Photosynthetic plants have a special place in living nature as they generate unbelievable amounts of carbohydrates, i.e., ca. 150 × 1012 kg on a global scale per year, and they are responsible for the production of the molecular oxygen in the Earth’s atmosphere. Thus, historically, studies of plants have been associated in the minds of scientists and laypeople alike with agriculture and related biotechnology fields. However, it is very far from reality. Already decades ago, plants joined the ranks of other higher model organisms, such as insects and animals, both from the standpoint of conceptual and technical sophistication of experimental approaches as well as their influence on our advances in the knowledge of biology.
Unlike most other organisms, plants as individuals have no capacity for travel and thus are unable to escape their surroundings. Along the eons of their evolution therefore plants have accumulated many biological capabilities to adapt to the changing biotic and abiotic environments. For example, plants can alter the degree of lipid unsaturation in their cellular membranes, and, therefore, alter the membrane fluidity, depending on the ambient temperature, i.e., increase it in the cold and decrease it in the heat [1]. Or, plants, in which the outside access to the cell membrane is limited due to the cell walls that encase the individual cells, have evolved intercellular connections, termed plasmodesmata [2], which are gateable and allow cell-to-cell transport of macromolecules and macromolecular structures, from nucleic acids to proteins to subviral particles [3–6]. Whereas the plasmodesmata were discovered more than 120 years ago [7], the concept of macromolecule/particle transport through intercellular connections emerged only 20 years ago with the discovery of membrane tunneling nanotubes (TNTs) [8], and since then TNTs have been shown to traffic large macromolecules and viruses [9–14], translating the concept developed in pants to the animal organisms. Even the discovery of viruses and, consequently, the science of virology began with the Tobacco mosaic virus (TMV), a plant RNA virus [15,16]. This cell-to-cell movement of plant viruses through plasmodesmata was one of the two biological (and, at the time, largely enigmatic) processes that attracted me to plant biology. Another process historically studied in plant systems is genetic engineering. In nature, diverse species of plants are genetically modified by a bacterial pathogen Agrobacterium tumefaciens which transfers to plants and integrates into their genome a segment of its plasmid DNA, termed T-DNA; the genes contained in the T-DNA induce neoplastic cell growth as well as biosynthesis of opines, amino acid derivatives that are secreted by the tumor cells into the environment and are used by Agrobacterium as carbon and nitrogen sources. In the laboratory, the native T-DNA is re- placed by the sequences of interest and used to produce transgenic plants [17–22]. Like Agrobacterium, many human pathogenic bacteria, e.g., Bartonella, Legionella, Helicobacter, and Shigella, have evolved to export their virulence effector proteins into the host cells using the type 4 secretion system (T4SS) machinery [23], and, at least one of these bacterial species, Bartonella henselae, was reported to export and integrate into a human cell genome a plasmid reporter DNA [24,25], although it remains unknown whether Bartonella exports its own, endogenous DNA and genetically transform its human host cells in vivo, during the course of infection.
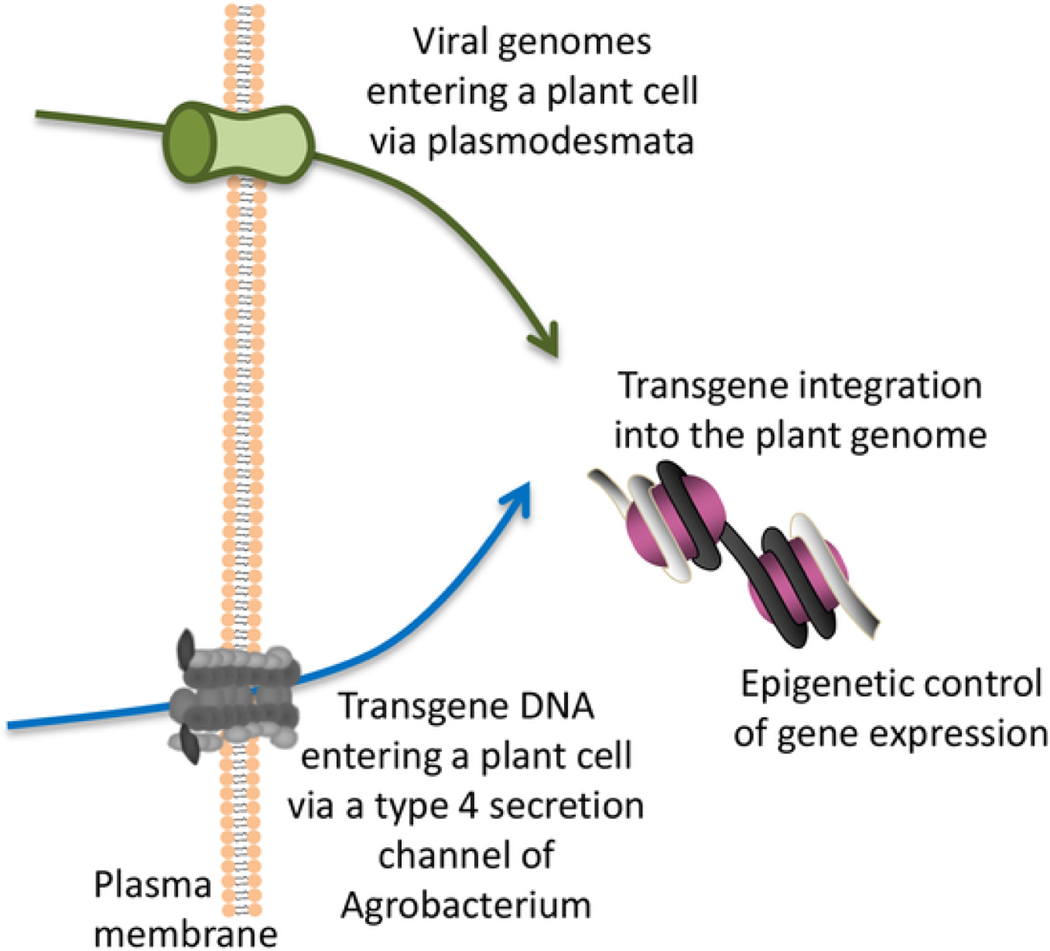
