Abstract
Background & Aims
Alcohol-associated liver disease (ALD) comprises a spectrum of disorders including steatosis, steatohepatitis, fibrosis, and cirrhosis. We aimed to study the role of protein arginine methyltransferase 6 (PRMT6), a new regulator of liver function, in ALD progression.
Methods
Prmt6-deficient mice and wild-type littermates were fed Western diet with alcohol in the drinking water for 16 weeks. Mice fed standard chow diet or Western diet alone were used as a control.
Results
We found that PRMT6 expression in the liver is down-regulated in 2 models of ALD and negatively correlates with disease severity in mice and human liver specimens. Prmt6-deficient mice spontaneously developed liver fibrosis after 1 year and more advanced fibrosis after high-fat diet feeding or thioacetamide treatment. In the presence of alcohol Prmt6 deficiency resulted in a dramatic increase in fibrosis development but did not affect lipid accumulation or liver injury. In the liver PRMT6 is primarily expressed in macrophages and endothelial cells. Transient replacement of knockout macrophages with wild-type macrophages in Prmt6 knockout mice reduced profibrotic signaling and prevented fibrosis progression. We found that PRMT6 decreases profibrotic signaling in liver macrophages via methylation of integrin α-4 at R464 residue. Integrin α-4 is predominantly expressed in infiltrating monocyte derived macrophages. Blocking monocyte infiltration into the liver with CCR2 inhibitor reduced fibrosis development in knockout mice and abolished differences between genotypes.
Conclusions
Taken together, our data suggest that alcohol-mediated loss of Prmt6 contributes to alcohol-associated fibrosis development through reduced integrin methylation and increased profibrotic signaling in macrophages.
Keywords: PRMT6, ALD, Inflammation, TGFβ, Macrophage
Abbreviations used in this paper: ALD, alcohol-associated liver disease; BMDM, bone marrow-derived macrophages; HCC, hepatocellular carcinoma; HSC, hepatic stellate cells; IL, interleukin; KC, Kupffer cells; KO, knockout; LSEC, liver sinusoidal endothelial cells; MACS, magnetic-activated cell sorting; PBS, phosphate-buffered saline; PRMT, protein arginine methyltransferase; TGF-β, transforming growth factor beta; TMT, tandem mass tag; WD, Western diet; WDA, Western diet alcohol; WT, wild-type
Summary.
Alcohol reduces liver PRMT6 levels, and loss of PRMT6 promotes fibrosis development. PRMT6 regulates fibrosis development via integrin α-4 methylation in macrophages. Proinflammatory and profibrotic signaling in macrophages exist in equilibrium determined by the activity of PRMT6.
Protein arginine methyltransferase 6 (PRMT6) is an enzyme that catalyzes the formation of mono and asymmetric dimethylarginine.1,2 PRMT6 was reported to methylate histones: histone H3 to form H3R2me2a,1 which acts as a repressive mark by blocking H3K4 methylation,1 and histones H2A and H4 to form H2AR3me and H4R3me leading to transcriptional activation.3
PRMT6 also methylates non-histone proteins; however, under normal conditions it has a narrow range of substrates compared with other methyltransferases such as PRMT1 or PRMT4.2,4 Despite its limited substrate scope, PRMT6 has been reported to regulate a wide range of cellular processes including cell cycle, cellular senescence, adipocyte differentiation, neuronal function, hematopoietic stem cell differentiation, and many others.3, 4, 5 Although PRMT6 is up-regulated in several types of cancer, it is down-regulated in hepatocellular carcinoma (HCC). PRMT6 down-regulation correlates with aggressive HCC behavior and contributes to tumorigenic properties of HCC by regulating glycolysis, stemness, and cell survival.5,6 Previous studies have identified targets AR, ERα, FOXO3, CRAF, and SIRT77, 8, 9 as potential targets linking PRMT6 to HCC progression.
Here we identified integrin α4 as a target of PRMT6 in alcohol-fed mouse liver. Integrin α4, or CD49d, is expressed on the surface of most leukocytes.10 It facilitates adhesion of the leukocytes to their receptors such as vascular cell adhesion molecule-1, mediating cell-cell and cell-matrix interactions, and involved in various processes including development, immune regulation, differentiation, and wound healing. In their inactive form, integrins are in a low-affinity, non-adhesive conformation. Upon stimulation either by ligand interaction or by signaling within the cell, integrins are converted to high-affinity conformations. Active integrins initiate intracellular signaling pathways that regulate migration, cell survival, and extracellular matrix homeostasis. Several integrins were reported to modulate matrix expression and degradation via regulation of the growth factor transforming growth factor beta (TGF-β) in a positive manner, like integrins α5, α11, and αv, or in a negative manner, like integrins α1β1, α2, or β3.11, 12, 13, 14, 15, 16, 17, 18
Mutations and posttranslational modifications of integrins are known to affect their activity and inside-out as well as outside-in signaling.19,20 Integrin α4 phosphorylation regulates paxillin binding to the integrin cytoplasmic domain, and this regulates cell migration.20 On the other hand, integrin α2β1 phosphorylation induced by adhesion affects intracellular Hippo pathway activation.21 Another example of integrin-regulated tumor functional change is TGF-β1–mediated phosphorylation of integrin β3, which results in a conformational change of the extracellular domains and promotes tumor invasion.22
In this work we identified that PRMT6 methylates integrin α4, which results in higher proinflammatory cytokine production but lower profibrotic signaling in macrophages. Prmt6-deficient mice developed more advanced liver fibrosis in several models of liver disease including high-fat diet feeding, thioacetamide-induced fibrosis, and alcohol-associated liver disease (ALD). Moreover, we found that Prmt6 expression is down-regulated in 2 models of ALD, suggesting that alcohol-mediated loss of Prmt6 contributes to alcohol-associated fibrosis development.
Results
Prmt6 Expression Is Down-regulated in Models of Alcohol-Associated Liver Disease
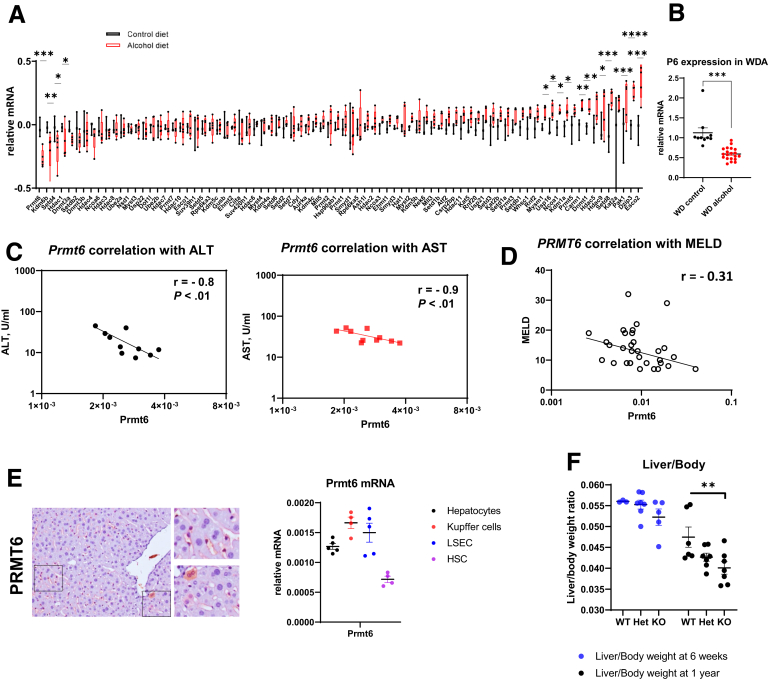
We examined gene expression of several epigenetic regulators in the livers of mice fed either control liquid diet or Lieber-DeCarli alcohol liquid diet for 4 weeks. We found that several epigenetic regulators were up-regulated and only a few were down-regulated by alcohol (Figure 1A). The top down-regulated gene was Prmt6 (Figure 1A). Prmt6 expression was down-regulated in another model of ALD as well, the Western diet alcohol (WDA) model23 (Figure 1B). In this model mice receive high-fat Western diet pellets and either water as a control (WD group) or alcohol in the drinking water (WDA group) for 16 weeks. Taken together, alcohol thus reduced Prmt6 gene expression in 2 different mouse ALD models.
Figure 1.
PRMT6 is down-regulated in liver disease. (A) Mice (male and female) were fed Lieber-DeCarli alcohol (N = 5) or control (N = 3) liquid diet for 4 weeks. Relative gene expression in whole liver mRNA. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. (B) Mice (male and female) were fed WD (control) or WD with alcohol in the drinking water (alcohol) for 16 weeks. Relative gene expression in whole liver mRNA. N = 10 and 20 per group, respectively. ∗∗∗P < .001. (C) Correlation between Prmt6 gene expression and serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) in alcohol-fed mice. (D) Correlation between PRMT6 gene expression and patients’ Model for End-stage Liver Disease (MELD) score. N = 48. (E, left) Representative images of PRMT6 protein staining in livers of mice. (Right) Prmt6 gene expression in isolated cells. N = 5 mice. (F) Liver to body weight ratios of wild-type (WT), Prmt6+/- (Het), and Prmt6-/- (KO) mice fed chow diet at 6 weeks and at 1 year of age. N = 3–6 mice per group. ∗∗P < .01.
We found that there was significant negative correlation between Prmt6 and serum levels of alanine aminotransferase and aspartate aminotransferase in alcohol-fed mice, suggesting that the Prmt6 decrease is greater in mice with more advanced liver disease (Figure 1C).
In humans, the PRMT6 gene is located in chromosome 1. Several polymorphisms in this gene were found as significant hits in multiple genome-wide association studies relevant to liver disease (genome-wide association study catalog). PRMT6 gene polymorphisms are associated with levels of low-density lipoprotein, high-density lipoprotein, and gamma-glutamyl transferase, an enzyme commonly elevated in ALD patients,24 suggesting that PRMT6 may have a role in human ALD as well. We further examined PRMT6 expression in human liver transplant explants and liver donor tissue obtained from KUMC Liver Bank. PRMT6 expression negatively correlated with Model for End-stage Liver Disease score (Figure 1D), suggesting that diminished PRMT6 expression correlates with more advanced liver disease.
To determine which cell types are relevant for PRMT6-mediated effects, we examined PRMT6 expression in the liver by immunohistochemical staining and by assessing mRNA expression in isolated cells (Figure 1E). We found that PRMT6 is expressed in a small subset of hepatocytes and in liver macrophages and endothelial cells and shows low expression in hepatic stellate cells (HSC) (Figure 1E). Data from single cell RNA sequencing analysis suggest that PRMT6 in healthy humans is enriched in immune cell clusters (HumanProteinAtlas25).
We obtained whole-body Prmt6 knockout (KO) mice from Jackson Laboratory (strain #028929, mixed C57BL/6J and C57BL/6N background; Bar Harbor, ME) and backcrossed them for 5 generations with C57BL/6J mice. We observed that Prmt6-deficient mice show no obvious phenotype, apart from livers that tend to be smaller in size than in their wild-type (WT) littermates. This difference was significant at 1 year of age (Figure 1F).
