Abstract
To evaluate the application of cycle threshold (Ct) values of coronavirus disease 2019 (COVID-19) patients in predicting epidemic dynamics and monitoring surface contamination. The Ct value of reverse transcriptase-polymerase chain reaction for SARS‑CoV-2 from COVID-19 patients inbound overseas in Xiamen, China was collected from October 2020 to December 2021, and the correlation of patients' Ct values with epidemic dynamics and surface contamination was evaluated. The results showed that there was an extreme inverse correlation of positivity rate in the current calendar month (ORF1ab, r = −0.692, P = 0.004; N,r = −0.629, P = 0.012) and the following calendar month (ORF1ab,r = −0.801, P = 0.001; N,r = −0.620, P = 0.018) with the median Ct values. Ct value showed better performance for monitoring surface contamination, with the area under the curve value 0.808(95 %CI: 0.748–0.869) for ORF1ab and 0.807(95 %CI:0.746–0.868) for the N gene. The patients’ ORF1ab Ct value< 29.09 or N Ct value< 28.03 were 11.25 times and 10.48 times more likely to result in surface contamination than those with ORF1ab Ct value ≥ 29.09 or N Ct value≥ 28.03 (OR:11.25,95 % CI: 5.52–22.35; OR:10.48,95 % CI:5.29–20.70). Ct values were associated with the positivity rate in the current or following calendar month and predicted the epidemic dynamics. The Ct values can be used as a predictor for monitoring surface contamination to develop public health responses to COVID-19.
Keywords: COVID-19, Cycle Threshold value, SARS-CoV-2, Epidemic dynamics, Surface contamination, RT-qPCR
Introduction
Reverse transcriptase-polymerase chain reaction (RT-PCR) is the gold standard for diagnosis of coronavirus disease 2019 (COVID-19) infection [1]. The demand for reporting Ct values has been justified to guide the administration of identifying reinfection, therapeutics, or removing isolation precautions [2]. On the contrary, a considerable number of studies have also shown some limitations in applying Ct values directly to individual patient levels and formulating treatment and prognosis schemes [3]. Despite the limitations of using Ct values for individual patient care, the population distributions of Ct values may be useful for forecasting COVID-19 trends [4]. These offer that there may be some benefits associated with the indications provided by Ct values, as they may provide insights into the epidemic dynamics.
Evidence confirms that surface samples taken from COVID-19 patients touch areas for SARS-CoV-2 with positive PCR results, the presence of a viable virus, and the implication for transmission through fomites contaminated by the settling of virus-laden particles onto the surface [5]. However, data on SARS-CoV-2 contamination in patient rooms correlating with patients' Ct values are scarce. This study incorporated Ct values of RT-PCR for SARS‑CoV-2 from COVID-19 patients among inbound overseas travelers in Xiamen, China, from October 2020 to December 2021, and evaluated the application of patients' Ct values in predicting epidemic dynamics and monitoring surface contamination, contributing to estimating epidemic dynamics and developing public health responses to COVID-19.
Material and methods
The participants in the study were the same group in our previous study [6]. A Daan 2019-nCoV RT-PCR Kit (Daan Gene, Guangzhou, China), targeting open reading frame 1a or 1b (ORF1ab) and the nucleocapsid protein (N) gene, was used to detect SARS-CoV-2 according to the manufacturer’s instructions. The surface samples were collected within 4 h when patients were confirmed positive for SARS-CoV-2. Surface contamination was defined as any surface sample in the patient’s room positive for SARS-CoV-2. The correlation of the results according to their r values was categorized as extreme (0.81–1.0), strong (0.61–0.8), moderate (0.41–0.6), and weak or poor (0–0.4). The χ 2 test was used for categorical variables, and odds ratios (OR) were calculated with a 95 % confidence interval (CI). Receiver operating characteristic (ROC) curves were used to evaluate the diagnostic values. The Youden index was calculated to determine the cutoff value. Statistical significance was defined as a two-sided P value of< 0.05.
This study was approved by the Ethics Committee of Zhongshan Hospital, Xiamen University(approval number,xmzsyyky2022–139), and the requirement for written informed consent was waived by the ethics committee owing to the emergent nature of COVID-19.
