of approvals can be found in the original publications.16, 17, 18, 19, 20, 21, 22, 23, 24, 25
Transcriptomics Data Generation and Processing
Table 1 shows the accession numbers for the published cohorts used in the study. The methods used for transcriptomics data generation and processing for all cohorts have been described in the Supplementary Material.
Clinical and Demographic Data
Meta-data available for the different cohorts is compiled in Table 1 and details are provided in the Supplementary Material. The sub-phenotypes’ meta-data in Table 1 include severe vs mild refractory in SB139, involved vs un-involved SB, and subsequent development of disease complication (B1 = inflammatory; B2 = stricturing, B3 = penetrating) in RISK, disease behavior in SB139 and Cedars100, disease recurrence in SB139, meta-data on active disease and Mayo endoscopy subscore for Cedars119 and need for oral steroid or anti–tumor necrosis factor (TNF) rescue therapy by week 52 in the PROTECT cohort. Further details of meta-data for various cohorts can be found in the Supplementary Material.
Methods for Data Set Downloaded Via Gene Expression Omnibus
Platform annotation, normalized gene expression, and phenotype meta-data were extracted using the R package GEOquery (GEO2R library).
Univariate and Multivariate Model Fits
Univariate and multivariate models were fitted with ACE2 or TMPRSS2 or TMPRSS4 as response and demographic or clinical data predictors where available in a given cohort.
Statistical Tools
Statistical package glm (R, version 3.5.1) was used for univariate and multivariate associations with significance cutoff of P < .05. Correlation analysis was done using Pearson or Spearman test based on data distribution (parametric or nonparametric). GraphPad Prism 7 (La Jolla, CA) was used to perform t test or Mann-Whitney test. Kruskal-Wallis test (nonparametric data) was used to compare the differences across multiple groups and adjusted P value (P adj) reported for pair-wise comparisons.
ACE2 Co-Expression Analysis
Co-expression analysis of ACE2 with genes of interest (Supplementary Table 1) involved in either IBD pathogenesis or high probability SARS-CoV-2 virus–host protein–protein interaction26 was performed using the SB139 and Cedars100 cohort. The details of the analysis can be found in the Supplementary Material.
Results
Differences in ACE2 Expression With Age, Body Mass Index, Disease, Smoking, and Sex
Univariate associations
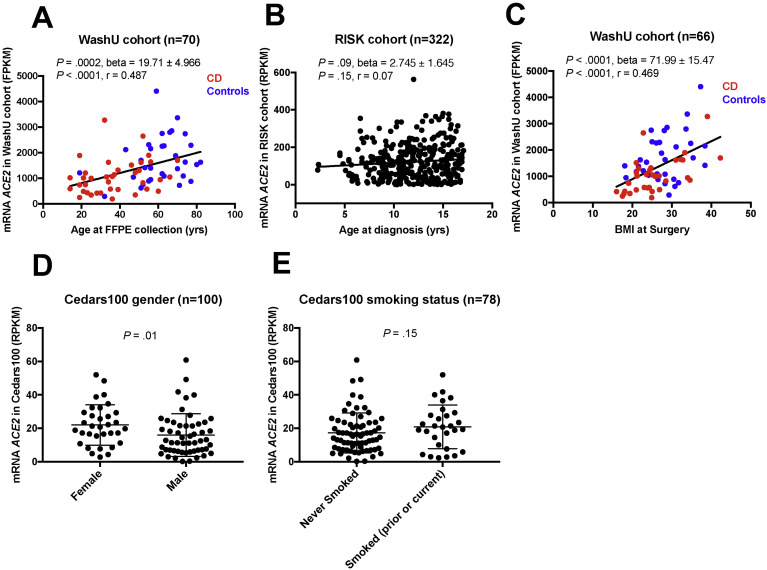
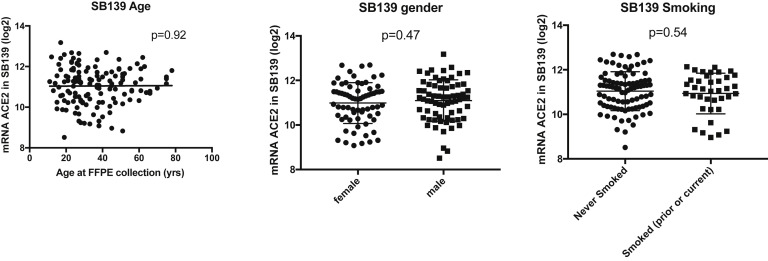
We examined ACE2 mRNA expression by subject age at the time of specimen collection when available. The expression of the most abundant ACE2 transcript isoform (ENST00000252519) was positively associated with age at collection in CD and non-IBD controls in the WashU cohort (Figure 1 A). The association with age trended toward significance in the pediatric RISK cohort (Figure 1 B). We did not see a statistically significant positive association with age in other SB cohorts, SB139 cohort (Supplementary Figure 1, Supplementary Table 2) and Cedars100 cohort (Supplementary Table 3) or colonic cohorts, PROTECT (Supplementary Table 4) and Cedars119 (Supplementary Table 5).
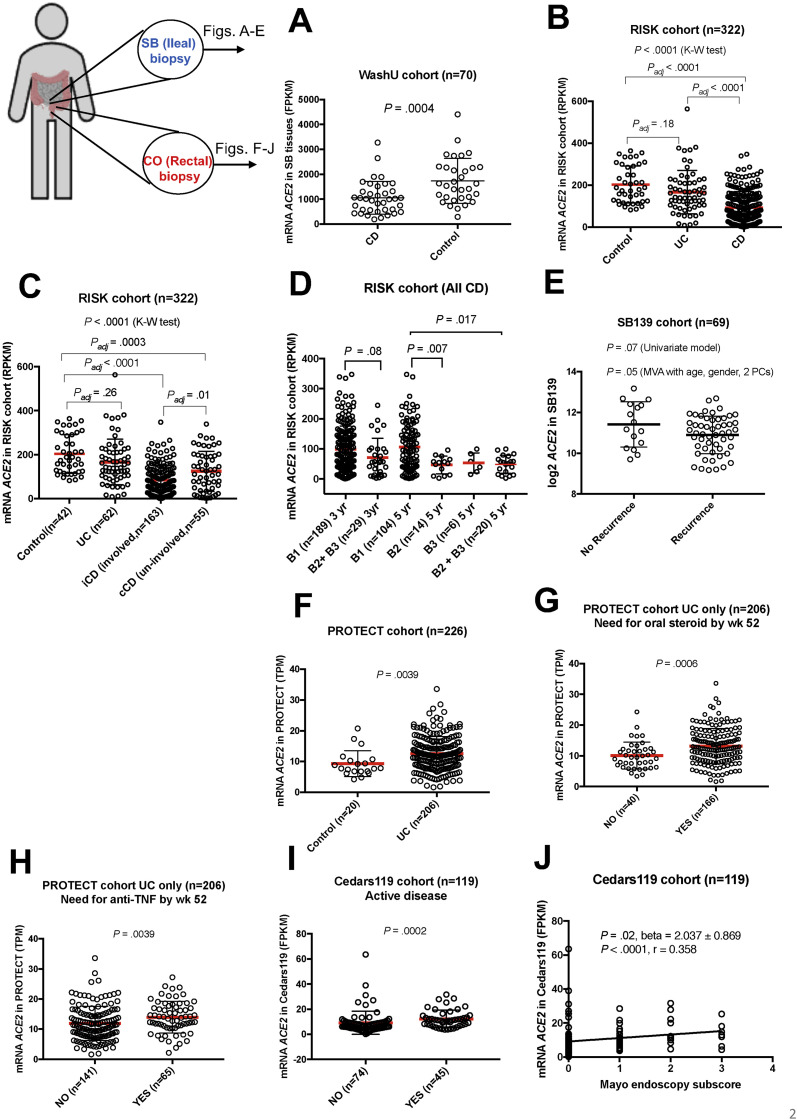
Figure 1.
Association of ACE2 expression with demographics. (A) WashU, age at sample collection (P = .0002, linear regression); (B) RISK, age at sample collection (P = .09, linear regression); (C) WashU, BMI (P < .0001, linear regression); (D) Cedars100, sex (P = .01, Mann-Whitney [M-W] test); and (E) Cedars100, smoking status (P = .15, M-W test).
Supplementary Figure 1.
Univariate association of ACE2 with age at specimen collection, gender and smoking status in SB139 cohort. FFPE, formalin-fixed, paraffin-embedded.
