Abstract
Background & Aims
Metastasis is found in most advanced hepatocellular carcinoma (HCC) patients, and it drives tumor recurrence and systemic failure. There is no effective treatment owing to its complex biological features. Many of the molecular drivers of metastasis are crucial players in normal physiology but behave unconventionally during cancer progression. Targeting these molecular drivers for therapy and differentiating them from a physiological background require a detailed examination of the novel mechanisms involved in their activation during metastasis.
Methods
Publicly available transcriptomic data such as that of TCGA-LIHC and Gene Expression Omnibus were utilized to identify novel targets upregulated in advanced and metastatic HCC. Validation of candidates was assisted by immunohistochemistry performed on tissue microarrays derived from more than 100 HCC patients. Expression of protein tyrosine kinase 7 (PTK7) was studied under the treatment of transforming growth factor-β1 and knockdown of SRY-Box Transcription Factor 9 (SOX9) to delineate upstream regulation, while CRISPR-mediated knockout and lentiviral overexpression of PTK7 in HCC cells were performed to study their functional and signaling consequences. Manipulated HCC cells were injected into mice models either by orthotopic or tail-vein injection to observe for any in vivo pro-metastatic effects.
Results
PTK7 was discovered to be the kinase most significantly upregulated in advanced and metastatic HCC, at both transcriptomic and proteomic level. Bioinformatic analyses and functional assays performed in HCC cell lines revealed transforming growth factor-β signaling and SOX9 to be important activators of PTK7 expression. Functionally, enrichment of PTK7 expression could positively regulate metastatic potential of HCC cells in vitro and in lung metastasis models performed in immunodeficient mice. The up-regulation of PTK7 recruited the epithelial–mesenchymal transition components, zinc finger protein SNAI2 (SLUG) and zinc finger E-box-binding homeobox 1 (ZEB1).
Conclusions
Our study proposes PTK7 as a novel molecular driver in metastatic HCC, particularly in a transforming growth factor-β–activated microenvironment. The preferential expression of PTK7 resulted in a previously unobserved regulatory effect on the recruitment of epithelial–mesenchymal transition components, which established PTK7 as a potential determinant of specific epithelial–mesenchymal transition status. Therefore, our data support the continual development of PTK7-targeted agents as antimetastatic therapies.
Keywords: PTK7, Liver Cancer, Metastasis, SOX9, TGF-β
Abbreviations used in this paper: APC, allophycocyanin; cDNA, complementary DNA; ChIP, chromatin immunoprecipitation; CRISPR-Cas9, Clustered regularly interspaced short palindromic repeats-associated 9; EMT, epithelial–mesenchymal transition; EV, empty vector; FACS, fluorescence-activated cell sorting; gRNA, guide RNA; GSEA, gene set enrichment analysis; HCC, hepatocellular carcinoma; IHC, immunohistochemistry; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; PTK7, protein tyrosine kinase 7; SLUG, Zinc finger protein SNAI2; SOX9, SRY-Box Transcription Factor 9; TCGA-LIHC, The Cancer Genome Atlas Liver Hepatocellular Carcinoma; TF, transcription factor; TGF-β, transforming growth factor-β; TMA, tissue microarray; Wnt, Wingless-related integration site; ZEB1, Zinc Finger E-box-binding Homeobox 1
Graphical abstract
Summary.
Findings from this study show protein tyrosine kinase 7 as a novel regulator of hepatocellular carcinoma metastasis. Specifically, we found protein tyrosine kinase 7 to be critical in driving transforming growth factor-β signaling activation, induction of metastatic phenotypes, and guiding of specific epithelial–mesenchymal transition status.
The frequent occurrence of intrahepatic metastasis in hepatocellular carcinoma (HCC) predisposes patients toward recurrence and compromises treatments. Extrahepatic metastasis of HCC often leads to systemic failure and further deteriorates the survival prospects of patients. Up to one fifth of early HCC patients experience intrahepatic metastasis, and as many as 90% of advanced HCC patients have extrahepatic metastasis into the lungs, lymph nodes, bones, and adrenal glands.1,2
Metastasis is a complex biological phenomenon that has been studied extensively, and it is best described by the invasion-metastasis cascade model.3, 4, 5 It now is widely accepted that the biological events (from invasion, intravasation, to extravasation) detailed in the model oversimplify the cellular changes of metastasizing cells. The molecular mechanisms underlying tumor cell dissemination often resemble physiological events, such as tissue development and wound healing.6,7 Understanding the novel strategies and regulatory elements adopted by cancer in the alternative recruitment of these signaling components is a top priority in the development of better therapeutic agents that are specific for metastatic cancer. Kinases are signaling modulators that respond to developmental cues and often are mobilized in unconventional manners during pathogenesis. These enzymes have long served to be a valuable repertoire for drug targets. Particularly in the context of HCC, regimens involving tyrosine kinase inhibitors in combination with immune checkpoint inhibitors are some of the few clinically studied options as potential targeted therapies.8 It would be of great value to build on these previous achievements by furthering our knowledge of how specific kinases participate in metastatic HCC.
Protein tyrosine kinase 7 (PTK7) is a membrane pseudokinase that resembles a receptor tyrosine kinase but lacks conserved residues in its intracellular kinase domain.9, 10, 11 Since its discovery in the 1990s, PTK7 has been characterized primarily in Drosophila and mouse models, in which the expression of PTK7 is associated with the proper formation of the neural crest and inner ear.12,13 The phenotypic resemblance of PTK7 knockout defects with Wingless-related integration site (Wnt) signaling deregulation, particularly planar cell polarity, showed its role in the complex membrane regulatory system of Wnt signaling.14, 15, 16, 17 However, the exact nature and properties of PTK7 interaction with Wnt signaling components are understudied. Despite the modest progress in understanding PTK7 in Wnt signaling, its involvement in cancer has quickly emerged over the past decades. Multiple studies have reported the up-regulation of PTK7 in lung,18 breast,19,20 esophageal,21,22 and colorectal cancers,23,24 and the evidence shows that PTK7 affects cell proliferation and cell mobility. However, the precise functional and mechanistic role of PTK7 in the complex molecular map and hallmark cancer features it primarily interacts with remain largely unexplored. Reports on the properties of PTK7 in HCC progression are comparably scant, which furthers our interest in its behavior in this model.
Our study established PTK7 as a novel regulator of HCC metastasis. The results indicate its role in transforming growth factor-β (TGF-β) signaling activation, induction of metastatic phenotypes, and driving of epithelial–mesenchymal transition (EMT) elements. The recent breakthroughs in the use of PTK7 as a receptor for therapeutic agents in other cancer types further support the relevance of PTK7 in the fight against HCC metastasis.
Results
PTK7 Is the Most Significantly Deregulated Kinase in Metastatic and Advanced HCC and Correlates Positively With Adverse Clinical Outcomes
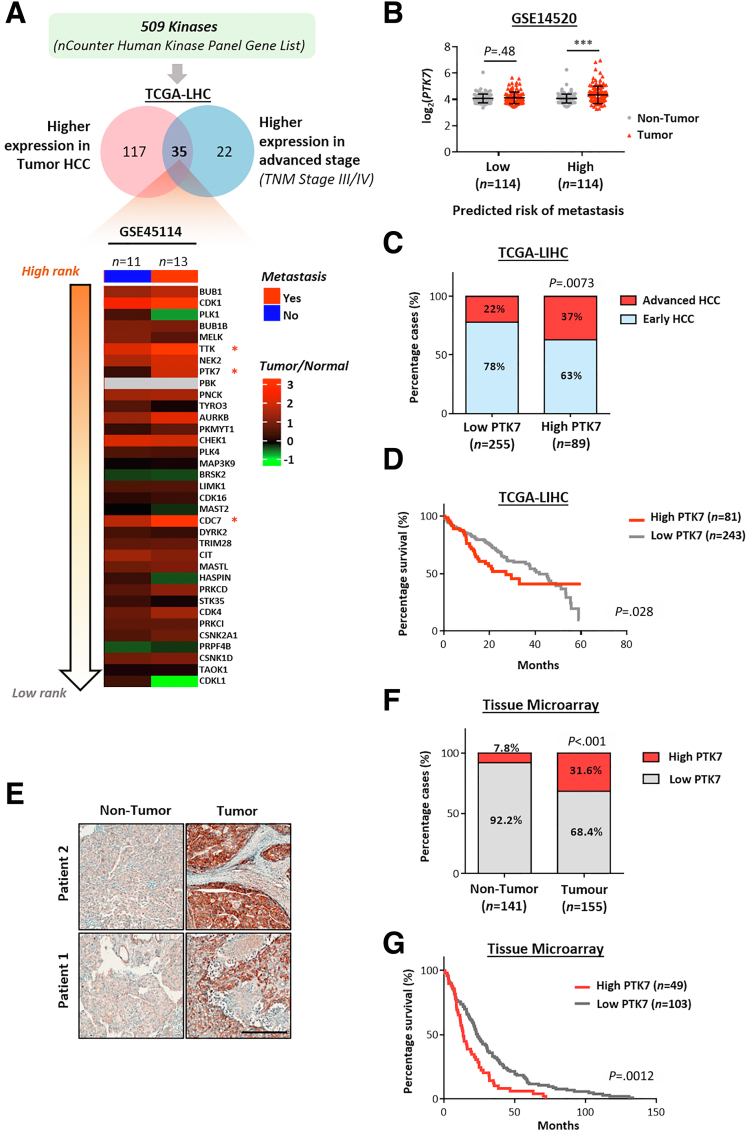
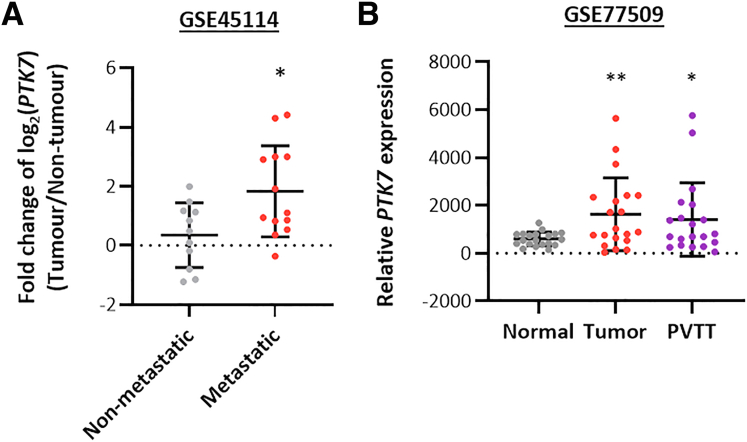
The nCounter Human Kinase Panel Gene List (nanoString) includes more than 500 kinases that commonly are deregulated in cancers. To understand the clinical relevance of these kinases in the setting of HCC metastasis, our initial bioinformatic analysis focused on their potential deregulation in advanced and metastatic HCC (Figure 1A, upper panel). Among the 509 kinases studied in The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) transcriptomic data, 152 showed significant up-regulation in HCC tumors compared with normal liver tissue. Only 35 kinases were significantly enriched in advanced HCC cases compared with early HCC. Notably, when these candidates were examined further for possible enrichment in metastatic HCC cases using transcriptomic data obtained from GSE45114, only 3 kinases (PTK7, dual specificity protein kinase (TTK), and cell division cycle 7-related protein kinase (CDC7)) showed significant up-regulation (Figures 1A, lower panel, and 2A). The up-regulation of PTK7 within the metastatic subset of HCC was further shown by the selective enrichment of PTK7 in tumor HCC predicted with high metastasis risk (Figure 1B)25 and the up-regulation of this protein in tumor and metastatic outgrowth of patients diagnosed with portal vein tumor thrombus, which is a representative event of intrahepatic metastasis (Figure 2B). Enrichment of PTK7 also was predictive of advanced-stage HCC (Figure 1C) and adverse prognosis of patients (median survival time, 27.2 months in the high-PTK7 group vs 41.8 months in the low-PTK7 group) (Figure 1D). To further investigate the clinical presentation of PTK7 at the proteomic level, immunohistochemistry (IHC) was performed on tissue microarray (TMA). High PTK7 staining was defined by the presence of discernible membrane expression or intense cytoplasmic expression (Figure 1E). Notably, although a subset of nontumor samples (∼8%) showed distinct cytoplasmic aggregation of PTK7 and was considered high-PTK7, no distinguishable membrane localization of the protein was observed in any of these samples. Conversely, more than 30% of tumor cases showed high levels of cytoplasmic PTK7 with frequent membrane localization of the protein (Figure 1F). Consistent with the transcriptomic data, the up-regulation of PTK7 in tumors was predictive of a poorer patient prognosis (median survival time, 15.7 months in the high-PTK7 group vs 26.1 months in the low-PTK7 group) (Figure 1G). Taken together, our bioinformatic analysis identified PTK7 as a novel protein that preferentially was enriched in metastatic and advanced HCC with clinical importance to disease outcome.
Figure 1.
PTK7 was the most significantly deregulated kinase in metastatic HCC and was enriched in advanced HCC tumors. (A) A total of 509 commonly deregulated kinases in cancer were extracted from the nCounter Human Kinase Panel Gene List, and their expression pattern was examined in the TCGA-LIHC patient cohort. Thirty-five candidate kinases were overexpressed in tumor and advanced HCC, and ranked (from top to bottom) according to their extent of up-regulation and evaluated against the GSE45114 data set, which described transcriptomic profiles of nontumor and tumor tissues harvested from metastatic HCC samples and their nonmetastatic counterparts. The heatmap was constructed to represent the fold-change in the expression of each candidate kinase in HCC tumors compared with nontumor samples under a metastatic or nonmetastatic background. Only 3 kinases (TTK, PTK7, and CDC7, marked with an asterisk) were significantly overexpressed in metastatic HCC, and PTK7 was the most significantly deregulated. (B) Statistical evaluation of the fold change in PTK7 expression (tumor vs nontumor) for 228 pairs of HCC samples evenly grouped by their predicted risk of metastasis as described in the GSE14520 cohort by paired t test between each tumor/nontumor comparison. The predicted risk of metastasis was defined by the integral signature score of 161 metastatic genes that robustly correlated with recurrence and adverse clinical outcome.25 (C) The distribution of early and advanced HCC cases in the high-PTK7 and low-PTK7 samples of the TCGA-LIHC data set compared using the chi-squared test. (D) The overall survival distribution of patients with high or low PTK7 expression in the TCGA-LIHC data set compared using the Gehan–Breslow–Wilcoxon test. (E) Representative images of high-PTK7 staining from IHC on TMA containing tumor and nontumor samples of HCC patients. Scale bar: 100 μm. (F) The distribution of high- and low-PTK7 cases in tumor and nontumor samples of TMA compared using the chi-squared test. (G) The overall survival distribution of TMA patients with high or low PTK7 expression compared using the log-rank test. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
Figure 2.
