Graphical abstract
Keywords: Antiviral, COVID-19, Physicochemical properties, SARS-CoV-2 Mpro, Molecular docking, Molecular dynamics
Abstract
COVID-19 and associated substantial inflammations continue to threaten humankind triggering death worldwide. So, the development of new effective antiviral and anti-inflammatory medications is a major scientific goal. Pyranopyrazoles have occupied a crucial position in medicinal chemistry because of their biological importance. Here, we report the design and synthesis of a series of sixteen pyranopyrazole derivatives substituted with two aryl groups at N-1 and C-4. The designed compounds are suggested to show dual activity to combat the emerging Coronaviruses and associated substantial inflammations. All compounds were evaluated for their in vitro antiviral activity and cytotoxicity against SARS-CoV infected Vero cells. As well, the in vitro assay of all derivatives against the SARS-CoV Mpro target was performed. Results revealed the potential of three pyranopyrazoles (22, 27, and 31) to potently inhibit the viral main protease with IC50 values of 2.01, 1.83, and 4.60 μM respectively compared with 12.85 and 82.17 μM for GC-376 and lopinavir. Additionally, in vivo anti-inflammatory testing for the most active compound 27 proved its ability to reduce levels of two cytokines (TNF-α and IL-6). Molecular docking and dynamics simulation revealed consistent results with the in vitro enzymatic assay and indicated the stability of the putative complex of 27 with SARS-CoV-2 Mpro. The assessment of metabolic stability and physicochemical properties of 27 have also been conducted. This investigation identified a set of metabolically stable pyranopyrazoles as effective anti-SARS-CoV-2 Mpro and suppressors of host cell cytokine release. We believe that the new compounds deserve further chemical optimization and evaluation for COVID-19 treatment.
1. Introduction
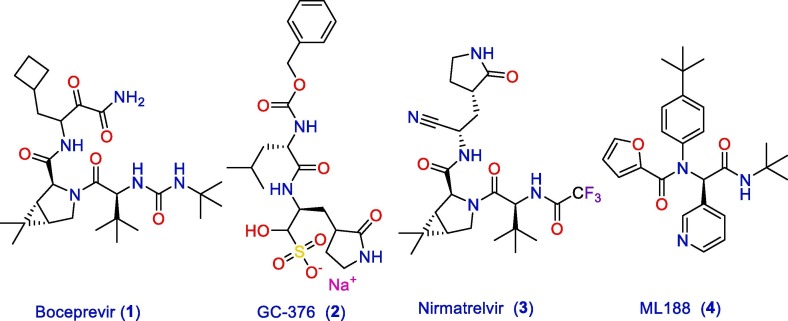
Coronavirus disease (COVID-19) and associated substantial inflammations can induce acute respiratory distress and even death in a considerable percentage of patients. By the end of June 2022, more than 534,000,000 confirmed cases of COVID-19, including more than 6,300,000 confirmed deaths, were recorded by the World Health Organization (WHO) [1]. To the best of our knowledge, all the treatment options for this critical infectious disorder are symptomatic to manage respiratory impairment and triggered inflammations. So, finding an effective remedy is urgent. The SARS-CoV-2 main protease (Mpro) is essential for viral replication and transcription. This functional importance made this protein a striking target for the development of potent antiviral agents [2]. So, researchers performed many attempts to develop Mpro inhibitors as antiviral medications. As a result, a number of molecules with prominent activity were identified (Fig. 1 ). For instance, boceprevir (1), GC-376 (2) nirmatrelvir (3), and ML188 (4) were verified as potential Mpro non-covalent inhibitors via binding to its catalytic active site [3], [4], [5], [6].
Fig. 1.
Prominent SARS-CoV-2 Mpro inhibitors reported in the literature.
Cytokine levels are usually elevated in coronavirus-infected patients and cause serious pathological changes in the lung [7], [8]. In particular, the level of IL-6 was significantly elevated in critically ill patients with acute respiratory distress syndrome (ARDS) compared to patients without ARDS and was statistically correlated with death. Both patients with mild or severe symptoms had pneumonia, and 29 % of them developed ARDS. The implication of inflammation in COVID-19 infection is the main cause of dementia [9]. ARDS-associated deaths were evidenced via accelerating the expression of pro-inflammatory cytokines such as TNF-α, IL-1β, NF-κβ, IL-6, and COX-II [10]. The induction of these inflammatory cytokines could disrupt the function of the neuronal cell [11]. Additionally, many studies have shown that severe COVID-19 patients may have hyperinflammatory syndrome [12], [13].
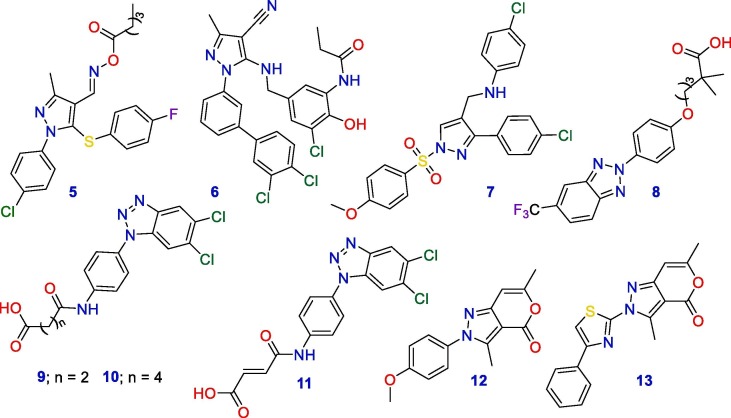
Pyranopyrazoles and their bioisosteric benzopyrazoles and benzotriazoles are a group of fused heterocyclic compounds that have gained precise importance in bioorganic chemistry because of their diverse biological activities [14], [15], [16], [17], [18]. Both pyran and pyrazole heterocycles have been individually studied as interesting classes of antiviral and anti-inflammatory drugs [19], [20], [21], [22], [23], [24], [25], [26], [27], [28]. A series of phenylpyrazole aldoxime ester derivatives displayed the good antiviral activity of the 4-carbaldehyde-O-propionyl oxime derivative 5 [29]. Structural optimization of this latter afforded a novel propenamide-based phenylpyrazole derivative bearing N terminal lipophilic fragments and nitrile group at C-4 of the pyrazole (6) as a potent anti-HIV agent [30]. Few years ago, phenylpyrazole derivative 7 has been verified as an attractive scaffold of antiviral activity [31]. Phenylpyrazoles have also been identified for many decades as a versatile promising scaffold for the development of anti-inflammatory medications [32], [33]. A number of commercially available anti-inflammatory drugs are belonging to this class of compound. These include Celecoxib, Ramifenazone, Lonazolac, and Phenylbutazone [34], [35]. In addition, pyran derivatives had been identified as promising motifs for designing antiviral agents [27], [28], [36], [37].
However, the investigation of pyranopyrazoles as dual antiviral and anti-inflammatory molecules has not yet been reported. To the best of our knowledge, few attempts were done for the development of certain bioisosteres of the pyranopyrazole heterocycle with either antiviral or anti-inflammatory activity (Fig. 2 ). In this context, a library of benzotriazoles was screened by a team of Italian researchers for their antiviral activity against twelve DNA and RNA viruses. Results revealed the potent antiviral effect and the superior selectivity of the carboxylic acid derivative 8 against Coxsackie Virus type-5 with an EC50 value of 0.15 μM and an excellent selectivity index [15]. In the past year, a library of fifty-six other benzotriazole derivatives was synthesized and tested for their antiviral activity. Among them, three compounds (9, 10, and 11) showed good antiviral activity against Picornaviruses and Coxsackievirus B5 [16]. On the other hand, pyranopyrazoles 12 and 13 exhibited good anti-inflammatory activity compared with Diclofenac sodium [14].
Fig. 2.
Structures of pyrazoles, pyranopyrazoles and bioisosteric antiviral and anti-inflammatory molecules.
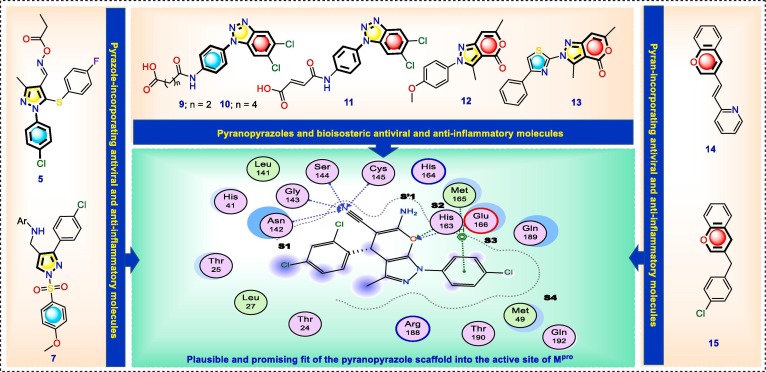
1.1. Compounds’ design strategy
The main protease, Mpro in SARS-CoV-2 is considered a viable drug target with an important role in the cleavage of the virus polypeptide [2], [38], [39]. Several Mpro inhibitors have been reported as covalent inhibitors that modify the catalytic Cys145 leading to protease inhibition and cellular antiviral activity, including GC376 containing pyrrolidone that mimics the glutamine at the P1 position of the natural Mpro substrate [40]. Based on the above verified scientific facts and as a continuation of our recent reports on identifying new bioactive agents [41], [42], [43], [44], [45], [46], [47], [48], we decided to get both privileged pharmacophoric fragments pyran and pyrazole in one molecular entity to get new SARS-CoV-2 Mpro inhibitor molecules with concomitant anti-inflammatory activity. Accordingly, a novel series of 6-amino-1-(4-chlorophenyl)-4-(4-aryl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile derivatives were designed and synthesized by joining structural aspects of the previously reported anti-inflammatory and SARS-CoV-2 Mpro inhibitors to evaluate their potential antiviral and anti-inflammatory activities. Different substitution patterns were introduced to the aryl ring at C-4 of the pyranopyrazole scaffold to assess the effect of such substitutions on biological activity. The inhibitor design approach of the present report is summarized in Fig. 3 . Initially, the pyranopyrazole scaffold was investigated by molecular docking of ligands into the active site of Mpro. Results revealed the putative interactions of the designed molecules with Mpro subsites through orientation of the pyranopyrazole toward the S1 pocket involving possible hydrogen bonding interactions. In addition, the warhead nitrile group was directed to the active-site Cys145 residue and the ligand’s position was stabilized through multiple hydrogen bonding interactions within the S1 pocket. Furthermore, the chlorophenyl residue of the pyranopyrazole extended into the S4 pocket as a rigid aromatic fragment. This overall plausible and promising fit of the pyranopyrazole scaffold into the active site of Mpro prompted us to synthesize sixteen potential inhibitors of the target protease based on this scaffold. All the synthesized compounds were evaluated as inhibitors of the SARS-CoV-2 Mpro and blockers of hyperinflammatory effects in host cells. So far, such derivatives have not been reported as antiviral agents for treating the COVID-19 infection. Additionally, the structure–activity relationship was explained guided by the results of biological activity evaluation. As well, the possible mechanism of action of the most potent pyranopyrazoles was further investigated in silico to understand their virtual binding affinity towards the active site of SARS-CoV-2 Mpro as a suggested target of their antiviral activity.
Fig. 3.
Molecular design of new pyranopyrazoles as suggested SARS-CoV-2 Mpro inhibitors and anti-inflammatory molecules.
2. Results and discussion
2.1. Chemistry
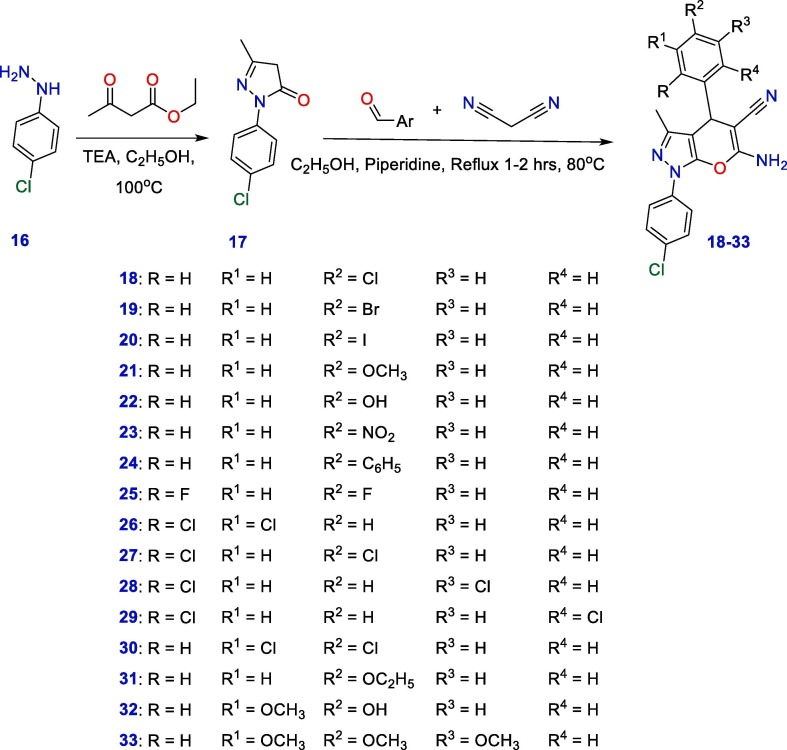
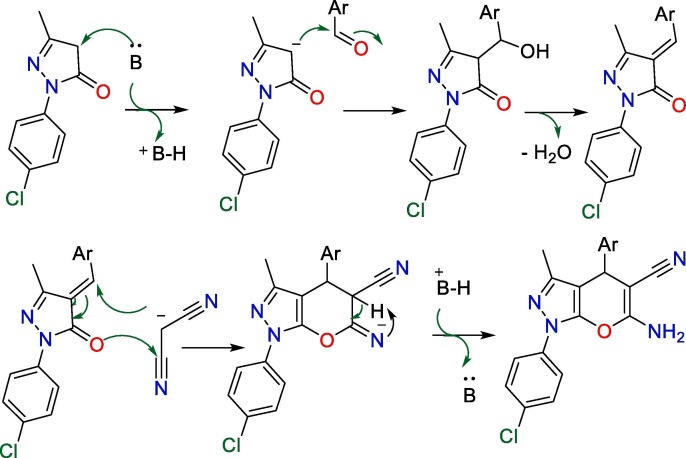
The adopted approach for the synthesis of the starting precursor 1-(4-chlorophenyl)-3-methyl-1H-pyrazol-5(4H)-one (17) is presented in Scheme 1 . Briefly, a solution of 4-chlorophenylhydrazine was treated with ethylacetoacetate in refluxing ethanol containing a catalytic amount of triethylamine. Our target 1-(4-chlorophenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole derivatives (18–33), were synthesized as represented in (Scheme 1). New pyranopyrazoles with substituted phenyl groups at the C-4 were easily acquired in a good yield by a one-pot, three-components reaction of 1 with malononitrile (2) and diversity of aromatic aldehydes in refluxing ethanolic containing piperidine as a catalyst [49].
Scheme 1.
