Abstract
The contribution of low affinity T cells to autoimmunity in the context of polyclonal T cell responses is understudied due to the limitations in their capture by tetrameric reagents and low level of activation in response to antigenic stimulation. As a result, low affinity T cells are often disregarded as non-antigen specific cells irrelevant to the immune response. Our study aimed to assess how the level of self-antigen reactivity shapes T cell lineage and effector responses in the context of spontaneous tissue specific autoimmunity observed in NOD mice. Using multi-color flow cytometry in combination with Nur77GFP reporter of TCR signaling we identified a dormant population of T cells that infiltrated the pancreatic islets of pre-diabetic NOD mice, which exhibited reduced level of self-tissue reactivity based on expression of CD5 and Nur77GFP. We showed that these CD5low T cells had a unique TCR repertoire, exhibited low activation and minimal effector function; however, induced rapid diabetes upon transfer. The CD4+CD5low T cell population displayed transcriptional signature of central memory T cells, consistent with the ability to acquire effector function post-transfer. Transcriptional profile of CD5low T cells was similar to T cells expressing a low affinity TCR, indicating TCR affinity to be the important factor in shaping CD5low T cell phenotype and function at the tissue site. Overall, our study suggests that autoimmune tissue can maintain a reservoir of undifferentiated central memory-like autoreactive T cells with pathogenic effector potential that might be an important source for effector T cells during long-term chronic autoimmunity.
Keywords: T cell, autoimmunity, TCR affinity, type 1 diabetes, CD5
Introduction
Low affinity T cells are biologically important, but hard to quantify [1,2]. Indeed, they dominate autoimmune responses, are more likely to escape exhaustion under conditions of chronic antigen stimulation, and are better at forming long-term memory [2-5]. Because low affinity T cells, especially CD4+ T cells, are not amenable to analysis by tetramer staining, their functional contribution to polyclonal autoimmune responses is understudied. Importantly, the full range of self-reactivity in autoimmune tissue, specifically pancreatic islet infiltration in type 1 diabetes, is still unknown.
Micropipette-based two-dimensional TCR affinity measurements provide a more sensitive alternative to tetramer staining and identify a higher frequency of antigen specific T cells in polyclonal T cell populations. Measurements performed on islet infiltrating T cells showed a broad range of T cell affinity for pancreatic antigens and captured about ten times the number of antigen specific cells that are normally observed using tetramer staining, exposing a large frequency of low affinity T cells in the tissue throughout the disease [6,7]. However, no study to date was able to fully resolve the total combined frequency of antigen specific T cells within autoimmune tissue response. A number of studies tracking T cells with known antigen specificities in mouse models of type 1 diabetes have shown that T cell accumulation in pancreatic islets is dependent on antigenic specificity; and few non-islet antigen reactive T cells are retained in the islets [8-10]. Although most of tissue infiltrating T cells seem to accumulate in antigen specific fashion, inflammation at the tissue site could make islets more permissive for entry of non-specific T cells [11]. A study using photoconvertible fluorescence protein to track cellular migration in NOD pancreatic islets observed that non-activated CD44lowCD62L+ and presumably non-antigen specific T cells had the ability to infiltrate pancreatic tissue [12]. In models of CNS autoimmunity and Lyme arthritis bystander CD4+ T cells activated in non-antigen dependent manner infiltrated the tissue and enhanced inflammation [13,14]. On the other hand, CD8+ bystander T cells have been shown to be protective in the model of autoimmune diabetes [15]. Therefore, there exists a potential for a subpopulation of islet-infiltrating T cells, especially at later stages of disease, that are non-antigen reactive bystander cells and might be inert or actively contribute to disease. In this study we performed ex vivo analysis of effector functions and tested intrinsic autoimmune capacity of cells that exhibited bystander phenotype based on low level of tissue reactivity.
In order to stratify islet infiltrating T cells based on their reactivity for tissue antigen in a model of spontaneous autoimmune diabetes, we used tetramer staining, in vivo reporter of TCR signaling (Nur77GFP) and the level of CD5 expression [2,16]. We observed that the level of tissue reactivity is associated with a distinct TCR repertoire, transcriptional profile, and effector function. Although phenotypically T cells with low reactivity exhibited characteristics common to naïve T cells, further analysis revealed their potential for acquiring effector function and inducing diabetes upon transfer. Our study uncovered a potential for a tissue resident T cell population to exhibit characteristics of stem-cell or central memory T cells and maintain undifferentiated status in the context of autoimmunity. These observations are consistent with recent identification of stem-like and central memory autoimmune T cells in periphery of patients with type 1 diabetes [17,18], and support their potentially unique and important role in long-term persistence of autoimmune T cell populations.
RESULTS
CD5 expression correlates with increased TCR signaling and effector potential of islet infiltrating T cells
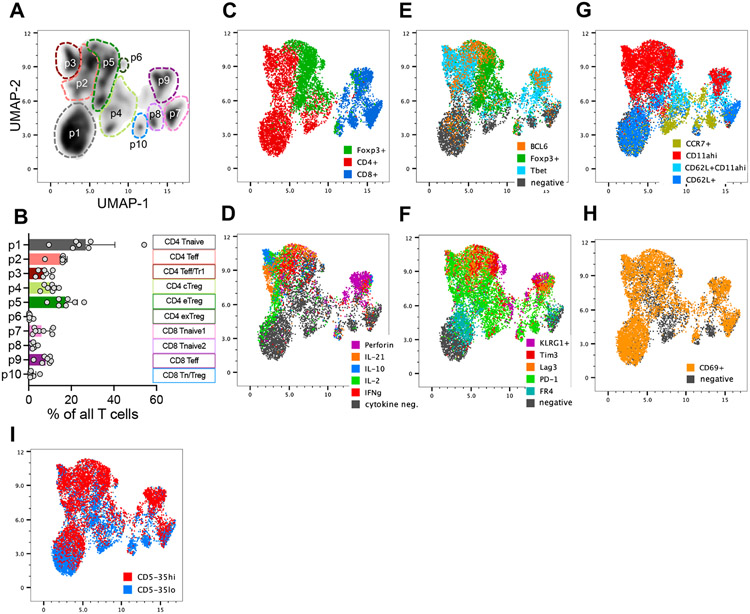
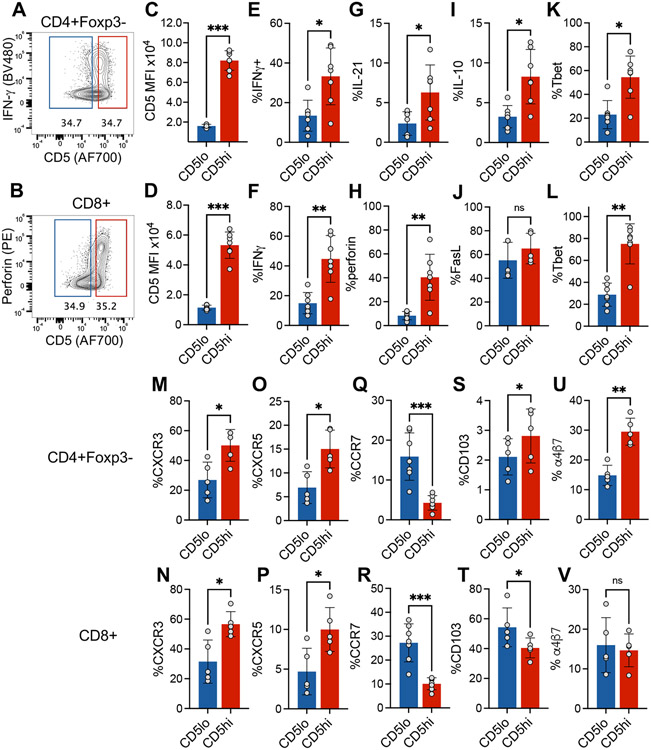
In order to observe the distribution of effector functional potential in polyclonal islet infiltrating CD4+ and CD8+ T cells we performed multi-parameter spectral cytometry analysis of T cells isolated from infiltrated pancreatic islets of pre-diabetic NOD female mice that were restimulated in vitro to elicit the full spectrum of effector functions. Unbiased UMAP analysis of flow cytometric data based on a combination of 22 T cell lineage markers and effector molecules revealed 10 major cell groups within TCR+ cells (Figure 1A, B, Supplemental Figure 1). The populations were broadly distinguished based on CD4 or CD8 expression, and more specifically based on their effector profile, such as cell surface markers of activation and cytokine production. These populations included CD4 and CD8 effector T cells characterized in part by the expression of Tbet and production of IFNγ (p2 at ~16% and p9 at ~8% respectively, Figure 1A-E). Expression of Foxp3 transcription factor was used to identify CD4+ Treg populations (p4, p5, and p6, Figure 1C). The three CD4+Foxp3+ populations were distinguished based on markers of central Tregs (cTreg - CD62L, CCR7), activated effector Tregs (eTreg – CD62L−CD11ahi), and a small subpopulation of terminally differentiated or exhausted Tregs (exTreg – KLRG1+) (Supplemental Figure 1B). Interestingly, population 3 consisted of Foxp3 negative CD4+ T cells with some characteristics of type 1 regulatory (Tr1) cells, including production of IL-10 and expression of Tim3 and Lag3 (Population 3, Figure 1D, F)[19]. CD8+ T cells also included a regulatory subpopulation characterized by the expression of Foxp3 and IL-10 (Population 10, Figure 1C, D, Supplemental Figure 1B). Perhaps most striking, was the presence of non-activated or naïve T cells (CD4+ p1, CD8+ p7 