Up-regulation of PTK7 in metastatic HCC. (A) The evaluation of PTK7 expression in metastatic vs nonmetastatic HCC samples in GSE45114, by Student t test. (B) The expression of PTK7 in primary tumor and portal vein tumor thrombus (PVTT) tissue of HCC patients was evaluated against that of paired normal tissue using the GSE77509 data set, by paired t test. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
The Expression Level and Transcriptomic Activity of SRY-Box Transcription Factor 9 (SOX9) Correlate Clinically and Functionally With PTK7
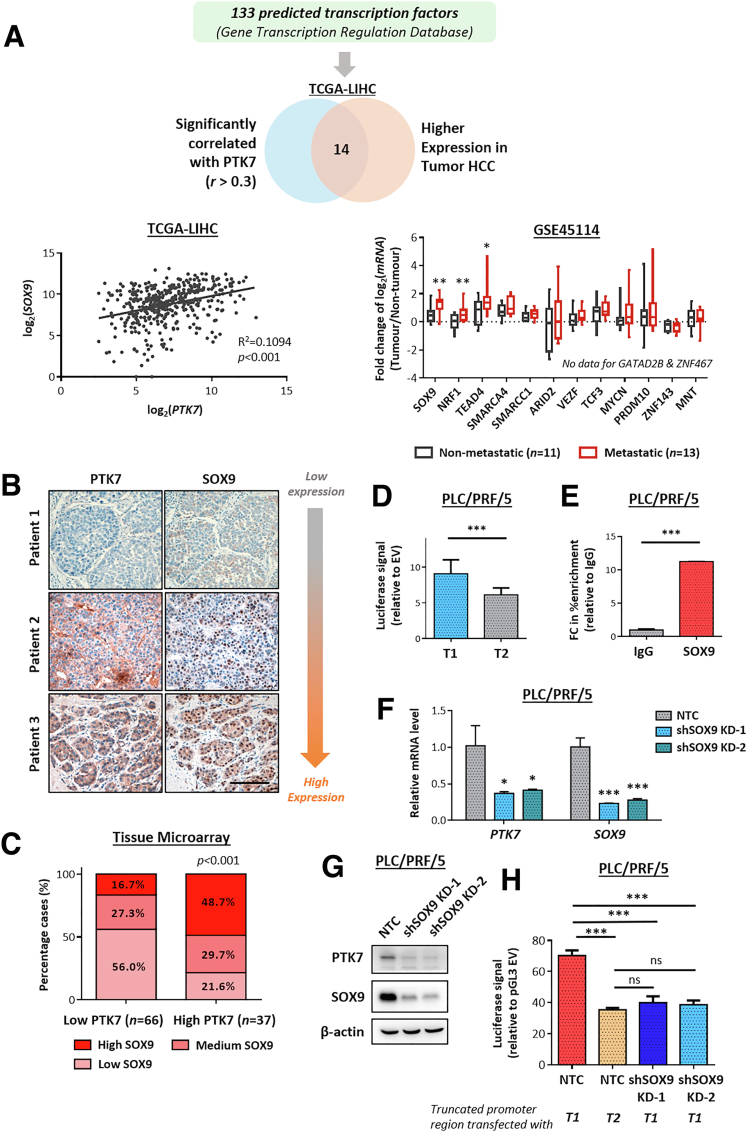
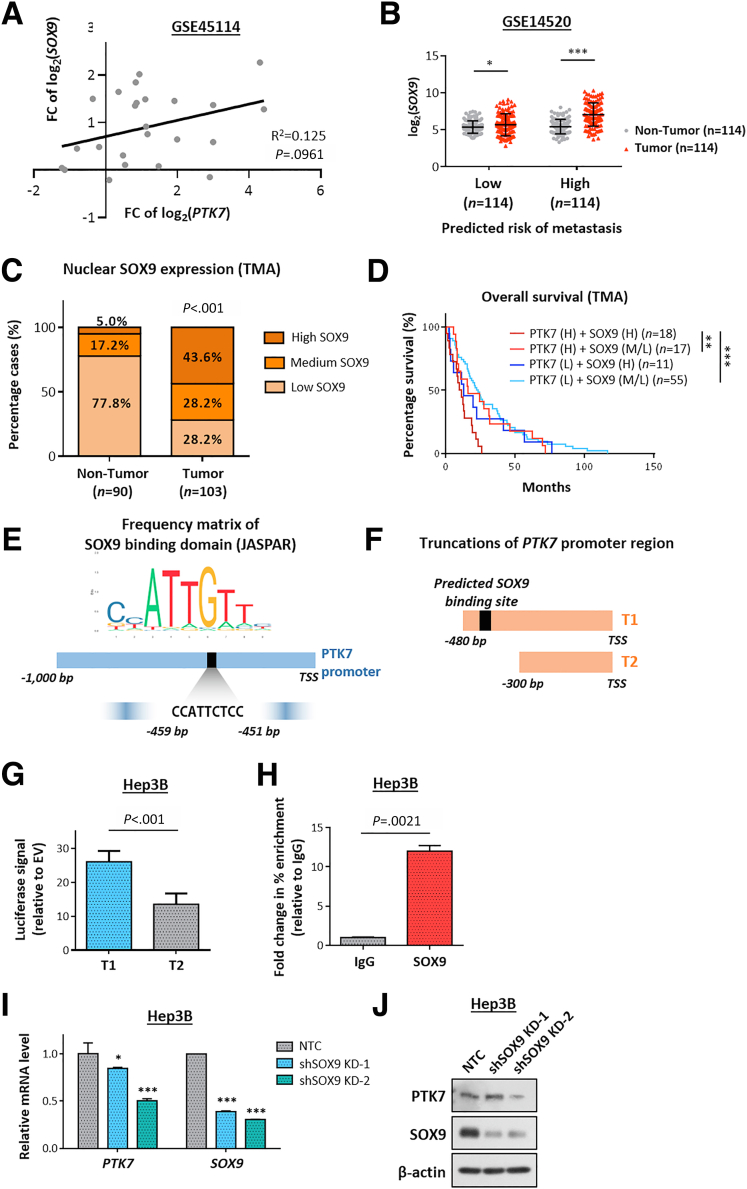
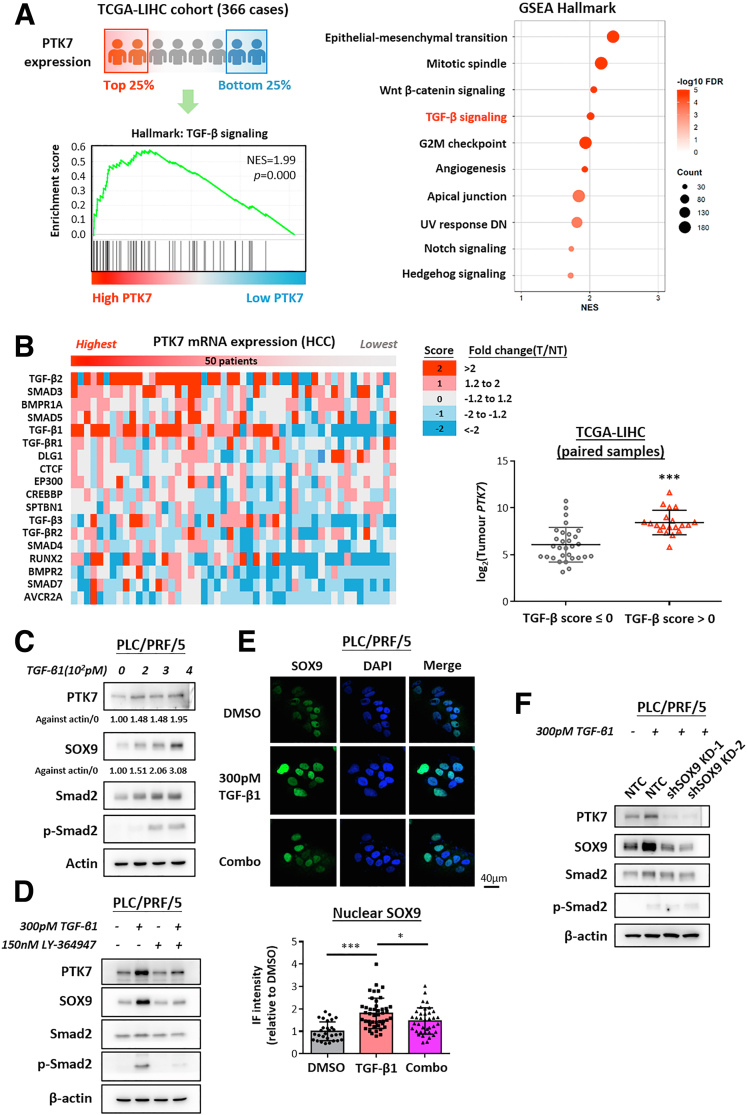
As shown in our initial bioinformatic analyses, transcriptional up-regulation of PTK7 occurred preferentially in advanced and metastatic HCC tumors. Therefore, it would be of great importance to understand the driving factors that initiate this stage-dependent enrichment. The focus was first placed on the potential transcription factors (TFs) binding to the PTK7 promoter and positively regulating promoter activity. To achieve this end, bioinformatic analysis was performed on the selected 1000–base pair region upstream of PTK7 using the publicly available database Gene Transcription Regulation Database to identify all reported and predicted binding TFs (Figure 3A). To limit the pool, only TFs that correlated with PTK7 expression (with r > 0.3) and were significantly up-regulated in HCC tumors were considered (Figure 3A, upper panel). Among the final candidates, SOX9 was the most significantly enriched TF in metastatic HCC (Figure 3A, lower right panel), and it highly correlated with PTK7 (Figures 3A, lower left panel, and 4A). The specific association of SOX9 expression with a more aggressive metastasis signature also was shown in the analysis of GSE14520, which showed a prominent tumor vs nontumor up-regulation of SOX9 for HCC cases with a high metastasis risk background, while the up-regulation for low metastasis risk samples was marginal (Figure 4B). At the proteomic level, a generally positive correlation between PTK7 and SOX9 levels was observed across HCC cell lines (Figure 5A). IHC of TMA showed that SOX9 was overexpressed significantly in HCC patient tumor tissue (Figure 4C), with a greater proportion of high-PTK7 patients having tumor SOX9 predominantly localized to the nucleus (Figure 3B and C). Clinically, the collective up-regulation of SOX9 and PTK7 in HCC patients reduced their survival prospects (Figure 4D). The JASPAR database was consulted for any putative SOX9-binding elements on the PTK7 promoter, and a single binding site was predicted (Figure 4E). Truncations of the PTK7 promoter were devised (Figure 4F) to study the importance of this binding site in regulating promoter activity. The exclusion of the region containing the predicted SOX9 binding site drastically reduced the ability of the promoter element to drive downstream luciferase expression in the reporter assay (Figure 3D). The physical binding of SOX9 to the predicted site also was validated using the chromatin immunoprecipitation (ChIP) assay (Figure 3E). The stable knockdown of SOX9 resulted in the down-regulation of endogenous PTK7 at the transcriptomic and proteomic levels, and SOX9 suppression inhibited the activity of the truncated PTK7 promoter harboring the SOX9 binding domain (Figure 3F–H). Similar findings were observed in Hep3B cells (Figure 4G–J). The clinical and in vitro data implicated SOX9 as an important upstream regulator of PTK7, and both factors are strongly implicated in HCC metastasis.
Figure 3.
The expression level and transcriptomic activity of SOX9 are clinically and functionally correlated with PTK7. (A) A total of 133 TFs potentially binding to the promoter region of PTK7, proximal to the transcriptional start site (between -1000 and +1), were compiled from the online Gene Transcription Regulation Database. Fourteen candidate TFs that highly correlated with PTK7 (r > 0.3, determined by Pearson correlation) and were overexpressed in tumor HCC at the messenger RNA (mRNA) level (analyzed using TCGA-LIHC data as a normalized count) were examined further for their specific enrichment in metastatic HCC using GSE45114 data. Only SOX9, nuclear respiratory factor 1 (NRF1), and TEA domain transcription factor (TEAD4) satisfied all of the tested criteria. The correlation of SOX9 and PTK7 expression at the mRNA level is shown for reference. (B) IHC was performed on TMA to investigate the extent of SOX9 nuclear localization in tumor HCC samples in which cases were designated as SOX9 high (>75% nuclei), medium (25%–75% nuclei), or low (<25% nuclei), depending on the amount of tumor cells showing apparent SOX9 nuclear staining. Graded expression of SOX9 was compared with PTK7 in the same TMA. Representative images of IHC are shown. Scale bar: 100 μm. (C) The distribution of cases with various nuclear SOX9 expression in the high-PTK7 and low-PTK7 samples of TMA was compared using the chi-squared test. (D) Two truncated versions of the PTK7 promoter region, one containing the predicted SOX9 binding site (T1) and one without (T2), were expressed in HCC cell lines via transient transfection. The ability of the promoter elements to drive protein expression was quantified using the Dual-Glo luciferase reporter assay and compared using the Student t test. (E) A ChIP assay was performed in HCC cell lines to examine the physical binding of SOX9 to the PTK7 promoter region. Targeted enrichment of the predicted binding sequence was quantified using reverse-transcription quantitative PCR and compared with mouse IgG immunoprecipitation control (IgG). (F) The messenger RNA (mRNA) levels of PTK7 and SOX9 were quantified and validated in HCC cells after successful knockdown of SOX9 by short hairpin RNA (shRNA). (G) Western blot validation of PTK7 suppression after SOX9 depletion by shRNA knockdown. (H) Two truncated PTK7 promoter elements, one harboring the predicted SOX9-binding motif (T1) and the other lacking the same motif (T2), were transfected into SOX9-knockdown (KD) cells and nontarget control (NTC) cells. The resultant luciferase signal induced by the 2 elements in the different cell lines was compared using the Student t test. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
Figure 4.