Prmt6-Deficient Mice Develop Similar Steatosis in the Presence of Alcohol
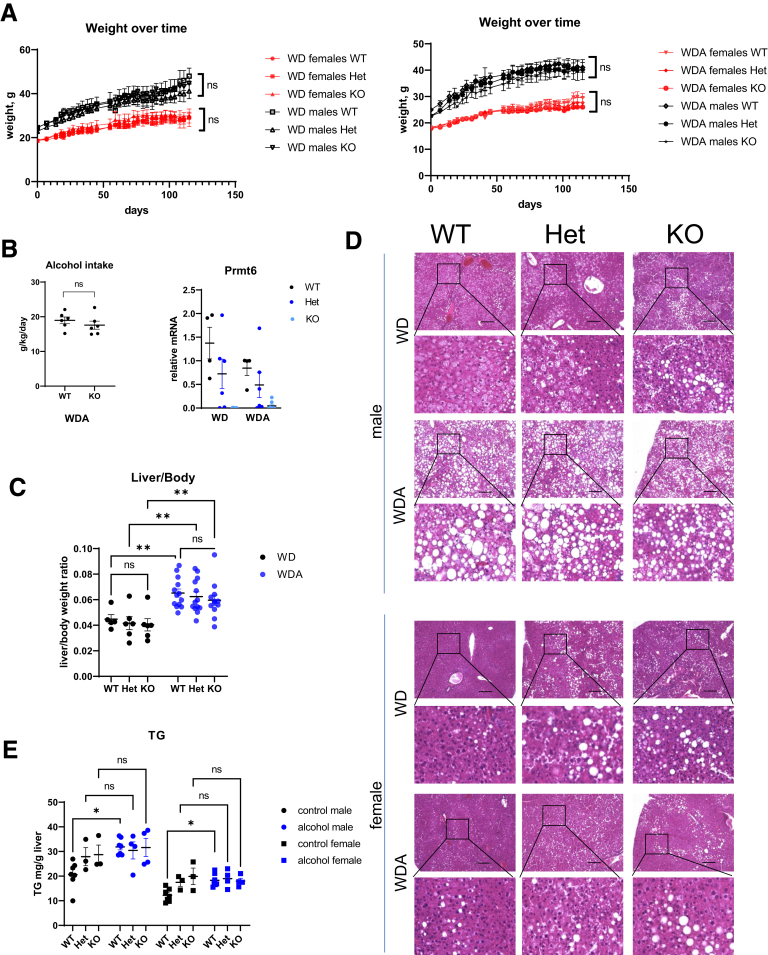
To assess the role of PRMT6 in ALD, we fed male and female WT Prmt6+/+, Prmt6+/- (Het), and Prmt6 -/- (KO) littermates WD and WDA diets for 16 weeks. We observed no difference in weight gain between genotypes (Figure 2A). We measured alcohol intake in WT and KO mice and found that Prmt6 -/- (KO) mice consumed similar amounts of alcohol as WT controls (Figure 2B). Prmt6 expression was reduced by alcohol in WT mice; Prmt6 expression in the livers of heterozygous Prmt6+/- mice was intermediate between WT and KO mice in both feeding groups (Figure 2B).
Figure 2.
PRMT6 loss does not affect lipid accumulation in alcohol-fed mice. (A–E) Wild-type (WT), Prmt6+/- (Het), and Prmt6-/- (KO) mice were fed western diet (WD, control) or WD with alcohol in the drinking water (WDA, alcohol) for 16 weeks. N = 3–8 mice per group. (A) Weight change over time in 12 groups of mice. ns, not significant between genotypes. (B) Alcohol intake in female mice and Prmt6 gene expression in livers of wild-type (WT), Prmt6+/- (Het), and Prmt6-/- (KO) mice fed WD or WDA. (C) Liver to body weight ratios. ∗∗P < .01. (D) Representative images of H&E staining from control and alcohol-fed mice. Bars, 200 μm. (E) Liver triglyceride (TG) levels in mice fed WD or WDA diets. ∗P < .05, ns, not significant. Two-way analysis of variance, males genotype (F2,22 = 1.299, P = .29); diet (F1,22 = 5.850, P = .024); interaction (F2,22 = 1.928, P = .16); females genotype (F2,22 = 3.909, P = .0353); diet (F1,22 = 2.187, P = .1534); interaction (F2,22 = 3.952, P = .034).
We observed that alcohol increased liver/body weight ratios in all genotypes, and although median liver weight was slightly lower in KO mice, we found no significant difference between genotypes in both feeding groups (Figure 2C). Similarly, we found comparable levels of lipid accumulation in the livers of mice after alcohol feeding (Figure 2D and E).
Interestingly, we observed that about half of the Prmt6-deficient mice had elevated lipid accumulation in the control WD fed group (Figure 2D and E); however, they did not have higher liver/body weight ratios. Because overall body weights were not increased, this could be due to other unidentified factors in the Prmt6-deficient mice that reduced non-lipid liver weight of the KO mice (Figure 1F) and compensated for increased lipid accumulation.
Importantly, our data show that Prmt6-deficient mice on WDA diet show no difference in weight gain, alcohol intake, or liver steatosis compared with WDA fed WT mice.
PRMT6 Protects From Fibrosis Development in Alcohol Fed Mice
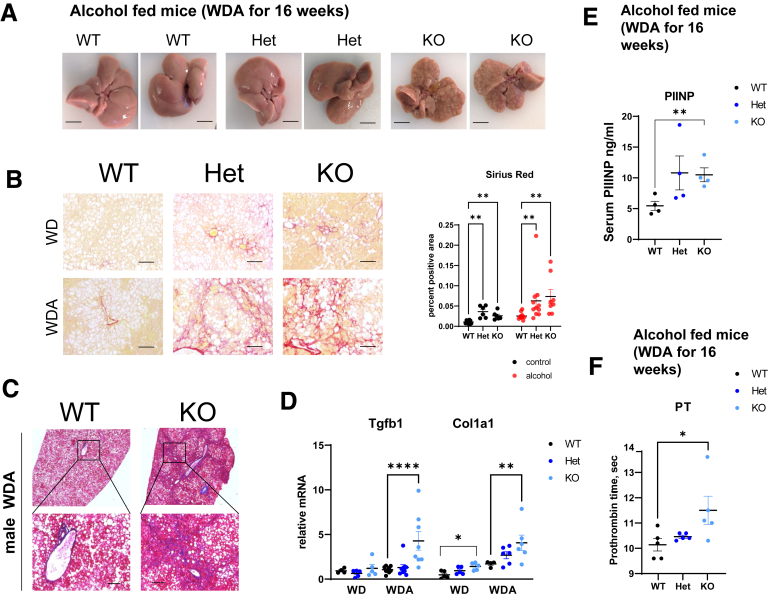
We found that Prmt6-deficient mice developed a dramatic liver disease phenotype after 16 weeks of WDA diet that did not occur in the WT mice. This was characterized by marked nodularity on gross liver appearance (Figure 3A) and more severe fibrosis as assessed by both Sirius red (Figure 3B) and trichrome (Figure 3C) staining of histologic sections (Figure 3A and B). We further observed a significant increase in total liver mRNA levels of Tgfb1 and Col1a1 (Figure 3D). Prmt6-/- mice on the WDA diet showed increased serum N-terminal propeptide of procollagen type III and increased prothrombin time, which agrees with more advanced fibrosis and more advanced liver disease in these animals (Figure 2E and F).
Figure 3.
PRMT6 protects against alcohol-induced fibrosis. (A–F) Wild-type (WT), Prmt6+/- (Het), and Prmt6-/- (KO) mice were fed western diet (WD, control) or WD with alcohol in the drinking water (WDA, alcohol) for 16 weeks. N = 3–8 mice per group. (A) Examples of liver gross appearance in mice after alcohol feeding. Bars, 0.5 cm. (B, left) Representative images of Sirius red staining. Bars, 200 μm. (Right) Positive area of Sirius red staining, N = 6–10 mice (male and female combined) per group. Area was measured by averaging 3 random images in a blinded manner. ∗∗P < .01. Two-way analysis of variance WT vs KO genotype (F1,32 = 15.37, P = .0004); diet (F1,32 = 14.88, P = .0005); interaction (F1,32 = 4.086, P = .05. (C) Masson trichrome staining of livers from WT and KO mice fed alcohol. Bars, 200 μm. (D) Relative liver mRNA in indicated groups, N = 3–8 mice per group. ∗∗P < .01, ∗∗∗∗P < .001. Two-way analysis of variance Tgfb1 genotype (F2,34 = 5.800, P = .0068); diet (F1,34 = 6.831, P = .0133); interaction (F2,34 = 3.419, P = .04); Col1a1 genotype (F2,25 = 5.814, P = .0084); diet (F1,25 = 22.61, P < .0001); interaction (F2,25 = 1.110, P = .34). (E) Serum PIIINP levels from mice fed alcohol, N = 4. ∗∗P < .01. (F) Prothrombin time (PT) in male mice fed alcohol. N = 5. ∗P < .05.
In contrast, Prmt6-deficient mice that were fed control WD showed only a small increase in Sirius red staining (Figure 3B) and an increase in liver mRNA levels of Col1a1 (Figure 3D). On the other hand, mRNA levels of Tgfb1 were not increased in WD fed KO mice. We found that for several parameters (Sirius red area, Tgfb1 expression) there is a significant diet-genotype interaction, suggesting that alcohol-induced factors are necessary to accelerate profibrotic signaling in KO mice.
Prmt6 Controls Fibrosis Development in Mice and Humans
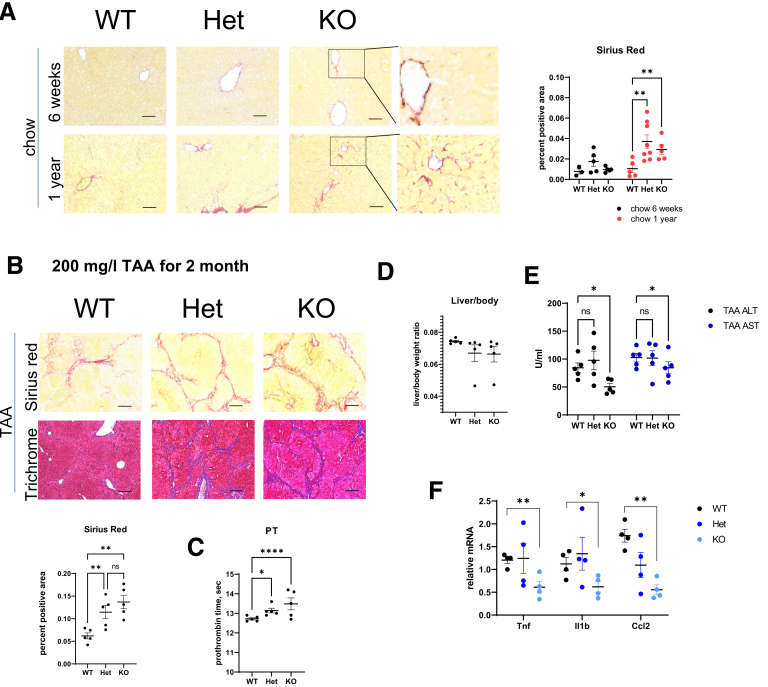
Because we observed an increase in fibrosis in Prmt6+/- and Prmt6-/- mice from all groups of treated animals, we next tested whether Prmt6-deficient mice develop liver fibrosis spontaneously. At 6 weeks of age Prmt6-/- mice did not show any significant fibrosis development (Figure 4A). In contrast at 1 year of age, both Prmt6+/- and Prmt6-/- mice showed an increase in Sirius red staining. Positive staining was present mostly around centrilobular hepatocytes (Figure 4A).
Figure 4.
