Abstract

It is known that oligomers of amyloid-β (Aβ) peptide are associated with Alzheimer’s disease. Aβ has two isoforms: Aβ40 and Aβ42. Although the difference between Aβ40 and Aβ42 is only two additional C-terminal residues, Aβ42 aggregates much faster than Aβ40. It is unknown what role the C-terminal two residues play in accelerating aggregation. Since Aβ42 is more toxic than Aβ40, its oligomerization process needs to be clarified. Moreover, clarifying the differences between the oligomerization processes of Aβ40 and Aβ42 is essential to elucidate the key factors of oligomerization. Therefore, to investigate the dimerization process, which is the early oligomerization process, Hamiltonian replica-permutation molecular dynamics simulations were performed for Aβ40 and Aβ42. We identified a key residue, Arg5, for the Aβ42 dimerization. The two additional residues in Aβ42 allow the C-terminus to form contact with Arg5 because of the electrostatic attraction between them, and this contact stabilizes the β-hairpin. This β-hairpin promotes dimer formation through the intermolecular β-bridges. Thus, we examined the effects of amino acid substitutions of Arg5, thereby confirming that the mutations remarkably suppressed the aggregation of Aβ42. Moreover, the mutations of Arg5 suppressed the Aβ40 aggregation. It was found by analyzing the simulations that Arg5 is important for Aβ40 to form intermolecular contacts. Thus, it was clarified that the role of Arg5 in the oligomerization process varies due to the two additional C-terminal residues.
Keywords: molecular dynamics simulation, generalized-ensemble algorithm, amyloid-β peptide, protein aggregation
Introduction
It is known that more than 40 proteins and peptides form aggregates associated with human diseases, such as Alzheimer’s disease (AD), Parkinson’s disease, and hemodialysis-related amyloidosis.1−4 An example of such a peptide is the amyloid-β (Aβ) peptide. Aβs form soluble oligomers and insoluble amyloid fibrils by their aggregation. These aggregates play a vital role in the pathogenesis of AD.5,6 Amyloid fibrils of Aβ are known to form cross-β-sheet structures.7,8 Additionally, their atomic-level structures have been reported by experiments using solid-state nuclear magnetic resonance (NMR) spectroscopy9−12 and cryogenic electron microscopy (cryo-EM).13,14 Although it has been shown by NMR experiments that oligomers also form β-sheet structures,15,16 their atomic-level structures have yet to be clarified.
Aβ is produced by the proteolytic processing of amyloid precursor protein.17 Although there are several Aβs with different numbers of amino acid residues, the main isoforms have 40 (Aβ40) and 42 (Aβ42) amino acid residues.18,19 The only difference between Aβ40 and Aβ42 is that Aβ42 has two additional residues at its C-terminus. However, Aβ42 is more toxic than Aβ4020 and is the major component of early senile plaques in the brains of AD patients.21−23 It is also known that in several early-onset familial ADs, the production of Aβ42 increases.24−27 Additionally, Aβ42 forms oligomers and amyloid fibrils more rapidly than Aβ40.28,29 The two C-terminal residues of Aβ42 play essential roles in the rapid aggregation, but the details have not been elucidated. Elucidating the difference between the oligomerization processes of Aβ40 and Aβ42 is essential to understand Aβ aggregation and overcome AD.
To investigate Aβ at the atomic level, all-atom molecular dynamics (MD) simulation is essential. Many studies have used MD simulation on aggregation and disaggregation of Aβ.30−51 For studies on the oligomerization process, most studies have employed Aβ fragments, such as Aβ(16–22),52−58 Aβ(10–35),59 and Aβ(29–42).60−63 Several studies have been reported for the oligomer formation of full-length Aβs.64−69 It has been shown that Aβ40 dimers form various secondary structures, such as intramolecular and intermolecular β-sheet structures and α-helix structures.64 Similar results were reported for Aβ42 dimers.65 Because Aβ oligomerization is a slow process for the all-atom MD simulation, most of these studies used the replica-exchange method (REM)70,71 to obtain efficient samplings of Aβ structures.66
Recently, replica-permutation method (RPM) has been proposed72 to enhance sampling efficiency more than REM. In this method, similar to REM, copies (replicas) of a target system are prepared. These replicas are assigned different temperatures to perform canonical simulations. During simulations, the assigned temperatures are permuted among more than two replicas, whereas they are exchanged between two replicas in REM. Additionally, instead of the Metropolis algorithm,73 the Suwa–Todo algorithm74 is used to minimize the rejection ratio for replica-permutation trials. Then, the Hamiltonian RPM (HPRM) was proposed to generalize RPM.75 In HRPM, the same temperature is assigned to all replicas. However, different values of a parameter introduced in the Hamiltonian are assigned and permuted among more than two replicas. The dimer and larger oligomer formation processes of Aβ fragments have been elucidated using this HRPM.62,63
It is necessary to clarify the difference in oligomerization between Aβ40 and Aβ42 at the atomic level to elucidate the key factors of the Aβ aggregation. However, no study has shown the differences in these oligomerization processes at the atomic level. Therefore, in this paper, we investigated the dimerization process, which is the early oligomerization process, for these Aβs using MD simulation. We employed the Coulomb RPM (CRPM)75 to enhance conformational sampling for Aβs in an explicit solvent. CRPM is a form of HRPM. It is a useful method for investigating the aggregation processes of biomolecules. As a result of the CRPM simulation, we identified a key residue in dimerization. We also conducted experiments on Aβ aggregation to verify whether the residue identified by our simulation is actually important for the aggregation process. Consequently, we obtained that Aβ aggregation is significantly suppressed by mutating the residue identified in the simulation.
Results and Discussion
Comparison of Aβ42 and Aβ40 Dimerizations
To investigate the dimerization process of Aβ40 and Aβ42, we applied the Coulomb replica-permutation MD (CRPMD) method75 to two Aβ40 molecules and two Aβ42 molecules. The initial conformations for these MD simulations are shown in Figure S1. We first calculated probabilities of dimer formation for Aβ42 and Aβ40 to see whether there is a difference in dimer formation. Here, the reweighting techniques76,77 were employed to calculate the physical quantities at the original parameter (λ = 1.00 for CRPMD). Figure 1 shows distributions of oligomer sizes. Here, when two Aβs formed more than one intermolecular β-bridge, it was regarded as these Aβs formed a dimer. The defined secondary structure of protein (DSSP) criteria78 were used to determine the secondary structures. As shown in the figure, Aβ42 has a slightly higher probability of dimer formation than Aβ40. Conversely, Aβ40 tends to be in a monomer state compared to Aβ42. This tendency and probabilities of the monomer states are consistent with experimental results that showed oligomer size distributions for Aβ42 and Aβ40.79
Figure 1.

Dimerization propensities of Aβ42 (green) and Aβ40 (red).
Next, we calculated the intermolecular contact probabilities of Cα atoms to see the dimer structure. Figure 2a,b shows the intermolecular contact probability for Aβ42 and Aβ40, respectively. Here, when the distance between a pair of Cα atoms was less than 6.5 Å, it was regarded as a contact. In the Aβ42 dimer, the β1 and β2 regions tended to form an intermolecular antiparallel β-sheet, as shown by the magenta ellipse in Figure 2a. Here, the β1 and β2 regions consist of residues 17–21 and C-terminal residues after residue 29, respectively. In Figure S1, the β1 and β2 regions in the initial conformations are represented by cyan and purple, respectively. These regions have hydrophobic cores, and these hydrophobic cores have been reported to form β-sheets in amyloid fibrils.9−14 An intermolecular parallel β-sheet was also formed moderately between the β1 regions (the black ellipse). In the Aβ40 dimer, an intermolecular antiparallel β-sheet was mainly formed between the β1 and β2 regions, as shown in Figure 2b (the magenta ellipse).
Figure 2.
Intermolecular contact probabilities of Cα atoms for (a) Aβ42 and (b) Aβ40. Probabilities with which the residues in (c) Aβ42 and (d) Aβ40 form β-strands with the corresponding length.
