Summary
Direct in vivo investigation of human placenta trophoblast’s susceptibility to SARS-CoV-2 is challenging. Here we report that human trophoblast stem cells (hTSCs) and their derivatives are susceptible to SARS-CoV-2 infection, which reveals heterogeneity in hTSC cultures. Early syncytiotrophoblasts (eSTBs) generated from hTSCs have enriched transcriptomic features of peri-implantation trophoblasts, express high levels of angiotensin-converting enzyme 2 (ACE2), and are productively infected by SARS-CoV-2 and its Delta and Omicron variants to produce virions. Antiviral drugs suppress SARS-CoV-2 replication in eSTBs and antagonize the virus-induced blockage of STB maturation. Although less susceptible to SARS-CoV-2 infection, trophoblast organoids originating from hTSCs show detectable viral replication reminiscent of the uncommon placental infection. These findings implicate possible risk of COVID-19 infection in peri-implantation embryos, which may go unnoticed. Stem cell-derived human trophoblasts such as eSTBs can potentially provide unlimited amounts of normal and genome-edited cells and facilitate coronavirus research and antiviral discovery.
Keywords: SARS-CoV-2, expanded potential stem cell, EPSC, human trophoblast, trophoblast organoid
Graphical abstract
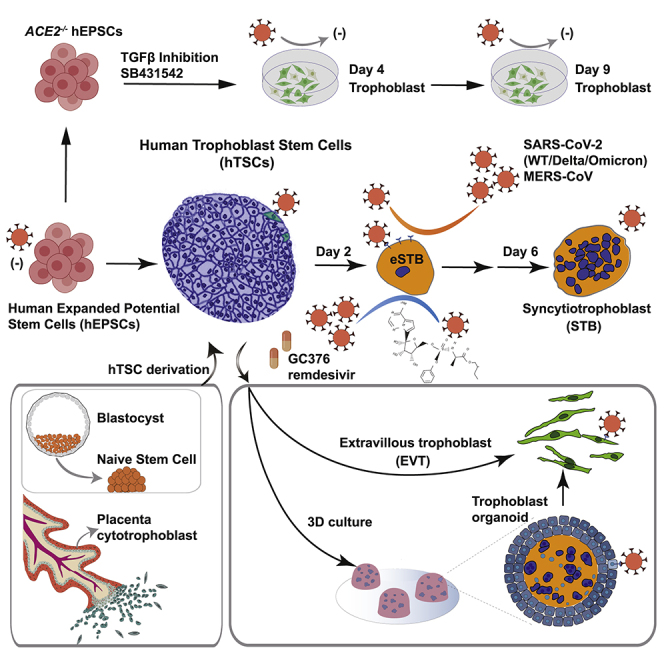
Highlights
-
•
SARS-CoV-2 infects human trophoblast stem cells (hTSCs) and their derivatives
-
•
ACE2+ early syncytiotrophoblasts (eSTBs) are susceptible to SARS-CoV-2 and MERS-CoV
-
•
eSTB virion production is comparable to that of Vero cells and eliminated by antivirals
-
•
Inefficient infection of trophoblast organoids models rare placental infection
Ruan et al. utilize human expanded potential stem cells (hEPSCs) to demonstrate the susceptibility of ACE2+ early syncytiotrophoblasts to SARS-CoV-2 variants. Productive viral infection blocks syncytiotrophoblast maturation, which is rescued by antivirals. Trophoblast organoids are inefficiently infected, reflecting uncommon placental infection in vivo.
Introduction
The human placenta protects the fetus by using multiple cellular and molecular defense mechanisms at the maternal-fetal interface to safeguard against infection during pregnancy, but certain viruses still replicate in the placenta and infect the fetus.1 SARS-CoV-2 infection of the human placenta and resultant damage, although relatively uncommon, have been reported.2,3,4,5,6,7,8,9 In rare cases, vertical transmission from mother to fetus has been reported.4,10,11
The human placenta consists of both maternal and fetal tissues.12 The extraembryonic ectoderm generates the proliferative mononucleated trophoblast progenitors known as villous cytotrophoblasts (vCTBs), which can differentiate into invasive extravillous trophoblasts (EVTs) in the anchoring villi that grow out into the maternal decidua and, by cell fusion, into non-proliferative multinucleated syncytiotrophoblasts (STBs) that form a physical barrier against pathogens.13
SARS-CoV-2 infects cells via its spike (S) protein binding to the host entry receptor ACE214,15 and being primed by the transmembrane serine protease 2 (TMPRSS2).16 Molecular assays and single-cell RNA sequencing (scRNA-seq) studies have identified ACE2 and TMPRSS2 co-expression in only a small number of first-trimester STBs and second-trimester EVTs,17,18,19 which are gradually decreased during pregnancy.17,20 ACE2 shedding may also help prevent SARS-CoV-2 infection from continuing to spread in the placenta.21 These molecular studies are in line with the overall low risk of COVID-19 to pregnant women. However, little is known about COVID-19’s risk to early pregnancy, since the impacts could be unnoticed and it is technically and ethically challenging to study normal trophoblasts of early pregnancy stages. Laboratory model organisms for SARS-CoV-2 have substantial differences from humans in trophoblast biology and placenta development. Therefore, 2D and 3D cellular models of normal human trophoblasts are needed to decipher SARS-CoV-2 infection in trophoblasts and in early pregnancy.
Expanded potential stem cells (EPSCs) derived from cleavage-stage preimplantation embryos retain developmental potential for both extraembryonic and embryonic cell lineages.22,23,24,25 In particular, human (h) EPSCs directly generated human trophoblast stem cells (hTSCs) in vitro.24,26 Standard human embryonic stem cells (hESCs) could also generate trophoblast-like cells27,28,29 and derive hTSC-like lines.30 hTSCs were recently established from human naive ESCs,26,31,32,33,34 which may reflect the property of human naive epiblast to regenerate trophoblasts.32
In the present study, we establish and validate a stem cell-based system to interrogate trophoblast susceptibility to SARS-CoV-2 infection by generating hTSCs from EPSCs and naive stem cells and hTSC derivatives. SARS-CoV-2 infected a small number of hTSCs expressing ACE2. In stark contrast, early STBs (eSTBs) generated from hTSCs expressed high levels of ACE2 and were highly efficient in supporting SARS-CoV-2 infection and virion production. Knockout of ACE2 abolished SARS-CoV-2 infection. eSTBs also expressed the DPP4 gene, which encodes the receptor for Middle East respiratory syndrome coronavirus (MERS-CoV), and were susceptible to the virus infection. Low concentrations (nanomolar) of the antiviral drugs remdesivir and GC376 effectively suppressed viral replication and rescued the developmental defects caused by the infection. In line with relatively uncommon placental infection, hTSC-derived trophoblast organoids expressed low ACE2 and TMPRSS2 and were inefficiently infected by SARS-CoV-2. Taken together, our stem cell-based 2D and 3D trophoblast infection results and the in vivo peri-implantation embryo gene expression profiles implicate a possible risk of COVID-19 infection in early pregnancy, but a relatively lower risk in late pregnancy. Importantly, human stem cells can provide normal and genome-edited cells such as eSTBs to advance coronavirus isolation, propagation, and production, which may help address some of the challenging technical issues of the currently used mammalian cells in virus research.
Results
Establishment of EPSC-TSCs and generation of STBs and EVTs for infection
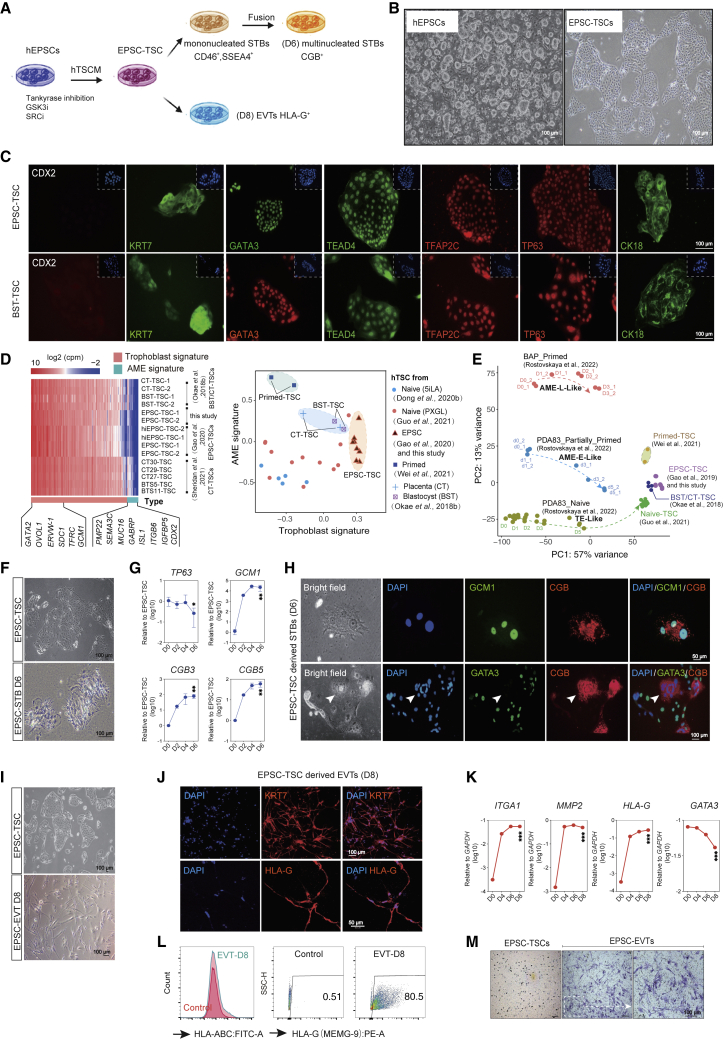
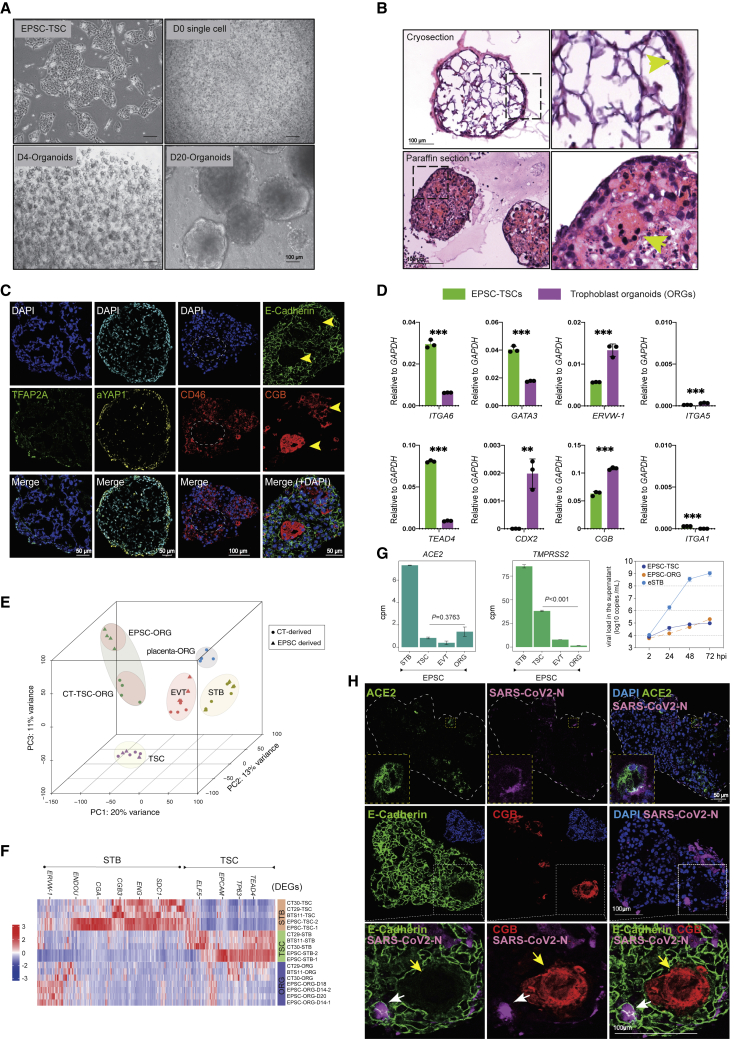
Based on the published culture conditions,35 we established several hTSC lines from M1 hEPSCs, which were converted from the primed human ESC line M124 (Figures 1A and 1B).
Figure 1.
Generation of human trophoblast stem cells (TSCs) and trophoblast subtypes from hEPSCs
(A) Schematic diagram of sequential generation of hTSCs, STBs, and EVTs from hEPSCs.
(B) Bright-field images of hEPSCs and EPSC-TSCs. Scale bars: 100 μm.
(C) Immunofluorescence-stained EPSC-TSCs and human blastocyst-derived hTSCs (BST-TSC).35 Scale bars, 100 μm.
(D) Left: RNA-seq analysis of human trophoblast and putative AME signature gene (Table S1) expression in EPSC- and BST/CT-TSC and five human placenta-derived CT-TSCs.36 Right: Scatterplot for gene set scores for human trophoblast and putative AME signature genes (Table S1) in hTSCs of various origins.
(E) PCA for comparison of hTSCs of in vivo (BST/CT) and in vitro origins (naive, primed, and EPSC). Datasets include in vitro differentiation of naive hPSCs to TSCs (D0–D5), primed hPSCs to AME-L (late)-like cells (D0–D3), and partially primed hPSCs to AME-E (early)-like (D0–D5).37 The dashed lines and arrows indicate the differentiation trajectories.
(F) Bright-field images of EPSC-TSCs differentiating toward STBs (STB-D6). Scale bars, 100 μm.
(G) Gene expression (qRT-PCR) during EPSC-TSC differentiation toward STBs. Data are mean ± SD; n = 3 biological replicates. Student’s t test, ∗p < 0.05, ∗∗p < 0.01.
(H) Immunostaining of STBs-D6 for GCM1 and CGB (top) and GATA3 (bottom). Arrows indicate the lack of GATA3 in a multinucleated CGB+ mature STB. Scale bars, 50 or 100 μm as indicated.
(I) Bright-field images of EPSC-TSCs differentiating toward EVT-D8. Scale bars, 100 μm.
(J) Immunostaining of EVT-D8 for KRT7 and HLA-G. Scale bars, 50 or 100 μm as indicated.
(K) Gene expression (qRT-PCR) during EPSC-TSC differentiation toward EVTs. Data are mean ± SD; n = 3 biological replicates. Student’s t test, ∗∗∗p < 0.001.
(L) Flow cytometric analysis of HLA-A, -B, -C and HLA-G on EVT-D8. EPSC-TSCs were used as the control.
(M) Invasiveness of EVT-D8 in the transwell assay. The invading/migrating cells on the lower surface were stained with crystal violet. Scale bar, 100 μm.
EPSC-TSCs formed cobblestone-shaped colonies and expressed typical hTSC markers (Figures 1B, 1C, and S1A) and trophoblast-specific C19MC miRNAs (has-miR-517c-3p, 517-5p, 525-3p, and 526b-3p)38 (Figure S1B) highly resembling human blastocyst-derived TSCs (BST-TSCs).35 They were low or negative for the classical human leukocyte antigen (HLA) class I molecules HLA-A and -B, like BST-TSCs (Figures S1C and S1D). EPSC-TSCs did not highly express putative amniotic epithelium (AME) signature genes such as CDX2, MUC16, GABRP, ITGB6, or VTCN139,40,41,42,43 (Figures 1D, S1E, and S1F; Table S1), similar to those derived from human blastocyst and placenta cytotrophoblasts (BST-TSCs, CT-TSCs, CT27-TSCs, CT29-TSCs, CT30-TSCs, BTS5-TSCs, and BTS-11-TSCs).35,36 We further transcriptomically compared hTSCs derived from EPSCs, primed, and naive stem cells with those of in vivo origins and AME-like cells24,30,31,32,35,36,37 (Table S2). EPSC-TSCs and naive-TSCs were more similar to each other than to “primed”-TSCs in the expression of putative AME signature genes (Figures 1D, 1E, and S1G).
EPSC-TSCs were induced to generate STBs (Figures 1F and S1H). The trophoblast progenitor marker TP63 was reduced, while STB genes such as GCM1, β-chorionic gonadotrophin 3 gene (CGB3), and CGB5 were quickly increased (Figure 1G). Immunofluorescence staining of day 6 differentiated cells (STB-D6) detected GCM1+ and CGB+ and multinucleated STBs (Figures 1H and S1I), some of which seemingly lost GATA3 expression (Figure 1H). Functionally, in the supernatant of STB-D6 cells, ELISA detected properly folded and secreted β-hCG hormone (Figure S1J). Under TGF-β inhibition, EPSC-TSCs efficiently generated EVTs with mesenchymal morphologies and were stained positively for KRT7, HLA-G, ITGA1, and IGTA5 (Figures 1I, 1J, and S1K), with ITGA1, MMP2, and HLA-G rapidly increased and GATA3 decreased or absent (Figures 1K and S1L). On day 8, most cells were positive for HLA-G, ITGA1, and ITGA5, but negative for the classical HLA class I molecules HLA-A and -B (Figures 1L and S1M). Functionally, EPSC-TSC-derived EVTs possessed potent invasiveness capability (Figure 1M).