The field of plant biology continues to provide excitement and intellectual and technical challenges to all involved. This is a very broad field of science that encompasses all aspects of living nature as they occur in plants. I am especially interested in three molecular aspects of the plant life cycle—genetic modification, intercellular communication, and epigenetic regulation—which include the transport of biologically active nucleic acids, i.e., transgenes and viral genomes, and transcriptional regulation of their expression (Fig. 1). In these areas, as in many others in plant research, I look forward to the increased involvement of synthetic biology. Generally, synthetic biology principles and techniques allow manipulation and refactoring, i.e., replacement of endogenous regulation of natural gene circuits with synthetic, orthogonal regulatory elements, many aspects of eukaryotic or prokaryotic cells, including orthogonal control of gene expression, genetic toggle switches, and logic gates as means to control gene expression with precision, and computationally designed control of cell and tissue-specific expression [26–29]. For example, orthogonal systems, i.e., engineered biologically active molecules that cooperate to provide a specific biological function without affecting or being affected by the corresponding endogenous cellular systems [27,28], would allow the conversion of living cells with particular natural capabilities into nanomachines dedicated solely to that specific set of capabilities, e.g., refactoring Agrobacterium into a dedicated genetic transformation nanomachine. Because Agrobacterium can genetically modify a wide range of eukaryotic cells, from plant to yeast to human [19], under laboratory conditions, the refactored Agrobacterium cells could be adapted to specific target cells/organisms, facilitating their genetic modification for medical, research, and biotechnological purposes. Furthermore, the optimal molecular composition of the refactored Agrobacterium would also shed new light on the cellular mechanisms involved in the genetic transformation process.
Fig. 1.
Schematic illustration of three biological processes—plant genetic engineering, plant virus cell-to-cell movement, and epigenetic regulation of plant gene expression—which involve the transmembrane transport of biologically active nucleic acids molecules and represent the focus of this Commentary.
Agrobacterium preferentially integrates its T-DNA into the double-stranded DNA breaks (DSBs) in the plant genome, presumably using the cellular DNA repair mechanisms [30–33]. Thus, our investigation of plant genetic transformation by Agrobacterium represents an aspect of the broader field of studies of the plant DNA damage response pathways. Collectively, future advances in these studies should facilitate elucidation of perhaps the most important of the remaining enigmatic steps of the Agrobacterium-mediated genetic transformation: the identity of the cellular proteins and the bacterial effectors that participate in T-DNA integration, the molecular interactions between these factors, and the temporal sequence of these interactions that culminates with the integration event. Similarly, I anticipate important advances in our understanding of the molecular reactions and sequential steps of targeting protein and nucleoprotein cargos to plasmodesmata, increase in plasmodesmal permeability, transit of the plasmodesmal channel by the cargo molecules, and the energy sources for this active transport. Detailed proteomic characterization of plasmodesmata, which has already begun [34–36], will facilitate the mechanistic studies by identifying and characterizing the complement of proteins associated with plasmodesmata. For example, by analogy to the dynamic composition of the nuclear pore complex [37], plasmodesmal proteins that are early in plasmodesmata assembly and stable in their residence time are likely structural in function whereas those that are late in the assembly and transient in the residence time are likely directly involved in the cell-to-cell transport process.
