Abstract
Lingxian white goose (LXW) is a goose breed indigenous to China that is famous for its meat quality and fast growth. However, the genomic evidence underlying such excellent breeding characteristics remains poorly understood. Therefore, we performed whole-genome resequencing of 141 geese from 3 indigenous breeds to scan for selection signatures and detect genomic regions related to breed features of LXW. We identified 5 reproduction-related genes (SYNE1, ESR1, NRIP1, CCDC170, and ARMT1) in highly differentiated regions and 11 notable genes in 26 overlapping windows, some of which are responsible for meat quality (DHX15), growth traits (LDB2, SLIT2, and RBPJ), reproduction (KCNIP4), and unique immunity traits (DHX15 and SLIT2). These findings provide insights into the genetic characteristics of LXW and identify genes affecting important traits in LXW, which extends the genetic resources and basis for facilitating genetic improvement in domestic geese breeds.
Key words: poultry, Lingxian white goose, selection signature, breeding
INTRODUCTION
Goose is an economically important agricultural animal. As one of the most important poultry, geese have been domesticated by humans for approximately 7,000 yr (Eda et al., 2022), which has led to a series of behavioral, morphological, and physiological changes. Artificial selection has been performed to improve agriculturally important traits, resulting in a variety of local breeds with different appearances and attributes. Among these geese, Lingxian white goose (LXW) is native to Yanling County, Hunan Province. It is a small, domesticated geese breed in China, with white feathers, orange beaks, and webs. Although small in body size, it shows rapid growth, precocious maturity and fattening, and good meat quality, as well as many excellent characteristics, such as resistance to rough feeding, strong adaptability, and disease resistance (Chen et al., 2011).
These breed characteristics are the result of natural and artificial selection pressures, and selective sweeps usually leave a footprint on the genome, leading to long haplotypes, high-frequency derived alleles, and highly differentiated alleles (Grossman et al., 2010). Identification of selection signatures at the whole-genome level is possible through the development of high-throughput sequencing technology. Recently, various methods have been applied to explore population selection signals. For example, Zhou et al. (2018) used F-statistics (Fst) and nucleotide diversity (π) methods to determine that MITF regulates the formation of white plumage in Pekin ducks. Jeong et al. (2015) employed cross-population-extended haplotype homozygosity (XP-EHH) and cross-population composite likelihood ratio test approaches to detect multiple genes that contribute to intramuscular fat in Berkshire pigs, which might affect the quality of pork. Bortoluzzi et al. (2020) found that the PITX1 and TBX5 genes are strongly associated with foot feathering in domestic chickens using a combination of genome-wide association studies and Z-transformed heterozygosity approaches. Although these selective sweeps have been widely used in breeding research and have accelerated the progress of genetic improvement, research on LXW is still lacking.
The physiological characteristics of LXW, an excellent goose breed in China, have received extensive attention. However, the genetic architecture of these important traits remains largely unknown, which may prevent full utilization of their genetic potential. In addition, no national conservation farm has been established for LXW, and the lack of effective protection will cause loss of genetic diversity and unique genomic information. Here, we sequenced 45 LXW individuals and compared them with 96 other Chinese geese (Guangfeng white goose [GFW] and Fengcheng gray goose [FCG]) to search for potential genomic evidence related to the breeding features of LXW. As efficient methods for detecting selection signals between 2 different groups, Fst (Weir and Cockerham, 1984), based on allele frequency, and XP-EHH (Sabeti et al., 2007), based on linkage disequilibrium, were used to detect selection effects. We aimed to identify genomic variants specific to LXW and a suite of promising genes that have undergone positive selection during domestication and breeding processes. Genetic research on breed characteristics will contribute to further genetic and breeding research on LXW and other geese breeds.
MATERIALS AND METHODS
Experimental Animals
A total of 141 geese from 3 Chinese indigenous geese breeds were used in this study: Lingxian white goose (LXW; n = 45), Fengcheng gray goose (FCG; n = 50), and Guangfeng white goose (GFW; n = 46). All samples were collected from Jiangxi Province, and LXW was collected from Lianhua County, Jiangxi Province. Whole blood samples were collected from 141 geese from veins under the wings. All experimental procedures were approved by the Animal Ethics Committee of Jiangxi Science and Technology Normal University and strictly followed the Guidelines of Animal Welfare, China.
Whole-Genome Resequencing
We used a standard phenol-chloroform extraction protocol to extract genomic DNA from whole blood of 141 geese and tested the integrity and purity of DNA by agarose gel electrophoresis and the A260/280 ratio (1.8–2.0), respectively. All qualified samples were used for library construction (Paired-end, 2 150 bp) and whole-genome resequencing was performed using the Illumina NovaSeq 6000 platform at Novogene (Beijing, China). The average sequencing depth of geese in this study was approximately 10-fold.
Genome-Wide Variant Calling and Annotation
To minimize artificial bias in the sequencing process, we applied Trimmomatic v0.32 (Bolger et al., 2014) to discard adaptors and low-quality reads (Q < 30). The clean reads were then mapped onto the Xingguo gray goose (XGG) reference genome (accession number: GWHBAAW00000000) using Burrows-Wheeler Aligner v0.7.17 (Li and Durbin, 2009) software with default parameters. The mapping results were further converted into BAM format files and sorted by SAMtools v1.10 (Li et al., 2009). Next, SNPs were detected using a custom version of Sentieon v201711.03 (Kendig et al., 2019) DNAseq pipeline following the best practice algorithms of GATK. LocusCollectora and Realigner functions from Sentieon were used to delete PCR duplicates and calibrate them according to the quality score. Finally, GATK v4.0.12 (McKenna et al., 2010) and SAMtools were employed to recalibrate the base quality for further analyses. After data filtering (minor allele frequency [MAF] > 1% and call rate > 90%), 10,459,469 SNPs were detected in all geese, and LXW yielded 9,756,715 SNPs (MAF > 1% and call rate > 90%). These variants were annotated using SnpEff v5.0 (Cingolani et al., 2012) software.
