Abstract
Neuropeptide release from dense-core vesicles in Caenorhabditis elegans is promoted by UNC-31, ortholog of the calcium-dependent activator protein for secretion (CAPS). Loss of UNC-31 causes multiple phenotypes in C. elegans including reduced motility, retention of late-stage eggs, and reduction in evoked synaptic release. However, the ability to analyze UNC-31 function over discrete timescales and in specific neurons is lacking. Here, we generated and validated a tool to enable UNC-31 expression and spatiotemporal functional analysis. We show that endogenously tagged UNC-31 is expressed in major ganglia and nerve cords from late embryonic stages through to adult. Using the auxin-inducible degradation system, we depleted UNC-31 postembryonically from the hermaphrodite nervous system and revealed defects in egg laying, locomotion, and vesicle release that were comparable to those in unc-31 null mutant animals. In addition, we found that depleting UNC-31 specifically from the BAG sensory neurons causes increased intestinal fat storage, highlighting the spatial sensitivity of this system. Together, this protein degradation tool may be used to facilitate studies of neuropeptide function at precise cellular and temporal scales.
SIGNIFICANCE STATEMENT Animal behavior and physiology is controlled by neuropeptides that are released from specific neuronal sources. The ability to dissect discrete neuropeptide functions requires precise manipulation of neuropeptide release. We have developed and validated a tool that enables precise spatiotemporal regulation of neuropeptide release that will enable researchers to examine neuropeptide function at exceptional resolution.
Keywords: behavior, Caenorhabditis elegans, CAPS, neuropeptide, physiology, UNC-31
Introduction
Neuronal stimulation causes the release of fast-acting chemical neurotransmitters and slower-acting neuropeptides. Calcium (Ca2+) entry into neurons precedes chemical neurotransmitter and neuropeptide exocytosis, with chemical neurotransmitters released from clear synaptic vesicles (SVs) and neuropeptides from dense-core vesicles (DCVs). UNC-31, the Caenorhabditis elegans ortholog of calcium-dependent activator protein for secretion (CAPS), is a phosphoinositide-binding protein that is critical for neuropeptide release from DCVs (Charlie et al., 2006; Gracheva et al., 2007; Sieburth et al., 2007; Speese et al., 2007). UNC-31 houses conserved C2 (coiled coil) and PH (pleckstrin homology) domains that coordinate tethering to the plasma membrane, a MUN (Munc13-homology) domain that interacts with syntaxin and a dense-core vesicle binding domain (DCVBD; Speese et al., 2007). Loss of UNC-31 causes synaptic DCV accumulation, reduced neuropeptide release, and defective behaviors associated with reduced neuropeptide signaling such as uncoordinated locomotion and retention of late-stage eggs (Charlie et al., 2006; Gracheva et al., 2007; Sieburth et al., 2007; Speese et al., 2007). In addition, unc-31 null mutant animals exhibit impaired evoked SV release that is likely an indirect effect of reduced neuropeptide action via the Gαs pathway (Gracheva et al., 2007). Genetic screens have previously generated multiple unc-31 mutant alleles to enable domain-specific functional analysis (Speese et al., 2007). However, the availability of tools to enable the manipulation of UNC-31 expression in a spatiotemporal manner is lacking. We endeavored to generate a genetic tool to enable (1) visualization of UNC-31 endogenous expression and localization and (2) temporal and spatial depletion of endogenous UNC-31.
The auxin-inducible degradation system in C. elegans is a powerful method that enables precise spatiotemporal manipulation of protein expression to facilitate functional studies (Zhang et al., 2015; Cao et al., 2021). The system leverages the ability of the Arabidopsis thaliana auxin hormone to regulate protein expression by activating the F-box transport inhibitor response 1 (TIR1) protein, the substrate recognition component of a Skp1-Cullin-F-box E3 ubiquitin ligase complex, which ubiquitinates degron-tagged proteins for proteasomal degradation (Ruegger et al., 1998; Gray et al., 1999; Dharmasiri et al., 2005). In C. elegans, the system requires the target protein to be endogenously tagged with the plant auxin-inducible degron (AID) sequence, expression of the plant TIR1 F-box protein in the tissue/cell of interest, and incubation with auxin (Zhang et al., 2015). Conditional depletion of C. elegans proteins can be precisely controlled by the spatial expression of TIR1, timing of auxin application, and degron tagging of specific protein isoforms (Zhang et al., 2015). In addition, depleted proteins can recover their expression when animals are removed from auxin. However, the rate at which this occurs can depend on the transcription/translation rates of the gene/protein of interest, the concentration of auxin used, and the developmental stage of treatment.
Here, we describe the generation and validation of a tool to enable UNC-31 conditional depletion. Endogenous tagging of UNC-31 with GFP-degron-TEV-3xFLAG reveals that expression commences during late embryogenesis and continues throughout adulthood. As reported previously using UNC-31 immunofluorescence (Gracheva et al., 2007), correct localization of endogenously tagged UNC-31 protein is dependent on the C. elegans kinesin UNC-104. We further show that addition of the GFP-degron-TEV-3xFLAG tag does not cause detectable phenotypes associated with unc-31 loss. We demonstrate the effectiveness of this conditional UNC-31 allele by analyzing the kinetics of auxin-induced UNC-31 depletion and confirming that postembryonic UNC-31 depletion phenocopies unc-31 null mutant phenotypes (egg laying, locomotion, and EPSCs). In addition, we show that depletion of UNC-31 from individual neurons (BAG sensory neurons) can reveal neuron-specific neuropeptide functions (regulation of intestinal fat storage). Together, we provide a tool that enables inhibition of neuropeptide release in a spatial and temporal manner, which will help accelerate dissection of neuropeptide functions.
Materials and Methods
Genetics
C. elegans strains were cultured on nematode growth medium (NGM) plates and fed with Escherichia coli OP50 bacteria at 20°C, unless otherwise stated (Brenner, 1974; Avery et al., 1993; Liu et al., 2009). Strains used were the following: Bristol N2 (wild type), RJP5269 rp166(unc-31-linker-GFP-TEV-AID-FLAG) IV, RJP5287 rp166 IV; him-5(e1490) V, RJP5296 reSi7(Prgef-1::TIR1::F2A::mTagBFP2::NLS::AID::tbb-2 3'UTR) I; rp166 IV, EG5096 oxIs364(unc-17p::channelrhodopsin::mCherry) X, RJP5382 reSi7(Prgef-1::TIR1::F2A::mTagBFP2::NLS::AID::tbb-2 3'UTR) I; rp166 IV; oxIs364 X, CB928 unc-31(e928) IV, RJP5601 unc-104(e1265) II; rp166 IV, and RJP5353 rpSi1(Pgcy-9::TIR1::F2A::mTagBFP2::NLS::AID::tbb-2 3'UTR) II; rp166(unc-31-linker-GFP-TEV-AID-FLAG) IV.
CRISPR-mediated generation of the gfp-tev-degron-flag-unc-31 strain
An N-terminal GFP-degron-FLAG-UNC-31 knock-in strain (rp166) was generated using CRISPR/Cas9-triggered homologous recombination (Dokshin et al., 2018). The crRNA used to target unc-31 (TTTTCAGGAGGATCATGATT) was designed and ordered using the online tool provided by https://sg.idtdna.com/pages. Asymmetric hybrid donor repair templates were amplified, which contain the GFP-TEV-degron-FLAG coding sequence from the pJW2086 plasmid (Ashley et al., 2021) as previously described (Dokshin et al., 2018). Ultramer primers used were as follows: forward (F), tttcttaaacatttactttttatgttttttttttcttttgcagtcttcgtttggaacatcATGTTAGGAGCAAGTAGTAGTGAAGAAGAAGACGACGATTTTCAGGAGGATCATGATTCTGGTGGCGGTGGATCGGGAGG; reverse (R), tgcagaggctctttagagcaaatttttggcagccaacaaaaatacatttcggcttaattttaccacctaattttattctccaaaaacaatatttacactttgaaataaaaaacttcttacCTTGTCATCGTCATCCTTGT). The following mix was then injected into wild-type animals: 4 µg repair template, 4 µg Cas9 protein, 2 µg universal tracrRNA, 1.1 µg crRNA, and myo-2::mCherry plasmid (3 ng/µl). F1 progeny were individually isolated and F2 progeny screened for degron::GFP knock-in by PCR. After confirmation by Sanger sequencing, the knock-in strain was backcrossed three times to N2 (wild type) before analysis.
