Summary
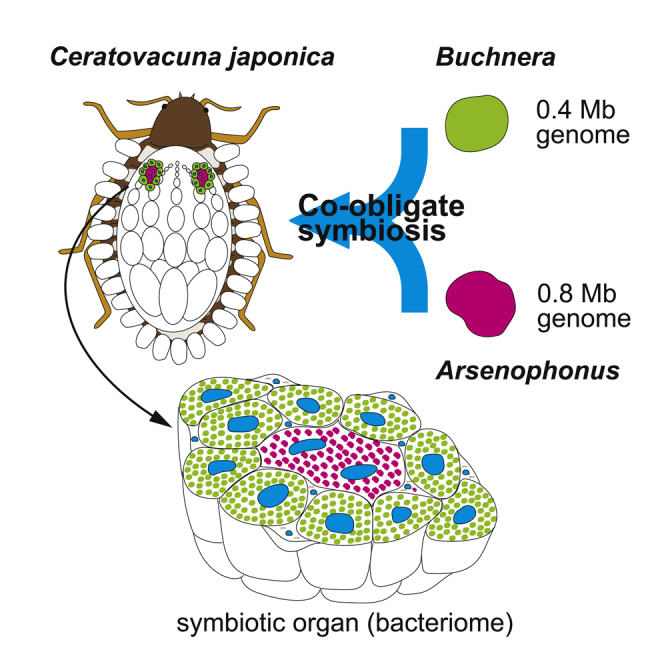
Some hemipteran insects rely on multiple endosymbionts for essential nutrients. However, the evolution of multi-partner symbiotic systems is not well-established. Here, we report a co-obligate symbiosis in the eusocial aphid, Ceratovacuna japonica. 16S rRNA amplicon sequencing unveiled co-infection with a novel Arsenophonus sp. symbiont and Buchnera aphidicola, a common obligate endosymbiont in aphids. Both symbionts were housed within distinct bacteriocytes and were maternally transmitted. The Buchnera and Arsenophonus symbionts had streamlined genomes of 432,286 bp and 853,149 bp, respectively, and exhibited metabolic complementarity in riboflavin and peptidoglycan synthesis pathways. These anatomical and genomic properties were similar to those of independently evolved multi-partner symbiotic systems, such as Buchnera–Serratia in Lachninae and Periphyllus aphids, representing remarkable parallelism. Furthermore, symbiont populations and bacteriome morphology differed between reproductive and soldier castes. Our study provides the first example of co-obligate symbiosis in Hormaphidinae and gives insight into the evolutionary genetics of this complex system.
Subject areas: Evolutionary biology, Genetics, Molecular biology
Graphical abstract
Highlights
-
•
A eusocial aphid evolved co-obligate symbiosis involving Buchnera and Arsenophonus
-
•
Two symbionts localize in the same host organ but within distinct specialized cells
-
•
Metabolic and developmental integration is observed among two symbionts and host
-
•
Symbiont densities and the bacteriome morphology differ between social castes
Evolutionary biology; Genetics; Molecular biology
Introduction
Endosymbionts, microorganisms that live inside the body or cells of host organisms, are widespread in eukaryotes, serving as important resources for evolutionary innovation in hosts (Moran and Baumann, 2000; Nowack and Melkonian, 2010; Pawlowska et al., 2018; Rosenberg et al., 2010). Endosymbionts have been shown to confer beneficial functions to hosts, such as provisioning essential nutrients that are not synthesized by hosts (Akman Gündüz and Douglas, 2012; Douglas et al., 2001; Shigenobu et al., 2000; The International Aphid Genomics Consortium, 2010; Wilson et al., 2010), providing resistance to heat stress (Chen et al., 2000; Russell and Moran, 2006) and protection against natural enemies via toxin production (Degnan et al., 2009; Nakabachi et al., 2013; Oliver et al., 2003). The acquisition of novel traits carried by endosymbionts contributes to niche divergence and lineage diversification of hosts (Janson et al., 2008; Sudakaran et al., 2017).
Aphids are good models for studying endosymbiosis. Almost all aphid species harbor the obligate mutualistic symbiont Buchnera aphidicola (Gammaproteobacteria) in specialized host cells known as bacteriocytes, and these symbionts are vertically transmitted to offspring (Braendle et al., 2003; Miura et al., 2003). Buchnera provides essential amino acids and riboflavin (vitamin B2), which aphids cannot synthesize and are deficient in their sole diet of phloem sap (Akman Gündüz and Douglas, 2012; Douglas et al., 2001; Nakabachi and Ishikawa, 1999; Shigenobu and Wilson, 2011). In addition to the primary symbiont Buchnera, aphids are often associated with facultative symbionts, which are not essential but can contribute to host fitness and phenotypes (Oliver et al., 2010). Buchnera genomes have experienced significant reductions down to ∼0.6 Mb with ∼600 protein-coding genes. Despite the drastic genome reduction, Buchnera genomes retain genes involved in the biosynthesis of amino acids and vitamins essential for the host aphids (Chong et al., 2019; Van Ham et al., 2003; Shigenobu et al., 2000). Facultative symbionts exhibit moderate genome reductions. For example, Serratia symbiotica, Hamiltonella defensa and Regiella insecticola of pea aphids have ∼2.0 Mb genomes encoding ∼2,000 genes (Burke and Moran, 2011; Degnan et al., 2009, 2010).
Recent genomic studies of aphid symbionts have revealed the dynamic evolution of coexisting bacterial partners that function collectively with the ancient and stable symbiont Buchnera (Shigenobu and Yorimoto, 2022). Novel symbionts, such as Sphingopyxis, Pectobacterium, Sodalis-related, and Erwinia-related bacteria, have been found by 16S rRNA amplicon sequencing in a wide variety of aphid lineages (McLean et al., 2019; Xu et al., 2020, 2021). In addition, co-obligate symbioses, where multiple species of symbionts are essential for host survival, have been found in several aphid lineages. Co-obligate symbiosis involving Buchnera and Serratia evolved repeatedly in multiple aphid lineages, including the subfamily Lachninae, genus Periphyllus, Microlophium carnosum, and Aphis urticata (Manzano-Marín et al., 2017; Monnin et al., 2020). Co-obligate Buchnera of Lachninae and Periphyllus (Chaitophorinae) aphids have ∼0.4 Mb genomes encoding ∼400 genes and have lost genes involved in the biosynthesis of some essential amino acids and vitamins that the co-obligate Serratia is presumed to compensate for (Lamelas et al., 2011a; Manzano-Marín et al., 2016; Monnin et al., 2020; Pérez-Brocal et al., 2006).
The tribe Cerataphidini (Hemiptera: Aphididae: Hormaphidinae) is an aphid clade that possesses a number of biologically interesting characteristics, including a complex life history, gall formation, and eusociality. They are able to alternate between primary host plants, where sexual reproduction occurs, and secondary host plants, where parthenogenesis occurs, seasonally (Aoki and Kurosu, 2010; Fukatsu et al., 1994). The fundatrix of Cerataphidini aphids hatches from an overwintering fertilized egg and induces morphologically diverse galls on the Styrax trees, their primary host plant (Aoki and Kurosu, 2010; Fukatsu et al., 1994). Cerataphidini aphids are eusocial; they produce sterile individuals known as a soldier caste to protect the colony from natural enemies (Stern and Foster, 1996). Fukatsu et al. (1994) reported that several species of Cerataphidini have secondary symbionts in addition to Buchnera (Fukatsu et al., 1994), although their identities and roles remain elusive. In several Cerataphidini aphids, the primary symbiont Buchnera has been replaced with extracellular yeast-like symbionts (Fukatsu et al., 1994).
In this study, we investigated bacterial symbionts of Ceratovacuna japonica in the Cerataphidini tribe (Figure 1A), taking an advantage of a recently established laboratory rearing system for this species (Hattori et al., 2013). We uncovered two endosymbionts of Ce. japonica by deep sequencing of 16S rRNA amplicons, Buchnera and an Arsenophonus-related bacterium, and these were consistently detected together. We sequenced the whole genomes of the two symbionts to assess the roles of both symbionts. We conducted microscopic observations to investigate their localization and dynamics in the host. We also compared symbiont populations between social castes. Our results suggest that the newly detected Arsenophonus-related symbiont is a novel species that has established an obligate relationship with the host Ce. japonica through collaboration with Buchnera.
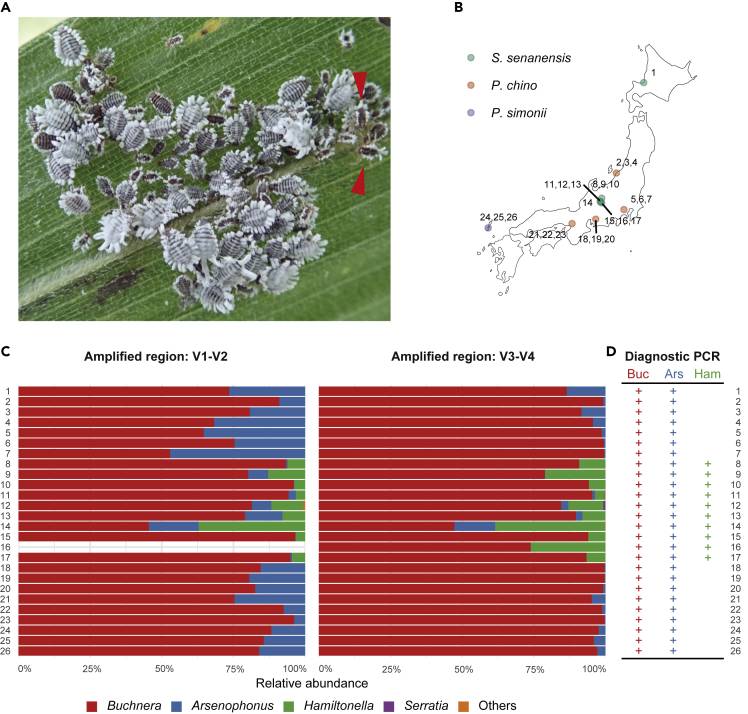
Figure 1.
Symbiont composition determined by deep sequencing of 16S rRNA amplicons of natural populations of Ceratovacuna japonica
(A) A colony of Ce. japonica on the bamboo grass P. chino. Red arrowheads indicate soldiers.
(B) Locations and host plant species of collected Ce. japonica.
(C) Relative abundance of bacterial symbionts in Ce. japonica. The V1–V2 region amplified from sample 16 was excluded owing to sequencing failure. “Others” includes unclassified sequences or those with low abundance.
(D) Summary of diagnostic PCR detection of Buchnera, Arsenophonus, and Hamiltonella in Ce. japonica. See also Figure S1 for agarose gel electrophoresis data. Each bacterial symbiont was detected using specific primers targeting a single-copy gene, dnaK. Each sample ID corresponds to those in Figures 1B, 1C, 1D, and S1. Detected bacterial symbionts are shown by the “+” character.
Results
Co-infection of Buchnera and Arsenophonus in natural populations of Ceratovacuna japonica
To assess the symbiont communities in Ce. japonica, we collected 26 colonies at nine geographically distinct populations from three different host plant species across Japan for high-throughput 16S rRNA amplicon sequencing (Figures 1B, 1C, Tables S1, and S2). The sequencing of 16S rRNA hypervariable regions, V1–V2 and V3–V4, yielded 332,656 and 432,594 reads after quality filtering. V1–V2 and V3–V4 reads were classified into 13 and 25 ASVs (amplicon sequence variants), respectively. The prevalent bacterial genera were identified as Buchnera (mean relative abundance ±SE of V1–V2: 81.48 ± 2.63%, V3–V4: 92.15 ± 2.22%), Arsenophonus (V1–V2: 14.89 ± 2.41%, V3–V4: 2.55 ± 0.76%), and Hamiltonella (V1–V2: 3.60 ± 1.58%, V3–V4: 5.25 ± 1.88%) (Figure 1C, Table S1, and S2). Buchnera, the nearly ubiquitous endosymbiont of aphids, was detected in all populations at an extremely high relative abundance (86.92 ± 1.86%) (Figure 1C). In addition, Arsenophonus was detected in all populations of Ce. japonica, regardless of host plant and geographical location (Figures 1B and 1C, Table S1 and S2). Hamiltonella was detected in specimens from 10 populations in the Nagano area (#8–17 in Figure 1) feeding on Sasa senanensis; however, it was not detected in the Hokkaido population (#1 in Figure 1) on the same host plant (Figures 1B, 1C, Table S1, and S2). Serratia was detected in only a single colony at a low level (Figures 1B and 1C, Table S2). We then performed PCR analyses to confirm the presence or absence of Buchnera, Arsenophonus, and Hamiltonella in the Ce. japonica populations with specific primers targeting a single-copy gene, dnaK, for each bacterium. Consistent with the result of 16S rRNA amplicon sequencing, PCR analyses showed that both Buchnera and Arsenophonus were detected in all populations, whereas Hamiltonella was detected only in populations in the central Japan area (Figures 1D and S1A). Next, we investigated the infection status at the individual level. Diagnostic PCR analyses of each of the 12 individuals in four geographically distinct populations detected both Buchnera and Arsenophonus in all cases (Figure S1B). Taken together, Ce. japonica was always co-infected with Buchnera and Arsenophonus, forming a multi-partner symbiosis. In addition, Ce. japonica could be infected with Hamiltonella as a facultative symbiont.
We established an isofemale line of the aphid Ce. japonica, named NOSY1, from a single individual collected at the foot of Mt. Norikura (location #14 in Figures 1, S1, Tables S1, and S2). It was cultured on bamboo shoots in the laboratory to undergo parthenogenetic reproduction for detailed molecular and microscopic analyses, as described below.
Molecular phylogeny indicates Arsenophonus of Ce. japonica is related to obligate insect endosymbionts with streamlined genomes
We performed molecular phylogenetic analyses based on the 16S rRNA sequences with our assembled data. Buchnera of Ce. japonica (hereafter referred to as Buchnera CJ for simplicity) was included within the Buchnera clade in Cerataphidini (Figure 2A). Buchnera CJ was closely related to the Buchnera strains of a species in the same aphid genus, Ceratovacuna nekoashi and Ce. lanigera, and the sister genus, Pseudoregma alexanderi, Ps. bambucicola, and Ps. panicola. Buchnera of Nipponaphidini, Hormaphidini, and Cerataphidini species were monophyletic with clear separation between clades. These phylogenetic relationships of Buchnera mirrored those of their host aphid species (Chen et al., 2014; Ortiz-Rivas and Martínez-Torres, 2010).
