Abstract
A single mutation from aspartate to glycine at position 614 has dominated all circulating variants of the severe acute respiratory syndrome coronavirus 2. D614G mutation induces structural changes in the spike (S) protein that strengthen the virus infectivity. Here, we use molecular dynamics simulations to dissect the effects of mutation and 630-loop rigidification on S-protein structure. The introduction of the mutation orders the 630-loop structure and thereby induces global structural changes toward the cryoelectron microscopy structure of the D614G S-protein. The ordered 630-loop weakens local interactions between the 614th residue and others in contrast to disordered structures in the wild-type protein. The mutation allosterically alters global interactions between receptor-binding domains, forming an asymmetric and mobile down conformation and facilitating transitions toward up conformation. The loss of salt bridge between D614 and K854 upon the mutation generally stabilizes S-protein protomer, including the fusion peptide proximal region that mediates membrane fusion. Understanding the molecular basis of D614G mutation is crucial as it dominates in all variants of concern, including Delta and Omicron.
Significance
The emergence of new variants of the severe acute respiratory syndrome coronavirus 2 continues to threaten global efforts to stop the pandemic despite the development of several vaccines and drugs. Mutations in spike (S) protein are central to the variants and might increase the viral infection rate. The mechanism that underlines various mutations and how they might alter the S-protein structure and viral infectivity is still illusive. Using all-atom molecular dynamics simulations, we elucidate structural changes in S-protein induced upon the D614G mutation. The loss of the anionic charge upon the mutation affects S-protein structure locally and globally. This includes the 630-loop rigidification, N-terminal subunit-outward rotation, and the break of symmetry in receptor-binding domains, which facilitate the transition to the angiotensin-converting enzyme-2-accessible up conformation. Dissecting the structural effects of the dominant mutation would impact the way that we target new severe acute respiratory syndrome coronavirus 2 variants.
Introduction
The emergence of new variants of the severe acute respiratory syndrome coronavirus 2 continues to threaten the ongoing efforts to stop the pandemic (1,2,3). Variants of concern usually show an enhanced virus fitness either by altering transmissibility and/or severity rate as well as their potential to evade natural/vaccine-acquired immunity, reducing the efficacy of the currently deployed vaccines (1,4). As of November 2022, several variants of concern have been reported so far (e.g., B1.1.7 [Alpha], B.1351 [Beta], B 1.1617 [Delta], and B.1.1.529 [Omicron]), while several others are continuously under investigation (e.g., Omicron BQ.1 and XBB) (1,4,5,6,7,8). Many of these variants share the presence of a limited number of advantageous mutations, while others might represent an antigenic drift (e.g., the Omicron variant has over 30 mutations in spike [S] protein) with potential serious consequences (9). Most of the functionally significant mutations are limited to specific variants, such as E484K and N501Y in Alpha, Beta, and Gamma, while L452R and T478K are in different classes of Delta and Omicron (1,6,8). In contrast, the D614G mutation is more universal in variants and has dominated all circulating strains worldwide within a month (10,11,12).
Mutations in S protein are at the heart of the severe acute respiratory syndrome coronavirus 2 variants’ evolution (13,14). S protein is an octadomain homotrimeric glycoprotein that decorates the virus surface, playing a central role in its cycle (15,16,17). It is formed of two subunits connected by a furin cleavage site (15,18). This includes the N-terminal subunit (S1), which mediates the virus binding to the host cell angiotensin-converting enzyme-2 (ACE2), and the C-terminal subunit (S2), which mediates membrane fusion upon cleavage from S1 (19,20,21). S1 consists of four domains, the N-terminal domain (NTD), the receptor-binding domain (RBD), and two subdomains (SD1 and SD2). The RBD undergoes an essential conformational change from ACE2 inaccessible (down) to ACE2 accessible (up), initiating the cell entry (20,21). Mutations in S protein have been reported in both subunits, whereas the majority of variations occur in S1 (1,4,13,14). This includes 1) RBD mutations such as N501Y, E484K, and L452R, with enhanced ACE2 and/or reduced antibody binding affinities (4,6,7,22); 2) NTD mutations/deletions (e.g., H69-, H70-, and R158G) with direct effects on NTD antibody binding (5,23); and 3) distal mutations in SD1 and SD2 with allosteric effects on the RBD structure and motion, such as the D614G and A570D mutations, which shift RBD down/up populations (12,24,25).
The D614G mutation is one of the early spotted dominant mutations in S1, as of March 2020 (10). It occurs as a result of a single nucleotide mutation (A to G) at the 23,403 position in the original Wuhan strain (26). G614 has become globally dominant and currently occurs in all variants of concern and interest including Alpha, Beta, Gamma, Delta, Lambda, Mu, and even Omicron, suggesting its convergent evolution and central role (27). Several studies investigated the effect of this mutation on the viral infectivity, severity, neutralization, and S-protein structure (2,11,12,24,28,29,30,31,32). Notably, D614G shows an enhanced transmissibility and infectivity rate not only in different cell lines but also in different species, which have been correlated to higher viral load (12). The mutation is neither associated with higher mortality rate nor reduced neutralization (27). The effect of mutation is mainly exerted at the membrane fusion step, where G614 S-protein (S-G614) conformational equilibrium is shifted toward the ACE2 accessible (up) conformation, as reported in several studies (24,28,29,32,33). However, contrasting reports exist on how the D614G mutation affects ACE2 binding and whether it causes weakened or stronger binding (11,30). The mutation has also been found to increase S-protein stability, where it prevents premature cleavage of the two subunits before ACE2 binding, while it enhances the cleavage upon binding (11,29).