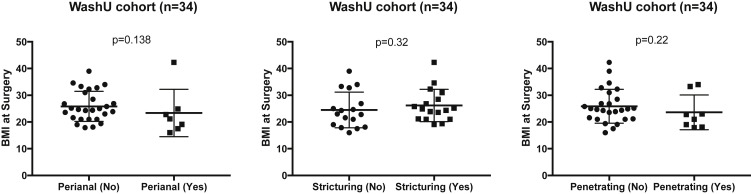
In the WashU cohort, we observed a strong positive association of ACE2 expression with BMI in both CD and non-IBD controls (P < .0001, linear regression) (Figure 1 C). No significant association of BMI with disease-severity phenotypes within CD (n = 34), such as presence of perianal disease, structuring, and penetrating disease, was observed (Supplementary Figure 2).
Supplementary Figure 2.
Correlation of BMI at surgery with disease severity in WashU cohort. No association of BMI at surgery with (A) perianal CD, (B) presence of stricturing disease, and (C) presence of penetrating disease was found among CD subjects in WashU cohort.
There was no significant association with sex among the SB139, WashU, RISK, PROTECT, and Cedars119 cohorts (Supplementary Figure 1, Tables 2 and 3 , Supplementary Tables 4 and 5). However, we observed higher ileal expression of ACE2 in female patients in the Cedars100 cohort (Figure 1 D, Supplementary Table 3), consistent with similar observations in GTEx.12
Table 2.
Univariate and Multivariate Models of ACE2 Messenger RNA Associations in the Washington University Cohort
| Response: ACE2 (FPKM)a | Univariate |
Multivariate |
Multivariate |
n | Multivariate |
n | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | P value | N | β | P value | n | β | P value | β | P value | |||
| BMI at surgery | 71.99 | .000017 | 66 | 51.37 | .002 | 51 | — | — | — | — | — | — |
| Age at collection | 19.71 | .000176 | 70 | 5.65 | .420 | 51 | 9.42 | .167 | 55 | 13.49 | .036 | 70 |
| Disease status (control) | 684.30 | .000515 | 70 | 487.7 | .052 | 51 | 550.6 | .039 | 55 | 369.78 | .120 | 70 |
| Sex (female) | –5.56 | .979007 | 55 | 78.47 | .672 | 51 | –30.08 | .873 | 55 | — | — | — |
| Smoking (yes) | 146.90 | .523000 | 35 | — | — | — | — | — | — | — | — | — |
FPKM, fragments per kilobase million.
Tested variables are indicated in parentheses.
Table 3.
Univariate and Multivariate Models of ACE2 Messenger RNA Associations in the RISK Cohort
| ACE2 (RPKM)a | Univariate |
Multivariate |
||
|---|---|---|---|---|
| β | P value | β | P value | |
| All (n = 322) | ||||
| Age at diagnosis | 2.745 | .0963 | 3.368 | .023 |
| Disease status (non-IBD) | 109.922 | 9.78e-14 | 113.091 | 2.14e-14 |
| Disease status (UC) | 73.518 | 3.13e-09 | 72.099 | 5.30e-09 |
| Sex (male) | –3.042 | .774 | –3.522 | .70886 |
| CD only (n = 218) | ||||
| Age at diagnosis | 1.464 | .388 | 1.1361 | .494 |
| Sex (male) | –0.196 | .985 | 0.9999 | .922 |
| CD type (iCD) | –41.12 | 4.86e-04 | –40.7184 | 5.93e-04 |
RPKM, reads per kilobase million.
Tested variables are indicated in parentheses.
We did not find significant association of smoking with ACE2 in most of the adult cohorts (Table 2, Supplementary Figure 1, Supplementary Table 5), although there was a trend toward higher expression in the Cedars100 cohort (Figure 1 E) (P = .15).
Data from ileal transcriptomics of non-IBD controls for comparison were only available for the WashU and RISK cohorts. In the WashU cohort (Figure 2 A), ileal ACE2 expression was lower in CD compared with controls (P = .0004). A univariate model with disease status as the predictor, was statistically significant for lower ileal ACE2 in CD vs control in the WashU cohort (Table 2).
Figure 2.
Association of ACE2 expression with disease status and sub-phenotypes in SB (A–E) and colon (CO) (F–J), error bars indicate mean ± SD, red bars indicate mean. (A) WashU, ACE2 in CD compared with control (non-IBD) (P = .0004, Mann-Whitney [M-W] test); (B) RISK, median ACE2 in CD, UC, and control (P < .0001, Kruskal-Wallis [K-W] test); (C) RISK, median ACE2 in control, UC, iCD, and cCD (P < .0001, K-W), iCD vs cCD (Padj = .01), iCD vs control (Padj < .0001); (D) RISK, ACE2 at diagnosis classified according to development of complicated disease (stricturing, B2 or penetrating, B3) or not (inflammatory, B1) at 3-year and 5-year follow-up (B2+B3 vs B1, P = .017; B2 vs B1, P = .007, adjusted for age and sex); (E) SB139, lower ACE2 expression associated with disease recurrence after surgery (P = .05, adjusted for age, sex, and 2 principal components [PCs]); (F) PROTECT, ACE2 was elevated in UC compared with control (P = .0039, M-W); (G) PROTECT, ACE2 was elevated in UC subjects that needed oral steroid by week 52 (P = .0006, M-W); (H) PROTECT, ACE2 was elevated in UC subjects that subsequently needed anti-TNF by week 52 (P = .0039, M-W); (I) Cedars119, ACE2 was elevated in UC subjects with active disease (P = .0002, M-W); (J) Cedars119, ACE2 was positively correlated with Mayo endoscopy score in UC (P < .0001, Spearman r = 0.358).
In the RISK cohort, median ACE2 expression in CD, UC, and controls was statistically different (P < .0001) (Figure 2 B). ACE2 levels in CD were lower compared with both UC (P adj < .0001) and controls (P adj < .0001) in ileal tissue. Univariate models of ileal ACE2 expression with disease status indicated ACE2 was lower in CD compared with controls (P = 9.78e-14) or UC (P = 3.13e-09) (Table 3).
Multivariate associations
Multivariate models with disease status as predictor, were statistically significant or trending toward significance for lower ileal ACE2 expression in CD vs control in the WashU cohort (Table 2). In this cohort, we observed BMI as the strongest predictor of ACE2 after adjusting for age at collection, disease status, and sex. In the RISK cohort, we again observed lower ileal ACE2 in CD compared with controls (P = 2.14e-14) or UC (P = 5.3e-09) after adjusting for age at diagnosis and sex (Table 3). Age at diagnosis was significantly associated with ileal ACE2 after adjusting for disease status and sex in the RISK cohort (Table 3). In contrast to SB, multivariate model of colonic ACE2 with disease status in the PROTECT cohort indicated elevated rectal ACE2 expression in UC compared with non-IBD (Supplementary Table 4).
Differences in Small Bowel ACE2 Expression by Disease Sub-Phenotype and Inflammation
In the RISK cohort, ileal ACE2 was lower in CD with SB involvement (iCD) compared with uninvolved CD (cCD) (P = .005, Figure 2 C and Table 3). We also found a trend toward association of ACE2 expression at diagnosis with the development of complicated disease by year 3, both without and with adjustment for age and sex (Figure 2 D, P = .08). This association of ACE2 expression at diagnosis and subsequent development of complicated disease became significant by year 5 of follow-up (Figure 2 D, B2+B3 vs B1; P = .017 and B2 vs B1; P = .007; after adjusting for age and sex).
We have previously reported transcriptomics-based sub-groups with varying disease severity in the SB139 cohort, where we found a severe-refractory sub-group (CD3) was associated with increased recurrence as well as faster time to both recurrence and second surgery compared with the mild-refractory (CD1) sub-group.15 In this SB139 cohort, ACE2 was lower in the CD3 vs the CD1 sub-group (fold-change [FC] = –3.23, corrected P < 1e-07). Using a multivariate model, we also found lower ACE2 in subjects with disease recurrence after surgery when corrected for age, sex, and first 3 principal components in genotype data (Figure 2 E; P = .05).
Differences in Colonic ACE2 Expression by Disease Sub-Phenotype and Inflammation
In the PROTECT cohort, colonic ACE2 was elevated in biopsies from UC subjects with varying disease severity and associated inflammation compared with controls (P = .004, Figure 2 F; Supplementary Table 4). In this cohort, we found elevated colonic ACE2 was predictive of UC patients requiring oral steroid by week 52 (Figure 2 G, P = .0006), as well as subjects that subsequently developed severe disease requiring the use of anti-TNF rescue therapy by week 52 (Figure 2 H, P = .004).
In the Cedars119 cohort, elevated colonic ACE2 was seen in subjects with active disease (Figure 2 I, P = .0002) and there was positive correlation with ACE2 and increasing Mayo score (Figure 2 J, P < .0001, r = 0.358, Spearman correlation).
