Abstract
The factors that influence survival during severe infection are unclear. Extracellular chromatin drives pathology, but the mechanisms enabling its accumulation remain elusive. Here, we show that in murine sepsis models, splenocyte death interferes with chromatin clearance through the release of the DNase I inhibitor actin. Actin-mediated inhibition was compensated by upregulation of DNase I or the actin scavenger gelsolin. Splenocyte death and neutrophil extracellular trap (NET) clearance deficiencies were prevalent in individuals with severe COVID-19 pneumonia or microbial sepsis. Activity tracing by plasma proteomic profiling uncovered an association between low NET clearance and increased COVID-19 pathology and mortality. Low NET clearance activity with comparable proteome associations was prevalent in healthy donors with low-grade inflammation, implicating defective chromatin clearance in the development of cardiovascular disease and linking COVID-19 susceptibility to pre-existing conditions. Hence, the combination of aberrant chromatin release with defects in protective clearance mechanisms lead to poor survival outcomes.
Keywords: DNA, degradation, proteomics, inflammation, sepsis, DNase I, NETs, actin, COVID-19, histone
Graphical abstract
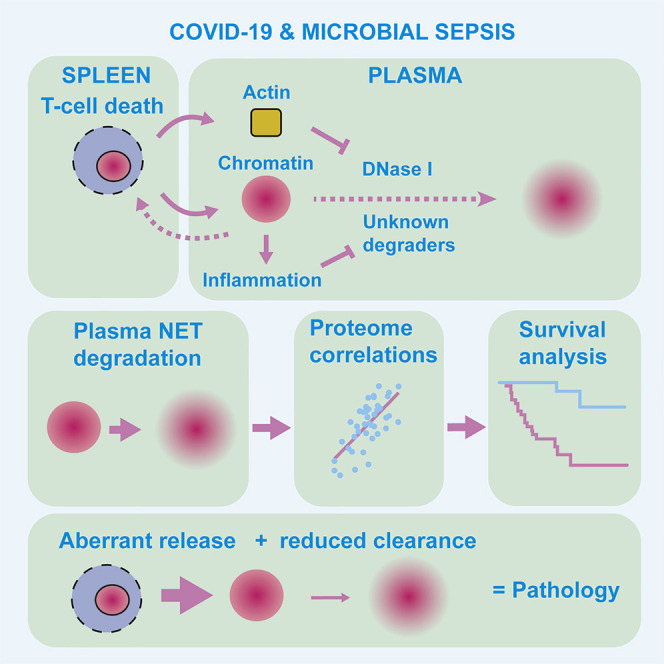
What determines survival during severe infection remains unclear. Aramburu, Hoving, et al. profile the plasma proteome of individuals with COVID-19 pneumonia or sepsis and link low extracellular chromatin clearance with increased mortality and inflammation that may pre-dispose healthy individuals to metabolic syndrome and pathology upon infection. Hence, poor survival results from a combination of aberrant release and low chromatin clearance.
Introduction
Microbial sepsis and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection cause high mortality despite available treatment options (Afzali et al., 2021; Finelli et al., 2021; Rudd et al., 2020). These conditions share common pathological features such as hyper-inflammation, coagulopathy, and immune dysfunction that can culminate in organ failure (Chousterman et al., 2017; Iba et al., 2020; Yang et al., 2021). Sepsis due to secondary co-infection is frequently encountered in severe COVID-19 infection (Langford et al., 2020). The underlying causes of pathologies such as coagulopathy and the mechanisms that counter them to reduce mortality during acute systemic infection are poorly understood.
Aberrant T cell death and excessive neutrophil extracellular trap (NET) release are major drivers of extracellular chromatin (Ioannou et al., 2022) that accumulates in microbial sepsis and SARS-CoV-2 infection and promotes hyperinflammation, coagulopathy, and immune dysfunction (McDonald et al., 2017; Tsourouktsoglou et al., 2020; Xu et al., 2009, 2011; Zuo et al., 2020). Whether these events must also be accompanied with a reduction in DNA clearance capacity for pathogenic extracellular chromatin to accumulate is unclear. Extracellular DNA degradation by serum endonucleases DNase I and DNase I L3 suppresses inflammation by decoupling the synergistic action of histones and DNA and exposes histones for degradation (Ioannou et al., 2022; Jimenez-Alcazar et al., 2017; Tsourouktsoglou et al., 2020). Given these clearance mechanisms, it is unclear why cell-free chromatin accumulates in the circulation upon acute infection.
Mice lacking both DNase I and DNase I L3 accumulate extracellular DNA in the circulation and develop excessive thrombosis upon sterile and microbial challenges (Jimenez-Alcazar et al., 2017). Deficiencies in extracellular DNA and NET clearance have been observed in individuals with autoimmune disease, coagulopathy, and thermal injury (Dinsdale et al., 2020; Dittmar et al., 2007; Hakkim et al., 2010; Jimenez-Alcazar et al., 2015; Martinez-Valle et al., 2009). Whether extracellular DNA clearance varies among healthy or infected individuals and its role in survival are unclear. Systemic actin release is also observed upon severe infection or injury (Belsky et al., 2018; Horvath-Szalai et al., 2017; Lee et al., 2007, 2008; Messner et al., 2020). F-actin scavenging systems control actin toxicity driven by aberrant polymerization in the circulation (Lee and Galbraith, 1992). Monomeric and filamentous actin inhibits DNase I, but its impact on extracellular chromatin clearance is poorly characterized (Lazarides and Lindberg, 1974; Morrison and Dawson, 2007; Podolski and Steck, 1988).
Here, we show that T cell-dependent actin release is associated with extracellular chromatin clearance deficiency and compensatory endonuclease upregulation. We then employ activity-based proteomic profiling to investigate the regulation and impact of extracellular chromatin clearance on SARS-CoV-2 pathology.
Results
T cell-dependent actin release modulates DNA clearance
We recently showed that T cell death promotes mortality by releasing chromatin in murine systemic candidiasis (Ioannou et al., 2022). Cell death was absent in the spleens of T cell receptor-α-deficient (TCRα−/−) mice lacking T cells that were infected intravenously with Candida albicans and stained for T cells (CD3) and apoptotic chromosomal fragmentation using terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) (Figures 1A and S1A). To understand the impact of T cell death on pathology, we employed mass spectrometry to analyze the plasma proteomes of naive or infected wild-type (WT) and TCRα−/− mice. Infected TCRα−/− mice exhibited a distinct proteomic profile compared with WT control animals (Figures 1B and 1C). Several actin peptides (Acta2, Actb, and Actg1) were among the plasma proteins whose abundance increased upon infection in WT mice but remained significantly lower in TCRα−/− mice (Figures 1D and S1B). T cell-dependent actin release was also detected in the plasma of mice infected intravenously with Staphylococcus aureus (Figure S1F). These results suggested that T cell death releases actin upon systemic infection. Major clotting components such as fibrinogen A, B, and G (FGA, FGB, and FGG) were also elevated in a T cell-dependent manner, suggesting that actin release correlates with an increased risk for thrombosis (Figures 1D and S1B).
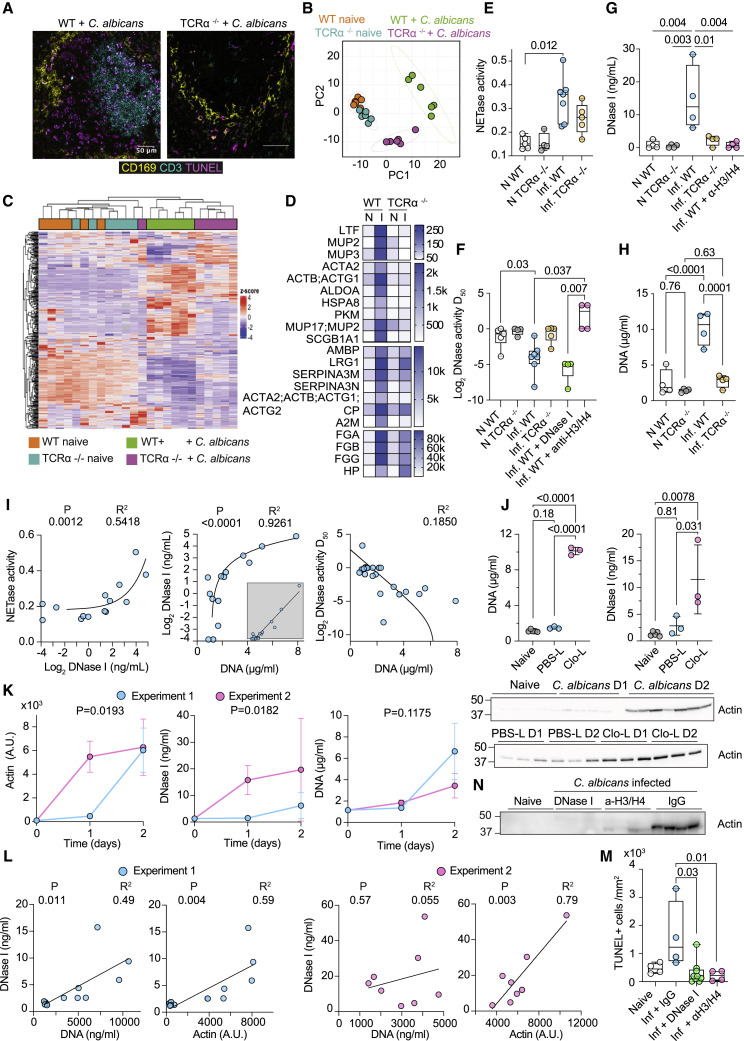
Figure 1.
T cell death promotes actin release and DNase I upregulation in systemic candidiasis
(A) Immunofluorescence confocal micrographs of spleens from WT and TCRα−/− mice infected intravenously with C. albicans, 3 days post-infection stained for CD169 (marginal-zone macrophages, yellow), CD3 (T cells, cyan), and TUNEL (apoptosis, magenta). Scale bars: 50 μm.
(B) Principal-component analysis of plasma proteomes from naive and infected WT and TCRα−/− mice analyzed by mass spectrometry.
(C) Heatmap depicting unbiased hierarchical clustering of plasma proteomes of mice in (B).
(D) Heatmap of plasma proteins that are upregulated in infected WT but not in infected TCRα−/− mice. N, naive; I, infected.
(E–I) Naive (N) or infected (Inf) WT or TCRα−/− mice untreated or treated with DNase I or anti-histone H3 and H4 antibodies.
(E) NET degradation (NETase) activity.
(F) DNase activity expressed as the dilution of murine plasmas required for 50% plasmid degradation (D50).
(G and H) DNase I and DNA concentrations in murine plasma.
(I) Correlations between plasma DNase I, DNA, DNase (D50), or NETase activities.
(J) Plasma DNA, DNase I, and actin concentrations in naive, control liposome (PBS-L)-, or clodronate liposome (Clo-L)-treated mice or infected with C. albicans, 1 or 2 days post-challenge.
(K) Kinetics of plasma DNA, DNase I, and actin in naive (T = 0) and C. albicans-infected (T = 1, 2 days) WT mice in two separate experiments.
(L) Correlation analysis between plasma actin and DNA or DNase I in experiment 1 (left panels, blue circles) and experiment 2 (right panels, pink circles).
(M and N) TUNEL+ splenic cells and plasma actin in naive or infected WT mice treated with control IgG antibody, anti-histone H3 and H4 antibodies, or DNase I. Statistics by Mann-Whitney or Kruskal-Wallis test for single comparisons, one-way ANOVA for multiple comparisons, simple linear or non-linear regression for correlations. See also Figure S1.
As actin inhibits DNase I, we hypothesized that actin release interfered with extracellular DNA clearance. Thus, we measured plasma DNA degradation against naked plasmid DNA or human NETs (Figures S1C and S1D). Plasmid DNA degradation by human healthy donor (HD) plasma was inhibited by G-actin, whereas NET degradation (NETase) was insensitive to G-actin inhibition (Figure S1E). These results suggested that plasmid DNA degradation reports on DNase I activity, whereas the dissolution of NETs involves the synergistic activity of multiple factors, like DNase I L3, which is resistant to actin, as well as proteases that degrade histones. We measured DNase activity at 0.01%, 0.05%, 0.1%, and 0.5% murine plasma dilutions and fitted the data to estimate the dilution required to reach 50% activity (D50). The plasma of infected symptomatic animals exhibited significantly higher NET and plasmid DNA degradation activity compared with naive mice (Figures 1E and 1F). Elevated activity was accompanied with the upregulation of plasma DNase I in infected WT mice, which was absent in infected TCRα−/− mice or WT mice treated with histone-blocking antibodies (Figure 1G). Higher DNase activity was also detected 72 h post-infection in WT mice receiving daily DNase I injections. These findings suggested that chromatin accumulation promoted the upregulation of DNase I as a compensatory mechanism. Consistently, DNA accumulated in infected WT but not in infected TCRα−/− plasma (Figure 1H). Moreover, chromatin degradation activities and DNA positively correlated with plasma DNase I in naive and infected WT and TCRα−/− mice (Figure 1I).