To investigate the lengths of β-strands composing the intermolecular β-sheet and identify residues in the β-strands, we calculated the probabilities of intermolecular β-bridge formation of residues at each length of the β-strand. Here, the β-strand length is the number of consecutive residues that form the β-strand. Figure 2c,d shows the probabilities for Aβ42 and Aβ40, respectively. As shown by the magenta ellipse in Figure 2c, the β-strand length with the highest probability was 3. As shown in Figure 2a,c, in the Aβ42 dimer, the intermolecular antiparallel β-sheet between the β1 and β2 regions was composed of the β-strands consisting of three residues. In contrast, in the Aβ40 dimer, the intermolecular antiparallel β-sheet had only one β-bridge (i.e., length of the β-strand is 1), as shown in Figure 2b,d. This means that the Aβ42 dimer had a more stable intermolecular β-sheet than the Aβ40 dimer because the formation of longer β-strands stabilizes the intermolecular β-sheet.
There were differences not only in the intermolecular structures but also in the intramolecular structures between Aβ42 and Aβ40. Figure 3a,b shows the intramolecular contact probabilities of Cα atoms. Aβ42 formed a β-hairpin between the β1 and β2 regions, as shown by the magenta ellipse in Figure 3a. However, in Aβ40, such β-hairpin was rarely formed (Figure 3b). This difference in β-hairpin formation affects the difference in intermolecular β-sheet formation between Aβ42 and Aβ40. This is because two of the authors (S.G.I. and H.O.) reported that a β-hairpin of an Aβ fragment readily formed intermolecular β-sheets with other Aβ fragments.62 In our simulation of full-length Aβs, we also observed that the β-hairpin formed intermolecular β-sheets with the other Aβ, as seen in the movies. In these movies, two Aβ42s that are spatially separated approach each other (Movie S1), and one Aβ42 forms the β-hairpin (Movie S2) and then forms the intermolecular β-sheet with the other Aβ (Movie S3). It is worth noting that the value of λ varies throughout the movies since these movies are the trajectory in one replica in the CRPMD simulation. Additionally, not only our works but also several experimental and computational works have shown that the β-hairpins accelerate the intermolecular β-sheet formation.80−82
Figure 3.
Intramolecular contact probabilities of Cα atoms for (a) Aβ42 and (b) Aβ40. Probability distributions with respect to the number of intramolecular and intermolecular β-bridges for (c) Aβ42 and (d) Aβ40.
To investigate the relationship between intramolecular and intermolecular β-bridges, we calculated the probability distributions with respect to the number of intramolecular and intermolecular β-bridges. Figure 3c,d shows the probability distributions for Aβ42 and Aβ40, respectively. In both systems, the probability of forming more intermolecular β-bridges is higher when more intramolecular β-bridges are formed, as shown by magenta circles. This indicates that an intermolecular β-sheet is readily formed in the presence of a β-hairpin. In other words, the β-hairpin stabilizes the intermolecular β-sheet. Therefore, the reason why Aβ42 forms a more stable intermolecular β-sheet is that Aβ42 tends to form the β-hairpin in comparison with Aβ40.
The mechanism by which the β-hairpin promotes the formation of the intermolecular β-sheet structure is as follows. The formation of the β-hairpin maintains the extended structures in the β1 and β2 regions and leaves their hydrophobic side chains exposed in the aqueous solution. These exposed hydrophobic side chains attract the hydrophobic side chains in the β1 and β2 regions of the other Aβ. Since the two Aβs are close and Aβ that forms the β-hairpin has the extended structures in the β1 and β2 regions, an intermolecular β-sheet is quickly formed when Aβ that does not form the β-hairpin forms an extended structure in the β1 or β2 regions.
Such a mechanism of β-sheet formation has been reported for other molecules, such as a designed peptide that forms α-helix and β-hairpin in equal proportions.83−86 This peptide was designed by adding seven residues to a fully helical peptide.83 The region consisting of the additional seven residues was hydrophobic and formed an extended structure. As the extended region approaches the helix region, the hairpin is formed when the helix region forms the extended structure.84−86 Consequently, this designed peptide can have both α-helix and β-hairpin.
Tertiary Structures of Aβ42 and Aβ40 Dimers
Principal component analysis (PCA)87 was used to observe the tertiary structures of Aβ42 and Aβ40. Here, to focus on the dimer structure, the conformations (snapshots obtained from our MD simulations) in which two Aβs formed more than one intermolecular β-bridge were employed for PCA. Additionally, only the coordinates of Cα atoms in the β1 and β2 regions were considered to perform PCA. More details of PCA are presented in the Supporting Information. Figure 4a shows the free-energy landscape for Aβ42 with respect to the first and second principal components. The representative tertiary structures in five local-minimum free-energy states (states A–E) are also shown in the figure.
Figure 4.
Free-energy landscapes for (a) Aβ42 and (b) Aβ40 with respect to the corresponding first and second principal components (PC1 and PC2). The local-minimum free-energy states are labeled as (a) states A–E for Aβ42 and (b) states A′–E′ for Aβ40. The units of the free-energy landscapes are kcal/mol. Representative dimer structures in (a) states A–E and (b) states A′–E′ are also shown.
Each representative structure is as follows (we focus on β-sheet structures of the β1 and β2 regions in two Aβs): (state A) the green Aβ42 forms a β-hairpin with intramolecular antiparallel β-bridges between the β1 and β2 regions. Intermolecular parallel β-bridges are also formed between the β1 region of this Aβ42 and that of the other Aβ42. (State B) The blue Aβ42 has a β-hairpin structure in which the β1 and β2 regions form intramolecular antiparallel β-bridges. A β-hairpin is also seen in the β2 region in the green Aβ42. These two β-hairpins have a β-sheet structure with intermolecular β-bridges between the β1 region in the blue Aβ42 and the β2 region in the green Aβ42. (State C) An intermolecular parallel β-bridge is formed between the β1 region in the green Aβ42 and the β2 region in the blue Aβ42. (State D) The two β2 regions in Aβ42s form intermolecular parallel β-bridges. (State E) The green Aβ42 has a β-hairpin with β-bridges between the β1 and β2 regions. An intermolecular parallel β-bridges between the two β2 regions are formed. A β-hairpin in the N-terminal region in the blue Aβ42 also forms intermolecular parallel β-bridges with the β1 region in the green Aβ42.
Figure 4b shows the free-energy landscape for Aβ40 with respect to the first and second principal components. The representative tertiary structure in five local-minimum free-energy states (states A′–E′) is also shown in this figure. The representative structures are as follows: (state A′) intermolecular antiparallel β-bridges are formed between the β1 region in the blue Aβ42 and the β2 region in the green Aβ42. (State B′) There are three β-strands in the β1 and β2 regions in the blue Aβ42. The β2 region in the green Aβ42 forms an intermolecular antiparallel β-sheet with the β1 region in the blue Aβ42. (State C′) An intermolecular β-bridge is formed between the β2 region in the green Aβ42 and the β1 region in the blue Aβ42. (State D′) The blue Aβ42 has a β-hairpin with intramolecular β-bridges between the β1 and β2 regions. The β2 region in the green Aβ42 forms intermolecular antiparallel β-bridges with the β1 region in the blue Aβ42. (State E′) An intermolecular β-bridge is formed between the two β2 regions.
From these representative tertiary structures, in Aβ42 and Aβ40 dimers, longer β-strand with intermolecular β-bridges tends to be formed when at least one Aβ has stable intramolecular β-bridges (i.e., β-hairpin or intramolecular β-sheet). This tendency is consistent with Figure 3c,d. Thus, the intramolecular β-bridges play an essential role in the formation of the intermolecular β-bridges.
Key Residue for the β-Hairpin of Aβ42
To investigate why Aβ42 forms more β-hairpins, we calculated the probability of intramolecular contacts, including side-chain atoms. Here, when the shortest distance between atoms included in two different residues was less than 5.0 Å, it was regarded as a contact between the two residues. Hydrogen atoms were not considered in calculating the contact probability. Figure 5a,b shows the contact probabilities. The contact patterns are almost the same as the intramolecular contacts between Cα atoms in Figure 3. However, as shown in Figure 5a, the contact peaks between the C-terminus and vicinity of residue 5 (Arg5) are more obvious in Aβ42, as indicated by the black circles. This is because these contacts were maintained by the electrostatic interaction between the negative charge of the carboxyl group (COO–) in the C-terminus and the positive charge of the guanidinium group in Arg5. Moreover, the positions of the peaks indicated by the black circles are located on the extension of the major axis of the magenta ellipse corresponding to the β-hairpin. Therefore, the contact between the C-terminus and Arg5 contributes to the stabilization of the β-hairpin. Conversely, no peak corresponds to the contact between the C-terminus and Arg5 in Aβ40 (Figure 5b).
Figure 5.
