Abstract
Background & Aims
NF-E2-related factor 2 (NRF2) is a transcription factor that regulates cytoprotective gene expression in response to oxidative and electrophilic stresses. NRF2 activity is mainly controlled by Kelch-like ECH-associated protein 1 (KEAP1). Constitutive NRF2 activation by NRF2 mutations or KEAP1 dysfunction results in a poor prognosis for esophageal squamous cell carcinoma (ESCC) through the activation of cytoprotective functions. However, the detailed contributions of NRF2 to ESCC initiation or promotion have not been clarified. Here, we investigated the fate of NRF2-activated cells in the esophageal epithelium.
Methods
We generated tamoxifen-inducible, squamous epithelium-specific Keap1 conditional knockout (Keap1-cKO) mice in which NRF2 was inducibly activated in a subset of cells at the adult stage. Histologic, quantitative reverse-transcription polymerase chain reaction, single-cell RNA-sequencing, and carcinogen experiments were conducted to analyze the Keap1-cKO esophagus.
Results
KEAP1-deleted/NRF2-activated cells and cells with normal NRF2 expression (KEAP1-normal cells) coexisted in the Keap1-cKO esophageal epithelium in approximately equal numbers, and NRF2-activated cells formed dysplastic lesions. NRF2-activated cells exhibited weaker attachment to the basement membrane and gradually disappeared from the epithelium. In contrast, neighboring KEAP1-normal cells exhibited accelerated proliferation and started dominating the epithelium but accumulated DNA damage that triggered carcinogenesis upon carcinogen exposure.
Conclusions
Constitutive NRF2 activation promotes the selective elimination of epithelial cells via cell competition, but this competition induces DNA damage in neighboring KEAP1-normal cells, which predisposes them to chemical-induced ESCC.
Keywords: Cell Competition, DNA Damage, Replication Stress
Abbreviations used in this paper: βTRCP, β-transducin repeat containing protein; cKO, conditional knockout; COL17A1, collagen 17a1; DEN, diethylnitrosamine; ESCC, esophageal squamous cell carcinoma; GSEA, gene set enrichment analysis; HPF, high-power fields; KEAP1, Kelch-like ECH-associated protein 1; K5CreERT2, keratin5-CreERT2; MPO, myeloperoxidase; NQO1, nicotinamide adenine dinucleotide plus hydrogen quinone oxidoreductase 1; NRF2, NF-E2-related factor 2; qPCR, quantitative polymerase chain reaction; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; RNA-seq, RNA-sequencing; Tam, tamoxifen; UMI, unique molecular identifier; 4NQO, 4-nitroquinoline 1-oxide
Graphical abstract
Summary.
A model of coexisting NRF2-activated and KEAP1-normal cells was established in the esophageal epithelium using Keap1 conditional knockout mice. NRF2-activated cells are selectively eliminated through cell competition, but loser NRF2-activated cells leave a memory for the remaining winner KEAP1-normal cells.
Esophageal cancer is one of the most aggressive tumors and the sixth leading cause of cancer-related death.1 Esophageal squamous cell carcinoma (ESCC) has been classified into 3 groups on the basis of the independent and integrated clustering of 4 genomic datasets,2 ie, somatic copy number alterations, DNA methylation, mRNA expression, and microRNA expression datasets. Among the 3 groups, ESCC1 is strongly correlated with genomic alterations in the NF-E2-related factor 2 (NRF2) pathway; these genomic alterations occur in 54% of ESCC1 cases. Genomic aberrations targeting the genes encoding NRF2 and Kelch-like ECH-associated protein 1 (KEAP1) enhance the cellular response to oxidative stress, and NRF2 activation is associated with a poor prognosis of ESCC.3,4 Therefore, the KEAP1-NRF2 system has been suggested to be among the most important therapeutic targets in ESCC.
NRF2 is a transcription factor that regulates a set of cytoprotective genes in response to oxidative and electrophilic stresses.5, 6, 7 NRF2 activity is controlled by ubiquitin-proteasomal degradation mediated by KEAP18,9 and β-transducin repeat containing proteins (βTRCP).10,11 In some cancer cells, NRF2 is constitutively activated because of abnormalities in multiple pathways such as dysregulation of the protein‒protein interaction of KEAP1 and NRF2.12,13 The resulting high level of NRF2 activation reinforces cytoprotection and confers a growth advantage or malignant phenotypes on cancer cells.3,14,15 We proposed that cancers with constitutive and high-level NRF2 expression should be referred to as NRF2-addicted cancers.12
NRF2 mutations in human cancers, including ESCC, accumulate at DLGex and ETGE motifs that interact with KEAP1 such that NRF2 is constitutively activated.12,14,16 We have studied NRF2 activation in cancer cells in vivo using mouse models with Keap1 deletion,17, 18, 19 because these mice are good experimental models in which to study NRF2-addicted cancers. In this regard, systemic Keap1 knockout in mice causes postnatal lethality due to hyperkeratosis in the esophagus and forestomach.20 Because these hyperkeratosis and lethal phenotypes are completely rescued in Keap1::Nrf2 double knockout mice, they most likely result from the constitutive activation of NRF2 induced by Keap1 deletion. Similarly, squamous epithelium-specific Keap1 knockout mice (Keratin5-Cre::Keap1floxA/floxA) die at a similar time point after birth because of hyperkeratosis in the upper digestive tract.19 These results clearly show that systemic Keap1 knockout causes lethality in mice via esophageal hyperkeratosis; thus, we cannot study esophageal carcinogenesis in adulthood in vivo using these Keap1 systemic knockout mice.
NRF2 also contributes to the proliferation and/or differentiation of hematopoietic stem cells and tissues.20, 21, 22 NRF2 activation drives the differentiation of hematopoietic stem cells toward the granulocyte-monocyte lineage. Constitutive activation of NRF2 in the small intestinal epithelium increases the tissue length and villus height because of the increase in enterocyte proliferation and differentiation.23 In addition, in the livers of Keap1::Pten double knockout mice, constitutive activation of NRF2 induced by the suppression of both the KEAP1 and βTRCP pathways causes cholangiocyte expansion.18 These results suggest that in certain tissues, NRF2 activation directs cells to proliferate and differentiate.
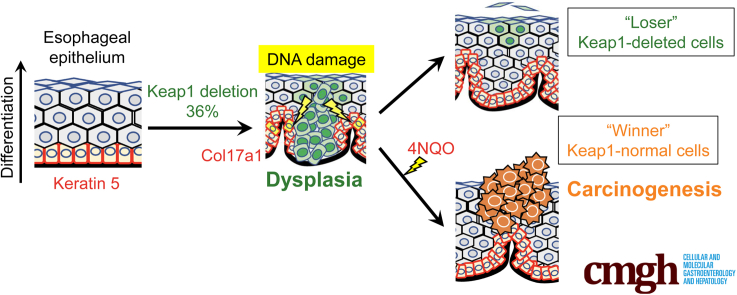
In the esophagus, where approximately equal numbers of NRF2-deleted cells and cells with normal NRF2 expression (NRF2-normal cells) are present,24 the NRF2-deleted cells are eliminated rapidly and selectively upon exposure to a chemical carcinogen in the esophagus.24 On the basis of these results, competition exists between NRF2-deleted and NRF2-normal cell populations. However, the behavior of constitutively NRF2-activated cells in a heterogeneous population containing NRF2-normal and NRF2-activated cells remains elusive. Therefore, we generated Keratin5-CreERT2 (K5CreERT2)::Keap1floxB/floxB (Keap1-cKO) mice with tamoxifen (Tam)-inducible Keap1 deletion/NRF2 activation in the squamous epithelium. Our results reveal that KEAP1-deleted/NRF2-activated cells in the esophageal epithelium are eliminated because of competition with KEAP1-normal/NRF2-normal cells. However, DNA damage accumulates in the KEAP1-normal/NRF2-normal cells that survive cell competition, and this damage appears to trigger chemical-induced esophageal carcinogenesis.
Results
Conditional Keap1 Knockout Induces Regional and Temporal Overexpression of NRF2 in the Esophageal Epithelium
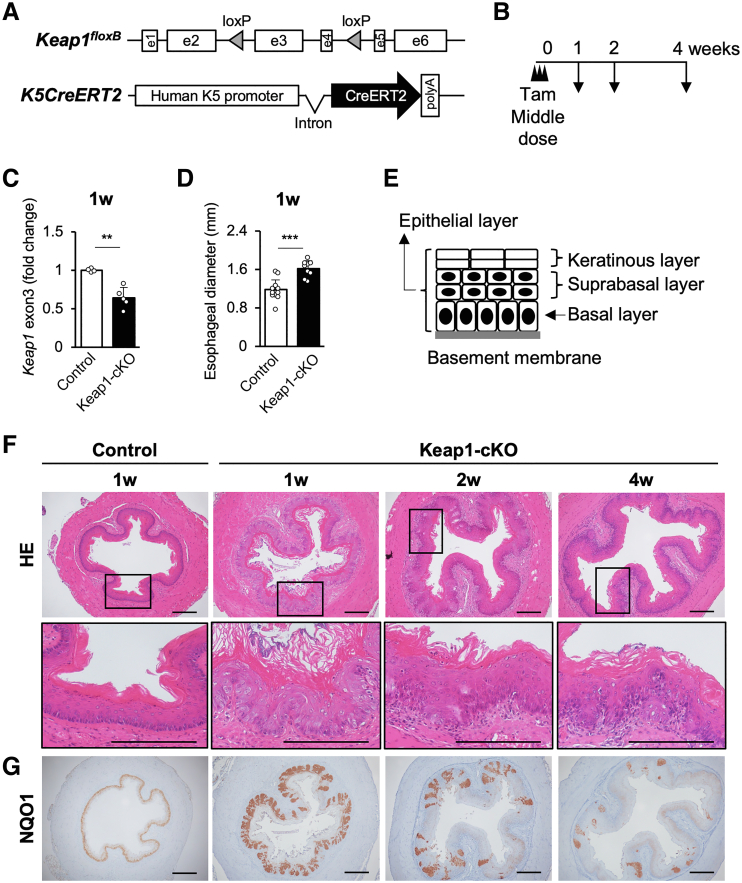
The fate of esophageal epithelial cells with inducible expression of NRF2 remains unknown. We used a squamous epithelium specific, Tam-inducible Keap1 knockout strategy to clarify the fate of NRF2-activated epithelial cells in the adult esophagus. For this experiment, we generated a K5CreERT2::Keap1floxB/floxB compound mutant mouse line, hereafter referred to as the Keap1-cKO mouse. Keap1-cKO mice were obtained by mating K5CreERT2 transgenic mice24 with Keap1floxB/floxB knock-in mutant mice.25 Maps of these modified genes are shown in Figure 1A.
Figure 1.
Partial Keap1 deletion causes dysplasia and thickening of the esophageal epithelium in Keap1-cKO mice. (A) Generation of K5CreERT2::Keap1floxB/floxB mice. (B) Experimental schedule. Tam (100 μg/g body weight) was injected 3 times. (C) Recombination rate of the Keap1 gene in the esophageal epithelium 1 week after Tam administration (n = 4–5 mice per group). The remaining level of Keap1 gene exon 3 was quantified. (D) Quantification of esophageal diameter 1 week after Tam administration (n = 12 control mice and n = 9 Keap1-cKO mice). (E) Schematic of the mouse esophageal epithelium. (F) HE staining of esophageal sections 1–4 weeks after Tam administration. The esophageal epithelium of Keap1-cKO mice became thicker than that of control mice and exhibited hyperkeratinization 1 week after Tam administration. Increase in thickness was reversed 4 weeks after Tam administration. (G) Immunohistochemical staining for NQO1 and time course of NQO1 expression in the Keap1-cKO mouse esophagus. Population of NQO1-positive cells was decreased 4 weeks after Tam administration. Scale bars: 200 μm. Data are presented as means ± standard deviations. ∗∗P < .01 and ∗∗∗P < .001 compared with control mice according to Welch t test.
First, Tam (100 μg/g body weight) was administered to Keap1-cKO and control (ie, Keap1floxB/floxB) mice at 6–9 weeks of age on 3 consecutive days. The mice were analyzed 1, 2, and 4 weeks after the last Tam administration (Figure 1B). Keap1-cKO mice treated in this manner survived for more than 25 weeks after Tam administration. This phenotype clearly contrasted with that of the systemic Keap1 knockout mice, which died before 3 weeks of age because of hyperkeratosis of the esophageal epithelium.20
Under this condition, the Keap1 gene recombination efficiency in the Keap1-cKO esophagus was approximately 36% 1 week after Tam administration (Figure 1C), indicating that approximately one-third of the epithelial cells lost the Keap1 gene.
A macroscopic inspection revealed a greater esophageal thickness in Keap1-cKO mice than in control mice. We next conducted histologic analyses and observed a significantly larger esophageal diameter in Keap1-cKO mice than in control mice 1 week after Tam treatment (Figure 1D and top panels in Figure 1F).
The esophageal epithelium consists of 3 layers (Figure 1E): the basal layer, which consists of Keratin5-expressing basal cells directly attached to the basement membrane, the suprabasal layer, and the keratinous layer. The esophageal epithelia were thicker in the Keap1-cKO mice 1 week after Tam treatment than those in the control mice and exhibited hyperkeratinization (Figure 1F). The thickening of the epithelial layer and hyperkeratinization were gradually reversed after the 1-week time point. The change in the body weights of Keap1-cKO mice was comparable with that in control mice treated with Tam during the observation period, indicating that the thickening and hyperkeratosis of the esophageal epithelium after Tam treatment did not affect the general condition of Keap1-cKO mice.