Both plant genetic transformation and, in many cases, plasmodesmal transport of viral and endogenous nucleoprotein complexes ultimately lead up to the expression of the transferred nucleic acid molecules, i.e., transgenes, viral genomic DNA or RNA, or non-cell-autonomous transcripts. One major factor in regulating the expression and/or formation of these molecules is the posttranslational modification of histones, which determines the active or inactive state of the chromatin. These modifications, which together determine the transcriptional outcomes [38,39], are dynamic, effected by writers and erasers, i.e., histone modifying enzymes that add or remove the specific functional groups [40]. In plant cells, one major class of important, yet relatively sparsely characterized, erasers are histone deubiquitinases that have been implicated in diverse aspects of physiology, growth, and development [41–47]. Only about 10% of all 50 deubiquitinases encoded by the Arabidopsis genome [48] have been proposed to target histones and participate in epigenetic regulation [46,47,49–56]. Of these, two histone deubiquitinases, UBP26 and OTLD1, participate both in transcriptional repression and activation of their target gene expression [46,47,51,53,54], with the dual function of OTLD1 likely to be direct [46,47]. Future understanding of how deubiquitylation of the same type of histone by the same histone deubiquitinase can elicit two opposing effects on transcription of the direct target genes may help will define a potential junction in chromatin remodeling pathways leading to transcriptional repression or activation.
This short Commentary reflects my own specific and relatively narrow scientific interests. Yet, it illustrates how plant-centered model systems contribute to our understanding of diverse and fundamental aspects of life.
Acknowledgment
The work in the V.C. laboratory is supported by grants from NIH, NSF, USDA/NIFA, and BARD to V.C.
Footnotes
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Vitaly Citovsky reports financial support was provided by National Institutes of Health. Vitaly Citovsky reports financial support was pro-vided by National Science Foundation. Vitaly Citovsky reports financial support was provided by United States Israel Binational Agricultural Research and Development Fund.
References
- [1].Falcone DL, Ogas JP, Somerville CR, Regulation of membrane fatty acid composition by temperature in mutants of Arabidopsis with alterations in membrane lipid composition, BMC Plant Biol 4 (2004) 17, 10.1186/1471-2229-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Robards AW, Lucas WJ, Plasmodesmata, Annu. Rev. Plant Physiol. Plant Mol. Biol. 41 (1990) 369–419. [Google Scholar]
- [3].Gibbs AJ, Viruses and plasmodesmata, in: Gunning BES, Robards AW (Eds.), Intercellular Communication in Plants: Studies on Plasmodesmata, Springer-Verlag, Berlin, 1976, pp. 149–164. [Google Scholar]
- [4].Oparka KJ, Plasmodesmata, Annual Plant Reviews, Blackwell Publishing, Oxford, 2005, p. 311. [Google Scholar]
- [5].Citovsky V, Probing plasmodesmal transport with plant viruses, Plant Physiol 102 (1993) 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tzfira T, Rhee Y, Chen MH, Citovsky V, Nucleic acid transport in plant-microbe interactions: the molecules that walk through the walls, Annu. Rev. Microbiol. 54 (2000) 187–219. [DOI] [PubMed] [Google Scholar]
- [7].Strasburger E, Ueber Plasmaverbindungen pflanzlicher Zellen, Jahrb. Wiss. Bot. 36 (1901) 493–601. [Google Scholar]
- [8].Ramirez-Weber FA, Kornberg TB, Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs, Cell 97 (1999) 599–607. [DOI] [PubMed] [Google Scholar]
- [9].Abounit S, Delage E, Zurzolo C, Identification and characterization of tunneling nanotubes for intercellular trafficking, Curr. Protoc. Cell Biol. 67 (2015) 12.10.11–12.10.21. [DOI] [PubMed] [Google Scholar]
- [10].Abounit S, Wu JW, Duff K, Victoria GS, Zurzolo C, Tunneling nanotubes: a possible highway in the spreading of tau and other prion-like proteins in neurodegenerative diseases, Prion 10 (2016) 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kumar A, Kim JH, Ranjan P, Metcalfe MG, Cao W, Mishina M, Gangappa S, Guo Z, Boyden ES, Zaki S, York I, Garcia-Sastre A, Shaw M, Sambhara S, Influenza virus exploits tunneling nanotubes for cell-to-cell spread, Sci. Rep. 7 (2017) 40360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Panasiuk M, Rychłowski M, Derewońko N, Bieńkowska-Szewczyk K, Tunneling nanotubes (TNT) as a novel route of cell-to-cell spread of herpesviruses, J. Virol. 92 (2018) e00090–00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cordero Cervantes D, Zurzolo C, Peering into tunneling nanotubes. The path forward, EMBO J 40 (2021) e105789, 10.15252/embj.2020105789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Haimovich G, Dasgupta S, Gerst JE, RNA transfer through tunneling nanotubes, Biochem. Soc. Trans. 49 (2021) 145–160, 10.1042/BST20200113. [DOI] [PubMed] [Google Scholar]