We next performed RNA-seq analysis of EPSC-TSCs and their derivative STBs (D2, D4, and D6) and EVTs (D4, D6, and D8) and compared them with human primary trophoblast-derived hTSCs (CT-TSC, BTS-TSC) and their derivatives.36 Principal-component analysis (PCA) showed that EPSC-TSCs again clustered closely to both CT-TSCs and BTS-TSCs, while EPSC-EVTs and EPSC-STBs clustered closely to CT-EVTs/BTS-EVTs and CT-STBs/BTS-STBs, respectively (Figure S1N). RNA-seq further confirmed low classical HLA class I molecules HLA-A and -B (Figure S1O) in all TSCs.36
Trophoblasts derived from hEPSCs molecularly resemble those in human peri-implantation embryos and placenta
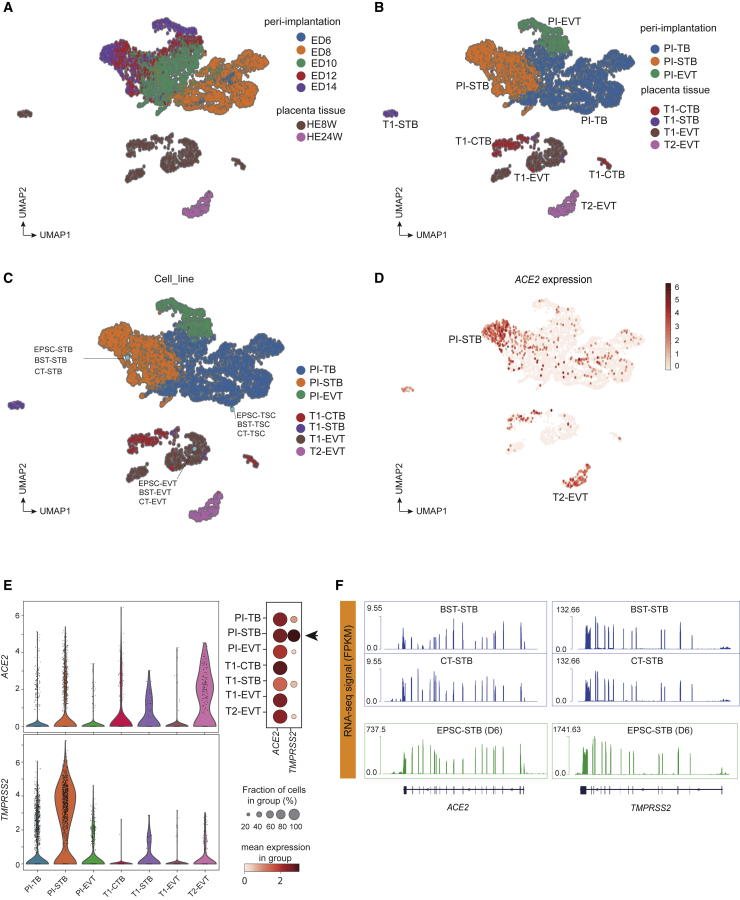
We next computationally compared EPSC-derived trophoblasts with those in human peri-implantation embryos and the placenta to validate the lineage identity of the in vitro-generated trophoblasts. We extracted scRNA-seq data of 4,041 peri-implantation extraembryonic cells (embryonic days 6–14) in a prolonged culture of human embryos in vitro44 and 952 placental cells from first- and second-trimester pregnancies45 and subsequently combined these scRNA-seq datasets to compute the joint uniform manifold approximation and projection (UMAP), with their developmental times highlighted (in embryonic day, or ED, or gestational week, or W) (Figure 2A) and developmental stages and subtypes annotated (Figure 2B). The peri-implantation sector contained the trophoblast cells (PI-TBs), which possessed stemness and potency to differentiate into EVTs (PI-EVTs) and STBs (PI-STBs) as shown in the same sector (Figure 2B). Correspondingly, the placenta cells were identified as first-trimester cytotrophoblast cells (T1-CTBs), T1-EVTs, and T1-STB or second-trimester placenta EVTs (T2-EVTs) (Figure 2B). The trophoblast markers were examined in each stage-specific subtype, which validated their trophoblast identities (Figure S2A). Notably, these human in vivo trophoblasts did not appear to highly express the reported putative amnion marker genes MUC16, GABRP, or CDX2, whereas some did express VTCN1, ITGB6, and ISL1 (Figure S2B), whose identity warrants further investigation.
Figure 2.
Trophoblasts derived from hEPSCs resemble those in human peri-implantation embryos and placenta
(A and B) UMAP analysis of scRNA-seq data of cells from in vitro-cultured peri-implantation (PI) embryo stages (top) and of cells from first (T1)- and second (T2)-trimester placenta (bottom). Cells are colored by developmental time points (ED6–14, embryonic day 6–14; HE8W/HE24W, placenta trophoblasts at 8 or 24 weeks of gestation, corresponding to first or second trimester) in (A) and trophoblast subtypes at each stage in (B).
(C) Mapping of in vitro human trophoblast cell bulk RNA-seq data to the peri-implantation and placental trophoblast clusters in (B). The in vitro cells are highlighted in light blue filled circles.
(D) Expression of ACE2 in peri-implantation and placenta trophoblast scRNA-seq clusters.
(E) Left: violin plots of ACE2 and TMPRSS2 expression (log-transformed transcripts per million [TPM]). Right: dot plot of ACE2 and TMPRSS2 expression levels in ACE2-positive cells, categorized according to stage and cell lineage. Dots are colored according to the mean expression value in each category and dot size indicates the percentage of ACE2- or TMPRSS2-positive cells from each category that expresses ACE2.
(F) RNA-seq signals of ACE2 and TMPRSS2 in BST/CT-STBs and EPSC-STBs. The ACE2 and TMPRSS2 genomic loci are plotted at the bottom, where each vertical bar represents an exon, and the transcription direction is from right to left.
We then projected and mapped EPSC-TSCs and blastocyst- or cytotrophoblast-derived TSCs (BST-TSCs, CT-TSCs) and their STBs/EVTs35 against the in vivo trophoblast clusters categorized by stage and lineage as described in Figures 2A and 2B. All hTSCs (EPSC-TSCs, BST-TSCs, CT-TSCs) were projected proximal to PI-TBs,33 whereas STBs were projected proximal to PI-STBs, and EVTs to T1-EVTs, regardless of cell line origin (Figure 2C). The resemblance between in vitro-generated trophoblasts and in vivo counterparts was supported by whole-transcriptome Pearson correlation (Figure S2C). Notably, none of the hTSCs or their STBs/EVTs expressed the putative AME genes at high levels (Figure S2D).
To explore the potential trophoblast susceptibility to SARS-CoV-2, we examined ACE2 in the in vivo trophoblasts and detected its high expression in the PI-STB cluster (Figures 2D and 2E). ACE2 and trophoblast subtype markers exhibit significant positive correlations with STB markers such as CD46, CGB5, ENG, and CSH2 (r = 0.245, 0.200, 0.207, and 0.248, respectively, p < 0.0001) (Figure S2E), but not EVT genes (Table S3). The PI-STB cluster also co-expressed the highest levels of ACE2 and TMPRSS2, whereas some co-expression was detected in PI-TBs, T1-STBs, and T2-EVTs (Figures 2E and S2F), in line with previous studies.17,19 Other reported SARS-CoV-2 receptors, like BSG46 and AXL,47 however, did not appear to be expressed in any specific cell cluster (Figure S2F).
In the in vitro-cultured trophoblasts, EPSC-TSCs, BST-TSCs, CT-TSCs, and their STBs and EVT derivatives, all expressed ACE2 and TMPRSS2, with STBs having the highest levels (Figures 2F, S2G, and S2H), whereas hEPSCs had barely detectable ACE2 or TMPRSS2 (Figure S2H). The host factor expression profiles in both in vivo and in vitro trophoblasts highlighted STBs’ potential susceptibility to SARS-CoV-2 infection.
hTSCs are susceptible to SARS-CoV-2 infection
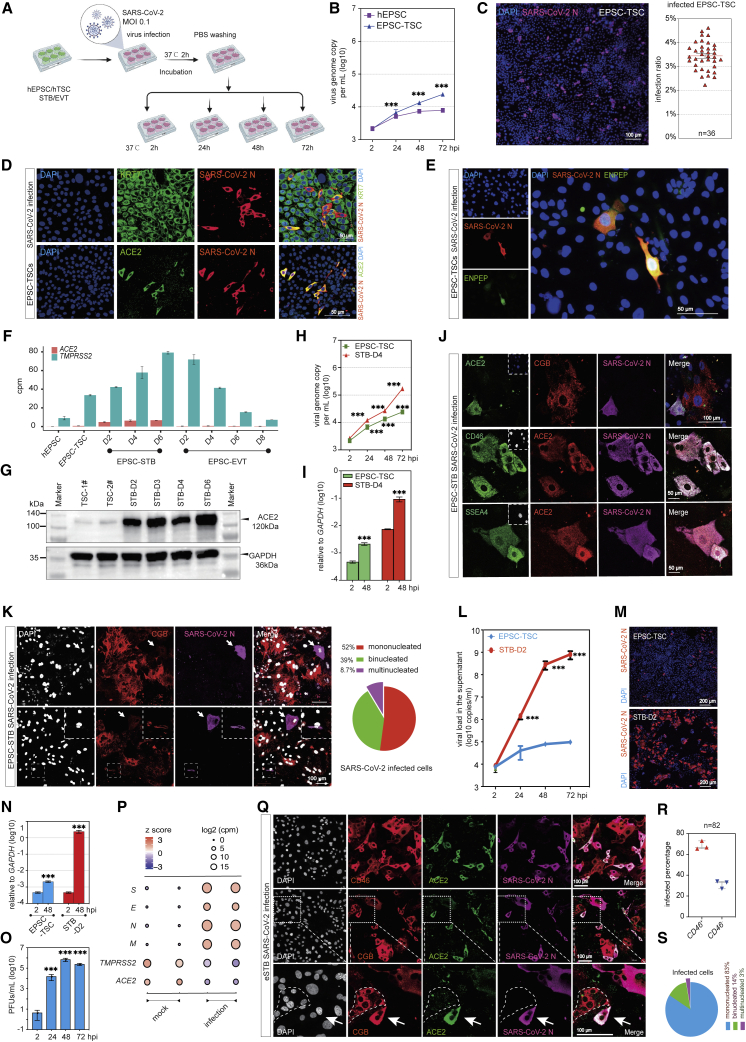
We next experimentally compared the viral replication kinetics among hEPSCs, EPSC-TSCs, STBs, and EVTs. Briefly, cells were infected with SARS-CoV-2 (SARS-CoV-2 HKU-001a strain; GenBank accession no. MT230940) for 2 h, followed by incubation in fresh medium for another 24, 48, or 72 h or hpi (hours post infection) (Figure 3A). The supernatant and cell lysates were collected for viral genome and antigen detection. hEPSCs did not express ACE2 (Figure S2H) and thus were poorly infected by SARS-CoV-2, as evidenced by viral genome detection in the supernatant or cell lysate and negative immunofluorescence staining of the viral N protein (Figures 3B, S3A, and S3B). In line with the relatively low ACE2 and TMPRSS2 expression levels (Figure S2H), SARS-CoV-2 infected EPSC-TSCs, but only about 3%–4% of the cells stained positive for the viral N protein (Figures 3B and 3C). Infected EPSC-TSCs were positively stained for ACE2 and the pan-trophoblast marker KRT7 (Figure 3D). Intriguingly, infected EPSC-TSCs expressed the blastocyst trophectoderm (TE) marker ENPEP34 (Figure 3E), which is a candidate co-receptor for SARS-CoV-2.48 Similar to EPSC-TSCs, about 0.5%–1% of BST-TSCs derived from human blastocysts35 were infected by SARS-CoV-2 and stained positive for KRT7, ACE2, and ENPEP (Figures S3C and S3D).
Figure 3.
eSTBs are highly susceptible to SARS-CoV-2 infection among trophoblasts
(A) Schematic diagram of SARS-CoV-2 infection of in vitro trophoblasts.
(B) qRT-PCR detection of SARS-CoV-2 genome copy numbers (per mL) in the supernatants of virus-co-cultured cells at different time points. Data are mean ± SD; n = 3 biological replicates. Student’s t test, ∗∗∗p < 0.001.
(C) Immunofluorescence image of 24 hpi EPSC-TSCs stained for SARS-CoV-2 N protein. Scale bar, 100 μm. Right: percentages of N-protein-positive cells. Data are mean ± SEM; n = 36, quantification of 36 random images.
(D) Immunofluorescence detection of KRT7, ACE2, and N protein in the infected EPSC-TSCs. Scale bars, 50 μm.
(E) Representative immunofluorescence images of N protein and ENPEP at 24 hpi in EPSC-TSCs. Scale bar, 50 μm.
(F) Bar plot for expression (cpm) of ACE2 and TMPRSS2 in hEPSCs, EPSC-TSCs, and STBs and EVTs differentiated from EPSC-TSCs at indicated time points.
(G) Detection of ACE2 protein in EPSC-TSCs and during their differentiation toward STBs.
(H) qRT-PCR detection of SARS-CoV-2 genome copy numbers (per mL) in the supernatants of virus-co-cultured EPSC-TSCs and (day 4) STBs. Data are mean ± SD; n = 3. Student’s t test, ∗∗∗p < 0.001.
(I) qRT-PCR analysis of SARS-CoV-2 genome copy number in cell lysates of 48 hpi EPSC-TSCs and STB-D4. Data are mean ± SD; n = 3. Student’s t test, ∗∗∗p < 0.001.
(J) Immunofluorescence staining of 48 hpi STB-D6 for ACE2, CGB, and N protein and for early STB markers CD46 and SSEA4. Scale bars, 50 or 100 μm as specified.
(K) Immunofluorescence staining of 48 hpi STB-D6 for ACE2, CGB, and N protein, and percentages of mono-, bi-, and multinucleated CGB+ cells in the infected STB-D6. Arrowheads indicate that some CGB-low or mono- or binucleated cells were infected by SARS-CoV-2. DAPI stains the nucleus. Scale bars, 100 μm.
(L) qRT-PCR detection of SARS-CoV-2 genome copy number (per mL) in the supernatants of virus-co-cultured EPSC-TSCs and eSTBs (STB-D2). Data are mean ± SD; n = 3. Student’s t test, ∗∗∗p < 0.001.
(M) Representative immunofluorescence images of 24 hpi EPSC-TSCs and eSTBs (STB-D2) stained for N protein. Scale bars, 200 μm.
(N) qRT-PCR detection of SARS-CoV-2 gene expression in cell lysates of the infected cells at 48 hpi. Data are mean ± SD; n = 3. Student’s t test, ∗∗∗p < 0.001.
(O) Plaque formation assay to detect SARS-CoV-2 virus in supernatants from the infected eSTBs (48 hpi). Data are mean ± SD; n = 3 three independent experiments; ∗∗∗p < 0.001 (Student’s t test). PFUs, plaque-forming units.
(P) Bubble plot for RNA-seq analysis of SARS-CoV-2 genes (E, M, N, S) in the cell lysates of 48 hpi eSTBs.
(Q) Representative immunofluorescence images of 24 hpi eSTBs stained for ACE2 and N protein and for STB markers CD46 and CGB. The dotted area shows an uninfected multinucleated CGB+ cell. Arrows indicate an infected binucleated STB. Scale bars, 100 μm.
(R) Quantification of the percentages of eSTB marker CD46 in 24 hpi eSTBs. Data are mean ± SEM.
(S) Quantification of the percentages of mono-, bi-, and multinucleated cells in 24 hpi eSTBs. SARS-CoV-2 preferentially infects mononucleated cells.
We next converted human primed ESCs (H1) into naive stem cells in the PXGL condition32 (Figures S3E-S3K) for deriving naive TSCs. Consistently, 2%–3% of the naive TSCs were infected by SARS-CoV-2 (Figure S3L) while expressing ACE2 (Figure S3M) and ENPEP (Figure S3M). The presence of a small number of ACE2+ and SARS-CoV-2-susceptible hTSCs revealed the heterogeneity of the current hTSC cultures, which warrants future investigation. The observation that hTSCs are transcriptionally similar to PI-TBs33 (Figure 2C) and that ACE2+ hTSCs were susceptible to SARS-CoV-2 indicates that human peri-implantation embryos are potentially at risk of SARS-CoV-2 infection.
eSTBs are highly susceptible to SARS-CoV-2 infection
STBs in peri-implantation embryos and those generated from hTSCs co-expressed ACE2 and TMPRSS2 (Figures 2D, 2E, 2F, and S2F). In hTSC differentiation toward STBs, expression of both ACE2 and TMPRSS2 was substantially increased starting from day 2 (STB-D2) (Figures 3F and 3G), in line with more efficient infection by SARS-CoV-2 in STBs than in hTSCs (Figures 3H and 3I). We noticed that the virus-infected cells were generally positive for early STB markers (SSEA4 and CD46)49,50 (Figure 3J). Indeed, multinucleated and CGB+ mature STBs did not express ACE2 highly and accounted for only a minor population of infected cells (Figure 3K). These results indicate that SARS-CoV-2 appears to preferentially infect immature or early STBs.
