Abstract
Infertility affects 10%–20% of the population in most countries, with approximately half of those cases resulting from male infertility. Although millions of infertile men are able to have children with the assistance of reproductive technology, individuals with non-obstructive azoospermia (NOA) syndrome are unable to do so because they lack functional sperm. Therefore, some other strategies for infertile NOA men are still urgently needed. Our current study uses an NOA-like mouse model to optimize microinjection and a subsequent electroporation method to test potential treatment strategies. We showed that the spermatogenetic process could be partially rescued in young Stra8-Rnf20−/− mice with microinjection and subsequent electroporation of Rnf20 plasmids into the testes. All meiotic prophase I stages could be identified, and programmed DNA double-strand break repair factors could successfully be recruited to Stra8-Rnf20−/− spermatocytes after electroporation. Moreover, by including an autophagy inhibitor in the treatment, electroporation significantly improved the spermatogenetic rescue efficiency of adult Stra8-Rnf20−/− mice. Most importantly, infertility caused by Rnf20 depletions could be overcome by electroporation coupled with intracytoplasmic sperm injection. Our studies establish a relative safe and efficient testis electroporation system and provide a promising therapeutic strategy for patients with NOA.
Keywords: MT: Delivery Strategies, male infertility, non-obstructive azoospermia, electroporation, autophagy, 3-methyladenine
Graphical abstract
Wang et al. report that testis electroporation coupled with autophagy inhibitor could rescue the spermatogenesis process of a non-obstructive azoospermia-like mouse model. These results establish a safe and efficient testes electroporation strategy to overcome male infertility, potentially providing a way to treat individuals with NOA.
Introduction
Fertility problems affect approximately 10%–20% of couples worldwide, and nearly half of the infertility cases are attributed to the male.1,2 Several factors contribute to male infertility, including spermatogenic disorder, obstruction of the seminal tract, abnormal sperm function, decreased sperm production, inflammation and some sexual disorders.3 According to pathogenesis, three ways to treat male infertility are available based on early diagnosis: medical treatment, surgical treatment or assisted reproduction technology (ART).4 ART has been used for decades and is steadily improving. The two most common methods are in vitro fertilization (IVF), a process by which an oocyte is fertilized by semen outside the body, and intracytoplasmic sperm injection (ICSI), a procedure in which a single sperm cell is directly injected into the cytoplasm of an oocyte.5,6,7 In recent years, ART has substantially improved birth rates, especially for infertility caused by genetic abnormalities.8 However, ART seems powerless with non-obstructive azoospermia (NOA) syndrome because functional haploid germ cells are absent. NOA results from spermatogenic failure that is defined by the absence of spermatozoa in seminal fluid and is the most severe form of male infertility.9,10 It has been reported that up to 35% of males with infertility problems were diagnosed with NOA. Currently, no strategy exists to restore spermatogenesis in the majority of individuals with NOA, apart from those with secondary testicular failure.11 Thus, the establishment of a method for restoring normal spermatogenesis to rescue male infertility is urgently needed, especially for men with NOA syndrome.
Given the genetic basis of NOA, delivering normal genes to affected spermatocytes or spermatids is the best way to overcome spermatogenic defects. Gene delivery to germ cells can be accomplished using genetically modified viruses, modified spermatogonial stem cells (SSC), and in vivo electroporation.12,13,14,15 Due to ethical issues, the integration of exogenous DNA into the genome of a human germ cell is currently not allowed; therefore, we can select to transiently express DNA delivered by microinjection and subsequent electroporation to treat NOA. Recent studies have shown that delivering DNA constructs into living mouse germ cells could be accomplished by electroporation after microinjecting DNA solutions into the seminiferous tubules. In mouse testes, microinjection of a DNA construct followed by electroporation led to the expression and localization of protein and to the observation of telomere movement during meiosis.15,16,17,18,19,20 Therefore, electroporation can be considered a promising method to deliver healthy genes into affected individuals, resulting in functional spermatozoa or even restoring male fertility.
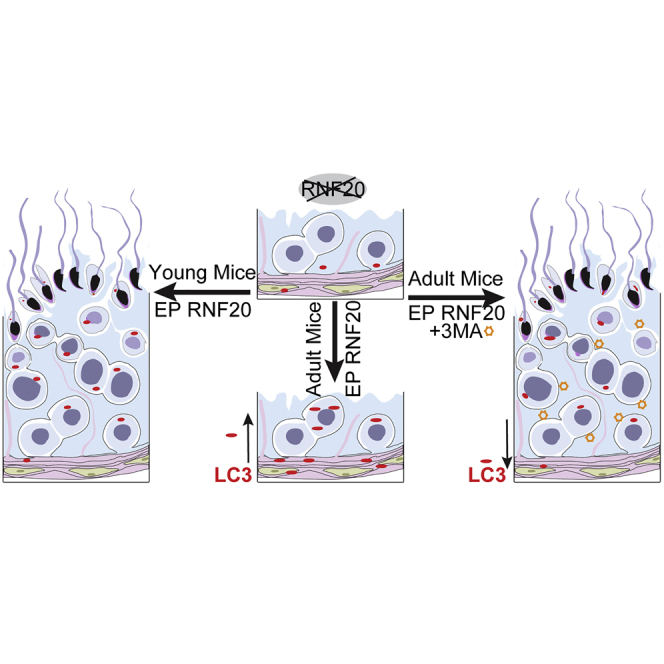
Previously, we developed an NOA-like mouse model with a germ-cell-specific Rnf20 knockout, resulting in almost a complete loss of spermatozoa in mice.9 Here, we use this model to test the possibility of microinjection and subsequent electroporation-based NOA treatment. After microinjecting and electroporating pCS2+-RNF20 plasmid into seminiferous tubules, the spermatogenetic defect of young, but not adult, Stra8-Rnf20−/− mice was shown to be partially rescued. The testes of rescued young Stra8-Rnf20−/− mice were bigger than those of young Stra8-Rnf20−/− mice. In electroporated young Stra8-Rnf20−/− mice, mature spermatozoa were found in some seminiferous tubules and the caudal epididymis. Immunostaining results further showed that the expression and localization of γH2AX and MRE11 were restored to some extent in the spermatocytes. Moreover, by including an autophagy inhibitor, we found that the spermatogenetic defect of adult Stra8-Rnf20−/− mice could also be rescued by electroporation. Most importantly, we found that healthy offspring were obtained from the spermatozoa of either electroporated young or adult Stra8-Rnf20−/− mice. Overall, these results establish a safe and efficient testes electroporation strategy to overcome male infertility, potentially providing a special way to treat individuals with NOA.
Results
In vivo microinjection and electroporation of mouse testes
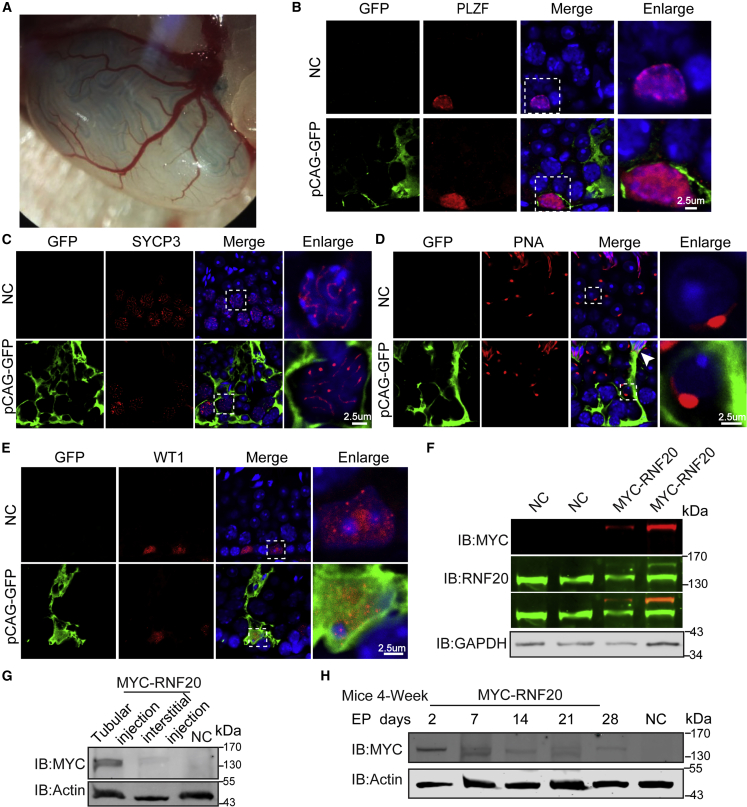
Testes DNA microinjection and electroporation have been well established to deliver exogenous DNA into the germ cells of live mice. Because the genes are only transiently expressed in the germ cells, genetic inheritance does not occur with progeny.15,16,18 However, this transitory technique could provide a unique opportunity as a male infertility treatment, circumventing ethical issues that accompany DNA integration. To test this possibility, we constructed GFP-tagged full-length cDNAs of RNF20 in pCAG-GFP plasmids. Next, pCAG-GFP and pCAG-GFP-RNF20 plasmids were solubilized in 1× HBS buffer to a final concentration of 7 μg/μL, respectively. Finally, 9 μL was mixed with 1 μL 0.1% FastGREEN (Figure 1A). The 10 μL mixture was then microinjected into the testes of live mice followed by electroporation. Two days post-injection, the testes were observed under microscopy, and GFP was found to be highly expressed (Figure S1A). GFP proteins were found to be localized in SSCs, spermatocyte cells, round spermatids, elongated spermatids, and Sertoli cells (Figures 1B–1E). However, the expression of GFP-RNF20 was much weaker than that of pCAG-GFP, and GFP-RNF20 was hardly detected by western blot (Figure S1B). We postulated that the constructs might impair GFP expression or be easily degraded, to get a robust and efficient transfection method, we constructed six MYC-tagged full-length cDNAs of RNF20 in pCS2+ plasmids, which were then subjected to electroporation. Western blot results showed that MYC-RNF20 was successfully expressed after electroporation (Figures 1F and S1C), and the efficiency of tubular microinjection was higher than that of interstitial injection (Figure 1G). The MYC-RNF20 protein could still be detected for a month after in vivo tubular microinjection and electroporation (Figure 1H). These results suggest that an exogenous gene can be successfully expressed in testes after in vivo microinjection and subsequent electroporation.