SOX9 as an important upstream regulator of PTK7 expression. (A) The correlation of the fold change (FC) in PTK7 and SOX9 expression of HCC tumor compared with nontumor tissue, analyzed using data from GSE45114. (B) Same statistical evaluation of GSE14520 data set as described in Figure 1B for the fold change in SOX9 expression of paired tumor/nontumor HCC samples with different predicted risk of metastasis, by paired t test. (C) The distribution of high-, medium-, and low-SOX9 cases in tumor or nontumor samples of TMA, compared by chi-squared test. (D) The overall survival distribution of TMA patients grouped according to their tumor PTK7 and SOX9 level, compared by log-rank test. (E) The frequency matrix of the DNA-binding motif of SOX9 as reported in JASPAR database. A single SOX9 binding site was predicted on the PTK7 promoter between the -450 and -460 region upstream of transcription start site. (F) Two truncations of the proximal PTK7 promoter region were synthesized and integrated into the pGL3-basic vector backbone for luciferase reporter assay. T1 truncation contained the predicted SOX9 binding site while T2 truncation did not. (G) The ability in driving protein expression of the 2 truncated promoter elements of PTK7 was quantified by the Dual-Glo luciferase reporter assay after transient transfection and compared by Student t test. (H) The physical binding of SOX9 to the PTK7 promoter region was examined by ChIP assay. Targeted enrichment of the predicted binding sequence was quantified by quantitative PCR and compared with mouse IgG immunoprecipitation control (IgG). (I) The messenger RNA (mRNA) level of PTK7 and SOX9 was quantified and validated in Hep3B cells after successful knockdown of SOX9 by short hairpin RNA (shRNA). (J) Western blot validation of the suppression of PTK7 level after SOX9 depletion by shRNA knockdown in Hep3B. H, high; L, low; M, medium; NTC, nontarget control. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
Figure 5.
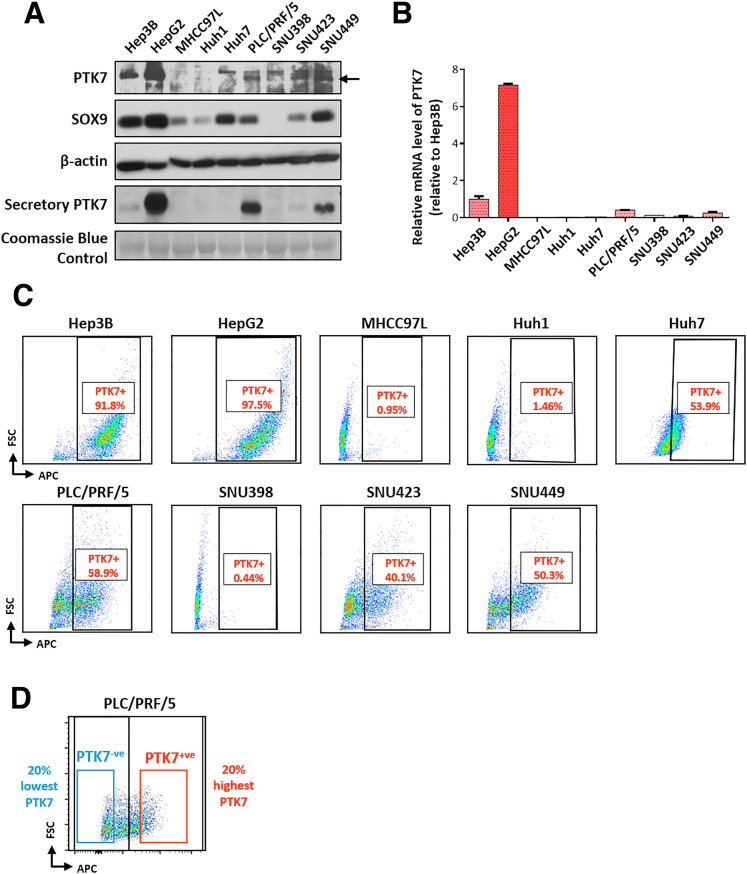
Expression of PTK7 and SOX9 in HCC cell lines. (A) A comparison of the expression of endogenous PTK7, SOX9, and secretory PTK7 in a panel of HCC cell lines. Coomassie Blue staining was used as a loading control for total secretory protein. (B) Quantitative PCR analysis on PTK7 messenger RNA (mRNA) level relative to that of the housekeeping gene β-actin across the HCC cell line panel. (C) Flow cytometry performed to evaluate the membrane expression of PTK7 across the HCC cell line panel. A mouse APC-conjugated IgG2a isotype control was used as the negative control. (D) A heterogenous population of PTK7-expressing and nonexpressing cells in PLC/PRF/5 as determined by flow cytometry. The 20% subsets with the highest- or lowest-membrane PTK7 expression were sorted out for functional and molecular assays. FSC, forward scatter.
The Enrichment of PTK7 in HCC Is Responsive to TGF-β Signaling Activation and May Be Mediated by Stimulated SOX9 Activity
The molecular landscape of the metastatic niche has important implications for the status of cancer cells and their readiness to migrate and invade into surrounding tissues. To understand the relationship of PTK7 with metastatic signatures, TCGA-LIHC cohort data were segregated to classify patients into high-PTK7 and low-PTK7 subpopulations (Figure 6A). Molecular signatures enriched under high-PTK7 conditions were identified using gene set enrichment analysis (GSEA) (Figure 6A, right panel). TGF-β and Wnt signaling were among the top deregulated pathways discovered in this bioinformatic analysis.
Figure 6.
The enrichment of PTK7 in HCC is responsive to the activation of TGF-β signaling and potentially mediated by stimulated SOX9 activity. (A) Patients in the TCGA-LIHC cohort were ranked according to the transcriptomic level of PTK7 in their tumors. Publicly available RNA sequencing data of the 25% highest-ranking patients were subjected to GSEA to identify molecular signatures and pathways deregulated under enriched PTK7 conditions compared with the 25% lowest-ranking patients. TGF-β signaling, which is highly relevant to metastasis, was identified as one of the top pathways significantly enriched under high-PTK7 conditions. The top 10 deregulated pathways under the high-PTK7 condition are arranged in a bubble diagram for reference. (B) Heatmap showing the correlation of the tumor PTK7 level with 18 TGF-β signaling signature genes in all of the paired tumor/nontumor (T/NT) cases of the TCGA-LIHC cohort. The T/NT fold change of all signature genes was calculated, and their activation status for each HCC case was based on a numeric scoring system, where a score of 2 means the most activated and a score of -2 is the most inactivated. A TGF-β score was assigned to each patient by integrating the scores of all 18 signature genes. An overall zero or negative TGF-β score was representative of a TGF-β–suppressive environment, and a positive overall score was representative of a TGF-β–activated environment. PTK7 expression was compared between HCC samples with a positive or negative TGF-β score using the Student t test. (C) Western blot validation of dose-dependent up-regulation of PTK7 and SOX9 expression in PLC/PRF/5 cells after a single treatment of recombinant TGF-β1 for 24 hours at various doses. Quantification of PTK7 and SOX9 expression was performed in ImageJ (National Institutes of Health, Bethesda, MD). (D) Western blot validation of PTK7 and SOX9 expression in PLC/PRF/5 cells after TGF-β1 single treatment or in combination with a TGF-βR1–specific inhibitor, LY-364947, for 24 hours. (E) Representative images of immunocytochemistry targeting SOX9 in PLC/PRF/5 cells after TGF-β1 single treatment or in combination with LY-364947 (Combo) for 24 hours. 4′,6-Diamidino-2-phenylindole (DAPI) staining was used to locate the nuclei of stained cells. The average nuclear localization of SOX9 was quantified in ImageJ and compared between treatment groups using the Student t test. (F) Western blot validation of PTK7 levels in SOX9-knockdown PLC/PRF/5 cells after TGF-β1 single treatment. DMSO, dimethyl sulfoxide; DN, down-regulated; FDR, false discovery rate; IF, immunofluorescence; mRNA, messenger RNA; NTC, nontarget control. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
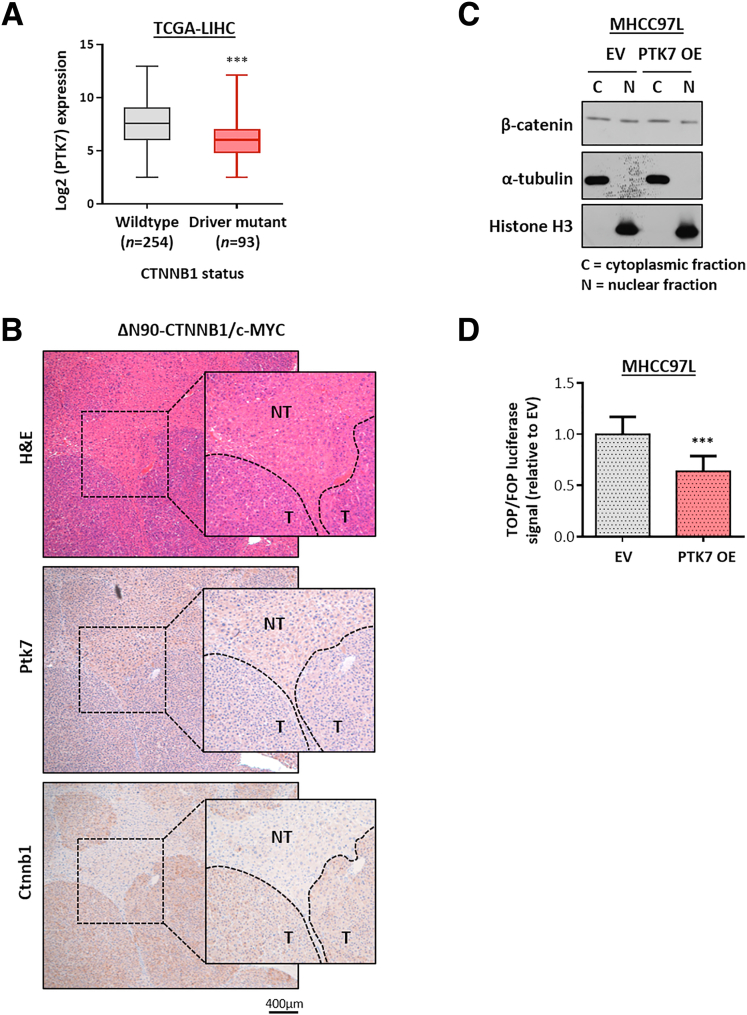
Because PTK7 has been widely implicated in the transduction of Wnt signaling,14, 15, 16, 17,26 we first focused on their potential association in the setting of HCC. Preliminary bioinformatic analysis based on HCC samples from the TCGA-LIHC cohort showed that HCC tumors driven by oncogenic CTNNB1 mutations co-expressed significantly lower levels of PTK7 than their counterparts harboring wild-type CTNNB1 (Figure 7A). Further examination of HCC derived from a mouse model driven by hydrodynamic tail-vein injection of constitutively active β-catenin (ΔN90-CTNNB1) and c-myelocytomatosis oncogene (c-MYC) overexpression plasmid also showed no noticeable accumulation of PTK7 in tumor regions compared with neighboring nontumor tissues (Figure 7B). The subsequent overexpression of PTK7 in the HCC cell line did not lead to increased nuclear localization of β-catenin (Figure 7C) or stimulation of its activity when activated by Wnt3a (Figure 7D). Taken together, we found no evidence suggesting that activated Wnt signaling is responsible for PTK7 accumulation and its prometastatic functionality in HCC.
Figure 7.
PTK7 enrichment in HCC tumor is not driven by and does not result in the activation of Wnt signaling. (A) A bioinformatic analysis was performed on TCGA-LIHC cohort to compare the transcriptomic level of PTK7 under HCC driven by different CTNNB1 status. Groups were compared by Student t test. (B) IHC was performed on formalin-fixed, paraffin-embedded tissue of terminal livers dissected from C57BL/6 mice transformed with a ΔN90-CTNNB1/c-MYC driver plasmid by hydrodynamic tail-vein injection. Representative IHC images showing PTK7 and β-catenin expression pattern at tumor boundaries. (C) The subcellular localization of β-catenin was investigated in MHCC97L cells overexpressing PTK7 by subcellular fractionation and Western blot. (D) The transcriptional activity of β-catenin in Wnt3a-stimulated MHCC97L cells overexpressing PTK7 was examined by TOP/FOP assay and compared with treated EV control by Student t test. NOT, nontumor; OE, overexpression; T, tumor. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
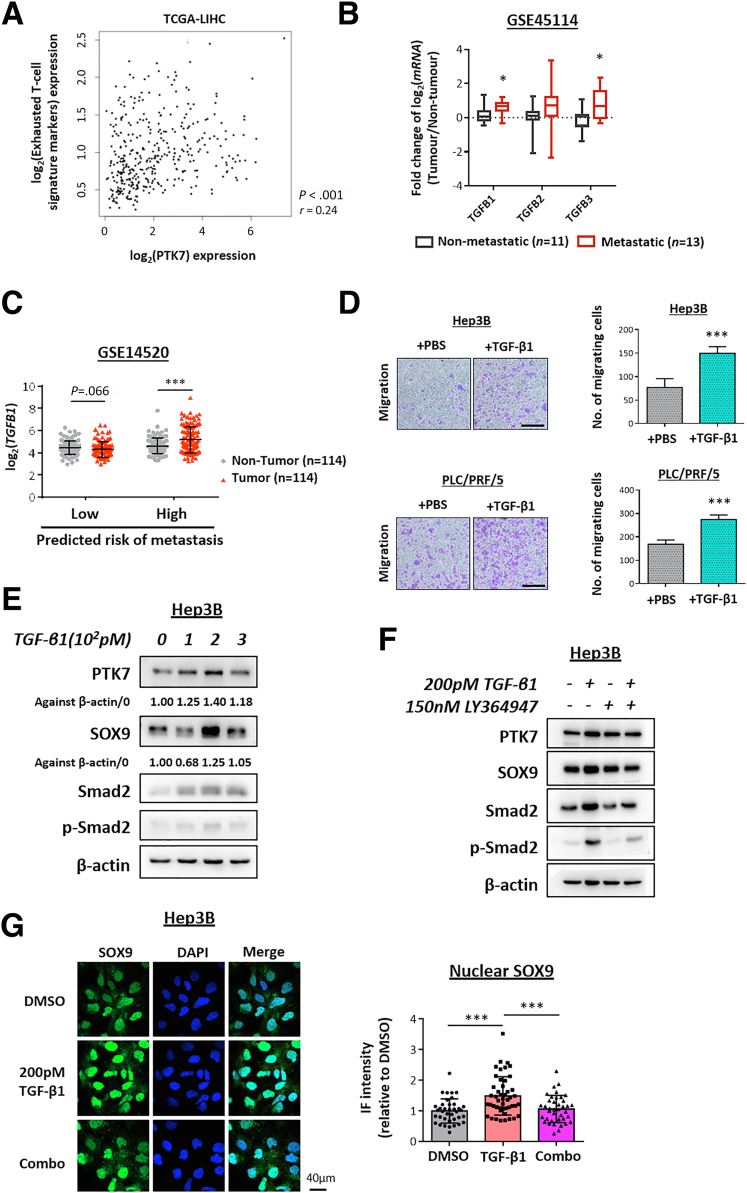
To validate the association between PTK7 and the activation status of TGF-β signaling in the TCGA patient cohort, 18 signature genes of the signaling pathway were selected as reported previously.27 The overall activation status of TGF-β signaling was determined by the collective enrichment of these signature genes in tumor HCC compared with paired nontumor tissue. Notably, higher tumor PTK7 expression corresponded with a general up-regulation of all these signature genes, and HCC tumors with activated TGF-β signaling (TGF-β score > 0) had a greater tendency to express PTK7 (Figure 6B). Increased TGF-β signaling is associated frequently with an inflammatory tumor microenvironment that is predisposed to T-cell exhaustion and suppression.28,29 Correlation analysis of PTK7 and the major markers of exhausted T cells using TCGA-LIHC data suggested a similar up-regulation of PTK7 in HCC tumors showing stronger signatures of T-cell exhaustion (Figure 8A). These results indicated that PTK7 enrichment is predictive of TGF-β signaling activation and is associated with biological features driven by this signaling pathway.