In STB-D2, most cells were mononucleated and negative for CGB, and only 3% of cells were multinucleated (Figures S3N and S3O). The early STB gene CD46 was transiently upregulated in STB-D2, whereas the mature STB gene CGB markedly increased after D2 (Figure S3P). Notably, ACE2 and TMPRSS2 expression was rapidly upregulated in STB-D2 (Figures 3F and 3G). We thus used STB-D2 cells for subsequent infection experiments and empirically named these cells as eSTBs.
Following infection of eSTBs with SARS-CoV-2 (MOI 0.1), supernatants were collected at 2, 24, 48, and 72 hpi and quantified for viral RNA load, which revealed that eSTBs produced high amounts of supernatant viral RNA at 48 and 72 hpi (Figure 3L). Immunofluorescence staining at 24 hpi showed substantially higher rates of infection than that of EPSC-TSCs (Figure 3M). In line with this observation, >3 log10 viral genome copies in the cell lyses was documented comparing eSTBs with hTSCs (Figure 3N). A continuous release of infectious virus particles from the infected eSTBs was detectable in plaque assays (Figure 3O). The robust production of SARS-CoV-2 was further revealed in RNA-seq, where abundant transcripts of viral Envelope (E), Membrane glycoprotein (M), Nucleocapsid (N), and S genes were detected (Figure 3P).51 Immunostaining for SARS-CoV-2 N protein revealed that the virus-infected cells were primarily CD46+, whereas those multinucleated cells were infrequently infected (Figures 3Q, 3R, and 3S). Similar results were obtained in eSTBs generated from naive TSCs (Figures S3Q and S3R).
EPSC-EVTs expressed low levels of ACE2 and TMPRSS2 (Figure S2H). Consistently, only 1%–2% of EVTs were infected by SARS-CoV-2 (Figures S3S, S3T, S3U, and S3V).
Susceptibility of human trophoblasts to SARS-CoV-2 variants of concern
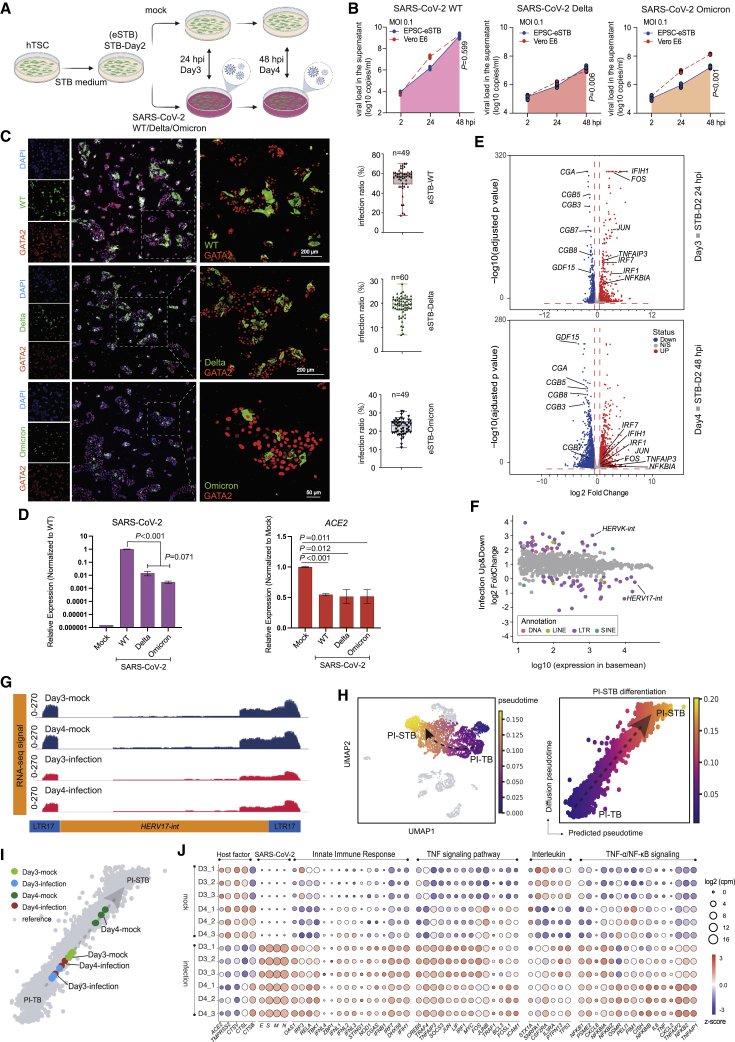
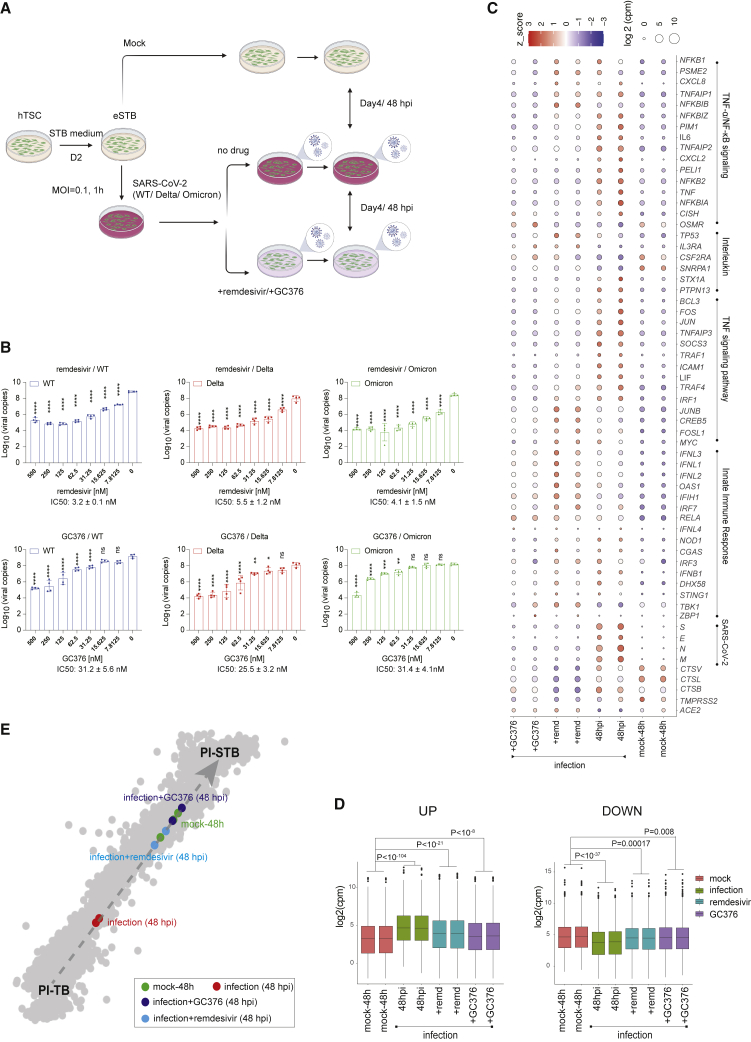
SARS-CoV-2 variants carrying various mutations have been found to influence virus replication, infectivity, transmission, and infection- and vaccine-induced immunity.52 The Delta variant was the previous predominant circulating SARS-CoV-2 strain, but was overtaken by the Omicron variants in late November 2021. Quantification of viral RNA genome copies generated over a period of 48 h demonstrated that both the Delta (B.1.617.2) and the Omicron (B.1.1.529) variants replicated robustly in EPSC-eSTBs and appeared to be as good as in Vero E6 cells (Figures 4A and 4B).
Figure 4.
SARS-CoV-2 variants robustly replicate in eSTBs
(A) Schematic diagram of eSTBs for SARS-CoV-2 wild-type (WT) and Delta or Omicron variant infection.
(B) Comparison of SARS-CoV-2 replication kinetics in EPSC-eSTBs and Vero E6 cells.
(C) Left: representative immunofluorescence images of N protein and GATA2 in EPSC-eSTBs infected by SARS-CoV-2 WT, Delta, and Omicron at 24 hpi. Right: percentages of N protein-positive EPSC-eSTBs. Error bars: mean and SEM; n = 49, 60, and 49 for quantification of random immunofluorescence images in WT-, Delta-, and Omicron-infected eSTBs, respectively.
(D) qRT-PCR analysis of ACE2 and virus gene expression levels in 48 hpi EPSC-eSTBs. Data are mean ± SD; n = 3. Student’s t test, exact p values are presented.
(E) Volcano plots of gene expression in infected STB-D3 and STB-D4. The horizontal red dashed line marks the adjusted p value (Wald test) 0.05, while the vertical red dashed lines marks expression fold change of 1.5.
(F) A scatter diagram of the transcriptomic analysis of TEs after virus infection. Differentially expressed TEs (p < 0.05) are in colors, while non-differentially expressed TEs are in gray. Expression levels of TEs used for the x axis are from DESeq2 results. Data from STB-D3 and STB-D4 are combined in this analysis.
(G) RNA-seq signal of HERV-W in infected STB-3 and STB-4 and the mock-infection control cells. Library size was used to normalize the read counts.
(H) Pseudotime analysis depicting PI-TB to PI-STB development trajectory. The black dashed arrow indicates the imputed direction of differentiation. PI-TB to PI-STB subpopulations are colored by machine learning predicted pseudotime. The x axis is the predicted pseudotime, and the y axis is the diffusion pseudotime computed using SCANPY. Gray dashed arrow describes the linear regression relationship between the predicted pseudotime and the diffusion pseudotime.
(I) Infected vs. normal (mock) STB-D3 and STB-D4 mapped against the PI-TB to PI-STB pseudotime trajectory (see STAR Methods).
(J) Bubble plot for host factors ACE2, TMPRSS2, CTSL, CTSV, and CTSB; infection-related signaling pathways; and virus genes in infected eSTBs. Bubble colors are in accordance with Z score and bubble sizes are proportional to expression levels.
Among the three lineages of SARS-CoV-2, less viral NP antigen in EPSC-eSTBs was found in Delta- (∼18.8%) and Omicron-infected cells (∼22%) compared with the wild-type (WT) SARS-CoV-2 (∼54%) (Figure 4C), as confirmed by viral genome quantification (Figure 4D). In line with a previous in vivo study,51 ACE2 expression was significantly downregulated in EPSC-eSTBs following either Delta or Omicron infection (Figure 4D). Similar phenotypes were observed in eSTBs generated from additional EPSC-TSC lines (EPSC-TSC 3# and hiEPSC-TSC24) and from naive-TSCs and BST-TSCs for their susceptibility to the WT, Delta, and Omicron variants (Figures S4A and S4B). Thus, eSTBs’ susceptibility to SARS-CoV-2 infection is independent of cell origin and genetic background.
SARS-CoV-2 infection impairs hTSC differentiation to STBs and induces a potent innate immune response
We next investigated global transcriptomic changes in infected EPSC-eSTBs (Figure 4E). Pearson correlation and cluster analysis revealed that the SARS-CoV-2-infected cells (24 hpi or STB-D3; 48 hpi or STB-D4) were clustered together and separated from the mock infection controls (Figure S4C). Comparative transcriptomic analysis identified 654 upregulated and 736 downregulated differentially expressed genes (DEGs) in the infected cells (Figure S4C).
Among the DEGs significantly decreased in SARS-CoV-2-infected cells were CGA, CGBs (CGB3, 5, 7, 8), GCM1, SDC1, and ENDOU and others that are highly expressed in mature and multinucleated STBs (Figures 4E and S4D), indicating that the infection caused a possible developmental blockage. Gene set enrichment analysis (GSEA) revealed a significant enrichment of cell-cycle, in particular, G2-M, related genes in the infected cells compared with the mock control (Figure S4E). The lack of mature STB signature and higher cell-cycle genes in infected cells are consistent, since mature and multinucleated STBs are known to have inactive cell cycles.
Interestingly, GSEA revealed that SARS-CoV-2 infection did not substantially affect the infected cells’ viability, as apoptosis-related genes were not significantly changed (Figure S4E). In contrast, infected Vero E6 cells had enriched expression of apoptosis-related genes53 (Figure S4E). Experimentally, Vero E6 cells mostly detached by 72 hpi due to cell death, whereas eSTBs did not show an obvious cytopathogenic effect (CPE) (Figure S4F). Furthermore, TUNEL cell apoptosis assay confirmed that, distinct from Vero E6 cells, eSTBs did not undergo appreciable apoptosis by 72 hpi (Figure S4F). Therefore, the lack of mature STBs in the infected cells is more likely caused by developmental block.
The multinucleate STB formation requires the endogenous retrovirus (HERV) proteins Syncytin-1, an envelope gene of HERV-W, and Syncytin-2, produced by HERV-FRD.54,55 SARS-CoV-2 infection of eSTBs resulted in lower levels of both HERV-W (HERV17-int, Syncytin-1) and HERV-FRD (Syncytin-2), but substantially increased HERV-K expression (Figures 4G and S4G). HERV-K is known to be exclusively expressed in cytotrophoblast cells (progenitors) in the human placenta.56
To further investigate eSTB development following the infection, we extracted the PI-TB and PI-STB differentiation process from the in vivo trophoblast scRNA-seq data44 to generate a pseudotime trajectory, which increased from immature PI-TB to relatively mature PI-STB (Figure 4H). Using machine learning, we mapped the mock- and virus-infected STB cells (STB-D3 and STB-D4) to this pseudotime trajectory and discovered that the infected cells were closer to PI-TB along the pseudotime trajectory compared with the mock cells (Figure 4I). SARS-CoV-2 infection thus impaired eSTB differentiation and maturation, implicating a potential COVID-19 risk in early pregnancy, which may be asymptomatic and unnoticed.57
SARS-CoV-2 infection caused a strong innate immunity response. Genes encoding interferon signaling components (IFNL1, IFIH1) and genes associated with TNF-α signaling via NF-κB, such as TNFAIP3 and NFKBIA, were upregulated in infected cells (Figures 4E and S4D). Gene ontology (GO) term analysis found enriched terms related to virus cellular response and immune response pathways in SARS-CoV-2-infected STBs (Figure S4H). As expected, KEGG pathway analysis revealed that the coronavirus disease COVID-19 pathway was among the overrepresented ones (Figure S4H). Bubble plot analysis further demonstrated an enrichment of pathways associated with innate immune response and interleukin and TNF signaling pathways in the upregulated genes (Figure 4J).
The double-stranded RNA (dsRNA) generated during coronavirus genome replication and transcription could be recognized by melanoma differentiation gene 5 (MDA5/IFIH1) in the cytoplasm to trigger innate immune activation upon coronavirus infection.58,59,60 IFIH1 was highly upregulated in the infected cells (Figure 4J). Meanwhile, DNA is not known to be involved in the SARS-CoV-2 life cycle. Consistently, genes encoding cGAS and STING1, both being components of the cGAS-STING pathway of the innate immune system detecting cytosolic DNA, were not substantially altered (Figure 4J).
In response to viral infections, interferons (IFNs) initiate signaling cascades that stimulate the expression of many genes and create an intracellular antiviral defense. Type III IFNs are important antiviral factors.61 Particularly, IFN-λ1 is known to be constitutively released from human placental trophoblasts to protect the fetus from viral infections.62,63 Type III IFNs have also been shown to restrict SARS-CoV-2 infection in airway and intestinal epithelia.64,65 Infected cells expressed high levels of genes encoding type III IFNs, including IFN-λ1 (IL-29 or IFNL1), IFN-λ2 (IL-28A or IFNL2), IFN-λ3 (IL-28B or IFNL3), and IFN-λ4 (IFNL4) (Figure 4J). 2′-5′-oligoadenylate synthetase 1 (OAS1) is a recently identified IFN-signaling downstream gene that stimulates RNase L and specifically inhibits the virus.66 It was highly expressed in the infected eSTBs (Figure 4J).
In line with Vero E6 cells being genetically defective in IFN signaling and TMPRSS2,67,68 they expressed ACE2 but not TMPRSS2 (Figure S4I).53 In the infected Vero E6 cells, the IFN genes IFNb1, IRF3, and TBK1 showed no changes and the antiviral effector OAS1 showed only mild upregulation (Figure S4I).
The relatively high Omicron replication in TMPRSS2-deficient Vero E6 cells, as presented in Figures 4B, S4A, and S4B, is possibly due to Omicron infection being more dependent on cathepsins or other endosomal proteases than other variants of concern.69,70,71,72 Vero E6 expressed Cathepsin L (CTSL) and Cathepsin L2 (CTSV), which were downregulated following the infection (Figure S4I). Similarly, eSTBs expressed both Cathepsin genes, and the infected cells showed decreased expression (Figure 4J), which may be a mechanism for Omicron replication in eSTBs.
