Abstract
Understanding the mechanisms responsible for the kidney's defense against ascending uropathogen is critical to devise novel treatment strategies against increasingly antibiotic resistant uropathogen. Growing body of evidence indicate Intercalated cells of the kidney as the key innate immune epithelial cells against uropathogen. The aim of this study was to find orthologous and differentially expressed bacterial defense genes in human versus murine intercalated cells. We simultaneously analyzed 84 antibacterial genes in intercalated cells enriched from mouse and human kidney samples. Intercalated cell “reporter mice” were exposed to saline versus uropathogenic Escherichia coli (UPEC) transurethrally for 1 h in vivo, and intercalated cells were flow sorted. Human kidney intercalated cells were enriched from kidney biopsy using magnetic‐activated cell sorting and exposed to UPEC in vitro for 1 h. RT2 antibacterial PCR array was performed. Mitogen‐activated protein kinase kinase kinase 7 (MAP3K7) messenger RNA (mRNA) expression increased in intercalated cells of both humans and mice following UPEC exposure. Intercalated cell MAP3K7 protein expression was defined by immunofluorescence and confocal imaging analysis, was consistent with the increased MAP3K7 mRNA expression profiles defined by PCR. The presence of the orthologous innate immune gene MAP3K7/TAK1 suggests that it may be a key regulator of the intercalated cell antibacterial response and demands further investigation of its role in urinary tract infection pathogenesis.
Keywords: intercalated cells, mitogen‐activated protein kinase, urinary tract infection, uropathogenic Escherichia coli
1. INTRODUCTION
An estimated 250 000 cases of pyelonephritis occur annually in the United States. 1 Multidrug resistance is emerging as a critical challenge regarding treatment of urinary tract infections (UTIs) and potential pyelonephritis complications include hypertension, chronic kidney disease, and urosepsis. 2 , 3 , 4 Uropathogenic Escherichia coli (UPEC) which is responsible for 70%–80% of acute pyelonephritis selectively localized to the apical surface of kidney intercalated cells (IC)s. 5 , 6 A body of emerging evidence indicates that ICs are critical bacterial defense cells in the kidneys. We have previously identified that human ICs express antimicrobial peptides (AMPs). 7 , 8 We and others have demonstrated that abnormal murine IC development results in increased susceptibility to UTIs and/or pyelonephritis. 9 , 10 Lastly, we have demonstrated innate immune and AMP gene expression in enriched murine ICs, and human ICs phagocytose UPEC. 11 , 12 , 13 Thus, defining the mechanisms that ICs use to defend the kidney will provide the foundation for future kidney based immunomodulation therapies. Differences and similarities between human and murine ICs will be important to delineate to contextualize mouse model findings into human innate immune pathophysiology. The objective of this study is to determine overlap and differences in the gene expression of human versus mouse IC response to ascending uropathogen.
2. MATERIALS AND METHODS
2.1. Study approval
Murine studies were approved by the Institutional Animal Care and Use Committees (IACUC) at the Indiana University School of Medicine protocol numbers 11333 and 20105 and adhered to the NIH Guide for the Care and Use of laboratory Animals. Uropathogen use was approved by Indiana biosafety committee protocol number IN‐891. Human studies were approved by Indiana University Institutional Review Board (protocol #1802253259).
2.2. Bacteria
UPEC strain CFT073 was used in the study (originally a gift from Dr. Matthew A. Mulvey, University of Utah). The bacteria were stored in 50% glycerol at −80°C until use. When needed for transurethral inoculation or in vitro exposure, an aliquot was placed in Luria broth then grown overnight at 37°C in a rotating shaker. The next day, bacterial stock was spun down and resuspended in sterile phosphate‐buffered saline (PBS) at a concentration of 1 × 108 CFT073 in 50 µl of sterile PBS for use in murine experimental UTI.
2.3. Mice
ICs were flow sorted from “IC reporter” mice as previously described. 13 Briefly, V‐ATPase B1‐cre mice (kindly provided by Dr. Raoul Nelson, University of Utah), in which 7‐kb B1 promoter drives the cre expression in ICs were crossed with tdTomato‐loxp homozygous mice (B6.Cg‐Gt(ROSA)26sortm9(CAG‐tdTomato) (Stock no. 007909; Jackson laboratory). The progeny of the crosses were genotyped for the presence of cre gene. Mice positive for the cre gene were used and termed “IC reporter” mice. ICs in this strain of mice are marked in situ with red fluorescence, which can be enriched by flowcytometry. To further confirm the messenger RNA (mRNA) data from “IC reporter” mice, c‐Kit+ IC cells were enriched from wild type C57BL/6 mice (Jackson laboratory) exposed to saline and UPEC in vivo. n = 3 mice/group were used, and targeted Map3k7 mRNA expression was measured in enriched IC cells.