PRMT6 protects against liver fibrosis in mice. (A, left) Representative images of Sirius red staining in chow fed mice (males and females). Bars, 200 μm. (Right) Positive area of Sirius red staining, N = 3–8 mice per group. Area was measured by averaging 3 random images in a blinded manner. ∗∗P < .01. (B–E) Wild-type (WT), Prmt6+/- (Het), and Prmt6-/- (KO) female mice were given 200 mg/L thioacetamide (TAA) in the drinking water for 2 months. (B, left) Representative images of Sirius red and trichrome staining. Bars, 200 μm. (Right) Positive area of Sirius red staining, N = 5 mice per group. Area was measured by averaging 3 random images in a blinded manner. ns, not significant. ∗∗P < .01. (C) Prothrombin time (PT) in treated mice. N = 5 mice per group. ∗P < .05, ∗∗∗∗P < .001. (D) Liver body weight ratio in these mice. (E) Serum alanine aminotransferase and aspartate aminotransferase levels. ns, not significant. ∗∗P < .01. (F) Relative liver mRNA in indicated groups, N = 4 mice per group. ∗∗P < .01, ∗P < .05.
We next evaluated whether PRMT6 controls fibrosis development in a different model of liver fibrosis, thioacetamide-induced liver fibrosis. WT, Prmt6 +/-, and Prmt6 -/- mice littermates were given thioacetamide in the drinking water for 2 months (Figure 4B–E). We found that Prmt6 +/- and Prmt6 -/- mice developed more severe fibrosis seen by Sirius red and trichrome staining (Figure 4B). In addition, Prmt6 -/- mice had higher prothrombin time (Figure 4C), suggesting a greater decrease in liver synthetic function. Mice had liver/body weight ratios similar to chow fed mice (Figure 4D). Interestingly, Prmt6 -/- mice had lower alanine aminotransferase and aspartate aminotransferase serum levels (Figure 4E) and lower levels of proinflammatory cytokines such as Tnf, Il1b, and Ccl2 (Figure 4F), suggesting that increased fibrosis was not due to a higher degree of liver injury or inflammation in these mice (Figure 4E).
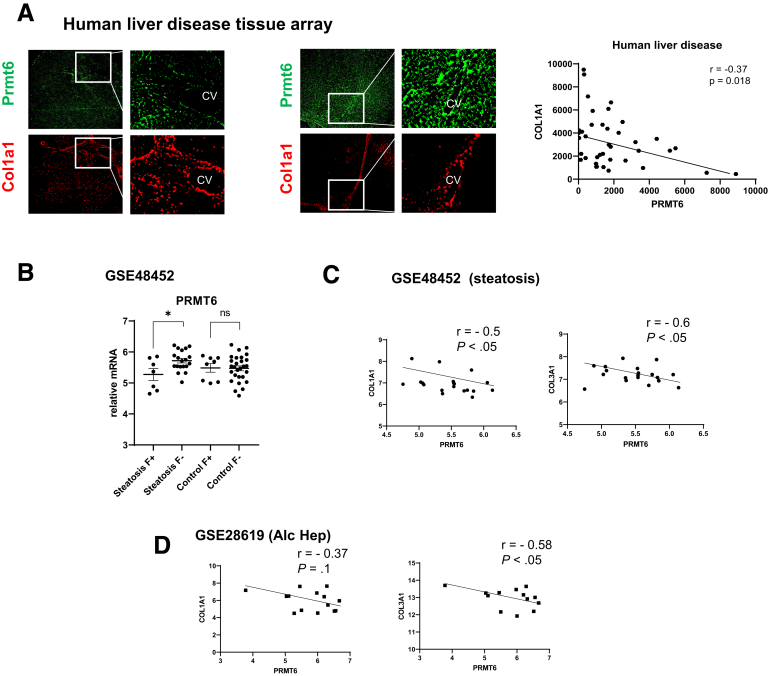
We next evaluated the role of PRMT6 in fibrosis development in human liver specimens using a tissue array. We analyzed PRMT6 protein staining and COL1A1 staining in human liver sections from patients with liver cirrhosis. We observed that PRMT6 levels and staining patterns (hepatocytes vs non-parenchymal cell staining) varied greatly between individuals (Figure 5A). There was a significant negative correlation between PRMT6 and COL1A1 staining (Figure 5A). We further assessed PRMT6 and COL1A1 gene expression in obese patients with normal livers or mild liver disease (steatosis and stage 0–2 fibrosis) and compared these with healthy, non-obese controls using a published dataset (GSE48452). We separated samples into groups on the basis of the presence of steatosis and fibrosis. We observed that PRMT6 gene expression was significantly reduced in livers of patients with steatosis and fibrosis (F+) compared with patients with steatosis but no fibrosis (F–). In contrast, we did not observe any difference in patients with fibrosis in the absence of steatosis (Figure 5B). This could be due to different etiology of fibrosis in these groups. Fibrosis in the absence of steatosis is likely due to causes such as viral hepatitis infection or biliary disease, and on the basis of these results, it is not mediated by PRMT6-dependent mechanism. Thus we excluded these patients from further analysis. We found that in patients with steatosis there was significant negative correlation between PRMT6 and COL1A1 and COL3A1 gene expression (Figure 5C), suggesting that PRMT6 loss promotes fibrosis development in patients with steatosis. In addition, we found a negative correlation between PRMT6 and COL1A1 and COL3A1 gene expression in patients with alcohol-associated hepatitis (GSE28619) (Figure 5D), suggesting that this mechanism is relevant in these patients as well.
Figure 5.
PRMT6 protects against liver fibrosis in humans. (A) Human liver samples were stained for PRMT6 and COL1A1. (Right) Correlation between PRMT6 and COL1A1 protein levels. N = 63. (B) PRMT6, COL1A1, and COL3A1 expression in human samples GSE48452. PRMT6 expression in patients with or without steatosis and fibrosis. (C and D) Correlation between PRMT6, COL1A1, and COL3A1 in patients with steatosis, N = 18 (C) and alcohol-associated hepatitis (N = 15, GSE28619).
PRMT6 Promotes Proinflammatory and Suppresses Profibrotic Signaling
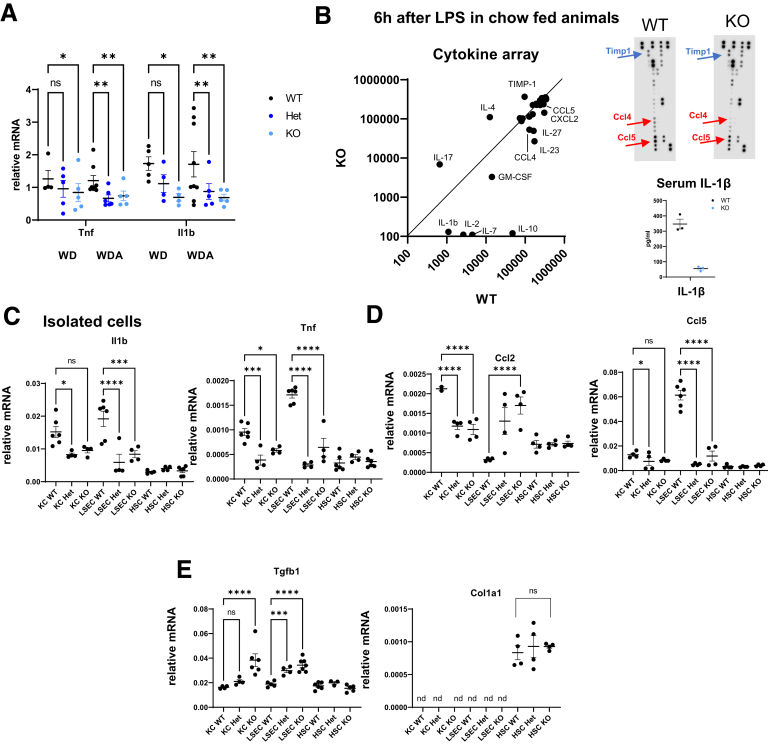
Fibrosis is often driven by an increase in liver inflammation. We evaluated proinflammatory cytokine gene expression in the livers of WT, Prmt6 +/-, and Prmt6 -/- mice fed WD or WDA (Figure 6A). We found that Prmt6 +/- and Prmt6 -/- mice have significantly lower levels of proinflammatory cytokines Tnf and Il1b (Figure 6A). We next tested whether PRMT6 regulates proinflammatory signaling in the absence of alcohol. We injected WT and Prmt6-/- mice with 2 mg/kg of lipopolysaccharide and collected serum samples after 6 hours (Figure 6B). We found that Prmt6-/- mice produce less proinflammatory cytokines and chemokines such as CCL5, CCL4, CXCL2, interleukin (IL)1β, and IL23. In contrast, the expression of the anti-inflammatory cytokine IL4 was increased. Prmt6-/- mice also showed a 3-fold increase in tissue inhibitor of metalloproteinases-1, an inhibitor of matrix metalloproteinases, and a 10-fold increase in IL17A, both known to be involved in fibrosis progression.
Figure 6.
PRMT6 promotes inflammation and suppresses fibrosis. (A) Prmt6-deficient mice have lower levels of proinflammatory cytokines. Wild-type (WT), Prmt6+/- (Het), and Prmt6-/- (KO) mice were fed Western diet (WD, control) or WD with alcohol in the drinking water (WDA, alcohol) for 16 weeks. Relative mRNA levels. N = 4–8 per group. ∗P < .05, ∗∗P < .01. (B) WT and Prmt6-/- (KO) mice were injected with 2 mg/kg lipopolysaccharide. Blood was collected 6 hours after injection, and cytokine levels in serum were measured using Proteome Profiler Cytokine Array. Relative dot densities in WT vs KO. (C–E) WT, Prmt6+/- (Het), and Prmt6-/- (KO) mice were used to isolate hepatic stellate cells (HSC), Kupffer cells (KC), and endothelial cells (LSEC). Relative mRNA expression levels. N = 3–6 mice. ns, not significant. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.
Next, we assessed proinflammatory and profibrotic signaling in isolated cells. We found that liver macrophages and endothelial cells from Prmt6+/- and Prmt6-/- mice show decreased expression of proinflammatory cytokines Tnf and Il1b (Figure 6C). Macrophages also showed a decrease in expression of chemokines Ccl2, whereas endothelial cells had a decrease in Ccl5 (Figure 6D). Both macrophages and endothelial cells had elevated expression levels of Tgfb1 (Figure 6E). In contrast, HSCs did not show genotype-dependent effects in gene expression for cytokines, chemokines, or fibrosis associated genes such as Tgfb1 and Col1a1 (Figure 6D and E). Taken together, these data suggest that proinflammatory and anti-fibrotic effects of PRMT6 are likely mediated by PRMT6 in macrophages and/or endothelial cells but not in HSC, which agrees with relative Prmt6 gene expression in these cell types (Figure 1E).