To address whether cell death promoted DNase I upregulation, we induced splenic macrophage cell death in uninfected mice using clodronate liposomes (Clo-Ls). Control liposomes (PBS-Ls) induced low actin release without DNA release or DNase I upregulation. In contrast, actin and DNA accumulation in Clo-L-treated mice was accompanied with a potent DNase I upregulation (Figure 1J). Therefore, cell death was sufficient to upregulate DNase I in the absence of infection, whereas lower actin concentrations did not promote DNA accumulation and DNase I upregulation. To further explore the relationship between DNA, actin, and DNase I, we examined their kinetics. In one experiment, actin release occurred 24 h post-infection, resulting in potent DNase I upregulation early post-infection and suppression of plasma DNA accumulation. By comparison, experiments where actin release occurred 48 h post-infection exhibited slower DNase I upregulation and higher DNA accumulation (Figure 1K). Actin and DNA correlated with DNase I in the samples associated with slow actin release, whereas DNase I correlated with actin but not with DNA in the experiment that exhibited faster actin release (Figure 1L). Hence, robust actin release was the prevailing factor in DNase I upregulation and subsequent DNA accumulation. Moreover, histone blockade reduced T cell death and actin release, explaining the lack of upregulation in DNase I and DNA clearance activity and exposing a feedback loop where histones promote further cell death and actin release (Figures 1F, 1G, 1M, and 1N). Therefore, actin release promoted chromatin accumulation and the subsequent upregulation of endonucleases to overcome endonuclease inhibition.
Chromatin clearance is reduced in sepsis and COVID-19 pneumonia
Considering that lymphopenia and T cell death are prominent features of severe COVID-19 pneumonia, we explored whether our observations in murine sepsis were relevant in SARS-CoV-2 infection. We stained post-mortem spleens for T cells, TUNEL, and myeloperoxidase (MPO) to document neutrophil infiltration. Cell death was evident in 6 out the 9 donors with a significant number of T cells being TUNEL+ (Figures 2A–2C). The TUNEL+ fraction included neutrophils but few CD163+ macrophages (Figure S1G). We also examined the relationship between plasma actin protein measured by mass spectrometry and lymphopenia in 465 blood samples from 63 CP (participants with severe COVID-19 pneumonia) who had reached maximum World Health Organization (WHO) ordinal severity scale grade 7 (WHO-7) (Figure S2A). We used the final samples collected from each participant because they exhibited the greatest degree of variance. Blood lymphocyte counts were inversely correlated with actin concentrations (Figure 2D). These findings linked cell death with actin release and are consistent with previous reports of lymphocytopenia and splenic cell death in severe CP (Ping et al., 2021; Xiang et al., 2021).
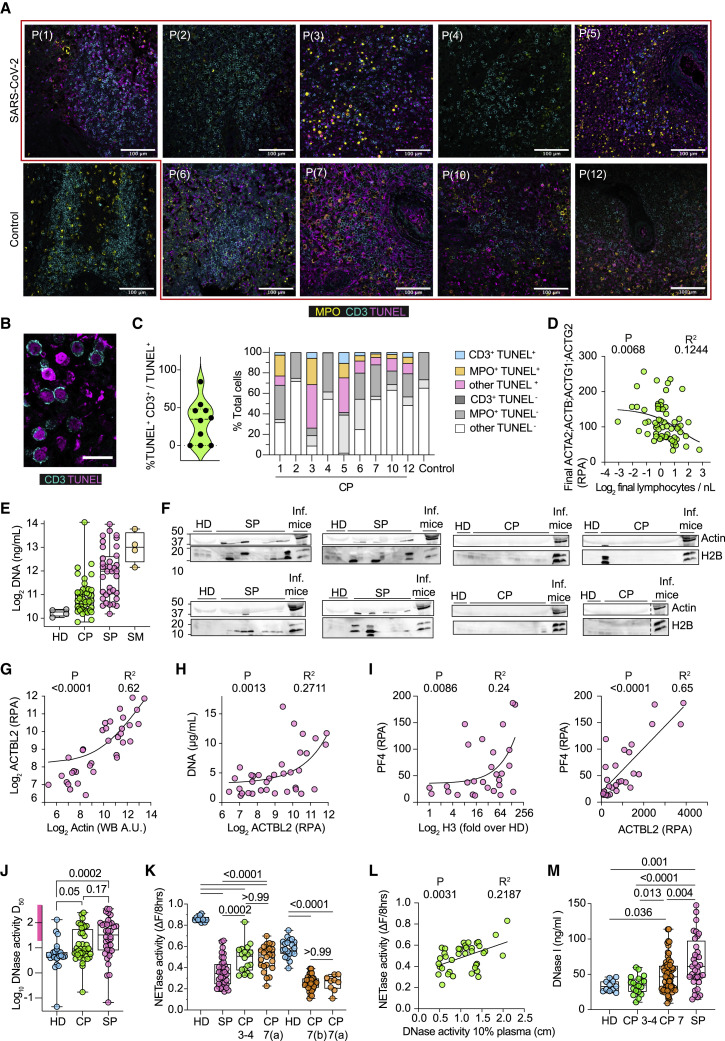
Figure 2.
Splenic cell death and reduced plasma DNA degradation capacity in sepsis and COVID-19 pneumonia
(A) Representative immunofluorescence confocal micrographs of post-mortem spleens from 1 control and 9 individuals with CP, stained for T cells (CD3, cyan), TUNEL (magenta), and myeloperoxidase (MPO, neutrophils; yellow). Scale bars: 100 μm.
(B) High-magnification micrographs from (A). Scale bars: 20 μm.
(C) Quantification of the percentage of TUNEL+ cells that are CD3+ in the splenic tissues shown in (A) (left) and distribution of TUNEL+ T cells (CD3+), neutrophils (MPO+), or other cells in (A) (right).
(D) Correlation between plasma actin (ACTA2; ACTB; ACTG1) relative protein abundance (RPA) and blood lymphocyte counts in the final sample obtained from 63 participants with CP who reached WHO-7.
(E) Plasma DNA in healthy (HD, gray), CP (green), and sepsis participants (SP, magenta) or mice infected with C. albicans (SM, yellow).
(F) Plasma samples from HD, SP, CP, and SM immunoblotted for actin and histone H2B.
(G) Correlation between actin abundance measured by densitometric analysis of the western immunoblots in (F) or mass spectrometry in SP plasma.
(H) Correlation between SP plasma actin and DNA.
(I) Correlation between histone H3 measured by western immunoblot (left) or actin (right) and platelet factor 4 (PF4) measured by mass spectrometry.
(J) Plasmid DNA degradation by 30 HD, 40 CP, and 36 SP plasmas. D50 values were calculated from raw data in Figures S2D and S2E. Low activity range (D50 > 15) depicted by the purple bar on the y axis.
(K) NETase activity in 3% plasma from 30 HD, 87 CP, and 36 SP measured by timelapse microscopy as the loss in human NET DNA intensity over 8 h.
(L) Correlation between DNase activity at 10% plasma dilution and NETase activity in CP plasmas.
(M) DNase I concentrations in HD (blue) or CP plasmas with WHO-3, -4 (green), or -7 (orange) and SP plasmas (pink). Statistics by Mann-Whitney or Kruskal-Wallis test for single comparisons, one-way ANOVA for multiple comparisons, simple linear or non-linear regression for correlations. See also Figure S2.
We also compared actin, DNA, and histone concentrations in CP samples with 36 plasma samples obtained from SP (participants with microbial sepsis). All SP had refractory septic shock with multi-organ failure as indicated by high norepinephrine doses, high necessity for invasive ventilation, and renal replacement therapy (Figure S2B). DNA concentrations were lower in CP plasma compared with SP plasma, and CP plasma actin and histones were not abundant at concentrations that could be detected by western immunoblotting, suggesting that CP and SP pathologies differ in severity (Figures 2E and 2F). As expected, DNA and histones positively correlated in SP plasma (Figure S2C). To increase the sensitivity of plasma protein detection, we analyzed CP and SP plasmas by mass spectrometry. Actin concentrations in SP plasma were consistent with measurements obtained by western immunoblot quantification and positively correlated with DNA (Figures 2G and 2H). Moreover, SP plasma histones and actin positively correlated with platelet factor 4 (PF4) (Figure 2I). PF4 is secreted by activated platelets and binds to heparin suppressing its anticoagulant effect. Elevated PF4 concentrations are an indicator of coagulopathy (Wegrzyn et al., 2021). Hence, SP and CP plasmas exhibited differences in actin and cell-free chromatin content, and high concentrations of these proteins in SP plasma correlated with coagulopathy markers.
To examine whether plasma actin affected DNA clearance capacity in 36 SP and 43 CP plasmas classified as WHO-3, -4, and -7, we measured DNase activity by plasmid degradation at 2.5%, 5%, and 10% plasma dilutions and NET degradation at a 3% plasma dilution (Figures S2A, S2D, and S2E). We employed the 95% confidence interval range in HD DNase activity to establish a D50 of 15 as the cutoff between low and high activity. Approximately 30% of CP samples and 60% of SP plasma samples exhibited reduced DNase activity compared with the HD group (Figures 2J, S2D, and S2E). Low NETase activity was observed in all SP and CP plasmas and in an additional set of 44 CP WHO-7 samples, including 9 WHO-7 samples from the first batch for validation (Figure 2K). A positive correlation between NETase and DNase activities suggested that loss of DNase activity exacerbates defects in NET dissolution. However, normal DNase activity alone was not sufficient for full NETase activity (Figure 2L). Therefore, low DNase activity was a contributing factor to the defects in NET degradation in a subset of the samples, reiterating that NET disassembly is a complex process mediated by multiple factors.
Next, we investigated which factors contributed to low DNA clearance. In contrast to mice, DNase I upregulation was mild in humans, with a mean increase of 0.3-fold in WHO-7 CP and 2-fold in SP plasmas, compared with the 12-fold increase in infected mice (Figure 2M). DNase activity was elevated in SP plasmas that contained high DNA and histone H3 concentrations and correlated with DNase I, indicating that accumulation of extracellular DNA results in high DNase activity in sepsis (Figures 3A and 3B and S3A). However, NETase activity did not correlate with DNase I (Figure 3B). Furthermore, DNase and NETase activities did not correlate with actin in SP plasmas (Figure 3C). Instead, correlation searches against the plasma proteomes of SP samples and the previously characterized CP samples uncovered a strong correlation with secreted actin scavenger gelsolin (GSN) (Figure 3D) (Messner et al., 2020). In contrast, DNase and NETase activities correlated with plasma actin in CP samples but did not correlate with DNase I or GSN (Figures 3E, 3F, and S3B). These findings suggested that the magnitude of actin release had a different impact on DNA clearance capacity. High actin concentrations in SP were compensated by DNase I and GSN upregulation, whereas compensation was absent in CP, allowing for a direct correlation between degradation activity and actin.
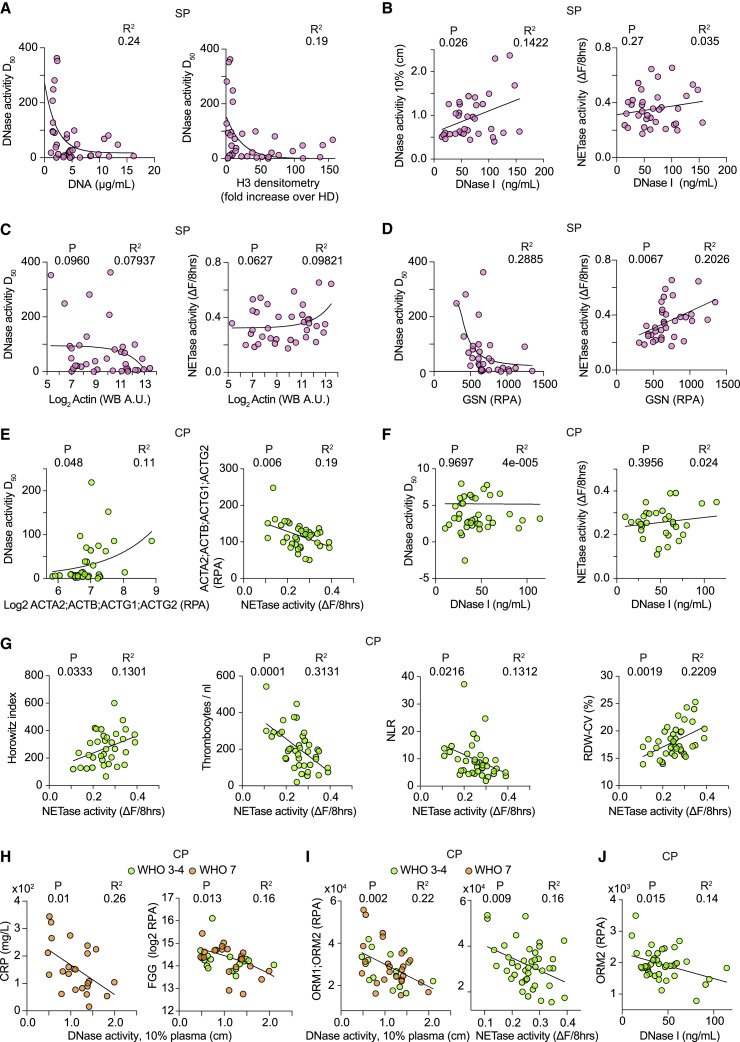
Figure 3.
DNA and NET clearance activities correlate with regulators and disease severity markers in sepsis and COVID-19 pneumonia
(A) Correlation between DNase activity (D50) and DNA (left) or histone H3 (right) in SP samples.
(B) Correlation between DNase I and DNase activity at 10% plasma (left) or NETase activity in SP samples (right).
(C) Correlation between SP plasma actin and DNase activity (D50) (left) or NETase activity (right).
(D) Correlation between SP plasma gelsolin (GSN) and DNase activity (D50) (left) or NETase activity (right).
(E) Correlation between CP plasma actin measured by mass spectrometry and DNase activity (D50) (left) or NETase activity (right).
(F) CP plasma DNase I plotted against DNase activity (D50) (left) or NETase activity (right).