We queried expression atlas27 to determine the impact of complicated CD (stricturing, penetrating, or disease recurrence) on colonic ACE2. We found that in the study by Peck et al28 (GSE66207), elevated levels of ACE2 in noninflamed colon tissue were associated with stricturing and penetrating disease compared with non-IBD (B2, FC = 2.1; P adj = .01; B3, FC = 1.5; P adj = .02). This is in contrast to the observations in noninflamed ileal tissue (SB139 cohort, lower ACE2 with disease recurrence, Figure 2 E) indicating discordant ACE2 signals (SB vs colon) with complicated disease in macroscopically normal tissue.
ACE2 Expression in Relation to Other COVID-19–Implicated Genes, Inflammatory Cytokines, and Known Inflammatory Bowel Disease Target
In addition to ACE2, we examined the relation of TMPRSS2 and TMPRSS4 that have also been implicated in viral entry in host cells in COVID-19,8 , 9 with various clinical parameters in all the cohorts (Supplementary Tables 2–7). Ileal TMPRSS2 expression was associated with age and positive smoking status in Cedars100 (Supplementary Table 3). Elevated expression of both TMPRSS2 and TMPRSS4 was associated with BMI in the WashU cohort. We found significantly elevated ileal TMPRSS2 in CD compared with controls in the RISK cohort (Supplementary Table 7).
We also examined the differential expression of ACE and SLC6A19 in non-IBD vs CD in WashU (Supplementary Table 8) and RISK cohorts (Supplementary Table 9). Similar to ACE2, expression of ACE was lower in CD vs controls in both WashU and RISK. We found lower ileal expression of SLC6A19 in CD compared with controls in the RISK cohort (Supplementary Table 9) and a similar trend in WashU cohort (Supplementary Table 8).
In the ACE2 co-expression analysis, we observed several genes that correlated with ACE2 expression in both SB139 and the Cedars100 CD cohorts (Supplementary Table 10) including SIGMAR1 (r = 0.6 to 0.43, P < .0001) and JAK1 (r = 0.34 to 0.25, P < .05), where r is the Spearman correlation coefficient. JAK3 was inversely correlated with ACE2 (r = –0.39 to –0.38, P < .0001) in both CD cohorts (Supplementary Table 10).
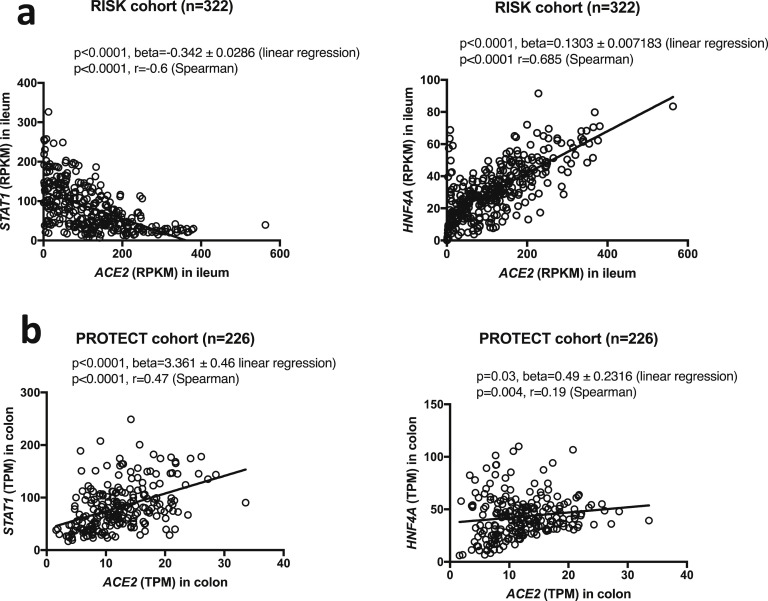
We observed that ileal ACE2 (RISK cohort) was negatively correlated with expression of transcription factor for interferon signaling, STAT1 (P < .0001, r = –0.6), while in colon ACE2 and STAT1 expression (PROTECT cohort) was positively correlated (P < .0001, r = 0.47) (Supplementary Figure 3, left). A stronger positive correlation was observed between ACE2 and HNF4A in ileum (P < .0001, r = 0.685) compared with that in colon (P = .004, r = 0.19) (Supplementary Figure 3, right).
Supplementary Figure 3.
ACE2 correlation with STAT1 and HNF4A in (A) ileal biopsies from CD subjects in RISK cohort and (B) colonic biopsies from UC subjects in PROTECT cohort. RPKM, reads per kilobase million; TPM, transcripts per kilobase million.
The Effect of Inflammation and Anti-Cytokine Therapy on ACE2 Expression in Small Bowel and Colon
We performed univariate analyses for trials where SB or colonic biopsy samples were collected pre- and post-exposure to anti-TNF (infliximab, IFX trial) and anti-IL12/23 (ustekinumab, CERTIFI and UNITI-2 trials) to query the effect of anti-cytokine monoclonal antibodies used in the treatment of IBD on intestinal ACE2 expression.
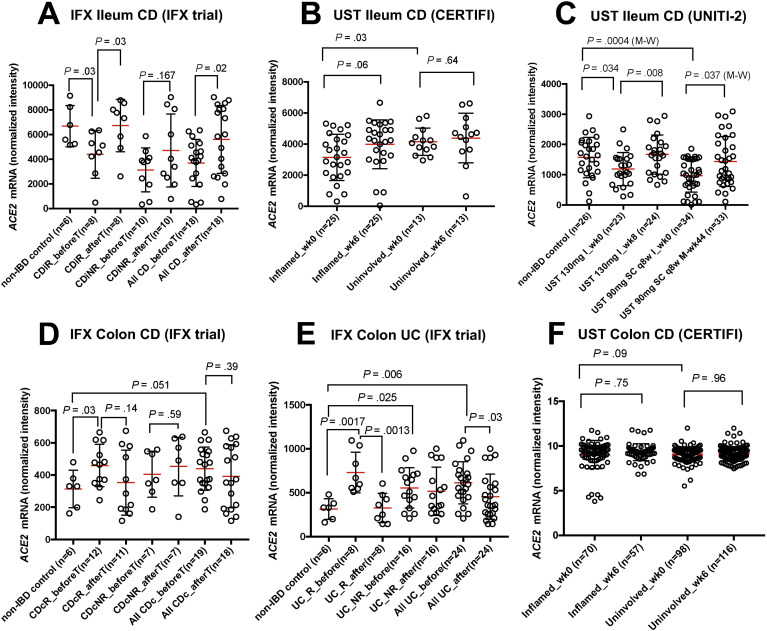
In the IFX trial, ileal ACE2 expression significantly increased after infliximab induction in CD subjects (P = .02). This phenomenon was significant in individuals who responded to treatment (P = .037), but not in nonresponders (Figure 3 A).
Figure 3.
Univariate analysis of association of ACE2 and IBD medication (infliximab, IFX trial or ustekinumab (UST), CERTIFI and UNITI-2 trials) in ileum (A–C) and colon (D–F). (A) IFX trial (ileum CD), ACE2 was elevated in non-IBD controls compared with CD responders pretreatment (CDiR_beforeT) (P = .03, t test). Post treatment, ACE2 was restored in responders (CDiR_afterT) compared with pre treatment (P = .03, t test); (B) CERTIFI (ileum CD), ACE2 pre- and post-treatment levels in inflamed and uninvolved samples; (C) UNITI-2 (ileum CD), lower ACE2 levels at baseline in CD compared with non-IBD in both UST induction group (I) (130 mg I_wk0, P = .034, t test) and maintenance group (M) (UST 90 mg subcutaneous [SC] q8w I_wk0, P = .0004, M-W test). Both post-induction therapy (130 mg I_wk8, P = .008, t test) and post-maintenance therapy (UST 90 mg SC q8w M-wk44, P = .037, M-W) , ACE2 levels are restored; (D) IFX trial (colon CD), lower ACE2 levels in non-IBD compared with Crohn’s colitis responders (P = .03, t test) pretreatment (CDcR_beforeT); (E) IFX trial (colon UC), ACE2 was lower in non-IBD compared with UC responders pretreatment (UC_R_before) (P = .0017, t test). Post treatment, the levels are restored to non-IBD in responders (UC_R_after, P = .0013, t test) as well as combined UC (P = .03, t test); and (F) CERTIFI (colon CD), ACE2 pre- and post-treatment levels in inflamed and uninvolved samples.