Population Structure and Genetic Diversity
After filtering, 10,459,469 SNPs were used for the phylogenetic and population structure analyses. A neighbor-joining (NJ) tree was constructed using PHYLIP v3.69 (Retief, 2000) based on the identical-by-state matrix calculated by PLINK v1.9 (Chang et al., 2015) with the parameter “–distance-matrix”, and then visualized via Figtree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). Principal component analysis (PCA) was performed with the parameter of “–make-grm” via GCTA v1.92 (Yang et al., 2011), and the first 2 principal components (PCs) were plotted using in-house R scripts. Admixture v1.3.0 (Alexander et al., 2009) was used to estimate the admixture proportions (from K = 2 to 3). Five parameters were calculated to evaluate the genetic diversity of the geese: the number of SNPs (NSNPs), runs of homozygosity (ROH), inbreeding coefficient (F), linkage disequilibrium (LD), and nucleotide diversity (π). ROH was estimated using the autosomal SNPs of each individual using PLINK. F was computed using the command “plink –het”. LD was quantified using the squared correlation coefficient () between pairs of SNPs as implemented in PLINK with the parameters “–r2 –ld-window-kb 1000 –ld-window-r2 0”. π was calculated using VCFtools v0.1.16 (Danecek et al., 2011) under 10 kb sliding windows.
Detection of Selective Signatures
To identify the positive signatures in LXW against other breeds (GFW and FCG), 2 statistical approaches (Fst and XP-EHH) were applied to improve detection efficacy. Only SNPs in the autosomes were preserved for selective sweeps. First, we estimated the pairwise Fst values in the groups of LXW and other geese using VCFtools v0.1.16 (Danecek et al., 2011), and the average Fst score was used as the summary statistics for each 10 kb non-overlapping window. Next, LXW was used as the experimental group and other geese as the reference to carry out the XP-EHH test with 10 kb non-overlapping sliding windows. The XP-EHH scores were then standardized across the entire genome. The results of the XP-EHH are directional, with a positive score indicating that the test population is highly likely to have experienced positive selection, whereas a negative score indicates that the reference population has experienced consistent selection during the evolutionary process.
For these two approaches, windows with less than 10 SNPs were excluded, and the top 1% was set as the significance threshold. To reduce the frequency of false-positive signals, the potential candidate regions detected by these 2 methods were considered to be significantly selective regions, and variants located within as more confident signals. Gene Ontology (GO) enrichment analysis was performed for genes overlapping with selective windows using the online Metascape (Zhou et al., 2019).
Haplotype Analysis and Functional Annotation of Candidate Genes
Genecard websites and literature from public databases (for example, NCBI) were used to further investigate the possible biological functions of candidate genes. The π and heterozygosity (Hp) in the target regions were calculated over 10 kb windows via VCFtools (Danecek et al., 2011) and PLINK (Chang et al., 2015), respectively. To further investigate the significantly positively selected genes associated with the breed features of LXW, the resequencing data of 300 individuals from 10 populations, including 89 wild geese and 211 domestic geese (Table S1), in our own bird database (unpublished) were used for haplotype analysis. The details were as follows: 1) First, XGG was used as a reference genome to detect SNPs and InDels (insertions and deletions of less than 50 bp) in candidate regions of 300 geese; 2) Next, we used the fastPhase function with 1,000 iterations in Beagle v5.1 (Browning et al., 2018) to infer haplotypes of candidate regions, and the haplotype network was constructed using pegas v1.1 (Paradis, 2010) in an R package. Additionally, the statistics for absolute relative allele frequencies (absRAFdif) within the tested populations were computed to verify the target regions, and SnpEff v5.0 (Cingolani et al., 2012) was used to predict the potential effects of SNPs.
RESULTS
Identification of Variants in LXW
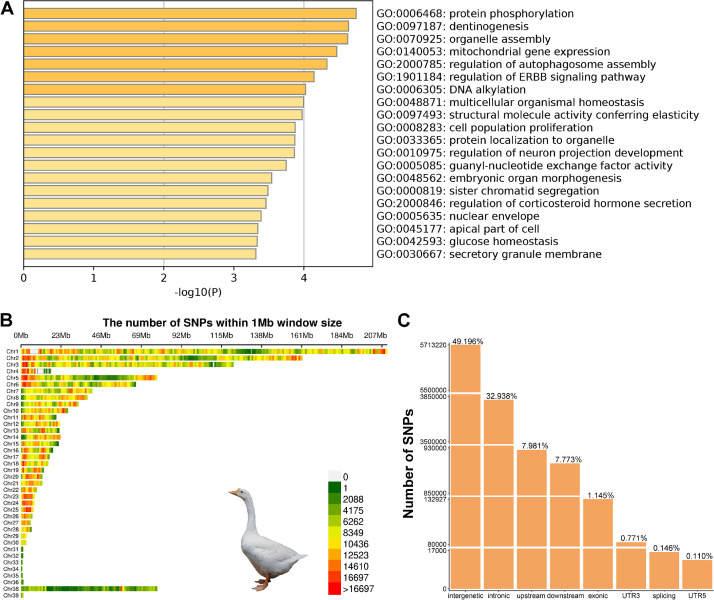
In the present study, 141 individuals, including 45 LXW, 50 FCG, and 46 GFW, were selected for whole-genome resequencing with an average depth of 10 ×. Aligning the sequencing data to the XGG reference genome (GWHBAAW00000000), we detected 10,459,469 SNPs (MAF > 1% and call rate > 90%). Most of these SNPs were located in intergenic regions (6,124,303; 49.196%), followed by 32.936% (4,100,134) of SNPs located in introns, 7.98% in upstream regions, 7.772% in downstream regions, and 1.149% in exons. From these SNPs, we found that 62,139 were specific to LXW (Table S2), with 415 missense mutations covering 278 protein-coding genes. GO enrichment of the SNPs indicated that these genes were involved in protein phosphorylation, dentinogenesis, organelle assembly, and embryonic organ morphogenesis (Figure 1A).
Figure 1.
Identification and annotation of SNPs in LXW population. (A) GO enrichment of the 278 protein coding genes where LXW-specific SNPs are located. (B) The SNPs density across the whole genome was estimated in each 1 Mb genome block. (C) The annotation of genome-wide SNPs according to SnpEff.