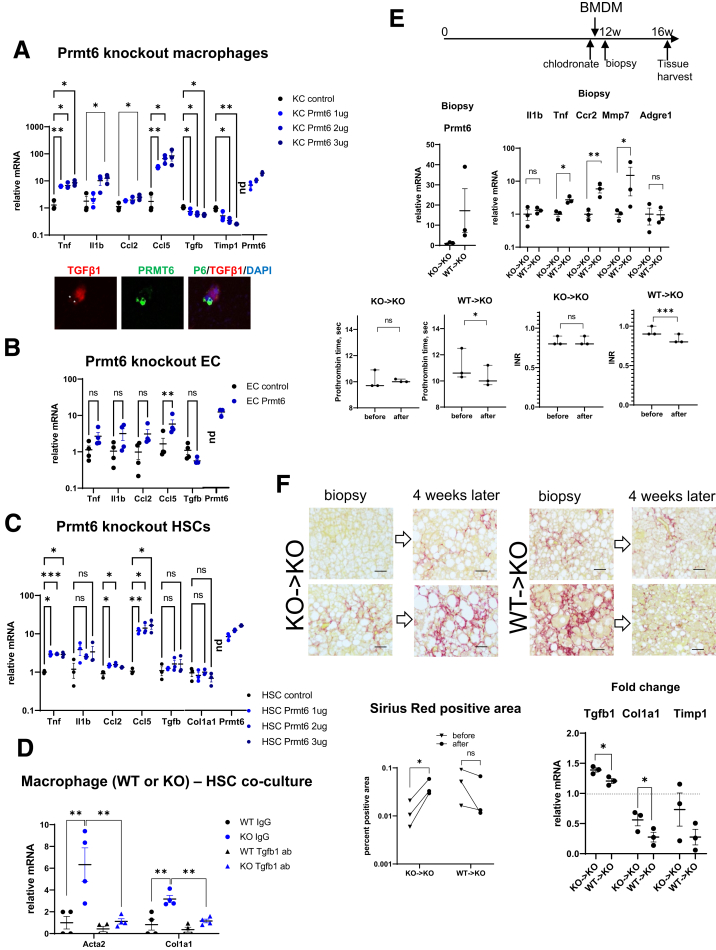
We then overexpressed Prmt6 in Prmt6-/- cells to confirm the role of PRMT6 in expression of proinflammatory and profibrotic genes (Figure 7A). In isolated liver macrophages we observed a dose-dependent increase in Tnf, Il1b, Ccl2, and Ccl5 in response to Prmt6 overexpression, whereas Tgfb1 and Timp1 gene expression was down-regulated 2-fold and 4-fold, respectively. We confirmed that PRMT6 overexpression resulted in a decrease of TGF-β1 expression at the protein level as well (Figure 7A). In isolated endothelial cells we observed that Prmt6 overexpression resulted only in a small increase in Ccl5, suggesting that Prmt6 loss in endothelial cells may not play as important role as macrophages in the phenotype of the KO mice (Figure 7B). Finally, in HSCs we observed an increase in Tnf, Ccl2, and Ccl5 in response to Prmt6 overexpression, although this increase was an order of magnitude lower than that of macrophages. In addition, Tgfb1 and Col1a1 gene expression was not affected (Figure 7C).
Figure 7.
PRMT6 in macrophages suppresses liver fibrosis. (A–C) Cells (liver macrophages (A), endothelial cells (EC) (B), or hepatic stellate cells (HSC) (C)) isolated from Prmt6-/- (KO) mice were treated with control vector or vector expressing Prmt6 at indicated concentrations. N = 3–4 mice. ns, not significant. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. Prmt6 expression is presented relative to WT cells. (A, bottom) TGF-β1 and PRMT6 protein levels in cells expressing Prmt6. Cells overexpressing PRMT6 are marked with asterisks. (D) Liver macrophages from WT or KO mice were used in a co-culture experiment with WT HSCs for 24 hours in the presence of 5 μg/mL of TGF-B1 antibodies or control immunoglobulin G. Relative gene expression in HSCs, ∗∗P < .01. N = 4. (E and F) Prmt6-/- (KO) mice were fed Western diet with alcohol in the drinking water (WDA, alcohol) for 12 weeks. Mice were treated with chlodronate liposomes and 1 day later were injected with 106 BMDM per mouse. Shortly after (within 1 hour after injection) mice liver biopsy was collected. Then mice were fed Western diet and alcohol for 4 more weeks. (E) Schematic of performed procedures. (Bottom left) Gene expression in liver biopsies from mice that received WT or KO macrophages as percent expression of a WT mouse. (Bottom right) International normalized ratio and prothrombin time in mice before chlodronate injection and after 16 weeks of feeding. N = 3 per group. ∗∗∗P < .001 paired t test. (F, left) Sirius red staining of biopsy and corresponding liver sections. Corresponding sections were placed on the same slide and stained simultaneously. Bars, 100 μm. (Right) Percent positive area of Sirius red staining. ns, not significant. ∗P < .05. (Bottom) Fold change in liver mRNA during last 4 weeks of feeding. ∗P < .05.
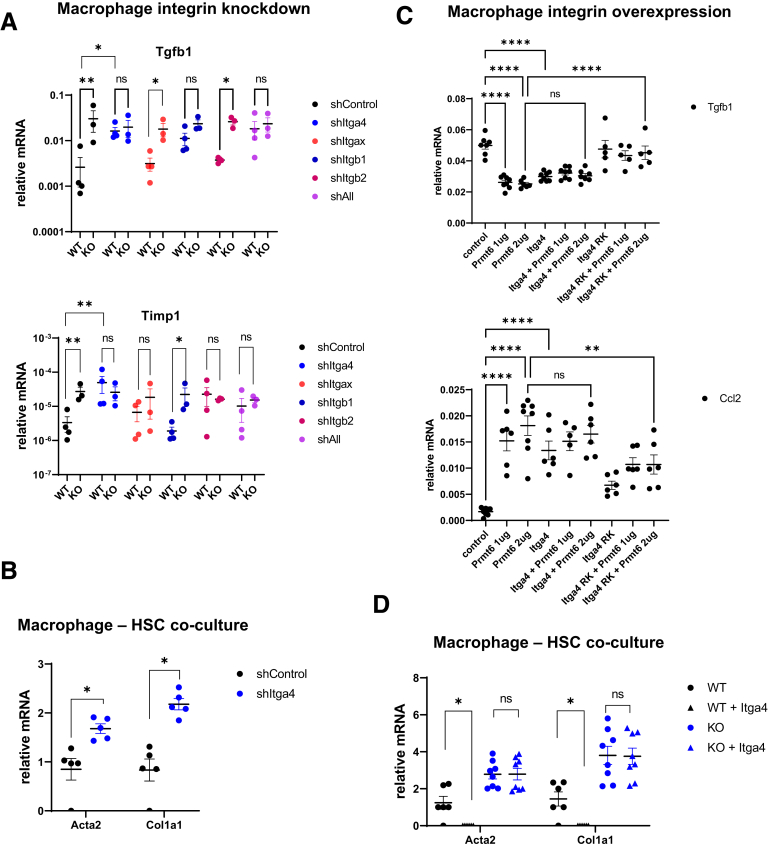
We confirmed that KO are responsible for profibrotic signaling using a co-culture experiment. WT and KO macrophages were co-cultured with HSCs in the presence of control immunoglobulin G or TGF-β1 antibodies. We found that Acta2 and Col1a1 expression in HSCs was greatly increased in the presence of KO macrophages in comparison with WT controls, anti-TGFβ1 antibodies reduced KO macrophage-mediated activation (Figure 7D).
To confirm that macrophages are the main cell type responsible for the Prmt6 role in fibrosis, we performed a macrophage transient replacement experiment (Figure 7E and F). We fed Prmt6 KO mice the WD and alcohol diet for 12 weeks and then treated with chlodronate liposomes to ablate liver macrophages. The clodronate-treated mice were subsequently injected with either Prmt6 knockout (KO->KO) or wild-type (WT->KO) bone marrow-derived macrophages (BMDM) 1 day after macrophage ablation. In addition, shortly after injection (within 1 hour) we collected liver biopsy samples, and mice continued alcohol diet for 4 more weeks (Figure 7E). We confirmed that biopsies from mice that received WT cells showed detectable Prmt6 expression, which was between 10% and 50% of Prmt6 levels in WT mice (Figure 7E). In addition, both groups of mice had similar liver mRNA expression of F4/80, suggesting that both groups received comparable number of macrophages (Figure 7E). Higher proinflammatory and fibrosis resolving (Mmp7) gene expression was observed after WT macrophages were injected. This was expected because WT macrophages are more proinflammatory than Prmt6 KO macrophages (Figure 6).
Four weeks after injection of WT macrophages into these KO mice, we observed lowered international normalized ratio/prothrombin time levels from those before the injection time point, whereas injection of KO macrophages resulted in no change in international normalized ratio/prothrombin time (Figure 7E). Moreover, we found that in mice that received KO macrophages, liver fibrosis measured by Sirius red staining was significantly increased during last 4 weeks of alcohol feeding (Figure 7F). In contrast, in the mice that received WT macrophages fibrosis was reduced, even though we did not detect original injected macrophages at this time point, suggesting that injection of WT cells compared with control KO cells was able to generate a fibrosis resolving rather than fibrosis promoting liver microenvironment. The effect of WT BMDM was similar to previously published studies showing that macrophage cell therapy improves liver synthetic function and reduces fibrosis area in an experimental fibrosis model.26 These data were confirmed by measuring changes in mRNA levels from time of the biopsy. We observed lower Tgfb1, Col1a1, and Timp1 in the WT macrophage treated group compared with the control KO macrophages treated group (Figure 7F).
PRMT6 Regulates Profibrotic Gene Expression Through Methylation of Integrin Alpha 4
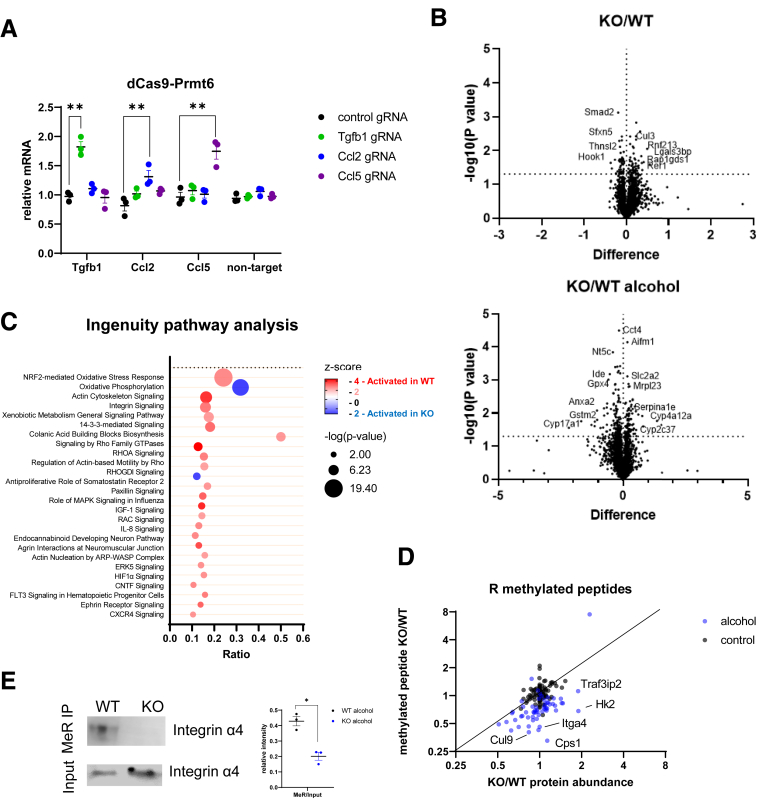
PRMT6 was initially reported as a transcriptional repressor.1 Thus, we hypothesized that PRMT6 can directly bind the promoter and suppress Tgfb1 gene expression. To test this hypothesis, we constructed a dCas9-PRMT6 fusion protein expression vector and used specific guide RNAs to guide PRMT6 to the promoters of its potential targets (Figure 8A). In contrast to our prediction, we found that PRMT6 specifically induces gene expression of Tgfb1, Ccl2, and Ccl5 in the presence of corresponding gRNA but not in the presence of other gRNAs. These data suggest that PRMT6 is a transcriptional activator and likely does not suppress Tgfb1 gene expression by direct promoter binding.
Figure 8.
