Abstract
Arsenic is a highly toxic metalloid of major concern for public safety. However, microorganisms have several resistance mechanisms, particularly the expression of arsenic pumps is a critical component for bacterial ability to expel it and decrease intracellular toxicity. In this study, we aimed to characterize the biochemical, structural, and genomic characteristics of the Acr3 pump among a group of Exiguobacterium strains isolated from different sites of the arsenic-rich Salar de Huasco (SH) ecosystem. We also determined whether the differences in As(III) resistance levels presented by the strains could be attributed to changes in the sequence or structure of this protein. In this context, we found that based on acr3 sequences the strains isolated from the SH grouped together phylogenetically, even though clustering based on gene sequence identity did not reflect the strain’s geographical origin. Furthermore, we determined the genetic context of the acr3 sequences and found that there are two versions of the organization of acr3 gene clusters, that do not reflect the strain’s origin nor arsenic resistance level. We also contribute to the knowledge regarding structure of the Acr3 protein and its possible implications on the functionality of the pump, finding that although important and conserved components of this family of proteins are present, there are several changes in the amino acidic sequences that may affect the interactions among amino acids in the 3D model, which in fact are evidenced as changes in the structure and residues contacts. Finally, we demonstrated through heterologous expression that the Exiguobacterium Acr3 pump does indeed improve the organisms As resistance level, as evidenced in the complemented E. coli strains. The understanding of arsenic detoxification processes in prokaryotes has vast biotechnological potential and it can also provide a lot of information to understand the processes of evolutionary adaptation.
Keywords: arsenic, ACR3, efflux pumps, resistance, Exiguobacterium
Introduction
Arsenic (As) is one of the most ubiquitous, non-essential, and highly toxic metalloids in the world; therefore, resistance mechanisms to its presence evolved very early and exist in all domains of life (Mateos et al., 2017). The release of arsenic from minerals into sediments and water can occur because of natural phenomena such as volcanic activities and erosion, but most arsenic pollution is due to human activities (Hoang et al., 2010; Zhu et al., 2014), such as mining and agriculture (Islam et al., 2004; Norra et al., 2005).
The inorganic forms arsenite [As(III)] and arsenate [As(V)] are more prevalent than the organic forms of arsenic in terrestrial environments, and these species can act as selective pressures on the ecosystems, especially for microorganisms. In bacteria, arsenic resistance ars genes are widespread and commonly found as arsenic resistant operons in two different configurations: consisting of three (arsRBC) or five (arsRDABC) genes, mostly organized into single transcriptional units (Mateos et al., 2017). Also, our research group found that an accessory arsenic efflux pump gene (acr3) is found outside the ars operon (Castro-Severyn et al., 2017), which also has been described for other species (Andres and Bertin, 2016).
The simplest ars operons have an As(III)-responsive trans-acting transcriptional regulator (ArsR), an arsenate reductase (ArsC), and the arsenite efflux pump (ArsB) (San Francisco et al., 1989, 1990; Wysocki et al., 1997; Ghosh et al., 1999; Meng et al., 2004). More complex forms appear to be later adaptations to enhance the activity of existing pumps, including an ATPase (ArsA) stimulated by As(III) that may interact with ArsB or Acr3 (Yang et al., 2012) and the metallochaperone/trans-acting repressor (ArsD). Other arsenic resistance genes have been characterized more recently, such as ArsM, ArsI, ArsP, and ArsH, some of which improve resistance to organic arsenicals (Ji and Silver, 1992; Gladysheva et al., 1994; Lin et al., 2006; Yang et al., 2012; Li et al., 2016; Serrato-Gamiño et al., 2018).
However, arsenic efflux is the most important arsenic detoxification mechanism; in this context, six types of arsenic efflux transporters have been characterized so far (ArsB, Acr3, ArsJ, ArsP, ArsK, and MSF1; Achour et al., 2007; Garbinski et al., 2019). The relative abundance of arsB and acr3 in microorganisms is very high, even in low arsenic-contaminated soils, and they show a significant correlation with arsenic concentrations (Wang et al., 2020; Castro-Severyn et al., 2021), providing an interesting biomarker for arsenic contamination (Cai et al., 2009; Poirel et al., 2013).
Even though the transporters ArcB and Acr3 share the same function, they evolved independently and show no sequence similarities (Mansour et al., 2007; Yang et al., 2015; Garbinski et al., 2019). ArsB is classified within the ion transporters superfamily and is found only in prokaryotes, while Acr3 proteins are smaller and belong to the bile/arsenite/riboflavin transporter (BART) superfamily and are found widespread in bacteria, archaea, fungi, and some plants (Mansour et al., 2007; Castillo and Saier, 2010; Yang et al., 2015). Shi et al. (2018) constructed a Neighbor-joining phylogenetic tree of arsenic efflux proteins and found that there was a clear association of proteins by function, as ArsB and Acr3 sequences formed distinct groups together, as did ArsK, ArsJ, and MFS1 sequences, but they were clearly divergent from each other at the subgroup level. ArsB and Acr3 are dominant in abundance compared to other As-pumps, but they are also the most ancient as it has been proposed that the minor types of pumps have a complementary function (Yang et al., 2015).
Resistance to As(III) by extrusion mediated by the Acr3 transporter is the most common tolerance mechanism found in prokaryotes and eukaryotes (Maciaszczyk-Dziubinska et al., 2012; Markowska et al., 2015; Yang et al., 2015). In yeast (Saccharomyces cerevisiae), arsenic resistance requires at least three genes: ACR1, ACR2, and ACR3, of which Acr3p is an oxyanion transporter that extrudes As(III) and is positively regulated by Acr1p (Wysocki et al., 1997). The impact of gene dose or multicopy on the ability to resist arsenic has also been previously addressed with variable results (Mateos et al., 2017).
The heterologous expression of Acr3 from yeast or plants in agronomical important species that are susceptible to arsenic is of a great biotechnological interest to generate safer and more resistant crops. Previous research has achieved increased arsenic tolerance, through enhanced arsenic efflux, in Escherichia coli AW3110 (Bandyopadhyay and Das, 2016), yeast (Bobrowicz et al., 1997; Wysocki et al., 1997; Ghosh et al., 1999), as well as Oryza sativa (rice), Arabidopsis thaliana, and Nicotiana tabacum plants (Ali et al., 2012; Duan et al., 2012; Chen et al., 2017).
Our research group has studied different strains of the Exiguobacterium bacterial genus isolated from the Chilean Altiplano and their genomes, focusing on their arsenic-resistance traits. These bacteria can thrive under several stressors, including high concentrations of arsenic in the sediments of Salar de Huasco (SH). This area is known for its high microbial biodiversity, as well as the complexity of different niches in a relatively small space, including salinity and arsenic gradients (Castro-Severyn et al., 2020). At metagenomic level, the most abundant genes related to arsenic in these bacterial communities of the SH are acr3 followed by arsB and arsJ, indicating that the As(V) reduction and subsequent As(III) expulsion mechanism is the most prevalent detoxification means used by bacteria in this ecosystem (Castro-Severyn et al., 2021).
Furthermore, we have studied the genome and physiology of the Exiguobacterium sp. SH31 strain that is able to grow in up to 10 mM arsenite and 100 mM arsenate, and we found that the expression of the ars operon and acr3 was strongly-induced in response to both toxics, suggesting that the arsenic efflux pump Acr3 mediates arsenic resistance in Exiguobacterium sp. SH31 (Castro-Severyn et al., 2017). Furthermore, the acr3 gene is not frequent among this bacterial genus; it was only found in strains S17 and SH31, both isolated from the Altiplano. Their protein sequences share 94% identity and the conservation of critical residues between both proteins, which could partially explain the high arsenic resistance of these strains (Ordoñez et al., 2015; Castro-Severyn et al., 2017). Interestingly, the Exiguobacterium strains isolated from SH possess four different arsenic pumps: ArsB, ArsK, ArsP and Acr3 (Figure 1; Castro-Severyn et al., 2020).
Figure 1.
Schematic representation of the phylogenetic relationships between the arsenic efflux pumps found in Exiguobacterium and their function inside the cell to face arsenic.
Moreover, our group has reported the differential expression of the pumps present in three Exiguobacterium strains (with different levels of arsenic resistance) at both transcriptional (Castro-Severyn et al., 2020) and proteomic (Castro-Severyn et al., 2019) levels induced by arsenic. In this study, we characterized the biochemical, structural, and genomic characteristics of the Acr3 pump among a group of Exiguobacterium strains isolated from different sites of the arsenic-rich SH ecosystem. In order to address whether possible changes in the protein sequence and structure among the strains could be the source of the displayed differences in arsenic resistance levels presented by the strains, something that has not been done previously nor reports are available.
Materials and methods
Similarities between the acr3 gene sequences
To make comparative analyses at the gene level, we extracted the acr3 sequences from the genomes of the 10 Exiguobacterium strains isolated from the SH, including the SH31 strain (Castro-Severyn et al., 2020), in addition to some reference sequences as shown in Table 1. A phylogeny was constructed to evaluate the evolutionary relationships between the acr3 genes. For this, the gene sequences were codon-aligned with RevTrans v2.0 (Rasmus and Pedersen, 2003), the alignment was used to infer phylogeny (GTR model) with FastTree v2.1.10 (Price et al., 2009) and FigTree v1.4.4 (Rambaut, 2009) was used to visualize the tree. Also, MAFFT v7 (Katoh and Standley, 2013) and PAL2NAL v14 (Suyama et al., 2006) tools were used to align the protein sequences, converting the multiple sequence alignment to the corresponding DNA codon alignment, which was used to calculate synonymous (dS) and non-synonymous (dN) substitution rates. Moreover, to analyze the degree of similarity and clustering between the different sequences, an identity matrix was calculated using pyANI v0.2.10 (Pritchard et al., 2016) using a multi-dimensional scaling (MDS) ordination with the R environment v4.0.3 (base and the packages stats v4.0.3, ggplot2 v3.3.5 and dplyr v1.0.7). Another phylogeny was constructed with the Acr3 amino acidic sequences, using MAFFT v7 (Katoh and Standley, 2013) with an L-INS-i strategy for iterative refinements, and a maximum-likelihood phylogeny tree was generated with IQ-TREE (Nguyen et al., 2015; 10,000 bootstrap) and visualized with FigTree v1.4.4 (Rambaut, 2009). The acr3 genomic context of the strains was visualized with Geneious v2022.1.1 (Kearse et al., 2012) and the homology of neighboring orthologous genes was evaluated using Blast (Altschul et al., 1990).
