Abstract
Mycobacterium avium causes disseminated disease in humans with AIDS, paratuberculosis in ruminants, lymphadenopathy in swine, and tuberculosis in birds. We constructed DNA vaccines expressing mycobacterial antigens as fusion proteins with enhanced green fluorescent protein (EGFP). Plasmids p65K-EGFP, p85A-EGFP, and p85B-EGFP expressed the M. avium 65-kDa antigen, the Mycobacterium bovis BCG 85A antigen, and the M. avium 85B antigen, respectively, as EGFP fusion proteins. We visualized protein expression directly in cultured murine fibroblasts and intact muscle. p65K-EGFP expressed fusion protein in a diffuse cytoplasmic pattern, and p85A-EGFP and p85B-EGFP produced a speckled pattern. We vaccinated C57BL/6 mice with three doses of plasmid DNA and then challenged them intraperitoneally with M. avium. Negative controls received saline, and positive controls received one dose of BCG vaccine. Mice in all groups developed disseminated infection with a high burden of organisms. Compared to negative controls, mice vaccinated with p85A-EGFP had an eightfold reduction in spleen M. avium CFU at 4 weeks after infection and a fourfold reduction at 8 weeks, reductions similar to those generated by BCG vaccine. Mice vaccinated with p65K-EGFP had a fourfold CFU reduction at 4 weeks and no effect at 8 weeks. This is the first report of DNA vaccines expressing foreign antigens as fusion proteins with EGFP and the first report of successful DNA vaccination against M. avium.
Mycobacterium avium is the most common bacterial infection in patients with AIDS in the United States, leading to substantial morbidity and mortality (6, 21, 24). M. avium is difficult to treat because it is naturally resistant to many antibiotics. It also causes paratuberculosis in ruminants, disseminated lymphadenopathy in swine, and tuberculosis in birds, resulting in significant economic losses. M. avium is closely related to other slowly growing mycobacterial pathogens, including Mycobacterium tuberculosis (the cause of human tuberculosis) and Mycobacterium leprae (the cause of leprosy). The development of effective vaccines against mycobacteria is an important public health goal. An effective M. avium vaccine would be useful in high-risk human and animal populations and might serve as a model for tuberculosis and leprosy vaccines.
DNA vaccination uses plasmids to express antigenic proteins in host cells. DNA vaccines produce both humoral and cell-mediated immune responses and have led to protective immunity in a wide variety of animal models of infectious diseases (10). DNA vaccines yield partial protection in murine models of M. tuberculosis infection (23, 39, 43).
We developed DNA vaccines and tested them in an established murine model of M. avium infection. We generated DNA vaccines which expressed mycobacterial antigens as fusion proteins with enhanced green fluorescent protein (EGFP). EGFP is a mutant form of the jellyfish GFP; it produces more intense fluorescence than the native protein (8). Expression of mycobacterial antigens as EGFP fusion proteins allowed us to monitor the amounts and localization of the expressed proteins in mammalian cells. We developed DNA vaccines expressing the M. avium 65-kDa antigen, the Mycobacterium bovis 85A antigen, and the M. avium 85B antigen as EGFP fusion proteins. Each of these antigens is secreted by pathogenic mycobacteria and generates cellular and humoral immune responses in natural infections. This report describes the development of these DNA vaccines, their expression in mammalian cells, and their protective efficacy in an animal model of M. avium infection.
MATERIALS AND METHODS
Plasmids used for DNA vaccination.
The plasmids used in this study are described in Table 1. pEGFP-N1 (pEGFP) (Clontech Laboratories, Inc., Palo Alto, Calif.) is a mammalian expression vector which contains a cytomegalovirus promoter active in mammalian cells, a multiple cloning site, the EGFP gene, RNA-stabilizing sequences, an Escherichia coli origin of replication, and antibiotic selection markers. Douglas B. Lowrie (National Institute for Medical Research, London, United Kingdom) kindly provided pCMV4.65 and pCMV7.36, which are optimized versions of the plasmids described by Tascon et al. (39).
TABLE 1.
Characteristics of plasmids used for DNA vaccinationa
| Plasmid name | Accession no. | Expressed protein | Synonyms for mycobacterial Ag | Sequence of insert | Protein expression in murine fibroblasts |
|---|---|---|---|---|---|
| pEGFP-N1 | U55762 | EGFP | None | No insert | Nucleus and cytoplasm, uniform |
| p65K-EGFP | U15989 | M. avium 65-kDa Ag–EGFP | WHO 2, CIE Ag 82, Hsp65, GroEL | Correct | Cytoplasm, uniform |
| p85A-EGFP | X53034 | M. bovis BCG Ag 85A-EGFP | WHO 10(a), MPB44, P32, 32-kDa Ag, FbpA | Correct | Cytoplasm, speckled |
| p85B-EGFP | X63437 | M. avium Ag 85B-EGFP | WHO 10(b), α-Ag, FbpB | Correct | Cytoplasm, speckled |
| pSodA-EGFP | U11550 | M. avium SodA-EGFP | WHO 4, CIE Ag 62, superoxide dismutase | Frameshift | No expression |
| pCMV4.65 | X65546 | M. leprae 65-kDa Ag | WHO 2, CIE Ag 82, Hsp65, GroEL | ND | ND |
| pCMV7.36 | U15183 | M. leprae 36-kDa Ag | WHO 8, proline-rich antigen | ND | ND |
Abbreviations: Ag, antigen; ND, not done.
Construction of plasmids expressing mycobacterial antigen-EGFP fusion proteins.
We inserted mycobacterial genes into pEGFP to generate genes expressing fusion proteins with EGFP at the carboxy terminus. We amplified each bacterial gene in its entirety by PCR with primers which modified both ends of the gene. Each forward primer inserted a specific restriction site and replaced the bacterial start codon with a Kozak consensus sequence (GCCACCATGG; positions −6 through +4) to enhance mammalian expression (27). Each reverse primer removed the stop codon and added a second restriction site. Each PCR product was then digested with two restriction enzymes and ligated into the multiple cloning site of pEGFP in frame with the EGFP gene. Each construct was designed to express a fusion protein comprising the complete mycobacterial antigen, a short amino acid spacer, and the complete EGFP. In some cases the second amino acid of the mycobacterial antigen was intentionally changed by the substitution of the Kozak consensus sequence G at nucleotide position 4.
p65K-EGFP.
