Abstract
Purpose
There are conflicting reports regarding the abundance of short-chain fatty acids producing bacteria in the gut microbiota in patients with type 1 and type 2 diabetes. We aimed to determine the amount of Akkermansia muciniphila, Anaerobutyricum hallii, Bifidobacterium adolescentis, Bifidobacterium longum, Collinsella aerofaciens, Faecalibacterium prausnitzii, Lacticaseibacillus rhamnosus, and Parabacteroides distasonis in the gut microbiota in patients with type1 and type2 diabetes, compared with the healthy controls and analyze the correlation between the gene expression levels of two short-chain fatty acids receptors GPR41 and GPR43.
Methods
Forty type 1, 40 type 2 stool and blood samples of diabetes patients, and 40 healthy control samples were studied. DNA and RNA were extracted, and bacteria were detected using a Microbial DNA qPCR Assay kit. Gene expressions were detected with GPR41 and GPR43 primers via in-house qPCR.
Results
Compared with healthy controls, B.longum and F.prausnitzii abundance were significantly decreased in patients with type1 and type2 diabetes, A.hallii abundance was increased in patients with type1 and decreased in type2 diabetes contrarily A.muciniphila abundance was decreased in patients with type1 and increased in type2 diabetes. GPR43 gene expression was upregulated in both patients group, however GPR41 was upregulated only in patients with type2 diabetes.
Conclusions
Elevated B. longum and F. prausnitzii abundances were detected in the gut microbiota of patients with type1 and type2 diabetes and compared with healthy controls. B. longum and F.prausnitzii abundances were also correlated with the GPR43 gene expression level in type1 diabetes patients. Extensive studies determining bacteria producing short-chain fatty acids in gut microbiota, and their contribution in the pathogenesis of diabetes, are needed to understand better the mechanism of these diseases.
Keywords: Type 1 diabetes, Type 2 diabetes, Gut microbiota, SCFAs receptor
Introduction
Diabetes mellitus is a complex metabolic disease that can create a risk factor for secondary diseases. It is characterized by hyperglycemia in patients [1]. Type 1 diabetes mellitus (T1D) and Type 2 diabetes mellitus (T2D) are the main subtypes of the disease [2]. While T1D is an autoimmune disease that develops as a result of an autoimmune response to pancreatic β cells, T2D is a disease associated with the defects of insulin secretion or insulin response [3, 4]. The increased prevalence of T1D and T2D are serious public health problems worldwide and the increase cannot be explained by the genetic mechanism of the diseases alone. Environmental factors such as microbiota and diet, have also been found to be associated with the risk factors of both T1D and T2D [5]. Short-chain fatty acids (SCFAs) play an important role in maintaining the metabolic health. SCFAs, such as acetate, butyrate, and propionate may contribute to mucosal immunity, glucose homeostasis, and immunomodulation by acting on tissue-specific mechanisms for metabolic health [6]. G protein-coupled receptor 41 (GPR41 or FFAR3: free fatty acid receptor 3) and G protein-coupled receptor 43 (GPR43 or FFAR2) have been known as G protein-coupled receptors (GPCRs) that can be expressed in different tissues and are activated by SCFAs. These receptors are involved in the chronic inflammatory response in many diseases such as obesity or asthma [7, 8]. Bacteria in gut microbiota such as Akkermansia muciniphila, Anaerobutyricum hallii (previously Eubacterium hallii), Bifidobacterium adolescentis, Bifidobacterium longum, Collinsella aerofaciens, Faecalibacterium prausnitzii, Lacticaseibacillus rhamnosus (previously Lactobacillus rhamnosus), and Parabacteroides distasonis have important biological effects in the host [9–11]. Lacticaseibacillus and Bifidobacterium species are known as classical probiotics. On the other hand, Akkermansia, Faecalibacterium, Anaerobutyricum, and Parabacteroides genera can be considered as food additives or biotherapeutics with their bioactive metabolites [12]. Changes in the abundances of these bacteria in the gut microbiota have been reported on many diseases and recommended to be used as the standard strains in fecal microbiota transplantation studies. These strains, that spesifically selected, have been taken into consideration in gut microbiome studies recently in the host due to their anti-inflammatory properties, probiotic qualities, probiotic potential or metabolite production properties such as SCFAs.[9–11]. The aim of this study, was to determine the amount of Akkermansia muciniphila, Anaerobutyricum hallii, Bifidobacterium adolescentis, Bifidobacterium longum, Collinsella aerofaciens, Faecalibacterium prausnitzii, Lacticaseibacillus rhamnosus, and Parabacteroides distasonis in the gut microbiota of T1D patients and T2D patients compared to healthy controls (HC). We also aimed to determine the GPR41 and GPR43 gene expression levels in the blood samples of our cases and controls and to analyze the correlation the amount of the bacteria that we studied.
Materials and methods
Study desing and participants.