Figure 1.
In vivo microinjection and electroporation of mouse testes
(A) Macroscopic appearance of a testis transfected with reporter plasmids. Microinjection involved efferent duct injection and filled 70%–80% of the tubules in each recipient testis. (B–E) The localization of GFP protein in testis. Immunofluorescence analysis of GFP localization in testis after electroporation. Promyelocytic leukemia zinc finger (PLZF) and SYCP3 served as markers of SSCs and spermatocyte cells, respectively. PNA is an established marker of the mammalian acrosome, and WT1 served as a marker for Sertoli cells. The white arrowhead indicates GFP signal in elongated spermatids. Scale bar, 2.5 μm. (F and G) Western blot analysis of testis extracts after electroporation. GAPDH and actin served as loading controls. (H) Western blot analysis of testis extracts after different times of electroporation. Actin served as a loading control.
Electroporation partially rescues spermatogenetic processes in Stra8-Rnf20−/− mice
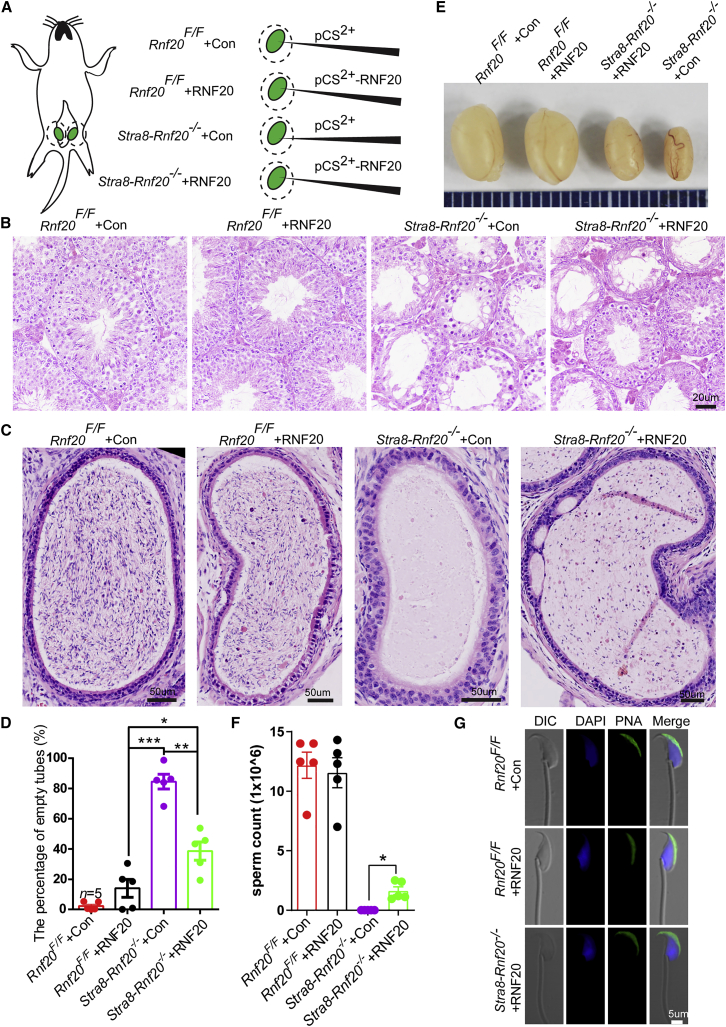
Previously, we found that germ-cell-specific Rnf20 knockout results in an NOA-like phenotype in mice.9 To study the potential of electroporation on NOA treatment, we turned to this mouse model. Since 17 to 30 days post-partum (dpp) testes have been reported to have higher electroporation efficiencies than 60 dpp testes,15 we carried out in vivo microinjection and electroporation on young mouse testes at 4 weeks of age. As shown in Figure 2A, pCS2+ plasmids were injected into wild-type (WT) and knockout mice (hereafter referred to as Rnf20F/F + Con and Stra8-Rnf20−/− + Con), and pCS2+-RNF20 plasmids were also injected into wild-type and Rnf20 knockout mice (hereafter referred to as Rnf20F/F + RNF20 and Stra8-Rnf20−/− + RNF20). At 37 days post-electroporation, histological examination of the testes and epididymal lumens showed that the pCS2+-RNF20 plasmids significantly improved the spermatogenetic processes in Stra8-Rnf20−/− mice (Figures 2B–2D). Most strikingly, the testes of the Stra8-Rnf20−/− + RNF20 mice after electroporation were larger than those of the Stra8-Rnf20−/− + Con mice (Figure 2E), and we could successfully harvest spermatozoa from Stra8-Rnf20−/− + RNF20 caudal epididymis (Figure 2F). To further test whether the spermatozoa of Stra8-Rnf20−/− + RNF20 mice have any impact on the acrosome, single-sperm immunofluorescence was performed using peanut agglutinin (PNA; staining the acrosome specifically) and 4′,6-diamidino-2-phenylindole (DAPI). Indeed, the spermatozoa of Stra8-Rnf20−/− + RNF20 mice were positive for both PNA and DAPI staining (Figure 2G). These results demonstrate that the spermatogenetic defect of Stra8-Rnf20−/− mice was partially rescued by microinjecting and electroporating pCS2+-RNF20 plasmid into seminiferous tubules.
Figure 2.
Electroporation partially rescues spermatogenetic processes in young Stra8-Rnf20−/− mice
(A) Diagram of in vivo electroporation procedures for mouse testes. (B) Histological analysis of the seminiferous tubules of the Rnf20F/F + Con, Rnf20F/F + RNF20, Stra8-Rnf20−/− + Con, and Stra8-Rnf20−/− + RNF20 mice after electroporation. Scale bar, 20 μm. (C) Histological analysis of the caudal epididymis of the Rnf20F/F + Con, Rnf20F/F + RNF20, Stra8-Rnf20−/− + Con, and Stra8-Rnf20−/− + RNF20 mice after electroporation. Scale bar, 50 μm. (D) Percentage of empty tubes. n = 5 biological replicates. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. (E) Testes of Stra8-Rnf20−/− + RNF20 mice were larger than those of the Stra8-Rnf20−/− + Con mice. (F) Sperm concentration of the Rnf20F/F + Con, Rnf20F/F + RNF20, and Stra8-Rnf20−/− + RNF20 mice after electroporation. n = 5 biological replicates. Data are presented as mean ± SEM. ∗p < 0.05. Statistical analysis was performed with two-tailed unpaired Student’s t test. (G) Morphological analysis of the spermatozoa in the Rnf20F/F + Con, Rnf20F/F + RNF20, and Stra8-Rnf20−/− + RNF20 mice after electroporation. Acrosomes were stained with PNA (green), and nuclei were stained with DAPI (blue). Scale bar, 5 μm.