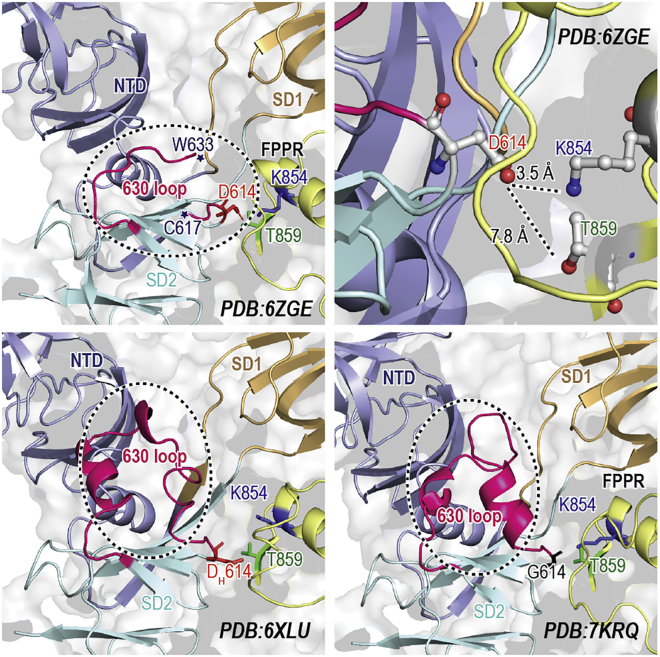
Cryoelectron microscopy (cryo-EM) studies on S-G614 have revealed several structural differences with respect to wild type (S-D614). This includes an outward rotation in S1 (away from S2), a more flexible RBD, a flexible down conformation, and slightly open conformations even in down (24,29,32). Likewise, a recent single-molecule Förster resonance energy transfer study suggests the formation of partially open or widely open conformations (34). Consequently, several mechanisms were proposed to explain structural basis of the abovementioned allosteric effects. Originally, such structural effects were attributed to break of a hydrogen bond with the adjacent S2 protomer Thr859, based on a wild-type cryo-EM structure (10). Although several studies advocated such hypothesis (28,33), this proposal was later challenged by suggesting the formation of a salt bridge with Lys854 as shown in wild-type cryo-EM structures (Fig. 1) (24,32). Note that D614 is located in SD2 at the interface with the adjacent protomer fusion peptide proximal region (FPPR); FPPR mediates membrane fusion after proteolysis at the TMPRSS2 cleavage site (29,32). Likewise, D614 is neighbored to a generally unresolved loop region, in the majority of S-protein cryo-EM structures, known as the 630 loop (Fig. 1). Notably, Zhange et al. (32) sheds more light on structural changes upon D614G mutation, showing the rigidification of this loop in S-G614. This loop, then, inserts into a gap between NTD and SD1, forming multiple hydrophobic contacts (Fig. 1). The authors also suggested that the insertion is hindered in S-D614 due to a smaller gap, forming a disordered 630 loop. In contrast, the rigidification was also observed in the wild-type S-D614 at low pH (D614 neutral), where it forms a helical structure (Fig. 1) (35). Although D614G has been extensively studied, the link between the mutation and structural changes are still elusive. Here, we use classical molecular dynamics (MD) simulations to scrutinize the atomistic basis on D614G mutation, ordered versus disordered 630 loops, and their structural ramification on S-protein conformation, stability, and, subsequently, infectivity.
Figure 1.
Ordered and disordered 630 loops in cryo-EM structures. Top: cartoon representations of the disordered 630 loop and D614 potential interactions in the S-D614 (PDB: 6ZGE). Bottom: cartoon representations of the ordered 630 loop observed in S-D614 at pH 4 (PDB: 6XLU) and S-G614 (PDB: 7KRQ). Three important residues, namely D614, K854, and T859, are highlighted and shown in the ball-and-stick representation in red, blue, and green, respectively. G614 is shown in black. The 630 loop is highlighted using a dotted black circle. To see this figure in color, go online.
Materials and methods
The wild-type S-D614 cryo-EM structure (PDB: 6ZGE) (36) was used as the initial model for all simulations as it has a 2.6 Å resolution and only lacks a small number of residues per protomer. The missing residues, namely 71–75, 677–688, and 941–943, were modeled using modeller 9.19 program (37). In addition, the 630 loop was partly resolved, with the only missing residues between C617 and W633. Consequently, the missing region was modeled as a flexible loop only in the D614loop simulation (disorder 630 loop). In all other simulations with an ordered 630 loop, the residues 610–650 were inserted from the wild-type cryo-EM structure at pH 4.0 (PDB: 6XLU) (35) upon protomer alignment with PDB: 6ZGE using the VMD program (38). The protonation state of D614 and mutation to G614 was performed using CHARMM-GUI (39). Like our previous study, 18 N-glycans and 1 O-glycan were added per protomer based on previous experimental and computational studies (40,41,42). The full list of added glycans is found in Fig. S2 of our previous study (42). In addition, 14 disulfide bonds were modeled in each protomer. CHARMM-GUI was also used to make the final model by adding 0.15 NaCl and solvation box. Four simulation models were made as listed in Table 1. The total numbers of atoms in the simulation models, D614SS, G614SS, DH614SS, and D614loop, are 652,308, 652,047, 652,266, and 652,371, respectively. Their average box lengths are 186.113, 186.228, 186.167, and 186.217 Å, respectively.
Table 1.
List of models and simulations performed
All simulations were performed using GENESIS 2.0 beta MD software on a Fugaku supercomputer (43,44) with an overall average performance of 55 ns/day using 128 nodes. Two independent simulations for 1 each were performed for each system with a total simulation time of 8 Protein and glycans were parametrized using the CHARMM 36m force field, while CHARMM TIP3P was used for water molecules (45). All simulations were first minimized for 5,000 steps while applying positional restraints of 1 kcal mol−1 Å−2 on the protein backbone and weak restraint of 0.1 kcal mol−1 Å−2 on all heavy atoms. Then, all systems were gradually heated to 310 K using the velocity Verlet integrator and stochastic velocity rescaling thermostat (46,47,48). Subsequently, all simulations were equilibrated in a step manner: 1) in an NVT ensemble using same integrator and thermostat; 2) in an NPT ensemble with stochastic velocity rescaling thermostat and MTK barostat (47); 3) in an NVT upon after removing all restraints; and 4) with a multiple time-step integrator with time steps of 2.5 and 5 fs for the fast and slow motions, respectively (49,50). Finally, a production run was performed in the NVT using a multiple time-step integrator and stochastic velocity rescaling thermostat. The first 200 ns of each production run was excluded from analysis since it was considered as a part of the equilibration process. Smooth particle mesh Ewald (51) was used to compute electrostatic interactions with 128 × 128 × 128 grids and the sixth-order B-spline function. The group-based approach was used to evaluate temperature, where the thermostat was applied every 10 steps (49). Bonds involving hydrogen atoms and water molecules were constrained with the SHAKE/RATTLE algorithm (52).