Table 1.
Exiguobacterium strains and other bacterial reference Acr3 sequences.
| Strains | GenBank accessions | MIC [mM] | ||
|---|---|---|---|---|
| Genes | Proteins | As(III) | As(V) | |
| Exiguobacterium sp. SH0S1 | GCA_004337165.1 | WP_131507397.1 | 7.5 | 200 |
| Exiguobacterium sp. SH0S2 | GCA_004337185.1 | WP_131489940.1 | 10 | 200 |
| Exiguobacterium sp. SH0S7 | GCA_004337195.1 | WP_131443547.1 | 20 | 150 |
| Exiguobacterium sp. SH3S1 | GCA_004337105.1 | WP_131469704.1 | 15 | 200 |
| Exiguobacterium sp. SH3S2 | GCA_004337285.1 | TCI59634.1 | 2.5 | 200 |
| Exiguobacterium sp. SH3S3 | GCA_004337115.1 | TCI44176.1 | 7.5 | 200 |
| Exiguobacterium sp. SH31 | GCA_001816105.1 | WP_084813073.1 | 10 | 100 |
| Exiguobacterium sp. SH4S7 | GCA_004336795.1 | TCI37681.1 | 10 | 200 |
| Exiguobacterium sp. SH5S4 | GCA_004337045.1 | WP_131458986.1 | 15 | 200 |
| Exiguobacterium sp. SH5S13 | GCA_004337085.1 | WP_131468614.1 | 15 | 100 |
| Exiguobacterium sp. SH5S32 | GCA_004336775.1 | TCI46015.1 | 15 | 100 |
| References | ||||
| Corynebacterium glutamicum ATCC 13032 | Cgl0262 | BAB97655.1 | 12 | >500 |
| Cgl1470 | BAB98863.1 | |||
| Cgl1510 | BAB98903.1 | |||
| Alkaliphilus metalliredigens QYMF | Amet_1828 | ABR47998.1 | – | – |
| Amet_0984 | ABR47201.1 | |||
| Amet_3996 | ABR50078.1 | |||
| Bacillus subtilis 168 | ASM904v1 | NP_390456.2 | – | – |
| Exiguobacterium sp. S17 | GCA_000411915.1 | WP_035385877.1 | 10 | 150 |
| Exiguobacterium mexicanum HUD | GCA_000763125.1 | WP_034781870.1 | – | – |
| Exiguobacterium aurantiacum PN47 | GCA_001766415.1 | WP_083274029.1 | – | – |
Acr3 protein modeling and structural analysis
Variations in Acr3 amino acid sequences from the eleven SH Exiguobacterium strains were discovered by alignment using clustalW (Madeira et al., 2019), and visualized with Jalview v2.11.1.4 (Waterhouse et al., 2009). The topology of Acr3 and transmembrane segments was predicted using TMHMM (Krogh et al., 2001) as well as PSIPRED (Jones, 1999) which predicts transmembrane helices, extracellular and cytoplasmic topologies. Moreover, all three-dimensional models of Acr3 were predicted using the ColabFold server (Mirdita et al., 2022), which uses the AlphaFold v2.0 (Jumper et al., 2021; Varadi et al., 2022) program, and the MMseqs2 (Steinegger and Söding, 2017) software for the generation of multiple sequence alignments and for template searching in order to construct the protein structure. Additionally, the predicted structures were relaxed to improve the geometry of the side chains by removing stereochemical violations and clashes. The relaxation of the structures was performed by an iterative restrained energy minimization procedure, with additional harmonic restraints that keep the system near its input structure. The minimization procedure was performed using the OpenMM v7.3.1 (Eastman et al., 2017) simulation package with the Amber99sb force field (Hornak et al., 2006), considering a tolerance of 2.39 kcal/mol and an unbounded maximum number of steps, which are OpenMM’s default values. The templates used for the AlphaFold2 predictions were the bile acid sodium transporters from Neisseria meningitidis and Yersinia frederiksenii bacteria (PDBs: 3ZUX, 3ZUY, 4N7W). Also, the per-residue confidence metric pLDDT value of AlphaFold was used to estimate how well the prediction of Acr3 proteins was performed, whilst the VMD program (Humphrey et al., 1996) was used to visualize the protein models, thus generating the structural superpositions through the Stamp Structural Alignment feature of MultiSeq (Roberts et al., 2006) and to measure the intra hydrogen bonds of proteins by the Hbonds plugin and default parameters. Furthermore, a contact map for the protein structures was created using CMView (Vehlow et al., 2011), selecting as contact type the alpha carbons of the protein, a cutoff distance of 8.0 angstroms and the Needleman-Wunsch sequence alignment. In order to understand the energy changes associated with the amino acids that vary between the Acr3 proteins and their contact network, the binding energy of each amino acid under study was calculated by the MM-GBSA method (Gohlke et al., 2003; Gohlke and Case, 2004) as follows:
where AB corresponds to the contact network formed by the amino acid that varies (for example, residue 10) and the residues in its environment that are at a distance of 5 angstrom. The term A corresponds only to the amino acid that varies, and B corresponds only to the residues of the environment without considering A. GTOTAL corresponds to the sum of HMM and Gsolv. HMM is the enthalpy, which corresponds to the molecular mechanics energies (bonds, angles, VDW, and electrostatic contribution). Gsolv represents the solvation free energy obtained by the Generalized Born approach and solvent accessible surface area (SASA).
Strains and culture conditions
The strains used in this study are listed in Table 2. Escherichia coli strains were grown routinely at 37°C in Luria Bertani (LB) medium with shaking (150 rpm). When required, LB was supplemented with ampicillin (Amp, 100 μg/ml), Tetracycline (Tet, 10 μg/ml) or Chloramphenicol (Cam, 25 μg/ml). The Exiguobacterium sp. SH31 strain was grown in LB medium with shaking (150 rpm) at 25°C.
Table 2.
Bacterial strains used in this study.
| Strain | Relevant characteristics | Source (Reference) |
|---|---|---|
| Exiguobacterium sp. SH31 | WT, arsenic resistant arsRDCBA + acr3 + | Our group (Castro-Severyn et al., 2017) |
| E. coli BW25113 | E. coli K-12 derivative arsRCB + acr3 - | CGSC* (Baba et al., 2006) |
| E. coli AW3110 | E. coli K-12 derivative Δars::Cam arsRCB - acr3 - | CGSC* (Carlin et al., 1995) |
| E. coli BW25113/pBR322-acr3 | BW25113 strain transformed with a derivative pBR322 vector carrying the Exiguobacterium sp. SH31 acr3 gene | This work |
| E. coli AW3110/pBR322-acr3 | AW3110 strain transformed with a derivative pBR322 vector carrying the Exiguobacterium sp. SH31 acr3 gene | This work |
The E. coli Genetic Stock Center (https://cgsc.biology.yale.edu/index.php).
Genetic complementation with acr3
To complement the BW25113 and AW3110 strains, the acr3 gene from Exiguobacterium sp. SH31 was amplified by PCR using the primers (F: 5′- CCCAAGCTTTTGAGTCGATTCGAGAAATT-3′ and R: 5′- CCCGGATCCCTAACCGAGTCGACGTTC-3′) which introduces the HindIII and BamHI restriction sites (underlined), respectively, using gDNA as a template. The PCR product and the pBR322 vector (Invitrogen) were then digested with both enzymes and the digested product was cloned between the vector HindIII – BamHI sites, interrupting the tet gene (losing the resistance). The generated pBR322-acr3 vector was transformed by electroporation into competent BW25113 and AW3110 cells. Correct transformants were selected by plating cells in LB medium (agar 10 gr/L) with the appropriate antibiotics.
Arsenic minimal inhibitory concentration
Minimal inhibitory concentration (MIC) assays for As(III) and As(V) were performed for all strains. Briefly, bacterial cultures in LB broth (supplemented with corresponding antibiotics) were grown at 37°C with constant agitation (150 rpm) until OD600 = 0.4. Subsequently, we set up a microplate with dilutions of As(III): NaAsO2 and As(V): Na3AsO4 to final concentrations ranging from 0.1 to 25 mM and from 1 to 200 mM, respectively. Controls were made by adding fresh medium to the corresponding well instead of arsenic. Each well was inoculated with the culture in 1:20 ratio in LB medium. Finally, the plates were incubated at 37°C for 12 h with constant agitation, and OD600 values were read with an Infinite 200 PRO Microplate Reader (Tecan).
Growth monitoring
The growth of all strains was monitored in LB media with different As(III) (0.5, 1, 1.5 and 2.5 mM) and As(V) (5, 10, 20, 50 mM) concentrations and the corresponding antibiotics. Cells were incubated at 37°C with constant agitation at 150 rpm for 24 h. The OD600 was measured every hour using an Infinite 200 PRO Microplate Reader (Tecan). Curves were plotted using the R package ggplot2 (Wickham, 2016).