Remdesivir and GC376 effectively eliminate eSTB SARS-CoV-2 and variant infection
The eSTBs permitted robust SARS-CoV-2 infection and thus provide normal and physiologically relevant cells for evaluating antiviral drugs. To this end, we tested the FDA-approved antiviral drug remdesivir73 and a veterinary drug, GC376,74 in our cell models (Figure 5A). Remdesivir effectively eliminated SARS-CoV-2 infection in Vero E6 cells at around 5 μM with an IC50 of 0.77 μM.75 In eSTBs, remarkably, remdesivir demonstrated an IC50 of 3.2 ± 0.1 nM for SARS-CoV-2 and 5.5 ± 1.2 nM for Delta and 4.1 ± 1.5 nM for Omicron variants (Figures 5B, S5A, and S5B).
Figure 5.
Antiviral drugs remdesivir and GC376 effectively inhibit replication of SARS-CoV-2 and its variants in eSTBs
(A) Schematic diagram of SARS-CoV-2 infection in an eSTB cell model.
(B) Dose-dependent inhibition of SARS-CoV-2 WT/Delta/Omicron by remdesivir/GC376 in EPSC-derived eSTBs. Viral copies in the supernatant were quantified by qRT-PCR. Data are mean ± SEM; n = 3. Two-way ANOVA; ns, not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(C) Bubble plot for genes of host factors, infection-related signaling pathways, and virus genes in infected eSTBs. Bubble color is in accordance with expression Z score and bubble size is expression level.
(D) Boxplots for upregulated and downregulated genes in 48 hpi EPSC-eSTBs, with or without SARS-CoV-2 infection, and in the presence or absence of remdesivir or GC376. Statistical significance was calculated by Wilcoxon test using mean of expression values.
(E) Mapping of 48 hpi EPSC-eSTBs in the presence or absence of remdesivir (10 μM) or GC376 (10 μM) against in vivo pseudotime trajectory.
GC376 is a repurposed SARS-CoV-2 main protease inhibitor that increases survival of mice with a fatal SARS-CoV-2 infection.76 GC376 suppressed the virus infection in eSTBs with an IC50 of 31.2 ± 5.6 nM against WT, 25.5 ± 3.2 nM against Delta variant, and 31.4 ± 4.1 nM against Omicron variant (Figures 5B and S5B), which were much lower than in Vero E6 cells (0.70 μM).77 The effectiveness of these two drugs against SARS-CoV-2 in eSTBs was further confirmed by the substantially reduced viral N antigen expression in immunofluorescence staining (Figure S5C). Similar results were obtained in naive eSTBs (Figure S5D).
Expression of the DPP4 gene, the host entry receptor of MERS-CoV, another highly pathogenic human coronavirus, is highly correlated with ACE2 and ENPEP in human preimplantation embryos48 and was detected in eSTBs (Figure S5E). MERS-CoV infected eSTBs derived from hEPSC-TSCs at an efficiency comparable to that in Vero E6 cells,78 and both remdesivir and GC376 could effectively block MERS-CoV replication in eSTBs with efficiencies comparable to those in Vero E6 cells (Figure S5F). These results indicate the potential of eSTBs for antiviral evaluation against SARS-CoV-2, MERS-CoV, and other coronaviruses.
We next examined the global transcriptome of infected and drug-treated cells. Both drug treatments drastically reduced expression of SARS-CoV-2 genes (E, M, N, and S) (Figure 5C). Neither drug appeared to cause substantial changes in host innate immune response genes such as MDA5, IFNL1-4, IFNB1, and OAS, but proinflammatory cytokine genes such as TNF, IL-6, and IL-8 were all downregulated in the drug-treated cells (Figures 5C and S5G).
We next asked whether remdesivir and GC376 treatment could mitigate the infection-caused developmental defect. Hierarchical clustering analysis confirmed a shift of the drug-treated cells toward the mock infection control STBs (Figure S5H), with substantially reduced upregulated and downregulated genes (Figure 5D). Furthermore, machine learning analysis based on global expression decomposition revealed that, along the PI-TB to PI-STB pseudotime trajectory, drug treatment induced a shift away from PI-TB and toward PI-STB, indicating a partial rescue of eSTB development defects (Figure 5E).
Direct derivation of 3D trophoblast organoids from EPSC-TSCs for SARS-CoV-2 infection
Human organoids, particularly of respiratory cells, have been extensively investigated for SARS-CoV-2 infection.79 Long-term and genetically stable trophoblast organoids are derived from first-trimester placenta tissues or blastocysts, which grow as complex structures closely recapitulating the organization of in vivo placental villi.80,81 It was recently reported that 3D or organoid culture of hTSCs more closely resembled the in vivo counterparts than 2D hTSCs.36 We thus investigated 3D trophoblast organoids in SARS-CoV-2 infection. From EPSC-TSCs, we directly established trophoblast organoids,82 where individual hTSCs self-aggregated and developed into 3D organoids (Figure 6A). The organoids were reminiscent of those derived from the placenta,80,81 where the basement membrane was on the outside in contact with the Matrigel substratum, whereas syncytial masses lined the central cavity (Figure 6B). Multinucleated mature STBs expressing CGB and ENDON were found at the organoids’ center and most of them did not highly express the stemness transcription factors GATA3, TFAP2A, and TFAP2C or the eSTB marker CD46 (Figures 6C, S6A, and S6B). Trophoblast organoids harbored both stem cells and STBs.80,81 They expressed genes of both hTSCs and STBs, although the levels were lower than those in hTSC or STB cultures (Figures 6D and S6C), and secreted full-length and properly folded β-hCG hormone (Figure S6D). Consistent with a recent study,36 hTSC-trophoblast organoids expressed higher levels of trophoblast-specific miRNAs (has-miR-517a and 525-3p) compared with the 2D TSCs (Figure S6E). These trophoblast organoids did not express appreciable levels of EVT genes but could be induced to robustly generate migrating HLA-G+ and ITGA5+ EVT cells (Figures S6F and S6G), confirming the presence of bipotential stem cells.
Figure 6.
Infection of EPSC-TSC trophoblast organoids (EPSC-ORG) by SARS-CoV-2
(A) Bright-field images of EPSC-ORGs derived from EPSC-TSCs. Scale bars, 100 μm.
(B) H&E staining of cryosections (top) and paraffin sections (bottom) of EPSC-ORGs. Arrowhead in the cryosection (top right) indicates densely packed cell clusters in the outer layer of EPSC-ORGs. Arrowhead in the paraffin section (bottom right) indicates the multinucleated STBs inside EPSC-ORG. Scale bars, 100 μm.
(C) Immunofluorescence staining of EPSC-ORG sections for cytotrophoblast markers TFAP2A and YAP1 and for STB markers CD46 and CGB. Yellow arrows point to the multinucleated STBs that express high CGB but are negatively stained for E-cadherin. The dashed line box areas are shown for the multinucleated STBs that are negatively stained for CD46. Scale bars, 100 or 50 μm as indicated.
(D) Changes in gene expression (qRT-PCR) in EPSC-TSCs and EPSC-ORGs. Data are mean ± SD; n = 3 independent replicates; ∗∗p < 0.01, ∗∗∗p < 0.001 (Student’s t test).
(E) 3D PCA for placenta-derived trophoblasts (CT-TSC/-STB/-EVT) and trophoblast organoids (CT-TSC-generated trophoblast organoids, CT-TSC-ORG; placenta-tissue-generated trophoblast organoids, placenta-ORG) and EPSC-derived trophoblasts (EPSC-TSC/STB/EVT) and EPSC-ORG. Cell sources are indicated by shape.
(F) Heatmap showing EPSC-ORG expression of both TSC and STB trophoblast genes, similar to trophoblast organoids generated from CT-TSCs (CT29, CT30, and BTS11). Z score of cpm was used and shown as heatmap signatures.
(G) Bar plots of expression (cpm) of ACE2 and TMPRSS2 in EPSC-TSC/STB/EVT and EPSC-ORG. Right: qRT-PCR detection of SARS-CoV-2 genome copy numbers (per mL) in the supernatants of virus-co-cultured EPSC-TSCs, eSTBs (STB-D2), and EPSC-ORGs. Data are mean ± SD; n = 3.
(H) Immunofluorescence images of paraffin sections of SARS-CoV-2-infected EPSC-ORG stained for N, ACE2, E-Cadherin, and CGB. White arrows indicate the mononucleated infected cells, whereas yellow arrows point to a large multinucleated STB that expresses high CGB. The dashed line box areas are shown in higher resolution. Scale bars, 50 or 100 μm as indicated.
RNA-seq analysis further unraveled the global gene expression similarities between trophoblast organoids derived from EPSC-TSCs (EPSC-ORGs) and from placental CT-TSCs (CT-TSC-ORGs) (Figure 6E).36 Both co-expressed markers for TSCs and STBs, similar to trophoblast organoids derived from primary villous placenta80,81 (Figure 6F and Table S4).
In EPSC-ORGs, ACE2 was expressed at levels comparable to those in EPSC-TSCs, but TMPRSS2 expression was much lower (Figure 6G), resembling that in the placenta.17,18,19 EPSC-ORGs were next infected with SARS-CoV-2 at an MOI of 10 based on the published condition for SARS-CoV-2 infection in organoids.83 SARS-CoV-2 RNA copy number in the supernatants from infected EPSC-ORGs was at levels comparable to those of EPSC-TSC infection (Figure 6G). Immunofluorescence staining revealed a small number of cells located along the periphery of the EPSC-ORGs positive for SARS-CoV-2 N, which varied substantially among organoids, possibly due to cellular heterogeneity. The infected cells tended to co-express ACE2, CD46, and E-cadherin but not CGB (Figures 6H and S6H), although the co-expression was much more clearly demonstrated in 2D-cultured eSTBs (Figure 3Q). The low SARS-CoV-2 infection in the trophoblast organoids in vitro supports clinical observations that opportunistic SARS-CoV-2 infection of the human placenta are uncommon.5,6
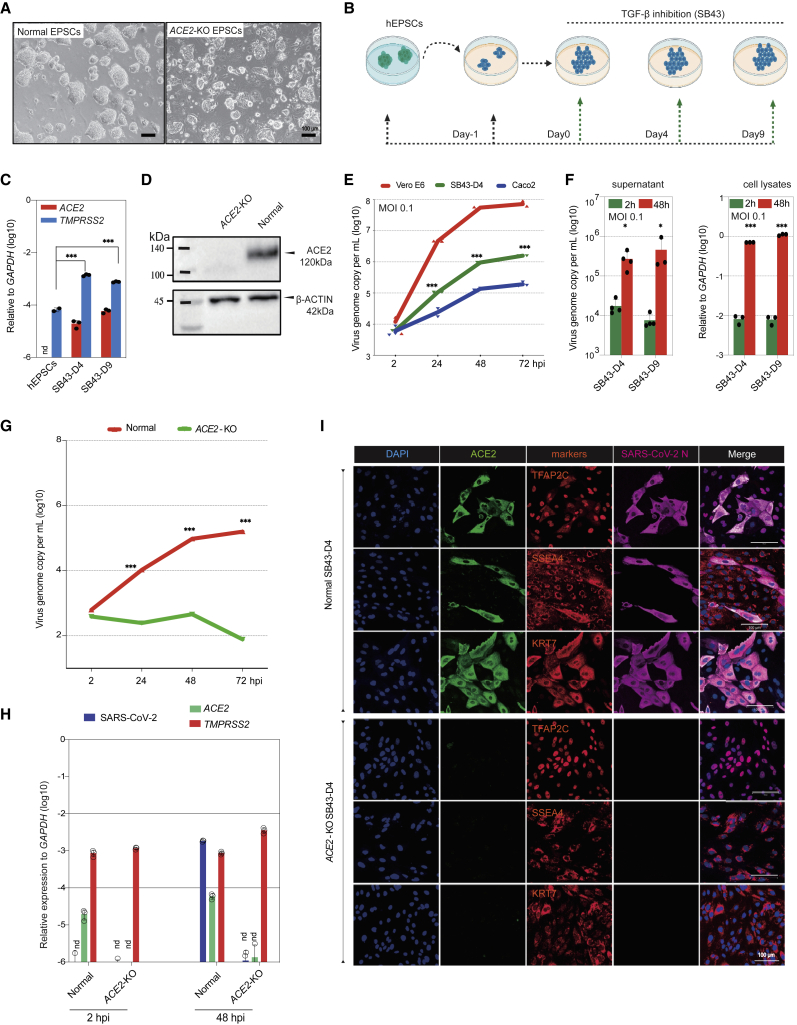
ACE2 is essential for SARS-CoV-2 infection in trophoblasts
To genetically validate the role of ACE2 in SARS-CoV-2 infection in human trophoblasts, we made homozygous deletions in ACE2 coding exon 2 in hEPSCs using CRISPR-Cas9 (Figures S7A–S7D). The ACE2 knockout hEPSCs (ACE2-KO) had normal morphology and expressed high levels of key pluripotency genes (OCT4 and NANOG) and markers (SSEA3 and TRA-1-60) but low levels of lineage genes (Figures 7A, S7E, and S7F), comparable to the normal parental hEPSCs.
Figure 7.
ACE2 is required for SARS-CoV-2 infection of trophoblasts differentiated from hEPSCs
(A) Normal and ACE2-KO hEPSCs. Scale bars, 100 μm.
(B) Experimental flow of hEPSC differentiation toward trophoblasts via TGF-β inhibitor SB-431542 (SB43) treatment.
(C) qRT-PCR analysis of ACE2 and TMPRSS2 in hEPSCs and the differentiated cells on days 4 and 9. nd, undetectable. Data are mean ± SD; n = 3; ∗∗∗p < 0.001 (Student’s t test).
(D) Western blotting confirms loss of ACE2 protein in the SB43-treated hEPSCs on day 9 (SB43D9) of differentiation.
(E) SARS-CoV-2 replication kinetics in cells differentiated from hEPSCs compared with Vero E6 and Caco2 cells. Data are mean ± SD; n = 3 biological replicates; ∗∗∗p < 0.001 (Student’s t test).
(F) Left: qRT-PCR of supernatant viral load in SB43-treated hEPSCs at 2 and 48 hpi. Right: qRT-PCR quantification of SARS-CoV-2 genome in cell lysates of SB43-treated hEPSCs on days 4 and 9. Data are mean ± SD; n = 3 biological replicates; ∗p < 0.05, ∗∗∗p < 0.001 (Student’s t test).
(G) Quantification of supernatant viral RNA loads of SB43-treated (day 4) normal and ACE2-KO EPSCs. Data are mean ± SD; n = 3 biological replicates; ∗∗∗p < 0.001 (Student’s t test).
(H) SARS-CoV-2 viral genome quantitation and expression of ACE2 and TMPRSS2 in normal vs. SB43-treated ACE2-KO EPSCs at day 4. Data are mean ± SD; n = 3 biological replicates; nd, not detectable.
(I) Representative immunofluorescence staining images for ACE2, trophoblast factors and markers, and SARS-CoV-2 N protein in normal (top) and ACE2-KO cells (bottom) (day 4 of SB43 treatment) at 48 hpi. Scale bars, 100 μm.
We induced normal and ACE2-KO hEPSCs to trophoblasts using a simple and efficient protocol via the TGF-β inhibitor SB431542 (Figures 7B and S7G).24 Normal hEPSCs did not express ACE2 (Figures 7C and S2H) and were not infected by SARS-CoV-2. The differentiated cells expressed ACE2, TMPRSS2s, and typical STB markers (Figures 7C, S7H, and S7I). In contrast, no ACE2 protein was detected in cells differentiated from the ACE2-KO hEPSCs (Figure 7D).
We next selected cells of days 4 and 9 differentiation for infection, as they expressed ACE2 and TMPRSS2 (Figure 7C), which produced substantial amounts of viral genome in the supernatant and cell lysates (Figures 7E and 7F). The infected cells, detected as SARS-CoV-2 nucleocapsid (N) protein positive, all expressed ACE2 and trophoblast markers TFAP2C, SSEA4, and KRT7 and were mostly mononucleated (Figure S7J). Loss of ACE2 abolished the infection, as indicated by the drastic decrease in viral genome in supernatants and cell lysates (Figures 7G and 7H). In immunofluorescence staining, no cells were stained positive for either SARS-CoV-2 N protein or ACE2 in the ACE2-KO cells (Figure 7I). Therefore, in trophoblasts, ACE2 is essential for SARS-CoV-2 infection.
Discussion
We report a stem cell-based system to study human trophoblasts’ susceptibility to SARS-CoV-2 and its variants. Specifically, eSTBs that were differentiated from hTSCs originating from EPSCs, naive stem cells, and human blastocysts were found to be highly susceptible to SARS-CoV-2.
An important issue about hTSCs is that they may possess AME gene signatures. We examined the transcriptome of hTSCs originating from human EPSCs and naive stem cells by comparing them with those of in vivo origins and found that these hTSCs were highly similar and expressed low or no putative AME cell genes.
hTSCs are thought to represent in vivo post-implantation cells.33 SARS-CoV-2 infected 0.5%–4% of hTSCs that were ACE2+ and co-expressed high levels of the TE marker ENPEP. These ACE2+ hTSCs warrant further investigation for their molecular properties and developmental potential.
eSTBs expressed high levels of ACE2 and TMPRSS2 and supported the replication of SARS-CoV-2 and the Delta and Omicron variants and MERS-CoV. The infection was effectively suppressed, and the infection-induced developmental delay was partially rescued by two known antivirals, remdesivir and GC376. hTSCs and eSTBs have enriched transcriptomic features of peri-implantation trophoblasts. Their susceptibility to coronaviruses and the resultant developmental defects imply that these early embryos may be at risk of the virus infection.