(G) From left to right, HI values, thrombocyte counts, neutrophil to lymphocyte ratio (NLR), and red cell blood distribution width (RDW-CV) plotted against their corresponding NETase activity in plasmas of individuals with COVID-19.
(H) Correlation between raw DNase activity measurements at 10% plasma dilution and CRP (left) or FGG protein (right) in WHO-7 (orange circle) and WHO-3 and -4 (green circle) CP samples.
(I) Correlation between ORM1;ORM2 and DNase activity measurements at 10% plasma dilution (left) or NETase activity (right) in WHO-7 and WHO-3 and -4 CP samples.
(J) Correlation between ORM2 (RPA) and DNase I measured in CP by ELISA. Statistics by Mann-Whitney or Kruskal-Wallis test for single comparisons, one-way ANOVA for multiple comparisons, simple linear or non-linear regression for correlations. See also Figure S3.
Defective NETase and DNase activities were associated with several clinical indications such as a lower Horowitz index (HI), which measures lung function as the ratio of the partial oxygen present in blood over the fraction of oxygen in inhaled air (Figures 3G and S3C). Normal HI values lie between 350–450, and a decrease toward 100 indicates poor lung function and low blood oxygen. Moreover, low NETase activity was associated with thrombophilia, which reflects the severity of coagulopathy, as well as a high neutrophil-to-lymphocyte ratio (NLR) that is linked to COVID-19 pathology and a poor prognosis (Figure 3G) (Hofstra et al., 2007). We also noted that higher NETase activity was associated with a higher red cell distribution width (RDW-CV), a clinical parameter that measures the variability in erythrocyte size (Figure 3G) (Mahmood et al., 2014). RDW-CV is elevated in many conditions and is associated with poor COVID-19 pneumonia outcomes (Jandaghian et al., 2021; Zinellu and Mangoni, 2021). However, we did not see a correlation with the associated mean corpuscular volume (MCV). Hence, low NETase activity correlated with elevated pathological markers but might also have beneficial impacts on specific clinical parameters such as lower RDW-CV readings.
Consistent with the ability of DNases to clear pro-inflammatory extracellular chromatin, low DNase activity in WHO-7 CP samples correlated with elevated C-reactive protein (CRP) and FGG (Figure 3H). Moreover, plasma proteomic profiling indicated that orosomucoid 1 and 2 (ORM1;OMR2) peptides that were elevated in microbial sepsis correlated with low DNase and NETase activities in SP and CP plasmas independently of the severity grade (Figures 3I, S3D, and S3E). High ORM protein concentrations were associated with high sepsis mortality and hinted that low DNA degradation capacity was detrimental (Kustan et al., 2017a, 2017b; Raju et al., 2016). Elevated ORM was also associated with higher rates of sepsis mortality in our study (Figure S3F). Several other proteins correlated with DNase activity in CP samples such as complement factor C8B, complement factor H and CFH related 1 (CFH;CFHR1), soluble CD44, and sex-hormone binding globulin (SHBG) that is elevated in sepsis survivors (Figure S3G) (Tsang et al., 2016). These correlations indicated that low chromatin degradation activity was associated with acute infection pathology.
ORM and actin correlate with COVID-19 pathology and mortality
The association between chromatin clearance and ORM1;ORM2 led us to use these proteins as surrogate markers to track DNA clearance capacity in a longitudinal set of 465 samples from 63 individuals with COVID-19 pneumonia who reached maximum WHO-7. We separated participants based on the survival outcome and plotted the cumulative daily average readings of all participants (Figure 4A). Average ORM1;ORM2 concentrations were high upon admission and decreased steadily thereafter in survivors, whereas they remained persistently high for up to 30 days post-admission in non-survivors. Direct DNase and NETase activity measurements confirmed an overall improvement in DNA clearance capacity over the course of hospitalization when examined collectively or individually (Figures 4B and 4C).
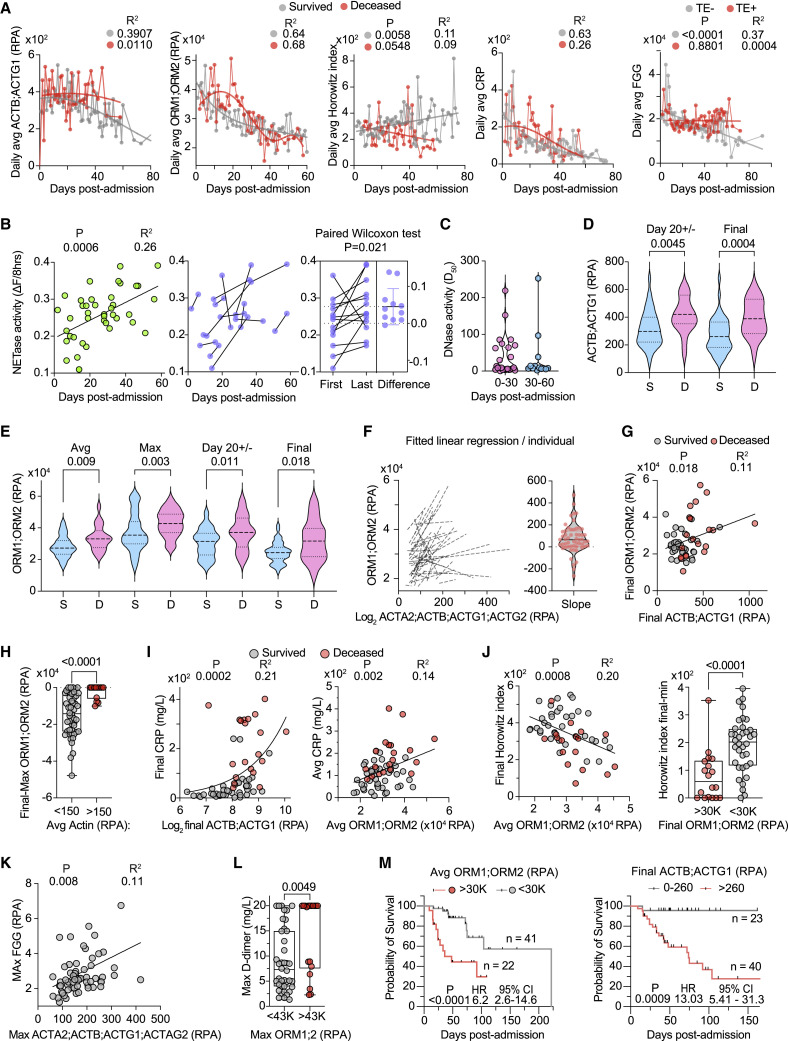
Figure 4.
Relationship between plasma actin, ORM proteins, and infection outcomes
Analysis of 465 samples from 63 CP with maximum WHO-7.
(A) Daily cumulative average relative to time post-admission for actin (ACTB;ACTG1), ORM1;ORM2, HI, and CRP separated by outcome: survivors (gray) or deceased (red). Right: Daily cumulative average FGG in individuals with (TE+ red) or without thromboembolism (TE− gray).
(B) Change in NETase activity over time in 44 CP samples (left). NETase activity in early and late samples connected by a line and the corresponding paired Wilcoxon test for the difference per participant (right).
(C) DNase activity (D50) values obtained from CP samples segregated by 0–30 and 30–60 days post-admission.
(D) Actin (ACTB;ACTG1) abundance in the sample closest to day 20 post-admission (D20+/−) and the final plasma sample obtained from each participant grouped by survival outcome (survived: S, blue; deceased: D, pink).
(E) Average, maximum, D20+/−, and final plasma ORM1;ORM2 per participant grouped by survival outcome.
(F) Fitted linear regression per participant of the correlation obtained from the values for ORM1;ORM2 and actin (ACTA2;ACTB;ACTG1) (left) and violin plot of the corresponding slopes (right).
(G) Correlation between the final values for ORM1;ORM2 and actin (ACTB;ACTG1) per participant who either survived (gray circles) or died (red circles).
(H) Difference between the final and longitudinal maximum ORM1;ORM2 abundance segregated by the corresponding longitudinal average actin values above (red circles) or below (gray circles)150 RPA per participant.
(I) Correlation between the actin and CRP abundance in the final plasma sample collected from each participant (left) or between the longitudinal average ORM1;ORM2 and CRP values (right).
(J) Correlation between the average ORM1;ORM2 and the final HI readings (left). Difference between the final and minimum HI values per participant segregated by whether their final ORM1;ORM2 values were above or below 30,000 RPA (right).
(K) Correlation between longitudinal maximum actin and FGG values.
(L) Longitudinal maximum D-dimer readings in participants with maximum ORM1;ORM2 values above or below 43,000 RPA (right).
(M) Probability of survival in CP clusters according to the longitudinal average values of ORM1;ORM2 (left) or the final actin values (right). Statistics by Mann-Whitney or Kruskal-Wallis test for single comparisons, one-way ANOVA for multiple comparisons, simple linear or non-linear regression for correlations, and Mantel-Cox survival analysis. See also Figure S4.
Similarly, actin abundance decreased 20 days post-admission in survivors but persisted in individuals that succumbed to the infection, alongside a declining daily HI average and higher CRP values between days 20–30 (Figure 4A). HI improved steadily in survivors from a daily average of 220 to above 400. Moreover, participants who experienced thromboembolisms maintained high cumulative daily average FGG levels (Figure 4A). These dynamics were reflected by the differences in the most proximal value to day 20 post-hospitalization (Day20+/−) and the maximum and average ORM and actin values in each participant for the entire hospitalization period (Figures 4D and 4E).
Consistent with the dependence of DNase activity on plasma actin, plasma ORM protein readings correlated with actin and remained high in most individuals with high average actin concentrations (Figures 4F–4H). Moreover, the average ORM and final actin concentrations positively correlated with the average or final CRP values per participant (Figure 4I). Individuals with high average or final ORM1;ORM2 readings exhibited poor lung function recovery as indicated by the low final HI values and smaller differences between the minimum and final readings per participant (Figure 4J). Furthermore, the maximum actin readings per participant positively correlated with the maximum FGG concentrations (Figure 4K). High maximum ORM1;ORM2 concentrations were also associated with elevated readings in the fibrinolysis degradation product D-dimer, consistent with the association of low DNase activity with coagulopathy (Figure 4L). In line with these clinical measurements, participants experienced higher rates of mortality if their longitudinal average ORM1;ORM2 reading surpassed 30,000 relative protein abundance (RPA) that corresponded to the median measured DNase and NETase activities (Figure 4M). Given that high mortality rates occurred approximately 20–30 days post-admission, we also examined mortality in relation to ORM1;ORM2 readings in samples most proximal to day 20 post-admission (Figure S4C). Deceased individuals exhibited higher actin and ORM concentrations during this critical interval (Figures 4D, 4E, S4A, and S4C). Survival differences were also apparent if segregated by the average, final, or D20+/− actin concentrations (Figures 4M, S4A, and S4C). These findings applied regardless of age and co-morbidities because individuals with different average ORM protein reading ranges had similar age ranges and rates of cardiovascular co-morbidities (Figure S4D). Hence, low DNA clearance capacity was associated with elevated plasma actin and lower survival in SARS-CoV-2-infected individuals.
Proteomic profiling links DNase activity with COVID-19 mortality
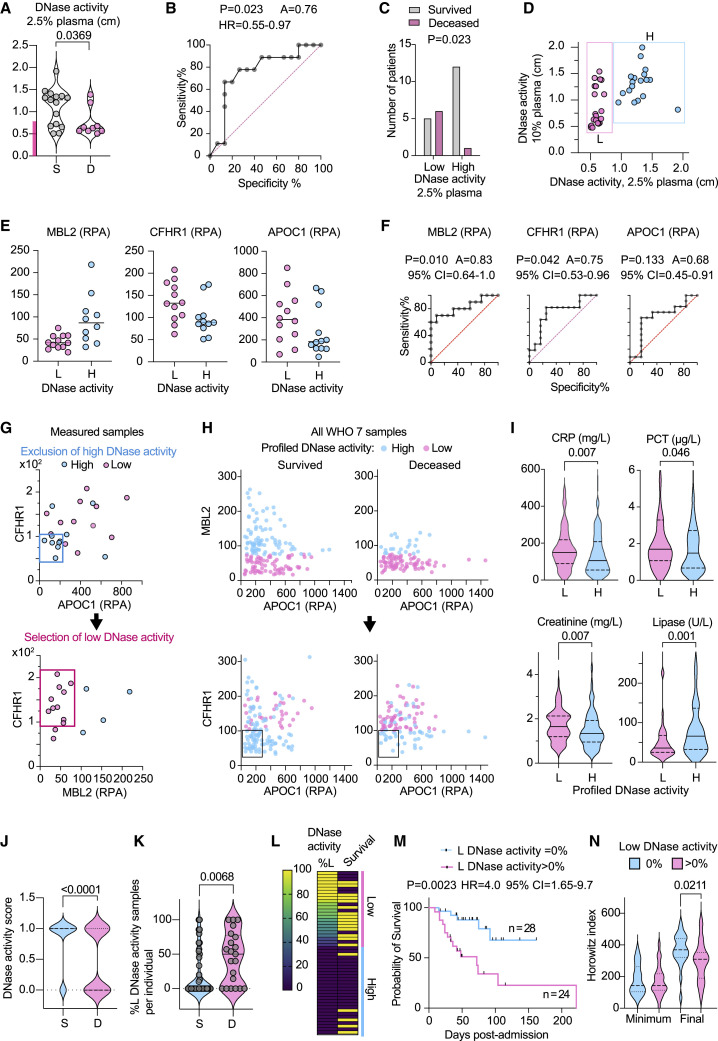
Next, we applied a gating strategy to assign each CP sample to different DNase activity categories based on multifactorial proteomic profiling. Using this digital approach, we analyzed the sample content per participant. Measurements obtained at 2.5% plasma dilution segregated the WHO-7 CP samples into high and low DNase activity clusters (Figure 5A). Plotting DNase activity at different plasma dilutions against the cumulative D50 indicated that the 2.5% dilution results were biphasic, whereas 10% plasma yielded higher continuity in activity variance (Figure S4E). Hence, 10% plasma readings worked better for correlations, whereas 2.5% plasma dilution results were more useful for clustering. At 2.5% plasma dilution, DNase activity could be used to predict mortality (p = 0.023) with an area under the curve (AUC) value of 0.76 (Figures 5B and 5C). Plotting the DNase measurements in 2.5% and 10% plasma dilutions highlighted different categories of samples (Figure 5D). We classified the samples that exhibited low activity at 2.5% dilution but higher activity at 10% dilution into a low activity category and the samples that had high activity at both dilutions into a high category. Proteome correlations identified mannose-binding lectin 2 (MBL2), CFHR1, and apolipoprotein C1 (APOC1) as markers that could segregate samples into different DNase activity groups (Figures 5E and 5F). These factors did not correlate with NETase activity and could be used to identify low DNase activity subsets independently of NETase activity assignments. A two-step gating approach of selecting low activity samples such as MBL2low;CHFR1high and correcting by excluding APOC1low;CHFR1low samples within this group assigned 300 WHO-7 CP samples into low and high DNase activity groups (Figures 5G and 5H). Profiled low DNase activity samples exhibited higher blood CRP, procalcitonin (PCT), and creatinine, indicative of elevated inflammation, bacterial co-infection, and kidney damage, respectively (Figure 5I). On the other hand, low activity samples were associated with reduced lipase concentrations, suggesting a lower incidence of pancreatitis. Moreover, low DNase activity samples were more frequent in non-survivors (Figure 5J) with most survivors either lacking or having only a small fraction of low DNase activity samples compared with the deceased group (Figure 5K). Individuals with low plasma DNase activity samples exhibited a 4.2-fold increase in mortality (Figures 5L and 5M) and lower lung-function recovery (Figure 5N). Hence, DNase activity assignments based on proteomic profiling identified individuals who exhibited increased pathology and mortality within a high-severity cohort.