Colony-forming unit determination
The number of viable microorganisms in control and arsenic conditions was estimated by the method described by Miles et al. (1938) and was expressed as CFU/ml. Briefly, cultures from all the strains were grown until their exponential phase (OD600 ~ 0.4). For the BW25113 strains the As(III) and As(V) concentrations were 1 and 10 mM respectively, while for the AW3110 strains, the corresponding concentrations were 0.5 and 5 mM. The cultures were then serially diluted, plated into LB medium and incubated for 12 h prior to the colony counting process. Plots were made using the R package ggplot2 (Wickham, 2016).
Results
Relationships among the acr3 gene sequences
We characterized at a genomic and structural level these sequences and their attributes, to identify differences among the data set. First, to evaluate the evolutive associations between known acr3 genes, we determined the phylogenetic relationships between the nucleotide sequences belonging to Exiguobacterium strains isolated from the SH; for comparisons we also included references of acr3 sequences belonging to other characterized Exiguobacterium strains as well as other bacterial species. Our results show that all Exiguobacterium strains are monophyletic and closely clustered together, belong to a different group compared to the other genera. In addition, the SH strains are grouped together, being the S17 strains the closest, which comes from a similar environment, also pressured by arsenic (Figure 2A). We also evaluated the mode and strength of selection pressures in the acr3 sequences by analyzing codon evolution and temporal dynamics through the calculations of dN/dS ratios, using the acr3 sequence from the S17 strain as reference. The S17 is a closer but external strain, providing proper resolution to evaluate relationships between SH strains sequences (Supplementary Table S1). We found that all the studied acr3 sequences are under negative selection (with similar dN/dS ratios ranging from 0.1202 to 0.1403) diverging from S17 sequence which seems to be from an independent branch. This is also reflected by the sequence identity values, which ranged from 92.5 to 94.4%. From the amino acidic point of view the phylogeny analysis broadly reflects the distribution observed at genomic level, separating the sequences into three main groups (Supplementary Figure S1), where the closest relationships are maintained.
Figure 2.
Genomic relationships among the studied acr3 sequences. (A) Phylogenetic reconstruction for all compared gene sequences (Exiguobacterium strains and references) and (B) MDS ordination using sequence identities as distance (for the Exiguobacterium strains and close related references).
Moreover, as these sequences are phylogenetically close to each other and to further characterize the relationships between the acr3 sequences in genomes isolated from SH, we calculated the identity matrix and used them as distances to determine relationships or clustering between the evaluated strains by MSD ordination. Our results show a clear pattern of association where the sequence from the S17 strain isolated from the Argentinian Altiplano is the outliner as expected, and another four clusters are formed within the sequences from the Exiguobacterium strains isolated from SH (Figure 2B). Furthermore, we determined that the associations between sequences are not correlated with the specific geographic site, as we found that one group is formed by sequences from strains isolated from site H5 and H3, another includes only strains from H3, also a third cluster includes sequences from sites H4 and H0 and the biggest group is formed by sequences associated with H0, H4, and H5 sites as well as the PN47 reference. Likewise, the clustering observed in the ordination plot corresponds to associations found on the phylogeny tree.
The genomic context of the acr3 gene is organized in two versions in the studied strains (Figure 3). The first corresponds to 9 genes - composed of two ABC transporters, two hypothetical proteins, the universal stress protein uspA, the sulfate transporter sulP, a SAM-dependent methyltransferase and the two arsenic transporters arsK and acr3 - in strains SH0S7, SH3S1, SH3S2, SH3S3, SH5S4, SH5S13 and the reference S17. The second version corresponds to 8 of the same genes with the same organization, with the absence of one of the two hypothetical proteins. Such a context was found in the SH31, SH0S1, SH0S2, SH4S7 and SH5S32 strains. The missing hypothetical protein does not have a known function but seems to belong to the DnaJ/Hsp40 cysteine-rich domain superfamily (SSF57938). The reference strain E. aurantiacum PN47 also has this second gene context organization, with the addition of a transposase related protein (TnpB), which could have been integrated afterwards. However, acr3 in E. mexicanum HUD strain is in a totally different context with genes related to metal resistance and transport with one hypothetical protein, one cation diffusion facilitator (CDF) family protein, bdbC (thiol-disulfide oxidoreductase), czrA (zinc, cobalt, nickel and cadmium transcriptional repressor), zupT (zinc transporter), czcD (Metal cation efflux), and cadA (Cadmium-transporting ATPase) genes. These organizations are mostly reflected in the phylogeny and ordination analyses. Consistently, the Acr3 from the HUD strain is phylogenetically closer to those of C. glutamicum ATCC13032, being outside the Exiguobacterium clade, which could be a hint of HGT in this case.
Figure 3.
Comparison of the acr3 gene cluster organization in the compared strains. Blue and orange arrows display the acr3 and arsK genes respectively; green arrows show the conserved flanking genes in the acr3 gene context and gray arrows display the uncommon genes. Shaded color connections represent gene conservation.
Molecular modeling of the Acr3 protein
To determine whether the differences and similarities at the genetic level are reflected in the 3D protein structure we characterize the Acr3 sequences of the Exiguobacterium studied strains and other sequences used as references trough molecular structural modeling, thus predict the protein structure and identify key amino acids (Figure 4). The results show that the predicted proteins have ten segments corresponding to transmembrane helices (Figure 4A). The model generated indicates a high confidence at each position, as can be seen in the E. sp. SH0S1 Acr3 protein model (Figure 4B). Moreover, these Acr3 protein models from Exiguobacterium and other species were structurally superimposed by using the crystal structure of the bile acid sodium transporter from Neisseria meningitidis (PDB: 3ZUX_A) as a template, which shares 23% identity (sd = 0.006) and an E-value of 9E-4 (sd = 0.0009) compared to the Acr3 modeled proteins. The RMSD (Root Mean Square Deviation) distance measurement between alpha carbons was calculated for all proteins against the template (Supplementary Table S2).
Figure 4.
Exiguobacterium Acr3 protein model. (A) Topology of the Acr3 protein. The ten transmembrane helices are shown, the eight continuous helices in light blue and discontinuous helices in purple and green. (B) Acr3 protein model from the SH0S1 strain, showing the three-dimensional conformation and the spatial orientation of the ten transmembrane helices; color corresponds to the modeling confidence level and green spheres represent the non-conserved amino acids.
Based on the predicted structure, the main transmembrane helix of Exiguobacterium proteins appears to have similar structural features and orientations, with small differences compared to the reference proteins (as can be seen in Supplementary Figure S2A). However, in the reference structures belonging to Bacillus subtilis 168, Corynebacterium glutamicum ATCC 13032 (BAB98903.1), Corynebacterium glutamicum ATCC 13032 (BAB97655.1), Alkaliphilus metalliredigens QYMF (ABR47998.1), Alkaliphilus metalliredigens QYMF (ABR47201.1) and Alkaliphilus metalliredigens QYMF (ABR50078.1) a beta sheet in the extracellular/transmembrane region interface (S2 segment) is observed, which is missing in the Acr3 proteins from Exiguobacterium strains. Also, when analyzing and comparing the structures of Acr3 proteins from Exiguobacterium strains, we found a clear diversification in three clusters (Supplementary Figure S2B). The first one (represented in orange) is formed by E. sp. SH0S1, the second one (in green) is comprised of E. sp. SH0S2, E. sp. SH0S7, E. sp. SH4S7 and E. sp. SH5S32, whilst the third one (in grey) is formed by E. sp. SH3S1, E. sp. SH3S2, E. sp. SH3S3, E. sp. SH5S4, E. sp. SH5S13 and E. sp. SH31. However, this clustering is not reflected in the phylogenetic analysis at the gene and protein level. In addition, the Acr3 proteins from E. aurantiacum PN47 and E. sp. S17 are structurally similar to the green cluster. Also, E. mexicanum HUD and C. glutamicum ATCC 13032 (BAB98863.1) Acr3 proteins are more structurally distant to the three clusters and do not possess the beta sheet.
Moreover, we found discontinuous membrane helices in the Acr3 modeled proteins belonging to the Exiguobacterium strains. Each structure contains a pair of discontinuous (an “α-helix – extended peptide – α-helix” motif) helix as described in Screpanti and Hunte (2007), which is associated with membrane proteins that transport ions in an active form. These discontinuous membrane helices are located between residues 93 and 119 (S4 Segment) and between residues 250 and 278 (S9 Segment; Supplementary Figure S2C). Both regions have conserved amino acids among the different Acr3 from Exiguobacterium strains studied, including a cysteine residue (position 107) of interest, which is conserved across the Acr3 family and has been described as essential for protein function (Fu et al., 2009). Furthermore, given the relevance of this residue, we searched for conformational interactions within a 5-angstroms range from the lateral chain of cysteine, and found that in all Acr3 proteins of the Exiguobacterium strains from SH and the reference S17 protein, the Cys-107 residue interacts with Thr-105, Pro-106, Thr-108, Asp-109, Trp-110, Tyr-111, Leu-129, Asn-132, Thr-260, Ala-263, Arg-264, Asn-265 and Glu-294. Nonetheless, there are some additional interactions: with Leu-136 in members from clusters II and III; with Ser-266 in members from clusters I and III; and with Pro-267 and Leu-298 exclusively in members from cluster III. Conversely, Glu-294 the other residue identified as key for Acr3 functions interacts with Leu-102, Cys-107, Trp-110, Phe-233, Leu-262, Ala-263, Arg-264, Ser-266, Ile-289, Gly-290, Pro-291, Leu-292, Ile-293, Leu-295, Pro-296, Val-297, Leu-298 and Val-299 in the Acr3 protein from all the studied strains. Interestingly, there are two more interaction with Tyr-111 and Leu-229 that only occur in cluster III members.
