Abstract
Background & Aims
Metastasis indicates a grave prognosis in patients with hepatocellular carcinoma (HCC). Our previous studies showed that RNA binding motif protein Y-linked (RBMY) is potentially a biomarker for poor survival in HCC patients, but its role in metastasis is largely unclear.
Methods
A total of 308 male patients with primary HCC were enrolled. RBMY expression was traced longitudinally by immunostaining from the manifestation of a primary HCC tumor to the formation of a distant metastasis, and its upstream regulators were screened with a protein microarray. A series of metastasis assays in mouse models and HCC cell lines were performed to explore new functional insights into RBMY.
Results
Cytoplasmic expression of RBMY was associated with rapid distant metastasis (approximately 1 year after resection) and had a predictive power of 82.4% for HCC metastasis. RBMY conferred high migratory and invasive potential upon phosphorylation by the provirus integration in Moloney 1 (PIM1) kinase. Binding of PIM1 to RBMY caused mutual stabilization and massive translocation of RBMY from nuclei to mitochondria, thereby preventing mitochondrial apoptosis and augmenting mitochondrial generation of adenosine triphosphate/reactive oxygen species to enhance cell motility. Depletion of RBMY suppressed Snail1/zinc finger E-box binding homeobox transcription factor 1–mediated epithelial–mesenchymal transition and dynamin-related protein 1–dependent mitochondrial fission. Inactivation and knockout of PIM1 down-regulated the expression of RBMY. In nude mice, cytoplasmic RBMY promoted liver-to-lung metastasis by increasing epithelial–mesenchymal transition, mitochondrial proliferation, and mitochondrial fission, whereas nuclear-restricted RBMY impeded the mitochondrial switch and failed to induce lung metastasis.
Conclusions
This study showed the regulation of HCC metastasis by PIM1-driven cytoplasmic expression of RBMY and suggested a novel therapeutic target for attenuating metastasis.
Keywords: Drp1, EMT, Mitochondria, Snail1, ZEB1
Abbreviations used in this paper: ATP, adenosine triphosphate; Bcl2, B-cell lymphoma 2; C+, cytoplasmic positive; CHX, cycloheximide; CIP, calf intestine phosphatase; CK1ε, casein kinase 1ε; Co-IP–WB, co-immunoprecipitation–Western blot; COX4, cyclooxygenase 4; CSC, cancer stem cell; DMEM, Dulbecco’s modified Eagle medium; Drp1, dynamin-related protein 1; EMT, epithelial-mesenchymal transition; FBS, fetal bovine serum; GFP, green fluorescent protein; GSK3β, glycogen synthase kinase 3β; HA, hemagglutinin; HCC, hepatocellular carcinoma; His, histidine; IFA, immunofluorescence assay; IHC, immunohistochemistry; IMM, inner mitochondrial membrane; MEK2, mitogen-activated protein/ERK kinase 2; Mix, mixed nuclear positive and cytoplasmic positive; mtROS, mitochondrial reactive oxygen species; N+, nuclear positive; NAC, N-acetyl cysteine; NESm, nuclear export signal mutant; NLSm, nuclear localization signal mutant; OMM, outer mitochondrial membrane; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; PIM1, provirus integration in Moloney 1; RBMY, RNA binding motif protein Y-linked family 1 member A1; ROS, reactive oxygen species; RRM, RNA recognition motif; sgPIM1, single-guide RNA against PIM1; sgRNA, single-guide RNA; shCtrl, short hairpin control; shRBMY, RNA binding motif protein Y-linked–specific short hairpin RNA; shRNA, short hairpin RNA; siCtrl, small interfering control; siPIM1, siRNA targeting provirus integration in Moloney 1; siRBMY, RNA binding motif protein Y-linked–specific small interfering RNA; siRNA, small interfering RNA; SRGY, serine-arginine-glycine-tyrosine repetitive segments; WT, wild-type; ZEB1, zinc finger E-box binding homeobox transcription factor 1; 3D, 3-dimensional
Graphical abstract

Summary.
We identified cytoplasmic RNA binding motif protein Y-linked (RBMY) as a predictive biomarker of hepatocellular carcinoma metastasis. Provirus integration in Moloney 1 phosphorylates RBMY. This stabilized form of RBMY may translocate to the cytoplasm and mitochondria, foster epithelial–mesenchymal transition, and induce mitochondrial dysregulation to result in distant metastasis of hepatocellular carcinoma.
A striking male predominance has been noted in a wide spectrum of human malignancies, including cancers of the lip, larynx, nasopharynx, esophagus, stomach, liver, and others.1, 2, 3 Although oncogenic roles of sex-related hormones have been proposed to explain the sex disparity, this is still insufficient to explain the mechanism of tumor development because the sex disparity is sometimes observed in pediatric malignancies.1,3 The involvement of an oncoprotein encoded by a gene on the Y chromosome seems to be a reasonable assumption. RNA binding motif protein Y-linked family 1 member A1 (RBMY) therefore is identified for its notable reactivation in male hepatocellular carcinomas (HCCs).4
Among tissues in adult human beings, RBMY is expressed exclusively in the testis (www.proteinatlas.org) and is assumed to be a male germline RNA splicing regulator.5, 6, 7, 8 It comprises an N-terminal RNA recognition motif (RRM) and 4 repeat segments rich in serine, arginine, glycine, and tyrosine residues (SRGY boxes) at the C-terminus.5 During early spermatogenesis, RBMY is detectable in spermatogonia throughout the maturation period.9,10 Spermatozoa motility is positively linked to the copy number of RBMY and decreases upon incubation with an anti-RBMY antibody,9,10 implying that RBMY may play a role in cell motility in addition to splicing.
During liver development, RBMY is present within hepatoblasts and cholangioblasts of human and rodent fetuses and then is silenced in mature hepatocytes.11 However, it is reactivated in childhood and adult HCC to confer key traits of cancer stem cells (CSCs), including self-renewal and chemoresistance.11 HCC generally shows a universal trend toward high mortality rates because of its frequent recurrence and metastasis even after curative resection.12 The high incidence of postoperative relapse is attributable to the presence of CSCs, which have a high capacity to transform into invasive and metastatic cancer cells via regulation of epithelial–mesenchymal transition (EMT).12 EMT is initially a critical morphogenetic event during embryonic development, allowing immotile epithelial cells to undergo a shift toward a motile mesenchymal phenotype; however, cancer cells switch on this embryonically restricted process to acquire migratory and invasive properties to trigger the metastatic cascade.13
Because developmental programs are reactivated in cancers, we presumed that the oncofetal protein, RBMY, may be re-expressed to mediate tumor aggressiveness. In this study, we proved this hypothesis from bedside to bench by validating patient data in the contexts of cancer cell lines and mouse models. Our findings showed that RBMY is indeed one of the metastatic signature genes and that its upstream serine/threonine kinase, provirus integration in Moloney 1 (PIM1), could enhance its phosphorylation-mediated activation and oncogenic function. PIM1 is expressed in a wide range of cancers and impacts various oncogenic pathways, including those mediating cell-cycle regulation, proliferation, apoptosis, drug resistance, stemness, and migration.14 This study further highlighted the prometastatic mechanism of PIM1 via phosphorylation of RBMY.
Results
Cytoplasmic RBMY Predicts Poor Prognosis in Patients With HCC
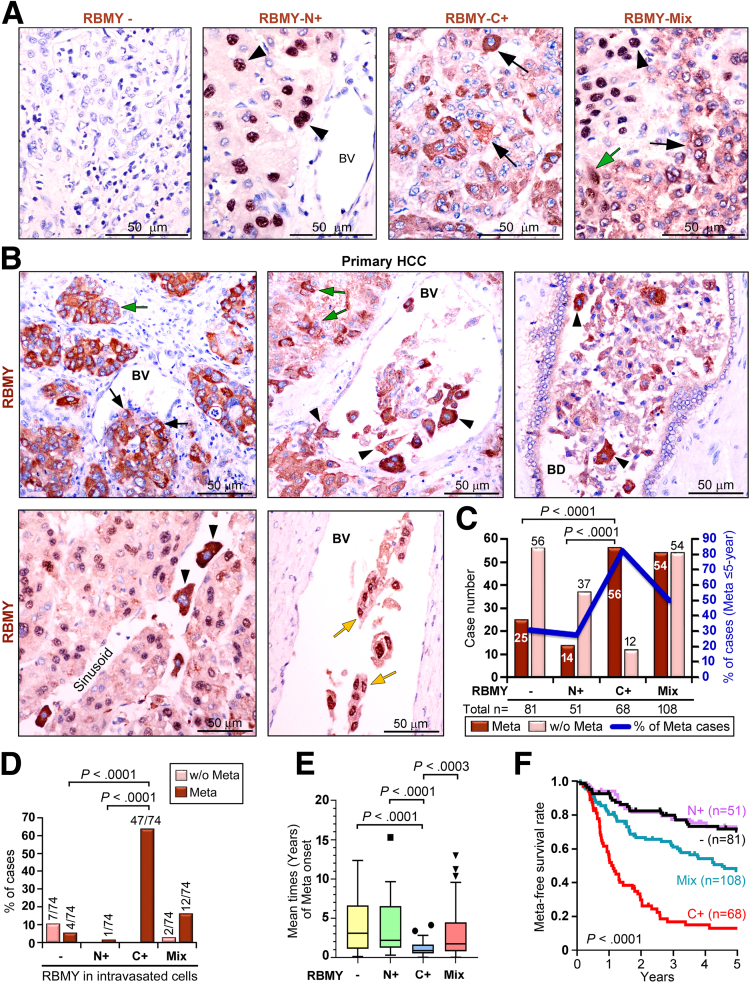
RBMY plays multiple oncogenic roles. In addition to driving HCC stemness,11 RBMY promoted xenograft tumor growth in nude mice, which were validated by the application of RBMY-specific small interfering RNA (siRNA) (siRBMY) or short hairpin RNA (shRNA) (shRBMY) and their parallel nontargeting controls, small interfering controls (siCtrls) and short hairpin controls (shCtrls), respectively (Figure 1A–C). In addition, RBMY enhanced colony formation on soft agar (Figure 1D), cell migration, and invasion (Figure 1E and F), suggesting that RBMY contributes to the malignancy of HCC. A large cohort (N = 308) of male HCC patients therefore was recruited, and the HCC specimens were analyzed by immunohistochemistry (IHC) to explore the expression of RBMY before and after metastasis formation. Four RBMY expression patterns, including negative (-), nuclear positive (N+), cytoplasmic positive (C+), and mixed N+ with C+ (Mix), were shown in the primary HCC tissues (Figure 2A). RBMY-C+ tumor cells not only were embedded in the hepatic parenchyma but also were spread along blood vessels and had successfully invaded into the blood vessels, bile ducts, and sinusoids as single cells or microemboli (Figure 2B). RBMY-Mix cells also were detected, albeit less frequently, among the intravasated cells (Figure 2B). Patients with the RBMY-C+ pattern were the most likely to experience relapse with distant metastasis because as many as 82.4% (56 of 68) of patients with the RBMY-C+ pattern developed distant metastasis within 5 years posthepatectomy (Figure 2C).
Figure 1.
RBMY plays multiple oncogenic functions in HCC cell lines. Subcutaneous HCC xenograft nude mice models engrafted with (A) siRNA-transfected and (B) shRNA-transduced Huh-7 cells were established. The endogenous RBMY was depleted with (A) siRBMY or (B) shRBMY. (B) Restoration of RBMY was achieved by overexpression of HA-RBMY. The xenograft tumor growth in nude mice was measured and compared with the control groups, (A) siCtrl and (B) shCtrl. The silencing efficacy of RBMY was assessed by immunoblotting. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) represents a loading control. (C) Tumor volume determined the xenograft growth and is plotted as means ± SDs. Statistical significance was evaluated with the Student t test. (D) Soft agar colony formation of stable Huh-7 transfectants was reported as means ± SDs (Student t test). (E) Scratch wound healing assay showed the migratory ability of indicated cells. Migration distance was assessed and depicted as means ± SDs (Student t test). (F) Transwell invasion assay showed the invasive capacity of the indicated cells. Bar graph (means ± SDs, Student t test) represents the number of cells migrating across Transwell membranes. RBMY-p, phosphorylated-RBMY.
Figure 2.
Expression of RBMY impacts the prognosis of patients with HCC. (A) RBMY-specific IHC was achieved on paraffin-embedded primary HCC tissue sections. Four different expression patterns of RBMY are shown: -, RBMY negative; N+, nuclear-positive (black arrowheads); C+, cytoplasmic-positive (black arrows); and Mix, positive signals in both nuclei and cytoplasm (green arrow). (B) IHC showed the distribution of RBMY in primary HCC tissues. Brown DAB-stained cells that expressed cytoplasmic RBMY were located in tumor nests (green arrows), migrated toward blood vessels (BV) (black arrows), and invaded into the blood vessels, bile duct (BD), and sinusoids (black arrowheads). RBMY sometimes was found in both the nuclei and cytoplasm of intravasated tumor cells (yellow arrows). (C) The number and percentage of cases with or without (w/o) distant metastasis (Meta) occurring within 5 years posthepatectomy were plotted according to the expression pattern of RBMY in primary HCC tissues. Statistical significance was estimated by the Mann‒Whitney U test. (D) Bar graphs show the percentage of primary HCC cases with or without distant metastasis with respect to the 4 expression patterns of RBMY in intravasated tumor cells (Mann‒Whitney U test). (E) Mean times (years, means ± SEMs) of metastasis onset posthepatectomy during the 15-year study were compared between the groups using the Student t test. (F) The metastasis-free survival rate of HCC patients based on the expression pattern of RBMY in primary HCC tissues was plotted by the Kaplan–Meier method with the log-rank test.
In 118 paraffin-embedded tissue sections, intravasated cells were clearly found in 74 primary HCC samples, and the RBMY-C+ pattern (in 63.5% [47 of 74] of patients) was distinctly associated with a higher prevalence of distant metastasis (Figure 2D). In addition, patients with the RBMY-C+ pattern showed an increased tendency toward earlier metastasis that occurred an average of 1 year posthepatectomy, and these patients had a significantly lower metastasis-free survival rate (Figure 2E and F). The level of RBMY was maintained in these HCC cells from patients who developed metastases in the distal organs (Figure 3A), as evidenced by the immunofluorescence assay (IFA) for detecting hepatocyte differentiation markers (hepatocyte paraffin-1 and hepatocyte nuclear factor 4α), which showed the RBMY-positive metastases in distant organs originated from HCC tissues (Figure 3B). Besides, RBMY was colocalized with cytokeratin 19, a biliary/hepatic progenitor cell marker (Figure 3A), confirming that RBMY was expressed not only in hepatocytes but also in progenitor cells within the metastases of HCC.
Figure 3.
RBMY is initially expressed in the primary HCC and the expression level is retained until the formation of distant metastases. (A) RBMY-specific IHC was performed on the paraffin-embedded tissue sections of primary tumors and paired metastases of HCC. Black arrows and arrowheads point to cells expressing RBMY-C+ and RBMY-N+, respectively. Green arrows indicate RBMY-Mix. (B) Double-staining IFA of RBMY and hepatic markers (Hep Par-1, hepatocyte nuclear factor 4α [HNF-4α] and cytokeratin 19 [CK19]) was conducted on the tissue sections of HCC metastases. Cells co-expressed RBMY and Hep Par-1, CK19, or HNF-4α are indicated by white arrows. Hep par-1 and CK19 are expressed in the cytoplasm, while HNF-4α is expressed in the nuclei. BV, blood vessel; DAPI, 4′,6-diamidino-2-phenylindole.
Phosphorylation of RBMY by PIM1 Enhances Its Oncogenicity
RBMY is a phosphoprotein.11 To reveal the regulatory control(s) that drove RBMY to become a prometastatic factor, a ProtoArray Human Protein Microarray (Invitrogen, Carlsbad, CA) was performed to identify RBMY-binding proteins. Several serine/threonine kinase candidates, including PIM1, mitogen-activated protein kinase kinase 2 (MEK2), glycogen synthase kinase 3β (GSK3β), casein kinase 1ε (CK1ε), and others, were revealed (Figure 4A). A subsequent in vitro kinase assay showed the ability of these kinases to phosphorylate histidine (His)-tagged RBMY was decreased in the order of PIM1 > CK1ε > MEK2, which was observable only in the presence of adenosine triphosphate (ATP) (Figure 4B). An enhanced phosphorylation of green fluorescent protein (GFP)-RBMY by hemagglutinin (HA)-tagged PIM1, MEK2, and CK1ε in the HCC cell line also was detected by co-immunoprecipitation coupled with Western blot (co-IP–WB) analysis (Figure 4C). Although GSK3β bound to RBMY, it did not phosphorylate RBMY (Figure 4B and C). Alkaline calf intestine phosphatase (CIP) treatment further verified that the distinct band shift patterns of His-RBMY and GFP-RBMY were produced by phosphorylation of RBMY catalyzed by PIM1, CK1ε, and MEK2 (Figure 4D and E). PIM1 also phosphorylated the endogenous RBMY, which was confirmed by immunoprecipitation and CIP assay (Figure 4F). To detail the interaction between kinase and substrate, co-IP–WB was achieved and showed that wild-type (WT) RBMY and the mutant (designated SRGY) containing 4 SRGY boxes showed a high capacity for binding to PIM1 and that the binding capacity was greatly reduced when SRGY boxes 1–3 were truncated simultaneously (Figure 4G).