Additionally, we independently extracted the SNPs of LXW and obtained 9,756,715 SNPs (MAF > 1%, call rate > 90%; Table S3). Among these SNPs, substitution mutations G > A and C > T occupied the largest number, followed by T > C and A > G (Table S4). The SNP density across the whole genome was presented in each 1 Mb genome block, among which Chr 23 and 24 showed the highest SNP density, and Chr 32 the lowest SNP density (Figure 1B). We found that the largest number of SNPs was enriched in the intergenic region (49.196%), followed by introns (32.938%), upstream regions (7.981%), downstream regions (7.773%), exons (1.145%), untranslated regions (0.722%), and splice site regions (0.146%) (Figure 1C). With respect to protein-coding genes, we identified 43,245 missense variants, 89,016 synonymous variants, 85 initiator-codon variants, 641 stop-lost variants, 113 start-lost variants, and 1,545 stop-gained variants (Table S5). These potential functional SNPs might be related to the altered traits in geese and would offer valuable genetic resources for in-depth research on the selective signatures of the LXW population.
Population Structure and Genetic Diversity of LXW
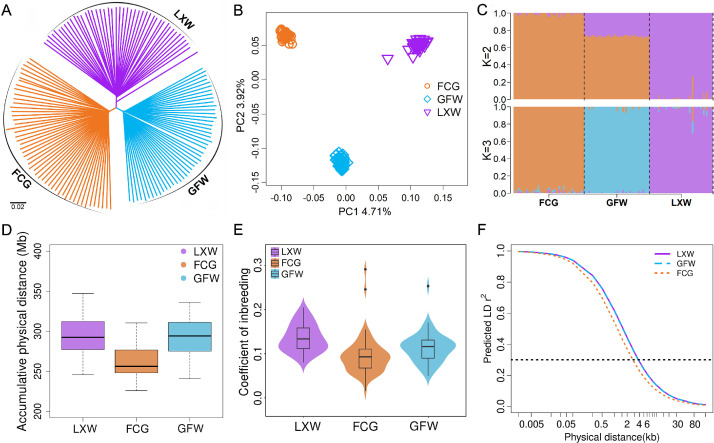
To infer the genetic structure of LXW, we used genome-wide variants (10,459,469 SNPs) to construct a phylogenetic tree and performed principal component analysis (PCA). All individuals were clustered together according to their breed, indicating that the individuals were genetically representative of the breeds (Figure 2A). Principal components 1 (PC1, 4.71% of the total variation) and 2 (PC2, 3.92% of the total variation) also clearly separated the samples into 3 distinct clusters corresponding to FCG, GFW, and LXW (Figure 2B). Additional ADMIXTURE analysis was conducted to estimate the admixture proportions by assuming K ancestral populations (Figure 2C). When K = 2, a clear division was observed between LXW and other geese breeds. When K = 3, there was a split between each breed that matched the phylogenetic tree and the PCA results. Notably, some individuals of LXW had a higher proportion of co-ancestry with others (Figure 2C), possibly due to complex introgression patterns among these geese breeds. Detailed PCA results for LXW also revealed the presence of several subgroups within the LXW population (Figure S1), which may require scientifically effective measures to maintain germplasm uniformity.
Figure 2.
Phylogeny and population genetics of three geese breeds. (A) The neighbor-joining phylogenetic tree of 141 geese was constructed based on 10,459,469 SNPs. (B) Two-dimensional principal component analysis (PCA) plot of three geese breeds. (C) Genetic structure of three geese breeds according to ADMIXTURE, with K = 2–3. (D) The total length of runs of homozygosity (ROH) of each breed. (E) The inbreeding coefficient (F) of each breed. (F) Linkage disequilibrium (LD) analysis for three geese breeds. Abbreviations: FCG, Fengcheng gray goose; GFW, Guangfeng white goose; LXW, Lingxian white goose.
We then estimated 5 parameters to evaluate the genetic diversity of LXW. Statistical results showed that the number of SNPs (NSNP) in LXW (9,634,706) was much lower than that in GFW (9,677,929) and FCG (10,087,193; Table S6), implying lower genetic diversity. As expected, low-diversity LXW showed high coverage by ROH (Figure 2D). Among these breeds, we also noticed that LXW (π = 2.51 × 10−3) had lower nucleotide diversity than GFW (π = 2.57 × 10−3) and FCG (π = 2.68 × 10−3), with the highest level of inbreeding (F = 0.138; Table S6 and Figure 2E). LD analysis also showed that the LXW genomes have relatively long LD distances () and a slower decay of the pairwise correlation coefficient () than the others (Figure 2F and Table S6).
Signatures of Selection in LXW
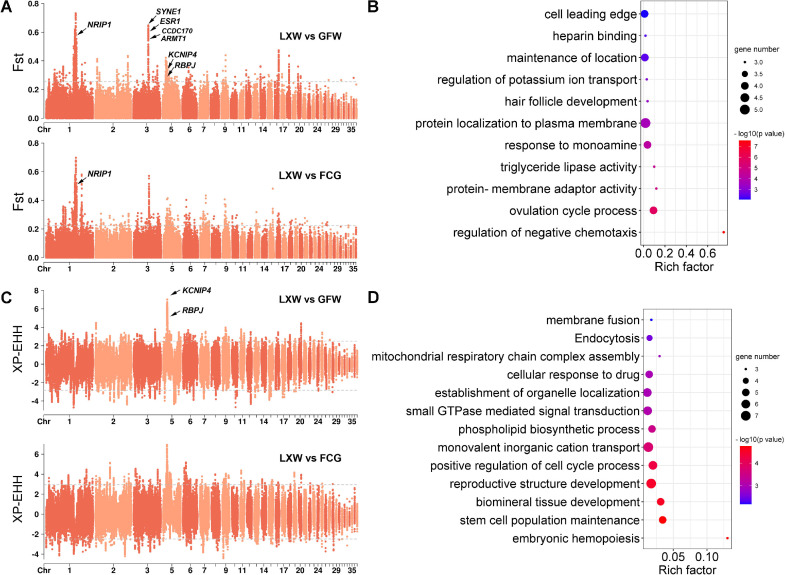
According to the population-scale genetic differences between LXW and other geese breeds, pairwise Fst was estimated to search for genomic regions where allele frequencies changed. We detected 1,059 and 861 regions (top 1% cut-off) with potential selection signatures of LXW against GFW (average Fst = 0.055) and FCG (average Fst = 0.057), respectively (Figure 3A). In the comparison of LXW and GFW, we identified 28 highly differentiated regions (Fst > 0.5) containing five reproduction-related genes, SYNE1, ESR1, NRIP1, CCDC170, and ARMT1 (Figure 3A and Table S7). Comparison of LXW and FCG also found that the region where NRIP1 gene is located had the strongest selective signal (Fst = 0.55; Figure 3A and Table S8). A total of 57 overlapping genes were found in the 2 comparisons, showing categories such as “ovulation cycle process” and “triglyceride catabolic process”, which are considered to be potentially involved in the response to reproductive activities and fat deposition processes in geese (Figure 3B). The XP-EHH test in LXW against GFW identified 997 positively selected regions (top 1% cut-off) harboring 154 genes, with the most remarkable signal in the KCNIP4 gene (Figure 3C and Table S9). A total of 998 potential regions (top 1% cut-off) harboring 133 genes were positively selected in LXW against FCG (Figure 3C and Table S10). The common selective regions of LXW against GFW and FCG contained 66 candidate genes enriched in GO terms associated with “embryonic hemopoiesis”, “reproductive structure development”, and “phospholipid biosynthetic process” (Figure 3D), which might also affect the reproductive and fattening characteristics of LXW.