Non-conserved amino acids
The non-conserved amino acids were identified by the alignment processes in their respective positions. These correspond to 24 amino acids across the Exiguobacterium strains Acr3 sequences, specifically at positions: 10, 17, 27, 31, 34, 87, 134, 164, 165, 181, 186, 187, 188, 190, 191, 198, 205, 226, 227, 238, 251, 304, 310 and 311 (Figure 5). The particular localization of each non-conserved amino acid in the three-dimensional protein can be seen in Figure 4B depicted as green spheres. Moreover, these non-conserved amino acids were identified and characterized according to the nature of their side chain. They correspond to amino acids with negatively-charged side chains (D, E), with positively-charged side chains (R, H, K), with polar-uncharged side chains (S, T, N, Q), with hydrophobic side chains (A, V, I, L, M, F, Y, W) and special cases (C, G, P). However, no correlation was found between the non-conserved amino acid positions and the resistance level presented by the strains to both As(III) and As(V). Even evaluating the type of mutation in each position, the nature of the amino acid, the number of non-conserved positions for each strain and differentiating between the three clusters, no association patterns were observed.
Figure 5.
Non-conserved amino acids among the Acr3 protein sequences of Exiguobacterium strains. The amino acid occurring in each variable position identified is shown for every sequence, as well as the amino acidic classification according to the side chain and how many variable positions each one has, with respect to the consensus sequence. The topological location of each position variable is also shown (E, M, C). The strains are sorted according to their As(III) MIC value.
Contact analysis
A representative protein from each cluster was selected to measure the distance between all amino acid residue pairs of the protein structures using a two-dimensional matrix comparing two contact maps. This contact analysis was performed to determine the associated changes in the amino acid environment when a mutation occurs. For this analysis, Acr3 proteins from E. sp. SH0S1, E. sp. SH0S7 and E. sp. SH3S2 were selected, representing each of the three clusters (Figure 6A). Additionally, the Acr3 proteins from the E. sp. SH0S7 and E. sp. SH3S2 strains were found to be the most resistant to As(III) and the least resistant, respectively. We carried out an inter-cluster pair-wise comparison and observed that the Acr3 protein of E. sp. SH0S1 has a total of 1,575 contacts, while Acr3 of E. sp. SH0S7 has 1,576 contacts. The number of unique contacts for E. sp. SH0S1 is 82 and for E. sp. SH0S7 is 83. Also, the number of contacts in common between these proteins is 1,493 (Figure 6A1). Comparing the second case, Acr3 of E. sp. SH0S7 has a total of 1,576 contacts and E. sp. SH3S2 has a total of 1,577 contacts. The number of unique contacts for E. sp. SH0S7 is 81 and for E. sp. SH3S2 is 82. The number of contacts in common between these proteins is 1,495 (Figure 6A2). In the third comparison, Acr3 of E. sp. SH0S1 has a total of 1,575 contacts and E. sp. SH3S2 has a total of 1,577 contacts. The number of unique contacts for E. sp. SH0S7 is 61 and for E. sp. SH3S2 is 63. The number of contacts in common between these proteins is 1,514 (Figure 6A3). Although the number of total contacts is similar between the proteins, the unique contacts are more variable, especially in the third case. Also, the differences between the protein contacts indicate that the amino acids at postions 10, 17, 27, 31, 87, 134, 165 and 188 stand out (indicated by the arrows in Figure 6A), as they generate a difference in the interactions of the amino acids in their close environment that can be reflected by the structural differences of the generated models. Finally, Figure 6B shows the localization of these eight-most-relevant identified residues in the structural 3D model of Acr3 from Exiguobacterium. The binding energy results indicate that the amino acids 10 and 165 of cluster 1, represented by the E. sp. SH0S1 Acr3 protein, have a higher affinity (lower binding energy) for surrounding amino acids, as well as residue 134 for cluster 2 and residue 165 for cluster 3 (Supplementary Table S3). Therefore, these amino acids offer an increased stabilization of the protein. In addition, some variants such as leucine at position 17 have a higher affinity than valine, isoleucine at position 31 than valine, or leucine at position 188 than arginine. Likewise, it was observed that residue 87 does not present variations in its affinity, independent of the nature of the amino acid. The protein from SH0S7 (strain with greater As(III) resistance) has a differential pattern with respect to the other two strains analyzed, regarding the binding energy values of residues 31, 134, 165 and 188.
Figure 6.
Contact analysis comparing selected Acr3 proteins from the Exiguobacterium strains. Contact maps: panels (A1–A3) shows the inter-cluster comparisons. The upper diagonal represents the pairwise comparison of the two contact maps: pink for the contacts that are unique to the first Acr3 protein of A1: E. sp. SH0S1, A2: E. sp. SH0S7, and A3: E. sp. SH0S1, and green the contacts unique to the second Acr3 protein of A1: E. sp. SH0S7, A2: E. sp. SH3S2, and A3: E. sp. SH3S2. The lower diagonal shows a heat-map of differences between the protein contacts, where red represents differences and blue represent similarities. The arrows indicate the positions of high variability and the amino acids for each case. (B) The position and localization of the eight residues with greatest variation are depicted by different colors on the structural model of the Acr3 protein.
To delve into these differences, hydrogen bonds were identified and measured within a 5-angstrom radius of the amino acids of interest, namely, which varied in the contact analysis. We selected the eight previously identified variable residues (10, 17, 27, 31, 87, 134, 165 and 188) and the results can be seen in Table 3.
Table 3.
Pairs of amino acids that form hydrogen bonds within 5 angstroms radius of residues with greatest variation.
| Residue position | E. sp. SH0S1 | E. sp. SH0S7 | E. sp. SH3S2 |
|---|---|---|---|
| 10 | Thr-18 – Leu-14 Leu-7 – Phe-4 | Ser-15 – Phe-11 Leu-13 – Pro-9 Leu-14 – Val-10 Thr-18 – Leu-14 | Phe-11 – Leu-7 Ser-15 – Phe-11 Ile-12 – Tyr-8 Leu-13 – Pro-9 Thr-18 – Leu-14 |
| 17 | Thr-18 – Leu-14 Leu-20 – Val-16 Phe-22 – Thr-18 Thr-24 – Ala-21 | Ser-15 – Phe-11 Leu-13 – Pro-9 Leu-20 – Val-16 Phe-22 – Thr-18 Thr-18 – Leu-14 Thr-24 – Ala-21 | Ile-12 – Tyr-8 Leu-13 – Pro-9 Thr-18 – Leu-14 Ser-15 – Phe-11 Leu-20 – Val-16 Phe-22 – Thr-18 Thr-24 – Ala-21 |
| 27 | Arg-30 – Glu-26 Ala-32 – Leu-28 Asp-33 – Arg-29 | Ala-32 – Leu-28 | Asp-33 – Arg-29 |
| 31 | Arg-30 – Glu-26 Ala-32 – Leu-28 Asp-33 – Arg-29 Leu-40 – Ile-36 | Ala-32 – Leu-28 Leu-40 – Ile-36 | Asp-33 – Arg-29 Leu-40 – Ile-36 |
| 87 | Ser-86 – Phe-82 Phe-82 – Pro-78 Phe-88 – Leu-84 Ala-85 – Ala-81 | Ser-86 – Phe-82 Phe-82 – Pro-78 Leu-84 – Phe-80 Ala-85 – Ala-81 Trp-96 – His-92 Ile-100 – Trp-96 | Phe-82 – Pro-78 Ser-86 – Phe-82 Leu-84 – Phe-80 Ala-85 – Ala-81 |
| 134 | Leu-135 – Leu-131 Asn-132 – Ile-128 Gln-136 – Asn-132 Tyr-143 – Leu-139 Ile-174 – Leu-170 | Leu-133 – Leu-129 Leu-135 – Leu-131 Gln-136 – Asn-132 Asn-132 – Ile-128 Tyr-143 – Leu-139 Ile-174 – Leu-170 | Leu-133 – Leu-129 Leu-135 – Leu-131 Asn-132 – Ile-128 Gln-136 – Asn-132 Tyr-143 – Leu-139 Leu-163 – Phe-159 Ala-167 – Leu-163 |
| 165 | Leu-160 – Val-156 Val-165 – His-161 His-161 – Ser-157 Ser-162 – Val-158 Ala-167 – Leu-163 Ser-172 – Ile-168 Thr-275 – Ala-271 Met-41 – Val-37 Leu-44 – Leu-40 Leu-40 – Ile-36 Phe-48 – Leu-44 | His-161 – Ser-157 Ala-167 – Leu-163 Ile-168 – Ala-164 Gly-171 – Ala-167 Ser-172 – Ile-168 Thr-275 – Ala-271 Ala-276 – Ile-272 Thr-274 – Leu-270 Met-41 – Val-37 Leu-44 – Leu-40 Leu-40 – Ile-36 Phe-48 – Leu-44 | Leu-160 – Val-156 Ala-165 – His-161 His-161 – Ser-157 Ser-162 – Val-158 Ile-168 – Ala-164 Ala-167 – Leu-163 Gly-171 – Ala-167 Ser-172 – Ile-168 Met-41 – Val-37 Phe-45 – Met-41 Leu-44 – Leu-40 Phe-48 – Leu-44 |
| 188 | Arg-187 – Aan-184 Leu-189 – Gln-185 Ala-190 – Phe-186 Arg-191 – Arg-187 | Leu-180 – Leu-176 Leu-188 – Gln-185 Ser-193 – Glu-190 | Leu-180 – Leu-176 Ser-193 – Glu-190 |
By analyzing the distribution of the non-conserved amino acids, we found that many are located in the area where the alpha helices are in contact with the membrane lipids (away from the channel) and many are hydrophobic (Figures 7A,B). On the other hand, the amino acids located towards the cytoplasm are negatively-or positively-charged, and those within the extracellular region are uncharged polar or hydrophobic. However, by analyzing the hydrogen bonds formed by non-conserved or key residues, we found that indeed there are relevant changes, especially in those residues that form these bonds in some structures but not in others (Figure 7C). We highlight residues 27 and 188 that could be key in the loss or gain of interactions, since they are not present in all cases. Residue 10 could also be key, since its interaction is maintained in the 3 clusters, regardless of the mutation of this position and at the same time presents four single cluster exclusive interactions.