Electroporation partially rescues meiotic pachytene arrest in Stra8-Rnf20−/− mice
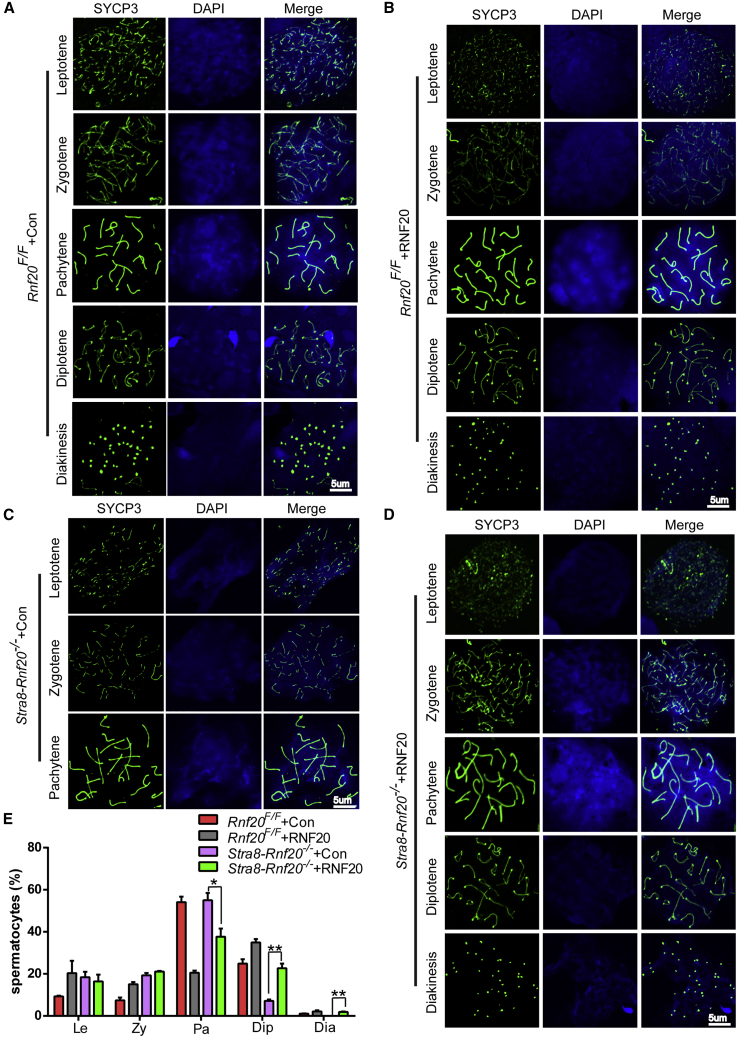
Spermatogenesis is a complex biological process that produces male haploid germ cells from diploid SSCs that undergo a series of developmental steps, including spermatogonial mitosis, spermatocytic meiosis, and spermiogenesis.21,22 Since Rnf20 knockout has been reported to lead to spermatocyte arrest at the pachytene and meiotic division stages,9 we speculated that electroporation might rescue meiotic pachytene arrest. To test this possibility, we observed various stages of meiotic prophase I in spermatocyte nucleus spreads by staining for a component of the synaptonemal complex, SYCP3.23 In the control group, all meiotic prophase I stages could be identified in the spermatocyte nuclei of Rnf20F/F + Con and Rnf20F/F + RNF20 testes (Figures 3A and 3B). As shown in Figure 3C, only spermatocytes during the leptotene to the pachytene stages were observed in Stra8-Rnf20−/− + Con testes, which is consistent with the previously reported NOA-like phenotype, whereas in the testes of the Stra8-Rnf20−/− + RNF20 mice, every prophase I stage could be identified (Figure 3D). Further quantification of the meiotic prophase I stage in testes indicated that the proportions of every prophase I-stage spermatocyte in Stra8-Rnf20−/− + RNF20 testes were similar to those of Rnf20F/F + Con and Rnf20F/F + RNF20 testes. The percentage of the pachytene stage cells was significantly increased, and zygotene stage cells decreased, in Stra8-Rnf20−/− + RNF20 testes compared with that of Stra8-Rnf20−/− + Con testes (Figure 3E). Together, these results suggest that electroporation partially rescues meiotic pachytene arrest in Stra8-Rnf20−/− mice.
Figure 3.
Electroporation partially rescues meiotic pachytene arrest in Stra8-Rnf20−/− mice
(A–D) Meiotic chromosome spreads from Rnf20F/F + Con, Rnf20F/F + RNF20, Stra8-Rnf20−/− + Con, and Stra8-Rnf20−/− + RNF20 male mice were stained with anti-SYCP3 antibody (green) and DAPI (blue). Scale bar, 5 μm. (E) Frequencies of meiotic stages in Rnf20F/F + Con, Rnf20F/F + RNF20, Stra8-Rnf20−/− + Con, and Stra8-Rnf20−/− + RNF20 testes suspensions. n = 3 biological replicates. Data are presented as mean ± SEM. ∗p < 0.05 and ∗∗p < 0.01. Statistical analysis was performed with two-tailed unpaired Student’s t test.
Programmed DSB repair factors can be successfully recruited to Stra8-Rnf20−/− spermatocytes after electroporation
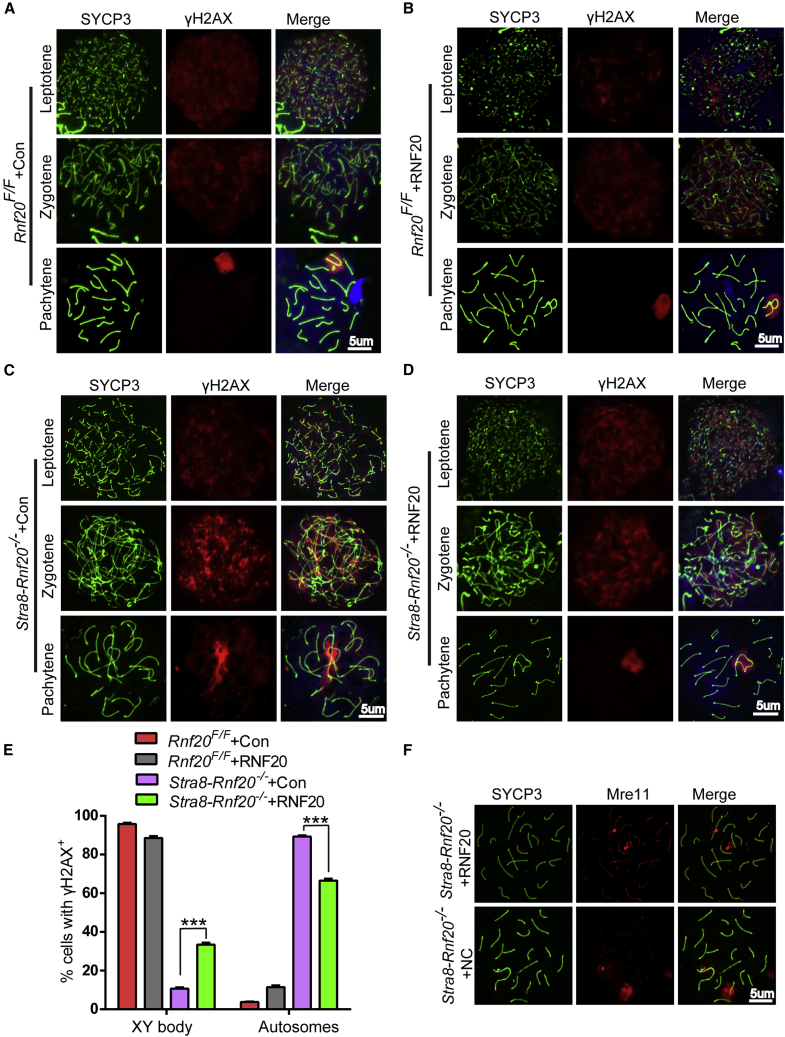
Recombination is a prominent feature of meiosis, playing an important role in increasing genetic diversity. Meiotic recombination is also essential for accurate pairing and segregation of homologous chromosomes.24,25 Meiotic programmed double-strand breaks (DSBs) recruit a series of DNA repair factors to form recombination foci and promote homologous chromosome synapsis and crossover/non-crossover.23,26,27 Rnf20 knockout has been demonstrated to affect the efficient repair of programmed DSBs due to insufficient recruitment of repair factors in Stra8-Rnf20−/− spermatocytes.9 To determine whether programmed DSB repair factors could be recruited to Stra8-Rnf20−/− spermatocytes after electroporation, we evaluated the localization of several repair factors by chromosome spreading coupled with immunofluorescent staining. γH2AX is a meiotic marker that distributes globally in the nucleus during the leptotene stage and begins to disappear during the zygotene stage. We found that the marker decreased in the autosomes and only concentrated in the XY body during the pachytene stage (Figures 4A and 4B).28 However, γH2AX signals in Stra8-Rnf20−/− + Con spermatocytes were modestly weaker than in Stra8-Rnf20−/− + RNF20 spermatocytes during the leptotene stage. During the pachytene stage, the γH2AX signals in Stra8-Rnf20−/− + Con spermatocytes were still diffused in the autosomes (Figures 4C and 4D). Further quantification of γH2AX outside of the sex chromosomes in Stra8-Rnf20−/− + Con and Stra8-Rnf20−/− + RNF20 spermatocytes during the pachytene stage also suggested that γH2AX could be recruited to Stra8-Rnf20−/− spermatocytes after electroporation (Figure 4E).