Figure 4.
Phosphorylation of RBMY by its upstream kinases. (A) Hierarchical clustering of RBMY-bound protein kinases, which were shown by analysis of a human protein array, based on Z-score classification. (B) An in vitro kinase assay followed by immunoblotting showed the phosphorylation of His-RBMY (His-RBMY-p) by the indicated kinases. Phosphorylation reactions were performed in the presence or absence of ATP. (C) Co-IP–WB on Huh-7 cells overexpressed with GFP-RBMY and HA-kinases are indicated. Ten percent input of total protein cell lysate used for co-IP–WB is shown. The blot showed clear bands of different phosphorylation patterns of RBMY by its upstream kinases. Blotting of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) verifies equal protein input. GAPDH was absent in the immunocomplex, confirming the specificity and purity of anti-GFP immunoprecipitation. (D) CIP dephosphorylated His-RBMY on Ser/Thr residues. Recombinant His-RBMY proteins were phosphorylated by His-PIM1 and His-CK1ε in an in vitro kinase assay. The phosphorylated His-RBMY proteins were purified using His-tagged protein purification spin columns, and the eluted proteins were subjected to CIP treatment and analyzed by Western blot using anti-RBMY antibody (SRGY clone 1). (E) Dephosphorylation of GFP-RBMY by CIP. Huh-7 cells cotransfected with GFP-RBMY and HA-kinases were harvested for CIP (20 U) treatment. The result was revealed by immunoblotting using SRGY clone 1 antibody. (F) Immunoprecipitation assay targeting the endogenous RBMY was performed on Huh-7 cells transfected with or without HA-PIM1. Anti-RBMY antibody (SRGY clone 1) was used to precipitate the phosphorylated RBMY (RBMY-p) mediated by HA-PIM1 (lane 5) and other endogenous kinases (lane 6). CIP treatment then was performed to dephosphorylate the immunoprecipitated RBMY-p. Western blot was accomplished with SRGY clone 1 monoclonal antibody, which recognized RBMY-p (lanes 5 and 6) and the nonphosphorylated RBMY (lanes 7 and 8). (G) Schematic illustration of domain structures of RBMY deletion mutants. These mutants bound (+) or did not bind (−) PIM1 p34 as indicated. Co-IP–WB using anti-HA and anti-GFP antibodies identified the RBMY domain essential for PIM1 interaction. Detection of GAPDH confirms equal protein input and the specificity of anti-HA and anti-GFP immunoprecipitation. The specific bands of interest are indicated by red arrowheads and the asterisk shows the nonspecific bands yielded by using anti-HA antibody for immunoprecipitation. Ab, antibody.
Compared with the HA controls, HA-MEK2 and HA-CK1ε, phosphorylation of GFP-RBMY by HA-PIM1 particularly augmented its oncogenic activity in terms of enhanced proliferation (Figure 5A), 3-dimensional (3D) spheroid invasion (Figure 5B), migration (Figure 5C), and Transwell invasion (Figure 5D). These observations were substantiated in a panel of male liver cancer cell lines, including Huh-7, PLC/PRF/5, SK-HEP-1, and HepG2 cells, as well as the RBMY-null female cell line (SNU-387) upon overexpression of GFP-RBMY and HA-PIM1 (Figure 5A–F). When RBMY was knocked-down by siRBMY, the migration and invasion of PLC/PRF/5 cells evidently were blocked (Figure 5G). Overexpression of HA-PIM1 moderately rescued the cell migratory ability of siRBMY cells but failed to induce invasion compared with those of cells expressing siCtrl + HA-PIM1 (Figure 5G), indicating that RBMY is one of the critical effectors of PIM1.
Figure 5.
Regulation of RBMY oncogenicity by its upstream kinases. (A) AlamarBlue cell viability assay evaluated the survival and proliferation ability of the indicated transfectants. Cell viability represented by the measured absorbance at 570 nm was plotted as the means ± SDs after normalization to the corresponding values at 0 hours. ∗P < .005 vs the GFP+HA control was determined by the Student t test. (B) 3D spheroid invasion assay of indicated transfectants. The invasiveness of spheroids grown in a 3D matrix was determined by the formation and enrichment of spindle-like protrusions (black arrows). Representative micrographs acquired before and after the formation of protrusions from spheroid bodies are shown. The mean percentage of the invaded spheroids was calculated and is shown in a bar graph. The error bars indicate the SDs of 3 wells in 2 independent experiments, and significance was evaluated by the Student t test. (C) A wound healing assay showed the migration efficacy of the indicated transfectants. A representative image of each group before (0 h) and after (48 h) cell migration is displayed. Cells expressing GFP and HA served as controls. The migration distances were measured and are reported as the means ± SDs (Student t test). The data shown are representative of 3 independent experiments. (D) Transwell invasion assay to determine the invasive potential of the indicated transfectants. The numbers of invaded cells are presented as the means ± SDs of 4 independent experiments, and significance was evaluated by the Student t test. (E) Scratch wound healing assay showed the migratory abilities of SNU-387 cells cotransfected with plasmids indicated. The migration distances were depicted as the means ± SDs (Student t test). (F) Transwell invasion assay on the indicated transfectants. The number of invaded cells was plotted as means ± SDs and evaluated by the Student t test for statistical significance. (G) Scratch wound healing and Transwell invasion assays on cells transfected twice with siCtrl/siRBMY, then once with HA/HA-PIM1 expression plasmids. Migration distance (means ± SDs) and number of invaded cells (means ± SDs) were analyzed 48 hours postseeding, and differences between groups were estimated by the Student t test.
PIM1 Increases the Cytoplasmic RBMY Level to Exacerbate HCC Malignancy
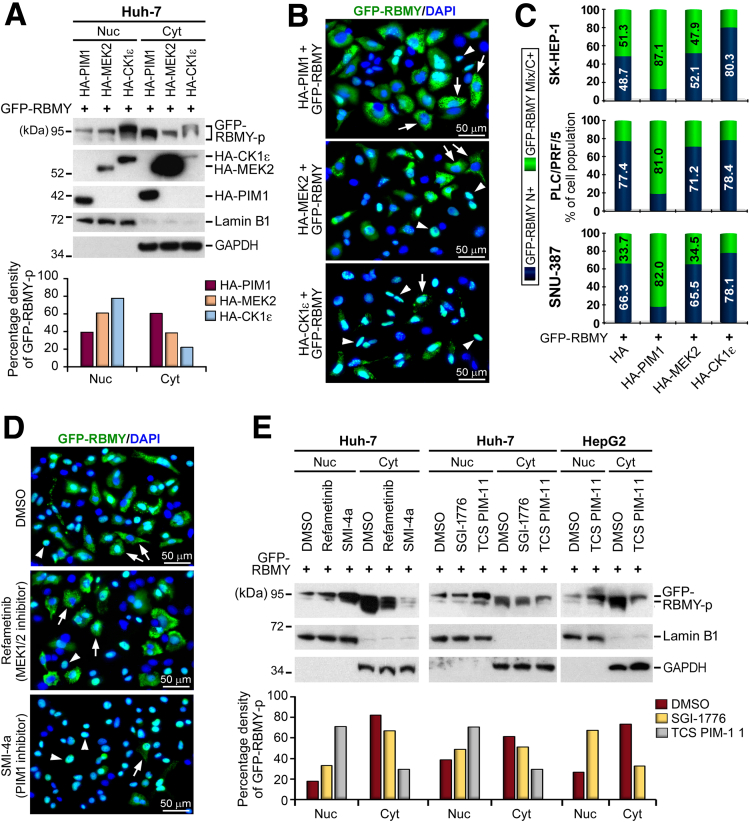
A prominent cytoplasmic pool of GFP-RBMY was observed in cells co-expressing HA-PIM1, whereas HA-CK1ε expression restricted GFP-RBMY to nuclei (Figure 6A and B). This phenomenon generally was detected in a variety of HCC cell lines (Figure 6C). Although neither overexpression of MEK2 nor inhibition of MEK2 by refametinib (a MEK1/2 inhibitor) affected the subcellular distribution of RBMY, blockade of PIM1 activity by a selective ATP-competitive kinase inhibitor (SMI-4a) restricted most of the GFP-RBMY protein to nuclei (Figure 6D and E). A similar result was obtained by treatment with another PIM1-specific inhibitor (TCS PIM-1 1), but not by treatment with a pan-PIM kinase inhibitor (SGI-1776) (Figure 6E), suggesting that PIM1 is the factor determining the cytoplasmic expression of RBMY.
Figure 6.
Increasing cytoplasmic accumulation of RBMY by PIM1. (A) Subcellular fractionation coupled with Western blot assay on Huh-7 cells co-expressed with proteins indicated. Detection of Lamin B1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was performed as for controls of the purities of nuclear (Nuc) and cytoplasmic (Cyt) fractions, respectively. Bar plot represents the densitometry analysis of GFP-RBMY-p. The densities of GFP-RBMY-p in nuclear and cytoplasmic fractions were normalized to the intensities of their corresponding markers, Lamin B1 and GAPDH, respectively, and then divided by the total nuclear + cytoplasmic densities of GFP-RBMY-p to obtain a percentage value. (B) Fluorescence imaging distinguished the impact of the indicated kinases on the distribution of GFP-RBMY in Huh-7 cells. The arrowheads and arrows indicate the RBMY-N+ and RBMY-C+/Mix patterns, respectively. (C) Comparison of the subcellular distribution of GFP-RBMY upon co-expression with HA-PIM1, HA-MEK2, or HA-CK1ε kinases in HCC cell lines. The microscopic signals of GFP-RBMY-Mix/C+ and N+ were plotted as a mean percentages of the cell population (±SDs). (D) Fluorescence microscopy analysis showed the N+ (arrowhead) and C+/Mix (arrow) patterns of GFP-RBMY in Huh-7 cells upon treatment of kinases inhibitors. (E) Western blot analysis exposed the expression level of GFP-RBMY in nuclear and cytoplasmic fractions of HCC cells treated with dimethyl sulfoxide (DMSO) solvent control or kinase inhibitors. Cells were incubated with the indicated inhibitors (30 μmol/L per well) after attachment. Transfection of GFP-RBMY was performed 24 hours post-treatment. Cells then were refreshed with medium containing the indicated inhibitors and cultured for another 24 hours before subcellular fractionation. Quantitative densitometry analysis was accomplished as mentioned previously. DAPI, 4′,6-diamidino-2-phenylindole; RBMY-p, phosphorylated RBMY.
To investigate the clinical significance of PIM1-RBMY regulation in patients with primary HCC, immunostaining of PIM1 and RBMY was performed. PIM1 showed the C+ and Mix patterns in primary HCC tissues (Figure 7A). A monoclonal antibody (SRGY clone 1) recognizing both PIM1-phosphorylated RBMY and nonphosphorylated RBMY (Figure 4F) was applied for subsequent double-stained IFA and IHC analysis. In the peritumoral tissues of primary HCC where PIM1 existed in C+ or Mix pattern, RBMY was either undetectable or co-expressed with PIM1 in nuclei (Figure 7B and C). However, RBMY tended to be expressed at higher levels and colocalized with PIM1-C+/Mix cells in tumors and intravasated cells, but restricted to the nuclei once PIM1 was absent (Figure 7B and C).
Figure 7.
Tumorigenic expression of PIM1 and RBMY in primary HCC tissues. (A) PIM1-specific IHC on paraffin-embedded tissue sections of primary HCC. PIM1-C+–expressing tissue showed PIM1-positive signal only in the cytoplasm. PIM1-Mix pattern comprised cells expressing both nuclear and cytoplasmic PIM1 (arrowheads) and also PIM1-C+ (arrows). (B) PIM1 and RBMY double-stained IFA was accomplished on primary HCC tissues using anti-PIM1 and SRGY clone 1 antibodies. Yellow arrows, PIM1-Mix/RBMY-N+; white arrows, PIM1-Mix/RBMY-Mix; and white arrowheads, PIM1-C+/RBMY-C+. (C) PIM1- and RBMY-specific IHC on serial paraffin-embedded tissue sections of primary HCC. BV, blood vessel; DAPI, 4′,6-diamidino-2-phenylindole.
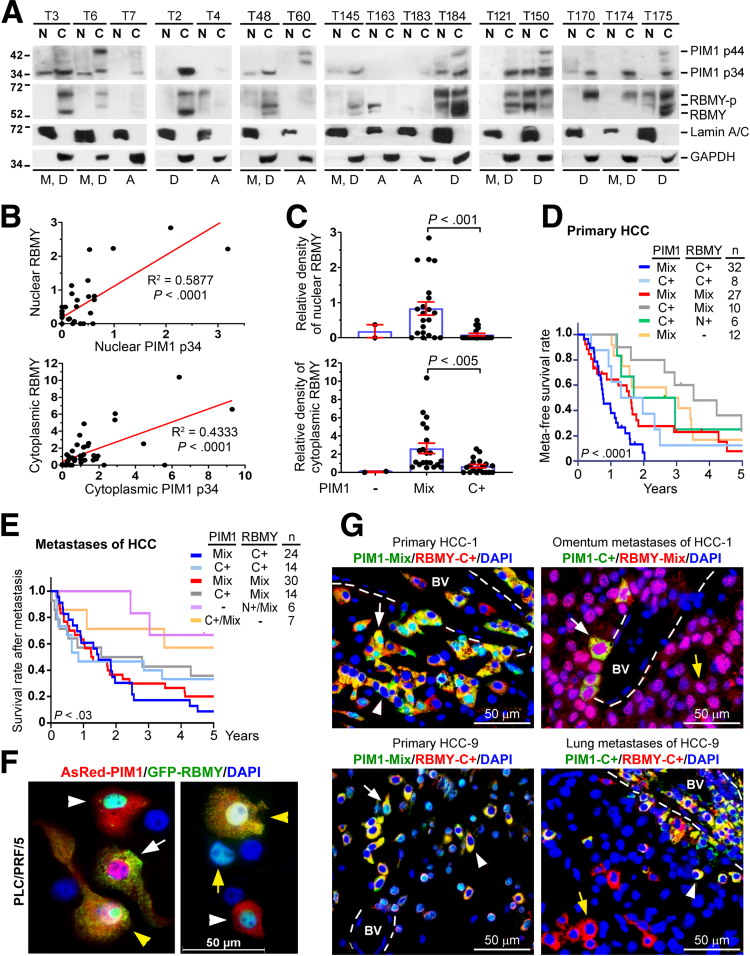
Subcellular fractionation of primary HCC tissues (tumor size, ≥25 mm3; total N = 42; 16 cases are shown) showed a greater positive rate of PIM1 p34 (85.7%; 36 of 42) than PIM1-p44 (31.0%; 13 of 42) (Figure 8A), which are the 34 and 44 kilodalton isoforms of PIM1, respectively.15 Notably, in patients without and with trivial amounts of PIM1 p34, RBMY was expressed at equivalent minute levels (T7, T60, and T183) or was retained in the nuclear fraction (T4 and T163) (Figures 8A ). These patients remained alive at the end of the study (Figure 8A). In contrast, patients who had abundant expression of PIM1 and RBMY were associated with a greater risk of tumor metastasis and death during the study period (Figure 8A). Densitometric measurements of PIM1 and RBMY in these 42 HCC tissues were performed. A strong positive correlation was found between PIM1 and RBMY in both nuclear (R2 = 0.5877; P < .0001) and cytoplasmic (R2 = 0.4333; P < .0001) fractions of HCC tissues (Figure 8B). Importantly, all patients with the PIM1-Mix pattern were positive for cytoplasmic RBMY, and showed higher levels of both nuclear (P < .001) and cytoplasmic (P < .005) RBMY than PIM1-C+ patients (Figure 8C). In addition, Kaplan‒Meier analysis showed that the patients with the PIM1-Mix/RBMY-C+ pattern in primary HCC tissues and metastases had significantly shorter metastasis-free survival times and the poorest survival rate after metastasis (Figure 8D and E).
Figure 8.