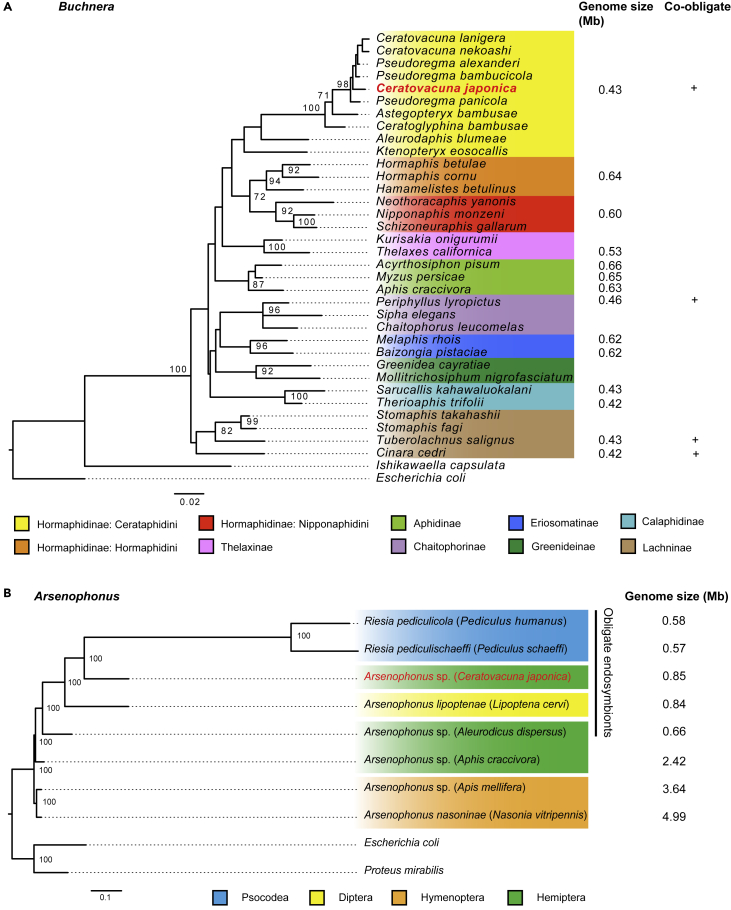
Figure 2.
Phylogenetic analysis of Buchnera and Arsenophonus
(A) Maximum likelihood (ML) tree of Buchnera aphidicola based on the length of 1,337 bp of 16S rRNA sequences. Escherichia coli and Ishikawaella capsulata were used as outgroups. The labels indicate the name of the host species.
(B) ML tree of Arsenophonus based on concatenated 204 single-copy orthologous protein sequences composed of 54,777 amino acid positions. E. coli and Proteus mirabilis were used as outgroups. Target symbionts in this study are highlighted in red on each tree. Bootstrap values no less than 70% are indicated on each node. Scale bars represent 0.02 and 0.1 substitutions per site.
We also built a phylogenetic tree of Arsenophonus of Ce. japonica (hereafter referred to as Arsenophonus CJ) and related bacteria based on the 16S rRNA sequences (Figure S2A). The tree indicated that Arsenophonus CJ was related to endosymbionts of hematophagous Diptera (the superfamily Hippoboscoidea), such as Lipoptena and Trichobius, and lice species (Group A in Figure S2A), whereas Arsenophonus CJ was not included in the clade of symbionts of Hemiptera and Hymenoptera (Group B in Figure S2A). This unusual clustering that does not reflect the phylogenetic relationship of the host insect might be an artifact caused by long-branch attraction and base-pair shift, which are often reported in the phylogenetic analyses of insect endosymbionts (Husník et al., 2011). To overcome this problem, we constructed the phylogeny of Arsenophonus using 204 single-copy orthologous protein sequences composed of 54,777 amino acid residues that are conserved among Arsenophonus and Riesia. The phylogenetic tree strongly supported the monophyly of the obligate endosymbionts of hematophagous insects and plant sap-feeding insects, all of which possess streamlined small genomes (Figure 2B). This phylogenetic position of Arsenophonus CJ was also supported by the phylogenetic trees based on Dayhoff6 recoded protein sequences, whose method minimized the heterogeneous composition and long-branch attraction (Figure S2B). The 16S rRNA sequence of Arsenophonus CJ showed the highest similarity (93.0%) to that of Candidatus Arsenophonus lipoptenae (Figure S3A), an obligate endosymbiont of the blood-sucking deer fly Lipoptena cervi with a role as a B vitamin provider (Nováková et al., 2016). This 93.0% identity was far lower than typical thresholds for assignment to the same species (e.g., ≤97%) (Stackebrandt and Goebel, 1994), indicating that the symbiont discovered in this study was a novel species in the genus Arsenophonus.
The 16S rRNA sequence of Hamiltonella of Ce. japonica (hereafter referred to as Hamiltonella CJ) was almost identical (99.8%) to that of Candidatus Hamiltonella defensa, a facultative symbiont of the pea aphid Acyrthosiphon pisum (Figure S3B).
Buchnera and Arsenophonus are intracellular symbionts housed inside distinct bacteriocytes
We inspected the internal morphology of Ce. japonica to uncover the localization of the three bacteria identified by 16S rRNA amplicon sequencing. Our histological observation identified a pair of symbiotic organs (bacteriomes) in the thorax of the adult parthenogenetic viviparous female (Figures 3A and 3C). Each bacteriome exhibited an oval-shaped structure with a major axis of approximately 191 μm composed of several large presumably polyploid nuclei surrounded by the cytoplasm, which was filled with bacterial cells (Figure 3B), a typical characteristic of aphid bacteriocytes. Of interest, HE staining and DAPI staining patterns allowed us to distinguish between two types of bacteriocytes. There were several uninucleate bacteriocytes on the surface of the bacteriome and a single double-nucleate bacteriocyte at the center of the bacteriome; the latter type of bacteriocytes showed less intense HE signals and more intense DAPI signals than those of the former type (Figures 3D and S4).
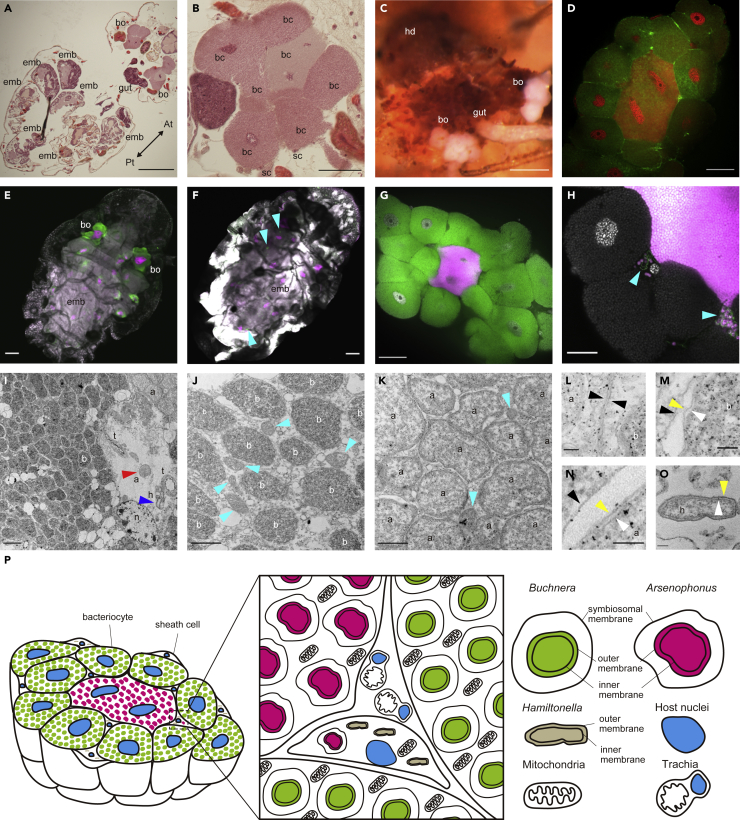
Figure 3.
Localization and morphology of Buchnera and Arsenophonus symbionts
(A and B) Light microscopic images of tissue sections stained with hematoxylin and eosin. The dorsal sections of an adult individual are shown. The whole body (A) and the magnified image of the bacteriome (B).
(C) Light microscopic image of dissected bacteriomes.
(D) Confocal image of the bacteriome structure. Nuclei (red) and F-actin (green) were stained by DAPI and phalloidin.
(E–H) Confocal images of Buchnera, Arsenophonus, and Hamiltonella stained with fluorescent probes specific to each bacterium. Whole bodies of adult individuals (E and F) and dissected bacteriomes (G and H). In (E and G), gray (DAPI), green (Cy5), and magenta (Cy3) signals indicate nuclei, Buchnera, and Arsenophonus, respectively. In (F and H), gray (DAPI), green (Cy5), and magenta (Cy3) signals indicate nuclei, Hamiltonella, and Arsenophonus, respectively. Cyan arrowheads indicate Hamiltonella in (F and H).
(I–O) Electron microscopy of a bacteriome dissected from an adult individual. (I) Low-magnification image of three types of symbionts in the bacteriome. Red and blue arrowheads indicate Arsenophonus and Hamiltonella in the space between bacteriocytes, respectively.
(J and K) Cytoplasm of a bacteriocyte harboring Buchnera (J) and Arsenophonus (K). Cyan arrowheads indicate mitochondria.
(L) Boundary of bacteriocytes harboring Buchnera and bacteriocytes harboring Arsenophonus. Black arrowheads indicate membranes of bacteriocytes.
(M−O) High magnification images of membranes of Buchnera (M), Arsenophonus (N), and Hamiltonella (O) In (M−O), black, yellow, and white arrowheads indicate the symbiosomal membrane, outer membrane, and inner membrane, respectively.
(P) Schematic diagram of the bacteriome morphology. Scale bars show 200 μm in (A and C), 100 μm in (E and F), 50 μm in (B, D, and G), 20 μm in (H), 2 μm in (I), 1 μm in (J and K), and 100 nm in (L–O). a, Arsenophonus; b, Buchnera; bc, bacteriocyte; bo, bacteriome; emb, embryo; h, Hamiltonella; n, host nucleus; sc, sheath cell; t, trachea.
We next conducted fluorescence in situ hybridization (FISH) using probes specifically targeting each bacterium, Buchnera CJ, Arsenophonus CJ, and Hamiltonella CJ. The FISH analysis revealed that Buchnera and Arsenophonus infected the same bacteriome but were localized in different bacteriocytes, consistent with the differential staining pattern of HE and DAPI. Buchnera was localized in the uninucleate bacteriocytes in the outer layer of the bacteriome, whereas Arsenophonus was localized in the single double-nucleate bacteriocyte at the center of the bacteriome (Figures 3E and 3G). Both Buchnera and Arsenophonus were observed exclusively in these specialized bacteriocytes and we had no evidence for the extracellular localization (e.g. hemolymph) of these endosymbionts (Figure 3E). In addition, ovarioles in the abdomen contained many embryos infected with both Buchnera and Arsenophonus (Figure 3E), suggesting that both symbionts are maternally and vertically transmitted to offspring. In contrast to the systematic localization of Buchnera and Arsenophonus in the bacteriocytes, sporadic Hamiltonella were found in the hemocoel and some were detected probably in the sheath cells located between bacteriocytes, where they often coexisted with Arsenophonus (Figures 3F, 3H, and 3I).
Electron microscopy also revealed three types of symbionts that differed in morphology and electron density (Figure 3I). Buchnera and Arsenophonus were round and resided within their specialized bacteriocytes, whereas Hamiltonella was rod-shaped and resided within sheath cells (Figures 3I–3K and 3O). Buchnera and Arsenophonus had a three-layered membrane structure (Figures 3M and 3N), indicating that both symbionts are packed inside the host-derived membranes called symbiosomal membranes, as reported in Buchnera of the pea aphid (Munson et al., 1991). In contrast, Hamiltonella had only a two-layered membrane, i.e., a bacterial inner membrane and outer membrane (Figure 3O).
Streamlined small genomes of Buchnera CJ and Arsenophonus CJ exhibit complementary metabolic capacity
We conducted a shotgun sequencing of the hologenome of the isofemale line of Ce. japonica NOSY1. A metagenomic assembly approach (see STAR Methods for details) yielded assemblies of the complete genomes of three bacterial symbionts, Buchnera CJ, Arsenophonus CJ, and Hamiltonella CJ (Figure S5). The Buchnera CJ genome consisted of one circular chromosome and two plasmids, pLeu and pTrp. (Figures 4A and 4D and Table 1). The Buchnera CJ chromosome had a length of 414,725 bp, G + C content of 20.0%, and coding density of 87.3%. The genome size of Buchnera CJ was significantly smaller than the typical size (approximately 600 kb) of Buchnera of many aphid species (Chong et al., 2019; Van Ham et al., 2003; Shigenobu and Yorimoto, 2022; Shigenobu et al., 2000; Tamas et al., 2002). It was similar to the sizes of genomes of Buchnera of aphids belonging to the genera Cinara, Tuberolachnus, and Periphyllus (Lamelas et al., 2011a; Manzano-Marín et al., 2016; Monnin et al., 2020; Pérez-Brocal et al., 2006), all of which coexist with another obligate symbiont, S. symbiotica. The chromosome of Buchnera CJ encoded 370 protein-coding genes (CDSs), 3 rRNAs (16, 5, and 23S), 30 tRNAs, and 6 pseudogenes. The plasmid pLeu consisted of at least two tandem repeats, the 7,001 bp units of six genes (Figure 4D). The plasmid pTrp consisted of at least two tandem repeats, the 10,560 bp units of one trpE, one pseudogenized trpG, and 10 trpG genes (Figure 4D).
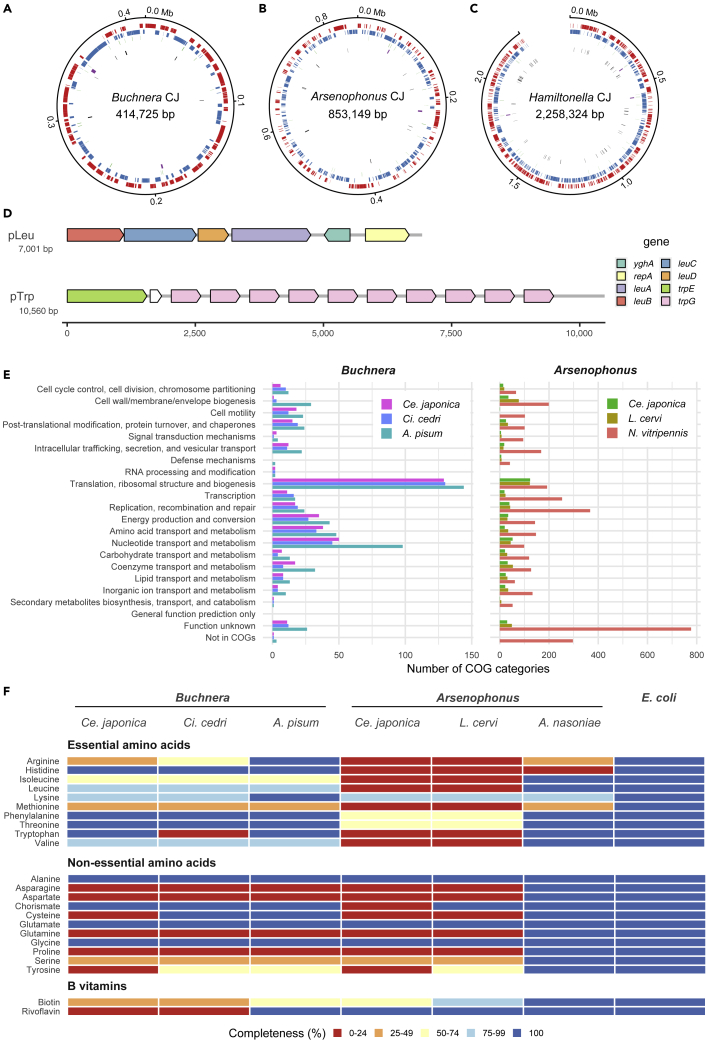
Figure 4.