CRISPR-mediated generation of the pgcy-9::TIR-1 strain
For Pgcy-9-TIR1-F2A-mTagBFP2-C1 vector and strain generation, the gcy-9 promoter (1918 bp) was amplified from C. elegans genomic DNA by PCR using primers F, AAACGACGGCCAGTGATGCGAGGAAAACTAATGGAGGC; R, TTCTCTTTTGCATCGTTATCATCTCATATGAGGGGCATT. Using In-Fusion HD Cloning (Takara Bio), the gcy-9 promoter was inserted into the pDD357(Psun-1-TIR1_F2A_mTagBFP2_C1) vector (a gift from Jordan Ward) that had been digested with SphI and ClaI. The resultant sequence-confirmed RJP737 Pgcy-9-TIR1-F2A-mTagBFP2-C1 vector was injected into wild-type C. elegans at 25 ng/µl with the following vectors: pCFJ2474(Cas9-expressing vector) at 25 ng/µl (Aljohani et al., 2020), pJW1883(ttTi5605 site targeting sgRNA (F + E) with R07E5.16 U6 promoter and 3′UTR vector) at 25 ng/µl, and the Pmyo-2::RFP vector at 5 ng/µl. Correct insertion of the Pgcy-9-TIR1-F2A-mTagBFP2-C1 sequence into the ttTi5605 transposon site on chromosome II was confirmed as described (Ashley et al., 2021). The resultant strain RJP5254 was backcrossed twice with N2 before crossing into rp166 to generate RJP5353.
AID plates
NGM plates were supplemented with 0.1 mm or 1 mm auxin (indole-3-acetic acid, catalog ALFA10556.14, Thermo Scientific) from a stock of 400 mm dissolved in ethanol. NGM plates supplemented with ethanol were used as a control.
Microscopy
Animals were anaesthetized with 20 mm NaN3 on 5% agarose pads, and images were obtained with a Axio Imager M2 fluorescence microscope and Zen software (Zeiss). Differential interference contrast (DIC) optics were used to image egg retention. Fluorescence levels were measured using FIJI software (ImageJ, version 1.47n).
Immunoprecipitation and Western blot
A packed volume (1 ml) of unc-31(rp166); Prgef-1::TIR1 1 d adult hermaphrodites were collected and washed with M9 buffer three times. Worms were pelleted by centrifugation; 3 ml of lysis buffer (50 mm Tris pH 7.4, 150 mm NaCl, 2% Triton X-100, and 1× protease inhibitor cocktail (cOmplete)) was added, and solution was sonicated using Bioruptor (Diagenode). Following centrifugation the supernatant was collected and anti-FLAG antibody (catalog #F1804, Sigma-Aldrich) binding Dynabeads were used to precipitate GFP-TEV-degron-FLAG-UNC-31. After mixing with 1X Bolt lithium dodecyl sulfate sample buffer (Invitrogen) and 1X Bolt Sample reducing Agent (Invitrogen), the input sample and postimmunoprecipitation Dynabeads were boiled at 95°C for 5 min, then loaded on an 8% polyacrylamide gel for Western Blot. The gel was blotted to a PVDF membrane using the iBlot semidry blot system (Thermo Fisher Scientific). After blocking with 5% BSA, the PVDF membranes were incubated with anti-FLAG antibody (catalog #F1804, Sigma-Aldrich), or anti-actin antibody (catalog #MAB1501, Sigma-Aldrich), followed by incubation with horseradish peroxidase-conjugated secondary antibody (catalog #61-6520, Thermo Scientific). ECL reagents (Thermo Fisher Scientific) and Bio-Rad ChemiDoc XRS+ System with CCD camera were used for imaging.
Locomotion assays
Locomotion speed was measured using the WormLab Imaging System (MBF Bioscience). NGM plates were seeded with 50 μl OP50 bacteria and dried for 1 h. Six 1-d-old adults, 20 h after mid-L4 larval stage, were picked from auxin or ethanol treatment plates to each assay plate and allowed to acclimate for 10 min. Videos were recorded for 1 min (3.75 frames/s). Speed was equal to distance traveled/time.
Egg-laying assay
Young adult hermaphrodites (36 h after the mid-L4 stage) were individually transferred to a drop of bleach solution (50% commercial bleach/50% 1 m NaOH) on a glass slide. The bleach solution dissolves the hermaphrodite to enable the number of eggs, which are protected by the eggshell, to be counted under DIC optics with an Axio Imager M2 fluorescence microscope.
Oil-Red O fat staining
For worm synchronization, five L4 larvae were picked per genotype to 8 NGM plates freshly coated with OP50, and incubated for 5 d. Worms were washed from the plates in 4 ml of M9 buffer into a 15 ml conical tube (Thermo Fisher Scientific), and eggs were isolated by adding 0.5 ml bleach (White King Premium) plus 0.5 ml 5 m NaOH, mixing and incubating for 4 min. M9 was added to fill the tube, then samples were centrifuged for 1 min at 1000 relative centrifugal force (RCF), and the supernatant was removed. Egg pellets were washed a further three times in M9 buffer, centrifuging for 1 min at 1000 RCF. The egg pellet was resuspended in ∼1 ml M9 and passed through a 40 µm filter to a fresh 15 ml tube. Samples were kept at room temperature, with gentle rocking for 24 h to allow all worms to hatch and reach the L1 stage. About 600 L1s were plated onto fresh auxin or ethanol plates (10 × 35 mm plates per condition, per genotype) and incubated at 20°C for 64 h to reach young adulthood. For staining, Oil-Red O (ORO) stock solution was prepared by adding 0.5 g Oil-Red O to 100 ml isopropanol. The solution was covered with foil and mixed at room temperature for 2 d. Immediately before commencing fat staining, Oil-Red O stock solution was diluted to 60% with sterile milli-Q filtered water, covered with foil, and rotated at room temperature until required. Worms were washed from plates with 10 ml PBS and placed into a 15 ml conical tube using a glass Pasteur pipette and centrifuged at 3300 RCF for 30 s. The supernatant was aspirated to 1 ml volume, and worms were resuspended and transferred with Pasteur pipette to a 1.5 ml microcentrifuge tube. Worms were centrifuged at 3300 RCF for 30 s, supernatant removed to 100 µl volume, and pellet resuspended in 1 ml PBS with a brief, gentle vortex mix. Samples were transferred to a 1.5 ml tube and centrifuged at 3300 RCF for 30 s. The supernatant was aspirated to 100 µl, and 1 ml PBS was added and then briefly vortexed. Wash steps were repeated for a total of three washes. After removing the final supernatant to 100 µl volume, 1 ml of 60% isopropanol was added to the worm pellet, and samples were incubated for 20 min with rotation at room temperature. Samples were centrifuged 3300 RCF for 30 s, and maximum supernatant was removed without disturbing the worm pellet. Oil-Red O working solution was filtered through a 0.22 µm filter, then 400 µl was added to each worm pellet. Samples were covered in foil, then incubated with rotation overnight at room temperature. The following day, the stained worms were washed twice in PBS plus 0.01% Triton X-100, then washed once in PBS, centrifuging at 3300 RCF for 30 s. The supernatant was removed, then worms were resuspended and mounted onto agarose pads for imaging. Oil-Red O staining was quantified in FIJI (ImageJ) software by tracing the first four intestinal cells proximal to the pharynx. The intensity in these cells was measured in the inverted green channel (where Oil-Red O absorbs the light), collecting area, mean gray value, and integrated density data. A region outside the worm was used as a background measurement for Oil-Red O staining and used to calculate the CTCF (corrected total cell fluorescence). CTCF is calculated as follows: Integrated Density – (area * mean gray of background).