Figure 4.
Programmed DSB repair factors can be recruited to Stra8-Rnf20−/− spermatocytes after electroporation
(A–D) Chromosome spreads of Rnf20F/F + Con, Rnf20F/F + RNF20, Stra8-Rnf20−/− + Con, and Stra8-Rnf20−/− + RNF20 spermatocytes. Chromosomes were immunostained with anti-SYCP3 antibodies (green) and anti-γ-H2AX antibodies (red). Scale bar, 5 μm. (E) Percentages of cells retaining γ-H2AX outside of the sex chromosomes in Rnf20F/F + Con, Rnf20F/F + RNF20, Stra8-Rnf20−/− + Con, and Stra8-Rnf20−/− + RNF20 pachytene spermatocytes. n = 3 biological replicates. Data are presented as mean ± SEM. ∗∗∗p < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. (F) Stra8-Rnf20−/− + Con and Stra8-Rnf20−/− + RNF20 chromosome spreads of spermatocytes were immunostained with anti-SYCP3 antibodies (green) and anti-MRE11 antibodies (red). Scale bar, 5 μm.
The MRE11-RAD50-NBS1 (MRN) complex is the major catalytic protein complex for initiating the DNA damage response and coordinating and sensing the DSB pathway.29 MRE11 cooperates with NBS1/Xrs2 and RAD50 to generate single strands at DSB sites.30 MRE11 has been shown to be insufficiently recruited to programmed DSB sites in Stra8-Rnf20−/− + Con spermatocytes, which is consistent with our current results. On the other hand, we showed that MRE11 could be recruited to programmed DSB sites in Stra8-Rnf20−/− + RNF20 spermatocytes (Figure 4F). These results further confirm that electroporation could restore the meiotic DSB repair process in Stra8-Rnf20−/− mice.
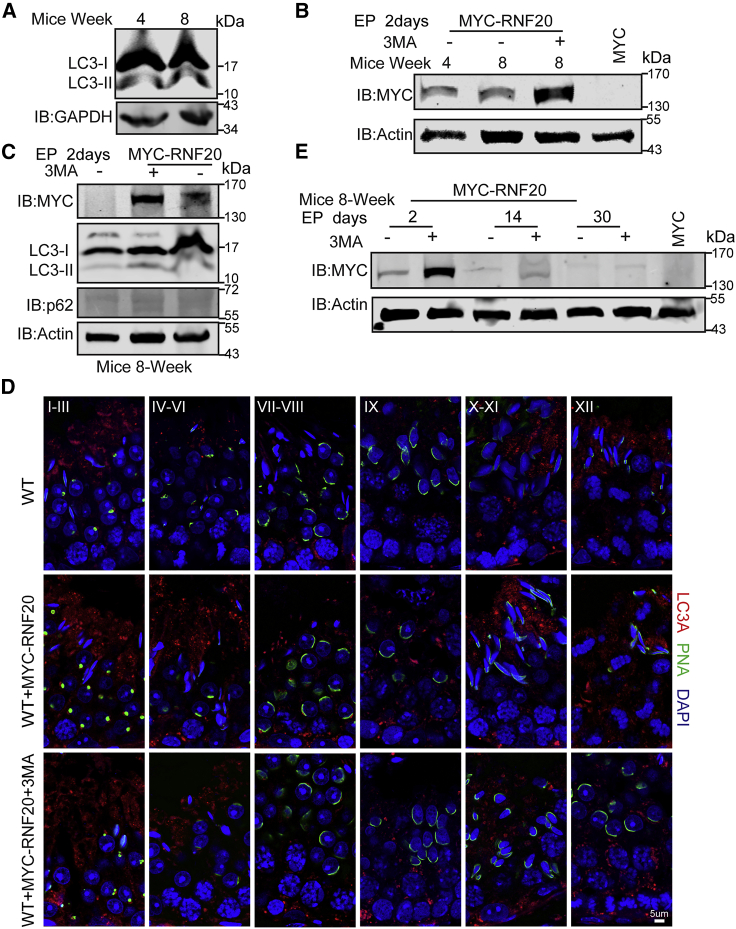
The efficiency of electroporation can be improved by inhibiting autophagy
Although electroporation works for delivering exogenous DNA to the germ cells of young male mice, delivering exogenous DNA to adult mice is more of a challenge. To improve delivery to adult mice, we considered autophagy, which has been shown to actively participate in spermatogenesis.31,32 Autophagy is a protein recycling system that delivers long-lived proteins and organelles to the lysosome for degradation. In addition to protein recycling, autophagy is also involved in other physiological processes, such as intracellular quality control, cell development and death, aging and infections.33 With infections, autophagy is reported to be an antiviral and antibacterial process of sorts that promotes the degradation of some proteins and DNA.34 As testis is an immune-exempt organ, testis may use autophagy against pathogenic microorganisms, thus preventing the expression of exogenous DNA. Microtubule-associated protein 1A/1B-light chain 3 (LC3) is an autophagosomal marker and could reflect autophagic activity.35 To test this hypothesis, we evaluated the expression of LC3 in the testes of 4- and 8-week-old male mice and found that the levels of LC3 were increased at 8 weeks compared with 4 weeks (Figures 5A and S2A). Because the efficiency of electroporation in an 8-week-old mouse is lower than that of a 4-week-old mouse (Figures 5B and S2B–S2D), autophagy might be directly involved in preventing exogenous DNA expression in the testis. To test this idea, we injected an autophagy inhibitor (3-methyladenine [3-MA]) together with RNF20 plasmids into the testis. The reduction of LC3 and the accumulation of autophagy substrate p62 suggests that the autophagic flux was blocked to some extent by 3-MA in 8-week-old mouse testes (Figures 5C, 5D, S2E, and S3A). Notably, the efficiency of electroporation was increased with injection of 3-MA into testes (Figures S2F and S2G), and expression levels of exogenous RNF20 were also increased with injection of 3-MA in 8-week-old mouse testes at different time points after electroporation (Figure 5E). These results demonstrate that autophagy may participate in preventing exogenous DNA expression in the testis, and electroporation efficiencies could be improved by inhibiting autophagy in the testis.
Figure 5.
The efficiency of electroporation is increased by adding autophagy inhibitor 3-MA in the testes of 8-week-old mice
(A) The expression levels of LC3 in the testes of 4- and 8-week-old mice. GAPDH served as a loading control. (B) Western blot analysis of testis extracts after electroporation. Actin served as a loading control. (C) Western blot analysis of the expression levels of LC3 and p62 in mouse testes after adding autophagy inhibitor 3-MA and electroporation. Actin served as a loading control. (D) The expression levels of LC3 are decreased in mouse testes that included an autophagy inhibitor 3-MA after electroporation. Scale bar, 5 μm. (E) Western blot results showed that the expression levels of exogenous RNF20 were increased after adding autophagy inhibitor 3-MA in mouse testes at different time points after electroporation. Actin served as a loading control.