Figure 8.
In vitro analyses of TGF-β1–treated HCC cells. (A) PTK7 correlation with 6 exhausted T-cell signature markers (HAVCR2, TIGIT, LAG3, PDCD1, CXCL13, and LAYN) was examined by Pearson correlation using TCGA-LIHC data set and aided by the publicly available GEPIA2 analysis platform with all expression in terms of transcript per million. (B) Statistical comparison of the expression of TGFB1/2/3 in metastatic and nonmetastatic HCC by GSE45114 and the Student t test. (C) Same statistical evaluation of the GSE14520 data set as described in Figure 1B for the fold change in TGFB1 expression of paired tumor/nontumor HCC samples with different predicted risk of metastasis, by paired t test. (D) Representative images and the quantification of migrating cells by Transwell assay after TGF-β1 single treatment of 24 hours’ duration. Statistical comparisons by the Student t test. (E) Western blot validation of dosage-dependent up-regulation of PTK7 and SOX9 expression in Hep3B cells after a single treatment of recombinant TGF-β1 for a duration of 24 hours at various dosages. Quantification of PTK7 and SOX9 expression was performed by ImageJ. (F) Western blot validation of PTK7 and SOX9 expression in Hep3B cells after TGF-β1 single treatment or in combination with LY-364947, a TGF-βR1–specific inhibitor, for a duration of 24 hours. (G) Representative images of immunocytochemistry targeting SOX9 in Hep3B cells after TGF-β1 single treatment or in combination with LY-364947 (Combo) for a duration of 24 hours. 4′,6-Diamidino-2-phenylindole (DAPI) staining was used to locate the nuclei of stained cells. The average nuclear localization of the SOX9 was quantified by ImageJ and compared between treatment groups by the Student t test. DMSO, dimethyl sulfoxide. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
The correlation between TGF-β signaling and HCC metastasis also was validated via bioinformatic analyses using Gene Expression Omnibus data sets (GSE45114 and GSE14520) to examine the possible enrichment of the 3 major TGF-β ligands (TGF-β1, TGF-β2, and TGF-β3) in the tumors of metastatic HCC or HCC predicted to have a high risk of metastasis (Figure 8B and C). TGF-β1 was the most pivotal TGF-β stimulatory factor up-regulated in metastatic HCC, and subsequent in vitro analyses confirmed its ability to induce the motility of HCC cells (Figure 8D). To consolidate the proposed response of PTK7 toward activated TGF-β signaling at a proteomic level, a TGF-β1–induced biological system was continually adopted. PLC/PRF/5 cells were treated with recombinant TGF-β1 at picomolar doses, followed by Western blot evaluation of PTK7 and SOX9 levels. There was an apparent dose-dependent accumulation of both proteins under stimulated TGF-β signaling (Figure 6C), and the simultaneous application of a TGF-βR1 inhibitor reversed this induction (Figure 6D). Several attempts have been reported previously that connected TGF-β signaling activation with enhanced SOX9 activity in terms of its transcription, nuclear localization, and phosphorylation.30, 31, 32, 33 To identify any TGF-β–mediated up-regulation of SOX9 functionality in HCC, immunocytochemistry was performed to visualize SOX9 localization. The results showed an increase in SOX9 nuclear translocation after TGF-β1 treatment (Figure 6E). TGF-β–activated PTK7 and SOX9 expression was validated simultaneously in Hep3B cells (Figure 8E–G). Stable knockdown of SOX9 directly reversed the up-regulation of proteomic PTK7 induced by TGF-β1 (Figure 6F). Taken together, our data suggested that PTK7 accumulation was a result of TGF-β signaling activation, and this process was highly dependent on SOX9 mediation.
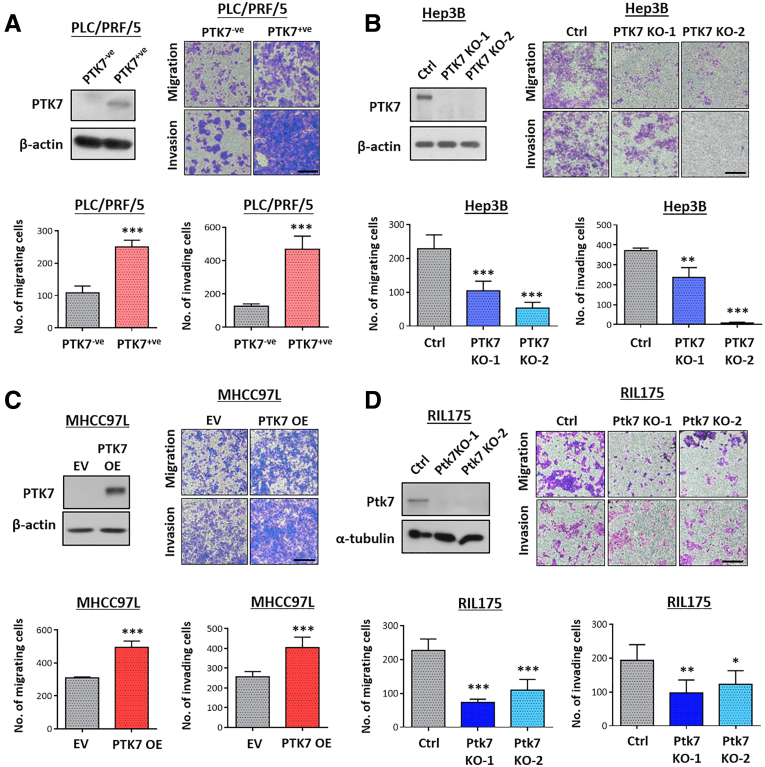
PTK7 Enrichment Leads to More Aggressive Metastatic Phenotypes of HCC Cells and PTK7 Knockout Reduces the Metastatic Capacity of Cells, While the Prometastatic Potential of PTK7 Is Mediated Primarily by its Secretory Domain
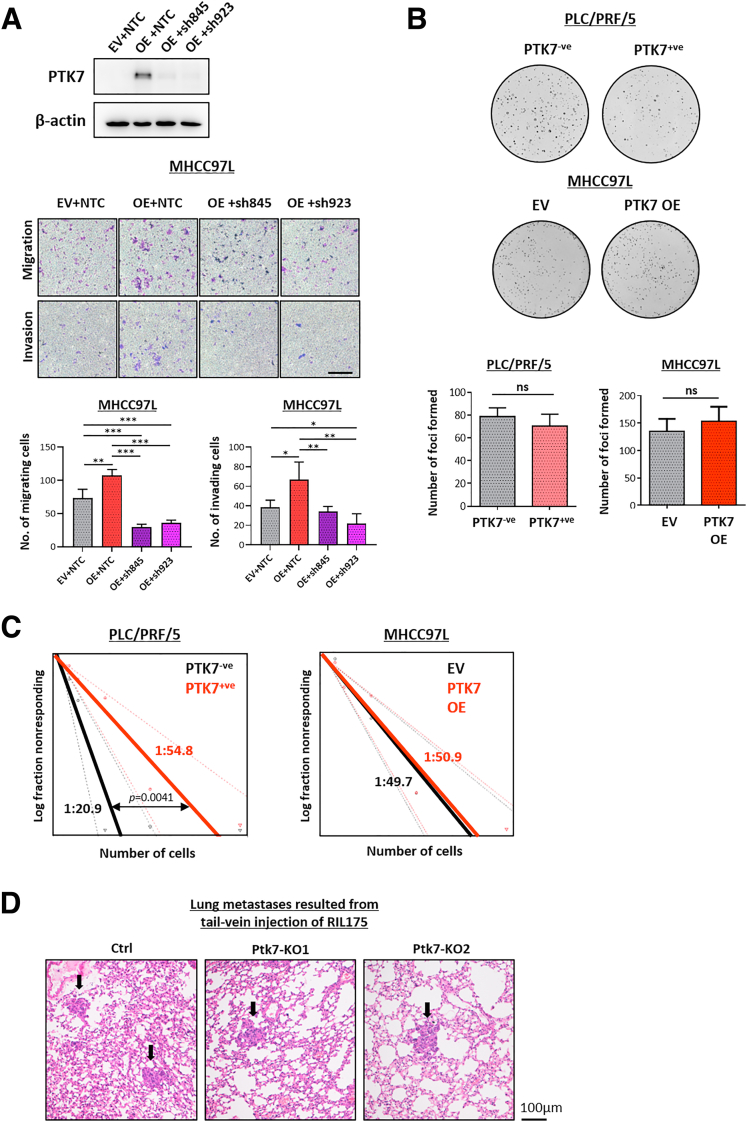
The endogenous and secretory levels of PTK7 and its membrane localization in various HCC cell lines were investigated using Western blot, quantitative polymerase chain reaction (PCR), and flow cytometry (Figure 5A–C). PLC/PRF/5 was observed in a heterogeneous population of PTK7-expressing and PTK7-nonexpressing cells. Fluorescence-activated cell sorting (FACS) was performed on this cell line to isolate PTK7-positive and PTK7-negative cell subsets (Figure 5D), and their metastatic potential was evaluated using Transwell migration and invasion assays (Figure 9A). Enhancing the PTK7-expressing population of PLC/PRF/5 drastically increased cell motility. In contrast, by stably suppressing PTK7 using CRISPR-Cas9–mediated knockout in Hep3B cells, a significant slowing of the migratory and invasive capacity of the cells was achieved (Figure 9B). Lentiviral-based overexpression and CRISPR-Cas9–mediated knockout of PTK7 were further performed on 2 other HCC cell lines, MHCC97L and murine RIL175, respectively. MHCC97L is one of the few HCC cell lines that potently metastasizes to the lung when orthotopically established in the liver, and RIL175 extravasates into the lung when injected through the tail vein.34,35, 36, 37 In vitro analyses indicated a positive correlation between the migratory and invasive properties of these cells and their PTK7 levels (Figure 9C and D). Lentiviral-based short hairpin RNA knockdown of PTK7 in PTK7-overexpressing MHCC97L successfully rescued the induced prometastatic properties (Figure 10A). Other oncogenic properties of PTK7 have been reported in other cancer types. Therefore, we performed foci formation and extreme limiting dilution assays on sorted PLC/PRF/5 cells and MHCC97L-overexpressing cells. The data showed that PTK7 enrichment exerted negligible or adverse effects on the proliferation and stemness of HCC cells depending on the cell line tested (Figure 10B and C).
Figure 9.
PTK7 enrichment and overexpression lead to more aggressive metastasis of HCC cells, and PTK7 knockout reduced the metastatic capacity of cells, and the prometastatic properties of PTK7 potentially are mediated by its secretory domain. (A) Low- and high-PTK7–expressing cells were isolated from the HCC cell line PLC/PRF/5 using FACS, and the separation was validated by Western blot. Sorted PLC/PRF/5 cells were subjected to Transwell migration and invasion assays. The number of migrating or invading cells was compared using the Student t test. (B) CRISPR-Cas9–mediated knockout of PTK7 was performed in Hep3B cells, followed by Western blot validation and Transwell migration and invasion assays. The number of migrating or invading cells was compared with the transfection control (Ctrl) using 1-way analysis of variance. (C) PTK7 was overexpressed in MHCC97L cells and validated using Western blot. The number of overexpressing cells migrating or invading in Transwell assays was compared with EV control using the Student t test. (D) CRISPR-Cas9–mediated knockout of Ptk7 was performed on murine RIL175 cells, followed by Western blot validation and Transwell migration and invasion assays. The number of migrating or invading cells was compared with the transfection control (Ctrl) using 1-way analysis of variance. All representative images of Transwell assays were taken under a light microscope at 40× magnification with at least 4 microscopic fields captured for statistical analyses. Scale bars: 100 μm. KO, knockout; OE, overexpression. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
Figure 10.