Synthetic strategy of new pyranopyrazoles.
The conversion of 17 into final target pyranopyrazoles (18–33) is suggested to be initiated by the condensation of benzaldehyde derivatives under the piperidine base catalyst with the dihydro-1H-pyrazole as an activated methylene derivative with concomitant elimination of a water molecule to form 4-aryliden-5-pyrazolone [45], [50], [51]. A Michael-type cyclization subsequently takes place with malononitrile accompanied by the recovery of the piperidine base catalyst to furnish the final 6-amino-3-methyl-1,4-diaryl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles [52] as presented in Scheme 2 . Both the spectroscopic data and the elemental analyses were consistent with the proposed structures of final pyranopyrazoles. The IR spectra of compounds 18–33 disclosed absorption bands around υ 3497–3450, 3388–3316, 3297–3164 cm -1 for NH2 in addition to the absorption bands of the cyano groups in the region υ 2204–2182 cm -1. On the other hand, the 1H NMR spectra of 18–33 divulged the signals of the amino, methine, and methyl protons in the range of δ 8.23–7.18, 5.66–4.62, and 1.86–1.77 ppm, respectively. Moreover, the 13C NMR spectra of 18–33 showed signals resonating in the range of δ 37.38–30.01 and 13.65–12.58 ppm, attributed to the pyran-CH and pyrazole-attached methyl carbons. In addition, the MS spectra and the 13C NMR-DEPT spectra at 135 ppm supported the structural elucidation (see supplementary materials).
Scheme 2.
Suggested mechanism for formation of 1,4-dihydropyranopyrazoles; B●● = Piperidine.
2.2. Biological evaluation
2.2.1. SARS-CoV-2 Mpro inhibition assay
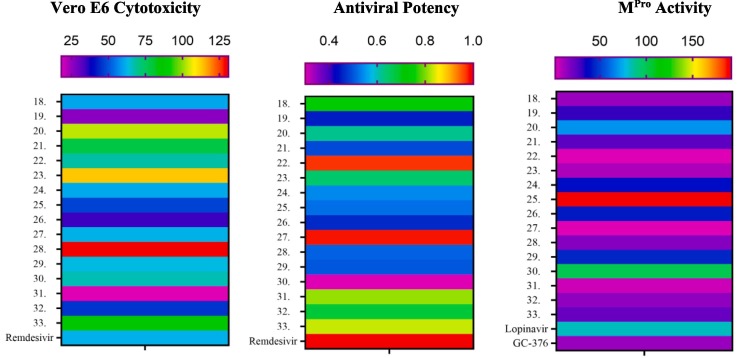
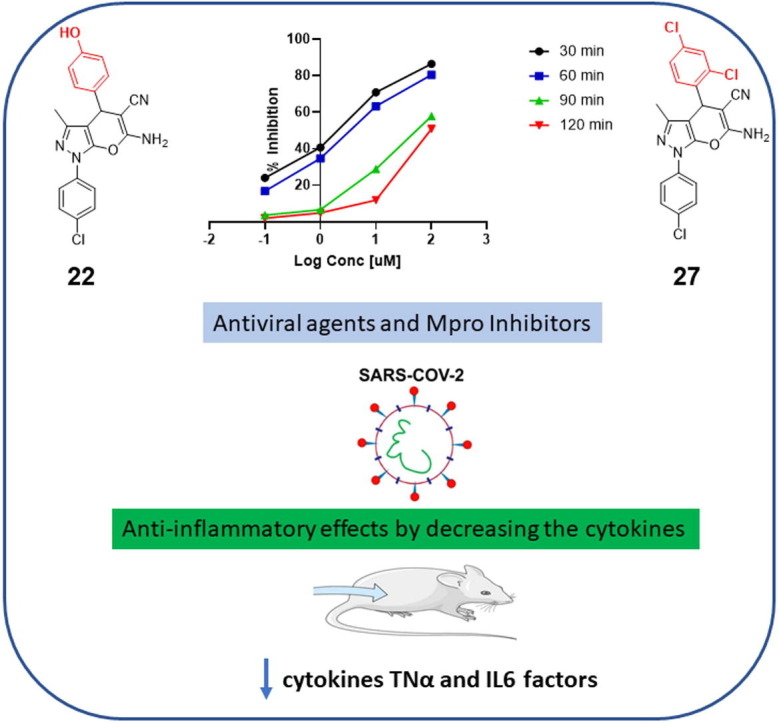
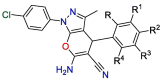
The SARS-CoV-2 Mpro has been validated as a viable target because of its importance in the cleavage of the viral polypeptide [53], [54]. A number of covalent and non-covalent SARS-CoV-2 Mpro inhibitors have been reported. Covalent inhibitors include GC-376 and carmofur. Each contains a nitrogenous heterocycle like pyrrolidone, and imidazole to mimic the glutamine P1 position [45], [55], [56]. Non-covalent inhibitors of SARS-CoV-2 Mpro were also developed and validated such as ML188 [5]. Here, the newly synthesized pyranopyrazoles, which are suggested to act as non-covalent inhibitors, were tested through an in vitro assay for inhibition of SARS-CoV-2 Mpro activity, and their results were compared with the results of two known antiviral agents lopinavir as non-specific and GC-376 as a specific Mpro inhibitor. The obtained results of in vitro MTT assay are shown in Table 1 . They indicate the importance of the phenyl substituents at the 2-position of the pyranopyrazole ring for the inhibition activity. We found that fourteen out of sixteen new derivatives inhibit the Mpro with IC50 values lower than that of the lopinavir while only five displayed better activity than that of the specific Mpro inhibitor GC-376. Among this series of analogs, three of the synthesized compounds were significantly more active than the rest of the other derivatives, i.e. 4‑hydroxy (22), 2,4‑dichloro (27), and 4‑ethoxy (31) substituents with IC50 values of 2.01, 1.83, and 4.60 μM, respectively which is more potent than the non-specific and specific inhibitors (IC50 values = 12.85, 82.17 μM for GC-376 and lopinavir).
Table 1.
Inhibitory data of the target compound 18–33 against SARS-CoV-2 Mpro.
|
IDs |
 |
|||||
|---|---|---|---|---|---|---|
| R | R1 | R2 | R3 | R4 | IC50 (µM) | |
| 18. | H | H | Cl | H | H | 12.77 ± 0.6 |
| 19. | H | H | Br | H | H | 30.93 ± 1.6 |
| 20. | H | H | I | H | H | 69.10 ± 3.50 |
| 21. | H | H | OCH3 | H | H | 24.55 ± 1.2 |
| 22. | H | H | OH | H | H | 2.01 ± 2.15 |
| 23. | H | H | NO2 | H | H | 9.50 ± 0.51 |
| 24. | H | H | C6H5 | H | H | 39.84 ± 2.2 |
| 25. | F | H | F | H | H | 191.7 ± 9.8 |
| 26. | Cl | Cl | H | H | H | 41.76 ± 2.1 |
| 27. | Cl | H | Cl | H | H | 1.83 ± 3.45 |
| 28. | Cl | H | H | Cl | H | 16.4 ± 0.8 |
| 29. | Cl | H | H | H | Cl | 44.64 ± 2.3 |
| 30. | H | Cl | Cl | H | H | 100 ± 1.70 |
| 31. | H | H | OC2H5 | H | H | 4.60 ± 0.25 |
| 32. | H | OCH3 | OH | H | H | 14.59 ± 0.7 |
| 33. | H | OCH3 | OCH3 | OCH3 | H | 22.09 ± 1.10 |
| Lopinavir | – | – | – | – | – | 82.17 ± 6.77 |
| GC-376 | – | – | – | – | – | 12.85 ± 0.74 |
2.3. SAR analysis
The obtained results in Table 1 showed that the replacement of the methoxy group at the para-position (R2) of the phenyl ring (21) with an ethoxy group (31) improved the inhibitory activity by 6-folds. However, both were less potent than the hydroxyl analog 22. Adding two additional groups as realized in the 3,4,5‑trimethoxy derivative 33, retained comparable inhibition (33 versus 21). The introduction of a bulky substituent at the para position (R2) as in the case 24 (biphenyl) had a bad impact on the Mpro inhibition (IC50 value = 39.84 μM). Introduction of different electron-withdrawing groups at the para position (R2) retained a relatively low micromolar inhibitory concentration (IC50 value = 9.50 μM) in case of the 4‑nitro compound 23 but increased the IC50 values for the chloro substituent (18, IC50 = 12.77 μM), bromo (19, IC50 = 30.93 μM). In contrast, the 2,4‑difluoro and 3,4‑dichloro compounds (25 and 30) showed a dramatic fall in the inhibitory activity (IC50 values are greater than 100.00 μM).
Relative to the activity of 18 with a para-chloro substituent, the introduction of an additional chloro substituent at different positions on the phenyl ring had a varying effect on the activity. For example, the 2,4‑dichloro substituent 27 displayed impressive potency (IC50 = 1.83 μM) compared to its corresponding 2,3‑dichloro (26, IC50 = 41.76 μM), 2,5‑dichloro (28, IC50 = 16.40 μM), 2,6‑dichloro (29, IC50 = 44.6 μM) and 3,4‑dichloro (30, IC50 = 100 μM) isomers. This result reflects the good impact of the 2,4‑dichloro substitution to get the optimum activity. This finding could be supported by the previously reported observation for analogs of the anti-epileptic drug perampanel, which stated that small substituents like chlorine at the ortho and meta positions yielded potent Mpro inhibitors with IC50 values of 0.018–0.037 μM [57]. Addition of a meta-methoxy group next to the para-hydroxy group of 22 to get compound 32 markedly reduced the inhibitory activity (IC50 changed from 2.01 to 14.59 µM). Overall, the 4‑hydroxy and the 2,4‑chloro substitution patterns turn out to be particularly advantageous for Mpro inhibition. A graphical presentation of the SAR study is given in Fig. 4 .
Fig. 4.
SAR analysis of target pyranopyrazoles correlated with Mpro inhibition activity.
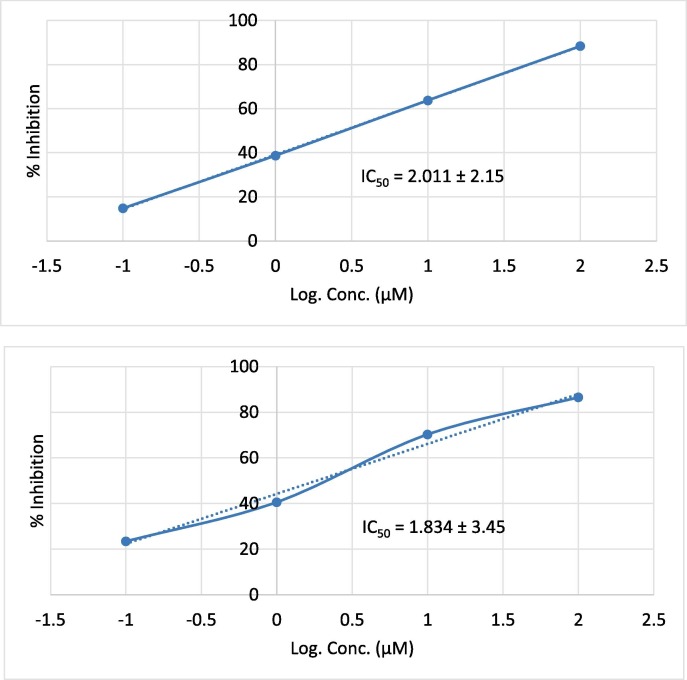
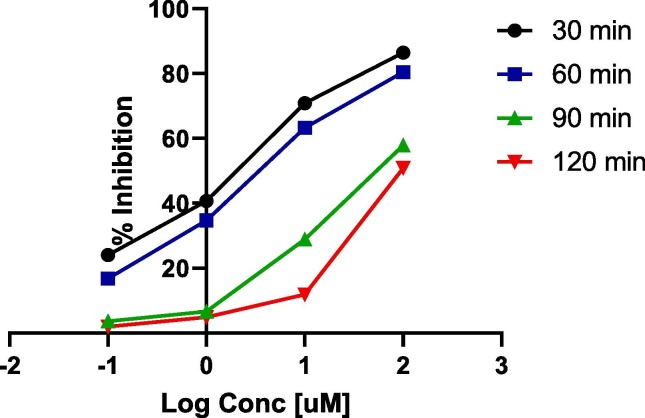
The kinetic analysis of the best effective derivatives (22 and 27) based upon the results shown in Fig. 5, Fig. 6 , revealed that they bind to the biological target reversibly and non-competitively with non-covalent interactions. Subjecting of the compound against the reaction with the protease at different time intervals showed increasing the inhibitory effect as IC50 values indicated, with the concentrations. Our kinetic rationale of their mode of action and binding for the formation of non-covalent-bound complexes is in good agreement with recently reported reference nitrile-incorporating drugs mechanistic studies [57], [58].
Fig. 5.
Dose-response curve and IC50 value of compounds 22 (upper panel) and 27 (lower panel) against SARS CoV-2-MPro.
Fig. 6.
Dose-response curve of compound 27 report with IC50 values at different time intervals.
2.3.1. Analysis of the Vero E6 cytotoxicity and correlation with SARS-CoV-2 Mpro activity
To test the ability of novel target compounds for their potential to inhibit the SARS-CoV-2 viral replication, a cellular antiviral assay on Vero E6 cells infected with SARS-CoV-2 at two different concentrations (1 and 10 μM) was performed using classical cell culture with real-time PCR [59]. The cell viability after treatment with pyranopyrazole target compounds or remdesivir was determined by MTT assay [60]. Remdesivir was used as a standard in the cytotoxicity assay to compare its cytotoxicity with that of our target molecules as it was reported as a highly selective antiviral agent with low off-target cytotoxicity [61]. CCL-8 Vero E6 cells of the American Type Culture Collection provided by the Virology Sector, VACSERA, Giza, Egypt and were infected later with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.01. The positive control remdesivir showed no cytotoxicity to Vero E6 cells at a concentration of 60 μM. Excluding five compounds; 19, 25, 26, 31, and 32, all the other eleven compounds were well tolerated and had IC50 values of over 60 μM. Although the para-ethoxy derivative (31) showed good inhibitory activity in the Mpro inhibitory activity, it was highly cytotoxic to Vero E6 cells with an IC50 value of 18.1 μM. Interestingly, the most potent couple of compounds (22 and 27) showed acceptable values of cytotoxicity with IC50 above 60 μM which are almost better than that of the positive control remdesivir. The pyranopyrazole compounds identified herein represent some of the most potent SARS-CoV-2 Mpro inhibitors with both enzymatic inhibition and cellular antiviral activity. Remdesivir was selected as a positive control in this study and the results showed that remdesivir potently inhibited viral replication, which reached 93.33 % and 89.61 % at 10 and 1 μM, respectively. Interestingly, 22 and 27 were potent inhibitors of SARS-CoV-2 Mpro (IC50: 2.01 and 1.83 μM, respectively) and were among the best performers in the viral replication assay which reached 90.45 % for 22 and 91.23 % for 27 at 10 μM, respectively. In general, none of the tested pyranopyrazoles reached 50 % inhibition of viral replication at the lowest concentration of 1 μM (Table 2 , Fig. 7 ). Moreover, some of the pyranopyrazole derivatives did not function positively in the cellular antiviral assay, and their relative abilities to inhibit viral replication did not always correlate directly with the in vitro inhibition parameters toward Mpro [58]. Compound 30 was the weakest inhibitor in cellular antiviral activity (27.8 % at 10 μM) which is comparable with its poor Mpro enzymatic inhibition (IC50 = 100 μM). Compounds 31 and 33 proved their capability of affecting the viability of cells and inhibiting viral replication with about 75 % at 10 μM for each but were not consistent with their Mpro enzymatic inhibition (IC50 = 4.60 μM for 31 and IC50 = 22.09 μM for 33). For compounds 18, 23, and 32, their Mpro inhibition IC50 values range from 9.50 to 14.59 μM, which were comparable to their inhibition of viral replication with around 65 % inhibition at 10 μM. Overall, the para-hydroxy and the ortho, para-dichloro derivatives (22 and 27) displayed the best combined results of SARS-CoV-2 Mpro inhibition, Vero E6 cytotoxicity assay, and as inhibitors of viral replication. They also demonstrated the highest antiviral potency indices compared with that of the positive control at 10 µM (0.97 and 0.98 respectively).