Intramolecular contact probabilities between residues for (a) Aβ42 and (b) Aβ40. Here, all atoms, including the side-chain atoms, except the hydrogen atoms, are considered in calculating the contact probabilities. (c) Schematic illustration where the β-hairpin of Aβ42 is stabilized by the contacts between the C-terminus and Arg5 and between E22 and K28.
Figure 5c shows a schematic illustration in which the β-hairpin of Aβ42 is stabilized by the contact between the C-terminus and Arg5. Here, a contact between residue 22 (E22) and residue 28 (K28) is also shown. This contact is formed with a high probability due to the electrostatic interaction between their side chains (i.e., a salt bridge) as in the contact between the C-terminus and Arg5. The intramolecular contact probabilities between E22 and K28 were calculated with and without the salt bridge between their side chains. Here, the DSSP criteria were employed for the hydrogen bond formation between the E22 and K28 side chains. The contact probability with and without the salt bridge was 0.37 and 0.12, respectively. It means that the salt bridge promotes the contact formation between E22 and K28. As long as the contacts between the C-terminus and Arg5 and between E22 and K28 are maintained, the distance between β1 and β2 regions is inevitably shortened. Additionally, because the number of residues between Arg5 and E22 and between K28 and A42 (C-terminus) are almost equal, both β1 and β2 regions can have extended structures simultaneously, as seen in Figure 5c. The β-hairpin is formed when the two extended regions are at a short distance. This is the mechanism of stabilizing the β-hairpin of Aβ42 by the contact between the C-terminus and Arg5. Regarding Aβ40, since the number of residues between K28 and V40 (C-terminus) is less than that between Arg5 and E22, these two regions cannot have extended structures simultaneously. Consequently, Aβ40 hardly forms the β-hairpin.
MD Simulations of Aβ42 Monomer
To investigate the effects of Arg5 on the stabilization of the β-hairpin, we performed MD simulations of an Aβ42 monomer and its mutants. We used R5G and R5E as the mutants. As mentioned in the previous subsection, the electrostatic interaction between Arg5 and C-terminus is expected to be essential to stabilize the β-hairpin. To decrease attractive electrostatic forces between residue 5 and C-terminus, we chose neutral and negatively charged residues, Gly and Glu. Figure 6a shows a typical time series of the shortest distance between residue 5 and C-terminal residue atoms for each Aβ monomer system, obtained from one of the MD simulations. In the wild type, the shortest distance tended to get trapped in the vicinity of 3 Å. This means that the C-terminus is often bound to Arg5, as in the structure shown on the left in Figure 6a. This structure is representative of the time indicated by the red circle. During this time, the β-hairpin was maintained while the side chain of Arg5 and C-terminus kept a short distance forming the salt bridge. In contrast, such bindings were not seen in the mutants. For R5G, however, we found that the distance between residue 5 and the C-terminus was maintained at about 10 Å during the last 60 ns. The structure at this distance is shown on the right side of Figure 6a. In this structure, the β-hairpin collapsed, forming a globular structure. Figure 6b shows the probability distribution of the shortest distance between residue 5 and C-terminal residue for each system. This distribution was calculated by averaging over 20 MD simulations. Here, for each MD simulation, the last 220 ns of the 320 ns trajectory data was used for this analysis. The distributions show that the binding between Arg5 and C-terminus was frequently formed in the wild type; however, no such binding was formed in the mutants.
Figure 6.
(a) Time series of distances between residue 5 and C-terminal residue for the wild type and mutants. Representative structures for the time periods indicated by red circles are also shown. Residues 5 and 42 are shown in the ball-and-stick model. (b) Probability distributions of distances between residue 5 and C-terminal residue for the wild type and mutants. (c) Time series of the average numbers of β-bridges between the β1 and β2 regions calculated from 20 MD simulations for each system.
To investigate the stability of the β-hairpin, the number of β-bridges between the β1 and β2 regions was counted at each MD step. Figure 6c shows the time series of the average numbers of β-bridges calculated from 20 MD simulations. In the wild type, the number of β-bridges decreased more slowly than in the mutants. This means that the β-bridges between the β1 and β2 regions were maintained in the wild type but were gradually broken in the mutants. Furthermore, the intramolecular contacts between Cα atoms were calculated to investigate whether the hairpin structures are maintained. Figure 7a–c shows the contact probabilities obtained from 20 MD simulations. Here, for each MD simulation, the last 220 ns of the 320 ns trajectory data was used for these analyses. As shown in these figures, the contact patterns corresponding to the hairpin structures in the mutants had lower probabilities than those in the wild type. It means that the hairpin structures of the mutants were gradually broken as the number of β-bridges decreased due to the lack of stable binding between residue 5 and C-terminus. Therefore, the interaction between Arg5 and C-terminus plays an essential role in the formation of the β-hairpin in Aβ42. Note that several computational studies reported that Arg5 tends to form intramolecular salt bridges with other negatively charged residues by performing MD simulations of an Aβ monomer or Aβ fragment.88−90 However, our study is the first to show that the intramolecular salt bridge between Arg5 and C-terminus is essential for β-hairpin formation.
Figure 7.
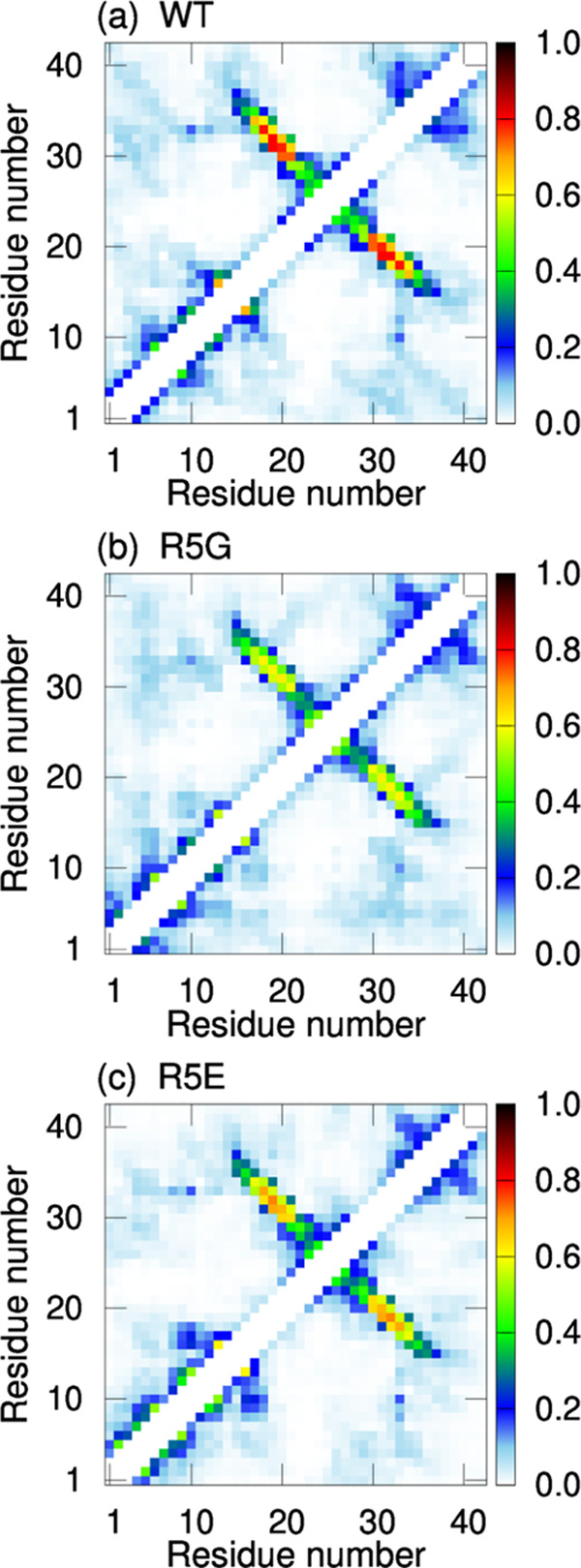
Intramolecular contact probabilities of Cα atoms in the three Aβ42 monomers: (a) the wild type, (b) R5G, and (c) R5E.