Response to treatment was unavailable for CERTIFI trial and we did not observe significant trends between pre and post treatment (Figure 3 B). The ileal ACE2 levels in UNITI-2 trial (Figure 3 C) were significantly lower at baseline in CD subjects compared with non-IBD controls for the 2 dosage groups (P = .034 and P = .0004). Post-ustekinumab induction, ACE2 levels were significantly restored compared with baseline (P = .008). In the maintenance-therapy group, ACE2 levels were significantly restored after 44 weeks compared with baseline (P = .037).
SB ACE2 expression was decreased in inflamed SB tissue compared with controls (Figures 3 A and C) and the severity of inflammation as measured by macroscopic and microscopic criteria (ileal Simple Endoscopic Score for Crohn’s Disease and Global Histology Activity Score) was negatively correlated with ACE2 expression in the UNITI-2 trial data set (Simple Endoscopic Score for Crohn’s Disease: week 0, P = .0007, (beta) β = –68.66; week 8, P = .0014, β = –68.3; Global Histology Activity Score: week 0, P < .0001, β = –80.75; week 8, P < .0001, β = –77.35) (Supplementary Figure 4).
Supplementary Figure 4.
Inverse correlation between ACE2 expression and increasing severity of inflammation as measured by macroscopic and microscopic criteria (ileal Global Histology Activity Score [GHAS] and Simple Endoscopic Score for Crohn’s Disease [SES-CD]).
In the IFX trial, colonic ACE2 levels (Figure 3 D) at baseline (pretreatment) were significantly elevated in Crohn’s colitis responders (P = .03). In the same trial, colonic ACE2 was significantly elevated in UC (both responders, P =.001 and nonresponders, P = .025) at baseline compared with non-IBD (Figure 3 E). After anti-TNF treatment, ACE2 levels were significantly reduced to non-IBD levels in UC responders (P = .0013), as well as combined UC cohort (P = .03). We did not observe a significant impact of treatment on colonic ACE2 levels in the CERTIFI ustekinumab trial (Figure 3 F).
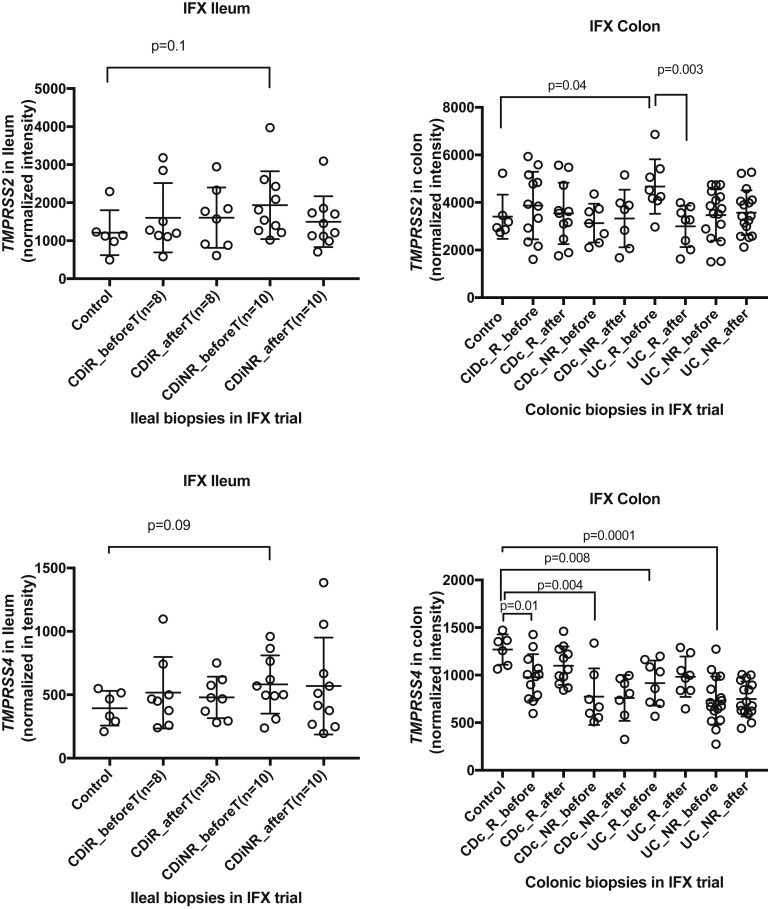
We did not find any modulation of TMPRSS2 or TMPRSS4 via anti-TNF therapy in ileal or colonic tissue, although colonic TMPRSS4 levels were reduced at baseline in both Crohn’s colitis as well as UC (Supplementary Figure 5).
Supplementary Figure 5.
Univariate analysis of association of TMPRSS2 and TMPRSS4 with anti-TNF (infliximab) IBD medication in ileal (left panel) and colonic (right panel) biopsies using IFX drug trial transcriptomics.
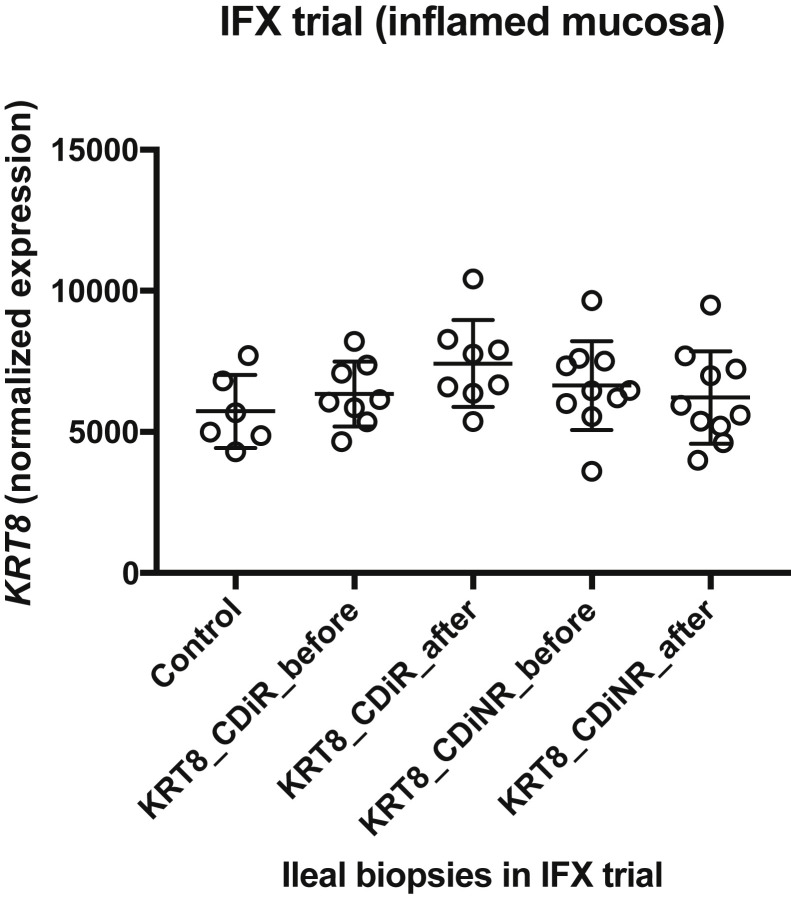
We evaluated whether the decrease in ACE2 before IFX therapy (Figure 3 A) was simply due to epithelial erosions by analyzing the mRNA expression of an epithelial marker,29 Keratin-8 (KRT-8) (Supplementary Figure 6). We found that KRT8 level in ileal biopsies pre and post treatment was fairly uniform, implying no substantial epithelial erosions were likely present at baseline in CD ileitis samples compared with controls. This indicated that the drop in ACE2 in CD ileum pretreatment is unlikely to be the result of epithelial cell loss in the areas sampled.
Supplementary Figure 6.
Altered ACE2 levels in ileum in baseline IBD compared with non-IBD controls may not be related to epithelial erosion: Expression of keratin 8 (KRT8) in IFX trial in ileal biopsies before and after treatment indicates no epithelial erosion at baseline CD ileitis compared with controls, indicating that the drop in ACE2 in CD ileum before treatment might not be the result of epithelial erosion.
Using the IFX trial colonic and ileal transcriptomics at baseline (pretreatment), we found that the direction of FC in IBD vs non-IBD for some canonical interferon-stimulated genes (ISGs) reported in the literature30 (STAT1, BST2, XAF1, IFI35, MX1, and GBP2) is the same as ACE2 in colon but not in ileum (Supplementary Tables 11 and 12). The expression of ACE2 itself in ileum was found to be 10 times that in colon in this data set (P < .0001, non-IBD control, ileum vs colon).
Discussion
Consistent with tissue- and cell-type–specific ACE2 expression reported in the literature,30, 31, 32 we found robust expression of ACE2 mRNA in SB tissue from both non-IBD as well as subjects with CD and UC. Our analysis of ACE2 expression in multiple cohorts uncovered several new observations in their relation to IBD.