Figure 3.
Genomic landscape of positive selection signatures by two statistical tests (XP-EHH and Fst). (A) Manhattan plot of Fst analyses with LXW against GFW or FCG. (B) Significant enriched GO terms (top 11) of the sharing common genes in Fst statistic. The y-axis in plot denotes –log10(P value). (C) Manhattan plot of XP-EHH analyses with LXW against GFW or FCG. The dashed line refers to the threshold level of top 1%. Each dot denotes the mean values in the 10-kb non-overlapping genomic region. (D) Significant enriched GO terms (top 13) of the sharing common genes in XP-EHH statistic. The y-axis in plot denotes –log10(P value).
Subsequently, we combined the candidate genes detected using the 2 approaches described above to obtain more confident selective signals. A total of 38 significantly selected genes were detected in LXW (top 1% cut-off for XP-EHH and Fst; Table S11), which were involved in “histone modification”, “cellular response to hormone stimulus”, and “smooth muscle contraction” (Figure S2). We checked the distribution of the selected regions within the genomic windows and found 11 overlapping genes (CCDC149, DHX15, KCNIP4, LDB2, NSG1, PACRGL, RBPJ, RPS6KA5, SEPSECS, SLIT2, and ZBTB49; Table 1). As expected, the haplotypes of these 11 genes also showed remarkable differences between LXW and other geese breeds as well as wild populations (Figure S3). The published literature was surveyed to further investigate possible biological functions (Table 1). DHX15 (Pattabhi et al., 2019) and SLIT2 (Altay et al., 2007) have been found to be involved in immunity; KCNIP4 (Fan et al., 2017), DHX15 (Xia et al., 2016), LDB2 (Wei et al., 2020), and NSG1 (Lee et al., 2016) were associated with the economic traits of livestock and poultry, such as reproductive capacity, meat quality, growth, and carcass traits; and RBPJ and SLIT2 were considered as potential candidate genes for chicken bone growth and development (Li et al., 2021).
Table 1.
The 11 positively selected genes were identified and shared in two comparisons.
| Chr | Start | End | Gene | Summary of gene function |
|---|---|---|---|---|
| 5 | 19,065,083 | 19,114,966 | CCDC149 | Beak length trait (Huang et al., 2022) |
| 5 | 18,966,684 | 19,013,336 | DHX15 | Meat tenderness (Xia et al., 2016) and immunity traits (Pattabhi et al., 2019) |
| 5 | 17,675,084 | 17,722,057 | KCNIP4 | Reproductive traits (Fan et al., 2017) |
| 5 | 16,093,124 | 16,312,302 | LDB2 | Body weight (Gu et al., 2011) |
| 5 | 13,660,999 | 13,680,953 | NSG1 | Milk traits (Lee et al., 2016) |
| 5 | 17,660,787 | 17,673,411 | PACRGL | Growth and carcass traits (Liu et al., 2022) |
| 5 | 19,477,656 | 19,537,056 | RBPJ | Bone traits (Li et al., 2021) |
| 5 | 19,177,151 | 19,203,929 | SEPSECS | Neurological disorders (Puppala et al., 2016) |
| 5 | 17,373,820 | 17,376,979 | SLIT2 | Immunity (Altay et al., 2007) and bone traits (Li et al., 2021) |
| 5 | 13,702,820 | 13,721,308 | ZBTB49 | - |
| 6 | 17,361,914 | 17,414,185 | RPS6KA5 | - |
Note: Pairwise test (XP-EHH and Fst) analyses were performed with LXW tested against GWF and FCG.
Abbreviations: FCG, Fengcheng gray goose; GFW, Guangfeng white goose; LXW, Lingxian white goose.