Genomic features of Buchnera CJ and Arsenophonus CJ
(A) Circular Buchnera CJ genome.
(B) Circular Arsenophonus CJ genome.
(C) Linear Hamiltonella CJ genome. Outer to innermost rings correspond to (i) genome coordinates in kilobases; (ii) predicted protein-coding genes on the plus strand (red); (iii) predicted protein-coding genes on the minus strand (blue); (iv) transfer RNAs (green); (v) ribosomal RNAs (purple); (vi) pseudogenes (black) in (A–C).
(D) Gene orders of plasmids of Buchnera CJ pLeu and pTrp. Arrows indicate the direction of transcription. White arrows indicate pseudogenes.
(E) COG classification of protein-coding genes of Buchnera and Arsenophonus.
(F) Comparison of gene repertoires responsible for nutrient synthesis by Buchnera and Arsenophonus. E. coli is shown as an example of a free-living bacterium. Color blocks indicate the completeness of the minimal gene set for metabolic pathways: red, orange, yellow, light blue, and blue mean 0–24%, 25–49%, 50–74%, 75%–99%, and 100% of the completeness, respectively. In the aphid-Buchnera symbiosis, a metabolic collaboration is known for amino acid syntheses, where the host complements the steps missing from Buchnera as observed in the pathways of leucine, isoleucine, valine, and methionine syntheses (Shigenobu and Wilson, 2011; The International Aphid Genomics Consortium, 2010).
Table 1.
General features of sequenced symbiont genomes of Ce. japonica and the closely related species
|
Buchnera |
Arsenophonus |
Hamiltonella |
|||||
|---|---|---|---|---|---|---|---|
| Ce. japonica | Ci. cedri | A. pisum | Ce. japonica | L. cervi | N. vitripennis | Ce. japonica | |
| Genome size (bp) | 432,286 | 422,434 | 655,725 | 853,149 | 836,724 | 4,987,107 | 2,258,324 |
| Chromosome size (bp) | 414,725 | 416,380 | 640,681 | 853,149 | 836,724 | 3,871,978 | 2,258,324 |
| Number of contigs | 3 | 2 | 3 | 1 | 1 | 18 | 1 |
| Plasmid | 2 | 1 | 2 | 0 | 0 | 17 | 0 |
| GC (%) | 20.0 | 20.1 | 26.3 | 18.5 | 24.9 | 40.2 | 41.2 |
| Coding density (%) | 87.3 | 84.6 | 86.0 | 59.9 | 73.1 | 75.7 | 82.8 |
| Protein-coding genes | 387 | 369 | 574 | 512 | 619 | 4,712 | 2,097 |
| tRNA | 30 | 31 | 31 | 32 | 35 | 70 | 42 |
| rRNA | 3 | 3 | 3 | 3 | 3 | 22 | 9 |
| Pseudogene | 7 | 2 | 13 | 11 | 17 | 485 | 72 |
Buchnera aphidicola of Cinara cedri (GenBank: GCA_000090965.1); B. aphidicola of Acyrthosiphon pisum (GCA_000009605.1); Candidatus Arsenophonus lipoptenae of Lipoptena cervi (GCA_001534665.1); Arsenophonus nasoniae of Nasonia vitripennis (GCA_004768525.1).
The Arsenophonus CJ genome consisted of one circular chromosome (Figures 4B and Table 1). The Arsenophonus CJ chromosome had a length of 853,149 bp and G + C content of 18.5%. The genome of Arsenophonus CJ was considerably smaller than the genome of Arsenophonus nasoniae (5.0 Mb), a son-killer bacterium of Nasonia vitripennis (Darby et al., 2010), and similar in size to the genome of Candidatus Arsenophonus lipoptenae (0.84 Mb), an obligate intracellular symbiont of the blood-feeding deer ked L. cervi (Nováková et al., 2016). Note that Arsenophonus lipoptenae was most closely related to Arsenophonus CJ based on the 16S rRNA sequences, as mentioned above (Figure S2A). In total, the Arsenophonus CJ genome encoded 512 proteins, 3 rRNAs (16, 5, and 23S), 32 tRNAs, and 11 pseudogenes. Notably, the Arsenophonus CJ genome exhibited a low coding density (59.9%), which may be a sign of ongoing genomic erosion.
The Hamiltonella CJ genome had a length of 2,258,324 bp, although we could not produce a circular contig probably because of the long repeats over 10,000 bp at both ends of the contig (Figures 4C and Table 1). The genome size of Hamiltonella CJ was similar to that of Hamiltonella of A. pisum (Degnan et al., 2009), which is a facultative symbiont of aphids, and was larger than that of Hamiltonella of Bemisia tabaci (Rao et al., 2015), which is a co-obligate endosymbiont of whiteflies. The Hamiltonella CJ genome encoded 2,097 proteins, 9 rRNAs (3 of 16S, 3 of 5S, and 3 of 23S), 42 tRNAs, and 72 pseudogenes.
Based on two observations (1) the systematic co-infection of two endosymbionts, Buchnera and Arsenophonus in Ce. japonica, and (2) the drastic genome reduction of these two bacteria, we hypothesized that the Ce. japonica aphid depends on Buchnera CJ and Arsenophonus CJ forming a co-obligate symbiosis. In several hemipteran insects, metabolic complementarity between co-symbionts has been observed (Husnik et al., 2013; Lamelas et al., 2011b; McCutcheon et al., 2009; Rao et al., 2015; Szabó et al., 2020; Wu et al., 2006). To determine if Buchnera CJ and Arsenophonus CJ form co-obligate associations with the host, we examined the genomic signature of metabolic complementarity by comparing the gene repertoires of these bacteria. We detected a complementarity gene repertoire in the metabolic pathways of riboflavin and peptidoglycan between Buchnera CJ and Arsenophonus CJ. Although all genes responsible for riboflavin biosynthesis were missing from the Buchnera CJ genome, these genes were retained in the Arsenophonus CJ genome (Figure 4F and Table S5), indicating a complementary riboflavin-related gene set between the two symbionts. Although Buchnera CJ lost all genes related to peptidoglycan synthesis, the Arsenophonus CJ genome retained genes to synthesize dap-type peptidoglycan (glmMSU, murABCDEFG, mraY, mrcB, ftsI, and dacA) (Table S6). This complementary gene repertoire related to nutrition and cell wall components represent a genomic signature of co-obligate symbiosis.
Although Arsenophonus-related bacteria infect some aphids as facultative symbionts, the genus has not been reported as obligate symbionts in aphids, to the best of our knowledge. As such, we further inspected the genome of Arsenophonus CJ to understand the evolution of the obligate symbiosis. Arsenophonus CJ had a streamlined 853 kb genome with an extremely low GC content (18.5%), a typical feature of obligatory endosymbionts. Notably, the Arsenophonus CJ genome exhibited a very low coding density (59.9%), suggesting ongoing genomic erosion. A COG analysis demonstrated that Arsenophonus CJ is missing many genes with broad functions, including essential housekeeping functions, such as “Replication, recombination and repair” (Figure 4E). For example, the Arsenophonus CJ genome lacked the SOS system genes recA, lexA, umuCD, and uvrABC, involved in DNA repair, as observed in Buchnera. The Arsenophonus CJ genome lacked a majority of genes involved in amino acid biosynthesis pathways; however, it retained all genes responsible for riboflavin biosynthesis (Figure 4F, Tables S4, and S5). Of interest, genes for lipid A biosynthesis were missing or pseudogenized in the Arsenophonus CJ genome. The lipid A biosynthesis pathway is highly conserved in free-living bacteria and facultative bacterial symbionts; however, obligate intracellular symbionts often lack all or some components of this pathway, which is interpreted as an adaptation to the host immune responses (Wu et al., 2006). Among nine key genes involved in lipid A biosynthesis, five genes were lost and four were found to be pseudogenes in the Arsenophonus CJ genome (Table S7). The analysis of lipid A-related gene repertoire of the Arsenophonus CJ provided two insights: (1) Arsenophonus CJ lost the ability to produce functional lipid A, as observed in other obligatory symbionts, and (2) the degenerative process occurred relatively recently, because four pseudogenes were detected. Taken together, the Arsenphonus CJ genome showed the features of obligatory symbionts and the signature of genomic erosion because of the ongoing obligate interaction.
Infection of Buchnera/Arsenophonus symbionts and host embryogenesis indicate developmental integration
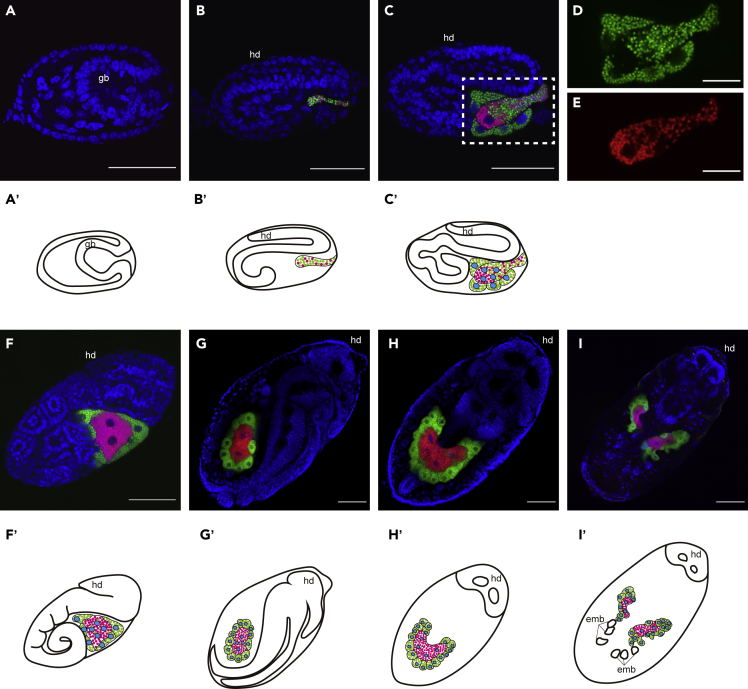
Obligate symbionts often exhibit “developmental integration” with hosts, where the processes of symbiont infection and host oogenesis or embryogenesis are well-coordinated (Braendle et al., 2003; Miura et al., 2003; Shigenobu and Wilson, 2011). To address the developmental integration in the context of multi-partner symbiosis in Ce. japonica, we analyzed the formation of the bacteriome and dynamics of symbionts during parthenogenetic embryogenesis. We dissected ovarioles from the third or fourth instar nymphs of viviparous aphids, within which a series of developing embryos can be observed. Until anatrepsis (stage 8), when the germband is invaginating from both the dorsal and ventral sides, no symbionts were detected (Figure 5A). Around the time of germband elongation and folding into an S shape (stage 11), we found a mixed population of Buchnera and Arsenophonus incorporated into the embryo from the posterior part (Figure 5B). At the later stage of the germband elongation, when it twisted with more abdominal segments (stage 12), we observed the first formation of bacteriocytes, where huge host nuclei and symbiont masses were cellularized into bacteriocytes (Figure 5C). At this stage, both Buchnera and Arsenophonus coexisted in the central bacteriocyte; however, only Buchnera cells were found in peripheral bacteriocytes (Figures 5C, 5D, and 5E). By the time of the initiation of limb bud formation (stage 13), several bacteriocytes aggregated into an organ-like structure (i.e., a bacteriome) located at the ventral part of the embryo. Within the bacteriome, Buchnera and Arsenophonus were segregated into different types of bacteriocytes: Arsenophonus resided in a single central syncytial bacteriocyte containing four host nuclei, whereas Buchnera resided in multiple peripheral uninucleate bacteriocytes surrounding the Arsenophonus-containing bacteriocyte (Figure 5F). After katatrepsis, the bacteriome was located in the dorsal abdomen of the embryo (Figures 5G and 5H). At this stage, the morphology of the bacteriome changed from a round shape to a U-shape, maintaining the orientation of the two types of bacteriocytes—outer Buchnera-containing bacteriocytes and inner Arsenophonus-containing bacteriocytes. In the late embryo before larviposition, the bacteriome divided laterally to form a pair of bacteriomes at the anterior abdomen (Figure 5I). After larviposition, the pair of bacteriomes was located at the anterior abdomen, maintaining the orientation of the two types of bacteriocytes (Figure 6E). The morphology and localization of the bacteriomes in first instar larvae were comparable to those observed in adults (Figure 3A).
Figure 5.
Infection and developmental integration of Buchnera and Arsenophonus symbionts during host embryogenesis
(A) Embryo during anatrepsis. Neither Buchnera nor Arsenophonus are observed at this stage.
(B) S-shape embryo. Both Buchnera and Arsenophonus begin to infect the embryo from the posterior part.
(C) Twisting embryo. Infection with both Buchnera and Arsenophonus is continuing. New bacteriocytes harboring Buchnera are formed.
(D and E) Only Cy5 (D) and Cy3 (E) signals in C are shown.
(F) Limb bud formation. Limb buds are formed in the thorax region and the germband is elongating. Symbiont transmission has finished at this stage.
(G and H) Germband retraction is completed after katatrepsis. G and H show the lateral view and dorsal view of the same individual. The germband is retracted to the posterior tip. One huge bacteriome exists in the abdomen.
(I) An embryo prior to larviposition. The bacteriome is divided and forms a pair of bacteriomes. (A–I) Blue (DAPI), green (Cy5), and red (Cy3) indicate nuclei, Buchnera, and Arsenophonus, respectively. (A′–I′) Schematic diagram of images in (A–I). Cyan, green, and magenta indicate nuclei of bacteriocytes, Buchnera, and Arsenophonus, respectively. (A–G): Lateral view, (H and I): Dorsal view. Scale bar: 20 μm for (D and E); 50 μm for (A–C and F–H); 100 μm for I. gb, germband; hd, head.
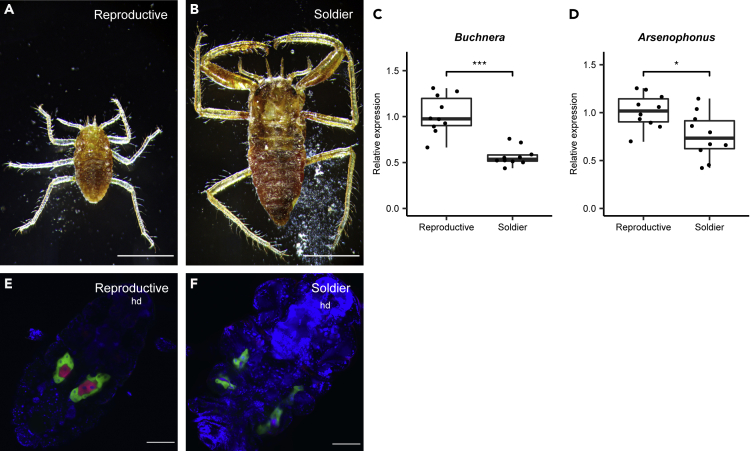
Figure 6.