Table 2.
Results of Vero E6 cytotoxicity and SARS-CoV-2 antiviral activity of novel pyranopyrazoles.
|
ID |
Vero E6 Cytotoxicity (IC50, µM) |
SARS-CoV-2 Antiviral Assay (%) |
Antiviral Potency Index (API)* |
|
|---|---|---|---|---|
| 1 µM | 10 µM | |||
| 18 | 60.5 ± 4.6 | 12.91 ± 0.06 | 69.21 ± 0.51 | 0.74 |
| 19 | 26.2 ± 20 | 10.21 ± 0.046 | 42.16 ± 0.23 | 0.45 |
| 20 | 103 ± 7.8 | 7.424 ± 0.034 | 58.13 ± 0.37 | 0.62 |
| 21 | 78.1 ± 5.9 | 1.515 ± 0.007 | 45.82 ± 0.27 | 0.49 |
| 22 | 67.7 ± 5.2 | 36.85 ± 0.19 | 90.45 ± 1.01 | 0.97 |
| 23 | 113 ± 8.6 | 18.65 ± 0.089 | 59.3 ± 0.31 | 0.64 |
| 24 | 60.7 ± 4.6 | 10.89 ± 0.05 | 50.31 ± 0.30 | 0.54 |
| 25 | 47.6 ± 3.6 | 0.758 ± 0.003 | 48.49 ± 0.29 | 0.52 |
| 26 | 34.8 ± 2.7 | 7.424 ± 0.09 | 42.99 ± 0.48 | 0.46 |
| 27 | 62 ± 4.70 | 20.46 ± 0.03 | 91.23 ± 0.25 | 0.98 |
| 28 | 131 ± 10 | 22.82 ± 0.11 | 47.75 ± 0.28 | 0.51 |
| 29 | 63.3 ± 4.8 | 5.303 ± 0.02 | 46.21 ± 0.26 | 0.50 |
| 30 | 66.5 ± 5.1 | 3.03 ± 0.01 | 27.84 ± 0.14 | 0.30 |
| 31 | 18.1 ± 1.4 | 21.06 ± 0.10 | 75.78 ± 0.61 | 0.81 |
| 32 | 45.7 ± 3.5 | 23.4 ± 0.11 | 63.37 ± 0.43 | 0.68 |
| 33 | 86.2 ± 6.6 | 28.38 ± 0.14 | 77.66 ± 0.65 | 0.83 |
| Remdesivir | 61.8 ± 4.7 | 89.61 ± 0.98 | 93.33 ± 1.17 | 1.00 |
API is the ratio between the % inhibition of tested compound to that of the remdesivir reference drug at 10 µM. SE represents standard deviation calculated from triplicate readings.
Fig. 7.
Heatmap correlation between the cytotoxicity effect, cellular antiviral effect (% inhibition), and SARS-CoV-2 Mpro inhibition of tested pyranopyrazoles and reference standards.
2.3.2. Effect on Pro-inflammatory Cytokine’s production
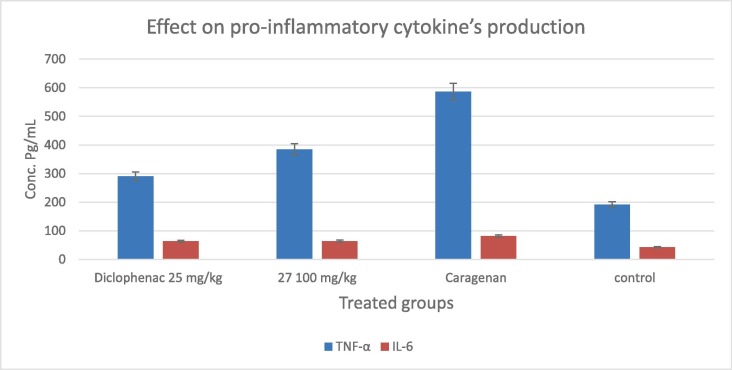
There is clear evidence from coronavirus-infected patients with both high cytokine levels and pathological changes in the lung [7], [8]. For example, in the plasma of COVID-19 patients, high concentrations of IL-2, IL-6, TNF-α, and IL-7 have been detected [7]. The induction of these inflammation mediators could disrupt the functions of neuronal cells [11]. Moreover, severe COVID-19 patients may have hyperinflammatory syndrome [12], [13]. In the current work, considering the proven scientific fact that phenylpyrazoles have the potential to show anti-inflammatory activity [34], [35], in vivo testing was performed to confirm the capacity of new compounds to lower the pro-inflammatory cytokines. As demonstrated in Fig. 8 , utilizing the most active chemical 27, the overproduction of cytokines in the carrageenan-treated group appears to be the highest when compared to the untreated group; 587 and 82 pg/mL for TNF-α and IL-6 respectively. When compared to the reference diclofenac medication, the target chemical 27 resulted in a considerable drop in cytokines’ levels 385 and 64 pg/mL for TNF-α and IL-6 respectively. These findings suggested that the new innovative scaffold-based derivative with good antiviral activity is also capable to show potent anti-inflammation effects; suggesting that it might be employed as a dual multitarget medicine for COVID-19 patients after further in vitro and in vivo optimization studies.
Fig. 8.
Effect of compound 27 in a dose of 100 mg/kg (triplicate ± SD) on serum levels of tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6), in male rats administrated Carrageenan compared to negative control and diclofenac-treated groups (25 mg/kg) in pg/mL.
2.4. Computational studies
2.4.1. Molecular docking
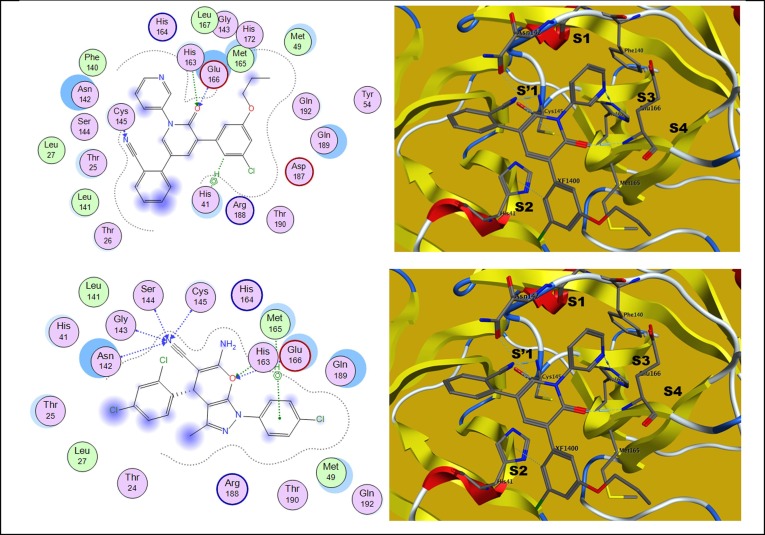
Using the proper in silico tool, researchers can reduce time, effort, and resources consumption in the journey of drug discovery [62], [63]. To gain a better understanding of the mode of action of novel compounds as antiviral and guide further SAR studies, we proceeded to examine the interaction of the most potent pyranopyrazole derivative with the SARS-CoV–2 Mpro protein target. Herein, the crystal structure of SARS-CoV–2 Mpro in complex with a potent bipyridine-benzonitrile inhibitor (XF1; PDB: 7L11; https://www.rcsb.org/structure/7l11) was selected as the binding model for nitrile-warhead containing non-covalent inhibitor [57]. This molecular docking study was performed using the default protocol associated with the glide module of the Schrodinger modelling suite. The molecular docking study showed that the promising compound 27 was appropriately oriented in the active site of SARS-CoV–2 Mpro protein with a significant docking score (-16.459 Kcal/mol) and binding free energy (-6.8 kcal/mol) compared to the reference bound ligand; docking score (-14.5 kcal/mol) and binding free energy (-4.8 kcal/mol). As appeared in Fig. 9 , the displayed interactions with amino acids of the catalytic residues His41 and Cys145 and accompanying pocket residues Glu166, His163, and Asn142 are mapped with the co-crystallized ligand and target compound 27. The binding affinity exhibited by compound 27 is attributed to significant bonded and non-bonded interactions with residues lining the active site of the SARS-CoV–2 Mpro. The binding pocket interaction analysis for the active compound 27 showed that the molecule is embedded into the active site through a series of significant van der Waals interactions. Fortunately, a prominent tetra-hydrogen bonding system was formed between the nitrile group and polar key residues; Cys145, Ser144, Gly143, and Asn142 (within a distance range of 2.61–3.05 Å) that strongly contribute to the stability of the compound. In addition, a pyrazole edge packs well against the imidazole ring of His41 in the S2 pocket with no indication of space for expansion (3.88 Å). Moreover, two strong hydrogen bonds are formed through the pyran oxygen and NH backbones of Glu166 (2.85 Å) and His163 residues at the S1 pocket. An edge-to-face-chlorophenyl interaction occurs with Met165 well contributes to the stabilization of new structure in the S2 pocket and might share in the non-covalent hydrophobic interaction. The docking score and interaction pattern of 27 show that this ligand may bind efficiently to the SARS-CoV–2 Mpro catalytic site with good affinity.
Fig. 9.
2D and 3D binding interactions of the co-crystalized ligand (upper row) compared to the active analog 27 (lower row) in the active site of SARS-CoV-2 Mpro.
2.4.2. Molecular dynamic simulation
Herein, we analyzed the protein's secondary structure of SARS-COV-2 19 Mpro protease bound to the novel active analog 27 before and after docking to understand the conformational changes and their stability. The protein's secondary structure of SARS-CoV–2 Mpro, when subjected to MD simulation studies, had a zero-net charge, 42,685 total atoms, 51.040 mM of Na+ ions, 53.956 mM of Cl− ions surrounded by 12,468 water molecules.
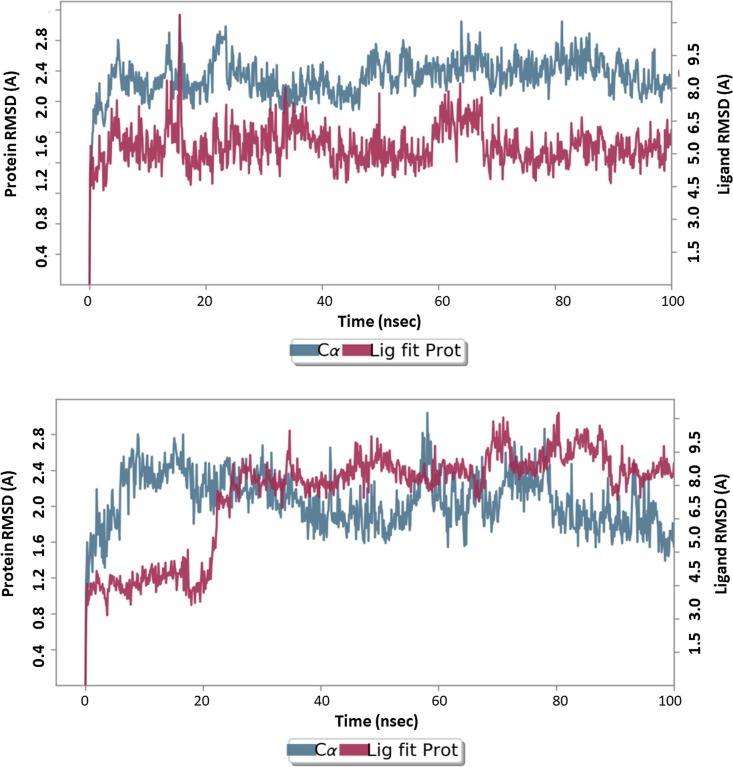
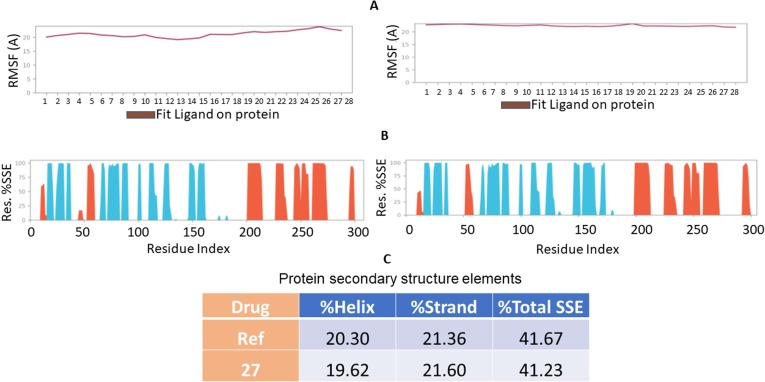
For an MD run of 100 ns, RMSD and RMSF were predicted for the complex to reference and 27 analog forms ligand’s interaction can ward off unfolding and stabilize the protein [64]. Hence, we analyzed the protein’s secondary structures before and after docking to understand the conformational changes due to ligand binding. Fig. 10 (up to reference drug, down for 27 shows the results of the RMSD record of the SARS-CoV–2 Mpro behavior before and after docking of pyranopyrazole derivative 27 and attained an equilibrium after 100 ns. The MD simulation showed a stable conformation was achieved by the target protein at RMSD 2 Å, an acceptable value for protein structures that disclosed the stability of the complex.
Fig. 10.
Results of MD of SARS-CoV-2 Mpro (PDB ID: 7L11) complex. RMSD of complex with reference compound (upper panel) and 27 (lower panel). The RMSDs were stable after 100 ns.
The plot of RMSF for SARS-CoV–2 Mpro with the reference ligand and 27 indicated that the N-terminal and C-terminal residues oscillated below 2.0 Å for the 27 and above 2.0 Å for the reference compound. The other secondary structures remained consistent throughout the trajectories, and the RMSF of the protein–ligand complex (Fig. 11 A) predicted was below 2.0 Å, indicating conformational stability during the simulation. The secondary structure of the SARS-CoV–2 Mpro complex with reference (Fig. 11 B and 11C) had 20.30 % α-helices and 21.36 % of β-strands, while the complex with 27 (Fig. 11 B left) had 20.04 % α-helices and 23.86 % β-strands. The negligible increase in the % of β-strands indicated the minimum unfolding of the α-helices during MD of the complex.