We next examined the distribution of KEAP1-deleted cells within the esophageal epithelium by detecting NRF2 activity. We performed immunostaining for the nicotinamide adenine dinucleotide plus hydrogen quinone oxidoreductase 1 (NQO1) protein, a representative NRF2 target gene product, because an anti-NQO1 antibody was previously shown to produce obvious staining and to constitute an excellent surrogate marker of NRF2 activity.24,26,27 We detected low NQO1 expression in the esophageal epithelium of control mice (Figure 1G, left panel). In contrast, we detected strongly stained NQO1-positive cells in the basal and suprabasal layers of the esophageal epithelium in Keap1-cKO mice. Consistent with the Keap1 gene analysis, these strongly NQO1-positive cells accounted for approximately one-third of the basal cells (Figure 1G, second to left panel). Consistent with the thickening and hyperkeratosis, the number and proportion of strongly NQO1-positive cells were markedly reduced over time after Tam treatment (Figure 1G, two right panels).
Squamous Epithelium-Specific Keap1 Knockout Induces Esophageal Dysplasia
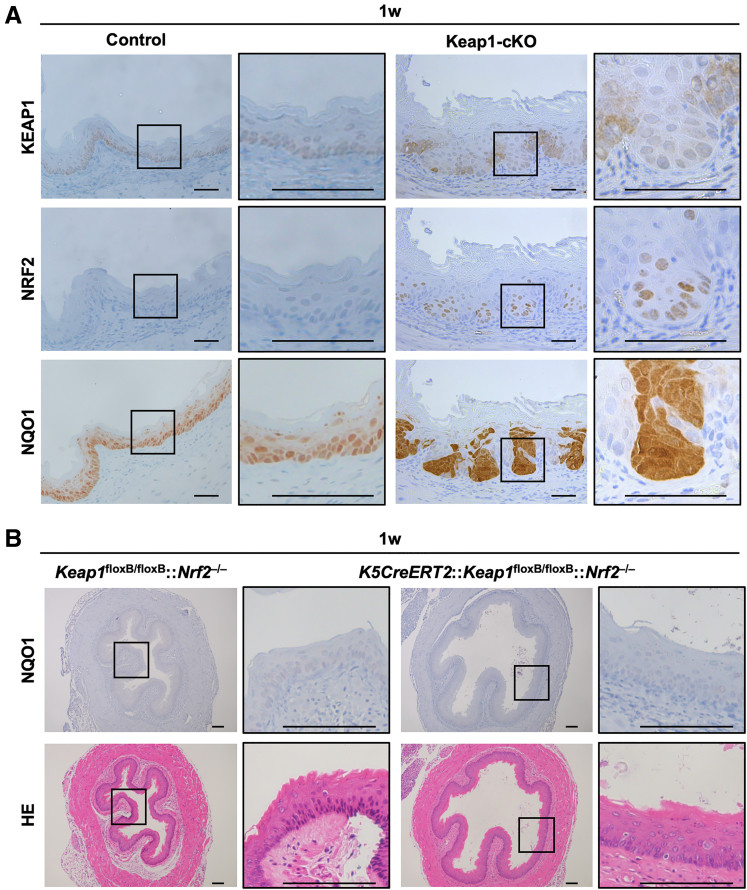
We next examined whether Keap1 deletion affects the cellular content and/or distribution of KEAP1 and NRF2 in the Keap1-cKO esophagus by performing an immunohistochemical analysis. In the control esophagus, KEAP1 was expressed in the cell layer, especially in a line of basal cells (Figure 2A, upper left panels). Although NRF2 expression was not detected in the esophagus of the control group, NQO1 expression was clearly detected in the basal and suprabasal layers of the esophagus in the control group (Figure 2A, middle and lower left panels, respectively). This NQO1 expression depended on NRF2 because NQO1 expression in control mice was substantially diminished by the simultaneous deletion of Nrf2 in the resulting Keap1floxB/floxB::Nrf2–/–mice (Figure 2B, upper left panels). Thus, although NRF2 expression was not detected because of the detection limit of the immunohistochemical analysis, NRF2 functioned at a basal level and activated its target gene.
Figure 2.
Esophageal dysplasia in Keap1-cKO mice was abolished by Nrf2 deletion. (A) Immunohistochemical staining for KEAP1, NRF2, and NQO1 at 1 week after Tam administration. In KEAP1-deleted cells, NRF2 accumulated in the nucleus, and NQO1 was expressed at high levels. Scale bars: 50 μm. (B) NQO1 immunohistochemistry and HE staining in Keap1floxB/floxB::Nrf2–/– and K5CreERT2::Keap1floxB/floxB::Nrf2–/– mouse esophagi. Scale bars: 100 μm.
Notably, Keap1 deletion induced by the middle dose of Tam occurred segmentally in the esophageal epithelium of Keap1-cKO mice (Figure 2A, upper right panels). KEAP1-deleted cells showed dysplastic features and proliferated toward the basal side, whereas KEAP1-normal cells proliferated toward the luminal side with attachment to the basement membrane. Importantly, under this condition, we detected marked NRF2 accumulation in the nucleus of KEAP1-deleted cells (Figure 2A, middle right panels). Consistent with the pattern of NRF2 expression, NQO1 expression was increased significantly and was observed in both the nucleus and cytoplasm of KEAP1-deleted cells in dysplastic lesions (Figure 2A, lower right panels).
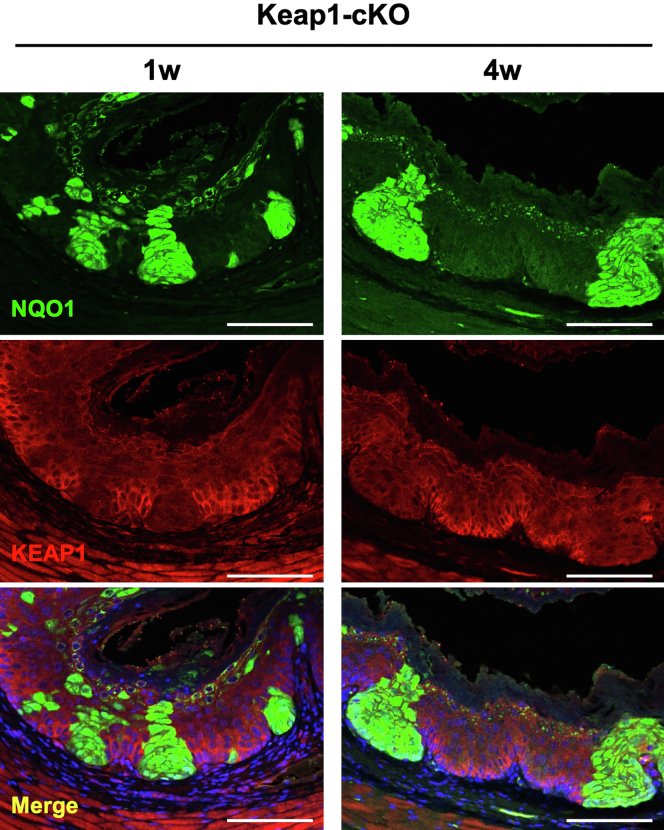
Intriguingly, NQO1 expression was reduced in the neighboring cells expressing the wild-type Keap1 gene (compare the lower left and lower right panels of Figure 2A). We do not have an immediate mechanistic explanation for this phenomenon, but this point is discussed in the single-cell RNA-sequencing (RNA-seq) section below. However, these results indicated very clear boundaries between the KEAP1-deleted/NQO1-positive dysplastic cells and the KEAP1-normal cells. Therefore, in this study, we used positive NQO1 staining as a surrogate marker of NRF2 expression in KEAP1-deleted cells. We performed immunofluorescence staining and confirmed that Keap1 deletion and the ensuing NQO1 up-regulation were detected at least 4 weeks after Tam administration (Figure 3).
Figure 3.
NQO1 expression was maintained in KEAP1-deleted cells until 4 weeks after Tam administration. Double immunofluorescence staining for NQO1 and KEAP1. NQO1 was markedly expressed in KEAP1-deleted cells in the esophageal epithelium of Keap1-cKO mice until 4 weeks after Tam administration. Scale bars: 100 μm.
Intriguingly, Keap1-cKO mice showed only mild esophageal hyperkeratosis in stark contrast to systemic Keap1 knockout mice,20 which exhibited severe esophageal hyperkeratosis and died before weaning. We concluded that inducible deletion at the adult stage in Keap1-cKO mice did not result in severe hyperkeratosis. However, in Keap1-cKO mice, the cellular arrangement in the basal layer was perturbed by NRF2 overexpression elicited by Keap1-cKO. The basal layer was elongated and convoluted as a result of the increase in the number of epithelial cells with enlarged nuclei, and these cells formed esophageal dysplastic lesions. Furthermore, the elongated basal layer appeared to be maintained for a long time (ie, at least 4 weeks) without shrinking (Figure 1F).
We generated Keap1-cKO::Nrf2–/– compound knockout mice and examined the esophageal epithelium in these mice to determine whether these epithelial phenotypes depended on the increase in NRF2 expression in the esophagus of Keap1-cKO mice. The diameter, keratosis, thickness, and elongation of the epithelial layer of the esophagus were within normal ranges in Keap1-cKO::Nrf2–/– mice (Figure 2B, lower panels), indicating that the increases in the esophageal diameter, hyperkeratinization of the cornified layer, thickness of the cell layer, and perturbation of the basal layer in Keap1-cKO esophagus were due to excessive NRF2 activation.
Studies of the Effects of the Dose and Time of Keap1 Conditional Knockout in Esophageal Epithelial Cells
We next sought to determine whether segmental deletion of the Keap1 gene depended on the specific Tam concentration used or whether graded segmental deletion would be achieved reproducibly over a wide range of Tam concentrations in this experimental system. Thus, we examined the proportion of NRF2-activated cells in mice with graded Keap1 deletion. We used 3 different doses of Tam, ie, 12.5 μg (low), 100 μg (middle; the dose used in the previous section), and 200 μg (high) per gram of body weight, and treated Keap1-cKO and control mice with these doses of Tam for 3 consecutive days.
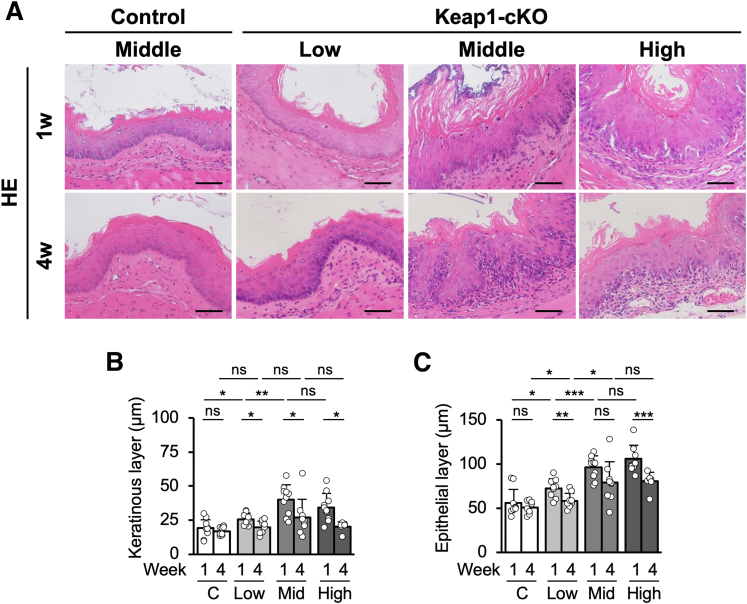
We did not identify a perturbation in the basal layer cell arrangement in the low-dose Keap1-cKO group or middle-dose control group 1 and 4 weeks after Tam treatment (Figure 4A, left panels). In contrast, Keap1-cKO mice treated with the middle dose of Tam showed marked perturbation of the basal layer cell arrangement 1 and 4 weeks after Tam treatment, and this perturbation did not differ substantially in mice treated with the high dose (Figure 4A, right panels). Similarly, the degree of hyperkeratinization showed Tam dose dependency, and again, no significant difference was observed between the middle- and high-dose groups (Figure 4A and B). The epithelial layers also exhibited thickening, depending on the dose of Tam (Figure 4). Consistent with the results described in the previous section, the keratinous and epithelial layers exhibited thinning 4 weeks after Tam administration (Figure 4).
Figure 4.
Increase in the esophageal epithelial thickness is gradually reversed 4 weeks after partial Keap1 deletion. (A) HE staining of esophageal sections after administration of low, middle, and high doses of Tam. (B and C) Quantification of keratinous and epithelial layer thicknesses. Both layers tended to exhibit thickening in a Tam dose-dependent manner. In all layers, the increase in thickness was reversed 4 weeks after Tam administration (n = 7–12 mice per group). Scale bars: 50 μm. Data are presented as means ± standard deviations. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 compared with control mice according to Welch t test.
Immunohistochemical Staining for NQO1 During the Dose and Time Studies
We also examined phenotypic changes occurring during the dose studies by performing NQO1 immunohistochemistry. One week after Tam treatment, the percentage of NQO1-positive cells in the esophageal epithelium increased in a Tam dose-dependent manner, but the difference between the middle- and high-dose groups was relatively moderate (Figure 5A, upper panels). The percentage of NQO1-positive cells was significantly decreased 4 weeks after Tam administration in all dose groups (Figure 5A, lower panels). We counted the strongly NQO1-positive cells in these immunostained sections by setting the staining level in the control esophageal epithelium as negative (Figure 5A, left panels) and found that the percentage of strongly NQO1-positive cells increased along with the Tam dose. However, the number of cells was significantly decreased 4 weeks after Tam administration (Figure 5B).
Figure 5.
NRF2 activation decreases 4 weeks after Tam treatment. (A) Immunohistochemical staining for NQO1. Population of NQO1-positive cells decreased over time in groups treated with each dose. Scale bars: 400 μm. (B) Quantification of NQO1-positive cells in the esophageal epithelium of mice treated with the 3 doses. Percentage of NQO1-positive cells was calculated as the NQO1-positive area within the total esophageal epithelial area in each mouse (n = 7–12 mice per group). (C) Positive correlations of percentage of NQO1-positive cells with thicknesses of keratinous and epithelial layers 1 week after administration of 3 doses of Tam (n = 29 mice). Line shows the two-tailed Pearson correlation. (D) Nqo1 and Gclc mRNA expression levels. Gene expression was decreased throughout the 4-week period in each group (n = 4–6 mice per group). Data are presented as means ± standard deviations. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 according to Welch t test.