We observed evidence of increased ACE2 mRNA in the ileum, with demographic features that have been associated with poor COVID-19 outcomes (age and elevated BMI). This age-related ACE2 expression might be one of the reasons for decreased COVID-19 susceptibility in children vs adults if these data, particularly from the non-IBD subjects, are reflective of ACE2 expression in other organs, such as the lung.33 Lower ACE2 expression in uninvolved SB tissue was associated with CD recurrence after surgery in an adult CD cohort. In the ileal biopsies from the RISK pediatric inception cohort, ACE2 levels at diagnosis were negatively associated with inflammation and disease severity (cCD vs iCD and UC vs CD) and, remarkably, the subsequent development of complicated disease at 5 years after diagnosis.
The demographic associations in non-IBD subjects and the relationship between ACE2 expression in macroscopically noninflamed tissue from CD patients point to systemic changes influencing ACE2 mechanisms. In the cases of aging and increased BMI, both conditions are associated with increased immune tone and myeloid skewing,34 , 35 as well as increased ACE2. Higher BMI has been linked with increased risk of infections.36 Increased ACE2 expression in lung has also been reported to be associated with age.37 There is speculation that the GI tract can serve as an alternate route for uptake of SARS-CoV-2, and our findings in the GI tract might take on increased relevance if this is confirmed.3 , 32 , 38 Furthermore, early but uncontrolled evaluations of the SECURE-IBD registry39 suggest that patients with IBD appear to be under-represented in those diagnosed with COVID-19 compared with what has been seen in the general populations in both Northern Italy and China.40 Our data suggesting reduced ACE2 expression in subsets of IBD can potentially contribute to this phenomenon, although additional work is needed to better understand these observations.
Recent findings have suggested that men are at risk of higher COVID-19 mortality, but we did not observe higher ACE2 expression in men—in fact, in 1 cohort we observed higher expression in women. This finding is in keeping with ACE2 expression in women (GTEx12). However, sex differences in ACE2 might be tissue-dependent and reflect tissue-specific escape from X-inactivation.41 Whether men are more susceptible to COVID-19, simply more likely to experience worse outcomes, or both, remains unknown. We saw a trend toward increased ACE2 expression in smokers in only 1 cohort, perhaps reflecting limited power, given the relatively low frequency of smokers in our populations, 2 of which included children only.
In contrast to the ileal tissue in CD, we found that there is elevated ACE2 expression in the colon in UC compared with non-IBD. These findings are consistent with a recent article42 studying tissue-specific (SB or colon) patterns of ACE2 expression. Furthermore, our findings suggest this ACE2 “compartmentalization” extends to disease phenotypes, including progression to complicated disease and disease recurrence in CD, with directionality of association with subsequent development of complicated disease (B2 or B3) dependent on SB (decreased) or colonic (increased) location. Consistent with this effect of location is the finding of increased ACE2 expression with increased Mayo score in UC. Overall, our analyses indicated discordant ACE2 signals in SB vs colon that are enhanced with inflammation but exist even in macroscopically normal tissue, where these discordant signals are associated with the development of complicated disease. These observations further emphasize SB/colon compartmentalization of ACE2-related immune responses.
In the colon (PROTECT pediatric UC inception cohort), we observed positive correlation between STAT1 (the reported transcription factor for interferon signaling and a canonical ISG30) and ACE2, consistent with recent reported literature of ACE2 being an ISG. However, in the ileum, STAT1 is negatively correlated with ACE2 (RISK pediatric inception cohort of CD subjects). We found strong correlation of ACE2 with HNF4A in ileum compared with colon consistent with a recent work,42 where HNF4A has been reported as an upstream regulator of ACE2 in ileum. Using the IFX trial colonic and ileal transcriptomics, we show that the direction of FC in IBD vs non-IBD for some canonical ISGs reported in literature30 is similar to ACE2 in colon but not in the ileum, consistent with ACE2 reported as an ISG in colon.42 What might account for this distinction of ileum vs colon? First, because the expression of ACE2 in ileum is 10 times of that in colon, we infer that the local tissue factors, distinct in different intestinal regions, set the homeostatic levels and direction of ACE2 response to inflammation. Second, we speculate that the threshold of biologic control for interferon signaling is surpassed in ileum compared with colon. Third, it is also possible that there are differences in the local RAAS in ileum vs colon, as we demonstrated discordant ACE2 signals in ileal and colonic inflammation.
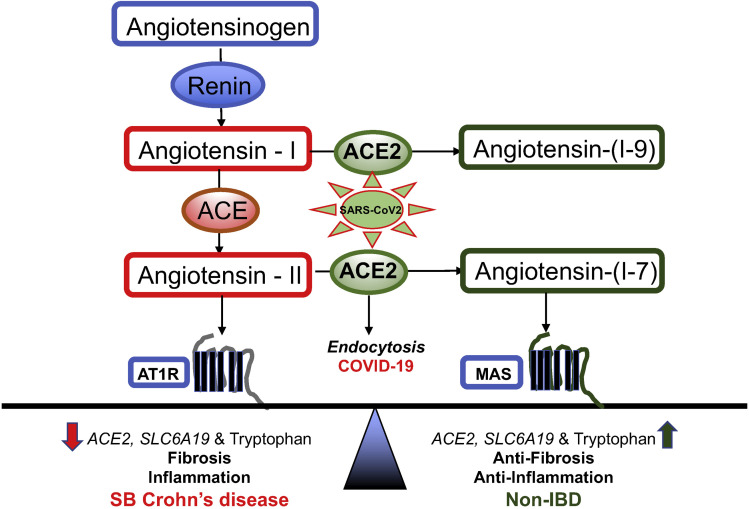
ACE2 may play a paradoxical role in disease progression of COVID-19.31 , 43 , 44 Although higher expression of ACE2 increases viral uptake by host, physiologically ACE2 has a significant anti-inflammatory role. This paradox is summarized in Figure 4 (adapted from the literature11 , 13 , 44) as it relates to SB CD and COVID-19. ACE2 is required to neutralize the pathologic effects of increased angiotensin-II in the classical RAAS by converting angiotensin II to angiotensin 1–7. Lung ACE2 expression is protective against diseases such as pulmonary fibrosis, lung injury, and asthma.45 Consistent with known anti-inflammatory role of ACE2, we report that within CD, reduced SB ACE2 expression was associated with inflammation, nonresponse to anti-cytokine therapy, and subsequent relapse of disease and development of complicated disease related to fibrosis.
Figure 4.
Main components of RAAS pathway in SB CD (left) compared with non-IBD (right). We hypothesize that ACE2 reduction in CD leads to low SLC6A19 levels and hence, low tryptophan, tipping the balance towards inflammation and fibrosis axis. Restoration of ACE2 leads to homeostasis, which can be disrupted by SARS-CoV-2 virus uptake in COVID-19. AT1R, angiotensin 1 receptor; MAS, MAS receptor.
How might down-regulation of ileal ACE2 in severe CD lead to inflammation and intestinal injury? ACE2 expression in the gut is necessary to maintain amino acid homeostasis, antimicrobial peptide expression, and “healthy” intestinal microbiome, and Ace2 –/– mice are more prone to developing colitis in induced models.10 , 14 , 31 Expression of amino acid transporter SLC6A19 (B(0)AT1) in SB is dependent on presence of ACE2,46 which acts as a chaperone for membrane trafficking of SLC6A19. 47 Accordingly, we find that expression of SLC6A19 is decreased in SB CD along with that of ACE2. Notably, lower SLC6A19 levels are selectively associated with lower tryptophan levels in SB CD.14 , 48 Dysregulated tryptophan metabolism has been linked to systemic inflammation.49 The biologic mechanisms that link levels of tryptophan to pathogenic intestinal inflammation and obesity are complex, including host and microbial production of bioactive tryptophan metabolites,48 the selective roles of these metabolites on molecular processes, such as energy checkpoint14 , 50 and transcriptional controls of inflammation pathways.51 Exploring these mechanisms in the ACE2 deficiency of SB CD can distill how the ACE2 network could serve as a protective pathway for IBD.