- [15].Citovsky V, Tobacco mosaic virus: a pioneer of cell-to-cell movement, Philos. Trans. R. Soc. Lond. B 354 (1999) 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Creager ANH, Scholthof KBG, Citovsky V, Scholthof HB, Tobacco mosaic virus: pioneering research for a century, Plant Cell 11 (1999) 301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nester EW, Agrobacterium: nature’s genetic engineer, Front. Plant Sci. 5 (2015) 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zupan J, Muth TR, Draper O, Zambryski PC, The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights, Plant J 23 (2000) 11–28. [DOI] [PubMed] [Google Scholar]
- [19].Lacroix B, Tzfira T, Vainstein A, Citovsky V, A case of promiscuity: Agrobacterium’s endless hunt for new partners, Trends Genet 22 (2006) 29–37. [DOI] [PubMed] [Google Scholar]
- [20].Lacroix B, Citovsky V, Pathways of DNA transfer to plants from Agrobacterium tumefaciens and related bacterial species, Annu. Rev. Phytopathol. 57 (2019) 231–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lacroix B, Citovsky V, Genetically modified organisms, plant transformation by Agrobacterium, in: Trefil J. (Ed.) Discoveries in Modern Science: Exploration, Invention, Technology, Farmington Hills: Macmillan; 2015, pp. 425–431. [Google Scholar]
- [22].Lacroix B, Citovsky V, Genetic factors governing bacterial virulence and host plant susceptibility during Agrobacterium infection, Adv. Genet. (2022), in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E, Biogenesis, architecture, and function of bacterial type IV secretion systems, Annu. Rev. Microbiol. 59 (2005) 451–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schröder G, Schuelein R, Quebatte M, Dehio C, Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae, Proc. Natl. Acad. Sci. USA 108 (2011) 14643–14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fernández-González E, de Paz HD, Alperi A, Agúndez L, Faustmann M, Sangari FJ, Dehio C, Llosa M, Transfer of R388 derivatives by a pathogenesis-associated type IV secretion system into both bacteria and human cells, J. Bacteriol. 193 (2011) 6257–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Endy D, Foundations for engineering biology, Nature 438 (2005) 449–453. [DOI] [PubMed] [Google Scholar]
- [27].Costello A, Badran AH, Synthetic biological circuits within an orthogonal central dogma, Trends Biotechnol 39 (2021) 59–71, 10.1016/j.tibtech.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu CC, Jewett MC, Chin JW, Voigt CA, Toward an orthogonal central dogma, Nat. Chem Biol 14 (2018) 103–106, 10.1038/nchembio.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Qian Y, Kong W, Lu T, Precise and reliable control of gene expression in Agrobacterium tumefaciens, Biotechnol. Bioeng. 118 (2021) 3962–3972, 10.1002/bit.27872. [DOI] [PubMed] [Google Scholar]
- [30].Puchta H, Repair of genomic double-strand breaks in somatic plant cells by one- sided invasion of homologous sequences, Plant J 13 (1998) 77–78. [Google Scholar]
- [31].Chilton MD, Que Q, Targeted integration of T-DNA into the tobacco genome at double-strand breaks: new insights on the mechanism of T-DNA integration, Plant Physiol 133 (2003) 956–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tzfira T, Frankmen L, Vaidya M, Citovsky V, Site-specific integration of Agrobacterium T-DNA via double-stranded intermediates, Plant Physiol 133 (2003) 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kralemann LEM, de Pater S, Shen H, Kloet SL, van Schendel R, Hooykaas PJJ, Tijsterman M, Distinct mechanisms for genomic attachment of the 5′ and 3′ ends of Agrobacterium T-DNA in plants, Nat. Plants 8 (2022) 526–534, 10.1038/s41477-022-01147-5. [DOI] [PubMed] [Google Scholar]
- [34].He R, Bernards MA, Wang A, Purification of plasmodesmata-enriched fraction for proteomic analyses, Methods Mol. Biol. 2400 (2022) 115–123, 10.1007/978-1-0716-1835-6_12. [DOI] [PubMed] [Google Scholar]