PIM1-associated cytoplasmic expression of RBMY is a risk factor for poor prognosis in HCC. (A) Subcellular fractionation and Western blot were performed to analyze the levels of PIM1 and RBMY in the nuclear and cytoplasmic fractions of primary HCC tissues (N = 42). Lamin A/C and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as the controls for the nuclear and cytoplasmic fractions, respectively, to confirm the purity and loading amount. (B and C) A significant positive correlation between PIM1 and RBMY in patients with HCC. (A) Densitometry analysis was conducted to measure the nuclear and cytoplasmic intensities of PIM1 p34 and RBMY and standardized with the intensities of Lamin A/C and GAPDH, respectively. (B) Linear correlations were established between the nuclear and cytoplasmic fractions of PIM1 p34 and RBMY. (C) The levels of nuclear and cytoplasmic RBMY were compared among cases expressing PIM1- (N = 2), PIM1-Mix (N = 21), and PIM1-C+ (N = 19). Data were analyzed by the Mann–Whitney U test for statistical significance. Kaplan–Meier analysis with the log-rank test was performed to estimate the (D) 5-year metastasis-free survival rate and the (E) 5-year survival rate after metastasis according to the expression patterns of PIM1 and RBMY in (D) primary tumors and (E) metastases of HCC tissues. (F) Fluorescence microscopy analysis of PLC/PRF/5 cells cotransfected with AsRed2-PIM1 and GFP-RBMY plasmids. 4′,6-diamidino-2-phenylindole (DAPI) indicates nuclear staining. White arrows point to cells expressing PIM1-Mix/RBMY-C+. Yellow arrowheads specify cells showing PIM1-Mix/RBMY-Mix. White arrowheads show cells expressing PIM1-C+/RBMY-N+. Yellow arrows indicate cell expressing RBMY-N+, but the absence of PIM1. (G) Double IFA staining of primary tumors and paired metastases of HCC. White arrows, PIM1-Mix/RBMY-C+ or PIM-C+/RBMY-Mix; white arrowheads, PIM1-C+/RBMY-C+. The yellow arrows show the dissociation of the PIM1-RBMY complex. A, alive; BV, blood vessel; C, cytoplasm; D, death; M, metastasis; N, nucleus.
PIM1-Mix caused poorer outcomes in HCC patients than PIM1-C+ (Figure 8D and E). To explain this observation, we examined the changes in the distribution of RBMY in response to the PIM1-Mix and C+ patterns. Fluorescence images of PLC/PRF/5 cells co-expressed AsRed-PIM1, and GFP-RBMY showed that AsRed-PIM1-Mix cells typically showed nuclear-to-cytoplasmic translocation of GFP-RBMY, whereas GFP-RBMY was retained in the nucleus of AsRed-PIM1-C+ cells, similar to the observation in cells without AsRed-PIM1 expression (Figure 8F). Time-lapse imaging of these live cells further elucidated this finding by comparing 2 daughter cells from 1 cell division, in which GFP-RBMY initially was expressed in the nuclei of both daughter cells (Video 1, white and black arrows). Once the cytoplasmic AsRed-PIM1 entered into the nucleus, a shift of the nuclear GFP-RBMY toward cytoplasm occurred and was spread to the cytoplasmic edge (Video 1, white arrows). However, in the other daughter cell, which had the AsRed-PIM1–C+ pattern, the nuclear export of GFP-RBMY was restricted (Video 1, black arrow). Although sorafenib (10 μmol/L) treatment conferred photosensitivity on AsRed-PIM1–expressing PLC/PRF/5 cells, the AsRed-PIM1–Mix pattern with GFP-RBMY expression effectively prevented cell death (Video 2), implying the increased aggressiveness of these cells. Thus, the oncogenicity of PIM1-Mix cells can be explained by their increased cytoplasmic RBMY level.
The colocalization signal of PIM1 and RBMY generally was found in primary HCC tissues and was maintained until these cells extravasated to distant organs, but was diminished when they colonized the metastatic sites (Figure 8G), suggesting that the formation of the PIM1-RBMY complex is necessary for metastatic spread before colonization. However, both PIM1 and RBMY are essential for the growth of metastases because patients with loss of either PIM1 or RBMY expression in distant metastases had longer survival times after metastasis (Figure 8E).
PIM1 Phosphorylates RBMY to Initiate EMT
The induction of EMT is a key driving force for metastasis development.13 Snail1, Slug (Snail2), twist, and zinc finger E-box binding homeobox transcription factor 1 (ZEB1) are the transcription factors of EMT that repress E-cadherin (epithelial marker) but enhance the expression of the mesenchymal genes encoding N-cadherin, fibronectin, and vimentin.16 The capability of PIM1 to modulate EMT has been suggested,17,18 we therefore hypothesized that RBMY may cooperate with PIM1 to enhance EMT and subsequently contribute to the metastasis of HCC. To this aim, we first tested the role of RBMY in EMT by isolating total RNA from HCC cells with overexpression of GFP-RBMY or depletion of RBMY (shRBMY) in parallel with the corresponding GFP and shCtrl cells. Reverse-transcription quantitative polymerase chain reaction (PCR) evaluating the expression levels of a panel of EMT markers was conducted. PLC/PRF/5 cells normally showed greater migration and invasion potential than HepG2 cells, yet Huh-7 cells showed the lowest potential (data not shown). Upon GFP-RBMY overexpression, both PLC/PRF/5 and HepG2 cells showed significantly increased amounts of Snail1, ZEB1, and N-cadherin accompanied by a decreased E-cadherin level (Figure 9A). On the other hand, silencing RBMY in HepG2 cells led to reductions in Snail1 and ZEB1 levels but an increase in E-cadherin (Figure 9A). Although increased levels of EMT transcripts were observed in Huh-7 cells in response to GFP-RBMY expression, these increases were not statistically significant (Figure 9A).
Figure 9.
RBMY regulates EMT at a transcriptional level. The EMT signature was evaluated by reverse-transcription quantitative PCR in (A) HCC cell lines transfected with GFP-RBMY or shRBMY and in (B) primary HCC tissues (N = 100). Up-regulation and down-regulation of EMT transcripts were quantified and reported as (A) average fold changes ± SDs and (B) 2-△Ct values. Statistical significance was evaluated with an unpaired Student t test. ∗P < .05, ∗∗P < .005, and ∗∗∗P < .0005. (C) The transcript–transcript correlations between RBMY and Snail1, ZEB1, E-cadherin, or N-cadherin were analyzed, and the significance of the linear regression analysis results is shown.
To further elaborate on the association between RBMY and EMT, 100 primary HCC tissues were subjected to RNA isolation and reverse-transcription quantitative PCR analysis. The positive rate of RBMY in these tissues was 80% (80 of 100), and the RBMY-positive tissues tended to express increased amounts of Snail1 and ZEB1 and a concomitantly reduced amount of E-cadherin, with statistically significant differences (Figure 9B). At the transcript level, RBMY expression in these primary HCC tissues was correlated significantly with those of Snail1 (R2 = 0.2663; P < .0001) and ZEB1 (R2 = 0.2605; P < .0001), but not with that of E-cadherin (R2 = 0.0058; P = .4513) or N-cadherin (R2 = 0.0265; P = .1060) (Figure 9C), hinting that RBMY may regulate EMT mainly through enhancing the transcription of Snail1 and ZEB1.
PIM1 has been noted to control EMT by modulating the activity of ZEB1.18 To reveal the mechanism by which RBMY contributes to EMT in comparison with that of PIM1, siRNAs targeting PIM1 (siPIM1), siRBMY, and siCtrl were transfected into HCC cell lines. PLC/PRF/5 cells harboring siPIM1 showed silencing of PIM1-p44/p34 and simultaneous reductions in Slug, ZEB1, and N-cadherin levels (Figure 10A). When the total PIM1 pool was rescued by overexpression of HA-PIM1, the levels of Slug and ZEB1 were increased, as was the level of RBMY-p (Figure 10A). Remarkably, knockdown of RBMY decreased the levels of Snail1, ZEB1, and PIM1 p34, while introduction of GFP-RBMY restored the levels of these proteins (Figure 10A). Of greater importance, RBMY replenishment reconstructed the EMT expression spectrum by decreasing the E-cadherin level and increasing the levels of Slug, N-cadherin, fibronectin, and vimentin, in addition to Snail1 and ZEB1 (Figure 10A). Likewise, this phenomenon was reproduced in Huh-7 cells upon RBMY depletion and restoration (Figure 10A).
Figure 10.
The PIM1–RBMY axis controls the translational activation of EMT. (A) Immunoblotting was performed to determine the levels of EMT proteins upon depletion of PIM1 and RBMY by siPIM1 and siRBMY, as well as restoration with HA-PIM1 and GFP-RBMY. The numbers under the blots of Snail1 and ZEB1 show the band densities of these proteins relative to α-tubulin and are presented as ratios with respect to the value of siCtrl, which is set as 1. (B) A schematic domain structure of RBMY is depicted. The mutation sites of NESm and NLSm are specially marked in pink characters and blue characters, respectively, showing the substitution of amino acids (a.a.) by alanine in comparison with the original sequence of RBMY-WT. Fluorescence imaging showed the distribution patterns of GFP-RBMY WT, NESm, and NLSm in Huh-7 cells. White arrowheads, white arrows, and yellow arrows indicate N+, C+, and Mix patterns of RBMY, respectively. Immunoblotting was performed to assess the expression of EMT proteins in cells overexpressing GFP-RBMY and its mutants with/without HA-PIM1 co-expression. (C) IFA showed the distribution of Snail1 and E-cadherin in Huh-7 cells overexpressing GFP-RBMY and GFP-NLSm. The white arrows, yellow arrows, and white arrowhead indicate GFP-RBMY-Mix, GFP-RBMY-C+, and GFP-RBMY-N+ cells, respectively. (D) Immunoblotting showed the expressions of GFP-RBMY-WT/mutants and EMT proteins upon SMI-4a treatment. Dimethyl sulfoxide (DMSO) served as the solvent control. (E) Immunoblotting evaluated the expressions of RBMY and EMT proteins in response to the knockout of PIM1. PLC/PRF/5 cells were transfected 3 times with sgPIM1 (clones 6, 4, and 3) and sgCtrl at 24-hour intervals. Part of these sgRNAs-expressed PLC/PRF/5 cells were transfected once with HA-RBMY (lower panel). All cells were harvested for measuring the levels of endogenous RBMY (upper) and HA-RBMY (lower), as well as PIM1 and EMT proteins, 24 hours after the last transfection. (F) Schematic diagram summarizing the findings of this study showing how PIM1 and RBMY are involved in the expression of EMT proteins. DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To test whether the subcellular distribution of RBMY affects EMT induction, a GFP-tagged nuclear export signal mutant (GFP-NESm) and a nuclear localization signal mutant (GFP-NLSm) of RBMY, which resulted in the RBMY-N+ and RBMY-C+ patterns, respectively, were generated (Figure 10B). Overexpression of GFP-RBMY-WT (Mix pattern), GFP-NESm, or GFP-NLSm alone was sufficient to increase the level of Snail1 and silence E-cadherin in both PLC/PRF/5 and Huh-7 cells (Figure 10B). In the absence of PIM1, nuclear RBMY (GFP-NESm) was able to up-regulate ZEB1, whereas cytoplasmic RBMY (GFP-NLSm) was more effective in inducing the expressions of Snail1 and vimentin (Figure 10B). Of note, co-expression of HA-PIM1 elicited full up-regulation of these EMT factors in PLC/PRF/5 cells and of all but N-cadherin in Huh-7 cells (Figure 10B). Double IFA staining showed that cells with the C+ and Mix patterns of GFP-RBMY showed a dramatic improvement in the nuclear entry of Snail1 compared with those with the N+ pattern or the absence of GFP-RBMY (Figure 10C). Expression of GFP-NLSm further proved the ability of cytoplasmic GFP-RBMY to drive the nuclear translocation of Snail1 (Figure 10C). In addition, loss of E-cadherin typically was found in GFP-RBMY C+ cells (Figure 10C).
Blockade of PIM1 kinase activity in PLC/PRF/5 and Huh-7 cells using the inhibitor SMI-4a led to diminished expressions of GFP-RBMY, Snail1, ZEB1, and vimentin, with an ensuing increase in E-cadherin expression (Figure 10D). Under this condition, the expression levels of N-cadherin and fibronectin were unaffected (Figure 10D), suggesting that at least one other factor is involved in regulating N-cadherin and fibronectin in addition to PIM1 and RBMY. Some kinase inhibitors exert off-target effects, and this phenomenon may occur with SMI-4a. Therefore, we knocked out the PIM1 gene to validate its regulation of RBMY and EMT by developing a clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-based gene editing system and engineering 3 single-guide RNAs (sgRNAs) against PIM1 (sgPIM1), designated as 6, 4 and 3 that specifically recognize the DNA sequence of PIM1. Three of these sgPIM1s effectively reduced the protein level of PIM1 and consequently reduced the levels of endogenous RBMY, HA-RBMY, Snail1, ZEB1, and vimentin, but increased the level of E-cadherin (Figure 10E). However, the expression levels of N-cadherin and fibronectin were not reduced consistently upon genetic ablation of PIM1 (Figure 10E), confirming that the PIM1–RBMY axis primarily controls the expressions of Snail1, ZEB1, vimentin, and E-cadherin, but has less influence on N-cadherin and fibronectin.
We concluded that after PIM1 phosphorylates RBMY, PIM1 cooperates with nuclear RBMY to drive the transcription of Snail1 and ZEB1 and with cytoplasmic RBMY to trigger the nuclear entry of Snail1 (Figure 10F). Both of these pathways synergistically support the transcription of ZEB1 and vimentin, as well as blockade of E-cadherin (Figure 10F).
Interaction Between PIM1 and RBMY Leads to Feedback Stabilization
Inhibition of the kinase activity of PIM1 blocked the expression of RBMY, and, in turn, depletion of RBMY suppressed the expression of PIM1, while restoration of the total RBMY pool resulted in rescued expression of PIM1 (Figure 10A, D, and E). It thus was likely that the relationship between PIM1 and RBMY is far more than a classic kinase–substrate relationship, which may additionally bridge other post-translational regulatory events. To further delineate the molecular interaction between PIM1 and RBMY, a cycloheximide (CHX)-chase assay was conducted to estimate the kinetic turnover rate of these proteins. In the presence of GFP-RBMY, the half-life of HA-PIM1 was prolonged, but it was reduced when endogenous RBMY was depleted (Figure 11A and B). Reciprocally, expression of HA-PIM1 improved the protein stability of GFP-RBMY (Figure 11C). In RBMY-null cells such as SNU-387 cells, the level of endogenous PIM1 p34 was unstable and was lost upon subculture (Figure 11D). In addition, a quick degradation of HA-PIM1 in SNU-387 cells was found in the CHX-chase assay unless GFP-RBMY was overexpressed to support the stabilization of HA-PIM1 (Figure 11E). This was evidenced further by female HCC tissues, which showed a decreased positivity rate of PIM1 p34 (33.3%; 2 of 6) as a result of intrinsic RBMY deficiency (Figure 11F). All these data suggested that PIM1 and RBMY indeed mutually stabilized each other when co-expressed. However, RBMY was not an obligate factor for the stable expression of PIM1 p34 because there still were 33.3% of female HCC tissues that expressed PIM1 p34 in the absence of RBMY.
Figure 11.
Positive feedback stabilization exists between PIM1 and RBMY. A CHX-chase assay followed by Western blot analysis was conducted to estimate the turnover rates of (A) HA-PIM1 upon GFP-RBMY co-expression and (B) RBMY depletion by siRBMY. Cells cotransfected with HA-PIM1 and (A) either GFP control or GFP-RBMY and (B) either siRBMY or siCtrl were harvested at the times indicated after CHX (300 μg/mL) treatment. The percentage of protein remaining was plotted by quantifying the HA-PIM1 intensities before normalization to their respective glyceraldehyde-3-phosphate dehydrogenase (GAPDH) signals. (C) CHX-chase assay assessing the half-life of GFP-RBMY upon HA-PIM1 overexpression was performed. Cell lysates harvested at the times indicated were analyzed by immunoblotting, and the percentage of protein remaining was calculated as described earlier. (D) Western blot analysis showed the absence of endogenous RBMY and PIM1 p34 in the HCC cell line SNU-387 derived from a female patient, as compared with the male HCC line PLC/PRF/5. (E) A CHX-chase assay was performed in SNU-387 cells to determine the half-life of HA-PIM1 in the presence or absence of GFP-RBMY. (F) Immunoblotting of PIM1 and RBMY expressions in male (N = 4) and female (N = 6) HCC tissue samples. Frozen tissues (20 mg) were homogenized in 400 μL RIPA lysis buffer containing 0.5% sodium dodecyl sulfate. After sonication, the protein lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis using Bolt 4%–20% Bis-Tris Plus gels, transferred onto a polyvinylidene difluoride membrane, and subsequently probed with antibodies recognizing the indicated proteins. (G) A CHX-chase assay was achieved before subcellular fractionation of cells cotransfected with HA-PIM1 and GFP-RBMY. The fractions were analyzed by Western blot. Protein degradation was estimated by quantifying the nuclear and cytoplasmic GFP-RBMY intensities before normalization to their respective markers, Lamin A/C and GAPDH. (H) CHX-chase assay determined the half-life of HA-PIM1 upon co-expression of GFP-RBMY-WT and its mutants.