Fouad A. K. El-Zaatari (Baylor College of Medicine, Houston, Tex.) kindly provided pMptb20, which contains the 65-kDa antigen gene from M. avium subsp. paratuberculosis ATCC 43015 (11). We amplified the mycobacterial gene from plasmid DNA by PCR. The forward primer introduced an EcoRI site and the Kozak consensus sequence (5′ CGGAATTCGC CACCATGGTG CTAGGTCGGG ACGGTGAGG 3′; the EcoRI site is underlined, and the Kozak consensus sequence is in italics). The reverse primer removed the stop codon and introduced a BamHI site (5′ CGGGATCCCA GAAGTCCATGCCGCCCATGC 3′; the BamHI site is underlined). The PCR product was digested and ligated into the EcoRI and BamHI sites of pEGFP. The resultant plasmid, p65K-EGFP, was predicted to express a fusion protein comprising the complete 65-kDa antigen, a 6-amino-acid spacer, and the complete EGFP.
p85A-EGFP.
The National Institutes of Health AIDS Research and Reference Reagent Program, Rockville, Md., kindly provided pαAg1, which contains the antigen 85A gene from M. bovis BCG (9). We amplified this gene by using the primers 5′ TGAGCGAGCT CGCCACCATG GAGCTTGTTG ACAGGGTTCG TG 3′ and 5′ ACGCGGATCC GCGCCCTGGG GCGCGGGCCC GGTG 3′ and subcloned the PCR product into the SacI and BamHI sites of pEGFP.
p85B-EGFP.
Naoya Ohara (Nagasaki University, Nagasaki, Japan) kindly provided pAASp56, which contains the antigen 85B gene from M. avium ATCC 15769 (31). We amplified this gene by using the primers 5′ TCCGCTCGAG CCACCATGGCAGATCTGAGC GAGAAGGTCC 3′ and 5′ CGGAATTCCG CCGCCGCCCG GGGACG 3′ and subcloned the PCR product into the XhoI and EcoRI sites of pEGFP.
pSodA-EGFP.
Vincent Escuyer (Institut National de la Santé et de la Recherche Médicale, Paris, France) kindly provided pSOD2, which contains the sodA gene from M. avium TMC 724 (13). We amplified this gene by using the primers 5′ GTCCGCTCGAGCCACCATGG CTGAATACAC CCTGCCCGAC 3′ and 5′ ACCGGAATTCTGCCGAAGAT CAGGCCTTGG 3′ and subcloned the PCR product into the XhoI and EcoRI sites of pEGFP.
Plasmid preparation and verification.
Plasmids were transformed by heat shock into competent E. coli INVαF′ cells (Invitrogen Corp., Carlsbad, Calif.). Plasmid DNA was prepared by using the EndoFree Plasmid Maxi Kit (Qiagen, Inc., Valencia, Calif.) and diluted in phosphate-buffered saline (PBS) to a final concentration of 1 mg/ml for vaccination. Each mycobacterial antigen-EGFP plasmid construct was verified by restriction digestion and by sequencing the complete insert DNA at the Duke University Sequencing Facility.
Cell culture and transfection.
Murine 3T3 fibroblasts (2 × 105/well) were grown in 35-mm-diameter tissue culture plates in Dulbecco’s medium (Life Technologies, Gaithersburg, Md.) with 10% fetal bovine serum (HyClone Laboratories, Inc., Logan, Utah), penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells in each well were transfected by using 2 μg of plasmid DNA and 5 μg of Lipofectin Reagent (Life Technologies) in 0.1 ml of Opti-MEM Reduced Serum Medium (Life Technologies) with a 12-h incubation at 37°C. Transfected cells were washed and incubated in complete Dulbecco’s medium for 48 h prior to analysis. Cells expressing fluorescent proteins were visualized by inverted fluorescence microscopy.
Colocalization experiments.
Murine 3T3 fibroblasts expressing fluorescent fusion proteins were fixed with paraformaldehyde, washed with PBS, and stained for 1 h at 37°C with 4′,6-diamidino-2-phenylindole (DAPI) or tetramethyl rhodamine isothiocyanate (TRITC)-labelled wheat germ agglutinin (both from Sigma Chemical Co., St. Louis, Mo.). Transferrin was labelled by indirect immunofluorescence with goat antitransferrin serum as the primary antibody and phycoerythrin-conjugated rabbit anti-goat immunoglobulin G as the secondary antibody (both from Sigma). TRAPα was labelled by using rabbit anti-TRAPα (previously designated SSRα [29]) as the primary antibody and phycoerythrin-conjugated goat anti-rabbit immunoglobulin G as the secondary antibody (Sigma). Cells were visualized by fluorescence microscopy.
Vaccination.
We vaccinated 4- to 6-week-old female C57BL/6 mice (National Cancer Institute, Frederick, Md.) in groups of 10. Mice were anesthetized with inhaled halothane and injected with 50 μg of plasmid DNA in 50 μl of PBS in each tibialis anterior muscle (100 μg DNA/mouse) by using an insulin syringe with a 28-gauge needle. Mice were vaccinated at week 0, week 3, and week 6 with plasmid DNA. Negative control mice were injected with PBS only. Positive control mice were injected intradermally once at week 0 with 50 μl of Tice BCG vaccine (Organon Teknika Corp., Durham, N.C.), containing 0.5 × 106 to 4 × 106 CFU of live mycobacteria. BCG vaccine is a live attenuated strain of M. bovis and is used in humans to prevent tuberculosis. Previous studies have demonstrated that BCG vaccine protects against M. avium in animal models (22, 33).
Mycobacterial infection.
Kevin A. Nash and Clark B. Inderlied (Children’s Hospital, Los Angeles, Calif.) kindly provided a spleen homogenate from a mouse infected with M. avium MAC 101. This serovar 1 strain was originally isolated from the blood of a human with AIDS and is widely used in mouse models (16). To maintain virulence, only transparent colonies from primary cultures of murine spleen tissue were used (16). Mycobacteria were grown from spleen homogenates for 10 days at 37°C on Middlebrook 7H11 agar with OADC (oleic acid-albumin-dextrose-catalase) (Difco, Detroit, Mich.) supplementation. Transparent colonies were selected, suspended in PBS, pooled, and stored in aliquots at −70°C at a concentration of 5 × 108 CFU/ml. Three weeks after the final injection of plasmid DNA (week 9), mice were anesthetized with inhaled methoxyflurane and injected intraperitoneally with 5 × 106 CFU of M. avium MAC 101. Vaccination groups of 10 were divided in half; these groups of 5 mice were sacrificed 4 and 8 weeks after infection (weeks 13 and 17).
Quantitative cultures of spleen.