Figure 5.
Low DNase activity is associated with increased COVID-19 mortality
(A) DNase activity in 25 WHO-7 CP samples measured at 2.5% dilution and segregated by survivors (S, gray) or deceased (D, pink). The pink bar in the y-axis indicates low DNase activity.
(B) AUC analysis of CP samples in (A) segregated by low and high DNase activity.
(C) Distribution of survivors and deceased CP in low and high DNase activity groups.
(D) DNase activity measured at 2.5% and 10% CP plasma dilution. The samples clustered as low degraders (L) are shown in pink, and the high degraders (H) are shown in blue.
(E) MBL2, CFHR1, APOC1 abundance in low (L, pink circles) and high (H, blue circles) DNase activity samples.
(F) AUC analysis of MBL2, CFHR1, APOC1 in low (L, pink circles) and high (H, blue circles) DNase activity samples.
(G) Gating strategy for segregation of measured DNase activity samples by proteomic measurements. In the first gating step CHFR1< 100 RPA; APOC1 < 200 RPA gating profiles a high DNase activity group. A second gating step is applied to the remaining samples profiling CHFR1 > 100 RPA; MBL2 < 80 RPA as low DNase activity samples and the excluded are grouped as high DNase activity samples.
(H) Illustration of how gating strategies in (G) apply on 300 WHO-7 CP samples segregated by survival and plotted for MBL2 and APOC1. The top plots illustrate the profiling of low DNase activity using a MBL2 < 80 RPA gate, and the plots below depict the second application of a CHFR1 > 100 RPA; APOC1 < 200 RPA correction filter to identify high DNase activity samples within the low DNase activity group from step one.
(I) CRP, procalcitonin (PCT), creatinine, and lipase values in 300 WHO-7 CP samples segregated by profiled high and low DNase activity.
(J) Distribution of low and high DNase activity assigned by proteomic profiling of 300 WHO-7 samples from 52 participants with maximum WHO-7 in survivors (blue circle) and deceased participants (pink circle).
(K) Fraction of low DNase activity samples per participant segregated by survival outcome.
(L) Heatmap of 52 participants ranked by their content of profiled DNase activity WHO-7 samples. Each row depicts one individual. The first column depicts the percentage of low DNase activity (%L) per participant and the second column depicts the survival outcome where yellow indicates deceased and purple indicates the survivors. Individuals containing only high profiled DNase activity samples (blue bar) or both high and low DNase activity samples (pink bar) are marked on the right.
(M) Probability of survival segregated by whether participants contained low profiled DNase activity samples (pink). The blue curve depicts participants that contained only high profiled DNase activity samples.
(N) Comparison of the longitudinal minimum and final HI values per participant segregated by the whether they contain low profiled DNase activity samples. Statistics by Mann-Whitney or Kruskal-Wallis test for single comparisons, one-way ANOVA for multiple comparisons, simple linear or non-linear regression for correlations, and Mantel-Cox survival analysis.
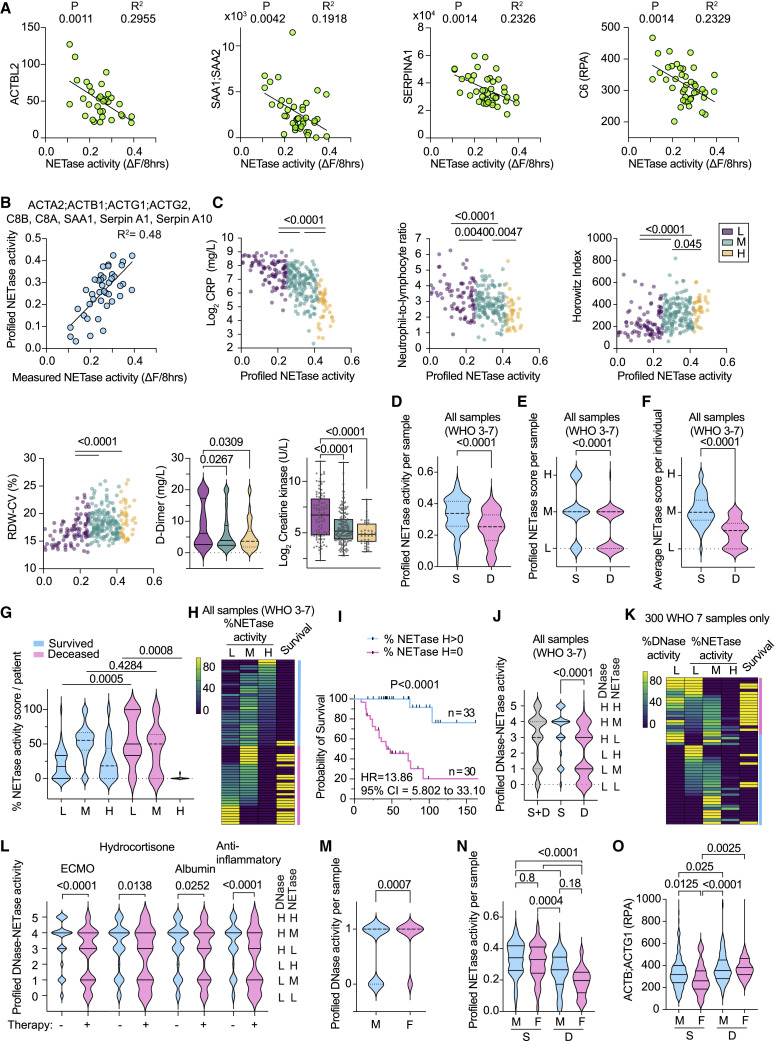
Proteomic profiling links NETase activity with COVID-19 outcomes
To profile NETase activity, we identified 18 correlating markers with high coefficient of determination (R2) values (Figures 6A and S5A). These included complement proteins (C6, C8A, C8B, C9, CP, and CPN1), actin peptides, vitronectin, haptoglobin (HP), serum amyloid A proteins, and serpins A1 and A10 that were upregulated with low NETase activity, as well as afamin (AFM), transthyretin (TTR), and serpin A4 that were downregulated. We employed the equations generated from the linear fitting of the proteomic measurements of each of these factors against the corresponding NETase activity measurements to calculate a predicted NETase value. Averaging these individual predictions reduced noise and improved accuracy as indicated by the R2 value obtained by plotting them against measured NETase activity (Figure S5B). Exploring marker combinations that varied with mortality and improved profiling accuracy when averaged yielded 5 peptides: ActA2;ActB;ActG1;ActG2, C8B, SAA1;SAA2, SeprinA1, and SeprinA10 (Figure 6B). We calculated the profiled NETase activity for all 465 CP samples and sub-divided them into low, medium, and high groups, corresponding to < 0.25, 0.25–0.4, and > 0.4 NETase activity, respectively. These ranges were determined by plotting profiled NETase values against their corresponding CRP readings and examining activity ranges that were associated with shifts in maximum CRP. Nevertheless, what we termed as high activity in CP samples was substantially lower than the average HD NETase activity.
Figure 6.
Low NETase activity is associated with increased COVID-19 mortality
(A) Correlation between NETase activity and ACTBL2, SAA1;SAA2, SERPINA1, and C6 protein measured by mass spectrometry in 41 WHO-7 CP plasmas. Significantly correlating proteins were identified by proteome-wide linear regression.
(B) Plot of measured and profiled NETase activity from averaging profiled NETase activity calculated from corresponding linear regression curves of ACTA2;ACTB1;ACTG1;ACTG2, SAA1;SAA2, SERPINA1, SERPINA10, C8A, and C8B.
(C) Graphs for CRP, NLR, HI, and RDW-CV against the corresponding profiled NETase activity in 300 WHO-7 CP plasmas, color coded by low (purple), medium (green), and high (yellow) profiled NETase activity. Violin plot of D-dimer and bar plot of creatinine kinase readings segregated by low, medium, or high profiled NETase activity.
(D–G) (D) Violin plot of the distribution of profiled NETase activity; (E) the profiled NETase low (L < 0.25), medium (M = 0.25–0.4), and high (H > 0.4) activity score or (F) the average NETase activity score per participant; or (G) the distribution of low (L), medium (M), and high (H) NETase activity samples per participant in 465 samples from 63 individuals with maximum WHO-7, segregated by survival outcome (S, survivors; D, deceased).
(H) Heatmap depicting the distribution of all L, M, and H NETase activity samples per participant in 63 individuals with maximum WHO-7, alongside the survival outcome where yellow indicates deceased and purple indicates survivors. Individuals containing high profiled NETase activity samples (blue bar) and individuals that contain only low and medium NETase activity samples (pink bar) are marked on the right. Each row represents 1 participant.
(I) Survival analysis of participants segregated by whether they contain high profiled NETase activity samples as shown in (H).
(J) Distribution of combined scoring for profiled DNase activity and NETase activity in 465 samples of all severity grades segregated by survival outcome.
(K) Heatmap displaying the low profiled DNase activity WHO-7 sample content and profiled NETase activity content per participant. In the last column, yellow indicates deceased and purple indicates survivors.
(L) Distribution of combined scoring for profiled DNase and NETase activities in 465 samples of all severities from 63 maximum WHO-7 participants segregated by whether they were receiving extracorporeal membrane oxygenation (ECMO), hydrocortisone, albumin, or anti-inflammatory therapies at the time of sample collection.
(M−O) Distribution of 465 samples of all WHO severity grades segregated by sex depicting (M) profiled DNase activity, (N) profiled NETase activity, or (O) plasma actin segregated by survival outcome.
Statistics by Mann-Whitney or Kruskal-Wallis test for single comparisons, one-way ANOVA for multiple comparisons, simple linear or non-linear regression for correlations, and Mantel-Cox survival analysis. See also Figure S5 and S6.
Several clinical parameters were improved with increasing profiled NETase activity, including markers for inflammation, such as CRP and IL-6, the NLR, HI, blood pH, which is indicative of acidosis due to the build-up of CO2, D-dimer, and creatine kinase, that reflect heart inflammation and are associated with a poor prognosis (Figure 6C). Correlations with lactate dehydrogenase (LDH), blood monocyte counts, and other clinical parameters are shown in Figure S6A. These results highlighted a link between profiled NETase activity and pathology in samples associated with the highest clinical severity. To increase longitudinal sample coverage, subsequent analyses were performed on all 465 samples, the majority of which were classified as WHO-6 and -7. The deceased group contained a larger proportion of low NETase activity and fewer high-activity samples compared with survivors (Figures 6D and 6E). This difference was also reflected in longitudinal traces as well as in the average NETase activity scoring per participant (Figures 6F and S6B). Differences between survivors and non-survivors were found in the upper and lower ends of the spectrum of the NETase activity sample content per participant (Figure 6G). A prevalence of low profiled NETase activity samples was associated with lower survival probability (Figures 6H and 6I).