PRMT6 loss results in global changes in liver proteome in the presence of alcohol. (A) Prmt6 knockout liver macrophages expressing fusion protein of catalytically inactive dCas9 and PRMT6 in the presence of indicated guide RNA. Relative mRNA expression of indicated genes or negative control (non-target, GFP). N = 3, ns, not significant. ∗P < .05, ∗∗P < .01. (B–D) Wild-type (WT) and Prmt6-/- (KO) mice were fed Western diet or WD with alcohol in the drinking water (alcohol) for 16 weeks. Livers from male mice (N = 3 mice per group) were analyzed by tandem mass tag mass spectrometry analysis. (B) Relative protein abundances in control and alcohol conditions. (C) Ingenuity pathway analysis of differentially regulated pathways in WT and KO mice fed alcohol. (D) Tandem mass tag mass spectrometry analysis of relative peptide and protein abundances in WT and Prmt6-/- (KO) mice fed Western diet (control) or WD with alcohol in the drinking water (alcohol) for 16 weeks. Fold change in peptide abundance was plotted versus fold change in corresponding protein abundance. (E) Methylated proteins were immunoprecipitated using anti-methyl arginine antibodies. Western blot analysis of integrin α4 protein abundance in precipitated fraction and in the input. Protein abundance ratios in N = 3 independent experiments. ∗P < .05.
To define the mechanism of PRMT6-dependent fibrosis development and to find PRMT6 methylation targets, we performed tandem mass tag (TMT) mass spectrometry analysis of whole liver protein extracts from male WT and Prmt6-/- mice fed WD and WD alcohol (Figure 8B, Supplementary Material). We observed that Prmt6 KO in alcohol-fed mice had a greater effect on the proteome that that in control-fed mice. Major pathways that were predicted to be regulated by PRMT6 in alcohol-fed mice were metabolic pathways, including xenobiotic metabolism, cell motility, and immune signaling related pathways, including integrin signaling (Figure 8C).
To identify methylation targets of PRMT6 we analyzed arginine methylated peptide abundance in WT and Prmt6-/- mouse livers. Figure 8D presents ratios of methylated peptide abundance between KO and WT mice vs ratio of total protein abundance. In the control condition, in the absence of alcohol, Prmt6 KO affects methylation of only a small number of proteins. In contrast, in alcohol-fed mice, Prmt6 KO results in a decrease in methylation for many proteins (Figure 8D, Supplementary Material). These data suggest that PRMT6 is important for protein methylation in alcohol-fed mice, whereas under control conditions PRMT6 is not an essential methyltransferase, possibly because PRMT6 loss is compensated by another methyltransferase such as PRMT1. Among proteins demethylated in Prmt6-/- mouse livers we identified integrin α4 (ITGA4). We found that integrin α4 is arginine methylated in WT livers, and its methylation is reduced in Prmt6-/- mouse livers (Figure 8E).
To test the role of integrin α4 in PRMT6-dependent profibrotic signaling in macrophages, we used short hairpin RNA mediated knockdown to reduce Itga4 and/or other integrin levels in macrophages isolated from WT and Prmt6-/- mice (Figure 9A). We found that Itga4 knockdown in WT increased Tgfb1 and Timp1 expression to the levels of Prmt6-/- cells (Figure 9A). In contrast, Itga4 knockdown in Prmt6-/- cells did not affect the expression of these genes. Knockdown of other integrins did not produce similar effects on both genes.
Figure 9.
PRMT6 regulates integrin α4 methylation. (A) Liver macrophages were isolated from wild-type (WT) and Prmt6-/- (KO) mice and treated with shRNA control vector, shRNA specific to Itga4, Itgax, Itgb1 or Itgb2, or a combination of all shRNA together (shAll). Relative mRNA expression. N = 3–4 mice per group. ns, not significant. ∗P < .05, ∗∗P < .01. (B) WT macrophages treated with shRNA control vector or shRNA specific to Itga4 were used in a co-culture experiment with WT HSCs. Relative gene expression in HSCs. ∗P < .05, N = 5. (C) Liver macrophages expressing PRMT6, ITGA4 WTe, or methylation deficient ITGA4 R464K where indicated. N = 5–8 independent experiments. ns, not significant. ∗∗P < .01, ∗∗∗∗P < .0001. (D) WT and KO macrophages expressing control vector or vector encoding Itga4 were used in a co-culture experiment with WT HSCs. Relative gene expression in HSCs. ∗P < .05, N = 6–8.
Next, we tested the role of integrin α4 in stellate cell activation. We found that Itga4 knockdown in WT macrophages increased their ability to induce HSC activation (Figure 9B). These data suggest that methylated integrin α4 (in WT cells) suppresses profibrotic gene expression and HSC activation, whereas demethylated integrin α4 (in KO cells) does not.
Mass spectrometry analysis identified arginine R464 as a site of integrin α4 methylation (Supplementary Material). R464 is located between the β-propeller and α-domains, regions involved in both β-integrin and ligand interactions. We mutated R464 to lysine and evaluated the role of WT and mutant integrin α4 in Tgfb1 and Ccl2 gene expression in macrophages in the presence of PRMT6 (Figure 9C). We found that overexpression of WT integrin α4 can suppress Tgfb1 gene expression and induce Ccl2 gene expression similar to PRMT6 overexpression. Overexpression of both proteins does not have additive effect, confirming that PRMT6 regulates gene expression through integrin α4 (Figure 9C). Overexpression of R464K mutant integrin did not suppress Tgfb1 gene expression and was less able to induce Ccl2 gene expression. In the presence of PRMT6 mutant integrin prevented PRMT6-induced suppression of Tgfb1 gene expression and induction of Ccl2 (Figure 9C).
Next, we overexpressed Itga4 in WT and KO macrophages and tested them in the HSC co-culture system. In contrast to Itga4 knockdown, Itga4 overexpression in WT macrophages reduced HSC activation (Figure 9D). In KO macrophages Itga4 overexpression had no effect on HSC activation ability, confirming that only the PRMT6-methylated form can prevent profibrotic signaling.
Monocyte Derived Macrophages Promote Liver Fibrosis in Prmt6-Deficient Mice
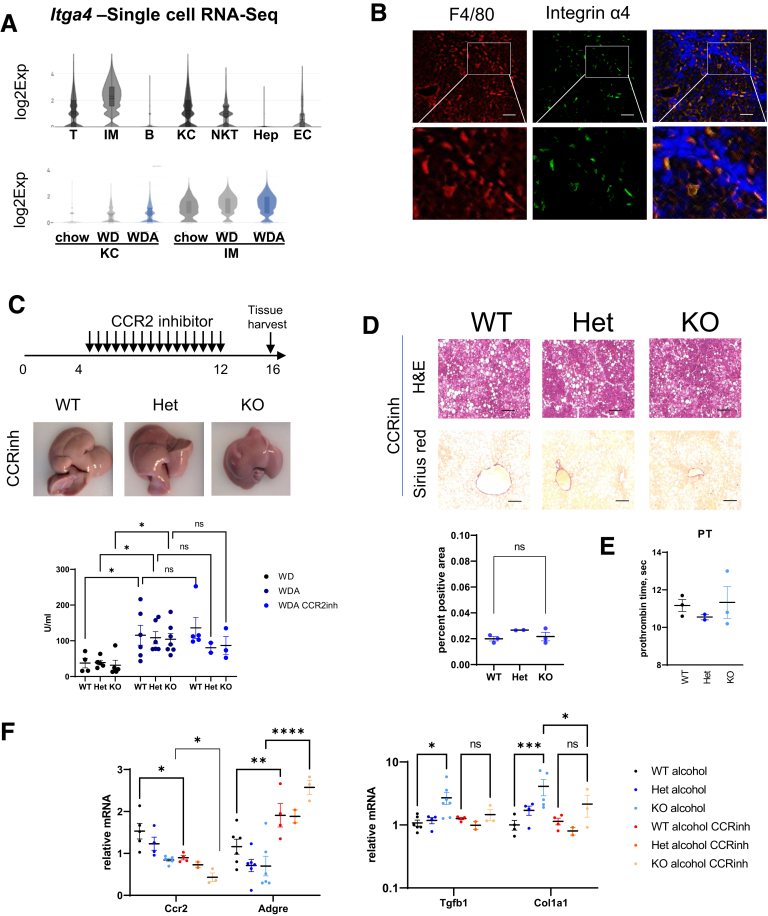
We used our previously reported small cytoplasmic RNA sequencing analysis to assess the expression of Itga4 in different cell types in the liver.23 Itga4 gene is predominantly expressed in infiltrating monocytes and macrophages (Figure 10A). Its expression is highest in infiltrating monocyte derived macrophages (Figure 10A). We confirmed that integrin α4 protein co-localizes with macrophage marker in fibrotic livers, specifically in the areas of fibrosis (Figure 10B).
Figure 10.
Infiltrating monocyte derived macrophages promote fibrosis in PRMT6-deficient mice. (A) Single cell RNA-sequencing analysis of liver cells from mice fed Western diet (WD) or WD with alcohol in the drinking water (WDA) for 16 weeks. (Top) Itga4 expression in combined cell clusters. (Bottom) Itga4 expression in myeloid cell clusters. ∗P < .05. (B) Immunofluorescence analysis of F4/80 and integrin α4 protein expression in fibrotic mouse livers. Bars, 100 μm. (C–F) Wild-type (WT), Prmt6+/- (Het), and Prmt6-/- (KO) mice were fed Western diet with alcohol in the drinking water (WDA, alcohol) for 16 weeks. Between weeks 4 and 12 of feeding mice received CCR2 inhibitor RS 504393 at 5 mg/kg twice a week. (C, top) Schematic of inhibitor treatment. (Middle) Gross liver appearance. (Bottom) Serum alanine aminotransferase levels in mice fed WD, WDA, and mice treated with the inhibitor. (D, top) H&E and Sirius red staining in treated mice. Bars, 200 μm. (Bottom) Sirius red positive area. ns, not significant. (E) Prothrombin time in treated mice. (F) Whole liver mRNA levels in mice fed WDA and WDA mice treated with the inhibitor. ns, not significant. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
Because integrin α4 is expressed predominantly on monocyte derived macrophages recruited to the liver during ALD progression, we tested whether blocking monocyte recruitment would rescue the phenotype of Prmt6-/- mice. To do so we injected mice fed WDA diet for 16 weeks with 2 weekly injections of 2 mg/kg of selective CCR2 inhibitor RS 504393 (inhibitory concentration of 50% = 330 nmol/L for CCL2 chemotaxis) starting week 4 of alcohol feeding (Figure 10C). We observed that inhibitor treatment abolished differences between genotypes in liver gross appearance (Figure 10C, compared with Figure 2A) but did not affect serum alanine aminotransferase levels (Figure 10C). Moreover, we found that that inhibitor treatment abolished differences in fibrosis development between genotypes (Figure 10D, compared with Figure 3B) and abolished differences in prothrombin time (Figure 10E, compared with Figure 3F).
Inhibitor treatment reduced Ccr2 levels, expressed predominantly in infiltrating monocytes and monocyte-derived macrophages, in WT and Prmt6-/- mice, whereas it increased levels of Adgre1 gene, expressed on Kupffer cells (KC) (Figure 10F). In addition, we found that treatment prevented the increase in Tgfb1 and Col1a1 levels in Prmt6-/- mice (Figure 10F).
Taken together, these data suggest that PRMT6 loss in CCR2-positive infiltrating monocyte-derived macrophages promotes the increase in profibrotic signaling in the liver.
Discussion
ALD is a complex disease that affects multiple pathways in the liver. Alcohol is known to have a big impact on epigenetic regulators that are involved in alcohol-induced steatosis, inflammation, and fibrosis development.27, 28, 29, 30 We have studied how alcohol affects 82 known epigenetic regulators. We found that about 15% of them were significantly up-regulated by alcohol, whereas only 4 were down-regulated. The most down-regulated enzyme was PRMT6, suggesting that PRMT6 loss might be involved in alcohol pathogenesis.