To gain insight into the subcellular localization of PIM1-stabilized RBMY, we subjected HA-PIM1– and GFP-RBMY–cotransfected cells to CHX-chase assay before separation of the nuclear and cytoplasmic fractions. GFP-RBMY was easily degraded in the nucleus but became stable when translocated into the cytoplasm (Figure 11G). Breakdown of nuclear GFP-RBMY was prevented by overexpression of HA-PIM1 (Figure 11G). In contrast, the half-life of nuclear HA-PIM1 was longer than that of cytoplasmic HA-PIM1; however, the presence of a large amount of GFP-RBMY in the cytoplasm could not protect HA-PIM1 from degradation (Figure 11G). To further investigate this finding, we sought to determine which subcellular localization mutant of GFP-RBMY—NLSm or NESm—stabilized PIM1. The subsequent CHX-chase assay indicated that expression of nuclear RBMY (NESm) but not cytoplasmic RBMY (NLSm) prolonged the half-life of HA-PIM1 (Figure 11H). In line with this observation, HCC tissues revealed a greater significance between PIM1 and RBMY expression in the nuclear extracts (Figure 8B). In addition, PIM1-C+ tissues had a lower amount of nuclear RBMY than those expressed by PIM1-Mix (Figure 8C), owing to a lack of nuclear PIM1 to stabilize the nuclear RBMY. Thus, feedback stabilization of PIM1-RBMY occurred mainly in the nuclear compartment.
PIM1 Induces Mitochondrial Translocation of RBMY to Prime the Metastatic Switch
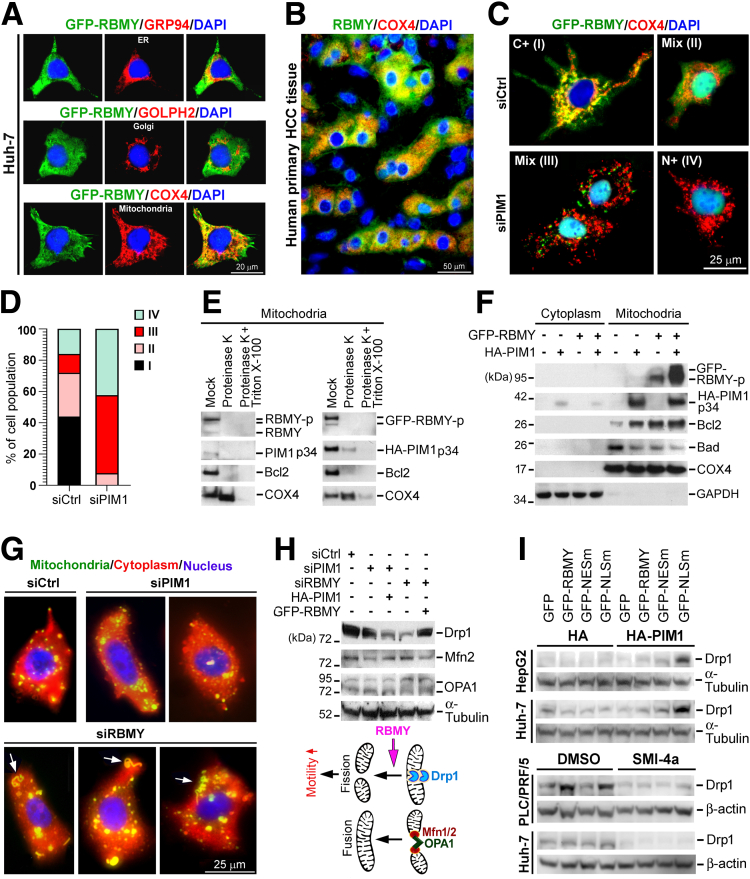
It is worthwhile to determine which cytoplasmic organelle is the site of RBMY activity because RBMY is functionally important in the cytoplasm. Costaining of RBMY and organelle markers showed that RBMY was localized mainly in mitochondria in Huh-7 cells and primary HCC tissues (Figure 12A and B). Depletion of PIM1 diminished the mitochondrial expression of RBMY and sequestered RBMY in nuclei (Figure 12C and D). Both endogenous RBMY and ectopic GFP-RBMY were localized at the outer mitochondrial membrane (OMM) because treating purified mitochondria with proteinase K degraded RBMY in the OMM as efficiently as the well-known OMM protein, B-cell lymphoma 2 (Bcl2, Figure 12E). Moreover, cyclooxygenase 4 (COX4), an inner mitochondrial membrane (IMM) protein, was retained in this fraction unless the integrity of the IMM was disrupted further by Triton X-100 in addition to proteinase K (Figure 12E). Endogenous PIM1 was expressed in the OMM, and PIM1 could enter the IMM when it was ectopically overexpressed (Figure 12E). PIM1 and RBMY synergistically regulated the expression of OMM proteins by increasing Bcl2 expression but suppressing Bcl2-associated agonist of cell death (Bad) level (Figure 12F), leading to enhanced apoptosis resistance and the preservation of mitochondrial integrity.
Figure 12.
PIM1 fosters the mitochondrial translocation of RBMY to regulate mitochondrial integrity and dynamics. Subcytoplasmic localization of RBMY was analyzed by double-stained IFA probing for the colocalization of RBMY and organelle markers in (A) GFP-RBMY–transfected Huh-7 cells and (B) primary HCC tissue. Glucose-regulated protein of 94 kDa (GRP94), Golgi phosphoprotein 2 (GOLPH2), and COX4 are markers of endoplasmic reticulum (ER), Golgi, and mitochondria, respectively. (C and D) IFA showed the distribution of GFP-RBMY in Huh-7 cells transfected 3 times with siCtrl or siPIM1. (D) The percentage of the cell population (means ± SDs, Student t test) was determined with respect to the different subcellular distribution patterns of GFP-RBMY, which are indicated as patterns I–IV. (E) Immunoblotting showed the submitochondrial localization of PIM1 and RBMY. Mitochondria isolated from Huh-7 cells 24 hours post-transfection were subjected to proteinase K and/or Triton X-100 treatment and analyzed by Western blot. Mock indicates the basal levels of mitochondrial proteins. Bcl2 and COX4 represent OMM and IMM proteins, respectively. (F) Mitochondrial levels of apoptosis-related proteins (Bcl2 and Bad) upon overexpression of HA-PIM1 and GFP-RBMY. After subcellular fractionation, the isolated cytoplasmic and mitochondrial fractions were analyzed by Western blot. COX4 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used to indicate the purity of the mitochondrial and cytoplasmic fractions, respectively. (G) Fluorescence staining of the mitochondria, cytoplasm, and nucleus of live PLC/PRF/5 cells with PIM1 or RBMY depletion using Mito-ID Green, CytoPainter Red, and Hoechst 33342, respectively. The white arrows indicate mitochondrial fusion. (H) Immunoblotting of PLC/PRF/5 cells transfected 3 times with the indicated siRNAs and once with HA-PIM1 or GFP-RBMY. A hypothetical model of the mechanism by which RBMY regulates mitochondrial dynamics is presented. (I) Western blot analyzed the Drp1 levels of the indicated cells expressing GFP-RBMY WT or mutants together with HA-PIM1 (upper) and kinase inhibitor SMI-4a (lower). Cells were treated with SMI-4a (30 μmol/L) or solvent control (dimethyl sulfoxide [DMSO]) 24 hours before transfecting with GFP-RBMY-WT or mutants. These cells were continuously supplied with SMI-4a and harvested after 24 hours post-transfection. DAPI, 4′,6-diamidino-2-phenylindole.
In addition to Bcl2, mitochondria integrity is critically controlled via continual fusion and fission.19 Silencing of RBMY caused mitochondrial fusion, an event impeding cell migration,20 yet this effect was absent in PIM1 knocked-down cells (Figure 12G). Depletion of RBMY but not PIM1 initiated evident down-regulation of the fission mediator (Drp1), but constitutive expression of the mitochondrial fusion proteins (mitofusin 2 and optic atrophy 1), thus rendering the cells prone to mitochondrial elongation (Figure 12H). Importantly, recovery of the RBMY level by overexpression of GFP-RBMY restored the amount of Drp1 (Figure 12H). Cytoplasmic RBMY was necessary for enhancing Drp1 expression, especially in the presence of active PIM1, and inhibition of PIM1 kinase activity by SMI-4a suppressed the RBMY-induced Drp1 expression (Figure 12I).
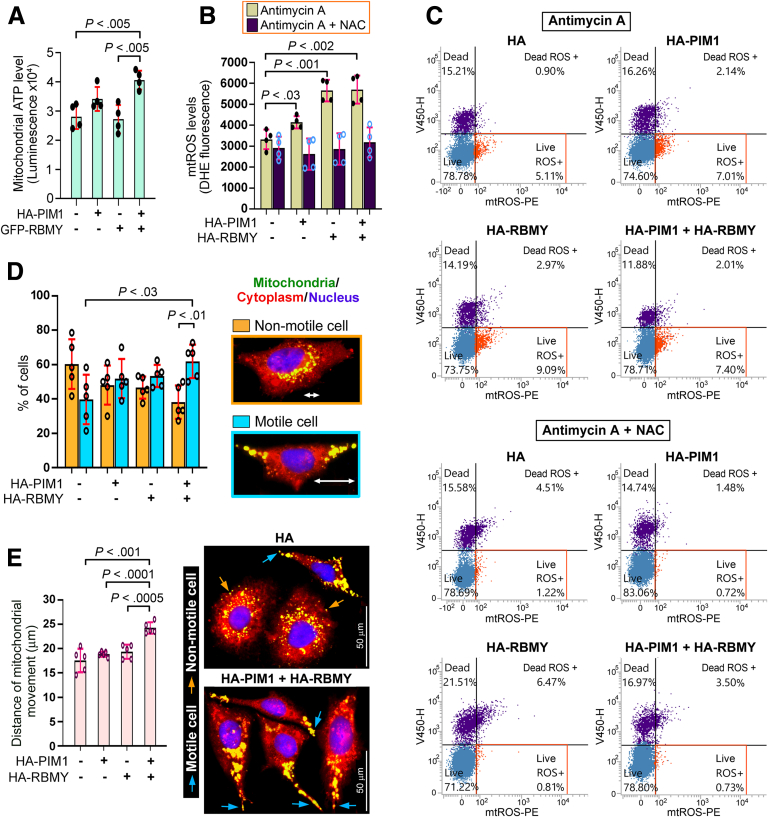
The production of high ATP and reactive oxygen species (ROS) levels allows cancer cells to achieve enhanced motility for invasion.21,22 Overexpression of HA-PIM1 and GFP-RBMY was sufficient to increase the mitochondrial ATP level; although GFP-RBMY alone failed to impact the mitochondrial ATP pool, this ability was gained by HA-PIM1 co-expression (Figure 13A). Antimycin A, an ATP-depleting agent and mitochondrial ROS (mtROS) inducer, then was applied to challenge the isolated mitochondria (Figure 13B) and intact PLC/PRF/5 cells (Figure 13C). The production of mtROS, which can be scavenged by the antioxidant N-acetylcysteine (NAC), was increased in mitochondria and cells expressing HA-RBMY and, to a lesser extent, in those expressing HA-PIM1 (Figure 13B and C). However, co-expressions of HA-RBMY and HA-PIM1 did not cause a further increase of mtROS (Figure 13B and C). Because high mtROS production is related to severe apoptosis,21 good control of mtROS level may protect cells from extensive damage.
Figure 13.
The PIM1–RBMY axis increases the mitochondrial motility. Bar plots showing the (A) luminescence signal of mitochondrial ATP in Huh-7 cells and the (B) fluorescence signal of mtROS in PLC/PRF/5 cells upon transfection with the indicated plasmids. The results shown are the means ± SDs from quadruplicate samples, and a representative data from 3 separate experiments is shown. The significance of differences was analyzed by the Student t test. (C) Flow cytometry analysis of mtROS in PLC/PRF/5 cells. Stable lines expressing HA-PIM1 and/or HA-RBMY were treated with antimycin A in the absence or presence of NAC. Cells were incubated with mtROS detection reagent before staining with SYTOX blue dead cell dye. For each sample, 10,000 cells were gated. The mtROS level was estimated in PE channel, and the live/dead cells were detected using V450 channel. (D) Mitochondrial locomotion in live Huh-7 transfectants. Representative fluorescence images of nonmotile and motile cells are shown. The percentage of these cells was depicted as means ± SDs and evaluated by the Student t test. (E) Traveling distance of mitochondria from the nuclear front in Huh-7 transfectants was plotted (means ± SDs, Student t test). Representative pictures showed staining of mitochondria, cytoplasm, and nucleus in live Huh-7 cells overexpressed with proteins indicated. Orange and blue arrows indicate nonmotile and motile cells, respectively.
A highly motile cell shows repositioning of mitochondria from the perinuclear region toward the distal edges of membrane protrusions, and this mitochondrial flux provides sufficient energy to activate membrane-cytoskeletal dynamics.23 Accordingly, the percentage of motile cells in comparison with that of nonmotile cells, which showed clustering of mitochondria around nuclei, was determined (Figure 13D). Co-expression of HA-PIM1 and GFP-RBMY triggered a larger increase in the number of motile cells and a longer distance of mitochondrial movement (Figure 13D and E). To further prove the role of mtROS in eliciting mitochondrial motility and contributing to cell migration and invasion, cell lines with stable HA-PIM1 and HA-RBMY expression were cultured in the presence and absence of NAC. Treatment with NAC not only caused defective mitochondrial locomotion but also abrogated the migration and invasion of HA-PIM1 and HA-RBMY cells (Figure 14), suggesting that mtROS are required for cell metastasis.
Figure 14.
ROS generation is crucial for PIM1/HA-RBMY-mediated mitochondrial motility and cellular migration/invasion. (A) PLC/PRF/5 cells stably expressing HA-PIM1 and/or HA-RBMY were treated with 30 μmol/L NAC and 1× PBS (mock) as solvent control. The distribution of mitochondria of these live cells were visualized by staining with Mito-ID Green. Staining of 4′,6-diamidino-2-phenylindole (DAPI) indicates the nucleus. Bar plots (means ± SDs) showed the distance of mitochondrial movement. The statistical significance was evaluated by the Student t test. (B) Scratch wound healing assay showed the migratory ability of cells indicated in the absence and presence of NAC. The migration distance was calculated and presented as means ± SDs. The Student t test was applied for the statistical analysis of the results. (C) Transwell invasion assay assessed the invasive potential of cells indicated with or without NAC treatment. The number of invaded cells was counted (means ± SDs) and the result between groups was compared by the Student t test. ∗P < .05, ∗∗P < .005, and ∗∗∗P < .0005.
PIM1 and RBMY Act as Metastatic Factors in the HCC Mouse Model
To establish in vivo evidence for PIM1–RBMY axis–mediated metastasis, PLC/PRF/5 cells stably expressing GFP-RBMY-WT/mutants (GFP-NLSm and GFP-NESm) with/without HA-PIM1 co-expression were implanted into the left hepatic lobes of nude mice and monitored for distant metastasis (Figure 15A). The expressions of HA-PIM1 and GFP-RBMY-WT/mutants in mouse liver tissues after death were confirmed by double IFA staining using anti-HA and anti-human RBMY antibodies (Figure 15B). Mice expressing GFP-NLSm with/without HA-PIM1 harbored the greatest number of metastatic nodules in the lungs (Figure 15C and D). However, the incidence (%) of lung metastasis was increased only with HA-PIM1 co-expression, and HA-PIM1+GFP-NLSm mice were the most susceptible to developing distant metastasis (Figure 15C). In the livers of these mice, we observed a large number of hepatocytes with the PIM1-Mix/RBMY-C+ pattern, which was the most malignant pattern found in HCC patients, which had surrounded and infiltrated into the blood vessels (Figure 15E). These cells grew into metastatic foci in the lungs, as confirmed by IHC staining of Hepatocyte Paraffin 1 (Her Par-1, Figure 15F).
Figure 15.