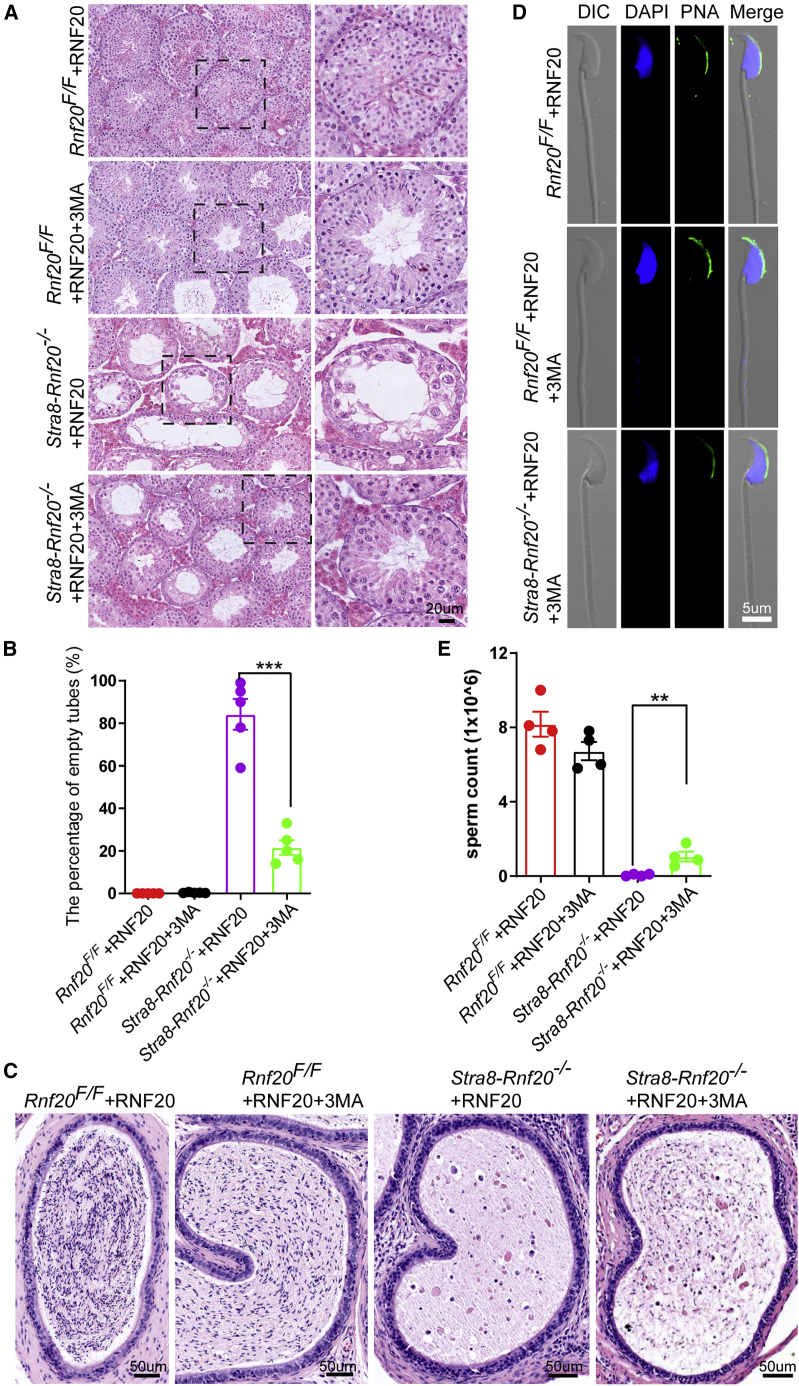
Electroporation plus 3-MA partially rescues spermatogenetic processes in adult Stra8-Rnf20−/− mice
Autophagy plays some key roles in spermatogenesis, such as spermatid differentiation, ectoplasmic specialization assembly, and acrosome biogenesis, and a higher concentration of 3-MA may result in teratospermia.31,32,36 It is required to optimize electroporation conditions to increase electroporation efficiency yet keep abnormal sperm levels low. To select the most optimized conditions, we used 3, 6, or 9 mM 3-MA solution directly injected into the testes of 8-week-old wild-type mice. We examined the spermatozoa released from the caudal epididymis and found that the injection of 3-MA into the testis resulted in teratospermia, which shows spermatozoa with coiled tails, aggregated spermatozoa, and spermatozoa with abnormal heads (Figure S4A). Moreover, the proportion of abnormal spermatozoa was dramatically increased, and total spermatozoa decreased, with high concentrations of 3-MA, whereas low (3 mM) concentrations of 3-MA had little effect on sperm count and sperm morphology (Figures S4B and S4C).
To further test the effect of the autophagy inhibitor on spermatogenesis after electroporation, we carried out microinjection and subsequent electroporation on the testes of 8-week-old adult mice. Microinjection delivered RNF20 plasmids together with or without low concentrations of 3-MA (3 mM) to wild-type and Rnf20 knockout mice (Figure 6A). At 37 days after electroporation, histological examination of the testis and epididymal lumens showed that Stra8-Rnf20−/− mice receiving RNF20 plasmids together with 3-MA clearly rescued the spermatogenetic processes (Figures 6A and 6C). Fewer empty tubules were observed in the testes of Stra8-Rnf20−/− + RNF20 + 3-MA mice than Stra8-Rnf20−/− + RNF20 mice (Figure 6B). As shown in Figure 6C, electroporation could not rescue spermatogenetic processes in 8-week-old adult Stra8-Rnf20−/− mice, which is consistent with the lower electroporation efficiencies in adult mice compared with that in young mice (Figure 5B). Notably, we successfully harvested mature sperm from Stra8-Rnf20−/− + RNF20 + 3MA mice. The spermatozoa within the Stra8-Rnf20−/− + RNF20 + 3MA caudal epididymis appeared to be morphologically normal compared with those in the control group (Figures 6D and 6E). These results demonstrate that electroporation together with an autophagy inhibitor can restore spermatogenetic processes in adult Stra8-Rnf20−/− mice.
Figure 6.
Electroporation partially rescues spermatogenetic processes in adult Stra8-Rnf20−/− mice with autophagy inhibitor 3-MA
(A) Histological analysis of the seminiferous tubules of the Rnf20F/F + RNF20, Rnf20F/F + RNF20+3-MA, Stra8-Rnf20−/− + RNF20, and Stra8-Rnf20−/− + RNF20+3-MA mice after electroporation. Scale bar, 20 μm. (B) Percentages of empty tubes. n = 5 biological replicates, Data are presented as mean ± SEM. ∗∗∗p < 0.001. Statistical analysis was performed with two-tailed unpaired Student’s t test. (C) Histological analysis of the caudal epididymis of the Rnf20F/F + RNF20, Rnf20F/F + RNF20 + 3-MA, Stra8-Rnf20−/− + RNF20, and Stra8-Rnf20−/− + RNF20 + 3-MA mice after electroporation. Scale bar, 50 μm. (D) Morphological analysis of the spermatozoa in the Rnf20F/F + RNF20, Rnf20F/F + RNF20 + 3-MA, and Stra8-Rnf20−/− + RNF20 + 3-MA mice after electroporation. Acrosomes were stained with PNA (green), and nuclei were stained with DAPI (blue). Scale bar, 5 μm. (E) Sperm concentration of the Rnf20F/F + RNF20, Rnf20F/F + RNF20 + 3-MA, and Stra8-Rnf20−/− + RNF20 + 3-MA mice after electroporation. n = 4 biological replicates, Data are presented as mean ± SEM. ∗∗p < 0.01. Statistical analysis was performed with two-tailed unpaired Student’s t test.
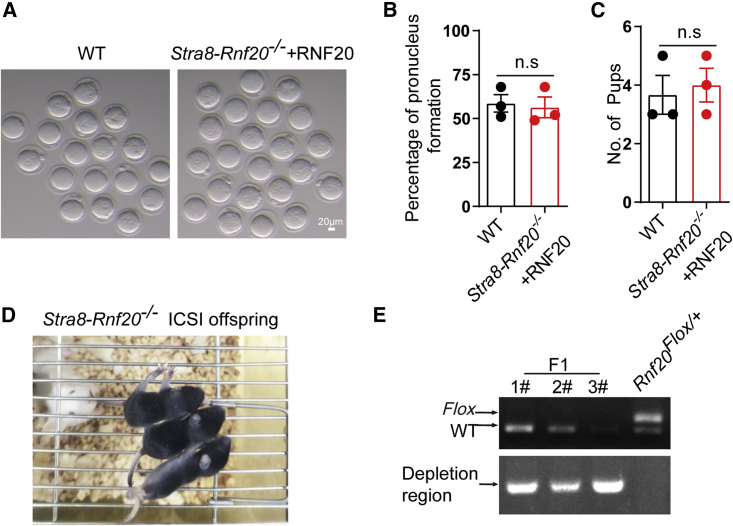
Infertility caused by Rnf20 knockout could be overcome by electroporation coupled with ICSI
To further evaluate Stra8-Rnf20−/− + RNF20 mice after electroporation, we performed a fertility test and found that the Stra8-Rnf20−/− + RNF20 male mice were still sterile, likely due to low concentrations of spermatozoa. To overcome the infertility of these NOA-like mice, we next harvested mature sperm from the Stra8-Rnf20−/− + RNF20 caudal epididymis and performed ICSI experiments, injecting spermatozoa into wild-type oocytes. There was no significant difference in the proportion of pronucleus formation and the average litter size between wild-type and Stra8-Rnf20−/− + RNF20 group (Figures 7A–7C). The ICSI resulted in healthy offspring (Figure 7D) with Rnf20+/− genotypes (Figure 7E). These results suggest that electroporation coupled with ICSI might provide an effective way to treat the infertility of individuals with NOA.
Figure 7.