The intensity of NQO1 staining in KEAP1-normal cells was markedly reduced, showing very good reproducibility with the results presented in Figure 2A. Thus, the difference in NQO1 staining between KEAP1-deleted cells and KEAP1-normal cells became very noticeable (Figure 5A, lower panels). We also examined the correlation between the percentage of NQO1-positive cells and the thickness of the keratinous and epithelial layers in the Keap1-cKO esophagus. We observed a significant positive correlation (Figure 5C), suggesting that the outgrowth of KEAP1-deleted cells induced dysplasia and perturbation of the esophageal epithelium.
We next sought to determine whether the changes in NQO1 expression identified by immunohistochemistry reflected the changes in NRF2 activity. We examined the mRNA expression levels of Nqo1 and Gclc, which are additional NRF2 target genes, at 1, 2, and 4 weeks after treatment with the 3 different doses of Tam. The expression of the Nqo1 and Gclc mRNAs was up-regulated 1 week after Tam treatment (Figure 5D). Consistent with the results of NQO1 immunohistochemistry, the expression levels of these mRNAs increased along with the dose of Tam and decreased gradually with increasing time after Tam administration in all dose groups.
On the basis of these results, treatment of Keap1-cKO mice with Tam generated KEAP1-deleted/NRF2-activated cells in a segmental pattern in the esophageal epithelium. The number of NRF2-activated cells initially increased but subsequently decreased after Tam administration. This decrease in the NRF2-activated cell population occurred regardless of the number of NRF2-activated cells in the epithelium. We surmise that complex mechanisms underlie the time-dependent changes (ie, the increase and decrease) in the number of KEAP1-deleted/NRF2-activated cells after Tam-induced Keap1 knockout. One mechanism is cell proliferation and the formation of dysplastic lesions, whereas the other is the competition between KEAP1-deleted/NRF2-activated cells and neighboring KEAP1-normal/NRF2-normal cells.
Competition Between KEAP1-Deleted/NRF2-Activated Cells and KEAP1-Normal Cells
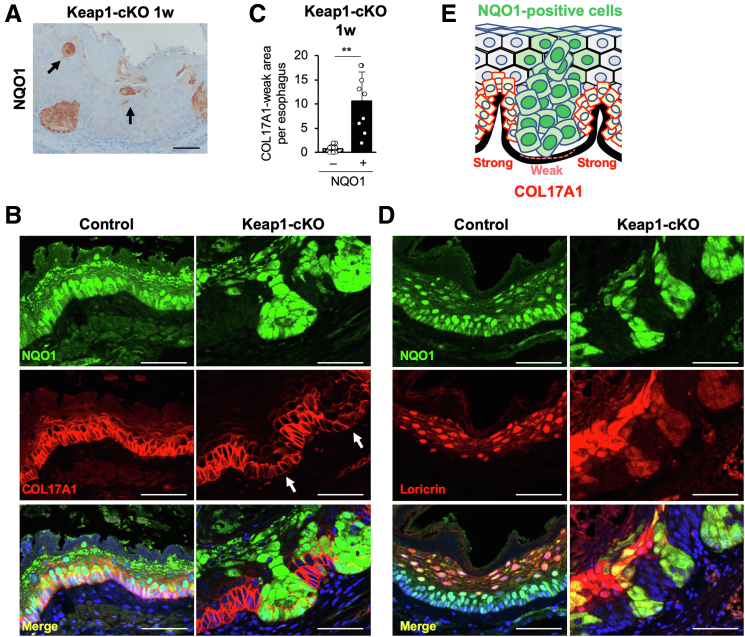
We next sought to verify that cell competition occurred between KEAP1-deleted/NRF2-activated cells and neighboring KEAP1-normal/NRF2-normal cells. To the best of our knowledge, the presence and physiological and/or pathologic contributions of cell competition in the esophageal epithelium have not been assessed. In contrast, reports indicate that in the skin, less-fit or “loser” cells exhibit weakened adhesion to the basement membrane and become floating cells, resulting in compensatory proliferation of the neighboring highly fit or “winner” cells.28,29 In fact, in our careful examination of NQO1 immunohistochemical sections, we observed floating NQO1-positive/NRF2-activated cells in the Keap1-cKO esophagus 1 week after Tam administration (Figure 6A). This result supports our hypothesis that KEAP1-deleted/NRF2-activated cells commit to differentiation, begin to lose their attachment to the basement membrane, and become losers in the esophageal epithelium.
Figure 6.
NRF2-activated cells are selectively eliminated and induce DNA damage in neighboring KEAP1-normal cells. (A) NQO1-positive floating cells were present in the Keap1-cKO mouse esophagus. (B) Immunofluorescence staining for NQO1 and COL17A1. White arrows indicate COL17A1-weak areas. NRF2-activated cells exhibited weak COL17A1 staining. (C) COL17A1-weak areas were counted in each section of esophageal epithelium from Keap1-cKO mice (n = 9). COL17A1-weak areas were particularly abundant in NQO1-positive basal cells. (D) Immunofluorescence staining for NQO1 and Loricrin. (E) Schematic of the esophageal epithelium of Keap1-cKO mice. NQO1-positive cells exhibited weaker COL17A1 staining than KEAP1-normal cells. Scale bars: 50 μm. Data are presented as means ± standard deviations. ∗∗P < .01, as compared using Welch t test.
Because collagen 17a1 (COL17A1) has been shown to play an important role in the attachment of basal cells to the basement membrane in the skin,28 we next examined COL17A1 expression in KEAP1-deleted/NRF2-activated cells and KEAP1-normal cells using immunofluorescence staining. In control mice, COL17A1 was expressed uniformly in basal layer cells of the esophageal epithelium along with NQO1 (Figure 6B, left panels). In contrast, COL17A1 expression appeared to be segmented and was visualized as clusters of cells with weakly positive and strongly positive regions in the basal layer of the esophageal epithelium in Keap1-cKO mice. The weakly positive and strongly positive regions of these cell clusters were arranged in a mirror image pattern relative to the NQO1 immunofluorescence-positive and NQO1-negative cells. Although KEAP1-normal cells exhibited strong positive staining with the anti-COL17A1 antibody and weak positive staining with the anti-NQO1 antibody, KEAP1-deleted/NRF2-activated cells strongly positive for NQO1 staining exhibited only weakly positive staining with the anti-COL17A1 antibody (Figure 6B, right panels). The KEAP1-deleted/NRF2-activated cells with weakly positive COL17A1 staining were clustered between the KEAP1-normal cells and were rarely detected in control mice (Figure 6C). In contrast, loricrin, a differentiated cell marker, was expressed in NRF2-activated cells of the basal layer (Figure 6D). Therefore, KEAP1-normal cells maintained their strong attachment to the basement membrane via hemidesmosomes, whereas the attachment of NRF2-activated cells was weakened and tended to differentiate from the basal cells (Figure 6E).
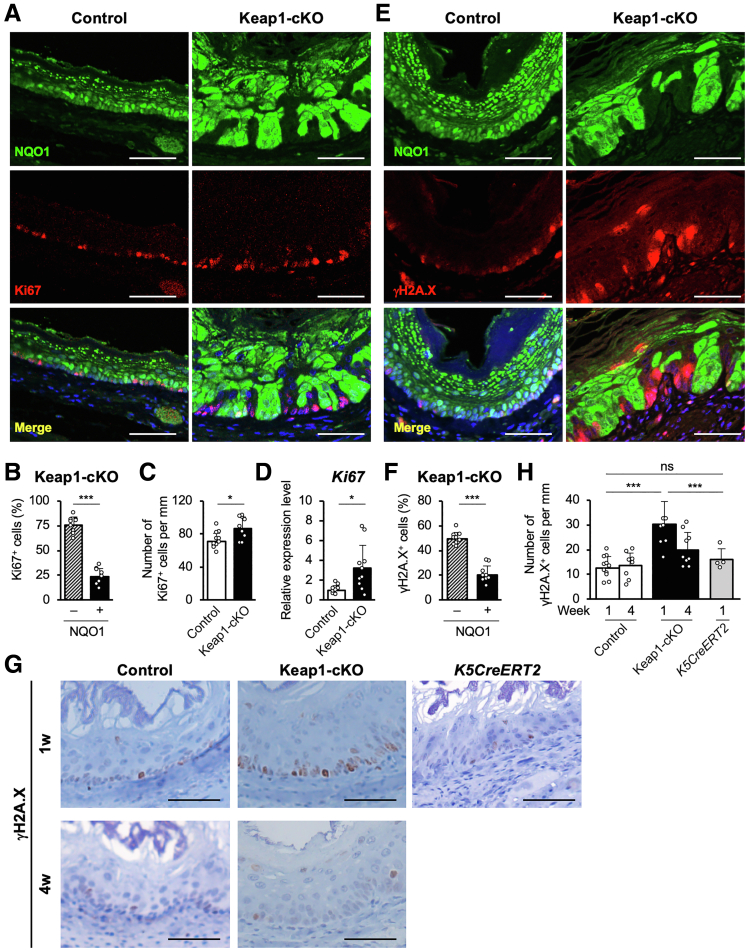
As a molecular mechanism that accelerates the elimination of NRF2-activated cells from the basal layer, compensatory proliferation of the neighboring KEAP1-normal cells was expected. We examined the expression of Ki67, a cell proliferation marker, in the esophagus 1 week after Tam administration to assess this hypothesis (Figure 7A). Ki67 expression was increased significantly in KEAP1-normal cells in the Keap1-cKO esophagus (Figure 7A and B). In addition, the total number of Ki67-positive cells in the esophagus was increased in Keap1-cKO mice compared with control mice (Figure 7C). Ki67 mRNA expression was also up-regulated in the esophageal epithelium of Keap1-cKO mice (Figure 7D), consistent with the results of histologic analyses. These results prompted us to hypothesize that NRF2 activation in esophageal epithelial cells drives these cells toward early proliferation to form dysplastic lesions; however, these cells stop proliferating and commit to differentiation, and NRF2 activation subsequently causes these cells to acquire a loser status. In contrast, the KEAP1-normal cells surrounding the KEAP1-deleted/NRF2-activated cells have acquired the ability to proliferate, and these cells begin expanding. Therefore, NRF2-activated cells are eliminated through cell competition, and neighboring KEAP1-normal cells undergo accelerated expansion as winners.
Figure 7.
NRF2-activated cells are selectively eliminated and induce DNA damage in neighboring KEAP1-normal cells. (A) Immunofluorescence staining for NQO1 and Ki67. The number of Ki67-positive cells was increased, especially among NRF2-activated cells, in the esophageal epithelium of Keap1-cKO mice. (B) Percentages of Ki67-positive cells among NQO1-negative and NQO1-positive cells in the Keap1-cKO mouse esophagus (n = 9). (C) The number of Ki67-positive cells in the basal layer was normalized to the length of the basement membrane (n = 12 control mice and n = 9 Keap1-cKO mice). (D) Ki67 mRNA expression level (n = 9 control mice and n = 10 Keap1-cKO mice). (E) Immunofluorescence staining for NQO1 and γH2A.X. The number of γH2A.X-positive cells was significantly increased, especially among NRF2-activated cells, in the Keap1-cKO mouse esophagus. (F) Percentages of γH2A.X-positive cells among NQO1-negative and NQO1-positive cells in the Keap1-cKO mouse esophagus. (G) Immunohistochemical staining for γH2A.X. (H) The number of γH2A.X-positive cells in the basal layer was normalized to the length of the basement membrane (n = 4–11 mice per group). Scale bars: 50 μm. Data are presented as means ± standard deviations. ∗P < .05 and ∗∗∗P < .001, as compared using Welch t test.
One of the remaining questions is how NRF2-activated cells communicate with KEAP1-normal neighboring cells. We examined the expression levels of γH2A.X, a marker of DNA double-strand breaks, in Keap1-cKO mice 1 week after Tam treatment to answer this question. Intriguingly, γH2A.X expression was increased significantly in KEAP1-normal cells in the esophageal epithelium of Keap1-cKO mice (Figure 7E), and DNA damage was induced in 49% of KEAP1-normal cells (Figure 7F).
Higher γH2A.X expression was observed in the esophagus of Keap1-cKO mice than in control mice 1 week after Tam administration, but the expression returned to the same level as that in control mice 4 weeks after Tam administration (Figure 7G, left two panels). Cre expression per se has been shown to induce DNA damage.30 Therefore, we examined the expression of γH2A.X in the K5CreERT2 mouse esophagus to exclude this possibility. However, the total number of γH2A.X-positive cells was not increased in K5CreERT2 mice treated with Tam for 1 week, indicating that the increase in γH2A.X-positive cells truly depended on the increase in NRF2 expression. Thus, we concluded that the coexistence of KEAP1-normal cells with NRF2-activated cells provoked DNA damage in KEAP1-normal cells in the esophagus (Figure 7G and H).
DNA replication stress is a DNA damage mechanism coupled with cell proliferation31,32 and induces genome instability and the formation of precancerous lesions.33 Therefore, the DNA damage in KEAP1-normal cells might also be caused by replication stress due to the coexistence and rapid elimination of neighboring NRF2-activated dysplastic cells in the esophageal epithelium.
Accelerated Elimination of NRF2-Activated Cells in Keap1-cKO::Nrf2SA Mice
Because a graded increase in the Tam dose resulted in no further increase in recombination, we were unable to examine the consequence of greater NRF2 activation in the esophageal epithelium by further increasing the Tam dose. In this regard, NRF2 ubiquitination has been shown to be accomplished via 2 ubiquitin ligases, ie, KEAP1 and βTRCP,5,10,11 and the NRF2 that accumulated as a result of Keap1 loss is degraded, at least in part, through the βTRCP pathway. βTRCP-mediated NRF2 degradation requires the phosphorylation of 2 serine residues in NRF2 at positions 335 and 338, and the Nrf2SA mutant in which these serine residues are replaced with alanine residues is resistant to βTRCP-mediated degradation. Therefore, we used Nrf2SA mutant knock-in mice34 to assess the consequence of high-level NRF2 activation.