Comparison of symbiosis status between soldier and reproductive castes
(A and B) Light microscopic images of first-instar nymphs of reproductive (A) and soldier (B) castes. (C and D) Comparison of symbiont titers of first instar nymphs between castes. (C) Buchnera and (D) Arsenophonus titers. Buchnera and Arsenophonus were measured by qPCR using the bacterial symbiont dnaK gene standardized by the host RpL7 gene. Asterisks indicate statistically significant differences (Welch’s t-test p< 0.001 in A, p< 0.05 in B).
(E and F) Confocal images of the localization of both symbionts in the first instar nymphs of reproductive (E) and soldier (F) castes. Blue (DAPI), green (Cy5), and red (Cy3) indicate nuclei, Buchnera, and Arsenophonus, respectively. Scale bars indicate 500 μm in (A and B) and 100 μm in (E and F). hd, head.
Hamiltonella CJ was also detected in the infecting bacterial mass together with Buchnera and Arsenophonus at the S-shape embryo stage (Figure S7A). However, unlike Buchnera and Arsenophonus, at later stages, Hamiltonella was scattered around the periphery of the bacteriome (Figure S7B) and was sometimes detected in or around unknown cells or organs, different from bacteriomes. Figure S7B shows an example of the localization near the tip of the germband.
Symbiosis status differs between soldier and reproductive castes
Ce. japonica is eusocial and produces sterile individuals known as a soldier caste specialized for colony protection (Figures 1A, 6A, and 6B). With our laboratory rearing system, asexual viviparous females cultured on the bamboo S. senanensis produce the normal (reproductive) caste and the soldier caste. Soldiers remain first-instar nymphs throughout their life on the bamboo. We compared symbiosis status between castes. First, we evaluated Buchnera and Arsenophonus titers in first-instar individuals of the reproductive caste and soldier caste in Ce. japonica by qPCR (Figures 6C and 6D). Buchnera was detected from both castes in Ce. japonica; however, the Buchnera titer was approximately 44% lower in the soldier caste than in the reproductive caste and the difference was statistically significant (Figure 6C; Welch’s t-test, p < 0.001). Arsenophonus was also detected in both castes in Ce. japonica; however, the Arsenophonus titer was approximately 25% lower in the soldier caste than in the reproductive caste and this difference was statistically significant (Figure 6D; Welch’s t-test, p < 0.05). Second, in an analysis of morphological differences, found that soldiers possess a pair of bacteriomes with a different morphology from that in the reproductive caste (Figures 6E and 6F). Taken together, soldiers exhibit a different symbiotic condition distinct from that of the reproductive caste, including fewer symbionts in apparently malformed bacteriomes.
Discussion
Arsenophonus is an obligate symbiont in Ce. japonica forming a dual symbiosis with Buchnera
We investigated bacterial symbionts of the eusocial aphid Ce. japonica by an integrative approach using molecular, genomic, and microscopic techniques. We found that two bacterial species, Buchnera CJ and Arsenophonus CJ, coexisted in all natural populations and individuals surveyed (Figures 1C and 1D, S1, Tables S1, and S2). The persistent presence of Buchnera in Ce. japonica is not surprising because the species is a common obligatory endosymbiont in aphids (Baumann et al., 1995; Chong et al., 2019; Shigenobu and Wilson, 2011; Shigenobu et al., 2000). Buchnera CJ showed typical characteristics of Buchnera of other aphids, i.e., a small genome, transovarial inheritance, intracellular localization in bacteriocytes, and a subcellular morphology exhibiting a round-shaped bacterial cell encapsulated by the host-derived membrane (symbiosomal membrane). On the other hand, the systematic infection of Arsenophonus CJ was an unexpected and novel finding in this study. Although Arsenophonus-related bacteria infect some aphids as facultative symbionts (Ayoubi et al., 2020; Wagner et al., 2015; Wulff et al., 2013), the consistent infection of Arsenophonus in Ce. japonica suggests that the symbiosis is obligatory for the host. Microscopic observation showed that Arsenophonus cells were localized inside bacteriocytes, similar to the intracellular symbiosis observed in Buchnera (Figures 3E and 3G). Each Arsenophonus cell was surrounded by the host-derived membrane (symbiosomal membrane), as observed in Buchnera (Figure 3N). Furthermore, Arsenophonus symbionts were maternally and vertically inherited in the host offspring (Figures 5B–5I). Arsenophonus CJ had a small, streamlined genome (Figures 3B and Table 1), typical of obligate bacterial symbionts. Taken together, we detected a dual obligate symbiosis involving Arsenophonus sp. and Buchnera in Ce. japonica.
The paraphyly of Arsenophonus genus and the origin of Arsenophonus CJ
The genus Arsenophonus is a cluster of bacterial symbionts found in taxonomically widespread insects (Nováková et al., 2009) and Arsenophonus-related bacteria have been reported to infect some aphids as facultative symbionts (Ayoubi et al., 2020; Jousselin et al., 2013; Wagner et al., 2015; Wulff et al., 2013). Our molecular phylogenetic analysis indicated that Arsenophonus is a paraphyletic genus sharing a common ancestor with Aschnera and Riesia. Based on the phylogenetic tree, obligate endosymbionts of hematophagous insects and plant sap-feeding insects were clustered into a monophyletic group (Figures 2B and S2), despite the distant host lineage. A. lipoptenae and Riesia bacteria are obligate endosymbionts of blood-feeding insects, Hippoboscoidea flies and Anoplura sucking lice, respectively (Boyd et al., 2017; Nováková et al., 2016). Arsenophonus of a whitefly Aleurodics dispersus is a co-obligate endosymbiont collaborating with the ancient obligate symbiont, Portiera aleyrodidarum (Santos-Garcia et al., 2018). Of interest, regardless of host lineages, the group of obligate endosymbionts including Arsenophonus CJ shares similar characteristics. These obligate endosymbionts have small genomes ranging from 0.53 to 0.84 Mb with AT-biased nucleotide composition and supply hosts with essential B vitamins that are deficient in the hosts’ diet (Boyd et al., 2017; Hosokawa et al., 2012; Nováková et al., 2016; Santos-Garcia et al., 2018). These features contrast facultative Arsenophonus symbionts that have relatively large genomes, e.g., the son-killer bacterium A. nasoniae (4.99 Mb) of N. vitripennis (Darby et al., 2010), Arsenophonus triatominarum (3.86 Mb) of Triatoma infestans, and Arsenophonus sp. (2.42 Mb) of Aphis craccivora. In sum, Arsenophonus CJ belonged to the group of obligatory endosymbionts, supporting the obligate nature of the observed symbiosis. Also, it should be noted that this is the first report of an obligate type of Arsenophonus endosymbiont found in aphid species.
The phylogenetic position of Arsenophonus CJ raises a question about its origin. Assuming the closest relative is the endosymbiont of hematophagous insects with currently available datasets, Arsenophonus CJ might be originated from symbionts of hematophagous insects by horizontal transfer. Alternatively, because Arsenophonus is widespread in Hormaphidinae aphids (Xu et al., 2021), the ancient acquisition of this symbiont by Ce. japonica is also possible. Extensive phylogenetic analyses of Arsenophonus in Hormaphidinae aphids with more extensive taxon sampling are needed to understand the origin of co-obligate Arsenophonus symbionts.
Buchnera and Arsenophonus as co-obligate symbionts
Our genome analysis revealed that the genome size of Buchnera CJ (Table 1) was significantly smaller than the majority of Buchnera genomes (Chong et al., 2019; Shigenobu and Yorimoto, 2022). It is possible that a loss of some functions in this Buchnera lineage was compensated by the other obligate symbiont Arsenophonus CJ. We examined the genomic signature of metabolic complementarity. All genes responsible for riboflavin biosynthesis were missing from the Buchnera CJ genome but present in the Arsenophonus CJ genome (Figure 4F and Table S5), indicating a complementary riboflavin-related gene set between two symbionts. Arsenophonus CJ may provide riboflavin to the host, instead of Buchnera, which provides riboflavin in the Aphid–Buchnera symbiosis (Nakabachi and Ishikawa, 1999). Similarly, peptidoglycan synthesis genes were lost in Buchnera CJ and present in the Arsenophonus CJ genome (particularly glmMSU, murABCDEFG, mraY, mrcB, ftsI, and dacA) (Table S6). These results provide evidence for complementary repertoires of genes involved in nutrition and cell wall production in Buchnera CJ and Arsenophonus CJ, further indicating that they represent a co-obligate symbiosis. Similar genomic complementarity was reported in the Buchnera-Serratia co-obligate symbioses found in Lachninae and Periphyllus aphids, as well as in the more recently evolved associations found in A. urticata and M. carnosum (Monnin et al., 2020) (See below for further discussion).
Our histological observation revealed a characteristic arrangement of the two symbionts within the same bacteriome. Both symbionts resided within the same bacteriome but in separate bacteriocytes; the central syncytial bacteriocytes harboring Arsenophonus CJ were morphologically different from the peripheral uninucleate bacteriocytes harboring Buchnera within the bacteriome of Ce. japonica (Figure 3). In bacteriome formation during embryogenesis, although both Buchnera CJ and Arsenophonus CJ were maternally transmitted simultaneously to the embryos, the symbionts were segregated into different types of bacteriocytes (Figures 5B–5F). These observations indicate a high level of anatomical and developmental integration of the two symbionts and the host.
Parallel evolution of co-obligate symbiosis in aphids
The co-obligate symbiosis of Buchnera and Serratia has occurred in aphid lineages, such as Cinara and Tuberolachnus in the subfamily Lachninae, Periphyllus in the subfamily Chaitophorinae, and A. urticata and M. carnosum in the subfamily Aphidinae (Lamelas et al., 2011a,2011b; Manzano-Marín and Latorre, 2014; Manzano-Marín et al., 2016; Monnin et al., 2020; Pérez-Brocal et al., 2006). In several Cinara species, co-obligate Serratia has been replaced by Erwinia, Fukatsuia, and Sodalis symbionts (Manzano-Marín et al., 2017; Manzano-Marín et al., 2019; Meseguer et al., 2017). Our study is the first report of co-obligate symbiosis in the subfamily Hormaphidinae and revealed a novel partner combination (i.e., Buchnera and Arsenophonus). Regardless of the aphid lineage or co-obligate symbiont species, there are amazing similarities among co-obligate symbioses ranging from genomic features to the morphology of bacteriocytes. Although typical Buchnera genome sizes are ∼0.6 Mb, those of co-obligate Buchnera in Ce. japonica, Lachninae, and Periphyllus aphids have ∼0.4 Mb genomes, regardless of phylogenetic positions (Table 1) (Chong et al., 2019; Shigenobu and Yorimoto, 2022). All of these ∼0.4 Mb Buchnera genomes lost genes in the riboflavin, ornithine, and peptidoglycan biosynthesis pathways, although the ornithine and peptidoglycan biosynthesis pathways were also lost in several Buchnera genomes of other subfamilies (Figure 4F, Tables S4, S5, and S6) (Chong et al., 2019; Lamelas et al., 2011a; Manzano-Marín et al., 2016, 2019; Monnin et al., 2020; Pérez-Brocal et al., 2006). The loss of nutritional functions in Buchnera to provide riboflavin to the host is presumably compensated by the partner obligate symbiont, as evidenced by the complementary gene repertoire. The loss of peptidoglycan pathway genes might be accounted for by the environment Buchnera resides where the obligate endosymbiont may not require cell walls inside the specialized host cells. In dual symbiosis, Buchnera and co-infected symbionts, such as Serratia, Erwinia, Fukatsuia, or Sodalis, are found in the same bacteriome but sorted separately into the distinct bacteriocytes in Lachninae aphids (Manzano-Marín et al., 2017, 2019). Mitochondrial density differs between Buchnera-containing bacteriocytes and co-obligate symbiont-containing bacteriocytes: Mitochondrial abundance was lower in bacteriocytes harboring Serratia in Ci. cedri (Gómez-Valero et al., 2004) and it seems to be the case with Arsenophonus-containing bacteriocytes in Ce. japonica (Figures 3J and 3K). These common characteristics are consistent with parallel evolution of the co-obligate symbiosis in aphid lineages, implying that there are evolutionary constraints on the aphid–Buchnera symbiosis.
In addition to the parallelism in co-obligate symbioses, there are significant variations across aphid lineages in the localization patterns of new symbionts. As we reported in this study, co-obligate Arsenophonus CJ resided inside the central syncytial bacteriocytes distinct from Buchnera’s one and exhibited no extracellular localization in Ce. japonica (Hormaphidinae) (Figure 3). In Lachninae aphids, co-obligate symbionts reside inside the distinct bacteriocytes, whereas the distribution of the bacteriocytes harboring co-obligate symbionts is diversified (Manzano-Marín et al., 2017, 2019). The co-obligate Serratia of Pe. lyropictus (Chaitophorinae) exhibits intracellular and extracellular localizations, such as in central syncytium bacteriocytes, sheath cells, hemolymph, and gut of the host (Renoz et al., 2022). The variations in anatomical integration in multi-partner symbioses seem to reflect the degree of the symbiotic association among the host and symbionts.
Roles of Arsenophonus CJ
Based on the genomic analysis, Arsenophonus CJ is likely involved in riboflavin provisioning to the host (Figure 4F, Tables S4, and S5). A similar function in B-vitamin provisioning has been reported for whiteflies, lice, and Hippoboscoidea flies (Nováková et al., 2015,2016; Santos-Garcia et al., 2018). An experimental study has demonstrated that a facultative Arsenophonus symbiont expands the dietary breadth in the aphid A. craccivora (Wagner et al., 2015). Arsenophonus CJ may also contribute to the usage of host plants, the snowbell tree Styrax japonicus and bamboo grasses, such as Pleioblastus chino, P. simonii and S. senanensis. However, the acquisition of new co-obligate symbiont species is not associated with adaptation to novel ecological niches in the evolution of co-obligate symbiosis of Cinara aphids (Meseguer et al., 2017). Further studies are needed to clarify the roles of the Arsenophonus symbiont in Ce. japonica.
Ongoing reductive evolution of the Arsenophonus CJ genome
Synteny analysis showed that Arsenophonus genomes have experienced many rearrangements and deletions (Figure S6C and S6D). In addition, the Arsenophonus CJ genome showed a low coding density and multiple pseudogenes (Table 1). Recently evolved symbionts are characterized by a low coding density, pseudogene formation, and many genome rearrangements and deletions (Burke and Moran, 2011; Ochman and Davalos, 2006). Accordingly, Arsenophonus CJ is a recently evolved co-obligate symbiont and gene inactivation is ongoing. An interesting example of ongoing genomic erosion in Arsenophonus CJ was the gene repertoire for lipid A biosynthesis. Lipid A is the hydrophobic anchor of lipopolysaccharide in the outer membrane of gram-negative bacteria (Raetz and Whitfield, 2002; Raetz et al., 2009), and elicits a strong immune response in host animals (Raetz and Whitfield, 2002). The key genes for lipid A biosynthesis were either missing or pseudogenes in the Arsenophonus CJ genome (Table S7). Losing the ability to produce lipid A in Arsenophonus CJ may be beneficial for the symbiotic association with host animals by accommodating the host immune response. The observed pseudogenization suggests an ongoing genomic erosion of Arsenophonus CJ, probably leading to a more streamlined obligatory endosymbiont specialized for the Arsenophonus–Buchnera dual symbiosis.