2.4. Experimental UTI
Under isoflurane anesthesia, Female “IC reporter” mice aged ~8–10 weeks were inoculated with 1 × 108 CFT073 in 50 µl sterile PBS with a transurethral injection. One hour later, mice were euthanized, and kidneys were processed for ICs enrichment. One‐hour UPEC exposure was chosen based on our previous data that showed kidney associated V‐ATPase subunit and AMP mRNA activation both in vitro and in vivo in mice. 12 , 13
2.5. Murine IC enrichment from “IC reporter” mice after in vivo UTI induction
Kidney single cell suspension was prepared from “IC reporter” mice after experimental UTI with Accumax (Innovative Cell Technology) enzymatic digestion solution with the help of GentleMACs dissociator (Miltenyi Biotec). Briefly, kidneys were placed in C tube (Miltenyi Biotec) with 5 ml Accumax solution and rapidly dissociated using program Lung_02_01 (45 s) then incubated for 15 min at room temp with gentle rotation. Cells were again placed on dissociator and Spleen_04_01 program (60 s) was used to further dissociate cells and cells were again incubated for 15 min at room temperature. After dissociation cells were mixed with pipetting up and down and filtered over 70 µm nylon mesh filter (BD Bioscience). Cells were pelleted by centrifugation at 300×g for 10 min. Red blood cells (RBCs) were lysed with RBC lysing buffer (Biolegend) and filtered and washed again with sterile RPMI1640 media. Dead cells were removed using Annexin V dead cell removal kit (Stem Cell Technology). After dead cell removal, cells were washed, and surface labeled with anti‐mouse CD45‐APC antibody (Biolegend) for 30 min at 4°C. After surface labeling, ICs were flow sorted after size and singlet gating as CD45− Tdt+ (IC) cells at the Indiana University School of Medicine (IUSM) flow cytometry core facility. We performed negative CD45 sorting to isolated IC from “IC reporter” mice because in our previous sorting experiments we noticed that these mice have tdtomato+ CD45+ cells in the kidney either due to leaky cre or more likely V‐ATPase B1 expressing immune cells. To avoid contamination from these resident immune cells, we added CD45 antibody to our cell suspension before IC sorting.
2.6. Enrichment of murine c‐Kit+ ICs and targeted Map3k7 mRNA expression
Single cell suspension was prepared from C57BL/6 mouse (saline and UPEC exposed) kidneys using Accumax enzymatic solution (Innovative Cell Technology) and GentleMACS dissociator (Miltenyi Biotec). The cell suspension was filtered with a 70‐μm basket filter (Fisher Scientific). Cells were centrifuged for 10 min at 2000 rpm. Red blood cells were lysed with red blood cell lysing buffer (Biolegend) and cells were then washed and resuspended in 5 ml fresh DMEM and filtered again with a 70‐μm basket filter. Dead cells were removed using Annexin V dead cell removal kit (Stem Cell Technology). From live cells, CD45+ immune cells were removed using anti‐mouse CD45 microbeads (Miltenyi Biotec) and then incubated with anti‐mouse CD117 (c‐Kit) microbeads (Miltenyi Biotec). After labeling, cells were washed and applied over an MS column (Miltenyi Biotec), and c‐Kit+ (IC) cells were flushed out of the column with 1 ml magnetic‐activated cell sorting (MACS) buffer (PBS containing 0.5% BSA and 2 mM EDTA). c‐Kit− cells were collected as non‐IC cells. RNA was prepared using the RNeasy plus Micro kit (Qiagen) and converted to complementary DNA (cDNA) using high‐capacity reverse transcription kit (Applied Biosystems). Map3k7 mRNA expression was analyzed on CFX connect Real‐time PCR system (Bio‐Rad). Predesigned and validated primer set for mouse Map3k7 were used (Sigma) and mouse Gapdh primers (Real Time Primers.com) were used to normalize gene expression. Map3k7 Forward primer sequence, CTTGTGATGGAATATGCAGAG, Map3k7 Reverse primer sequence, CTTGGGAACACTGTAAACAC and Gapdh Forward primer sequence, CTGGAGAAACCTGCCAAGTA and Reverse primer sequence, TGTTGCTGTAGCCGTATTCA.