We attempted to generate a compound knockout mouse line, K5CreERT2::Keap1floxB/floxB::Nrf2SA/SA, by mating Keap1-cKO mice with Nrf2SA/SA mice; this line is hereafter referred to as the Keap1-cKO::Nrf2SA mouse line. Because the Nrf2SA mutation was systemic, NQO1 expression in the esophageal epithelium was moderately higher in Nrf2SA mice than in control mice (Figure 8A, left panels).
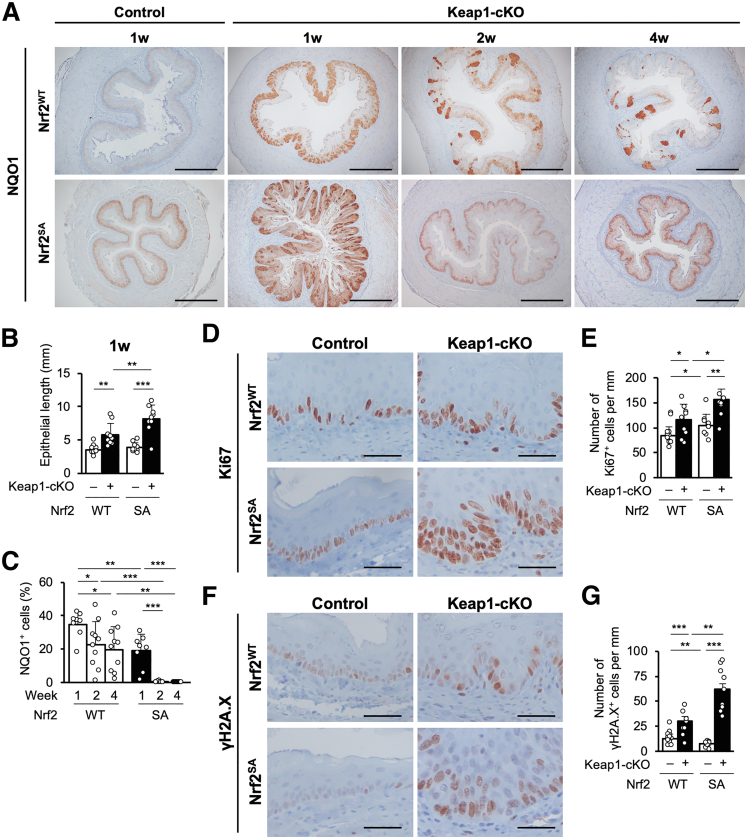
Figure 8.
Additional NRF2 activation in Keap1-cKO::Nrf2SAmice promotes elimination of NRF2-activated cells. (A) Immunohistochemical staining for NQO1. NQO1-positive cells were distributed segmentally in the esophageal epithelium of both Keap1-cKO::Nrf2SA mice and Keap1-cKO mice. Scale bars: 400 μm. (B) Quantification of esophageal epithelial length. The epithelial length was greatest in Keap1-cKO::Nrf2SA mice (n = 9–12 mice per group). (C) Quantification of NQO1-positive cells in the esophageal epithelium. The number of NQO1-positive cells decreased earlier in Keap1-cKO::Nrf2SA mouse esophagus than in Keap1-cKO mouse esophagus (n = 8–10 mice per group). (D and E) Immunohistochemical staining for Ki67. A greater increase in number of Ki67-positive cells was observed in the Keap1-cKO::Nrf2SA mouse esophagus than in the Keap1-cKO mouse esophagus. Scale bars: 50 μm. (F and G) Immunohistochemical staining for γH2A.X. Greater increase in number of γH2A.X-positive cells was observed in Keap1-cKO::Nrf2SA mouse esophagus than in Keap1-cKO mouse esophagus. Scale bars: 50 μm. Data are presented as means ± standard deviations. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 according to Welch t test.
One week after the middle-dose Tam administration, we detected much stronger activation of NRF2 in the esophageal epithelium of Keap1-cKO::Nrf2SA mice than in Keap1-cKO mice. As observed in Keap1-cKO mice, NRF2-activated cells were distributed segmentally in Keap1-cKO::Nrf2SA mice (Figure 8A, second left panels). Surprisingly, the number of cells with high NRF2 activation in Keap1-cKO::Nrf2SA mice decreased much more rapidly than the number of cells with lower NRF2 activation in Keap1-cKO mice 2 and 4 weeks after Tam administration (Figure 8A, right panels). The basal layer was much more substantially elongated in Keap1-cKO::Nrf2SA mice than in Keap1-cKO mice (Figure 8B). We counted the strongly NQO1-positive cells and found that the number of NRF2-activated cells in Keap1-cKO::Nrf2SA mice had already decreased to a level lower than that in Keap1-cKO mice at 1 week after Tam administration (Figure 8C). The decrease in the number of strongly NQO1-positive cells was quite rapid, and only 0.4% of the total epithelial cell population exhibited strong positive staining for NQO1 2 weeks after Tam administration. These results support the hypothesis that the substantial increase in NRF2 activation resulting from the inhibition of both KEAP1 and βTRCP pathways markedly promotes a rapid decrease in the number of NRF2-activated cells during basal cell proliferation.
We next examined Ki67 expression in the Keap1-cKO::Nrf2SA esophagus. Ki67-positive cells were observed in the basal layer of the esophageal epithelium in Nrf2 wild-type control mice (Figure 8D, upper left panel), and the appearance of the esophageal epithelium was generally similar in Nrf2SA mice (Figure 8D, lower left panel). The number of Ki67-positive cells increased in the Keap1-cKO esophagus (Figure 8D, upper right panel), but in stark contrast, the number of Ki67-positive cells was substantially increased in the esophageal epithelium of Keap1-cKO::Nrf2SA mice 1 week after Tam administration (Figure 8D, lower right panel). In Keap1-cKO mice, the Ki67-positive cells appeared to expand from the basal layer into multiple layers of cells, which overlapped with the KEAP1-normal cells. We counted the Ki67-positive cells and found that the number of cells increased more significantly in Keap1-cKO::Nrf2SA mice than in Keap1-cKO mice 1 week after Tam treatment (Figure 8E).
We also examined the presence of γH2A.X-positive cells in the esophagus of these mice. A moderate number of γH2A.X-positive cells and very weak γH2A.X expression were observed in the esophagus of both KEAP1-normal Nrf2WT and KEAP1-normal Nrf2SA mice (Figure 8F, left panels). However, along with the increase in Ki67-positive cells, the number of γH2A.X-positive cells and the expression of γH2A.X were increased in the Keap1-cKO esophagus (Figure 8F, right panels). In both mice, γH2A.X expression overlapped with that of Ki67 in the esophagus, and γH2A.X appeared to be expressed in KEAP1-normal cells. Notably, Keap1-cKO::Nrf2SA mice showed much stronger expression of γH2A.X than Keap1-cKO::Nrf2WT mice.
Therefore, additional NRF2 activation resulting from the Nrf2SA mutation markedly accelerated the proliferation and turnover of basal epithelial cells in Keap1-cKO::Nrf2SA mice. NRF2-hyperactivated cells in the esophagus of Keap1-cKO::Nrf2SA mice were eliminated much faster than NRF2-activated cells in Keap1-cKO esophagus, supporting the hypothesis that NRF2 hyperactivation in the esophageal epithelium worsens the outcome of cell competition and results not only in rapid elimination of loser cells but also in damage to the surrounding KEAP1-normal winner cells. We surmise that the DNA damage in KEAP1-normal winner cells is elicited by replication stress, because the replication rate in these cells is very high.
Accelerated Proliferation of KEAP1-Normal Cells in Keap1-cKO Esophagus
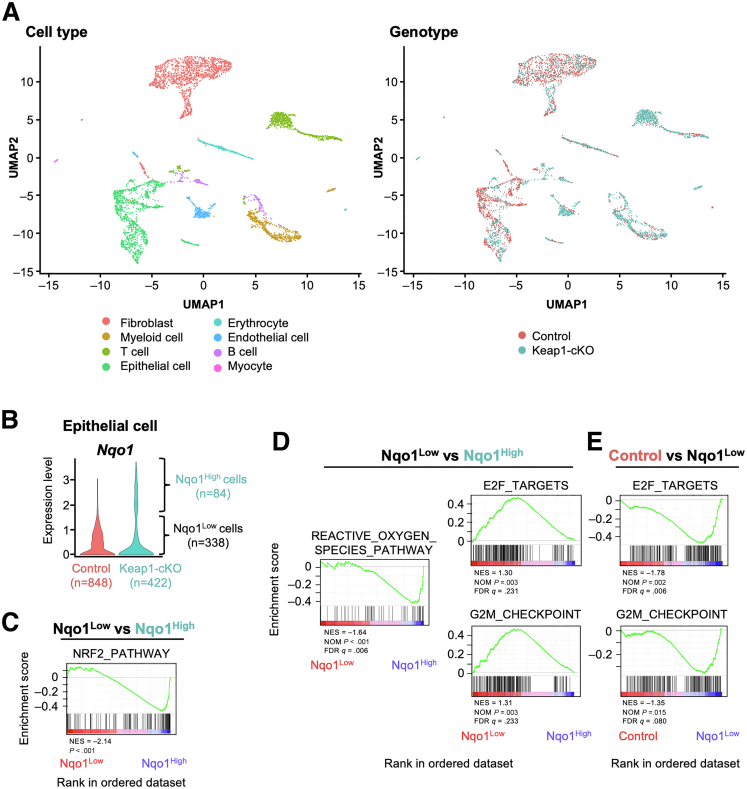
We further characterized KEAP1-normal/NRF2-normal cells and KEAP1-deleted/NRF2-activated cells in the esophageal epithelium of Keap1-cKO mice by conducting a single-cell RNA-seq analysis. One week after Tam administration, the esophageal epithelia of Keap1-cKO and control mice were dispersed into single cells and subjected to a single-cell RNA-seq analysis. After sequencing, the esophageal cells were classified into 8 types (Figure 9A and Figure 10).
Figure 9.
Accelerated proliferation of KEAP1-normal cells. (A) UMAP visualization of cells isolated from control and Keap1-cKO mouse esophagi. Left panel, 4853 cells are colored on basis of cell type. Right panel, 2472 and 2381 cells are colored on basis of their origin from control and Keap1-cKO mice, respectively. (B) Nqo1 expression in epithelial cells. (C) GSEA enrichment plot comparing Nqo1Low and Nqo1High epithelial cells using the NRF2 pathway gene set obtained from Wiki Pathways. (D) GSEA enrichment plot comparing Nqo1Low and Nqo1High epithelial cells using hallmark gene sets. Left panel shows the gene sets enriched in Nqo1High cells. Only the reactive oxygen species pathway was significantly enriched among the hallmark gene sets. The right two panels show the hallmark gene sets enriched in Nqo1Low cells. (E) GSEA enrichment plot comparing control and Nqo1Low epithelial cells. The gene sets E2F targets and G2/M checkpoint were significantly enriched in Nqo1Low cells.
Figure 10.
Cell type annotation of single-cell RNA-seq analyses. Expression of cell type marker genes in 8 cell types.
A total of 848 and 422 epithelial cells were detected in control and Keap1-cKO mice, respectively (Table 1), and we determined the expression level of the Nqo1 mRNA using the LogNormalize method. Nqo1 expression showed a bimodal distribution in the epithelial cells of Keap1-cKO mice but a unimodal distribution in the epithelium of control mice (Figure 9B). According to the Nqo1 expression level, we classified the epithelial cells from the Keap1-cKO esophagus into 2 types. Cells with an Nqo1 expression level greater than 1.6 were considered Nqo1High cells (n = 84), whereas those with a level less than 1.6 were considered Nqo1Low cells (n = 338). The Nqo1Low cells corresponded to the KEAP1-normal cells. Among the differentially expressed genes between Nqo1Low and Nqo1High cells, Nqo1 expression exhibited the most significant change (Supplementary Table 1). An intriguing observation was that the expression level of Nqo1 in Nqo1Low cells was substantially lower than that in epithelial cells from control mice. Upon closer examination, this lower Nqo1 expression level in Keap1-normal cells was reproduced in the NQO1 immunostaining results (Figure 1G, leftmost panel vs right panels). These observations suggest that Nqo1 expression and other functions of Nqo1Low cells were affected by the surrounding Nqo1High (ie, NRF2-activated) cells.
Table 1.
Numbers of the 8 Cell Types Detected in Single-Cell RNA-Sequencing Analyses
| Cell type | Control | Keap1-cKO | Total |
|---|---|---|---|
| Fibroblast | 1001 | 428 | 1429 |
| Myeloid cell | 173 | 412 | 585 |
| T cell | 141 | 589 | 730 |
| Epithelial cell | 848 | 422 | 1270 |
| Erythrocyte | 178 | 120 | 298 |
| Endothelial cell | 80 | 218 | 298 |
| B cell | 34 | 178 | 212 |
| Myocyte | 17 | 14 | 31 |
| Total | 2472 | 2381 | 4853 |
We then conducted a gene set enrichment analysis (GSEA) on Nqo1High and Nqo1Low epithelial cells from Keap1-cKO mice. We first tried to validate the enrichment of the NRF2 pathway gene set by searching WikiPathways.35 The NRF2 pathway gene set was indeed highly enriched in Nqo1High cells compared with Nqo1Low cells (Figure 9C).
We next analyzed hallmark gene sets (50 gene sets) to explore differentially expressed gene sets between Nqo1High and Nqo1Low cells.36 The reactive oxygen species pathway, which includes well-known NRF2 target genes, was enriched in Nqo1High cells from Keap1-cKO mice (Figure 9D and Table 2). In contrast, 4 gene sets were enriched in Nqo1Low cells compared with Nqo1High cells (Table 2). Among the gene sets enriched in Nqo1Low cells, the gene sets G2/M checkpoint and E2F targets, which are responsible for cell cycle regulation, were important because enrichment of these pathways indicates accelerated proliferation of Nqo1Low cells compared with Nqo1High cells (Figure 9D). A similar comparison of Nqo1Low cells with cells from control mice revealed enrichment of 1 gene set in control mouse cells and 15 gene sets in Nqo1Low cells (Table 3). Consistent with the aforementioned findings, the gene sets G2/M checkpoint and E2F targets were enriched in Nqo1Low cells (Figure 9E). These results support the assertion that KEAP1-normal cells exhibit accelerated compensatory proliferation, consistent with the results of our histologic analyses.