Mice were anesthetized with methoxyflurane and euthanized by cervical dislocation. Spleens were removed by aseptic dissection, weighed, homogenized, and serially diluted in Middlebrook 7H9 broth (Difco). Aliquots of the suspension were plated onto Middlebrook 7H11 agar with OADC supplementation. Colonies counts were expressed as log10 CFU per spleen. The logarithmic transformation was used because the CFU counts were not normally distributed. The spleen weight and log10 CFU in each experimental group were compared to those in the negative control group by using a two-tailed t test. A P value of <0.05 was considered statistically significant.
RT-PCR of cytokine mRNA.
We amplified mRNAs for β-actin, gamma interferon (IFN-γ), and interleukin-4 (IL-4) from spleen tissues by using reverse transcriptase PCR (RT-PCR). Spleen samples were frozen in liquid nitrogen at the time of euthanasia. Spleen RNA was purified according to the manufacturer’s protocol by using Trizol reagent (Life Technologies). cDNA was synthesized in a 20-μl volume containing 1 μg of total RNA, 5 mM random hexamers, 0.5 mM (each) four deoxynucleoside triphosphates, 20 U of RNase inhibitor (Promega, Madison, Wis.), 50 mM Tris-HCl, 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, and 100 U of Moloney murine leukemia virus RT (Life Technologies). This mixture was incubated for 1 h at 37°C. PCR was conducted in a 50-μl volume containing 1 μl of cDNA, 5 μl of GeneAmp 10× PCR Buffer II (Perkin-Elmer Cetus, Emeryville, Calif.), 1.5 mM MgCl2, 100 nM each primer, 200 μM (each) four deoxynucleoside triphosphates, and 1.25 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer Cetus). An initial denaturation of 12 min at 95°C was followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 1 min, and extension at 72°C for 2 min, followed by a final extension at 72°C for 10 min. Primers were previously published (35). IFN-γ and IL-4 were amplified by using undiluted cDNA, and β-actin was amplified by using a 1:100 dilution. These cDNA quantities were selected empirically such that PCR amplification did not reach a plateau in 30 cycles. Each PCR product was subjected to electrophoresis in 3% agarose, stained with ethidium bromide, and visualized by UV transillumination. The intensity of each fluorescent band was compared to those of DNA standards by using the EDAS 120 digital imaging system (Kodak, Rochester, N.Y.).
Animal care.
All animal protocols were approved by the Institutional Animal Care and Use Committees of both the Durham Veterans Affairs Medical Center and Duke University. The method of euthanasia followed the recommendations of the American Veterinary Medical Association. Death was not an experimental end point.
RESULTS
We constructed plasmids to express mycobacterial antigens as EGFP fusion proteins. We confirmed the expression of the fusion proteins in vitro in murine fibroblasts and in vivo in murine skeletal muscle. We then tested the plasmid DNA vaccines in a murine model of M. avium infection. After completing the vaccination series, mice were injected intraperitoneally with live M. avium bacteria. Protective immunity was determined by spleen weight and spleen CFU at 4 and 8 weeks after infection.
Plasmid sequencing.
Plasmid constructs were verified by restriction enzyme digestion and sequencing of the insert (Table 1). Three plasmid constructs had the predicted insert sequences (p65K-EGFP, p85A-EGFP, and p85B-EGFP). However, plasmid pSodA-EGFP had a frameshift error at the site of the forward PCR primer. It was predicted to express a 39-amino-acid polypeptide with only the initial five amino acids corresponding to the M. avium SodA protein. This construct was used in expression and vaccination studies as a negative control plasmid.
Protein expression in murine fibroblasts.
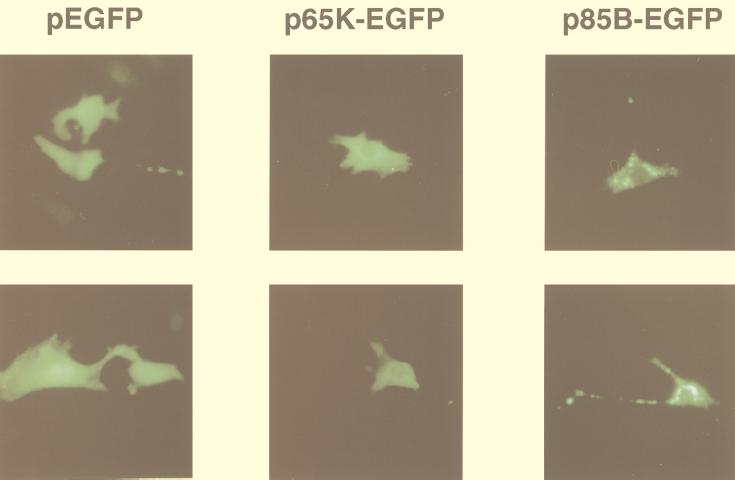
We transfected murine 3T3 fibroblasts with pEGFP and each of the four mycobacterial antigen-EGFP constructs. Fluorescent protein was visualized after transfection with pEGFP, p65K-EGFP, p85A-EGFP, and p85B-EGFP but not after transfection with the pSodA-EGFP plasmid containing the frameshift mutation. Fluorescence indicates the production of full-length mycobacterial antigen-EGFP fusion proteins, since the EGFP component is at the carboxy terminus. Three distinct patterns were observed (Fig. 1). EGFP was uniformly distributed in the nucleus and cytoplasm. The 65K-EGFP protein was uniformly distributed in the cytoplasm, with little or no nuclear expression. The 85A-EGFP and the 85B-EGFP fusion proteins were distributed in a speckled cytoplasmic pattern. These patterns of cellular localization were reproducible in at least three independent transfections for each plasmid. The mycobacterial antigen-EGFP fusion proteins were visible in a smaller proportion of cells and fluoresced with a reduced intensity compared to EGFP. This may be due to decreased transfection efficiency (larger plasmid), decreased translation efficiency (larger coding sequence or mycobacterial coding preferences), decreased fluorescence of the fusion proteins, or some combination of these factors.
FIG. 1.
Expression and localization of EGFP and mycobacterial antigen-EGFP fusion proteins in vitro. Murine 3T3 fibroblasts were transiently transfected with pEGFP, p65K-EGFP, p85A-EGFP, or p85B-EGFP and visualized 3 days later by fluorescence microscopy. Two photographs are shown for each plasmid. EGFP produced a homogenous pattern in the nucleus and cytoplasm. The 65K-EGFP fusion protein produced a homogenous cytoplasmic pattern with little or no protein in the nucleus. The 85A-EGFP (not shown) and 85B-EGFP fusion proteins produced speckled cytoplasmic patterns.
We performed colocalization experiments in fibroblasts expressing 85A-EGFP and 85B-EGFP by using DAPI (nucleus and mitochondria), TRITC-labelled wheat germ agglutinin (Golgi apparatus), and antibodies directed against the endosomal marker transferrin and the resident endoplasmic reticulum membrane protein TRAPα. However, the fluorescent fusion proteins did not localize to any of these compartments (not shown).