Infertility caused by RNF20 depletion could be bypassed by electroporation coupled with ICSI
(A) Representative images of oocytes at 6 h after ICSI. Scale bar, 20 μm. (B) Statistical analysis of pronucleus formation of wild-type (totaling 81 injected oocytes) and Stra8-Rnf20−/− + RNF20 (totaling 125 injected oocytes) mice. n = 3 biological replicates. Data are presented as mean ± SEM. n.s., non-significant. Statistical analysis was performed with two-tailed unpaired Student’s t test. (C) The average litter size of WT and Stra8-Rnf20−/− + RNF20 mice. n = 3 biological replicates. Data are presented as mean ± SEM. n.s., non-significant. Statistical analysis was performed with two-tailed unpaired Student’s t test. (D) Representative images of Stra8-Rnf20−/− offspring resulting from electroporation coupled with ICSI. (E) Representative genotypes of the Stra8-Rnf20−/− offspring resulting from electroporation coupled with ICSI.
Discussion
Our previous studies established an NOA-like mouse model resulting from germ-cell-specific Rnf20 knockout that leads to impaired meiotic recombination and spermatocyte arrest during the pachytene stage.9 Using this mouse model, we optimized a microinjection and subsequent electroporation method that partially restored spermatogenetic processes in young Stra8-Rnf20−/− mice. To extend the method to adult mice, we disrupted autophagy, which might inhibit exogenous DNA expression in the immune-exempt testes. By inhibiting autophagy with 3-MA, we improved the electroporation efficiency and bypassed the challenge of testis transfection in adult mice (Figures 5E and 6A). Most importantly, infertility caused by RNF20 depletion could be overcome by electroporation coupled with ICSI to produce healthy offspring. Our studies establish a safe and efficient testis electroporation strategy to overcome male infertility, potentially providing a special way to treat individuals with NOA.
NOA constitutes approximately 10%–15% of all infertile men with obstructive azoospermia (OA).37 A number of genes have already been found to be associated with NOA, such as ATM, BRCA2, TEX11, MLH1, and WT1.38 Although many patients who are infertile are able to have children with the assistance of reproductive technology, no therapeutic options are available for most patients with NOA because they lack functional haploid germ cells. SSC transplantation, in vitro spermatogenesis, and gene therapy have been proposed as three potential ways to treat the infertility of individuals with NOA,39 yet practical application is impeded by either technical or ethical issues. For individuals with NOA who have received a genetic diagnosis, targeted gene therapy should be the best treatment and could be delivered with genetically modified viruses or electroporation.17,40 Studies have demonstrated that microinjection of lentiviral vectors into seminiferous tubules or inter-tubular space results in transgenic mice.14,41,42,43 Lentivirus-based in vivo gene knockdown and complementation in male germ cells have also been shown to work in some cases.44,45 However, the potential for random gene integration and other side effects with lentivirus-based delivery calls this method into question for gene therapy.
Electroporation is another useful gene delivery system that has been developed for many years, but it has only recently been translated as a testis transfection system.15,16,46 Moreover, plasmid DNA is safe for gene transfer therapy and rarely integrates with the host genome upon introduction into target cells, and use of plasmid DNA is not associated with inducing host immune responses.47,48 For example, in vivo electroporation-based SUN1-MYC expression has been shown to partially restore telomere attachment defects in Sun1−/− spermatocytes.15 In our study, we conducted microinjection and subsequent electroporation experiments in our NOA-like mouse model and found that this method could partially rescue spermatogenetic processes in Stra8-Rnf20−/− mice. Coupled with ICSI, NOA-like male mouse infertility was overcome. We also compared the efficiency between tubular injection and interstitial injection and found that tubular injection resulted in greater efficiency (Figure 1G), and MYC-RNF20 expression was also more efficient in 4-week-old mice than in 8-week-old mice (Figure 5B). These findings are consistent with previous reports showing electroporation efficiencies are correlated with age, DNA concentration and voltage of electroporation.15
To uncover the underlying mechanism, we considered that autophagy might be the major factor that inhibits exogenous DNA expression in the germ cells of testis. As a membrane-trafficking process for degrading recycling long-lived proteins and organelles, autophagy also participates as an antiviral and antibacterial process of sorts that promotes the degradation of some proteins and DNA.34 We found that the autophagic flux is negatively correlated to electroporation efficiency (Figures 5C, 5D, and S3), and electroporation efficiency is increased by adding an autophagy inhibitor (3-MA) to the testes of 8-week-old mice (Figures 5B and 5E). It is possible that exogenous DNA that is electroporated into germ cells are also degraded by the autophagy-lysosome pathway, thus preventing expression in the germ cells. A fine balance must be achieved because although an autophagy inhibitor could improve electroporation efficiency, it might also affect spermatogenesis. By considering the role of autophagy in infertility treatments, we have identified a special strategy, electroporation coupled with 3-MA, to successfully deliver exogenous DNA into adult male germ cells. This method coupled with ICSI establishes a promising way to treat the infertility of individuals with NOA, and provides a reliable platform for infertile men to have children with the assistance of reproductive technology.
Materials and methods
Generation of Rnf20 conditional knockout mice
The Rnf20Flox/Flox mice (C57BL/6J) been reported previously.9 The Rnf20Flox/Flox; Stra8-Cre mice were bred from intercrosses of Rnf20Flox/Flox mice and Stra8-Cre mice.49 The genotyping primers of Rnf20Flox/Flox mice were as follows: forward: 5′-GCTGTAAGAGTTCTTAATGTATG-3′, and reverse: 5′-GGCTTGTCACACAAGCATGAGCATC-3′. All animal studies were carried out in accordance with the protocols approved by the Institutional Animal Care and Use Committee (IACUC) protocols (#2021-002) of the Institute of Zoology, Chinese Academy of Sciences.
Plasmid DNA preparation for electroporation
pCAG-GFP and pCS2+ plasmids were used for in vivo testis microinjection and electroporation. Gene-specific cDNA sequences were inserted into these vectors. Plasmids containing the cDNA of specific genes were extracted with a MAXI-Kit (Macherey-Nagel, 740420.50), and endotoxins were removed so that in vivo immune reactions could be avoided. DNA pellets were solubilized in 1× HBS buffer (2% HEPES, 0.8% NaCl, 5 M KCl, 0.7 mM Na2HPO4, 0.1% glucose), and final plasmid concentrations were adjusted to 7 μg/μL. Prior to injection, 9 μL plasmid (7 μg/μL) was mixed together with 1 μL 0.1% FastGREEN (Sigma-Aldrich, F7258).
DNA injection into mouse testes
For performing gene transfer into mouse testes, microinjection pipettes were prepared by heating glass capillaries that were then pulled with a micropipette puller. Surgical scissors were used to break capillary tips to approximately 50–70 μm in diameter and about 1 cm long, ensuring the microinjection pipette was stiff enough to penetrate tissue. Male mice were anesthetized with 2,2,2-tribromoethanol (Sigma-Aldrich, T48402-5G) and processed for surgery. During surgery, the testes were pulled from the abdominal cavity, and plasmid DNA was injected into the rete testis using a microcapillary pipettes (Sigma-Aldrich, A5177-5EA) equipped with a glass capillary by breath pressure under a stereomicroscope. The entire injection process was monitored by observing testes filling status with the help of the FastGREEN. The testis was only partially filled up to 2/3 of its volume, and 1 h after injection, the testes were held between a pair of tweezer-type electrodes, which had been soaked in PBS (1×) to ensure effective electroporation. Electric pulses were applied four times in one direction, followed by four times in the reverse direction, at 30 V for 50 ms at 950 ms intervals per pulse using an electroporator. When electroporation was finished, the testes were placed back into the abdominal cavity, and the abdominal wall and skin were closed with sutures. In the rescue assay using Stra8-Rnf20−/− mice, 3-MA (Sigma-Aldrich, M9281, 30 mM) was dissolved in 0.9% NaCl and stored at −20°C, and 1 μL was mixed with 9 μL plasmids to a final concentration of 3 mM. Thirty-seven days after electroporation, phenotypic analysis was performed.
Tissue collection and histological analysis
For histological examination, the testes and caudal epididymis from at least three mice for each genotype were dissected and fixed with 4% paraformaldehyde (PFA; Solarbio, P1110) for up to 24 h. Next, the testes were dehydrated using graded ethanol and embedded in paraffin. Then, the testes were cut into 5 μm sections using microtome-cryostat (Leica, CM1950) and mounted on glass slides. After deparaffinization, the slides were stained with H&E for histological analysis. Images were collected with a Nikon inverted microscope with a charge-coupled device (CCD) (Nikon, Eclipse Ti-S, Japan).
Epididymal sperm count
The caudal epididymis was dissected immediately from treated mice after euthanasia. Sperm were released from the cauda epididymis and incubated at 37°C for 30 min under 5% CO2. The incubated sperm medium was then diluted 1:500 and transferred to a hemocytometer for counting.