Table 2.
Enriched Gene Sets in Epithelial Cells Identified by GSEA Using Hallmark Gene Sets (Nqo1Low vs Nqo1High)
| Enriched in Nqo1High (Nqo1Low vs Nqo1High) | |||
|---|---|---|---|
| Gene set | NES | NOM P value | FDR q value |
| REACTIVE_OXYGEN_SPECIES_PATHWAY | –1.64 | <.001 | .003 |
| Enriched in Nqo1Low (Nqo1Low vs Nqo1High) | |||
|---|---|---|---|
| Gene set | NES | NOM P value | FDR q value |
| INTERFERON_ALPHA_RESPONSE | 1.43 | .005 | .107 |
| COAGULATION | 1.37 | .002 | .144 |
| G2M_CHECKPOINT | 1.31 | .003 | .233 |
| E2F_TARGETS | 1.30 | .003 | .231 |
FDR, false discovery rate.
Table 3.
Enriched Gene Sets in Epithelial Cells Identified by GSEA Using Hallmark Gene Sets (Control vs Nqo1Low)
| Enriched in control (control vs Nqo1Low) | |||
|---|---|---|---|
| Gene set | NES | NOM P value | FDR q value |
| KRAS_SIGNALING_DN | 1.73 | <.001 | .006 |
| Enriched in Nqo1Low (Nqo1Low vs control) | |||
|---|---|---|---|
| Gene set | NES | NOM P value | FDR q value |
| INFLAMMATORY_RESPONSE | –1.97 | <.001 | <.001 |
| ALLOGRAFT_REJECTION | –1.93 | <.001 | .001 |
| E2F_TARGETS | –1.78 | <.001 | .002 |
| KRAS_SIGNALING_UP | –1.75 | <.001 | .003 |
| IL2_STAT5_SIGNALING | –1.66 | <.001 | .008 |
| MTORC1_SIGNALING | –1.51 | .003 | .033 |
| IL6_JAK_STAT3_SIGNALING | –1.47 | .012 | .045 |
| ESTROGEN_RESPONSE_EARLY | –1.47 | .001 | .041 |
| INTERFERON_GAMMA_RESPONSE | –1.46 | .003 | .037 |
| COMPLEMENT | –1.46 | <.001 | .035 |
| APICAL_SURFACE | –1.46 | .026 | .033 |
| HEDGEHOG_SIGNALING | –1.43 | .043 | .037 |
| TNFA_SIGNALING_VIA_NFKB | –1.37 | .007 | .068 |
| G2M_CHECKPOINT | –1.35 | .015 | .080 |
| ESTROGEN_RESPONSE_LATE | –1.29 | .037 | .117 |
FDR, false discovery rate.
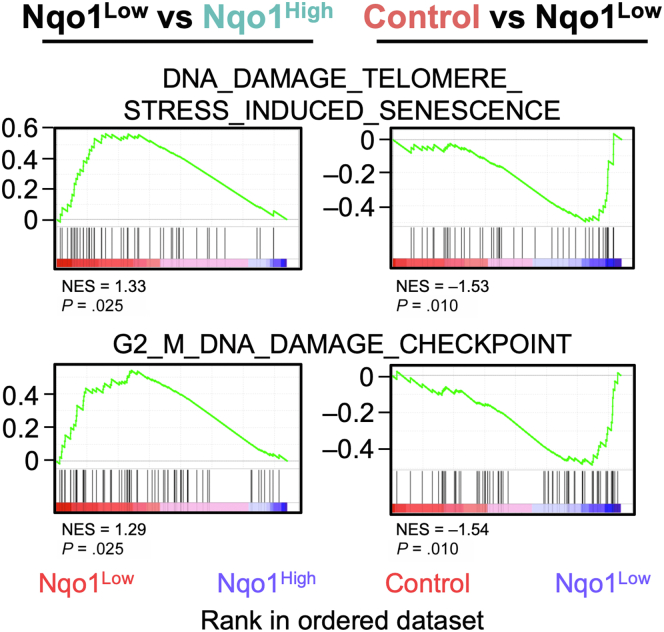
We next examined 7 DNA damage-related gene sets in the Reactome pathway database and found that 2 of the 7 gene sets were significantly enriched in Nqo1Low cells compared with Nqo1High cells (Table 4). The enriched pathways were “DNA damage telomere stress induced senescence” and “G2/M DNA damage checkpoint” (Figure 11, left panels). We also compared the 7 gene sets between Nqo1Low cells and control cells. We found that 4 of the 7 gene sets were significantly enriched in Nqo1Low cells compared with control cells (Table 4), namely “DNA damage bypass”, “Recognition of DNA damage by PCNA containing replication complex”, and the 2 gene sets listed above (Table 4 and Figure 11, right panels). The 7 DNA damage-related gene sets were not enriched in Nqo1High or control cells. Notably, the 2 gene sets commonly enriched in Nqo1Low cells are responsible for cell cycle progression, suggesting the considerable accumulation of DNA damage associated with rapid cell cycle progression in KEAP1-normal cells surrounding NRF2-activated cells.
Table 4.
DNA Damage-Related Gene Sets in the Reactome Pathway Database
| Gene set | Nqo1Low vs Nqo1High | Nqo1Low vs control |
|---|---|---|
| DNA_DAMAGE_BYPASS | ns | P = .012 |
| DNA_DAMAGE_RECOGNITION_IN_GG_NER | ns | ns |
| DNA_DAMAGE_TELOMERE _STRESS_INDUCED_SENESCENCE |
P = .025 | P = .010 |
| G1_S_DNA_DAMAGE_CHECKPOINTS | ns | ns |
| G2_M_DNA_DAMAGE_CHECKPOINT | P = .025 | P = .010 |
| RECOGNITION_OF_DNA_DAMAGE_BY_PCNA _CONTAINING_REPLICATION_COMPLEX |
ns | P = .016 |
| SUMOYLATION_OF_DNA_DAMAGE _RESPONSE_AND_REPAIR_PROTEINS |
ns | ns |
ns, not significant.
Figure 11.
DNA damage in KEAP1-normal cells. GSEA enrichment plots showing DNA damage-related gene sets obtained from Reactome that were significantly enriched in Nqo1Low cells.
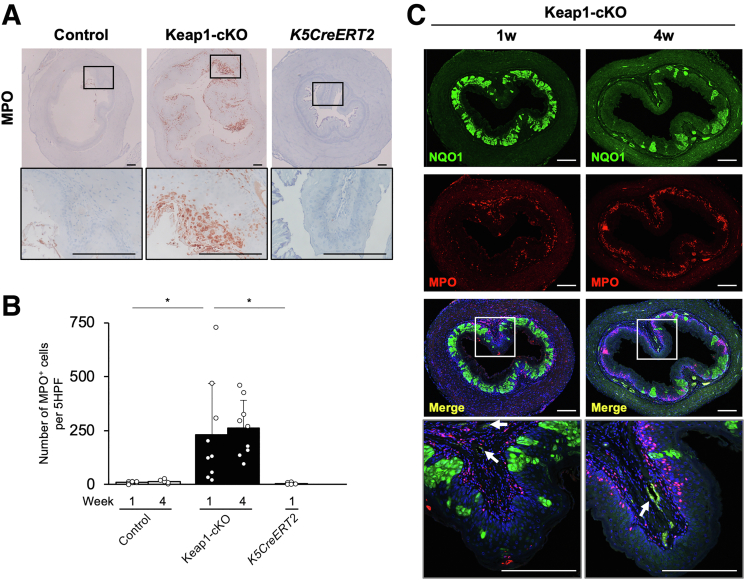
We then addressed the origins of the cell populations in the Keap1-cKO mouse esophagus (Figure 9A, right panel and Table 1). The populations of myeloid cells, T cells, and B cells were markedly increased in the esophagus of Keap1-cKO mice compared with control mice. We performed immunohistochemistry for myeloperoxidase (MPO), a myeloid cell marker, to validate the increased number of myeloid cells. The population of MPO-positive cells was increased in the submucosal and intraepithelial layers of the Keap1-cKO esophagus (Figure 12A). In contrast, the control and K5CreERT2 esophagi contained very few MPO-positive cells (Figure 12A and B).
Figure 12.
Inflammatory response in the Keap1-cKO mouse esophagus. (A) Immunohistochemical staining for MPO. The number of MPO-positive cells was increased in Keap1-cKO mouse esophagus 1 week after Tam administration. (B) Number of MPO-positive cells per 5 HPFs in the esophagus (n = 5–10 mice per group). (C) Immunofluorescence staining for NQO1 and MPO. White arrows indicate the small vessels surrounding the esophageal epithelium. Scale bars: 100 μm. Data are presented as means ± standard deviations. ∗P < .05, as compared using Welch t test.
The MPO-positive cells persisted for at least 4 weeks after Tam administration (Figure 12C). The red MPO-positive cells become purple when nuclei are simultaneously stained with DAPI. These MPO-positive cells appeared to migrate from the region surrounding small vessels (Figure 12C, lower left panels) into the intraepithelial region, particularly to the region surrounding the KEAP1-normal cells, ie, the region not surrounding the green NQO1-positive cell region (Figure 12C, lower right panels). The MPO-positive cells migrated into the luminal side of the basement membrane. These results support the hypothesis that the replication stress induced by compensatory proliferation in KEAP1-normal cells triggers inflammatory cell infiltration.
DNA Damage in Neighboring KEAP1-Normal Cells Promotes Esophageal Carcinogenesis
We next sought to determine whether the marked DNA damage in the neighboring KEAP1-normal esophageal cells surrounding the KEAP1-deleted cells in Keap1-cKO mice contributed to esophageal carcinogenesis. We used a chemical carcinogenesis approach using 4-nitroquinoline 1-oxide (4NQO) as a carcinogen. We achieved 4NQO-induced esophageal carcinogenesis using a standard procedure.24,37,38
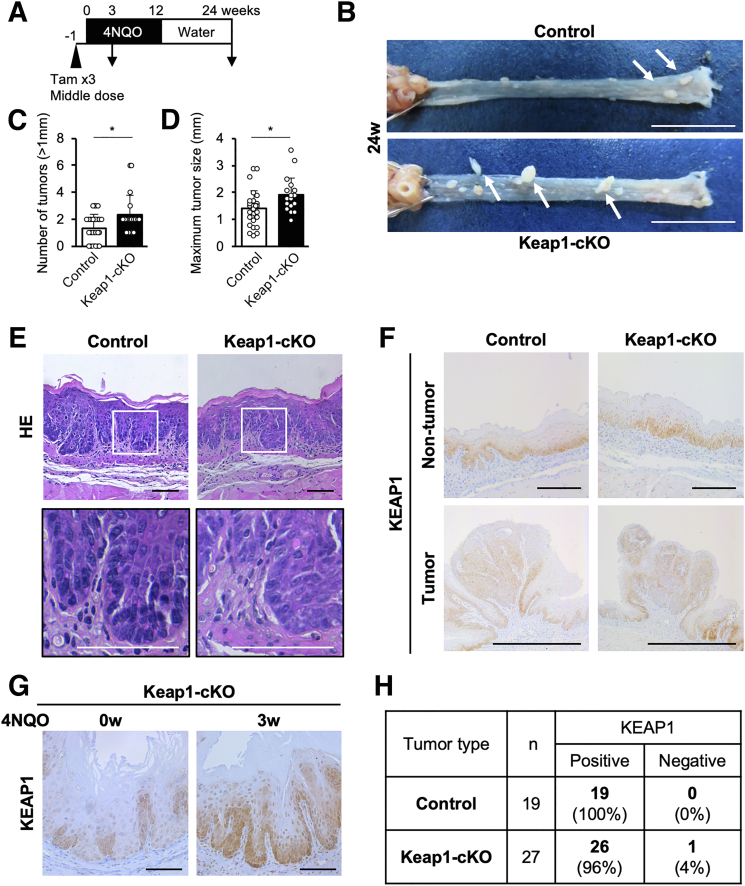
We treated both Keap1-cKO and control mice with the middle dose of Tam for 3 consecutive days to delete Keap1 and started administering 4NQO in the drinking water 1 week later. After 12 weeks of treatment, the mice were supplied 4NQO-free water for 12 weeks (Figure 13A). At 24 weeks, we detected esophageal tumors in all Keap1-cKO and control mice (Figure 13B, white arrows). Keap1-cKO mice developed many more tumors with a diameter greater than 1 mm than the corresponding control mice (Figure 13C). In addition, the maximum tumor sizes in Keap1-cKO mice were significantly greater than those in control mice (Figure 13D).
Figure 13.
KEAP1-normal cells constitute an origin of 4NQO-induced esophageal tumors in Keap1-cKO mice. (A) Experimental schedule of 4NQO and Tam administration (n = 28 control mice and n = 19 Keap1-cKO mice). (B) Esophageal tumors were observed in both control and Keap1-cKO mice 24 weeks after start of 4NQO administration. White arrows indicate tumors (<1 mm). Scale bars: 1 cm. (C) Quantification of tumors greater than 1 mm in diameter. Keap1-cKO mice had more tumors than control mice. (D) Quantification of maximum tumor size. Keap1-cKO mice had larger tumors than control mice. (E) HE staining of esophageal sections 24 weeks after start of 4NQO administration. Both Keap1-cKO and control mice exhibited atypical esophageal epithelial cells. Scale bars: 50 μm. (F) Immunohistochemical staining for KEAP1. Most tumors originated from KEAP1-positive cells. Scale bars: 100 μm. (G) Immunohistochemical staining for KEAP1. KEAP1-deleted cells were eliminated from Keap1-cKO mouse esophagus within 3 weeks of 4NQO treatment compared with 0 weeks (before 4NQO treatment). Scale bars: 50 μm. (H) Numbers of KEAP1-positive and KEAP1-negative tumors. Data are presented as means ± standard deviations. ∗P < .05, as compared using Welch t test.