Fig. 11.
Results of MD of SARS-CoV-2 Mpro (PDB ID: 7L11) complexes. (A) Ligand Root Mean Square Fluctuation (l-RMSF) for reference complexed with SARS-CoV-2 Mpro (left) and for 27 complexed with SARS-CoV-2 Mpro (right). (B) Secondary structure elements (SSE) of the complex SARS-CoV-2 Mpro with reference compound (left) and with 27 (right). (C) % SSE of the two complexes of SARS-CoV-2 Mpro with reference and 27 compounds. No significant change in the percentage of SSE between the apo and bound proteins indicates conformational stability.
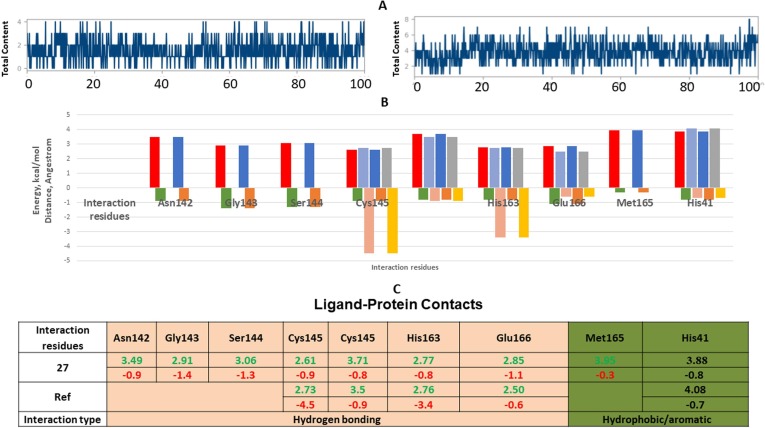
Analysis of intermolecular interactions of the SARS-CoV–2 Mpro complexes during the MD simulation, shown in Fig. 12 A-C, indicated that analyzed trajectories exhibited different promising contacts of polar and non-polar interactions confirming the molecular docking results. Additionally, the plot of interaction fractions against the binding site residues confirmed that Cys106 conserved residue of the furnished multiple contacts (interaction fraction greater than 1.0). Results of molecular docking were consistent with the MD results, as the plot indicated that the binding site residues of the SARS-CoV–2 Mpro complex Cys145, His163, and Glu166 were involved in hydrogen bonding for different simulation times, Fig. 10 C with consistent binding energies. The hydrophobic interactions of 27 with SARS-CoV-2 19 Mpro complex residues Met165, and catalytic His41 were also prominent through the pyrazole, linker, and terminal chromene fragments. These interactions contributed to the stability of the complex for 50 % of the simulation time.
Fig. 12.
Results of MD of SARS-CoV 2 Mpro (PDB ID: 7L11) complexes. (A) Timeline of the total contacts of the complex SARS-CoV 2 Mpro with reference compound (right) and 27 (left). (B) Stacked bar chart represents the normalized interactions throughout the trajectory. Y-axis represents the % of simulation time the specific interaction was maintained of the complex SARS-CoV–2 Mpro with reference (right) and 27 (left). (C) Ligand-Protein Contacts of the two complexes of SARS-CoV–2 Mpro with reference compound and 27.
2.5. Initial assessment of the physicochemical properties and pharmacokinetic parameters
One of the most critical characteristics of drug candidates must be evaluated early in the drug development process is the physicochemical properties [65], [66], [67], [68]. Herein, the physicochemical properties of 27 as the most potent analog were explored. First, the aqueous solubility was analyzed using a turbidimetric assay in phosphate-buffered saline [69]. The importance of solubility as a pharmacokinetic parameter is known to directly affects the pharmacokinetic profiles of drugs, routes of administration and the formulation of compounds into suitable dosage forms [70]. Compound 27 showed moderate aqueous solubility compared with two FDA-approved drugs rifampicin and tamoxifen (Table 3 ).
Table 3.
Evaluation of solubility in phosphate-buffered saline (PBS).
| Compound ID | Solubility limit (μM)* | c Log P | Note |
|---|---|---|---|
| Rifampicin | 179.4 | 4.24 | Reference with good solubility |
| Tamoxifen | 2.15 | 6.35 | Reference with limited solubility |
| 27 | 22.4 | 4.80 | Moderate aqueous solubility |
The solubility limit corresponds to the highest concentration of test compound at which no precipitate was detected (OD540).
2.6. Initial assessment of the enzymatic metabolic stability
The antiviral activity of the synthesized pyranopyrazoles is not the sole factor for them to be considered successful candidates. They must possess a set of drug-likeness properties as the metabolic stability and rate of clearance [71], [72]. Therefore, a certain stability analysis of compound 27 was initially investigated using in vitro assays. The most promising derivative in this series, compound 27, showed a high degree of metabolic stability with an intrinsic half-life (t1/2) of more than 2 h (Table 4 ). This value was increased in the absence of NADPH, the cofactor of CYP-450, indicating that 27 is mainly metabolized by this class of metabolic oxidases. Furthermore, this dichlorophenylpyrazole derivative 27 has an intermediate clearance rate between those of the reference drugs propranolol and terfenadine. Despite the increase in its half-life in the absence of NADPH suggesting that the compound is mainly metabolized by the co-factor CYP-450, compound 27 also exhibited pronounced stability to hepatic metabolism, with a clearance rate of 54.6 µL/min/mg. This result endows the new series of pyranopyrazoles to be metabolically stable and suggested to have a relatively long duration of action.
Table 4.
Enzymatic metabolic stability of the active derivative 27 compared with Propranolol and Terfenadine.
| Compound ID | Mean CLint (μL/min/mg) | Mean t1/2 (min) | Metabolic rate |
|---|---|---|---|
| Terfenadine | 570.30 | 8.41 | High |
| Propranolol | 46.20 | 143.80 | Low |
| 27 | 54.60 | 133.51 | Intermediate |
3. Conclusion
In conclusion, we have introduced novel pyranopyrazole-based derivatives and evaluated their antiviral activity against COVID-19 infection. Most of the novel compounds exhibited potent antiviral activity. Analog 27 showed potent anti-SARS-CoV-2 activity at 1 and 10 µM by 95 % inhibition of the viral growth. These novel derivatives were subjected to SARS-CoV-2 Mpro inhibitory assay to assess their possible antiviral mechanism. Compound 27 strongly inhibited the activity of Mpro with an IC50 value of 1.83 µM compared to 8.12 and 82.17 µM for the positive controls GC-376 and Lopinavir. Additionally, in vivo testing was also conducted to evaluate the capacity of 27 to lower the levels of pro-inflammatory cytokines compared with diclofenac sodium. Compound 27 significantly lowered the cytokines TNF-α and IL-6 levels. Finally, extensive molecular docking and dynamics simulation proved the prominent interaction behavior and stability of 27 within the binding pocket through the conserved His41, Glu166, and Cys145 residues. Docking results exhibited that some of the new pyranopyrazoles might work through the non-covalent interaction with the SARS-CoV main protease. SAR analysis of the novel scaffold was estimated based on biological and theoretical results obtained for these compounds. In addition, physicochemical and in vivo pharmacokinetic profiles of the most active derivative 27 suggested its promising ADME properties and metabolic stability. Overall, the target compounds behaved as non-competitive non-covalent protease inhibitors.
4. Materials and method
4.1. Chemistry
All chemicals were purchased from Sigma-Aldrich Chemical Co. (Sigma-Aldrich Corp., St. Louis, MO, USA). All the melting points were measured with a Stuart Scientific Co. ltd apparatus, which are uncorrected. The IR spectra were recorded on a KBr disc on a Jasco FT/IR 460 plus spectrophotometer. The 1H (850 MHz),13C (214 MHz), and 13C DEPT/APT (214 MHz) NMR spectra were measured on a BRUKER AV 850 MHz spectrometer in DMSO‑d 6, a solvent, using the tetramethylsilane (TMS) as an internal standard. HPLC-Mass Spectrometry was performed on Agilent 1100 / ZQ MSD including C18 column and diod-array UV detector. The mobile phase (containing 0.01 M ammonium acetate) was gradient starting from 20 % acetonitrile/80 % water to 80 % acetonitrile/20 % water. The elemental analysis was carried out at the Regional Centre for Mycology and Biotechnology (RCMP), Al-Azhar University, Cairo, Egypt, and the results were within ± 0.25 %. The reaction courses and product mixtures were routinely monitored by the thin layer chromatography (TLC) on silica gel precoated F254 Merck plates.
4.2. General procedure for synthesis of 6-Amino-1-(4-chlorophenyl)-4-(aryl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile derivatives (18–33)
A reaction mixture of 1-(4-chlorophenyl)-3-methyl-1H-pyrazol-5(4H)-one (17) [73] (0.01 mol), malononitrile (0.01 mol), the appropriate aromatic aldehydes (0.01 mol), and piperidine (0.5 ml) in an ethanol solution (30 ml) was heated under reflux for 1–2 hr at temperature 80 °C. After the reaction reached completion, the reaction mixture was cooled to room temperature, and the precipitated solid was filtered off, washed with methanol, and recrystallized from ethanol or ethanol/benzene. The physical and spectral data of compounds (18–33) are as follows:
4.2.1. 6-Amino-1-(4-chlorophenyl)-4-(4-chlorophenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (18)
Colorless crystals; yield 95 %; m. p. 205-206οC; IR (KBr) υ (cm−1): 3447, 3388, 3246 (NH2), 2182 (CN); 1H NMR (DMSO‑d 6, 850 MHz) δ: 7.82 (d, 2H, J = 8.5 Hz, Ar), 7.55 (d, 2H, J = 8.5 Hz, Ar), 7.32 (d, 2H, J = 8.5 Hz, Ar1), 7.30 (s, 2H, NH2), 7.19 (d, 2H, J = 8.5 Hz, Ar1), 4.72 (s, 1H, H-4), 1.78 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 159.77 (C-6),146.19 (C-7a), 144.39 (C-3), 140.20 (C-1, Ar), 136.79 (C-1, Ar1), 130.71 (C-4, Ar, Ar1), 130.26, 130.22 (C-2,6, Ar1), 129.73 (C-3,5, Ar), 121.92 (C-3,5, Ar1), 120.38 (CN), 115.81, 115.71 (C-2,6, Ar), 99.19 (C-3a), 58.44 (C-5), 36.32 (C-4), 13.03 (CH3); 13C NMR-DEPT spectrum at 135°CH, CH3 (positive, up), CH2 (negative, down), revealed the following signals at δ: 130.26, 130.22 (C-2,6, Ar1↑), 129.73 (C-3,5, Ar↑), 121.92 (C-3,5, Ar1↑), 115.81, 115.71 (C-2,6, Ar↑), 36.32 (C-4↑), 13.03 (CH3↑); LC-MS (ESI) RT = 4.99 min, m/z 396.9 [M++H]; Anal. Calcd for C20H14Cl2N4O (397.26): C, 60.47; H, 3.55; N, 14.10. Found: C, 60.54; H, 3.61; N, 14.16 %.
4.2.2. 6-Amino-4-(4-bromophenyl)-1-(4-chlorophenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (19)
Colorless crystals; yield 94 %; m. p. 231-232οC; IR (KBr) υ (cm−1): 3482, 3385, 3231 (NH2), 2185 (CN); 1H NMR (DMSO‑d 6, 850 MHz) δ: 7.82 (d, 2H, J = 8.5 Hz, Ar), 7.54 (d, 2H, J = 8.5 Hz, Ar), 7.54 (d, 2H, J = 8.5 Hz, Ar1), 7.31 (s, 2H, NH2), 7.25 (d, 2H, J = 8.5 Hz, Ar1), 4.71 (s, 1H, H-4), 1.79 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 159.85 (C-6),146.17 (C-7a), 144.45 (C-3), 143.42 (C-1, Ar), 136.76 (C-1, Ar1), 131.94 (C-2,6, Ar1), 130.75 (C-4, Ar), 130.58 (C-3,5, Ar1), 130.52 (C-4, Ar1), 129.74 (C-3,5, Ar), 121.93 (C-2,6, Ar), 120.66 (CN), 98.84 (C-3a), 58.05 (C-5), 36.51 (C-4), 13.05 (CH3);13C NMR-DEPT spectrum at 135°CH, CH3 (positive, up), CH2 (negative, down), revealed the following signals at δ: 131.94 (C-2,6, Ar1 ↑), 130.98 (C-3,5, Ar1 ↑), 129.74 (C-3,5, Ar ↑), 121.93 (C-2,6, Ar ↑), 36.51 (C-4 ↑), 13.05 (CH3 ↑); LC-MS (ESI) RT = 5.51 min, m/z 440.8 [M++H]; Anal. Calcd for C20H14BrClN4O (441.71): C, 54.38; H, 3.19; N, 12.68. Found: C, 54.30; H, 3.12; N, 12.62 %.
4.2.3. 6-Amino-1-(4-chlorophenyl)-4-(4-iodophenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (20)
Colorless crystals; yield 94 %; m. p. 228-229οC; IR (KBr) υ (cm−1): 3497, 3366, 3231 (NH2), 2187 (CN); 1H NMR (DMSO‑d 6, 850 MHz) δ: 7.82 (d, 2H, J = 8.5 Hz, Ar), 7.72 (d, 2H, J = 8.5 Hz, Ar), 7.54 (d, 2H, J = 8.5 Hz, Ar1), 7.31 (s, 2H, NH2), 7.10 (d, 2H, J = 8.5 Hz, Ar1), 4.68 (s, 1H, H-4), 1.79 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 159.85 (C-6),146.17 (C-7a), 144.45 (C-3), 143.82 (C-1, Ar), 137.79 (C-3,5, Ar1), 136.77 (C-1, Ar1), 130.74 (C-4, Ar), 130.71 (C-2,6, Ar1), 129.74 (C-3,5, Ar), 121.91 (C-2,6, Ar), 120.33 (CN), 98.83 (C-3a), 93.64 (C-4, Ar1), 58.04 (C-5), 36.63 (C-4), 13.06 (CH3); 13C NMR-DEPT spectrum at 135°CH, CH3 (positive, up), CH2 (negative, down), revealed the following signals at δ: 137.79 (C-3,5, Ar1 ↑), 130.71 (C-2,6, Ar1 ↑), 129.74 (C-3,5, Ar ↑), 121.91 (C-2,6, Ar ↑), 36.63 (C-4 ↑), 13.06 (CH3 ↑); LC-MS (ESI) RT = 6.97 min, m/z 388.8 [M++H]; Anal. Calcd for C20H14ClIN4O (488.71): C, 49.15; H, 2.89; N, 11.46. Found: C, 49.09; H, 2.82; N, 11.40 %.