Simulation trajectories was analyzed using GENESIS 1.6 analysis tools. Principal component analysis (PCA) was performed using all the available cryo-EM S-protein structures with down conformation in the PDB released by the end of September 2021. Structures that include other molecules, such as neutralizing antibodies, were excluded from the selection to avoid any induced conformational changes due to the binding. Also, structures that lack resolutions at the regions involved in the coarse-grained (CG) model preparation (discussed below) were also excluded. A total of 52 structures were selected for PCA including 156 protomers. Similar to our previous work, we represent all selected structures using a CG beads model, while we use 11 beads instead of 9 per protomer to represent S2 better. Our model consists of two beads for the RBD (core and top), three beads for the NTD (core, base and top), one bead for SD1 and SD2 each, and four beads for S2. We performed PCA for both the monomeric structure (11 beads) and the trimeric one (33 beads) using the selected 156 and 52 structures, respectively. Root-mean-square deviation (RMSD) analyses on S1 were performed using only the Cα atoms upon fitting S2 (residues 689–827 and 854–1134). Likewise, only rigid regions in the RBD (328–444, 462–468, 489–501, 503–533) and the NTD (14–69, 80–143, 165–172, 186–245, and 263–306) were used for the analysis. Stride in VMD was used to assign secondary structures in the 630-loop region. Average interresidue contact was also calculated with the iTRAj plugin in VMD (38). Electrostatic potential was performed with ABPS in PyMOL (53). Solvent-accessible surface areas were also calculated using PyMOL with a probe radius of 7.2 Å. The distance between the two RBDs, the hinge angle, and solvent-accessible surface area (SASA) analyses were performed using the last 200 ns of the simulation to characterize changes after the conformational shift of S protein. The definition of the hinge angle is the same as that in our previous work. All structural figures were made using PyMOL (53), and VMD was used for trajectories visualization (38). In contact analysis, a “contact” pair is defined as those with an atomic distance less than 2.5 Å, and the selected pairs with a 30% occurrence threshold in any protomer are discussed.
Results and discussion
MD simulations of three S-D614 and one S-G614 systems (Table 1; Fig. S1) were carried out. These systems include disordered 630 loops (in D614loop) or an ordered loop (in D614SS, DH614SS, and G614SS) as well as different types of the 614th residue, namely anionic Asp (in D614loop and D614SS), neutral Asp (in DH614SS), and Gly (in G614SS). Taking advantage of the trimeric nature of S protein, we analyzed six protomers per system from two independent 1 simulations; summing up to simulation data corresponds to 6 per system (24 in total). In the main text, we discuss the comparison of one of the replicas between different molecular models as for the 614th residue and the 630-loop structure. Due to the limitation of conformational sampling in classical MD simulations, two replicas in each system do not show completely identical results, including monomer and trimer PCA projections (Figs. 2 and S2), RMSD analysis (Fig. S3), RMS fluctuation (RMSF) analysis (Figs. 4 d and S12), and analysis of main distances (Fig. S7). However, the general trend in the effect on the mutation and the 630-loop structure is kept in the replicas. It is also difficult to obtain completely the same results for each protomer in each simulation, although the same molecular model was used. This is also caused by the insufficient conformational sampling. To understand molecular mechanisms upon the mutation and relate one feature to another, the variation of results in each protomer might be useful instead of performing significantly longer MD simulations.
Figure 2.
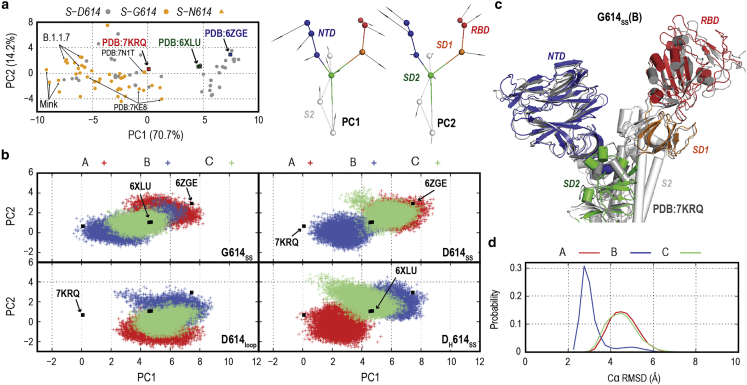
D614G mutation and ordered 630 loop drive S-protein structural changes toward a S-G614 cryo-EM-like structure. (a) PCA for 156 protomers from 52 S-protein cryo-EM structures with down form using the CG model with 11 CG beads per protomer. The lowest two modes (PC1 and PC2) representing an outward motion of S1 from S2 are shown. The projections of S-D614 and G-614 cryo-EM structures on the two-dimensional PC map are shown in gray and orange circles, respectively. Three important PDB structures, S-D614 (PDB: 6ZGE), S-D614 at low pH (PDB: 6XLU), and S-G614 (PDB: 7KRQ), are highlighted with blue, green, and red boxes, respectively. (b) Projections of MD simulations (run 1) for the four systems, G614SS, D614SS, DH614SS, and D614loop, on the first and second PC maps. Different protomers are shown as A, B, and C. Projections of other MD simulations (run 2) are shown in Fig. S2a. (c) A cartoon representation of the superposition of S-G614 protomer cryo-EM structure (shown in white) with the last structure from the G614SS simulation. Only protomer B is shown upon fitting S2, wherein the NTD, RBD, SD1, SD2, and part of S2 are colored in red, blue, orange, green, and gray, respectively. (d) Probability distribution of root-mean-square deviation of part of S1 (NTD, RBD, and SD1) upon fitting S2, with respect to S-G614 cryo-EM structure (PDB: 7KRQ). To see this figure in color, go online.
Figure 4.