Figure 7.
Distribution and possible effects of relevant residues in the Exiguobacterium Acr3 model. The location of non-conserved amino acids in the protein structure are shown (A) in a longitudinal view and (B) in a top-down view. Panel (C) shows the number of H-bonds that the non-conserved residues as well as the key Cys-107 and Glu-294 can form with proteins from the three clusters or exclusively with some of them.
Complemented bacteria arsenic minimal inhibitory concentration
The ability of the Exiguobacterium sp. SH31 (model strain) Acr3 pump to impact the arsenic resistance was tested through heterologous expression in susceptible E. coli strains. We determined the minimal inhibitory concentration (MIC) of each strain (Table 2) and found that as expected the complemented E. coli BW25113/pBR322-acr3 strain has 2X and 5X increased resistance for As(III) and As(V) respectively, given that this strain possesses both the ars genes and the accessory acr3 pump. Furthermore, the E. coli BW25113 strain with the ars genome (without the acr3 pump) is 50% less resistant to As(III) and 37.5% less resistant to As(V) than the E. coli BW25113/pBR322-acr3 strain. Moreover, the E. coli AW3110/pBR322-acr3 strain (that does not harbor the ars operon but was complemented with the Acr3 pump) increased its ability to resist 2X more As(III) and 4X more As(V), compared to the complete sensitivity displayed by E. coli AW3110 (Table 4).
Table 4.
Genotype of each strain used and their minimal inhibitory concentration against As(III) and As(V).
| Strain | Genotype of interest | MIC (mM) | |
|---|---|---|---|
| As(III) | As(V) | ||
| E. coli BW25113 | arsRCB + / acr3 − | 1.25 | 15 |
| E. coli BW25113/pBR322-acr3 | arsRCB + / acr3 + | 2.5 | 40 |
| E. coli AW3110 | arsRCB − / acr3 − | 0.2 | 1.25 |
| E. coli AW3110/ pBR322-acr3 | arsRCB − / acr3 + | 1 | 5 |
Growth monitoring
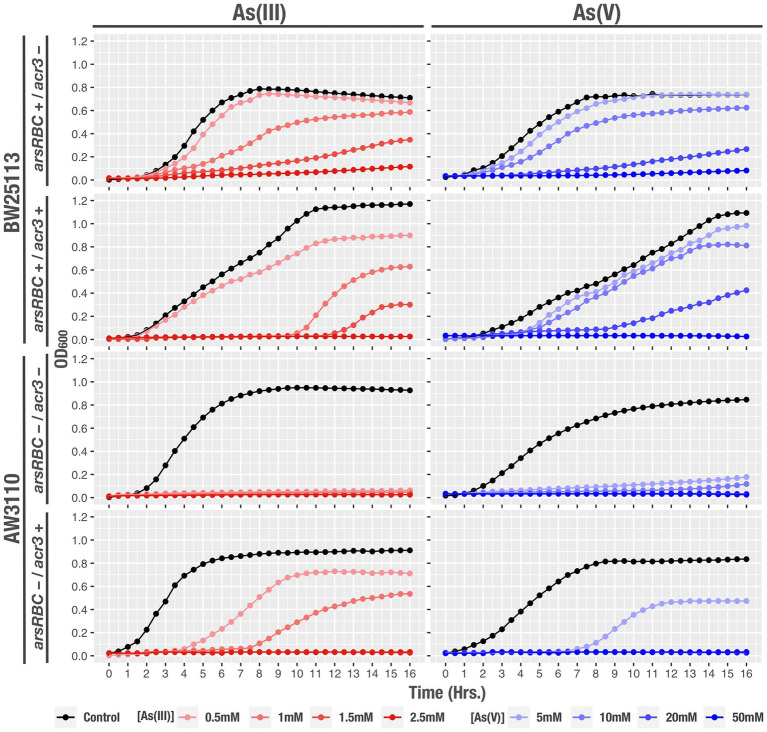
In order to further characterize the resistance of each strain to arsenic and determine the role of Acr3 in their ability to grow and develop in the presence of the toxic compound, we measured by spectrophotometry the bacterial growth curves at increasing concentrations of the metalloid. As expected, the strain with both the ars operon and the acr3 gene had the ability to grow and reach exponential phase more quickly and efficiently than the other strains, despite the high As(III) and As(V) concentrations. E. coli BW25113 containing the classical resistance genes (ars), and the strain that only possessed acr3 were able to reach exponential growth under conditions of 0.5 and 1 mM of As(III), and 5 mM of As(V), whereas the strain that lacked either mechanism did not grown in any of the conditions tested (Figure 8).
Figure 8.
Growth curves of the studied strains, under the tested conditions: control, As(III) and As(V) at different concentrations. OD600 readings were recorded every hour during 16 h. Data represents an average of three independent experiments with three technical replicates each.
Colony-forming unit determination
Additionally, to determine the amount of viable bacteria that were able to survive the presence of arsenic, we measured the Colony Forming Units (CFU) that were generated after growth in media containing As(III) and As(V) for the same strains. Our results show statistically significant differences in cell viability with the strains AW3110 arsRBC−/acr3+ after As(III) exposure and BW25113 arsRBC+/acr3-in the presence of As(III) compared to the control condition. Bacteria that lacked the ars operon (AW3110 arsRBC-acr3+) had significantly decreased cell viability in the presence of As(V) and the strains without acr3 (BW25113 ars + acr3-) had lower recovery under As(III) treatment (Figure 9).
Figure 9.
Viable cell recovery after arsenic treatment [control, As(III) and As(V)] of the studied strains. Bars represents an average of three independent experiments with three technical replicates each. * p<0.05; ** p<0.01; *** p<0.001.
Discussion
The prevalence of arsenite transporters in contaminated and non-contaminated soils has always attracted research. Acr3 transporter sequences are more common than those of ArsB, and have been found across every phylogenetic domain conferring resistance to As(III) (Achour et al., 2007; Yang et al., 2012). This is also reflected by a wider distribution of Acr3 in sequenced bacterial genomes (Shi et al., 2018). The evolution of several means of arsenic detoxification is the result of its environmental prevalence. Arsenic efflux is the most common detoxification mechanism and a considerable diversity of transporter families has been described (Ghosh et al., 1999; Yang et al., 2015). Our group came to the same conclusion, by evaluating at the metagenomic level the abundance and distribution of arsenic efflux pumps in the arsenic rich sediment from SH (Castro-Severyn et al., 2021). ArsB is an antiporter [exchanging As(III) for H], where counter-transport is driven by the protonmotive force, and some experiments have indicated that arsenite efflux via Acr3 could also be coupled to protonmotive force (Ghosh et al., 1999; Villadangos et al., 2012). On the other hand, there is also evidence that demostrate that Acr3 is a uniporter, with As(III) efflux coupled to the membrane potential (Fu et al., 2009). Therefore, despite the widespread distribution of Acr3, its transport mechanism remains unclear.
To identify the genomic basis of these phenotypic capabilities, we addressed the genomic context, phylogenic relationships, as well as protein structure and amino acidic interactions, further elucidating the underlying mechanisms that influence the functionality of Acr3 and its contribution to the variety of resistance levels observed among the studied Exiguobacterium strains. Hence, the phylogeny of the acr3 gene sequences show a clear grouping of all strains from the SH, indicating their close evolutionary history. This could be attributed to the isolation, uniqueness, and diversity of the ecosystem, where bacterial communities change and adapt, but basic functions, such as resistance to metal(oids) is critical given the geochemical characteristics of the area (Risacher et al., 2003; Dorador et al., 2013; Hernández et al., 2016). Therefore, the results of the dN/dS ratio indicate that the acr3 genes of these bacteria are under negative selection, that could be attributed to the mechanism in which the selective forces tend to maintain an improved structure/function, thus contributing to the stability of the trait without generating further mutations. The dN/dS ratio remains one of the most popular and reliable measures of evolutionary pressures on protein-coding regions, with a simple and intuitive interpretation of dN/dS < 1 as negative selection, dN/dS = 1 as neutrality, and dN/dS > 1 as positive selection (Kryazhimskiy and Plotkin, 2008).
Moreover, the study of the genetic context of the acr3 gene could shed light on associated functions, evolution and regulatory networks. Among the Exiguobacterium strains from SH we found two distinct versions of the gene cluster, differing mainly due to the presence of one small hypothetical protein, with an unknown function but belonging to a cysteine-rich domain superfamily that could be important for arsenic resistance; this protein thus requires further biochemical and functional studies. However, it is relevant to highlight that strains S17 and PN47, used as references, are closely related to the ones isolated from the SH, as they belong to nearby places in the Altiplano-Puna Region, where arsenic concentrations are also high (Ordoñez et al., 2013; Strahsburger et al., 2018; Castro-Severyn et al., 2020). This differs from the situation of the HUD strain, which is totally different and whose arsenic resistance has not been experimentally demonstrated.
To study a transport protein at the functional level, it is vital to have a basic understanding of its structure. Hence, the first ever experimentally derived structural study of the Acr3 family concluded that the Acr3 protein from Bacillus subtilis has 10 transmembrane helices, short loops connecting the membrane spanning segments and cytoplasmic loops longer than the extracellular ones (Aaltonen and Silow, 2008). Other Acr3 proteins from organisms such as C. glutamicum, Exiguobacterium sp., S. cerevisiae and A. metalliredigens share the same protein structure (Maciaszczyk-Dziubinska et al., 2012; Ordoñez et al., 2015; Hwang et al., 2016; Mateos et al., 2017). Our findings of predicted topology agree with those previous studies indicating that acr3 encodes a transport protein with transmembrane characteristics (ten transmembrane segments), and structural similarities between the sequences studied also show the formation of three distinct groups. Additionally, we determined that the discontinuous alpha-helices are in the same area as the conserved residue Cys-107, which is distinctive of the Sodium/Bile Acid symporter family (Ordoñez et al., 2015).