Electrophysiology recording
NGM plates (60 mm) were seeded with 250 μl OP50 mixed with 5 μl 100 mm all-trans retinal (Sigma-Aldrich) as previously described (Piggott et al., 2011). Seeded retinal plates were stored in the dark at 4°C for up to 1 week. C. elegans hermaphrodites were transferred to retinal plates and grown in the dark for an additional 6 h before electrophysiological recording. All patch-clamp electrophysiology was performed with dissected adult hermaphrodites under an Olympus microscope (BX51WI) with an EPC-10 amplifier and Patchmaster software (HEKA), using a protocol previously described (Richmond et al., 1999). Each worm was glued on a SYLGARD-coated coverslip along the dorsal side with a medical-grade, cyanoacrylate-based glue (Dermabond Topical Skin Adhesive, Ethicon). A small piece of cuticle on the worm body was carefully cut and glued down to the coverslip to expose the body wall muscles (BWMs) for recording. Current data were filtered at 2 kHz and sampled at 20 kHz. Series resistance and membrane capacitance were both compensated. Recording pipettes were pulled from thick-walled, borosilicate glass (catalog #BF150-86-10, Sutter Instruments) and had resistances of 3–5 MΩ. The regular bath solution contained the following (in mm): 140 NaCl, 5 KCl, 5 MgCl2, 1 CaCl2, 11 glucose, and 10 HEPES (330 mOsm, pH adjusted to 7.3). The pipette solution contained the following (in mm): 120 KCl, 20 KOH, 4 MgCl2, 5 EGTA, 0.25 CaCl2, 10 HEPES, and 5 Na2ATP (325 mOsm, pH adjusted to 7.2 using NaOH). BWMs were clamped at −60 mV. Channelrhodopsin was excited with a blue light pulse (3 ms) generated with a blue-light-emitting diode (LED) source (catalog #M00552407, Thorlabs). The light switch was controlled by transistor–transistor logic signals from a HEKA EPC-10 double amplifier. All experiments were performed at room temperature (20–22°C).
Experimental design and statistical analyses
Experiments were performed in three independent replicates with the experimenter blinded to genotype/condition. Statistical analysis was performed in GraphPad Prism 9 software using either one-way ANOVA when more than two samples were being compared or Welch's t test when two samples were being compared. Values are expressed as mean ± SEM. Differences with a p value < 0.05 were considered significant. The number of animals analyzed, along with exact p values resulting from defined statistical tests, is reported in each figure legend.
Results
Generation of a conditional degradation allele for UNC-31/CAPS
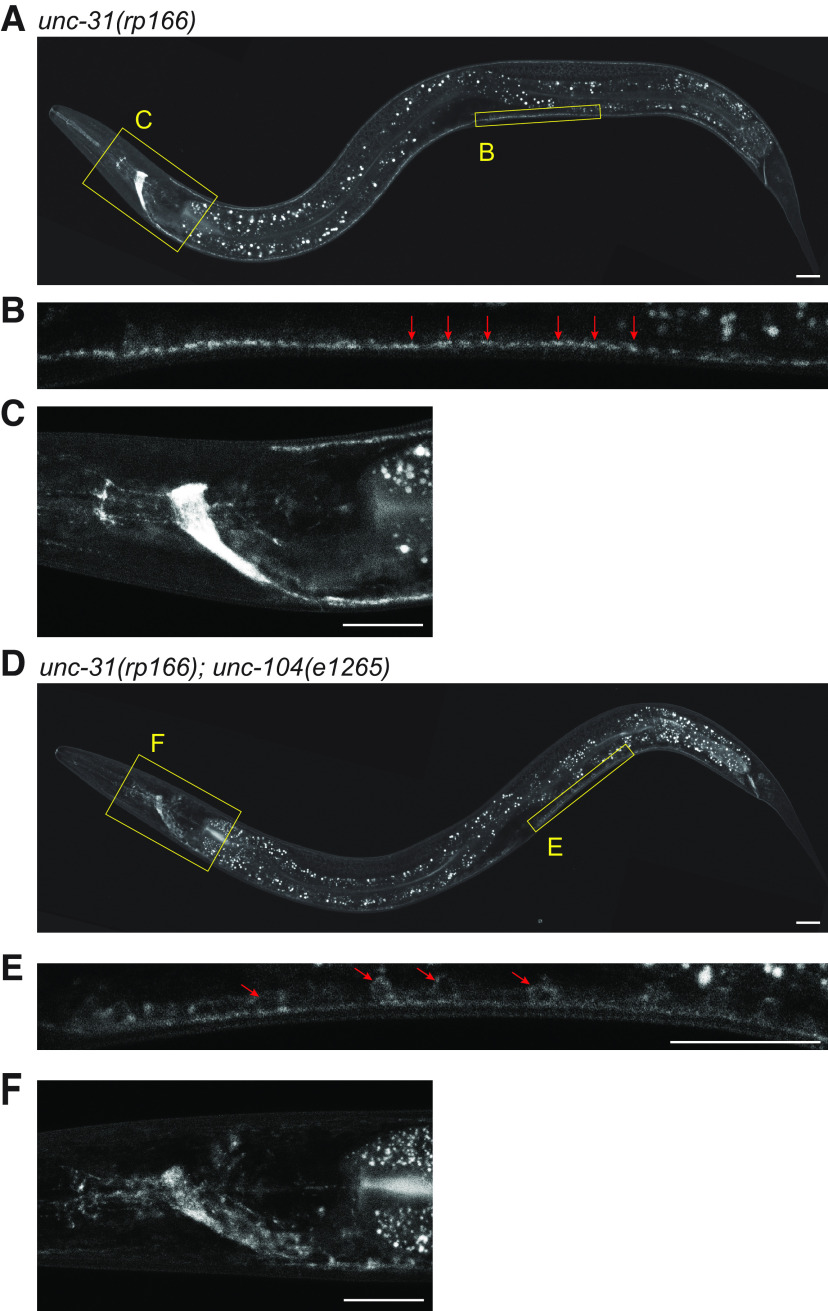
The unc-31 gene encodes multiple isoforms. We elected to tag the longest isoforms that contain all the recognized functional domains (Fig. 1A,B; Speese et al., 2007). Expression of N-terminally tagged UNC-31 was previously shown to support transgenic rescue of unc-31 mutant phenotypes (Speese et al., 2007). Therefore, we used CRISPR-Cas9 engineering to insert a GFP-degron-TEV-3xFLAG cassette at the N terminus (30 amino acids downstream of the ATG codon) of UNC-31 (Dokshin et al., 2018). The resultant unc-31(rp166) knock-in strain was sequenced to confirm the insertion, backcrossed three times with wild-type animals, and reisolated. Next, we analyzed GFP fluorescence in unc-31(rp166) hermaphrodites and unc-31(rp166); him-5(e1490) males (Fig. 1C). We first detected expression in the nerve ring of threefold embryos just before hatching (Fig. 1C). In adults, strong fluorescence was maintained in the nerve ring, and expression was also detected in the ventral nerve cord (VNC) and dorsal nerve cord (DNC). In males, a similar expression pattern was observed, albeit with additional expression in the male-specific postcloacal sensilla (Fig. 1C). Focusing on unc-31(rp166) expression in VNC motor neurons, we observed punctate expression reminiscent of UNC-31 immunostaining representing synapses (Fig. 2; Gracheva et al., 2007). UNC-31 synaptic localization is dependent on the UNC-104 kinesin motor protein (Gracheva et al., 2007). We found that unc-31(rp166) expression in VNC motor neurons and the nerve ring is also disrupted in unc-104(e1265) mutant animals, with GFP accumulation in cell bodies (Fig. 2). Thus, unc-31(rp166) expression is detected in the nervous system of both sexes, with the most prominent expression in the nerve ring.
Figure 1.