PIM1 and RBMY prime HCC cells for lung metastasis in a mouse model. (A) A schematic illustration of the establishment of the liver cancer lung metastasis model is shown. (B) Double-stained IFA with anti-HA and anti-human RBMY (SRGY clone 1) antibodies was performed to assess the expression of transfected HA-PIM1 and/or GFP-RBMY WT/mutants in the liver tissue sections. A representative figure from each group is shown. (C) The number of pulmonary metastatic nodules and the percentage incidence of lung metastasis in each group were plotted (means ± SEMs, Student t test ). (D) H&E staining assessed the number of pulmonary HCC metastases (black arrows). A representative image from each group is presented. (E) Double IFA staining of PIM1 and RBMY in liver tissues of HA-PIM1+GFP-NLSm mice. Positive immunoreactivity for PIM1-Mix/RBMY-C+ is indicated by white arrows. (F) IHC staining of the indicated proteins in serial lung sections from HA-PIM1+GFP-NLSm mice. The black arrows indicate the metastatic nodule in serial sections. BV, blood vessel; DAPI, 4′,6-diamidino-2-phenylindole.
The results of microscopic and macroscopic analyses showed that HA-PIM1+GFP-NESm mice were predisposed to the formation of multiple tumor nodules in their livers but were highly resistant to lung metastasis because no mice in this group developed lung nodules (Figures 15C and D and 16A). Nuclear restriction of RBMY did not abrogate EMT induction in these mice because increased expression of Snail1 and Vimentin were observed in their liver tissues (Figure 16B). However, they did not show a mitochondrial proliferative response, given that the COX4 levels in these mice were as low as those in control (Vector) mice, and far lower than those in HA-PIM1+GFP-NLSm mice (Figure 16B and C). In mice bearing prometastatic cells such as those expressing GFP-RBMY or GFP-NLSm with/without HA-PIM1, their mitochondria showed a prominent change—in particular, bursts of mitochondrial mass and increased Drp1 levels, with concomitant loss of mitofusin 2 expression (Figure 16B and C). This observation indicated that mitochondrial proliferation and fission occurred in the livers of mice bearing metastases.
Figure 16.
Increasing mitochondrial mass and fission is essential for PIM1 and RBMY-mediated distance metastasis of HCC. (A) Macroscopic views of livers and lungs harvested from the indicated mice. The black arrows show hepatic and pulmonary nodules. No metastatic tumors were found in the lungs of any control vector and HA-PIM1+GFP-NESm mice. (B) Immunoblotting of the protein lysates harvested from liver tissues of the indicated mice after death. A cartoon diagram illustrating that PIM1/RBMY-C+ and PIM1/RBMY-N+ impact different mechanisms is shown. (C) Double IFA staining of COX4 and Na+/K+ adenosine triphosphatase (ATPase), a plasma membrane marker, in liver tissue sections from the indicated mice. The white arrows show cells with mitochondria-surrounding nuclei. The yellow arrows indicate cells with increased mitochondrial mass and increased mitochondrial trafficking to the leading edge. DAPI, 4′,6-diamidino-2-phenylindole.
HA-PIM1 mice expressed trivial amounts of Snail1 and vimentin compared with those in control mice, but had higher levels of COX4 than control mice, and lung metastasis occurred in HA-PIM1 mice but not in control mice (Figure 16B and C). On the contrary, HA-PIM1+GFP-NESm mice showed evident expression of Snail1 and vimentin, but low COX4, yet lung metastasis was completely suppressed (Figure 16B and C), indicating mitochondrial changes had the greatest influence on distant metastasis, while EMT played a supportive role.
To further corroborate our finding that cytoplasmic RBMY activates a mitochondrial switch promoting distant metastasis, which is impeded by inhibition of nuclear RBMY export, a more rigorous mouse model of metastasis was established via tail-vein injection of HCC cells (Figure 17A). PLC/PRF/5 cells expressing HA-PIM1+GFP-RBMY-WT/mutants (N = 10 mice/group) were prepared. Before injection, Western blot was performed to evaluate the expression of EMT and mitochondrial proteins. High levels of Snail1, ZEB1, COX4, and Drp1, accompanied by down-regulation of E-cadherin, were observed in cells stably expressing HA-PIM1+GFP-RBMY-WT and HA-PIM1+GFP-NLSm (Figure 17B). These data confirmed that both cell lines possessed mesenchymal features and contained large amounts of fragmented mitochondria, with a metastatic advantage. This was prohibited in HA-PIM1+GFP-NLSm cells because of low expression of COX4 and Drp1 (Figure 17B). After injection, all tumor cells were allowed to circulate in the harsh microenvironment of the bloodstream (subject to apoptosis and immune attack), from which they could reach the capillary beds of distant organs, invade the endothelial cells lining blood vessels, and extravasate into tissues to form metastases. Only cells expressing HA-PIM1+GFP-RBMY-WT and HA-PIM1+GFP-NLSm successfully developed metastases in the lungs (30%; 3 of 10) (Figure 17C–E). Cells expressing HA-PIM1+GFP-NLSm even formed multiple large nodules after extravasation from pulmonary blood vessels (Figure 17C–E). Moreover, none of the mice expressing vector or HA-PIM1+GFP-NESm developed lung metastases (Figure 17C–E). HA-PIM1+GFP-NESm cells underwent EMT but contained less mitochondrial mass, reduced Drp1-mediated mitochondrial fission, and did not establish distant metastases (Figure 17B–E). These data were consistent with the observations shown in Figures 15 and 16, verifying that loss of cytoplasmic RBMY expression and mitochondrial changes attenuated the metastatic potential regardless of the presence of PIM1.
Figure 17.
Metastasis of cells expressing HA-PIM1 and GFP-RBMY-WT/mutants in a tail-vein injection mouse model. (A) Schematic diagram shows the steps by which a tail-vein injection metastasis mouse model was generated. (B) Western blot monitored the levels of EMT and mitochondrial proteins achieved on PLC/PRF/5 stable lines expressing proteins indicated before subjecting to tail-vein injection. (C) Number of lung metastatic masses and percentage of the metastatic occurrence in each group were counted and reported as means ± SEMs (Student t test). (D) H&E staining was performed on the lung tissues resected from mice indicated, and a representative image from each group is shown. Black arrowheads show pulmonary nodules. (E) Representative macroscopic views of lungs excised from each group at 10 weeks postinjection. Black arrowheads denote pulmonary nodules. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
In summary, we present evidence of RBMY as a new substrate of PIM1. The interaction between PIM1 and RBMY triggered not only the phosphorylation and stabilization of RBMY but also the mitochondrial translocation of RBMY. This resulted in EMT induction, a switch to mitochondrial fission, and increased synthesis of ATP and ROS for mitochondrial movement as a mechanism to fuel tumor metastasis and influence poor prognosis in patients with HCC.
Discussion
Previously, we showed a notable increase of cytoplasmic RBMY in the CSCs of primary HCC tissues, which correlated significantly with more advanced Barcelona Clinic Liver Cancer stage, larger tumor size, earlier recurrence, and poorer survival in HCC patients.11 Herein, we further proved that patients with cytoplasmic RBMY expression tended to experience metastasis. RBMY was reactivated in primary HCC tumors before metastasis. These RBMY-positive tumor cells were shed from the primary HCC tumor and infiltrated into blood vessels, bile ducts, and sinusoids, from which they migrated into the circulatory system. After extravasation at the metastatic organs, the initial expression level of RBMY was maintained in these distant metastases of HCC for priming metastatic seeding and proliferation into metastatic lesions. The observation that HCC patients without RBMY expression had longer metastasis-free survival times supported this hypothesis.
PIM1-mediated RBMY phosphorylation was the driving force to increase the metastatic potential of HCC. Moreover, PIM1 induced cytoplasmic accumulation of RBMY to cause metastasis formation; in contrast, HCC cells harboring nuclear RBMY failed to rapidly metastasize. In line with this, Kido et al24 reported that nuclear RBMY neither support the proliferation of HCC cells nor the development of HCC in myristoylated-protein kinase B (AKTmyr) and mutant neuroblastoma Ras viral oncogene homolog (NRASV12)-stimulated liver cancer in mice. A similar phenomenon was shown in our earlier study, in which expression of the nuclear-restricted RBMY mutant inhibits the proliferation of HCC cells in 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and xenograft tumor growth assays.11 In addition, the RBMY-transgenic mice that we generated in the FVB/N strain generally express nuclear RBMY and develop steatosis instead of liver tumors.11,25 Fifteen months after breeding, nuclear RBMY begins to translocate to the cytoplasm, showing the Mix pattern of RBMY, and adenomas are found in the livers of these mice.11,25 When these mice express an increasing RBMY-C+ pattern in their livers, spontaneous formation of HCC occurs in the absence of carcinogenic stimulation.11 All of these findings highlighting the nucleus-to-cytoplasm translocation of RBMY-conferred hepatic oncogenicity.
According to previous works identifying the putative substrate recognition sites of PIM1, amino acid sequences with 2–3 Arg residues adjacent to the phosphorylation site Ser/Thr are important for PIM1 binding.26,27 These typical sequences are found particularly in SRGY boxes 1–3 but not SRGY box 4, for example, 257RNHRS261 in SRGY box 1, 284RRDES288 and 294RNRRS298 in SRGY box 2, and 321RRHES325 in SRGY box 3, where the S residues are the probable sites of PIM1-mediated phosphorylation. Consequently, serial deletion of amino acids in SRGY boxes 1–3 abolished the ability of these mutants to bind PIM1.
We herein identified a formerly unrecognized pool of RBMY in mitochondria and found that PIM1 is dispensable for mitochondrial recruitment of RBMY. The mitochondrial roles of the PIM1 are significant not only in cancer cells but also in cardiomyocytes and hematopoietic cells, in which it increases the Bcl2 level to prevent mitochondrial apoptosis and maintain the mitochondrial transmembrane potential and mitochondrial integrity.28, 29, 30, 31, 32 RBMY may enhance these mitochondrial roles of PIM1 via mutual stability control. In addition, we found that PIM1 and RBMY synergistically increased the mitochondrial mass and ROS/ATP production and stimulated the movement of these energetically active mitochondria toward the leading edge of HCC cells to promote cell motility.
RBMY supported mitochondrial fission, a feature that differentiated it from PIM1. PIM1 is found to preferentially induce mitochondrial fusion in lung cancer cells,32 and this was also the case in our present study because knockdown of PIM1 tended to result in more mitochondrial fission, in contrast to the effect of RBMY depletion. However, highly metastatic HCC cells normally show excessive mitochondrial fission.20 Although activation of the Ca2+/calmodulin-dependent protein kinase II (CaMKII)/ERK/focal adhesion kinase (FAK) and interleukin 2/c-Jun N-terminal kinase (JNK)/F-actin pathways is known to control mitochondrial fission in liver cancer cells,20,33 RBMY was identified as another factor underpinning this mechanism. The key player in mitochondrial fission, Drp1, is controlled by PIM1 and GSK3β, which phosphorylate Drp1 on Ser637 and Ser693, respectively, to decrease the guanosine triphosphatase activity and total pool of Drp1 to attenuate mitochondrial fragmentation.34,35 Our present and previous studies showed the ability of RBMY to bind to both kinases. RBMY interacts with GSK3β and promotes its phospho-Ser9–dependent autoinhibition, which consequently abolishes its kinase activity and disrupts the phosphorylation of its downstream substrates.11 We speculated that RBMY bound to PIM1 and GSK3β, imposing a competitive constraint on the activity of these kinases toward Drp1, which therefore is released from the interaction with PIM1 and GSK3β to initiate mitochondrial fission.
Both nuclear and cytoplasmic RBMY regulated EMT by increasing Snail1 and ZEB1 levels. GSK-3β first binds and phosphorylates Snail1 in the nucleus, leading to its nuclear export and triggering its proteasomal degradation in the cytoplasm.36 Both nuclear and cytoplasmic RBMY can suppress the activity of GSK-3β toward Snail1 by preventing its phosphorylation-mediated inactivation in the nucleus and prolonging its half-life in the cytoplasm. Then, the stabilized cytoplasmic Snail1 may enter the nucleus to drive the transcription of EMT genes. On the other hand, RBMY inhibits GSK-3β activity to promote nuclear entry of β-catenin and enhances the interaction of the β-catenin/transcription factor 4 (TCF4) complex,11 which therefore may gain control and bind to the promoters of Snail and ZEB1 to trigger β-catenin–dependent transcription of Snail and ZEB1.37,38
During spermatogenesis, RBMY adapts a mechanistic scheme similar to that in hepatocarcinogenesis by translocating from the nucleus of spermatogonia to the cytoplasm of spermatozoa.9 Thereafter, RBMY is relocated to the mitochondria-rich midpiece of mature spermatozoa,9,10 similar to its nuclear localization in the peritumoral regions followed by cytoplasmic/mitochondrial translocation in the tumor nests of HCC. The expression of PIM1 in male germ cells also has been reported.39 It is possible that in spermatozoa, similar to cancer cells, motility is managed through RBMY and PIM1 and is fully powered by mitochondria.
The RBMX gene, an X-homolog of RBMY that encodes a protein with an organization identical to RBMY at the N-terminal RRM region but different at the SRGY box domain, is expressed in all male and female tissues.4,40,41 RBMX facilitates chromosome morphogenesis and DNA repair.42,43 Depletion of RBMX increases chromosome segregation defects and alters cell-cycle progression.44 The role of RBMX in cancers is controversial, whereby it acts as a tumor suppressor in bladder and breast cancers but contributes to poor prognosis in both male and female patients with HCC by enhancing HCC cell proliferation and sorafenib resistance.45, 46, 47 Because of the ubiquitous expression and beneficial physiological functions of RBMX in human tissues, it may not be a good therapeutic target, which is entirely different from RBMY.
In summary, our findings provide a glimpse into the expression patterns of RBMY before and after HCC metastasis. RBMY had the greatest gain of function upon phosphorylation by PIM1, which is an indispensable kinase for cell survival and is expressed throughout the human body. For years, PIM1 has been a focus in cancer therapy, and some of its small-molecule inhibitors are currently in clinical trials.14 However, all of these inhibitors are ATP competitors that lack specificity among kinases and whose use is accompanied by unfavorable side effects. Alternatively, selective targeting of PIM1 substrate binding may provide a feasible option in anticancer treatment. RBMY expression is normally absent in adult livers but is reactivated after hepatocarcinogenesis,4,11,24 indicating the diagnostic potential and therapeutic targeting value of RBMY. In particular, expression of cytoplasmic RBMY is a novel biomarker for predicting the risk of metastasis and mortality in HCC.
Materials and Methods
Human Tissue Collection
Primary HCC tissues of male (aged 20∼95 y; N = 308) and female (aged 3∼76 y; N = 6) patients were collected from the Departments of Surgery and Pathology at the National Taiwan University Hospital. Ninety-five patients provided tissue sections of both primary tumors and paired metastases of HCC, and the others provided only primary HCC tissues at the time of surgery. Patient recruitment was from March 2005 to September 2015. All patients were followed up for more than 5 years after surgery, and ended in September 2020. Paraffin-embedded tissue sections of primary HCC were obtained from 118 cases and the other 190 cases were optimal cutting temperature (OCT)-embedded frozen tissue sections. During the study period, 56.2% (173 of 308) of cases were diagnosed with distant metastasis, 82.6% (143 of 173) of them occurred within 5 years posthepatectomy, and 67.9% (209 of 308) of patients died of the disease. Distant metastasis was diagnosed by clinical manifestations, physical examination, contrast-enhanced computed tomography, magnetic resonance imaging, and histologic findings of biopsies and/or surgical specimens. Extrahepatic metastases of HCC were found in omentum, bone, lung, diaphragm, pelvic cavity, adrenal gland, tonsil, brain, skin, and soft tissues in the abdomen. Lung and bone are the most frequent metastatic sites. The time of metastasis onset was calculated from the date of surgery to the date of first diagnosis of distant metastasis. This study was approved by the Institutional Review Board of the National Taiwan University Hospital Research Ethics Committee (approval numbers: 9361701139, 201003073R, and 201512093RINC).
Human Cell Lines
Human HCC cell lines, including Huh-7 [Japanese Cancer Research Resources Bank (JCRB), SK-HEP-1 (ATCC), Hep3B (ATCC), PLC/PRF/5 (ATCC), and SNU-387 (ATCC), as well as human hepatoblastoma cell line HepG2 (ATCC) all were obtained from the commercial sources as indicated. Cells were grown in high-glucose Dulbecco’s modified Eagle medium (DMEM) supplied with 10% fetal bovine serum (FBS), 2 mmol/L L-glutamate (Gibco, Grand Island, NY), 1 mmol/L sodium pyruvate (Gibco), 1× MEM nonessential amino acids (Gibco), and antibiotics (MicroZap Plus-CL; Lonza, Basel, Switzerland). HA-RBMY/GFP-RBMY and HA-PIM1 stable lines, as well as their paired control and mutants, established in Huh-7, Hep3B, and PLC/PRF/5, were maintained in G418-selective complete DMEM.