Using Prmt6 KO mice we found that PRMT6 is involved in multiple aspects of alcohol-induced liver disease; however, the most striking result was the effect of Prmt6 deficiency on alcohol-induced fibrosis development. We observed that PRMT6 is anti-fibrotic and functions as part of a mechanism that prevents fibrosis development, both in the absence of alcohol and especially during alcohol exposure.
PRMT6 regulates fibrosis development via integrin α4 methylation in macrophages. Integrin methylation suppresses profibrotic signaling. In contrast, proinflammatory signaling is induced. Thus, in Prmt6-deficient mice increased fibrosis is accompanied by decreased inflammation and decreased liver injury markers, suggesting that this mechanism of fibrosis development is independent of proinflammatory cytokine production and hepatocyte injury induced by alcohol. On the other hand, we found that Prmt6 down-regulation correlates with injury markers in WT mice fed alcohol (Figure 1). These data suggest that liver injury promotes Prmt6 down-regulation, which in turn promotes fibrosis development.
We observed that in the presence of alcohol, Prmt6 deficiency results in a greater increase in fibrosis compared with other disease models (high-fat diet feeding, thioacetamide). This effect could not be due to alcohol-induced reduction in Prmt6 levels, because then the effect of KO would be less in the presence of alcohol. This apparent contradiction is due to complex role of arginine methyltransferases in the liver. We observed a similar situation in our previous studies of another methyltransferase, PRMT1. We found that PRMT1 is inhibited by alcohol31; however, Prmt1 KO in alcohol-fed mice resulted in a dramatic increase in liver injury.27 Our data suggested that alcohol-induced PRMT1 inhibition is a protective mechanism, because it promoted its anti-inflammatory and pro-survival properties. Similarly, our data suggest that reduction in Prmt6 can prevent excessive inflammation induced by alcohol, which might be important in early ALD development. On the other hand, reduction in Prmt6 accelerates fibrosis development in later stages of ALD (Figure 11). Our data on significant diet-genotype interactions suggest that other alcohol-induced factors are necessary to accelerate profibrotic signaling in KO mice.
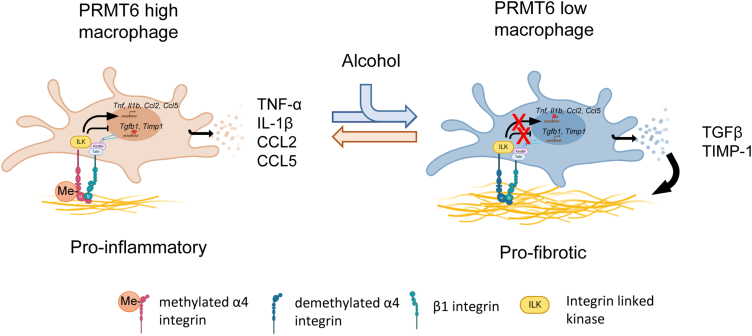
Figure 11.
Model of PRMT6-dependent fibrosis regulation. PRMT6 regulates fibrosis development via integrin α4 methylation in macrophages. Proinflammatory and profibrotic signaling in macrophages exist in equilibrium determined by activity of PRMT6. Methylated integrin α4β1 promotes downstream expression of proinflammatory cytokines and chemokines such as Tnf, Il1b, and Ccl2, Ccl5. On the other hand, methylated integrin α4β1 inhibits profibrotic gene expression of genes such as Tgfb1 and Timp1. Alcohol reduces PRMT6 levels, and in the presence of alcohol integrin α4β1 is demethylated. Demethylated integrin is less able to promote proinflammatory signaling or suppress profibrotic gene expression. As a result, loss of PRMT6 promotes fibrosis development in the liver.
PRMT6 was first identified as an epigenetic regulator that is involved in histone H3 methylation, depositing R2 methylation, a repressive mark.1 Later, PRMT6 was identified to induce an activating histone methylation mark on H2A that promotes gene expression. In our study we confirmed that PRMT6 can promote gene expression when recruited to gene promoters of genes involved in proinflammatory signaling. This mechanism likely contributes to PRMT6-mediated up-regulation of these genes in multiple cell types including macrophages, endothelial cells, and HSCs. In contrast, we did not find that PRMT6 can suppress gene expression when recruited to the promoters of its target genes, suggesting that suppressive histone methylation might not be playing a role in PRMT6 function in the liver.
Previous studies indicated that PRMT6 has very narrow substrate specificity and methylates very few targets outside the nucleus.4 When we analyzed PRMT6 non-histone targets, we found that protein methylation was most affected in alcohol-fed mice. In contrast, in control mice PRMT6 KO had less effect on protein methylation, suggesting that in the absence of alcohol, PRMT6 loss can be compensated by other methyltransferases such as PRMT1. These other methyltransferases are likely also inhibited by alcohol, perhaps to an even greater degree, making PRMT6 essential for protein methylation in alcohol-fed mice (Figure 8). These data agree with our previous work on alcohol-induced reduction in protein arginine methylation and PRMT1 inhibition.27,32,33
One of the top pathways that were dysregulated in PRMT6 KO mice was integrin signaling. Moreover, we found that PRMT6 methylates integrin α4, and this modification controls its downstream signaling and its role in proinflammatory and profibrotic gene expression. A PRMT6-dependent methylation site is located in the region responsible for both β-integrin interaction in its inactive form and for ligand interaction in its active ligand-bound form.19,34, 35, 36, 37 Arginine methylation does not affect protein charge, unlike many other posttranslational modifications such as phosphorylation or lysine acetylation. However, methylation interferes with the formation of hydrogen bonds, thus affecting protein conformation and/or protein-protein interactions. Our data suggest that integrin methylation might affect integrin-mediated intracellular signaling, ie, when integrin is activated. Thus, it is likely that methylation affects a conformation switch between active and inactive forms. However, further studies are necessary to study the role of integrin methylation in its conformation changes.
We identified integrin α4 as a regulator of profibrotic signaling in macrophages. Our data using integrin gene knockdowns suggest that other integrins might also be involved (Figure 9A), including integrins αx, β1, and β2. These data agree with previous studies on the role of these integrins in extracellular matrix expression and regulation of TGF-β.14,18,21 Integrins α4β1 and αxβ2 are predominantly expressed on monocyte derived infiltrating macrophages, which are known for their proinflammatory and profibrotic roles. Our results suggest that proinflammatory and profibrotic functions in these macrophages exist in equilibrium, and the precise setting of this equilibrium state is determined by the activity of PRMT6 (Figure 11). These data explain contradicting findings on the role of infiltrating macrophages in fibrosis progression and fibrosis resolution. Our data suggest that Prmt6 WT macrophages can promote fibrosis resolution, whereas Prmt6-deficient macrophages are profibrotic.
Interestingly we found that in the liver, PRMT6 is primarily expressed in non-parenchymal cells, macrophages, and endothelial cells. However, we detected PRMT6 protein expression in a subset of hepatocytes. Although we found that macrophage PRMT6 is involved in inflammation and fibrosis, hepatocyte PRMT6 could be involved in regulation of liver size (Figure 1F) and lipid accumulation (Figure 2E). These functions could be mediated by PRMT6-dependent metabolic pathways and insulin-like growth factor-1 signaling (Figure 8C). Further studies are necessary to identify targets of PRMT6 in hepatocytes that are involved in these processes.
Taken together, we found that PRMT6 regulates fibrosis development in an ALD model as well as other models such as high-fat diet, thioacetamide treatment, and aging. Targeting PRMT6 and its downstream targets such as integrins can be a promising strategy for reducing or preventing liver fibrosis.
Materials and Methods
Mice
Prmt6 KO mice were purchased from Jackson Laboratory (strain #028929, mixed C57BL/6J and C57BL/6N background) and backcrossed them for 5 generations with C57BL/6J mice. Prmt6 +/+, Prmt6 +/-, and Prmt6 -/- littermates were used for experiments at 6–8 weeks of age.
All mice were housed in a temperature-controlled, specific pathogen-free environment with 12-hour light-dark cycles. All animal handling procedures were approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center (Kansas City, KS).
Feedings
Lieber-DeCarli liquid diet feeding was performed as previously described.38
For the previously described WDA model,23 both male and female mice were fed ad libitum Western diet (Research Diets, Inc, New Brunswick, NJ; cat# D12079B), and alcohol was given ad libitum in water. Mice received progressively increasing amount of alcohol in water (1%, 3%, 10%, 15%, and 20% for 3 days each). After reaching 20%, mice were then alternated between 20% (4 days) and 10% (3 days) to achieve maximum alcohol intake.
Tandem Mass Tag Labeling and Mass Spectrometric Analysis
Whole liver extracts were lysed in 20 mmol/L HEPES (pH 8.0), 150 mmol/L NaCl, 0.5% NP-40, and 0.1% sodium dodecyl sulfate. Trypsin digestion and TMT labeling were performed following the manufacturer’s instructions using the TMT-sixplex Mass tagging kit (Thermo Fisher Scientific, Waltham, MA). Briefly, 100 μg of protein per condition was digested with 2.5 μg of trypsin (Promega, Madison, WI). After trypsin digestion, TMT label reagent was added. Equal amounts of labeled samples were combined, and sample was then loaded into a high pH reverse phase spin column previously conditioned following the manufacturer’s instructions (Thermo Fisher Scientific). The peptides were eluted in 9 fractions. Eluted samples were injected into the high-performance liquid chromatography coupled with the Orbitrap Fusion Lumos spectrometer (Thermo Fisher Scientific). For data analysis all MSMS scans were searched using Protein Discoverer v.2.4 running Sequest HT and a mouse database downloaded from the NCBI NR repository. Protein quantification was done using unique peptides only.
Cytokine Array and Enzyme-linked Immunosorbent Assay
Proteome Profiler Mouse Cytokine Array Kit (R&D Systems, Minneapolis, MN) detecting 111 mouse cytokines and mouse IL1β ELISA kit (R&D Systems) was used according to manufacturer’s instructions.
Small Cytoplasmic RNA Sequencing
Liver CD45 positive cells were used to generate barcoded cDNA libraries using a 10x Genomics Chromium platform with a total input of 10,000 cells per condition as previously described.39 Libraries were sequenced with an Illumina Novoseq (San Diego, CA) sequencer, and data were analyzed with the 10x Genomics Cell Ranger and Loupe Cell Browser software.
Human Samples
Liver tissue microarray containing de-identified human liver cirrhosis samples was purchased from US Biolab (Rockville, MD).
De-identified human specimens were obtained from the Liver Center Tissue Bank at the University of Kansas Medical Center. All studies using human tissue samples were approved by the Human Subjects Committee of the University of Kansas Medical Center.
Macrophage Replacement
Mice were injected with 150 μL of chlodronate liposomes (anionic Clophosome; Formumax, Sunnyvale, CA) 1 day before macrophage injections. BMDM were prepared as previously described40 and injected intravenously under isoflurane anesthesia.
Liver Biopsy
Mice were anesthetized with isoflurane, a small incision was made on the upper right side of the abdomen, and the liver was exposed. One lobe of liver was carefully lifted, and small piece of liver (3–5 mm) was removed. The tissue was immediately placed in zinc-formalin and RNA-later for further processing. The gap in the liver was closed with an absorbable hemostatic gelatin sponge (Vetspon #96002; Novartis, Cambridge, MA) to stop all eventual bleeding. The incision was closed with 5-0 absorbable surgical suture (Redilene Redisorb Fast Pro #VF493-M) and 7-mm wound-clips (Reflex 7 #203-1000). Mice were then injected with 1 mL saline (subcutaneously) and 1.0 mg/kg SR Buprenorphine (subcutaneously). Mice were placed on a heating pad and monitored until fully awake from anesthesia; thereafter mice were monitored daily for the next 7 days. The wound-clips were removed on day 8 after surgery.