Symbiosis in the sterile soldier in eusocial aphids
Eusocial aphid species have been found in only two subfamilies, Hormaphidinae and Eriosomatinae (Pike and Foster, 2008; Stern and Foster, 1996). The eusocial aphid Ce. japonica provides a unique opportunity to study how symbiosis operates in a caste system. In a comparison of normal (reproductive) nymphs and sterile soldiers, we detected significantly fewer symbionts in soldiers than in normal nymphs (Figures 6C and 6D) as well as a distorted bacteriome shape (Figures 6E and 6F), indicating that the symbiotic condition differs between social castes in Ce. japonica. Soldiers of eusocial aphids are sterile and do not grow after birth (Chung and Shigenobu, 2022; Stern and Foster, 1996). Such differences in nutritional conditions between castes may be related to the difference in symbiotic status. In the eusocial aphid Colophina arma of the subfamily Eriosomatinae, the soldier caste lacks endosymbionts and bacteriomes entirely (Fukatsu and Ishikawa, 1992). In the eusocial ant tribe Camponotini, the obligate symbiont Blochmannia enhances nutrition, thereby influencing the size of worker ants (Feldhaar et al., 2007; Sinotte et al., 2018; Zientz et al., 2006). The mechanisms and biological significance of caste-dependent symbiosis in social insects are intriguing subjects for future exploration.
Limitations of the study
This study found a dual obligate symbiosis involving Arsenophonus sp. and Buchnera in Ce. japonica with molecular, genomic, and microscopic approaches. First, the role of Arsenophnus CJ remains elusive. Our genomic analysis predicted that Arsenophonus CJ likely has a role involved in riboflavin provisioning to the host, but it should be further validated experimentally. We do not exclude the possibility of other functions the symbionts exert. For the experimental assessment, a technique of selective and specific elimination of each symbiont, which is currently unfeasible for this system, by using antibiotics or some chemicals should be developed. Second, the origin of Arsenophonus CJ is unclear. Our molecular phylogenetic analysis indicated that Arsenophonus CJ was positioned in a clade that includes obligate endosymbionts of diverse host insects in origin such as hematophagous Diptera and sap-sacking Hemiptera. To understand the origin of Arsenophonus CJ and the evolutionary trajectory of Arsenophonus–Buchnera dual symbiosis, further genomic analysis of symbionts with more extensive taxon sampling in the subfamily Hormaphidinae is needed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Buchnera aphidicola strain CjNOSY1 | This study | N/A |
| Arsenophonus sp. strain CjNOSY1 | This study | N/A |
| Hamiltonella defensa strain CjNOSY1 | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RNase A (17,500 U) | Qiagen | Cat#19101 |
| Proteinase K (2 mL) | Qiagen | Cat#19131 |
| DAPI | Dojindo Laboratories | Cat#340-07971 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat#H3570 |
| Alexa Fluor™ 594 phalloidin | Thermo Fisher Scientific | Cat#A12381 |
| FM™ 4-64FX | Thermo Fisher Scientific | Cat#F34653 |
| Critical commercial assays | ||
| DNeasy Blood & Tissue Kit (250) | Qiagen | Cat#69506 |
| Quick-16S NGS Library Prep Kit | Zymo Research | Cat#D6400 |
| TruSeq DNA PCR-Free Low Throughput Library Prep Kit (24 samples) | Illumina | Cat#20015962 |
| Genomic-tip 20/G | Qiagen | Cat#10223 |
| Ligation Sequencing Kit | Oxford Nanopore Technologies | Cat#SQK-LSK110 |
| Deposited data | ||
| Ceratovacuna japonica strain NOSY1 Nanopore genome sequencing data | This study | Accession number: DRR379965 |
| Ceratovacuna japonica strain NOSY1 Illumina genome sequencing data | This study | Accession number: DRR379966 |
| Ceratovacuna japonica strain NOSY1 mtDNA, complete genome | This study | Accession number: LC722794 |
| Buchnera aphidicola (Ceratovacuna japonica) strain CjNOSY1 genome | This study | Accession number: AP026065 (chromosome), AP026066 (pLeu), AP026067 (pTrp) |
| Arsenophonus sp. (Ceratovacuna japonica) strain CjNOSY1 genome | This study | Accession number: AP026064 |
| Candidatus Hamiltonella defensa (Ceratovacuna japonica) strain CjNOSY1 | This study | Accession number: AP026068 |
| 16S rRNA amplicon sequencing data of Ceratovacuna japonica | This study | Accession number: DRA014323 |
| Predicted protein-coding genes, amino acid sequences, and annotations | This study | Figshare: 10.6084/m9.figshare.c.6026333 |
| Scripts for analyses | This study | GitHub: https://github.com/shigenobulab/21shunta_cjsym_msprep |
| Experimental models: Organisms/strains | ||
| Ceratovacuna japonica strain NOSY1 | This study | N/A |
| Oligonucleotides | ||
| For primer sequences, please see Table S8 | This study | N/A |
| For probe sequences, please see Table S8 | This study | N/A |
| Software and algorithms | ||
| MAFFT version 7.490 | Katoh et al. (2002) | https://mafft.cbrc.jp/alignment/software/ |
| QIIME2 version 2020.8 | Bolyen et al. (2019) | https://qiime2.org/ |
| dada2 | Callahan et al. (2016) | https://benjjneb.github.io/dada2/dada-installation.html |
| naïve Bayes classifier | Bokulich et al. (2018) | N/A |
| Primer3Plus | Untergasser et al. (2007) | https://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi |
| Guppy version 4.3.4 | Oxford Nanopore Technologies | https://github.com/nanoporetech |
| Raven version 1.5.0 | Vaser and Šikić (2021) | https://github.com/lbcb-sci/raven |
| BLAST version 2.12.0 | Altschul et al. (1990) | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastDocs&DOC_TYPE=Download |
| DIAMOND version 2.0.5 | Buchfink et al. (2015) | https://github.com/bbuchfink/diamond |
| BlobTools version version 1.1.1 | Laetsch and Blaxter (2017) | https://github.com/DRL/blobtools |
| minimap2 version 2.17-r941 | Li (2018) | https://github.com/lh3/minimap2 |
| Racon version 1.4.20 | Vaser et al. (2017) | https://github.com/isovic/racon |
| medaka version 1.4.1 | Oxford Nanopore Technologies | https://github.com/nanoporetech/medaka |
| cutadapt version 2.10 | Martin (2011) | https://github.com/marcelm/cutadapt |
| Bowtie2 version 2.4.2 | Langmead and Salzberg (2012) | https://github.com/BenLangmead/bowtie2 |
| Pilon version 1.24 | Walker et al. (2014) | https://github.com/broadinstitute/pilon |
| JBrowse2 version 2.1.0 | Hershberg et al. (2021) | https://jbrowse.org/jb2/download/ |
| Prokka version 1.14.6 | Seemann (2014) | https://github.com/tseemann/prokka |
| eggNOG-mapper version 2.1.3 | Huerta-Cepas et al. (2017), 2019 | https://github.com/eggnogdb/eggnog-mapper |
| KEGG mapper | Kanehisa and Sato (2020) | https://www.genome.jp/kegg/mapper/ |
| circlize version 0.4.15 | Gu et al. (2014) | https://cran.r-project.org/web/packages/circlize/index.html |
| gggenes version 0.4.1 | Wilkins and Kurtz (2019) | https://github.com/wilkox/gggenes |
| TrimAL version 1.4.rev15 | Capella-Gutiérrez et al. (2009) | https://vicfero.github.io/trimal/ |
| ModelTest-NG version 0.1.6 | Darriba et al. (2020) | https://github.com/ddarriba/modeltest |
| RAxML-NG version 1.0.3 | Kozlov et al. (2019) | https://github.com/amkozlov/raxml-ng |
| OrthoFinder version 2.5.2 | Emms and Kelly (2019) | https://github.com/davidemms/OrthoFinder |
| FigTree version 1.4.4 | Institute of Evolutionary Biology, University of Edinburgh, Edinburgh | http://tree.bio.ed.ac.uk/software/figtree/ |
| Jalview version 2.11.0 | Waterhouse et al. (2009) | http://www.jalview.org/getdown/release/ |
| FIJI | Schindelin et al. (2012) | https://imagej.net/software/fiji/ |
| R version 4.2.0 | R Core Team (2021) | https://www.r-project.org/ |
| ggplot2 version 3.3.6 | Wickham, 2016 | https://ggplot2.tidyverse.org/ |
| Python version 3.9.1 | Python Software Foundation | https://www.python.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Shuji Shigenobu (shige@nibb.ac.jp).
Materials availability
The isofemale NOSY1 strain of Ce. japonica established in this study has been maintained at the Laboratory of Evolutionary Genomics, National Institute of Basic Biology.
Experimental model and subject details
Aphid rearing
An isofemale strain of the aphid Ce.japonica, strain NOSY1, was established from a single female collected from a secondary host plant, bamboo grass S. senanensis, at the foot of Mt. Norikura, Nagano Prefecture, Japan (1525.7 m above the sea level; 36°07′09.0″N, 137°37′18.4″E). The strain has been maintained in the asexual viviparous phase on S. senanensis at 20°C with a long-day condition (16 h light/8 h dark cycle) and 60% relative humidity in our laboratory since September 2017. S. senanensis was grown singly in 0.6L pots filled with potting soil at 25°C before use for feeding aphids. For species identification, a partial cytochrome c oxidase I (COI) sequence (658 bp) of Ce. japonica registered in NCBI (GenBank: EU701571.1) and the mitochondrial genome sequence obtained from our de novo assembly data of Ce. japonica strain NOSY1 (see “Symbiont genome assembly and annotation” section) were aligned using MAFFT (v7.490) (Katoh et al., 2002) with the default options, which resulted in only one nucleotide mismatch.
Method details
Aphid collection
Twenty-six colonies of Ceratovacuna japonica (Takahashi, 1958) were collected at nine geographically distinct populations from three different host plant species across Japan from 2006 to 2019. The host plants and locations of sampling are listed in Tables S1 and S2. These aphid samples were preserved in 100% ethanol at 4°C until 16S ribosomal DNA amplicon sequencing was performed.
16S ribosomal DNA amplicon sequencing analysis
The genome DNA (gDNA) of Ce.japonica colonies was extracted using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction. Aphids were rinsed with fresh 70% ethanol with vigorous agitation to remove wax on the surface of their bodies. Each gDNA sample was extracted from 6 to 20 individuals of the reproductive caste. gDNA samples with low concentration were concentrated by ethanol precipitation with Ethachinmate (Nippon Gene, Tokyo, Japan). The 16S rDNA libraries were prepared using Quick-16S NGS Library Prep Kit (Zymo Research, Irvine, CA, USA) targeting hypervariable 16S rRNA regions, V1–V2 and V3–V4 regions, with one negative (ZymoBIOMICS DNase/RNaseFree Distilled Water, Zymo Research) and one positive control (ZymoBIOMICS Microbial Community DNA Standard, Zymo Research). Although the selection of the target hypervariable region in 16S rDNA amplicon sequencing is a controversial issue, both primer sets (V1–V2 and V3–V4) provided in the kit were used in this study to achieve both wider coverage of detectable bacterial groups (V1–V2) and better resolution in classifying sequences of Proteobacteria (V3-V4). Concentrations of the libraries were quantified with KAPA SYBR FAST qPCR Kit (Kapa Biosystems, Wilmington, MA, USA) by Applied Biosystems 7500 Real-Time PCR Systems (Applied Biosystems, Foster City, CA, USA). The libraries were sequenced using the Illumina MiSeq platform (Illumina, Foster City, CA, USA), and 250 bp of paired-end reads were generated. The sequencing of 16S rRNA hypervariable regions, V1–V2 and V3–V4, yielded 360,704 and 484,288 raw reads, respectively. The Illumina raw reads were deposited in the DDBJ DRA database under accession number DRA014323.
Raw paired-end reads were analyzed using QIIME 2 (version 2020.8) (Bolyen et al., 2019) with the following plugins: dada2 (Callahan et al., 2016) for quality filtering, trimming length, merge paired reads and removing chimeric sequences; naïve Bayes classifier (Bokulich et al., 2018) for taxonomy assignment against the Silva132 database (Quast et al., 2013). Forward and reverse primer regions were trimmed from the raw reads as follows: 19 bp from 5′ end of forward reads and 16 bp from 5′ end of reverse reads in the V1–V2 amplified data, 16 bp from 5′ end of forward reads and 24 bp from 5′ end of reverse reads in the V3–V4 amplified data. After quality filtering and removing chimeric sequences, 332,656 and 432,594 reads from the V1–V2 and V3–V4 regions were remained, respectively.
Diagnostic PCR
Genomic DNA was prepared from Ce. japonica colonies in the same way as described in the “16S ribosomal DNA amplicon sequencing analysis” section. Same gDNA samples corresponding to natural populations of Ce. japonica (#1–#26) were used to confirm the presence or absence of symbionts. In addition, gDNA from each of the 12 adult individuals (populations of #2–4, #14, #18–20, and #24–26) was used to confirm the infection status at the individual level. To detect each symbiont specifically, we designed the PCR primers as follows. A single-copy gene, dnaK, is often used in di-symbiotic aphids infected with Buchnera and Serratia to detect each symbiont specifically (Koga et al., 2003; Monnin et al., 2020). Three dnaK genes of Buchnera, Arsenophonus, and Hamiltonella of Ce. japonica were aligned using MAFFT (v7.490) (Katoh et al., 2002) and the variable regions were manually selected. Multiple candidate primers targeting dnaK of Buchnera, Arsenophonus, and Hamiltonella were designed using Primer3Plus (Untergasser et al., 2007) with a setting where the selected variable regions were specified in “included region” option. To confirm the specificity of the designed primers, BLASTN (version 2.12.0) (Altschul et al., 1990) was performed with the “-task blastn-short” option against the assembled genome sequences of Buchnera, Arsenophonus, Hamiltonella, and the host aphid Ce. japonica (see “Hologenome sequencing” section). The amplification and the product size of the PCR products generated with candidate primers were validated using the specimen #14 in Figure 1. Based on these considerations, the following primers were finally established: CjBucDnaK_FW2 (5′- CAGCAGATTCATCTGGACCTAAAC-3′) and CjBucDnaK_RV2 (5′- CCATAGGCATTCTAGTTTGACCAC-3′) primers for Buchnera; CjArsDnaK_FW2 (5′- TGGAATTCAAGCAGCACCAC-3′) and CjArsDnaK_RV2 (5′- TCTGCATTTGCTTCTGCATC-3′) primers for Arsenophonus; CjHamDnaK_F3 (5′- ATGCACTGACGATGGTTTCTGC-3′) and CjHamDnaK_R3 (5′- ACTCAGCATCAACAGCATCTGC-3′) for Hamiltonella. PCR reaction mixtures were comprised of 10 μL KOD SYBR qPCR Mix (Toyobo, Osaka, Japan), 0.6 μL of each primer (0.3 μM), 2 μL gDNA, and 6.8 μL UltraPure DNase/RNase-Free Distilled Water (Invitrogen, Carlsbad, CA, USA). PCR was performed on LightCycler 96 (Roche, Basel, Switzerland) with the following program: 98°C for 2 min, followed by 30 cycles consisting of 98°C for 10 s, 60°C for 10 s, and 68°C for 30 s. The PCR products and FastGene 100 bp DNA Marker (Nippon Genetics) were loaded on 1.8% UltraPure™ Agarose (Invitrogen) gels containing SYBR Safe DNA Gel Stain (Invitrogen) with 100V for 25 min and imaged on the Molecular Imager ChemiDoc™ XRS+ system using UV transillumination (Bio-Rad, Hercules, CA, USA).