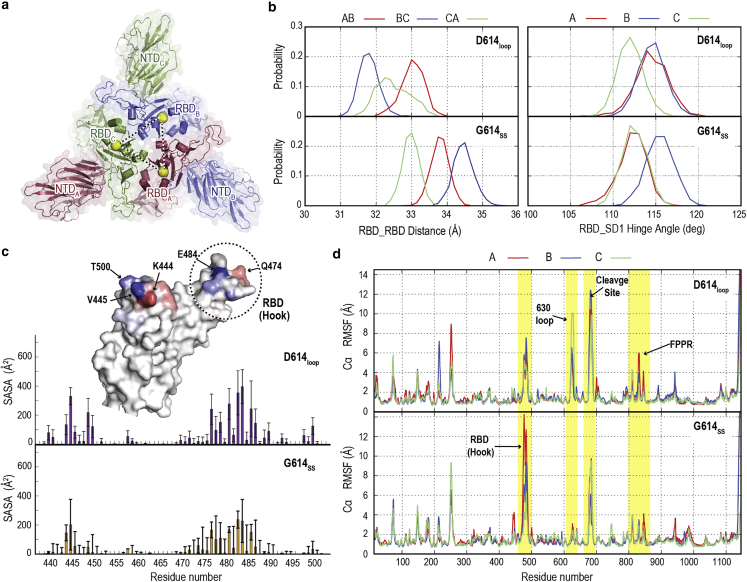
Effect of mutation on protein stability and dynamics. (a) Top view of S protein showing the organization of RBDs and NTDs in cartoon and surface representations. The center of mass of the RBD is highlighted by a yellow sphere. (b) Left: interdomain RBD center-of-mass distance probability distribution in the last 200 ns of the S-G614SS and S-D614loop. Right: RBD hinge angle with respect to SD1. (c) SASA per residue the RBM (residues 410–510) in the absence of glycan of S-D614loop (top) and S-G614SS (bottom), which was calculated using a probe radius of 7.2 Å. The surface representation shows the difference in RBM SASAs between S-G614 and S-D614 in the presence of glycan. Colors from blue to red indicate the difference in SASA from −100 and 100 (). Residues with large differences are highlighted with black arrows. The RBD hook region is also highlighted by a dash black circle. (d) Root-mean-square fluctuation in S-protein protomers (residues 14–1146) of the S-D614loop (top) and S-G614SS (bottom). Four regions with large changes in root-mean-square fluctuation are highlighted by yellow shades. They include the RBD hook, 630 loop, furin cleavage site, and protomer fusion peptide proximal region. To see this figure in color, go online.
The effects of 630-loop secondary structure on S-protein conformation
To assign induced conformational changes by the mutation, we utilize the vast number of S-protein cryo-EM structures, guided by the work of Henderson et al. (54) and our previous study (42). We constructed a CG model with 11 beads per protomer (Fig. 2 a; Table S1) of the 52 S-protein three RBD down structures (156 promoters). Next, we performed PCA for the monomeric (11 beads) as well as trimeric (33 beads) models. Fig. 2 a shows that the first and second PCs both represent an outward motion of S1 with respect to S2, with a total contribution of over 84% in the observed motions. Similar PCs were also obtained for the 33-bead trimeric model (Fig. S2 b). The projection of the 156 protomers on the PC1-PC2 map in Fig. 2 a roughly separates S-G614 (orange) from S-D614 (gray) structures along PC1, though some protomers of S-D614 are found in the S-G614 distribution. Notably, the PDB structures of S-D614 with a disordered 630 loop (PDB: 6ZGE) (36) and those with an ordered loop (PDB: 7KRQ) (32) are distinctive, while S-D614 at low pH with an ordered loop (PDB: 6XLU) (35) lays in the middle between them. They are separated similarly in the trimeric PCA (Fig. S2 c).
The last 800 ns MD simulations of all six promoters per system were projected along the same PC1-PC2 space (Figs. 2 b and S2 a). A clear difference can be seen between the simulations of D614SS, G614SS, and DH614SS (start from an ordered 630 loop) and D614loop (from a disordered 630 loop), where D614loop projection overlaps only with wild-type cryo-EM structures. In contrast, all the other simulations (D614SS, G614SS, and DH614SS) show projections toward the S-G614 structures. Projections of two G614SS protomers (Chains B and C) cover the objective S-G614 (PDB: 7KRQ). In Fig. 2 c, the final structure of G614SS (B) aligns almost perfectly with the cryo-EM structure (PDB: 7KRQ) upon fitting S2. RMSD analysis of individual and combined S1 domains suggests the differences of structure ensembles between the wild-type and mutant structures (Figs. 2 d and S3). Few protomers in D614SS (chain B) and DH614SS (chain A) could cover the S-G614 structure (Figs. 2 b and S3). PCA for the trimeric model (Fig. S2 d) shows more clear differences upon the D614G mutation, where only G614SS samples toward the cryo-EM structure (PDB: 7KRQ) in both runs, while D614SS and DH614SS projections only align with wild-type structure.
The predicted outward motion in S1 was further analyzed via the interdomain angles in S1, which is defined using the center of mass of adjacent domains, namely SD1, SD2, and the base of NTD (NTD(b)). Fig. S4 shows that the angle increases in all simulations with an ordered 630 loop (D614SS, G614SS, and DH614SS). The angle in D614loop distributes between 67 and 85, while D614SS and G614SS show angle distributions between 74 and 90. Note that in G614SS, chain B also shows the largest shift, which is in consistent with the monomeric PCA. We also calculate the angle between nonadjacent domains, namely the RBD, SD2, and the core of NTD. Even using this definition, increases of the angles in D614SS, G614SS, and DH614SS (with ordered 630 loops) could be seen. Fig. S5 a shows the interdomain angles in cryo-EM structures, wherein G614 conformations have an Sd1_SD2_NTD(b) angle larger than 84, while D614 shows a much wider distribution with the lowest minimum around 80 Fig. S5 b shows the angle distributions obtained from all MD simulations, which align with PCA results in Fig. 2 b. The D614loop has a distinct distribution from the other simulations with a rigidified 630 loop.
The 630-loop rigidification drives an insertion and the interdomain angle in S1 increases (Fig. S4) regardless of the nature of 614 residue. The motions do not necessary shift the S-protein conformation to a mutant one (Fig. S3). Trimeric PCAs (Fig. S2 d) may suggest that both an ordered 630 loop and a D614G mutation are required to reproduce key features of global structure in S-G614. To a lesser extent, the 630-loop rigidification in S-D614 could drive changes in one protomer (Fig. 2 b), raising the question of if the loop rigidification could happen in S-D614. A previous study suggested that an ordered loop is not observed in wild-type S-D614 due to a limited gap between the NTD and SD1 (32). In contrast, our simulation suggests the formation of this gap as a consequence of the 630-loop rigidification. In addition, projection of S-D614 cryo-EM structures shows a wider distribution along PC1, which might suggest that the mutation shifts the conformational ensemble to specific orientations.