Forty patients with T1D, 40 patients with T2D, and 40 HC were included to this observational case-control study. T1D and T2D cohorts were diagnosed according to American Diabetes Association (ADA) criteria [13]. Criteria for the diagnosis of T1D and T2D were used the fasting blood sugar (FBS) levels (≥ 126 mg/dL) after the not eat anything for at least 8 h, HbA1c rate (%) (≥ 6.5%), and 2 h plasma glucose levels during 75-gr oral glucose tolerance test (≥ 200 mg/dL), human anti glutamic acid decarboxylase antibodies on the ELISA results (MyBioSource, San Diego, CA, USA). It has been used for the diagnosis of T1D in clinical information such as age at diagnosis (< 35 years), lower BMI (< 25 kg/m2), unintentional weight loss and ketoacidosis [13]. T2D patients consisted of newly diagnosed patients who had not started drug therapy yet. Ethical approvals were obtained from the Clinical Research Ethical Committee of Istanbul University-Cerrahpasa, Cerrahpasa Medical Faculty (Approval numbers: 83,045,809/60,401 and 83,045,809/30,339). To prevent bias, after the patient groups were determined, age- and sex-matched healthy controls were included to our study by the matching method. While designing the groups, individuals were checked for the following inclusion criteria; (I) Being an adult (age > 18 years), (II) Not to have used any antibiotics and probiotics in last one month, (III) To not have any disease other than T1D and T2D for the patient groups, (IV) To not have any active infection, (V) To not have any allergy history. Recent probiotic and antibiotic use, active infection and presence of other diseases were determined as an exclusion criteria. The patient and control groups were selected from individuals with similar nutritional habits and similar economic levels. Individuals with different eating habits such as being a vegan, vegetarian or following a specific diet program such as the dukan diet were not included to the study.
Sample collection and storage.
Stool and whole blood samples were collected from individuals of patients and control groups admitted to the endocrinology clinic of Istanbul University-Cerrahpasa, Cerrahpasa Medical Faculty between January 2014 and April 2017. All stool samples were classified to use Bristol Stool Form Scale (BSFS) and found as Type 3–5 [14]. Stool samples were stored in the Stool Transport and Recovery solution (S.T.A.R. buffer) (Roche Diagnostics, Mannheim, Germany), and whole blood samples were stored in the PAXgene blood RNA tube (Becton-Dickinson, Franklin Lakes, NJ) at − 80 °C until the DNA and RNA extraction respectively [15].
DNA and RNA extraction from the stool and whole blood samples.
DNA and RNA extraction was performed with the Magna Pure 96 DNA/Viral NA Small volume kit and MagNA Pure 96 Cellular RNA Large Volume Kit (Roche Diagnostics GmBH, Mannheim, Germany) respectively using the MagNA Pure 96 (Roche Diagnostics, Mannheim, Germany) system according to the manufacturer’s instructions [15]. Quantitative polymerase chain reaction (qPCR or real-time PCR) reactions for bacterial quantification and for the detection of GPR41 and GPR43 gene expression levels were performed in the Medical Microbiology Department of Kirklareli University School of Medicine.
Bacterial quantification by qPCR.
The Microbial DNA qPCR assay kit (Qiagen GmbH, Hilden, Germany) for Akkermansia muciniphila (GeneGlobe ID: BBID00026A), Anaerobutyricum hallii (previously Eubacterium hallii) (BBID00147A), Bifidobacterium adolescentis (BBID00063A), Bifidobacterium longum (BBID00067A), Collinsella aerofaciens (BBID00118A), Faecalibacterium prausnitzii (BBID00154A), Lacticaseibacillus rhamnosus (previously Lactobacillus rhamnosus) (BBID00195A), and Parabacteroides distasonis (BBID00257A) were utilized to detect specific gut bacteria on LightCycler 480 II system (Roche Diagnostics GmBH, Mannheim, Germany) according to the manufacturer’s instructions. qPCR reactions were performed in triplicate in three different runs. Absolute quantification was done via qPCR protocols and the amount of bacteria in the gut microbiota was presented as log10copy/gr [10].
GPR41 and GPR43 gene expression levels by qPCR.
Transcriptor First Strand cDNA synthesis kit (Roche Diagnostics, Mannheim, Germany) was used for complementary DNA (cDNA) synthesis from 50 ng of template RNA [15]. GPR41 (Forw: GTT GGC ATC CTG GCT GTT and Rev: CCT CTT CTT CAC CAC CGT CTA), GPR43 (Forw: CGC TAC CTG GGA GTG GCT T and Rev: CGG CCT TCT GGG TTG AGT T) and GAPDH (Forw: TTT GCG TCA GTG TCA TCG and Rev: TGC TCT GCC TTG GGT AAT) primers were used to detect GPR41 and GPR43 [16]. GAPDH gene was used as a housekeeping gene for normalization of gene expression results of GPR41 and GPR43. FastStart Essential DNA Green Master kit (Roche Diagnostics, Mannheim, Germany) was used for qPCR runs according to the manufacturer’s instructions. qPCR reactions were performed in triplicate in three different runs. The relative quantification analysis was used to detect GPR41 and GPR43 gene expression levels and analyzed by Livak and Schmittgen’s 2−ΔΔCT (delta-delta Ct) method. Relative ratio values were presented as GPR41 and GPR43 gene expressions [17].