- [35].Kirk P, Amsbury S, German L, Gaudioso-Pedraza R, Benitez-Alfonso Y, A comparative meta-proteomic pipeline for the identification of plasmodesmata proteins and regulatory conditions in diverse plant species, BMC Biol 20 (2022) 128, 10.1186/s12915-022-01331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Salmon MS, Bayer EM, Dissecting plasmodesmata molecular composition by mass spectrometry-based proteomics, Front. Plant Sci. 3 (2012) 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Raices M, D’Angelo MA, Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions, Nat. Rev. Mol. Cell Biol. 13 (2012) 687–699, 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- [38].Lee JS, Smith E, Shilatifard A, The language of histone crosstalk, Cell 142 (2010) 682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Suganuma T, Workman JL, Crosstalk among histone modifications, Cell 135 (2008) 604–607. [DOI] [PubMed] [Google Scholar]
- [40].Yun M, Wu J, Workman JL, Li B, Readers of histone modifications, Cell Res 21 (2011) 564–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bourbousse C, Ahmed I, Roudier F, Zabulon G, Blondet E, Balzergue S, Colot V, Bowler C, Barneche F, Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis, PLOS Genet 8 (2012) e1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fleury D, Himanen K, Cnops G, Nelissen H, Boccardi TM, Maere S, Beemster GT, Neyt P, Anami S, Robles P, Micol JL, Inzé D, Van Lijsebettens M, The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth, Plant Cell 19 (2007) 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gu X, Jiang D, Wang Y, Bachmair A, He Y, Repression of the floral transition via histone H2B monoubiquitination, Plant J 57 (2009) 522–533. [DOI] [PubMed] [Google Scholar]
- [44].Himanen K, Woloszynska M, Boccardi TM, De Groeve S, Nelissen H, Bruno L, Vuylsteke M, Van Lijsebettens M, Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis, Plant J 72 (2012) 249–260. [DOI] [PubMed] [Google Scholar]
- [45].Liu Y, Koornneef M, Soppe WJJ, The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy, Plant Cell 19 (2007) 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Keren I, Citovsky V, The histone deubiquitinase OTLD1 targets euchromatin to regulate plant growth, Sci. Signal. 9 (2016) ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Keren I, Citovsky V, Activation of gene expression by histone deubiquitinase OTLD1, Epigenetics 12 (2017) 584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Isono E, Nagel MK, Deubiquitylating enzymes and their emerging role in plant biology, Front. Plant Sci. 5 (2014) 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Derkacheva M, Liu S, Figueiredo DD, Gentry M, Mozgova I, Nanni P, Tang M, Mannervik M, Köhler C, Hennig L, H2A deubiquitinases UBP12/13 are part of the Arabidopsis polycomb group protein system, Nat. Plants 2 (2016) 16126. [DOI] [PubMed] [Google Scholar]
- [50].Suen DF, Tsai YH, Cheng YT, Radjacommare R, Ahirwar RN, Fu H, Schmidt W, The deubiquitinase OTU5 regulates root responses to phosphate starvation, Plant Physiol 176 (2018) 2441–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Schmitz RJ, Tamada Y, Doyle MR, Zhang X, Amasino RM, Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis, Plant Physiol 149 (2009) 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, Hasegawa PM, Bressan RA, Zhu JK, Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination, Nature 447 (2007) 735–738. [DOI] [PubMed] [Google Scholar]
- [53].Luo M, Luo MZ, Buzas D, Finnegan J, Helliwell C, Dennis ES, Peacock WJ, Chaudhury A, UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis, Genetics 180 (2008) 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Krichevsky A, Lacroix B, Zaltsman A, Citovsky V, Involvement of KDM1C histone demethylase-OTLD1 otubain-like histone deubiquitinase complexes in plant gene repression, Proc. Natl. Acad. Sci. USA 108 (2011) 11157–11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Phan MSV, Keren I, Tran PT, Lapidot M, Citovsky V, Arabidopsis LSH10 transcription factor interacts with the co-repressor histone deubiquitinase OTLD1 to recruit it to the target promoters, bioRxiv (2022), 10.1101/2022.07.30.502139. [DOI] [Google Scholar]
- [56].Keren I, Lacroix B, Kohrman A, Citovsky V, Histone deubiquitinase OTU1 epigenetically regulates DA1 and DA2, which control Arabidopsis seed and organ size, iScience 23 (2020) 100948. [DOI] [PMC free article] [PubMed] [Google Scholar]