Signatures of Selection in Two Well-Characterized Growth Genes
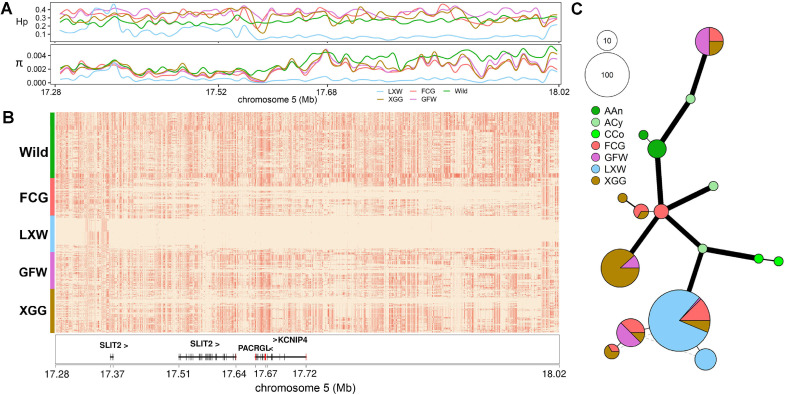
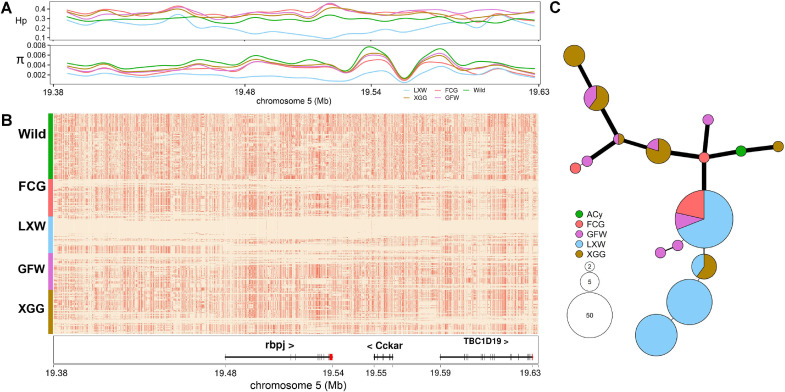
LXW is small in body size and is characterized by early maturity. KCNIP4 (Fan et al., 2017) and RBPJ (Li et al., 2021) have been identified as targets for the selection of reproductive and bone traits in poultry, respectively. We calculated the π and Hp values for each group in the region containing KCNIP4. The results showed that LXW was strongly selected in this gene region (Figure 4A), which was verified in haplotype heatmap analysis as having nearly fixed haplotypes (Figure 4B). A haplotype network of KCNIP4 showed that haplotypes in domestic geese were completely different from those in wild geese. Among these domestic geese, LXW had the least haplotype type (n = 2) and the smallest difference (nucleotide difference = 1) within the different haplotypes (Figure 4C and Figure S4). We also noted that 5 loci had extreme differences in absolute relative allele frequencies (absRAFdif > 0.9) between LXW and wild geese in KCNIP4. Two of these loci were found only in domestic geese, and the frequency in LXW was remarkably higher than that in other domestic geese breeds (absRAFdif > 0.5; Table S12). Evidence of strong selection for LXW was also observed in the RBPJ region (Figure 5A). The haplotypes of RBPJ were constructed and four main haplotypes were found in LXW. The most common haplotypes appeared 42 times in the 3 geese breeds, with 29 times in LXW, 9 times in FCG, and 4 times in GFW (Figures 5B and 5C). Furthermore, we noted that 2 main haplotypes were only found in LXW but were absent in GFW and FCG. Given the function of these genes, we speculated that this may explain the differences in body size and reproductive performance among distinct domestic geese breeds.
Figure 4.
Population differentiation of KCNIP4 gene (chr5: 17,675,084–17,722,057 bp) between LXW and other geese. (A) The Nucleotide diversity (π) and heterozygosity (Hp) values are plotted surrounding KCNIP4 gene. (B) Haplotypes comparison between LXW and other geese. Major and minor alleles are labeled in beige and coral, respectively. (C) The haplotype network diagram of KCNIP4 gene. Each circle denotes a haplotype, the circle size and haplotype frequency are proportional, and the width and length of the line represent the difference between haplotypes. Abbreviations: FCG, Fengcheng gray goose; GFW, Guangfeng white goose; LXW, Lingxian white goose; XGG, Xingguo gray goose; Wild, wild species (ACy: Anser cygnoides, AAn: Anser anser, AAl: Anser albifrons, AEr: Anser erythropus, AFa: Anser fabalis, CCo: Cygnus columbianus).
Figure 5.
Population differentiation of RBPJ gene (chr5: 19,477,656–19,537,056 bp) between LXW and other geese. (A) The Nucleotide diversity (π) and heterozygosity (Hp) values are plotted surrounding RBPJ gene. (B) Haplotypes comparison between LXW and other geese. Major and minor alleles are labeled in beige and coral, respectively. (C) The haplotype network diagram of RBPJ gene. Each circle denotes a haplotype, the circle size and haplotype frequency are proportional, and the width and length of the line represent the difference between haplotypes. Abbreviations: FCG, Fengcheng gray goose; GFW, Guangfeng white goose; LXW, Lingxian white goose; XGG, Xingguo gray goose; Wild, wild species (ACy: Anser cygnoides, AAn: Anser anser, AAl: Anser albifrons, AEr: Anser erythropus, AFa: Anser fabalis, CCo: Cygnus columbianus).
DISCUSSION
LXW is an invaluable component of the genetic resources of geese in China. It is recognized for its small body size, rapid growth, strong disease resistance, tender meat, and excellent reproductive performance (Chen et al., 2011). Dissection of the genetic architecture of these traits is conducive to the genetic improvement of domestic geese, conservation of germplasm resources, and development of the local economy. Here, whole-genome resequencing data with more comprehensive and reliable information was used for further selective sweeps. We adopted a feasible strategy that combines the pairwise tests of Fst, which depend on allele frequency, and XP-EHH based on linkage disequilibrium patterns across genomes to explore candidate genes associated with breed characteristics at the genome-wide level. These 2 approaches are necessary and helpful for revealing the genetic mechanisms of phenotypic diversity.
Selection can increase beneficial allele frequency over time and fix it within a population. Our 2 comparative analyses of Fst statistics (LXW vs. GFW and LXW vs. FCG) identified extremely significant genetic differentiation (Fst > 0.5) of LXW in some regions that contained 5 important functional genes. Of these, NRIP1 has been reported to be essential for ovulation and female fertility (White et al., 2000). ESR1 is an estrogen receptor that participates in ovarian follicular development and has a potential effect on egg production traits in laying hens (Niu et al., 2017) and litter size in pigs (Munoz et al., 2007). CCDC170 and SYNE1 are involved in sex steroid hormone pathways (Sapkota et al., 2017). ARMT1 is highly homologous to the dominantly expressed genes in Drosophila testes (Tang et al., 2018). All these genes and the pathways involved directly or indirectly play an important role in reproductive function, which provides strong evidence for the excellent reproductive performance of LXW.
Genomic regions identified by Fst and XP-EHH (top 1% cut-off) identified only 11 common genes that potentially harbored selective signals. One possible explanation for this is that these 2 methods are based on different principles. Fst is a detection method based on population differentiation, which is mainly affected by migration and genetic drift (Weir and Cockerham, 1984). However, when the mutation is subjected to artificial or natural selection, its frequency increases to improve the level of population differentiation, and the Fst values also increase. Therefore, the Fst method is sensitive to signature selection between different populations. The XP-EHH test aims to assess haplotype differences between 2 populations and is more effective for some artificially selected signatures, considering the change in haplotypes (Simonson et al., 2010). Combining the results of 2 different statistical analyses provides more powerful information than the results of a single test. Furthermore, stringent screening criteria were used to obtain high-quality data; therefore, less stringent criteria may yield better results. Despite these limitations, the results partially explain the selective pressures that LXW underwent during evolution.