Statistical analysis
Variables data were presented as mean ± standard deviation. Mann-Whitney U test and Spearman Correlation analysis were used to perform statistical analysis using IBM SPSS version 20.0 software (SPSS Inc., Chicago, IL). p values below 0.05 (p < 0.05) were considered statistically significant. For Spearman’s correlation value (rs), rs < 0.25 was evaluated as not statistically correlated, rs = 0.25–0.5 was evaluated as a weak correlation, rs = 0.5–0.75 was evaluated as a moderate correlation, rs = 0.76–0.85 was evaluated as a strong correlation and rs > 0.85 was evaluated as a very strong correlation. Only significant correlation links (rs-values) were shown.
Results
This study was conducted with 120 individuals including 40 T1D patients (20 male, 20 female), 40 T2D patients (20 male, 20 female), and 40 HC (20 male, 20 female). The mean age of the T1D, T2D, and HC groups were 31.23 ± 5.12, 31.83 ± 3.53, and 31.05 ± 4.86 years respectively and a statistically significant difference was not found between the groups in terms of the mean age (p > 0.05). The mean body mass index (BMI) values, the mean fasting blood sugar (FBS) levels, and the mean HbA1c rate (%) were significantly different between the groups (Table 1). The mean age of the male cohorts of T1D, T2D, and HC groups were 30.63 ± 5.19, 31.77 ± 3.65, and 30.52 ± 4.46 years respectively and the mean age of female cohorts of T1D, T2D, and HC groups were 32.13 ± 5.03, 31.85 ± 3.54, and 32.15 ± 5.64 years respectively.
Table 1.
Demographic characteristics of studied cohorts
| Type 1 Diabetes (T1D) | Type 2 Diabetes (T2D) |
Healthy Controls (HC) | p* | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | T1D vs. HC | T2D vs. HC | T1D vs. T2D | |
| Gender (F/M) | 20/20 | 20/20 | 20/20 | 1,000 | 1,000 | 1,000 | |||
| Age | 31.23 | 5.12 | 31.83 | 3.53 | 31.05 | 4.86 | 0.996 | 0.448 | 0.451 |
| BMI (kg/m2) | 21.84 | 2.36 | 28.17 | 4.29 | 20.86 | 1.64 | 0.020 | 0.001 | 0.001 |
| FBS (mg/dL) | 125.03 | 39.63 | 141.43 | 31.55 | 96.88 | 7.66 | 0.001 | 0.001 | 0.001 |
| HbA1c (%) | 7.72 | 0.84 | 8.55 | 1.35 | 4.68 | 0.53 | 0.001 | 0.001 | 0.009 |
*Calculated with Mann Whitney U
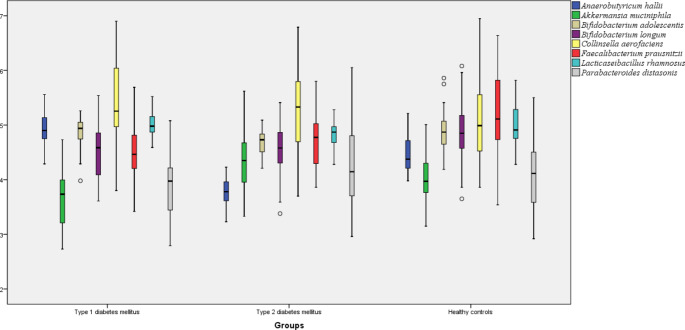
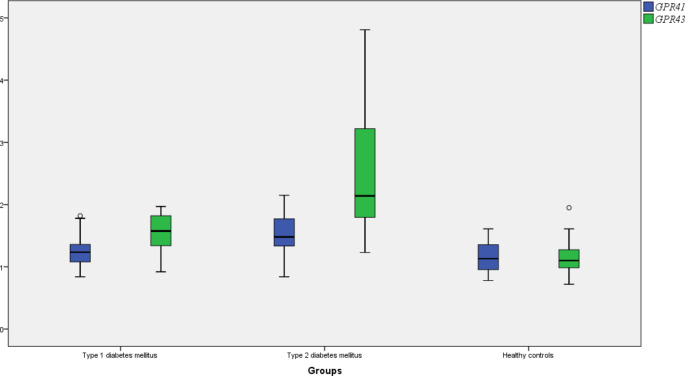
In the gut microbiota, the amount of A.muciniphila, B.longum, F.prausnitzii, and P.distasonis were significantly reduced and the amounts of C.aerofaciens and A.hallii were significantly increased in T1D patients compared to HC. There was no significant change in the amount of B. adolescentis and L. rhamnosus in the gut microbiota of T1D patients compared to HC (p > 0.05). In the mean time, a significant upregulation was found in the circulating GPR43 gene expression level of T1D patients compared to HC (p < 0.05). Comparing the gut microbiota of T2D patients with HC, the amount of A.muciniphila was significantly increased and contrarily the amount of B. adolescentis, B. longum, (A) hallii, and F.prausnitzii were significantly reduced (p < 0.05). There was no significant change in the amount of P.distasonis and C.aerofaciens. Both GPR41 and GPR43 gene expression levels were significantly upregulated in the blood of T2D patients compared to HC (p > 0.05). The amount of (B) adolescentis, A. hallii, and L. rhamnosus were significantly increased and the amount of A. muciniphila, F.prausnitzii, and P. distasonis were significantly reduced in the gut microbiota of T1D patients compared to T2D patients (p < 0.05). In addition, GPR41 and GPR43 gene expression levels were significantly downregulated in the blood of T1D patients compared to T2D patients (p < 0.05) (Table 2). The distribution of the amount of gut bacteria and GPR gene expression levels between the groups were shown in Figs. 1 and 2 respectively.