PTK7 in vitro functional assays performed on various HCC cell lines. (A) PTK7 was overexpressed in MHCC97L cells and subsequently suppressed by lentiviral-based knockdown using 2 independent short hairpin RNA (shRNA) structures (sh845 and sh923). The number of cells migrating or invading in Transwell assays were compared with EV + nontarget control (NTC) by 1-way analysis of variance with multiple t tests. The successful knockdown of PTK7 was validated by Western blot. (B) Representative images and the comparison of the number of foci formed by sorted PLC cells and MHCC97L cells overexpressing PTK7. Statistical comparisons were performed by the Student t test. (C) The predicted tumor-initiating cell frequency of sorted PLC cells and MHCC97L overexpression cells was determined by extreme limiting dilution assay and analyzed with a publicly available algorithm. The frequency was represented by the estimated population of cells required for the detection of 1 tumor-initiating cell. (D) Representative H&E staining of a lung tissue section harvested from mice injected with RIL175 through the tail vein. Metastases are indicated with black arrows. Ctrl, control; OE, overexpression. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
PTK7 Overexpression Promotes Extrahepatic Metastasis to the Lung of Liver-Engrafted HCC Cells, and PTK7 Knockout Suppresses the Ability of Cells to Extravasate Into the Lung
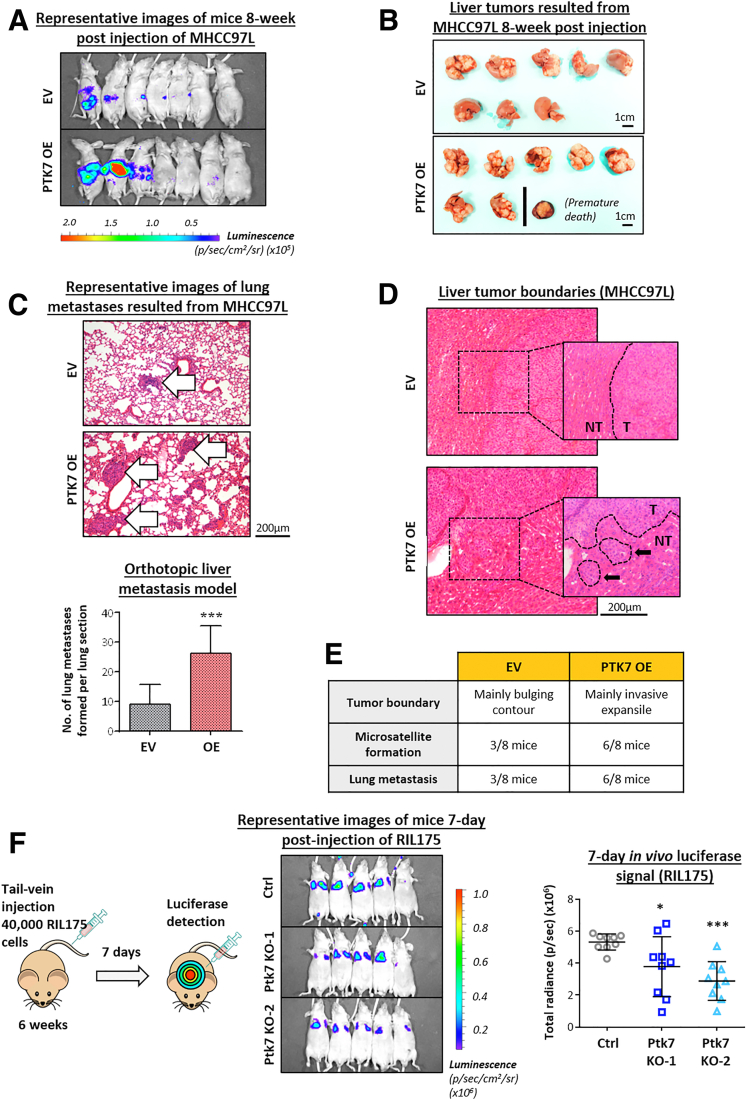
Metastasis is a highly contextual biological event and must be studied in greater detail using animal models. MHCC97L is a human HCC cell line with reliable metastatic potential in immunodeficient mice. MHCC97L cells overexpressing PTK7 and stably expressing a luciferase reporter gene were injected into the livers of nude mice, and empty vector (EV) cells were used as a control. The luciferase signal induced by descendent cells was monitored until 8 weeks after orthotopic injection, and the mice were killed for further examination of the formation of lung metastases. According to the intensity of the luciferase signal induced (Figure 11A) and the morphology of liver tumors formed (Figure 11B), overexpression of PTK7 resulted in noticeably greater tumor nodules in size and number compared with the EV control, which indicated more aggressive cancer development and intrahepatic metastasis. Histologic examination of dissected lung and liver tissues showed that more frequent lung metastases were induced after PTK7 enrichment (Figure 11C), and the boundaries of PTK7-overexpressing tumors were more invasive in terms of morphology and microsatellite formation than EV tumors (Figure 11D and E). To summarize, our PTK7-overexpression animal model echoed the in vitro data and reinforced the potential of PTK7 in driving intrahepatic and extrahepatic metastasis. To study in greater detail the role of PTK7 in specific biological stages of the metastasis cascade, particularly extravasation and the establishment of distal metastases, Ptk7 was stably knocked out in luciferase-tagged RIL175 murine HCC cells using the CRISPR-Cas9 system. Ptk7-knockout RIL175 cells were injected into the blood stream of nude mice through the tail vein to simulate the circulating tumor cell population. Establishment of circulating RIL175 cells in the lungs of mice was visualized by their luciferase signal intensity and the formation of metastatic outgrowths ( Figure 10D and Figure 11F left panel). After 7 days of incubation, the knockout cells settled in the lung at a significantly lower rate than the control cells (Figure 11F, middle and right panels), which indicated a reduced capacity of these cells to extravasate into foreign tissues.
Figure 11.
PTK7 overexpression promotes extrahepatic metastasis to the lung of liver-engrafted HCC cells, and PTK7 knockout suppresses the ability of cells to extravasate into the lung. (A) In vivo detection of luciferase-induced luminescence of tumor nodules and images of livers dissected from nude mice 8 weeks after orthotopic liver injection of MHCC97L overexpressing cells. (B) External morphology of HCC nodules formed in the livers of nude mice 8 weeks after orthotopic liver injection of MHCC97L-overexpressing cells. (C) H&E-stained tissue sections of lungs dissected from the orthotopic injection model showing the formation of lung metastases (indicated by white arrows). The number of discernible metastases formed in the lungs of mice injected with MHCC97L-overexpressing cells was counted and compared using the Student t test. (D) H&E-stained tissue sections of livers dissected from the orthotopic injection model showing different features of the tumor boundary. The presence of microsatellites is indicated by black arrows. (E) A summary table of the parameters related to metastatic aggressiveness of the tumors in the orthotopic injection model. (F) Murine RIL175 cells with Ptk7 depleted by CRISPR-Cas9–mediated knockout were injected into nude mice through the tail vein to observe lung metastasis formation. Luciferase-induced luminescence of tumor nodules developed from RIL175 cells was detected using in vivo imaging 7 days after tail vein injection. The total detectable radiance was compared between the control and Ptk7-knockout groups using 1-way analysis of variance. Ctrl, control; OE, overexpression. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
The Up-Regulation of PTK7 Resulted in the Enrichment of the EMT Components Zinc Finger Protein SNAI2 (SLUG) and Zinc Finger E-box-binding Homeobox 1 (ZEB1), Which May Confer the Prometastatic Properties of PTK7
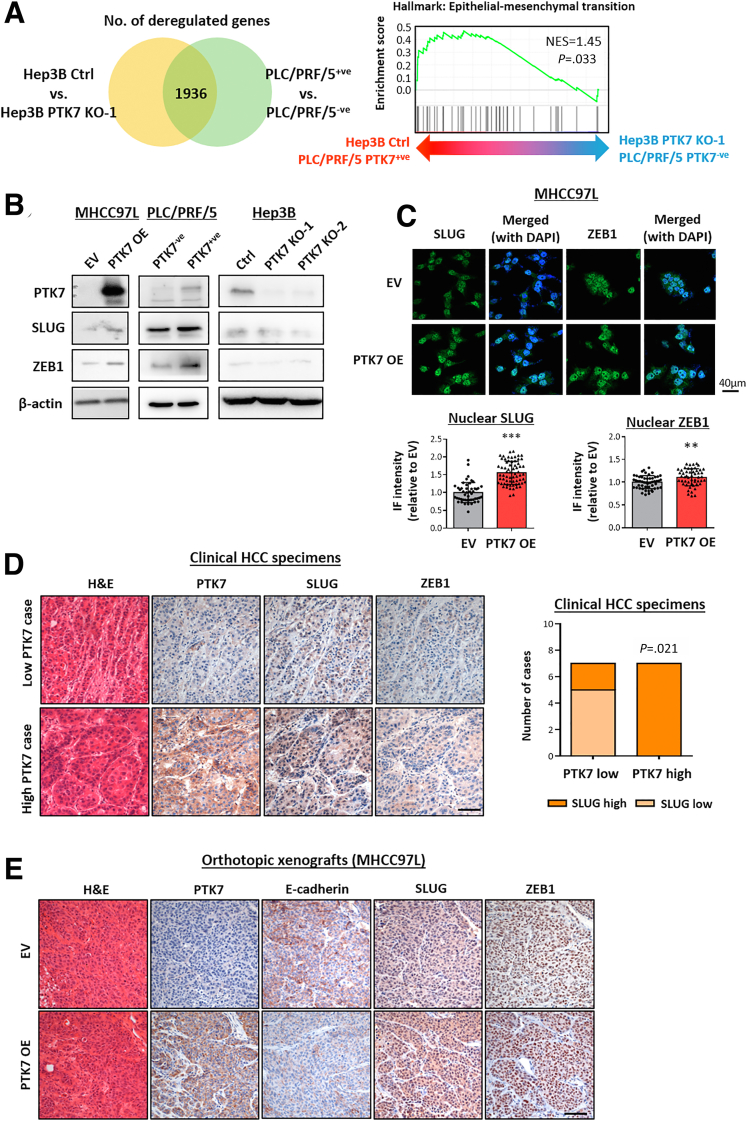
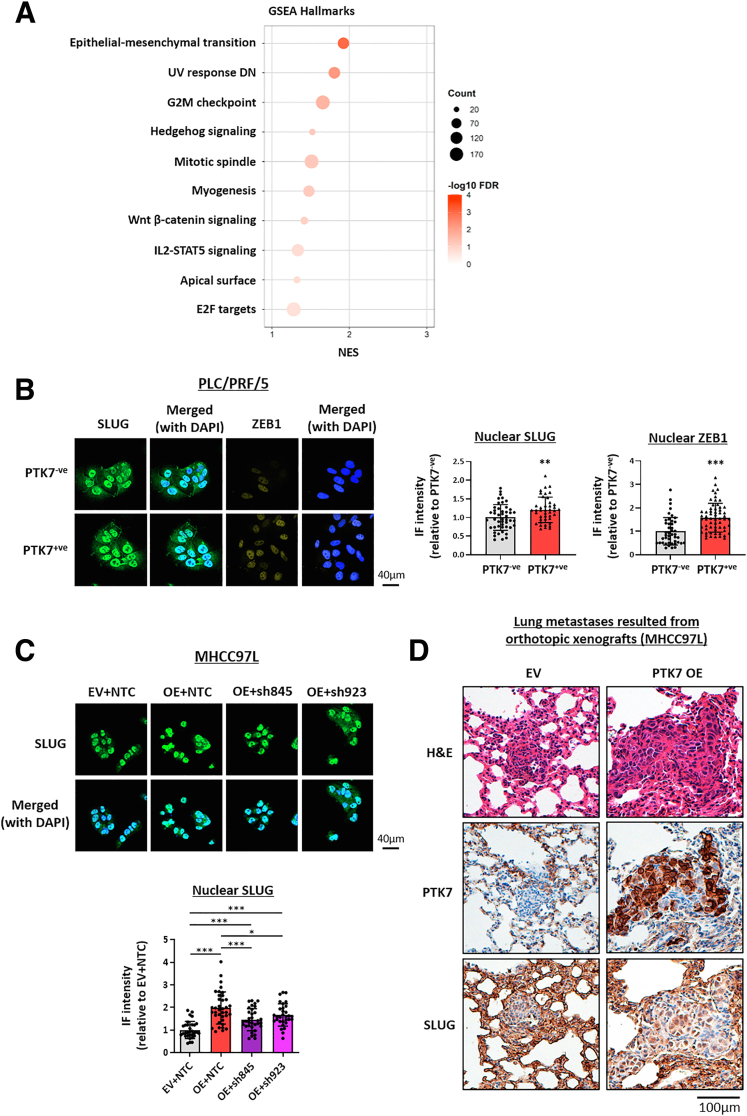
RNA sequencing was performed on the sorted PLC/PRF/5 cells and Hep3B knockout vs control cells to help gain further insights into the potential signaling pathways driven by PTK7 (Figure 12A). Among the top deregulated Hallmark pathways identified by GSEA (Figure 13A), EMT was one of the top deregulated signaling events under PTK7 enrichment. Notably, the EMT players SLUG and ZEB1 were deregulated consistently in the manipulated HCC cell lines (Figure 12B), and their nuclear localization was significantly induced upon PTK7 overexpression or enrichment (Figures 12C and 13B). Lentiviral-based short hairpin RNA knockdown of PTK7 in PTK7-overexpressing MHCC97L cells partially rescued and reversed the increase in nuclear SLUG expression (Figure 13C). By reviewing a collection of HCC patient-derived tumor tissues using IHC, higher PTK7 expression was associated with denser SLUG and ZEB1 staining (Figure 12D). SLUG protein localized preferentially to the nuclei of PTK7-enriched HCC cells, as quantified by the greater proportion of cells showing noticeable nuclear 3,3'-diaminobenzidine (DAB) staining per microscopic field. ZEB1 was only marginally detectable by IHC in the same set of HCC samples, which dissuaded more detailed quantification. The accumulation of SLUG and ZEB1 also was evident in metastasizing tumors developed from our orthotopic liver injection model. More intense cellular and nuclear signals of these proteins were found by IHC on PTK7-overexpressing tumors obtained from the mouse model (Figure 12E), which also was accompanied by a decrease in the epithelial marker E-cadherin. Similarly, a more profound nuclear expression of SLUG was noticed in the lung metastases resulting from PTK7-overexpressing cells (Figure 13D), which suggested that the potential activation and up-regulation of SLUG resulting from PTK7 enrichment was a conserved feature during the course of the metastatic cascade and may have played a central role in driving the process.
Figure 12.
The up-regulation of PTK7 resulted in enrichment of the EMT components SLUG and ZEB1, which may confer the prometastatic properties of PTK7. (A) RNA sequencing was performed on HCC cells with differential PTK7 expression, and Hep3B control was compared against one of the PTK7-knockout clones. FACS-sorted PLC/PRF/5 cells were compared against each other. Commonly deregulated genes (total, 1936 genes) were identified in the 2 comparison groups with a threshold of transcript per million greater than 1 and relative fold-change difference of 1.2 or greater, and then subjected to GSEA. EMT was one of the top enriched pathways under PTK7 enrichment. (B) The proteomic levels of SLUG and ZEB1 in PTK7-overexpressing MHCC97L cells, sorted PLC/PRF/5 cells, and PTK7-knockout Hep3B cells were evaluated using Western blot. (C) Representative images of immunocytochemistry (ICC) targeting SLUG and ZEB1 in PTK7-overexpressing MHCC97L cells. 4′,6-Diamidino-2-phenylindole (DAPI) staining was used to locate the nuclei of stained cells. The average nuclear localization of SLUG and ZEB1 was quantified in ImageJ and compared between EV and overexpressing cells using the Student t test. (D) IHC was performed on formalin-fixed, paraffin-embedded (FFPE) tissue sections derived from the livers of 14 HCC patients to detect SLUG and ZEB1 expression. Representative images of tumors with high or low PTK7 expression are shown. The proportion of HCC nuclei observed with SLUG expression was evaluated further using light microscopy and compared between PTK7-low and PTK7-high patient groups with at least 4 microscopic fields captured for statistical analyses. High-SLUG nuclear expression was defined by >50% nuclei observed with intense DAB staining, and vice versa. Group comparisons were performed using the Fisher exact test. (E) IHC was performed on FFPE tissue sections of liver tumors developed after orthotopic injection of MHCC97L cells to compare E-cadherin, SLUG, and ZEB1 expression patterns between EV control and PTK7-overexpressing xenografts. Scale bars: 100 μm. Ctrl, control; IF, immunofluorescence. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
Figure 13.