2.7. Human c‐Kit+ IC enrichment
Human kidney biopsy normal margins were obtained from cooperative human tissue network (CHTN) (https://www.CHTN.org), in Dulbecco's modified eagle medium (DMEM) in cold packs in 2–4 mm pieces, immediately upon arrival tissue was chopped into further smaller pieces and transferred to a C tube (Miltenyi Biotec) containing 5 ml Liberase TL (Sigma; final conc. 500 µg/ml) in DMEM media containing 100 µg/ml DNase I (Sigma). Single cell suspension was prepared using GentleMACS tissue dissociator (Miltenyi Biotec) using Lung 02_01 program (45 s program). C tubes were then incubated at 37°C for 15 min and then program spleen 04_01 (60 s program) was used. Cells were incubated for another 15 min at 37°C. To stop the dissociation, DMEM containing 10% Fetal bovine serum (FBS) was added. Cell suspension was filtered through 70 µm basket filter (Fisher Scientific). Cells were centrifuged for 10 min at 2000 rpm and RBCs were lysed with 1× RBC lysing buffer (Biolegend) by incubation on ice for 5 min. Cells were then washed with DMEM and re‐suspended in 5 ml fresh DMEM and were filtered again with 70 µm basket filter. Dead cells were removed from cell suspension using dead cell removal microbeads (Cat No. 130‐090‐101; Miltenyi Biotec) over LS column (Miltenyi Biotec). Cells were counted over hemocytometer for viability testing with trypan blue dye exclusion test (at this time viability was ~90%) then CD45+ immune cells were removed from cell suspension using anti‐human CD45 microbeads (Miltenyi Biotec). Cells were centrifuged at 300×g for 10 min and counted with hemocytometer and then were incubated with 100 µl FcR blocking reagent and 100 µl anti‐human CD117 (c‐Kit) microbeads for 15 min (Miltenyi Biotec). Cells were applied over MS column (Miltenyi Biotec, CA) and c‐Kit + (CD117+) enriched cells were flushed out of the column with 1 ml MACS buffer. Enriched C‐Kit+ cells (presumed IC) were tested for mRNA expression of V‐ATPase B1 subunit with real‐time PCR to confirm relative IC enrichment. 12
2.8. In vitro UPEC exposure of enriched human c‐Kit+ ICs
UPEC (strain CFT073) originally provided by Dr. Matthew A. Mulvey (University of Utah) was grown overnight in Luria broth at 300 rpm and 37°C. An aliquot of bacterial broth was pelleted and suspended in sterile PBS. Optical density of the culture was measured at 600 nm. Based on this calculation; OD at 600 of 1 = 8 × 108 cells/ml, 1 × 105 bacterial cells were estimated. Enriched IC cells were equally divided in two wells of 96‐well U‐bottom plate and incubated for 1 h at 37°C and 5% CO2 environment in DMEM containing 10% FBS with 1 × 105 UPEC cells or equal volume of sterile PBS alone. After incubation, cells were centrifuged at 1200 rpm for 10 min and media was carefully removed and cell pellets were washed again with sterile PBS, and RNA was prepared using RNeasy plus micro kit (Qiagen).
2.9. RT2 profiler PCR array
RNA was obtained from enriched MACS sorted human and flow sorted murine ICs using RNeasy plus micro kit (Qiagen). Following quantification with spectrometry, quality was evaluated using Agilent Bioanalyzer (Agilent). A 260/280 nm ratio with 2.0 ± 0.1 was considered adequate quality. RT2 PreAMP cDNA synthesis kit (Qiagen) in combination with pathway specific primers was used to generate cDNA. RT2 Profiler Antibacterial Response Arrays (mouse GeneGlobe ID PAMM‐148Z and human GeneGlobe ID PAHS‐148Z) were performed using Bio‐Rad CFX96 PCR System (Bio‐Rad). Only results that passed quality checks in PCR array reproducibility, RT efficiency, and genomic DNA contamination were included. Gene expression was normalized using a panel of five housekeeping genes β‐actin (Actb), β2‐microglobulin (β2m), glyceraldehyde 3‐phosphate dehydrogenase (Gapdh), glucuronidase‐β (Gusb), and heat shock protein 90 kDa α (cytosolic) class B member 1 (Hsp90ab1). Data were analyzed on GeneGlobe data analysis center and presented as volcano plot (Qiagen).
2.10. Immunofluorescence
To localize MAP3K7 in mice IC, we used wild type C57BL/6 mice (rather “IC reporter”) and human kidney biopsy sections. Seven‐micrometer thick paraffin embedded human and C57BL/6 mice kidney tissue sections were stained with polyclonal chicken anti‐V‐ATPase E1 antibody (Sigma) to localize collecting duct IC and rabbit anti‐MAP3K7 antibody (Sigma) at 1:200 dilution for overnight at 4°C. To visualize, MAP3K7, anti‐Rabbit AF488 (1:600 final dilution) and to visualize V‐ATPase E1, anti‐chicken Cy3 (1:600 final dilution) secondary antibodies were used (Jackson Immunoresearch). Sections were visualized using a Keyence BZ9000 microscope (Keyence Corporation). Red images were pseudo‐colored to magenta to allow better visualization for individuals with color blindness.
2.11. Confocal microscopy
Single plane tile‐imaging of the entire C57BL/6 kidney sections was conducted with a Leica TCS SP8 confocal/2P microscope (Leica Microsystems Inc.) available at the ICBM imaging facility, using a Leica HC PL APO CS2 20×/0.75IMM objective lens adjusted to oil immersion. Confocal images (12bit) were acquired at 1024 × 1024 pixels, 400 Hz, 1 AU, bidirectional X scan, 1.2 zoom, line averaging of 2, and using a sequential laser illumination mode set up to three sequences: (1) Nuclei/DAPI (405 nm laser illumination, PMT1 adjusted to collect 415–485 nm emission), (2) MAP3K7/Alexa Fluor 488 (488 nm laser illumination, PMT2 adjusted to collect 500–500 nm emission), (3) ICs/Cyanin‐3 (552 nm laser illumination, PMT3 adjusted to collect 560–630 nm emission light). The hardware scanning set up was kept constant for all samples. Mosaic images were generated using automatic stitching in Leica LAS X software v.3.5.7.
2.12. Image analysis
Image quantification was performed on mosaic images using Imaris image analysis software v. 9.82 (Bitplane). The “Surfaces” segmentation module was used to evaluate percentage of ICs expressing MAP3K7. First, the red channel was segmented to obtain total number of ICs (represented by total number of segmented voxels) and then filtering was applied to extract voxels with green signal above threshold (MAP3K7 positive cells). The % of MAP3K7 positive cells was calculated based on the total voxels values. The same segmentation parameters were applied to all images.