Expression in mouse muscle.
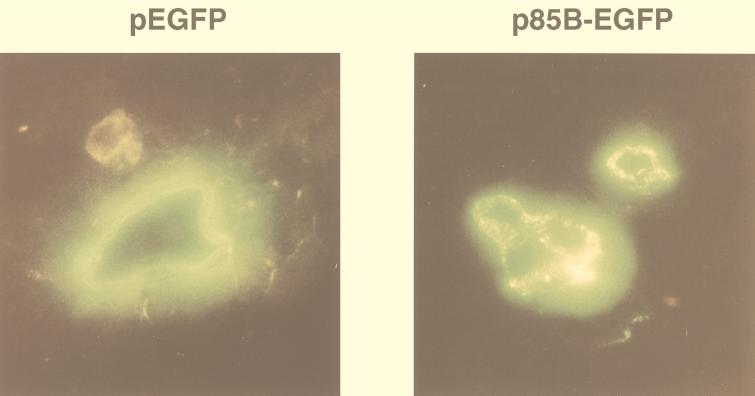
We injected skeletal muscle to confirm in vivo protein expression from the mycobacterial antigen-EGFP constructs. We injected 50 μg of pEGFP, p65K-EGFP, and p85B-EGFP into the tibialis anterior muscles of C57BL/6 mice. Four days later, the mice were euthanized, and frozen muscle sections were examined by fluorescence microscopy (Fig. 2). All three plasmids expressed fluorescent protein throughout the length of individual muscle cells. Both EGFP and 65K-EGFP were distributed in a uniform pattern, but 85B-EGFP had a speckled localization, similar to that seen in cultured fibroblasts. Green fluorescence was not observed in muscle fibers injected with PBS.
FIG. 2.
Expression and localization of EGFP and 85B-EGFP fusion protein in vivo. Tibialis anterior muscles of C57BL/6 mice were injected with pEGFP, p65K-EGFP, or p85B-EGFP. Four days later, the mice were euthanized, and frozen sections of the muscle were visualized by fluorescence microscopy. EGFP and 65K-EGFP (not shown) produced homogenous fluorescence, but 85B-EGFP produced a speckled pattern. The clear areas inside the muscle cells are frozen-section artifacts.
Protection against experimental M. avium infection.
Mice were vaccinated with plasmid DNA at weeks 0, 3, and 6, challenged by intraperitoneal M. avium injection at week 9, and euthanized at week 13 or 17. Injection of PBS alone served as a negative control. A single intradermal injection of BCG vaccine at week 0 served as a positive control. Two independent experiments (experiments 1 and 2) were conducted with the same methods but different DNA vaccines. Mice in all groups developed disseminated M. avium infection with high bacterial loads in the spleen. All mice developed splenomegaly. A typical spleen weight in uninfected mice of this age is 0.1 g. All mice survived the infection, gained weight, and displayed normal activity.
Vaccination experiment 1.
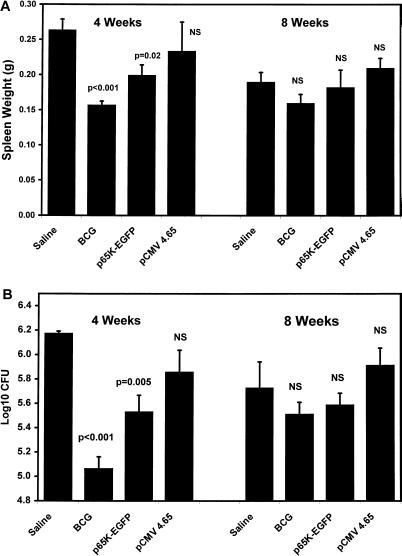
Experiment 1 included two DNA vaccines (p65K-EGFP and pCMV4.65), each of which expressed a mycobacterial 65-kDa antigen (Fig. 3). Vaccination with either BCG or p65K-EGFP yielded a significant reduction in the degree of splenomegaly at 4 weeks after infection compared to the saline control group (Fig. 3A), but there was no difference among the four groups at 8 weeks. Similarly, vaccination with either BCG or p65K-EGFP yielded a reduction in M. avium CFU in the spleen 4 weeks after challenge (Fig. 3B) but had no effect at 8 weeks. p65K-EGFP generated a 0.6-log-unit (fourfold) reduction in CFU at 4 weeks, which was less than that produced by BCG. Mice vaccinated with pCMV4.65 did not differ from negative controls in spleen weight or M. avium CFU at either 4 or 8 weeks after infection.
FIG. 3.
Spleen weights (A) and spleen CFU (B) after M. avium infection in vaccination experiment 1. C57BL/6 mice were vaccinated with three intramuscular injections of saline, p65K-EGFP, or pCMV4.65 or with one intradermal injection of BCG vaccine and then challenged with intraperitoneal M. avium, as described in Materials and Methods. All groups contained five mice, and all data represent means ± standard errors. Each experimental group was compared to the saline group by using a two-tailed t test. NS, not significant.
Vaccination experiment 2.
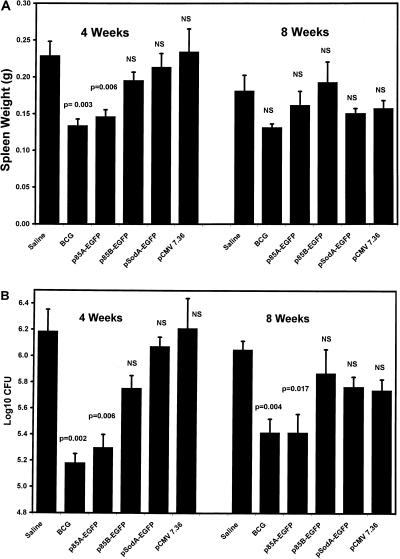
Experiment 2 included four DNA vaccines (p85A-EGFP, p85B-EGFP, pSodA-EGFP, and pCMV7.36) (Table 1). The results are shown in Fig. 4. Compared to negative controls, vaccination with either BCG or p85A-EGFP yielded a significant reduction in the degree of splenomegaly at 4 weeks after infection, but there was no significant effect at 8 weeks (Fig. 4A). Vaccination with either BCG or p85A-EGFP yielded a reduction in spleen CFU at both 4 and 8 weeks after challenge (Fig. 4B). p85A-EGFP generated a 0.9-log-unit (eightfold) reduction in CFU at 4 weeks and a 0.6-log-unit (fourfold) reduction at 8 weeks, both of which were similar to the reduction generated by BCG. Mice vaccinated with p85B-EGFP, pSodA-EGFP, or pCMV7.36 did not differ from negative controls in spleen weight or CFU at either 4 or 8 weeks after infection.