RNA sequencing and representative images of IHC conducted on lung sections harvested from the orthotopic injection mice model. (A) A bubble plot showing the top 10 enriched Hallmark pathways under PTK7-enriched conditions as determined by GSEA analysis. (B) Representative images of immunocytochemistry (ICC) targeting SLUG and ZEB1 in PLC/PRF/5 cells sorted by their PTK7 expression. 4′,6-Diamidino-2-phenylindole (DAPI) staining was used to locate the nucleus of stained cells. The average nuclear localization of the SLUG and ZEB1 was quantified by ImageJ and compared between PTK7-ve and PTK7+ve cells by the Student t test. (C) Representative images of ICC targeting SLUG in MHCC97L, of which PTK7 was overexpressed and subsequently suppressed by lentiviral-based knockdown using 2 independent short hairpin RNA structures (sh845 and sh923). DAPI staining was used to locate the nucleus of stained cells. The average nuclear localization of SLUG was quantified by ImageJ and compared between the 4 transfected lines by 1-way analysis of variance with multiple t tests. (D) Images showing IHC staining of PTK7 and SLUG performed on lung metastases resulted from orthotopic liver injection of MHCC97L cells with or without PTK7 overexpression. DN, down-regulated; E2F, E2F transcription factor; FDR, false discovery rate; G2M, G2 to M phase cell cycle transition; IF, immunofluorecence; IL, interleukin; NTC, nontarget control; OE, overexpression; STAT5, signal transducer and activator of transcription 5. P values (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
Discussion
Kinases are one of the largest repertoires of drug targets exploited in cancer therapies. More than 100 kinase inhibitors are under development and in clinical trials, with dozens approved and routinely prescribed to cancer patients with various disease conditions.38 Receptor tyrosine kinases are favorable targets for inhibition owing to their exposure to the extracellular environment and their functional importance in mediating cell‒cell signal transduction. Therefore, understanding the specific recruitment of these kinases during different stages of cancer will immensely benefit targeted therapy and drug delivery.
The present study sought to discover novel kinases representative of metastatic and advanced-stage HCC to aid in the development of unique inhibitors for this subset of patients and provide alternatives to the broadly prescribed first-line medications. Our bioinformatic analyses found that the pseudokinase PTK7 was the most relevant to metastasis. The lack of an active kinase domain in pseudokinases was once deemed unfavorable for its inhibition, but increasing evidence has suggested that pseudokinases drive oncogenic pathways in cancer via novel, noncatalytic mechanisms, with new avenues for inhibitor design.39,40 Similarly, our functional analyses confirmed that apart from being a functional component driving metastasis, the preferential expression of PTK7 also resulted in the activation of selective prometastatic factors, such as SLUG and ZEB1, which showed novel mechanisms regulating these central EMT drivers.
Insights also were gained into the transcriptional mechanism and functional role of PTK7 in the cancer-specific molecular landscape. Previous studies in developmental models positioned PTK7 as a crucial member of Wnt signaling and neuronal development. However, other reports that focused on PTK7 in the cancer context failed to address the nature of its up-regulation, whether it was associated similarly with Wnt signaling or promoted by other oncogenic signatures. Our bioinformatic analyses and in vitro assays identified an intimate link between TGF-β signaling activation status, SOX9 transcriptional activity, and the presentation of PTK7 in HCC patients. This observed correlation established PTK7 as a potential reporter of activated TGF-β signaling, which has been widely implicated in cancer metastasis. The functional role of SOX9 as a stemness marker in HCC has been proposed by several recent studies.41, 42, 43 We further suggested that SOX9 was preferentially expressed in metastatic HCC, and PTK7 was its potential effector in potentiating metastasis. The observed contribution of PTK7 to the selective up-regulation of SLUG and ZEB1 levels reported here elucidate a possible regulatory pathway in which the activity of a pseudokinase influences the determination of EMT states under the more global TGF-β signaling cascade. However, the exact nature of this mediation requires closer inspection because preliminary data have shown that SLUG and ZEB1 up-regulation by PTK7 enrichment was not a result of transcriptional activation.
The potential proteolysis of PTK7 in HCC cells was not overlooked during our investigation. Earlier studies showed that PTK7 is subjected to extensive cleavage by several extracellular matrix (ECM)-remodeling enzymes upon its membrane localization.44, 45, 46 The release of PTK7 N-terminal extracellular and C-terminal intracellular domains after cleavages may promote distinct oncogenic properties. When our team examined the expression pattern of PTK7 across a panel of HCC cell lines, we found that all PTK7-expressing cell lines showed a tendency to secrete PTK7. However, the extent of PTK7 secretion did not strictly follow the endogenous level or membrane presentation of PTK7, which shows a great dependence of this event on other cellular components, such as the intrinsic activity of various ECM-remodeling enzymes. As a result, this study focused solely on the endogenous behavior of PTK7. A detailed depiction of PTK7 secretion in HCC would promote the prognostic value of this novel protein. Therefore, to further our current findings, it is important to identify the presence of secretory PTK7 in patient serum via the development of an enzyme-linked immunosorbent assay platform and to observe the unique functional autocrine and paracrine consequences of PTK7 secretion with the assistance of cleavage-resistant mutants.
Efforts are gradually building toward the use of PTK7 as a drug delivery platform for specific cancer types or cell populations. The secretory nature of PTK7 and its probable absence in normal hepatic function further enhance the diagnostic and therapeutic value of PTK7 for HCC treatment. These therapeutic platforms ranged from small interfering RNA–mediated PTK7 silencing,47 PTK7-specific antibody‒drug conjugates,48 PTK7-targeting CAR T-cell therapy,49 and PTK7-targeting DNA aptamers.50 The increasing potential of neutralizing antibodies targeting surface epitopes also introduces the tremendous possibility of PTK7-mediated treatment. Our collaborator generously provided PTK7-specific antibody‒drug conjugates for preliminary testing, and our team compared the cytotoxicity of the conjugate in PTK7-rich or PTK7-depleted HCC cells. The results showed that the drug conjugate only marginally induced apoptosis in HepG2 cells, which have the highest PTK7 expression. Whether this finding was a result of cell line–specific behavior or a general insensitivity of most HCC cells toward the microtubule inhibitor conjugated to the antibody must be further determined.
In conclusion, the present study discovered a novel regulatory axis of HCC metastasis that was initiated by the global enrichment of TGF-β–activating signals, mediated by the transcriptional activity of SOX9, and depended on the accumulation of PTK7, which eventually led to the activation of EMT drivers. We hope our investigation and findings will shed light on the importance and possibility of identifying drug targets specific to biological abnormalities as complex as metastasis and contribute to the collective attempt to enrich the repertoire of targetable kinases, particularly pseudokinases, by discovering novel strategies performed by these molecules.
Materials and Methods
Cell Lines
The HCC cell lines Hep3B and PLC/PRF/5 were purchased from American Type Culture Collection (Manassas, VA). The HCC cell line MHCC97L was kindly provided by the Chinese Academy of Medical Sciences (Beijing, China). The murine HCC cell line RIL175 was kindly provided by Dr Judy W. P. Yam (University of Hong Kong, Hong Kong, SAR). HEK293T and HEK293FT cells were purchased from Invitrogen and American Type Culture Collection, respectively.
Reagents for Cell Culture Treatment
Recombinant TGF-β1 was purchased from R&D Systems and TGF-βR1–specific inhibitor LY364947 was purchased from Sigma-Aldrich.
Cell Treatment
HCC cell lines were treated with recombinant TGF-β1, alone or in combination with LY364947, for 24 hours before processing for proteomic analyses or IHC.
TMA
A total of 3 sets of TMAs representative of 53 patients per set were investigated in this study. IHC of PTK7 was performed on all 3 sets of TMAs, and SOX9 staining was performed on only 2 sets for correlation studies because of the limited quantity of available TMA. TMAs were obtained from Professor Jing-Ping Yun (Sun Yat-sen University Cancer Centre, Guangzhou, China), with approval by the Institutional Review Board for ethical review from the university. The procurement of all clinical information received consent from patients.
Bioinformatic Analyses
TCGA-LIHC data set was used to analyze the expression pattern of PTK7, its correlation with potential transcription regulators, and the enrichment of TGF-β signaling signature genes in HCC patients. Expression data in relation to metastatic HCC and HCC with described metastatic potential were extracted from Gene Expression Omnibus (GSE45114 and GSE14520) of the National Centre for Biotechnology Information. All statistical significance was calculated in PRISM (GraphPad) and presented as P values unless stated otherwise.
In Vivo Metastasis Models
MHCC97L cells overexpressing PTK7 were orthotopically injected into the left lateral lobe of nude mice (400,000 cells) supplemented with 50% Matrigel. The luciferase signal of the injected cells and the formation of tumors and metastases were recorded 8 weeks postinjection. Livers and lungs were obtained for histologic procedures at the end of the experiment. To examine the functional consequence of PTK7 depletion in the colonization of lung tissue, PTK7-knockout RIL175 cells were injected via the tail vein as previously described.34 The establishment of circulating RIL175 cells in lungs was visualized by luciferase signal detection 7 days after injection. Lungs were obtained for histologic procedures afterward. All animal work was subjected to the supervision of and followed the regulations for animal ethics as set out by the Committee of the Use of Live Animals in Teaching and Research at The University of Hong Kong and the Animals (Control of Experiments) Ordinance of Hong Kong.
Hydrodynamic Tail-Vein Injection of Constitutively Active β-Catenin+Protein Kinase B (Akt) Plasmids to Induce HCC in a Mouse Model
A total of 10 μg of 2 plasmids each encoding a proto-oncogene, the β-catenin mutant with N-terminal deletion before the α-catenin binding site (ΔN90-CTNNB1) and human c-MYC, respectively, together with sleeping beauty transposase at a 25:1 ratio, were diluted and filtered as 2 mL saline solution. Injection of plasmid solution into 6- to 8-week-old male C57BL/6 wild-type mice was performed through the lateral tail vein, with a 5- to 7-second purge. Mice were monitored for tumor growth and their livers were harvested at week 8 postinjection for histologic processing and evaluation of PTK7 expression by IHC.
Cloning and Transfection of PTK7 Overexpression, PTK7 Knockout, and PTK7 and SOX9 Knockdown Plasmids
Human PTK7 complementary DNA (cDNA) representative of the full-length protein (1070 amino acids) was subcloned from a pcDNA3 backbone (supplied by Dr Seung-Taek Lee of Yonsei University in South Korea)51 into pDONR 201 entry clone (Addgene) by PCR amplification and recombination reaction using Gateway BP clonase II reaction (Invitrogen). Shuttling of target cDNAs into a lentiviral-based system using pEZ-Lv199 destination vector (GeneCopoeia) was further performed by Gateway LR clonase II reaction (Invitrogen). For human HCC cell lines, PTK7 knockout was performed by a lentiviral-based CRISPR-Cas9 system using plentiCRISPR-v2 vector (Addgene) as backbone. The annealed oligos encoding scaffold guide RNA sequence were ligated with the predigested backbone by Quick ligation reaction (NEB) at their Esp3I blunt ends. An untransformed backbone was used as empty vector control. For murine RIL175 cells, CRISPR-Cas9–mediated knockout was performed by a 3-plasmid system composed of a pGFP (Addgene) plasmid for cell labeling, pCas9 (Addgene), and 2 independent pgRNA-Cloning vectors (Addgene) harboring the guide RNA sequences. Guide RNAs were cloned into pgRNA-Cloning vectors by Gibson’s Assembly (NEB) at their AflII-digested ends. Guide RNA sequences for PTK7-knockout are as follows, human guide RNA 1: forward: GGGTCCCATCTCGGAACCAT, reverse: ATGGTTCCGAGATGGGACCCC; human guide RNA 2: forward: ACGGAGCGGCGTTTCGCCCA, reverse: TGGGCGAAACGCCGCTCCGTC; mouse guide RNA 1: forward: CGTCTACCGTTGCATCG, reverse: GACCGATGCAACGGTAGACG; and mouse guide RNA 2: forward: CAAACCGTCGCTCCGTG, reverse: GGACACGGAGCGACGGTTTG.
Lentiviral-based PTK7 knockdown was performed with the cloning of the following short-hairpin RNAs sequences (underlined) into pLKO.1-blast vector (Addgene) by T4 ligase (Promega) at the AgeI and EcoRI overhangs. Oligo sequences for the synthesis of short-hairpin structure are as follows,
sh845: forward: CCGGCCTTGAGCATTGCTGATGAAACTCGAGTTTCATCAGCAATGCTCAAGGTTTTTG, reverse: AATTCAAAAACCTTGAGCATTGCTGATGAAACTCGAGTTTCATCAGCAATGCTCAAGG; and sh923: forward: CCGGCCATGTTCCATTGCCAGTTCTCTCGAGAGAACTGGCAATGGAACATGGTTTTTG, reverse: AATTCAAAAACCATGTTCCATTGCCAGTTCTCTCGAGAGAACTGGCAATGGAACATGG.
Lentiviral-based SOX9 knockdown was performed with the cloning of the following short-hairpin RNAs sequences (underlined) into pLKO.1-puro vector (Addgene) by T4 ligase (Promega) at the AgeI and EcoRI overhangs. Oligo sequences for the synthesis of short-hairpin structure are as follows, short hairpin RNA1: forward: CCGGCTCCACCTTCACCTACATGAACTCGAGTTCATGTAGGTGAAGGTGGAGTTTTTG, reverse: AATTCAAAAACTCCACCTTCACCTACATGAACTCGAGTTCATGTAGGTGAAGGTGGAG; and short hairpin RNA2: forward: CCGGACTTCTGAACGAGAGCGAGAACTCGAGTTCTCGCTCTCGTTCAGAAGTTTTTTG, reverse: AATTCAAAAAACTTCTGAACGAGAGCGAGAACTCGAGTTCTCGCT CTCGTTCAGAAGT.