We also examined the distribution of a dual classification comprised of a combined DNase and NETase score where 0, 1, and 2 corresponded to low DNase activity with low, medium, and high NETase activity, and 3, 4, and 5 denoted the same NETase activity scoring in high DNase activity samples (Figure 6J). The frequency of these combinations highlighted the interdependence of the two activities because samples with high NETase activity and low DNase activity under group 2 were rare. Moreover, samples scored as 0 and 1 were overrepresented in individuals who died compared with survivors. Hence, defects in NET clearance associated with low DNase activity correlated with poor survival. Consistently, CRP was higher in low and medium NETase activity WHO-7 samples that exhibited a low DNase activity profile (Figure S6C). Furthermore, a high mortality frequency was observed in individuals containing a significant proportion of WHO-7 samples with low profiled DNase and NETase activities, as opposed to participants with low NETase activity samples in the absence of low DNase activity (Figure 6K). Here, we compared WHO-7 samples to eliminate a clinical severity bias. This analysis suggested that low NET degradation associated with DNase activity deficiency was linked to high mortality.
We also explored the impact of therapies on chromatin clearance capacity. These participants were recruited during the first wave of the pandemic in March 2020 and did not receive the subsequently approved regimens such as dexamethasone or anti-interleukin 6 (IL-6) therapy. We anticipated that individuals who received therapy would exhibit improved DNA clearance capacity. However, the data indicated a higher frequency of low profiled NETase and DNase activity in samples of individuals who were being actively treated with either extracorporeal membrane oxygenation (ECMO), intravascular hydrocortisone, albumin, or anti-inflammatory therapies (Figures 6L and S6D). This difference was also reflected in measured NETase activity in WHO-7 samples from individuals who received active anti-inflammatory therapy (Figure S6E). These findings suggested that low chromatin clearance was associated with a higher requirement for therapeutic intervention.
Sexual dimorphism in DNase activity in SARS-CoV-2-infected individuals
In addition, we examined whether DNA clearance capacity varied in samples from males and females (44 male, n = 336 samples and 19 female, n = 129 samples). Low DNase activity was associated predominately with male individuals (Figure 6M), whereas NETase activity did not display a sex bias (Figure 6N). The difference in DNase activity was also reflected in the dual DNase-NETase activity classification when considering all WHO severity grade samples. However, female WHO-6 samples displayed a higher incidence of low DNase activity, as these samples were over-represented in female participants compared with males, reflecting the lower maximum severity in females (Figures S6F and S6G). Consistently, plasma actin concentrations were lower in female compared with male survivors but did not differ in the deceased groups (Figure 6O). Thus, differences in DNase activity between males and females may contribute to differences in survival.
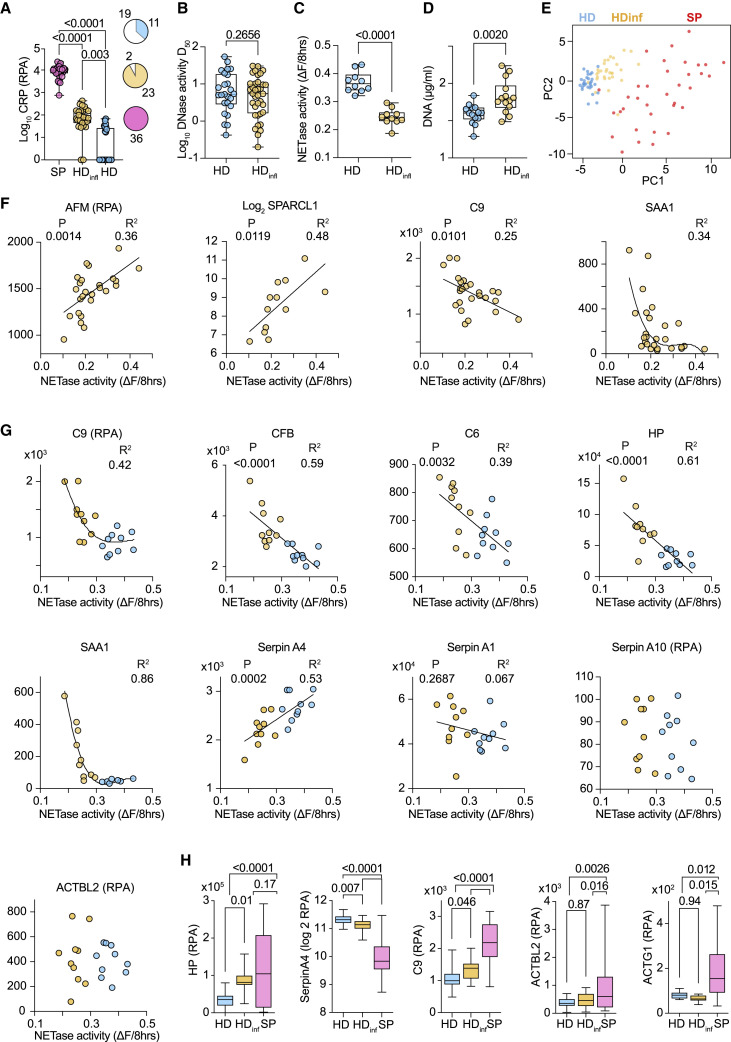
Low NET degradation in HDs with inflammation
We also explored whether chronic inflammation affected chromatin clearance capacity. We analyzed plasma from participants in the Avon Longitudinal Study of Parents and Children (ALSPAC) (Boyd et al., 2013; Fraser et al., 2013; Northstone et al., 2019). ALSPAC participants are tracked from childhood, and elevated plasma concentrations of the novel inflammatory marker GlycA at 15 years of age correlated with the appearance of cardiovascular disease risk indicators after a 10-year period (Chiesa et al., 2022). We segregated participant baseline samples into high and low GlycA containing groups designated as HD and HDinfl. CRP was elevated in one third of HD samples (11 out of 30) and in nearly all HDinfl plasmas (28 out of 30) (Figure 7A). The two groups exhibited comparably high DNase activity, but NETase activity was significantly lower in all tested HDinfl samples, alongside a higher DNA content (Figures 7B–7D). These results prompted us to investigate the connection between profiled activities and cardiovascular co-morbidities in the COVID-19 cohort. Low DNAse activity profiles were more prominent in individuals with cardiovascular co-morbidities, but NETase activity exhibited a comparable distribution (Figure S7A).
Figure 7.
Low NETase activity is associated with chronic inflammation in healthy individuals
(A) CRP in plasma from sepsis participants (SP) or healthy ALSPAC donors segregated by low (HD) or high (HDinfl) plasma concentrations of the inflammatory marker GlycA. Pie chart represents the number of high (colored) and low (white) CRP abundance samples.
(B–D) DNase activity (D50), NETase activity, and plasma DNA in ALSPAC HD and HDinfl plasma samples.
(E) Principal-component analysis of SP and ALSPAC HD and HDinfl plasma proteomes measured simultaneously by mass spectrometry.
(F and G) Correlation between measured NETase activity in HD (blue) and HDinfl (yellow) and plasma proteins selected based on CP correlation analysis.
(H) Comparison of plasma haptoglobin (HP), serpinA4, complement factor C9, and actin (ACTBL2 or ACTG1) protein abundance in HD (blue), HDinfl (yellow), and SP (pink) samples. Statistics by Mann-Whitney or Kruskal-Wallis test for single comparisons, one-way ANOVA for multiple comparisons, simple linear or non-linear regression for correlations. See also Figure S7.
Principal-component analysis and hierarchical clustering exposed significant differences in HD, HDinfl, and SP plasma proteomes that were measured concomitantly. (Figures 7E and S7B). Nearly all the NETase activity correlating proteins in CP plasma exhibited comparable correlations in HD and HDinfl samples, except for serpin A10 and actin (Figures 7F and 7G). Moreover, a shared set of factors could be used to trace these activities in different conditions (Figure S7C). While correlating proteins shifted in HDinfl samples, actin peptides remained low or undetectable when compared with SP plasmas (Figure 7H). Consistently, there were no defects in DNase activity among ALSPAC participants. Thus, low NETase activity was linked to chronic inflammation but could not be attributed to low DNase activity, substantiating further the multifactorial nature of NET degradation.
Discussion
The critical role for chromatin clearance in the survival of individuals with severe SARS-CoV-2 infection highlights the pathogenic significance of extracellular chromatin. The combined release of actin enabled the systemic accumulation of chromatin despite the presence of plasma endonucleases. Actin exacerbates defects in chromatin clearance driven by inflammation. DNase I and DNase I L3 have different sensitivities to actin and specificities for free and micelle-bound chromatin. In our experiments, G-actin did not inhibit NET dissolution, suggesting potential contributions from DNase I L3 or other plasma factors. Past work has pointed to the importance of synergy between endonucleases and proteases in chromatin clearance (Napirei et al., 2009; Papayannopoulos et al., 2011; Xu et al., 2009). However, actin correlated with reduced DNA and NET degradation capacity in SARS-CoV-2-infected individuals and triggered compensatory mechanisms in sepsis that underline the stress that actin exerts on these pathways. These findings are consistent with the notion that while DNA degradation comprises one of several enzymatic activities that contribute to chromatin clearance, it plays an important role in pathology and survival.
T cell apoptosis plays a critical pathogenic role through systemic chromatin accumulation. Consistently, in human sepsis, T cells upregulate pro-apoptotic Bim and downregulate anti-apoptotic Bcl-2 and Bcl-xL, promoting apoptosis (Hotchkiss et al., 1999; Schwulst et al., 2008; Weber et al., 2008). Moreover, PD-1-mediated T cell exhaustion, reactive oxygen species (ROS)-mediated endoplasmic reticulum (ER) stress, and mitochondrial dysfunction are also implicated in sepsis (Chang et al., 2014; Jimbo et al., 2003; Lee et al., 2015; Qu et al., 2013). Splenic T cell death is likely to be triggered by histone-mediated activation of adjacent marginal-zone macrophages, and macrophage-derived FasL induces lymphocyte death in other contexts (Daigneault et al., 2012; Ioannou et al., 2022; Roth et al., 2021). Although apoptosis is considered a silent process, it can be immunogenic and release chromatin (Choi et al., 2004; Jahr et al., 2001; Jiang et al., 2007; Montico et al., 2018; Radic et al., 2004; Reich and Pisetsky, 2009). Nevertheless, systemic chromatin and actin release by other splenocytes may occur in human infections.
Cell death drove DNase I expression, resulting in plasma chromatin positively correlating with the enzyme that degrades it. Therefore, endonuclease expression matches substrate abundance via unknown mechanisms that depend on the magnitude of actin release. These compensatory responses suggest that actin-resistant DNase I L3 was not sufficient to clear systemic DNA upon acute challenge, but the increase in degradation activity indicated that elevated DNase I overcame actin inhibition. The delay in circulating chromatin reduction explains the effectiveness of prophylactic DNase I administration and emphasizes the role of amplification loops. Consistently, therapeutic pulmozyme lowered circulating DNA and inflammation in participants with SARS-CoV-2 (Porter et al., 2022). Targeting extracellular chromatin may prove more beneficial than anti-cytokine monotherapies because chromatin induces multiple inflammatory cytokines and immune dysfunction (Ioannou et al., 2022).
The link between chronic inflammation and low NETase activity may explain the association of CP susceptibility with inflammatory conditions. However, the impact of predispositions is difficult to ascertain during the acute infection phase. The decoupling between defects in DNase and NETase activities in HDinf plasma provides further evidence for a role of additional enzymes in chromatin disassembly beyond DNase I. Correlating proteins may play regulatory roles or denote downstream consequences of differential chromatin clearance. ORM proteins may play a functional role because a recent genome-wide association study reported a correlation between ORM1 genetic variants and circulating DNA (Lopez et al., 2022). The similar trends with plasma proteome correlations between SARS-CoV-2-infected and HDinfl individuals suggested that low NETase activity had a similar impact on physiology in both groups. As NET degradation measurements were robust enough to sub-stratify samples, plasma proteomics enabled the characterization of samples that were otherwise difficult to distinguish by conventional clinical approaches, identifying high-risk individuals within clinically indistinguishable cohorts and pre-clinical states that may predispose individuals to acute or chronic disease.
Limitations of the study
We acknowledge that the associations introduced in the study do not provide tangible mechanistic insight on the chromatin degradation defects. Further work is needed to elucidate the machinery regulating DNase I expression.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD169-AlexaFluor647 clone 3D6.112 | BioLegend | Cat# 142408; RRID:AB_2563621 |
| Anti-mouse CD3-AlexaFluor488 clone 17A2 | BioLegend | Cat# 100210; RRID:AB_389301 |
| Anti-MPO | R&D Systems | Cat# AF3667; RRID:AB_2250866 |
| Anti-actin clone C4 | Millipore | Cat# MAB1501; RRID:AB_2223041 |
| Anti-CD163 | Abcam | Cat# ab182422; RRID:AB_2753196 |
| Bacterial and virus strains | ||
| Staphylococcus aureus 8325-4 | Provided by R. Novick, New York University, New York, NY | N/A |
| Candida albicans | clinical isolate SC5314 | N/A |
| Biological samples | ||
| Plasma samples from PA-COVID-19 study group | Charite obtained from Markus Ralser | N/A |
| Plasma samples from sepsis participants | Provided by Andrew Aswani | N/A |
| plasma from participants of the Avon Longitudinal Study of Parents and Children (ALSPAC) study | Obtained from Scott T. Chiesa and John E. Deanfield | N/A |
| Post-mortem COVID-19 spleens | Provided by Jana Ihlow (Charité, Institute of Pathology) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Deoxyribonuclease I from bovine pancreas (DNase I) | Sigma | Cat# D5025 |
| Actin from rabbit muscle | Sigma | Cat# A2522 |
| PMA (Phorbol 12-myristate 13-acetate) | Sigma | P8139 |
| Hoechst 33342 | Cell Signalling | 4082 |
| Sytox green | Invitrogen | S7020 |
| Critical commercial assays | ||
| Click-iT TUNEL Alexa 594 imaging assay | Invitrogen | Cat# C10246 |
| Deoxyribonuclease-1 (DNase1) (Human) ELISA Kit | BioVision | Cat# E4500 |
| Deposited data | ||
| Human sepsis plasma samples | This paper | PRIDE: PXD034391 |
| A time-resolved proteomic and prognostic map of COVID-19 | PMC8201874 | PRIDE: PXD025752 |
| Plasma proteomics in mouse sepsis | This paper | PRIDE: PXD037676 |
| ALSPAC human inflammation plasma proteomics | This paper | PRIDE: PXD037682 |
| Experimental models: Organisms/strains | ||
| WT C57BL/6J | The Francis Crick institute | N/A |
| C57BL/6J TCRα−/− (Tcratm1Phi) | The Francis Crick institute | N/A |
| Recombinant DNA | ||
| pBabe plasmid 6250 bp substrate of DNase assay | This paper | N/A |
| Software and algorithms | ||
| Imagej2 (v2.3.0/1.53f) | Courtesy of Spike Walker | https://imagej.net/ |
| Prism 9 Version 9.1.2 (225) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Jupyter notebook 6.4 | https://jupyter.org/ | N/A |
| NETase analysis | This paper | https://zenodo.org/badge/latestdoi/563418622 |
| Other | ||
| Nikon eclipse T2 inverted wide-field microscope | Imaging core facility | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Venizelos Papayannopoulos (veni.p@crick.ac.uk).