3. RESULTS
3.1. RT2 PCR array quality control
Only results that passed quality checks in PCR array reproducibility, RT efficiency, and genomic DNA contamination were included. All samples passed the quality control test Gene expression was normalized using a panel of five housekeeping genes β‐actin (Actb), β2‐microglobulin (β2m), glyceraldehyde 3‐phosphate dehydrogenase (Gapdh), glucuronidase‐β (Gusb), and heat shock protein 90 kda α (cytosolic) class B member 1 (Hsp90ab1) (see Supporting Information: S1 and S2).
3.2. Murine IC antibacterial PCR array
To investigate the response of murine ICs exposed to UPEC pyelonephritis strain CFT073, “IC reporter” mice were subjected to saline or UPEC transurethral challenge in vivo for 1 h and ICs were flow sorted and subjected to robust antibacterial RT2 PCR array that detects 84 genes simultaneously. Out of 84 genes investigated, 15 genes were found to be significantly upregulated that include, Map3k7/Tak1 (9.5‐fold, p = 0.000014), Mapk1 (6.21‐fold, p = 0.000031), Mapk3 (3.75‐fold, p = 0.02), Mapk8 (3.11‐fold, p = 0.00031), Sugt1 (11.5‐fold, p = 0.0006), Il12a (50.36‐fold, p = 0.04), Il18 (5.7‐fold, p = 0.007), Fadd (13.5‐fold, p = 0.005), Ikbkb (13.7‐fold, p = 0.005), Ly96 (4.28‐fold, p = 0.02), Pik3ca (2.57‐fold, p = 0.03), Pycard (26.57‐fold, p = 0.025), Xiap (7.45‐fold, p = 0.04), Tlr5 (22.82‐fold, p = 0.04), and Tollip (41.53‐fold, p = 0.04). Five genes were found to be significantly downregulated such as Casp8 (−5.71‐fold, p = 0.01), Myd88 (−5.38‐fold, p = 0.01), Ripk1 (−2.47‐fold, p = 0.01), Tnfrsf1a (−2.58‐fold, p = 0.009) and Traf6 (−5.54‐fold, p = 0.01). Some other notable genes that were upregulated but missed the significance were Map2k4 (3.39‐fold, p = 0.05), Irak1 (3.86‐fold, p = 0.07), and Lbp (12.44, p = 0.08) and downregulated genes Ripk2 (−2.65‐fold, p = 0.05), Nfkbia (−3.81‐fold, p = 0.06) (Figure 1, Supporting Information: datasheet 1 and S1). Increased Map3k7 mRNA expression in mouse ICs, was confirmed with enriched c‐Kit+ ICs from wild type C57BL/6 mice exposed to UPEC versus saline in vivo for 1 h and Map3k7 mRNA was measured by targeted RT‐PCR. c‐Kit+ ICs upregulated Map3k7 upon UPEC exposure in vivo (p = 0.039) (Supporting Information: material S3). These data strengthen the finding that ICs upregulate Map3k7 upon UPEC exposure in vivo.
Figure 1.
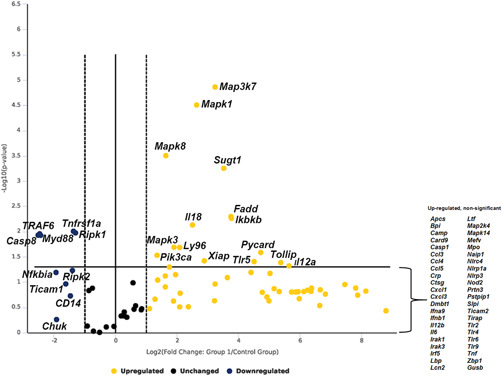
Volcano plot of murine IC RT2 profiler antibacterial PCR array. Murine ICs were enriched by flowsorting from “IC reporter” mice following exposure to UPEC versus saline in vivo for 1‐h. Data are presented as volcano plot to identify significant gene expression changes in UPEC exposed ICs as compared to saline. Thresholds for the volcano plot were a fold change of >2 and p‐value of <0.05. Dotted lines represent significance. Upper right quadrant next to dotted lines has genes which are significant and upregulated. Genes listed on lower right are upregulated but not significant. Genes listed on upper left quadrant are significantly downregulated and genes listed in lower left quadrant are downregulated but not significant. n = 3 “IC reporter” mice/group. IC, intercalated cell; UPEC, uropathogenic Escherichia coli.
3.3. Human IC antibacterial PCR array
To investigate the clinical relevance of IC function, we performed antibacterial array on human enriched ICs exposed to saline versus UPEC strain CFT073. Out of 84 genes analyzed, 12 genes increased expression upon UPEC exposure while Interferon regulatory factor 5 (IRF5) (2.05‐fold, p = 0.014) and mitogen‐activated protein kinase kinase kinase 7 (MAP3K7) (2.42‐fold, p = 0.009) were found to be significantly elevated similar to murine ICs. Six genes were found to be downregulated but failed to reach significance which include CARD9, IL12A, MPO, PIK3CA, SUGT1, and TNFRSF1A (Figure 2 and Supporting Information: datasheet 2 and S1).
Figure 2.