Table 2.
Comparison of gut bacteria amount and gene expression levels between studied cohorts
| Type 1 Diabetes (T1D) | Type 2 Diabetes (T2D) | Healthy Controls (HC) | p* | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | T1D vs. HC | T2D vs. HC | T1D vs. T2D | |
| Akkermansia muciniphila** | 3.66 | 0.56 | 4.33 | 0.57 | 4.00 | 0.45 | 0.007 | 0.009 | 0.001 |
| Anaerobutyricum hallii** | 4.91 | 0.33 | 3.78 | 0.25 | 4.47 | 0.31 | 0.001 | 0.000 | 0.000 |
| Bifidobacterium adolescentis** | 4.87 | 0.26 | 4.69 | 0.23 | 4.88 | 0.36 | 0.675 | 0.012 | 0.001 |
| Bifidobacterium longum** | 4.53 | 0.54 | 4.59 | 0.44 | 4.89 | 0.49 | 0.003 | 0.006 | 0.541 |
| Collinsella aerofaciens** | 5.38 | 0.77 | 5.27 | 0.72 | 5.08 | 0.71 | 0.047 | 0.186 | 0.551 |
| Faecalibacterium prausnitzii** | 4.51 | 0.43 | 4.76 | 0.51 | 5.15 | 0.79 | 0.001 | 0.009 | 0.039 |
| Lacticaseibacillus rhamnosus ** | 4.99 | 0.20 | 4.81 | 0.23 | 4.98 | 0.36 | 0.535 | 0.085 | 0.003 |
| Parabacteroides distasonis** | 3.91 | 0.58 | 4.30 | 0.75 | 4.07 | 0.62 | 0.225 | 0.211 | 0.038 |
| GPR41*** | 1.25 | 0.25 | 1.53 | 0.31 | 1.15 | 0.23 | 0.087 | 0.000 | 0.001 |
| GPR43*** | 1.55 | 0.31 | 2.42 | 0.89 | 1.14 | 0.25 | 0.001 | 0.001 | 0.001 |
*Calculated with Mann Whitney U. **log10copy/gr. ***relative ratio
Fig. 1.
The distribution of the amounts of gut bacteria between the groups
Fig. 2.
The distribution of GPR41 and GPR43 gene expression levels between the groups
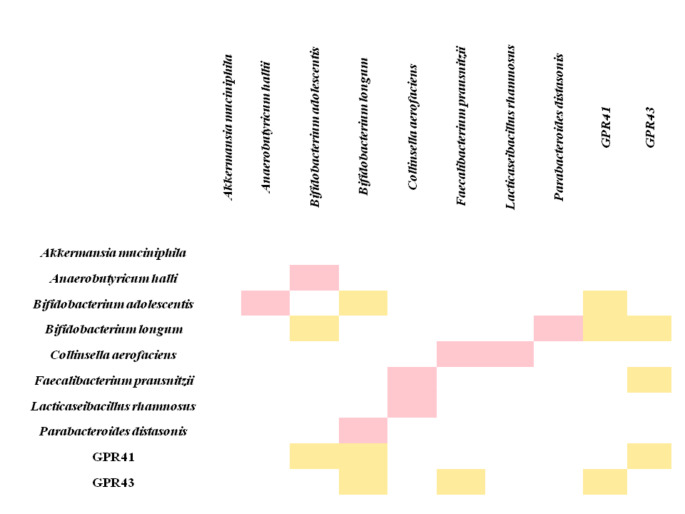
In T1D patients, a statistically weak positive correlation was found between, B. adolescentis and B. longum amounts in the gut microbiota and blood GPR41 gene expression levels (rs=0.314, rs=0.319 respectively), also between the amount of F.prausnitzii in the gut microbiota and the blood GRP43 gene expression levels (rs=0.324).
A statistically weak positive link was also found between GPR41 and GPR43 gene expression in the blood (rs=0.336) and between the amount of B. adolescentis and B. longum in the gut microbiota of T1D patients (rs=0.296). There was a statistically weak negative correlation between the amount of B. adolescentis and A.hallii (rs=-0.286), B. longum and P. distasonis (rs=-0,316), C.aerofaciens and F.prausnitzii (rs=-0.319), C.aerofaciens and L. rhamnosus (rs=-0.299) in the gut microbiota of T1D patients (Fig. 3). There was also a statistically weak positive correlation between the FBS and GPR43 gene expression (rs=0.266) in the blood of T1D patients.
Fig. 3.