Lentiviral packaging of PTK7 overexpression plasmids was performed in 293T cells using Lipofectamine 3000 transfection reagent (Thermo Scientific) and a third-generation lentiviral packaging system (pMDL/pRRE, pRSV-Rev, VSV.G; Addgene). Packaging of PTK7 knockout plasmids was performed in 293T cells using Lipofectamine 3000 transfection reagent and a second-generation lentiviral packaging system (pMD2.G, psPAX2; Addgene). Packaging of SOX9 knockdown plasmids was performed in 293T cells using the FuGENE HD Transfection Reagent (Promega) and a packaging mix of pGAG, pREV, and VSV.G (provided by Dr Terence Lee, The Hong Kong Polytechnic University, Hong Kong, SAR). HCC cells were infected by viruses with 8 μg/mL polybrene (Sigma-Aldrich) followed by recovery and selection using puromycin. Transient transfection of the 3-plasmid CRISPR-Cas9 system into murine HCC was performed using Lipofectamine 3000 transfection reagent, and isolation of positive clones was performed by FACS and single-cell selection in 96-well plates. GFP-only transfected cells were used as control in this transient knockout model.
Flow Cytometry and FACS
Membrane PTK7 expression of various HCC cell lines was assessed by flow cytometry. Marking of surface PTK7 was performed using an APC-conjugated anti-human PTK7 mouse antibody (130-099-660; Miltenyi Biotec) with an allophycocyanin (APC)-conjugated mouse IgG2a (130-113-831; Miltenyi Biotec) stained as negative control. Flow cytometer analyses were performed on the FACS Canto II (BD Biosciences) with a red laser channel (633 nm) to activate the APC signal. Gating, compensation, and analysis of PTK7-stained cell populations were performed with FlowJo software (FlowJo, LLC). PLC/PRF/5 cells were stained for their membrane PTK7 expression to identify populations differentially expressing PTK7. FACS was performed on a FACS Aria SORP (BD Biosciences). The subpopulation with the 20% highest APC signal was sorted out as PTK7+ve, while the lowest 20% was sorted out as PTK7-ve. All cells were harvested in complete medium for further functional assays and validation.
RNA Extraction, cDNA Synthesis, and Quantitative PCR
RNA was extracted from cells by RNAiso Plus (TaKaRa) followed by standard chloroform phase separation and alcohol precipitation. cDNA was synthesized from total RNA by the PrimeScript RT Master Mix kit (TaKaRa) according to the standard protocol. BlasTaq quantitative PCR MasterMix (ABM) and primers listed in Table 1 were used to amplify specific target cDNA under the LightCycler 480 II (384-well) system (Roche). Relative expression differences were calculated using the 2-ΔΔCt method.
Table 1.
Primers Used in This Study
| Target | Sequence | Target size, base pair |
|---|---|---|
| qPCR | ||
| Human PTK7 | Forward: GTAGTAGCGAGGTATGAGGAGG Reverse: TGCGGTTAGTGATGGGAGTCT |
106 |
| Human SOX9 | Forward: AGCGAACGCACATCAAGAC Reverse: CTGTAGGCGATCTGTGGGG |
85 |
| Human ACTB (β-actin) | Forward: CATCCACGAAACTACCTTCAACTC Reverse: GAGCCGCCGATCCAGACG |
213 |
| For ChIP-qPCR | ||
| SOX9-binding site on PTK7 promoter | Forward: ATTCTGGGGGAACTGTTGGC Reverse: AGTGCAGGGGACCTGAGTAT |
202 |
qPCR, quantitative PCR.
Total Protein Extraction, Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis, and Western Blot
All cell lysis was performed with RIPA buffer (Cell Signaling Technology) with added cOmplete protease inhibitor cocktail (25× dilution; Roche), 1 mmol/L phenylmethylsulfonyl fluoride (Sigma-Aldrich), and phosphatase inhibitor cocktails 2 and 3 (100× dilution; Sigma-Aldrich). Total protein extracted was quantified by the Bradford assay with the Bio-Rad protein dye reagent (Bio-Rad). Quantified protein samples, denatured by heating, were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Merck Millipore). Primary antibodies used for immunoblotting against various targets were listed as follows: PTK7 (1:1000, AF4499; R&D Systems), SOX9 (1:1000, AB553; Merck Millipore), Smad2 (1:1000, 5339; Cell Signaling Technology), pSmad2 (1:1000, 3108; Cell Signaling Technology), SLUG (1:1000, MA5-26385; Thermo Scientific), ZEB1 (1:500, ab203829; Abcam), β-catenin (1:1000, 9587; Cell Signaling Technology), α-tubulin (1:1000, T9026; Sigma-Aldrich), histone H3 (1:2000, ab24834; Abcam), and β-actin (1:10,000, A5316; Sigma-Aldrich).
Immunocytochemistry
The nuclear localization of various proteins was assessed by immunocytochemistry in HCC cells. Cells were preseeded on glass coverslips, fixed in 4% paraformaldehyde (Sigma-Aldrich), and permeabilized with 0.1% Triton X-100 solution (Sigma-Aldrich). Nonspecific binding sites were blocked with 5% bovine serum albumin solution in phosphate-buffered saline (PBS). Primary antibodies used for immunoblotting against various targets were listed as follows: SOX9 (1:500, AB553; Merck Millipore), SLUG (1:100, 9585; Cell Signaling Technology), and ZEB1 (1:200, 3396; Cell Signaling Technology). Cells were counterstained with antifade 4′,6-diamidino-2-phenylindole (Invitrogen) and visualized by a fluorescent confocal microscope (LSM 880; Carl Zeiss).
IHC
After initial deparaffinization, tissue sections were immersed in xylene and rehydrated through a gradient of ethanol mixture from 100% down to distilled water. Antigen retrieval was performed by heating at sub-boiling temperature and submersion in antigen-retrieval solutions (PTK7/E-cadherin/SLUG/ZEB1: Tris-EDTA solution with 0.05% Tween-20, pH 9.0; SOX9: sodium citrate solution with 0.05% Tween-20, pH 6.0). Blocking was performed with 3% hydrogen peroxidase solution and incubation with primary antibodies diluted with Dako antibody diluent (Agilent) was performed overnight at 4ºC. Primary antibodies used for immunoblotting against various targets were listed as follows: PTK7 (1:1000, provided by AbbVie), SOX9 (1:1000, AB5535; Merck Millipore), E-cadherin (1:400, 3195; Cell Signaling Technology), SLUG (1:200, MA5-26385; Thermo Scientific), and ZEB1 (1:150, ab203829; Abcam). The reaction was developed with secondary antibody blotting and DAB+Substrate-Chromogen System (Dako). Slides were counterstained with Mayer’s hematoxylin.
Transwell Migration and Invasion Assays
HCC cell lines with differential PTK7 expression were resuspended in serum-free culture medium and deposited into the chamber of Millicell 8.0-μm polyethylene terephthalate hanging cell culture inserts (Merck Millipore). The invasion assay was performed with an additional layer of 10% Matrigel coated in the inner surface of the chambers. Cells were allowed to migrate or invade through Matrigel toward complete medium in the outer 24-well space for 48–72 hours. Cells that migrated or invaded were washed with PBS, fixed with 4% paraformaldehyde for 15 minutes, and stained with 2% crystal violet.
Foci Formation Assay
Cells were seeded in a 6-well plate at a density of 1000 cells/well and incubated at 37ºC for 10–14 days depending on the cell lines. Medium was replenished every 3 days. After incubation, colonies were washed with PBS, fixed with 4% paraformaldehyde, and stained with 2% crystal violet. All visible colonies were counted by the naked eye and compared among cell lines.
Extreme Limiting Dilution Assay
Extreme limiting dilution assay was performed to evaluate the ability of HCC cells to multiply and proliferate into spheroid at very low cell densities (5, 10, 20, 50, 100, 200 cells). In a 96-well plate precoated with polyhydroxyethylmethacrylate (Sigma-Aldrich), cells were seeded at the desired densities in a specialized spheroid medium, containing Gibco B27 supplement (Thermo Scientific), human epidermal growth factor (PeproTech), human fibroblast growth factor (PeproTech), and insulin (Sigma-Aldrich) mixed with 0.25% methylcellulose (Sigma-Aldrich) dissolved in fetal bovine serum–free medium. Any spheroid formation in each well was interpreted as a single event for the given cell density and the number of positive events over the total number of wells was considered as the spheroid-initiating probability. Data from all of the tested cell densities were compiled and analyzed with the extreme limiting dilution assay online software (http://bioinf.wehi.edu.au/software/elda)52 to yield the final spheroid-initiating frequency.
Luciferase Reporter Assays
Two truncations (T1 and T2) of the PTK7 promoter region (representative of -480 to +1 and -300 to +1 DNA sequence upstream of PTK7) were synthesized and subcloned into pGl3-basic vector (service provided by Hitrobio.tech, Beijing, China). The pGl3 vectors harboring PTK7 promoter were transiently transfected into HCC cells using Lipofectamine 3000 transfection reagent (Thermo Scientific). A pRL-CMV construct (Addgene) was delivered simultaneously into the same HCC cells as a transfection efficiency control. Transfected cells were seeded into 96-well plates to perform luciferase signal detection by the Dual-Glo luciferase assay system (Promega), according to the manufacturer’s instructions.
ChIP
Potential binding of SOX9 to the predicted binding site on the PTK7 promoter was validated by ChIP performed using the Magna ChIP G kit (Merck Millipore), according to the suggested protocol by the manufacturer. A total of 1 × 107 cells were collected for each experiment. Cells were washed with PBS and cross-linked by the addition of 1% formaldehyde (Sigma-Aldrich) (by diluting 550 μL 37% formaldehyde in 20 mL culture medium) followed by glycine neutralization. Cells were harvested by scraping and subjected to cellular and nuclear lysis following the manufacturer’s instructions. Sonication was performed to shear genomic DNA to a desirable size of 200 to 1000 base pairs. All procedures were performed in the presence of protease inhibitor cocktail II. Sheared DNA (50 μL) was incubated overnight at 4ºC with either 5 μg of an anti-SOX9 antibody (AB5535; Merck Millipore) or a purified rabbit IgG (P120-101; Bethyl Laboratories) as negative control, in the presence of protein G magnetic beads. A separate 5 μL sheared DNA was stored for input control without being subjected to any immunoprecipitation. Protein G magnetic beads with any chromatin bound were pelleted using a magnet and consecutively washed by various buffers supplied by the kit following standard protocol. After the last wash, beads were pelleted and incubated with ChIP Elution Buffer and proteinase K to release the protein-bound DNA. The same incubation was performed on the input control. All pulldown DNA fragments were purified further by spin filter columns, eluted in diethyl pyrocarbonate-treated water, and analyzed by quantitative PCR using primers specified in Table 1.
Total RNA Extraction and RNA Sequencing
Total RNA of FACS-sorted PLC/PCF/5 cells, Hep3B control cells, and Hep3B PTK7-knockout cells was harvested using the RNeasy Plus Mini Kit (Qiagen). The extracted RNA was sent to the University of Hong Kong Centre for PanorOmic Sciences for quality control, library construction, RNA-sequencing, postsequencing quality control, genome mapping, and gene expression analyses. Differentially expressed genes were first ranked according to P value and false discovery rate, then selected based on a threshold of transcript per million >1 and fold-change >1.2. Selected genes were subjected to pathway enrichment analyses by GSEA where PLC PTK7+ve cells and Hep3B control cells were grouped under high-PTK7, while PLC PTK7-ve and Hep3B knockout cells were grouped under low-PTK7.
Statistical Analyses
GraphPad Prism 7.0 (GraphPad Software) was mainly used to perform statistical analyses in this study. All graphs were generated using GraphPad Prism 7.0 and data in graphs were expressed as the means ± SD unless otherwise specified. The mean values of 2 groups were compared by an unpaired Student t test unless specified otherwise. Correlation of PTK7 and SOX9 expression in clinical tissue sections and TMA was analyzed by the chi-squared test. Correlation of PTK7 expression with advanced HCC in TCGA-LIHC was analyzed by the chi-squared test. To calculate and compare the overall survival of HCC patients between PTK7-high and PTK7-low conditions, Kaplan–Meier survival plot, Gehan–Breslow–Wilcoxon, and log-rank tests were used. The threshold for statistical significance was expressed as P values unless stated otherwise.
Acknowledgments
The authors thank the Centre for PanorOmic Sciences (The University of Hong Kong) for providing and maintaining the equipment and technical support needed for transcriptome sequencing, animal imaging, and confocal microscopy studies. The authors also thank the Centre for Comparative Medicine (The University of Hong Kong) for supporting our animal work studies.
CRediT Authorship Contributions
Matthew Tsz Lam Wong (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Tin-Lok Wong (Investigation: Supporting; Methodology: Supporting; Validation: Supporting; Writing – review & editing: Supporting)
Lei Zhou (Investigation: Supporting; Methodology: Supporting)
Kwan Man (Resources: Supporting)
James Purcell (Resources: Supporting)
Terence K Lee (Resources: Supporting)
Jing-Ping Yun (Resources: Supporting)
Stephanie Ma, PhD (Conceptualization: Equal; Formal analysis: Equal; Funding acquisition: Lead; Project administration: Lead; Resources: Lead; Validation: Equal; Writing – original draft: Equal; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the Health and Medical Research Fund from the Food and Health Bureau of the Hong Kong Government (05161756) and the Research Grants Council of Hong Kong - Research Fellow Scheme (RFS2122-7S05). Also supported by the Guangdong Science and Technology Department (2020B1212030004).