Immunofluorescence
Testis sections were fixed with 4% PFA for 15 min and washed in PBS three times (pH 7.4). Then, the slides were treated with 0.5% Triton X-100 for 10 min and washed in PBS three times. After blocking with 5% BSA for 60 min, the slides were incubated overnight at 4°C with a primary antibody in 5% BSA containing 0.3% Triton X-100. Primary antibodies included anti-LC3 (Cell Signaling Technology, 4599); anti-P62 (Cell Signaling Technology, 5114); anti-MRE11 (Novus, NB100-142); anti-SYCP3 (Abcam, ab150292); anti-γH2AX (EMD Millipore, 05-636); anti-PLZF (Novus, BAF2944); anti-WT1 (Abcam, ab89901); anti-GFP (Abmart, M20004L); and anti-PNA (Thermo Fisher, L21409). The next day, samples were washed three times with PBS, incubated with fluorescein isothiocyanate (FITC)- or tetramethylrhodamine (TRITC)-conjugated secondary antibody at a dilution of 1:200 for 1 h at 37°C. Next, the nuclei were stained with DAPI. Goat anti-rabbit FITC-, goat anti-mouse FITC-, goat anti-rabbit TRITC-, and goat anti-mouse TRITC-conjugated secondary antibodies were purchased from Zhong Shan Jin Qiao (China). Images were collected immediately using a confocal microscope (LSM 880 Meta plus Zeiss Axiovert Zoom; Zeiss) equipped with a 63× (numerical aperture [NA] = 1.40) oil-immersion objective lens (Plan-Apochromat; Zeiss) and acquired by highly sensitive photomultiplier tube (PMT) at room temperature (RT). Images were analyzed by using ZEN software (2011_Lite_x64) and were aligned by Adobe Illustrator (CS4). Stages of seminiferous epithelium cycle were determined as previously described.22
Chromosome spreads of spermatocyte
Spermatocyte surface spreading was conducted according to the drying-down technique as previously described.50 Briefly, the testes were dissected from treated mice after euthanasia. The seminiferous tubules were washed in PBS. Subsequently, seminiferous tubules were placed in a hypotonic extraction buffer for 40 min. Then, the seminiferous tubules were torn to pieces in 100 mM sucrose (pH 8.2) on a clean glass slide. The cell suspensions were placed on slides containing 1% PFA (pH 9.2) and 0.15% Triton X-100. The slides were dried for at least 2 h and washed three times with PBS for immunofluorescence experiment according to the standard protocols mentioned above.
Immunoblotting
Immunoblotting was performed as previously described.51 Briefly, protein lysates were separated by SDS-PAGE and electro-transferred onto a nitrocellulose membrane. The membrane was incubated with corresponding primary antibodies. Primary antibodies included anti-MYC (Abmart, M20002M) and anti-actin (Abmart, M20011). The next day, the proteins on the membrane were hybridized with Alexa Fluor 680-conjugated goat anti-mouse or Alexa Fluor 800-conjugated goat anti-rabbit secondary antibodies and scanned using the ODYSSEY Sa Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA).
ICSI
ICSI was performed as previously described.52 Briefly, 8-week-old C57BL/6 mice were used to prepare mature oocyte donors. Stra8-Rnf20−/− sperm heads were collected by centrifugation in 70% Percoll (Solarbio, 65455-52-9), followed by three washes of M2 medium (Sigma-Aldrich, M7167-100ML). At RT, single sperm heads were picked from the sperm suspension and injected into WT oocytes using a micromanipulator with a piezoelectric actuating pipette. The injected oocytes were transferred into the oviducts of pseudo-pregnant ICR females. Full-term pups derived from ICSI embryos were obtained through natural labor.
Statistical analysis
All data are presented as the mean ± SEM. The statistical significance of the difference between the mean values for the different genotypes was examined using a Student’s t test with a paired two-tailed distribution. Statistical analysis and figures were made with GraphPad Prism 9. The data were considered significant when ∗p < 0.05, ∗∗p < 0.01, or ∗∗∗p < 0.001.
Data availability
The authors declare that all other data supporting the findings of this study are available within the article and its supplemental information or from the corresponding author upon reasonable request.
Acknowledgments
We thank Shiwen Li, Xili Zhu, and Yue Wang for their help with confocal laser scanning microscopy. This work was supported by National Key Research and Development Program of China (2022YFC2702604) and the National Science Fund for Distinguished Young Scholars (81925015).
Author contributions
Liying Wang, C.L., and H.W. performed most of the experiments and analyzed the data. R.Z., Lina Wang, and Y.C. performed some experiments and analyzed the data. Y.O. and M.D. performed ICSI experiments. Y.M. and M.G. constructed plasmids. Y.Y. conceived the project. W.L. and Q.-Y.S. wrote the manuscript, designed the research, and supervised the project.
Declaration of interests
The authors declare that there is no conflict of interest.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.10.022.
Contributor Information
Qing-Yuan Sun, Email: sunqy@gd2h.org.cn.
Wei Li, Email: leways@gwcmc.org.
Supplemental information
References
- 1.Boivin J., Bunting L., Collins J.A., Nygren K.G. International estimates of infertility prevalence and treatment-seeking: potential need and demand for infertility medical care. Hum. Reprod. 2007;22:1506–1512. doi: 10.1093/humrep/dem046. [DOI] [PubMed] [Google Scholar]
- 2.Safarinejad M.R. Infertility among couples in a population-based study in Iran: prevalence and associated risk factors. Int. J. Androl. 2008;31:303–314. doi: 10.1111/j.1365-2605.2007.00764.x. [DOI] [PubMed] [Google Scholar]
- 3.Namiki M. Genetic aspects of male infertility. World J. Surg. 2014;24:1176–1179. doi: 10.1007/s002680010198. [DOI] [PubMed] [Google Scholar]
- 4.O'Flynn N. Assessment and treatment for people with fertility problems: NICE guideline. Br. J. Gen. Pract. 2014;64:50–51. doi: 10.3399/bjgp14X676609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luke B. Pregnancy and birth outcomes in couples with infertility with and without assisted reproductive technology: with an emphasis on US population-based studies. Am. J. Obstet. Gynecol. 2017;217:270–281. doi: 10.1016/j.ajog.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palermo G.D., O'Neill C.L., Chow S., Cheung S., Parrella A., Pereira N., Rosenwaks Z. Intracytoplasmic sperm injection: state of the art in humans. Reproduction. 2017;154:F93–F110. doi: 10.1530/REP-17-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abou-Setta A.M., Peters L.R., D'Angelo A., Sallam H.N., Hart R.J., Al-Inany H.G. Post-embryo transfer interventions for assisted reproduction technology cycles. Cochrane Database Syst. Rev. 2014:CD006567. doi: 10.1002/14651858.CD006567.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adamson D., Baker V. Multiple births from assisted reproductive technologies: a challenge that must be met. Fertil. Steril. 2004;81:517–522. doi: 10.1016/j.fertnstert.2003.09.041. discussion: 526. [DOI] [PubMed] [Google Scholar]
- 9.Xu Z., Song Z., Li G., Tu H., Liu W., Liu Y., Wang P., Wang Y., Cui X., Liu C., et al. H2B ubiquitination regulates meiotic recombination by promoting chromatin relaxation. Nucleic Acids Res. 2016;44:9681–9697. doi: 10.1093/nar/gkw652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua R., Wei H., Liu C., Zhang Y., Liu S., Guo Y., Cui Y., Zhang X., Guo X., Li W., Liu M. FBXO47 regulates telomere-inner nuclear envelope integration by stabilizing TRF2 during meiosis. Nucleic Acids Res. 2019;47:11755–11770. doi: 10.1093/nar/gkz992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiba K., Enatsu N., Fujisawa M. Management of non-obstructive azoospermia. Reprod. Med. Biol. 2016;15:165–173. doi: 10.1007/s12522-016-0234-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano M., Brinster C.J., Orwig K.E., Ryu B.Y., Avarbock M.R., Brinster R.L. Transgenic mice produced by retroviral transduction of male germ-line stem cells. Proc. Natl. Acad. Sci. USA. 2001;98:13090–13095. doi: 10.1073/pnas.231473498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe S., Kanatsu-Shinohara M., Ogonuki N., Matoba S., Ogura A., Shinohara T. In vivo genetic manipulation of spermatogonial stem cells and their microenvironment by Adeno-associated viruses. Stem Cell Rep. 2018;10:1551–1564. doi: 10.1016/j.stemcr.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin J., Xu H., Zhang P., Zhang C., Zhu Z., Qu R., Qin Y., Zeng W. An efficient strategy for generation of transgenic mice by lentiviral transduction of male germline stem cells in vivo. J. Anim. Sci. Biotechnol. 2015;6:59. doi: 10.1186/s40104-015-0058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibuya H., Morimoto A., Watanabe Y. The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLoS Genet. 2014;10:e1004821. doi: 10.1371/journal.pgen.1004821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michaelis M., Sobczak A., Weitzel J.M. In vivo microinjection and electroporation of mouse testis. J. Vis. Exp. 2014:51802. doi: 10.3791/51802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coward K., Kubota H., Parrington J. In vivo gene transfer into testis and sperm: developments and future application. Arch. Androl. 2007;53:187–197. doi: 10.1080/01485010701426455. [DOI] [PubMed] [Google Scholar]