Cell Isolation
Liver cells were isolated by a modification of the method described by Troutman et al.41 Mouse livers were digested by retrograde perfusion with liberase via the inferior vena cava. The dissociated cell mixture was placed into a 50-mL conical tube and centrifuged twice at 50g for 2 minutes to pellet hepatocytes. The NPC-containing cell supernatant was further used to isolate KC, liver sinusoidal endothelial cells (LSEC), and HSC. The cell suspension was pelleted by centrifugation (700g, 10 minutes, 4°C) and resuspended in phosphate-buffered saline (PBS) and OptiPrep (Sigma-Aldrich, St Louis, MO) to a final concentration of 17%. Afterwards, 5 mL of the indicated suspension was placed in a 15-mL polystyrene conical centrifuge tube (BD Biosciences) and overlaid with 5 mL of 9% Optiprep solution, followed by 2 mL PBS. After centrifugation at 1400g for 20 minutes at 4°C with decreased acceleration and without breaks, the various cell types were arranged according to their density. HSC were enriched in the upper cell layer, whereas KC and LSEC were separated as a second layer of higher density. Cell fractions were collected separately by pipetting. HSC purity over 99% was confirmed by retinoid based fluorescence-activated cell sorter sorting. The KC/LSEC fraction was pelleted, and KCs and ECs were isolated with F4/80+ and CD146+MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany), respectively, according to the manufacturer’s instructions. Cells were applied onto LS magnetic-activated cell sorting (MACS) columns (Miltenyi Biotec), which were placed within the magnetic field of a MACS separator and washed 3 times with MACS buffer (Miltenyi Biotec). Cells were eluted and then seeded into culture dishes. ECs were seeded on dishes coated with collagen-I.
Transwell Co-culture
For co-culture experiments, primary macrophages were placed in cell inserts of 24-well Transwell (Corning Incorporated, Acton, MA; 0.4-μm pore size) at a seeding density of 5 × 104/well. Cells were treated as indicated and incubated for 24 hours. Freshly isolated HSC were seeded in bottom well at a seeding density of 1 × 104/well. The cells were then cultured for additional 24 hours, and HSCs were harvested for RNA isolation.
Immunohistochemistry/Immunofluorescence
Liver tissue sections (5 μm thick) were prepared from formalin-fixed, paraffin-embedded samples. The quantification of Sirius red stained sections was performed in a blinded manner. Immunostaining on formalin-fixed sections was performed by deparaffinization and rehydration, followed by antigen retrieval by heating in a pressure cooker (121°C) for 5 minutes in 10 mmol/L sodium citrate, pH 6.0 as described previously.42 Peroxidase activity was blocked by incubation in 3% hydrogen peroxide for 10 minutes. Sections were rinsed 3 times in PBS/PBS-T (0.1% Tween-20) and incubated in Dako Protein Block (Dako, Carpinteria, CA) at room temperature for 1 hour. After removal of blocking solution, slides were placed into a humidified chamber and incubated overnight with a primary antibody, diluted 1:300 in Dako Protein Block at 4°C. Antigen was detected using the SignalStain Boost IHC detection reagent (catalogue #8114; Cell Signaling Technology, Beverly, MA), developed with diaminobenzidene (Dako), counterstained with hematoxylin (Sigma-Aldrich), and mounted.
Alternatively, slides were incubated with fluorescent Alexa Fluor-conjugated secondary antibodies diluted 1:300 in 0.1 μg/ml DAPI in Dako Protein Block for 1 hour at room temperature. Slides were then washed with PBS and mounted with FluorSave Reagent (Calbiochem, La Jolla, CA). Signal intensity for both immunohistochemistry and immunofluorescence was analyzed by Aperio ImageScope 12.1. N = 5 random field were quantified in a blinded way to obtain average signal intensity.
Reverse Transcription Polymerase Chain Reaction
RNA was extracted from livers using the RNeasy Mini Kit (Qiagen, Hilden, Germany). cDNA was generated using the RNA reverse transcription kit (Applied Biosystems, Waltham, MA; cat. no. 4368814). Quantitative real-time reverse transcription polymerase chain reaction was performed in a CFX96 Real time system (Bio-Rad, Hercules, CA) using specific sense and antisense primers combined with iQ SYBR Green Supermix (Bio-Rad) for 40 amplification cycles: 5 seconds at 95°C, 10 seconds at 57°C, 30 seconds at 72°C. mRNA concentrations were calculated relative to Actb.
| Actb: forward | ATGTCACGCACGATTTCCCT |
| Actb: reverse | CGGGACCTGACAGACTACCT |
| Col1a1: forward | TGGCCAAGAAGACATCCCTG |
| Col1a1: reverse | GGGTTTCCACGTCTCACCAT |
| Tgfb1: forward | TACGTCAGACATTCGGGAAGC |
| Tgfb1: reverse | TTTAATCTCTGCAAGCGCAGC |
| Timp1: forward | GTAAGGCCTGTAGCTGTGCC |
| Timp1: reverse | AGCCCTTATGACCAGGTCCG |
| mCcl2 forward | ACCTGGATCGGAACCAAATGAG |
| mCcl2: reverse | GCTGAAGACCTTAGGGCAGAT |
| mCcl5: forward | GGATTACTGAGTGGCATCCCC |
| mCcl5: reverse | TCTGACCCTGTATAGCTTCCCT |
| m-Tnf: forward | CTGAGACATAGGCACCGCC |
| m-Tnf: reverse | CAGAAAGCATGATCCGCGAC |
| m-IL1b:forward | ACGGGAAAGACACAGGTAGC |
| m-IL1b: reverse | AGCTTCAGGCAGGCAGTATC |
| mPrmt6: forward | CACCGGCTCGTTCAAGTAGA |
| mPrmt6: reverse | AAACCTCTGGTGCTGTCCAC |
Vectors
The pCMV6-PRMT6 vector was from OriGene (Rockville, MD). Plasmid expressing Itga4 ORF was from Sino Biological US (Chesterbrook, PA; cat# MG50049-ACG). The R464K point mutation was generated by Q5 mutagenesis kit from NEB.
The shRNA plasmids were from Sigma-Aldrich:
Cat# Target
| TRCN0000348624 | Itgb1 |
| TRCN0000066375 | Itgax |
| TRCN0000066043 | Itga4 |
| TRCN0000055311 | Itgb2 |
Antibodies
PRMT6 Santa Cruz #sc-271744
COL1A1 Cell Signaling #77397
F4/80 Cell Signaling #70076
Integrin α4 Cell Signaling #4749
Methyl-R Cell Signaling #8015
Deoxyuride-5´-Triphosphate Nick End Labeling Assay
Deoxyuride-5´-triphosphate nick end labeling assay was performed using the DeadEND Colorimetric TUNEL System (Promega) according to the manufacturer’s instructions.
Immunoprecipitation
RIPA (20 mmol/L HEPES pH 7.5, 150 mmol/L NaCl, 1% NP-40, 0.25% sodium deoxycholate, 10% glycerol) extracts were pre-cleared using 10 μg/mL of pre-immune rabbit or mouse immunoglobulin G (Millipore, Burlington, MA) for 2 hours at 4°C. One μL of protein G magnetic beads (Millipore; LSKMAGG10) per 10 μg of antibody was added, and the sample was incubated for another 1 hour at 4°C. After removal of beads, proteins were immunoprecipitated using 10 μg/mL of the IP antibody overnight at 4°C. Ten μL of protein G magnetic beads per 1 μg of antibody was added, and the sample incubated for 4 hours at 4°C. The beads were magnetically separated and washed 2 times in RIPA buffer. Beads were then resuspended in 10 μL of 4x sodium dodecyl sulfate loading buffer (0.25 mol/L Tris pH 6.8, 40% glycerol, 20% β-mercaptoethanol, 4% sodium dodecyl sulfate), and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Western Blots
Protein extracts (15 μg) were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, electrophoretically transferred to nitrocellulose membranes (Amersham Hybond ECL; GE Healthcare, Chicago, IL), and blocked in 3% bovine serum albumin/PBS at room temperature for 1 hour. Primary antibodies were incubated overnight at manufacturer’s recommended concentrations. Immunoblots were detected with the ECL Plus Western Blotting Detection System (Amersham Biosciences, Piscataway, NJ) or using near-infrared fluorescence with the ODYSSEY Fc, Dual-Mode Imaging system (Li-COR, Lincoln, NE). Expression levels were evaluated by quantification of relative density of each band normalized to that of the corresponding β-actin or GAPDH band density.
Statistics
Results are expressed as mean ± standard deviation. The Student t test, paired t test, Pearson correlation, or one-way analysis of variance with Bonferroni post hoc test were used for statistical analyses. P value <.05 was considered significant.
Acknowledgments
CRediT Authorship Contributions
Michael Schonfeld (Investigation: Lead; Methodology: Lead)
Maria T. Villar (Formal analysis: Equal; Investigation: Equal; Methodology: Equal)
Antonio Artigues (Formal analysis: Equal; Investigation: Equal; Methodology: Equal)
Steven A. Weinman (Formal analysis: Supporting; Funding acquisition: Supporting; Methodology: Supporting; Writing – review & editing: Equal)
Irina Tikhanovich, PhD (Formal analysis: Equal; Funding acquisition: Lead; Investigation: Supporting; Methodology: Supporting; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by grants AA027586 and AA012863 from the National Institute on Alcoholism and Alcohol Abuse and VA Merit Award I01BX004694.