Elevated ACE2 levels can promote tissue propagation of virus and, in theory, could promote COVID-19 disease severity. However, the secondary cytokine storm likely promotes tissue injury via mechanisms independent of viral propagation, and this process might be independent of ACE2. Alternatively, ACE2, with its anti-inflammatory properties, can play a role in protection from the secondary cytokine storm. Due to the SARS-CoV-2–ACE2 interaction, there has been interest in treatments for COVID-19 that modulate ACE2. A study examining ACE2 with TNF-α production found that viral entry modulated TNF-α–converting enzyme via the ACE2 cytoplasmic domain and caused tissue damage through increased TNF-α production.52 We identified that ACE2 levels were restored after infliximab therapy and that this was significant in anti-TNF responders. We also observed a significant increase in ileal ACE2 expression with both ustekinumab induction and maintenance therapies. The inverse relationship of ACE2 with inflammatory cytokines and restoration of enhanced ileal ACE2 levels after response to anti-cytokine therapy point toward the anti-inflammatory function of ACE2 in SB. It has been reported that fecal calprotectin is elevated and correlates with serum IL-6 in COVID-19, linking gut inflammation and systemic cytokines in patients infected with SARS-CoV-2.53 However, additional work will be needed to delineate the anti-inflammatory function of ACE2 in COVID-19 and determine whether anti-cytokine therapies could be effective in modulating the secondary cytokine storm associated with COVID-19.54 , 55
Consistent with our findings, a recent study by Suárez-Fariñas et al56 also reported compartmentalization of intestinal ACE2 in IBD with inflammation and recognized a potential role of anti-cytokine therapy for COVID-19 treatment. Using gene regulatory networks, they also dissected overlapping molecular signals in IBD and COVID-19. Independently, we report ACE2 association with other demographics (elevated BMI); significant differences in ileal ACE2 levels in UC and CD subjects in the RISK cohort; and that reduced ileal ACE2 at diagnosis were predictive of development of complicated CD at 5-year follow-up in RISK cohort and also associated with severe refractory CD in the SB139 cohort. We also extended the region-specific discordant ACE2 signals in IBD inflammation to both CD and UC disease sub-phenotypes, prognosis and need for therapy.
Our study looked at intestinal ACE2 mRNA expression in colon and SB in a relatively large sample size (n = 976). In order to understand the functional relevance of altered ACE2 levels in IBD, as well as COVID-19, large sample studies of ACE2 protein levels on the surface of enterocytes and in the lumen will be needed in the future. There is also the possibility of unknown post-transcriptional and post-translational mechanisms that occur in IBD. ACE2 is known to be cleaved by proteases at the cell surface and shed apically, further complicating any protein analysis.57 A careful study of ACE2 at the protein level will require further work beyond the scope of this study.
Finally, we examined ileal ACE2 co-expression with a set of candidate genes as potential targets for novel or repurposed drugs. For example, our analysis revealed SIGMAR1 (candidate target for the drug hydroxychloroquine) to be consistently co-expressed with ACE2. The use of hydroxychloroquine in treating COVID-19 remains controversial. In addition, we identified that JAK1 was consistently co-expressed with ACE2 in contrast to JAK3. Selective JAK inhibitors are available and in development. Baricitinib (a JAK1/2 inhibitor) is being tested in COVID-19 based on both its anti-inflammatory properties and its possible role in inhibiting endocytosis and viral entry.58 Our observation of co-occurrence of ileal ACE2 and JAK1 provides some support for the testing of this compound in COVID-19.
To summarize, we observed association of ACE2 with various demographics (associated with worse outcomes from COVID-19) and clinical factors in multiple IBD transcriptomic data sets. We significantly extended the observation of discordant ACE2 signals in SB and colonic inflammation to prognosis and response to therapy. We report an impaired ileal ACE2 expression that leads to worse outcomes in CD and evidence that implicates ACE2 pathway as a protective, tryptophan-dependent anti-inflammatory mechanism in severe IBD. Anti-TNF and anti-IL12/23 might restore ACE2 levels in the context of inflammation reduction, suggesting that restoration of the ACE2 pathway can be a mechanism by which these drugs promote recovery in IBD. Our work supports the potential paradoxical function of ACE2 in inflammation and COVID-19. Individuals with higher ACE2 expression might be at increased risk of infection with SARS-CoV-2, but ACE2 likely has anti-inflammatory and anti-fibrotic functions in SB CD and can play an important role in preventing the secondary cytokine storm seen in COVID-19, as well as preventing the development of complicated disease in IBD. How anti-cytokine, as well as other immune targeting therapies, might modulate these processes requires urgent investigation.
Acknowledgments
The authors are thankful to all clinicians, coordinators, and especially the patients who have contributed time, data, and samples to the MIRIAD (Material and Information Resources for Inflammatory and Digestive Diseases) Biobank.
CRediT Authorship Contributions
Alka A. Potdar, PhD (Conceptualization: Equal; Data curation: Lead; Formal analysis: Lead; Investigation: Equal; Methodology: Equal; Project administration: Supporting; Software: Equal; Validation: Equal; Writing – original draft: Lead; Writing – review & editing: Equal).
Shishir Dube, PhD (Data curation: Equal; Formal analysis: Equal; Methodology: Supporting; Software: Equal; Validation: Equal; Writing – original draft: Supporting; Writing – review & editing: Equal; Equal contribution first author: Equal).
Takeo Naito, PhD (Data curation: Supporting; Methodology: Supporting).
Katherine Li, PhD (Data curation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting).
Gregory Botwin, BS (Data curation: Supporting; Methodology: Supporting).
Talin Haritunians, PhD (Data curation: Supporting; Investigation: Supporting; Resources: Supporting; Writing – review & editing: Supporting).
Dalin Li, PhD (Conceptualization: Supporting; Formal analysis: Supporting; Methodology: Supporting). David Casero, PhD (Methodology: Supporting).
Shaohong Yang, PhD (Data curation: Supporting; Methodology: Supporting; Writing –review & editing: Supporting).
Janine Bilsborough, PhD (Funding acquisition: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting).
Jacqueline G. Perrigoue, PhD (Data curation: Supporting; Methodology: Supporting).
Lee A. Denson, MD (Data curation: Supporting; Resources: Supporting). Mark Daly, PhD (Data curation: Supporting; Writing – review & editing: Supporting).
Stephan R. Targan, MD (Data curation: Supporting; Funding acquisition: Supporting; Project administration: Supporting; Supervision: Supporting; Writing – review & editing: Supporting).
Phillip Fleshner, MD (Data curation: Supporting; Resources: Supporting; Writing – review & editing: Supporting).
Jonathan Braun, MD, PhD (Conceptualization: Supporting; Data curation: Supporting; Resources: Supporting; Supervision: Supporting; Writing – review & editing: Supporting).
Subra Kugathasan, MD (Data curation: Supporting; Resources: Supporting).
Thaddeus S. Stappenbeck, MD, PhD (Data curation: Supporting; Investigation: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting).
Dermot P. B. McGovern, MD, PhD, FRCP(Lon) (Conceptualization: Lead; Data curation: Supporting; Funding acquisition: Lead; Investigation: Lead; Methodology: Supporting; Project administration: Lead; Resources: Lead; Software: Supporting; Supervision: Lead; Validation: Supporting; Writing – original draft: Equal; Writing – review & editing: Equal).
Footnotes
Conflicts of interest These authors disclose the following: Dermot P. B. McGovern, Janine Bilsborough, and Stephan R. Targan own stock in Prometheus Biosciences Inc. Alka A. Potdar, Dalin Li, Janine Bilsborough, Stephan R. Targan, and Dermot P. B. McGovern are consultants for Prometheus Biosciences, Inc. Dermot P. B. McGovern has consulted for Gilead, Pfizer, Boehringer Ingelheim, Qu Biologics, and Bridge Biotherapeutics, and received grant support from Janssen. Thaddeus S. Stappenbeck has consulted for Janssen, Boehringer Ingelheim, Genentech, and Takeda. Mark Daly is a founder of Maze Therapeutics. Lee A. Denson has received grant support from FrieslandCampina, Glycosyn, and Janssen. Subra Kugathasan consults for Janssen, steering committee for DEVELOP registry and is Takeda DSMB chair. Katherine Li and Jacqueline G. Perrigoue are employees of Janssen Research and Development, LLC. The remaining authors disclose no conflicts. Cedars-Sinai has financial interests in Prometheus Biosciences, Inc., a company that has access to the data and specimens in Cedars-Sinai’s MIRIAD Biobank (including the data and specimens used in this study) and seeks to develop commercial products.
Funding This work was supported by internal funds from the F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute. The Cedars-Sinai MIRIAD IBD Biobank is supported by the F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute, National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (grants P01 DK046763 and U01 DK062413], and The Leona M. and Harry B. Helmsley Charitable Trust.
Author names in bold designate shared co-first authorship.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://doi.org/10.1053/j.gastro.2020.10.041.
Supplementary Methods
Tissue Samples and Study Subjects for Drug Trial Data Sets
The transcriptomics for the IFX trial21 were generated using Affymetrix Human Genome U133 Plus 2.0 microarray platform using biopsies from inflamed mucosa (n = 61 IBD subjects) before and 4–6 weeks after first infliximab infusion and in normal mucosa from 12 control patients (6 colon and 6 ileum). The patients were classified as responders or nonresponders for treatment based on endoscopic and histologic findings at 4–6 weeks after infliximab induction treatment.