The syncytialized trophoblasts generated from human ESCs (primed) by BAP treatment (BMP4, TGF-β inhibitor A83-01, and FGF2 signaling inhibitor PD173074) were found to co-express ACE2 and TMPRSS2 and supported replicative and persistent infection by SARS-CoV-2 57. These results differ from our observation that eSTBs derived from hTSCs expressed ACE2 and were highly susceptible to SARS-CoV-2 infection, whereas the syncytialized or more mature STBs were much less susceptible. The discrepancy could be due to either technical reasons, as eSTBs are expected to be transiently present in hTSC differentiation, or the difference between BAP cells (putative trophoblasts) and hTSCs/eSTBs. Indeed, our discovery of eSTBs was made possible by the stem cell-based in vitro system, since eSTB-like cells likely exist only transiently along placental trophoblast development before they differentiate to the non-proliferative multinucleated STBs. Neither trophoblastic cell lines nor primary trophoblasts are representative of these eSTBs.
Several human cell lines have been used in coronavirus research. However, they suffer from genetic and/or innate immune defects and have some long-standing technical challenges such as cell transfection and genetic manipulation. Vero E6 cells, which originated from African green monkey, are commonly used for isolation and propagation of SARS-CoV-215 and have been approved for use in vaccine manufacturing. They have genetic defects, including large genomic deletions encompassing the IFN gene clusters (IFN-a and -b) and the CDKN2A/B loci, and are thus deficient in IFN response,67 which has prevented their application in the study of antiviral response. Furthermore, the lack of TMPRSS2 in Vero E6 cells results in the selection of SARS-CoV-2 variants that have lost the polybasic Furin cleavage site at the S1-S2 junction.68 Meanwhile, the current dominant SARS-CoV-2 Omicron variant, which relies more on Cathepsins and other endosomal proteases, replicates well in Vero E6 cells and the upper airway, in which ACE2 and Cathepsins are more abundant, but not in Calu3 and Caco2 cells or in lung cells.71,72
eSTBs are normal human cells with an intact innate immune system and no major known genetic or epigenetic defects and support highly productive propagation of SARS-CoV-2 and its variants. SARS-CoV-2 infection causes massive cell death in Vero E6 cells but not in eSTBs. The lack of cell death in infected eSTBs is different from recent reports that primitive human trophoblasts generated from human ESCs were susceptible to and lysed by an African lineage strain Zika virus,84,85 but may provide a practical advantage for viral production.
The hTSC-derived trophoblast organoids expressed low levels of ACE2 and TMPRSS2 and were much less susceptible to the virus infection, reminiscent of the placenta. Similar findings on stem cell-derived trophoblast organoids and their use in SARS-CoV-2 study were recently reported.86
Human EPSCs and naive stem cells permit efficient genome editing and possess differentiation potential for all embryonic and extraembryonic lineages.24,32 In addition to trophoblasts, these stem cells can be explored to derive diverse cell types and to functionally assess their susceptibility to SARS-CoV-2 and variants, like airway epithelium, gastrointestinal epithelium, and neurons. These qualities grant them potential in overcoming the long-standing challenges in cell transfection and genetic manipulation of various target cells. Cells generated from these stem cells, in particular eSTBs, may thus represent a genetically normal human cell source for investigating host-virus interactions in current and future pandemics, performing genetic screens to identify biological determinants of infections, and evaluating and discovering antivirals.
Limitations of this study
The current study lacks experimental and clinical data on SARS-CoV-2 infection in human peri-implantation pregnancy, particularly regarding any in vivo counterparts of eSTBs. The eSTB model in vitro also requires further characterization at the single-cell level to delineate culture heterogeneity and compare their identity with in vivo counterparts. The current in vitro eSTBs are transient in culture and thus require enrichment for performing high-throughput antiviral drug screens and large-scale viral production. Meanwhile, the scRNA-seq finding of some peri-implantation embryonic trophoblasts expressing ACE2 and TMPRSS2 requires proteomic validation in human embryos. The mechanistic role of ACE2 in trophoblast development also remains to be dissected.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Oct-3/4 | R and D Systems | Cat#AF1759; RRID:AB_354975 |
| Anti-TEAD4 | Abcam | Cat#ab58310; RRID:AB_945789 |
| ENPEP | Abcam | Cat#ab155991; RRID:AB_2916048 |
| Purified anti-human CD75 Antibody | BioLegend | Cat#326901; RRID:AB_2194435 |
| Anti-KLF17 antibody | Sigma-Aldrich | Cat#HPA024629; RRID:AB_1668927 |
| Recombinant Anti-active YAP1 antibody | Abcam | Cat#ab205270; RRID:AB_2813833 |
| Anti-CDX2 antibody [EPR2764Y] | Abcam | Cat#ab76541; RRID:AB_1523334 |
| GCM1 Antibody | Novus | Cat#NBP2-48520; RRID:AB_2916049 |
| PE anti-human CD49e Antibody | BioLegend | Cat#328009; RRID:AB_893368 |
| Purified anti-human/mouse CD49f Antibody | BioLegend | Cat#313602; RRID:AB_345296 |
| FITC anti-human CD49a Antibody | BioLegend | Cat#328307; RRID:AB_1236430 |
| Alexa Fluor® 488 anti-human HLA-A,B,C Antibody | BioLegend | Cat#311413; RRID:AB_493133 |
| ENDOU Antibody | Novus biologicals | Cat#NBP2-55877; RRID:AB_2916050 |
| Anti-E Cadherin [EP700Y] | Abcam | Cat#ab40772; RRID:AB_731493 |
| Anti-Transcription factor AP-2-alpha [EPR2688(2)] | Abcam | Cat#ab108311; RRID:AB_10861200 |
| Purified anti-human CD49e Antibody | BioLegend | Cat#328002; RRID:AB_8933 |
| Anti-GATA-3 | R and D Systems | Cat#MAB6330; RRID:AB_10640512 |
| Anti-TRA-1-60 | STEMCELL Technologies | Cat#60064; RRID:AB_2686905 |
| Anti-ACE2 | Thermo Fisher Scientific | Cat# MA5-32307; RRID:AB_2809589 |
| ACE2 Rabbit mAb | ABclonal | Cat#A4612; RRID:AB_2863309 |
| Anti-SSEA-4 | STEMCELL Technologies | Cat#60062; RRID:AB_2721031 |
| Anti-hCGB | Thermo Fisher Scientific | Cat#14-6508-82; RRID:AB_10853505 |
| Anti-KRT7 | Santa Cruz Biotechnology | Cat# sc-70936; RRID:AB_2234501 |
| Anti-AP-2 gamma | R and D Systems | Cat#AF5059; RRID:AB_2255891 |
| Anti-HLA-G | Abcam | Cat#ab7759; RRID:AB_306053 |
| Anti-CD46 | BioLegend | Cat#352403; RRID:AB_10897448 |
| Anti-ZO-1 | ABclonal | Cat#A0659; RRID:AB_2757321 |
| Anti-KRT18 | R and D Systems | Cat#MAB7619; RRID:AB_2893116 |
| Alexa Fluor® 488 anti-human/mouse SSEA-3 Antibody | BioLegend | Cat#330305; RRID:AB_1279441 |
| Alexa Fluor® 488 anti-human/mouse TRA-1-60 Antibody | BioLegend | Cat#330613; RRID:AB_2295395 |
| Anti-p63/TP73L | R and D Systems | Cat#AF1916; RRID:AB_2207174 |
| Donkey anti-Mouse 555 | Thermo Fisher Scientific | Cat#A-31570; RRID:AB_2536180 |
| Donkey anti-Mouse 488 | Thermo Fisher Scientific | Cat#A-21202; RRID:AB_141607 |
| Donkey anti-Goat 555 | Thermo Fisher Scientific | Cat#A-21432; RRID:AB_2535853 |
| Donkey anti-Goat 488 | Thermo Fisher Scientific | Cat#A-11055; RRID:AB_2534102 |
| Donkey anti-Rabbit 594 | Thermo Fisher Scientific | Cat#A-21207; RRID:AB_141637 |
| Donkey anti-Rabbit 488 | Thermo Fisher Scientific | Cat#A-21206; RRID:AB_2535792 |
| Goat anti-Guinea Pig 647 | Thermo Fisher Scientific | Cat#A-21450; RRID:AB_141882 |
| Bacterial and virus strains | ||
| SARS-CoV-2 (HKU-001a) | In house | GenBank: MT230904 |
| SARS-CoV-2 Omicron | hCoV-19/Hong Kong/HKU-344/2021 | GISAID: EPI_ISL_7357684 |
| SARS-CoV-2 Delta | hCoV-19/Hong Kong/HKU-210804-001/2021 | GISAID: EPI_ISL_3221329 |
| MERS-CoV (EMC/2012) | Erasmus Medical Center, Netherlands | GenBank:JX869059.2 |
| Top10 Competent Cell | TIANGEN | CB104-02 |
| Chemicals, peptides, and recombinant proteins | ||
| GC376 | MedChemExpress | HY-100721 |
| CTS™ (Cell Therapy Systems) N-2 Supplement | Thermo Fisher Scientific | A1370701 |
| B-27™ Supplement (50X), serum free | Thermo Fisher Scientific | 17504044 |
| XAV939 | MedChemExpress | HY-15147 |
| PD0325901 | MedChemExpress | HY-10254 |
| Gö 6983 | MedChemExpress | HY-13689 |
| Neurobasal Medium | Thermo Fisher Scientific | 21103049 |
| remdesivir | MedChemExpress | HY-104077 |
| DMEM/F-12, no glutamine | Thermo Fisher Scientific | 21331020 |
| Y-27632 dihydrochloride | Tocris | 1254/10 |
| FBS | Thermo Fisher Scientific | 10270 |
| LIF Recombinant Human Protein | Thermo Fisher Scientific | PHC9484 |
| DPBS, powder, no calcium, no magnesium | Thermo Fisher Scientific | 21600010 |
| FastQuant RT Super Mix | TIANGEN | KR108-01 |
| PowerUp™ SYBR™ Green Master Mix | Thermo Fisher Scientific | A25776 |
| SB-431542 | Tocris | 1614 |
| A83-01 | Tocris | 2939 |
| CHIR-99021 | Tocris | 4423 |
| A 419259 trihydrochloride | MedChemExpress | HY-15764A |
| Geltrex™ LDEV-Free Reduced Growth Factor Basement Membrane Matrix | Thermo Fisher Scientific | A1413201 |
| MicroAmp™ Fast Optical 96-Well Reaction Plate with Barcode, 0.1 mL | Thermo Fisher Scientific | 4346906 |
| MicroAmp™ Optical Adhesive Film | Thermo Fisher Scientific | 4360954 |
| Forskolin | Sigma-Aldrich | F3917 |
| Matrigel Matrix. GFR | Corning | 354230 |
| EGF Recombinant Human Protein | Thermo Fisher Scientific | PHG0311 |
| Prostaglandin E2 | MedChemExpress | HY-101952 |
| HGF Protein, Human, Recombinant | Sino biological | Cat: 10463-HNAS |
| RSPO1 Protein, Human, Recombinant | Sino biological | Cat: 11083-HNAS |
| DMSO, Anhydrous | Thermo Fisher Scientific | D12345 |
| BSA solution | Sigma-Aldrich | A7979 |
| Recombinant Human NRG1-beta 1/HRG1-beta 1 EGF Domain Protein | R and D Systems | 396-HB-050 |
| Normal Donkey Serum | Abcam | ab7475 |
| Knockout serum replacement | Thermo Fisher Scientific | 10828010 |
| Valproic Acid (Sodium Salt) | Stem cell technology | 72292 |
| Cell Recovery Solution | Corning | 354253 |
| Recombinant Human FGF basic/FGF2/bFGF (146 aa) Protein | R and D Systems | 233-FB-500/CF |
| μ-Slide 8 Well high Glass Bottom | ibidi | 80807 |
| b-Mercaptoethanol | Thermo Fisher Scientific | 31350010 |
| TryPLE-Express | Thermo Fisher Scientific | 12605036 |
| Trypsin/EDTA | Thermo Fisher Scientific | 25200072 |
| Penicillin-Streptomycin-Glutamine (100X) | Thermo Fisher Scientific | 10378016 |
| Minimum Essential Medium (MEM) Vitamin Solution | Thermo Fisher Scientific | 11120052 |
| Insulin-Transferrin-Selenium-Ethanolamine (ITS-X) (100X) | Thermo Fisher Scientific | 51500056 |
| Triton X-100 | Sigma-Aldrich | T9284 |
| 4% Paraformaldehyde | Sigma-Aldrich | P6148 |
| 2-phospho-L-ascorbic-acid | Sigma-Aldrich | A8960-5G |
| Critical commercial assays | ||
| CellTiter-Glo® Luminescent Cell Viability Assay | Promega | G7570 |
| miRcute Plus miRNA cDNA First-Strand cDNA Kit | TIANGEN | KR211-01 |
| miRcute Plus miRNA qPCR Kit (SYBR Green) | TIANGEN | FP411-01 |
| QuantiNova Probe RT-PCR kit | QIAGEN | 208354 |
| Transwell invasion assay | Corning | 354480 |
| TUNEL cell apoptosis detection kit | Servicebio | G1501 |
| Deposited data | ||
| RNA-seq data | (Okae et al., 2018)35 | DDBJ: JGA00000000074 |
| RNA-seq data | (Okae et al., 2018)35 | DDBJ: JGA00000000117 |
| RNA-seq data | (Okae et al., 2018)35 | DDBJ: JGA00000000122 |
| RNA-seq data | (Gao, ,2019)24 | ARRAYEXPRESS: E-MTAB-7253 |
| RNA-seq data | (Sheridan, 2021) | ARRAYEXPRESS: E-MTAB-10429 |
| scRNA-seq data | (Zhou, et al., 2019)44 | GEO: GSE109555 |
| scRNA-seq data | (Liu, et al., 2018)45 | GEO: GSE89497 |
| RNA-seq data | This paper | GEO: GSE190432 |
| scRNA analysis | This paper | https://github.com/leeyoyohku/EPSC_COVID |
| Experimental models: Cell lines | ||
| Monkey: Vero E6 cells | ATCC | CCL-81 |
| Human embryonic stem cell (hESC) line: Man-1/M1 | (Camarasa, 2010) | N/A |
| Caco-2 [Caco2] | ATCC | HTB-37 |
| TSC-BST, Human Blastocyst derived hTSCs | (Okae et al., 2018)35 | N/A |
| Oligonucleotides | ||
| Primers used, see Table S5 | This paper | N/A |
| Software and algorithms | ||
| ImageJ Fiji (2.0.0) | NIH | https://imagej.net/Fiji |
| GraphPad Prism 8.0 | GraphPad | https://www.graphpad.com/scientific- software/prism/ |
| Microsoft | Microsoft | https://www.microsoft.com/de-at/microsoft- 365/excel |
| Synthego | N/A | https://ice.synthego.com/#/ |
| BioRender | N/A | https://biorender.com/ |
| Adobe Illustrator | Adobe | https://www.adobe.com/at/products/illustrator.html |
| FlowJo | BD Life Sciences | https://www.flowjo.com/ |
| Zeiss Zen (Blue edition) | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen-lite.html |
| FeatureCounts v2.0.1 | N/A | http://subread.sourceforge.net |
| SCANPY (v1.7.2, scRNA/integrative analysis) | N/A | https://scanpy-tutorials.readthedocs.io/en/latest/index.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Pentao Liu (pliu88@hku.hk).
Materials availability
All unique/stable reagents generated in this study are available from the Lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Human expanded potential stem cells (hEPSCs)
The primed human embryonic stem cells (hESCs) Man-1/M1 line (female, XX) was converted to hEPSCs (M1 hEPSCs). C5-hiEPSCs (female, XX) are derived from fetal neural stem cell line BRC1019 (NSCs, a gift from Dr. Colin Watts) and were established by episomal (integration-free) six-factor reprogramming.24,87 The six factors were human complementary DNAs of OCT4, cMYC, KLF4, SOX2, RARG, and LRH1 linked by 2A peptides. Transfected cells (2×105; GM00013, Coriell Institute; Amaxa Nucleofector) were seeded on feeder cells (10-cm dish) in M15 (Knockout DMEM, 15% FBS, 1×Glutamine Penicillin-Streptomycin and 1×NEAA, 50 μM 2-mercaptoethanol) and supplemented with 50 μg mL−1 vitamin C and Dox (1.0 μg mL−1). Dox was removed at days 12–14 and the media was switched to hEPSCM. The survival colonies were picked at day 21 to hEPSCM and expanded to stable hiEPSC lines. The maintenance and expansion of hEPSCs were routinely performed on SNL feeder cells (SNL76/7). Gamma irradiation-inactivated SNL feeder cells were plated on 0.1% gelatinised plates 3–4 days before seeding hEPSCs at a density of ∼3.125×104 cells/cm2.