Plasmid Construction
The coding regions of full-length RBMY and its serial deletion mutants including 2860-9, 2860-10, 1212-1, SRGY, and RRM were generated from childhood HCC or hepatoblastoma tissues by reverse-transcription PCR amplification and cloned into the vectors designated pcDNA3.1-HA (with 1 copy of HA tag at N-terminal; a gift from Dr Li-Min Huang, Department of Pediatrics, National Taiwan University Hospital) and pEGFP-c2 (Clontech, Carlsbad, CA). The amino acid sequence of this full-length RBMY construct matched with the NCBI reference sequence NP_005049.1 of human RBMY1A1. The open reading frames of PIM1 p34, MEK2, CK1ε, and GSK3β were amplified from the full-length MGC complementary DNA clones (Thermo Fisher Scientific Dharmacon, Lafayette, CO) and inserted into the pcDNA3.1-3HA (encoding 3 copies of HA epitope at the N-terminal, a gift from Dr Li-Min Huang, Department of Pediatrics, National Taiwan University Hospital) and pAsRed2-N1 (Clontech) expression vectors to produce fusion proteins. Plasmid carrying GFP-NESm, which has a mutated nuclear export signal at the N-terminus of RBMY, was constructed by mutating the I34 and V37 codons of RBMY to alanine using the QuikChange Lightening Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA). Adopting a similar approach, GFP-NLSm, which harbored double mutations on both bipartite nuclear localization signals, was created by replacing K151, R152, R284, and R285 residues of RBMY to alanine. Mutant V5-SRGY, which spans RBMY amino acid residues 227–374 and comprises 4 repeated SRGY box domains, was cloned into a mammalian expression vector pEF6/V5-His TOPO (Thermo Fisher Scientific, Waltham, MA).
The siRNA duplexes siCtrl (ON-TARGETplus Non-Targeting Control Pool), siRBMY (On-TARGETplus Human RBMY1A1 [5940] siRNA-SMARTpool), and siPIM1 (On-TARGETplus Human PIM1 [5292] siRNA-SMARTpool) were purchased from Thermo Fisher Scientific Dharmacon. Human RBMY1 shRNA lentiviral particles (sc-270225-V) and control shRNA lentiviral particles-A (sc-108080) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The sgRNAs of PIM1 (sgPIM1) and control (sgCtrl) cloned in the enhanced specificity Cas9-2A-green fluorescent protein (eSpCas9-2A-GFP) vector, which expressed GFP protein for monitoring the transfection efficiency, were purchased from Genscript Biotech (Piscataway, NJ). The sequences of these sgRNAs are as follows: sgCtrl (CGCTTCCGCGGCCCGTTCAA), sgPIM1 6 (ATCTCTTCGTCATGCTCGAA), sgPIM1 4 (ACCATCGAAGTCCGTGTAGA), and sgPIM1 3 (AGAAGGACCGGATTTCCGAC).
Transfection and Transduction
The expression plasmids and siRNA duplexes were reverse-transfected into cell lines using Lipofectamine 3000 (Life Technologies, Carlsbad, CA) and DharmaFECT (Thermo Scientific Dharmacon) transfection reagents, respectively, following the manufacturers’ protocols. Reverse-transfection of siRNA duplexes was performed repeatedly (3 times) at 24-hour intervals. The transduction of shRNA lentivirus particles was performed using Polybrene (sc-134220; Santa Cruz Biotechnology) according to the manufacturer's instructions. The shRNAs were transduced twice, once on day 2 and once on day 3, for a total multiplicity of infection of 20. To efficiently knockout the target gene, the sgRNAs were transfected 3 times at 24-hour intervals using Lipofectamine 3000.
Generation of Nude Mouse Model of Xenograft Tumor
Male BALB/c athymic nude mice (BALB/cAnN.Cg-Foxn1nu/CrlNarl; age, 5 wk; N = 5/group) were purchased from the National Laboratory Animal Center (Taipei, Taiwan) and bred in the Animal Center of the College of Medicine, National Taiwan University. Mice were housed in ventilated caging units under positive pressure in a specific pathogen-free facility with a standard 12-hour light-dark cycle and free access to mouse chow (LabDiet 5053; Purina Mills, Richmond, IN) and sterile water. These mice were adapted for 1 week before xenograft implantation. Huh-7 cells (1 × 106 cells/mouse) were transfected 3 times with siCtrl and siRBMY, as well as transduced twice with shCtrl, shRBMY, and shRBMY+HA-RBMY, and were resuspended in 150 μL FBS-free DMEM/F12 (Thermo Fisher Scientific) plus Matrigel (BD Biosciences, San Jose, CA) in a 1:1 ratio. Cells were inoculated subcutaneously into the right and left flanks of nude mice. Mice were monitored for the growth of implanted cells and killed after 90 days. All animal procedures were in compliance and approved by the Institutional Animal Care and Use Committee of National Taiwan University, College of Medicine (Institutional Animal Care and Use Committee approval 20150409).
Establishment of Nude Mouse Model of Pulmonary Metastasis
BALB/c nude mice (male; age, 3 wk; N = 6∼10/group) purchased from the National Laboratory Animal Center of Taiwan were adapted for 1 week in the Animal Center of the College of Medicine, National Taiwan University. To prepare cells for inoculation, PLC/PRF/5 was reverse-transfected with HA-PIM1 and/or GFP-RBMY-WT/mutants using Lipofectamine 3000. These cells were replated in medium containing G418 (1 mg/mL) 24 hours post-transfection, and continued passage 3 times per week for 2 weeks. For each mouse injection, these PLC/PRF/5 transfectants (1 × 106 cells/mouse) were resuspended in 50 μL FBS-free DMEM/F12 and mixed with 25 μL Matrigel (Matrigel Basement Membrane Matrix High Concentration; Corning, Inc, Corning, NY). Mice were anesthetized intramuscularly with an injection of ketamine/xylazine cocktail (75 mg/kg ketamine + 5 mg/kg xylazine), and then received a subxyphoid laparotomy of 1.5∼2 cm in length to expose the livers. Cell suspensions were injected directly into the left lobe of livers using a 29-gauge needle. To prevent bleeding, the puncture sites were gently compressed by cotton gauze for 15 seconds, and the wound was sutured in layers. Six weeks after surgery, nude mice were killed for lung metastasis analysis.
BALB/c nude mice (male; age, 3 wk) also were applied to create a tail-vein injection metastasis mouse model after a 1-week adaptation at the Animal Center of the College of Medicine, National Taiwan University. Cells were thoroughly washed 3 times with 1× phosphate-buffered saline (PBS), resuspended in 200 μL 1× PBS, and pipetted to single-cell suspension. These cells (1 × 106 cells/mouse) then were injected into the tail veins of nude mice. Mice were killed 10 weeks after injection and analyzed for the occurrence of lung metastasis. All experimental procedures were approved by the Institutional Animal Care and Use Committee of National Taiwan University College of Medicine (Institutional Animal Care and Use Committee approval 20150409).
IHC
Deparaffinization, rehydration, and antigen unmasking were consecutively achieved on paraffin-embedded HCC tissue sections with Trilogy reagent (Cell Marque, Rocklin, CA) via microwave heating at 100ºC for 10 minutes. Once cooled to room temperature, the endogenous peroxidase activity was inactivated with UltraVision Hydrogen Peroxide Block (Thermo Fisher Scientific) for 10 minutes at room temperature, and the reaction was eliminated by washing 3 times with 1× PBS. OCT-embedded HCC tissue sections were fixed with acetone for 5 minutes at -20°C. All sections were blocked with UltraVision Protein Block (Thermo Fisher Scientific) at room temperature for 10 minutes. Primary antibodies (Table 1) were incubated with tissue sections at 4°C overnight. Subsequent immunostaining was conducted using the UltraVision Quanto Detection System Horseradish Peroxidase kit (Thermo Fisher Scientific) for human tissue sections, and N-Histofine Simple Stain Mouse MAX PO or MOUSESTAIN KIT (Nichirei Biosciences, Tokyo, Japan) for mouse tissue sections, following the manufacturer’s instructions. Positive signals were revealed by staining for 1 minute with 3, 3’-diaminobenzidine (DAB Quanto; Thermo Fisher Scientific). Nuclei were counterstained with hematoxylin (Merck, Darmstadt, Germany). After air-drying, sections were cleared in Neo-clear (Merck) and cover-slipped using Neo-Mount (Merck).
Table 1.
Detailed Information of Antibodies Used in IHC and IFA Analysis
| Antibody (clone/catalog number) | Manufacturer | Host species | Dilution and incubation conditions |
|---|---|---|---|
| RBMY (SRGY clone 1) | Homemade | Mouse | 5×, 4°C, Overnight (O/N) |
| RBMY1A1 (GTX110098) | GeneTex | Rabbit | 150×, 4°C, O/N |
| Her Par-1 (OCH1E5, ab75677) | Abcam | Mouse | 1×, 4°C, O/N |
| PIM1 (TA354029) | OriGene | Rabbit | 150×, 4°C, O/N |
| COX4 (GTX101499) | GeneTex | Rabbit | 100×, 4°C, O/N |
| c-MET (8198) | Cell Signalling Technology | Rabbit | 100×, 4°C, O/N |
| Fibronectin (GTX112794) | GeneTex | Rabbit | 100×, 4°C, O/N |
| E-cadherin (ab1416) | Abcam | Mouse | 50×, 4°C, O/N |
| Snail1 (sc-271977) | Santa Cruz Biotechnology | Mouse | 50×, 37°C, 2 h |
| GRP94 (GTX103232) | GeneTex | Rabbit | 100×, 4°C, O/N |
| Na+/K+-ATPase α (sc-48345) | Santa Cruz Biotechnology | Mouse | 500×, 4°C, O/N |
| GOLPH2 (GTX107702) | GeneTex | Rabbit | 100×, 4°C, O/N |
| CK19 (TA313117) | OriGene | Rabbit | 100×, 4°C, O/N |
| HNF4α (C11F12) | Cell Signalling Technology | Rabbit | 100×, 4°C, O/N |
| HA.11 epitope tag (901501) | BioLegend | Mouse | 100×, 4°C, O/N |
IFA
Cultured cells plated on 8-well chamber slides (Millipore, Billerica, MA) were fixed with 4% paraformaldehyde for 12 minutes, except those used for Snail1 staining, which were fixed with ice-cold methanol at -20ºC for 5 minutes. Then, cells were permeabilized with 0.1% Triton X-100/PBS for 5 minutes and blocked with UltraVision Protein Block for 10 minutes at room temperature. Paraffin-embedded tissue sections were immersed in blocking buffer as mentioned previously after Trilogy treatment. All samples were subsequent to incubation with primary antibodies (Table 1) overnight at 4ºC, followed by extensive washing with 1× PBS. Thereafter, incubation with Alexa Fluor 488– or 594–conjugated secondary antibodies (1:200; Invitrogen) was performed at room temperature for 1 hour and mounted with ProLong Gold Antifade Mountant with 4′,6-diamidino-2-phenylindole (Thermo Fisher Scientific).
Western Blot
Total protein lysates were extracted with RIPA lysis buffer (50 mmol/L Tris-HCl, pH 7.4, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 150 mmol/L NaCl, 1 mmol/L EDTA, pH 8.0, 1 mmol/L phenylmethylsulfonyl fluoride (PMSF) and 1X HaltTM protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Each sample (20 μg/reaction) was dissolved in 1× Novex LDS Sample Buffer and 1× Novex sample reducing agent, heated at 70°C for 10 minutes, and run on precast Bolt 10% Bis-Tris Plus gels (Thermo Fisher Scientific). The separated proteins were transferred to polyvinylidene difluoride membranes before blocking with 0.5% skim milk at 37°C for 30 minutes. The membranes were probed with primary antibodies (Table 2) at 4ºC overnight. Successive washing with Tris-buffered saline (pH 7.5) containing 0.05% Tween-20 and incubation of horseradish-peroxidase–conjugated secondary antibodies (Jackson Immunoresearch, West Grove, PA) were accomplished using the SNAP id Protein Detection System (Millipore), in accordance with the manufacturer’s instructions. Peroxidase signals were developed with WesternBright ECL (Advansta, Menlo Park, CA). Densitometry analysis was achieved using ImageJ software (National Institutes of Health, Bethesda, MD).
Table 2.
Information of the Antibodies Used in Western Blot Analysis
| Antibody (clone/catalog number) | Manufacturer | Host | Blotting conditions | Molecular weight, kilodaltons |
|---|---|---|---|---|
| RBMY (SRGY clone 1) | Homemade | Mouse | 10×, room temperature, 2 h | 56–68 |
| RBMY1A1 (GTX110098) | GeneTex | Rabbit | 1000×, 4°C, Overnight (O/N) | 56–68 |
| His (MAB050H) | R&D Systems | Mouse | 1500×, 4°C, O/N | Depends |
| GFP (PT-LA-GFP-RB) | Protech | Rabbit | 1000×, 4°C, O/N | Depends |
| HA.11 Epitope Tag (901501) | BioLegend | Mouse | 1000×, 4°C, O/N | Depends |
| GAPDH (GT239) | GeneTex | Mouse | 2000×, room temperature, 2 h | 38 |
| Lamin B1 (ab16048) | Abcam | Rabbit | 5000×, room temperature, 2 h | 68 |
| Lamin A/C (ab108922) | Abcam | Rabbit | 5000×, room temperature, 2 h | 74 |
| α-Tubulin (GTX115141) | GeneTex | Rabbit | 5000×, room temperature, 2 h | 55 |
| PIM1 (2907) | Cell Signalling Technology | Rabbit | 200×, 4°C, O/N | 34, 44 |
| Bcl2 (GTX100064) | GeneTex | Rabbit | 500×, 4°C, O/N | 26 |
| Bad (GTX130108) | GeneTex | Rabbit | 500×, 4°C, O/N | 22 |
| COX4 (GTX101499) | GeneTex | Rabbit | 1000×, 4°C, O/N | 16 |
| Snail1 (sc-271977) | Santa Cruz Biotechnology | Mouse | 100×, 37°C, 2 h | 29 |
| Slug (9585) | Cell Signalling Technology | Rabbit | 1000×, 4°C, O/N | 30 |
| Twist1 | GeneTex | Rabbit | 1000×, 4°C, O/N | 29 |
| ZEB1 (ab203829) | Abcam | Rabbit | 500×, 4°C, O/N | 210 |
| E-cadherin (ab1416) | Abcam | Mouse | 50×, 4°C, O/N | 110 |
| N-cadherin (GTX127345) | GeneTex | Rabbit | 500×, 4°C, O/N | 140 |
| Fibronectin (GTX112794) | GeneTex | Rabbit | 1000×, 4°C, O/N | 263 |
| Vimentin (ab16700) | Abcam | Rabbit | 100×, 4°C, O/N | 53 |
| Drp1 (ab184247) | Abcam | Rabbit | 1000×, 4°C, O/N | 83 |
| OPA1 (sc-393296) | Santa Cruz Biotechnology | Mouse | 100×, 4°C, O/N | 80-100 |
| Mfn2 (ab56889) | Abcam | Mouse | 1000×, 4°C, O/N | 86 |
| β-actin | Abcam | Rabbit | 2000×, 4°C, O/N | 45 |
H&E Staining
Tissue sections were deparaffinized, rehydrated, and unmasked antigen sites with Trilogy reagent and stained with Mayer's Hematoxylin (BioGenex, Fermont, CA) for 5 minutes at room temperature. Sections then were washed thoroughly with double-distilled water (ddH2O) and dehydrated with an ascending ethanol series (50%, 70%, 80%, 95%, and 99%). After staining with eosin (Merck) at room temperature for 30 seconds, these sections were washed with 99% ethanol, cleared in Neo-clear, and coverslipped by Neo-Mount after air drying.
Subcellular Fractionation
The adherent cells grown in 6-cm dishes were trypsinized and pelleted down by centrifugation at 500 × g for 5 minutes to remove cell debris. Human HCC frozen tissues (≈20 mg) were washed with 1× PBS and homogenized using a sterile disposable micropestle (VWR, Radnor, PA). Nuclear and cytoplasmic proteins of the adherent cells and HCC tissues were extracted using NE-PER Nuclear and Cytoplasmic Extraction reagents (Thermo Fisher Scientific) in strict accordance with the manufacturer’s instructions.