and p8). These represented a high frequency of all CD4+Foxp3- T cells (35% of CD4+Foxp3−) and CD8+ T cells (50% of CD8 T cells). The naïve-like populations were negative for most all other markers associated with T cell activation and differentiation, including IL-21, perforin, PD-1, Lag3, TIGIT, Tim3, and Bcl6. Although, these cells upregulated CD69 in response to in vitro stimulation, a marker of early T cell activation, suggesting that they are functional (Figure 1H). Given the observed heterogeneity of T cell differentiation and effector function in the infiltrated T cells, we wondered how these phenotypes reflected the level of T cell self-reactivity for pancreatic antigens and the downstream TCR signaling. CD5 is upregulated during thymic selection in direct proportion to the strength of TCR activation by self-ligands and serves as a relatively stable marker of T cell reactivity to self-antigens after T cell exit from the thymus [16]. We have previously shown that insulin B:9-23 reactive T cells in single TCR mice maintain levels of CD5 set during thymic selection after activation in the draining lymph nodes and in the pancreatic tissue, further supporting CD5 as a useful stable surrogate of TCR self-reactivity[2]. When we overlaid CD5 expression onto the population distribution plot, we observed that CD5low cells were preferentially located within naïve-like populations 1 and 8, while CD5high T cells populated highly activated effector populations 2 and 9 (Figure 1I). To directly assess how effector function correlated to the level of CD5 expression, we performed standard gating analysis to compare effector molecule expression in CD5high (top 35%) and CD5low (bottom 35%) CD4+ and CD8+ T cells (Supplemental Figure 2A,B). In both T cell compartments effector molecule expression (IFNγ, IL-21 and perforin) was associated with higher level of CD5 expression, suggesting that CD5high T cell compartment encompasses pathogenic effectors (Figure 2A-H). However, further analysis revealed similar high-level expression of FasL on both CD5high and CD5low CD8+ T cells, suggesting that CD5low T cells are not completely void of effector function (Figure 2J). Regulatory cytokine IL-10 was increased in CD5high CD4+ T cells, (Figure 2I), while TGFβ was expressed at equally low levels in both CD5high and CD5low T cells compared to high expression in Foxp3+ Tregs (Supplemental Figure 2C), excluding the possibility that CD4+CD5low T cells are primarily of a regulatory phenotype. The effector functions correlated with the increase in markers of differentiated T cells including Tbet, CXCR3, and CXCR5 in CD5high cells, while naïve or central memory associated CCR7 was primarily expressed in CD5low cells (Figure 2K-R). Analysis of integrins CD103 and α4β7 associated with tissue infiltration showed an overall higher expression of CD103 on CD8+ T cells, and partial preference for α4β7 expression on CD4+ T cells (Figure 2S-V). Interestingly, CD4+CD5high T cells maintained higher level of both integrins compared to CD5low T cells, suggesting their competitive advantage in tissue accumulation and residence compared to CD5low T cells that preferentially expressed lymph node homing marker CCR7. Interestingly, CD8+CD5low T cells expressed higher levels of CD103 compared to CD5high T cells (Figure 2T). CD103 has been used as a marker of tissue resident memory T cells and could be an indicator of resident cells within the CD8+CD5low compartment.
Figure 1. Functionally distinct T cell subpopulations infiltrate the pancreas during autoimmune diabetes.
Islet infiltrating T cells from pancreata of 8–12-week-old pre-diabetic female NOD mice were analyzed by flow cytometry after 5-hour restimulation in vitro with PMA and ionomycin. Analysis is gated on TCR+ T cells. (A) UMAP analysis based on a concatenated sample of TCR+ cells infiltrating the pancreatic islets of 7 individual female mice (see supplemental Figure 1). (B) Average frequency of each cluster among mice. Each point represents a single mouse, (n=7). (C-H) Indicated markers are backgated onto the UMAP populations. (I) CD5high (top 35%) and CD5low (bottom 35%) were determined by standard flow gating for CD8, CD4, and Foxp3+ T cell populations and backgated onto the UMAP plot. Error bars designate mean ± SD, n=7 mice, data from one experiment.
Figure 2. Increased activation of CD5high islet infiltrating T cells.
(A and B) Representative CD5 gating strategy. (C and D) CD5 MFI for CD4+ (C) and CD8+ T cells (D) after gating on CD5high or CD5low cells (top and bottom 35%). (E, G, I, K, M, O, Q, S, U) Analysis of CD5high or CD5low CD4+ T cells. (F, H, J, L, N, P, R, T, V) Analysis of CD5high or CD5low CD8+ T cells. Statistical analysis was performed using Mann-Whitney test (n=5-7). Error bars designate mean ± SD, n=5-7 mice, each point represents a single mouse, data from one experiment. *p<0.05, **p<0.01, ***p<0.001.
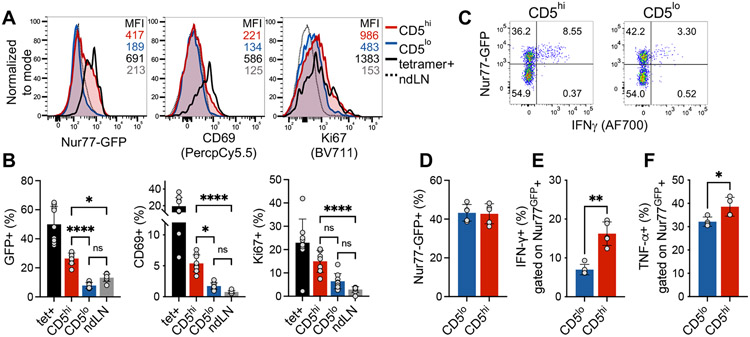
Since CD5 is correlated with self-reactivity, we postulated that the CD5low population did not accumulate sufficient TCR signal in response to self-antigens in the pancreas to elicit a functional response. We have previously observed a direct correlation between CD5 expression and TCR signaling at the pancreatic tissue site, as measured by Nur77GFP reporter [20]. Using insulin tetramer+ CD4+ T cells as a positive control for tissue specificity, we compared the proportion of activated T cells in islet infiltrating CD5high, CD5low, and non-draining lymph node (LN) derived CD4+Foxp3− T cells. As expected, islet infiltrating tetramer+ T cells exhibited high levels of Nur77GFP, CD69 and Ki67 compared to cells in non-draining LN (Figure 3A, B). While CD5high T cell population contained T cells with high levels of Nur77GFP and CD69 expression, CD5low cells exhibited a low level of TCR signaling on par with non-draining LN T cells. However, Ki67 was increased CD5high and to some level in CD5low cells, although the difference did not reach significance for CD5low cells. These observations suggest that even though TCR signaling was not detectable in CD5low cells using Nur77GFP reporter, there remained a possibility that CD5low T cells were in their early stages of activation or partially activated due to lower TCR affinity. To assess whether equivalent high level of TCR stimulation would reveal reserved functional capacity of CD5low cells we stimulated islet infiltrating T cells with plate bound anti-CD3. A comparable frequency of CD5high and CD5low T cells from infiltrated islets of NOD.Nur77GFP mice upregulated GFP in response to TCR cross-linking (Figure 3C,D). However, within the activated (GFP+) T cells, CD5low cells again exhibited inferior effector potential as defined by their reduced ability to produce IFNγ (Figure 3E). In order to confirm that CD5low population was indeed functional, we assessed TNFα expression after TCR stimulation. TNFα is expressed by naïve T cells and does not require full differentiation into Th1 type effectors, unlike IFNγ [21]. Although CD5high T cells exhibited significantly higher levels of TNFα production, TNFα was still expressed by a high frequency of CD5low cells (38% vs 32%, Figure 3F), supporting functional potential and competency of CD5low cells. Overall, these data suggest that CD5low T cells possess reduced levels of effector function and might represent a naïve or bystander T cell population in infiltrated pancreatic islets.
Figure 3. Increased in vivo TCR signaling and effector potential of CD5high islet infiltrating T cells.