Real-time qPCR
Genome DNA was extracted from first-instar nymphs of the reproductive caste and soldier caste in the strain NOSY1 in the same way as described in the “16S ribosomal DNA amplicon sequencing analysis” section. Each sample contained the gDNA of three fresh individuals reared on S. senanensis at 20°C. We used the primers targeting the dnaK gene: CjBucDnaK_FW2 and CjBucDnaK_RV2 for Buchnera and CjArsDnaK_FW2 and CjArsDnaK_RV2 for Arsenophonus. We additionally designed primers targeting the RpL7 gene in the host Ce. japonica as an internal control: Cjap_TR33288_RpL7_F1 (5′- GGCCTTTCAAATTAAACACCCCAAC-3′) and Cjap_TR33288_RpL7_R1 (5′- ATCTTCCCGGTTTCCAAAGTCG-3′) primers. PCR reaction mixtures were composed of 10 μL KOD SYBR qPCR Mix (Toyobo), 0.6 μL of each primer (0.3 μM), 2 μL gDNA, and 6.8 μL UltraPure DNase/RNase-Free Distilled Water (Invitrogen). Real-time quantitative PCR was performed in 40 cycles using LightCycler 96 Instrument (Roche) with the following program: 98°C for 2 min, followed by 40 cycles consisting of 98°C for 10 s, 60°C for 10 s, and 68°C for 30 s. The amount of the dnaK genes of bacterial symbionts was normalized by the RpL7 gene of the host and the relative amount was calculated using the ΔΔCt methodology (Livak and Schmittgen, 2001).
Hologenome sequencing
Nanopore long reads and Illumina short reads were generated to achieve high-quality symbiont genome assemblies. For Nanopore library preparation, 19.69 mg of 25 fresh adult individuals of the strain NOSY1 reared on S. senanensis at 20°C was used. Aphids were rinsed with fresh 70% ethanol with vigorous agitation to remove wax on the surface of their bodies. To prepare the high molecular weight (HMV) DNA of aphids, frozen aphids were transferred to a mortar and gently ground into a fine powder with liquid nitrogen. Frozen powdery QIAGEN G2 buffer (Qiagen), which was generated by adding 2-mercaptoethanol to QIAGEN G2 buffer and spraying the mixed buffer into liquid nitrogen in a glass beaker, was added to the sample and blended quickly. Letting the mixture thaw in a tube, RNaseA (Qiagen) and Proteinase K (Qiagen) were added, and the sample was incubated at 40°C for 3.0 h without agitation. The sample was centrifuged at 9,500 rpm at 4°C for 20 min and the supernatant was subjected to DNA extraction with a QIAGEN Genomic-tip 20/G column. The gDNA was eluted with 800 μL of Buffer QF twice, a 0.7-fold volume of isopropanol was added, and then the gDNA was centrifuged at 9,500 rpm at 4°C for 20 min. The pellet was washed twice using fresh 70% ethanol and centrifuged at 15,000 rpm at 4°C for 5 min. The pellet was dried for a few minutes and then the gDNA was eluted with 51μLTE buffer at room temperature overnights. Note that the extracted gDNA includes genomes derived from aphids and the symbionts, i.e., hologenome. The quantity of extracted gDNA was measured using Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) and Qubit 2.0 Fluorometer (Thermo Fisher Scientific). The quality of extracted gDNA was assessed using Nanodrop ND-2000C (Thermo Fisher Scientific). The integrity of the HMW genome was assessed by a pulsed-field gel electrophoresis using CHEF Mapper (Bio-Rad). With this HMW genome, a Nanopore sequencing library was prepared using the SQL-LSK110 Ligation Sequencing Kit (Oxford Nanopore Technologies, Oxford, UK) according to the manufacturer’s instruction and sequenced using the R10.3 flow cell on the GridION system. Reads were basecalled using GUPPY (version 4.3.4). The total number of raw Nanopore reads was 646,913. The raw Nanopore reads were deposited in the DDBJ database under accession number DRR379965.
For Illumina library preparation, 10.93 mg of 15 fresh adult individuals of the strain NOSY1 reared on S. senanensis at 20°C was used. Aphids were rinsed with fresh 70% ethanol with vigorous agitation to remove wax on the surface of their bodies. Genomic DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen) and then the gDNA was purified using the Genomic DNA Clean & Concentrator Kit (Zymo Research) according to the manufacturer’s instruction. The gDNA was fragmented into 200–500 bp (peak at 350 bp) using Covaris Focused-ultrasonicator M220. A library for whole genome sequencing was prepared using the TruSeq DNA PCR-Free Library Prep Kit (Illumina) according to the manufacturer’s instruction. Quality of the library was validated by the TapeStation HS D5000 (Agilent Technologies, Santa Clara, CA, USA). A concentration of the library was quantified by Applied Biosystems 7500 Real-Time PCR Systems (Applied Biosystems). The Illumina library was sequenced using the Illumina HiSeq X Ten platform (Illumina) at Macrogen Japan (Tokyo, Japan) with the 2x 150 bp paired-end sequencing protocol. The total number of raw Illumina paired-end reads was 204,892,576. The raw Illumina reads were deposited in the DDBJ database under accession number DRR379966.
Genome assembly and annotation
Raw Nanopore reads obtained from hologenomic samples (see above the section “Hologenome sequencing”) were used to assemble symbiont genomes and a mitochondrial genome of the host using raven (version 1.5.0) (Vaser and Šikić, 2021). The metagenome-assembled contigs were subjected to BlobTools (version 1.1.1) (Laetsch and Blaxter, 2017) for metagenomic binning. All assembled contigs were aligned against the RefSeq protein database (last accessed November 23, 2020) using DIAMOND (version 2.0.5) (Buchfink et al., 2015) with the blastx mode and “--range-culling--evalue 1e-25 ” options. One hundred million Illumina raw reads were mapped to the contigs using Bowtie2 (version 2.4.2) (Langmead and Salzberg, 2012). Then, a blobplot was created based on the DIAMOND results and sequencing depth using BlobTools (version 1.1.1) (Laetsch and Blaxter, 2017), which allowed us to identify contigs corresponding to chromosomes of Buchnera, Arsenophonus, Hamiltonella, and the host mitochondrion. Plasmids were not well-assembled probably due to the repetitive sequences. Plasmid sequences of Buchnera were extracted from the raw Nanopore reads based on the results of BLASTN (version 2.12.0) (Altschul et al., 1990) search using plasmid sequences of Buchnera of A. pisum (pLeu: AP001071.1, pTrp: AP001070.1) as queries. To confirm the circularity, the Nanopore reads were mapped to the symbiont genome sequences using minimap2 (version 2.17-r941) (Li, 2018) and the presence of overlapping reads at both ends was checked. We finally obtained closed circular sequences of chromosomes and plasmids for Buchnera and Arsenophonus, while the chromosomal contig of Hamiltonella was not closed probably due to the long repeats over 10,000 bp at both ends of the contig.
The raw Nanopore reads were mapped to the plasmid sequences using minimap2 (version 2.17-r941) (Li, 2018), and the sequences were polished once using Racon (version 1.4.20) (Vaser et al., 2017). Then, assembled symbiont genomes and the plasmids are polished using medaka (version 1.4.1, https://github.com/nanoporetech/medaka). Adapter trimming and quality filtering were performed on raw Illumina paired-reads using cutadapt (version 2.10) (Martin, 2011) and 182,821,759 (89.2%) paired-reads were passed. The cleaned Illumina reads were mapped to the assembled genomes and the plasmids using Bowtie2 (version 2.4.2) (Langmead and Salzberg, 2012) and then Pilon (version 1.24) (Walker et al., 2014) was used for assembly polishing. This polishing step with Bowtie2 and Pilon was repeated three times. In de novo genome assembling, 17,120 (read depth: 126.6 x), 13,819 (39.7 x), 44,970 (97.3 x), and 10,873 (813.4 x) of 646,913 raw Nanopore long reads were used for the Buchnera, Arsenophonus, Hamiltonella, and mitochondrial genomes, respectively. For polishing the plasmid sequences with Racon, 335 (83.9 x) and 668 (130.6 x) of 646,913 raw Nanopore reads were used for pLeu and pTrp, respectively. For polishing the genome assembly with Illumina reads with pilon, 5,401,205 (1,819.4 x), 2,508,501 (413.9 x), 11,614,114 (702.9 x), 1,223,783 (8,126.6x), 180,211 (1,882.0 x), and 304,557 (2,228.9 x) of 182,821,759 cleaned Illumina reads were used for the genomes of Buchnera, Arsenophonus, Hamiltonella, and mitochondrion, and the plasmids of pLeu and pTrp, respectively. The assembled genome and the plasmid sequences were deposited in the DDBJ database under accession numbers: LC722794.1 for mitochondrion, AP026065–AP026067 for Buchnera, AP026064 for Arsenophonus, and AP026068 for Hamiltonella. The genome browser was built using JBrowse2 (version 2.1.0) (Hershberg et al., 2021) and is available at https://shigenobulab-bkt1.s3.ap-northeast-1.amazonaws.com/jb-CerJaSym-220622a/index.html.
Gene predictions were performed using Prokka (version 1.14.6) (Seemann, 2014). Pseudogene candidates were predicted using Pseudofinder (version 1.0) (Syberg-Olsen et al., 2022) and determined these as pseudogenes according to the following criteria: genes less than 70% of the average length of DIAMOND BlastP hits and/or interrupted by pre-mature terminal codons. Functional annotations were performed using eggNOG-mapper (version 2.1.3) (Huerta-Cepas et al., 2017, 2019), the annotations include Cluster of Orthologous Genes (COG) category tags (Tatusov et al., 2000) and Kyoto Encyclopedia of Genes and Genomes (KEGG) IDs (Kanehisa and Goto, 2000). Metabolic pathways were reconstructed using the KEGG mapper tool (Kanehisa and Sato, 2020). Three symbiont genomes and two plasmids were visualized with annotations using circlize (version 0.4.15) (Gu et al., 2014) and gggenes (version 0.4.1) (Wilkins and Kurtz, 2019), respectively. To observe genome synteny, symbiont chromosomes are compared using BLASTN with dc-megablast task option (Altschul et al., 1990). The gene annotation files and scripts are available on Figshare (https://doi.org/10.6084/m9.figshare.C.6026333) and GitHub (https://github.com/shigenobulab/21shunta_cjsym_msprep), respectively.
Molecular phylogenetic analysis
To reconstruct phylogenetic trees with symbiont sequences of Ce. japonica obtained in this study, 16S rRNA sequences of relatives and the outgroup were downloaded from NCBI Database on January 5, 2022 (See Table S3). Multiple alignments of 16S rRNA sequences were performed using MAFFT with the L-INS-i algorithm (version 7.490) (Katoh et al., 2002). Gaps in the alignments were stripped using TrimAL with the “-gt 1.0” option (version 1.4.rev15) (Capella-Gutiérrez et al., 2009), resulting in the length of 1,337 and 1,451 bp of 16S rRNA sequences to build Buchnera and Arsenophonus phylogenies, respectively. To find the best models for phylogenetic analysis, ModelTest-NG (version 0.1.6) (Darriba et al., 2020) was used; all of the bayesian information criterion (BIC), Akaike information criterion (AIC), and Akaike information criterion corrected for small samples (AICs) supported consistently the GTR + I + G4 model and TVM + I + G4 model for Buchnera and Arsenophonus, respectively, for the Maximum likelihood (ML) trees. ML trees were reconstructed using RAxML-NG (version 1.0.3) (Kozlov et al., 2019) with the aforementioned models. Bootstrap values for ML phylogeny were obtained by 1,000 replicates. Phylogenetic trees were visualized using FigTree (version 1.4.4) (http://tree.bio.ed.ac.uk/software/figtree/). A multiple alignment of Arsenophonus of Ce. japonica, L. cervi, and T. parasiticus and a pairwise alignment of Hamiltonella of Ce. japonica and A. pisum were performed using MAFFT with the L-INS-i algorithm (version 7.490) and visualized using Jalview (version 2.11.0) (Waterhouse et al., 2009).
For better resolution of Arsenophonus phylogeny, a core set of single-copy protein sequences was also used to build it. All protein sequences derived from complete genomes of five Arsenophonus, two Riesia, and the outgroup (Proteus mirabilis and Escherichia coli) were downloaded from the NCBI Assembly Database on August 11, 2022 (See Table S3). Orthology analysis using OrthoFinder (version 2.5.2) (Emms and Kelly, 2019) identified 204 single-copy orthologous groups universally conserved among all species analyzed. For each orthologous group, member orthologs were aligned using MAFFT with the L-INS-i algorithm (version 7.490) (Katoh et al., 2002), trimmed using TrimAL with the “-gt 1.0” option (version 1.4.rev15) (Capella-Gutiérrez et al., 2009), and then concatenated by our custom Python script, resulting in a multiple alignment sequences composed of 54,777 amino acid residues. An ML tree was built using RAxML-NG (version 1.0.3) (Kozlov et al., 2019) with the CPREV + I + G4 + F model, supported by BIC, AIC, and AICs on the ModelTest-NG (version 0.1.6) (Darriba et al., 2020). In addition, to minimize adverse effect caused by heterogeneous composition and long-branch attraction, which were often observed in the phylogenetic analyses of endosymbiont genomes (Husník et al., 2011), the Dayhoff6 (A, G, P, S, T) (D, E, N, Q) (H, K, R) (F, Y, W) (I, L, M, V) (C) recoded dataset was used to build the Arsenophonus phylogeny. The concatenated amino acid alignments were converted based on the Dayhoff6 matrix with our custom Python script and an ML tree was reconstructed using RAxML-NG (version 1.0.3) (Kozlov et al., 2019) with the MULTI6_GTR + I + G4 +M{SNPHAC}{X} model. Bootstrap values for ML phylogeny were obtained by 1,000 replicates and the phylogenetic trees were visualized using FigTree (version 1.4.4) (http://tree.bio.ed.ac.uk/software/figtree/).