Effects of Amino Acid Substitutions of Arg5 on Aβ Aggregation
From our MD simulations, Arg5 is expected to promote the Aβ42 aggregation. Conversely, in the mutations of Arg5 to Gly or Glu, their aggregations are expected to be suppressed. To confirm this prediction from our MD simulations, we conducted experiments on the aggregations of the wild type and mutants. Figure 8a–c shows the aggregation of these Aβ42s monitored by thioflavin T (ThT) fluorescence. As expected from the MD simulations, the Aβ42 aggregation is suppressed by the mutations of Arg5 to Gly or Glu. The effect of the mutations is remarkable. Thus, in the experiments, it was confirmed that Arg5 plays an essential role in the Aβ42 aggregation. To the best of our knowledge, no fibril model with the salt bridge between Arg5 and C-terminus has been reported so far. This suggests that this intramolecular salt bridge might be transient in the early stages of amyloidogenesis.
Figure 8.
Aggregation of (a) the wild type, (b) R5G, and (c) R5E of Aβ42s monitored by ThT assay in 20 mM sodium phosphate buffer, pH 7.4. Aggregation of (d) the wild type, (e) R5G, and (f) R5E of Aβ40s monitored by ThT assay in 20 mM sodium phosphate buffer, pH 7.4.
Additionally, we investigated whether Arg5 affects the Aβ40 aggregation. Figure 8d–f shows the experimental results on the aggregation of the Aβ40 wild type and mutants using ThT assay. Interestingly, mutations of Arg5 also affect the Aβ40 aggregation. However, the effect of suppressing aggregation seems to be weaker than that of Aβ42. For instance, for R5G of Aβ40, although the start of aggregation is delayed, the aggregation is not suppressed much. Therefore, the role of Arg5 in the Aβ40 aggregation may be different from that in the Aβ42 aggregation. In fact, in our MD simulations for Aβ40, the contact between Arg5 and C-terminus seen in Aβ42 is hardly formed (Figure 5b).
To investigate the role of Arg5 in the Aβ40 aggregation, we calculated the probability of intermolecular contacts, including side-chain atoms, from our MD simulations. For comparison, we also calculated those for Aβ42. Figure 9 shows the contact probabilities for both Aβs. In both Aβs, as with the intramolecular contacts, the contact patterns are similar to the intermolecular contacts between Cα atoms in Figure 2a,b. In Aβ42, there is a contact peak between the C-terminus and Arg5 residues, as seen in the intramolecular contacts in Figure 5a. However, this contact probability is lower than that of the intramolecular contacts in Figure 5a. This indicates that the intramolecular contact between Arg5 and the C-terminus is more dominant than the intermolecular contact between them. For Aβ40, Arg5 has intermolecular contacts with residues in the N-terminal regions, as shown in Figure 9b (the magenta ellipse). The reason why Arg5 and N-terminal residues form contacts is that there are several negatively charged residues in the N-terminal region, such as Asp1, Glu3, Asp7, and Glu11. In contrast, Arg5 is the only positively charged residue in the N-terminal region, except for the N-terminus (NH3+). This contact between Arg5 and N-terminal region plays an essential role in the dimer formation of Aβ40. This is because when there is such a contact, the distance between the two Aβ40s is shorter, thereby forming a dimer.
Figure 9.
Intermolecular contact probabilities between residues for (a) Aβ42 and (b) Aβ40. Here, all atoms, including the side-chain atoms, except the hydrogen atoms, are considered in calculating the contact probabilities.
From these simulation results, the experimental results of Aβ40 in Figure 8 can be explained as follows. The total negative charge in the N-terminal region increases by mutating Arg5 to a neutral or negatively charged residue. The larger the total negative charge, the less the N-terminal regions form contacts with each other. Consequently, the Aβ40 aggregation is suppressed.
Known Mutations in the Vicinity of Residue 5
Several Aβ mutants, where the vicinity of residue 5 is mutated, are known in association with AD. For example, rodent Aβ has three mutations (R5G, Y10T, and H13R) in the N-terminal region, and it has been shown that age-associated amyloid plaques do not accumulate in rodents.91 It was reported that this mutant aggregates more slowly than human Aβ.92,93 However, it was reported that single mutations for Y10 and H13 promote Aβ aggregation.92,94 Therefore, the mutation of Arg5 is important in suppressing Aβ aggregation, as shown in our study. Additionally, the English (H6R) and Tottori (D7N) mutations are associated with familial AD. They are known as mutations that accelerate the Aβ aggregation. Since these mutations increase the positive charges or decrease the negative charges in the region near residue 5, a β-hairpin is considered to be more readily formed in Aβ42 (Figure 5c). For Aβ40, Aβ40 molecules may easily form intermolecular contacts due to an increase in the positive charge in the N-terminal region. As mentioned in the previous subsection, such intermolecular contacts in the N-terminal region are essential for Aβ oligomerization.
Conclusions
It is known that Aβ42 forms oligomers more rapidly than Aβ40. To investigate the role of the two additional C-terminal residues of Aβ42 in accelerating the oligomer formation, we performed the Coulomb replica-permutation molecular dynamics (CRPMD) simulation for two Aβ42 molecules in explicit water. We also conducted the CRPMD simulation for two Aβ40 molecules to clarify the difference in oligomerization processes between Aβ42 and Aβ40.
We showed that the probability of the dimer formation for Aβ42 was slightly higher than that of Aβ40. In the dimer structures for both Aβ systems, the β1 and β2 regions tended to form the intermolecular antiparallel β-sheets. Additionally, we observed that the Aβ42 dimer forms a stable intermolecular β-sheet with longer β-strands than the Aβ40 dimer. For the intramolecular structures, Aβ42 formed the β-hairpin with a higher probability than Aβ40. The β-hairpin formation is essential in forming a stable intermolecular β-sheet. In fact, more intermolecular β-bridges were formed with more intramolecular β-bridges in both Aβs.
Aβ42 forms more β-hairpins because of the following reasons. The contacts between the C-terminus and Arg5 and between E22 and K28 are maintained by their electrostatic interactions in Aβ42. Due to these contacts, the distance between the β1 and β2 regions is inevitably shortened. The region from Arg5 to E22 and that from K28 to A42 have almost the same number of residues. These regions can have extended structures simultaneously. The β-hairpin can easily be formed between these close extended structures. To see whether Arg5 is needed to stabilize the β-hairpin, we performed additional molecular dynamics (MD) simulations of the wild type and mutants (R5G and R5E). Consequently, the β-hairpin was maintained in the wild type; however, it was gradually broken in the mutants.
These simulation results show that Arg5 plays an essential role in the Aβ42 aggregation. We conducted experiments on Aβ aggregations to confirm the accuracy of the prediction from the simulation. The experimental results show that the mutation of Arg5 suppresses the Aβ42 aggregation. We also obtained that the Arg5 mutation suppresses not only the Aβ42 aggregation but also the Aβ40 aggregation. The MD simulations elucidated that Arg5 is essential for the intramolecular contact in Aβ42, whereas it is essential for the intermolecular contact in Aβ40.
In this study, we have successfully identified the key residue, Arg5, for the Aβ42 and Aβ40 oligomerizations. For the Aβ42 oligomerization, we predicted this key residue prior to the experiments using the Hamiltonian replica-permutation method. This shows that the MD simulation with efficient conformational sampling is useful for elucidating oligomerization processes of proteins. By performing the MD simulations and experiments, we obtained that the key residue plays different roles in the Aβ42 and Aβ40 oligomerizations. Such a collaborative approach between simulation and experiments is essential in understanding protein oligomerization. Through the simulations, we investigated dimerization, which is the smallest unit of oligomerization. Consequently, we observed that there is a difference between Aβ42 and Aβ40. The fact that we could predict the experimental results from the simulation results means that the differences seen in the formation of dimers make a difference in the formation of much larger aggregates, such as amyloid fibrils observed in experiments. Thus, it is essential to elucidate the process of small oligomer formation to fully understand the Aβ aggregation.