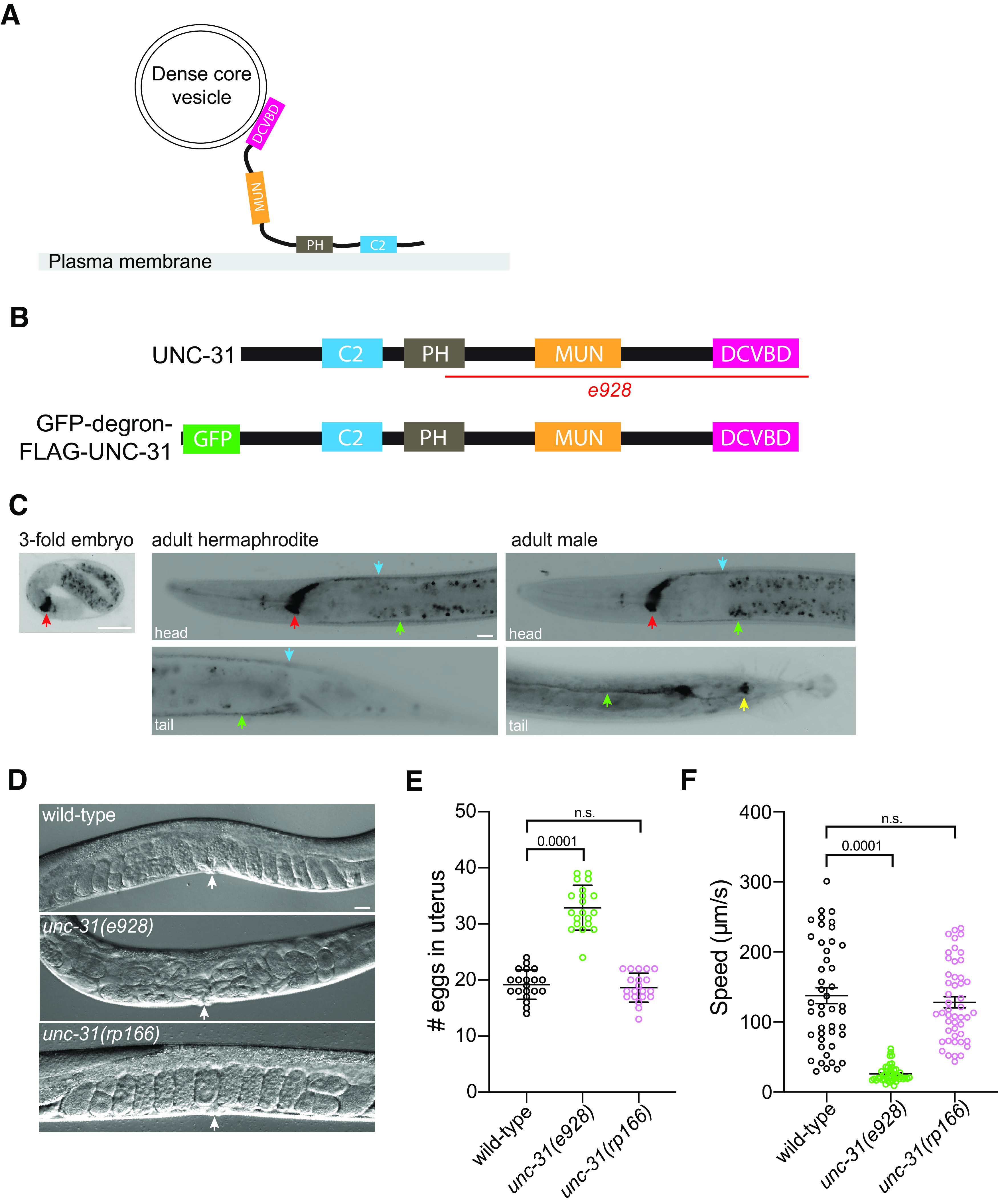
Generation of an UNC-31 conditional degradation allele. A, Schematic of the UNC-31 protein. B, Schematic of wild-type UNC-31 protein (top) and GFP-degron-FLAG-UNC-31 protein (bottom). Deleted region e928 in unc-31 null mutant (red line). C, Fluorescent micrographs of unc-31(rp166) knock-in GFP expression in a threefold embryo (left), adult hermaphrodite (center), and adult male (right). Red arrows indicate nerve ring; green arrows, ventral nerve cord; blue arrows, dorsal nerve cord; yellow arrow, postcloacal sensilla. Lateral views, anterior to the left (except male tail image, ventral view). Scale bars: 20 µm. D, E, DIC images (D), quantification (E) of egg accumulation in wild-type, unc-31(e928), and unc-31(rp166) adult hermaphrodites (36 h after mid-L4). White arrows indicate vulva (n = 20). Data expressed as mean ± SEM. Statistical significance was assessed by one-way ANOVA multiple comparison with Dunnett's multiple-comparisons test (F(2,57) = 132.8, p = 0.0001), n.s, Not significant. Scale bar, 20 µm. F, Quantification of locomotory speed (distance traveled/time) of wild-type, unc-31(e928), and unc-31(rp166) adult hermaphrodites (20 h after mid-L4; n = 45–48). Data expressed as mean ± SEM. Statistical significance was assessed by one-way ANOVA multiple comparison with Tukey's multiple-comparisons test (F(2,137) = 59.98, p = 0.0001), n.s.
Figure 2.
Localization of endogenously tagged UNC-31 is dependent on the kinesin UNC-104. A–F, Fluorescent confocal images of unc-31(rp166) knock-in GFP expression in wild-type (A–C) and unc-104(e1265) (D–F) adult hermaphrodites. Yellow boxes in A, D expanded in B, C and E, F. Arrows show punctate localization (B) compared with cell body accumulation (E). Lateral views, anterior to the left. Scale bars: 20 µm.
Fusion of the GFP-degron-TEV-3xFLAG tag to the endogenous UNC-31 protein may have a detrimental effect on UNC-31 function. We did not observe any overt phenotype in unc-31(rp166) animals; however, to examine this possibility further, we analyzed two well-studied paradigms of UNC-31 function, egg laying and locomotion (Avery et al., 1993). We first examined the steady-state accumulation of eggs in the uterus of wild-type, unc-31(e928) null mutant, and unc-31(rp166) 1 d adult hermaphrodites. We found that unc-31(rp166) animals exhibit wild-type levels of egg retention (Fig. 1D,E). This contrasts to unc-31(e928) null mutant animals, which accumulate significantly more eggs than wild-type animals (Fig. 1D,E). Next, we examined locomotion of wild-type, unc-31(e928), and unc-31(rp166) 1 d adult hermaphrodites using WormLab automated tracking (MBF Bioscience). We found that the locomotory speed (distance traveled/time) of unc-31(rp166) animals is not significantly different from that of wild type (Fig. 1F). In contrast, unc-31(e928) null mutant animals exhibit significantly reduced locomotion (Fig. 1F). Together, our data show that the CRISPR-Cas9 generated unc-31(rp166) knock-in strain reveals expression in the nervous system and that in vivo tagging of UNC-31 does not cause detectable defects in egg laying or locomotion.