Materials availability
This study did not generate new, unique reagents.
Experimental model and subject details
Animal studies
All mice were bred and maintained in a pathogen free, 12-hour light-dark cycle environment. All experiments were conducted with age-matched and cage-controlled, 8 to 12-week-old female WT C57BL/6J (RRID:IMSR_JAX:000664) and TCRα−/− (Tcra tm1Phi) (MGI:2180883) mice, according to local guidelines and UK Home Office regulations under the Animals Scientific Procedures Act 1986 (ASPA). Mice were infected with Candida albicans (clinical isolate SC5314) or Staphylococcus aureus (serotype 8325-4).
Human studies
For the in vitro human neutrophil experiments, peripheral blood was isolated from consenting healthy adult volunteers, according to approved protocols of the ethics board of the Francis Crick Institute and the Human Tissue act. Sepsis participant samples (Sepsis plasma sample size:36) were provided by the Hannover Medical School approved the ethics committee under the study protocol (No. 2786-2015). Written informed consent was obtained from participants or authorized representatives. The study was performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. No funding specific to this project was received. COVID Sampling was performed as part of the Pa-COVID-19 study, a prospective observational cohort study assessing pathophysiology and clinical characteristics of participants with COVID-19 at Charité Universitätsmedizin Berlin (Kurth et al., 2020). All participants with SARS-CoV-2 infection proven by positive PCR from respiratory specimens and willing to provide written informed consent are eligible for inclusion. Exclusion criteria are refusal to participate in the clinical study by participant or legal representative or clinical conditions that do not allow for blood sampling. The study assesses epidemiological and demographic parameters, medical history, clinical course, morbidity and quality of life during hospital stay of COVID-19 participants. Moreover, serial high-quality bio-sampling consisting of various sample types with deep molecular, immunological and virological phenotyping is performed. Treatment and medical interventions follow standard of care as recommended by current international and German guidelines for COVID-19. Severity of illness in the present study follows the WHO ordinal outcome scale. The Pa-COVID-19 study is carried out according to the Declaration of Helsinki and the principles of Good Clinical Practice (ICH 1996) where applicable and was approved by the ethics committee of Charité- Universitätsmedizin Berlin (EA2/066/20) (COVID plasma samples=87). Plasmas of healthy donors with and without inflammation signatures were selected from participants in the Avon Longitudinal Study of Parents and Children (ALSPAC) (Boyd et al., 2013; Fraser et al., 2013; Northstone et al., 2019) (ALSPAC plasma sample = 50). All ethical approvals conformed to the Declaration of Helsinki and all biological samples used in the study were collected in accordance with the Human Tissue Act (2004).
Method details
Murine sepsis models
Wild-type C. albicans was cultured overnight in yeast extract peptone dextrose (YEPD; Sigma) medium, while shaking at 190RPM at 37°C. On the subsequent day, overnight culture was sub-cultured in YEPD and allowed to grow for approximately 4 hours, to an optical density (A600) of 0.4–0.8. Subcultures were microscopically examined for lack of hyphae, washed in sterile phosphate-buffered saline (PBS) and spun for 10 min at 1000xg at RT. S. aureus were plated on LB agar plates, cultured overnight in LB media and sub-cultured and grown to an optical density (A600) of 0.8. Bacteria were washed and resuspended in PBS.
Mice were intravenously injected with 5x105 yeast or 1x106 S. aureus bacteria per mouse via the tail vein. The weight and rectal temperature of the mice were recorded daily over the course of infection to track health status. A body temperature below 32°C, a weight loss superior to 80% of initial weight accompanied by slow movement and non-responsiveness were considered collectively as septic shock and the humane endpoint for the mice. The mice were culled via cervical dislocation or by lethal dose of pentobarbital (600 mg/kg) with mepivacaine hydrochloride (20 mg/ml).
For degradation of circulating DNA in vivo, mice were treated with deoxyribonuclease I (DNase I) from bovine pancreas (Sigma; 2000 units/mouse) daily via intraperitoneal injection, starting on the day prior to infection (D-1). For in vivo histone neutralisation, the mice received combined anti-histone 3 (Merck Millipore; 07-690) and anti-histone 4 antibodies (Merck Millipore; 04-858) or control polyclonal rabbit IgG (BioXCell). Anti-histone antibodies were dialysed and injected intraperitoneally, starting on D-1 (200μg/mouse) and daily afterward (200μg H3 and 100μg H4). Treatments were given daily until completion of the experiment. Macrophage depletion was performed via the administration of 1 mg Clodronate liposomes (Clo-L) or 1 mg PBS liposomes (PBS-L) as control (Liposoma) intravenously at 1 day prior to infection.
Histology and immunofluorescence imaging
For mouse tissue staining, freshly extracted spleens were directly embedded in optimal cutting temperature (OCT) compound cryo-embedding media (VWR Chemicals BDH) and frozen. Sections were cut to 8μm thickness on positively charged glass slides. To prepare samples for staining, the glass slides were air dried for 20 min at RT, washed in PBS and fixed for 10min with 4% paraformaldehyde (PFA; Sigma) at RT.
For staining of paraffinized and cut COVID participant spleens, slides were first baked at 60°C for 1hr. To remove paraffin, samples received 3 sequential baths of 5mins in Neo Clear and were then rehydrated with sequential 100%, 96%, 80%, 70% and 50% EtOH baths, 5min each, and washed. Antigen retrieval was performed with Dako Target Retrieval Solution pH9, for 45 min at 97°C. All tissues were permeabilized with 0.5% Triton X-100 in PBS for 20 min at RT. For terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), the Click-iT TUNEL Alexa Fluor 594 Imaging Assay kit (Invitrogen) was used and instructions from the manufacturer were followed. After TUNEL staining, non-specific binding was blocked with 2% BSA (Sigma) and 2% donkey serum (Sigma) in PBS for 1 hr at RT. Samples were then stained overnight in a dark humified chamber with the following primary antibodies in blocking buffer: anti-mouseCD3 (BioLegend; clone 17A2), anti-CD3 (Abcam; ab5690), anti-CD169 (BioLegend; clone 3D6.112) and anti-MPO (R&D Systems; AF3667) anti-CD163 (Abcam clone EPR19518). For secondary staining, tissues were stained for 2hrs in a humidified dark chamber at RT with the following labelled secondary antibodies in blocking buffer: donkey anti-rabbit IgG (Invitrogen) and donkey anti-goat IgG (Invitrogen). Nuclear staining with 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI; Invitrogen) was added with the secondary staining. All stained tissue sections were mounted in ProLong Gold (Molecular Probes). Images were taken using the Leica TCS SP5 inverted confocal microscope (20x, 40x, 63x original magnification) and analysis was performed using Fiji/ImageJ version 2.0.0 software.
Neutrophil isolation from peripheral human blood
Peripheral venous blood collected in EDTA tubes was layered on Histopaque 1119 (Sigma-Aldrich) and centrifuged for 20 min at 800xg. The plasma was collected and centrifuged for a second time. The neutrophil layer was collected and washed in Hyclone Hank's Balanced Salt Solution (HBSS) -Ca, -Mg, -Phenol red (GE Healthcare) supplemented with 10mM HEPES (Invitrogen) 0.1% plasma and further purified with a discontinuous Percoll (GE Healthcare) gradient consisting of layers with densities of 85%, 80%, 75%, 70% and 65% and centrifugated for 20 min at 800xg. Neutrophil enriched layer were collected and washed. Neutrophil purity was assessed via flow cytometry.
Plasmid DNA degradation assay (DNase assay)
1μg of a 6250 bp plasmid was incubated with 10 mM Tris, 2.5 mM MgCl2 and 2.5 mM CaCl2 at a pH 7.4 with 2.5/5 or 10% plasma in a final volume of 20μL for 2h at 37°C. The sample was then run in 1% agarose gel electrophoresis for 30 min at 120V. The images acquired from the gel were then analysis using Fiji. For the analysis; line profiles of each well were measured and the migrated distance of the DNA was quantified. The distance corresponding to the maximum intensity point (representing the most abundant fragment size) was plotted at the different plasma % for each plasma sample.
Time-lapse imaging and NET degradation quantification
5x104 human neutrophils from healthy volunteers were seeded in a black 96-well plate (PerkinElmer) in HyClone HBSS + Ca, +Mg, - Phenol red (GE Healthcare) containing 10mM HEPES (Invitrogen) and 3% plasma from healthy, inflammatory, septic or COVID-19 donors. Neutrophils from healthy volunteers were incubated for 45 mins at 37°C and CO2 (5%) prior to stimulation with 100 nM PMA (Phorbol 12-myristate 13-acetate) (Sigma). DNA of life cells was labelled with 4μg/mL Hoechst (membrane permeable; Thermo Scientific) and of dead cells with 0.2μM Sytox-green (membrane impermeable; Invitrogen). The cells were imaged on an inverted Nikon wide-field microscope system at 37°C and CO2 (5%). Four fields of view, containing around 100 cells per field of view, were acquired per well every 10 mins for 10–15 hrs using a 40x objective.
Quantification of NET degradation was performed by automatically identifying Regions of interest (ROI) containing individual neutrophils in the four different fields of view per well, each well containing a different plasma sample. ROIs were determined by using Otsu intensity-based thresholding on the Hoechst channel. Sytox signal was measured on individual ROI over time during the course of the acquisition. Each field of view contained 60-80% of NETotic cells and 40-20% of necrotic cells. Discrimination between NETotic and necrotic cells was performed based on the sytox intensity trace (Figure S1D). Continuously increasing sytox intensity traces with the maximum intensity values within the last 10% of the total measurement time were cataloged as necrotic. NETotic cells were characterized by reaching the maximum intensity sytox peak within the first 25% of the acquisition time post-stimulation followed by a decrease of signal over time. Comparison of the NET degradation capacity of different plasmas from human cohorts was achieved by calculating the difference of sytox intensity between the maximum point and the end of the acquisition for each individual NET. The median values of Δintensity for all the NETs detected on each field were used to used to obtain the average and standard deviation of the four fields of view per donor sample, values used to represent the NET degradation capacity per donor.
Western blot analysis
Samples were boiled in a sodium dodecyl sulphate (SDS) buffer containing dithiothreitol (DTT) and resolved by polyacrylamide gel electrophoresis (SDS-PAGE) on a Criterion TGX precast gel (Any-KD; Bio-Rad Laboratories). Proteins were then transferred to a PVDF membrane (Bio-Rad Laboratories) via semi-dry transfer. The membrane was blocked with 5% bovine serum albumin (BSA; Fisher Scientific) in Tris-buffered saline with 0.1% Tween 20 (TBS-T). H3 was detected with anti-histone 3 (Milipore) and HRP–conjugated goat anti-rabbit (Thermo Scientific). Actin was detected with anti-actin (Milipore) and HRP–conjugated donkey anti-mouse (Thermo Scientific) antibodies.
DNA quantitation
Circulating DNA in plasma was quantified using the Quant-iTTM PicoGreen dsDNA assay kit (Thermo Fisher Scientific). The fluorescent signal (excitation at 488nm) was measured using a spectrophotometric microplate reader (Fluostar Omega, BMG labtech).
Plasma sample preparation for proteomic analysis
Healthy donor and participant plasma samples were randomised and plated in a 96-well plate (Eppendorf). Protein/peptide extraction and proteomics analysis was performed following the previously described protocol (Messner et al., 2020). 5μL of plasma was denatured in 50μL 8M Urea (Honeywell Research Chemicals), 100mM ammonium bicarbonate (ABC, Honeywell Research Chemicals) and reduced with 5μL of 50mM dithiothreitol (DTT, Sigma Aldrich) at 30°C for 1 hour. Followed by alkylation with 5μL of 100mM iodoacetamide (IAA, Sigma Aldrich) at 23°C for 30 min in the dark. The samples were diluted with 340μL of 100mM ABC and 220μL was added to trypsin solution (Promega) for protein digestion at a trypsin/protein ratio of 1/40 and incubated at 37°C overnight (17h). Quenching of digestion was done by the addition of 25μL of 10% v/v formic acid (FA, Thermo Fisher Scientific). Rounds of solid phase extraction clean-up steps were performed with the use of C18 96-well plates (BioPureSPE Macro 96-Well, 100mg PROTO C18, The Nest Group) as described previously (Messner et al., 2020). Methanol (Fisher Chemicals), 50% v/v acetonitrile (ACN, Fisher Chemicals) or 0.1% v/v FA was used at each centrifugation step as required. After final elution, the collected peptide material was dried by a vacuum concentrator (Eppendorf Concentrator Plus) and redissolved in 50μL 0.1% v/v FA, to be processed by liquid chromatography-mass spectrometry.