References
- 1.Reynolds A.R., Furlan A., Fetzer D.T., Sasatomi E., Borhani A.A., Heller M.T., Tublin M.E. Infiltrative hepatocellular carcinoma: what radiologists need to know. Radiographics. 2015;35:371–386. doi: 10.1148/rg.352140114. [DOI] [PubMed] [Google Scholar]
- 2.Katyal S., Oliver J.H., III Extrahepatic metastases of hepatocellular carcinoma. Radiology. 2000;216:698–703. doi: 10.1148/radiology.216.3.r00se24698. [DOI] [PubMed] [Google Scholar]
- 3.Fidler I.J. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 4.Gupta G.P., Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Valastyan S., Weinberg R.A. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fares J., Fares M.Y., Khachfe H.H., Salhab H.A., Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther. 2020;5:1–17. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambert A.W., Pattabiraman D.R., Weinberg R.A. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang A., Yang X.R., Chung W.Y., Dennison A.R., Zhou J. Targeted therapy for hepatocellular carcinoma. Signal Transduct Target Ther. 2020;5:1–13. doi: 10.1038/s41392-020-00264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung J.W., Shin W.S. Cloning and characterization of the full-length mouse Ptk7 cDNA encoding a defective receptor protein tyrosine kinase. Gene. 2004;328:75–84. doi: 10.1016/j.gene.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Murphy J.M., Zhang Q., Young S.N., Reese M.L., Bailey F.P., Eyers P.A., Ungureanu D., Hammaren H., Silvennoinen O., Varghese L.N., Chen K., Tripaydonis A., Jura N., Fukuda K., Qin J., Nimchuk Z., Mudgett M.B., Elowe S., Gee C.L., Liu L., Daly R.J., Manning G., Babon J.J., Lucet I.S. A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem J. 2014;457:323–334. doi: 10.1042/BJ20131174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park S.K., Lee H.S. Characterization of the human full-length PTK7 cDNA encoding a receptor protein tyrosine kinase-like molecule closely related to chick KLG. J Biochem. 1996;119:235–239. doi: 10.1093/oxfordjournals.jbchem.a021228. [DOI] [PubMed] [Google Scholar]
- 12.Lu X., Borchers A.G. PTK7/CCK-4 is a novel regulator of planar cell polarity in vertebrates. Nature. 2004;430:93–98. doi: 10.1038/nature02677. [DOI] [PubMed] [Google Scholar]
- 13.Shnitsar I., Borchers A. PTK7 recruits DSH to regulate neural crest migration. Development. 2008;135:4015–4024. doi: 10.1242/dev.023556. [DOI] [PubMed] [Google Scholar]
- 14.Berger H., Breuer M. PTK7 localization and protein stability is affected by canonical Wnt ligands. J Cell Sci. 2017;130:1890–1903. doi: 10.1242/jcs.198580. [DOI] [PubMed] [Google Scholar]
- 15.Bin-Nun N., Lichtig H. PTK7 modulates Wnt signaling activity via LRP6. Development. 2014;141:410–421. doi: 10.1242/dev.095984. [DOI] [PubMed] [Google Scholar]
- 16.Martinez S., Scerbo P., Giordano M., Daulat A.M., Lhoumeau A.C., Thome V., Kodjabachian L., Borg J.P. The PTK7 and ROR2 protein receptors interact in the vertebrate WNT/planar cell polarity (PCP) pathway. J Biol Chem. 2015;290:30562–30572. doi: 10.1074/jbc.M115.697615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peradziryi H., Kaplan N.A., Podleschny M., Liu X., Wehner P., Borchers A., Tolwinski N.S. PTK7/Otk interacts with Wnts and inhibits canonical Wnt signaling. EMBO J. 2017;30:3729–3740. doi: 10.1038/emboj.2011.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen R., Khatri P., Mazur P.K., Polin M., Zheng Y., Vaka D., Hoang C.D., Shrager J., Xu Y., Vicent S., Butte A.J., Sweet-Cordero E.A. A meta-analysis of lung cancer gene expression identifies PTK7 as a survival gene in lung adenocarcinoma. Cancer Res. 2014;74:2892–2902. doi: 10.1158/0008-5472.CAN-13-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ataseven B., Angerer R., Kates R., Gunesch A., Knyazev P., Hogel B., Becker C., Eiermann W., Harbeck N. PTK7 expression in triple-negative breast cancer. Anticancer Res. 2013;33:3759–3763. [PubMed] [Google Scholar]
- 20.Gärtner S., Gunesch A., Knyazeva T., Wolf P., Hogel B., Eiermann W., Ullrich A., Knyazev P., Ataseven B. PTK7 is a transforming gene and prognostic marker for breast cancer and nodal metastasis involvement. PLoS One. 2014;9 doi: 10.1371/journal.pone.0084472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin W.S., Gim J. Biphasic regulation of tumorigenesis by PTK7 expression level in esophageal squamous cell carcinoma. Sci Rep. 2018;8:1–9. doi: 10.1038/s41598-018-26957-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin W.S., Na H.W. Biphasic effect of PTK7 on KDR activity in endothelial cells and angiogenesis. Biochim Biophys Acta Mol Cell Res. 2015;1853:2251–2260. doi: 10.1016/j.bbamcr.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 23.Lhoumeau A.C., Martinez S., Boher J.M., Monges G., Catellano R., Goubard A., Doremus M., Poizat F., Lelong B., de Chaisemartin C., Bardin F., Viens P., Raoul J.L., Prebet T., Aurrand-Lions M., Borg J.P., Goncalves A. Overexpression of the promigratory and prometastatic PTK7 receptor is associated with an adverse clinical outcome in colorectal cancer. PLoS One. 2015;10 doi: 10.1371/journal.pone.0123768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian X., Yan L., Zhang D., Guan X., Dong B., Zhao M., Hao C. PTK7 overexpression in colorectal tumors: clinicopathological correlation and prognosis relevance. Oncol Rep. 2016;36:1829–1836. doi: 10.3892/or.2016.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roessler S., Jia H.L., Budhu A., Forgues M., Ye Q.H., Lee J.S., Thorgeirsson S.S., Sun Z., Tang Z.Y., Qin L.X., Wang X.W. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger H., Wodarz A., Borchers A. PTK7 faces the Wnt in development and disease. Front Cell Dev Biol. 2017;5:31. doi: 10.3389/fcell.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J., Zaidi S., Rao S., Chen J.S., Phan L., Farci P., Su X., Shetty K., White J., Zamboni F., Wu X., Rashid A., Pattabiraman N., Mazumder R., Horvath A., Wu R.C., Li S., Xiao C., Deng C.X., Wheeler D.A., Mishra B., Akbani R., Mishra L. Analysis of genomes and transcriptomes of hepatocellular carcinomas identifies mutations and gene expression changes in the transforming growth factor-β pathway. Gastroenterology. 2018;154:195–210. doi: 10.1053/j.gastro.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas D.A., Massagué J. TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8:369–380. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Gunderson A.J., Yamazaki T., McCarty K., Fox N., Phillips M., Alice A., Blair T., Whiteford M., O’Brien D., Ahmad R., Kiely M.X., Hayman A., Crocenzi T., Gough M.J., Crittenden M.R., Young K.H. TGFβ suppresses CD8+ T cell expression of CXCR3 and tumor trafficking. Nat Commun. 2020;11:1–13. doi: 10.1038/s41467-020-15404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Cai H., Deng J., Tu X., Sun Y., Huang Z., Ding Z., Dong L., Chen J., Zang Y., Zhang J. TGF-β-mediated upregulation of Sox9 in fibroblast promotes renal fibrosis. Biochim Biophys Acta Mol Basis Dis. 2018;1864:520–532. doi: 10.1016/j.bbadis.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 31.Coricor G., Serra R. TGF-β regulates phosphorylation and stabilization of Sox9 protein in chondrocytes through p38 and Smad dependent mechanisms. Sci Rep. 2016;6:1–11. doi: 10.1038/srep38616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chavez R.D., Coricor G., Perez J., Seo H.S., Serra R. SOX9 protein is stabilized by TGF-β and regulates PAPSS2 mRNA expression in chondrocytes. Osteoarthr Cartil. 2017;25:332–340. doi: 10.1016/j.joca.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirano T., Saito D., Yamada H., Ishisaki A., Kamo M. TGF-β1 induces N-cadherin expression by upregulating Sox9 expression and promoting its nuclear translocation in human oral squamous cell carcinoma cells. Oncol Lett. 2020;20:474–482. doi: 10.3892/ol.2020.11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao X., Tey S.K., Yeung C.L., Kwong E.M., Fung Y.M., Chung C.Y., Mak L.Y., Wong D.K., Yuen M.F., Ho J.C., Pang H., Wong M.P., Leung C.O., Lee T.K., Ma V., Cho W.C., Cao P., Xu X., Gao Y., Yam J.W. Nidogen 1-enriched extracellular vesicles facilitate extrahepatic metastasis of liver cancer by activating pulmonary fibroblasts to secrete tumor necrosis factor receptor 1. Adv Sci. 2020;7 doi: 10.1002/advs.202002157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loh J.J., Li T.W., Zhou L., Wong T.L., Liu X., Ma V.W., Lo C.M., Man K., Lee T.K., Ning W., Tong M., Ma S. FSTL1 secreted by activated fibroblasts promotes hepatocellular carcinoma metastasis and stemness. Cancer Res. 2021;81:5692–5705. doi: 10.1158/0008-5472.CAN-20-4226. [DOI] [PubMed] [Google Scholar]
- 36.Luk S.T., Ng K.Y., Zhou L., Tong M., Wong T.L., Yu H., Lo C.M., Man K., Guan X.Y., Lee T.K., Ma S. Deficiency in embryonic stem cell marker REX1 activates MKK6-dependent p38 MAPK signaling to drive hepatocarcinogenesis. Hepatology. 2019;72:183–197. doi: 10.1002/hep.31020. [DOI] [PubMed] [Google Scholar]
- 37.Ng K.Y., Chan L.H., Chai S., Tong M., Guan X.Y., Lee N.P., Yuan Y.F., Xie D., Lee T.K., Dusetti N.J., Carrier A., Ma S. TP53INP1 downregulation activates a p73-dependent DUSP10/ERK signaling pathway to promote metastasis of hepatocellular carcinoma. Cancer Res. 2017;77:4602–4612. doi: 10.1158/0008-5472.CAN-16-3456. [DOI] [PubMed] [Google Scholar]
- 38.Bhullar K.S., Lagarón N.O., McGowan E.M., Parmar I., Jha A., Hubbard B.P., Rupasinghe H.P. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol Cancer. 2018;17:1–20. doi: 10.1186/s12943-018-0804-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kung J.E., Jura N. Prospects for pharmacological targeting of pseudokinases. Nat Rev Drug Discov. 2019;18:501–526. doi: 10.1038/s41573-019-0018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheetz J.B., Mathea S., Karvonen H., Malhotra K., Chatterjee D., Niininen W., Perttila R., Preuss F., Suresh K., Stayrook S.E., Tsutsui Y., Radhakrishnan R., Ungureanu D., Knapp S., Lemmon M.A. Structural insights into pseudokinase domains of receptor tyrosine kinases. Mol Cell. 2020;79:390–405. doi: 10.1016/j.molcel.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawai T., Yasuchika K., Ishii T., Miyauchi Y., Kojima H., Yamaoka R., Katayama H., Yoshitoshi E.Y., Ogiso S., Kita S., Yasuda K., Fukumitsu K., Komori J., Hatano E., Kawaguchi Y., Uemoto S. SOX9 is a novel cancer stem cell marker surrogated by osteopontin in human hepatocellular carcinoma. Sci Rep. 2016;6:1–11. doi: 10.1038/srep30489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu C., Liu L., Chen X., Cheng J., Zhang H., Shen J., Shan J., Xu Y., Yang Z., Lai M., Qian C. Sox9 regulates self-renewal and tumorigenicity by promoting symmetrical cell division of cancer stem cells in hepatocellular carcinoma. Hepatology. 2016;64:117–129. doi: 10.1002/hep.28509. [DOI] [PubMed] [Google Scholar]
- 43.Richtig G., Aigelsreiter A., Schwarzenbacher D., Ress A.L., Adiprasito J.B., Stiegelbauer V., Hoefler G., Schauer S., Kiesslich T., Kornprat P., Winder T., Eisner F., Gerger A., Stoeger H., Stauber R., Lackner C., Pichler M. SOX9 is a proliferation and stem cell factor in hepatocellular carcinoma and possess widespread prognostic significance in different cancer types. PLoS One. 2017;12 doi: 10.1371/journal.pone.0187814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Golubkov V.S., Prigozhina N.L., Zhang Y., Stoletov K., Lewis J.D., Schwartz P.E., Hoffman R.M., Strongin A.Y. Protein-tyrosine pseudokinase 7 (PTK7) directs cancer cell motility and metastasis. J Biol Chem. 2014;289:24238–24249. doi: 10.1074/jbc.M114.574459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Golubkov V.S., Strongin A.Y. Downstream signaling and genome-wide regulatory effects of PTK7 pseudokinase and its proteolytic fragments in cancer cells. Cell Commun Signal. 2014;12:15. doi: 10.1186/1478-811X-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Na H.W., Shin W.S. The cytosolic domain of protein-tyrosine kinase 7 (PTK7), generated from sequential cleavage by a disintegrin and metalloprotease 17 (ADAM17) and γ-secretase, enhances cell proliferation and migration in colon cancer cells. J Biol Chem. 2012;287:25001–25009. doi: 10.1074/jbc.M112.348904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Messerli S.M., Hoffman M.M. Therapeutic targeting of PTK7 is cytotoxic in atypical teratoid rhabdoid tumors. Mol Cancer Res. 2017;15:973–983. doi: 10.1158/1541-7786.MCR-16-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Damelin M., Bankovich A., Bernstein J., Lucas J., Chen L., Williams S., Park A., Aguilar J., Ernstoff E., Charati M., Dushin R., Aujay M., Lee C., Ramoth H., Milton M., Hampl J., Lazetic S., Pulito V., Rosfjord E., Sun Y., King L., Barletta F., Betts A., Guffroy M., Falahatpisheh H., O'Donnell C.J., Stull R., Pysz M., Escarpe P., Liu D., Foord O., Gerber H.P., Sapra P., Dylla S.J. A PTK7-targeted antibody-drug conjugate reduces tumor-initiating cells and induces sustained tumor regressions. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aag2611. [DOI] [PubMed] [Google Scholar]
- 49.Jie Y., Liu G., Feng L., Li Y., E M., Wu L., Li Y., Rong G., Li Y., Wei H., Gu A. PTK7-targeting CAR T-cells for the treatment of lung cancer and other malignancies. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.665970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sicco E., Mónaco A. Metastatic and non-metastatic melanoma imaging using Sgc8-c aptamer PTK7-recognizer. Sci Rep. 2021;11:1–12. doi: 10.1038/s41598-021-98828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin W.S., Maeng Y.S., Jung J.W., Min J.K., Kwon Y.G., Lee S.T. Soluble PTK7 inhibits tube formation, migration, and invasion of endothelial cells and angiogenesis. Biochem Biophys Res Commun. 2008;371:793–798. doi: 10.1016/j.bbrc.2008.04.168. [DOI] [PubMed] [Google Scholar]
- 52.Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]