Islet infiltrating T cells from pancreata of 8-12 week old pre-diabetic female NOD.Nur77GFP mice were analyzed by flow cytometry directly ex vivo (A and B) and after in vitro stimulation with plate-bound anti-CD3 (C-F). (A) Representative histograms. Analysis is gated on TCR+CD4+, followed by gating on insulin tetramer+ T cells (PE/APC double tetramer gate), or CD5high or CD5low cells (CD5high - top 35%; CD5low - bottom 35%). Dotted line - TCR+CD4+ T cells from non-draining lymph nodes (ndLN). MFI number colors correspond with histogram colors; black is tetramer, grey is ndLN. (B) Cumulative analysis of 9 individual mice, (n=9), data from one experiment; (C-F) n=5 mice, data from two independent experiments. Each point represents a single mouse. Statistical analysis was performed using Kruskal-Wallis (B) and Mann-Whitney (C-F) tests. Error bars designate mean ± SD, *p<0.05; **p<0.01; ****p<0.0001.
Level of CD5 expression distinguishes islet infiltrating populations with unique TCR repertoires
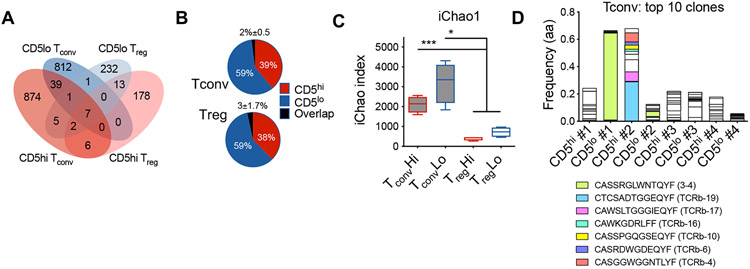
In order to assess whether CD5high and CD5low expressing T cell populations were composed of distinct clonotypes, we compared their TCR repertoires. Since CD4+ T cells are more heterogeneous in their TCR repertoire and antigen specificity in autoimmune diabetes compared to CD8+ T cells, a large proportion of which respond to a single IGRP epitope in NOD islets [22,23], we focused our analysis on CD4+ T cells to identify potential divergent clonotypes in CD5high and CD5low T cell responses. We have previously shown that high expression of CD5 can identify functionality superior Foxp3+ islet infiltrating Tregs [20], which provided us with an internal control of two functionally distinct populations (CD5high vs CD5low Tregs) where we expected limited TCR overlap. To that end, we evaluated TCR repertoires of CD5high and CD5low conventional (Foxp3−) and regulatory (Foxp3+) CD4 T cells in pancreatic islets of pre-diabetic NOD.Foxp3GFP mice. High-throughput TCR-beta sequencing revealed few shared TCR sequences between CD5high and CD5low CD4+Foxp3GFP− conventional (Tconv) and CD4+Foxp3GFP+ regulatory (Treg) cells (Tconv, 2±0.5%, MH=0.0778; Treg 3±1.7%, MH=0.1198, Figure 4A, B, Supplemental Figure 3A, B). This observation confirmed that the level of CD5 expression is set in individual T cell clones during development and is maintained in periphery [2,16]. Moreover, the data supported previous observations of negligible conversion between effector and regulatory T cell populations in autoimmune pancreatic infiltration [24]. Importantly, the minimal overlap between CD5high and CD5low populations indicated that CD5 level separates unique T cell clones.
Figure 4. Distinct TCR repertoires of CD5high and CD5low T cells in pancreatic islets.
TCR-beta sequences were obtained from CD4+ Tconv (Foxp3GFP−) and Treg (Foxp3GFP+) cells sorted based on the level of CD5 expression from the islets of NOD.Foxp3GFP+ mice. (A) A representative analysis of TCR-beta amino acid (a.a.) sequence overlap among T cell populations of a single mouse. (B) Average of TCR-beta a.a. sequence overlap between CD5high and CD5low populations (n=4 mice, ±SD). (C) Sample TCR-beta diversity was calculated using iChao index. (D) Frequencies of top 10 T cell clones in Tconv populations. Colored bars designate CDR3 sequences shared with TCRs of known specificity (insulin B:9-23). The results are from one experiment (n=4 mice). Statistical analysis was performed using Two-way ANOVA. Error bars designate mean ± SD, *p<0.05; ***p<0.001.
Since we have previously observed that CD5high Tregs exhibited increased activation and proliferation in the pancreas of NOD mice [20], we expected increased clonal expansion within the CD5high Treg population. As expected, there was a trend toward increased clonality and reduced diversity within the CD5high Tregs, consistent with our hypothesis (Figure 4C, Supplemental Figure 3C). Surprisingly, CD5high and CD5low Tconv populations showed overall similar levels of clonality, with some mice exhibiting increased clonal expansion within CD5high population and others in CD5low population (Supplemental Figure 3D). Moreover, several highly expanded clones within the CD5low Tconv compartment shared CDR3 sequence motifs with previously characterized insulin specific T cell clones (Figure 4D)[2]. Although, we did not find CDR3 motifs associated with specific beta cell antigen specificities within the top Treg TCR sequences, one of the highest frequency clones was shared among CD5low Treg populations of three separate mice, suggesting similar antigenic targets and public TCR usage (Supplemental Figure 3F). In general, Tconv cells displayed both decreased clonality and increased repertoire diversity compared to Tregs (Figure 4C, Supplemental Figure 3E), suggesting that Tregs possess a more restricted TCR repertoire in the pancreatic tissue, and their expansion at the tissue site is less clonally focused than Tconvs. In the case of Tconv cells, these results demonstrate that clonal expansion can be observed within either high or low affinity tissue infiltrating effector populations.
Transcriptional characteristic of CD5low islet infiltrating T cells
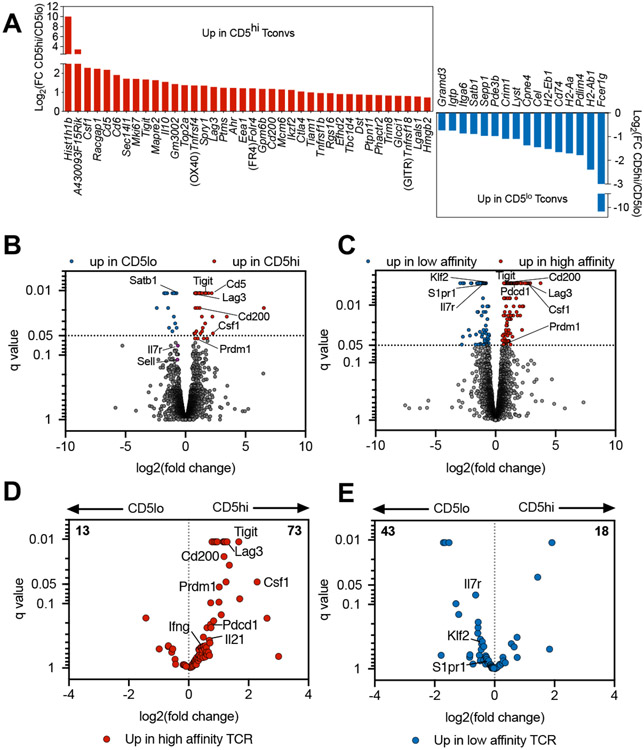
In order to assess whether functional differences observed in restimulated CD5high and CD5low CD4+ islet infiltrating T cells could be observed directly ex vivo, we analyzed the transcriptional profile of CD5high and CD5low CD4+Foxp3− Tconv cells sorted from the islets of NOD female mice. A total of 55 genes were significantly different (q<0.05) between the two populations (Figure 5A, B). Genes involved in proliferation and activation of T cells were expressed at a higher level in CD5high subset (Ki67, Csf1, Tigit, Tnfrsf4 (OX40), Ctla4, Tnfrsf18 (GITR)), which showed that TCR reactivity for tissue antigen correlated with an overall higher level of activation. In order to confirm that TCR affinity for self-antigens was the determinant factor in driving transcriptional differences between CD5high and CD5low cells, we compared the genes to a previously obtained data set from T cells expressing TCRs with defined affinities. The dataset was derived from a two-TCR chimera from which we obtained transcriptional profiles of islet infiltrating high affinity T cells expressing insulin B:9-23 specific TCR 4-8 and low affinity InsB:9-23 specific 12-4.4m1 T cells sorted from the same pancreatic islets [25]. Importantly, unlike the majority of insulin reactive T cells that show direct correlation between CD5 and TCR affinity for insulin epitope, the low affinity 12-4.4m1 T cells developed with a high level of CD5 expression that did not correlate with TCR reactivity for insulin [2]. The elevated levels of CD5 in the low affinity 12-4.4m1 T cells were likely due to strong reactivity for an unrelated self-antigen during thymic selection. Using 12-4.4m1 TCR allowed us to exclude the possibility that the transcriptional differences were induced by different levels of CD5 expression, rather than TCR activation. We observed similar genes up or down regulated in high and low affinity cells in the two-TCR system and between polyclonal populations sorted on CD5 high or low expression (Figure 5B, C). Transcripts upregulated in both CD5high and high affinity TCR expressing cells included Tigit, Lag3, Csf1 – genes associated with T cell activation. When we focused our analysis on transcripts that were significantly upregulated in high-affinity 4-8 T cells from the two-TCR comparison (q<0.05, total 86), we observed that many of the genes (73 out of 86) were preferentially associated with CD5high cells (Figure 5D). Similarly, transcripts significantly increased in T cells expressing low affinity insulin 12-4.4m1 TCR (q<0.05, total 61) were primarily associated with CD5low T cells (43 out of 61; Figure 5E). These observations confirmed that TCR affinity for antigen is an important factor in driving distinct T cell phenotype of CD5high and CD5low cells.