Culture of human trophoblast stem cells (hTSCs)
Human trophoblast stem cells (hTSCs) (the BST-TSC line provided from Dr. T. Arima35; EPSC-TSCs derived from M1 hEPSCs; naive TSCs derive from H1 PXGL naive stem cells) were cultured with the TSC medium in Matrigel/Geltrex-coated plates at 37°C and 5% CO2 conditions. The medium was changed daily. Human TSCs were treated with TrypLE and then passaged onto new Matrigel/Geltrex-coated plates every 3–4 days. All cells were maintained in a 5% CO2 incubator at 37°C and routinely tested negative for mycoplasma.
Cell lines
The SNL cell line is an immortalized subclone of the STO line created to stably express the neomycin resistance and leukemia inhibitory factor (LIF) genes. SNL feeder cells were maintained in regular M10 medium: Knockout DMEM, 10% FBS, 1×Glutamine Penicillin-Streptomycin and 1× NEAA. Human colon Caco-2 cells (epithelial cells isolated from colon tissue, ATCG, HTB-37) and monkey Vero E6 cells (kidney epithelial cells from female African green monkey, ATCC, CCL-81) were maintained in DMEM culture medium supplemented with 10% heat-inactivated FBS, 50 Uml−1 penicillin and 50 μgml−1 streptomycin. All cells were maintained in a 5% CO2 incubator at 37°C and routinely tested negative for mycoplasma.
Culturing hEPSC-TSC derived trophoblast organoids
Dissociated hEPSC-TSCs were resuspended into single-cells in ice-cold trophoblast organoid medium (TOM) containing 80 ng/mL R-spondin-1, 1.5 μM CHIR99021, 500 nM A83-01, 50 ng/mL human EGF, 100 ng/mL FGF2, 50 ng/mL human HGF, 2.5 μM prostaglandin E2, and Y-27632 5.0 μM. A final concentration of 60% growth factor-reduced Matrigel (GFR-M, Corning) was added. 40 μL drops of cell/Matrigel mixture) with 1 × 105 EPSC-TSCs each were pipetted into the well centers of 24-well plates. After 15 min solidification at 37°C for the drops to form domes, 500 μL prewarmed TOM was added to overlay the domes. Trophoblast organoid formation require around 4–6 days at P0. They were passaged after reaching 200–300 μm in diameter.
Virus
The wildtype SARS-CoV-2 HKU-001a strain (GenBank accession number: MT230904) was isolated from the nasopharyngeal aspirate specimen of a patient who was laboratory-confirmed to have COVID-19 in Hong Kong. The SARS-CoV-2 Delta variant isolate (B.1.617.2) (hCoV-19/Hong Kong/HKU-210804-001/2021; GISAID accession number EPI_ISL_3221329) has been described previously.88 The Omicron variant strain was isolated from the nasopharyngeal throat swab of a COVID-19 patient in Hong Kong (hCoV-19/Hong Kong/HKU-344/2021; GISAID accession number EPI_ISL_7357684). The MERS-CoV strain (HCoV-EMC/2012) was a gift from R. Fouchier. All experiments involving live SARS-CoV-2 and MERS-CoV were performed following the approved standard operating procedures of the biosafety level 3 facility in Queen Mary Hospital at The University of Hong Kong.89
Method details
Culture of hEPSCs and naive stem cells
Human EPSC cells were maintained on SNL 76/7 feeder layers and enzymatically passaged (1:10) every 3–5 days by a brief PBS wash followed by treatment with TrypLE for 3 min. Cells were dissociated and centrifuged (300g for 3 min) in 10% fetal bovine serum (FBS)-containing medium (M10 medium). After removing the supernatant, human EPSCs were resuspended and seeded in hEPSC Medium (EPSCM) supplemented with 5.0 μM Y27632. The hEPSCM is a N2B27-based media supplement with 3 small molecules 5 μM XAV939, 1 μM CHIR99021 and 0.1 μM A-419259 as previously published.24 N2B27 basal media [1:1 of DMEM/F12 (Thermo, Cat.21331020) and Neurobasal Medium (Thermo, Cat. 21103049); 200x N2 supplement and 100x B27 supplement, 100x ITS-X, 50.0 μM β-mercaptoethanol, 1% Penicillin-Streptomycin-Glutamine, 100x Non-essential amino acid solution, 50 μg/mL Vitamin C].
Naive stem cells were maintained on inactivated MEF (mouse embryonic fibroblasts) feeder layers in PXGL medium. The PXGL medium is prepared by supplementing N2B27 basal medium with PD0325901(1 μM), XAV-939(2 μM), Gö 6983 (2 μM), 0.45% BSA and human leukemia inhibitory factor (hLIF, 10 ng/mL) as previously reported.32
Differentiation of hEPSCs to trophoblast lineages by TGF-β inhibitor SB431542 treatment
Human EPSCs were dissociated with TrypLE and seeded in 100x Geltrex coated six-well plates at a density of 1×105 cells per well. Cells were cultured (pre-treatment) in 20% KSR media supplemented with 10 μM Y27632 for one day. From the second day, 10 μM SB431542 was added into 20% KSR media to start the differentiation. Cells were collected at the indicated time points for analysis.
Derivation of human trophoblast stem cells (hTSCs) from hEPSCs and naive stem cells
Single cell-dissociated hEPSCs were plated on 6-well plates pre-coated with 100x Geltrex at a density of 2,000 cells per well and cultured in the published hTSC media with modifications.35 DMEM/F12 supplemented with 50.0 μM β mercaptoethanol, 0.2% FBS, 0.5% Penicillin-Streptomycin-Glutamine, 0.3% BSA, 1.0% ITS-X supplement, 50.0 μg/mL Vc, 50.0 ng/mL EGF, 2.0 μM CHIR99021, 0.5 μM A83-01, 1.0 μM SB431542, 10.0 μM VPA and 5.0 μM Y27632. After 12–14 days of culture, the colonies with TSC-like morphologies were picked, dissociated in TrypLE, and replated on a plate pre-coated with 100x Geltrex. After 10 passages, the cells were collected for syncytiotrophoblast (STB) and extravillous trophoblast (EVT) differentiation.
Single cell-dissociated naive stem cells were plated on 6-well plates pre-coated with 100x Geltrex and cultured in PD0325901 (1 μM) + A83-01(1 μM) for 3 days then induced in hTSCM as described.32
Differentiation of EPSC-TSCs to STBs
Wells of a six-well plate were coated with 100× Matrigel for at least 1h. 2.0×105 hTSCs were seeded per well in 2 mL STB medium [DMEM/F12 supplemented with 50 μM β-mercaptoethanol, 0.5% Penicillin-Streptomycin-Glutamine, 0.3% BSA, 1% ITS-X, 2.5 μM Y-27632, 2 μM Forskolin and 4% KnockOut Serum Replacement. Media was changed on day 3, and the cells were ready for downstream analysis on day6. The eSTBs correspond to day 2 cells differentiating in the STB medium.
Differentiation of EPSC-TSCs to EVTs
For EVT differentiation, wells of a 6-well plate were coated with 100× Matrigel for at least 1h. 2.0 ×105 hTSCs were seeded per well in 3.0 mL EVT basal medium [DMEM/F12 supplemented with 50uM β-mercaptoethanol, Penicillin-Streptomycin-Glutamine, 0.3% BSA, 1%ITS-X, 7.5 μM A83-01, 10 μM Y27632] supplemented with 4% KSR, 100 ng/mL NRG1 and 2% Matrigel. On day 3, the media were replaced with 2 mL EVT basal medium supplemented with 4% KSR and 0.5% Matrigel. On day 6, they were dissociated with TrypLE for 10–15 min and passaged to a fresh matrigel-coated 6-well plate. The media were replaced with 2 mL EVT basal medium and 0.5% Matrigel. On day 8, the cells were ready for downstream analysis.
Generation of trophoblast organoids from EPSC-TSCs
EPSC-TSCs were digested with TrypLE and dissociated into single cells by pipetting. After centrifugation, growth factor-reduced Matrigel (GFR-M) was added to reach a final concentration of 60% and the remaining 40% was made up of trophoblast organoid medium (TOM). The Matrigel/TOM mixture (40 μL) containing 1.0 × 105 hTSCs was seeded into the wells’ center of a 24-well plates. After 3 min at 37°C in a CO2 incubator, the plates were turned upside down to ensure equal spreading of the cells in the well. After 15 min at 37°C in an incubator, the solidifying GFR-M formed domes and were carefully overlaid with 500 μL TOM. Trophoblast organoids were allowed to form for 4–6 days at P0. After 20 days of culturing, trophoblast organoids were collected by dissolving the Matrigel with recovery solution and submitted to SARS-CoV-2 infection. TOM is composed of N2B27 basal media, recombinant human EGF 50 ng/mL, CHIR99021 1.5μM, recombinant human R-spondin-1 80 ng/mL, recombinant human FGF-2 100 ng/mL, recombinant human HGF 50 ng/mL, A83-01 500 nM, Prostaglandin E2 2.5 μM, Y-27632 5 μM. All medium was stored at 4°C for up to 2 weeks.
SARS-CoV-2 infection and viral copy detection
SARS-CoV-2 stock was propagated using Vero E6 cells, and the titer of supernatant was assessed by plaque assay as previously described.90
For viral detection, the supernatant of the cultured cells challenged by SARS-CoV-2 was harvested at various time points. A total of 140 μL of culture supernatant was lysed with 560 μL of AVL buffer, which was subsequently extracted for total RNA with the QIAamp® Viral RNA Mini Kit. The extracted RNA was quantified with the one-step QuantiNova Probe RT-PCR kit. Each 20 μL reaction mixture contained 10 μL of 2× QuantiNova Probe RT-PCR Master Mix, 0.2 μL of QuantiNova Probe RT-Mix, 1.6 μL each of 10μM forward and reverse primer, 0.4 μL of 10 μM probe, 5 μL of extracted RNA as template, and 1.2 μL of RNase-free water. Reactions were incubated at 45°C for 10 min for reverse transcription, 95°C for 5 min for denaturation, 45 cycles of 95°C for 5 s and 55°C for 30 s, followed by a cooling step at 40°C for 30 s. The primers and probe sequences were against the RNA-dependent RNA polymerase/helicase (RdRP/Hel) gene region of SARS-CoV-2 were listed in Table S5.
Plaque assay
Plaque assay was performed as previously described.91 Briefly, Vero E6 cells were seeded at 300,000 cells/well in 12-well tissue culture plates on the day before carrying out the assay. After 24 h of incubation, a serial dilution of supernatant was added to the cell monolayer and the plates were further incubated for 1 h at 37°C in 5% CO2 before removal of unbound viral particles by aspiration of the media and washing once with DMEM. Monolayers were then overlaid with media containing 1% low melting agarose (Cambrex Corporation, East Rutherford, NJ, USA) in DMEM, inverted and incubated as above for another 72 h. The wells were then fixed with 10% formaldehyde (BDH, Merck, Darmstadt, Germany) overnight. After removal of the agarose plugs, the monolayers were stained with 0.7% crystal violet (BDH, Merck) and the plaques were counted. The plaque assay experiments were performed in triplicates.
Antiviral evaluation in eSTBs
The EPSC-TSCs were dissociated into single cells and rinsed with STB medium. Then cells were seeded into 96 well plate at the density of 10000 cells per well and cultured for two days (eSTBs). The eSTBs were infected with SARS-CoV-2 WT, Delta, Omicron variant or MERS-CoV with 0.1 MOI inoculum at 100 μL per well. After 1 h, the inoculum was removed, and the eSTBs were washed 3 times with PBS. The infected eSTBs were then cultured in 100ul of STB medium with remdesivir or GC376 at indicated concentrations. Supernatants were collected at 48 h after inoculation for downstream assays.
Guide RNA design and plasmid DNA preparation
The human ACE2 exon 2 sequence was analyzed by the online CRISPR tool for designing a pair of highly specific gRNAs. Chemically synthesized ssDNA oligos were incubated at 95°C for 10 min for annealing into dsDNA, which were then ligated into a linearized empty gRNA vector using the DNA Ligation Kit, Mighty Mix. The ligation product was transformed into Chemically Competent DH 5α Cell (KT Health) and spread onto an LB agar plate with ampicillin for selection. The next day, single colonies were picked and cultured in LB broth with ampicillin for plasmid miniprep using the TIANprep Rapid Mini Plasmid Kit. After confirmation of ligated gRNA sequences by Sanger Sequencing, gRNA plasmids with the correct target sequence were amplified and extracted by the Endo-free Plasmid Kit II to purify a large amount of endotoxin-free gRNA plasmid DNA for electroporation. Plasmid DNA of BSD-Cas9 was similarly prepared as described previously.24
Electroporation and selection
Electroporation of hEPSCs was performed when the cells reached 70–80% confluence. Plasmid DNA including Cas9 and double sgRNA at a ratio of 2:1:1 (4μg Cas9, 2μg sgRNA1, 2μg sgRNA2 per 1millon cells as one group) were added into 300 μL Opti-MEM. hEPSCs were washed twice with PBS, then dissociated into single cells using 0.05% Trypsin-EDTA. M10 medium (DMEM with 10% FSB) was added to neutralize the trypsin. Single-cell hEPSCs were resuspended in an Opti-MEM medium containing the plasmid DNA mixture. Electroporation was performed using the Bio-Rad Gene Pulser Xcell Electroporation Systems in 0.4cm cuvettes at 230V, 500 μF. After electroporation, cells were seeded with a recovery medium (500 μL EPSCM + 10% KSR+10μM Y27632). After incubation overnight, cells were switched to a typical hEPSC medium. One day after electroporation, cells were selected by 10 μg/mL Blasticidin S HCl. Two days after electroporation, cells were selected by 10 μg/mL Blasticidin S HCl and 1 μg/mL Puromycin Dihydrochloride for another 2 days. After 7–10 days, single colonies were picked, expanded, and genotyped.
Genotyping and sanger sequencing
Single hEPSC colonies were picked and digested into single cells in a 96-well plate using 0.05% trypsin-EDTA for 3–5 min. Half of these cells were transferred into a 48-well plate with EPSCM medium and SNL feeders for culturing. The other half cells of the same colony were collected for genotyping using primers designed to amplify the targeted as well as the wildtype bands. The genotyping primer sequences for ACE2 were as follows: Forward: 5′-GTGGCCTGGTCACTCTTAAC-3′; Reverse: 5′-CAAATAAAGGCAGCTGCTGTG-3′. The mutant PCR band was gel-purified and confirmed by Sanger sequencing for a 148-bp deletion.
Reverse transcription- quantitative polymerase chain reaction (RT-qPCR)
Total RNA extraction was performed using the RNeasy Mini Kit as per the manufacturer’s instructions. The isolated RNA was reverse transcribed into complementary (cDNA) using the Fastking gDNA Dispelling RT Super-Mix on a thermal cycler. The PowerUp™ SYBR™ Green Master Mix was used for amplifying intracellular SARS-CoV-2 and gene cDNA. Primer sequences are listed in Table S5. All qPCR experiments were performed using The StepOnePlus™ Real-Time PCR System (Applied Biosystems). Gene expression levels were normalized to GAPDH using the ΔCt method. Data were analyzed using one/two-tailed Student’s t test in Prism 8 (GraphPad).
Immunofluorescence staining
Samples were fixed in 4% paraformaldehyde at room temperature for 15 min, permeabilized with 0.3% Triton X-100 for 30 min, and blocked for 3h with 5% donkey serum as well as 1% BSA. This was followed by incubation with primary antibodies in a 4°C cold room overnight. After washing in PBS, fluorophore-conjugated secondary antibodies were incubated with the samples at room temperature for 1 h. After another 3 rounds of washing in PBS, samples were counterstained with 10 μg/mL DAPI for 10 min to mark nuclei and were imaged under a fluorescence microscope or confocal LSM900.
Western blotting
Proteins were separated in 7.5% polyacrylamide gels and transferred to PVDF membranes using the Bio-Rad transblot turbo system according to the manufacturer’s guidance. The following primary antibodies were used for Western blot: rabbit ACE2 (1:500, Abclonal. Cat. A4612), rabbit β-actin (1:5000, Abmart. Cat. P30002M). Goat anti-rabbit IgG H&L (HRP) (1:10000, Abcam. Cat. ab205718) was used as the secondary antibody. Images were developed and analyzed by the ChemiDoc Imaging System.
Flow cytometry
Cells were digested with 0.25% trypsin/EDTA for 2–3 min at 37°C and dissociated to single cells by pipetting. The dissociated cells were filtered through a 40-μm nylon mesh to remove cell clumps. After centrifugation, the cells were fixed using Fixation Medium according to the manufacturer’s protocol and the washed cells were stored at 4°C in PBS supplemented with 0.1% NaN3 and 5% FBS before analysis with flow cytometry. All the samples were assayed by the ACEA NovoCyte Quanteon. 488nm (530/30 bandpass filter) and 561nm (610/20 bandpass filter) channels were used to detect FITC and excluded autofluorescence. 405nm (445/45 bandpass filter) channel was used to detect DAPI positive cells. FACS data were analyzed by the Flowjo software.