Correlations between SCFAs receptor genes expressions and the amount of gut bacteria in T1D patients (Only significant correlation links (rs-values) are shown. rs: Spearman correlation value. rs < 0.25 was evaluated as not statistically correlated, rs = 0.25–0.5 was evaluated as a weak correlation, rs = 0.5–0.75 was evaluated as a moderate correlation, rs = 0.76–0.85 was evaluated as a strong correlation and rs > 0.85 was evaluated as a very strong correlation. Yellow color: Positive weak correlation; Pink color: Negative weak correlation)
For T2D patients, there was a statistically weak negative correlation between the amount of C. aerofaciens in the gut microbiota and both GPR41 (rs=-0.258) and GPR43 gene expressions (rs=-0.321) in the blood samples of T2D patients. A statistically weak positive correlation was also found between the amount of C. aerofaciens and the amount of L. rhamnosus in the gut microbiota (rs=-0.375) (Fig. 4). There was also a statistically weak positive correlation between the FBS and GPR43 gene expression (rs=0.385), BMI and GPR43 gene expression (rs=0.279) in the blood of T2D patients.
Fig. 4.
Correlations between SCFAs receptor genes expressions in blood and the amount of gut bacteria of T2D patients (Only significant correlation links (rs-values) are shown. rs: Spearman correlation value. rs < 0.25 was evaluated as not statistically correlated, rs = 0.25–0.5 was evaluated as a weak correlation, rs = 0.5–0.75 was evaluated as a moderate correlation, rs = 0.76–0.85 was evaluated as a strong correlation and rs > 0.85 was evaluated as a very strong correlation. Pink color: Negative weak correlation)
Discussion
Diabetes mellitus is an important health problem. It is estimated the number of cases will increase over the years worldwide [18]. The pathogenesis of diabetes is still complex and unresolved. In addition to host genetics, many factors including microorganisms, diet, and immune system problems, have been reported to play a role in the pathogenesis of diabetes [19]. It has been reported that specific composition changes in the gut microbiota affect specific mechanisms in the metabolic pathways and immune system that may cause the development of T1D and T2D [20]. SCFAs are microbiota-derived metabolites and circulating SCFAs are known to affect glucose metabolism in critical tissues of diabetes patients [21]. Therefore, in our study, we focused on the amounts of some critical SCFA-producing bacteria in the gut microbiota of T1D and T2D cohorts and the determination of SCFAs receptor gene expression levels in the blood of these patients.
Leiva-Gea et al. [22] reported that the abundance of Bifidobacterium and Faecalibacterium genera in the gut microbiota of patients with T1D decreased compared to HC. In this study, decreases in these strains were negatively correlated with serum IL-1β levels. Similarly, in our study, we found a significant decrease in these bacterial strains. Matos et al. [23] reported that P.distasonis was detected only in T1D patients gut microbiota, but not detected in healthy controls. Additionally, researchers did not detect any changes in the abundance of Bifidobacterium spp. in the gut microbiota of patients with T1D compared to healthy controls, however they mentioned a significant decrease in Lactobacillus spp. They also reported that Lactobacillus spp. inhibited the invasion ability of P.distasonis. In our study, contradictory data was obtained compared to the study. We found no significant change between P.distasonis and L. rhamnosus abundance in T1D patients compared to HC. In the correlation analysis, we did not detect any relationship between P.distasonis and L. rhamnosus amounts. On the other hand, in our study, a significant negative correlation was found between the amounts of P.distasonis and B. longum in the gut microbiota of the patients with T1D. It was thought that, the difference between the results might be about the study was conducted in pediatric patients and the number of patients was low. Liu et al. [24] reported that C.aerofaciens and A.hallii were significantly enriched but P.distasonis amount was significantly decreased in the gut microbiota of T1D patients compared to HC. In this study, the correlation between the amounts of A.hallii and FBS was also determined. In our study, similar results were found on the amounts of bacteria other than P.distasonis in the gut microbiota of T1D patients compared to HC patients. We also found a decrease in the amount of P.distasonis, but this decrease was not significant. The difference was thought to be due to the age and BMI differences of the study groups. Groele et al. [25] reported that no significant effect on the beta cells (β-cells) function was observed in children newly diagnosed with T1D and receiving L. rhamnosus. Similarly, in the gut microbiota, no significant difference was found on the abundance of L. rhamnosus of T1D patients compared to HC. Bell et al. [26] reported that B. adolescentis and B. longum amounts were correlated with HbA1c reduction in adult patients with T1D who were followed up with SCFAs supplementation. We did not find a similar correlation for these bacteria in our study; the fact that we couldn’t follow the patients prospectively may have caused this different result. Similar to our results, Talukdar et al. [27] reported elevated abundance on C.aerofaciens amounts in the gut microbiota of patients with T1D. However, in contrast to our results, they reported an increased abundance on B. longum amounts. This difference may due to the small number of patients used in their study when compared to our patient numbers. Fassatoui et al. [28] reported a significantly decreased F. prausnitzii and A.muciniphila amounts and no significantly decreased abundance of B. longum in the gut microbiota of T1D and T2D patients compared to HC. Similar results were seen on our study for F.prausnitzii and A.muciniphila amounts in the gut microbiota of T2D patients compared to the HC. However we also found a significant decreased abundance of B. longum and B. adolescentis. These differences may due to the small number of patients used in their study. Similar to our findings, Talukdar et al. [27] showed a decreased abundance of F.prausnitzii and Wang et al. [29] reported a declined abundance for B. longum in the gut microbiota of T2D patients compared to HC. In our study, we also found elevated abundances of A. hallii in the gut microbiota of T2D patients compared to HC. However on the literature, a contradictory data to our study as A. hallii treatment at db mice improved insulin sensitivity and energy consumption was reported by Udayappan et al. [30].