To study the effects of D614G mutation and the 630 loop on structural changes, we first focus on the two protomer Bs in G614SS (in Fig. 2 b) compared with the D614SS structure. Fig. S6 a shows that the loop insertion in D614SS breaks the backbone hydrogen bonding between the linker regions connecting the NTD to SD1 (residues 315–321) and SD1 to SD2 (residues 590–595). In contrast, protomer B in G614ss maintains such interactions. Fig. S6 b emphasizes this result. The breaking of hydrogen bonding was found to be directly correlated to the formation of stronger hydrophobic interactions between 630-loop and NTD_SD1 linker regions, as shown in Fig. S6 c and d. Such a stronger 630-loop/linker interaction was correlated to the formation of a sharp angle in the 630 loop nearby the mutation site (Fig. S6 c and e). Notably, the protomer in G614SS, which lacks hydrogen-bonding interactions between linker regions, is the only protomer that shows the formation of a sharp angle in the 630 loop and, subsequently, strong hydrophobic interactions between 630-loop and linker residues.
To date, not much consideration has been given to the modeling of the missing 630-loop conformation and its effect on global S-protein structure. In the previous models, the loop was simply treated as helical or disordered (40,42,55). Ironically, the modeling of such a small region (less than 30 residues) has a great effect on the S-protein structures and motions, suggesting that more careful modeling of S variants is required, especially if the mutation site is located near an experimentally unresolved region.
Order versus disorder 630 loop in S-D614
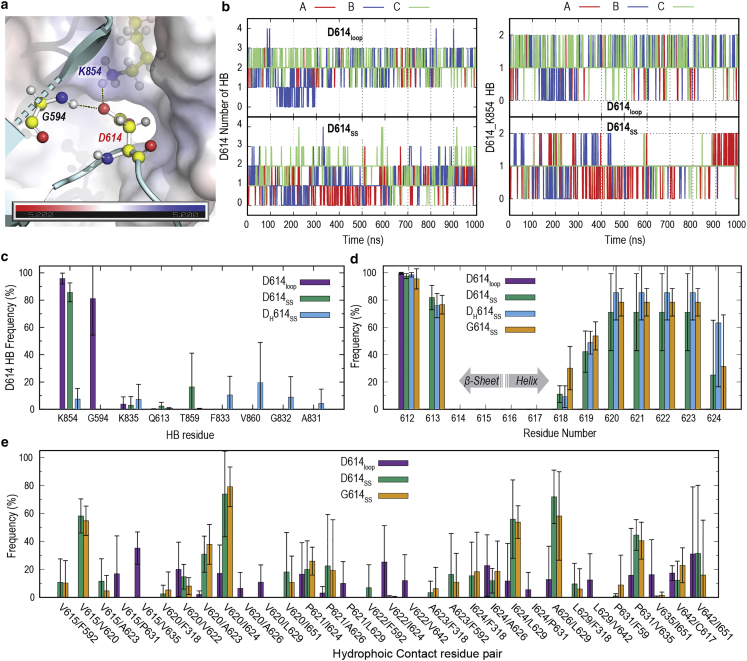
The correlation between D614G mutation and 630-loop rigidification as well as the possibility of secondary structure formation in the 630 loop were uncertain. It was also unclear how the presence of the anionic residue (D614) affects the loop conformation nearby. To answer these questions, we compare simulation results of S-D614 with ordered (D614SS) and disordered (D614loop) 630 loops. Structural and electrostatic potential analyses show that D614 is located at the interface of a hydrophobic pocket (Fig. 3 a) with one charged residue in its vicinity (K854). D614 was previously proposed to be stabilized by H-bonding with T859 based on a cryo-EM structure (PDB: 6VSB) (10,28). However, the PDB structure shows that the orientation of both residues does not reflect direct interaction. Fig. 3 a also shows that, in PDB: 6ZGE, D614 forms two possible hydrogen bonds (H-bonds): one with K854 and the other with the main chain of G594. Accordingly, we first analyze the total number of H-bonds that could stabilize the side chain of anionic D614. Figs. 3 b and S8 signify differences of D614 stabilization in the presence of an ordered (D614SS) and disordered 630 loop (D614loop), where the loop rigidification reduces the total number of H-bonds in all six protomers in D614SS simulations. Similarly, the probability of H-bond formation between D614 and K854 is also reduced in D614SS. Note that the salt bridge between D614 and K845 was formed just after the equilibration regardless of the 630-loop structure. The weakening of the D614 interaction with K854 is reflected in their C distance (Fig. S7 a), which increases from 8.5–11.5 Å in D614loop to 7–13 Å in D614SS. Note that the C distances further increased in G614SS upon the mutation. Fig. 3 c shows the H-bond formation of a D614 side chain with any residues in MD simulations. D614loop reflects the dominancy of two main H-bonds with K854 and G594, in agreement with the cryo-EM structure (Fig. 3 a). In contrast, the formation of secondary structure in the 630 loop of D614SS diminishes the interaction with G594, while the interaction with K854 is maintained with a lesser number of H-bonds (Figs. 3 b and S8). Such a reduction in stabilization might originate from the reorientation of D614 due to loop rigidification. The interaction between D614 and K854 was drastically reduced in neutral D614 in DH614SS. Only few H-bond partners are identified (in Figs. 3 c and S8). In summary, our data show that the anionic D614 is better stabilized in the presence of a disordered 630 loop, where rigidification weakens its interactions.
Figure 3.
Contacts and secondary structure analysis in 630 loop. (a) An electrostatic potential map of D614-adjacent protomer interface in S-D614 (PDB: 6ZGE). Three residues are highlighted as balls and sticks with yellow carbon atoms including G594, D614, and the adjacent protomer K854. D614 hydrogen bonding and salt bridge are shown as black dash lines. (b) Time series of D614 side-chain total number of H-bonds (left) and the salt bridge between D614 and K854 (right) in all protomers from MD simulations (run 1). Results from MD simulations (run 2) are shown in Fig. S8. Three protomers are shown as A, B, and C in red, blue, and green, respectively. Top and bottom represent S-D614 with an ordered and disordered 630 loop, respectively. (c) Hydrogen-bonding formations with the 614th residue in the MD simulations of D614SS, D614loop, and DH614SS. (d) Secondary structure formations (α-helix and β-sheet) in residues near to the mutation point (residues 612–625) in all simulated systems. Fig. S13 shows probability of helicity per protomer. (e) Hydrophobic contacts in residues from V615 to V642 in the MD simulations of D614SS, D614loop, and G614SS. All reported values in (c)–(e) are the averages of six protomers (three protomer × two runs), while error bars represent the standard deviations. To see this figure in color, go online.