Histology
Heads, tails, and appendages of adult insects were removed in a Bouin’s fixative (saturated picric acid: formaldehyde: glacial acetic acid = 15:5:1) to facilitate infiltration of reagents into the insect tissues at room temperature. The insects were fixed in fresh Bouin’s fixative at 4°C overnight, washed with 90% ethanol added lithium carbonate, and preserved in 90% ethanol at 4°C until use. Subsequently, the insects were dehydrated and cleared through an ethanol-butanol series. The cleared insects were immersed and embedded in paraffin at 60°C. The paraffin blocks were polymerized at 4°C and cut into 5 μm thick sections using the rotary microtome Leica RM 2155 (Leica, Wetzlar, Germany). The sections were mounted on microscopic slides coated with egg white-glycerin and deparaffinized by Lemosol A (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan). The deparaffinized sections were hydrated through an ethanol-water series and stained with Delafield’s hematoxylin. The stained sections were washed with 1% hydrochloric acid-ethanol for 40 s and then stained with eosin. The stained sections were dehydrated through a water-ethanol series and cleaned by Lemosol A. The sections were mounted into Canada balsam and the images were taken using the digital camera DS-FIL-U2 (Nikon, Tokyo, Japan).
Fluorescence in situ hybridization
Adults and first-instar nymphs were fixed by Carnoy’s fixative (ethanol: chloroform: acetic acid = 6:3:1) overnight and decolorized by the alcoholic 6% H2O2 solution for several weeks at room temperature. Late embryos were fixed by Carnoy’s fixative for 30 min and decolorized by the alcoholic 6% H2O2 solution overnight. H2O2 solution was exchanged every two or three days during incubation. Young embryos and dissected bacteriomes were fixed by 4% paraformaldehyde dissolved 1xPBS (pH 7.4). The fixed samples were treated with 100% methanol for 30 min except for bacteriome samples. The samples were hydrated in a graded series of 0.3% Triton X-100 in 1x PBS prior to the hybridization. Hybridization was performed in 20 mM Tris-HCl (pH 8.0), 0.9 M NaCl, 0.01% SDS, 30% (v/v) formamide containing 100 nM each of fluorescent-labeled oligonucleotide DNA probes: Cy5_CjapBuc16S_1 (5’-Cy5-CCT CTT CTA AGT AGA TCC-3′), Cy3_CjapArs16S_2 (5′-Cy3-CCC GAC CGA ATC GAT GGC-3′), and Cy5_CjapHam16S_V1V2 (5’-Cy5-CTC AGT AAA CTG CGC TCA C-3′), which are complementary to 16S rRNA of Buchnera, Arsenophonus, and Hamiltonella, respectively, at room temperature for 2 h. Excess probes were washed out with 0.3% Triton X-100 in 1x PBS. DNA was counterstained with DAPI (Dojindo Laboratories, Kumamoto, Japan) or Hoechst 33,342 (Thermo Fisher Scientific). F-actin was stained with Alexa Fluor 594 phalloidin (Thermo Fisher Scientific) in 1x PBS at room temperature for 2 h. Membranes were stained with FM 4-64FX (Thermo Fisher Scientific) in 1x PBS for 10 min on ice; then fixed by 4% paraformaldehyde dissolved 1x PBS for 1 h on ice. The specimens were mounted with VECTASHIELD (Vector Laboratories, Burlingame, CA, USA) and analyzed underOlympus FLUOVIEW FV1000 confocal laser scanning microscope (Olympus, Tokyo, Japan). The sizes of bacteriomes were measured using FIJI (Schindelin et al., 2012).
Transmission electron microscopy (TEM)
Bacteriomes were dissected from adult aphids in 2.5% glutaraldehyde in 1x PBS (pH 7.4). The fixed samples were then postfixed with 1% osmium tetroxide for 1 h. After washing with the same buffer, the specimens were dehydrated in a graded ethanol series at room temperature. The samples were treated with propylene oxide and infiltrated with propylene oxide-Epon (Epon 812 resin; TAAB Laboratories, Aldermaston, UK) solution (propylene oxide-Epon resin, 1:1 [v/v]) overnight. The samples were then embedded in Epon resin that was allowed to polymerize at 60°C for 72 h. Ultrathin sections were cut on an ultramicrotome (Leica) and mounted on nickel grids. The sections were then stained with 4% uranyl acetate and lead citrate. After staining, all sections were examined under a transmission electron microscope (model JEM1010; JEOL, Tokyo, Japan) operated at 80 kV.
Quantification and statistical analysis
The symbiont titers between castes were evaluated using Welch’s t-test on the ΔΔCt values. All statistical analyses were performed using R version 4.2.0 (R Core Team, 2021).
Acknowledgments
We appreciate Dr. Katsushi Yamaguchi and Dr. Asaka Akita in the Functional Genomics Facility, NIBB Core Research Facilities for technical support with Illumina library preparation and sequencing. We appreciate Dr. Yuuki Kobayashi, Dr. Wen Hsin-i, and Mika Ikeda for the technical support with Nanopore library preparation and sequencing. We appreciate Dr. Chen-yo Chung, Dr. Yasuhiko Chikami, Dr. Naoki Minamino, and Dr. Yasuhiro Kamei for technical advices. We appreciate Dr. Shinichi Yoda, Dr. Yasuhiko Chikami, and Kathrine Tan for support and discussion. Computational resources were provided by the Data Integration and Analysis Facility, National Institute for Basic Biology. This work was funded by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant-in-Aid for Scientific Research (B) (no. JP17H03717), (A) (no. JP20H00478), and Grant-in-Aid for Scientific Research on Innovative Areas (Research in a Proposed Research Area) (no. JP17H06384) to S.S., and Grant-in-Aid for JSPS Fellows (no. JP20J11981) and SOKENDAI Publication Grant for Research Papers to S.Y..
Author contributions
S.Y. and S.S. designed the research. S.Y. and M.H. collected aphid samples. S.Y. and M.K. performed TEM. S.Y. performed molecular biology experiments and microscopic analyses. S.Y. and S.S. performed next-generation sequencing and bioinformatics analyses. S.Y. and S.S. wrote the manuscript. All authors provided comments on revisions and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: December 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105478.
Supplemental information
Buchnera of Ac. pisum (GCA_000009605.1); Buchnera of Ci.cedri (GCA_000090965.1); Arsenophonus of L. cervi (GCA_001534665.1); Arsenophonus of N. vitripennis (GCA_004768525.1); Escherichia coli (GCA_000005845.2).
Buchnera of Ac. pisum (GCA_000009605.1); Buchnera of Ci.cedri (GCA_000090965.1); Arsenophonus of L. cervi (GCA_001534665.1); Arsenophonus of N. vitripennis (GCA_004768525.1); Escherichia coli (GCA_000005845.2).
Buchnera of Ac. pisum (GCA_000009605.1); Buchnera of Ci.cedri (GCA_000090965.1); Arsenophonus of L. cervi (GCA_001534665.1); Arsenophonus of N. vitripennis (GCA_004768525.1); Escherichia coli (GCA_000005845.2).
Buchnera of Ac. pisum (GCA_000009605.1); Arsenophonus of L. cervi (GCA_001534665.1); Arsenophonus of Ap. craccivora (GCA_013460135.1); Arsenophonus of N. vitripennis (GCA_004768525.1); Serratia of T. salginus (GCA_900016775.1); Serratia of Ci. cedri (GCA_000238975.1); Serratia of Ac. pisum (GCA_008370165.1); Serratia marcescens (GCA_000336425.1); Blochmannia floridanus (GCA_000043285.1); Wigglesworthia glossinidia (GCA_000008885.1); Baumannia cicadellinicola (GCA_000013185.1); Escherichia coli (GCA_000005845.2). Φ: pseudogenes.
Data and code availability
-
•
All sequencing data and assembled genome data have been deposited at the DDBJ under the bioProject ID: PRJDB13695. Accession numbers are listed in the Key resources table.
-
•
Original codes are available on GitHub (https://github.com/shigenobulab/21shunta_cjsym_msprep). Annotation data are available through Figshare (https://doi.org/10.6084/m9.figshare.c.6026333).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Akman Gündüz E., Douglas A.E. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc. R. Soc. B Biol. Sci. 2012;276:987–991. doi: 10.1098/rspb.2008.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aoki S., Kurosu U. Psyche (Stuttg); 2010. A Review of the Biology of Cerataphidini (Hemiptera, Aphididae, Hormaphidinae), Focusing Mainly on Their Life Cycles, Gall Formation, and Soldiers. [Google Scholar]
- Ayoubi A., Talebi A.A., Fathipour Y., Mehrabadi M. Coinfection of the secondary symbionts, Hamiltonella defensa and Arsenophonus sp. contribute to the performance of the major aphid pest, Aphis gossypii (Hemiptera: Aphididae) Insect Sci. 2020;27:86–98. doi: 10.1111/1744-7917.12603. [DOI] [PubMed] [Google Scholar]
- Baumann P., Baumann L., Lai C.Y., Rouhbakhsh D., Moran N.A., Clark M.A. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu. Rev. Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen E., Knight R., Huttley G.A., Gregory Caporaso J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd B.M., Allen J.M., Nguyen N.P., Vachaspati P., Quicksall Z.S., Warnow T., Mugisha L., Johnson K.P., Reed D.L. Primates, lice and bacteria: speciation and genome evolution in the symbionts of hominid lice. Mol. Biol. Evol. 2017;34:1743–1757. doi: 10.1093/molbev/msx117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braendle C., Miura T., Bickel R., Shingleton A.W., Kambhampati S., Stern D.L. Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol. 2003;1:E21. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Burke G.R., Moran N.A. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol. Evol. 2011;3:195–208. doi: 10.1093/gbe/evr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B.J., Mcmurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.Q., Montllor C.B., Purcell A.H. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid. Entomol. Exp. Appl. 2000;95:315–323. [Google Scholar]
- Chen J., Jiang L.Y., Qiao G.X. A total-evidence phylogenetic analysis of Hormaphidinae (Hemiptera: Aphididae), with comments on the evolution of galls. Cladistics. 2014;30:26–66. doi: 10.1111/cla.12024. [DOI] [PubMed] [Google Scholar]
- Chong R.A., Park H., Moran N.A. Genome evolution of the obligate endosymbiont Buchnera aphidicola. Mol. Biol. Evol. 2019;36:1481–1489. doi: 10.1093/molbev/msz082. [DOI] [PubMed] [Google Scholar]
- Chung Chen-yo, Shigenobu Shuji. Reproductive constraint in the social aphid Ceratovacuna japonica: Sterility regulation in the soldier caste of a viviparous insect. Insect Biochem Molec. 2022 doi: 10.1016/j.ibmb.2022.103756. [DOI] [PubMed] [Google Scholar]
- Darby A.C., Choi J.H., Wilkes T., Hughes M.A., Werren J.H., Hurst G.D.D., Colbourne J.K. Characteristics of the genome of Arsenophonus nasoniae, son-killer bacterium of the wasp Nasonia. Insect Mol. Biol. 2010;19:75–89. doi: 10.1111/j.1365-2583.2009.00950.x. [DOI] [PubMed] [Google Scholar]
- Darriba D., Posada D., Kozlov A.M., Stamatakis A., Morel B., Flouri T. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020;37:291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan P.H., Yu Y., Sisneros N., Wing R.A., Moran N.A. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc. Natl. Acad. Sci. USA. 2009;106:9063–9068. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan P.H., Leonardo T.E., Cass B.N., Hurwitz B., Stern D., Gibbs R.A., Richards S., Moran N.A. Dynamics of genome evolution in facultative symbionts of aphids. Environ. Microbiol. 2010;12:2060–2069. doi: 10.1111/j.1462-2920.2009.02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas A.E., Minto L.B., Wilkinson T.L. Quantifying nutrient production by the microbial symbionts in an aphid. J. Exp. Biol. 2001;204:349–358. doi: 10.1242/jeb.204.2.349. [DOI] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238–314. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldhaar H., Straka J., Krischke M., Berthold K., Stoll S., Mueller M.J., Gross R. Nutritional upgrading for omnivorous carpenter ants by the endosymbiont Blochmannia. BMC Biol. 2007;5:48. doi: 10.1186/1741-7007-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T., Ishikawa H. Soldier and male of an eusocial aphid Colophina arma lack endosymbiont: implications for physiological and evolutionary interaction between host and symbiont. J. Insect Physiol. 1992;38:1033–1042. [Google Scholar]
- Fukatsu T., Aoki S., Kurosu U., Ishikawa H. Phylogeny of cerataphidini aphids revealed by their symbiotic microorganisms and basic structure of their galls: implications for host-symbiont coevolution and evolution of sterile soldier castes. Zoolog. Sci. 1994;11:613–623. [Google Scholar]
- Gómez-Valero L., Soriano-Navarro M., Pérez-Brocal V., Heddi A., Moya A., García-Verdugo J.M., Latorre A., Moya A., García-Verdugo J.M. Coexistence of Wolbachia with Buchnera aphidicola and a secondary symbiont in the aphid Cinara cedri. J. Bacteriol. 2004;186:6626–6633. doi: 10.1128/JB.186.19.6626-6633.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Gu L., Eils R., Schlesner M., Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- Hattori M., Kishida O., Itino T. Buying time for colony mates: the anti-predatory function of soldiers in the eusocial aphid Ceratovacuna japonica (Homoptera, Hormaphidinae) Insectes Soc. 2013;60:15–21. [Google Scholar]
- Hershberg E.A., Stevens G., Diesh C., Xie P., De Jesus Martinez T., Buels R., Stein L., Holmes I. JBrowseR: an R interface to the JBrowse 2 genome browser. Bioinformatics. 2021;37:3914–3915. doi: 10.1093/bioinformatics/btab459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T., Nikoh N., Koga R., Satô M., Tanahashi M., Meng X.Y., Fukatsu T. Reductive genome evolution, host-symbiont co-speciation and uterine transmission of endosymbiotic bacteria in bat flies. ISME J. 2012;6:577–587. doi: 10.1038/ismej.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Forslund K., Coelho L.P., Szklarczyk D., Jensen L.J., Von Mering C., Bork P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen L.J., et al. EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik F., Nikoh N., Koga R., Ross L., Duncan R.P., Fujie M., Tanaka M., Satoh N., Bachtrog D., Wilson A.C.C., et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell. 2013;153:1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- Husník F., Chrudimský T., Hypša V. Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 2011;9:87. doi: 10.1186/1741-7007-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson E.M., Stireman J.O., Singer M.S., Abbot P. Phytophagous insect-microbe mutualisms and adaptive evolutionary diversification. Evolution. 2008;62:997–1012. doi: 10.1111/j.1558-5646.2008.00348.x. [DOI] [PubMed] [Google Scholar]
- Jousselin E., Cœur d'Acier A., Vanlerberghe-Masutti F., Duron O. Evolution and diversity of Arsenophonus endosymbionts in aphids. Mol. Ecol. 2013;22:260–270. doi: 10.1111/mec.12092. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29:28–35. doi: 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K.i., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R., Tsuchida T., Fukatsu T. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. Biol. Sci. 2003;270:2543–2550. doi: 10.1098/rspb.2003.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetsch D.R., Blaxter M.L. BlobTools: interrogation of genome assemblies. F1000Research. 2017;6:1287. [Google Scholar]