Lê et al. [31] reported significantly increased abundance of L. rhamnosus amounts and significantly decreased abundance of B. adolescentis amounts in the gut microbiota of T2D patients. They also showed decreased abundances of B. longum but it was not significant. Our results for the B. adolescentis amounts were similar to that study. It was thought that the opposite results were due to the patients in that study were on medication.
GPR41 and GPR43 are known as SCFA receptors expressed in different tissues and they are activated by SCFAs [7, 8]. Acetate, butyrate, and propionate are the most common SCFAs and these are also the most potent effector against GPR41 and GPR43 [7]. They are important in the regulation of insulin secretion from pancreatic islet cells and regulation of metabolic homeostasis [32]. It has been reported that the GPR43 levels contributes to inflammation by inducing M1 and M2 macrophages after stimulation by SCFAs, and the gene expression is upregulated on obesity and diabetes in adipose tissues. However, it has been stated that the mechanism of GPR43-related inflammation is complex [33]. Shi et al. [34] reported that GPR43 (FFAR2) gene expression levels were upregulated in T1D patients and this was induced by NFκB activation. Similarly, Fang et al. [35] showed that GPR41 and GPR43 gene expression levels were increased in T2D as well. In particular, it has been reported that GPR43 has a critical role in microorganism-host communication [36]. Lu et al. [37] reported that SCFAs supplementation upregulated the GPR41 and GPR43 gene expressions, and these upregulations were correlated with some bacteria such as Lactobacillus spp. They also reported that these receptors are involved in a complex inflammation mechanism [37]. In vivo and in vitro studies on co-culture of SCFA-producing bacteria or adding them to the diet show that SCFA production increases in co-existence [38, 39]. In accordance with these reports, we found some weak positive correlations in some SCFA-producing bacteria in the gut microbiota of T1D and T2D patients in our study.
Our study has also some limitations like the inability to define the gut microbiota profile with the metagenomic approach, the gene expression profiles with the transcriptomic approach, and the SCFAs profiles with the metabolomics approach. These methods are expensive and our budget was not enough. In addition, not measuring the amount of SCFAs in patient and control groups was a deficiency, but it was sufficient to show the correlation of GPR41 and GPR43 levels which were the main SCFAs receptors.
In conclusion, it was determined that there were similar signatures on the abundance changes of B. longum and F.prausnitzii strains in the gut microbiota of T1D and T2D patients. In particular, these compositional changes were also found to be correlated with the GPR43 gene expression levels in the blood of T1D patients. The increase abundances of C.aerofaciens in the gut microbiota of patients with T2D was correlated with GPR43 gene expression upregulation in the blood. Diabetes is a complex metabolic disease and it is clear that this complex mechanism also exists in terms of the host-microorganism relationship. It has been concluded that there were changes in the composition of SCFAs-producing microorganisms with probiotic properties or probiotic potential and these changes also affected the circulating SCFA receptors and contributed to the inflammation that causes diabetes. To understand the mechanism of these diseases, it is necessary to carry out comprehensive studies that determine the contribution of SCFAs-producing microorganisms in the gut microbiota.
Acknowledgements.
Statements and Declarations.
.
List of abbreviations
- ADA
American Diabetes Association.
- BMI
Body mass index.
- FBS
Fasting blood sugar.
- FFAR2
Free fatty acid receptor 2.
- FFAR3
Free fatty acid receptor 3.
- GPCRs
G protein-coupled receptors.
- GPR41
G protein-coupled receptor 41.
- GPR43
G protein-coupled receptor 43.
- HC
Healthy controls.
- rs
Spearman correlation value.
- SCFAs
Short-chain fatty acids.
- T1D
Type 1 diabetes mellitus.
- T2D
Type 2 diabetes mellitus.
Contribution
All authors have made substantial contributions to and have approved the final manuscript. HBT, MD, FEK, ZT, MO, NK and BSK have been involved in the conception and design of the study. MD, ZT, FEK, HBT and MO have contributed to the acquisition and analysis of data. HBT, MD, FEK, ZT, MO, NK and BSK have contributed to the interpretation of data.
Funding
No external funding was received for this study.
Declarations
Conflict of interest
All authors state that we don’t have any potential conflict of interest including any financial activities, additional affiliations, personal or other relationships with other people or organizations that could influence, or be perceived to influence, their work, such as employment, consultancies, stock ownership, honoraria, patent applications/registrations, grants or other funding.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zaccardi F, Webb DR, Yates T, Davies MJ. Pathophysiology of type 1 and type 2 diabetes mellitus: a 90-year perspective. Postgrad Med J. 2016;92(1084):63–9. doi: 10.1136/postgradmedj-2015-133281. [DOI] [PubMed] [Google Scholar]
- 2.Sapra A, Bhandari P. Diabetes Mellitus. [Updated 2021 Sep 18]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2022 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK551501/.