Although unfolding of the 630 loop is beyond the scope of this study, we compare the relative stability of the helical structure adjacent to the mutation site. Fig. 3 d illustrates that the stability of the helical structure between residues V620 and A623 might be reduced in D614SS, as indicated by an average of 71.1% with large standard deviations (SDs) of 28.2%, with a minimum probability of 25% in protomer B2. The protonation of D614 (DH614SS) increases this average to 85.4% while reducing the SD to 20. Notably, the D614G mutation further enhances the stability in the same region considering the reduction of SD to 10 with an average of 78.4% and a minimum population of 66.1%. Indeed, a much longer simulation and a probably enhanced sampling approach are necessary to test secondary structure formation in both wild-type and mutant structures. Furthermore, all three systems with an ordered loop (D614SS, DH614SS, and G614SS) show the extension of the adjacent -sheet to include Q613. Such extension is diminished in D614loop, probably due to the kink region formed by a stronger salt bridge between D614 and K845 (Fig. 3 a) as well as an interaction with G594. These results align with the experimental observation of the shortened distance between G614 and A647 to 2.7 Å in the S-G614 cryo-EM structure (PDB: 6XS6) (28), indicating the elongation of the -sheet.
The 630-loop rigidification in S-G614 was previously suggested, in part, due to the formation of hydrophobic interactions upon the insertion between SD1 and NTD (32). In fact, the 630-loop region (615–642) is highly hydrophobic and is formed of five Val, two Pro, two Ala, Leu, Ile, and Trp residues. Fig. 3 e shows hydrophobic contacts between one of the residues in the 630 loop with rest of the S protein, wherein a switch of interactions is observed upon the change from disordered to ordered loop. For instance, V615 interacts with V635 in D614loop, while it interacts with V620 in D614SS. The shift in the interactions aligns with the calculated C distance between V615 and V635, which increases the average distances from 8 Å in D614loop to over 16 Å in all other systems with an ordered loop (Fig. S7 b). The formation of an ordered 630 loop forms several interactions including V620/V624, A626/L629, I624/L629, and P631/V635, as well as interactions with NTD(b), SD1, and the linker region reflecting a loop insertion. No significant difference is observed comparing the results in D614SS with G614SS (Figs. 3 e and S9). Thus, the formation of hydrophobic contacts is a consequence of 630-loop rigidification and not directly related to the mutation. In fact, the disordered 630 loop also shows the formation of fluctuating hydrophobic interactions, which could compensate for stability.
A comparison of H-bonding, hydrophobic interactions, and secondary structure stability suggests that structural changes mainly originate from the breaking of the salt bridge between D614 and K854 upon the mutation. In the Wild-type S-D614 the formation of a flexible 630 loop is preferred to stabilize the anionic charge of D614. The salt bridges between D614 and K854 and between D614 and G594 play important roles in forming a kink structure around Q613. This hinders the formation of a -sheet and, subsequently, allows for a different pattern of hydrophobic interactions, including those between V615 and V635. In contrast, the D614G mutation sets the loop region free from this interaction, extending the -sheet to include Q613, which increases the distance between V615 and V635, leading to the formation of different forms of hydrophobic contacts. In addition, the loss of interaction between D614 and K854 sets the loop region free, as indicated by their C distance, which permits an insertion between SD1 and the NTD. Our results explain the possibility of observing an ordered loop in S-D614 at low pH (35) due to the breaking of the salt bridge. This hypothesis can be confirmed experimentally upon mutating the K854 in wild type or by reducing the hydrophobicity at the D614 S2 interface.
Structural ramifications of D614G mutation on RBD
Two main mechanisms have been proposed to explain superior transmission rate in D614G, which includes a regulated shedding mechanism of S1/S2 depending on the absence or presence of ACE2, and the shift in conformational equilibrium toward the RBD up (29,32). To understand the allosteric effect of the mutation, we compare our simulations results of wild type with disordered loop (D614loop) with G614SS. Fig. 4 a shows that a change in one RBD due to a mutation is expected to alter the neighboring RBD organization. In Fig. 4 b, we calculate the center-of-mass distances between all three RBDs. The interdomain RBD distances were increased in S-G614, wherein RBD (B) shows the largest distances. Furthermore, the hinge angle defined by RBD with respect to SD1 in G614SS reflects a higher hinge angle in one of the RBDs compared with wild type (D614loop). These results suggest that a S-G614-like structure leads to more spacing between asymmetric RBDs, which forms a slightly more open conformation, in agreement with the cryo-EM structure (32). Fig. S10, a and b, shows that a similar effect is observed upon the rigidification of the 630 loop in S-D614 (D614SS) but to a lesser extent, supporting our finding that S-D614 would prefer a flexible loop.
The effect of RBD rearrangement on the exposure of the receptor-binding motif (RBM; residues 410–510) and glycans covered in down was examined. In the presence of glycan, the RBM average SASA value was slightly reduced from 1,635.2 Å2 in D614loop to 1,530.9 Å2 in G614SS with SDs of 225.7 and 246.8, respectively. Similarly, in the absence of glycan, the total SASA value was reduced more significantly, from 3,812.3 to 3,576.4 Å2 with SDs of 243.1 and 460.8, respectively. This difference was scrutinized by calculating the SASA per residue, which indicates an overall reduction in the absence of glycan (Fig. 4 c). Plotting the difference in the accessibility on the RBM surface reflects a shift of residue exposure due to the RBD rearrangements. K444 has become relatively more solvent exposed in S-G614, while V445 has higher accessibility in S-D614. Furthermore, E484, an important point of mutation in S variants, is found to be less exposed in G614SS. Comparison of SASA values in the absence of glycans (Fig. 4 c) clearly shows large differences in accessibility between G614SS and D614SS, especially in residues between 470 and 503, suggesting that the RBD rearrangement can alter the RBM accessibility in down.