FIG. 4.
Spleen weights (A) and spleen CFU (B) after M. avium infection in vaccination experiment 2. C57BL/6 mice were vaccinated with three intramuscular injections of saline, p85A-EGFP, p85B-EGFP, pSodA-EGFP, or pCMV7.36 or with one intradermal injection of BCG vaccine and then challenged with intraperitoneal M. avium, as described in Materials and Methods. All groups contained five mice, and all data represent means ± standard errors. Each experimental group was compared to the saline group by using a two-tailed t test. NS, not significant.
Reproducibility.
The vaccine model used in these experiments has been used successfully by other researchers. The reproducibility of the vaccine model was demonstrated by the saline and BCG groups, for which the procedure was repeated in the two experiments (Fig. 3 and 4). When the results of experiment 1 and experiment 2 were compared, there were no significant differences in either spleen weight or spleen CFU in either the saline or BCG groups at either 4 or 8 weeks after infection (P > 0.05 for each of the eight comparisons).
Side effects.
Some mice vaccinated with intradermal BCG vaccine developed local ulcers as described for humans receiving this vaccine (5). Mice receiving saline or plasmid DNA vaccines had no local signs and had normal gait.
Cytokine mRNAs in spleens after M. avium infection.
As an indicator of local Th1/Th2 responses to infection, we amplified cytokine mRNAs from spleen tissue after infection by RT-PCR with primers directed at IL-4, IFN-γ, and β-actin (control). We determined the quantity of each PCR product by using a digital imaging system. IL-4 PCR products were absent or faint at all time points (data not shown). IFN-γ PCR products increased at 4 weeks after infection and declined toward baseline levels at 8 weeks after infection. However, the quantities of IFN-γ RT-PCR products in vaccinated mice did not differ from those in the mock-vaccinated controls at either 4 or 8 weeks after infection. This assay had a power of over 80% to detect threefold differences in the amounts of RT-PCR products between groups (alpha = 0.05) (data not shown). Data from vaccination experiment 1 are displayed in Fig. 5. Vaccination experiment 2 yielded similar results.
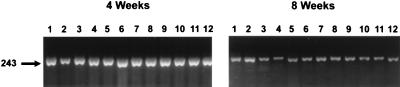
FIG. 5.
Spleen IFN-γ mRNA after M. avium infection as determined by RT-PCR. Total RNA was extracted from spleen tissue at 4 and 8 weeks after infection. Murine IFN-γ mRNA was amplified by RT-PCR. PCR products were subjected to electrophoresis on a 3% agarose gel and stained with ethidium bromide. Each lane shows the 243-bp IFN-γ PCR product from a single mouse. PCR products from mice vaccinated with pCMV4.65 (lanes 1 to 3), p65K-EGFP (lanes 4 to 6), BCG (lanes 7 to 9), and saline (lanes 10-12) are shown. IFN-γ mRNA was increased at 4 weeks in all groups and declined toward baseline levels at 8 weeks. There were no significant differences among the groups.
DISCUSSION
Expression of mycobacterial antigens as EGFP fusion proteins.
The visualization of EGFP provides rapid confirmation of foreign protein expression in mammalian cells in vitro and in vivo (3). Western blot analysis can also be used for this purpose, but it requires the availability of specific antibodies (23, 39, 43). EGFP fusion proteins can be used to confirm expression of antigens for which specific antibodies are not available. In contrast to many expression markers, EGFP does not require the addition of substrate. EGFP expression in tissue can be quantitated with a fluorometer. Quantitative expression analysis will be important when DNA vaccines are combined with adjuvants, because some adjuvants reduce protein expression by DNA vaccines (40).
Our work is the first report on the use of antigen-EGFP fusion proteins in a DNA vaccine. The EGFP component of these proteins could diminish immune responses by competing with the intended antigen, or it could enhance immune responses by acting as a carrier for a poor immunogen. Neither of these effects appeared to dominate in our experiments, since the levels of protective immunity that we observed are similar to those in previous reports of mycobacterial DNA vaccination. However, we did not directly compare immune responses to the same DNA vaccine with and without the EGFP component.
Cellular localization of mycobacterial antigen-EGFP fusion proteins.
We observed two patterns of localization of mycobacterial antigen-EGFP fusion proteins. The 65K-EGFP protein was distributed uniformly in the cytoplasm, but the 85A-EGFP and 85B-EGFP proteins had a speckled pattern. These patterns were similar in cultured fibroblasts and intact murine muscles. These antigens may contain signal sequences recognized by mammalian cells, although we were unable to identify a specific cellular compartment. Alternatively, the speckled pattern of 85A-EGFP and 85B-EGFP may simply represent intracellular aggregation due to poor solubility of the fusion proteins. Mycobacterial pathogens replicate in vivo inside host macrophages and secrete large amounts of these antigens. Intracellular localization of these antigens in host macrophages may modulate the host immune response in natural infection.
The intracellular localization of antigens expressed by DNA vaccines may influence the nature and intensity of the immune responses generated. However, Huygen et al. (23) found similar immune responses with DNA vaccines which expressed a secreted and a nonsecreted form of antigen 85A. We observed a modest level of protective immunity with both 65K-EGFP (uniform pattern) and 85A-EGFP (speckled pattern). Our experiments do not directly address the role of intracellular localization in protective immunity. The EGFP marker will be useful in future experiments on the intracellular localization of antigens expressed by DNA vaccines.
Protection from M. avium challenge.
Two plasmid DNA vaccines (p85A-EGFP and p65K-EGFP) generated protective immunity in this animal model of M. avium infection, as demonstrated by reduced spleen weight and reduced spleen CFU (up to a 0.9-log-unit CFU reduction). Several studies have reported similar CFU reductions with DNA vaccines in mycobacterial infection models. Tascon et al. (39) used DNA vaccines expressing the 65-kDa antigen and the 36-kDa antigen and found about a 1.0-log-unit CFU reduction in inbred mice and up to a 2-log-unit CFU reduction in crossbred or outbred mice after intraperitoneal M. tuberculosis challenge. Huygen et al. (23) used DNA vaccines expressing antigen 85A to generate a 0.7- to 1.0-log-unit CFU reduction after aerosol challenge with M. tuberculosis. Zhu et al. (43) used a DNA vaccine expressing the 38-kDa antigen to produce a 0.5-log-unit CFU reduction in spleen after intraperitoneal challenge with M. tuberculosis. Erb et al. (12) demonstrated no protection from intranasal BCG challenge with DNA vaccines expressing the 19-kDa antigen and the AhpC protein.