Temporal degradation of UNC-31
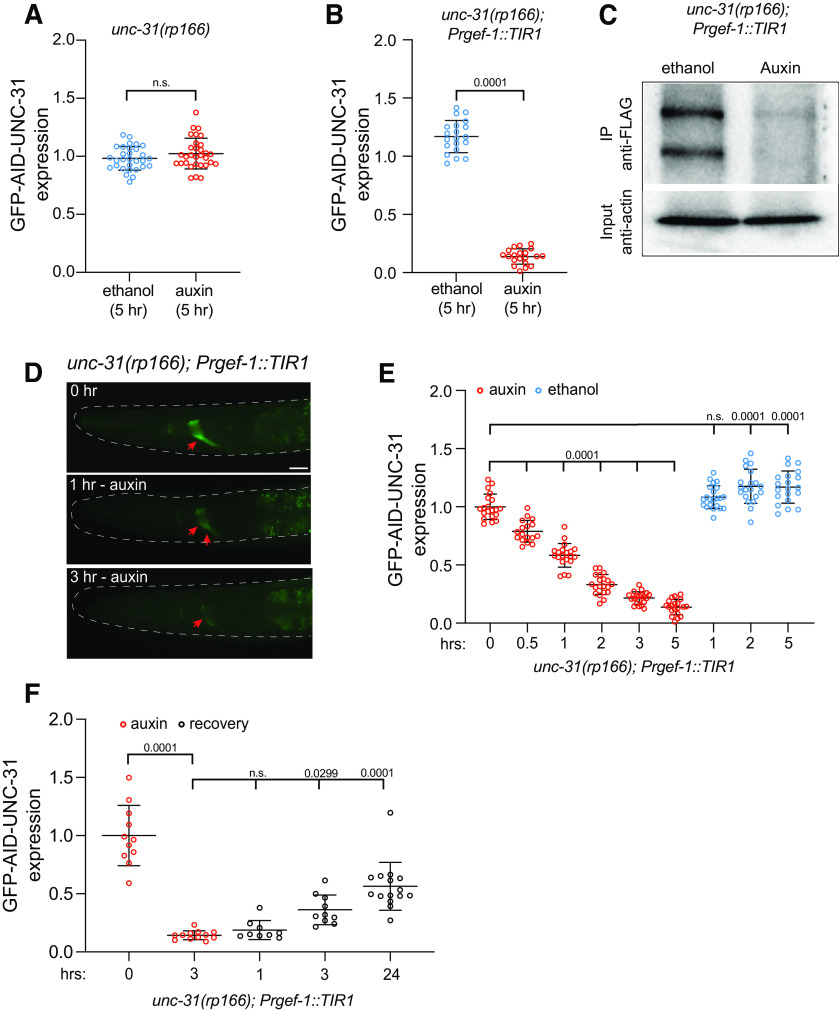
To determine whether UNC-31 can be efficiently depleted by the auxin-inducible degradation system, we crossed a transgene into unc-31(rp166) animals that expresses TIR1 in the nervous system under control of the rgef-1 promoter (Fig. 3; Ashley et al., 2021). We exposed either unc-31(rp166) or unc-31(rp166); Prgef-1::TIR1 animals to auxin for 5 h and measured GFP fluorescence in the nerve ring (Fig. 3A,B). We found that GFP levels were unchanged in unc-31(rp166) animals exposed to auxin compared with the ethanol control (Fig. 3A). In contrast, auxin robustly depleted GFP fluorescence in unc-31(rp166); Prgef-1::TIR1 animals (Fig. 3B). We confirmed auxin-induced depletion of unc-31(rp166); Prgef-1::TIR1 animals by probing for the FLAG tag by Western blot, finding that GFP-degron-FLAG-UNC-31 protein was undetectable after 24 h of auxin treatment (Fig. 3C).
Figure 3.
Temporal knockdown of UNC-31 expression with auxin. A, B, Quantification of GFP expression in the nerve ring of unc-31(rp166) (A) and unc-31(rp166); Prgef::TIR1 (B) mid-L4 larvae exposed to 1 mm auxin or ethanol for 5 h, relative to untreated controls; n = 30–31 in A, 20 in B. Data expressed as mean ± SEM. Statistical significance was assessed by unpaired t test, A (t(1.330) = 59, n.s., Not significant); B (t(30.09) = 38, p = 0.0001). C, Western blot of immunoprecipitated GFP-degron-FLAG-UNC-31 protein from unc-31(rp166); Prgef::TIR1 animals exposed to ethanol or auxin for 24 h. Two major UNC-31 bands were detected. D, Fluorescent micrographs of GFP expression in unc-31(rp166); Prgef::TIR1 L4 hermaphrodites following 1 mm auxin exposure for 0, 60, and 180 min. Red arrows indicate nerve ring; dotted line shows worm outline. Scale bar, 20 µm. E, Quantification of GFP expression in the nerve ring of unc-31(rp166); Prgef::TIR1 in mid-L4 larvae exposed to 1 mm auxin or ethanol for the specified time periods in hours; n = 17–21. Data expressed as mean ± SEM. Statistical significance was assessed by one-way ANOVA multiple comparison with Tukey's multiple-comparisons test (F(8,170) = 327.4), p = 0.0001, n.s. F, Quantification of GFP expression in the nerve ring of unc-31(rp166); Prgef::TIR1 animals exposed to 0.1 mm auxin for 3 h and then recovered for 1, 3, and 24 h without auxin; n = 10–15. Data expressed as mean ± SEM. Statistical significance was assessed by one-way ANOVA multiple comparison with Tukey's multiple-comparisons test (F(4,52) = 46.94), p = 0.0001, n.s.
The successful use of unc-31(rp166); Prgef-1::TIR1 animals in behavioral experiments may depend on the rapidity of depletion. We therefore analyzed the kinetics of auxin-mediated depletion of UNC-31 over a 5 h period by measuring nerve ring fluorescence in L4 larvae (Fig. 3D,E). We found that within 30 min of auxin exposure, UNC-31 expression reduced by ∼20% (Fig. 3E) and observed a gradual reduction of UNC-31 expression over time to ∼90% of control levels after 5 h of auxin exposure (Fig. 3D,E). We found that ethanol, the diluent for auxin that is used as a control, caused a slight but significant increase in UNC-31 expression after 1 h (Fig. 3E). This should be considered when performing experiments with this strain.
In certain experiments, it may be desired to examine the effects of restoring UNC-31 expression following auxin-induced degradation. We therefore determined whether UNC-31 expression could recover following removal of animals from auxin (Fig. 3F). We first exposed unc-31(rp166); Prgef-1::TIR1 L4 hermaphrodites to 1 mm auxin for 3 h and measured UNC-31 levels in the nerve ring following recovery on standard NGM growth plates. We found that UNC-31 levels did not recover under these conditions, even after a 24 h recovery period (data not shown). Minimal recovery from 1 mm auxin exposure has also been shown in other contexts (Zhang et al., 2015). Therefore, we repeated the experiment using a 10-fold reduction of auxin (0.1 mm). We found that exposure to 0.1 mm auxin robustly reduced UNC-31 levels after 3 h and that expression was restored to ∼50% of untreated levels after a 24 h recovery period (Fig. 3F). The slow restoration of UNC-31 expression may be because of the residual presence of auxin and potentially low rates of unc-31 transcription in L4/adult animals. Optimization of auxin levels should be considered in specific experiments where reversibility is required.
Conditional depletion of UNC-31 causes defects in egg laying and locomotion
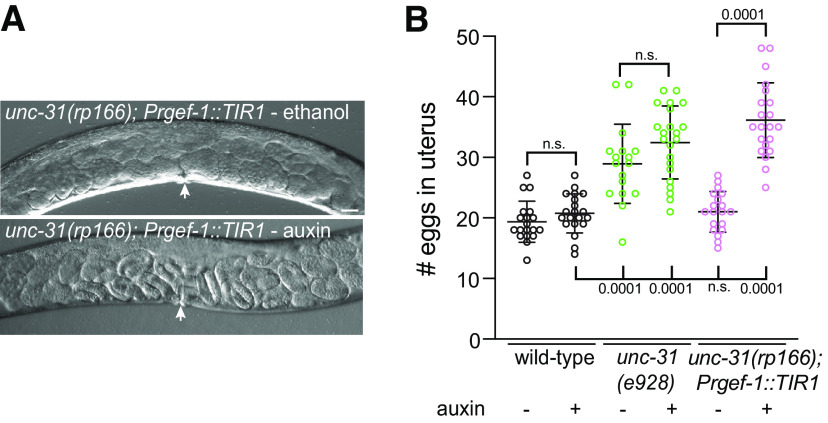
To determine the utility of the unc-31(rp166) tool for functional studies, we assayed egg laying and locomotion (Figs. 3, 4). We exposed wild-type, unc-31(rp166), and unc-31(rp166); Prgef-1::TIR1 mid-L4 stage hermaphrodites to auxin or ethanol for 24 h and counted egg retention in the uterus (Fig. 4). We found that the accumulation of eggs in wild-type and unc-31(e928) animals is not significantly affected by auxin (Fig. 4B). In contrast, auxin caused significant accumulation of eggs in unc-31(rp166); Prgef-1::TIR1 animals (Fig. 4A,B).