Liquid chromatography-mass spectrometry
1μg of protein digest (peptides) was injected and analysed on a nanoAcquity Liquid Chromatograph (Waters) coupled to a TripleTOF 6600 Mass Spectrometer (Sciex) at a flow-rate of 5 μL/min. This was followed by a separation using a Waters HSS T3 column (150 mm × 300 μm, 1.8μm particles) in 20-min non-linear gradients starting with 3% B up to 40% B (Buffer A: 0.1% v/v FA; Buffer B: ACN/0.1% v/v FA). A data independent acquisition (DIA/SWATH) method was used, with MS1 scan from m/z 400 to m/z 1250 and 50ms accumulation time followed by 40 MS2 scans of 35ms accumulation time with variable precursor isolation width covering the mass range from m/z 400 to m/z 1250. Ion source gas 1 (nebulizer gas), ion source gas 2 (heater gas) and curtain gas were set to 30, 15 and 25 respectively. The source temperature was set to 450°C and the ion spray voltage to 5500V. Injections of samples took place in a random order.
The “gas-phase fractionation” methodology was used to generate the library out of pooled peptide digest of all plasma samples with the use of a LC-MS/MS method as mentioned before. 3μg of protein digest was separated with a 60-min linear gradient (3% B to 40% B). Injections were performed at the mass ranges of: 400–500 m/z, 495–600 m/z, 595–700 m/z, 695–800 m/z, 795–900 m/z, 895–1000 m/z, 995–1100 m/z, 1095–1250 m/z. The precursor selection windows were 2 m/z (1 m/z overlap). DIA-NN 1.7.10 proteomics analysis software (Demichev et al., 2020) was used for the library preparation with Mus musculus (mouse) UniProt (UniProt Consortium, 2019) isoform sequence database (UP000000589) to annotate the library. To quantify proteins, raw data acquired were processed with DIA-NN 1.7.10 with the “robust LC (high precision)” mode with MS2, MS1 and scan window size set to 20ppm, 12ppm and 8 respectively.
Quantification and statistical analysis
Correlation and statistical analysis
All correlation and fitting analysis were performed using GraphPad Prism software aided by sorting and grouping samples using Microsoft Excel. Thresholds for identifying and grouping individuals into groups were determined using frequency analysis in Excel. Single comparison statistical significance was assessed by an unpaired, two-tailed Student’s t-test. Mann-Whitney test for single comparisons, Fisher’s exact test for contingency, simple linear regression, or non-linear exponential or Gaussian distribution fitting. Statistical analysis was performed on GraphPad Prism software.
Acknowledgments
We are extremely grateful to the study participants and their families. We thank Andrew Aswani for sourcing sepsis participant samples and Zak Morgan for NETase analysis scripts. For the purpose of Open Access, the authors have applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission. This work was supported by the Francis Crick Institute, which receives its core funding from the UK Medical Research Council (FC0010129 and FC001134), Cancer Research UK (FC0010129 and FC001134), and the Wellcome Trust (FC0010129 and FC001134). I.V.A was funded by an EMBO LTF (ALTF 113-2019) and a Wellcome Trust fellowship (SHWF 222825/Z/21/Z). S.T.C. was funded by BHF project grant PG/18/45/33814. Special thanks to the Charite PA-COVID-19 study group: Sebastian Albus, Florian Alius, Tim Andermann, Stefan Angermair, Uwe D. Behrens, Laure Bosquillon de Jarcy, Markus Brack, Astrid Breitbart, Felix Bremer, Felix Bremer, Dana Briesemeister, Victor M. Corman, Thomas Cronen, Chantip Dang-Heine, Sophy Denker, Phillip van Dijck, Jan-Moritz Doehn, Christian Drosten, Kai-Uwe Eckardt, Marius A. Eckart, Andreas Edel, Lucas Elbert, Matthias Endres, Philipp Enghard, Matthias Felten, Carolin Ferse, Simon Fraumann, Nikolaj Frost, Carmen Garcia, Dominik Geus, Gisele J. Godzick-Njomgang, Daniel Grund, Sascha S. Haenel, Jens K. Haumesser, Julia Heeschen, Friederike L. Hefele, Katrin M. Heim, Anne Herholz, Vera Hermanns, Moritz Hilbrandt, David Hillus, Stefan Hippenstiel, Andreas Hocke, Johannes Hodes, Ralf-Harto Hübner, Michael Hummel, Till Jacobi, Linda Jürgens, Christof von Kalle, Ute Kellermann, Ye-Ji Kim, Malte Kleinschmidt, Philipp Knape, Samuel Knauß, Roland Körner, Alexander Krannich, Philipp M. Krause, Lucie Kretzler, Jan M. Kruse, Liron Lechtenberg, Lukas Lehner, Yaosi Li, Linna Li, Emma Lieker, Tilman Lingscheid, Felix Machleidt, Elena Madlung, Josef Mang, Luise Martin, Nikolai Menner, Lil Meyer-Arndt, Agata Mikolajewska, Belén Millet Pascual-Leone, Mirja Mittermaier, Martin Möckel, Luisa Mrziglod, Nadine Muller, Holger Müller-Redetzky, Sergej Münzenberg, Christopher Neumann, Michaela Niebank, Nadine Olk, Bastian Opitz, Eva Pappe, Panagiotis Pergantis, Frieder Pfäfflin, Lennart Pfannkuch, Moritz Pfeiffer, Thuy N. Pham, Peter Radünzel, Matthias Raspe, Nicola Reck, Johannes Rein, Jana Riecke, Teresa Ritter, Anne Ritter, Maria Rönnefarth, Christoph Ruwwe-Glösenkamp, Jacopo Saccomanno, Leif-Erik Sander, Birgit Sawitzki, Laura K. Schmalbrock, Sein Schmidt, Solveig Schönberger, Rosa C. Schuhmacher, Dirk Schürmann, Mariana Schürmann, Georg Schwanitz, Dominik Soll, Claudia Spies, Miriam S. Stegemann, Fridolin Steinbeis, Britta Stier, Paula Stubbemann, Norbert Suttorp, Christoph Tabeling, Frank Tacke, Bettina Temmesfeld-Wollbrück, Charlotte Thibeault, Pinkus Tober-Lau, Sascha Treskatsch, Denise Treue, Alexander Uhrig, Markus Vogtmann, Nadia A. de Vries, Lisa-Marie Wackernagel, Susanne Weber, Steffen Weber-Carstens, Anne Wetzel, Isabelle Wirsching, Marcel Wittenberg, Martin Witzenrath, Christian Wollboldt, Alexander Wree, Yinan Wu, Berna Yedikat, Tamar Zhamurashvili, Daniel Zickler, Christian Zobel, Thomas Zoller, Saskia Zvorc, and Selina Greuel.
Author contributions
I.V.A., D.H., M.I., M.I.T., M.C.F.T., and N.M.D.V. performed experiments and analysis. S.I.V., V.D., and M.W. performed mass spectrometry. E.T.H., J.I., H.R., L.L., E.T.H., and F.K. supervised the SARS-CoV-2 study. S.D., K.S., and C.B. supervised the microbial sepsis study. S.T.C. and J.E.D. provided HD samples. M.R. directed the proteomic analysis. V.P. directed the study, performed analysis, and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: November 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2022.11.007.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository and are publicly available from the date of publication. Dataset identifiers are listed in the key resources table.
-
•
The code originated to analyze the NETase assay has been deposited in Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Afzali B., Noris M., Lambrecht B.N., Kemper C. The state of complement in COVID-19. Nat. Rev. Immunol. 2021;22:77–84. doi: 10.1038/s41577-021-00665-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky J.B., Rivers E.P., Filbin M.R., Lee P.J., Morris D.C. Thymosin beta 4 regulation of actin in sepsis. Expert Opin. Biol. Ther. 2018;18:193–197. doi: 10.1080/14712598.2018.1448381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A., Golding J., Macleod J., Lawlor D.A., Fraser A., Henderson J., Molloy L., Ness A., Ring S., Davey Smith G. Cohort profile: the 'children of the 90s'--the index offspring of the avon longitudinal study of parents and children. Int. J. Epidemiol. 2013;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K., Svabek C., Vazquez-Guillamet C., Sato B., Rasche D., Wilson S., Robbins P., Ulbrandt N., Suzich J., Green J., et al. Targeting the programmed cell death 1: programmed cell death ligand 1 pathway reverses T cell exhaustion in patients with sepsis. Crit. Care. 2014;18:R3. doi: 10.1186/cc13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa S.T., Charakida M., Georgiopoulos G., Roberts J.D., Stafford S.J., Park C., Mykkänen J., Kähönen M., Lehtimäki T., Ala-Korpela M., et al. Glycoprotein acetyls: a novel inflammatory biomarker of early cardiovascular risk in the young. J. Am. Heart Assoc. 2022;11 doi: 10.1161/JAHA.121.024380. e024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.J., Reich C.F., 3rd, Pisetsky D.S. Release of DNA from dead and dying lymphocyte and monocyte cell lines in vitro. Scand. J. Immunol. 2004;60:159–166. doi: 10.1111/j.0300-9475.2004.01470.x. [DOI] [PubMed] [Google Scholar]
- Chousterman B.G., Swirski F.K., Weber G.F. Cytokine storm and sepsis disease pathogenesis. Semin. Immunopathol. 2017;39:517–528. doi: 10.1007/s00281-017-0639-8. [DOI] [PubMed] [Google Scholar]
- Daigneault M., De Silva T.I., Bewley M.A., Preston J.A., Marriott H.M., Mitchell A.M., Mitchell T.J., Read R.C., Whyte M.K.B., Dockrell D.H. Monocytes regulate the mechanism of T-cell death by inducing Fas-mediated apoptosis during bacterial infection. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002814. e1002814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demichev V., Messner C.B., Vernardis S.I., Lilley K.S., Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods. 2020;17:41–44. doi: 10.1038/s41592-019-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale R.J., Hazeldine J., Al Tarrah K., Hampson P., Devi A., Ermogenous C., Bamford A.L., Bishop J., Watts S., Kirkman E., et al. Dysregulation of the actin scavenging system and inhibition of DNase activity following severe thermal injury. Br. J. Surg. 2020;107:391–401. doi: 10.1002/bjs.11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar M., Poppe R., Bischofs C., Fredenhagen G., Kanitz M., Kahaly G.J. Impaired deoxyribonuclease activity in monoglandular and polyglandular autoimmunity. Exp. Clin. Endocrinol. Diabetes. 2007;115:387–391. doi: 10.1055/s-2007-967082. [DOI] [PubMed] [Google Scholar]
- Finelli L., Gupta V., Petigara T., Yu K., Bauer K.A., Puzniak L.A. Mortality among US patients hospitalized with SARS-CoV-2 infection in 2020. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.6556. e216556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A., Macdonald-Wallis C., Tilling K., Boyd A., Golding J., Davey Smith G., Henderson J., Macleod J., Molloy L., Ness A., et al. Cohort Profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int. J. Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkim A., Fürnrohr B.G., Amann K., Laube B., Abed U.A., Brinkmann V., Herrmann M., Voll R.E., Zychlinsky A. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc. Natl. Acad. Sci. USA. 2010;107:9813–9818. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstra J.J., Schouten M., Levi M. Thrombophilia and outcome in severe infection and sepsis. Semin. Thromb. Hemost. 2007;33:604–609. doi: 10.1055/s-2007-985757. [DOI] [PubMed] [Google Scholar]
- Horváth-Szalai Z., Kustán P., Mühl D., Ludány A., Bugyi B., Kőszegi T. Antagonistic sepsis markers: serum gelsolin and actin/gelsolin ratio. Clin. Biochem. 2017;50:127–133. doi: 10.1016/j.clinbiochem.2016.10.018. [DOI] [PubMed] [Google Scholar]
- Hotchkiss R.S., Swanson P.E., Knudson C.M., Chang K.C., Cobb J.P., Osborne D.F., Zollner K.M., Buchman T.G., Korsmeyer S.J., Karl I.E. Overexpression of Bcl-2 in transgenic mice decreases apoptosis and improves survival in sepsis. J. Immunol. 1999;162:4148–4156. [PubMed] [Google Scholar]
- Iba T., Levy J.H., Levi M., Thachil J. Coagulopathy in COVID-19. J. Thromb. Haemost. 2020;18:2103–2109. doi: 10.1111/jth.14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou M., Hoving D., Aramburu I.V., Temkin M.I., De Vasconcelos N.M., Tsourouktsoglou T.D., Wang Q., Boeing S., Goldstone R., Vernardis S., et al. Microbe capture by splenic macrophages triggers sepsis via T cell-death-dependent neutrophil lifespan shortening. Nat. Commun. 2022;13:4658. doi: 10.1038/s41467-022-32320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahr S., Hentze H., Englisch S., Hardt D., Fackelmayer F.O., Hesch R.D., Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- Jandaghian S., Vaezi A., Manteghinejad A., Nasirian M., Vaseghi G., Haghjooy Javanmard S. Red blood cell distribution width (RDW) as a predictor of in-hospital mortality in COVID-19 patients; a cross sectional study. Arch. Acad. Emerg. Med. 2021;9:e67. doi: 10.22037/aaem.v9i1.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Bell C.W., Pisetsky D.S. The relationship between apoptosis and high-mobility group protein 1 release from murine macrophages stimulated with lipopolysaccharide or polyinosinic-polycytidylic acid. J. Immunol. 2007;178:6495–6503. doi: 10.4049/jimmunol.178.10.6495. [DOI] [PubMed] [Google Scholar]