Among the 11 commonly selected genes, we found that DHX15 (Pattabhi et al., 2019) and SLIT2 (Altay et al., 2007) play roles in immune regulation. The cell-intrinsic innate immune response serves as the first line of defense against viral infection, and RNA helicases play an important role in the response to microbial infection. DHX15 is a pre-mRNA processing factor that participates in the disassembly of spliceosomes after the release of mature mRNA. In human cells, DHX15 is a co-receptor required for innate immune responses to regulate and control RNA viral infection (Pattabhi et al., 2019). SLIT2 has been previously identified as a ligand for the Robo family of immunoglobulin receptors. A study in mice found that SLIT2 reduces leukocyte migration to cortical venules after global cerebral ischemia (Altay et al., 2007). Accordingly, both genes may be of great importance for the innate immunity of LXW.
We also identified a set of genes related to economic traits. For example, NSG1 is associated with milk-related traits (milk yield, fat, and protein) of Holstein (Lee et al., 2016). DHX15 plays an essential role in the shear force measured by the tenderness of Simmental beef cattle (Xia et al., 2016); thus, it may also affect the meat tenderness of LXW. Compared to the GFW and FCG populations, LXW displayed a faster growth rate and excellent reproductive performance. Our findings also illustrate these differences. KCNIP4 has an important effect on egg weight in 300-day-old Jinghai Yellow chickens (Fan et al., 2017). We noted that the haplotype in KCNIP4 had absolute differences between wild and domestic geese, and the genetic diversity of LXW around this gene was significantly reduced in comparison to other domestic geese. Given that the age of LXW at the first egg was earlier than that of the other 2 varieties (Chen et al., 2011), we speculated that the gene may play a critical role in the reproductive traits of LXW. These findings further confirmed that breeding LXW in the direction of egg and meat production might have achieved some results, which would lay a foundation for future research on the genomic characteristics of LXW and other poultry to develop improved varieties with excellent production performance.
Several genes have been suggested to be associated with growth traits. For example, the LDB2 gene was found to be related to the body weight of chickens at wk 7 to 12 and average daily gain at wk 6 to 12 (Gu et al., 2011) and has been repeatedly reported to be associated with growth and carcass traits in chickens (Liu et al., 2013; Zhang et al., 2021). In addition, SLIT2 participates in bone metabolism and inhibits osteoclastogenesis and bone resorption in vitro (Park et al., 2019). RBPJ is a key regulator of osteoclastogenesis and bone resorption and plays an important upstream negative regulatory role in osteoclast formation (Li et al., 2014). A recent genome-wide study suggested that SLIT2 and RBPJ may be predominantly selected for bone traits in chickens (Li et al., 2021). Bones and the skeletal system play an important role in supporting and protecting the animal body and are key factors that affect the body size of animals. LXW is known for its small body size, whereas GFW, FCG, and XGG are medium-sized goose breeds. The strong selection of bone-related genes may explain, to a certain extent, the genetic basis of the difference in body size between LXW and other geese.
CONCLUSIONS
In summary, natural and artificial selection have remarkably shaped variation in the goose genome during breed formation. We integrated Fst and XP-EHH tests to detect alleles that rapidly increased to a high frequency in the LXW population. A series of potential genes associated with meat quality, reproductive performance, and other functions, which could have strong impacts on the breed characteristics of LXW, were identified. To our knowledge, this study is one of the few to identify the selection signatures in LXW and could help better understand the selection mechanisms and breeding practices of LXW and other goose breeds.
ACKNOWLEDGMENTS
This study is supported by the National Natural Science Foundation of China (31860622 and 32060735) and the technology research project of Jiangxi Provincial Education Department (GJJ180595)
Data availability statement: The Xingguo gray goose genome used in this study has been deposited in the Genome Warehouse in BIG Data Center (https://bigd.big.ac.cn/gwh/) under accession number GWHBAAW00000000. The genome resequencing data has been deposited in the NCBI database as BioProject No. PRJNA678815.
DISCLOSURES
The authors declare that they have no competing financial interests.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.102269.
Appendix. Supplementary materials
REFERENCES
- Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altay T., McLaughlin B., Wu J.Y., Park T.S., Gidday J.M. Slit modulates cerebrovascular inflammation and mediates neuroprotection against global cerebral ischemia. Exp. Neurol. 2007;207:186–194. doi: 10.1016/j.expneurol.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortoluzzi C., Megens H.J., Bosse M., Derks M.F.L., Dibbits B., Laport K., Weigend S., Groenen M.A.M., Crooijmans R. Parallel genetic origin of foot feathering in birds. Mol. Biol. Evol. 2020;37:2465–2476. doi: 10.1093/molbev/msaa092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning B.L., Zhou Y., Browning S.R. A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 2018;103:338–348. doi: 10.1016/j.ajhg.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.W., Yang N., Wang G.Y., Wang Z.Y., Wang K.H., Wang J.Y., Wang J.W., Wen J., Lu L.Z., Li H.F., Zhang L., Zhang X.Y., Zhang X.Q., Chen Y.T., Chen J.L., Gong G.F., Xu G.Y., Kang X.T. China Agriculture Press; China: 2011. Animal Genetic Resources in China: Poultry. [Google Scholar]
- Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., McVean G., Durbin R., Genomes Project Analysis G. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda M., Itahashi Y., Kikuchi H., Sun G., Hsu K.H., Gakuhari T., Yoneda M., Jiang L., Yang G., Nakamura S. Multiple lines of evidence of early goose domestication in a 7,000-y-old rice cultivation village in the lower Yangtze River, China. Proc. Natl. Acad. Sci. U S A. 2022;119 doi: 10.1073/pnas.2117064119. e2117064119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q.C., Wu P.F., Dai G.J., Zhang G.X., Zhang T., Xue Q., Shi H.Q., Wang J.Y. Identification of 19 loci for reproductive traits in a local Chinese chicken by genome-wide study. Genet. Mol. Res. 2017;16:1–8. doi: 10.4238/gmr16019431. [DOI] [PubMed] [Google Scholar]
- Grossman S.R., Shlyakhter I., Karlsson E.K., Byrne E.H., Morales S., Frieden G., Hostetter E., Angelino E., Garber M., Zuk O., Lander E.S., Schaffner S.F., Sabeti P.C. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010;327:883–886. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- Gu X., Feng C., Ma L., Song C., Wang Y., Da Y., Li H., Chen K., Ye S., Ge C., Hu X., Li N. Genome-wide association study of body weight in chicken F2 resource population. PLoS One. 2011;6:e21872. doi: 10.1371/journal.pone.0021872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wang C., Ouyang J., Tang H., Zheng S., Xiong Y., Gao Y., Wu Y., Wang L., Yan X., Chen H. Identification of key candidate genes for beak length phenotype by whole-genome resequencing in geese. Front. Vet. Sci. 2022;9 doi: 10.3389/fvets.2022.847481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Song K.D., Seo M., Caetano-Anolles K., Kim J., Kwak W., Oh J.D., Kim E., Jeong D.K., Cho S., Kim H., Lee H.K. Exploring evidence of positive selection reveals genetic basis of meat quality traits in Berkshire pigs through whole genome sequencing. BMC Genet. 2015;16:104. doi: 10.1186/s12863-015-0265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendig K.I., Baheti S., Bockol M.A., Drucker T.M., Hart S.N., Heldenbrand J.R., Hernaez M., Hudson M.E., Kalmbach M.T., Klee E.W., Mattson N.R., Ross C.A., Taschuk M., Wieben E.D., Wiepert M., Wildman D.E., Mainzer L.S. Sentieon DNASeq variant calling workflow demonstrates strong computational performance and accuracy. Front. Genet. 2019;10:736. doi: 10.3389/fgene.2019.00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., Shin D., Lee W., Taye M., Cho K., Park K.D., Kim H. The prediction of the expected current selection coefficient of single nucleotide polymorphism associated with holstein milk yield, fat and protein contents. Asian-Australas. J. Anim. Sci. 2016;29:36–42. doi: 10.5713/ajas.15.0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Miller C.H., Giannopoulou E., Hu X., Ivashkiv L.B., Zhao B. RBP-J imposes a requirement for ITAM-mediated costimulation of osteoclastogenesis. J. Clin. Invest. 2014;124:5057–5073. doi: 10.1172/JCI71882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.D., Liu X., Li Z.W., Wang W.J., Li Y.M., Cao Z.P., Luan P., Xiao F., Gao H.H., Guo H.S., Wang N., Li H., Wang S.Z. A combination of genome-wide association study and selection signature analysis dissects the genetic architecture underlying bone traits in chickens. Animal. 2021;15 doi: 10.1016/j.animal.2021.100322. [DOI] [PubMed] [Google Scholar]
- Liu R., Sun Y., Zhao G., Wang F., Wu D., Zheng M., Chen J., Zhang L., Hu Y., Wen J. Genome-wide association study identifies Loci and candidate genes for body composition and meat quality traits in Beijing-You chickens. PLoS One. 2013;8:e61172. doi: 10.1371/journal.pone.0061172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Bai C., Shi L., He Y., Hu M., Sun H., Peng H., Lai W., Jiao S., Zhao Z. Detection of selection signatures in South African Mutton Merino sheep using whole-genome sequencing data. Anim. Genet. 2022;53:224–229. doi: 10.1111/age.13173. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz G., Ovilo C., Estelle J., Silio L., Fernandez A., Rodriguez C. Association with litter size of new polymorphisms on ESR1 and ESR2 genes in a Chinese-European pig line. Genet. Sel. Evol. 2007;39:195–206. doi: 10.1186/1297-9686-39-2-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X., Tyasi T.L., Qin N., Liu D., Zhu H., Chen X., Zhang F., Yuan S., Xu R. Sequence variations in estrogen receptor 1 and 2 genes and their association with egg production traits in Chinese Dagu chickens. J. Vet. Med. Sci. 2017;79:927–934. doi: 10.1292/jvms.17-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- Park S.J., Lee J.Y., Lee S.H., Koh J.M., Kim B.J. SLIT2 inhibits osteoclastogenesis and bone resorption by suppression of Cdc42 activity. Biochem. Biophys. Res. Commun. 2019;514:868–874. doi: 10.1016/j.bbrc.2019.05.046. [DOI] [PubMed] [Google Scholar]
- Pattabhi S., Knoll M.L., Gale M., Jr, Loo Y.M. DHX15 Is a coreceptor for RLR signaling that promotes antiviral defense against RNA virus infection. J. Interferon Cytokine Res. 2019;39:331–346. doi: 10.1089/jir.2018.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppala A.K., French R.L., Matthies D., Baxa U., Subramaniam S., Simonovic M. Structural basis for early-onset neurological disorders caused by mutations in human selenocysteine synthase. Sci. Rep. 2016;6:32563. doi: 10.1038/srep32563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retief J.D. Phylogenetic analysis using PHYLIP. Methods Mol. Biol. 2000;132:243–258. doi: 10.1385/1-59259-192-2:243. [DOI] [PubMed] [Google Scholar]
- Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R., Schaffner S.F., Lander E.S., International HapMap C., Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., Pasternak S., Wheeler D.A., Willis T.D., Yu F., Yang H., Zeng C., Gao Y., Hu H., Hu W., Li C., Lin W., Liu S., Pan H., Tang X., Wang J., Wang W., Yu J., Zhang B., Zhang Q., Zhao H., Zhao H., Zhou J., Gabriel S.B., Barry R., Blumenstiel B., Camargo A., Defelice M., Faggart M., Goyette M., Gupta S., Moore J., Nguyen H., Onofrio R.C., Parkin M., Roy J., Stahl E., Winchester E., Ziaugra L., Altshuler D., Shen Y., Yao Z., Huang W., Chu X., He Y., Jin L., Liu Y., Shen Y., Sun W., Wang H., Wang Y., Wang Y., Xiong X., Xu L., Waye M.M., Tsui S.K., Xue H., Wong J.T., Galver L.M., Fan J.B., Gunderson K., Murray S.S., Oliphant A.R., Chee M.S., Montpetit A., Chagnon F., Ferretti V., Leboeuf M., Olivier J.F., Phillips M.S., Roumy S., Sallee C., Verner A., Hudson T.J., Kwok P.Y., Cai D., Koboldt D.C., Miller R.D., Pawlikowska L., Taillon-Miller P., Xiao M., Tsui L.C., Mak W., Song Y.Q., Tam P.K., Nakamura Y., Kawaguchi T., Kitamoto T., Morizono T., Nagashima A., Ohnishi Y., Sekine A., Tanaka T., Tsunoda T., Deloukas P., Bird C.P., Delgado M., Dermitzakis E.T., Gwilliam R., Hunt S., Morrison J., Powell D., Stranger B.E., Whittaker P., Bentley D.R., Daly M.J., de Bakker P.I., Barrett J., Chretien Y.R., Maller J., McCarroll S., Patterson N., Pe'er I., Price A., Purcell S., Richter D.J., Sabeti P., Saxena R., Schaffner S.F., Sham P.C., Varilly P., Altshuler D., Stein L.D., Krishnan L., Smith A.V., Tello-Ruiz M.K., Thorisson G.A., Chakravarti A., Chen P.E., Cutler D.J., Kashuk C.S., Lin S., Abecasis G.R., Guan W., Li Y., Munro H.M., Qin Z.S., Thomas D.J., McVean G., Auton A., Bottolo L., Cardin N., Eyheramendy S., Freeman C., Marchini J., Myers S., Spencer C., Stephens M., Donnelly P., Cardon L.R., Clarke G., Evans D.M., Morris A.P., Weir B.S., Tsunoda T., Johnson T.A., Mullikin J.C., Sherry S.T., Feolo M., Skol A., Zhang H., Zeng C., Zhao H., Matsuda I., Fukushima Y., Macer D.R., Suda E., Rotimi C.N., Adebamowo C.A., Ajayi I., Aniagwu T., Marshall P.A., Nkwodimmah C., Royal C.D., Leppert M.F., Dixon M., Peiffer A., Qiu R., Kent A., Kato K., Niikawa N., Adewole I.F., Knoppers B.M., Foster M.W., Clayton E.W., Watkin J., Gibbs R.A., Belmont J.W., Muzny D., Nazareth L., Sodergren E., Weinstock G.M., Wheeler D.A., Yakub I., Gabriel S.B., Onofrio R.C., Richter D.J., Ziaugra L., Birren B.W., Daly M.J., Altshuler D., Wilson R.K., Fulton L.L., Rogers J., Burton J., Carter N.P., Clee C.M., Griffiths M., Jones M.C., McLay K., Plumb R.W., Ross M.T., Sims S.K., Willey D.L., Chen Z., Han H., Kang L., Godbout M., Wallenburg J.C., L'Archeveque P., Bellemare G., Saeki K., Wang H., An D., Fu H., Li Q., Wang Z., Wang R., Holden A.L., Brooks L.D., McEwen J.E., Guyer M.S., Wang V.O., Peterson J.L., Shi M., Spiegel J., Sung L.M., Zacharia L.F., Collins F.S., Kennedy K., Jamieson R., Stewart J. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota Y., Steinthorsdottir V., Morris A.P., Fassbender A., Rahmioglu N., De Vivo I., Buring J.E., Zhang F., Edwards T.L., Jones S., O D., Peterse D., Rexrode K.M., Ridker P.M., Schork A.J., MacGregor S., Martin N.G., Becker C.M., Adachi S., Yoshihara K., Enomoto T., Takahashi A., Kamatani Y., Matsuda K., Kubo M., Thorleifsson G., Geirsson R.T., Thorsteinsdottir U., Wallace L.M., S.I.B.G.i P.-S., Yang J., Velez Edwards D.R., Nyegaard M., Low S.K., Zondervan K.T., Missmer S.A., D'Hooghe T., Montgomery G.W., Chasman D.I., Stefansson K., Tung J.Y., Nyholt D.R. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat. Commun. 2017;8:15539. doi: 10.1038/ncomms15539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson T.S., Yang Y., Huff C.D., Yun H., Qin G., Witherspoon D.J., Bai Z., Lorenzo F.R., Xing J., Jorde L.B., Prchal J.T., Ge R. Genetic evidence for high-altitude adaptation in Tibet. Sci. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- Tang J.B., Cao H.W., Xu R., Zhang D.D., Huang J. [Mutant generation of the testis genes and phenotype analyses in Drosophila] Yi Chuan. 2018;40:478–487. doi: 10.16288/j.yczz.18-055. [DOI] [PubMed] [Google Scholar]
- Wei C., Hou D., Feng Y., Li T., Jing Z., Li W., Han R., Li G., Sun G., Tian Y., Liu X., Kang X., Li Z. Molecular characterization and a duplicated 31-bp indel within the LDB2 gene and its associations with production performance in chickens. Gene. 2020;761 doi: 10.1016/j.gene.2020.145046. [DOI] [PubMed] [Google Scholar]
- Weir B.S., Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- White R., Leonardsson G., Rosewell I., Ann Jacobs M., Milligan S., Parker M. The nuclear receptor co-repressor nrip1 (RIP140) is essential for female fertility. Nat. Med. 2000;6:1368–1374. doi: 10.1038/82183. [DOI] [PubMed] [Google Scholar]
- Xia J., Qi X., Wu Y., Zhu B., Xu L., Zhang L., Gao X., Chen Y., Li J., Gao H. Genome-wide association study identifies loci and candidate genes for meat quality traits in Simmental beef cattle. Mamm. Genome. 2016;27:246–255. doi: 10.1007/s00335-016-9635-x. [DOI] [PubMed] [Google Scholar]
- Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Li Y., Wu J., Wang X., Bian C., Tian Y., Sun G., Han R., Liu X., Jiang R., Wang Y., Li G., Li W., Hu X., Kang X. Genome-wide association study reveals the genetic determinism of growth traits in a Gushi-Anka F2 chicken population. Heredity (Edinb) 2021;126:293–307. doi: 10.1038/s41437-020-00365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Li M., Cheng H., Fan W., Yuan Z., Gao Q., Xu Y., Guo Z., Zhang Y., Hu J., Liu H., Liu D., Chen W., Zheng Z., Jiang Y., Wen Z., Liu Y., Chen H., Xie M., Zhang Q., Huang W., Wang W., Hou S., Jiang Y. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat. Commun. 2018;9:2648. doi: 10.1038/s41467-018-04868-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.