Immunoprecipitation
Protein lysates extracted with RIPA lysis buffer were sonicated before performing immunoprecipitation. To precipitate the GFP-RBMY, the GFP-Trap Magnetic Agarose Kit (Chromotek GmbH, Planegg-Martinsried, Germany) was used. Briefly, 200 μL (≈500 μg) cell lysates were mixed with 300 μL kit-provided dilution buffer. In addition, GFP-Trap_MA magnetic agarose beads (25 μL/reaction) were washed 3 times with 500 μL dilution buffer, and the supernatant then was replaced with diluted cell lysates. The reactions were equilibrated at 4ºC for 1 hour, and subsequently washed 3 times with dilution buffer by magnetically separating the beads. The resulting complex-bound beads were resuspended in 30 μL of 1× Novex LDS-sample buffer containing 1× PBS and 1× Novex sample reducing agent (Thermo Fisher Scientific). To precipitate the HA-PIM1 and endogenous PIM1, cell lysates were harvested as mentioned previously and precleared with protein G Sepharose beads (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) for 1 hour at 4°C. After centrifugation at 13,000 rpm for 2 minutes, the supernatant was incubated with 2 μg anti-HA (BioLegend, San Diego, CA) or anti-PIM1 p34 antibodies (Santa Cruz Biotechnology) overnight at 4°C. Protein G Sepharose beads subsequently were added to the reactions and mixed by rotation for 2 hours at 4°C. The resulting Sepharose bead–bound immunocomplexes were washed once with RIPA lysis buffer and twice with 1× PBS, and dissolved in 1× Novex LDS sample buffer containing 1× PBS and Novex sample reducing agent. All the immunocomplexes were finally analyzed by Western blot.
Time-Lapse Live Imaging
HCC cell lines (4 × 104 cell/well) seeded onto chambered coverslips (μ-slide 8-well ibitreat; Ibidi, Gräfelfing, Germany) were grown overnight at 37ºC. Five hours after transfection with pEGFP-RBMY and pAsRed2-PIM1, live cells were subjected to time-lapse digital fluorescent imaging. Images were captured at 15-minute intervals and recorded for 18 hours using an Axiovert 200M fluorescence microscope (Zeiss, Ltd, Hertfordshire, UK) equipped with a CoolSNAP HQ2 CCD camera (Photometrics, Tucson, AZ) and under the control of MetaMorph imaging software (Universal Imaging Corp, Downingtown, PA).
Protein Microarray Analysis
V5-SRGY expression plasmid was transfected into 293T cells to yield a 23.5-kilodalton fusion protein. After purification by Ni-NTA His Bind Resins (QIAGEN, Valencia, CA) and ultrafiltration on an Amicon Ultra centrifugal filter (Millipore), the resulting V5-SRGY protein was used as a probe in the following ProtoArray Human Protein Microarray (Invitrogen) analysis. Briefly, the array was blocked for 1 hour with 1% bovine serum albumin blocking buffer before incubation with V5-SRGY in probing buffer at 4ºC for 90 minutes. Then, the array was washed with probing buffer. The V5-SRGY probe that bound on the array was fluorescently labeled using anti-V5–AlexaFluor 647 antibody, and the reaction was performed for 30 minutes on ice. After thoroughly washing with probing buffer, the array was dried in the dark at room temperature and subsequent to fluorescent microarray scanning. The pixel intensity data were analyzed by ProtoArray Prospector software, which identified statistically significant interactors by calculating the P value and Z-score. A Z-score threshold of ≥3.0 is considered a significant interaction. Solvent control containing a relatively high concentration of bovine serum albumin (10 mg/mL) was included in the assay to rule out nonspecific interactions. The identified binding partners of RBMY, sorting with the descending order of Z-score, included PIM1, MEK2, CK1ε, GSK3β, ephrin type-A receptor 3, p21-activated kinase 6, neurotrophic tyrosine kinase receptor type 3, feline sarcoma/Fujinami poultry sarcoma proto-oncogene, carbonic anhydrase 1, N-methylpurine-DNA glycosylase, SH3 and cysteine-rich domain, and chromosome 7 open reading frame 50.
In Vitro Kinase (Phosphorylation) Assay
Recombinant human proteins including His-RBMY (as a substrate; MyBioSource, San Diego, CA) and kinases such as His-PIM1, His-MEK2, His-CK1ε, and His-GSK3β (Thermo Fisher Scientific) were obtained from commercial sources. These proteins were incubated in phosphorylation reaction containing kinase buffer (25 mmol/L Tris-HCl [pH 7.5], 5 mmol/L b-glycerophosphate, 2 mmol/L dithiothreitol, 0.1 mmol/L Na3VO4, and 10 mmol/L MgCl2; Cell Signaling Technology, Beverly, MA) and 0.2 mmol/L nonradioactive ATP (Cell Signaling Technology) at 30ºC for 30 minutes. The resulting phosphorylated RBMY proteins were analyzed by Western blot.
Alkaline CIP Treatment
A total of 30 μL dephosphorylation reaction was prepared by adding 2 μL CIP (20 U; New England Biolabs, Beverly, MA), 15 μg cell lysate, and 1× NEBufferTM 3 (New England Biolabs). The reactions were incubated at 37°C for 2 hours, and stopped by mixing with 1× sodium dodecyl sulfate sample buffer and then heated to 70ºC for 10 minutes. The patterns of protein dephosphorylation were analyzed by immunoblotting.
RNA Isolation and RT-PCR
Total RNAs were isolated from GFP-RBMY–/shRBMY-transfected HCC cell lines and primary HCC tissues using TRIzol reagent (Thermo Fisher Scientific). Complementary DNAs were synthesized with the High-Capacity Complementary DNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Real-time quantitative PCR was achieved using SYBR Green PCR Master Mix (Applied Biosystems) on an ABI Prism 7500 Fast System (Applied Biosystems), following the manufacturer’s instructions. The primer sequences are provided in Table 3.
Table 3.
Primer Sequences for RT-QPCR Amplification
| Primer name | Sequences |
|---|---|
| RBMY_E4B | 5’-GGTGGCTTCCCTCACATGAAGG-3’ |
| RBMY_E5I | 5’-ACTATTGCTTCTTGCCACAGCAG-3’ |
| Snail1_F | 5’-GCACATCCGAAGCCACAC-3’ |
| Snail1_R | 5’-GGAGAAGGTCCGAGCACAC-3’ |
| Slug_F | 5’-ATACCACAACCAGAGATCCTCA-3’ |
| Slug_R | 5’-GACTCACTCGCCCCAAAGATG-3’ |
| Twist_F | 5’-AATTGGGATGCATTCGAGTCTGTAA-3’ |
| Twist_R | 5’-TTCTGTCCGATGTCACTGCTGTC-3’ |
| ZEB1_F2 | 5’-GCCAATAAGCAAACGATTCTG-3’ |
| ZEB1_R2 | 5’-TTTGGCTGGATCACTTTCAAG-3’ |
| E-cadherin_F | 5’-CCCGGGACAACGTTTATTAC-3’ |
| E-cadherin_R | 5’-GCTGGCTCAAGTCAAAGTCC-3’ |
| N-cadherin_F | 5’-ACAGTGGCCACCTACAAAGG-3’ |
| N-cadherin_R | 5’-CCGAGATGGGGTTGATAATG-3’ |
| Fibronectin_F | 5’-TCCCTCGGAACATCAGAAAC-3’ |
| Fibronectin_R | 5’-CAGTGGGAGACCTCGAGAAG-3’ |
| Vimentin_F | 5’-TACAGGAAGCTGCTGGAAGG-3’ |
| Vimentin_R | 5’-ACCAGAGGGAGTGAATCCAG-3’ |
| hGAPDH_F | 5’-GAAGGTGAAGGTCGGAGTCAAC-3’ |
| hGAPDH_R | 5’-CAGAGTTAAAAGCAGCCCTGGT-3’ |
F, forward; R, reverse.
CHX-Chase Assay
Cells cultured in 6-well plates were transfected with plasmids of interest. For each well, CHX (300 μg/mL) was administered to the culture media 18 hours post-transfection. Cells were harvested in a time series at 0, 2, 4, and 8 hours post-treatment using RIPA lysis buffer. Western blot was performed to monitor protein degradation and ImageJ software was applied to determine the relative intensities of protein bands.
Soft Agar Colony Formation Assay
The 60-mm plates were layered with solidified modified Eagle's medium comprising 10% FBS, 10% tryptose phosphate broth (Merck), and 0.6% Bacto agar (BD Biosciences). Cells (1 × 104) resuspended in DMEM/F12 containing 0.3% Bacto agar and 1% FBS then were seeded on top of the base agar for anchorage-independent growth. The forming colonies were counted after being cultured for 1 month at 37ºC under humidified 5% CO2 conditions.
Scratch Wound Healing Assay
Cultured cells grown into confluent monolayers in 12-well plates were scraped with a sterilized SPLScar Scratcher (SPL Life Sciences, Pocheon, Korea), and then washed extensively with 1× PBS to remove nonadherent cells. The remaining attached cells were continuously cultured in DMEM supplemented with 1% FBS at 37ºC, and monitored for cell migration from 24 to 48 hours.
Transwell Invasion Assay
The trypsinized cells were washed 3 times with FBS-free DMEM/F12 medium and resuspended in 100 μL of 0.1% FBS containing DMEM/F12 at a density of 5 × 104 cells/well (for SK-HEP-1) and 1 × 105 cells/well (for PLC/PRF/5 and HepG2). Cell suspensions were seeded onto each upper chamber of a Transwell plate [24-well insert, 8.0-μm pores, polyester (PET) membrane; Corning, Inc] that was freshly precoated with Matrigel Basement Membrane Matrix Growth Factor Reduced (50 μL; Corning, Inc). Medium containing 10% FBS was used as a chemoattractant and loaded in the lower chamber. After a 24-hour culture at 37ºC, the noninvaded cells on the top of the Transwell insert were scraped off with a cotton swab. Cells that invaded to the lower surface of the insert were washed twice with 1× PBS, fixed with 4% paraformaldehyde for 12 minutes at room temperature, permeabilized for 20 minutes with 100% methanol, and stained purple with KaryoMAX Giemsa dye (Gibco/Fisher Scientific, Auckland, Germany).
Cell Viability Assay
Cells were seeded onto 96-well plates (1 × 104 cells/well) at 24 hours post-transfection. The viability of live cells was monitored by the metabolic colorimetric Alamar blue assay (Life Technologies, Thermo Fisher Scientific), following the manufacturer's instructions. The absorbance at 570 nm was measured with a microplate spectrophotometer.
3D Spheroid Invasion Assay
After washing twice with 1× PBS, cells (3 × 103/well) resuspended in 50 μL FBS-free DMEM were mixed with 5 μL Cultrex 10× Spheroid Formation ECM (Trevigen, Gaithersburg, MD), and seeded onto each well of a Cultrex 3D culture 96-well plate (Trevigen). After 3 days postculture, cells were assembled into compact spheroids, and this 3D culture 96-well plate was placed on ice for 15 minutes. Cultrex spheroid invasion matrix (50 μL) was added to each well, and incubated at 37ºC for 1 hour to enhance matrix gel formation. Next, 100 μL warm DMEM containing chemoattractants [10% FBS, 20 ng/mL epidermal growth factor (EGF), and 25 ng/mL basic fibroblast growth factor (bFGF)] was supplied to each well. Spheroids then were cultured for another 1 week.
Mitochondrial Isolation
Cells grown in 60-mm dishes were subjected to mitochondrial purification using the Mitochondrial Isolation Kit for Cultured Cells (Thermo Fisher Scientific). Briefly, the attached cells (1 × 107) were incubated with 800 μL reagent A on ice for 2 minutes. Then, cell lysates were transferred to Eppendorf tube, followed by treatment with 10 μL reagent B on ice and the reactions were vortexed every minute at maximum speed for a total of 5 minutes. Reagent C (800 μL) was successively added to the reactions, which then were centrifuged at 700 × g, 4ºC for 10 minutes. Supernatant was saved and continuously centrifuged at 12,000 × g, 4ºC for 15 minutes. The resulting supernatant is the cytosolic fraction. The pellet (mitochondrial fraction) was washed with 500 μL reagent C at 12,000 × g, 4ºC for 5 minutes. After discarding the supernatant, the pelleted mitochondria are active and were used in the following study of mitochondrial bioactivity. Alternatively, the mitochondria were lyzed with RIPA lysis buffer for subsequent Western blot analysis.
Differentiation between OMM and IMM Proteins
In order to break down OMM, the isolated mitochondria were subjected to proteolysis by proteinase K (50 ng/μL, Sigma-Aldric, St. Louis, MO) in 100 μL M buffer (210 mM mannitol, 70 mM sucrose, 10 mM HEPES and 1 mM EDTA, pH 7.5] for 30 min at room temperature. To digest IMM, the purified mitochondria were treated with proteinase K (50 ng/μL) and 1% Triton X-100 (Sigma-Aldric) in 100 μL M buffer at room temperature for 30 min. The reactions were stopped by adding RIPA lysis buffer containing 1 mmol/L PMSF and 1X HaltTM protease and phosphatase inhibitor cocktail. The OMM and IMM proteins were then analyzed by western blotting.
Mitochondrial ATP Detection
Cells initially were grown in high-glucose DMEM. After plasmid transfection, the culture media were replaced with glucose-free, galactose-contained DMEM, and continuously cultured for 24 hours before mitochondrial purification. The ATP contents of these mitochondrial isolates were measured with the ATP detection reagent Mitochondrial ToxGlo Assay kit (Promega, Madison, WI), which lysed the intact mitochondria and produced luminescent signal proportional to the level of ATP. The procedure was performed in accordance with the manufacturer's instructions.
ROS Detection
This assay was accomplished by applying the ROS Detection Cell-Based Assay kit (Cayman, Ann Arbor, MI). Dihydroethidium, which can be oxidized by ROS, was used as a redox-sensitive fluorescent probe for the detection of ROS generation. Antimycin A, an inhibitor of complex III of the mitochondrial electron transport chain, was included to stimulate ROS generation. NAC was used as an antioxidant control. After isolating mitochondria from the transfected cells, the pelleted mitochondria were mixed with 130 μL ROS staining buffer. In addition, NAC (30 μmol/L) was added to the antioxidant control reactions. Incubation was performed at 37ºC for 30 minutes and all of the reactions were protected from light. Then, antimycin A (15 μmol/L) was administered to each reaction and incubated at 37ºC in the dark for another 1 hour. Thereafter, the reaction was centrifuged at 400 × g to pellet down the mitochondria. These pellets were mixed with 100 μL cell-based assay buffer and transferred to a black 96-well plate for measuring the fluorescence signal at 570 nm.
Measurement of mtROS in viable cells was performed using a mitochondrial ROS detection assay kit (Cayman). For each reaction, 5 × 105 cells were washed with cell-based assay buffer and centrifuged at 400 × g to remove the assay buffer. Cells were incubated with 0.5 μmol/L mtROS detection reagent for 20 minutes at 37ºC. The stained cells were repeatedly washed 3 times with Hank’s balanced salt solution containing Ca2+ and Mg2+. NAC (500 μmol/L) treatment was performed on these stained cells in Hank’s balanced salt solution at 37ºC for 1 hour, followed by antimycin A (10 μmol/L) at 37ºC for another 1 hour. Each reaction was stained with SYTOX blue dead cell stain solution (1 μmol/L; Life Technologies) to distinguish live cells from dead cells. Finally, 10,000 cells from each reaction were analyzed by flow cytometry (BD FACSLyric; Becton Dickinson Biosciences, San Jose, CA) using the phycoerythrin (PE) channel for mtROS and the V450 channel for live/dead cell detection.
Mitochondrial Staining
Live cells transfected with expression plasmids or siRNAs were grown on 8-well chamber slides. Mitochondrial staining was performed using the Mito-ID Green Detection Kit (Enzo Life Sciences, Lausen, Switzerland). To clarify the distribution of mitochondria, the cytoplasm and nucleus of each cell were labeled simultaneously with Cytopainter Cell Tracking Dye Kit-Red (Abcam, Cambridge, MA) and Hoechst 33342 (Enzo Life Sciences), respectively. Briefly, 1 μL Mito-ID green detection reagent, 20 μL Cytopainter, and 1 μL Hoechst 33342 were mixed in 1 mL of 1× assay buffer, and 100 μL of the mixture was added to each well and incubated at 37ºC for 30 minutes. After 3 repeated washes with 1× PBS, cells were mounted and observed with a fluorescence microscope.
Statistical Analysis
Statistical analysis was conducted using Microsoft Excel 2016 software (Microsoft), SPSS v16.0 software (SPSS Inc, Chicago, IL), and Prism software v8.0 (GraphPad Software, La Jolla, CA). Kaplan–Meier survival curves were generated based on clinical follow-up data to describe survival rates. The log-rank test and the Gehan–Breslow–Wilcoxon test were executed to find statistical differences. The Student t test was used for statistical comparisons, and the Mann–Whitney U test was applied for analyzing the significant differences in categoric variables among groups. A P value less than .05 was considered statistically significant.