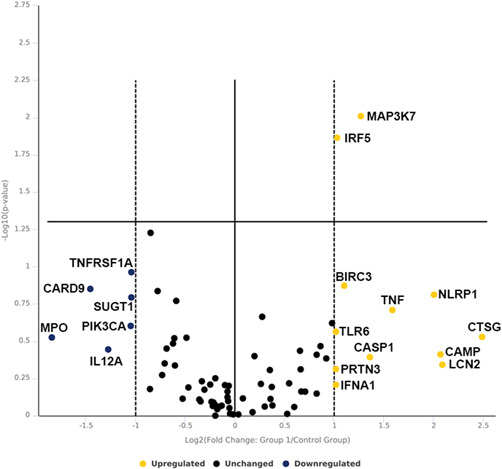
Volcano plot of human IC RT2 antibacterial PCR array. Human ICs were enriched by MACS from kidney biopsy and exposed to UPEC versus saline for 1‐h in vitro. Data are presented as volcano plot to identify significant gene expression changes in UPEC exposed ICs as compared to saline. Thresholds for the volcano plot were a fold change of >2 and p‐value of <0.05. Dotted lines represent significance. Upper right quadrant next to dotted lines has genes which are significant and upregulated. Genes listed on lower right are upregulated but not significant. Genes listed in lower left quadrant are downregulated but nonsignificant. n = 3 kidney biopsy/group. IC, intercalated cell; MACS, magnetic‐activated cell sorting; UPEC, uropathogenic Escherichia coli.
3.4. Confocal imaging reveal increase in MAP3K7 expressing ICs
To quantitatively analyze MAP3K7 expression in ICs, we performed confocal imaging and whole kidney imaris analysis in C57BL/6 mice challenged with UPEC in vivo for 1 h. Kidney sections from uninfected and infected mice were labeled with V‐ATPase E1 (Red) to identify IC and MAP3K7 (Green) and acquired on confocal imaging microscope at 20× magnification. Whole kidney image stitch section was analyzed for MAP3K7 expression in ICs (Figure 3A–F). Imaris quantification was performed to analyze the % of MAP3K7 expressing ICs (Green positive Red voxels) which revealed that UPEC exposure result in increased number of ICs expressing MAP3K7 in two individual mice with varying degree (Figure 3G).
Figure 3.
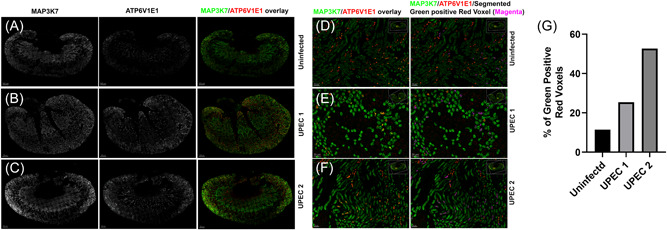
MAP3K7 confocal imaging. C57BL/6 mice were challenged with UPEC in vivo for 1 h or remain uninfected. Kidney sections were stained with MAP3K7 (Green) and ATP6V1E1 (Red) to locate ICs and acquired on confocal imaging microscope. 20× whole kidney image stitch was performed, and quantification was done using imaris software. (A–C) MAP3K7 (left panel) and ATP6V1E1 (middle panel) is shown in greyscale and overlay of MAP3K7 (Green) expressing IC (Red) (right panel). (D–F) Enlarged inset images of MAP3K7 and ATP6V1E1 overlay (left panel) and overlay plus segmented red plus green expressing IC in pseudocolor magenta (right panel). (G) Imaris quantification of % MAP3K7 expressing IC. Scale bar (A–C) 500 µm, Scale bar (D–F) 50 µm. IC, intercalated cell; UPEC, uropathogenic Escherichia coli.
3.5. MAP3K7 is expressed in human and murine pyelonephritis
To confirm the mRNA finding of common expression of MAP3K7 at the protein level, we also performed immunofluorescence on human and wild type C57BL/6 mouse kidney sections. In non‐pyelonephritis patient, few non‐IC (presumed principal cells) collecting duct cells were positive for MAP3K7 with apical staining pattern while several ICs in pyelonephritis patient showed positivity for MAP3K7 and staining pattern was more cytoplasmic upon UPEC infection in vivo (Supporting Information: S4). Interestingly, human kidney also had strong staining in several tubules likely resembling thick ascending limbs (TAL) in pyelonephritis. TAL staining in human kidney is absorbed or induced protein need to be further investigated.
4. DISCUSSION
Understanding the mechanism ICs use to defend kidney from ascending uropathogen is essential to develop novel treatment methods pyelonephritis. ICs of kidney are located at the collecting duct where they are among the first cells to encounter ascending uropathogen. In an attempt to identify mechanisms used by ICs to defend against ascending uropathogen, we simultaneously investigated a set of 84 antibacterial genes by robust PCR array in both mice and humans ICs. The results revealed MAP3K7 as a common upregulated antibacterial gene in both mice and humans ICs exposed to UPEC.