- Jimbo A., Fujita E., Kouroku Y., Ohnishi J., Inohara N., Kuida K., Sakamaki K., Yonehara S., Momoi T. ER stress induces caspase-8 activation, stimulating cytochrome c release and caspase-9 activation. Exp. Cell Res. 2003;283:156–166. doi: 10.1016/s0014-4827(02)00033-2. [DOI] [PubMed] [Google Scholar]
- Jiménez-Alcázar M., Napirei M., Panda R., Köhler E.C., Kremer Hovinga J.A., Mannherz H.G., Peine S., Renné T., Lämmle B., Fuchs T.A. Impaired DNase1-mediated degradation of neutrophil extracellular traps is associated with acute thrombotic microangiopathies. J. Thromb. Haemost. 2015;13:732–742. doi: 10.1111/jth.12796. [DOI] [PubMed] [Google Scholar]
- Jiménez-Alcázar M., Rangaswamy C., Panda R., Bitterling J., Simsek Y.J., Long A.T., Bilyy R., Krenn V., Renné C., Renné T., et al. Host DNases prevent vascular occlusion by neutrophil extracellular traps. Science. 2017;358:1202–1206. doi: 10.1126/science.aam8897. [DOI] [PubMed] [Google Scholar]
- Kurth F., Roennefarth M., Thibeault C., Corman V.M., Müller-Redetzky H., Mittermaier M., Ruwwe-Glösenkamp C., Heim K.M., Krannich A., Zvorc S., et al. Studying the pathophysiology of coronavirus disease 2019: a protocol for the Berlin prospective COVID-19 patient cohort (Pa-COVID-19) Infection. 2020;48:619–626. doi: 10.1007/s15010-020-01464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustán P., Horváth-Szalai Z., Mühl D. Nonconventional markers of sepsis. EJIFCC. 2017;28:122–133. [PMC free article] [PubMed] [Google Scholar]
- Kustán P., Szirmay B., Horváth-Szalai Z., Ludány A., Kovács G.L., Miseta A., Kőszegi T., Mühl D. Urinary orosomucoid: a novel, early biomarker of sepsis with promising diagnostic performance. Clin. Chem. Lab. Med. 2017;55:299–307. doi: 10.1515/cclm-2016-0840. [DOI] [PubMed] [Google Scholar]
- Langford B.J., So M., Raybardhan S., Leung V., Westwood D., MacFadden D.R., Soucy J.P.R., Daneman N. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin. Microbiol. Infect. 2020;26:1622–1629. doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarides E., Lindberg U. Actin is the naturally occurring inhibitor of deoxyribonuclease I. Proc. Natl. Acad. Sci. USA. 1974;71:4742–4746. doi: 10.1073/pnas.71.12.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.S., Patel S.R., Christiani D.C., Bajwa E., Stossel T.P., Waxman A.B. Plasma gelsolin depletion and circulating actin in sepsis: a pilot study. PLoS One. 2008;3:e3712. doi: 10.1371/journal.pone.0003712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.S., Waxman A.B., Cotich K.L., Chung S.W., Perrella M.A., Stossel T.P. Plasma gelsolin is a marker and therapeutic agent in animal sepsis. Crit. Care Med. 2007;35:849–855. doi: 10.1097/01.CCM.0000253815.26311.24. [DOI] [PubMed] [Google Scholar]
- Lee W.M., Galbraith R.M. The extracellular actin-scavenger system and actin toxicity. N. Engl. J. Med. 1992;326:1335–1341. doi: 10.1056/NEJM199205143262006. [DOI] [PubMed] [Google Scholar]
- Lee W.S., Sung M.S., Lee E.G., Yoo H.G., Cheon Y.H., Chae H.J., Yoo W.H. A pathogenic role for ER stress-induced autophagy and ER chaperone GRP78/BiP in T lymphocyte systemic lupus erythematosus. J. Leukoc. Biol. 2015;97:425–433. doi: 10.1189/jlb.6A0214-097R. [DOI] [PubMed] [Google Scholar]
- Lopez S., Martinez-Perez A., Rodriguez-Rius A., Viñuela A., Brown A.A., Martin-Fernandez L., Vilalta N., Arús M., Panousis N.I., Buil A., et al. Integrated GWAS and gene expression suggest ORM1 as a potential regulator of plasma levels of cell-free DNA and thrombosis risk. Thromb. Haemost. 2022;122:1027–1039. doi: 10.1055/s-0041-1742169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood N.A., Mathew J., Kang B., DeBari V.A., Khan M.A. Broadening of the red blood cell distribution width is associated with increased severity of illness in patients with sepsis. Int. J. Crit. Illn. Inj. Sci. 2014;4:278–282. doi: 10.4103/2229-5151.147518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Valle F., Balada E., Ordi-Ros J., Bujan-Rivas S., Sellas-Fernandez A., Vilardell-Tarres M. DNase 1 activity in patients with systemic lupus erythematosus: relationship with epidemiological, clinical, immunological and therapeutical features. Lupus. 2009;18:418–423. doi: 10.1177/0961203308098189. [DOI] [PubMed] [Google Scholar]
- McDonald B., Davis R.P., Kim S.J., Tse M., Esmon C.T., Kolaczkowska E., Jenne C.N. Platelets and neutrophil extracellular traps collaborate to promote intravascular coagulation during sepsis in mice. Blood. 2017;129:1357–1367. doi: 10.1182/blood-2016-09-741298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner C.B., Demichev V., Wendisch D., Michalick L., White M., Freiwald A., Textoris-Taube K., Vernardis S.I., Egger A.S., Kreidl M., et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020;11:11–24.e4. doi: 10.1016/j.cels.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montico B., Nigro A., Casolaro V., Dal Col J. Immunogenic apoptosis as a novel tool for anticancer vaccine development. Int. J. Mol. Sci. 2018;19:e594. doi: 10.3390/ijms19020594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.S., Dawson J.F. A high-throughput assay shows that DNase-I binds actin monomers and polymers with similar affinity. Anal. Biochem. 2007;364:159–164. doi: 10.1016/j.ab.2007.02.027. [DOI] [PubMed] [Google Scholar]
- Napirei M., Ludwig S., Mezrhab J., Klöckl T., Mannherz H.G. Murine serum nucleases--contrasting effects of plasmin and heparin on the activities of DNase1 and DNase1-like 3 (DNase1l3) FEBS J. 2009;276:1059–1073. doi: 10.1111/j.1742-4658.2008.06849.x. [DOI] [PubMed] [Google Scholar]
- Northstone K., Lewcock M., Groom A., Boyd A., Macleod J., Timpson N., Wells N. The Avon Longitudinal Study of Parents and Children (ALSPAC): an update on the enrolled sample of index children in 2019. Wellcome Open Res. 2019;4:51. doi: 10.12688/wellcomeopenres.15132.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulos V., Staab D., Zychlinsky A. Neutrophil elastase enhances sputum solubilization in cystic fibrosis patients receiving DNase therapy. PLoS One. 2011;6:e28526. doi: 10.1371/journal.pone.0028526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D.J., Prakash A., Frericks-Zipper A., Eisenacher M., et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping H., Zhang K., Wang Y., Tong X., Chen Z., Cai C., Lu Z., Gui X., Liu L., Wang X., Ke H. Cell death and pathological findings of the spleen in COVID-19 patients. Pathol. Res. Pract. 2021;227:153610. doi: 10.1016/j.prp.2021.153610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolski J.L., Steck T.L. Association of deoxyribonuclease I with the pointed ends of actin filaments in human red blood cell membrane skeletons. J. Biol. Chem. 1988;263:638–645. [PubMed] [Google Scholar]
- Porter J.C., Inshaw J., Solis V.J., Denneny E., Evans R., Temkin M.I., De Vasconcelos N., Aramburu I.V., Hoving D., Basire D., et al. Anti-inflammatory therapy with nebulised dornase alfa in patients with severe COVID-19 pneumonia a randomised clinical trial. medRxiv. 2022 doi: 10.7554/eLife.87030. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu K., Shen N.Y., Xu X.S., Su H.B., Wei J.C., Tai M.H., Meng F.D., Zhou L., Zhang Y.L., Liu C. Emodin induces human T cell apoptosis in vitro by ROS-mediated endoplasmic reticulum stress and mitochondrial dysfunction. Acta Pharmacol. Sin. 2013;34:1217–1228. doi: 10.1038/aps.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radic M., Marion T., Monestier M. Nucleosomes are exposed at the cell surface in apoptosis. J. Immunol. 2004;172:6692–6700. doi: 10.4049/jimmunol.172.11.6692. [DOI] [PubMed] [Google Scholar]
- Raju M S., J. V., Kamaraju R.S., Sritharan V., Rajkumar K., Natarajan S., Kumar A.D., Burgula S. Continuous evaluation of changes in the serum proteome from early to late stages of sepsis caused by Klebsiella pneumoniae. Mol. Med. Rep. 2016;13:4835–4844. doi: 10.3892/mmr.2016.5112. [DOI] [PubMed] [Google Scholar]
- Reich C.F., Pisetsky D.S. The content of DNA and RNA in microparticles released by Jurkat and HL-60 cells undergoing in vitro apoptosis. Exp. Cell Res. 2009;315:760–768. doi: 10.1016/j.yexcr.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Roth S., Cao J., Singh V., Tiedt S., Hundeshagen G., Li T., Boehme J.D., Chauhan D., Zhu J., Ricci A., et al. Post-injury immunosuppression and secondary infections are caused by an AIM2 inflammasome-driven signaling cascade. Immunity. 2021;54:648–659.e8. doi: 10.1016/j.immuni.2021.02.004. [DOI] [PubMed] [Google Scholar]
- Rudd K.E., Johnson S.C., Agesa K.M., Shackelford K.A., Tsoi D., Kievlan D.R., Colombara D.V., Ikuta K.S., Kissoon N., Finfer S., et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the global burden of disease study. Lancet. 2020;395:200–211. doi: 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwulst S.J., Muenzer J.T., Peck-Palmer O.M., Chang K.C., Davis C.G., McDonough J.S., Osborne D.F., Walton A.H., Unsinger J., McDunn J.E., Hotchkiss R.S. Bim siRNA decreases lymphocyte apoptosis and improves survival in sepsis. Shock. 2008;30:127–134. doi: 10.1097/shk.0b013e318162cf17. [DOI] [PubMed] [Google Scholar]
- Tsang G., Insel M.B., Weis J.M., Morgan M.A.M., Gough M.S., Frasier L.M., Mack C.M., Doolin K.P., Graves B.T., Apostolakos M.J., Pietropaoli A.P. Bioavailable estradiol concentrations are elevated and predict mortality in septic patients: a prospective cohort study. Crit. Care. 2016;20:335. doi: 10.1186/s13054-016-1525-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsourouktsoglou T.D., Warnatsch A., Ioannou M., Hoving D., Wang Q., Papayannopoulos V. Histones, DNA, and citrullination promote neutrophil extracellular trap inflammation by regulating the localization and activation of TLR4. Cell Rep. 2020;31:107602. doi: 10.1016/j.celrep.2020.107602. [DOI] [PubMed] [Google Scholar]
- Weber S.U., Schewe J.C., Lehmann L.E., Müller S., Book M., Klaschik S., Hoeft A., Stüber F. Induction of Bim and Bid gene expression during accelerated apoptosis in severe sepsis. Crit. Care. 2008;12:R128. doi: 10.1186/cc7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn G., Walborn A., Rondina M., Fareed J., Hoppensteadt D. Biomarkers of platelet activation and their prognostic value in patients with sepsis-associated disseminated intravascular coagulopathy. Clin. Appl. Thromb. Hemost. 2021;27 doi: 10.1177/1076029620943300. 1076029620943300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q., Feng Z., Diao B., Tu C., Qiao Q., Yang H., Zhang Y., Wang G., Wang H., Wang C., et al. SARS-CoV-2 induces lymphocytopenia by promoting inflammation and decimates secondary lymphoid organs. Front. Immunol. 2021;12:661052. doi: 10.3389/fimmu.2021.661052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang X., Monestier M., Esmon N.L., Esmon C.T. Extracellular histones are mediators of death through TLR2 and TLR4 in mouse fatal liver injury. J. Immunol. 2011;187:2626–2631. doi: 10.4049/jimmunol.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang X., Pelayo R., Monestier M., Ammollo C.T., Semeraro F., Taylor F.B., Esmon N.L., Lupu F., Esmon C.T. Extracellular histones are major mediators of death in sepsis. Nat. Med. 2009;15:1318–1321. doi: 10.1038/nm.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xie X., Tu Z., Fu J., Xu D., Zhou Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Target. Ther. 2021;6:255. doi: 10.1038/s41392-021-00679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinellu A., Mangoni A.A. Red blood cell distribution width, disease severity, and mortality in hospitalized patients with SARS-CoV-2 infection: a systematic review and meta-analysis. J. Clin. Med. 2021;10:e286. doi: 10.3390/jcm10020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y., Yalavarthi S., Shi H., Gockman K., Zuo M., Madison J.A., Blair C., Weber A., Barnes B.J., Egeblad M., et al. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5:138999. doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository and are publicly available from the date of publication. Dataset identifiers are listed in the key resources table.
-
•
The code originated to analyze the NETase assay has been deposited in Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.