4.2.4. 6-Amino-1-(4-chlorophenyl)-4-(4-methoxyphenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (21)
Colorless crystals; yield 94 %; m. p. 278–280 οC; IR (KBr) υ (cm−1): 3492, 3377, 3232 (NH2), 2182 (CN); 1H NMR (DMSO‑d 6, 850 MHz) δ: 7.82 (d, 2H, J = 8.5 Hz, Ar), 7.54 (d, 2H, J = 8.5 Hz, Ar), 7.20 (s, 2H, NH2), 7.17 (d, 2H, J = 8.5 Hz, Ar1), 6.90 (d, 2H, J = 8.5 Hz, Ar1), 4.62(s, 1H, H-4), 3.74 (s, 3H, OCH3), 1.78 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 159.62 (C-6),158.66 (C-4, Ar1), 146.32 (C-7a), 144.31 (C-3), 136.82 (C-1, Ar), 135.94 (C-4, Ar), 130.64 (C-1, Ar1), 129.74 (C-2,6, Ar1), 129.33 (C-3,5, Ar), 121.83 (C-2,6, Ar), 120.50 (CN), 114.33 (C-3,5, Ar1), 99.58 (C-3a), 58.96 (C-5), 55.48 (CH3), 36.32 (C-4), 13.04 (CH3); 13C NMR-DEPT spectrum at 135°CH, CH3 (positive, up), CH2 (negative, down), revealed the following signals at δ: 129.74 (C-2,6, Ar1 ↑), 129.33 (C-3,5, Ar ↑), 121.83 (C-2,6, Ar ↑), 114.33 (C-3,5, Ar1 ↑), 55.48 (CH3 ↑), 36.32 (C-4 ↑), 13.04 (CH3 ↑); LC-MS (ESI) RT = 3.91 min, m/z 396.9 [M++H]; LC-MS (ESI) RT = 6.99 min, m/z 392.9 [M++H]; Anal. Calcd for C21H17ClN4O2 (392.84): C, 64.21; H, 4.36; N, 14.26. Found: C, 64.15; H, 4.30; N, 14.21 %.
4.2.5. 6-Amino-1-(4-chlorophenyl)-4-(4-hydroxyphenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (22)
Colorless crystals; yield 94 %; m. p. 219–220 οC; IR (KBr) υ (cm−1): 3479, 3369, 3208 (NH2), 2200 (CN); 1H NMR (DMSO‑d 6, 850 MHz) δ: 9.40 (s, 1H, OH), 7.82 (d, 2H, J = 8.5 Hz, Ar), 7.54 (d, 2H, J = 8.5 Hz, Ar), 7.18 (s, 2H, NH2), 7.04 (d, 2H, J = 8.5 Hz, Ar1), 6.72 (d, 2H, J = 8.5 Hz, Ar1), 4.56 (s, 1H, H-4), 1.78 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 159.57 (C-6),156.75 (C-4, Ar1), 146.34 (C-7a), 144.28 (C-3), 136.86 (C-1, Ar), 134.27 (C-4, Ar), 130.58 (C-1, Ar1), 129.73 (C-2,6, Ar1), 129.27 (C-3,5, Ar), 121.78 (C-2,6, Ar), 120.55 (CN), 115.67 (C-3,5, Ar1), 99.76 (C-3a), 59.14 (C-5), 36.37 (C-4), 13.06 (CH3); 13C NMR-DEPT spectrum at 135°CH, CH3 (positive, up), CH2 (negative, down), revealed the following signals at δ: 129.73 (C-2,6, Ar1 ↑), 129.27 (C-3,5, Ar ↑), 121.78 (C-2,6, Ar ↑), 115.67 (C-3,5, Ar1 ↑), 36.37 (C-4 ↑), 13.06 (CH3 ↑); LC-MS (ESI) RT = 4.2 min, m/z 378.9 [M++H]; Anal. Calcd for C20H15ClN4O2 (378.81): C, 63.41; H, 3.99; N, 14.79. Found: C, 63.34; H, 3.91; N, 14.72 %.
4.2.6. 6-Amino-1-(4-chlorophenyl)-4-(4-nitrophenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (23)
Colorless crystals; yield 90 %; m. p. 221-222οC; IR (KBr) υ (cm−1): 3487, 3363, 3246 (NH2), 2189 (CN); 1H NMR (DMSO‑d 6, 850 MHz) δ: 8.24 (d, 2H, J = 8.5 Hz, Ar1), 7.83 (d, 2H, J = 8.5 Hz, Ar1), 7.59 (d, 2H, J = 8.5 Hz, Ar1), 7.56 (d, 2H, J = 8.5 Hz, Ar1), 7.42 (s, 2H, NH2), 4.92 (s, 1H, H-4), 1.79 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 160.09 (C-6),151.53 (C-4, Ar1), 147.09 (C-7a), 145.14 (C-3), 144.53 (C-1, Ar1), 136.09 (C-1, Ar), 130.87 (C-4, Ar), 129.76 (C-2,6, Ar1), 129.72 (C-3,5, Ar), 124.38 (C-3,5, Ar1), 122.03 (C-2,6, Ar), 120.17 (CN), 98.31 (C-3a), 57.28 (C-5), 36.75 (C-4), 13.02 (CH3); 13C NMR-DEPT spectrum at 135°CH, CH3 (positive, up), CH2 (negative, down), revealed the following signals at δ: 129.76 (C-2,6, Ar1↑), 129.72 (C-3,5, Ar↑), 124.39 (C-3,5, Ar1↑), 122.03 (C-2,6, Ar↑), 36.75 (C-4↑), 13.02 (CH3↑); LC-MS (ESI) RT = 5.27 min, m/z 407.9 [M++H]; Anal. Calcd for C20H14ClN5O3 (407.81): C, 58.90; H, 3.46;N, 17.17. Found: C, 58.82; H, 3.40;N, 17.11 %.
4.2.7. 4-([1,1′-Biphenyl]-4-yl)-6-amino-1-(4-chlorophenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (24)
Colorless crystals; yield 89 %; m. p. 289–290 οC; IR (KBr) υ (cm−1): 3491, 3380, 3249 (NH2), 2204 (CN); 1H NMR (DMSO‑d 6, 850 MHz) δ: 7.84 (d, 2H, J = 8.5 Hz, Ar), 7.68 (d, 2H, J = 8.5 Hz, Ar1), 7.66 (d, 2H, J = 8.5 Hz, Ar), 7.55 (d, 2H, J = 8.5 Hz, Ar1), 7.48 (t, 1H, J = 8.5 Hz, Ar1), 7.37(d, 2H, J = 8.5 Hz, Ar1), 7.35 (d, 2H, J = 8.5 Hz, Ar1), 7.30 (s, 2H, NH2), 4.75 (s, 1H, H-4), 1.83 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 159.88 (C-6),146.27 (C-7a), 144.45 (C-3), 143.21 (C-1′, Ar1), 140.20 (C-1, Ar1), 139.35 (C-1, Ar), 136.82 (C-4, Ar1), 130.71 (C-4, Ar),129.75 (C-3,5, Ar1), 129.43 (C-3,5, Ar), 128.87 (C-3′,5′, Ar1), 127.91 (C-2′,6′, Ar1), 127.34 (C-2,6, Ar1), 127.10 (C-4′, Ar1), 121.91 (C-2,6, Ar), 120.51 (CN), 99.28 (C-3a), 58.42 (C-5), 36.74 (C-4), 13.12 (CH3); 13C NMR-DEPT spectrum at 135°CH, CH3 (positive, up), CH2 (negative, down), revealed the following signals at δ: 129.75 (C-3,5, Ar1 ↑), 129.43 (C-3,5, Ar ↑), 128.87 (C-3′,5′, Ar1 ↑), 127.91 (C-2′,6′, Ar1 ↑), 127.34 (C-2,6, Ar1 ↑), 127.10 (C-4′, Ar1 ↑), 121.91 (C-2,6, Ar ↑), 36.74 (C-4 ↑), 13.12 (CH3 ↑); LC-MS (ESI) RT = 7.45 min, m/z 439 [M++H]; Anal. Calcd for C26H19ClN4O (438.91): C, 71.15; H, 4.36; N, 12.77. Found: C, 71.20; H, 4.40; N, 12.81 %.
4.2.8. 6-Amino-1-(4-chlorophenyl)-4-(2,4-difluorophenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (25)
Colorless crystals; yield 88 %; m. p. 227-228οC; IR (KBr) υ (cm−1): 3485, 3385, 3239 (NH2), 2189 (CN); 1H NMR (DMSO‑d 6, 850 MHz)δ: 7.82 (d, 2H, J = 8.5 Hz, Ar), 7.55 (d, 2H, J = 8.5 Hz, Ar), 7.38 (dt, 1H, J = 11.3, 5.6 Hz, Ar1), 7.35 (s, 2H, NH2), 7.26–7.23 (m, 1H, Ar1), 7.09 (td, 1H, J = 8.4, 2.2 Hz, Ar1), 4.96 (s, 1H, H-4), 1.81 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 161.27 (C-6),160.27 (C-2, Ar1), 158.78 (C-4, Ar1), 145.88 (C-7a), 144.68 (C-3), 136.72 (C-1, Ar), 132.03 (C-6, Ar1), 130.80 (C-4, Ar), 129.76 (C-3,5, Ar), 126.76 (C-1, Ar1), 121.91 (C-2,6, Ar), 120.19 (CN), 112.90 (C-5, Ar1), 104.50 (C-3, Ar1), 98.01 (C-3a), 56.70 (C-5), 30.60 (C-4), 12.69 (CH3); 13C NMR-DEPT spectrum at 135°CH, CH3 (positive, up), CH2 (negative, down), revealed the following signals at δ: 132.03 (C-6, Ar1 ↑), 129.76 (C-3,5, Ar ↑), 121.91 (C-2,6, Ar↑), 112.90 (C-5, Ar1 ↑), 104.50 (C-3, Ar1 ↑), 30.60 (C-4 ↑), 12.69 (CH3 ↑); LC-MS (ESI) RT = 5.85 min, m/z 398.9 [M++H]; Anal. Calcd for C20H13ClF2N4O (398.79): C, 60.24; H, 3.29; N, 14.05. Found: C, 60.31; H, 3.36; N, 14.11 %.
4.2.9. 6-Amino-1-(4-chlorophenyl)-4-(2,3-dichlorophenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (26)
Colorless crystals; yield 91 %; m. p. 195–196 οC; IR (KBr) υ (cm−1): 3460, 3330, 3291 (NH2), 2195 (CN), 1660 (C N); 1H NMR (DMSO‑d 6, 850 MHz) δ: 8.23 (s, 2H, NH2), 7.96–7.26 (m, 7H, Ar and Ar1), 5.00 (s, 1H, H-4), 1.77 (s, 3H, CH3);13C NMR (DMSO‑d 6, 214 MHz) δ: 159.14 (C-6),146.64 (C-7a), 145.76 (C-3), 144.84 (C-1, Ar1), 139.69 (C-1, Ar), 136.73 (C-3, Ar), 131.62 (C-4, Ar), 130.04 (C-2, Ar1), 129.76 (C-3,5, Ar), 129.15 (C-5, Ar1), 128.01 (C-4, Ar1), 127.40 (C-6, Ar1), 122.00 (C-2,6, Ar), 120.82 (CN), 101.66 (C-3a), 56.49 (C-5), 30.01 (C-4), 13.65 (CH3); LC-MS (ESI) RT = 5.11 min, m/z 432 [M++H];Anal. Calcd for C20H13Cl3N4O (431.70): C, 55.64; H, 3.04; N, 12.98. Found: C, 55.69; H, 3.10; N, 13.02 %.
4.2.10. 6-Amino-1-(4-chlorophenyl)-4-(2,4-dichlorophenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (27)
Colorless crystals; yield 91 %; m. p. 200-201οC; IR (KBr) υ (cm−1): 3487, 3378, 3241 (NH2), 2188 (CN); 1H NMR (DMSO‑d 6, 850 MHz)δ: 7.82 (d, 2H, J = 8.7 Hz, Ar), 7.64 (s, 1H, Ar1), 7.55 (d, 2H, J = 8.7 Hz, Ar), 7.43 (d, 2H, J = 8.1 Hz, Ar1), 7.39 (s, 2H, NH2), 5.15 (s, 1H, H-4), 1.77 (s, 3H, CH3);13C NMR (DMSO‑d 6, 214 MHz) δ: 160.32 (C-6),145.80 (C-7a), 144.82 (C-3), 142.20 (C-1, Ar1), 138.81 (C-2, Ar1), 136.71 (C-1, Ar), 133.02 (C-6, Ar1), 130.84 (C-4, Ar, Ar1), 129.76 (C-3,5, Ar), 128.61 (C-3,5, Ar1), 121.98 (C-2,6, Ar), 119.97 (CN), 98.01 (C-3a), 56.70 (C-5), 30.60 (C-4), 12.81 (CH3); 13C NMR-DEPT spectrum at 135°CH, CH3 (positive, up), CH2 (negative, down), revealed the following signals at δ: 132.02 (C-6, Ar1 ↑), 129.76 (C-3,5, Ar ↑), 128.60 (C-3,5, Ar1 ↑), 121.98 (C-2,6, Ar ↑), 30.60 (C-4 ↑), 12.81 (CH3 ↑); LC-MS (ESI) RT = 7.41 min, m/z 430.8 [M++H];Anal. Calcd for C20H13Cl3N4O (431.70): C, 55.64; H, 3.04; N, 12.98. Found: C, 55.58; H, 3.00; N, 12.92 %.
4.2.11. 6-Amino-1-(4-chlorophenyl)-4-(2,5-dichlorophenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (28)
Colorless crystals; yield 91 %; m. p. 215–216 οC; IR (KBr) υ (cm−1): 3438, 3317, 3194 (NH2), 2189 (CN), 1660 (C N); 1H NMR (DMSO‑d 6, 850 MHz)δ: 7.96–7.36 (m, 9, Ar, Ar1andNH2), 5.16 (s, 1H, H-4), 1.78 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 160.44 (C-6),146.61 (C-7a), 145.70 (C-3), 144.93 (C-1, Ar1), 136.76 (C-1, Ar), 133.82 (C-5, Ar1), 130.82 (C-3,6 Ar1), 129.70 (C-3,5, Ar), 129.55 (C-4, Ar), 128.80 (C-2, Ar1), 122.08 (C-4, Ar1), 120.91 (C-2,6, Ar),120.00 (CN), 99.55 (C-3a), 56.49 (C-5), 32.81 (C-4), 12.84 (CH3); LC-MS (ESI) RT = 5.01 min, m/z 431.9 [M++H];Anal. Calcd for C20H13Cl3N4O (431.70): C, 55.64; H, 3.04; N, 12.98. Found: C, 55.70; H, 3.11; N, 13.04 %0.4.2.12 6-Amino-1-(4-chlorophenyl)-4-(2,6-dichlorophenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (29).