Protection across mycobacterial species.
Antigens from different mycobacteria often have substantial sequence similarity, and monoclonal antibodies directed at these antigens often react with multiple mycobacterial species (42). DNA vaccines expressing these antigens sometimes provide cross-species protection. The DNA vaccine with the highest level of protection against M. avium in our study (p85A-EGFP) actually expressed the M. bovis BCG 85A antigen, which has 79% amino acid identity with its M. avium homologue (32). It is also paradoxical that p85A-EGFP was protective, while p85B-EGFP was not, since M. avium strains express antigen 85A at lower levels than antigen 85B (32). Tascon et al. used a DNA vaccine expressing the M. leprae 65-kDa antigen (pCMV3.65) to protect against M. tuberculosis challenge (39). We used a modified version of this DNA vaccine (pCMV4.65) but achieved no protection against M. avium. The M. leprae and M. avium 65-kDa antigens have 87% amino acid identity. We did observe modest protection with p65K-EGFP, which expresses the M. avium 65-kDa antigen. However, p65K-EGFP also differs from pCMV4.65 in the plasmid backbone and the presence of the EGFP marker.
There is no obvious way to predict which DNA vaccines will protect across mycobacterial species. However, the demonstration of cross-species protection in some cases supports the expectation that DNA vaccines should protect against multiple strains of a single mycobacterial species. Although M. avium strains can be distinguished serologically based on surface glycopeptidolipids, protein antigens from diverse strains have highly conserved sequences. For example, Swanson et al. (38) sequenced a 360-bp portion of the 65-kDa antigen gene in 16 M. avium strains belonging to 11 serovars and found identical amino acid sequences in all 16 strains. M. avium subsp. paratuberculosis also has an identical amino acid sequence in this segment of the 65-kDa antigen (11). Similarly, M. tuberculosis strains from diverse geographic locations have highly conserved amino acid sequences (37).
Limitations of protective immunity results.
Our results should be interpreted with several limitations in mind. First, the reductions in spleen weight and CFU which we observed were highly statistically significant, but they represent only modest levels of protection. Second, we do not know the duration of protective immunity, since all mice were challenged 3 weeks after the vaccination series. Most studies of DNA vaccines against mycobacteria have also used short intervals (1 to 4 weeks) between the completion of the vaccination series and the challenge (12, 28, 39, 43). However, Huygen et al. (23) demonstrated similar levels of protective immunity when mice were challenged at either 4 weeks or 10 weeks after completion of the vaccination series. Third, the level of protective immunity in our experiments decreased between 4 and 8 weeks after challenge for both BCG and DNA vaccines. Similar results were obtained with a mouse model of aerosol infection with M. tuberculosis (36). BCG-vaccinated mice had lower numbers of CFU from 3 to 7 weeks after infection, but there was no difference from 8 to 15 weeks. Fourth, each of the protective DNA vaccines was tested only once in a single animal model. Further experiments are needed to determine the reproducibility and generalizability of our results.
Subsets of T-cell responses.
Two subsets of CD4+ T cells (Th1 and Th2) are identified by their patterns of cytokine production. A variety of lines of evidence support the protective role of a Th1 response in mycobacterial infection. Mice lacking the IFN-γ gene and humans with mutations in the IFN-γ receptor are highly susceptible to mycobacterial infection (7, 30). Exogenous administration of Th1 cytokines in humans and mice enhances resistance to mycobacterial infection (20, 26, 30). Th1 cytokine production at the site of infection is associated with control of mycobacterial infection. Yamamura et al. (41) demonstrated that leprosy patients with low M. leprae burdens express Th1 cytokines in their skin lesions, whereas patients with high M. leprae burdens express Th2 cytokines.
Splenocytes cultured after mycobacterial DNA vaccination have normal basal IFN-γ production (23) and produce high levels of IFN-γ after stimulation with specific antigens (23, 28, 39, 43). We did not repeat these analyses but instead measured spleen cytokine production after M. avium challenge as a measure of T-cell responses at a site of infection. Based on the quantity of RT-PCR products, IFN-γ mRNA increased 4 weeks after infection and decreased toward the baseline at 8 weeks. However, the groups receiving DNA vaccines, BCG, or saline had no differences in IFN-γ production at either 4 or 8 weeks after challenge. Our results are consistent with previous studies using these methods (2, 15). Appelberg et al. (2) used RT-PCR to measure IFN-γ mRNA after intravenous M. avium challenge in BCG-vaccinated and unvaccinated mice. At baseline, both groups of mice had similar levels of splenic IFN-γ mRNA. At 2 and 14 days after M. avium challenge, these levels rose sharply in the BCG-vaccinated mice but were unchanged in the unvaccinated mice. At 28 and 60 days after challenge, IFN-γ levels were elevated to similar degrees in vaccinated and unvaccinated mice. Protective immunity generated by BCG was associated with enhanced IFN-γ production at early time points after infection (2). However, neither BCG nor the DNA vaccines in our study led to a long-term change in IFN-γ production at the site of infection.
Specific versus nonspecific vaccine effects.
The host response to mycobacterial infections includes macrophages and natural killer cells, components of the innate immune system (1, 4, 14, 19). Bacterial DNA (including plasmid DNA) has prominent immunostimulatory effects, including Th1 cytokine secretion, macrophage activation, and enhancement of natural killer cell activity (34). Further experiments are needed to determine whether nonspecific immune stimulation contributes to the protection generated by DNA vaccines. These experiments could include trials with a longer delay between vaccination and challenge, cytokine measurement after vaccination, and determination of macrophage and natural killer cell activation after vaccination.
Relevance to human M. avium infections.
The M. avium complex is an important cause of disseminated disease in AIDS patients. An effective DNA vaccine would provide a clinical benefit for these patients. Although pulmonary disease is caused by both M. avium and M. intracellulare, disseminated disease in AIDS patients is nearly always caused by M. avium (6, 21, 38). M. avium strains isolated from AIDS patients belong to several serovars based on surface glycopeptidolipids, but they share highly conserved protein antigens (38).
The work should not be extrapolated to support a clinical vaccine trial against M. avium infection. In our model, DNA vaccines reduced spleen weight and CFU, but all mice developed chronic M. avium infections. We used an intraperitoneal challenge, but natural M. avium infection is acquired by the gastrointestinal or respiratory route. We used EGFP expression to monitor the expression of mycobacterial antigens. Until more is known about immune responses to EGFP, it would not be an appropriate vaccine component for a clinical trial. Finally, mycobacterial heat shock proteins have close human homologues and may produce autoimmunity (17, 18, 25).