Figure 5. Transcriptional analysis reveals increased activation and differentiation of CD4+ CD5high islet infiltrating T cells.
(A and B) CD4+Foxp3− Tconvs were isolated from islets of 4 pre-diabetic female NOD.Foxp3GFP mice based on CD5 expression (CD5high - top 35%; CD5low - bottom 35%) and subjected to RNAseq analysis. (A) Genes significantly different between CD5low and CD5high T cells (q<0.05). (B) Volcano plot showing comparison between CD5low and CD5high T cells. (C) Islet infiltrating T cells from two-TCR retrogenic mice expressing a high and a low affinity TCR were sorted based on fluorescent reporters GFP or Ametrine of the two TCR vectors and subjected to RNAseq analysis. (D, E) Transcriptional analysis of CD5low and CD5high T cells is shown for the selected genes that were significantly increased in high affinity (D) or low affinity (E) islet infiltrating cells from two-TCR mice. The results are from one experiment, n=4 mice (CD5hi/lo) and n=3 mice (TCRhigh/low). Data was analyzed by Cuffinks Assembly & DE, with differently expressed genes defined by q<0.05 with Benjamini-Hochberg correction for multiple testing.
Intriguingly, CD5low subset showed higher expression of genes associated with undifferentiated naïve or central memory T cells, while CD5high population exhibited a signature of terminal differentiation or exhaustion. CD5low T cells expressed higher levels of Satb1, a transcriptional regulator involved in repression of PD-1 locus and maintaining T cell responsiveness to antigen in autoimmunity [26,27]. Consistent with this observation, CD5low cells had reduced levels of PD-1 expression (Supplemental Fig. 4). In addition, CD5high T cells exhibited increased expression of inhibitory receptors Lag3, Tigit and the marker of anergy Folr4, characteristic of terminally differentiated T cells (Figure 5A, B). This transcriptional increase was confirmed at the protein level (Supplemental Fig. 4). In contrast, CD5low cells expressed a number of genes reminiscent of naïve or central memory T cell phenotype, including higher levels of Ccr7, Sell (CD62L) and Il7r (Figure 5B, E). This observation was also confirmed at the protein level (Figure 2, Supplemental Fig. 1 and 4). Overall, our data showed that CD4+ T cells infiltrating pancreatic islets of pre-diabetic NOD mice are heterogeneous in their level of differentiation, effector function, and TCR repertoire, but it was unclear how these differences affect their capacity to cause disease.
CD4+ CD5low islet-infiltrating T cells induce diabetes upon transfer
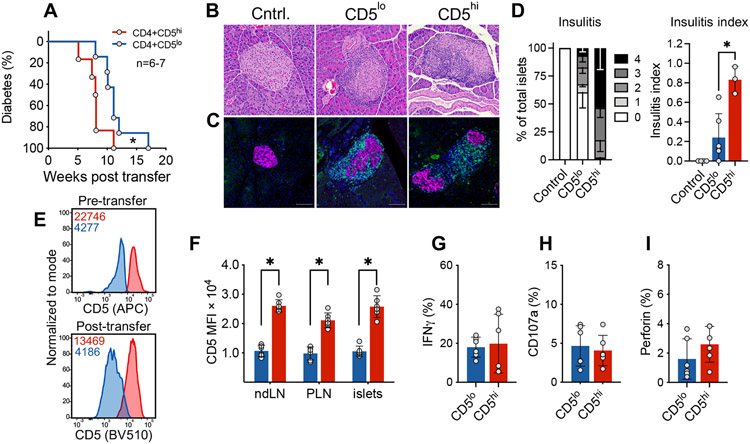
Since CD5high cells exhibited all the characteristics of activated effector T cells, while CD5low cells had a non-activated, potentially bystander, phenotype we wanted to compare intrinsic autoimmune potential of the two populations in a transfer model of diabetes. Given that CD5low cells exhibited reduced TCR activation at the tissue site indicating their low reactivity for tissue antigen, we predicted that this cell population would have a reduced ability to home to the pancreas and cause beta cell damage. We sorted CD5high and CD5low CD4+ Tconvs from infiltrated islets of pre-diabetic female NOD.Foxp3GFP mice, and transferred cells into NOD.TCRα−/− recipients. Mice that received CD5high subset exhibited accelerated diabetes development starting at 5 weeks after transfer, compared to the group that received CD5low T cells, in which diabetes onset was delayed by 3 weeks (Figure 6A). Strikingly, both of the T cell subsets induced diabetes in 100% of mice within 18 weeks post transfer. Histological analysis confirmed that both CD5high and CD5low cells infiltrated the pancreatic islets and induced insulitis, which could be observed as early as 6 weeks post transfer (Figure 6B, C, D). Consistent with the delayed diabetes induction, insulitis was reduced in CD5low recipient mice at this early time-point. Nevertheless, about 40% of islets in CD5low recipient mice exhibited some level of insulitis, and about 18% of islets had lymphocytic penetration of more than 75% of the islet (Figure 6D). These observations further confirm the ability of CD5low cells to infiltrate the pancreatic islets and induce islet damage. Importantly, the differences in the levels of CD5 expression of the two sorted populations were maintained post-transfer and were consistent among the examined organs irrespective of antigen availability (Figure 6E, F). These results show that although CD5low population seemed “dormant”, it had a strong pathogenic potential.
Figure 6. CD5low islet-infiltrating CD4+ T cells induce diabetes upon transfer.
CD4+Foxp3GFP− T cells were isolated from the islets of pre-diabetic 11-20 week old NOD.Foxp3GFP mice. Sorted cells were transferred into 4-8 week old NOD.TCRα−/− recipients. (A) Diabetes development, n=6-7. (B) Representative islet H&E from non-transferred (Cntrl.), CD5low cell recipient and CD5high recipient mice. (C) Immunofluorescent detection of transferred T cells (green) infiltrating the pancreatic islets (magenta) in control and transferred mice at 6 weeks post transfer. DAPI is depicted in blue; green is anti-CD3; magenta is BSRP-A marker of endocrine cells. (D) Cumulative histological (H&E) analysis of insulitis at 6 weeks post transfer (n=3-6 mice). Scale bar is 100um. (E) Representative flow plot analysis of CD5 expression on cells immediately after sorting and in the infiltrated pancreatic islets 5 weeks post-transfer. Numbers within flow plots represent MFI of the two populations; red is CD5high, blue is CD5low MFI. (F) Analysis shows the maintenance of high/low CD5 expression at the diabetes endpoint; ndLN = non-draining lymph nodes, PLN = pancreatic draining lymph nodes; n=6. (G-I) Transferred CD5high and CD5low CD4+ T cells were isolated from the islets of NOD.TCRα−/− recipients 5 weeks post transfer. Cells were restimulated with PMA/ionomycin; n=6. (A, E-I) are pooled from at least two independent experiments with n=6-7. (B, C) are representative of 3 independent experiments (Cntrl., n=4; CD5lo, n=6; CD5hi, n=3). (D) Insulitis scoring were performed on data pooled from 3 independent experiments (n=3-6). For each mouse, >100 islets from 5 consecutive layers (100μm apart) were scored. Statistical analysis was performed using Mantel-Cox (A) or Mann-Whitney (D-I). Error bars designate mean ± SEM in D, and mean ± SD in F-I, *p<0.05.
We next assessed whether the delay in disease development in our transfer model was due to reduced infiltration, expansion, or effector function of CD5low cells. There was no difference in the number of CD4+ T cells in the spleens or in the islets of the two groups at endpoint, suggesting that the two cell subsets survive, expand, and can reach similar levels of islet infiltration (Supplemental Figure 5A). CD69, PD-1 and Lag3 activation markers were equivalently upregulated on both CD5high and CD5low cells post transfer (Supplemental Figure 5B-D). Previous studies have shown that CD5high T cells were more equipped at differentiating into Foxp3+ cells [28]. Although, the proportion of Foxp3+ regulatory T cells in CD5high and CD5low subsets was variable (0-12%), there was no significant difference between the two populations in either spleens or islets (Supplemental Figure 5E). Intriguingly, we observed that CD5low cells acquired markers of anergy (FR4 and CD73) in the draining PLN; however, the anergic phenotype was lower at the tissue site (Supplemental Figure 5F). Thus, CD5high and a significant proportion of CD5low Tconvs were able to avoid anergy and ultimately cause diabetes.