Materials and Methods
CRPMD Simulations of Two Aβ Molecules
CRPMD was applied to a system of two Aβ40 molecules and that of two Aβ42 molecules. The N-termini and C-termini were left uncapped for these Aβs. The two Aβ molecules were put in a cubic unit cell with explicit water molecules and counterions. The side lengths of the cubic unit cells were 101.7 Å in both Aβ systems (i.e., a system containing two Aβ40 molecules and that containing two Aβ42 molecules). Periodic boundary conditions were utilized. The Amber parm99SB force field95 and TIP3P rigid-body model96 were employed for the Aβ and water molecules, respectively. The SHAKE algorithm was used to constrain bond lengths with the hydrogen atoms of Aβ and fix the water molecule structures during the simulations. The cutoff distance for the Lennard-Jones potential energy was 12.0 Å. The electrostatic potential energy was calculated using the particle mesh Ewald method.97 The temperature was controlled by the Nosé–Hoover thermostat.98−101 The multiple time-step method102 was employed, and the time steps were taken to be 4.0 fs for interactions between the water molecules and 1.0 fs for other interactions. Initial conformations for these CRPMD simulations were the same for all replicas; they are prepared as presented in the Supporting Information. The number of replicas was 18. The values of the parameter λ for the CRPM were set as 0.82, 0.84, 0.86, 0.88, 0.90, 0.92, 0.94, 0.96, 0.98, 1.00, 1.01, 1.02, 1.03, 1.04, 1.05, 1.06, 1.07, and 1.08. Here, we employed λ only for the intermolecular electrostatic interactions between the Aβ atoms. The values of λ less than 1 result in weaker electrostatic interactions between Aβs, and the values greater than 1 result in stronger electrostatic interactions between them. Therefore, the dimerization of Aβs is inhibited or promoted by changing the value of λ to be smaller or larger than 1. The details of CRPM are described in the Supporting Information. The 18 replicas were divided into three subsets, and each subset had six replicas and six parameter values (see ref (72) for more details). Each CRPMD simulation was performed for 1.12 μs at 350 K per replica, including an equilibration run performed for 100 ns. The production run of each Aβ system was conducted for 18.36 μs in total. The trajectory data were stored every 2.0 ps, and trials of replica permutations were performed every 4.0 ps.
We employed the reweighting techniques76,77 to obtain the physical quantities at the original parameter (λ = 1.00) in the Results and Discussion section. The errors were estimated using the jackknife method.103 The number of bins for the jackknife method was 20.
MD Simulations of the Aβ42 Monomer
We performed MD simulations of the Aβ42 monomer. We employed two different β-hairpins as the initial structures, as shown in Figure S2. The Aβ42 molecule was put in a cubic unit cell. The side length of the cubic unit cell was 64.2 Å. Ten different initial velocities were employed for each initial structure, meaning that we employed 20 different initial conditions. For each initial condition, an MD simulation was performed for 320 ns. The other simulation conditions were the same as in the previous subsection. For comparison, we performed MD simulations of two Aβ42 mutants: R5G and R5E. Two initial structures were also employed for each mutant, and their structures were the same as those for the wild type, except for the side chain of residue 5. For each initial structure, 10 different initial velocities were also employed. In other words, 20 MD simulations for 320 ns were commonly performed for WT and the mutants. The other simulation conditions were the same as for the wild type.
Aggregation Assays
The synthetic wild-type Aβ42 and Aβ40 peptides were purchased from Toray Research Center, Inc. The synthetic Aβ42 and Aβ40 peptides with a substitution of Arg5 to Gly (R5G) or Glu (R5E) were purchased from Abclonal. The peptides were dissolved in 6 M guanidine hydrochloride and purified using a Superdex 75 Increase 10/300 column (Cytiva) at a 0.4 mL/min flow rate with 20 mM sodium phosphate buffer, a pH of 7.4, to remove from potential aggregated species. The obtained monomer was diluted with 20 mM sodium phosphate buffer, a pH of 7.4, to the desired concentration and supplemented with 0.1 mM ThT from a 2 mM stock solution. Then, each sample was pipetted into multiple wells of a 96-well half-area, low-binding polyethylene glycol coating plate with a clear bottom (Corning 3881) at 0.1 mL per well. Aggregation assays were initiated by placing the 96-well plate at 37 °C under quiescent conditions in a plate reader (Infinite 200Pro, TECAN). The ThT fluorescence was measured through the bottom of the plate with a 430 nm excitation filter and a 485 nm emission filter. The ThT fluorescence was followed for three repeats of each sample.
Acknowledgments
This work used supercomputers at the Research Center for Computational Science, Okazaki Research Facilities, the National Institutes of Natural Sciences (Projects 16-IMS-C127, 17-IMS-C144, 18-IMS-C152, 19-IMS-C172, 20-IMS-C155, 21-IMS-C172, 22-IMS-C186), and the Supercomputer Center, the Institute for Solid State Physics, the University of Tokyo. This work was supported by JSPS KAKENHI Grant Numbers JP21K06040, JP16K18531, JP19K07041, and JP21K06118. This work was also supported by the ExCELLS Research for Young Scientists and Grant-in-Aid for Research in Nagoya City University Grant Numbers 2212008 and 2222004.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschemneuro.2c00358.
Description of CRPM, preparation of the initial conformations for the CRPMD simulations and for the MD simulations of the Aβ monomers, and details of PCA (PDF)
Two Aβ42s that are spatially separated approach each other (Movie S1) (AVI)
One Aβ42 forms the β-hairpin (Movie S2) (AVI)
An intermolecular β-sheet is formed with the other Aβ (Movie S3) (AVI)
Author Contributions
S.G.I. and H.O. designed the research. S.G.I. modeled the initial conformations, performed simulations, and analyzed the simulation results. M.Y.-U. and K.K. prepared samples and conducted experiments. S.G.I. and M.Y.-U. wrote the paper. All authors discussed the results and revised the paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Chiti F.; Dobson C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Chiti F.; Dobson C. M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- Iadanza M. G.; Jackson M. P.; Hewitt E. W.; Ranson N. A.; Radford S. E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 755–773. 10.1038/s41580-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picken M. M. The Pathology of Amyloidosis in Classification: A Review. Acta Haematol. 2020, 143, 322–334. 10.1159/000506696. [DOI] [PubMed] [Google Scholar]
- Sipe J. D. Amyloidosis. Annu. Rev. Biochem. 1992, 61, 947–975. 10.1146/annurev.bi.61.070192.004503. [DOI] [PubMed] [Google Scholar]