However, the grouping of the proteins in three clusters does not reflect the phylogenetic relationships. Therefore, these differences are associated with structural traits that could influence the efficiency of the protein. For instance, the protein crystal used as reference has a beta-sheet that was not present in the Exiguobacterium Acr3 proteins; this common motif could modify the interaction with the membrane or the accessibility to the extracellular space. Nonetheless, our predicted topology is consistent with previous proposals for B. subtilis and A. metalliredigens (Aaltonen and Silow, 2008; Fu et al., 2009).
The Acr3 from S. cerevisiae is the best characterized member of the family, conferring resistance to high concentrations of As(III) and cells lacking this protein are hypersensitive and accumulate more As(III) compared to wild type. A complete loss of transport activity was evident in mutant cells in which the highly conserved cysteine residue was eliminated (Cys151Ala; Maciaszczyk-Dziubinska et al., 2012). The same conclusion was reached in another study where a modification of the same cysteine residue that is conserved in all analyzed Acr3 homologues resulted in loss of transport activity (Fu et al., 2009). This residue is the only cysteine in the Acr3 protein. It is predicted to be in the fourth transmembrane segment (a hydrophobic region), located right in the middle of a clear channel in the structure, and is proposed to be the catalytic residue that binds arsenite (Aaltonen and Silow, 2008), which strongly suggests that the thiol group is required to interact with the As(III) for translocation and thus activation of transport. Also, as single thiols bind trivalent arsenic with low affinity, the binding to As(III) might be weak on one side of the membrane, so the low affinity would facilitate release of As(III) from the carrier on the other side of the membrane (Fu et al., 2009).
The Acr3 proteins of the studied Exiguobacterium strains and S17, share the presence of an intracellular region with a preponderance of positively charged residues (Arg-3, Arg-57, Arg-63, Arg-177, Arg-191, Arg-241, Arg-242, Arg-249, Lys-6, Lys-119, Lys-182, Lys-245, Lys-246), compared with the number of negative residues (Asp-121, Asp-251, Glu-5, Glu-190). Meanwhile, on the extracellular side, there are more negative residues (Asp-33, Asp-91, Glu-26, Glu-217, Glu-250, Glu-280) than positive ones (Arg-29, Arg-30, Arg-213). As the pKa of arsenite is 9.2 and the Exiguobacterium genus is known to be alkaliphile, a low percentage of arsenite would be ionized on the cytosolic side of the membrane, where positively charged residues would interact electrostatically with arsenite due to its negative charge. Interestingly, there are three charged residues (Arg-264, Asp-109, Glu-294) in the core of the transmembrane segments, which are close to Cys-107; all four of them are placed in the catalytic area, and so could be involved in arsenite binding (Ordoñez et al., 2015).
Although the relationship between individual residues with the function of Acr3 proteins is not completely clear, the binding of As(III) by cysteine residues is common for a number of arsenic resistance proteins, including the ArsR repressor, the ArsD metallochaperone, and the ArsA ATPase (Villadangos et al., 2012). In addition, another residue that has been proposed as possibly having great relevance in proton translocation during As(III)/H+ antiport by C. glutamicum Acr3 is Glu-305, whose mutation also causes loss of transport capacity (Maciaszczyk-Dziubinska et al., 2012; Villadangos et al., 2012). The involvement of glutamic acid in proton translocation has been previously described for proteins such as cytochromes and haem-copper oxidases (Riistama et al., 1997; Verkhovskaya et al., 1997; Backgren et al., 2000). Only substitutions of Acr3 Cys and Glu altered As(III) resistance in homologous (C. glutamicum) and heterologous (E. coli) expression analyses, supporting the key role for these two amino acids in As(III) translocation and cellular release (Mateos et al., 2017). Also, as both amino acids are located in the middle of transmembrane segments it has been speculated that both could serve as selectivity filters for As(III).
In this context we evaluated the interaction between these key amino acids (Cys-107 and Glu-294; part of the active site of the protein) and with others in each strain, finding that Cys-107 can interact with a total of 17 residues in the 3D structure, of which 13 interactions occur in all the studied strains, leaving 4 interactions exclusive to some strains. On the other hand, Glu-294 can interact with a total of 20 amino acids in the 3D structure, of which 18 interactions occur in all the studied strains whilst only 2 are exclusive to some strains. All amino acids that are part of these interactions are fully conserved in all of the Acr3 proteins studied; thus the differences or the inability to form some interactions in some sequences may be due to the influence exerted on the 3D structure by the variation level of 24 amino acids positions that are not conserved in each particular sequence.
There is a potential impact of conserved amino acids over Acr3 functionality. As this level of conservation is observed in our studied proteins, we aimed to elucidate the role or participation of variable amino acids in the different arsenic resistance levels observed among the SH Exiguobacterium strains. We found variation within the proteins based on non-conserved amino acids by identifying changes and their position. Despite not finding any association patterns between the non-conserved positions and the arsenic resistance level of each strain, we found that the variations are associated with the more flexible loops; but not within the channel nor the motifs that interact with the membrane. Moreover, the amino acids that vary in the extracellular region are uncharged polar or hydrophobic, while the residues that change in the membrane are hydrophobic as well as those residues found in the helices that are in contact with the membrane lipids on the outside of the channel. However, the amino acids that are located towards the cytoplasm are negatively-or positively-charged, possibly forming a charged channel. Finally, the conservation or variation of this composition directly influences the critical electrostatic interactions of the influx/efflux of ions, as can be seen in Figure 7.
Considering the level of variation observed between the sequences (24 of the 316 positions), we aimed to evaluate if these changes could derive into modifications in interactions or contacts, thus impacting the protein structure or function. We found that residues 27 and 31, located in the predicted membrane/extracellular interface are key as they could be important for the entry of ions into the cell, as well as residue 186 located in the part of the cytoplasm/membrane interface that is vertical to residues 27 and 31, thus possibly affecting the entry or exit of arsenic. Also, it is well known that hydrogen bonds stabilize alpha-helices (Senes et al., 2001; Jarosch, 2005). In the case of Acr3, we detected important changes in these interactions between the different clusters, mainly due to non-conserved amino acids, which clearly exert an effect on the pump structure and dynamics, that could derive into functional differences. Finally, the analysis of the contact maps shows that interactions between clusters 1 and 3 behave similarly, as there are minimal hot-spots that are only found in the charged amino acids of the cytoplasmic region. However, comparing the contact maps of cluster 1 vs. cluster 2 and cluster 2 vs. cluster 3, we detected major differences. There appears to be a pattern where amino acids from intra or inter cellular regions are key for pump function, as they modify the contacts around them, whilst conservative substitutions of hydrophobic amino acids in the transmembrane regions, do not seem to alter the surroundings. Measurement of binding energy shows a distinctive pattern in some Acr3 residues for the most As(III) resistant strain; however these possible relationships need to be experimentally demonstrated.
Finally, the acquisition of the Acr3 pump increases the organism’s tolerance to arsenic exposure, as has been shown before (Ali et al., 2012; Chen et al., 2017; Yang et al., 2022). Of particular relevance is the successful cloning and heterologous expression of a proteorhodopsin gene from a halotolerant, UV-and arsenic-resistant Exiguobacterium sp. S17 isolated from a high-altitude Andean Lake in E. coli XL1 (Albarracín et al., 2016). Our results with the heterologous expression of acr3 indicate that, as expected, the presence of the gene that encodes for the Acr3 pump increase the ability of the strains to withstand greater concentrations of arsenite and arsenate, by enhancing resistance to arsenic (Table 4), growth capability (Figure 8) and cell viability (Figure 9) when propagated in high concentrations of the toxic compounds.
Conclusion
Our results indicate that the arsenic pump Acr3 plays an active role in the ability to detoxify the cell and resist higher concentrations of arsenic in bacteria under laboratory-controlled conditions. Heterologous acquisition of Arc3 provides augmented resistance to this toxic compound. At the genetic level the acr3 sequences from the SH strains have a close evolutionary history, which can be influenced by environmental conditions. Genetic cluster analyses show that one hypothetical protein with an unknown function marks the difference within the genetic context of the acr3 gene. At the protein structural level, observed differences segregate the pumps into three different clusters, where the non-conserved amino acids could impact the function of the protein. Consequently, we are able to observe changes in many interactions such as 5-angstrom contacts and hydrogen bonds. Indeed, the Cys-107 and Glu-294 residues seem to be in critical positions, and are key for the functionality. In any case, further molecular dynamics and structural studies are necessary to elucidate whether these interactions and changes within the active site are relevant for protein efficiency and the ability of the strain to eliminate arsenic.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found at: DDBJ/ENA/GenBank under the Bioproject: PRJNA319980.
Author contributions
JC-S and CS conceived and designed the study. JC-S and VG performed all experimental procedures. JC-S and IA-D performed all bioinformatic analyzes. JC-S, IA-D, and CP-E analyzed interpreted all data and wrote the first manuscript draft. CS and FR contributed with reagents, materials, and analysis. All authors read and approved the final manuscript.
Funding
This research was sponsored by ANID (Agencia Nacional de Investigación y Desarrollo de Chile) grants. CS was funded by ANID-FONDECYT Regular 1210633 and ECOS-ANID 170023. JC-S was funded by ANID 2021 Post-Doctoral FONDECYT 3210156. FR was funded by ANID-FONDECYT Regular 1220902. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the illustrator Florence Gutzwiller for Figure 1 (https://spideryscrawl-illustration.webnode.com/).