Next, we compared the functional profile of the two subsets post-transfer. After 5 weeks post-transfer, we detected comparable levels of IFNγ (Figure 6G), which was contrary to our observations prior to transfer, where CD5high cells expressed higher levels of IFNγ (Figure 2E). In addition, levels of CD107a and perforin did not differ between these two subsets post-transfer (Figure 6H, I), demonstrating similar level of activation, Th1 differentiation, and effector mechanisms utilized by the two populations. Collectively, our results demonstrate that CD4+ Tconvs with high- and low-reactivity have comparable capacity for Th1 differentiation and IFNγ production, indicating similar effector mechanisms in the pancreas.
DISCUSSION
Our analysis of islet infiltrating T cells has uncovered a population of cells with reduced effector function. The phenotype was associated with reduced TCR signaling and lower expression of CD5. Although, the level of CD5 expression can be modulated in certain situations, for example downregulated in MHC knock out mice [29], and upregulated in response to exceedingly high concentrations of the peptide/MHC ligand [30], both conditions are outside physiologically relevant environment experienced by T cells in vivo. We have previously tracked CD5 expression on insulin specific T cells in single TCR retrogenic system and did not observe changes in CD5 at the site of antigen in pancreatic islets and draining lymph nodes, although Nur77GFP was induced in those sites indicating T cell activation [2]. In the current study, relative levels of CD5 on islet-derived T cells remained stable post-transfer in lymphoid organs and in the pancreatic tissue (Figure 6E, F), suggesting that CD5 expression is stable in vivo and can serve as a valuable readout of self-reactivity at the autoimmune tissue site.
A number of studies suggest that the level of self-reactivity can influence T cell helper lineage development and T cell responses in periphery. T cells with higher affinity for self-ligands exhibit increased basal level of TCR signaling in periphery, presumably due to continuous interactions with self-peptides in secondary lymphoid organs and are poised to respond quickly to agonist foreign antigens [31]. As a result, T cells with increased self-reactivity are more prevalent among the polyclonal population recruited into the immune response, while T cells with lower level of self-reactivity are considered less effective. These observations seem to suggest that autoimmune responses should be similarly dominated by higher affinity T cells. However, contrary to these expectations, autoimmune T cells recruited to the targeted tissue span a wide range of TCR affinities [7,32]. Accordingly, our previous study using single TCR retrogenic model has demonstrated that pathogenic effector T cells can span a range of TCR affinities [2]. The robust pathogenic potential of CD4+CD5low T cells post-transfer is consistent with the idea of broad TCR affinities that can support autoimmune T cell responses.
The dynamics of lymphocyte populations and their functional heterogeneity in autoimmune tissue lesions have been a focus of continuous investigation and debate [8,9,11,12]. In our study we identified a subpopulation of tissue infiltrating cells with reduced level of TCR signaling, effector function, but enriched in markers of naïve T cells. While this population exhibited many characteristics of bystander or naïve T cells, transfer experiments showed that these cells have near equal pathogenic capacity. This led us to postulate that a proportion of islet infiltrating CD4+ T cells are partially differentiated, central memory-like cells. Accumulating evidence suggests an important pathogenic role for memory T cells in the development of autoimmune disorders, including T1D. T cells expressing memory markers and reactive to pancreatic antigens have been identified in peripheral blood of individuals with T1D [17,33-36]. While memory T cells can be targeted therapeutically, and some approaches are showing promising results in models of autoimmune diabetes [37,38], the role of long-lived autoimmune T cell memory vs. continuous recruitment of effector T cells from naïve repertoire in T1D disease pathology is unknown. Our data suggests a presence of stem-cell or central memory phenotype T cells within pancreatic islets that have significant pathogenic potential. It is not clear whether this memory-like population is dynamically recruited into inflammatory response via differentiation into effector T cells, or is maintained as long-lasting tissue resident effector precursors, and requires further investigation.
The ultimate contribution of CD5low T cells to diabetes development is still unclear. CD4+CD5low T cells have the capacity to produce TNFα and CD8+CD5low T cells express FasL supporting their pathogenic potential. Importantly, CD5low T cell population contain expanded clones and some of these share CDR3 motifs with known insulin reactive TCRs. CD5low T cells preferentially express CCR7, which promotes migration to lymph nodes via recognition of CCL21 on high endothelial venules (HEVs). However, chronic inflammation results in the formation of HEV venules and tertiary lymphoid organs (TLOs) in peripheral tissues, including pancreatic islets. Inflamed islets express CCL21 and recruit CCR7+ T cells [39,40]. CCR7/CCL21 axis is not limited to T cells and has been implicated in recruitment and activation of macrophages and dendritic cells in chronic autoimmunity [41,42]. Importantly, naïve T cells themselves can be a source of lymphotoxins and further enhance TLO formation [43-45]. Presence of CD5low or naïve T cells in the tissue could make islets permissive for infiltration of additional stem-like CCR7+ cells, perpetuating the persistence of autoimmune T cell infiltrate. Perhaps surprisingly, transfer of CD62L+ cells or enhanced early recruitment of CD62L+ T cells in response to ectopic over-expression of CCL21 in the islets was associated with reduction in diabetes[46,47]. Therefore, we cannot exclude the possibility that additional recruitment of CD62L+CCR7+ T cells could have a regulatory function early in disease onset. However, the proportion of antigen specific T cells within CD62L+ T cell populations or their TCR affinities in these studies were not defined and might be the key underlying difference between regulatory or pathogenic CD62L+ T cells. Alternatively, the ability of self-reactive T cells to escape from regulation, rather than their affinity for antigen, might be more critical for their pathogenic potential and might explain the persistence and critical pathogenic role for low affinity CD5low T cells in autoimmunity [2].
In conclusion, we show that islet infiltrating CD4+ T cells with a low level of TCR signaling characteristic of bystander or naïve T cells are pathogenic and are capable of inducing beta cell destruction. Our study suggests that islet infiltrate in pre-diabetes is composed of several functionally and phenotypically distinct T cell populations that could have unique contribution to disease pathology. Lastly, the findings presented here raise questions about the longevity and memory potential of high vs. low reactive T cells in autoimmune diabetes.
Materials and methods
Mice
NOD/ShiLtJ (NOD), NOD/ShiLt-Tg(Foxp3-EGFP/cre)1cJbs/J (NOD.Foxp3GFP), NOD.129P2(C)-Tcratm1Mjo/DoiJ (NOD.TCRα−/−), and NOD.CB17-Prkdcscid/J (NOD.scid) mice were purchased from Jackson Laboratories and maintained at our facility. NOD.Nur77GFP mice were previously backcrossed to NOD and maintained in our mouse facility [2,20]. Mice were housed in specific pathogen free conditions and maintained on standard diet. Experimental mice were monitored weekly for diabetes development by urine glucose testing (Diastix; Bayer); positive readings were confirmed by blood glucose testing (Breeze2 glucometer; Bayer). Mice were considered diabetic after two consecutive blood glucose readings >300mg/dl or a single reading >400mg/dl. The protocols were approved by the Baylor College of Medicine and University of Utah Institutional Animal Care and Use Committees.
Isolation of islet infiltrating cells
T cells were isolated from pancreatic islets of NOD, NOD.Foxp3GFP, or NOD.Nur77GFP mice as described previously [20]. Briefly, pancreata were perfused by injecting 600u/mL collagenase 4 (Worthington) 5%FBS/HBSS solution into the bile duct, excised, and digested at 37°C for 30min. After tissue disruption and washing, single islets were picked using microdissection scope with a p10 pipette. Islets were dissociated into single cell suspension using enzyme-free cell dissociation buffer (Gibco®). Islet cells were directly analyzed by flow cytometry or sorted for further analysis.
T cell stimulation
Dissociated islets were stimulated for five hours with plate bound anti-CD3 (1μg/ml) or 10 ng/mL phorbol myristic acid (PMA) (Sigma) and 1 μmol/L ionomycin (Sigma) in the presence of brefeldin A and monensin.