The histologic analysis of mice at 24 weeks revealed dysplastic lesions with atypical cells in the esophagus of both control and Keap1-cKO mice treated with 4NQO (Figure 13E). Immunohistochemical analysis of mice at 24 weeks showed that KEAP1 was expressed uniformly in the esophageal epithelium in both control mice (Figure 13F, upper left panel) and Keap1-cKO mice (Figure 13F, upper right panel). We interpreted this result to indicate that KEAP1-deleted/NRF2-activated cells had disappeared by this time point. Indeed, KEAP1-deleted cells in Keap1-cKO mice had disappeared 3 weeks after the start of 4NQO administration (Figure 13G).
After a careful inspection, we found that most of the 4NQO-induced tumors expressed KEAP1, regardless of the Keap1 genotype (Figure 13F, lower panels). We counted esophageal tumors in multiple 4NQO-treated control (n = 11) and Keap1-cKO (n = 12) mice and found that 100% and 96% of the tumors in control and Keap1-cKO mice, respectively, were KEAP1-positive; only 1 tumor was KEAP1-negative (Figure 13H). Thus, after 4NQO treatment, almost all tumors originated from KEAP1-normal cells, and KEAP1-deleted/NRF2-activated cells were selectively eliminated during 4NQO-induced tumorigenesis.
On the basis of these results, we concluded that the coexistence of KEAP1-deleted/NRF2-activated cells and KEAP1-normal/NRF2-normal cells in the esophageal epithelium stimulated 4NQO-induced tumorigenesis in the latter type of cells. Although KEAP1-deleted/NRF2-activated cells were eliminated through cell competition, these cells left a hidden message, ie, DNA damage that accumulated in neighboring KEAP1-normal cells. Our results suggested that this message contributed intimately to 4NQO-induced tumorigenesis.
Discussion
We developed Tam-induced, squamous epithelium-specific Keap1-cKO mice to determine the fate of NRF2-activated cells in the esophageal epithelium and their contribution to esophageal carcinogenesis. We found that 2 types of epithelial cell populations coexist in the esophagus of mice with Tam-induced Keap1-cKO, KEAP1-deleted cells with NRF2 activation and KEAP1-normal cells. Importantly, we found that the esophageal epithelium contains a mixture of these 2 types of cells at 1 week after Tam administration. Capitalizing on this condition, specifically the finding that approximately 36% of the cells exhibit Keap1 deletion and 64% have an intact Keap1 gene, we discovered the mechanism underlying the selective elimination of KEAP1-deleted/NRF2-activated cells from the esophageal epithelium. KEAP1 deletion/NRF2 activation strongly directs initial dysplasia formation and the subsequent differentiation of squamous epithelial cells; thus, NRF2-activated cells become loser cells and are eliminated through competition with KEAP1-normal cells. Importantly, the eliminated NRF2-activated cells leave a memory for the remaining KEAP1-normal winner cells. We found that DNA damage is frequently induced in KEAP1-normal cells located near NRF2-activated cells. In fact, in the Keap1-cKO esophagus, the cells in 4NQO-induced tumors are almost exclusively KEAP1-normal cells. As shown in our previous study, Keap1 knockdown mice in which Keap1 expression is systematically decreased in the esophageal epithelium are resistant to 4NQO-induced esophageal carcinogenesis.38 In contrast to that study, Keap1-cKO mice showed the coexistence of KEAP1-normal and KEAP1-deleted cells in the esophageal epithelium, resulting in the formation of many more tumors than in control mice. Taken together, these results indicate that genetic NRF2 activation in the esophageal epithelium induces the differentiation of epithelial cells and renders these cells the losers; however, these loser cells leave a hidden message for the neighboring KEAP1-normal winner cells (ie, cryptic DNA damage), and these cells eventually promote esophageal carcinogenesis upon exposure to chemical carcinogens.
Although our laboratory has extensively studied systemic Keap1 knockout mice and squamous epithelium-specific Keap1 knockout mice such as K5Cre::Keap1floxA/floxA mice,19,20 the esophagus in these mice uniformly contains KEAP1-deleted squamous epithelial cells; thus, we were unable to detect any competition between KEAP1-deleted and KEAP1-normal epithelial cells. In these mice, we observed a substantial increase in squamous cell differentiation in the esophagus, resulting in severe hyperkeratosis.20 In contrast, in the present study, we examined Tam-induced Keap1-cKO mice that harbor both KEAP1-deleted and KEAP1-normal cells in a patchwork pattern, which conveniently allowed us to analyze the competition between these types of cells. At 1 week after Tam administration, we observed the formation of dysplastic lesions by cells with high NRF2 expression, which suggests the outgrowth of KEAP1-deleted cells in the esophageal epithelium. However, surprisingly, these KEAP1-deleted cells proliferated only briefly and gradually disappeared. In contrast, the KEAP1-normal cells surrounding the KEAP1-deleted cells actively proliferated and eventually filled the region occupied by the KEAP1-deleted cells. To the best of our knowledge, this study is the first to identify the accelerated differentiation of KEAP1-deleted cells and their elimination from the esophageal epithelium.
Cell competition was discovered in 1975 between minute mutant cells and normal cells in Drosophila imaginal discs.29 Thereafter, the concept of cell competition was thoroughly adopted in the study of mammalian development.39, 40, 41 In this concept, highly fit cells, or “winners”, undergo clonal expansion and outcompete the surrounding less-fit cells, or “losers”, which are finally eliminated from the cell population. Cell competition seems to be a homeostatic mechanism that eliminates abnormal cells with mutations.42 Moreover, cell competition has emerged as an important mechanism in the selection of cells during carcinogenesis, because cancer progression is affected by competitive interactions between cancer cells and the surrounding cells.17 In the present study, we showed that NRF2-activated cells commit to differentiation and acquire a loser status in the esophageal epithelium. In this regard, recent reports suggest that somatic mutant clones colonize the human esophagus during aging, and various somatic mutant clones then form a patchwork pattern and compete.43,44 In this scenario, when the winner cells commit to clonal expansion, the loser clones exit the cell cycle.42,45,46 The attachment of the loser cells to the basement membrane via hemidesmosomes is attenuated, and these cells start differentiating and migrating from the basal layer to the luminal layer. COL17A1 has been shown to be a marker of the esophageal stem/progenitor cell compartment,47 and a decrease in COL17A1 expression has been shown to be related to skin cell differentiation.28 Consistent with this mechanism, we observed decreased COL17A1 expression in NRF2-activated basal cells, along with weaker attachment of these cells to the basement membrane.
In a similar setting study, we previously found that conditional NRF2-deleted cells coexist with KEAP1-normal cells in esophageal epithelium under normal, unstressed conditions, whereas these NRF2-deleted cells are selectively eliminated from the epithelium upon treatment with a chemical carcinogen.24 This observation strongly argues that because NRF2 expression is usually suppressed under unstressed conditions, a significant difference should not be observed between KEAP1-normal cells and NRF2-deleted cells under normal conditions. However, upon exposure to carcinogens or strong chemical stress, NRF2-deleted cells become vulnerable to chemical stress and are eliminated from the esophageal epithelium. Thus, a double-edged sword exists in which too much NRF2 and too little NRF2, ie, NRF2-activated cells and NRF2-deleted cells, both lead to the elimination of the respective esophageal epithelial cells.
Intriguingly, in analyses of experimental hepatocellular carcinoma, the gradual loss of NRF2-activated cells that harbor Nrf2 mutations was observed during tumorigenesis induced by diethylnitrosamine (DEN).48 The number of cells harboring Nrf2 mutations was decreased in preneoplastic lesions, accompanied by the progression of DEN-induced hepatocarcinogenesis in rats. This observation is consistent with our current observation in ESCC induced by 4NQO. We thus propose that the decrease in the number of NRF2-activated cells in the hepatocellular carcinoma model may also be due to the selective elimination of NRF2-activated cells.
Intriguingly, the mechanism by which DNA damage is induced in KEAP1-normal cells is unknown. KEAP1-normal cells in the Keap1-cKO esophagus showed considerably increased proliferation compared with those in the control esophagus, indicating that cell competition accelerates the proliferation of these cells. Actively proliferating cells tend to be subjected to replication stress, including DNA damage.49,50 Therefore, one plausible explanation for the accumulation of DNA damage is replication stress. Replication stress often triggers immune responses51,52 in addition to DNA damage. Consistent with these findings, our single-cell RNA-seq analysis revealed an increase in the number of inflammatory cells in the Keap1-cKO esophagus. This increase in inflammatory cells resembles the observations in reflux esophagitis, in which neutrophils and other inflammatory cells infiltrate the submucosa and intraepithelial region of the esophagus.53 In the Keap1-cKO mouse esophagus, the infiltration of inflammatory cells continues for at least 4 weeks after Tam administration. Therefore, the appearance of KEAP1-deleted cells likely provokes the infiltration of inflammatory cells; consequently, DNA damage is induced in surrounding KEAP1-normal cells because of inflammation.
Alternatively, alterations in metabolic activities54 in KEAP1-deleted cells might induce the production of toxic metabolites. In fact, we detected the secretion of several toxic metabolites in the culture medium of NRF2-activated lung cancer cells.55 We also surmise that mechanical stresses generated by the initial dysplasia-like growth of KEAP1-deleted cells might elicit DNA damage in KEAP1-normal cells.56,57 Finally, paracrine signaling from NRF2-activated cells might also be a plausible cause of this DNA damage. Thus, the mechanisms by which DNA damage is induced in KEAP1-normal cells may be numerous and await further evaluation.
In the wing discs of Drosophila, competitive stress induces apoptotic damage in loser cells, and these cells are subsequently eliminated.58 However, previous studies have shown that apoptosis is not related to cell competition in the squamous epithelium of the esophagus and skin.28,42,46 Our study indicates that cell competition instead induces DNA damage in the neighboring cells in the esophageal epithelium. In fact, accumulating reports have shown that DNA damage constantly accumulates in normal cells in the human esophagus and skin.43,44,59,60 We hypothesize that DNA damage in winner cells may be one cause of the accumulation of mutations in aged epithelial tissues.
In conclusion, this study reveals that constitutively NRF2-activated cells are eliminated from the esophageal epithelium as the losers of cell competition. Thus, other mechanisms, eg, Tp53 mutations, might be necessary for the initiation of NRF2-addicted cancers. NRF2-activated cells leave DNA damage in the remaining winner cells that express KEAP1 and exhibit a normal level of NRF2 activity. This DNA damage serves as a cellular memory, which becomes an important predisposing factor for carcinogenesis. We propose that the identification of this pathway is important for elucidating the mechanisms underlying the initiation and promotion of ESCC.
Methods
Mice
K5CreERT2 mice,24 systemic Nrf2 knockout mice,7 and Nrf2SA mice34 have been described previously. Keap1floxB mice were provided by Dr Sham Biswal (Johns Hopkins University).25 These mice were maintained in the animal facility at Tohoku University. All animal experiments were approved by the Animal Care Committee at Tohoku University.
Gene Deletion in the Esophagus Using K5CreERT2 Mice
K5CreERT2 mice were crossed with Keap1floxB mice on a hybrid C57BL/6J and BALB/c background, and Keap1floxB/floxB mice and K5CreERT2::Keap1floxB/floxB mice were obtained. For induction of Cre expression, Tam (T5648; Sigma‒Aldrich, St Louis, MO) dissolved in corn oil (032-17016; Wako Pure Chemical Industries, Osaka, Japan) was intraperitoneally administered to 6- to 9-week-old mice (5 μL/g body weight) on 3 consecutive days. The following doses of Tam were used: low (12.5 μg/g body weight), middle (100 μg/g body weight), and high (200 μg/g body weight).
Histologic Analyses
Esophagi were opened longitudinally or transversely and fixed with Mildform 10N (131-10317; Wako Pure Chemical Industries). The fixed esophagi were embedded in paraffin, sliced into 4-μm-thick sections, and stained with hematoxylin-eosin. Histologic images were acquired with a Leitz DMRD microscope (Leica, Wetzlar, Germany) using cellSens Standard version 2.3 software (Olympus, Tokyo, Japan). The thicknesses of the cell layer and keratinous layer were measured at 3 points on each slide using ImageJ/Fiji software.61 The epithelial thickness was calculated by summing the thicknesses of the cell layer and keratinous layer. In addition, the epithelial length was measured using ImageJ/Fiji software.
Immunohistochemistry
Paraffin sections were rehydrated, autoclaved in 10 mmol/L sodium citrate buffer (pH 6.0) for antigen retrieval, treated with 3% H2O2, blocked with Protein Block Serum-free (X0909; Dako, Glostrup, Denmark), and sequentially incubated with primary antibodies (Table 5) for 16 hours at 4°C and the corresponding secondary antibodies. The positive cells were counted in the immunohistochemical sections, and the percentage of positive cells in the epithelium was calculated using ImageJ/Fiji software.61 In addition, the NQO1-positive area and total area of the cell layer were determined using ImageJ software, and the proportion of the NQO1-positive area was calculated. MPO-positive cells were counted in 5 high-power fields (HPFs).
Table 5.
Antibodies Used for Immunohistochemistry and Immunofluorescence
| Antibody | Clone | Company | Catalog no. | Dilution for IHC | Dilution for IF |
|---|---|---|---|---|---|
| KEAP1 | Rabbit polyclonal | Proteintech | 10503-2-AP | 1:200 | 1:50 |
| NRF2 (D9J1B) | Rat monoclonal | Cell Signaling Technology | 14596 | 1:400 | — |
| γH2A.X (Ser139; 20E3) | Rabbit monoclonal | Cell Signaling Technology | 9718 | 1:300 | 1:100 |
| NQO1 | Goat polyclonal | Abcam | ab2346 | 1:500 | 1:200 |
| Collagen XVII | Rabbit monoclonal | Abcam | ab184996 | — | 1:100 |
| MPO | Rabbit monoclonal | Abcam | ab208670 | 1:1000 | 1:100 |
| Ki67 | Rat monoclonal | BioLegend | 652402 | 1:1000 | 1:100 |
| Loricrin | Rabbit polyclonal | BioLegend | 905101 | 1:200 | 1:200 |
IF, immunofluorescence; IHC, immunohistochemistry.