DNA vaccines produced modest levels of protective immunity against M. avium challenge. Future studies are needed to determine which immune responses correlate with vaccine-mediated protective immunity and to enhance the modest levels of protection observed. Promising approaches include the use of DNA vaccine mixtures expressing multiple antigens and the combination of DNA vaccines with immune adjuvants.
ACKNOWLEDGMENTS
We thank Elizabeth A. Talbot for assistance in the development of the cytokine RT-PCR assay. We thank Douglas B. Lowrie, Fouad A. K. El-Zaatari, Naoya Ohara, Vincent Escuyer, and the NIH AIDS Research and Reference Reagent Program for providing plasmids. We thank Kevin A. Nash and Clark B. Inderlied for providing a spleen homogenate from a mouse infected with M. avium MAC 101. We thank Christopher V. Nicchitta (Duke University Medical Center) for advice regarding the colocalization experiments and for providing antiserum to TRAPα. We thank Gerald A. Olson and the staff of the Durham VA Medical Center Animal Research Facility for their excellent care of the mice used in this research.
This work was supported by the Department of Veterans Affairs, by NIH grants AI-07392 (to M. Velaz-Faircloth) and GM-07184 (to A. L. Horstman), and by the North Carolina Biotechnology Center (to R. Frothingham).
REFERENCES
- 1.Andersen P. Host responses and antigens involved in protective immunity to Mycobacterium tuberculosis. Scand J Immunol. 1997;45:115–131. doi: 10.1046/j.1365-3083.1997.d01-380.x. [DOI] [PubMed] [Google Scholar]
- 2.Appelberg R, Castro A G, Pedrosa J, Silva R A, Orme I M, Minóprio P. Role of gamma interferon and tumor necrosis factor alpha during T-cell-independent and -dependent phases of Mycobacterium avium infection. Infect Immun. 1994;62:3962–3971. doi: 10.1128/iai.62.9.3962-3971.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett R J, Secore S L, Singer J T, Bodo M, Sharma K, Ricordi C. Long-term expression of a fluorescent reporter gene via direct injection of plasmid vector into mouse skeletal muscle: comparison of human creatine kinase and CMV promoter expression levels in vivo. Cell Transplant. 1996;5:411–419. doi: 10.1177/096368979600500308. [DOI] [PubMed] [Google Scholar]
- 4.Bermudez L E. Immunobiology of Mycobacterium avium infection. Eur J Clin Microbiol Infect Dis. 1994;13:1000–1006. doi: 10.1007/BF02111501. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. The role of BCG vaccine in the prevention and control of tuberculosis in the United States. Morbid Mortal Weekly Rep. 1996;45(RR-4):1–18. [PubMed] [Google Scholar]
- 6.Chin D P, Reingold A L, Stone E N, Vittinghoff E, Horsburgh C R, Jr, Simon E M, Yajko D M, Hadley W K, Ostroff S M, Hopewell P C. The impact of Mycobacterium avium complex bacteremia and its treatment on survival of AIDS patients—a prospective study. J Infect Dis. 1994;170:578–584. doi: 10.1093/infdis/170.3.578. [DOI] [PubMed] [Google Scholar]
- 7.Cooper A M, Dalton D K, Stewart T A, Griffin J P, Russell D G, Orme I M. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 9.De Wit L, de la Cuvellerie A, Ooms J, Content J. Nucleotide sequence of the 32 kDa-protein gene (antigen 85A) of Mycobacterium bovis BCG. Nucleic Acids Res. 1990;18:3995. doi: 10.1093/nar/18.13.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnelly J J, Ulmer J B, Shiver J W, Liu M A. DNA vaccines. Annu Rev Immunol. 1997;15:617–648. doi: 10.1146/annurev.immunol.15.1.617. [DOI] [PubMed] [Google Scholar]
- 11.El-Zaatari F A K, Naser S A, Lars E, Burch P E, Hachem C Y, Whipple D L, Graham D Y. Nucleotide sequence analysis and seroreactivities of the 65K heat shock protein from Mycobacterium paratuberculosis. Clin Diagn Lab Immunol. 1995;2:657–664. doi: 10.1128/cdli.2.6.657-664.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erb K J, Kirman J, Woodfield L, Wilson T, Collins D M, Watson J D, LeGros G. Identification of potential CD8+ T-cell epitopes of the 19 kDa and AhpC proteins from Mycobacterium tuberculosis. No evidence for CD8+ T-cell priming against the identified peptides after DNA-vaccination of mice. Vaccine. 1998;16:692–697. doi: 10.1016/s0264-410x(97)00253-3. [DOI] [PubMed] [Google Scholar]
- 13.Escuyer V, Haddad N, Frehel C, Berche P. Molecular characterization of a surface-exposed superoxide dismutase of Mycobacterium avium. Microb Pathog. 1996;20:41–55. doi: 10.1006/mpat.1996.0004. [DOI] [PubMed] [Google Scholar]
- 14.Fenton M J, Vermeulen M W. Immunopathology of tuberculosis: roles of macrophages and monocytes. Infect Immun. 1996;64:683–690. doi: 10.1128/iai.64.3.683-690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florido M, Appelberg R, Orme I M, Cooper A M. Evidence for a reduced chemokine response in the lungs of beige mice infected with Mycobacterium avium. Immunol. 1997;90:600–606. doi: 10.1046/j.1365-2567.1997.00206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gangadharam P R J. Beige mouse model for Mycobacterium avium complex disease. Antimicrob Agents Chemother. 1995;39:1647–1654. doi: 10.1128/aac.39.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruber R, Lederer S, Bechtel U, Lob S, Riethmuller G, Feucht H E. Increased antibody titers against mycobacterial heat-shock protein 65 in patients with vasculitis and arteriosclerosis. Int Arch Allergy Immunol. 1996;110:95–98. doi: 10.1159/000237318. [DOI] [PubMed] [Google Scholar]
- 18.Handley H H, Yu J, Yu D T, Singh B, Gupta R S, Vaughan J H. Autoantibodies to human heat shock protein hsp60 may be induced by Escherichia coli groEL. Clin Exp Immunol. 1996;103:429–435. doi: 10.1111/j.1365-2249.1996.tb08298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harshan K V, Gangadharam P R J. In vivo depletion of natural killer cell activity leads to enhanced multiplication of Mycobacterium avium complex in mice. Infect Immun. 1991;59:2818–2821. doi: 10.1128/iai.59.8.2818-2821.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland S M, Eisenstein E M, Kuhns D B, Turner M L, Fleisher T A, Strober W, Gallin J I. Treatment of refractory disseminated nontuberculous mycobacterial infection with interferon gamma. N Engl J Med. 1994;330:1348–1355. doi: 10.1056/NEJM199405123301904. [DOI] [PubMed] [Google Scholar]