Results
Association of the Ct values with the positivity rate of the COVID-19 in the calendar month
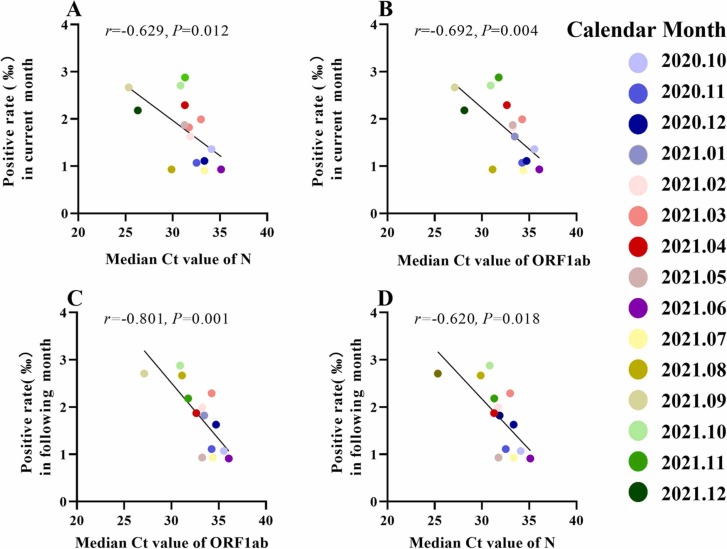
The trend of positivity rate over calendar month was showed in the Fig. S1. The median Ct values of ORF1ab and N had similar trajectories over the calendar month from October 2020 to December 2021. (Fig. S2). There was a strong inverse correlation between the positivity rate in the current calendar month and the median Ct values (ORF1ab, r = −0.692, P = 0.004; N, r = −0.629, P = 0.012). Interestingly, a strong, even extreme inverse correlation was found between the positivity rate and median Ct values in the following calendar month (ORF1ab, r = −0.801, P = 0.001; N, r = −0.620, P = 0.018) ( Fig. 1). These results showed that Ct values were associated with the positivity rate in the current calendar month and even predicted the trend of positivity rate in the following calendar month.
Fig. 1.
Association of the positivity rate of COVID-19 with median Ct values. A-B: Associations of the positivity rate of COVID-19 in the current month with median Ct values of ORF1ab (A) or the N gene (B); C-D: Associations of the positivity rate of COVID-19 in the following month with median Ct values of ORF1ab (C) or the N gene (D). Spearman analysis was performed to determine the correlation between the positivity rate of COVID-19 and the median Ct values. Each dot represents the median Ct value (n = 15 for the current month and n = 14 for the following month). The straight line in the graph is the result of the linear regression analysis. Spearman's correlation coefficients (r) and P values are shown.
Monitoring surface contamination by the Ct value
In this study, 188 surface samples were collected from 519 COVID-19 patients to assess surface contamination, 104 (55.32 %) with surface contamination, and 84 (44.68 %) with no surface contamination. ORF1ab and N genes had a high predictive performance for surface contamination, with an area under the curve of 0.808 (95 % CI:0.748–0.869), 0.807 (95 % CI:0.746–0.868), respectively(Fig. S3).
The cut-off of Ct value was recommended by Youden index values (29.09 for ORF1ab and 28.03 for N gene). Patients’ ORF1ab gene Ct value< 29.09 or N gene Ct value< 28.03 were 11.25 times and 10.48 times more likely to result in surface contamination than those with ORF1ab Ct value ≥ 29.09 or N Ct value ≥ 28.03 (OR: 11.25, 95 % CI: 5.52–22.35; OR: 10.48, 95 % CI: 5.29–20.70) ( Table 1).
Table 1.
Monitoring surface contamination by the Ct value.
| Gene | Ct value | Surface contamination, n | No surface contamination, n | Sensitivity (%) | Specificity (%) | OR (95 % CI) | P for OR |
|---|---|---|---|---|---|---|---|
| ORF | ≥29.09 | 14 | 72 | 1 | |||
| <29.09 | 70 | 32 | 83.3 (73.9–89.8) | 69.2 (59.8–77.3) | 11.25(5.52–22.35) | < 0.001 | |
| N | ≥28.03 | 16 | 74 | 1 | |||
| <28.03 | 68 | 30 | 81.0 (71.3–87.9) | 71.1 (61.8–78.9) | 10.48(5.29–20.70) | < 0.001 |
Ct, threshold cycle; OR, odds ratio.
The chi-square test was used to explore the odds ratio of Ct value for predicting surface contamination.
Ct, threshold cycle; OR, odds ratio.
Discussion
Estimating the epidemic is crucial for developing public health responses to COVID-19 [7]. The study showed that the median Ct values of both ORF1ab and N genes had a strong inverse relationship with the positivity rate of inbound overseas in the current calendar month and even had an extreme inverse relationship with the following calendar month. The results confirmed that the distribution of Ct values coincided with changes in the epidemic trajectory. Using Ct data collected would help estimate an epidemic's trend.