Immunofluorescence Staining
Paraffin sections subjected to antigen retrieval were initially incubated with primary antibodies (Table 5) for 16 hours at 4°C, followed by appropriate secondary antibodies conjugated to Alexa Fluor 488, 546, or 647 (Molecular Probes, Eugene, OR) for 1 hour at room temperature. After washes with Tris-buffered saline, 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; D1306, Invitrogen, Waltham, MA) was added for nuclear counterstaining. Coverslips were mounted on glass slides with fluorescent mounting medium (S3023; Dako). All images were acquired with an LSM780 confocal microscope (Zeiss, Oberkochen, Germany) using ZEN software. The COL17A1-weak area was calculated, the Ki67-positive cells and γH2A.X-positive cells were counted, and the proportion of positive cells in the epithelium was calculated using ImageJ/Fiji software.61
Quantitative Reverse Transcription-Polymerase Chain Reaction
Harvested esophagi were opened longitudinally and treated with 0.5 g/L trypsin/0.53 mmol/L EDTA solution (32778-05; Nacalai Tesque, Kyoto, Japan) for 16 hours at 4°C. The epithelial layer was separated from the submucosal layer. Total RNA was extracted from the esophageal epithelium using Sepasol-RNA I Super G (09379-97; Nacalai Tesque). The RNA concentration was measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). RNA was reverse transcribed into cDNAs using ReverTra Ace qPCR RT Master Mix with gDNA Remover (FSQ-301; Toyobo, Osaka, Japan) according to the manufacturer’s instructions. The obtained templates were used for quantitative reverse transcription-polymerase chain reaction (qRT‒PCR) with a KAPA SYBR Fast qPCR Master Mix (2x) Kit (KK4602; Kapa Biosystems, London, UK) and a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). rRNA was used as the internal control. The primers used for qRT‒PCR are listed in Table 6.
Table 6.
Primers Used for Quantitative Reverse-Transcription Polymerase Chain Reaction
| Mouse gene | Primer sequence (5′ => 3′) | |
|---|---|---|
| Gclc | Forward Reverse |
ATCTGCAAAGGCGGCAAC ACTCCTCTGCAGCTGGCTC |
| Ki67 | Forward Reverse |
CATCCATCAGCCGGAGTCA TGTTTCGCAACTTTCGTTTGTG |
| Nqo1 | Forward Reverse |
AGCTGGAAGCTGCAGACCTG CCTTTCAGAATGGCTGGCA |
| rRNA | Forward Reverse |
CGGCTACCACATCCAAGGAA GCTGGAATTACCGCGGCT |
Analysis of the DNA Recombination Rate
The DNA recombination rate was analyzed using quantitative PCR (qPCR). Harvested esophagi were opened longitudinally and treated with 2.5 g/L trypsin/1 mmol/L EDTA solution (32777-15; Nacalai Tesque) for 3 hours at 37°C. The epithelial layer was separated from the submucosal layer. DNA was extracted from the esophageal epithelium. The concentrations of the obtained templates were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific), and qPCR was performed as described above. The β-Actin gene was used as the internal control. The primers used for qPCR are listed in Table 7.
Table 7.
Primers Used for Quantitative Reverse-Transcription Polymerase Chain Reaction Analysis of the DNA Recombination Rate
| Mouse gene | Primer sequence (5′ => 3′) | |
|---|---|---|
| β-Actin | Forward Reverse |
CCATAGGCTTCACACCTTCCTG GCACTAACACTACCTTCCTCAACCG |
| Keap1 exon3 | Forward Reverse |
GCGTGAGCTCCTGGAATATC TGCATCGACTGGGTCAAATA |
Single-Cell RNA-Sequencing Analysis
Single-cell RNA-seq libraries were prepared from control (n = 9) and Keap1-cKO (n = 8) esophagi 1 week after Tam administration. The whole esophagus was opened longitudinally, and the muscle layer was removed with forceps. All esophageal epithelia were pooled into a single sample from mice of the same genotype. The epithelia were cut into small pieces with scissors and incubated with 2.5 g/L trypsin/1 mmol/L EDTA solution (32777-15; Nacalai Tesque) for 10 minutes at 37°C. The digested samples were dispersed into single cells with a gentleMACS dissociator (Miltenyi Biotec, Gladbach, Germany). After adding Dulbecco modified Eagle medium, the suspension was filtered through a mesh 3 times and centrifuged twice at 300g and 4°C for 5 minutes each. Debris was removed using debris removal solution (130-109-398; Miltenyi Biotec), and the cell concentration was adjusted to 1 × 106 cells/mL. Single-cell RNA-seq libraries were prepared using Chromium Single Cell 3’ Reagent Kits (v3.1 chemistry; 10x Genomics, Pleasanton, CA) and sequenced with a DNBSEQ-G400 instrument (MGI Tech Co, Ltd, Shenzen, China). Two independent experiments were performed using different samples.
The raw gene expression matrix was obtained with CellRanger software (10x Genomics, ver. 6.0.1), and the single-cell RNA-seq data were analyzed with the Seurat R package (ver. 4.0.5).62 Before the next threshold was reached, 2472 and 2381 cells were selected from the control and Keap1-cKO samples, respectively. Genes detected in 3 or fewer cells were ignored. Cells expressing 200–8000 genes and <5% mitochondrial reads were selected. We used the LogNormalize method to eliminate the effects of different cell library sizes, in which the number of unique molecular identifiers (UMIs) of the genes was divided by the total UMI count in each cell, the quotient was multiplied by 10,000, the logarithm of the product was calculated, and the resulting value was considered the expression level. On the basis of gene expression, 19 clusters were identified by uniform manifold approximation and projection. The clusters were manually classified into 8 known biological cell types63 using the corresponding marker genes (Figure 10).
GSEA was performed using GSEA 4.1.0 software to characterize epithelial cells: Nqo1Low (n = 84) and Nqo1High cells (n = 338) in Keap1-cKO samples and control cells (n = 848). An NRF2 pathway gene set in WikiPathways35 was used to validate NRF2 activation in Nqo1High cells. Hallmark gene sets36 were used for a comprehensive analysis. A P value <.05 and a false discovery rate q value <0.25 were considered to indicate a statistically significant difference.
Induction of Esophageal Carcinogenesis by 4NQO
4NQO (N8141; Sigma‒Aldrich) was administered as previously described24 1 week after Tam-induced Keap1 knockout. Tam (middle dose) was administered intraperitoneally to Keap1-cKO and control (Keap1floxB/floxB) mice. One week after the last Tam administration, these mice were treated with 4NQO for 12 weeks and then provided 4NQO-free drinking water for 12 weeks after 4NQO administration. 4NQO was dissolved in dimethyl sulfoxide (09659-85; Nacalai Tesque) at 10 mg/mL and diluted in drinking water to 0.1 mg/mL. All mice were allowed ad libitum access to water in all stages of the experiment. The mice were weighed once weekly, and the drinking water was replaced with fresh water. At 24 weeks, these mice were euthanized with isoflurane, and the esophagus was harvested. The whole esophagus was opened longitudinally and photographed. Tumors larger than 1 mm were counted, and the maximum diameter of every tumor was measured using ImageJ/Fiji software.61
Statistical Analyses
The average values were calculated, and the error bars indicate the standard deviations. Differences in continuous data were analyzed using two-tailed Welch t test. P <.05 was considered statistically significant.
Acknowledgments
The authors thank Prof. Dr. Sham Biswal (Johns Hopkins University) for providing the Keap1floxB mice, Prof. Toru Furukawa and Dr. Fumiyoshi Fujishima (Tohoku University) for determining the pathologic diagnosis, Keiko Tateno and Nanae Osanai (Tohoku University) for the sequencing analysis, and the Biomedical Research Core of Tohoku University Graduate School of Medicine for providing technical support in pathologic analyses.
CRediT Authorship Contributions
Wataru Hirose (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead)
Makoto Horiuchi (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Supporting; Investigation: Equal)
Donghan Li (Data curation: Equal; Formal analysis: Equal; Visualization: Equal; Writing – original draft: Supporting)
Ikuko N. Motoike (Supervision: Equal)
Lin Zhang (Supervision: Supporting)
Hafumi Nishi (Supervision: Supporting)
Yusuke Taniyama (Supervision: Supporting)
Takashi Kamei (Supervision: Supporting)
Mikiko Suzuki (Resources: Equal)
Kengo Kinoshita (Supervision: Equal)
Fumiki Katsuoka (Data curation: Supporting; Supervision: Supporting)
Keiko Taguchi (Funding acquisition: Lead; Project administration: Lead; Supervision: Equal; Writing – review & editing: Equal)
Masayuki Yamamoto (Conceptualization: Equal; Funding acquisition: Lead; Project administration: Lead; Supervision: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by funding from MEXT/JSPS KAKENHI [19H05649 (M.Y.), 19K07395 (K.T.), and 21K16466 (M.H.)] and AMED-P-CREATE [JP20cm0106101 (M.Y.)].
Note: to access the supplementary materials accompanying this article, visit the online version of Cellular and Molecular Gastroenterology and Hepatology at www.cmghjournal.org.
Supplementary Material
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC, Brigham Women’s H, Broad I, Brown U, Case Western Reserve U, Dana-Farber Cancer I, Duke U, Greater Poland Cancer C, Harvard Medical S, Institute for Systems B, Leuven KU, Mayo C, Memorial Sloan Kettering Cancer C, National Cancer I, Nationwide Children’s H, Stanford U, University of A, University of M, University of North C, University of P, University of R, University of Southern C, University of Texas MDACC, University of W, Van Andel Research I, Vanderbilt U, Washington U, Genome Sequencing Center: Broad I, Washington University in St L, Genome Characterization Centers BCCA, Broad I, Harvard Medical S, Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins U, University of North C, University of Southern California Epigenome C, University of Texas MDACC, Van Andel Research I, Genome Data Analysis Centers: Broad I, Brown U, Harvard Medical S, Institute for Systems B, Memorial Sloan Kettering Cancer C, University of California Santa C, University of Texas MDACC, Biospecimen Core Resource: International Genomics C, Research Institute at Nationwide Children’s H, Tissue Source Sites: Analytic Biologic S, Asan Medical C, Asterand B, Barretos Cancer H, BioreclamationIvt, Botkin Municipal C, Chonnam National University Medical S, Christiana Care Health S, Cureline, Duke U, Emory U, Erasmus U, Indiana University School of M, Institute of Oncology of M, International Genomics C, Invidumed, Israelitisches Krankenhaus H, Keimyung University School of M, Memorial Sloan Kettering Cancer C, National Cancer Center G, Ontario Tumour B, Peter MacCallum Cancer C, Pusan National University Medical S, Ribeirao Preto Medical S, St Joseph’s H, Medical C, St Petersburg Academic U, Tayside Tissue B, University of D, University of Kansas Medical C, University of M, University of North Carolina at Chapel H, University of Pittsburgh School of M, University of Texas MDACC, Disease Working Group: Duke U, Memorial Sloan Kettering Cancer C, National Cancer I, University of Texas MDACC, Yonsei University College of M, Data Coordination Center CI, Project Team: National Institutes of H. Integrated genomic characterization of oesophageal carcinoma. Nature 2017;541:169–175. [DOI] [PMC free article] [PubMed]
- 3.Kitano Y., Baba Y., Nakagawa S., Miyake K., Iwatsuki M., Ishimoto T., Yamashita Y.I., Yoshida N., Watanabe M., Nakao M., Baba H. Nrf2 promotes oesophageal cancer cell proliferation via metabolic reprogramming and detoxification of reactive oxygen species. J Pathol. 2018;244:346–357. doi: 10.1002/path.5021. [DOI] [PubMed] [Google Scholar]
- 4.Xia D., Zhang X.R., Ma Y.L., Zhao Z.J., Zhao R., Wang Y.Y. Nrf2 promotes esophageal squamous cell carcinoma (ESCC) resistance to radiotherapy through the CaMKIIalpha-associated activation of autophagy. Cell Biosci. 2020;10:90. doi: 10.1186/s13578-020-00456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol Rev. 2018;98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baird L., Yamamoto M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol Cell Biol. 2020;40 doi: 10.1128/MCB.00099-20. e00099-00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., Yamamoto M., Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katoh Y., Iida K., Kang M.I., Kobayashi A., Mizukami M., Tong K.I., McMahon M., Hayes J.D., Itoh K., Yamamoto M. Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Arch Biochem Biophys. 2005;433:342–350. doi: 10.1016/j.abb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rada P., Rojo A.I., Evrard-Todeschi N., Innamorato N.G., Cotte A., Jaworski T., Tobon-Velasco J.C., Devijver H., Garcia-Mayoral M.F., Van Leuven F., Hayes J.D., Bertho G., Cuadrado A. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/beta-TrCP axis. Mol Cell Biol. 2012;32:3486–3499. doi: 10.1128/MCB.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taguchi K., Yamamoto M. The KEAP1-NRF2 system in cancer. Front Oncol. 2017;7:85. doi: 10.3389/fonc.2017.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cloer E.W., Goldfarb D., Schrank T.P., Weissman B.E., Major M.B. NRF2 activation in cancer: from DNA to protein. Cancer Res. 2019;79:889–898. doi: 10.1158/0008-5472.CAN-18-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibata T., Kokubu A., Saito S., Narisawa-Saito M., Sasaki H., Aoyagi K., Yoshimatsu Y., Tachimori Y., Kushima R., Kiyono T., Yamamoto M. NRF2 mutation confers malignant potential and resistance to chemoradiation therapy in advanced esophageal squamous cancer. Neoplasia. 2011;13:864–873. doi: 10.1593/neo.11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taguchi K., Yamamoto M. The KEAP1-NRF2 system as a molecular target of cancer treatment. Cancers (Basel) 2020;13 doi: 10.3390/cancers13010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma S., Paiboonrungruan C., Yan T., Williams K.P., Major M.B., Chen X.L. Targeted therapy of esophageal squamous cell carcinoma: the NRF2 signaling pathway as target. Ann N Y Acad Sci. 2018;1434:164–172. doi: 10.1111/nyas.13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi M., Kuga A., Suzuki M., Panda H., Kitamura H., Motohashi H., Yamamoto M. Microenvironmental activation of Nrf2 restricts the progression of Nrf2-activated malignant tumors. Cancer Res. 2020;80:3331–3344. doi: 10.1158/0008-5472.CAN-19-2888. [DOI] [PubMed] [Google Scholar]