- Hayden E. Y.; Teplow D. B. Amyloid beta-protein oligomers and Alzheimer’s disease. Alzheimer’s Res. Ther. 2013, 5, 60 10.1186/alzrt226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner D. A.; Abraham C.; Selkoe D. J. X-ray diffraction from intraneuronal paired helical filaments and extraneuronal amyloid fibers in Alzheimer disease indicates cross-beta conformation. Proc. Natl. Acad. Sci. U.S.A. 1986, 83, 503–507. 10.1073/pnas.83.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye H.; Fraser P. E.; Kirschner D. A. Structure of beta-crystallite assemblies formed by Alzheimer beta-amyloid protein analogues: analysis by x-ray diffraction. Biophys. J. 1993, 64, 502–519. 10.1016/S0006-3495(93)81393-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravastu A. K.; Leapman R. D.; Yau W. M.; Tycko R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 18349–18354. 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.; Ma B.; McElheny D.; Parthasarathy S.; Long F.; Hoshi M.; Nussinov R.; Ishii Y. Abeta(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat. Struct. Mol. Biol. 2015, 22, 499–505. 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin M. T.; Silvers R.; Ni Q. Z.; Can T. V.; Sergeyev I.; Rosay M.; Donovan K. J.; Michael B.; Wall J.; Linse S.; Griffin R. G. Atomic Resolution Structure of Monomorphic Abeta42 Amyloid Fibrils. J. Am. Chem. Soc. 2016, 138, 9663–9674. 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhrs T.; Ritter C.; Adrian M.; Riek-Loher D.; Bohrmann B.; Dobeli H.; Schubert D.; Riek R. 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 17342–17347. 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremer L.; Scholzel D.; Schenk C.; Reinartz E.; Labahn J.; Ravelli R. B. G.; Tusche M.; Lopez-Iglesias C.; Hoyer W.; Heise H.; Willbold D.; Schroder G. F. Fibril structure of amyloid-beta(1-42) by cryo-electron microscopy. Science 2017, 358, 116–119. 10.1126/science.aao2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M.; Rohou A.; Lasker K.; Yadav J. K.; Schiene-Fischer C.; Fandrich M.; Grigorieff N. Peptide dimer structure in an Abeta(1-42) fibril visualized with cryo-EM. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 11858–11863. 10.1073/pnas.1503455112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.; Edalji R.; Harlan J. E.; Holzman T. F.; Lopez A. P.; Labkovsky B.; Hillen H.; Barghorn S.; Ebert U.; Richardson P. L.; Miesbauer L.; Solomon L.; Bartley D.; Walter K.; Johnson R. W.; Hajduk P. J.; Olejniczak E. T. Structural characterization of a soluble amyloid beta-peptide oligomer. Biochemistry 2009, 48, 1870–1877. 10.1021/bi802046n. [DOI] [PubMed] [Google Scholar]
- Sarkar B.; Mithu V. S.; Chandra B.; Mandal A.; Chandrakesan M.; Bhowmik D.; Madhu P. K.; Maiti S. Significant structural differences between transient amyloid-beta oligomers and less-toxic fibrils in regions known to harbor familial Alzheimer’s mutations. Angew. Chem., Int. Ed. 2014, 53, 6888–6892. 10.1002/anie.201402636. [DOI] [PubMed] [Google Scholar]
- Kang J.; Lemaire H. G.; Unterbeck A.; Salbaum J. M.; Masters C. L.; Grzeschik K. H.; Multhaup G.; Beyreuther K.; Muller-Hill B. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature 1987, 325, 733–736. 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- Jakob-Roetne R.; Jacobsen H. Alzheimer’s disease: from pathology to therapeutic approaches. Angew. Chem., Int. Ed. 2009, 48, 3030–3059. 10.1002/anie.200802808. [DOI] [PubMed] [Google Scholar]
- Zheng L.; Cedazo-Minguez A.; Hallbeck M.; Jerhammar F.; Marcusson J.; Terman A. Intracellular distribution of amyloid beta peptide and its relationship to the lysosomal system. Transl. Neurodegener. 2012, 1, 19 10.1186/2047-9158-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A. M.; Kowall N. W.; Ferrante R. J. Neurotoxicity and oxidative damage of beta amyloid 1-42 versus beta amyloid 1-40 in the mouse cerebral cortex. Ann. N. Y. Acad. Sci. 1999, 893, 314–320. 10.1111/j.1749-6632.1999.tb07845.x. [DOI] [PubMed] [Google Scholar]
- Miller D. L.; Papayannopoulos I. A.; Styles J.; Bobin S. A.; Lin Y. Y.; Biemann K.; Iqbal K. Peptide compositions of the cerebrovascular and senile plaque core amyloid deposits of Alzheimer’s disease. Arch. Biochem. Biophys. 1993, 301, 41–52. 10.1006/abbi.1993.1112. [DOI] [PubMed] [Google Scholar]
- Iwatsubo T.; Odaka A.; Suzuki N.; Mizusawa H.; Nukina N.; Ihara Y. Visualization of Aβ42(43) and Aβ40 in senile plaques with end-specific Aβ monoclonals: Evidence that an initially deposited species is Aβ42(43). Neuron 1994, 13, 45–53. 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- Gravina S. A.; Ho L.; Eckman C. B.; Long K. E.; Otvos L. Jr.; Younkin L. H.; Suzuki N.; Younkin S. G. Amyloid beta protein (A beta) in Alzheimer’s disease brain. Biochemical and immunocytochemical analysis with antibodies specific for forms ending at A beta 40 or A beta 42(43). J. Biol. Chem. 1995, 270, 7013–7016. 10.1074/jbc.270.13.7013. [DOI] [PubMed] [Google Scholar]
- Borchelt D. R.; Thinakaran G.; Eckman C. B.; Lee M. K.; Davenport F.; Ratovitsky T.; Prada C. M.; Kim G.; Seekins S.; Yager D.; Slunt H. H.; Wang R.; Seeger M.; Levey A. I.; Gandy S. E.; Copeland N. G.; Jenkins N. A.; Price D. L.; Younkin S. G.; Sisodia S. S. Familial Alzheimer’s disease-linked presenilin 1 variants elevate Abeta1-42/1-40 ratio in vitro and in vivo. Neuron 1996, 17, 1005–1013. 10.1016/S0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- Kumar-Singh S.; Theuns J.; Van Broeck B.; Pirici D.; Vennekens K.; Corsmit E.; Cruts M.; Dermaut B.; Wang R.; Van Broeckhoven C. Mean age-of-onset of familial alzheimer disease caused by presenilin mutations correlates with both increased Abeta42 and decreased Abeta40. Hum. Mutat. 2006, 27, 686–695. 10.1002/humu.20336. [DOI] [PubMed] [Google Scholar]
- Baldassarre M.; Baronio C. M.; Morozova-Roche L. A.; Barth A. Amyloid beta-peptides 1-40 and 1-42 form oligomers with mixed beta-sheets. Chem. Sci. 2017, 8, 8247–8254. 10.1039/C7SC01743J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi C.; Bi S.; Li B. Processing of Mutant beta-Amyloid Precursor Protein and the Clinicopathological Features of Familial Alzheimer’s Disease. Aging Dis. 2019, 10, 383–403. 10.14336/AD.2018.0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett J. T.; Berger E. P.; Lansbury P. T. Jr. The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer’s disease. Biochemistry 1993, 32, 4693–4697. 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- El-Agnaf O. M.; Mahil D. S.; Patel B. P.; Austen B. M. Oligomerization and toxicity of beta-amyloid-42 implicated in Alzheimer’s disease. Biochem. Biophys. Res. Commun. 2000, 273, 1003–1007. 10.1006/bbrc.2000.3051. [DOI] [PubMed] [Google Scholar]
- Sgourakis N. G.; Merced-Serrano M.; Boutsidis C.; Drineas P.; Du Z.; Wang C.; Garcia A. E. Atomic-Level Characterization of the Ensemble of the Aβ(1–42) Monomer in Water Using Unbiased Molecular Dynamics Simulations and Spectral Algorithms. J. Mol. Biol. 2011, 405, 570. 10.1016/j.jmb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez-Vega C.; Escobedo F. A. Characterizing the structural behavior of selected Abeta-42 monomers with different solubilities. J. Phys. Chem. B 2011, 115, 4900–4910. 10.1021/jp1086575. [DOI] [PubMed] [Google Scholar]
- Olubiyi O. O.; Strodel B. Structures of the Amyloid β-Peptides. Aβ1–40 and Aβ1–42 as Influenced by pH and a d-Peptide. J. Phys. Chem. B 2012, 116, 3280. 10.1021/jp2076337. [DOI] [PubMed] [Google Scholar]
- Rosenman D. J.; Connors C. R.; Chen W.; Wang C.; Garcia A. E. Aβ Monomers Transiently Sample Oligomer and Fibril-Like Configurations: Ensemble Characterization Using a Combined MD/NMR Approach. J. Mol. Biol. 2013, 425, 3338. 10.1016/j.jmb.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenman D. J.; Wang C.; Garcia A. E. Characterization of Aβ Monomers through the Convergence of Ensemble Properties among Simulations with Multiple Force Fields. J. Phys. Chem. B 2016, 120, 259. 10.1021/acs.jpcb.5b09379. [DOI] [PubMed] [Google Scholar]
- Itoh S. G.; Yagi-Utsumi M.; Kato K.; Okumura H. Effects of a Hydrophilic/Hydrophobic Interface on Amyloid-β Peptides Studied by Molecular Dynamics Simulations and NMR Experiments. J. Phys. Chem. B 2019, 123, 160–169. 10.1021/acs.jpcb.8b11609. [DOI] [PubMed] [Google Scholar]