Flow cytometry and antibodies
Flow cytometry analyses were performed on Cytek Aurora or BD LSRFortessa II, and data were analyzed with FlowJo software (Tree Star Inc.). Antibodies used in this study are as follows: anti-Foxp3 (FJK-16s, BioLegend or eBioscience), anti-Bcl6 (7D1), anti-CD3 (145-2C11, BioLegend or BD), anti-CD4 (GK1.5 or RM4-5), anti-CD8a (53-6.7), anti-CD8b (H35-17.2, eBioscience), anti-CD5 (53-7.3), anti-Ki67 (B56), anti-CD11a (M17/4, BD), anti-CTLA-4 (UC10-4B9), anti-PD1 (29F.1A12), anti-LAG3 (C9B7W, BD), anti-TIGIT (1G9, BD or GIGD7, eBioscience), anti-CD73 (TY/11.8), anti-FR4 (12A5), anti-CD44 (IM7), anti-CD62L (MEL-14), anti-IFNγ (XMG1.2, BioLegend or BD), anti-perforin (S16009A), anti-Tbet (4B10 BioLegend or eBiosciences), anti-CD69 (H1.2F3, BD), anti-CCR7 (4B12, BD), anti-KLRG1 (2F1, BD), anti-IL-10 (JESS-16E3), anti-Tim3 (RMT3-23), anti-IL-2 (JES6-5H4, BD), anti-IL-21 (mhalx21, eBiosciences), anti-CD103 (2E7), anti-FasL (MFL3, eBioscience), anti-CXCR3 (CXCR3-173), anti-CXCR5 (L138D7), anti-integrinα4β7 (DATK32), anti-LAP (pro-TGFβ, TW7-16B4), anti-TNFα (MP6-XT22), anti-CD107a (1D4B). Antibodies were obtained from BioLegend, unless indicated otherwise. Double staining with insulin B:10-23/I-Ag7 tetramers (HVEALYLVCGGEG) conjugated to PE and APC was used to identify antigen specific T cells. Flow cytometry and sorting studies adhere to the ‘Guidelines for the use of flow cytometry and cell sorting in immunological studies’[48].
Sorting and transfer of islet infiltrating CD4+ T cells
Islet-derived CD4+ Foxp3− conventional T cells (Tconvs) were sorted based on CD5 expression (CD4+Foxp3GFP− CD5high/low) from 11-20 week old pre-diabetic NOD.Foxp3GFP mice. Sorting was performed on a FACSAria Fusion flow cytometer (BD Biosciences). The sort purity was above 90% for CD5high Tconvs, and 95% for CD5low Tconvs. CD5high and CD5low CD4+ Tconvs were adoptively transferred (i.p.) into female NOD.TCRα−/− recipient mice. Diabetes was induced with 20,000 cells per mouse. Insulitis and islet infiltration were assessed after transfer of 10,000 cells.
Histological analysis of islet infiltration
Five step sections from each mouse were stained with H&E. An average of 90–100 islets per mouse were scored in a blinded manner. Two methods of insulitis measurement were used. First, insulitis and peri-insulitis were determined based on the percentage of the islets that possessed leukocyte infiltrate using the following metric: no insulitis (score 0: normal islet and no infiltration), peri-insulitis (score 1: infiltration on edges of islet); and insulitis (score 2: up to 25% of islet, score 3: 25-75% of islet, score 4: >75% of islet). Second, a defined insulitis score was determined using the method outlined in Current Protocols in Immunology[49].
Immunofluorescence
Pancreatic sections were stained with anti-CD3 (CD3-12, Thermo Fisher), anti-BSRP-A (polyclonal sheep IgG, R&D), and DAPI. Images were obtained using Nikon Ti-E widefield microscope at x20 magnification. BSRP-A was used to identify endocrine cells [50,51]. Nikon NIS-Elements scope/software was used to obtain the images. Background autofluorescence was removed using a previously published method in Fiji [52].
TCR sequence analysis
TCR sequence analysis was performed on individual mice. Genomic DNA was extracted from sorted Foxp3GFP+ and Foxp3GFP− CD4+TCR+ T cells isolated from pancreatic islets of pre-diabetic NOD.Foxp3GFP mice at 14-16 weeks of age. Purification of gDNA was performed using QIAamp DNA Micro Kit. TCRβ repertoires of Tconv and Treg populations were assessed by ImmunoSEQ Survey assay (Adaptive Biotechnologies). Data were analyzed by ImmunoSEQ Analyzer version 2.0.
RNAseq analysis
RNA was isolated from CD4+ T cells sorted from single cell suspensions of the pancreatic islets of NOD.Foxp3GFP mice stained with anti-CD4 and anti-CD5 antibodies. All samples were sorted with >97% purity. cDNA was synthesized using the SMARTer Ultra Low Input RNA Kit (Clonetech). Library preparation was performed with the Illumina Nextera XT kit before paired-end RNA-sequencing using the Illumina NextSeq500 platform for 150 cycles (NextSeq500 Mid Output Kit). Sequencing reads were aligned to the mouse genome (RefSeq mm10) using TopHat Alignment (version 1.0.0; [53]) and gene expression was quantified by FPKM. Cufflinks Assembly & DE (version 1.1.0; [54]) was used to compute differential expression between groups, with differentially expressed genes defined by q<0.05 with Benjamini-Hochberg correction for multiple testing. Gene ontology analysis was performed using Metascape. Two-TCR RNAseq data files were previously published in [25] . Data Resources: The accession number for the raw data reported in this paper are GSE102231, GSE106467, and GSE131345.
Statistical Analysis
Statistical analyses were performed using Prism, except TCR sequencing analysis, which was performed using ImmunoSEQ Analyzer.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the BCM Cytometry and Cell Sorting Core for assistance with cell sorting, and the NIH tetramer core for providing tetramer reagents. This work was supported by the National Institutes of Health [R01 AI125301 to M.B, and R01 DK114456 to M.L.B.], and The Robert and Janice McNair Foundation.
Footnotes
Conflict of interests:
The authors declare no commercial or financial conflict interests.
Data availability statement:
The RNAseq datasets generated and analyzed during the current study are available in the GEO repository, and can be found under these identifiers: GSE102231, GSE106467, and GSE131345. The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- [1].Martinez RJ, Evavold BD, Lower Affinity T Cells are Critical Components and Active Participants of the Immune Response, Front Immunol. 6 (2015) 468. 10.3389/fimmu.2015.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bettini M, Blanchfield L, Castellaw A, Zhang Q, Nakayama M, Smeltzer MP, Zhang H, Hogquist KA, Evavold BD, Vignali DAA, TCR Affinity and Tolerance Mechanisms Converge To Shape T Cell Diabetogenic Potential, J Immunol. 193 (2014) 571–579. 10.4049/jimmunol.1400043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Snook JP, Kim C, Williams MA, TCR signal strength controls the differentiation of CD4+ effector and memory T cells., Sci Immunol. 3 (2018) eaas9103. 10.1126/sciimmunol.aas9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fiege JK, Stone IA, Fay EJ, Markman MW, Wijeyesinghe S, Macchietto MG, Shen S, Masopust D, Langlois RA, The Impact of TCR Signal Strength on Resident Memory T Cell Formation during Influenza Virus Infection., J Immunol Baltim Md 1950. 203 (2019) 936–945. 10.4049/jimmunol.1900093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schober K, Voit F, Grassmann S, Müller TR, Eggert J, Jarosch S, Weißbrich B, Hoffmann P, Borkner L, Nio E, Fanchi L, Clouser CR, Radhakrishnan A, Mihatsch L, Lückemeier P, Leube J, Dössinger G, Klein L, Neuenhahn M, Oduro JD, Cicin-Sain L, Buchholz VR, Busch DH, Reverse TCR repertoire evolution toward dominant low-affinity clones during chronic CMV infection, Nat Immunol. 21 (2020) 434–441. 10.1038/s41590-020-0628-2. [DOI] [PubMed] [Google Scholar]
- [6].Bettini M, Scavuzzo MA, Liu B, Kolawole E, Guo L, Evavold BD, Borowiak M, Bettini ML, A Critical Insulin TCR Contact Residue Selects High-Affinity and Pathogenic Insulin-Specific T Cells., Diabetes. 69 (2019) 392–400. 10.2337/db19-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu B, Hood JD, Kolawole EM, Woodruff DM, Vignali DA, Bettini M, Evavold BD, A Hybrid Insulin Epitope Maintains High 2D Affinity for Diabetogenic T Cells in the Periphery., Diabetes. 69 (2019) 381–391. 10.2337/db19-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lennon GP, Bettini M, Burton AR, Vincent E, Arnold PY, Santamaria P, Vignali DAA, T Cell Islet Accumulation in Type 1 Diabetes Is a Tightly Regulated, Cell-Autonomous Event, Immunity. 31 (2009) 643–653. 10.1016/j.immuni.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang J, Tsai S, Han B, Tailor P, Santamaria P, Autoantigen Recognition Is Required for Recruitment of IGRP206–214-Autoreactive CD8+ T Cells but Is Dispensable for Tolerance, J Immunol. 189 (2012) 2975–2984. 10.4049/jimmunol.1201787. [DOI] [PubMed] [Google Scholar]