Supplementary Material
References
- 1.Guccione E., Bassi C., Casadio F., Martinato F., Cesaroni M., Schuchlautz H., Luscher B., Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 2.Bedford M.T., Clarke S.G. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Andaloussi N., Valovka T., Toueille M., Steinacher R., Focke F., Gehrig P., Covic M., Hassa P.O., Schar P., Hubscher U., Hottiger M.O. Arginine methylation regulates DNA polymerase beta. Mol Cell. 2006;22:51–62. doi: 10.1016/j.molcel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Cheng D., Gao G., Di Lorenzo A., Jayne S., Hottiger M.O., Richard S., Bedford M.T. Genetic evidence for partial redundancy between the arginine methyltransferases CARM1 and PRMT6. J Biol Chem. 2020;295:17060–17070. doi: 10.1074/jbc.RA120.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong T.L., Ng K.Y., Tan K.V., Chan L.H., Zhou L., Che N., Hoo R.L.C., Lee T.K., Richard S., Lo C.M., Man K., Khong P.L., Ma S. CRAF methylation by PRMT6 regulates aerobic glycolysis-driven hepatocarcinogenesis via ERK-dependent PKM2 nuclear relocalization and activation. Hepatology. 2020;71:1279–1296. doi: 10.1002/hep.30923. [DOI] [PubMed] [Google Scholar]
- 6.Chan L.H., Zhou L., Ng K.Y., Wong T.L., Lee T.K., Sharma R., Loong J.H., Ching Y.P., Yuan Y.F., Xie D., Lo C.M., Man K., Artegiani B., Clevers H., Yan H.H., Leung S.Y., Richard S., Guan X.Y., Huen M.S.Y., Ma S. PRMT6 regulates RAS/RAF binding and MEK/ERK-mediated cancer stemness activities in hepatocellular carcinoma through CRAF methylation. Cell Reports. 2018;25:690–701 e8. doi: 10.1016/j.celrep.2018.09.053. [DOI] [PubMed] [Google Scholar]
- 7.Yan W.W., Liang Y.L., Zhang Q.X., Wang D., Lei M.Z., Qu J., He X.H., Lei Q.Y., Wang Y.P. Arginine methylation of SIRT7 couples glucose sensing with mitochondria biogenesis. EMBO Rep. 2018;19 doi: 10.15252/embr.201846377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scaramuzzino C., Casci I., Parodi S., Lievens P.M.J., Polanco M.J., Milioto C., Chivet M., Monaghan J., Mishra A., Badders N., Aggarwal T., Grunseich C., Sambataro F., Basso M., Fackelmayer F.O., Taylor J.P., Pandey U.B., Pennuto M. Protein arginine methyltransferase 6 enhances polyglutamine-expanded androgen receptor function and toxicity in spinal and bulbar muscular atrophy. Neuron. 2015;85:88–100. doi: 10.1016/j.neuron.2014.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y., Chung H.H., Woo A.R., Lin V.C. Protein arginine methyltransferase 6 enhances ligand-dependent and -independent activity of estrogen receptor alpha via distinct mechanisms. Biochim Biophys Acta. 2014;1843:2067–2078. doi: 10.1016/j.bbamcr.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 10.O’Doherty C., Roos I.M., Antiguedad A., Aransay A.M., Hillert J., Vandenbroeck K. ITGA4 polymorphisms and susceptibility to multiple sclerosis. J Neuroimmunol. 2007;189:151–157. doi: 10.1016/j.jneuroim.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Han Z., Ma Y., Cao G., Ma Z., Chen R., Cvijic M.E., Cheng D. Integrin alphaVbeta1 regulates procollagen I production through a non-canonical transforming growth factor beta signaling pathway in human hepatic stellate cells. Biochem J. 2021;478:1689–1703. doi: 10.1042/BCJ20200749. [DOI] [PubMed] [Google Scholar]
- 12.Bon H., Hales P., Lumb S., Holdsworth G., Johnson T., Qureshi O., Twomey B.M. Spontaneous extracellular matrix accumulation in a human in vitro model of renal fibrosis is mediated by alphaV integrins. Nephron. 2019;142:328–350. doi: 10.1159/000499506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cha B.H., Shin S.R., Leijten J., Li Y.C., Singh S., Liu J.C., Annabi N., Abdi R., Dokmeci M.R., Vrana N.E., Ghaemmaghami A.M., Khademhosseini A. Integrin-mediated interactions control macrophage polarization in 3D hydrogels. Adv Healthc Mater. 2017;6 doi: 10.1002/adhm.201700289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X., Wang H., Liao H.J., Hu W., Gewin L., Mernaugh G., Zhang S., Zhang Z.Y., Vega-Montoto L., Vanacore R.M., Fassler R., Zent R., Pozzi A. Integrin-mediated type II TGF-beta receptor tyrosine dephosphorylation controls SMAD-dependent profibrotic signaling. J Clin Invest. 2014;124:3295–3310. doi: 10.1172/JCI71668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson N.C., Sheppard D. Integrin-mediated regulation of TGFbeta in fibrosis. Biochim Biophys Acta. 2013;1832:891–896. doi: 10.1016/j.bbadis.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girgert R., Martin M., Kruegel J., Miosge N., Temme J., Eckes B., Muller G.A., Gross O. Integrin alpha2-deficient mice provide insights into specific functions of collagen receptors in the kidney. Fibrogenesis Tissue Repair. 2010;3:19. doi: 10.1186/1755-1536-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Annes J.P., Rifkin D.B., Munger J.S. The integrin alphaVbeta6 binds and activates latent TGFbeta3. FEBS Lett. 2002;511:65–68. doi: 10.1016/s0014-5793(01)03280-x. [DOI] [PubMed] [Google Scholar]
- 18.Sheppard D. Integrin-mediated activation of transforming growth factor-beta(1) in pulmonary fibrosis. Chest. 2001;120(Suppl):49S–53S. doi: 10.1378/chest.120.1_suppl.s49. [DOI] [PubMed] [Google Scholar]
- 19.Ripamonti M., Liaudet N., Azizi L., Bouvard D., Hytonen V.P., Wehrle-Haller B. Structural and functional analysis of LIM domain-dependent recruitment of paxillin to alphavbeta3 integrin-positive focal adhesions. Commun Biol. 2021;4:380. doi: 10.1038/s42003-021-01886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dikeman D.A., Rivera Rosado L.A., Horn T.A., Alves C.S., Konstantopoulos K., Yang J.T. Alpha4 beta1-integrin regulates directionally persistent cell migration in response to shear flow stimulation. Am J Physiol Cell Physiol. 2008;295:C151–C159. doi: 10.1152/ajpcell.00169.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong K.F., Liu A.M., Hong W., Xu Z., Luk J.M. Integrin alpha2beta1 inhibits MST1 kinase phosphorylation and activates Yes-associated protein oncogenic signaling in hepatocellular carcinoma. Oncotarget. 2016;7:77683–77695. doi: 10.18632/oncotarget.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galliher A.J., Schiemann W.P. Beta3 integrin and Src facilitate transforming growth factor-beta mediated induction of epithelial-mesenchymal transition in mammary epithelial cells. Breast Cancer Research. 2006;8:R42. doi: 10.1186/bcr1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schonfeld M., O’Neil M., Villar M.T., Artigues A., Averilla J., Gunewardena S., Weinman S.A., Tikhanovich I. A Western diet with alcohol in drinking water model recapitulates features of alcohol-associated liver disease in mice. Alcohol Clin Exp Res. 2021 doi: 10.1111/acer.14700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irie M., Suzuki N., Sohda T., Anan A., Iwata K., Takeyama Y., Watanabe H., Fischer P., Scherberich J.E., Sakisaka S. Hepatic expression of gamma-glutamyltranspeptidase in the human liver of patients with alcoholic liver disease. Hepatol Res. 2007;37:966–973. doi: 10.1111/j.1872-034X.2007.00151.x. [DOI] [PubMed] [Google Scholar]
- 25.Karlsson M., Zhang C., Mear L., Zhong W., Digre A., Katona B., Sjostedt E., Butler L., Odeberg J., Dusart P., Edfors F., Oksvold P., von Feilitzen K., Zwahlen M., Arif M., Altay O., Li X., Ozcan M., Mardinoglu A., Fagerberg L., Mulder J., Luo Y., Ponten F., Uhlen M., Lindskog C. A single-cell type transcriptomics map of human tissues. Sci Adv. 2021;7 doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas J.A., Pope C., Wojtacha D., Robson A.J., Gordon-Walker T.T., Hartland S., Ramachandran P., Van Deemter M., Hume D.A., Iredale J.P., Forbes S.J. Macrophage therapy for murine liver fibrosis recruits host effector cells improving fibrosis, regeneration, and function. Hepatology. 2011;53:2003–2015. doi: 10.1002/hep.24315. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J., Adams A., Weinman S.A., Tikhanovich I. Hepatocyte PRMT1 protects from alcohol induced liver injury by modulating oxidative stress responses. Scientific Reports. 2019;9:9111. doi: 10.1038/s41598-019-45585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Argemi J., Latasa M.U., Atkinson S.R., Blokhin I.O., Massey V., Gue J.P., Cabezas J., Lozano J.J., Van Booven D., Bell A., Cao S., Vernetti L.A., Arab J.P., Ventura-Cots M., Edmunds L.R., Fondevilla C., Starkel P., Dubuquoy L., Louvet A., Odena G., Gomez J.L., Aragon T., Altamirano J., Caballeria J., Jurczak M.J., Taylor D.L., Berasain C., Wahlestedt C., Monga S.P., Morgan M.Y., Sancho-Bru P., Mathurin P., Furuya S., Lackner C., Rusyn I., Shah V.H., Thursz M.R., Mann J., Avila M.A., Bataller R. Defective HNF4alpha-dependent gene expression as a driver of hepatocellular failure in alcoholic hepatitis. Nature Communications. 2019;10:3126. doi: 10.1038/s41467-019-11004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curtis B.J., Zahs A., Kovacs E.J. Epigenetic targets for reversing immune defects caused by alcohol exposure. Alcohol Research: Current Reviews. 2013;35:97–113. [PMC free article] [PubMed] [Google Scholar]
- 30.Gao B., Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141:1572–1585. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J., Adams A., Roberts B., O’Neil M., Vittal A., Schmitt T., Kumer S., Cox J., Li Z., Weinman S.A., Tikhanovich I. Protein arginine methyl transferase 1- and Jumonji C domain-containing protein 6-dependent arginine methylation regulate hepatocyte nuclear factor 4 alpha expression and hepatocyte proliferation in mice. Hepatology. 2018;67:1109–1126. doi: 10.1002/hep.29587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tikhanovich I., Zhao J., Long A., Roberts B.R., Weinman S.A. Alcohol increases hepatocyte proliferation by inhibition of histone arginine methylation. Hepatology. 2016;64(Suppl):262a–a. [Google Scholar]
- 33.Tikhanovich I., Kuravi S., Campbell R.V., Kharbanda K.K., Artigues A., Villar M.T., Weinman S.A. Regulation of FOXO3 by phosphorylation and methylation in hepatitis C virus infection and alcohol exposure. Hepatology. 2014;59:58–70. doi: 10.1002/hep.26618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J., Teh M., Kim J., Eva M.M., Cayrol R., Meade R., Nijnik A., Montagutelli X., Malo D., Jaubert J. A loss-of-function mutation in the integrin alpha L (Itgal) gene contributes to susceptibility to Salmonella enterica Serovar Typhimurium infection in collaborative cross strain CC042. Infect Immun. 2019;88 doi: 10.1128/IAI.00656-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L., Dong Y., Dong Y., Cheng J., Du J. Role of integrin-beta3 protein in macrophage polarization and regeneration of injured muscle. J Biol Chem. 2012;287:6177–6186. doi: 10.1074/jbc.M111.292649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margadant C., Sonnenberg A. Integrin-TGF-beta crosstalk in fibrosis, cancer and wound healing. EMBO Rep. 2010;11:97–105. doi: 10.1038/embor.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bittner M., Gosslar U., Luz A., Holzmann B. Sequence motifs in the integrin alpha 4 cytoplasmic tail required for regulation of in vivo expansion of murine lymphoma cells. J Immunol. 1998;161:5978–5986. [PubMed] [Google Scholar]
- 38.Guo F., Zheng K., Benede-Ubieto R., Cubero F.J., Nevzorova Y.A. The Lieber-DeCarli diet: a flagship model for experimental alcoholic liver disease. Alcohol Clin Exp Res. 2018;42:1828–1840. doi: 10.1111/acer.13840. [DOI] [PubMed] [Google Scholar]
- 39.Schonfeld M., Averilla J., Gunewardena S., Weinman S.A., Tikhanovich I. Alcohol-associated fibrosis in females is mediated by female-specific activation of lysine demethylases KDM5B and KDM5C. Hepatol Commun. 2022 doi: 10.1002/hep4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z., Zhao J., Tikhanovich I., Kuravi S., Helzberg J., Dorko K., Roberts B., Kumer S., Weinman S.A. Serine 574 phosphorylation alters transcriptional programming of FOXO3 by selectively enhancing apoptotic gene expression. Cell Death Differ. 2016;23:583–595. doi: 10.1038/cdd.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Troutman T.D., Bennett H., Sakai M., Seidman J.S., Heinz S., Glass C.K. Purification of mouse hepatic non-parenchymal cells or nuclei for use in ChIP-seq and other next-generation sequencing approaches. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J., Adams A., Roberts B., O’Neil M., Vittal A., Schmitt T., Kumer S., Cox J., Li Z., Weinman S.A., Tikhanovich I. PRMT1 and JMJD6 dependent arginine methylation regulate HNF4alpha expression and hepatocyte proliferation. Hepatology. 2018;67:1109–1126. doi: 10.1002/hep.29587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.