- 18.Taguchi K., Hirano I., Itoh T., Tanaka M., Miyajima A., Suzuki A., Motohashi H., Yamamoto M. Nrf2 enhances cholangiocyte expansion in Pten-deficient livers. Mol Cell Biol. 2014;34:900–913. doi: 10.1128/MCB.01384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taguchi K., Maher J.M., Suzuki T., Kawatani Y., Motohashi H., Yamamoto M. Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol Cell Biol. 2010;30:3016–3026. doi: 10.1128/MCB.01591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wakabayashi N., Itoh K., Wakabayashi J., Motohashi H., Noda S., Takahashi S., Imakado S., Kotsuji T., Otsuka F., Roop D.R., Harada T., Engel J.D., Yamamoto M. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 21.Murakami S., Shimizu R., Romeo P.H., Yamamoto M., Motohashi H. Keap1-Nrf2 system regulates cell fate determination of hematopoietic stem cells. Genes Cells. 2014;19:239–253. doi: 10.1111/gtc.12126. [DOI] [PubMed] [Google Scholar]
- 22.Murakami S., Motohashi H. Roles of Nrf2 in cell proliferation and differentiation. Free Radic Biol Med. 2015;88(Pt B):168–178. doi: 10.1016/j.freeradbiomed.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 23.Yagishita Y., McCallum M.L., Kensler T.W., Wakabayashi N. Constitutive activation of Nrf2 in mice expands enterogenesis in small intestine through negative regulation of Math1. Cell Mol Gastroenterol Hepatol. 2021;11:503–524. doi: 10.1016/j.jcmgh.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horiuchi M., Taguchi K., Hirose W., Tsuchida K., Suzuki M., Taniyama Y., Kamei T., Yamamoto M. Cellular Nrf2 levels determine cell fate during chemical carcinogenesis in esophageal epithelium. Mol Cell Biol. 2021;41 doi: 10.1128/MCB.00536-20. e00536–00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blake D.J., Singh A., Kombairaju P., Malhotra D., Mariani T.J., Tuder R.M., Gabrielson E., Biswal S. Deletion of Keap1 in the lung attenuates acute cigarette smoke-induced oxidative stress and inflammation. Am J Respir Cell Mol Biol. 2010;42:524–536. doi: 10.1165/rcmb.2009-0054OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nioi P., McMahon M., Itoh K., Yamamoto M., Hayes J.D. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J. 2003;374(Pt 2):337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirotsu Y., Katsuoka F., Funayama R., Nagashima T., Nishida Y., Nakayama K., Engel J.D., Yamamoto M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012;40:10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu N., Matsumura H., Kato T., Ichinose S., Takada A., Namiki T., Asakawa K., Morinaga H., Mohri Y., De Arcangelis A., Geroges-Labouesse E., Nanba D., Nishimura E.K. Stem cell competition orchestrates skin homeostasis and ageing. Nature. 2019;568:344–350. doi: 10.1038/s41586-019-1085-7. [DOI] [PubMed] [Google Scholar]
- 29.Morata G., Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- 30.Bersell K., Choudhury S., Mollova M., Polizzotti B.D., Ganapathy B., Walsh S., Wadugu B., Arab S., Kuhn B. Moderate and high amounts of tamoxifen in alphaMHC-MerCreMer mice induce a DNA damage response, leading to heart failure and death. Dis Model Mech. 2013;6:1459–1469. doi: 10.1242/dmm.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hills S.A., Diffley J.F. DNA replication and oncogene-induced replicative stress. Curr Biol. 2014;24:R435–R444. doi: 10.1016/j.cub.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Piazzolla D., Palla A.R., Pantoja C., Canamero M., de Castro I.P., Ortega S., Gomez-Lopez G., Dominguez O., Megias D., Roncador G., Luque-Garcia J.L., Fernandez-Tresguerres B., Fernandez A.F., Fraga M.F., Rodriguez-Justo M., Manzanares M., Sanchez-Carbayo M., Garcia-Pedrero J.M., Rodrigo J.P., Malumbres M., Serrano M. Lineage-restricted function of the pluripotency factor NANOG in stratified epithelia. Nat Commun. 2014;5:4226. doi: 10.1038/ncomms5226. [DOI] [PubMed] [Google Scholar]
- 33.Gorgoulis V.G., Vassiliou L.V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A., Jr., Kastrinakis N.G., Levy B., Kletsas D., Yoneta A., Herlyn M., Kittas C., Halazonetis T.D. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 34.Kuga A., Tsuchida K., Panda H., Horiuchi M., Otsuki A., Taguchi K., Katsuoka F., Suzuki M., Yamamoto M. The β-TrCP-mediated pathway cooperates with the Keap1-mediated PPathway in Nrf2 degradation in vivo. Mol Cell Biol. 2022 doi: 10.1128/mcb.00563-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martens M., Ammar A., Riutta A., Waagmeester A., Slenter D.N., Hanspers K., R AM, Digles D., Lopes E.N., Ehrhart F., Dupuis L.J., Winckers L.A., Coort S.L., Willighagen E.L., Evelo C.T., Pico A.R., Kutmon M. WikiPathways: connecting communities. Nucleic Acids Res. 2021;49:D613–D621. doi: 10.1093/nar/gkaa1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang X.H., Knudsen B., Bemis D., Tickoo S., Gudas L.J. Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin Cancer Res. 2004;10(Pt 1):301–313. doi: 10.1158/1078-0432.ccr-0999-3. [DOI] [PubMed] [Google Scholar]
- 38.Ohkoshi A., Suzuki T., Ono M., Kobayashi T., Yamamoto M. Roles of Keap1-Nrf2 system in upper aerodigestive tract carcinogenesis. Cancer Prev Res (Phila) 2013;6:149–159. doi: 10.1158/1940-6207.CAPR-12-0401-T. [DOI] [PubMed] [Google Scholar]
- 39.Simpson P., Morata G. Differential mitotic rates and patterns of growth in compartments in the Drosophila wing. Dev Biol. 1981;85:299–308. doi: 10.1016/0012-1606(81)90261-x. [DOI] [PubMed] [Google Scholar]
- 40.Clayton E., Doupe D.P., Klein A.M., Winton D.J., Simons B.D., Jones P.H. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 41.Kon S., Ishibashi K., Katoh H., Kitamoto S., Shirai T., Tanaka S., Kajita M., Ishikawa S., Yamauchi H., Yako Y., Kamasaki T., Matsumoto T., Watanabe H., Egami R., Sasaki A., Nishikawa A., Kameda I., Maruyama T., Narumi R., Morita T., Sasaki Y., Enoki R., Honma S., Imamura H., Oshima M., Soga T., Miyazaki J.I., Duchen M.R., Nam J.M., Onodera Y., Yoshioka S., Kikuta J., Ishii M., Imajo M., Nishida E., Fujioka Y., Ohba Y., Sato T., Fujita Y. Cell competition with normal epithelial cells promotes apical extrusion of transformed cells through metabolic changes. Nat Cell Biol. 2017;19:530–541. doi: 10.1038/ncb3509. [DOI] [PubMed] [Google Scholar]
- 42.Colom B., Herms A., Hall M.W.J., Dentro S.C., King C., Sood R.K., Alcolea M.P., Piedrafita G., Fernandez-Antoran D., Ong S.H., Fowler J.C., Mahbubani K.T., Saeb-Parsy K., Gerstung M., Hall B.A., Jones P.H. Mutant clones in normal epithelium outcompete and eliminate emerging tumours. Nature. 2021;598:510–514. doi: 10.1038/s41586-021-03965-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martincorena I., Fowler J.C., Wabik A., Lawson A.R.J., Abascal F., Hall M.W.J., Cagan A., Murai K., Mahbubani K., Stratton M.R., Fitzgerald R.C., Handford P.A., Campbell P.J., Saeb-Parsy K., Jones P.H. Somatic mutant clones colonize the human esophagus with age. Science. 2018;362:911–917. doi: 10.1126/science.aau3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yokoyama A., Kakiuchi N., Yoshizato T., Nannya Y., Suzuki H., Takeuchi Y., Shiozawa Y., Sato Y., Aoki K., Kim S.K., Fujii Y., Yoshida K., Kataoka K., Nakagawa M.M., Inoue Y., Hirano T., Shiraishi Y., Chiba K., Tanaka H., Sanada M., Nishikawa Y., Amanuma Y., Ohashi S., Aoyama I., Horimatsu T., Miyamoto S., Tsunoda S., Sakai Y., Narahara M., Brown J.B., Sato Y., Sawada G., Mimori K., Minamiguchi S., Haga H., Seno H., Miyano S., Makishima H., Muto M., Ogawa S. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature. 2019;565:312–317. doi: 10.1038/s41586-018-0811-x. [DOI] [PubMed] [Google Scholar]
- 45.Doupe D.P., Alcolea M.P., Roshan A., Zhang G., Klein A.M., Simons B.D., Jones P.H. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science. 2012;337:1091–1093. doi: 10.1126/science.1218835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colom B., Alcolea M.P., Piedrafita G., Hall M.W.J., Wabik A., Dentro S.C., Fowler J.C., Herms A., King C., Ong S.H., Sood R.K., Gerstung M., Martincorena I., Hall B.A., Jones P.H. Spatial competition shapes the dynamic mutational landscape of normal esophageal epithelium. Nat Genet. 2020;52:604–614. doi: 10.1038/s41588-020-0624-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Busslinger G.A., Weusten B.L.A., Bogte A., Begthel H., Brosens L.A.A., Clevers H. Human gastrointestinal epithelia of the esophagus, stomach, and duodenum resolved at single-cell resolution. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108819. [DOI] [PubMed] [Google Scholar]
- 48.Orru C., Perra A., Kowalik M.A., Rizzolio S., Puliga E., Cabras L., Giordano S., Columbano A. Distinct mechanisms are responsible for Nrf2-Keap1 pathway activation at different stages of rat hepatocarcinogenesis. Cancers (Basel) 2020;12:2305. doi: 10.3390/cancers12082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haynie J.L., Bryant P.J. The effects of X-rays on the proliferation dynamics of cells in the imaginal wing disc of Drosophila melanogaster. Wilehm Roux Arch Dev Biol. 1977;183:85–100. doi: 10.1007/BF00848779. [DOI] [PubMed] [Google Scholar]
- 50.Fan Y., Bergmann A. Apoptosis-induced compensatory proliferation: the cell is dead—long live the cell. Trends Cell Biol. 2008;18:467–473. doi: 10.1016/j.tcb.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ragu S., Matos-Rodrigues G., Lopez B.S. Replication stress, DNA damage, inflammatory cytokines and innate immune response. Genes (Basel) 2020;11 doi: 10.3390/genes11040409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gasser S., Orsulic S., Brown E.J., Raulet D.H. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zand Irani M., Talley N.J., Ronkainen J., Aro P., Andreasson A., Agreus L., Vieth M., Jones M.P., Walker M.M. Neutrophils, eosinophils, and intraepithelial lymphocytes in the squamous esophagus in subjects with and without gastroesophageal reflux symptoms. Hum Pathol. 2021;115:112–122. doi: 10.1016/j.humpath.2021.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 55.Saigusa D., Motoike I.N., Saito S., Zorzi M., Aoki Y., Kitamura H., Suzuki M., Katsuoka F., Ishii H., Kinoshita K., Motohashi H., Yamamoto M. Impacts of NRF2 activation in non-small-cell lung cancer cell lines on extracellular metabolites. Cancer Sci. 2020;111:667–678. doi: 10.1111/cas.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levayer R. Solid stress, competition for space and cancer: the opposing roles of mechanical cell competition in tumour initiation and growth. Semin Cancer Biol. 2020;63:69–80. doi: 10.1016/j.semcancer.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayr M., Hu Y., Hainaut H., Xu Q. Mechanical stress-induced DNA damage and rac-p38MAPK signal pathways mediate p53-dependent apoptosis in vascular smooth muscle cells. FASEB J. 2002;16:1423–1425. doi: 10.1096/fj.02-0042fje. [DOI] [PubMed] [Google Scholar]
- 58.de la Cova C., Abril M., Bellosta P., Gallant P., Johnston L.A. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- 59.Martincorena I., Campbell P.J. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 60.Yizhak K., Aguet F., Kim J., Hess J.M., Kubler K., Grimsby J., Frazer R., Zhang H., Haradhvala N.J., Rosebrock D., Livitz D., Li X., Arich-Landkof E., Shoresh N., Stewart C., Segre A.V., Branton P.A., Polak P., Ardlie K.G., Getz G. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science. 2019;364 doi: 10.1126/science.aaw0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han L., Chaturvedi P., Kishimoto K., Koike H., Nasr T., Iwasawa K., Giesbrecht K., Witcher P.C., Eicher A., Haines L., Lee Y., Shannon J.M., Morimoto M., Wells J.M., Takebe T., Zorn A.M. Single cell transcriptomics identifies a signaling network coordinating endoderm and mesoderm diversification during foregut organogenesis. Nat Commun. 2020;11:4158. doi: 10.1038/s41467-020-17968-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.