- 3.Kawasaki E. Type 1 diabetes and autoimmunity. Clin Pediatr Endocrinol. 2014;23(4):99–105. doi: 10.1297/cpe.23.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galicia-Garcia U, Benito-Vicente A, Jebari S, et al. Pathophysiology of Type 2 Diabetes Mellitus. Int J Mol Sci. 2020;21(17):6275. doi: 10.3390/ijms21176275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skyler JS, Bakris GL, Bonifacio E, Darsow T, Eckel RH, Groop L, et al. Differentiation of Diabetes by Pathophysiology, Natural History, and Prognosis. Diabetes. 2017;66(2):241–55. doi: 10.2337/db16-0806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaak EE, Canfora EE, Theis S, Frost G, Groen AK, Mithieux G, et al. Short chain fatty acids in human gut and metabolic health. Benef Microbes. 2020;11(5):411–55. doi: 10.3920/BM2020.0057. [DOI] [PubMed] [Google Scholar]
- 7.Ang Z, Ding JL. GPR41 and GPR43 in Obesity and Inflammation - Protective or Causative? Front Immunol. 2016;7:28. doi: 10.3389/fimmu.2016.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nøhr MK, Pedersen MH, Gille A, Egerod KL, Engelstoft MS, Husted AS, et al. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology. 2013;154(10):3552–64. doi: 10.1210/en.2013-1142. [DOI] [PubMed] [Google Scholar]
- 9.Gagliardi A, Totino V, Cacciotti F, Iebba V, Neroni B, Bonfiglio G, et al. Rebuilding the Gut Microbiota Ecosystem. Int J Environ Res Public Health. 2018;15(8):1679. doi: 10.3390/ijerph15081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demirci M, Tokman HB, Uysal HK, Demiryas S, Karakullukcu A, Saribas S, et al. Reduced Akkermansia muciniphila and Faecalibacterium prausnitzii levels in the gut microbiota of children with allergic asthma. Allergol Immunopathol (Madr) 2019;47(4):365–71. doi: 10.1016/j.aller.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Bolsega S, Bleich A, Basic M. Synthetic Microbiomes on the Rise-Application in Deciphering the Role of Microbes in Host Health and Disease. Nutrients. 2021;13(11):4173. doi: 10.3390/nu13114173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrade JC, Almeida D, Domingos M, Seabra CL, Machado D, Freitas AC, et al. Commensal Obligate Anaerobic Bacteria and Health: Production, Storage, and Delivery Strategies. Front Bioeng Biotechnol. 2020;8:550. doi: 10.3389/fbioe.2020.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.American Diabetes Association. 2. Classification and Diagnosis of Diabetes. Diabetes Care. 2017 Jan;40(Suppl 1):S11-S24. doi: 10.2337/dc17-S005. PMID: 27979889. [DOI] [PubMed]
- 14.Blake MR, Raker JM, Whelan K. Validity and reliability of the Bristol Stool Form Scale in healthy adults and patients with diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther. 2016;44(7):693–703. doi: 10.1111/apt.13746. [DOI] [PubMed] [Google Scholar]
- 15.Demirci M, Bahar Tokman H, Taner Z, Keskin FE, Çağatay P, Ozturk Bakar Y, et al. Bacteroidetes and Firmicutes levels in gut microbiota and effects of hosts TLR2/TLR4 gene expression levels in adult type 1 diabetes patients in Istanbul, Turkey. J Diabetes Complications. 2020;34(2):107449. doi: 10.1016/j.jdiacomp.2019.107449. [DOI] [PubMed] [Google Scholar]
- 16.Li G, Su H, Zhou Z, Yao W. Identification of the porcine G protein-coupled receptor 41 and 43 genes and their expression pattern in different tissues and development stages. PLoS ONE. 2014;9(5):e97342. doi: 10.1371/journal.pone.0097342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Faisal K, Tusiimire J, Yadesa TM. Prevalence and Factors Associated with Non-Adherence to Antidiabetic Medication Among Patients at Mbarara Regional Referral Hospital, Mbarara, Uganda. Patient Prefer Adherence. 2022;16:479–91. doi: 10.2147/PPA.S343736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Chu J, Hao W, Zhang J, Li H, Yang C, et al. Gut Microbiota and Type 2 Diabetes Mellitus: Association, Mechanism, and Translational Applications. Mediators Inflamm. 2021;2021:5110276. doi: 10.1155/2021/5110276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li WZ, Stirling K, Yang JJ, Zhang L. Gut microbiota and diabetes: From correlation to causality and mechanism. World J Diabetes. 2020;11(7):293–308. doi: 10.4239/wjd.v11.i7.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim CH. Microbiota or short-chain fatty acids: which regulates diabetes? Cell Mol Immunol. 2018;15(2):88–91. doi: 10.1038/cmi.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leiva-Gea I, Sánchez-Alcoholado L, Martín-Tejedor B, Castellano-Castillo D, Moreno-Indias I, Urda-Cardona A, et al. Gut Microbiota Differs in Composition and Functionality Between Children With Type 1 Diabetes and MODY2 and Healthy Control Subjects: A Case-Control Study. Diabetes Care. 2018;41(11):2385–95. doi: 10.2337/dc18-0253. [DOI] [PubMed] [Google Scholar]