Figure 4.
Auxin-induced UNC-31 knockdown causes egg accumulation. A, DIC images of unc-31(rp166); Prgef::TIR1 1 d adult hermaphrodites exposed to ethanol or auxin for 24 h from the mid-L4 larval stage. White arrows indicate vulva. Scale bar, 20 µm. B, Quantification of egg accumulation in wild-type, unc-31(e928), and unc-31(rp166); Prgef::TIR1 1 d adult hermaphrodites. Auxin and ethanol treatments were for 24 h from the mid-L4 larval stage; n = 17–22. Data expressed as mean ± SEM. Statistical significance was assessed by one-way ANOVA multiple comparison with Dunnett's multiple-comparisons test (F(5,112) = 40.3, p = 0.0001), n.s., Not significant.
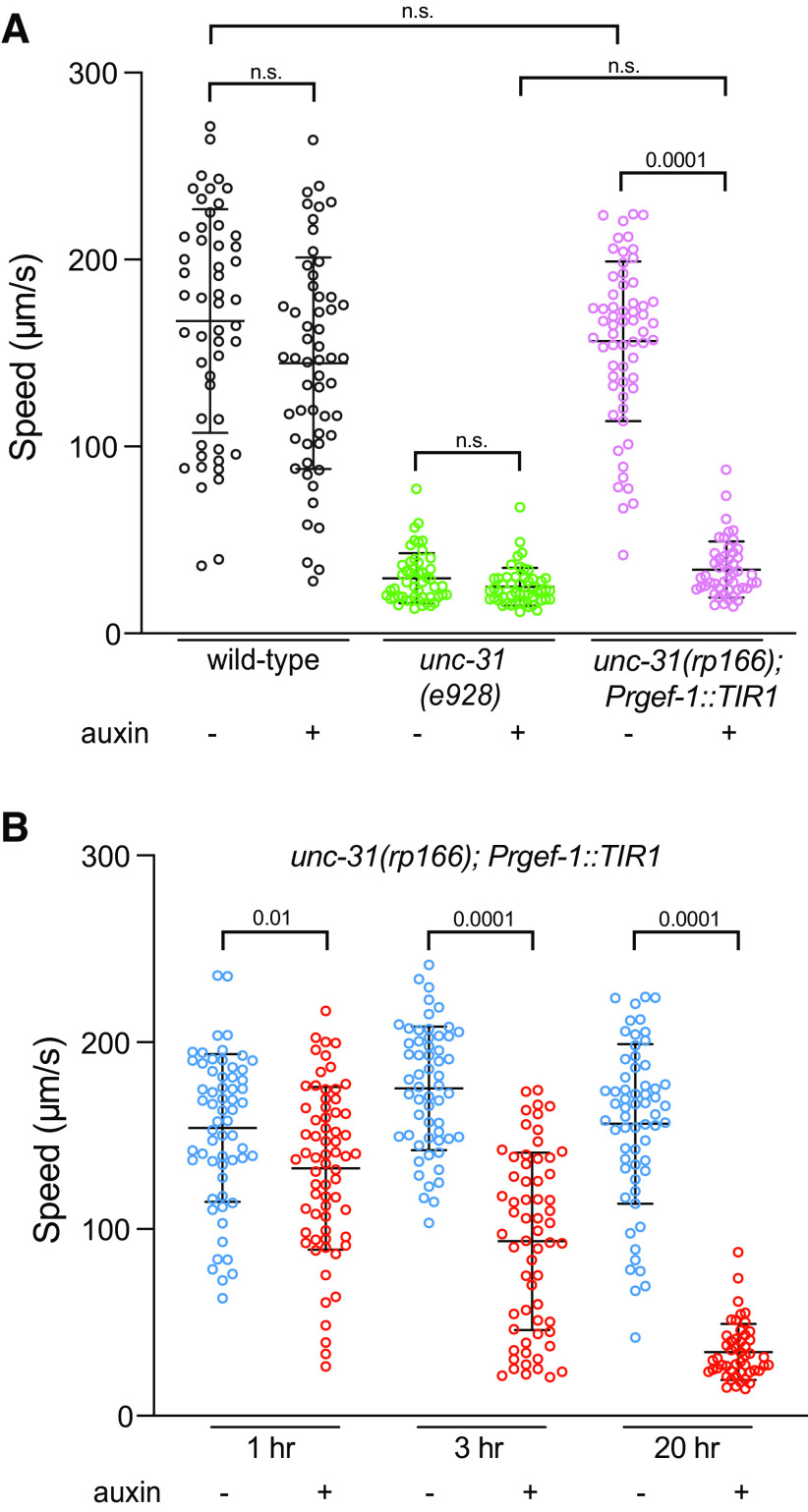
Next, we examined locomotion of wild-type, unc-31(e928), and unc-31(rp166); Prgef-1::TIR1 1 d adult hermaphrodites that were exposed to auxin for 20 h from the mid-L4 stage (Fig. 5). Using WormLab automated tracking (MBF Bioscience) we found that the locomotory speed of wild-type and unc-31(e928) animals was not affected by auxin exposure (Fig. 5A). In contrast, unc-31(rp166); Prgef-1::TIR1 exposed to auxin exhibited locomotion similar to the unc-31(e928) null mutant (Fig. 5A). This shows that UNC-31 is continually required for C. elegans locomotion. To examine the kinetics of locomotory decline following auxin-induced UNC-31 depletion, we measured the locomotion speed of unc-31(rp166); Prgef-1::TIR1 adult hermaphrodites exposed to auxin for 1 h, 3 h, and 20 h (Fig. 5B). We found that within 1 h of auxin exposure, when ∼40% of UNC-31 protein is depleted (Fig. 3E), there is a significant decrease in locomotion speed compared with control animals (Fig. 5B). After 3 h of auxin exposure, the locomotory speed of unc-31(rp166); Prgef-1::TIR1 animals reduces by ∼60%, with a further decrease to levels of unc-31 null mutant locomotion within 20 h (Fig. 5A,B). Together, these data show that unc-31(rp166) enables temporal depletion of UNC-31 protein to generate robust behavioral phenotypes.
Figure 5.
Auxin-induced UNC-31 knockdown causes defects in locomotion. A, Quantification of locomotory speed (distance traveled/time) of wild-type, unc-31(e928), and unc-31(rp166); Prgef::TIR1 adult hermaphrodites exposed to 1 mm auxin or ethanol for 20 h from the mid-L4 stage; n ≥ 50. Data expressed as mean ± SEM. Statistical significance was assessed by one-way ANOVA multiple comparison with Tukey's multiple-comparisons test (F(5,313) = 165.4, p = 0.0001), n.s., Not significant. B, Quantification of locomotory speed (distance traveled/time) of unc-31(rp166); Prgef::TIR1 adult hermaphrodites exposed to 1 mm auxin or ethanol for 1 h, 3 h, or 20 h; n ≥ 50. Data expressed as mean ± SEM. Statistical significance was assessed by one-way ANOVA multiple comparison with Tukey's multiple-comparisons test (F(5,351) = 96.02, p = 0.0001), n.s.