The CERTIFI trial consists of microarray (Affymetrix HT HG-U133+ PM Array Plate) transcriptomics of human blood and intestinal biopsy samples from a phase 2b, double-blind, placebo-controlled study of ustekinumab in CD.23 The cohort contained gene expression on 329 biopsies from multiple regions in the intestine of 87 subjects with CD. Response outcomes to ustekinumab were not available.
The UNITI-225 induction and maintenance trial consists of microarray (Affymetrix HT HG-U133+ PM Array Plate) transcriptomics of terminal ileum biopsy samples collected at baseline, 8 weeks after induction (ustekinumab or placebo), and 44 weeks after maintenance (ustekinumab 90 mg subcutaneous q12w, ustekinumab 90 mg subcutaneous q8w, or placebo) from patients with moderate-to-severe CD who participated in phase 3 studies. Ileal biopsy specimens were taken from patients with ileal or ileocolonic CD (n = 110), as well as non-IBD controls (n = 26). Ileal histologic activity was quantified based on modified Global Histology Activity Score and endoscopic activity was quantified by Simple Endoscopic Score for Crohn’s Disease.16 , 24
Transcriptomics Data Generation and Processing
Table 1 shows the accession numbers for the published cohorts used in the study. The Genome Technology Access Center at Washington University (St Louis, MO) generated data sets in the SB139, WashU, and Cedars100 cohorts. The methods used to generate microarray SB139 cohort data have been described previously.15 For the WashU cohort, RNA-seq library preparation, sequencing, and read alignment were performed and sequencing done on an Illumina HiSeq2000 SR42 (Illumina, San Diego, CA) using single reads extending 42 bases.
For the Cedars100 cohort, total RNAs were processed with Sigma Seqplex to create amplified double-stranded complementary DNA, followed by traditional Illumina library preparation with unique dual indexing. One hundred libraries were run on NovaSeq6000, S2 flow cell, using single-end 100 base reads. The run generated approximately 4.2B reads passing filter, thus an average of 42 million reads per library were generated.
The Cedars119 RNA-seq data set was generated by EA genomics, Q2 solutions. Briefly, RNA samples were converted into complementary DNA libraries using the Illumina TruSeq stranded mRNA sample preparation kit and hiSeq-Sequencing-2 × 50-bp paired end sequencing performed on an Illumina sequencing platform. Across all samples, the median number of actual reads was 24.8 million with 23.6 million on-target reads, after removal of various sequencing artifacts and normalized data in fragments per kilobase million generated.
The data generation methods for the other cohorts (RISK, PROTECT, IFX, CERTIFI, UNITI-2) have been published previously.16, 17, 18, 19, 20, 21 , 23
The methods used to process microarray data from SB139 cohort have been described previously.15 The pipeline used for RNA-seq data processing and normalizing for the Cedars100 cohort was similar to the one used for the WashU cohort, as described previously.16 For Cedars100, RNA-seq data was normalized and resultant reads per kilobase million values were generated for analysis, and for WashU, normalized data were generated in fragments per kilobase million. The methods used to process the RNA-seq data from RISK cohort have also described previously.17 , 18
Normalized processed data for some cohorts and trials (RISK, PROTECT, IFX, and CERTIFI) were downloaded using accession numbers available at Gene Expression Omnibus (GEO) in series matrix files, which were cleaned and annotated with geneids. Clean, processed data for SB139, Cedars100, and WashU along with respective meta-data were available in-house at Cedars-Sinai. UNITI-2 trial data were analyzed at Janssen.
Clinical and Demographic Data
Meta-data available for the different transcriptomics cohorts is compiled in Table 1. Clinical phenotype data available for SB139 included age at collection, sex, smoking status, disease status, disease sub-phenotypes, and disease recurrence after surgery. The Cedars100 cohort included age at collection, sex, smoking status, disease status, and CD sub-phenotypes. The Cedars119 cohort included age at collection, sex, BMI at collection, smoking, disease status, and sub-phenotypes.
For the WashU cohort, data were extracted from the clinical charts and include age at collection, sex, disease status, smoking, and BMI at collection. Some meta-data for RISK cohort were downloaded from the National Center for Biotechnology Information (GEO/Sequence Read Archive), such as age at collection, sex, and disease diagnosis, including information for involved vs unaffected CD, but complication data were available from the prospective follow-up. Meta-data for IFX, CERTIFI, and UNITI-2 trials were downloaded from their respective GEO accession numbers. Some meta-data for PROTECT cohort were downloaded from the National Center for Biotechnology Information (GEO), including age at collection, sex, and diagnosis, but data on need for rescue medication were available from the prospective follow-up.
The sub-phenotypes meta-data in Table 1 include severe vs mild refractory in SB139; involved vs un-involved SB and subsequent development of disease complication in RISK; disease behavior (B1 = inflammatory; B2 = stricturing, B3 = penetrating) in SB139, Cedars100, and RISK; meta-data on active disease and Mayo endoscopy subscore for Cedars119; and need for oral steroid or anti-TNF rescue therapy by week 52 in the PROTECT cohort.
Methods for Data Sets Downloaded Via Gene Expression Omnibus
Platform annotation, normalized gene expression, and phenotype meta-data were extracted using the R package GEOquery (GEO2R library). The phenotype meta-data table was used to identify categories such as tissue type (noninvolved/inflamed biopsy tissue samples), disease status (control, CD, UC), time points for treatment, and treatment type, as available per cohort.
Univariate and Multivariate Model Fits
Univariate models were fitted with ACE2 or TMPRSS2 or TMPRSS4 as response and each available demographic data (age, sex, BMI at collection, and smoking status) as a predictor in each cohort. A similar pipeline was followed for clinical predictors, such as disease status, CD severity sub-groups, recurrence, and treatment, where available in a given cohort. This was followed by fitting multivariate models with ACE2 expression as response and all available predictors within each cohort.
In the WashU and RISK cohorts, multivariate models were also fitted for expression of other COVID-19 relevant genes, such as ACE and SLC6A19. We examined the relationship between ACE2 expression and disease recurrence (only available in SB139) through a multivariate model with age, sex, and first 2 principal components in genotype data calculated using genetic data published previously.15 We also examined association of ACE2 with CD disease behavior B1, B2, and B3 (available in SB139, Cedars100, and RISK) using age and sex as covariates.
ACE2 Gene Co-Expression Analysis
Co-expression analysis of ACE2 with multiple genes of interest involved in either IBD pathogenesise1 or high probability SARS-CoV-2 virus–host protein–protein interaction26 was performed using the SB139 and Cedars100 cohorts. Genomic annotations for candidate genes of interest were extracted at the probe/transcript level from the platform annotation file for SB13915 and Cedars100 (R-based GenomicFeatures package in Bioconductor). The statistical package glm was used to fit a multivariate linear regression model on the gene pairs and included covariates, such as age at collection and sex (when available) with a P < .05 cutoff for statistical significance. The full list of genes examined in the co-expression analysis are available in Supplementary Table 1.
Supplementary Material
References
- 1.Han C., Duan C., Zhang S., et al. Digestive symptoms in COVID-19 patients with mild disease severity: clinical presentation, stool viral RNA testing, and outcomes. Am J Gastroenterol. 2020;115:916–923. doi: 10.14309/ajg.0000000000000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan L., Mu M., Yang P., et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am J Gastroenterol. 2020;115:766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention Coronvirus disease 2019 (COVID-19). People at increased risk. https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-at-higher-risk.html Available at: Updated November 20, 2020. Accessed April 19, 2020.