- 23.Matos J, Matos I, Calha M, Santos P, Duarte I, Cardoso Y, et al. Insights from Bacteroides Species in Children with Type 1 Diabetes. Microorganisms. 2021;9(7):1436. doi: 10.3390/microorganisms9071436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Cheng YW, Shao L, Sun SH, Wu J, Song QH, et al. Gut microbiota dysbiosis in Chinese children with type 1 diabetes mellitus: An observational study. World J Gastroenterol. 2021;27(19):2394–414. doi: 10.3748/wjg.v27.i19.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groele L, Szajewska H, Szalecki M, Świderska J, Wysocka-Mincewicz M, Ochocińska A, et al. Lack of effect of Lactobacillus rhamnosus GG and Bifidobacterium lactis Bb12 on beta-cell function in children with newly diagnosed type 1 diabetes: a randomised controlled trial. BMJ Open Diabetes Res Care. 2021;9(1):e001523. doi: 10.1136/bmjdrc-2020-001523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell KJ, Saad S, Tillett BJ, McGuire HM, Bordbar S, Yap YA, et al. Metabolite-based dietary supplementation in human type 1 diabetes is associated with microbiota and immune modulation. Microbiome. 2022;10(1):9. doi: 10.1186/s40168-021-01193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talukdar R, Sarkar P, Jakkampudi A, Sarkar S, Aslam M, Jandhyala M, et al. The gut microbiome in pancreatogenic diabetes differs from that of Type 1 and Type 2 diabetes. Sci Rep. 2021;11(1):10978. doi: 10.1038/s41598-021-90024-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fassatoui M, Lopez-Siles M, Díaz-Rizzolo DA, Jmel H, Naouali C, Abdessalem G, et al. Gut microbiota imbalances in Tunisian participants with type 1 and type 2 diabetes mellitus. Biosci Rep. 2019;39(6):BSR20182348. doi: 10.1042/BSR20182348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Yu X, Xu X, Ming J, Wang Z, Gao B, et al. The Fecal Microbiota Is Already Altered in Normoglycemic Individuals Who Go on to Have Type 2 Diabetes. Front Cell Infect Microbiol. 2021;11:598672. doi: 10.3389/fcimb.2021.598672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Udayappan S, Manneras-Holm L, Chaplin-Scott A, Belzer C, Herrema H, Dallinga-Thie GM, et al. Oral treatment with Eubacterium hallii improves insulin sensitivity in db/db mice. NPJ Biofilms Microbiomes. 2016;2:16009. doi: 10.1038/npjbiofilms.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lê KA, Li Y, Xu X, Yang W, Liu T, Zhao X, et al. Alterations in fecal Lactobacillus and Bifidobacterium species in type 2 diabetic patients in Southern China population. Front Physiol. 2013;3:496. doi: 10.3389/fphys.2012.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veprik A, Laufer D, Weiss S, Rubins N, Walker MD. GPR41 modulates insulin secretion and gene expression in pancreatic β-cells and modifies metabolic homeostasis in fed and fasting states. FASEB J. 2016;30(11):3860–9. doi: 10.1096/fj.201500030R. [DOI] [PubMed] [Google Scholar]
- 33.Nakajima A, Nakatani A, Hasegawa S, Irie J, Ozawa K, Tsujimoto G, et al. The short chain fatty acid receptor GPR43 regulates inflammatory signals in adipose tissue M2-type macrophages. PLoS ONE. 2017;12(7):e0179696. doi: 10.1371/journal.pone.0179696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi G, Sun C, Gu W, Yang M, Zhang X, Zhai N, et al. Free fatty acid receptor 2, a candidate target for type 1 diabetes, induces cell apoptosis through ERK signaling. J Mol Endocrinol. 2014;53(3):367–80. doi: 10.1530/JME-14-0065. [DOI] [PubMed] [Google Scholar]
- 35.Tang C, Ahmed K, Gille A, Lu S, Gröne HJ, Tunaru S, et al. Loss of FFA2 and FFA3 increases insulin secretion and improves glucose tolerance in type 2 diabetes. Nat Med. 2015;21(2):173–7. doi: 10.1038/nm.3779. [DOI] [PubMed] [Google Scholar]
- 36.Bindels LB, Dewulf EM, Delzenne NM. GPR43/FFA2: physiopathological relevance and therapeutic prospects. Trends Pharmacol Sci. 2013;34(4):226–32. doi: 10.1016/j.tips.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y, Fan C, Li P, Lu Y, Chang X, Qi K. Short Chain Fatty Acids Prevent High-fat-diet-induced Obesity in Mice by Regulating G Protein-coupled Receptors and Gut Microbiota. Sci Rep. 2016;6:37589. doi: 10.1038/srep37589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de los Reyes-Gavilan CG. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol Lett. 2015;362(21):fnv176. doi: 10.1093/femsle/fnv176. [DOI] [PubMed] [Google Scholar]
- 39.Kim H, Jeong Y, Kang S, You HJ, Ji GE. Co-Culture with Bifidobacterium catenulatum Improves the Growth, Gut Colonization, and Butyrate Production of Faecalibacterium prausnitzii: In Vitro and In Vivo Studies. Microorganisms. 2020;8(5):788. doi: 10.3390/microorganisms8050788. [DOI] [PMC free article] [PubMed] [Google Scholar]