- O’Brien E. P.; Okamoto Y.; Straub J. E.; Brooks B. R.; Thirumalai D. Thermodynamic perspective on the dock-lock growth mechanism of amyloid fibrils. J. Phys. Chem. B 2009, 113, 14421–14430. 10.1021/jp9050098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub J. E.; Thirumalai D. Toward a molecular theory of early and late events in monomer to amyloid fibril formation. Annu. Rev. Phys. Chem. 2011, 62, 437–463. 10.1146/annurev-physchem-032210-103526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwierz N.; Frost C. V.; Geissler P. L.; Zacharias M. Dynamics of Seeded Abeta40-Fibril Growth from Atomistic Molecular Dynamics Simulations: Kinetic Trapping and Reduced Water Mobility in the Locking Step. J. Am. Chem. Soc. 2016, 138, 527–539. 10.1021/jacs.5b08717. [DOI] [PubMed] [Google Scholar]
- Sasmal S.; Schwierz N.; Head-Gordon T. Mechanism of Nucleation and Growth of Abeta40 Fibrils from All-Atom and Coarse-Grained Simulations. J. Phys. Chem. B 2016, 120, 12088–12097. 10.1021/acs.jpcb.6b09655. [DOI] [PubMed] [Google Scholar]
- Bacci M.; Vymetal J.; Mihajlovic M.; Caflisch A.; Vitalis A. Amyloid β Fibril Elongation by Monomers Involves Disorder at the Tip. J. Chem. Theory Comput. 2017, 13, 5117. 10.1021/acs.jctc.7b00662. [DOI] [PubMed] [Google Scholar]
- Buchete N. V.; Tycko R.; Hummer G. Molecular dynamics simulations of Alzheimer’s beta-amyloid protofilaments. J. Mol. Biol. 2005, 353, 804–821. 10.1016/j.jmb.2005.08.066. [DOI] [PubMed] [Google Scholar]
- Baumketner A.; Krone M. G.; Shea J. E. Role of the familial Dutch mutation E22Q in the folding and aggregation of the 15-28 fragment of the Alzheimer amyloid-beta protein. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 6027–6032. 10.1073/pnas.0708193105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemkul J. A.; Bevan D. R. Assessing the stability of Alzheimer’s amyloid protofibrils using molecular dynamics. J. Phys. Chem. B 2010, 114, 1652–1660. 10.1021/jp9110794. [DOI] [PubMed] [Google Scholar]
- Okumura H.; Itoh S. G. Structural and Fluctuational Difference Between Two Ends of Aβ Amyloid Fibril: MD Simulations Predict Only One End has Open Conformations. Sci. Rep. 2016, 6, 38422 10.1038/srep38422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R. A.; Chen L. Y.; Plascencia-Villa G.; Perry G. Thermodynamics of Amyloid-beta Fibril Elongation: Atomistic Details of the Transition State. ACS Chem. Neurosci. 2018, 9, 783–789. 10.1021/acschemneuro.7b00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson D. S.; Brown A. M.; Lemkul J. A. Insights into Stabilizing Forces in Amyloid Fibrils of Differing Sizes from Polarizable Molecular Dynamics Simulations. J. Mol. Biol. 2018, 430, 3819–3834. 10.1016/j.jmb.2018.05.020. [DOI] [PubMed] [Google Scholar]
- Ilie I. M.; Caflisch A. Disorder at the Tips of a Disease-Relevant Aβ42 Amyloid Fibril: A Molecular Dynamics Study. J. Phys. Chem. B 2018, 122, 11072. 10.1021/acs.jpcb.8b05236. [DOI] [PubMed] [Google Scholar]
- Okumura H.; Itoh S. G. Amyloid fibril disruption by ultrasonic cavitation: nonequilibrium molecular dynamics simulations. J. Am. Chem. Soc. 2014, 136, 10549–10552. 10.1021/ja502749f. [DOI] [PubMed] [Google Scholar]
- Hoang Viet M.; Derreumaux P.; Nguyen P. H. Nonequilibrium all-atom molecular dynamics simulation of the bubble cavitation and application to dissociate amyloid fibrils. J. Chem. Phys. 2016, 145, 174113 10.1063/1.4966263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang Viet M.; Derreumaux P.; Li M. S.; Roland C.; Sagui C.; Nguyen P. H. Picosecond dissociation of amyloid fibrils with infrared laser: A nonequilibrium simulation study. J. Chem. Phys. 2015, 143, 155101 10.1063/1.4933207. [DOI] [PubMed] [Google Scholar]
- Okumura H.; Itoh S. G.; Nakamura K.; Kawasaki T. Role of Water Molecules and Helix Structure Stabilization in the Laser-Induced Disruption of Amyloid Fibrils Observed by Nonequilibrium Molecular Dynamics Simulations. J. Phys. Chem. B 2021, 125, 4964–4976. 10.1021/acs.jpcb.0c11491. [DOI] [PubMed] [Google Scholar]
- Nguyen P. H.; Li M. S.; Stock G.; Straub J. E.; Thirumalai D. Monomer adds to preformed structured oligomers of Abeta-peptides by a two-stage dock-lock mechanism. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 111–116. 10.1073/pnas.0607440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimov D. K.; Thirumalai D. Dissecting the Assembly of Aβ16–22 Amyloid Peptides into Antiparallel β Sheets. Structure 2003, 11, 295–307. 10.1016/S0969-2126(03)00031-5. [DOI] [PubMed] [Google Scholar]
- Hwang W.; Zhang S.; Kamm R. D.; Karplus M. Kinetic control of dimer structure formation in amyloid fibrillogenesis. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 12916–12921. 10.1073/pnas.0402634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanakaran S.; Nussinov R.; Garcia A. E. Atomic-level description of amyloid beta-dimer formation. J. Am. Chem. Soc. 2006, 128, 2158–2159. 10.1021/ja0548337. [DOI] [PubMed] [Google Scholar]
- Nguyen P. H.; Li M. S.; Derreumaux P. Effects of all-atom force fields on amyloid oligomerization: replica exchange molecular dynamics simulations of the Abeta(16-22) dimer and trimer. Phys. Chem. Chem. Phys. 2011, 13, 9778–9788. 10.1039/c1cp20323a. [DOI] [PubMed] [Google Scholar]
- Okumura H.; Itoh S. G. Molecular dynamics simulations of amyloid-beta(16-22) peptide aggregation at air-water interfaces. J. Chem. Phys. 2020, 152, 095101 10.1063/1.5131848. [DOI] [PubMed] [Google Scholar]
- Ngoc L. L. N.; Itoh S. G.; Sompornpisut P.; Okumura H. Replica-permutation molecular dynamics simulations of an amyloid-β(16–22) peptide and polyphenols. Chem. Phys. Lett. 2020, 758, 137913 10.1016/j.cplett.2020.137913. [DOI] [Google Scholar]
- Tarus B.; Straub J. E.; Thirumalai D. Probing the initial stage of aggregation of the Abeta(10-35)-protein: assessing the propensity for peptide dimerization. J. Mol. Biol. 2005, 345, 1141–1156. 10.1016/j.jmb.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Itoh S. G.; Okamoto Y. Amyloid-β(29-42) dimer formations studied by a multicanonical-multioverlap molecular dynamics simulation. J. Phys. Chem. B 2008, 112, 2767–2770. 10.1021/jp712170h. [DOI] [PubMed] [Google Scholar]
- Lu Y.; Wei G.; Derreumaux P. Effects of G33A and G33I mutations on the structures of monomer and dimer of the amyloid-beta fragment 29-42 by replica exchange molecular dynamics simulations. J. Phys. Chem. B 2011, 115, 1282–1288. 10.1021/jp110269a. [DOI] [PubMed] [Google Scholar]
- Itoh S. G.; Okumura H. Dimerization process of amyloid-β(29-42) studied by the Hamiltonian replica-permutation molecular dynamics simulations. J. Phys. Chem. B 2014, 118, 11428–11436. 10.1021/jp505984e. [DOI] [PubMed] [Google Scholar]
- Itoh S. G.; Okumura H. Oligomer Formation of Amyloid-β(29-42) from Its Monomers Using the Hamiltonian Replica-Permutation Molecular Dynamics Simulation. J. Phys. Chem. B 2016, 120, 6555–6561. 10.1021/acs.jpcb.6b03828. [DOI] [PubMed] [Google Scholar]
- Tarus B.; Tran T. T.; Nasica-Labouze J.; Sterpone F.; Nguyen P. H.; Derreumaux P. Structures of the Alzheimer’s Wild-Type Abeta1-40 Dimer from Atomistic Simulations. J. Phys. Chem. B 2015, 119, 10478–10487. 10.1021/acs.jpcb.5b05593. [DOI] [PubMed] [Google Scholar]
- Man V. H.; Nguyen P. H.; Derreumaux P. Conformational Ensembles of the Wild-Type and S8C Abeta1-42 Dimers. J. Phys. Chem. B 2017, 121, 2434–2442. 10.1021/acs.jpcb.7b00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilie I. M.; Caflisch A. Simulation Studies of Amyloidogenic Polypeptides and Their Aggregates. Chem. Rev. 2019, 119, 6956–6993. 10.1021/acs.chemrev.8b00731. [DOI] [PubMed] [Google Scholar]