Transwell invasion assay
The invasion ability of hTSCs-derived EVTs was determined by cell invasion according to manufacturers' instructions. Briefly, the invasion chambers were incubated with a warm DMEM base medium at 37°C for 1 h. After rehydration, the medium was removed. EPSC-EVTs (1 × 105 cells/well) was mixed with DMEM basal medium, and the mixture was added into an invasion chamber and placed into a 24 well culture plate, with the lower chamber filled with DMEM containing 10% FBS. The cells were allowed to pass through the chamber and attached to the lower bottom of the polycarbonate membrane for 22 h. After that, the medium from the top insert was aspirated, and non-invasive/-migratory cells on the upper surface were wiped away with a cotton bud. The invaded/migrated cells on the lower surface were stained with crystal violet for 15 min. The membrane was observed under a light microscope.
Quantification and statistical analysis
Statistical analysis and reproducibility
The statistical analysis was conducted with Microsoft Excel or Prism 8 (GraphPad). p values were calculated using (un)paired Student’s t test and ANOVA for comparisons of multiple groups. The threshold for statistical significance was p < 0.05. The exact number of measurements, the number of independent experiments, and the statistical test used for each analysis performed were listed in the figure legends. Experiments were repeated independently with similar results obtained.
RNA-seq analysis
Cutadapt was used to remove adapter sequences and low-quality 3′ end sequences. Processed reads were mapped to the human hg38 genome assembly by hisat2. Gene annotation form Ensembl was used. FeatureCounts was used to quantify gene expression. Genes with mean count number <5 were filtered out. Transposable element annotations were from UCSC Genome Browser (RepeatMasker). SQuIRE with “total” mode was used to quantify TE expression. DESeq2 was used to analyze differentially expressed genes and TEs. Genes and TEs with expression fold change >1.5 and adjusted p-value < 0.05 were considered significant differential expressed. R package clusterprofiler was used for gene ontology (GO) and KEGG analysis. GSEA (Gene Set Enrichment Analysis) was performed by GSEApy, and the gene sets were downloaded directly from https://www.gsea-msigdb.org. Bigwig files for RNA-seq signal were generated by bamCoverage from Deeptools and IGV was used for visualization. For the RNA-seq signal on endogenous retrovirus, GenBank: AY101582.1, BC068585.1, JN675077.1 were used to inquire about the sequence of HERV-W, HERV-FRD, and HERV-K and the reads were mapped by hisat2. GenBank accession number MN985325 was used to inquire about the sequence of SARS-CoV-2 genes, then the reads were mapped to sequences by hisat2 and quantified. For data deposited in E-MTAB-10429, the processed count table was used directly. For the comparison between E-MTAB-10429 and our data, we used ComBat-seq to remove the batch effect.
For the PCA analysis for hEPSC-TSCs, naive hPSC-hTSCs, primed-hTSCs and early/late AMEs, data processing were followed by another study92: the expression of genes are normalized by DESeq2 and the top 1000 most variable protein coding genes were used in the analysis.
scRNA preprocessing
scRNA preprocessing was performed according to the SCANPY pipelines.93 Briefly, Transcripts Per Million (TPM) of pre-to post-implantation cells were extracted from Zhou’s dataset44 (GEO: GSE109555), and placenta cells were extracted from Liu’s dataset45(GEO: GSE89497). In the original studies, cells have been categorized by their stages and lineages into 7 coarse clusters (3 in pre-to post-implantation, 3 in the first-trimester placenta, and 1 in the second-trimester placenta; N.B., Zhou’s dataset annotation was obtained from Castel. et al.).33 Here, we performed pseudotime analysis on these two datasets jointly. First, we pre-processed the combined dataset by 1) log-transformation of the TPM counts, 2) scaling to 10 on each gene, and 3) regressing out on the sequencing depth. Then, we calculated the diffusion pseudotime by DPT using scanpy.tl.dpt with default parameters.94 Cells with inconsistent stage-lineage annotations and pseudotime values have been removed, and 4041 and 952 cells were retained for downstream analysis, respectively. Based on log counts, Pearson correlations were computed for selected markers with ACE2, and visualized by scatterplots. Batch effects among bulk RNA-seq have also been regressed out before comparing in vitro cells to in vivo reference.
Integration of in vitro and in vivo extra-embryonic datasets
For integrative analysis, we mapped the in vitro bulk RNA-seq to the in vivo scRNA reference by using singular value decomposition (SVD) modeling,95,96 based on the assumption that top components could capture cell identity and biological variations, regardless of sequencing types. First, the batch effects between the two scRNA datasets were regressed out to construct an extra-embryonic landscape containing peri-implantation to placenta stages. Then, we fitted the SVD model using the whole transcriptome of the combined in vivo scRNA reference and generated a 50-component decomposition result for the in vivo reference. Next, we applied the fitted model to project the in vitro cells (bulk RNA-seq) to the corresponding 50 components in the same space. After concatenating the bulk RNA and scRNA datasets as 50-component samples, UMAP visualization could be generated. As a proof of the effectivity of the SVD modeling in segregating cell types, for the in vivo sector, inter-cell type variations were captured by each cluster in the UMAP.
Whole-transcriptome correlation analysis between in vitro cell line bulk RNA-seq and in vivo scRNA-seq
With the assumption that bulk RNA-seq reflects additive effects of scRNA levels, we generated pseudo-bulk sets from the in vivo scRNA-seq. For each in vivo subtype, we randomly selected two sets of 50 cells as two “pseudo-bulk cells” and calculated an average to simulate two bulk samples for each subtype, hence we have 14 pseudo-bulk samples. After merging the 14 pseudo-bulk samples with in vitro cells (19 samples, 10 types), we calculated the pairwise Pearson coefficients across the whole transcriptome between every two samples. The coefficients were visualized in the cluster map to impute the corresponding in vivo stage of each in vitro cell line.
Integrative pseudotime analysis
First, diffusion pseudotime was computed for the whole in vivo reference using SCANPY (scanpy.tl.dpt, default parameters). We then extracted the PI-TB to PI-STB subsets as the pseudotime reference set for downstream analysis of in vitro ST differentiation cells. There are 2295 PI-TB cells and 1282 PI-STB cells, constituting a unidirectional differentiation trajectory. Similar to the process described in the integration of bulk RNA and scRNA datasets part, we fitted a 50-component SVD model using the scRNA reference to reduce the dimensions of both scRNA and bulk RNA to a common 50-component space. We hypothesized that these 50 components could predict the pseudotime calculated by SCANPY. The machine learning prediction process was achieved using sklearn.linear_model. LinearRegression. Briefly, the 50 components and the pseudotime of 1721 PI-TB cells and 961 PI-STB cells (75% of each subtype) were used to train the linear regression model, and the remaining 25% test set of each subtype was used to validate the model. The validation result was visualized by the density plot. After confirming the effectivity of the model, we applied the model to predict the pseudotime of in vitro differentiated cells. We also calculated the theoretical “observed pseudotime” for in vitro cells by the following linear equation between the predicted and observed pseudotime in the in vivo reference: Observed pseudotime = 1.022 ∗ Predicted pseudotime - 0.0032. Then pseudotime of each dataset was then merged to visualize the relative differentiation stage of each cell.
Acknowledgments
This work was financially supported by the National Key Research and Development Program of China (no. 2018YFA0902702); ITC Health@InnoHK; National Natural Science Foundation of China (nos. 81570202, 32100639, 82071856, 81671579, and 31370904 to L.L.); the Health and Medical Research Fund, the Food and Health Bureau, The Government of the HKSAR (no. COVID190114 to D.-Y.J. and no. 09202056 to C.-L.L.) and Hong Kong Research Grant Council (no. C7142-20GF to D.-Y.J.; GRF 17127219, 17126421, and 17112422 to P.L. and GRF 17112822 to P.C.N.C.); China Postdoctoral Science Foundation (2021M702280); High-Level-Hospital Program, Health Commission of Guangdong Province (no. HKUSZH201902025, HKUSZH201902037, HKUSZH201902008 to W.Y.); Program for Scientific and Technological Innovation from the Science and Technology Commission of Shanghai Municipality (22490760400, L.L.); and HKU and HKU Shenzhen Hospital internal funding (P.L. and D.R.). Instrument or analysis software was supported by the Imaging and Flow Cytometry Core of HKU Centre for PanorOmic Sciences (CPOS).
Author contributions
P.L., D.J., F.L., and D.R. conceived the project and designed the experiments. The co-first authors, P.L. and D.J., wrote the manuscript together. F.L. and T.T.K.K.T. performed the SB43 differentiation-related experiments and ACE2-KO to establish mutant hEPSCs. J.G., Y.H., and Y.X. contributed to the CRISPR technology and western blotting. Z.Y. and C.P.O. performed all infection studies, including viral load analysis. S.Y. and K.T. helped with viral infection and antiviral drug evaluation. D.R. performed the hTSC generation and differentiation, immunofluorescence staining, and FACS assays. D.R., Q.Z., and C.L. contributed to trophoblast organoid generation and characterization. W.Z. and Z.L. performed RNA-seq data analysis. P.C.N.C. and W.S.B.Y. provided advice.
Declaration of interests
A patent application related to the data presented here has been filed on behalf of the Centre for Translational Stem Cell Biology.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100849.
Contributor Information
Fang Liu, Email: fang.liu@hkstemcell.hk.
Dong-Yan Jin, Email: dyjin@hku.hk.
Pentao Liu, Email: pliu88@hku.hk.
Supplemental information
Data and code availability
-
•
RNA-seq data generated in this study have been deposited at NCBI Gene Expression Omnibus (GEO): GEO: GSE190432.
-
•
All original code has been deposited (https://github.com/leeyoyohku/EPSC_COVID) and is publicly available as of date of publication.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead contact upon request.
-
•
Additional Supplemental Items are available from Mendeley Data: https://doi:10.17632/rnsbwdr7gw.2.
References
- 1.Pereira L. Congenital viral infection: traversing the uterine-placental interface. Annu. Rev. Virol. 2018;5:273–299. doi: 10.1146/annurev-virology-092917-043236. [DOI] [PubMed] [Google Scholar]
- 2.Algarroba G.N., Rekawek P., Vahanian S.A., Khullar P., Palaia T., Peltier M.R., Chavez M.R., Vintzileos A.M. Visualization of severe acute respiratory syndrome coronavirus 2 invading the human placenta using electron microscopy. Am. J. Obstet. Gynecol. 2020;223:275–278. doi: 10.1016/j.ajog.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hosier H., Farhadian S.F., Morotti R.A., Deshmukh U., Lu-Culligan A., Campbell K.H., Yasumoto Y., Vogels C.B., Casanovas-Massana A., Vijayakumar P., et al. SARS-CoV-2 infection of the placenta. J. Clin. Invest. 2020;130:4947–4953. doi: 10.1172/JCI139569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivanti A.J., Vauloup-Fellous C., Prevot S., Zupan V., Suffee C., Do Cao J., Benachi A., De Luca D. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020;11:3572. doi: 10.1038/s41467-020-17436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts D.J., Edlow A.G., Romero R.J., Coyne C.B., Ting D.T., Hornick J.L., Zaki S.R., Das Adhikari U., Serghides L., Gaw S.L., et al. A standardized definition of placental infection by SARS-CoV-2, a consensus statement from the national institutes of health/eunice Kennedy Shriver national Institute of child Health and human development SARS-CoV-2 placental infection workshop. Am. J. Obstet. Gynecol. 2021;225:593.e1–593.e9. doi: 10.1016/j.ajog.2021.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Male V. SARS-CoV-2 infection and COVID-19 vaccination in pregnancy. Nat. Rev. Immunol. 2022;22:277–282. doi: 10.1038/s41577-022-00703-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz D.A., Avvad-Portari E., Babál P., Baldewijns M., Blomberg M., Bouachba A., Camacho J., Collardeau-Frachon S., Colson A., Dehaene I., et al. Placental tissue destruction and insufficiency from COVID-19 causes stillbirth and neonatal death from hypoxic-ischemic injury: a study of 68 cases with SARS-CoV-2 placentitis from 12 countries. Arch. Pathol. Lab. Med. 2022 doi: 10.5858/arpa.2022-0029-SA. [DOI] [PubMed] [Google Scholar]
- 8.Valdespino-Vázquez M.Y., Helguera-Repetto C.A., León-Juárez M., Villavicencio-Carrisoza O., Flores-Pliego A., Moreno-Verduzco E.R., Díaz-Pérez D.L., Villegas-Mota I., Carrasco-Ramírez E., López-Martínez I.E., et al. Fetal and placental infection with SARS-CoV-2 in early pregnancy. J. Med. Virol. 2021;93:4480–4487. doi: 10.1002/jmv.26965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baud D., Greub G., Favre G., Gengler C., Jaton K., Dubruc E., Pomar L. Second-trimester miscarriage in a pregnant woman with SARS-CoV-2 infection. JAMA. 2020;323:2198–2200. doi: 10.1001/jama.2020.7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allotey J., Chatterjee S., Kew T., Gaetano A., Stallings E., Fernández-García S., Yap M., Sheikh J., Lawson H., Coomar D., et al. SARS-CoV-2 positivity in offspring and timing of mother-to-child transmission: living systematic review and meta-analysis. BMJ. 2022;376:e067696. doi: 10.1136/bmj-2021-067696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong L., Tian J., He S., Zhu C., Wang J., Liu C., Yang J. Possible vertical transmission of SARS-CoV-2 from an infected mother to her newborn. JAMA. 2020;323:1846–1848. doi: 10.1001/jama.2020.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burton G.J., Fowden A.L. The placenta: a multifaceted, transient organ. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20140066. doi: 10.1098/rstb.2014.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeldovich V.B., Clausen C.H., Bradford E., Fletcher D.A., Maltepe E., Robbins J.R., Bakardjiev A.I. Placental syncytium forms a biophysical barrier against pathogen invasion. PLoS Pathog. 2013;9:e1003821. doi: 10.1371/journal.ppat.1003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashary N., Bhide A., Chakraborty P., Colaco S., Mishra A., Chhabria K., Jolly M.K., Modi D. Single-cell RNA-seq identifies cell subsets in human placenta that highly expresses factors driving pathogenesis of SARS-CoV-2. Front. Cell Dev. Biol. 2020;8:783. doi: 10.3389/fcell.2020.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pique-Regi R., Romero R., Tarca A.L., Luca F., Xu Y., Alazizi A., Leng Y., Hsu C.D., Gomez-Lopez N. Does the human placenta express the canonical cell entry mediators for SARS-CoV-2? Elife. 2020;9:e58716. doi: 10.7554/eLife.58716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W., Yuan P., Yang M., Yan Z., Kong S., Yan J., Liu X., Chen Y., Qiao J., Yan L. SARS-CoV-2 entry factors: ACE2 and TMPRSS2 are expressed in peri-implantation embryos and the maternal–fetal interface. Engineering. 2020;6:1162–1169. doi: 10.1016/j.eng.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colson A., Depoix C.L., Dessilly G., Baldin P., Danhaive O., Hubinont C., Sonveaux P., Debiève F. Clinical and in vitro evidence against placenta infection at term by severe acute respiratory syndrome coronavirus 2. Am. J. Pathol. 2021;191:1610–1623. doi: 10.1016/j.ajpath.2021.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taglauer E.S., Wachman E.M., Juttukonda L., Klouda T., Kim J., Wang Q., Ishiyama A., Hackam D.J., Yuan K., Jia H. Acute severe acute respiratory syndrome coronavirus 2 infection in pregnancy is associated with placental angiotensin-converting enzyme 2 shedding. Am. J. Pathol. 2022;192:595–603. doi: 10.1016/j.ajpath.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y., Liu B., Xu J., Wang J., Wu J., Shi C., Xu Y., Dong J., Wang C., Lai W., et al. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell. 2017;169:243–257.e25. doi: 10.1016/j.cell.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J., Ryan D.J., Wang W., Tsang J.C.H., Lan G., Masaki H., Gao X., Antunes L., Yu Y., Zhu Z., et al. Establishment of mouse expanded potential stem cells. Nature. 2017;550:393–397. doi: 10.1038/nature24052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao X., Nowak-Imialek M., Chen X., Chen D., Herrmann D., Ruan D., Chen A.C.H., Eckersley-Maslin M.A., Ahmad S., Lee Y.L., et al. Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 2019;21:687–699. doi: 10.1038/s41556-019-0333-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao L., Gao X., Zheng Y., Wang Z., Zhao G., Ren J., Zhang J., Wu J., Wu B., Chen Y., et al. Establishment of bovine expanded potential stem cells. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2018505118. e2018505118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cinkornpumin J.K., Kwon S.Y., Guo Y., Hossain I., Sirois J., Russett C.S., Tseng H.-W., Okae H., Arima T., Duchaine T.F., et al. Naive human embryonic stem cells can give rise to cells with a trophoblast-like transcriptome and methylome. Stem Cell Rep. 2020;15:198–213. doi: 10.1016/j.stemcr.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amita M., Adachi K., Alexenko A.P., Sinha S., Schust D.J., Schulz L.C., Roberts R.M., Ezashi T. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc. Natl. Acad. Sci. USA. 2013;110:E1212–E1221. doi: 10.1073/pnas.1303094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu R.-H., Chen X., Li D.S., Li R., Addicks G.C., Glennon C., Zwaka T.P., Thomson J.A. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 29.Horii M., Li Y., Wakeland A.K., Pizzo D.P., Nelson K.K., Sabatini K., Laurent L.C., Liu Y., Parast M.M. Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease. Proc. Natl. Acad. Sci. USA. 2016;113:E3882–E3891. doi: 10.1073/pnas.1604747113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei Y., Wang T., Ma L., Zhang Y., Zhao Y., Lye K., Xiao L., Chen C., Wang Z., Ma Y., et al. Efficient derivation of human trophoblast stem cells from primed pluripotent stem cells. Sci. Adv. 2021;7:eabf4416. doi: 10.1126/sciadv.abf4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong C., Beltcheva M., Gontarz P., Zhang B., Popli P., Fischer L.A., Khan S.A., Park K.-m., Yoon E.-J., Xing X., et al. Derivation of trophoblast stem cells from naïve human pluripotent stem cells. Elife. 2020;9:e52504. doi: 10.7554/eLife.52504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo G., Stirparo G.G., Strawbridge S.E., Spindlow D., Yang J., Clarke J., Dattani A., Yanagida A., Li M.A., Myers S., et al. Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell. 2021;28:1040–1056.e6. doi: 10.1016/j.stem.2021.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castel G., Meistermann D., Bretin B., Firmin J., Blin J., Loubersac S., Bruneau A., Chevolleau S., Kilens S., Chariau C., et al. Induction of human trophoblast stem cells from somatic cells and pluripotent stem cells. Cell Rep. 2020;33:108419. doi: 10.1016/j.celrep.2020.108419. [DOI] [PubMed] [Google Scholar]