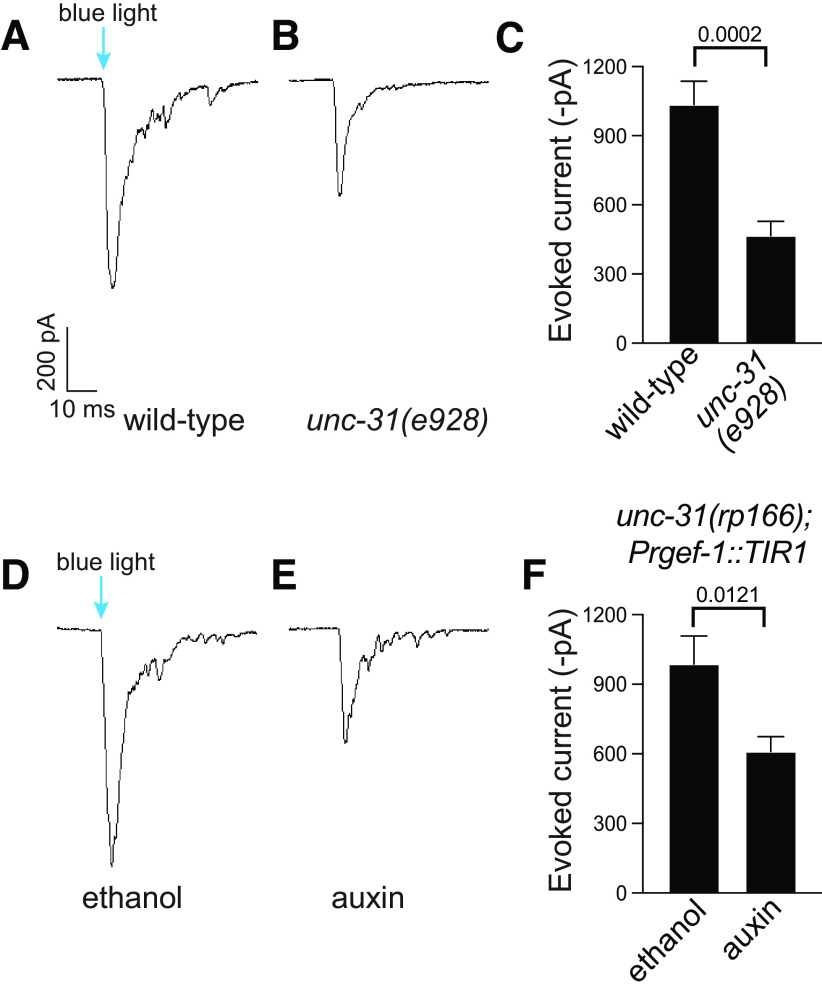
Conditional depletion of UNC-31 causes reduced channelrhodopsin-induced currents at the neuromuscular junction
UNC-31 is a known regulator of DCV release (Gracheva et al., 2007; Sieburth et al., 2007). However, it was previously shown that unc-31 null mutants also exhibit a smaller evoked response to electrical stimulation (Gracheva et al., 2007) and blue-light-induced evoked currents in animals expressing channelrhodopsin (Lindsay et al., 2011). To examine whether auxin-induced depletion of UNC-31 causes defects in vesicle release, we performed electrophysiological recording at the cholinergic neuromuscular junctions (Liu et al., 2018). We expressed channelrhodopsin in cholinergic motor neurons (oxIs364 [Punc-17::ChR2]) and examined evoked currents following blue-light stimulation (Liewald et al., 2008; Liu et al., 2009). As previously reported, we found that unc-31(e928) null mutant animals expressing channelrhodopsin in cholinergic motor neurons exhibit smaller evoked currents compared with wild-type animals (Fig. 6A–C; Gracheva et al., 2007). Next, we examined blue-light-induced inward currents in unc-31(rp166); Prgef-1::TIR1; oxIs364 animals exposed to auxin or the ethanol control for 6 h. We found that unc-31(rp166); Prgef-1::TIR1; oxIs364 animals exposed to ethanol exhibit a response similar to that of wild-type animals (Fig. 6). In contrast, unc-31(rp166); Prgef-1::TIR1; oxIs364 animals exposed to auxin show a significant decrease in evoked amplitudes, which is comparable to unc-31(e928) null mutant animals (Fig. 6). These data show that auxin-induced depletion of UNC-31 causes defects in vesicle release.
Figure 6.
Auxin-induced UNC-31 knockdown reduces channelrhodopsin-induced currents at the neuromuscular junction. A–C, Sample traces (A, B) and quantification (C) of blue-light-stimulated amplitude of evoked EPSCs of wild-type (A) and unc-31(e928) (B) adult hermaphrodites; n = 9–11. Data expressed as mean ± SEM. Statistical significance was assessed by unpaired t test (t(4.548) = 18, p = 0.0002). D–F, Sample traces (D, E) and quantification (F) of blue-light-stimulated amplitude of evoked EPSCs of ethanol-treated (D) and auxin-treated (E) unc-31(rp166); Prgef::TIR1 adult hermaphrodites; n = 6–8. Data expressed as mean ± SEM. Statistical significance was assessed by unpaired t test (t(2.951) = 12, p = 0.0121).
BAG neuron-specific depletion of UNC-31 causes intestinal fat accumulation
Next, we examined the utility and spatial resolution of the UNC-31 conditional depletion tool. The BAG sensory neurons are key regulators of C. elegans behavior and physiology (Zimmer et al., 2009; Brandt et al., 2012; Carrillo et al., 2013; Juozaityte et al., 2017; Handley et al., 2022). We previously showed that animals lacking the BAG neuron fate-determining transcription factor ETS-5 store excess intestinal fat (Juozaityte et al., 2017). In addition, BAG-specific unc-31 RNAi or loss of the INS-1 peptide from the BAGs results in increased fat storage (Juozaityte et al., 2017; Handley et al., 2022). To examine whether the conditional auxin system could be used to confer neuron-specific phenotypes, we expressed TIR1 specifically in the BAG neurons using the gcy-9 promoter (Brandt et al., 2012) and crossed this strain into unc-31(rp166). We exposed wild-type and unc-31(rp166); Pgcy-9::TIR1 animals to ethanol or auxin from L1 to adult and measured fat levels by ORO staining (Fig. 7). We found that BAG-specific UNC-31 depletion increases intestinal fat storage levels (Fig. 7). These data confirm the overall inhibitory effect of BAG-derived neuropeptides on intestinal fat levels.
Figure 7.
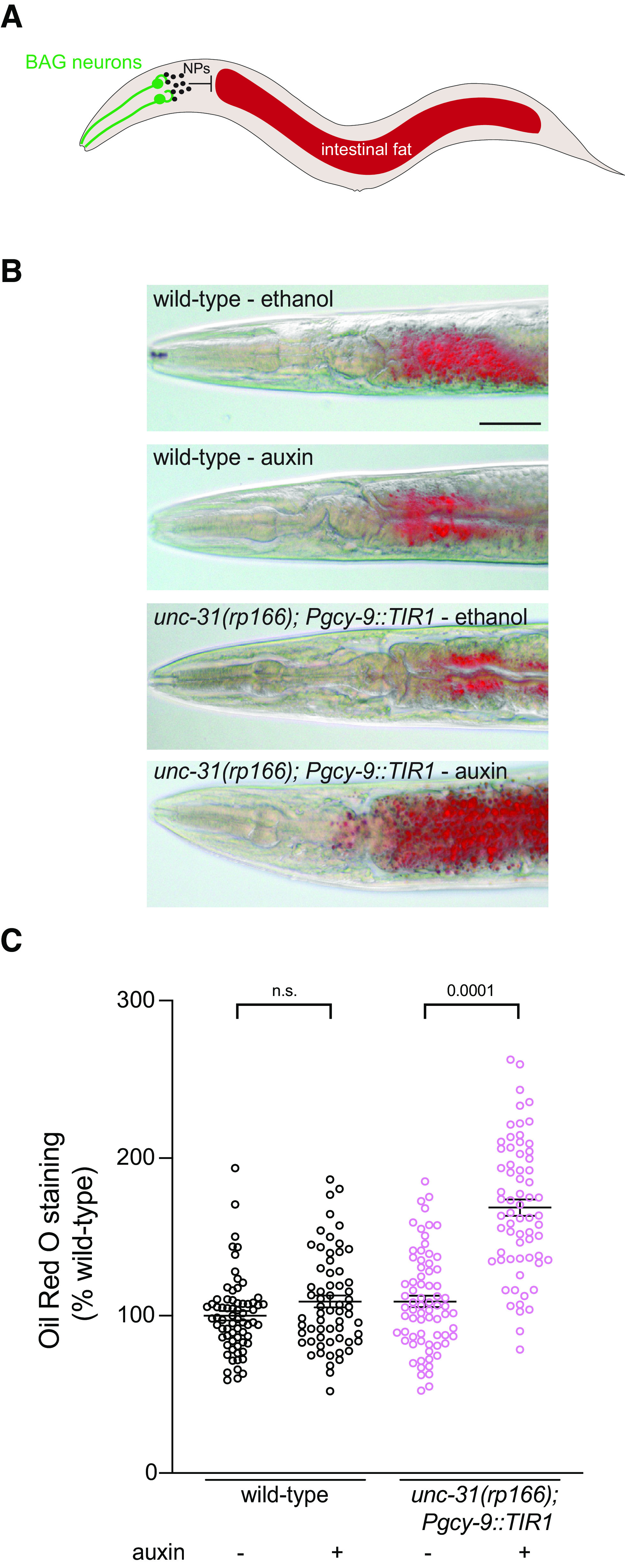
BAG neuron-specific UNC-31 knockdown increases intestinal fat levels. A, Schematic of C. elegans showing that neuropeptides (NPs) released from the BAG neurons inhibit intestinal fat storage (Juozaityte et al., 2017; Handley et al., 2022). B, C, Representative images (B) and quantification (C) of ORO staining in wild-type and unc-31(rp166); Pgcy-9::TIR1 adult hermaphrodites exposed to 1 mm auxin or ethanol from L1 larvae to adult; n ≥ 62. Data presented as ORO intensity as percentage of wild type. Data expressed as mean ± SEM. Statistical significance was assessed by one-way ANOVA multiple comparison with Tukey's multiple-comparisons test (F(3,264) = 63, p = 0.0001), n.s., Not significant. Scale bar, 50 µm.