- Barz B.; Liao Q.; Strodel B. Pathways of Amyloid-beta Aggregation Depend on Oligomer Shape. J. Am. Chem. Soc. 2018, 140, 319–327. 10.1021/jacs.7b10343. [DOI] [PubMed] [Google Scholar]
- Liao Q.; Owen M. C.; Bali S.; Barz B.; Strodel B. Aβ under stress: the effects of acidosis, Cu2+-binding, and oxidation on amyloid β-peptide dimers. Chem. Commun. 2018, 54, 7766–7769. 10.1039/C8CC02263A. [DOI] [PubMed] [Google Scholar]
- Fatafta H.; Khaled M.; Owen M. C.; Sayyed-Ahmad A.; Strodel B. Amyloid-β peptide dimers undergo a random coil to β-sheet transition in the aqueous phase but not at the neuronal membrane. Proc. Natl. Acad. Sci. U.S.A. 2021, 118, e2106210118 10.1073/pnas.2106210118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukushima K.; Nemoto K. Exchange Monte Carlo method and application to spin glass simulations. J. Phys. Soc. Jpn. 1996, 65, 1604–1608. 10.1143/JPSJ.65.1604. [DOI] [Google Scholar]
- Sugita Y.; Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999, 314, 141–151. 10.1016/S0009-2614(99)01123-9. [DOI] [Google Scholar]
- Itoh S. G.; Okumura H. Replica-Permutation Method with the Suwa-Todo Algorithm beyond the Replica-Exchange Method. J. Chem. Theory Comput. 2013, 9, 570–581. 10.1021/ct3007919. [DOI] [PubMed] [Google Scholar]
- Metropolis N.; Rosenbluth A. W.; Rosenbluth M. N.; Teller A. H.; Teller E. Equation of State Calculations by Fast Computing Machines. J. Chem. Phys. 1953, 21, 1087–1092. 10.1063/1.1699114. [DOI] [Google Scholar]
- Suwa H.; Todo S. Markov chain Monte Carlo method without detailed balance. Phys. Rev. Lett. 2010, 105, 120603 10.1103/PhysRevLett.105.120603. [DOI] [PubMed] [Google Scholar]
- Itoh S. G.; Okumura H. Hamiltonian replica-permutation method and its applications to an alanine dipeptide and amyloid-β(29-42) peptides. J. Comput. Chem. 2013, 34, 2493–2497. 10.1002/jcc.23402. [DOI] [PubMed] [Google Scholar]
- Ferrenberg A. M.; Swendsen R. H. Optimized Monte Carlo data analysis. Phys. Rev. Lett. 1989, 63, 1195–1198. 10.1103/PhysRevLett.63.1195. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Rosenberg J. M.; Bouzida D.; Swendsen R. H.; Kollman P. A. THE weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992, 13, 1011–1021. 10.1002/jcc.540130812. [DOI] [Google Scholar]
- Kabsch W.; Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Bitan G.; Kirkitadze M. D.; Lomakin A.; Vollers S. S.; Benedek G. B.; Teplow D. B. Amyloid beta -protein (Abeta) assembly: Abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 330–335. 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abelein A.; Abrahams J. P.; Danielsson J.; Graslund A.; Jarvet J.; Luo J.; Tiiman A.; Warmlander S. K. The hairpin conformation of the amyloid beta peptide is an important structural motif along the aggregation pathway. JBIC, J. Biol. Inorg. Chem. 2014, 19, 623–634. 10.1007/s00775-014-1131-8. [DOI] [PubMed] [Google Scholar]
- Maity S.; Hashemi M.; Lyubchenko Y. L. Nano-assembly of amyloid beta peptide: role of the hairpin fold. Sci. Rep. 2017, 7, 2344 10.1038/s41598-017-02454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang H. L.; Chen C. J.; Okumura H.; Hu C. K. Transformation between alpha-helix and beta-sheet structures of one and two polyglutamine peptides in explicit water molecules by replica-exchange molecular dynamics simulations. J. Comput. Chem. 2014, 35, 1430–1437. 10.1002/jcc.23633. [DOI] [PubMed] [Google Scholar]
- Araki M.; Tamura A. Transformation of an alpha-helix peptide into a beta-hairpin induced by addition of a fragment results in creation of a coexisting state. Proteins 2007, 66, 860–868. 10.1002/prot.21263. [DOI] [PubMed] [Google Scholar]
- Itoh S. G.; Tamura A.; Okamoto Y. Helix-Hairpin Transitions of a Designed Peptide Studied by a Generalized-Ensemble Simulation. J. Chem. Theory Comput. 2010, 6, 979–983. 10.1021/ct9005932. [DOI] [Google Scholar]
- Okumura H.; Itoh S. G. Transformation of a design peptide between the alpha-helix and beta-hairpin structures using a helix-strand replica-exchange molecular dynamics simulation. Phys. Chem. Chem. Phys. 2013, 15, 13852–13861. 10.1039/c3cp44443k. [DOI] [PubMed] [Google Scholar]
- Chakraborty D.; Chebaro Y.; Wales D. J. A multifunnel energy landscape encodes the competing alpha-helix and beta-hairpin conformations for a designed peptide. Phys. Chem. Chem. Phys. 2020, 22, 1359–1370. 10.1039/C9CP04778F. [DOI] [PubMed] [Google Scholar]
- Karplus M.; Kushick J. N. Method for Estimating the Configurational Entropy of Macromolecules. Macromolecules 1981, 14, 325–332. 10.1021/ma50003a019. [DOI] [Google Scholar]
- Ball K. A.; Phillips A. H.; Nerenberg P. S.; Fawzi N. L.; Wemmer D. E.; Head-Gordon T. Homogeneous and Heterogeneous Tertiary Structure Ensembles of Amyloid-β Peptides. Biochemistry 2011, 50, 7612–7628. 10.1021/bi200732x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise-Scira O.; Xu L.; Kitahara T.; Perry G.; Coskuner O. Amyloid-β peptide structure in aqueous solution varies with fragment size. J. Chem. Phys. 2011, 135, 205101 10.1063/1.3662490. [DOI] [PubMed] [Google Scholar]
- Coskuner O.; Wise-Scira O.; Perry G.; Kitahara T. The Structures of the E22Δ Mutant-Type Amyloid-β Alloforms and the Impact of E22Δ Mutation on the Structures of the Wild-Type Amyloid-β Alloforms. ACS Chem. Neurosci. 2013, 4, 310–320. 10.1021/cn300149j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B.; Simons M.; Multhaup G.; Van Leuven F.; Beyreuther K.; Dotti C. G. Production of intracellular amyloid-containing fragments in hippocampal neurons expressing human amyloid precursor protein and protection against amyloidogenesis by subtle amino acid substitutions in the rodent sequence. EMBO J. 1995, 14, 4932–4938. 10.1002/j.1460-2075.1995.tb00176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduslo J. F.; Gilles E. J.; Ramakrishnan M.; Howell K. G.; Wengenack T. M.; Curran G. L.; Kandimalla K. K. HH domain of Alzheimer’s disease Abeta provides structural basis for neuronal binding in PC12 and mouse cortical/hippocampal neurons. PLoS One 2010, 5, e8813 10.1371/journal.pone.0008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroutanpay B. V.; Kumar J.; Kang S. G.; Danaei N.; Westaway D.; Sim V. L.; Kar S. The Effects of N-terminal Mutations on beta-amyloid Peptide Aggregation and Toxicity. Neuroscience 2018, 379, 177–188. 10.1016/j.neuroscience.2018.03.014. [DOI] [PubMed] [Google Scholar]
- Dai X.; Chang P.; Liu W.; Xu K.; Sun Y.; Zhu S.; Jiang Z. Abeta-40 Y10F increases betafibrils formation but attenuates the neurotoxicity of amyloid-beta peptide. Int. J. Mol. Sci. 2012, 13, 5324–5337. 10.3390/ijms13055324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V.; Abel R.; Okur A.; Strockbine B.; Roitberg A.; Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984, 52, 255–268. 10.1080/00268978400101201. [DOI] [Google Scholar]
- Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. 10.1063/1.447334. [DOI] [Google Scholar]
- Hoover W. G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695–1697. 10.1103/PhysRevA.31.1695. [DOI] [PubMed] [Google Scholar]
- Okumura H.; Itoh S. G.; Okamoto Y. Explicit symplectic integrators of molecular dynamics algorithms for rigid-body molecules in the canonical, isobaric-isothermal, and related ensembles. J. Chem. Phys. 2007, 126, 084103 10.1063/1.2434972. [DOI] [PubMed] [Google Scholar]
- Allen M. P.; Tildesley D. J.. Computer Simulation of Liquids: Second Edition, 2nd ed.; Oxford University Press: Oxford, 2017. [Google Scholar]
- Berg B. A.Markov Chain Monte Carlo Simulations and Their Statistical Analysis; World Scientific, 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.