- 18.Tu Z., Bayazit M.B., Liu H., Zhang J., Busayavalasa K., Risal S., Shao J., Satyanarayana A., Coppola V., Tessarollo L., et al. Speedy A-Cdk2 binding mediates initial telomere-nuclear envelope attachment during meiotic prophase I independent of Cdk2 activation. Proc. Natl. Acad. Sci. USA. 2017;114:592–597. doi: 10.1073/pnas.1618465114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M., Huang T., Li M.J., Zhang C.X., Yu X.C., Yin Y.Y., Liu C., Wang X., Feng H.W., Zhang T., et al. The histone modification reader ZCWPW1 is required for meiosis prophase I in male but not in female mice. Sci. Adv. 2019;5:eaax1101. doi: 10.1126/sciadv.aax1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibuya H., Watanabe Y. Live-cell microscopy of meiosis in spermatocytes. Methods Cell Biol. 2018;145:269–277. doi: 10.1016/bs.mcb.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 21.Roosen-Runge E.C. The process of spermatogenesis in mammals. Biol. Rev. 1962;37:343–376. doi: 10.1111/j.1469-185x.1962.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 22.Hess R.A., Renato de Franca L. Spermatogenesis and cycle of the seminiferous epithelium. Adv. Exp. Med. Biol. 2008;636:1–15. doi: 10.1007/978-0-387-09597-4_1. [DOI] [PubMed] [Google Scholar]
- 23.Zickler D., Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 24.Zickler D., Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harbor Perspect. Biol. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harbor Perspect. Biol. 2015;7:a016618. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guillon H., Baudat F., Grey C., Liskay R.M., de Massy B. Crossover and noncrossover pathways in mouse meiosis. Mol. Cell. 2005;20:563–573. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 27.Kohl K.P., Sekelsky J. Meiotic and mitotic recombination in meiosis. Genetics. 2013;194:327–334. doi: 10.1534/genetics.113.150581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanco-Rodríguez J. Programmed phosphorylation of histone H2AX precedes a phase of DNA double-strand break-independent synapsis in mouse meiosis. Reproduction. 2012;144:699–712. doi: 10.1530/REP-12-0326. [DOI] [PubMed] [Google Scholar]
- 29.Situ Y., Chung L., Lee C.S., Ho V. MRN (MRE11-RAD50-NBS1) complex in human cancer and prognostic implications in colorectal cancer. Int. J. Mol. Sci. 2019;20:816. doi: 10.3390/ijms20040816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 2007;15:551–563. doi: 10.1007/s10577-007-1147-9. [DOI] [PubMed] [Google Scholar]
- 31.Wang H., Wan H., Li X., Liu W., Chen Q., Wang Y., Yang L., Tang H., Zhang X., Duan E., et al. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 2014;24:852–869. doi: 10.1038/cr.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang Y., Wang H., Jia P., Zhao H., Liu C., Liu W., Song Z., Xu Z., Yang L., Wang Y., Li W. Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy. 2016;12:1575–1592. doi: 10.1080/15548627.2016.1192750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 34.Niedźwiedzka-Rystwej P., Tokarz-Deptuła B., Deptuła W. Autophagy in physiological and pathological processes--selected aspects. Pol. J. Vet. Sci. 2013;16:173–180. doi: 10.2478/pjvs-2013-0026. [DOI] [PubMed] [Google Scholar]
- 35.Tanida I., Ueno T., Kominami E. LC3 and autophagy. Methods Mol. Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 36.Liu C., Wang H., Shang Y., Liu W., Song Z., Zhao H., Wang L., Jia P., Gao F., Xu Z., et al. Autophagy is required for ectoplasmic specialization assembly in sertoli cells. Autophagy. 2016;12:814–832. doi: 10.1080/15548627.2016.1159377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cocuzza M., Alvarenga C., Pagani R. The epidemiology and etiology of azoospermia. Clinics. 2013;68:15–26. doi: 10.6061/clinics/2013(Sup01)03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagirnaja L., Aston K.I., Conrad D.F. Genetic intersection of male infertility and cancer. Fertil. Steril. 2018;109:20–26. doi: 10.1016/j.fertnstert.2017.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vij S.C., Sabanegh E., Jr., Agarwal A. Biological therapy for non-obstructive azoospermia. Expert Opin. Biol. Ther. 2018;18:19–23. doi: 10.1080/14712598.2018.1380622. [DOI] [PubMed] [Google Scholar]
- 40.Park F. Lentiviral vectors: are they the future of animal transgenesis? Physiol. Genom. 2007;31:159–173. doi: 10.1152/physiolgenomics.00069.2007. [DOI] [PubMed] [Google Scholar]
- 41.Kanatsu-Shinohara M., Toyokuni S., Shinohara T. Transgenic mice produced by retroviral transduction of male germ line stem cells in vivo. Biol. Reprod. 2004;71:1202–1207. doi: 10.1095/biolreprod.104.031294. [DOI] [PubMed] [Google Scholar]
- 42.Kim T.S., Choi H.S., Ryu B.Y., Gang G.T., Kim S.U., Koo D.B., Kim J.M., Han J.H., Park C.K., Her S., Lee D.S. Real-time in vivo bioluminescence imaging of lentiviral vector-mediated gene transfer in mouse testis. Theriogenology. 2010;73:129–138. doi: 10.1016/j.theriogenology.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 43.Sehgal L., Thorat R., Khapare N., Mukhopadhaya A., Sawant M., Dalal S.N. Lentiviral mediated transgenesis by in vivo manipulation of spermatogonial stem cells. PLoS One. 2011;6:e21975. doi: 10.1371/journal.pone.0021975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai J., Voloshin O., Potapova S., Camerini-Otero R.D. Meiotic knockdown and complementation reveals essential role of RAD51 in mouse spermatogenesis. Cell Rep. 2017;18:1383–1394. doi: 10.1016/j.celrep.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai P., Wang X., Gou L.-T., Li Z.-T., Wen Z., Chen Z.-G., Hua M.-M., Zhong A., Wang L., Su H., et al. A translation-activating function of MIWI/piRNA during mouse spermiogenesis. Cell. 2019;179:1566–1581.e16. doi: 10.1016/j.cell.2019.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen M., Yao C., Qin Y., Cui X., Li P., Ji Z., Lin L., Wu H., Zhou Z., Gui Y., et al. Mutations of MSH5 in nonobstructive azoospermia (NOA) and rescued via in vivo gene editing. Signal Transduct. Targeted Ther. 2022;7:1. doi: 10.1038/s41392-021-00710-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ledwith B.J., Manam S., Troilo P.J., Barnum A.B., Pauley C.J., Griffiths T.G., 2nd, Harper L.B., Beare C.M., Bagdon W.J., Nichols W.W. Plasmid DNA vaccines: investigation of integration into host cellular DNA following intramuscular injection in mice. Intervirology. 2000;43:258–272. doi: 10.1159/000053993. [DOI] [PubMed] [Google Scholar]
- 48.Tomizawa M., Shinozaki F., Motoyoshi Y., Sugiyama T., Yamamoto S., Sueishi M. Sonoporation: gene transfer using ultrasound. World J. Methodol. 2013;3:39–44. doi: 10.5662/wjm.v3.i4.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sadate-Ngatchou P.I., Payne C.J., Dearth A.T., Braun R.E. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46:738–742. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters A.H., Plug A.W., van Vugt M.J., de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- 51.Wang L., Xu Z., Wang L., Liu C., Wei H., Zhang R., Chen Y., Wang L., Liu W., Xiao S., et al. Histone H2B ubiquitination mediated chromatin relaxation is essential for the induction of somatic cell reprogramming. Cell Prolif. 2021;54:e13080. doi: 10.1111/cpr.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shang Y., Zhu F., Wang L., Ouyang Y.C., Dong M.Z., Liu C., Zhao H., Cui X., Ma D., Zhang Z., et al. Essential role for SUN5 in anchoring sperm head to the tail. Elife. 2017;6:e28199. doi: 10.7554/eLife.28199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all other data supporting the findings of this study are available within the article and its supplemental information or from the corresponding author upon reasonable request.