The effect of D614G mutation on S-protein stability is examined by calculating the RMSF in all protomers. G614SS shows an overall stabilizing effect upon the mutation in all domains (Fig. 4 d), in comparison to D614loop. Remarkably, the FPPR has enhanced the conformational stability in S-G614. Despite the ordered loop in D614ss, FPPR has larger fluctuations that suggest general destabilizing effects in S-D614 regardless of the 630-loop conformation. Neutral D614 (DH614SS), increases FPPR stability, confirming the role of salt bridge between D614 and K854 in the observed instability. The furin cleavage site is also found to be relatively stable in G614SS and DH614SS while the presence of anionic D614 shows higher fluctuations. Fig. 4 d also shows that the rearrangement of RBDs loosens its hook region, leading to higher fluctuations and disorders in this region. The disordering of the region has been observed in other S-protein variants of concern that includes D614G (1).
In summary, comparison of wild type with a disordered loop and S-G614 with an ordered 630 loop describes the allosteric effect of the mutation. The 630-loop rigidification and insertion between SD1 and the NTD alter the RBD organization, forming asymmetric down due to their larger RBD-RBD distances. The increase in the RBD-RBD distances allows the formation of a mobile RBD hook region as shown from the RMSF results, which might help initiate the transition to up state. The change of RBD organization was also found to reduce RBM accessibility, as indicated by calculated SASA values in the absence of glycan as well as a shift of residue exposure in the presence of glycans. Finally, RMSF analysis also shows a global effect of the mutation where the loss of a salt bridge stabilizes FPPR.
Protomer B in G614SS, which shows the closest RMSD with respect to the cryo-EM structure of the mutant, indicates that the conversion between wild-type and mutant structures is accompanied by several structural features. This includes the 630-loop insertion between the NTD and SD1 and the increased interdomain angles leading to outward motion in S1 while maintaining linker/linker backbone interactions. The increase of RBD-RBD distances with slightly larger hinge angles might facilitate down to up transition.
The allosteric effect of a D614G mutation alters S-protein conformation at a different level that goes beyond the S1 rotation observed in cryo-EM structures. First, the break of the symmetry and the formation of flexible down is a prerequisite for N343 glycan contact changes that initiate transition from down to up, as suggested previously (42). In addition, a higher disorder in the RBD hook region might also help loosen the interaction between different RBDs and enhance the transition toward up, which aligns with the previous proposal for E484K mutant (1). These results might explain the origin of the observed higher population in up compared with wild type. Likewise, Gobeil et al. (1) previously suggested that the increase in RBD mobility in down reduces the barrier for up transition in B.1.1.17. Second, the rearrangement of the RBD is also found to alter the RBD interface and residue solvent exposure, as indicated by the SASA per residue. Such a reduction in accessibility might partially compensate for the larger exposure to neutralization due to the conformational shift toward up. Indeed, S-G614 was found to be moderately more sensitive to neutralization despite a large shift in the up population (30). Third, our simulations also show the stabilizing effect upon the mutation, especially on FPPR and the furin cleavage site. It also increases conformational stability upon the protonation of D614. Note that the cryo-EM structure also suggests a change from disorder to order upon mutation. Gobeil et al. (1) suggested the regulatory role of the FPPR region and 630-loop order/disorder on S-protein stability and structural rearrangement, based on their cryo-EM study of different variants of concern.
Conclusion
In this study, we performed classical MD simulations to examine the effect of D614G mutation and 630-loop rigidification starting from a wild-type S-protein cryo-EM structure. Projection of all simulations along the cryo-EM-based PC1-PC2 space shows the role of the ordered 630 loop in inducing an outward motion in S1. G614ss simulation shifts the S-protein conformation toward the mutant cryo-EM structure. Analysis of H-bonding patterns in wild type with the ordered and disordered loops indicates a weaker stabilization of anionic D614 in the presence of an ordered 630 loop. Likewise, secondary structure analysis suggests that instability of the 630 loop in the presence of anionic D614 explains the disordered loop in wild-type cryo-EM structures. The loss of the salt bridge between the 614th residue and K854 and the H-bond with G594 mainly causes the observed structural changes in S-G614, wherein an ordered 630 loop inserts between SD1 and the NTD. The loop insertion allosterically reorganizes the RBD arrangements and interactions at the interface, forming a mobile asymmetric down state with a lesser barrier toward up. The breaking of the salt bridge between D614 and K854 alters not only the 630-loop conformation but also has a general stabilizing effect on FPPR and the furin cleavage region, which, in part, explains the experimentally observed stabilized prefusion state in S-G614. In summary, our results dissect the observed structural transition in D614G, showing how a single mutation could have a drastic structural effect. It also points out the importance of careful modeling of S-protein structures in the upcoming emerging variants. Notably, understanding the molecular basis and consequence of mutation is crucial for vaccine and antiviral drug development.
Author contributions
H.M.D. and Y.S. designed and initiated the research. H.D.M. performed simulations and analyzed trajectories. H.M.D. and Y.S wrote the manuscript.
Acknowledgments
This research used computational resources of the supercomputer Fugaku (the evaluation environment in the trial phase) provided by the RIKEN Center for Computational Science. The results obtained on the evaluation environment in the trial phase do not guarantee the performance, power, and other attributes of the supercomputer Fugaku at the start of its public use operation. The computer resources of Oakforest-PACS were also provided through HPCI System Research project (Project IDs: hp200028, hp200135, hp200153, hp210107, hp210177, and hp220087). The research was supported, in part, by MEXT as “Program for Promoting Researches on the Supercomputer Fugaku” (Biomolecular dynamics in a living cell/MD-driven Precision Medicine) (to Y.S.) and JSPS KAKENHI (grant numbers 19H05645 and 21H05249 to Y.S. and 20K15737 to H.M.D.); RIKEN Pioneering Research Projects (Glycolipidologue Initiative/Biology of Intracellular Environments) (to Y.S.); and by RIKEN Incentive Research Projects (the effects of mutations on the structure of spike protein and SARS-CoV-2 infectivity) (to H.M.D.). Finally, we would like to thank Dr. Suyong Re for helpful discussion.
Declaration of interests
The authors declare no competing interests.
Editor: Chris Chipot.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.11.025.