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1047283/full#supplementary-material
References
- Aaltonen E. K., Silow M. (2008). Transmembrane topology of the Acr3 family arsenite transporter from Bacillus subtilis. BBA-Biomembranes 1778, 963–973. doi: 10.1016/j.bbamem.2007.11.011, PMID: [DOI] [PubMed] [Google Scholar]
- Achour A. R., Bauda P., Billard P. (2007). Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res. Microbiol. 158, 128–137. doi: 10.1016/j.resmic.2006.11.006, PMID: [DOI] [PubMed] [Google Scholar]
- Albarracín V. H., Kraiselburd I., Bamann C., Wood P. G., Bamberg E., Farias M. E., et al. (2016). Functional green-tuned proteorhodopsin from modern stromatolites. PLoS One 11:e0154962. doi: 10.1371/journal.pone.0154962, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali W., Isner J. C., Isayenkov S. V., Liu W., Zhao F. J., Maathuis F. J. (2012). Heterologous expression of the yeast arsenite efflux system ACR3 improves Arabidopsis thaliana tolerance to arsenic stress. New Phytol. 194, 716–723. doi: 10.1111/j.1469-8137.2012.04092.x, PMID: [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Andres J., Bertin P. N. (2016). The microbial genomics of arsenic. FEMS Microbiol. Rev. 40, 299–322. doi: 10.1093/femsre/fuv050 [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006–0008. doi: 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backgren C., Hummer G., Wikström M., Puustinen A. (2000). Proton translocation by cytochrome c oxidase can take place without the conserved glutamic acid in subunit I. Biochemistry 39, 7863–7867. doi: 10.1021/bi000806b, PMID: [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S., Das S. K. (2016). Functional analysis of ars gene cluster of Pannonibacter indicus strain HT23 T (DSM 23407 T) and identification of a proline residue essential for arsenate reductase activity. Appl. Microbiol. Biotechnol. 100, 3235–3244. doi: 10.1007/s00253-016-7390-2, PMID: [DOI] [PubMed] [Google Scholar]
- Bobrowicz P., Wysocki R., Owsianik G., Goffeau A., Ulaszewski S. (1997). Isolation of three contiguous genes, ACR1, ACR2 and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisiae. Yeast 13, 819–828. doi: , PMID: [DOI] [PubMed] [Google Scholar]
- Cai L., Liu G., Rensing C., Wang G. (2009). Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol. 9, 1–11. doi: 10.1186/1471-2180-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin A., Shi W., Dey S., Rosen B. P. (1995). The ars operon of Escherichia coli confers arsenical and antimonial resistance. J. Bacteriol. 177, 981–986. doi: 10.1128/jb.177.4.981-986.1995, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo R., Saier M. H. (2010). Functional promiscuity of homologues of the bacterial ArsA ATPases. Int. J. Microbiol. 2010:187373. doi: 10.1155/2010/187373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Severyn J., Pardo-Esté C., Mendez K. N., Fortt J., Marquez S., Molina F., et al. (2021). Living to the high extreme: unraveling the composition, structure, and functional insights of bacterial communities thriving in the arsenic-rich Salar de Huasco altiplanic ecosystem. Microbiol. Spectr. 9:e0044421. doi: 10.1128/Spectrum.00444-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Severyn J., Pardo-Esté C., Mendez K. N., Morales N., Marquez S. L., Molina F., et al. (2020). Genomic variation and arsenic tolerance emerged as niche specific adaptations by different Exiguobacterium strains isolated from the extreme Salar de Huasco environment in Chilean–Altiplano. Front. Microbiol. 11:1632. doi: 10.3389/fmicb.2020.01632, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Severyn J., Pardo-Esté C., Sulbaran Y., Cabezas C., Gariazzo V., Briones A., et al. (2019). Arsenic response of three altiplanic Exiguobacterium strains with different tolerance levels against the metalloid species: a proteomics study. Front. Microbiol. 10:2161. doi: 10.3389/fmicb.2019.02161, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Severyn J., Remonsellez F., Valenzuela S. L., Salinas C., Fortt J., Aguilar P., et al. (2017). Comparative genomics analysis of a new Exiguobacterium strain from Salar de Huasco reveals a repertoire of stress-related genes and arsenic resistance. Front. Microbiol. 8:456. doi: 10.3389/fmicb.2017.00456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Hua C. Y., Jia M. R., Fu J. W., Liu X., Han Y. H., et al. (2017). Heterologous expression of Pteris vittata arsenite antiporter PvACR3; 1 reduces arsenic accumulation in plant shoots. Environ. Sci. Technol. 51, 10387–10395. doi: 10.1021/acs.est.7b03369, PMID: [DOI] [PubMed] [Google Scholar]
- Dorador C., Vila I., Witzel K. P., Imhoff J. F. (2013). Bacterial and archaeal diversity in high altitude wetlands of the Chilean Altiplano. Fundam. Appl. Limnol. 182, 135–159. doi: 10.1127/1863-9135/2013/0393 [DOI] [Google Scholar]
- Duan G., Kamiya T., Ishikawa S., Arao T., Fujiwara T. (2012). Expressing ScACR3 in rice enhanced arsenite efflux and reduced arsenic accumulation in rice grains. Plant Cell Physiol. 53, 154–163. doi: 10.1093/pcp/pcr161, PMID: [DOI] [PubMed] [Google Scholar]
- Eastman P., Swails J., Chodera J. D., McGibbon R. T., Zhao Y., Beauchamp K. A., et al. (2017). OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comp. Biol. 13:e1005659. doi: 10.1371/journal.pcbi.1005659, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H. L., Meng Y., Ordóñez E., Villadangos A. F., Bhattacharjee H., Gil J. A., et al. (2009). Properties of arsenite efflux permeases (Acr3) from Alkaliphilus metalliredigens and Corynebacterium glutamicum. J. Biol. Chem. 284, 19887–19895. doi: 10.1074/jbc.M109.011882, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbinski L. D., Rosen B. P., Chen J. (2019). Pathways of arsenic uptake and efflux. Environ. Int. 126, 585–597. doi: 10.1016/j.envint.2019.02.058, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Shen J., Rosen B. P. (1999). Pathways of as (III) detoxification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 96, 5001–5006. doi: 10.1073/pnas.96.9.5001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladysheva T. B., Oden K. L., Rosen B. P. (1994). Properties of the arsenate reductase of plasmid R773. Biochemistry 33, 7288–7293. doi: 10.1021/bi00189a033, PMID: [DOI] [PubMed] [Google Scholar]
- Gohlke H., Case D. A. (2004). Converging free energy estimates: MM-PB (GB) SA studies on the protein–protein complex Ras–Raf. J. Comp. Chem. 25, 238–250. doi: 10.1002/jcc.10379, PMID: [DOI] [PubMed] [Google Scholar]
- Gohlke H., Kiel C., Case D. A. (2003). Insights into protein–protein binding by binding free energy calculation and free energy decomposition for the Ras–Raf and Ras–RalGDS complexes. J. Mol. Biol. 330, 891–913. doi: 10.1016/S0022-2836(03)00610-7, PMID: [DOI] [PubMed] [Google Scholar]
- Hernández K. L., Yannicelli B., Olsen L. M., Dorador C., Menschel E. J., Molina V., et al. (2016). Microbial activity response to solar radiation across contrasting environmental conditions in Salar de Huasco, Northern Chilean Altiplano. Front. Microbiol. 7:1857. doi: 10.3389/fmicb.2016.01857, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. H., Bang S., Kim K. W., Nguyen M. H., Dang D. M. (2010). Arsenic in groundwater and sediment in the Mekong River delta, Vietnam. Environ. Pollut. 158, 2648–2658. doi: 10.1016/j.envpol.2010.05.001, PMID: [DOI] [PubMed] [Google Scholar]
- Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. (2006). Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725. doi: 10.1002/prot.21123, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. (1996). VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38. doi: 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Hwang C., Copeland A., Lucas S., Lapidus A., Barry K., Detter J. C., et al. (2016). Complete genome sequence of Alkaliphilus metalliredigens strain QYMF, an alkaliphilic and metal-reducing bacterium isolated from borax-contaminated leachate ponds. Genome Announc. 4:e01226-16. doi: 10.1128/genomeA.01226-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. R., Jahiruddin M., Rahman G. K. M. M., Miah M. A. M., Farid A. T. M., Panaullah G. M. (2004). Assessment of arsenic in the water–soil–plant systems in gangetic flood plains of Bangladesh. Asian J. Plant Sci. 3, 489–493. doi: 10.3923/ajps.2004.489.493 [DOI] [Google Scholar]
- Jarosch R. (2005). The alpha-helix, an overlooked molecular motor. Protoplasma 227, 37–46. doi: 10.1007/s00709-005-0136-0, PMID: [DOI] [PubMed] [Google Scholar]
- Ji G., Silver S. (1992). Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. PNAS 89:9474e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202. doi: 10.1006/jmbi.1999.3091, PMID: [DOI] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., Von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315, PMID: [DOI] [PubMed] [Google Scholar]