Discussion
In conclusion, we describe here the generation and validation of a genetic tool that enables the function of the UNC-31/CAPS protein to be dissected in a spatiotemporal manner. In combination with neuron-specific expression of the A. thaliana TIR1 protein, this tool will enable investigators to inhibit neuropeptide release in a highly controlled manner. This will facilitate functional studies of individual postmitotic neurons, which will help to determine the roles of individual neurons and circuits in controlling behavior and physiology. However, the kinetics of UNC-31 knockdown and recovery will need to be considered when performing functional studies.
Footnotes
This work was supported by National Health and Medical Research Council Grant GNT1105374 to R.P. and Veski Innovation Fellowship VIF23 to R.P. Some Caenorhabditis elegans strains were provided by the Caenorhabditis Genetics Center at University of Minnesota, which is funded by National Institutes of Health–Office of Research Infrastructure Programs Grant P40 OD010440. We thank members of the Pocock laboratory for comments on the manuscript and Jordan Ward for providing the pPD357 vector prior to publication.
The authors declare no competing financial interests.
References
- Aljohani MD, El Mouridi S, Priyadarshini M, Vargas-Velazquez AM, Frøkjær-Jensen C (2020) Engineering rules that minimize germline silencing of transgenes in simple extrachromosomal arrays in C. elegans. Nat Commun 11:6300. 10.1038/s41467-020-19898-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley GE, Duong T, Levenson MT, Martinez MAQ, Johnson LC, Hibshman JD, Saeger HN, Palmisano NJ, Doonan R, Martinez-Mendez R, Davidson BR, Zhang W, Ragle JM, Medwig-Kinney TN, Sirota SS, Goldstein B, Matus DQ, Dickinson DJ, Reiner DJ, Ward JD (2021) An expanded auxin-inducible degron toolkit for Caenorhabditis elegans. Genetics 217:iyab006. 10.1093/genetics/iyab006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Bargmann CI, Horvitz HR (1993) The Caenorhabditis elegans unc-31 gene affects multiple nervous system-controlled functions. Genetics 134:455–464. 10.1093/genetics/134.2.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt JP, Aziz-Zaman S, Juozaityte V, Martinez-Velazquez LA, Petersen JG, Pocock R, Ringstad N (2012) A single gene target of an ETS-family transcription factor determines neuronal CO2-chemosensitivity. PLoS One 7:e34014. 10.1371/journal.pone.0034014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77:71–94. 10.1093/genetics/77.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Tran C, Archer SK, Gopal S, Pocock R (2021) Functional recovery of the germ line following splicing collapse. Cell Death Differ 29:772–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo MA, Guillermin ML, Rengarajan S, Okubo RP, Hallem EA (2013) O2-sensing neurons control CO2 response in C. elegans. J Neurosci 33:9675–9683. 10.1523/JNEUROSCI.4541-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlie NK, Schade MA, Thomure AM, Miller KG (2006) Presynaptic UNC-31 (CAPS) is required to activate the G alpha(s) pathway of the Caenorhabditis elegans synaptic signaling network. Genetics 172:943–961. 10.1534/genetics.105.049577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435:441–445. 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Dokshin GA, Ghanta KS, Piscopo KM, Mello CC (2018) Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics 210:781–787. 10.1534/genetics.118.301532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracheva EO, Burdina AO, Touroutine D, Berthelot-Grosjean M, Parekh H, Richmond JE (2007) Tomosyn negatively regulates CAPS-dependent peptide release at Caenorhabditis elegans synapses. J Neurosci 27:10176–10184. 10.1523/JNEUROSCI.2339-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13:1678–1691. 10.1101/gad.13.13.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley A, Wu Q, Sherry T, Cornell R, Pocock R (2022) Diet-responsive transcriptional regulation of insulin in a single neuron controls systemic metabolism. PLoS Biol 20:e3001655. 10.1371/journal.pbio.3001655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juozaityte V, Pladevall-Morera D, Podolska A, Nørgaard S, Neumann B, Pocock R (2017) The ETS-5 transcription factor regulates activity states in Caenorhabditis elegans by controlling satiety. Proc Natl Acad Sci U S A 114:E1651–E1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liewald JF, Brauner M, Stephens GJ, Bouhours M, Schultheis C, Zhen M, Gottschalk A (2008) Optogenetic analysis of synaptic function. Nat Methods 5:895–902. 10.1038/nmeth.1252 [DOI] [PubMed] [Google Scholar]
- Lindsay TH, Thiele TR, Lockery SR (2011) Optogenetic analysis of synaptic transmission in the central nervous system of the nematode Caenorhabditis elegans. Nat Commun 2:306. 10.1038/ncomms1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li L, Wang W, Gong J, Yang X, Hu Z (2018) Spontaneous vesicle fusion is differentially regulated at cholinergic and GABAergic synapses. Cell Rep 22:2334–2345. 10.1016/j.celrep.2018.02.023 [DOI] [PubMed] [Google Scholar]
- Liu Q, Hollopeter G, Jorgensen EM (2009) Graded synaptic transmission at the Caenorhabditis elegans neuromuscular junction. Proc Natl Acad Sci U S A 106:10823–10828. 10.1073/pnas.0903570106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggott BJ, Liu J, Feng Z, Wescott SA, Xu XZ (2011) The neural circuits and synaptic mechanisms underlying motor initiation in C. elegans. Cell 147:922–933. 10.1016/j.cell.2011.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Davis WS, Jorgensen EM (1999) UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci 2:959–964. 10.1038/14755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12:198–207. 10.1101/gad.12.2.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth D, Madison JM, Kaplan JM (2007) PKC-1 regulates secretion of neuropeptides. Nat Neurosci 10:49–57. 10.1038/nn1810 [DOI] [PubMed] [Google Scholar]
- Speese S, Petrie M, Schuske K, Ailion M, Ann K, Iwasaki K, Jorgensen EM, Martin TF (2007) UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in Caenorhabditis elegans. J Neurosci 27:6150–6162. 10.1523/JNEUROSCI.1466-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Ward JD, Cheng Z, Dernburg AF (2015) The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C-elegans. Development 142:4374–4384. 10.1242/dev.129635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer M, Gray JM, Pokala N, Chang AJ, Karow DS, Marletta MA, Hudson ML, Morton DB, Chronis N, Bargmann CI (2009) Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron 61:865–879. 10.1016/j.neuron.2009.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]