Colorless crystals; yield 91 %; m. p. 253–254 οC; IR (KBr) υ (cm−1): 3472, 3316, 3297 (NH2), 2198 (CN), 1655 (C N); 1H NMR (DMSO‑d 6, 850 MHz)δ: 7.81–7.38 (m, 9, Ar, Ar1andNH2), 5.66 (s, 1H, H-4), 1.78 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 161.16 (C-6),145.46 (C-7a), 145.24 (C-3),136.70 (C-1, Ar1), 135.74 (C-1, Ar), 135.23 (C-2, Ar1), 135.15 (C-6 Ar1), 131.49 (C-4, Ar), 130.86 (C-4, Ar1), 130.54 (C-3,5, Ar), 129.81 (C-3 Ar1), 129.18 (C-5, Ar1), 121.87 (C-2,6, Ar), 119.79 (CN), 96.42 (C-3a), 54.10 (C-5), 33.45 (C-4), 12.58 (CH3); LC-MS (ESI) RT = 4.81 min, m/z 431.8 [M++H];Anal. Calcd for C20H13Cl3N4O (431.70): C, 55.64; H, 3.04; N, 12.98. Found: C, 55.71; H, 3.12; N, 13.05 %.
4.2.12. 6-Amino-1-(4-chlorophenyl)-4-(3,4-dichlorophenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (30)
Colorless crystals; yield 91 %; m. p. 216–217 οC; IR (KBr) υ (cm−1): 3450, 3324, 3205 (NH2), 2199 (CN), 1655 (C N); 1H NMR (DMSO‑d 6, 850 MHz)δ: 7.83–7.30 (m, 9, Ar, Ar1andNH2), 4.78 (s, 1H, H-4), 1.81 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 159.99 (C-6),146.04 (C-7a), 145.26 (C-3), 144.58 (C-1, Ar1), 136.78 (C-1, Ar), 131.62 (C-3 Ar1), 131.30 (C-4, Ar), 130.78 (C-2, Ar1), 130.33 (C-5, Ar1), 130.17 (C-4, Ar1), 129.69 (C-3,5, Ar), 128,85 (C-6 Ar1), 122.06 (C-2,6, Ar), 120.26 (CN), 98.31 (C-3a), 57.53 (C-5), 36.18 (C-4), 13.08 (CH3); LC-MS (ESI) RT = 5.04 min, m/z 432 [M++H];Anal. Calcd for C20H13Cl3N4O (431.70): C, 55.64; H, 3.04; N, 12.98. Found: C, 55.57; H, 2.95; N, 12.91 %.
4.2.13. 6-Amino-1-(4-chlorophenyl)-4-(4-ethoxyphenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (31)
Colorless crystals; yield 91 %; m.p.211-212οC; IR (KBr) υ (cm-1): 3459, 3361, 3230 (NH2), 2189 (CN); 1H NMR (DMSO‑d 6, 500 MHz) δ: 7.98 (d, 2H, J = 8.5 Hz, Ar), 7.72 (d, 2H, J = 8.5 Hz, Ar1), 7.54 (d, 2H, J = 8.5 Hz, Ar), 7.31 (s, 2H, NH2), 7.18 (d, 2H, J = 8.5 Hz, Ar1), 4.68 (s, 1H, H-4), 4.17 (q, 2H, J = 8.5 Hz, CH2), 1.79 (s, 3H, CH3), 1.36 (t, 3H, J = 8.5 Hz, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 160.93 (C-6), 151.53 (C-4, Ar1), 147.09 (C-7a), 146.14 (C-3), 136.09 (C-1, Ar), 133.92 (C-2,6, Ar1), 130.87 (C-4, Ar), 129.76 (C-3,5, Ar), 124.44 (C-1, Ar1), 122.03 (C-2,6, Ar), 120.17 (CN), 116.03 (C-3,5, Ar1), 98.56 (C-3a), 64.56 (CH2), 57.92 (C-5), 36.75 (C-4), 14.84 (CH3), 13.02 (CH3); 13C NMR-DEPT spectrum at 135o CH, CH3 (positive, up), CH2 (negative, down), revealed the following signals at δ: 133.92 (C-2,6, Ar1 ↑), 129.76 (C-3,5, Ar ↑), 122.03 (C-2,6, Ar ↑), 116.03 (C-3,5, Ar1 ↑), 64.56 (CH2 ↓), 36.75 (C-4 ↑), 14.84 (CH3 ↑), 13.02 (CH3 ↑); LC-MS (ESI) RT = 5.2 min, m/z 406.9 [M++H]; Anal. Calcd. for C22H19ClN4O2 (406.86): C, 64.94; H, 4.71; N, 13.77. Found: C, 64.88; H, 4.64; N, 13.70 %.
4.2.14. 6-Amino-1-(4-chlorophenyl)-4-(4-hydroxy-3-methoxyphenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (32)
Colorless crystals; yield 91 %; m. p. 181-282οC; IR (KBr) υ (cm−1): 3492, 3360, 3245 (NH2), 2190 (CN); 1H NMR (DMSO‑d 6, 850 MHz) δ: 8.97 (s, 1H, OH), 7.83 (d, 2H, J = 8.5 Hz, Ar), 7.54 (d, 2H, J = 8.5 Hz, Ar), 7.18 (s, 2H, NH2), 6.81 (d, 1H, J = 2.0 Hz, Ar1), 6.75–6.72 (m, 1H, Ar1), 6.62 (dd, 1H, J = 8.1, 2.0 Hz, Ar1), 4.58 (s, 1H, H-4), 3.73 (s, 3H, OCH3), 1.82 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 159.64 (C-6),147.86 (C-3, Ar1), 146.39 (C-7a), 145.94 (C-4, Ar1), 144.26 (C-3), 136.89 (C-1, Ar), 134.87 (C-4, Ar1), 130.56 (C-1, Ar1), 129.71 (C-3,5, Ar),121.75 (C-2,6, Ar), 120.60 (C-6, Ar1), 120.75 (CN), 115.94 (C-5, Ar1),112.36 (C-2, Ar1), 99.62 (C-3a), 58.98 (C-5), 56.13 (CH3), 36.75 (C-4), 13.15 (CH3); 13C NMR-DEPT spectrum at 135°CH, CH3 (positive, up), CH2 (negative, down), revealed the following signals at δ: 129.71 (C-3,5, Ar ↑),121.75 (C-2,6, Ar ↑), 120.60 (C-6, Ar1 ↑), 115.94 (C-5, Ar1 ↑),112.36 (C-2, Ar1 ↑), 56.13 (CH3 ↑), 36.75 (C-4 ↑), 13.15 (CH3 ↑); LC-MS (ESI) RT = 5.97 min, m/z 408.9 [M++H];Anal. Calcd for C21H17ClN4O3 (408.84): C, 61.69; H, 4.19; N, 13.70. Found: C, 61.63; H, 4.14; N, 13.65 %.
4.2.15. 6-Amino-1-(4-chlorophenyl)-4-(3,4,5-trimethoxyphenyl)-3-methyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile (33)
Colorless crystals; yield 89 %; m. p. 192-193οC; IR (KBr) υ (cm−1): 3495, 3386, 3240 (NH2), 2187 (CN); 1H NMR (DMSO‑d 6, 850 MHz) δ: 7.82 (d, 2H, J = 8.5 Hz, Ar), 7.54 (d, 2H, J = 8.5 Hz, Ar), 7.25 (s, 2H, NH2), 6.55 (s, 1H, Ar1), 4.66 (s, 1H, H-4), 3.73 (s, 6H, 2OCH3), 3.64 (s, 3H, OCH3), 1.86 (s, 3H, CH3); 13C NMR (DMSO‑d 6, 214 MHz) δ: 159.95 (C-6),153.31 (C-3,5, Ar1), 146.40 (C-7a), 144.32 (C-3), 139.58 (C-1, Ar1), 136.81 (C-1, Ar), 136.77 (C-4, Ar), 130.64 (C-4, Ar1), 129.72 (C-3,5, Ar), 121.84 (C-2,6, Ar), 120.54 (CN), 105.41 (C-2,6, Ar1), 99.08 (C-3a), 60.47 (CH3), 58.26 (CH3), 56.35 (C-5), 37.38 (C-4), 13.24 (CH3); 13C NMR-DEPT spectrum at 135°CH, CH3 (positive, up), CH2 (negative, down), revealed the following signals at δ: 129.72 (C-3,5, Ar ↑), 121.84 (C-2,6, Ar ↑), 105.41 (C-2,6, Ar1 ↑), 60.47 (CH3 ↑), 58.26 (CH3 ↑), 37.38 (C-4 ↑), 13.24 (CH3 ↑); LC-MS (ESI) RT = 4.47 min, m/z 453 [M++H];Anal. Calcd for C23H21ClN4O4 (452.89): C, 61.00; H, 4.67; N, 12.37. Found: C, 61.08; H, 4.75; N, 12.44 %.
4.3. Mpro protease inhibition assay
The SARS-CoV-2 Mpro enzyme assay was developed in 384-well black, medium binding microplates (Greiner Bio-One, Monroe, NC, USA) with a total volume of 20 μL and then miniaturized to 1536-well format. In a 384-well plate format, 10 μL enzyme in reaction buffer was added into each well, followed by the addition of 10 μL Mpro self-quenching 14-mer fluorogenic (FRET) peptide containing cleavage site indicated by arrow (Dabcyl-KTSAVLQ↓SGFRKME-Edans) (BPS Bioscience)) using a counter- screen assay for elimination of the fluorescence quenching effect of inhibitors. Fluorescent intensity was measured at different time points on a PHERAstar FSX plate reader (BMG Labtech, Cary, NC, USA) with Ex = 340 nm/Em = 460 nm after the addition of substrate. Briefly, 10 μL of drugs (250 μM) were incubated for 1 h with 4 ng Mpro-MBP tagged enzyme in 30 μL of assay buffer. By the end of the incubation time, 10 μL fluorescent substrate (250 μM) were added to initiate the enzymatic reaction. The final reaction volume was 50 μL and the final concentration of drugs and substrate in the reaction mixture was 50 μM. After a second incubation period of 16–18 h, fluorescence was measured at 360/40 nm excitation and 460/40 nm emission using Synergy HT fluorescent plate reader. Drugs were screened from 0 to 1000 μM dose range to calculate the IC50. To prepare positive control wells, 1 % DMSO with 4 ng of enzyme and 50 μM of substrate were added without enzyme inhibition. The standard lopinavir inhibitor was used at 50 μM which also served as a negative control. Wells containing 1 % DMSO with 50 μM of substrate without enzyme served as blank. All the values were subtracted from blank values (see supplementary part for manufacturer’s kit (BPS Biosciences (CA)). The experiment was conducted at both room temperature (RT) and 37 °C. Steady-state kinetic parameters were evaluated using 50 nM 3CLpro and different concentrations of substrate. In brief, 10 μL/well enzyme was added to the 384-well plate. The reaction was then initialized by adding the substrate solutions at different concentrations. The substrate stock solution was serially diluted at 1:2 to obtain seven concentrations. The final concentrations used in this test were 160, 80, 40, 20, 10, 5, and 2.5 μM. The fluorescent intensity was measured for incubated hydrolysate of substrate reaction at 5, 10, 15, and 30 min (see supplementary part). Experiment was performed in triplicate and the error from global fit with variable hill slope to obtain IC50 value is reported, see supporting materials.
4.4. SARS-CoV-2 antiviral assay
To determine the effect of different treatments of target compounds on SARS-CoV-2 viral load (SARS-CoV-2 isolate EGY/WAT-2 VACCERA), qualitative detection of SARS-CoV-2 viral RNA was carried out using a Real-Time PCR assay [74], [75]. Total RNA was extracted using genesig® Coronavirus SARS-CoV-2 Real-Time PCR Assay kit (Primer design TM ltd, Southampton, United Kingdom) according to the manufacturer’s protocol. This kit contains a master mix, primers, and probe designed for reverse transcription of the extracted RNA and Real-Time PCR for the detection of SARS-CoV-2. The assay was performed using the Rotor-Gene Q instrument (Qiagen, Germany) under the following amplification conditions: 10 min Reverse Transcription at 55 °C, 2 min initial activation at 95 °C, followed by 45 cycles of 10 s denaturation at 95 °C and 60 s annealing and extension at 60 °C (see supplementary kit).
4.5. Carrageenan-induced paw edema in rats
Twelve Sprague Dawley Rat (200 g each) were randomly divided into four groups, group 1: healthy control receiving 100 µL DMSO, group 2: positive control group receiving 1 % carrageen an, group 3: diclofenac sodium group receiving 25 mg/kg PO in 100 µL DMSO, and group 4: treatment group receiving 100 mg/kg 27 compound dissolved in in 100 µL DMSO. After 1hr of initiating the different treatments, animals of group 2, 3 and 4 received a subcutaneous injection of 100 µL carrageen an (1 % in saline) in the plantar region of the left hind paw to induce inflammation. Six hours after carrageenan treatment animals were scarified to collect blood for determination of inflammatory markers IL-6&TNF [76], [77]. Results were expressed as mean ± SD compared with the control group. Statistical significance was determined by one-way analysis of variance (ANOVA) at the 95 % confidence interval.
4.6. Molecular docking study
Molecular docking simulation was carried out using combination of the program AutoDock 4.0.1.34 (version 4.0, Molecular graphics laboratories, La Jolla, CA, USA) with the graphical user interface AutoDock tools (ADT) [78] and Molecular Operating Environment (MOE 2014) [79]. The active compounds were docked into (3D) crystal structure of SARS-COV-19 Mpro (PDB: 7L11; https://www.rcsb.org/structure/7L11) in complex with 2-[3-(3,5-dichlorophenyl)-2-oxo[2H–[1,3′-bipyridine]]-5-yl]benzonitrile inhibitor at 1.63 Å resolution. The protocol of docking is fully described prevously [80], [81], [82].
4.7. Molecular dynamics simulation
MD simulations were carried out using templates of the 3D structure crystal of SARS-COV 19 Mpro protease (PDB ID: 7L11) complexed with the target active compound 27 compared to the placement of the reference bound drug. The simulation was carried out using the Desmond ver. 3.8 package [83] with the OPLS2005 forcefield. SGI Rackalbe RP2 Standard-Depth Servers C2108-RP2 (Intel Xeon Processor E5-2670, 16CPU/node) at AIST were used as the computational hardware in this simulation. The initial docked model structure was refined using the protein preparation wizard in Maestro (Schrödinger, LLC) and placed into TIP3P water molecules solvated with 0.15 M NaCl. After minimization and relaxation of the model, the production molecular dynamics phase was performed for three independent 100 ns simulations in an isothermal-isobaric (NpT) ensemble at 300 K and 1 bar using Langevin dynamics. Long-range electrostatic interactions were computed using the Smooth Particle Mesh Ewald method. All system setups were performed using Maestro. Trajectory coordinates were recorded every 100 ps. The simulation trajectories were then analyzed for the sampled conformations in all structural models. The obtained trajectory was processed utilizing the AMBER11 tool ptraj for the calculations of RMSD and PMSF for protein RMSD. All molecular figures were generated using PyMOL (Schrödinger, LLC), see supplementry data.
4.8. PBS solubility screening
The solubility screen was conducted as described previously [84].