- Kryazhimskiy S., Plotkin J. B. (2008). The population genetics of dN/dS. PLoS Genet. 4:e1000304. doi: 10.1371/journal.pgen.1000304, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Pawitwar S. S., Rosen B. P. (2016). The organoarsenical biocycle and the primordial antibiotic methylarsenite. Metallomics 8, 1047–1055. doi: 10.1039/C6MT00168H, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. F., Walmsley A. R., Rosen B. P. (2006). An arsenic metallochaperone for an arsenic detoxification pump. PNAS 103, 15617–15622. doi: 10.1073/pnas.0603974103, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciaszczyk-Dziubinska E., Wawrzycka D., Wysocki R. (2012). Arsenic and antimony transporters in eukaryotes. Int. J. Mol. Sci. 13, 3527–3548. doi: 10.3390/ijms13033527, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F., Park Y. M., Lee J., Buso N., Gur T., Madhusoodanan N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour N. M., Sawhney M., Tamang D. G., Vogl C., Saier M. H., Jr. (2007). The bile/arsenite/riboflavin transporter (BART) superfamily. FEMS J. 274, 612–629. doi: 10.1111/j.1742-4658.2006.05627.x [DOI] [PubMed] [Google Scholar]
- Markowska K., Maciaszczyk-Dziubinska E., Migocka M., Wawrzycka D., Wysocki R. (2015). Identification of critical residues for transport activity of Acr3p, the Saccharomyces cerevisiae as (III)/H+ antiporter. Mol. Microbiol. 98, 162–174. doi: 10.1111/mmi.13113, PMID: [DOI] [PubMed] [Google Scholar]
- Mateos L. M., Villadangos A. F., Alfonso G., Mourenza A., Marcos-Pascual L., Letek M., et al. (2017). The arsenic detoxification system in Corynebacteria: basis and application for bioremediation and redox control. Adv. Appl. Microbiol. 99, 103–137. doi: 10.1016/bs.aambs.2017.01.001, PMID: [DOI] [PubMed] [Google Scholar]
- Meng Y. L., Liu Z., Rosen B. P. (2004). As (III) and Sb (III) uptake by GlpF and efflux by ArsB in Escherichia coli. J. Biol. Chem. 279, 18334–18341. doi: 10.1074/jbc.M400037200, PMID: [DOI] [PubMed] [Google Scholar]
- Miles A. A., Misra S. S., Irwin J. O. (1938). The estimation of the bactericidal power of the blood. Epidemiol. Infect. 38, 732–749. doi: 10.1017/S002217240001158X, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. (2022). ColabFold: making protein folding accessible to all. Nature Methods, 1–4. doi: 10.1038/s41592-022-01488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T., Schmidt H. A., Von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norra S., Berner Z. A., Agarwala P., Wagner F., Chandrasekharam D., Stuben D. (2005). Impact of irrigation with as-rich ground-water on soil and crops: a geochemical case study in West Bengal delta plain, India. Appl. Geochem. 20, 1890–1906. doi: 10.1016/j.apgeochem.2005.04.019 [DOI] [Google Scholar]
- Ordoñez O. F., Lanzarotti E., Kurth D., Cortez N., Farías M. E., Turjanski A. G. (2015). Genome comparison of two Exiguobacterium strains from high altitude andean lakes with different arsenic resistance: identification and 3D modeling of the Acr3 efflux pump. Front. Environ. Sci. 3:50. doi: 10.3389/fenvs.2015.00050 [DOI] [Google Scholar]
- Ordoñez O. F., Lanzarotti E., Kurth D., Gorriti M. F., Revale S., Cortez N., et al. (2013). Draft genome sequence of the polyextremophilic Exiguobacterium sp. strain S17, isolated from hyperarsenic lakes in the Argentinian Puna. Genome Announc. 1:e00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel J., Joulian C., Leyval C., Billard P. (2013). Arsenite-induced changes in abundance and expression of arsenite transporter and arsenite oxidase genes of a soil microbial community. Res. Microbiol. 164, 457–465. doi: 10.1016/j.resmic.2013.01.012, PMID: [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard L., Glover R. H., Humphris S., Elphinstone J. G., Toth I. K. (2016). Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Analytical Methods 8, 12–24. doi: 10.1039/C5AY02550H [DOI] [Google Scholar]
- Rambaut A. (2009). FigTree. Tree figure drawing tool. Available at: http://tree.bio.ed.ac.uk/software/figtree/-ci.nii.ac.jp
- Rasmus W., Pedersen A. G. (2003). RevTrans—constructing alignments of coding DNA from aligned amino acid sequences. Nucl. Acids Res. 31, 3537–3539. doi: 10.1093/nar/gkg609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riistama S., Hummer G., Puustinen A., Dyer R. B., Woodruff W. H., Wikström M. (1997). Bound water in the proton translocation mechanism of the haem-copper oxidases. FEMS Lett. 414, 275–280. [DOI] [PubMed] [Google Scholar]
- Risacher F., Alonso H., Salazar C. (2003). The origin of brines and salts in Chilean salars: a hydrochemical review. Earth-Sci. Rev. 63, 249–293. doi: 10.1016/S0012-8252(03)00037-0 [DOI] [Google Scholar]
- Roberts E., Eargle J., Wright D., Luthey-Schulten Z. (2006). MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformat. 7, 1–11. doi: 10.1186/1471-2105-7-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Francisco M. J., Hope C. L., Owolabi J. B., Tisa L. S., Rosen B. P. (1990). Identification of the metalloregulatory element of the plasmid-encoded arsenical resistance operon. Nucleic Acids Res. 18, 619–624. doi: 10.1093/nar/18.3.619, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Francisco M. J., Tisa L. S., Rosen B. P. (1989). Identification of the membrane component of the anion pump encoded by the arsenical resistance operon of R-factor R773. Mol. Microbiol. 3, 15–21. doi: 10.1111/j.1365-2958.1989.tb00098.x, PMID: [DOI] [PubMed] [Google Scholar]
- Screpanti E., Hunte C. (2007). Discontinuous membrane helices in transport proteins and their correlation with function. J. Struct. Biol. 159, 261–267. doi: 10.1016/j.jsb.2007.01.011, PMID: [DOI] [PubMed] [Google Scholar]
- Senes A., Ubarretxena-Belandia I., Engelman D. M. (2001). The Cα—Ḥ·· O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc. Natl. Acad. Sci. U. S. A. 98, 9056–9061. doi: 10.1073/pnas.161280798, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrato-Gamiño N., Salgado-Lora M. G., Chávez-Moctezuma M. P., Campos-García J., Cervantes C. (2018). Analysis of the ars gene cluster from highly arsenic-resistant Burkholderia xenovorans LB400. World J. Microbiol. Biotechnol. 34, 1–10. doi: 10.1007/s11274-018-2526-4 [DOI] [PubMed] [Google Scholar]
- Shi K., Li C., Rensing C., Dai X., Fan X., Wang G. (2018). Efflux transporter ArsK is responsible for bacterial resistance to arsenite, antimonite, trivalent roxarsone, and methylarsenite. Appl. Environ. Microbiol. 84, e01842–e01818. doi: 10.1128/AEM.01842-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinegger M., Söding J. (2017). MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028. doi: 10.1038/nbt.3988, PMID: [DOI] [PubMed] [Google Scholar]
- Strahsburger E., Zapata F., Pedroso I., Fuentes D., Tapia P., Ponce R., et al. (2018). Draft genome sequence of Exiguobacterium aurantiacum strain PN47 isolate from saline ponds, known as “Salar del Huasco”, located in the Altiplano in the North of Chile. Braz. J. Microbiol. 49, 7–9. doi: 10.1016/j.bjm.2017.03.011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612. doi: 10.1093/nar/gkl315, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., et al. (2022). AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444. doi: 10.1093/nar/gkab1061, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vehlow C., Stehr H., Winkelmann M., Duarte J. M., Petzold L., Dinse J., et al. (2011). CMView: interactive contact map visualization and analysis. Bioinformatics 27, 1573–1574. doi: 10.1093/bioinformatics/btr163, PMID: [DOI] [PubMed] [Google Scholar]
- Verkhovskaya M. L., Garcìa-Horsman A., Puustinen A., Rigaud J. L., Morgan J. E., Verkhovsky M. I., et al. (1997). Glutamic acid 286 in subunit I of cytochrome bo 3 is involved in proton translocation. Proc. Natl. Acad. Sci. U. S. A. 94, 10128–10131. doi: 10.1073/pnas.94.19.10128, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadangos A. F., Fu H. L., Gil J. A., Messens J., Rosen B. P., Mateos L. M. (2012). Efflux permease CgAcr3-1 of Corynebacterium glutamicum is an arsenite-specific antiporter. J. Biol. Chem. 287, 723–735. doi: 10.1074/jbc.M111.263335, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yin Z., Jing C. (2020). Metagenomic insights into microbial arsenic metabolism in shallow groundwater of Datong basin, China. Chemosphere 245:125603. doi: 10.1016/j.chemosphere.2019.125603, PMID: [DOI] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009). Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016). “Data analysis” in ggplot2 (Cham: Springer; ), 189–201. [Google Scholar]
- Wysocki R., Bobrowicz P., Ułaszewski S. (1997). The Saccharomyces cerevisiae ACR3 gene encodes a putative membrane protein involved in arsenite transport. J. Biol. Chem. 272, 30061–30066. [DOI] [PubMed] [Google Scholar]
- Yang H. C., Fu H. L., Lin Y. F., Rosen B. P. (2012). Pathways of arsenic uptake and efflux. Curr. Top. Membr. 69, 325–358. doi: 10.1016/B978-0-12-394390-3.00012-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Li Y., Feng R., Chen J., Alwathnani H. A., Xu W., et al. (2022). Characterization of two highly arsenic-resistant Caulobacteraceae strains of Brevundimonas nasdae: discovery of a new arsenic resistance determinant. Intl. J. Mol. Sci. 23:5619. doi: 10.3390/ijms23105619, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wu S., Lilley R. M., Zhang R. (2015). The diversity of membrane transporters encoded in bacterial arsenic-resistance operons. PeerJ 3:e943. doi: 10.7717/peerj.943, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. G., Yoshinaga M., Zhao F. J., Rosen B. P. (2014). Earth abides arsenic biotransformations. Annu. Rev. Earth Planet. Sci. 42, 443–467. doi: 10.1146/annurev-earth-060313-054942, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found at: DDBJ/ENA/GenBank under the Bioproject: PRJNA319980.