Supporting material
References
- 1.Gobeil S.M.C., Janowska K., et al. Acharya P. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science. 2021;373:eabi6226. doi: 10.1126/science.abi6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniloski Z., Jordan T.X., et al. Sanjana N.E. The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. Elife. 2021;10:e65365. doi: 10.7554/eLife.65365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weisblum Y., Schmidt F., et al. Bieniasz P.D. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey W.T., Carabelli A.M., et al. Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCallum M., Bassi J., et al. Veesler D. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373:648–654. doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J., Xiao T., et al. Chen B. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science. 2021;374:1353–1360. doi: 10.1126/science.abl9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang P., Nair M.S., et al. Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 8.Ulrich L., Halwe N.J., et al. Benarafa C. Enhanced fitness of SARS-CoV-2 variant of concern Alpha but not Beta. Nature. 2022;602:307–313. doi: 10.1038/s41586-021-04342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X., Wu S., et al. He X. SARS-CoV-2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduct. Target. Ther. 2021;6:430. doi: 10.1038/s41392-021-00852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korber B., Fischer W.M., et al. Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L., Jackson C.B., et al. Choe H. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plante J.A., Liu Y., et al. Shi P.Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q., Wu J., et al. Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greaney A.J., Starr T.N., et al. Bloom J.D. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., Xiao T., et al. Chen B. Structure of SARS-CoV-2 spike protein. Curr. Opin. Virol. 2021;50:173–182. doi: 10.1016/j.coviro.2021.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.V'Kovski P., Kratzel A., et al. Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai Y., Zhang J., et al. Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrapp D., Wang N., et al. McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang J., Wan Y., et al. Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan R., Zhang Y., et al. Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q., Zhang Y., et al. Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan M., Huang D., et al. Wilson I.A. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373:818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCallum M., Walls A.C., et al. Veesler D. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science. 2021;374:1621–1626. doi: 10.1126/science.abl8506. [DOI] [PubMed] [Google Scholar]
- 24.Benton D.J., Wrobel A.G., et al. Gamblin S.J. The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2022586118. e2022586118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ray D., Le L., Andricioaei I. Distant residues modulate conformational opening in SARS-CoV-2 spike protein. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2100943118. e2100943118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valdes-Balbin Y., Santana-Mederos D., et al. Verez-Bencomo V. Molecular aspects concerning the use of the SARS-CoV-2 receptor binding domain as a target for preventive vaccines. ACS Cent. Sci. 2021;7:757–767. doi: 10.1021/acscentsci.1c00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C., Zheng Y., et al. Sun Q. The virological impacts of SARS-CoV-2 D614G mutation. J. Mol. Cell Biol. 2021;13:712–720. doi: 10.1093/jmcb/mjab045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yurkovetskiy L., Wang X., et al. Luban J. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gobeil S.M.C., Janowska K., et al. Acharya P. D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2020.108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozono S., Zhang Y., et al. Tokunaga K. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12:848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansbach R.A., Chakraborty S., et al. Gnanakaran S. The SARS-CoV-2 Spike variant D614G favors an open conformational state. Sci. Adv. 2021;7:eabf3671. doi: 10.1126/sciadv.abf3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J., Cai Y., et al. Chen B. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science. 2021;372:525–530. doi: 10.1126/science.abf2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang X., Zhang Z., et al. Sun Q. Bimodular effects of D614G mutation on the spike glycoprotein of SARS-CoV-2 enhance protein processing, membrane fusion, and viral infectivity. Signal Transduct. Target. Ther. 2020;5:268. doi: 10.1038/s41392-020-00392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z., Han Y., et al. Lu M. SARS-CoV-2 variants increase kinetic stability of open spike conformations as an evolutionary strategy. mBio. 2022;13 doi: 10.1128/mbio.03227-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou T., Tsybovsky Y., et al. Kwong P.D. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe. 2020;28:867–879.e5. doi: 10.1016/j.chom.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wrobel A.G., Benton D.J., et al. Gamblin S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 38.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 39.Lee J., Cheng X., et al. Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woo H., Park S.J., et al. Im W. Developing a fully glycosylated full-length SARS-CoV-2 spike protein model in a viral membrane. J. Phys. Chem. B. 2020;124:7128–7137. doi: 10.1021/acs.jpcb.0c04553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe Y., Allen J.D., et al. Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dokainish H.M., Re S., et al. Sugita Y. The inherent flexibility of receptor binding domains in SARS-CoV-2 spike protein. Elife. 2022;11:e75720. doi: 10.7554/eLife.75720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung J., Kobayashi C., et al. Sugita Y. New parallel computing algorithm of molecular dynamics for extremely huge scale biological systems. J. Comput. Chem. 2021;42:231–241. doi: 10.1002/jcc.26450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobayashi C., Jung J., et al. Sugita Y. Genesis 1.1: a hybrid-parallel molecular dynamics simulator with enhanced sampling algorithms on multiple computational platforms. J. Comput. Chem. 2017;38:2193–2206. doi: 10.1002/jcc.24874. [DOI] [PubMed] [Google Scholar]
- 45.Huang J., Rauscher S., et al. MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126 doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 47.Bussi G., Zykova-Timan T., Parrinello M. Isothermal-isobaric molecular dynamics using stochastic velocity rescaling. J. Chem. Phys. 2009;130 doi: 10.1063/1.3073889. [DOI] [PubMed] [Google Scholar]
- 48.Jung J., Kobayashi C., Sugita Y. Kinetic energy definition in velocity Verlet integration for accurate pressure evaluation. J. Chem. Phys. 2018;148 doi: 10.1063/1.5008438. [DOI] [PubMed] [Google Scholar]
- 49.Jung J., Sugita Y. Group-based evaluation of temperature and pressure for molecular dynamics simulation with a large time step. J. Chem. Phys. 2020;153 doi: 10.1063/5.0027873. [DOI] [PubMed] [Google Scholar]
- 50.Tuckerman M., Berne B.J., Martyna G.J. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992;97:1990–2001. doi: 10.1063/1.463137. [DOI] [Google Scholar]
- 51.Essmann U., Perera L., et al. Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 52.Miyamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. doi: 10.1002/jcc.540130805. [DOI] [Google Scholar]
- 53.Schrodinger, LLC The PyMOL Molecular Graphics System, Version 2.4, Schrodinger, LLC.
- 54.Henderson R., Edwards R.J., et al. Acharya P. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27:925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sztain T., Ahn S.H., et al. Amaro R.E. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat. Chem. 2021;13:963–968. doi: 10.1038/s41557-021-00758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.