Mitogen‐activated protein kinase kinase kinase 7 (human: MAP3K7, mouse Map3k7), also known as Transforming growth factor‐β activated kinase‐1 (TAK1), was initially described to be involved with transcriptional regulation of TGFβ and later as a member of the mitogen‐activated protein kinase kinase kinase (MAP3K) family. MAP3K7 is essential for innate and adaptive immune signaling cascades. 14 , 15 , 16 , 17 , 18 , 19 Recent studies have defined an essential role of Map3k7 in T cell receptor (TCR) and B cell receptor (BCR)‐induced activation of NF‐kB and in the survival and development of immune cells, including mature B and T cells. 17 , 20 , 21 , 22 , 23 , 24 Importantly, multilineage hematopoietic cell survival requires Map3k7 as its deletion triggered uncontrolled apoptosis in hematopoietic cells and bone marrow and liver failure. 23
Myeloid cells such as neutrophils and macrophages are critical cells in controlling microbial invasion. Interestingly, in neutrophils, Map3k7 negatively regulates NF‐kB and p38 activation and renders mice susceptible to septic shock. 25 Map3k7 role in these cells, which frequently reside in tissue such as kidney for microbial surveillance, indicate a stringent control of cells' pro‐inflammatory response. An unchecked pro‐inflammatory response can damage the tissue by hyperactivation and cause tissue damage. We speculate that Map3k7 may also be playing a similar role in ICs regulating the crosstalk with immune cells such as neutrophils and remains to be studied. In a similar role, gut epithelial cell specific deletion of a protein kinase Map3k7 depletes Paneth cells and increases ROS production in mouse models. 26
Of note, we detected upregulation of Map3k7 mRNA following 1 h of UPEC exposure. At this timepoint, we do not detect UPEC in kidney by culture. Likely, ICs sensed pathogen‐associated molecular pattern or bacterial‐derived lipopolysaccharide (LPS). Similar to our finding, previous studies have reported that MAP3K7 phosphorylates RIPK1 within 5 min of TNF‐α exposure. In another study, mice receiving intraperitoneal injection of LPS activated, guanylate‐binding protein (Gbp1) within 1 h of exposure. 27 In another study, bacterial LPS induced NF‐kB in bone marrow derived macrophages within minutes with peak activation in 5 min. 28 Viruses also show rapid activation of signaling molecules, such as extracellular signal‐regulated kinase activity is sustained within 15min of cytomegalovirus infection. 29 Increased expression of Map3k7 mRNA in both human and mice may indicate rapid sensing and priming of ICs to dampen ascending UPEC‐induced inflammation and maintenance of kidney homeostasis. In addition to its tissue homeostasis role, MAP3K7 has been implicated in protection of lysosomal integrity and thereby regulating macrophage population. 30 Therefore, we speculate that recently described ICs phagocytic ability 12 may also be regulated by Map3k7. In addition to Map3k7, a notable upregulated gene was Fas‐associated protein with death domain (Fadd) in murine ICs which is shown to be a key gene involved in virus induced innate immune response including production of type 1 interferon in mammalian cells and protection against Gram‐negative bacteria and AMP production in Drosophila. 31 , 32
Another noteworthy gene that was significantly upregulated was toll‐interacting protein (Tollip) in murine ICs. This upregulation may suggests negative regulation of UPEC derived LPS and TLR4 signaling and control inflammation by ICs. 33 , 34 Interestingly, among significantly downregulated genes in murine ICs, was caspase 8 (Casp8), whose downregulation has been shown for sustained release of AMP in skin infection, regulation of cell death versus survival decisions and antiviral innate immunity. 35 , 36 , 37 Interferon regulatory factor 5 (IRF5) is a transcription factor known to be a stress sensor and plays critical role in innate immune response and bacterial clearance 38 , 39 , 40 was significantly elevated in human ICs but only elevated in murine ICs after UPEC infection. Collectively, the results of this study indicate that ICs rapidly act to encounter ascending uropathogen while balancing pro‐inflammatory signaling and preserving tissue integrity.
This study does have limitations as the number of significantly elevated and downregulated genes in human ICs and murine ICs were not comparable. We speculate that the primary reason for this difference could be that human IC response could be due to the different bacterial exposure methods between murine and human studies. It is plausible that intact collecting duct epithelium is more responsive to UPEC infections due to crosstalk with other collecting duct epithelial and immune cells. Another possibility is that the human biopsy samples we obtained were from different kidney locations that may behave differently such as cortical IC versus medullary ICs response may vary while murine ICs preparations were a consistent mix of both locations. We will need direct human to mouse comparisons using a range of UPEC doses and timepoints. C57BL6/J mice are less susceptible to UTIs than humans which is why the relatively high transurethral inoculums are needed. For the direct human to mouse comparison, we would need to use a range of bacterial doses and timepoints which is beyond the scope of this study.
Nonetheless, presence of a common IC innate immune gene upon UPEC infection such as MAP3K7 is promising. Our future studies will also focus on the role of MAP3K7 in vivo using conditional knockout mice and the selective MAP3K7 antagonist, such as Takinib, in mouse models of UTI to understand clinical significance of MAP3K7. We will also study the MAP3K7 activation by upon UPEC exposure in vivo and in vitro by looking at its phosphorylation and its downstream signaling molecules such as NFkB and AP‐1.