Interpersonal SARS-CoV-2 transmission primarily occurs by respiratory droplets or contact [8]. The Ct values of the PCR reaction correlate inversely with the viral load; low Ct values indicate high viral loads and vice versa [9]. Increased viral loads facilitate spread by increasing the risk of infection transmission. The results showed that the Ct value had a better predictive performance, with an area under the curve of 0.807 for the ORF1ab gene and 0.808 for the N gene, to monitor surface contamination. The results also showed that surface contamination was significantly more likely in participants with low Ct values. A recent study about stability of SARS-CoV-2 indicated that infectious aerosols persist for several hours and on surfaces for as long as two days [10]. SARS-CoV-2 transmission may occur via direct contact with an infected person or by settling virus-laden particles on the surface [11]. Knowledge regarding the information on SARS-CoV-2 is continuously evolving as new evidence accumulates [12]. Due to the high surface contamination rate (55.32 %, 104/188) of COVID-19 patients and the possibility of the presence of SARS-CoV-2 on inanimate surfaces, regular disinfection practices and washing hands would reduce the likelihood of coronavirus transmission by this potential route of infection, especially in patients with low Ct values. This study only predicted the Ct value for surface contamination.
Conclusion
Ct values were associated with the positivity rate in the current or following calendar month and predicted the epidemic dynamics. The Ct values can be used as a predictor for monitoring surface contamination to develop public health responses to COVID-19.
Funding
This work was supported by the National Natural Science Foundation of China [grant numbers 82172331, 81972028, 81973104, 81772260, 81672094], the Key Projects for Province Science and Technology Program of Fujian Province, China [grant number 2020D017], the Natural Science Foundation of Fujian Province, China [grant number 2021J02055] and the Guiding Projects of Xiamen Science and Technology Program [grant number 3502Z20214ZD1056]. The funders played no role in the study design, data collection, or analyses, the decision to publish, or manuscript preparation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jiph.2022.11.012.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Stokes W., Kanji J., Hu J., Zelyas N., Berenger B.M. Wide variation in cycle threshold values cloud the interpretation of COVID-19 infectiousness. Clin Chem. 2021:1–8. doi: 10.1093/clinchem/hvab145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ade C., Pum J., Abele I., Raggub L., Bockmuhl D., Zollner B. Analysis of cycle threshold values in SARS-CoV-2-PCR in a long-term study. J Clin Virol. 2021;138:1–10. doi: 10.1016/j.jcv.2021.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez M.J., Basile L., Siso-Almirall A., Cristino V., Cuesta G., Carlos Hurtado J., et al. Lack of prognostic value of SARS-CoV2 RT-PCR cycle threshold in the community. Infect Dis Ther. 2022;11:587–593. doi: 10.1007/s40121-021-00561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin N., Dellicour S., Daubie V., Franco N., Wautier M., Faes C., et al. Leveraging of SARS-CoV-2 PCR cycle thresholds values to forecast COVID-19 trends. Front. Med. 2021;8 doi: 10.3389/fmed.2021.743988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanamori H. An overview of research on air and environmental contamination with severe acute respiratory coronavirus virus 2 (SARS-CoV-2) in healthcare settings. Infect Control Hosp Epidemiol. 2022;43:130–133. doi: 10.1017/ice.2020.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y.-J., Xue J.-H., Fang Z.-X., Xie J.-W., Niu J.-J., Yang T.-C., et al. A 14+7 day quarantine period and a dual nucleic acid testing reagent strategy detect potentially indiscoverable Coronavirus disease 2019 infections in Xiamen, China. Clin Chim Acta. 2022;532:89–94. doi: 10.1016/j.cca.2022.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson E.J., McKune S.L., Ryan K.A., Shapiro J., Mott-Young A.H., Myers P.D., et al. Antigen vs RT-PCR tests for screening quarantined students in Florida during the COVID-19 pandemic SARS-CoV-2 delta variant surge. JAMA Pediatr. 2022:1–10. doi: 10.1001/jamapediatrics.2022.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinoi A., Feltracco M., Chirizzi D., Trabucco S., Conte M., Gregoris E., et al. A review on measurements of SARS-CoV-2 genetic material in air in outdoor and indoor environments: Implication for airborne transmission. Sci Total Environ. 2022;809 doi: 10.1016/j.scitotenv.2021.151137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabaan A.A., Tirupathi R., Sule A.A., Aldali J., Al Mutair A., Alhumaid, Muzaheed S., et al. Viral dynamics and real-time RT-PCR Ct values correlation with disease severity in COVID-19. Diagnostics (Basel) 2021;11:1–10. doi: 10.3390/diagnostics11061091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phuong Mai Le. Thi, Tran V.D., Mai Phuong H.V., Thi Hong Trang U., Thanh L.T., Vu Son N., et al. Investigation of SARS-CoV-2 presence on environmental surfaces and waste in healthcare and non-healthcare facilities. Environ. Challenges. 2022;7 doi: 10.1016/j.envc.2022.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stiefel U., Bhullar D., Zabarsky T.F., Palmieri N.F., Diaz K.D., Torres-Teran M.M., et al. Healthcare personnel frequently have positive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigen tests 5 days or more after diagnosis of coronavirus disease 2019 (COVID-19) Infect Control Hosp Epidemiol. 2022:1–8. doi: 10.1017/ice.2022.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material