- [10].Calderon B, Carrero JA, Miller MJ, Unanue ER, Cellular and molecular events in the localization of diabetogenic T cells to islets of Langerhans, Proc National Acad Sci. 108 (2011) 1561–1566. 10.1073/pnas.1018973108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Calderon B, Carrero JA, Miller MJ, Unanue ER, Entry of diabetogenic T cells into islets induces changes that lead to amplification of the cellular response, Proc National Acad Sci. 108 (2011) 1567–1572. 10.1073/pnas.1018975108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Magnuson AM, Thurber GM, Kohler RH, Weissleder R, Mathis D, Benoist C, Population dynamics of islet-infiltrating cells in autoimmune diabetes, Proc National Acad Sci. 112 (2015) 1511–1516. 10.1073/pnas.1423769112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Whiteside SK, Snook JP, Ma Y, Sonderegger FL, Fisher C, Petersen C, Zachary JF, Round JL, Williams MA, Weis JJ, IL-10 Deficiency Reveals a Role for TLR2-Dependent Bystander Activation of T Cells in Lyme Arthritis, J Immunol. 200 (2018) 1457–1470. 10.4049/jimmunol.1701248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee H-G, Lee J-U, Kim D-H, Lim S, Kang I, Choi J-M, Pathogenic function of bystander-activated memory-like CD4+ T cells in autoimmune encephalomyelitis, Nat Commun. 10 (2019) 709. 10.1038/s41467-019-08482-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Christoffersson G, Chodaczek G, Ratliff SS, Coppieters K, von Herrath MG, Suppression of diabetes by accumulation of non–islet-specific CD8+ effector T cells in pancreatic islets, Sci Immunol. 3 (2018) eaam6533. 10.1126/sciimmunol.aam6533. [DOI] [PubMed] [Google Scholar]
- [16].Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE, CD5 Expression Is Developmentally Regulated By T Cell Receptor (TCR) Signals and TCR Avidity, J Exp Medicine. 188 (1998) 2301–2311. 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Vignali D, Cantarelli E, Bordignon C, Canu A, Citro A, Annoni A, Piemonti L, Monti P, Detection and Characterization of CD8+ Autoreactive Memory Stem T Cells in Patients with Type 1 Diabetes, Diabetes. 67 (2018) db171390. 10.2337/db17-1390. [DOI] [PubMed] [Google Scholar]
- [18].Abdelsamed HA, Zebley CC, Nguyen H, Rutishauser RL, Fan Y, Ghoneim HE, Crawford JC, Alfei F, Alli S, Ribeiro SP, Castellaw AH, McGargill MA, Jin H, Boi SK, Speake C, Serti E, Turka LA, Busch ME, Stone M, Deeks SG, Sekaly R-P, Zehn D, James EA, Nepom GT, Youngblood B, Beta cell-specific CD8+ T cells maintain stem cell memory-associated epigenetic programs during type 1 diabetes, Nat Immunol. 21 (2020) 578–587. 10.1038/s41590-020-0633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Anderson AC, Joller N, Kuchroo VK, Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation, Immunity. 44 (2016) 989–1004. 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sprouse ML, Scavuzzo MA, Blum S, Shevchenko I, Lee T, Makedonas G, Borowiak M, Bettini ML, Bettini M, High self-reactivity drives T-bet and potentiates Treg function in tissue-specific autoimmunity, Jci Insight. 3 (2018) e97322. 10.1172/jci.insight.97322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brehm MA, Daniels KA, Welsh RM, Rapid production of TNF-alpha following TCR engagement of naive CD8 T cells., J Immunol Baltim Md 1950. 175 (2005) 5043–9. 10.4049/jimmunol.175.8.5043. [DOI] [PubMed] [Google Scholar]
- [22].Lieberman SM, Evans AM, Han B, Takaki T, Vinnitskaya Y, Caldwell JA, Serreze DV, Shabanowitz J, Hunt DF, Nathenson SG, Santamaria P, DiLorenzo TP, Identification of the β cell antigen targeted by a prevalent population of pathogenic CD8+ T cells in autoimmune diabetes, Proc National Acad Sci. 100 (2003) 8384–8388. 10.1073/pnas.0932778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fuchs YF, Eugster A, Dietz S, Sebelefsky C, Kühn D, Wilhelm C, Lindner A, Gavrisan A, Knoop J, Dahl A, Ziegler A-G, Bonifacio E, CD8+ T cells specific for the islet autoantigen IGRP are restricted in their T cell receptor chain usage, Sci Rep-Uk. 7 (2017) 44661. 10.1038/srep44661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wong J, Mathis D, Benoist C, TCR-based lineage tracing: no evidence for conversion of conventional into regulatory T cells in response to a natural self-antigen in pancreatic islets, J Exp Medicine. 204 (2007) 2039–2045. 10.1084/jem.20070822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sprouse ML, Shevchenko I, Scavuzzo MA, Joseph F, Lee T, Blum S, Borowiak M, Bettini ML, Bettini M, Cutting Edge: Low-Affinity TCRs Support Regulatory T Cell Function in Autoimmunity, J Immunol. 200 (2017) 909–914. 10.4049/jimmunol.1700156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Akiba Y, Kuwabara T, Mukozu T, Mikami T, Kondo M, Special AT - rich sequence binding protein 1 is required for maintenance of T cell receptor responsiveness and development of experimental autoimmune encephalomyelitis, Microbiol Immunol. 62 (2018) 255–268. 10.1111/1348-0421.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Stephen TL, Payne KK, Chaurio RA, Allegrezza MJ, Zhu H, Perez-Sanz J, Perales-Puchalt A, Nguyen JM, Vara-Ailor AE, Eruslanov EB, Borowsky ME, Zhang R, Laufer TM, Conejo-Garcia JR, SATB1 Expression Governs Epigenetic Repression of PD-1 in Tumor-Reactive T Cells, Immunity. 46 (2017) 51–64. 10.1016/j.immuni.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Henderson JG, Opejin A, Jones A, Gross C, Hawiger D, CD5 Instructs Extrathymic Regulatory T Cell Development in Response to Self and Tolerizing Antigens, Immunity. 42 (2015) 471–483. 10.1016/j.immuni.2015.02.010. [DOI] [PubMed] [Google Scholar]
- [29].Smith K, Seddon B, Purbhoo MA, Zamoyska R, Fisher AG, Merkenschlager M, Sensory Adaptation in Naive Peripheral CD4 T Cells, J Exp Medicine. 194 (2001) 1253–1262. 10.1084/jem.194.9.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ryan KR, McCue D, Anderton SM, Fas - mediated death and sensory adaptation limit the pathogenic potential of autoreactive T cells after strong antigenic stimulation, J Leukocyte Biol. 78 (2005) 43–50. 10.1189/jlb.0205059. [DOI] [PubMed] [Google Scholar]
- [31].Fulton RB, Hamilton SE, Xing Y, Best JA, Goldrath AW, Hogquist KA, Jameson SC, The TCR’s sensitivity to self peptide-MHC dictates the ability of naive CD8+ T cells to respond to foreign antigens, Nat Immunol. 16 (2015) 107–117. 10.1038/ni.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sabatino JJ, Huang J, Zhu C, Evavold BD, High prevalence of low affinity peptide–MHC II tetramer–negative effectors during polyclonal CD4+ T cell responses, J Exp Medicine. 208 (2011) 81–90. 10.1084/jem.20101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Luce S, Lemonnier F, Briand J-P, Coste J, Lahlou N, Muller S, Larger E, Rocha B, Mallone R, Boitard C, Single Insulin-Specific CD8+ T Cells Show Characteristic Gene Expression Profiles in Human Type 1 Diabetes, Diabetes. 60 (2011) 3289–3299. 10.2337/db11-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Orban T, Beam CA, Xu P, Moore K, Jiang Q, Deng J, Muller S, Gottlieb P, Spain L, Peakman M, the T 1 D.T.A.S. Group, Reduction in CD4 Central Memory T-Cell Subset in Costimulation Modulator Abatacept-Treated Patients With Recent-Onset Type 1 Diabetes Is Associated With Slower C-Peptide Decline, Diabetes. 63 (2014) 3449–3457. 10.2337/db14-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Narsale A, Davies JD, Memory T Cells in Type 1 Diabetes: the Devil is in the Detail, Curr Diabetes Rep. 17 (2017) 61. 10.1007/s11892-017-0889-9. [DOI] [PubMed] [Google Scholar]