- Lamelas A., Gosalbes M.J., Moya A., Latorre A. New clues about the evolutionary history of metabolic losses in bacterial endosymbionts, provided by the genome of Buchnera aphidicola from the aphid Cinara tujafilina. Appl. Environ. Microbiol. 2011;77:4446–4454. doi: 10.1128/AEM.00141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamelas A., Gosalbes M.J., Manzano-Marín A., Peretó J., Moya A., Latorre A. Serratia symbiotica from the aphid Cinara cedri: a missing link from facultative to obligate insect endosymbiont. PLoS Genet. 2011;7:e1002357. doi: 10.1371/journal.pgen.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Manzano-Marín A., Latorre A. Settling down: the genome of Serratia symbiotica from the aphid Cinara tujafilina zooms in on the process of accommodation to a cooperative intracellular life. Genome Biol. Evol. 2014;6:1683–1698. doi: 10.1093/gbe/evu133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A., Simon J.C., Latorre A. Reinventing the wheel and making it round again: evolutionary convergence in Buchnera-Serratia symbiotic consortia between the distantly related Lachninae aphids Tuberolachnus salignus and Cinara cedri. Genome Biol. Evol. 2016;8:1440–1458. doi: 10.1093/gbe/evw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A., Szabó G., Simon J.C., Horn M., Latorre A. Happens in the best of subfamilies: establishment and repeated replacements of co-obligate secondary endosymbionts within Lachninae aphids. Environ. Microbiol. 2017;19:393–408. doi: 10.1111/1462-2920.13633. [DOI] [PubMed] [Google Scholar]
- Manzano-Marín A., Coeur d’acier A., Clamens A.L., Orvain C., Cruaud C., Barbe V., Jousselin E. Serial horizontal transfer of vitamin-biosynthetic genes enables the establishment of new nutritional symbionts in aphids’ di-symbiotic systems. ISME J. 2019;14:259–273. doi: 10.1038/s41396-019-0533-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. j. 2011;17:10–12. [Google Scholar]
- McCutcheon J.P., McDonald B.R., Moran N.A. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. USA. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean A.H.C., Godfray H.C.J., Ellers J., Henry L.M. Host relatedness influences the composition of aphid microbiomes. Environ. Microbiol. Rep. 2019;11:808–816. doi: 10.1111/1758-2229.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meseguer A.S., Manzano-Marín A., Coeur d’Acier A., Clamens A.L., Godefroid M., Jousselin E. Buchnera has changed flatmate but the repeated replacement of co-obligate symbionts is not associated with the ecological expansions of their aphid hosts. Mol. Ecol. 2017;26:2363–2378. doi: 10.1111/mec.13910. [DOI] [PubMed] [Google Scholar]
- Miura T., Braendle C., Shingleton A., Sisk G., Kambhampati S., Stern D.L. A comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: aphidoidea) J. Exp. Zool. B Mol. Dev. Evol. 2003;295:59–81. doi: 10.1002/jez.b.3. [DOI] [PubMed] [Google Scholar]
- Monnin D., Jackson R., Kiers E.T., Bunker M., Ellers J., Henry L.M. Parallel evolution in the integration of a Co-obligate aphid symbiosis. Curr. Biol. 2020;30:1949–1957.e6. doi: 10.1016/j.cub.2020.03.011. [DOI] [PubMed] [Google Scholar]
- Moran N.A., Baumann P. Bacterial endosymbionts in animals. Curr.Opin.Microbiol. 2000;3:270–275. doi: 10.1016/s1369-5274(00)00088-6. [DOI] [PubMed] [Google Scholar]
- Munson M.A., Baumann P., Kinsey M.G. Buchnera gen. Nov. And Buchnera aphidicola sp. nov., a taxon consisting of the mycetocyte-associated, primary endosymbionts of aphids. Int. J. Syst. Bacteriol. 1991;41:566–568. [Google Scholar]
- Nakabachi A., Ishikawa H. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J. Insect Physiol. 1999;45:1–6. doi: 10.1016/s0022-1910(98)00104-8. [DOI] [PubMed] [Google Scholar]
- Nakabachi A., Ueoka R., Oshima K., Teta R., Mangoni A., Gurgui M., Oldham N.J., Van Echten-Deckert G., Okamura K., Yamamoto K., et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 2013;23:1478–1484. doi: 10.1016/j.cub.2013.06.027. [DOI] [PubMed] [Google Scholar]
- Nováková E., Hypša V., Moran N.A. Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 2009;9:143. doi: 10.1186/1471-2180-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková E., Husník F., Šochová E., Hypša V. Arsenophonus and Sodalis symbionts in louse flies: an analogy to the Wigglesworthia and Sodalis system in tsetse flies. Appl. Environ. Microbiol. 2015;81:6189–6199. doi: 10.1128/AEM.01487-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková E., Hypša V., Nguyen P., Husník F., Darby A.C. Genome sequence of Candidatus Arsenophonus lipopteni, the exclusive symbiont of a blood sucking fly Lipoptena cervi (Diptera: hippoboscidae) Stand. Genomic Sci. 2016;11:1–7. doi: 10.1186/s40793-016-0195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack E.C.M., Melkonian M. Endosymbiotic associations within protists. Phil. Trans. R. Soc. B. 2010;365:699–712. doi: 10.1098/rstb.2009.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Davalos L.M. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- Oliver K.M., Russell J.A., Moran N.A., Hunter M.S. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. USA. 2003;100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver K.M., Degnan P.H., Burke G.R., Moran N.A. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 2010;55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- Ortiz-Rivas B., Martínez-Torres D. Combination of molecular data support the existence of three main lineages in the phylogeny of aphids (Hemiptera: Aphididae) and the basal position of the subfamily Lachninae. Mol. Phylogenet. Evol. 2010;55:305–317. doi: 10.1016/j.ympev.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Pawlowska T.E., Gaspar M.L., Lastovetsky O.A., Mondo S.J., Real-Ramirez I., Shakya E., Bonfante P. Biology of fungi and their bacterial endosymbionts. Annu. Rev. Phytopathol. 2018;56:289–309. doi: 10.1146/annurev-phyto-080417-045914. [DOI] [PubMed] [Google Scholar]
- Pérez-Brocal V., Gil R., Ramos S., Lamelas A., Postigo M., Michelena J.M., Silva F.J., Moya A., Latorre A. A small microbial genome: the end of a long symbiotic relationship? Science. 2006;314:312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- Pike N., Foster W. The ecology of altruism in a clonal insect. Ecol. Soc. Evol. 2008:37–57. [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . Vol. 2. R Core Team; 2021. R: A Language and Environment for Statistical Computing; pp. 1–12. [Google Scholar]
- Raetz C.R.H., Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz C.R.H., Guan Z., Ingram B.O., Six D.A., Song F., Wang X., Zhao J. Discovery of new biosynthetic pathways: the lipid A story. J. Lipid Res. 2009;50:S103–S108. doi: 10.1194/jlr.R800060-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Q., Rollat-Farnier P.A., Zhu D.T., Santos-Garcia D., Silva F.J., Moya A., Latorre A., Klein C.C., Vavre F., Sagot M.F., et al. Genome reduction and potential metabolic complementation of the dual endosymbionts in the whitefly Bemisia tabaci. BMC Genom. 2015;16:226–313. doi: 10.1186/s12864-015-1379-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renoz F., Lopes M.R., Gaget K., Duport G., Eloy M.-C., Geelhand de Merxem B., Hance T., Calevro F. Compartmentalized into bacteriocytes but highly invasive: the puzzling case of the Co-obligate symbiont Serratia symbiotica in the aphid Periphyllus lyropictus. Microbiol. Spectr. 2022;10:e0045722. doi: 10.1128/spectrum.00457-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E., Sharon G., Atad I., Zilber-Rosenberg I. The evolution of animals and plants via symbiosis with microorganisms. Environ. Microbiol. Rep. 2010;2:500–506. doi: 10.1111/j.1758-2229.2010.00177.x. [DOI] [PubMed] [Google Scholar]
- Russell J.A., Moran N.A. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc. Biol. Sci. 2006;273:603–610. doi: 10.1098/rspb.2005.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Garcia D., Juravel K., Freilich S., Zchori-Fein E., Latorre A., Moya A., Morin S., Silva F.J. To B or not to B: comparative genomics suggests Arsenophonus as a source of B vitamins in whiteflies. Front. Microbiol. 2018;9:1–16. doi: 10.3389/fmicb.2018.02254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Shigenobu S., Wilson A.C.C. Genomic revelations of a mutualism: the pea aphid and its obligate bacterial symbiont. Cell. Mol. Life Sci. 2011;68:1297–1309. doi: 10.1007/s00018-011-0645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S., Yorimoto S. Aphid hologenomics: current status and future challenges. Curr.Opin. Insect Sci. 2022;50 doi: 10.1016/j.cois.2022.100882. [DOI] [PubMed] [Google Scholar]
- Shigenobu S., Watanabe H., Hattori M., Sakaki Y., Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Sinotte V.M., Freedman S.N., Ugelvig L.V., Seid M.A. Camponotus floridanus ants incur a trade-off between phenotypic development and pathogen susceptibility from their mutualistic endosymbiont Blochmannia. Insects. 2018;9:E58. doi: 10.3390/insects9020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E., Goebel B.M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 1994;44:846–849. [Google Scholar]
- Stern D.L., Foster W.A. The evolution of soldiers in aphids. Biol. Rev. Camb. Philos. Soc. 1996;71:27–79. doi: 10.1111/j.1469-185x.1996.tb00741.x. [DOI] [PubMed] [Google Scholar]
- Sudakaran S., Kost C., Kaltenpoth M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 2017;25:375–390. doi: 10.1016/j.tim.2017.02.014. [DOI] [PubMed] [Google Scholar]
- Syberg-Olsen M.J., Garber A.I., Keeling P.J., McCutcheon J.P., Husnik F. Pseudofinder: detection of pseudogenes in prokaryotic genomes. Mol. Biol. Evol. 2022;39:msac153. doi: 10.1093/molbev/msac153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó G., Schulz F., Manzano-Marín A., Toenshoff E.R., Matthias H. ISME 2020. 2020. Evolutionary recent dual obligatory symbiosis among adelgids indicates a transition between fungus and insect associated lifestyles. 10.16.342642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas I., Klasson L., Canbäck B., Näslund A.K., Eriksson A.S., Wernegreen J.J., Sandström J.P., Moran N.A., Andersson S.G.E. 50 Million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:71–74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ham R.C.H.J., Kamerbeek J., Palacios C., Rausell C., Abascal F., Bastolla U., Fernández J.M., Jiménez L., Postigo M., Silva F.J., et al. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA. 2003;100:581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R., Šikić M. Time- and memory-efficient genome assembly with Raven. Nat. Comput. Sci. 2021;1:332–336. doi: 10.1038/s43588-021-00073-4. [DOI] [PubMed] [Google Scholar]
- Vaser R., Sović I., Nagarajan N., Šikić M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S.M., Martinez A.J., Ruan Y., Kim K.L., Lenhart P.A., Dehnel A.C., Oliver K.M., White J.A. Facultative endosymbionts mediate dietary breadth in a polyphagous herbivore. Funct. Ecol. 2015;29:1402–1410. [Google Scholar]
- Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K., Earl A.M. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins D., Kurtz Z. R Package Version 0.4. 0 342. 2019. gggenes: draw gene arrow maps in ‘ggplot2’. [Google Scholar]
- Wilson A.C.C., Ashton P.D., Calevro F., Charles H., Colella S., Febvay G., Jander G., Kushlan P.F., MacDonald S.J., Schwartz J.F., et al. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol. Biol. 2010;19:249–258. doi: 10.1111/j.1365-2583.2009.00942.x. [DOI] [PubMed] [Google Scholar]
- Wu D., Daugherty S.C., Van Aken S.E., Pai G.H., Watkins K.L., Khouri H., Tallon L.J., Zaborsky J.M., Dunbar H.E., Tran P.L., et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 2006;4:e188. doi: 10.1371/journal.pbio.0040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff J.A., Buckman K.A., Wu K., Heimpel G.E., White J.A. The endosymbiont Arsenophonus is widespread in soybean aphid, Aphis glycines, but does not provide protection from parasitoids or a fungal pathogen. PLoS One. 2013;8:e62145. doi: 10.1371/journal.pone.0062145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T.T., Jiang L.Y., Chen J., Qiao G.X. Host plants influence the symbiont diversity of Eriosomatinae (Hemiptera: Aphididae) Insects. 2020;11:217–316. doi: 10.3390/insects11040217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T.T., Chen J., Jiang L.Y., Qiao G.X. Diversity of bacteria associated with Hormaphidinae aphids (Hemiptera: Aphididae) Insect Sci. 2021;28:165–179. doi: 10.1111/1744-7917.12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientz E., Beyaert I., Gross R., Feldhaar H. Relevance of the endosymbiosis of Blochmannia floridanus and carpenter ants at different stages of the life cycle of the host. Appl. Environ. Microbiol. 2006;72:6027–6033. doi: 10.1128/AEM.00933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Buchnera of Ac. pisum (GCA_000009605.1); Buchnera of Ci.cedri (GCA_000090965.1); Arsenophonus of L. cervi (GCA_001534665.1); Arsenophonus of N. vitripennis (GCA_004768525.1); Escherichia coli (GCA_000005845.2).
Buchnera of Ac. pisum (GCA_000009605.1); Buchnera of Ci.cedri (GCA_000090965.1); Arsenophonus of L. cervi (GCA_001534665.1); Arsenophonus of N. vitripennis (GCA_004768525.1); Escherichia coli (GCA_000005845.2).
Buchnera of Ac. pisum (GCA_000009605.1); Buchnera of Ci.cedri (GCA_000090965.1); Arsenophonus of L. cervi (GCA_001534665.1); Arsenophonus of N. vitripennis (GCA_004768525.1); Escherichia coli (GCA_000005845.2).
Buchnera of Ac. pisum (GCA_000009605.1); Arsenophonus of L. cervi (GCA_001534665.1); Arsenophonus of Ap. craccivora (GCA_013460135.1); Arsenophonus of N. vitripennis (GCA_004768525.1); Serratia of T. salginus (GCA_900016775.1); Serratia of Ci. cedri (GCA_000238975.1); Serratia of Ac. pisum (GCA_008370165.1); Serratia marcescens (GCA_000336425.1); Blochmannia floridanus (GCA_000043285.1); Wigglesworthia glossinidia (GCA_000008885.1); Baumannia cicadellinicola (GCA_000013185.1); Escherichia coli (GCA_000005845.2). Φ: pseudogenes.
Data Availability Statement
-
•
All sequencing data and assembled genome data have been deposited at the DDBJ under the bioProject ID: PRJDB13695. Accession numbers are listed in the Key resources table.
-
•
Original codes are available on GitHub (https://github.com/shigenobulab/21shunta_cjsym_msprep). Annotation data are available through Figshare (https://doi.org/10.6084/m9.figshare.c.6026333).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.