- 34.Io S., Kabata M., Iemura Y., Semi K., Morone N., Minagawa A., Wang B., Okamoto I., Nakamura T., Kojima Y., et al. Capturing human trophoblast development with naive pluripotent stem cells in vitro. Cell Stem Cell. 2021;28:1023–1039.e13. doi: 10.1016/j.stem.2021.03.013. [DOI] [PubMed] [Google Scholar]
- 35.Okae H., Toh H., Sato T., Hiura H., Takahashi S., Shirane K., Kabayama Y., Suyama M., Sasaki H., Arima T. Derivation of human trophoblast stem cells. Cell Stem Cell. 2018;22:50–63.e6. doi: 10.1016/j.stem.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Sheridan M.A., Zhao X., Fernando R.C., Gardner L., Perez-Garcia V., Li Q., Marsh S.G.E., Hamilton R., Moffett A., Turco M.Y. Characterization of primary models of human trophoblast. Development. 2021;148 doi: 10.1242/dev.199749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rostovskaya M., Andrews S., Reik W., Rugg-Gunn P.J. Amniogenesis occurs in two independent waves in primates. Cell Stem Cell. 2022;29:744–759.e6. doi: 10.1016/j.stem.2022.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee C.Q.E., Gardner L., Turco M., Zhao N., Murray M.J., Coleman N., Rossant J., Hemberger M., Moffett A. What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Rep. 2016;6:257–272. doi: 10.1016/j.stemcr.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng Y., Xue X., Shao Y., Wang S., Esfahani S.N., Li Z., Muncie J.M., Lakins J.N., Weaver V.M., Gumucio D.L., Fu J. Controlled modelling of human epiblast and amnion development using stem cells. Nature. 2019;573:421–425. doi: 10.1038/s41586-019-1535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang L., Yin Y., Zheng Y., Ma Y., Li Y., Zhao Z., Guo J., Ai Z., Niu Y., Duan K., et al. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature. 2020;577:537–542. doi: 10.1038/s41586-019-1875-y. [DOI] [PubMed] [Google Scholar]
- 41.Zhao C., Reyes A.P., Schell J.P., Weltner J., Ortega N.M., Zheng Y., Björklund Å.K., Rossant J., Fu J., Petropoulos S., Lanner F. Reprogrammed blastoids contain amnion-like cells but not trophectoderm. bioRxiv. 2021 doi: 10.1101/2021.05.07.442980. Preprint at. [DOI] [Google Scholar]
- 42.Chhabra S., Warmflash A. BMP-treated human embryonic stem cells transcriptionally resemble amnion cells in the monkey embryo. Biol. Open. 2021;10:bio058617. doi: 10.1242/bio.058617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyser R.C.V., Mahammadov E., Nakanoh S., Vallier L., Scialdone A., Srinivas S. Single-cell transcriptomic characterization of a gastrulating human embryo. Nature. 2021;600:285–289. doi: 10.1038/s41586-021-04158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou F., Wang R., Yuan P., Ren Y., Mao Y., Li R., Lian Y., Li J., Wen L., Yan L., et al. Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature. 2019;572:660–664. doi: 10.1038/s41586-019-1500-0. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y., Fan X., Wang R., Lu X., Dang Y.L., Wang H., Lin H.Y., Zhu C., Ge H., Cross J.C., Wang H. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 2018;28:819–832. doi: 10.1038/s41422-018-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang K., Chen W., Zhang Z., Deng Y., Lian J.-Q., Du P., Wei D., Zhang Y., Sun X.-X., Gong L., et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal. Transduct. Target. Ther. 2020;5:283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S., Qiu Z., Hou Y., Deng X., Xu W., Zheng T., Wu P., Xie S., Bian W., Zhang C., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gamage T.K., Chamley L.W., James J.L. Stem cell insights into human trophoblast lineage differentiation. Hum. Reprod. Update. 2016;23:77–103. doi: 10.1093/humupd/dmw026. [DOI] [PubMed] [Google Scholar]
- 50.Holmes C.H., Simpson K.L., Okada H., Okada N., Wainwright S.D., Purcell D.F., Houlihan J.M. Complement regulatory proteins at the feto-maternal interface during human placental development: distribution of CD59 by comparison with membrane cofactor protein (CD46) and decay accelerating factor (CD55) Eur. J. Immunol. 1992;22:1579–1585. doi: 10.1002/eji.1830220635. [DOI] [PubMed] [Google Scholar]
- 51.Banu N., Panikar S.S., Leal L.R., Leal A.R. Protective role of ACE2 and its downregulation in SARS-CoV-2 infection leading to Macrophage Activation Syndrome: therapeutic implications. Life Sci. 2020;256:117905. doi: 10.1016/j.lfs.2020.117905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeGrace M.M., Ghedin E., Frieman M.B., Krammer F., Grifoni A., Alisoltani A., Alter G., Amara R.R., Baric R.S., Barouch D.H., et al. Defining the risk of SARS-CoV-2 variants on immune protection. Nature. 2022;605:640–652. doi: 10.1038/s41586-022-04690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mi S., Lee X., Li X., Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.Y., Edouard P., Howes S., et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 55.Dupressoir A., Marceau G., Vernochet C., Bénit L., Kanellopoulos C., Sapin V., Heidmann T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. USA. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kämmerer U., Germeyer A., Stengel S., Kapp M., Denner J. Human endogenous retrovirus K (HERV-K) is expressed in villous and extravillous cytotrophoblast cells of the human placenta. J. Reprod. Immunol. 2011;91:1–8. doi: 10.1016/j.jri.2011.06.102. [DOI] [PubMed] [Google Scholar]
- 57.Zhou J., Choi S., Liu H., Zhang J., Tian Y., Edlow A.G., Ezashi T., Roberts R.M., Ma W., Schust D.J. Is SARS-CoV-2 infection a risk factor for early pregnancy loss? ACE2 and TMPRSS2 coexpression and persistent replicative infection in primitive trophoblast. J. Infect. Dis. 2021;224:S660–S669. doi: 10.1093/infdis/jiab309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kindler E., Thiel V., Weber F. Interaction of SARS and MERS coronaviruses with the antiviral interferon response. Adv. Virus Res. 2016;96:219–243. doi: 10.1016/bs.aivir.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J., Liu Y., Zhang X. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J. Virol. 2010;84:6472–6482. doi: 10.1128/JVI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin X., Riva L., Pu Y., Martin-Sancho L., Kanamune J., Yamamoto Y., Sakai K., Gotoh S., Miorin L., De Jesus P.D., et al. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep. 2021;34:108628. doi: 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye L., Schnepf D., Staeheli P. Interferon-λ orchestrates innate and adaptive mucosal immune responses. Nat. Rev. Immunol. 2019;19:614–625. doi: 10.1038/s41577-019-0182-z. [DOI] [PubMed] [Google Scholar]
- 62.Chen J., Liang Y., Yi P., Xu L., Hawkins H.K., Rossi S.L., Soong L., Cai J., Menon R., Sun J. Outcomes of congenital Zika disease depend on timing of infection and maternal-fetal interferon action. Cell Rep. 2017;21:1588–1599. doi: 10.1016/j.celrep.2017.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bayer A., Lennemann N.J., Ouyang Y., Bramley J.C., Morosky S., Marques E.T.D.A., Jr., Cherry S., Sadovsky Y., Coyne C.B. Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe. 2016;19:705–712. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanderheiden A., Ralfs P., Chirkova T., Upadhyay A.A., Zimmerman M.G., Bedoya S., Aoued H., Tharp G.M., Pellegrini K.L., Manfredi C., et al. Type I and type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J. Virol. 2020;94 doi: 10.1128/jvi.00985-20. e009855–e01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stanifer M.L., Kee C., Cortese M., Zumaran C.M., Triana S., Mukenhirn M., Kraeusslich H.G., Alexandrov T., Bartenschlager R., Boulant S. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32:107863. doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wickenhagen A., Sugrue E., Lytras S., Kuchi S., Noerenberg M., Turnbull M.L., Loney C., Herder V., Allan J., Jarmson I., et al. A prenylated dsRNA sensor protects against severe COVID-19. Science. 2021;374:eabj3624. doi: 10.1126/science.abj3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Osada N., Kohara A., Yamaji T., Hirayama N., Kasai F., Sekizuka T., Kuroda M., Hanada K. The genome landscape of the african green monkey kidney-derived vero cell line. DNA Res. 2014;21:673–683. doi: 10.1093/dnares/dsu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sasaki M., Uemura K., Sato A., Toba S., Sanaki T., Maenaka K., Hall W.W., Orba Y., Sawa H. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 2021;17:e1009233. doi: 10.1371/journal.ppat.1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koch J., Uckeley Z.M., Doldan P., Stanifer M., Boulant S., Lozach P.-Y. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J. 2021;40:e107821. doi: 10.15252/embj.2021107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beumer J., Geurts M.H., Lamers M.M., Puschhof J., Zhang J., van der Vaart J., Mykytyn A.Z., Breugem T.I., Riesebosch S., Schipper D., et al. A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses. Nat. Commun. 2021;12:5498. doi: 10.1038/s41467-021-25729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shuai H., Chan J.F.-W., Hu B., Chai Y., Yuen T.T.-T., Yin F., Huang X., Yoon C., Hu J.-C., Liu H., et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022 doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 72.Hui K.P.Y., Ho J.C.W., Cheung M.-c., Ng K.-c., Ching R.H.H., Lai K.-l., Kam T.T., Gu H., Sit K.-Y., Hsin M.K.Y., et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022;603:715–720. doi: 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 73.Rubin D., Chan-Tack K., Farley J., Sherwat A. FDA approval of remdesivir - a step in the right direction. N. Engl. J. Med. 2020;383:2598–2600. doi: 10.1056/NEJMp2032369. [DOI] [PubMed] [Google Scholar]
- 74.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dampalla C.S., Zheng J., Perera K.D., Wong L.-Y.R., Meyerholz D.K., Nguyen H.N., Kashipathy M.M., Battaile K.P., Lovell S., Kim Y., et al. Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2101555118. e2101555118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., et al. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W.A.L., Posthuma C.C., van der Meer Y., Bárcena M., Haagmans B.L., et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Han Y., Yang L., Lacko L.A., Chen S. Human organoid models to study SARS-CoV-2 infection. Nat. Methods. 2022;19:418–428. doi: 10.1038/s41592-022-01453-y. [DOI] [PubMed] [Google Scholar]
- 80.Turco M.Y., Gardner L., Kay R.G., Hamilton R.S., Prater M., Hollinshead M.S., McWhinnie A., Esposito L., Fernando R., Skelton H., et al. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature. 2018;564:263–267. doi: 10.1038/s41586-018-0753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haider S., Meinhardt G., Saleh L., Kunihs V., Gamperl M., Kaindl U., Ellinger A., Burkard T.R., Fiala C., Pollheimer J., et al. Self-renewing trophoblast organoids recapitulate the developmental Program of the early human placenta. Stem Cell Rep. 2018;11:537–551. doi: 10.1016/j.stemcr.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sheridan M.A., Fernando R.C., Gardner L., Hollinshead M.S., Burton G.J., Moffett A., Turco M.Y. Establishment and differentiation of long-term trophoblast organoid cultures from the human placenta. Nat. Protoc. 2020;15:3441–3463. doi: 10.1038/s41596-020-0381-x. [DOI] [PubMed] [Google Scholar]
- 83.Zhang B.-Z., Chu H., Han S., Shuai H., Deng J., Hu Y.-f., Gong H.-r., Lee A.C.-Y., Zou Z., Yau T., et al. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020;30:928–931. doi: 10.1038/s41422-020-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sheridan M.A., Yunusov D., Balaraman V., Alexenko A.P., Yabe S., Verjovski-Almeida S., Schust D.J., Franz A.W., Sadovsky Y., Ezashi T., Roberts R.M. Vulnerability of primitive human placental trophoblast to Zika virus. Proc. Natl. Acad. Sci. USA. 2017;114:E1587–E1596. doi: 10.1073/pnas.1616097114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sheridan M.A., Balaraman V., Schust D.J., Ezashi T., Roberts R.M., Franz A.W.E. African and Asian strains of Zika virus differ in their ability to infect and lyse primitive human placental trophoblast. PLoS One. 2018;13:e0200086. doi: 10.1371/journal.pone.0200086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Karvas R.M., Khan S.A., Verma S., Yin Y., Kulkarni D., Dong C., Park K.M., Chew B., Sane E., Fischer L.A., et al. Stem-cell-derived trophoblast organoids model human placental development and susceptibility to emerging pathogens. Cell Stem Cell. 2022;29:810–825.e8. doi: 10.1016/j.stem.2022.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilkinson A.C., Ryan D.J., Kucinski I., Wang W., Yang J., Nestorowa S., Diamanti E., Tsang J.C.-H., Wang J., Campos L.S. Expanded potential stem cell media as a tool to study human developmental hematopoiesis in vitro. Exp. Hematol. 2019;76:1–12.e5. doi: 10.1016/j.exphem.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao H., Lu L., Peng Z., Chen L.L., Meng X., Zhang C., Ip J.D., Chan W.M., Chu A.W.H., Chan K.H., et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microbes Infect. 2022;11:277–283. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan S., Chu H., Huang J., Zhao X., Ye Z.-W., Lai P.-M., Wen L., Cai J.-P., Mo Y., Cao J., et al. Viruses harness YxxØ motif to interact with host AP2M1 for replication: a vulnerable broad-spectrum antiviral target. Sci. Adv. 2020;6:eaba7910. doi: 10.1126/sciadv.aba7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yuan S., Wang R., Chan J.F.-W., Zhang A.J., Cheng T., Chik K.K.-H., Ye Z.-W., Wang S., Lee A.C.-Y., Jin L., et al. Metallodrug ranitidine bismuth citrate suppresses SARS-CoV-2 replication and relieves virus-associated pneumonia in Syrian hamsters. Nat. Microbiol. 2020;5:1439–1448. doi: 10.1038/s41564-020-00802-x. [DOI] [PubMed] [Google Scholar]
- 91.Yuan S., Yin X., Meng X., Chan J.F.-W., Ye Z.-W., Riva L., Pache L., Chan C.C.-Y., Lai P.-M., Chan C.C.-S., et al. Clofazimine broadly inhibits coronaviruses including SARS-CoV-2. Nature. 2021;593:418–423. doi: 10.1038/s41586-021-03431-4. [DOI] [PubMed] [Google Scholar]
- 92.Rostovskaya M., Andrews S., Reik W., Rugg-Gunn P.J. Amniogenesis occurs in two independent waves in primates. Cell Stem Cell. 2022;29:744–759.e6. doi: 10.1016/j.stem.2022.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haghverdi L., Büttner M., Wolf F.A., Buettner F., Theis F.J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 95.Halko N., Martinsson P.-G., Tropp J.A. Finding structure with randomness: probabilistic algorithms for constructing approximate matrix decompositions. arXiv. 2009 doi: 10.48550/arXiv.0909.4061. Preprint at. 0909.4061. [DOI] [Google Scholar]
- 96.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-learn: machine learning in Python. J.Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNA-seq data generated in this study have been deposited at NCBI Gene Expression Omnibus (GEO): GEO: GSE190432.
-
•
All original code has been deposited (https://github.com/leeyoyohku/EPSC_COVID) and is publicly available as of date of publication.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead contact upon request.
-
•
Additional Supplemental Items are available from Mendeley Data: https://doi:10.17632/rnsbwdr7gw.2.