4.9. Analysis of the metabolic stability
The metabolic stability of compound 27 was analyzed as described in a previous study [85].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements:
This research was funded by Institutional Fund Projects under grant No. (IFPHI-182-166-2020), The authors, therefore, gratefully acknowledge technical and financial support from the Ministry of Education and King Abdulaziz University, the Deanship of Scientific Research (DSR), Jeddah, Saudi Arabia. In addition, authors would like to thank Prof. Michael Gütschow and Prof. Jürgen Bajorath for their help in manuscript revision.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioorg.2022.106255.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.The World Health Organization, WHO Coronavirus (COVID-19) Dashboard, (2022). https://covid19.who.int/ (accessed July 7, 2022).
- 2.M.I.A. Hamed, K.M. Darwish, R. Soltane, A. Chrouda, A. Mostafa, N.M. Abo Shama, S.S. Elhady, H.S. Abulkhair, A.E. Khodir, A.A. Elmaaty, A.A. Al-karmalawy, β-Blockers bearing hydroxyethylamine and hydroxyethylene as potential SARS-CoV-2 Mpro inhibitors: rational based design, in silico , in vitro , and SAR studies for lead optimization, RSC Adv. 11 (2021) 35536–35558. 10.1039/D1RA04820A. [DOI] [PMC free article] [PubMed]
- 3.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19. Science (80-.) 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh A.K., Brindisi M., Shahabi D., Chapman M.E., Mesecar A.D. Drug Development and Medicinal Chemistry Efforts toward SARS-Coronavirus and Covid-19 Therapeutics. ChemMedChem. 2020;15:907–932. doi: 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.N. Kitamura, M.D. Sacco, C. Ma, Y. Hu, J.A. Townsend, X. Meng, F. Zhang, X. Zhang, M. Ba, T. Szeto, A. Kukuljac, M.T. Marty, D. Schultz, S. Cherry, Y. Xiang, Y. Chen, J. Wang, Expedited Approach toward the Rational Design of Noncovalent SARS-CoV-2 Main Protease Inhibitors., J. Med. Chem. (2021). 10.1021/acs.jmedchem.1c00509. [DOI] [PMC free article] [PubMed]
- 7.X. Chen, B. Zhao, Y. Qu, Y. Chen, J. Xiong, Y. Feng, D. Men, Q. Huang, Y. Liu, B. Yang, J. Ding, F. Li, Detectable Serum Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load (RNAemia) Is Closely Correlated With Drastically Elevated Interleukin 6 Level in Critically Ill Patients With Coronavirus Disease 2019, Clin. Infect. Dis. 71 (2020) 1937–1942. 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed]
- 8.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou X., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyne J.D., Brickman A.M. The Impact of the COVID-19 Pandemic on Dementia Risk: potential pathways to cognitive decline. Neurodegener. Dis. 2021;21:1–23. doi: 10.1159/000518581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayden K.M., Beavers D.P., Steck S.E., Hebert J.R., Tabung F.K., Shivappa N., Casanova R., Manson J.E., Padula C.B., Salmoirago-Blotcher E., Snetselaar L.G., Zaslavsky O., Rapp S.R. The association between an inflammatory diet and global cognitive function and incident dementia in older women: the women’s health initiative memory study. Alzheimer’s Dement. 2017;13:1187–1196. doi: 10.1016/j.jalz.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A.P. Muller, G.K. Ferreira, A.J. Pires, G. de Bem Silveira, D.L. de Souza, J. de A. Brandolfi, C.T. de Souza, M.M.S. Paula, P.C.L. Silveira, Gold nanoparticles prevent cognitive deficits, oxidative stress and inflammation in a rat model of sporadic dementia of Alzheimer’s type, Mater. Sci. Eng. C. 77 (2017) 476–483. 10.1016/j.msec.2017.03.283. [DOI] [PubMed]
- 12.Cully M. Immune status could determine efficacy of COVID-19 therapies. Nat. Rev. Drug Discov. 2020;19:431–434. doi: 10.1038/d41573-020-00110-3. [DOI] [PubMed] [Google Scholar]
- 13.Mehta P., Fajgenbaum D.C. Is severe COVID-19 a cytokine storm syndrome: a hyperinflammatory debate. Curr. Opin. Rheumatol. 2021;33:419–430. doi: 10.1097/BOR.0000000000000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar A., Lohan P., Aneja D.K., Gupta G.K., Kaushik D., Prakash O. Design, synthesis, computational and biological evaluation of some new hydrazino derivatives of DHA and pyranopyrazoles. Eur. J. Med. Chem. 2012;50:81–89. doi: 10.1016/j.ejmech.2012.01.042. [DOI] [PubMed] [Google Scholar]
- 15.Loddo R., Novelli F., Sparatore A., Tasso B., Tonelli M., Boido V., Sparatore F., Collu G., Delogu I., Giliberti G., La Colla P. Antiviral activity of benzotriazole derivatives. 5-[4-(Benzotriazol-2-yl)phenoxy]-2,2-dimethylpentanoic acids potently and selectively inhibit Coxsackie Virus B5. Bioorg. Med. Chem. 2015;23:7024–7034. doi: 10.1016/j.bmc.2015.09.035. [DOI] [PubMed] [Google Scholar]
- 16.Ibba R., Piras S., Corona P., Riu F., Loddo R., Delogu I., Collu G., Sanna G., Caria P., Dettori T., Carta A. Synthesis, antitumor and antiviral in vitro activities of new benzotriazole-dicarboxamide derivatives. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.660424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A.A. Gaber, A.M. El‐Morsy, F.F. Sherbiny, A.H. Bayoumi, K.M. El‐Gamal, K. El‐Adl, A.A. Al‐Karmalawy, R.R. Ezz Eldin, M.A. Saleh, H.S. Abulkhair, Pharmacophore‐linked pyrazolo[3,4‐ d ]pyrimidines as EGFR‐TK inhibitors: Synthesis, anticancer evaluation, pharmacokinetics, and in silico mechanistic studies, Arch. Pharm. (Weinheim). (2021). 10.1002/ardp.202100258. [DOI] [PubMed]
- 18.Ezzat H.G., Bayoumi A.H., Sherbiny F.F., El-Morsy A.M., Ghiaty A., Alswah M., Abulkhair H.S. Design, synthesis, and molecular docking studies of new [1,2,4]triazolo[4,3-a]quinoxaline derivatives as potential A2B receptor antagonists. Mol. Divers. 2021;25:291–306. doi: 10.1007/s11030-020-10070-w. [DOI] [PubMed] [Google Scholar]
- 19.McCord R.S., Breinig M.K., Morahan P.S. Antiviral Effect of Pyran Against Systemic Infection of Mice with Herpes Simplex Virus Type 2. Antimicrob. Agents Chemother. 1976;10:28–33. doi: 10.1128/AAC.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Costa E.C.B., Amorim R., da Silva F.C., Rocha D.R., Papa M.P., de Arruda L.B., Mohana-Borges R., Ferreira V.F., Tanuri A., da Costa L.J., Ferreira S.B. Synthetic 1,4-Pyran Naphthoquinones Are Potent Inhibitors of Dengue Virus Replication. PLoS One. 2013;8:e82504. doi: 10.1371/journal.pone.0082504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J.-J., Liao H.-R., Chen K.-S., Cheng M.-J., Shu C.-W., Sung P.-J., Lim Y.-P., Wang T.-C., Kuo W.-L. A New 2H-Pyran-2-One Derivative and Anti-inflammatory Constituents of Alpinia zerumbet. Chem. Nat. Compd. 2017;53:40–43. doi: 10.1007/s10600-017-1906-6. [DOI] [Google Scholar]
- 22.Mohareb R.M., Zaki M.Y., Abbas N.S. Synthesis, anti-inflammatory and anti-ulcer evaluations of thiazole, thiophene, pyridine and pyran derivatives derived from androstenedione. Steroids. 2015;98:80–91. doi: 10.1016/j.steroids.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Mantzanidou M., Pontiki E., Hadjipavlou-Litina D. Pyrazoles and Pyrazolines as Anti-Inflammatory Agents. Molecules. 2021;26:3439. doi: 10.3390/molecules26113439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surendra Kumar R., Arif I.A., Ahamed A., Idhayadhulla A. Anti-inflammatory and antimicrobial activities of novel pyrazole analogues, Saudi. J. Biol. Sci. 2016;23:614–620. doi: 10.1016/j.sjbs.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Sabbagh O.I., Baraka M.M., Ibrahim S.M., Pannecouque C., Andrei G., Snoeck R., Balzarini J., Rashad A.A. Synthesis and antiviral activity of new pyrazole and thiazole derivatives. Eur. J. Med. Chem. 2009;44:3746–3753. doi: 10.1016/j.ejmech.2009.03.038. [DOI] [PubMed] [Google Scholar]
- 26.Ouyang G., Cai X.-J., Chen Z., Song B.-A., Bhadury P.S., Yang S., Jin L.-H., Xue W., Hu D.-Y., Zeng S. Synthesis and antiviral activities of pyrazole derivatives containing an oxime moiety. J. Agric. Food Chem. 2008;56:10160–10167. doi: 10.1021/jf802489e. [DOI] [PubMed] [Google Scholar]
- 27.Conti C., Proietti Monaco L., Desideri N. Synthesis and anti-rhinovirus activity of novel 3-[2-(pyridinyl)vinyl]substituted -2H-chromenes and -4H-chromen-4-ones. Bioorg. Med. Chem. 2014;22:1201–1207. doi: 10.1016/j.bmc.2013.11.054. [DOI] [PubMed] [Google Scholar]
- 28.Conti C., Proietti Monaco L., Desideri N. Design, synthesis and in vitro evaluation of novel chroman-4-one, chroman, and 2H-chromene derivatives as human rhinovirus capsid-binding inhibitors. Bioorg. Med. Chem. 2011;19:7357–7364. doi: 10.1016/j.bmc.2011.10.060. [DOI] [PubMed] [Google Scholar]
- 29.Ouyang G., Chen Z., Cai X.-J., Song B.-A., Bhadury P.S., Yang S., Jin L.-H., Xue W., Hu D.-Y., Zeng S. Synthesis and antiviral activity of novel pyrazole derivatives containing oxime esters group. Bioorg. Med. Chem. 2008;16:9699–9707. doi: 10.1016/j.bmc.2008.09.070. [DOI] [PubMed] [Google Scholar]
- 30.Mizuhara T., Kato T., Hirai A., Kurihara H., Shimada Y., Taniguchi M., Maeta H., Togami H., Shimura K., Matsuoka M., Okazaki S., Takeuchi T., Ohno H., Oishi S., Fujii N. Structure–activity relationship study of phenylpyrazole derivatives as a novel class of anti-HIV agents. Bioorg. Med. Chem. Lett. 2013;23:4557–4561. doi: 10.1016/j.bmcl.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 31.Desideri N., Fioravanti R., Proietti Monaco L., Atzori E.M., Carta A., Delogu I., Collu G., Loddo R., Design S. Antiviral Evaluation, and SAR Studies of New 1-(Phenylsulfonyl)-1H-Pyrazol−4-yl-Methylaniline Derivatives. Front. Chem. 2019;7 doi: 10.3389/fchem.2019.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menozzi G., Mosti L., Schenone P., Donnoli D., Schiariti F., Marmo E. 1-phenyl-1H-pyrazole derivatives with antiinflammatory, analgesic and antipiretic activities. Farmaco. 1990;45:167–186. http://www.ncbi.nlm.nih.gov/pubmed/2133993 [PubMed] [Google Scholar]
- 33.Menozzi G., Mosti L., Schenone P., D’Amico M., Falciani M., Filippelli W. 1-Aryl-1H-pyrazole-5-acetic acids with antiinflammatory, analgesic and other activities. Farmaco. 1994;49:115–119. http://www.ncbi.nlm.nih.gov/pubmed/8003179 [PubMed] [Google Scholar]
- 34.Penning T.D., Talley J.J., Bertenshaw S.R., Carter J.S., Collins P.W., Docter S., Graneto M.J., Lee L.F., Malecha J.W., Miyashiro J.M., Rogers R.S., Rogier D.J., Yu S.S., Anderson G.D., Burton E.G., Cogburn J.N., Gregory S.A., Koboldt C.M., Perkins W.E., Seibert K., Veenhuizen A.W., Zhang Y.Y., Isakson P.C. Synthesis and Biological Evaluation of the 1,5-Diarylpyrazole Class of Cyclooxygenase-2 Inhibitors: Identification of 4-[5-(4-Methylphenyl)-3- (trifluoromethyl)-1 H -pyrazol-1-yl]benzenesulfonamide (SC-58635, Celecoxib) J. Med. Chem. 1997;40:1347–1365. doi: 10.1021/jm960803q. [DOI] [PubMed] [Google Scholar]
- 35.Reed G.A., Griffin I.O., Eling T.E. Inactivation of prostaglandin H synthase and prostacyclin synthase by phenylbutazone. Requirement for peroxidative metabolism. Mol. Pharmacol. 1985;27:109–114. http://www.ncbi.nlm.nih.gov/pubmed/3917545 [PubMed] [Google Scholar]
- 36.Conti C., Proietti Monaco L., Desideri N. 3-Phenylalkyl-2 H -chromenes and -chromans as novel rhinovirus infection inhibitors. Bioorg. Med. Chem. 2017;25:2074–2083. doi: 10.1016/j.bmc.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Raj V., Lee J.-H., Shim J.-J., Lee J. Antiviral activities of 4H-chromen-4-one scaffold-containing flavonoids against SARS–CoV–2 using computational and in vitro approaches. J. Mol. Liq. 2022;353 doi: 10.1016/j.molliq.2022.118775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahmoud A., Mostafa A., Al-Karmalawy A.A., Zidan A., Abulkhair H.S., Mahmoud S.H., Shehata M., Elhefnawi M.M., Ali M.A. Telaprevir is a potential drug for repurposing against SARS-CoV-2: computational and in vitro studies. Heliyon. 2021;7:e07962. doi: 10.1016/j.heliyon.2021.e07962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amin S.A., Banerjee S., Ghosh K., Gayen S., Jha T. Protease targeted COVID-19 drug discovery and its challenges: Insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg. Med. Chem. 2021;29 doi: 10.1016/j.bmc.2020.115860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 41.Hammoud M.M., Khattab M., Abdel-Motaal M., Van der Eycken J., Alnajjar R., Abulkhair H., Al-Karmalawy A.A. Synthesis, structural characterization, DFT calculations, molecular docking, and molecular dynamics simulations of a novel ferrocene derivative to unravel its potential antitumor activity. J. Biomol. Struct. Dyn. 2022 doi: 10.1080/07391102.2022.2082533. [DOI] [PubMed] [Google Scholar]
- 42.Othman E.M., Fayed E.A., Husseiny E.M., Abulkhair H.S. Rationale design, synthesis, cytotoxicity evaluation, and in silico mechanistic studies of novel 1,2,3-triazoles with potential anticancer activity. New J. Chem. 2022;46:12206–12216. doi: 10.1039/d2nj02061k. [DOI] [Google Scholar]
- 43.Othman E.M., Fayed E.A., Husseiny E.M., Abulkhair H.S. Apoptosis induction, PARP-1 inhibition, and cell cycle analysis of leukemia cancer cells treated with novel synthetic 1,2,3-triazole-chalcone conjugates. Bioorg. Chem. 2022;123 doi: 10.1016/j.bioorg.2022.105762. [DOI] [PubMed] [Google Scholar]
- 44.Othman E.M., Fayed E.A., Husseiny E.M., Abulkhair H.S. The effect of novel synthetic semicarbazone- and thiosemicarbazone-linked 1,2,3-triazoles on the apoptotic markers, VEGFR-2, and cell cycle of myeloid leukemia. Bioorg. Chem. 2022;127 doi: 10.1016/j.bioorg.2022.105968. [DOI] [PubMed] [Google Scholar]
- 45.Abul-Khair H., Elmeligie S., Bayoumi A., Ghiaty A., El-Morsy A., Hassan M.H. Synthesis and evaluation of some new (1,2,4) triazolo(4,3-a)quinoxalin- 4(5h)-one derivatives as AMPA receptor antagonists. J. Heterocycl. Chem. 2013;50:1202–1208. doi: 10.1002/jhet.714. [DOI] [Google Scholar]
- 46.El-Adl K., Ibrahim M.K., Khedr F., Abulkhair H.S., Eissa I.H. Design, synthesis, docking, and anticancer evaluations of phthalazines as VEGFR-2 inhibitors. Arch. Pharm. (Weinheim). 2022;355:2100278. doi: 10.1002/ardp.202100278. [DOI] [PubMed] [Google Scholar]
- 47.Abulkhair H.S., Turky A., Ghiaty A., Ahmed H.E.A., Bayoumi A.H. Novel triazolophthalazine-hydrazone hybrids as potential PCAF inhibitors: Design, synthesis, in vitro anticancer evaluation, apoptosis, and molecular docking studies. Bioorg. Chem. 2020;100 doi: 10.1016/j.bioorg.2020.103899. [DOI] [PubMed] [Google Scholar]
- 48.Aljuhani A., Ahmed H.E.A., Ihmaid S.K., Omar A.M., Althagfan S.S., Alahmadi Y.M., Ahmad I., Patel H., Ahmed S., Almikhlafi M.A., El-Agrody A.M., Zayed M.F., Turkistani S.A., Abulkhair S.H., Almaghrabi M., Salama S.A., Al-Karmalawy A.A., Abulkhair H.S. In vitro and computational investigations of novel synthetic carboxamide-linked pyridopyrrolopyrimidines with potent activity as SARS-CoV-2-M Pro inhibitors. RSC Adv. 2022;12:26895–26907. doi: 10.1039/D2RA04015H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wakodkar R.R., Sanap G., Farooqui M., Kayande D.D. A Facile Synthesis of 6-Amino-4-Aryl-3-Methyl-2,4- Dihydropyrano [2,3-C] Pyrazole-5-Carbonitriles in Aqueous Medium. J. Ultra Chem. 2018;14:50–56. doi: 10.22147/juc/140201. [DOI] [Google Scholar]
- 50.Otto H.-H. Darstellung einiger 4H-Pyrano[2.3-c]pyrazolderivate. Arch. Pharm. 1974;307:444–447. doi: 10.1002/ardp.19743070609. [DOI] [PubMed] [Google Scholar]
- 51.Mamaghani M., Hossein Nia R. A Review on the Recent Multicomponent Synthesis of Pyranopyrazoles, Polycycl. Aromat. Compd. 2021;41:223–291. doi: 10.1080/10406638.2019.1584576. [DOI] [Google Scholar]
- 52.H.-H. Otto, H. Schmelz, Heterocyclen durch Michael-Reaktionen, 5. Mitt. Nucleophile Additionen an 4-Aryliden-pyrazolone, Arch. Pharm. (Weinheim). 312 (1979) 478–486. 10.1002/ardp.19793120604.