In summary, we evaluated 84 human and murine IC for antibacterial gene expression at baseline and following experimental pyelonephritis. Using a validated commercial antibacterial PCR array in human and mouse enriched ICs we found that both mice and human ICs significantly increased MAP3K7 mRNA expression upon 1 h of UPEC exposure. Confocal imaging and imaris quantification analysis verified that more IC express MAP3K7 upon UPEC exposure. Given the critical role of MAP3K7 in innate and adaptive immunity, we hypothesize that MAP3K7 protects the kidney's cellular integrity by regulating the organ's pro‐inflammatory response.
AUTHOR CONTRIBUTIONS
Vijay Saxena, Andrew Schwaderer, and David S. Hains made substantial contribution to the research design and interpretation of data. Vijay Saxena performed and analyzed most of the experiments, drafted the manuscript, and revised it critically with inputs from all the authors. Samuel Arregui was instrumental in performing in vivo murine UPEC infections. Malgorzata Maria Kamocka performed statistical analysis of the confocal imaging data. All authors have read and approved the final submitted manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
The authors thank the members of the Indiana University Melvin and Bren Simon Cancer Center Flow Cytometry Resource Facility for their outstanding technical support in murine IC enrichment. This study was supported in part by, The Indiana University Melvin and Bren Simon Comprehensive Cancer Center Flow Cytometry Resource Facility, which is funded in part by NIH, National Cancer Institute (NCI) grant P30 CA082709 and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant U54 DK106846. The FCRF is supported in part by NIH instrumentation grant 1S10D012270. This study was also funded in part by the National Institute of Diabetes and Digestive and Kidney Diseases R01DK106286, Eli Lily Foundation (A. S. and D. S. H.).
Saxena V, Arregui S, Kamocka MM, Hains DS, Schwaderer A. MAP3K7 is an innate immune regulatory gene with increased expression in human and murine kidney intercalated cells following uropathogenic Escherichia coli exposure. J Cell Biochem. 2022;123:1817‐1826. 10.1002/jcb.30318
DATA AVAILABILITY STATEMENT
The raw data files for RT2 PCR array are included as supplementary datasheets and materials.
REFERENCES
- 1. Medina M, Castillo‐Pino E. An introduction to the epidemiology and burden of urinary tract infections. Ther Adv Urol. 2019;11:1756287219832172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roberts JA. Etiology and pathophysiology of pyelonephritis. Am J Kidney Dis. 1991;17:1‐9. [DOI] [PubMed] [Google Scholar]
- 3. Schneeberger C, Holleman F, Geerlings SE. Febrile urinary tract infections: pyelonephritis and urosepsis. Curr Opin Infect Dis. 2016;29:80‐85. [DOI] [PubMed] [Google Scholar]
- 4. Zowawi HM, Harris PN, Roberts MJ, et al. The emerging threat of multidrug‐resistant Gram‐negative bacteria in urology. Nat Rev Urol. 2015;12:570‐584. [DOI] [PubMed] [Google Scholar]
- 5. Chassin C, Goujon JM, Darche S, et al. Renal collecting duct epithelial cells react to pyelonephritis‐associated Escherichia coli by activating distinct TLR4‐dependent and ‐independent inflammatory pathways. J Immunol. 2006;177:4773‐4784. [DOI] [PubMed] [Google Scholar]
- 6. Ronald A. The etiology of urinary tract infection: traditional and emerging pathogens. Am J Med. 2002;113(suppl 1A):14S‐19S. [DOI] [PubMed] [Google Scholar]
- 7. Spencer JD, Hains DS, Porter E, et al. Human alpha defensin 5 expression in the human kidney and urinary tract. PLoS One. 2012;7:e31712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spencer JD, Schwaderer AL, Dirosario JD, et al. Ribonuclease 7 is a potent antimicrobial peptide within the human urinary tract. Kidney Int. 2011;80:174‐180. [DOI] [PubMed] [Google Scholar]
- 9. Hains DS, Chen X, Saxena V, et al. Carbonic anhydrase 2 deficiency leads to increased pyelonephritis susceptibility. Am J Physiol Renal Physiol. 2014;307:F869‐F880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Paragas N, Kulkarni R, Werth M, et al. Alpha‐intercalated cells defend the urinary system from bacterial infection. J Clin Invest. 2014;124:2963‐2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saxena V, Fitch J, Ketz J, et al. Whole transcriptome analysis of renal intercalated cells predicts lipopolysaccharide mediated inhibition of retinoid X receptor alpha function. Sci Rep. 2019;9:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saxena V, Gao H, Arregui S, et al. Kidney intercalated cells are phagocytic and acidify internalized uropathogenic Escherichia coli . Nat Commun. 2021;12:2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saxena V, Hains DS, Ketz J, et al. Cell‐specific qRT‐PCR of renal epithelial cells reveals a novel innate immune signature in murine collecting duct. Am J Physiol Renal Physiol. 2018;315:F812‐F823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arthur JS, Ley SC. Mitogen‐activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13:679‐692. [DOI] [PubMed] [Google Scholar]
- 15. Ishitani T, Ninomiya‐Tsuji J, Nagai S, et al. The TAK1‐NLK‐MAPK‐related pathway antagonizes signalling between beta‐catenin and transcription factor TCF. Nature. 1999;399:798‐802. [DOI] [PubMed] [Google Scholar]