- 21.Horsburgh C R., Jr Mycobacterium avium complex infection in the acquired immunodeficiency syndrome. N Engl J Med. 1991;324:1332–1338. doi: 10.1056/NEJM199105093241906. [DOI] [PubMed] [Google Scholar]
- 22.Hubbard R D, Flory C M, Collins F M. T-cell immune responses in Mycobacterium avium-infected mice. Infect Immun. 1992;60:150–153. doi: 10.1128/iai.60.1.150-153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huygen K, Content J, Denis O, Montgomery D L, Yawman A M, Deck R R, Dewitt C M, Orme I M, Baldwin S, D’Souza C, Drowart A, Lozes E, Vandenbussche P, Van Vooren J-P, Liu M A, Ulmer J B. Immunogenicity and protective efficacy of a tuberculosis DNA vaccine. Nat Med. 1996;2:893–898. doi: 10.1038/nm0896-893. [DOI] [PubMed] [Google Scholar]
- 24.Inderlied C B, Kemper C A, Bermudez L E M. The Mycobacterium avium complex. Clin Microbiol Rev. 1993;6:266–310. doi: 10.1128/cmr.6.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karopoulos C, Rowley M J, Handley C J, Strugnell R A. Antibody reactivity to mycobacterial 65 kDa heat shock protein: relevance to autoimmunity. J Autoimmun. 1995;8:235–248. doi: 10.1006/jaut.1995.0018. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi K, Yamazaki J, Kasama T, Katsura T, Kasahara K, Wolf S F, Shimamura T. Interleukin (IL)-12 deficiency in susceptible mice infected with Mycobacterium avium and amelioration of established infection by IL-12 replacement therapy. J Infect Dis. 1996;174:564–573. doi: 10.1093/infdis/174.3.564. [DOI] [PubMed] [Google Scholar]
- 27.Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981;9:5233–5262. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozes E, Huygen K, Content J, Denis O, Montgomery D L, Yawman A M, Vandenbussche P, Van Vooren J-P, Drowart A, Ulmer J B, Liu M A. Immunogenicity and efficacy of a tuberculosis DNA vaccine encoding the components of the secreted antigen 85 complex. Vaccine. 1997;15:830–833. doi: 10.1016/s0264-410x(96)00274-5. [DOI] [PubMed] [Google Scholar]
- 29.Migliaccio G, Nicchitta C V, Blobel G. The signal sequence receptor, unlike the signal recognition particle receptor, is not essential for protein translocation. J Cell Biol. 1992;117:15–25. doi: 10.1083/jcb.117.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newport M J, Huxley C M, Huston S, Hawrylowicz C M, Oostra B A, Williamson R, Levin M. A mutation in the interferon-γ-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 31.Ohara N, Matsuo K, Yamaguchi R, Yamazaki A, Tasaka H, Yamada T. Cloning and sequencing of the gene for alpha antigen from Mycobacterium avium and mapping of B-cell epitopes. Infect Immun. 1993;61:1173–1179. doi: 10.1128/iai.61.4.1173-1179.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohara N, Ohara-Wada N, Kitaura H, Nishiyama T, Matsumoto S, Yamada T. Analysis of the genes encoding the antigen 85 complex and MPT51 from Mycobacterium avium. Infect Immun. 1997;65:3680–3685. doi: 10.1128/iai.65.9.3680-3685.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orme I M, Collins F M. Prophylactic effect in mice of BCG vaccination against nontuberculous mycobacterial infections. Tubercle. 1985;66:117–120. doi: 10.1016/0041-3879(85)90076-5. [DOI] [PubMed] [Google Scholar]
- 34.Pisetsky D S. Immune activation by bacterial DNA: a new genetic code. Immunity. 1996;5:1996. doi: 10.1016/s1074-7613(00)80256-3. [DOI] [PubMed] [Google Scholar]
- 35.Platzer C, Richter G, Uberla K, Muller W, Blocker H, Diamantstein T, Blankenstein T. Analysis of cytokine mRNA levels in interleukin-4-transgenic mice by quantitative polymerase chain reaction. Eur J Immunol. 1992;22:1179–1184. doi: 10.1002/eji.1830220511. [DOI] [PubMed] [Google Scholar]
- 36.Schell R F, Ealey W F, Harding G E, Smith D W. The influence of vaccination on the course of experimental airborne tuberculosis in mice. J Reticuloendothel Soc. 1974;16:131–138. [PubMed] [Google Scholar]
- 37.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanson D S, Kapur V, Stockbauer K, Pan X, Frothingham R, Musser J M. Subspecific differentiation of Mycobacterium avium complex strains by automated sequencing of a region of the gene (hsp65) encoding a 65-kilodalton heat shock protein. Int J Syst Bacteriol. 1997;47:414–419. doi: 10.1099/00207713-47-2-414. [DOI] [PubMed] [Google Scholar]
- 39.Tascon R E, Colston M J, Ragno S, Stavropoulos E, Gregory D, Lowrie D B. Vaccination against tuberculosis by DNA injection. Nat Med. 1996;2:888–892. doi: 10.1038/nm0896-888. [DOI] [PubMed] [Google Scholar]
- 40.Weeratna R, Millan C L B, Krieg A M, Davis H L. Reduction of antigen expression from DNA vaccines by coadministered oligodeoxynucleotides. Antisense Nucleic Acid Drug Dev. 1998;8:351–356. doi: 10.1089/oli.1.1998.8.351. [DOI] [PubMed] [Google Scholar]
- 41.Yamamura M, Uyemura K, Deans R J, Weinberg K, Rea T H, Bloom B R, Modlin R L. Defining protective responses to pathogens: cytokine profiles in leprosy lesions. Science. 1991;254:277–279. doi: 10.1126/science.254.5029.277. [DOI] [PubMed] [Google Scholar]
- 42.Young D B, Kaufmann S H, Hermans P W, Thole J E. Mycobacterial protein antigens: a compilation. Mol Microbiol. 1992;6:133–145. doi: 10.1111/j.1365-2958.1992.tb01994.x. [DOI] [PubMed] [Google Scholar]
- 43.Zhu X, Venkataprasad N, Thangaraj H S, Hill M, Singh M, Ivanyi J, Vordermeier H M. Functions and specificity of T cells following nucleic acid vaccination of mice against Mycobacterium tuberculosis infection. J Immunol. 1997;158:5921–5926. [PubMed] [Google Scholar]