- [36].Yeo L, Woodwyk A, Sood S, Lorenc A, Eichmann M, Pujol-Autonell I, Melchiotti R, Skowera A, Fidanis E, Dolton GM, Tungatt K, Sewell AK, Heck S, Saxena A, Beam CA, Peakman M, Autoreactive T effector memory differentiation mirrors β-cell function in type 1 diabetes, J Clin Invest. 128 (2018) 3460–3474. 10.1172/jci120555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Penaranda C, Kuswanto W, Hofmann J, Kenefeck R, Narendran P, Walker LSK, Bluestone JA, Abbas AK, Dooms H, IL-7 receptor blockade reverses autoimmune diabetes by promoting inhibition of effector/memory T cells, Proc National Acad Sci. 109 (2012) 12668–12673. 10.1073/pnas.1203692109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee L-F, Logronio K, Tu GH, Zhai W, Ni I, Mei L, Dilley J, Yu J, Rajpal A, Brown C, Appah C, Chin SM, Han B, Affolter T, Lin JC, Anti–IL-7 receptor-α reverses established type 1 diabetes in nonobese diabetic mice by modulating effector T-cell function, Proc National Acad Sci. 109 (2012) 12674–12679. 10.1073/pnas.1203795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Weninger W, Carlsen HS, Goodarzi M, Moazed F, Crowley MA, Baekkevold ES, Cavanagh LL, von Andrian UH, Naive T Cell Recruitment to Nonlymphoid Tissues: A Role for Endothelium-Expressed CC Chemokine Ligand 21 in Autoimmune Disease and Lymphoid Neogenesis, J Immunol. 170 (2003) 4638–4648. 10.4049/jimmunol.170.9.4638. [DOI] [PubMed] [Google Scholar]
- [40].Shan Z, Xu B, Mikulowska-Mennis A, Michie SA, CCR7 directs the recruitment of T cells into inflamed pancreatic islets of nonobese diabetic (NOD) mice, Immunol Res. 58 (2014) 351–357. 10.1007/s12026-014-8500-9. [DOI] [PubMed] [Google Scholar]
- [41].Raemdonck KV, Umar S, Palasiewicz K, Volkov S, Volin MV, Arami S, Chang HJ, Zanotti B, Sweiss N, Shahrara S, CCL21/CCR7 signaling in macrophages promotes joint inflammation and Th17-mediated osteoclast formation in rheumatoid arthritis, Cell Mol Life Sci. 77 (2020) 1387–1399. 10.1007/s00018-019-03235-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bouma G, Coppens JMC, Mourits S, Nikolic T, Sozzani S, Drexhage HA, Versnel MA, Evidence for an enhanced adhesion of DC to fibronectin and a role of CCL19 and CCL21 in the accumulation of DC around the pre - diabetic islets in NOD mice, Eur J Immunol. 35 (2005) 2386–2396. 10.1002/eji.200526251. [DOI] [PubMed] [Google Scholar]
- [43].Ohshima Y, Yang LP, Avice MN, Kurimoto M, Nakajima T, Sergerie M, Demeure CE, Sarfati M, Delespesse G, Naive human CD4+ T cells are a major source of lymphotoxin alpha., J Immunol Baltim Md 1950. 162 (1999) 3790–4. [PubMed] [Google Scholar]
- [44].Gramaglia I, Mauri DN, Miner KT, Ware CF, Croft M, Lymphotoxin alphabeta is expressed on recently activated naive and Th1-like CD4 cells but is down-regulated by IL-4 during Th2 differentiation., J Immunol Baltim Md 1950. 162 (1999) 1333–8. [PubMed] [Google Scholar]
- [45].Luther SA, Bidgol A, Hargreaves DC, Schmidt A, Xu Y, Paniyadi J, Matloubian M, Cyster JG, Differing activities of homeostatic chemokines CCL19, CCL21, and CXCL12 in lymphocyte and dendritic cell recruitment and lymphoid neogenesis., J Immunol Baltim Md 1950. 169 (2002) 424–33. 10.4049/jimmunol.169.1.424. [DOI] [PubMed] [Google Scholar]
- [46].You S, Slehoffer G, Barriot S, Bach J-F, Chatenoud L, Unique role of CD4+CD62L+ regulatory T cells in the control of autoimmune diabetes in T cell receptor transgenic mice., P Natl Acad Sci Usa. 101 Suppl 2 (2004) 14580–5. 10.1073/pnas.0404870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Badillo FEG, Tegou FZ, Abreu MM, Masina R, Sha D, Najjar M, Wright SH, Bayer AL, Korpos É, Pugliese A, Molano RD, Tomei AA, CCL21 Expression in β-Cells Induces Antigen-Expressing Stromal Cell Networks in the Pancreas and Prevents Autoimmune Diabetes in Mice, Diabetes. 68 (2019) 1990–2003. 10.2337/db19-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cossarizza A, Chang H, Radbruch A, Abrignani S, Addo R, Akdis M, Andrä I, Andreata F, Annunziato F, Arranz E, Bacher P, Bari S, Barnaba V, Barros - Martins J, Baumjohann D, Beccaria CG, Bernardo D, Boardman DA, Borger J, Böttcher C, Brockmann L, Burns M, Busch DH, Cameron G, Cammarata I, Cassotta A, Chang Y, Chirdo FG, Christakou E, Čičin - Šain L, Cook L, Corbett AJ, Cornelis R, Cosmi L, Davey MS, Biasi SD, Simone GD, del Zotto G, Delacher M, Rosa FD, Santo JD, Diefenbach A, Dong J, Dörner T, Dress RJ, Dutertre C, Eckle SBG, Eede P, Evrard M, Falk CS, Feuerer M, Fillatreau S, Fiz - Lopez A, Follo M, Foulds GA, Fröbel J, Gagliani N, Galletti G, Gangaev A, Garbi N, Garrote JA, Geginat J, Gherardin NA, Gibellini L, Ginhoux F, Godfrey DI, Gruarin P, Haftmann C, Hansmann L, Harpur CM, Hayday AC, Heine G, Hernández DC, Herrmann M, Hoelsken O, Huang Q, Huber S, Huber JE, Huehn J, Hundemer M, Hwang WYK, Iannacone M, Ivison SM, Jäck H, Jani PK, Keller B, Kessler N, Ketelaars S, Knop L, Knopf J, Koay H, Kobow K, Kriegsmann K, Kristyanto H, Krueger A, Kuehne JF, Kunze - Schumacher H, Kvistborg P, Kwok I, Latorre D, Lenz D, Levings MK, Lino AC, Liotta F, Long HM, Lugli E, MacDonald KN, Maggi L, Maini MK, Mair F, Manta C, Manz RA, Mashreghi M, Mazzoni A, McCluskey J, Mei HE, Melchers F, Melzer S, Mielenz D, Monin L, Moretta L, Multhoff G, Muñoz LE, Muñoz - Ruiz M, Muscate F, Natalini A, Neumann K, Ng LG, Niedobitek A, Niemz J, Almeida LN, Notarbartolo S, Ostendorf L, Pallett LJ, Patel AA, Percin GI, Peruzzi G, Pinti M, Pockley AG, Pracht K, Prinz I, Pujol - Autonell I, Pulvirenti N, Quatrini L, Quinn KM, Radbruch H, Rhys H, Rodrigo MB, Romagnani C, Saggau C, Sakaguchi S, Sallusto F, Sanderink L, Sandrock I, Schauer C, Scheffold A, Scherer HU, Schiemann M, Schildberg FA, Schober K, Schoen J, Schuh W, Schüler T, Schulz AR, Schulz S, Schulze J, Simonetti S, Singh J, Sitnik KM, Stark R, Starossom S, Stehle C, Szelinski F, Tan L, Tarnok A, Tornack J, Tree TIM, van Beek JJP, van de Veen W, van Gisbergen K, Vasco C, Verheyden NA, von Borstel A, Ward - Hartstonge KA, Warnatz K, Waskow C, Wiedemann A, Wilharm A, Wing J, Wirz O, Wittner J, Yang JHM, Yang J, Guidelines for the use of flow cytometry and cell sorting in immunological studies (third edition), Eur J Immunol. 51 (2021) 2708–3145. 10.1002/eji.202170126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Leiter EH, Current Protocols in Immunology, Curr Protoc Immunol Ed Coligan John E et al. Chapter 15 (2001) 15.9.1–15.9.23. 10.1002/0471142735.im1509s24. [DOI] [PubMed] [Google Scholar]
- [50].Hald J, Galbo T, Rescan C, Radzikowski L, Sprinkel AE, Heimberg H, Ahnfelt-Rønne J, Jensen J, Scharfmann R, Gradwohl G, Kaestner KH, Stoeckert C, Jensen JN, Madsen OD, Pancreatic islet and progenitor cell surface markers with cell sorting potential, Diabetologia. 55 (2012) 154–165. 10.1007/s00125-011-2295-1. [DOI] [PubMed] [Google Scholar]
- [51].Ito Y, Ashenberg O, Pyrdol J, Luoma AM, Rozenblatt-Rosen O, Hofree M, Christian E, de Andrade LF, Tay RE, Teyton L, Regev A, Dougan SK, Wucherpfennig KW, Rapid CLIP dissociation from MHC II promotes an unusual antigen presentation pathway in autoimmunity, J Exp Med. 215 (2018) jem.20180300. 10.1084/jem.20180300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Baharlou H, Canete NP, Bertram KM, Sandgren KJ, Cunningham AL, Harman AN, Patrick E, AFid: A tool for automated identification and exclusion of autofluorescent objects from microscopy images, Biorxiv. (2019) 566315. 10.1101/566315. [DOI] [PubMed] [Google Scholar]
- [53].Trapnell C, Pachter L, Salzberg SL, TopHat: discovering splice junctions with RNA-Seq, Bioinformatics. 25 (2009) 1105–1111. 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks, Nat Protoc. 7 (2012) 562. 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNAseq datasets generated and analyzed during the current study are available in the GEO repository, and can be found under these identifiers: GSE102231, GSE106467, and GSE131345. The data that support the findings of this study are available from the corresponding author upon reasonable request.