- 16. Ninomiya‐Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. The kinase TAK1 can activate the NIK‐I kappaB as well as the MAP kinase cascade in the IL‐1 signalling pathway. Nature. 1999;398:252‐256. [DOI] [PubMed] [Google Scholar]
- 17. Sato S, Sanjo H, Takeda K, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6:1087‐1095. [DOI] [PubMed] [Google Scholar]
- 18. Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin‐dependent kinase of MKK and IKK. Nature. 2001;412:346‐351. [DOI] [PubMed] [Google Scholar]
- 19. Yamaguchi K, Shirakabe K, Shibuya H, et al. Identification of a member of the MAPKKK family as a potential mediator of TGF‐beta signal transduction. Science. 1995;270:2008‐2011. [DOI] [PubMed] [Google Scholar]
- 20. Liu HH, Xie M, Schneider MD, Chen ZJ. Essential role of TAK1 in thymocyte development and activation. Proc Natl Acad Sci USA. 2006;103:11677‐11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato S, Sanjo H, Tsujimura T, et al. TAK1 is indispensable for development of T cells and prevention of colitis by the generation of regulatory T cells. Int Immunol. 2006;18:1405‐1411. [DOI] [PubMed] [Google Scholar]
- 22. Schuman J, Chen Y, Podd A, et al. A critical role of TAK1 in B‐cell receptor‐mediated nuclear factor kappaB activation. Blood. 2009;113:4566‐4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang M, Wei X, Guo Y, et al. TAK1 is required for the survival of hematopoietic cells and hepatocytes in mice. J Exp Med. 2008;205:1611‐1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wan YY, Chi H, Xie M, Schneider MD, Flavell RA. The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nat Immunol. 2006;7:851‐858. [DOI] [PubMed] [Google Scholar]
- 25. Ajibade AA, Wang Q, Cui J, et al. TAK1 negatively regulates NF‐kappaB and p38 MAP kinase activation in Gr‐1+CD11b+ neutrophils. Immunity. 2012;36:43‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simmons AN, Kajino‐Sakamoto R, Ninomiya‐Tsuji J. TAK1 regulates Paneth cell integrity partly through blocking necroptosis. Cell Death Dis. 2016;7:e2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qiu X, Guo H, Yang J, Ji Y, Wu CS, Chen X. Down‐regulation of guanylate binding protein 1 causes mitochondrial dysfunction and cellular senescence in macrophages. Sci Rep. 2018;8:1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bagaev AV, Garaeva AY, Lebedeva ES, Pichugin AV, Ataullakhanov RI, Ataullakhanov FI. Elevated pre‐activation basal level of nuclear NF‐kappaB in native macrophages accelerates LPS‐induced translocation of cytosolic NF‐kappaB into the cell nucleus. Sci Rep. 2019;9:4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodems SM, Spector DH. Extracellular signal‐regulated kinase activity is sustained early during human cytomegalovirus infection. J Virol. 1998;72:9173‐9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sakamachi Y, Morioka S, Mihaly SR, et al. TAK1 regulates resident macrophages by protecting lysosomal integrity. Cell Death Dis. 2017;8:e2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Balachandran S, Thomas E, Barber GN. A FADD‐dependent innate immune mechanism in mammalian cells. Nature. 2004;432:401‐405. [DOI] [PubMed] [Google Scholar]
- 32. Naitza S, Rosse C, Kappler C, et al. The Drosophila immune defense against gram‐negative infection requires the death protein dFADD. Immunity. 2002;17:575‐581. [DOI] [PubMed] [Google Scholar]
- 33. Didierlaurent A, Brissoni B, Velin D, et al. Tollip regulates proinflammatory responses to interleukin‐1 and lipopolysaccharide. Mol Cell Biol. 2006;26:735‐742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang G, Ghosh S. Negative regulation of toll‐like receptor‐mediated signaling by Tollip. J Biol Chem. 2002;277:7059‐7065. [DOI] [PubMed] [Google Scholar]
- 35. Bhatt T, Bhosale A, Bajantri B, et al. Sustained secretion of the antimicrobial peptide S100A7 is dependent on the downregulation of Caspase‐8. Cell Rep. 2019;29:2546‐2555. [DOI] [PubMed] [Google Scholar]
- 36. Chen H, Ning X, Jiang Z. Caspases control antiviral innate immunity. Cell Mol Immunol. 2017;14:736‐747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. DeLaney AA, Berry CT, Christian DA, et al. Caspase‐8 promotes c‐Rel‐dependent inflammatory cytokine expression and resistance against Toxoplasma gondii . Proc Natl Acad Sci USA. 2019;116:11926‐11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ban T, Sato GR, Tamura T. Regulation and role of the transcription factor IRF5 in innate immune responses and systemic lupus erythematosus. Int Immunol. 2018;30:529‐536. [DOI] [PubMed] [Google Scholar]
- 39. Hedl M, Yan J, Witt H, Abraham C. IRF5 is required for bacterial clearance in human M1‐Polarized macrophages, and IRF5 immune‐mediated disease risk variants modulate this outcome. J Immunol. 2019;202:920‐930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao GN, Jiang DS, Li H. Interferon regulatory factors: at the crossroads of immunity, metabolism, and disease. Biochim Biophys Acta. 2015;1852:365‐378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Data Availability Statement
The raw data files for RT2 PCR array are included as supplementary datasheets and materials.