- 53.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.-H. An Overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL Protease Inhibitors: Peptidomimetics and Small Molecule Chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoffman R.L., Kania R.S., Brothers M.A., Davies J.F., Ferre R.A., Gajiwala K.S., He M., Hogan R.J., Kozminski K., Li L.Y., Lockner J.W., Lou J., Marra M.T., Mitchell L.J., Murray B.W., Nieman J.A., Noell S., Planken S.P., Rowe T., Ryan K., Smith G.J., Solowiej J.E., Steppan C.M., Taggart B. Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19. J. Med. Chem. 2020;63:12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma C., Hu Y., Townsend J.A., Lagarias P.I., Marty M.T., Kolocouris A., Wang J. Ebselen, Disulfiram, Carmofur, PX-12, Tideglusib, and Shikonin Are Nonspecific Promiscuous SARS-CoV-2 Main Protease Inhibitors. ACS Pharmacol. Transl. Sci. 2020;3:1265–1277. doi: 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vuong W., Khan M.B., Fischer C., Arutyunova E., Lamer T., Shields J., Saffran H.A., McKay R.T., van Belkum M.J., Joyce M.A., Young H.S., Tyrrell D.L., Vederas J.C., Lemieux M.J. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang C.-H., Stone E.A., Deshmukh M., Ippolito J.A., Ghahremanpour M.M., Tirado-Rives J., Spasov K.A., Zhang S., Takeo Y., Kudalkar S.N., Liang Z., Isaacs F., Lindenbach B., Miller S.J., Anderson K.S., Jorgensen W.L. Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations. ACS Cent. Sci. 2021;7:467–475. doi: 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osipiuk J., Azizi S.-A., Dvorkin S., Endres M., Jedrzejczak R., Jones K.A., Kang S., Kathayat R.S., Kim Y., Lisnyak V.G., Maki S.L., Nicolaescu V., Taylor C.A., Tesar C., Zhang Y.-A., Zhou Z., Randall G., Michalska K., Snyder S.A., Dickinson B.C., Joachimiak A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021;12:743. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gunther S., Asper M., Roser C., Luna L., Drosten C., Beckerziaja B., Borowski P., Chen H., Hosmane R. Application of real-time PCR for testing antiviral compounds against Lassa virus, SARS coronavirus and Ebola virus in vitro. Antiviral Res. 2004;63:209–215. doi: 10.1016/j.antiviral.2004.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 61.Xu Y., Barauskas O., Kim C., Babusis D., Murakami E., Kornyeyev D., Lee G., Stepan G., Perron M., Bannister R., Schultz B.E., Sakowicz R., Porter D., Cihlar T., Feng J.Y. Off-target in vitro profiling demonstrates that remdesivir is a highly selective antiviral agent. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.02237-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khadke S.K., Lee J.-H., Kim Y.-G., Raj V., Lee J. Assessment of Antibiofilm Potencies of Nervonic and Oleic Acid against Acinetobacter baumannii Using In Vitro and Computational Approaches. Biomedicines. 2021;9:1133. doi: 10.3390/biomedicines9091133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raj V., Park J.G., Cho K.-H., Choi P., Kim T., Ham J., Lee J. Assessment of antiviral potencies of cannabinoids against SARS-CoV-2 using computational and in vitro approaches. Int. J. Biol. Macromol. 2021;168:474–485. doi: 10.1016/j.ijbiomac.2020.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mazal H., Aviram H., Riven I., Haran G. Effect of ligand binding on a protein with a complex folding landscape. Phys. Chem. Chem. Phys. 2018;20:3054–3062. doi: 10.1039/C7CP03327C. [DOI] [PubMed] [Google Scholar]
- 65.El-Adl K., El-Helby A.G.A., Sakr H., Ayyad R.R., Mahdy H.A., Nasser M., Abulkhair H.S., El-Hddad S.S.A. Design, synthesis, molecular docking, anticancer evaluations, and in silico pharmacokinetic studies of novel 5-[(4-chloro/2,4-dichloro)benzylidene]thiazolidine-2,4-dione derivatives as VEGFR-2 inhibitors. Arch. Pharm. (Weinheim). 2021;354:2000279. doi: 10.1002/ardp.202000279. [DOI] [PubMed] [Google Scholar]
- 66.El-Shershaby M.H., Ghiaty A., Bayoumi A.H., Ahmed H.E.A., El-Zoghbi M.S., El-Adl K., Abulkhair H.S. 1,2,4-Triazolo[4,3- c ]quinazolines: a bioisosterism-guided approach towards the development of novel PCAF inhibitors with potential anticancer activity. New J. Chem. 2021;45:11136–11152. doi: 10.1039/d1nj00710f. [DOI] [PubMed] [Google Scholar]
- 67.El-Adl K., Sakr H., El-Hddad S.S.A., El-Helby A.G.A., Nasser M., Abulkhair H.S. Design, synthesis, docking, ADMET profile, and anticancer evaluations of novel thiazolidine-2,4-dione derivatives as VEGFR-2 inhibitors. Arch. Pharm. (Weinheim). 2021;354:2000491. doi: 10.1002/ardp.202000491. [DOI] [PubMed] [Google Scholar]
- 68.Khedr F., Ibrahim M.K., Eissa I.H., Abulkhair H.S., El-Adl K. Phthalazine-based VEGFR-2 inhibitors: Rationale, design, synthesis, in silico, ADMET profile, docking, and anticancer evaluations. Arch. Pharm. (Weinheim). 2021;354:202100201. doi: 10.1002/ardp.202100201. [DOI] [PubMed] [Google Scholar]
- 69.Mancy A., Abutaleb N.S., Elsebaei M.M., Saad A.Y., Kotb A., Ali A.O., Abdel-Aleem J.A., Mohammad H., Seleem M.N., Mayhoub A.S. Balancing Physicochemical Properties of Phenylthiazole Compounds with Antibacterial Potency by Modifying the Lipophilic Side Chain. ACS Infect. Dis. 2020;6:80–90. doi: 10.1021/acsinfecdis.9b00211. [DOI] [PubMed] [Google Scholar]
- 70.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- 71.Turky A., Sherbiny F.F., Bayoumi A.H., Ahmed H.E.A., Abulkhair H.S. Novel 1,2,4-triazole derivatives: design, synthesis, anticancer evaluation, molecular docking, and pharmacokinetic profiling studies. Arch. Pharm. (Weinheim). 2020;353:2000170. doi: 10.1002/ardp.202000170. [DOI] [PubMed] [Google Scholar]
- 72.Abulkhair H.S., Elmeligie S., Ghiaty A., El-Morsy A., Bayoumi A.H., Ahmed H.E.A., El-Adl K., Zayed M.F., Hassan M.H., Akl E.N., El-Zoghbi M.S. In vivo- and in silico-driven identification of novel synthetic quinoxalines as anticonvulsants and AMPA inhibitors. Arch. Pharm. (Weinheim). 2021;354:2000449. doi: 10.1002/ardp.202000449. [DOI] [PubMed] [Google Scholar]
- 73.Naik C.G., Malik G.M. Synthesis of pyrazolone derivatives and their biological activities. Orient. J. Chem. 2010;26:113–116. http://www.orientjchem.org/vol26no1/synthesis-of-pyrazolone-derivatives-and-their-biological-activities/ [Google Scholar]
- 74.W. Zhu, M. Xu, C.Z. Chen, H. Guo, M. Shen, X. Hu, P. Shinn, C. Klumpp-Thomas, S.G. Michael, W. Zheng, Identification of SARS-CoV-2 3CL Protease Inhibitors by a Quantitative High-throughput Screening, 2020. 10.1101/2020.07.17.207019. [DOI] [PMC free article] [PubMed]
- 75.Kutkat O., Moatasim Y., Al-Karmalawy A.A., Abulkhair H.S., Gomaa M.R., El-Taweel A.N., Abo Shama N.M., GabAllah M., Mahmoud D.B., Kayali G., Ali M.A., Kandeil A., Mostafa A. Robust antiviral activity of commonly prescribed antidepressants against emerging coronaviruses: in vitro and in silico drug repurposing studies. Sci. Rep. 2022;12:12920. doi: 10.1038/s41598-022-17082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Al-Majed A.A., Khattab M., Raza M., Al-Shabanah O.A., Mostafa A.M. Potentiation of diclofenac-induced anti-inflammatory response by aminoguanidine in carrageenan-induced acute inflammation in rats: The role of nitric oxide. Inflamm. Res. 2003;52:378–382. doi: 10.1007/s00011-003-1189-1. [DOI] [PubMed] [Google Scholar]
- 77.Winter C.A., Risley E.A., Nuss G.W. Carrageenin-Induced Edema in Hind Paw of the Rat as an Assay for Antiinflammatory Drugs. Exp. Biol. Med. 1962;111:544–547. doi: 10.3181/00379727-111-27849. [DOI] [PubMed] [Google Scholar]
- 78.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- 79.Integrated Computer-Aided Molecular Design Platform, (2014). https://www.chemcomp.com/Products.htm (accessed May 12, 2022).
- 80.Ahmed H.E.A., Ihmaid S.K., Omar A.M., Shehata A.M., Rateb H.S., Zayed M.F., Ahmed S., Elaasser M.M. Design, synthesis, molecular docking of new lipophilic acetamide derivatives affording potential anticancer and antimicrobial agents. Bioorg. Chem. 2018;76:332–342. doi: 10.1016/j.bioorg.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 81.Rezki N., Almehmadi M.A., Ihmaid S., Shehata A.M., Omar A.M., Ahmed H.E.A., Aouad M.R. Novel scaffold hopping of potent benzothiazole and isatin analogues linked to 1,2,3-triazole fragment that mimic quinazoline epidermal growth factor receptor inhibitors: synthesis, antitumor and mechanistic analyses. Bioorg. Chem. 2020;103 doi: 10.1016/j.bioorg.2020.104133. [DOI] [PubMed] [Google Scholar]
- 82.Zaki A.A., Kaddah M.M.Y., Abulkhair H.S., Ashour A. Unravelling the antifungal and antiprotozoal activities and LC-MS/MS quantification of steroidal saponins isolated from Panicum turgidum. RSC Adv. 2022;12:2980–2991. doi: 10.1039/D1RA08532H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.K.J. Bowers, D.E. Chow, H. Xu, R.O. Dror, M.P. Eastwood, B.A. Gregersen, J.L. Klepeis, I. Kolossvary, M.A. Moraes, F.D. Sacerdoti, J.K. Salmon, Y. Shan, D.E. Shaw, Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters, in: ACM/IEEE SC 2006 Conf., IEEE, 2006: pp. 43–43. 10.1109/SC.2006.54.
- 84.Seleem M.A., Disouky A.M., Mohammad H., Abdelghany T.M., Mancy A.S., Bayoumi S.A., Elshafeey A., El-Morsy A., Seleem M.N., Mayhoub A.S. Second-generation phenylthiazole antibiotics with enhanced pharmacokinetic properties. J. Med. Chem. 2016;59:4900–4912. doi: 10.1021/acs.jmedchem.6b00233. [DOI] [PubMed] [Google Scholar]
- 85.Ihmaid S.K., Alraqa S.Y., Aouad M.R., Aljuhani A., Elbadawy H.M., Salama S.A., Rezki N., Ahmed H.E.A. Design of molecular hybrids of phthalimide-triazole agents with potent selective MCF-7/HepG2 cytotoxicity: synthesis, EGFR inhibitory effect, and metabolic stability. Bioorg. Chem. 2021;111 doi: 10.1016/j.bioorg.2021.104835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.