- 5.Brake S.J., Barnsley K., Lu W., et al. Smoking upregulates angiotensin-converting enzyme-2 receptor: a potential adhesion site for novel coronavirus SARS-CoV-2 (Covid-19) J Clin Med. 2020;9:841–847. doi: 10.3390/jcm9030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du R.-H., Liang L.-R., Yang C.-Q., et al. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. European Respir J. 2020;382:2000524. doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrilli C.M., Jones S.A., Yang J., et al. Factors associated with hospitalization and critical illness among 4,103 patients with COVID-19 disease in New York City. BMJ. 2020;369:m1966. doi: 10.1136/bmj.m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zang R., Gomez Castro M.F., McCune B.T., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Science Immunol. 2020;5(47) doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vuille-dit-Bille R.N., Camargo S.M., Emmenegger L., et al. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids. 2014;47:693–705. doi: 10.1007/s00726-014-1889-6. [DOI] [PubMed] [Google Scholar]
- 11.Oakes J.M., Fuchs R.M., Gardner J.D., et al. Nicotine and the renin-angiotensin system. Am J Physiol Regul Integr Comp Physiol. 2018;315:R895–R906. doi: 10.1152/ajpregu.00099.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguet F., Barbeira A.N., Bonazzola R., et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Preprint. Posted online October. 2019;3 doi: 10.1101/787903. bioRxiv 787903. [DOI] [Google Scholar]
- 13.Garg M., Royce S.G., Tikellis C., et al. Imbalance of the renin-angiotensin system may contribute to inflammation and fibrosis in IBD: a novel therapeutic target? Gut. 2020;69:841–851. doi: 10.1136/gutjnl-2019-318512. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto T., Perlot T., Rehman A., et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487(7408):477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potdar A.A., Li D., Haritunians T., et al. Ileal gene expression data from Crohn’s Disease small bowel resections indicate distinct clinical subgroups. J Crohns Colitis. 2019;13:1055–1066. doi: 10.1093/ecco-jcc/jjz021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanDussen K.L., Stojmirovic A., Li K., et al. Abnormal small intestinal epithelial microvilli in patients with Crohn’s disease. Gastroenterology. 2018;155:815–828. doi: 10.1053/j.gastro.2018.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haberman Y., Tickle T.L., Dexheimer P.J., et al. Pediatric Crohn’s disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kugathasan S., Denson L.A., Walters T.D., et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet. 2017;389:1710–1718. doi: 10.1016/S0140-6736(17)30317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyams J.S., Davis S., Mack D.R., et al. Factors associated with early outcomes following standardised therapy in children with ulcerative colitis (PROTECT): a multicentre inception cohort study. Lancet Gastroenterol Hepatol. 2017;2:855–868. doi: 10.1016/S2468-1253(17)30252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haberman Y., Karns R., Dexheimer P.J., et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat Commun. 2019;10:38. doi: 10.1038/s41467-018-07841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arijs I., De Hertogh G., Lemaire K., et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0007984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandborn W.J., Gasink C., Gao L.-L., et al. Ustekinumab Induction and maintenance therapy in refractory Crohn’s disease. N Engl J Med. 2012;367:1519–1528. doi: 10.1056/NEJMoa1203572. [DOI] [PubMed] [Google Scholar]
- 23.Peters L.A., Perrigoue J., Mortha A., et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet. 2017;49:1437–1449. doi: 10.1038/ng.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li K., Friedman J.R., Chan D., et al. Effects of ustekinumab on histologic disease activity in patients with Crohn’s disease. Gastroenterology. 2019;157:1019–1031. doi: 10.1053/j.gastro.2019.06.037. [DOI] [PubMed] [Google Scholar]
- 25.Feagan B.G., Sandborn W.J., Gasink C., et al. Ustekinumab as induction and maintenance therapy for Crohn’s disease. N Engl J Med. 2016;375:1946–1960. doi: 10.1056/NEJMoa1602773. [DOI] [PubMed] [Google Scholar]
- 26.Gordon D.E., Jang G.M., Bouhaddou M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papatheodorou I., Moreno P., Manning J., et al. Expression Atlas update: from tissues to single cells. Nucleic Acids Res. 2020;48:D77–D83. doi: 10.1093/nar/gkz947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peck B.C.E., Weiser M., Lee S.E., et al. MicroRNAs classify different disease behavior phenotypes of Crohnʼs disease and may have prognostic utility. Inflamm Bowel Dis. 2015;21:2178–2187. doi: 10.1097/MIB.0000000000000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miao Q., Xu Y., Yin H., et al. KRT8 phosphorylation regulates the epithelial-mesenchymal transition in retinal pigment epithelial cells through autophagy modulation. J Cell Mol Med. 2020;24:3217–3228. doi: 10.1111/jcmm.14998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziegler C.G.K., Allon S.J., Nyquist S.K., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J., Zhao S., Liu M., et al. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism. Preprint. Posted online February. 2020;7 doi: 10.1101/2020.02.05.20020545. medRxiv 2020.02.05.20020545. [DOI] [Google Scholar]
- 32.Zhang H., Kang Z., Gong H., et al. Digestive system is a potential route of COVID-19: an analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut. 2020;69:1010–1018. [Google Scholar]
- 33.Gudbjartsson D.F., Helgason A., Jonsson H., et al. Spread of SARS-CoV-2 in the Icelandic population. N Engl J Med. 2020;382:2302–2315. doi: 10.1056/NEJMoa2006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagareddy P.R., Kraakman M., Masters S.L., et al. Adipose tissue macrophages promote myelopoiesis and monocytosis in obesity. Cell Metab. 2014;19:821–835. doi: 10.1016/j.cmet.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J., Yoon S., Choi I., et al. Causes and mechanisms of hematopoietic stem cell aging. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20061272. 1272–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winter-Jensen M., Afzal S., Jess T., et al. Body mass index and risk of infections: a Mendelian randomization study of 101,447 individuals. European J Epidemiol. 2020;35:347–354. doi: 10.1007/s10654-020-00630-7. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y, Shan K, Qian W. Asians and other races express similar levels of and share the same genetic polymorphisms of the SARS-CoV-2 cell-entry receptor. Preprint. Posted on line February 25, 2020. Preprints 2020020258. doi: 10.20944/preprints202002.0258.v1.
- 38.Wu Y., Guo C., Tang L., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.SECURE-IBD Database Updates on COVID-19 and IBD. http://www.covidibd.org Available at: Accessed April 19, 2020.
- 40.Higgins P., Ng S., Danese S., et al. The Risk of SARS-CoV-2 in immunosuppressed IBD patients. Crohns Colitis 360. 2020;2(2) doi: 10.1093/crocol/otaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tukiainen T., Villani A.-C., Yen A., et al. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verstockt B, Verstockt S, Adbu Rahiman S, et al. Intestinal receptor of SARS-CoV-2 in inflamed IBD tissue is downregulated by HNF4A in ileum and upregulated by interferon regulating factors in colon [Epub ahead of print Sep 11, 2020]. J Crohns Colitis doi: 10.1093/ecco-jcc/jjaa185/5904222. [DOI] [PMC free article] [PubMed]
- 43.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med. 2020;8(4):e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaduganathan M., Vardeny O., Michel T., et al. Renin-angiotensin-aldosterone system inhibitors in patients with Covid-19. N Engl J Med. 2020;382:1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richards E.M., Raizada M.K. ACE2 and pACE2: a pair of aces for pulmonary arterial hypertension treatment? Am J Respir Crit Care Med. 2018;198:422–423. doi: 10.1164/rccm.201803-0569ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camargo S.M.R., Singer D., Makrides V., et al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology. 2009;136:872–882. doi: 10.1053/j.gastro.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan R., Zhang Y., Li Y., et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikolaus S., Schulte B., Al-Massad N., et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology. 2017:1504–1516. doi: 10.1053/j.gastro.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 49.Cussotto S., Delgado I., Anesi A., et al. Tryptophan metabolic pathways are altered in obesity and are associated with systemic inflammation. Front Immunol. 2020;11:95–97. doi: 10.3389/fimmu.2020.00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Virtue A.T., McCright S.J., Wright J.M., et al. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aav1892. eaav1892-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lamas B., Richard M.L., Leducq V., et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haga S., Yamamoto N., Nakai-Murakami C., et al. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc Natil Acad Sci U S A. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Effenberger M., Grabherr F., Mayr L., et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut. 2020;69:1543–1544. doi: 10.1136/gutjnl-2020-321388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feldmann M., Maini R.N., Woody J.N., et al. Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet. 2020;395(10234):1407–1409. doi: 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon D., Tascilar K., Krönke G., et al. Patients with immune-mediated inflammatory diseases receiving cytokine inhibitors have low prevalence of SARS-CoV-2 seroconversion. Nat Commun. 2020;11:3774. doi: 10.1038/s41467-020-17703-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suárez-Fariñas M, Tokuyama M, Wei G, et al. Intestinal inflammation modulates the expression of ACE2 and TMPRSS2 and potentially overlaps with the pathogenesis of SARS-CoV-2 related disease [Epub ahead of print Sep 25, 2020]. Gastroenterology doi: 10.1053/j.gastro.2020.09.029. [DOI] [PMC free article] [PubMed]
- 57.Lambert D.W., Yarski M., Warner F.J., et al. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J Biol Chem. 2005;280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richardson P., Griffin I., Tucker C., et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary Reference
- Cheng C., Hua J., Tan J., et al. Identification of differentially expressed genes, associated functional terms pathways, and candidate diagnostic biomarkers in inflammatory bowel diseases by bioinformatics analysis. Exp Ther Med. 2019:278–288. doi: 10.3892/etm.2019.7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.