Acknowledgments
The authors thank Yu-Chuan Hong (Department of Pediatrics, National Taiwan University Hospital) for technical support in animal experiments and flow cytometry, and also the staff of the Second Core Laboratory (Department of Medical Research, National Taiwan University Hospital) for providing imaging technical support.
CRediT Authorship Contributions
Huey-Huey Chua (Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Project administration: Lead; Software: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead)
Mei-Hwei Chang (Conceptualization: Lead; Funding acquisition: Equal; Methodology: Supporting; Project administration: Lead; Resources: Equal; Visualization: Lead; Writing – review & editing: Equal)
Ya-Hui Chen (Investigation: Supporting; Methodology: Equal; Project administration: Supporting; Validation: Supporting)
Daw-Jen Tsuei (Conceptualization: Equal; Data curation: Equal; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Project administration: Supporting; Software: Supporting; Validation: Supporting)
Yung-Ming Jeng (Conceptualization: Supporting; Methodology: Supporting; Resources: Equal; Validation: Supporting)
Po-Huang Lee (Funding acquisition: Supporting; Resources: Equal)
Yen-Hsuan Ni, MD, PhD (Conceptualization: Lead; Funding acquisition: Lead; Investigation: Equal; Methodology: Equal; Project administration: Lead; Resources: Lead; Supervision: Lead; Validation: Equal; Visualization: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was financially supported by the Ministry of Science and Technology, Executive Yuan, Taiwan (MOST105-2314-B-002-137-MY3 and MOST109-2634-F-002-043), and the Center of Precision Medicine from the Featured Areas Research Center Program within the Framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan (109L9014).
Supplementary Material
References
- 1.Chang M.H., You S.L., Chen C.J., Liu C.J., Lee C.M., Lin S.M., Chu H.C., Wu T.C., Yang S.S., Kuo H.S., Chen D.S., Taiwan Hepatoma Study Group Decreased incidence of hepatocellular carcinoma in hepatitis B vaccinees: a 20-year follow-up study. J Natl Cancer Inst. 2009;101:1348–1355. doi: 10.1093/jnci/djp288. [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Clocchiatti A., Cora E., Zhang Y., Dotto G.P. Sexual dimorphism in cancer. Nat Rev Cancer. 2016;16:330–339. doi: 10.1038/nrc.2016.30. [DOI] [PubMed] [Google Scholar]
- 4.Tsuei D.J., Hsu H.C., Lee P.H., Jeng Y.M., Pu Y.S., Chen C.N., Lee Y.C., Chou W.C., Chang C.J., Ni Y.H., Chang M.H. RBMY, a male germ cell-specific RNA-binding protein, activated in human liver cancers and transforms rodent fibroblasts. Oncogene. 2004;23:5815–5822. doi: 10.1038/sj.onc.1207773. [DOI] [PubMed] [Google Scholar]
- 5.Venables J.P., Elliott D.J., Makarova O.V., Makarov E.M., Cooke H.J., Eperon I.C. RBMY, a probable human spermatogenesis factor, and other hnRNP G proteins interact with Tra2beta and affect splicing. Hum Mol Genet. 2000;9:685–694. doi: 10.1093/hmg/9.5.685. [DOI] [PubMed] [Google Scholar]
- 6.Heinrich B., Zhang Z., Raitskin O., Hiller M., Benderska N., Hartmann A.M., Bracco L., Elliott D., Ben-Ari S., Soreq H., Sperling J., Sperling R., Stamm S. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J Biol Chem. 2009;284:14303–14315. doi: 10.1074/jbc.M901026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreumont N., Bourgeois C.F., Lejeune F., Liu Y., Ehrmann I.E., Elliott D.J., Stevenin J. Human RBMY regulates germline-specific splicing events by modulating the function of the serine/arginine-rich proteins 9G8 and Tra2-{beta. J Cell Sci. 2010;123:40–50. doi: 10.1242/jcs.055889. [DOI] [PubMed] [Google Scholar]
- 8.Elliott D.J., Millar M.R., Oghene K., Ross A., Kiesewetter F., Pryor J., McIntyre M., Hargreave T.B., Saunders P.T., Vogt P.H., Chandley A.C., Cooke H. Expression of RBM in the nuclei of human germ cells is dependent on a critical region of the Y chromosome long arm. Proc Natl Acad Sci U S A. 1997;94:3848–3853. doi: 10.1073/pnas.94.8.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abid S., Sagare-Patil V., Gokral J., Modi D. Cellular ontogeny of RBMY during human spermatogenesis and its role in sperm motility. J Biosci. 2013;38:85–92. doi: 10.1007/s12038-012-9281-8. [DOI] [PubMed] [Google Scholar]
- 10.Yan Y., Yang X., Liu Y., Shen Y., Tu W., Dong Q., Yang D., Ma Y., Yang Y. Copy number variation of functional RBMY1 is associated with sperm motility: an azoospermia factor-linked candidate for asthenozoospermia. Hum Reprod. 2017;32:1521–1531. doi: 10.1093/humrep/dex100. [DOI] [PubMed] [Google Scholar]
- 11.Chua H.H., Tsuei D.J., Lee P.H., Jeng Y.M., Lu J., Wu J.F., Su D.S., Chen Y.H., Chien C.S., Kao P.C., Lee C.N., Hu R.H., Ni Y.H., Chang M.H. RBMY, a novel inhibitor of glycogen synthase kinase 3beta, increases tumor stemness and predicts poor prognosis of hepatocellular carcinoma. Hepatology. 2015;62:1480–1496. doi: 10.1002/hep.27996. [DOI] [PubMed] [Google Scholar]
- 12.Wang N., Wang S., Li M.Y., Hu B.G., Liu L.P., Yang S.L., Yang S., Gong Z., Lai P.B.S., Chen G.G. Cancer stem cells in hepatocellular carcinoma: an overview and promising therapeutic strategies. Ther Adv Med Oncol. 2018;10 doi: 10.1177/1758835918816287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nieto M.A. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342 doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 14.Panchal N.K., Sabina E.P. A serine/threonine protein PIM kinase as a biomarker of cancer and a target for anti-tumor therapy. Life Sci. 2020;255 doi: 10.1016/j.lfs.2020.117866. [DOI] [PubMed] [Google Scholar]
- 15.Saris C.J., Domen J., Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 1991;10:655–664. doi: 10.1002/j.1460-2075.1991.tb07994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nieto M.A., Huang R.Y., Jackson R.A. Thiery JP. Emt. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. 2016. [DOI] [PubMed] [Google Scholar]
- 17.Zhao B., Liu L., Mao J., Zhang Z., Wang Q., Li Q. PIM1 mediates epithelial-mesenchymal transition by targeting Smads and c-Myc in the nucleus and potentiates clear-cell renal-cell carcinoma oncogenesis. Cell Death Dis. 2018;9:307. doi: 10.1038/s41419-018-0348-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X., Zou Y., Liu Y., Cao Y., Zhu J., Zhang J., Chen X., Zhang R., Li J. Inhibition of PIM1 kinase attenuates bleomycin-induced pulmonary fibrosis in mice by modulating the ZEB1/E-cadherin pathway in alveolar epithelial cells. Mol Immunol. 2020;125:15–22. doi: 10.1016/j.molimm.2020.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Adebayo M., Singh S., Singh A.P., Dasgupta S. Mitochondrial fusion and fission: the fine-tune balance for cellular homeostasis. FASEB J. 2021;35 doi: 10.1096/fj.202100067R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun X., Cao H., Zhan L., Yin C., Wang G., Liang P., Li J., Wang Z., Liu B., Huang Q., Xing J. Mitochondrial fission promotes cell migration by Ca(2+)/CaMKII/ERK/FAK pathway in hepatocellular carcinoma. Liver Int. 2018;38:1263–1272. doi: 10.1111/liv.13660. [DOI] [PubMed] [Google Scholar]
- 21.Cheung E.C., Vousden K.H. The role of ROS in tumour development and progression. Nat Rev Cancer. 2022;22:280–297. doi: 10.1038/s41568-021-00435-0. [DOI] [PubMed] [Google Scholar]
- 22.Bergers G., Fendt S.M. The metabolism of cancer cells during metastasis. Nat Rev Cancer. 2021;21:162–180. doi: 10.1038/s41568-020-00320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunniff B., McKenzie A.J., Heintz N.H., Howe A.K. AMPK activity regulates trafficking of mitochondria to the leading edge during cell migration and matrix invasion. Mol Biol Cell. 2016;27:2662–2674. doi: 10.1091/mbc.E16-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kido T., Tabatabai Z.L., Chen X., Lau Y.C. Potential dual functional roles of the Y-linked RBMY in hepatocarcinogenesis. Cancer Sci. 2020;111:2987–2999. doi: 10.1111/cas.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuei D.J., Lee P.H., Peng H.Y., Lu H.L., Su D.S., Jeng Y.M., Hsu H.C., Hsu S.H., Wu J.F., Ni Y.H., Chang M.H. Male germ cell-specific RNA binding protein RBMY: a new oncogene explaining male predominance in liver cancer. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullock A.N., Debreczeni J., Amos A.L., Knapp S., Turk B.E. Structure and substrate specificity of the Pim-1 kinase. J Biol Chem. 2005;280:41675–41682. doi: 10.1074/jbc.M510711200. [DOI] [PubMed] [Google Scholar]
- 27.Friedmann M., Nissen M.S., Hoover D.S., Reeves R., Magnuson N.S. Characterization of the proto-oncogene pim-1: kinase activity and substrate recognition sequence. Arch Biochem Biophys. 1992;298:594–601. doi: 10.1016/0003-9861(92)90454-5. [DOI] [PubMed] [Google Scholar]
- 28.Braso-Maristany F., Filosto S., Catchpole S., Marlow R., Quist J., Francesch-Domenech E., Plumb D.A., Zakka L., Gazinska P., Liccardi G., Meier P., Gris-Oliver A., Cheang M.C., Perdrix-Rosell A., Shafat M., Noel E., Patel N., McEachern K., Scaltriti M., Castel P., Noor F., Buus R., Mathew S., Watkins J., Serra V., Marra P., Grigoriadis A., Tutt A.N. PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat Med. 2016;22:1303–1313. doi: 10.1038/nm.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borillo G.A., Mason M., Quijada P., Volkers M., Cottage C., McGregor M., Din S., Fischer K., Gude N., Avitabile D., Barlow S., Alvarez R., Truffa S., Whittaker R., Glassy M.S., Gustafsson A.B., Miyamoto S., Glembotski C.C., Gottlieb R.A., Brown J.H., Sussman M.A. Pim-1 kinase protects mitochondrial integrity in cardiomyocytes. Circ Res. 2010;106:1265–1274. doi: 10.1161/CIRCRESAHA.109.212035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lilly M., Sandholm J., Cooper J.J., Koskinen P.J., Kraft A. The PIM-1 serine kinase prolongs survival and inhibits apoptosis-related mitochondrial dysfunction in part through a bcl-2-dependent pathway. Oncogene. 1999;18:4022–4031. doi: 10.1038/sj.onc.1202741. [DOI] [PubMed] [Google Scholar]
- 31.Wen Q.L., Yi H.Q., Yang K., Yin C.T., Yin W.J., Xiang F.Y., Bao M., Shuai J., Song Y.W., Ge M.H., Zhu X. Role of oncogene PIM-1 in the development and progression of papillary thyroid carcinoma: Involvement of oxidative stress. Mol Cell Endocrinol. 2021;523 doi: 10.1016/j.mce.2020.111144. [DOI] [PubMed] [Google Scholar]
- 32.Chauhan S.S., Toth R.K., Jensen C.C., Casillas A.L., Kashatus D.F., Warfel N.A. PIM kinases alter mitochondrial dynamics and chemosensitivity in lung cancer. Oncogene. 2020;39:2597–2611. doi: 10.1038/s41388-020-1168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji K., Lin K., Wang Y., Du L., Xu C., He N., Wang J., Liu Y., Liu Q. TAZ inhibition promotes IL-2-induced apoptosis of hepatocellular carcinoma cells by activating the JNK/F-actin/mitochondrial fission pathway. Cancer Cell Int. 2018;18:117. doi: 10.1186/s12935-018-0615-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou C.H., Lin C.C., Yang M.C., Wei C.C., Liao H.D., Lin R.C., Tu W.Y., Kao T.C., Hsu C.M., Cheng J.T., Chou A.K., Lee C.I., Loh J.K., Howng S.L., Hong Y.R. GSK3beta-mediated Drp1 phosphorylation induced elongated mitochondrial morphology against oxidative stress. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Din S., Mason M., Volkers M., Johnson B., Cottage C.T., Wang Z., Joyo A.Y., Quijada P., Erhardt P., Magnuson N.S., Konstandin M.H., Sussman M.A. Pim-1 preserves mitochondrial morphology by inhibiting dynamin-related protein 1 translocation. Proc Natl Acad Sci U S A. 2013;110:5969–5974. doi: 10.1073/pnas.1213294110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou B.P., Deng J., Xia W., Xu J., Li Y.M., Gunduz M., Hung M.C. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez-Tillo E., de Barrios O., Siles L., Cuatrecasas M., Castells A., Postigo A. beta-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci U S A. 2011;108:19204–19209. doi: 10.1073/pnas.1108977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song J. Targeting epithelial-mesenchymal transition pathway in hepatocellular carcinoma. Clin Mol Hepatol. 2020;26:484–486. doi: 10.3350/cmh.2020.0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jimenez-Garcia M.P., Lucena-Cacace A., Robles-Frias M.J., Narlik-Grassow M., Blanco-Aparicio C., Carnero A. The role of PIM1/PIM2 kinases in tumors of the male reproductive system. Sci Rep. 2016;6 doi: 10.1038/srep38079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delbridge M.L., Lingenfelter P.A., Disteche C.M., Graves J.A. The candidate spermatogenesis gene RBMY has a homologue on the human X chromosome. Nat Genet. 1999;22:223–224. doi: 10.1038/10279. [DOI] [PubMed] [Google Scholar]
- 41.Ehrmann I., Dalgliesh C., Tsaousi A., Paronetto M.P., Heinrich B., Kist R., Cairns P., Li W., Mueller C., Jackson M., Peters H., Nayernia K., Saunders P., Mitchell M., Stamm S., Sette C., Elliott D.J. Haploinsufficiency of the germ cell-specific nuclear RNA binding protein hnRNP G-T prevents functional spermatogenesis in the mouse. Hum Mol Genet. 2008;17:2803–2818. doi: 10.1093/hmg/ddn179. [DOI] [PubMed] [Google Scholar]
- 42.Matsunaga S., Takata H., Morimoto A., Hayashihara K., Higashi T., Akatsuchi K., Mizusawa E., Yamakawa M., Ashida M., Matsunaga T.M., Azuma T., Uchiyama S., Fukui K. RBMX: a regulator for maintenance and centromeric protection of sister chromatid cohesion. Cell Rep. 2012;1:299–308. doi: 10.1016/j.celrep.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Adamson B., Smogorzewska A., Sigoillot F.D., King R.W., Elledge S.J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munschauer M., Nguyen C.T., Sirokman K., Hartigan C.R., Hogstrom L., Engreitz J.M., Ulirsch J.C., Fulco C.P., Subramanian V., Chen J., Schenone M., Guttman M., Carr S.A., Lander E.S. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 2018;561:132–136. doi: 10.1038/s41586-018-0453-z. [DOI] [PubMed] [Google Scholar]
- 45.Yan Q., Zeng P., Zhou X., Zhao X., Chen R., Qiao J., Feng L., Zhu Z., Zhang G., Chen C. RBMX suppresses tumorigenicity and progression of bladder cancer by interacting with the hnRNP A1 protein to regulate PKM alternative splicing. Oncogene. 2021;40:2635–2650. doi: 10.1038/s41388-021-01666-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Arribas F., Agudo D., Pollan M., Gomez-Esquer F., Diaz-Gil G., Lucas R., Schneider J. Positive correlation between the expression of X-chromosome RBM genes (RBMX, RBM3, RBM10) and the proapoptotic Bax gene in human breast cancer. J Cell Biochem. 2006;97:1275–1282. doi: 10.1002/jcb.20725. [DOI] [PubMed] [Google Scholar]
- 47.Song Y., He S., Ma X., Zhang M., Zhuang J., Wang G., Ye Y., Xia W. RBMX contributes to hepatocellular carcinoma progression and sorafenib resistance by specifically binding and stabilizing BLACAT1. Am J Cancer Res. 2020;10:3644–3665. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.