Abstract
A murine model of disseminated candidiasis was utilized to determine whether Candida albicans Als proteins are produced in vivo. The kidneys, spleen, heart, liver, and lungs were collected from mice inoculated with one of three C. albicans strains (SC5314, B311, or WO-1). Immunohistochemical analysis of murine tissues by using a rabbit polyclonal anti-Als serum indicated that Als proteins were produced by each C. albicans cell in the tissues examined. Patterns of staining with the anti-Als serum were similar among the C. albicans strains tested. These data indicated that Als protein production was widespread in disseminated candidiasis and that, despite strain differences in ALS gene expression previously noted in vitro, Als protein production in vivo was similar among C. albicans strains. The extensive production of Als proteins in vivo and their presence on the C. albicans cell wall position these proteins well for a role in host-pathogen interaction.
The fungus Candida albicans can cause a variety of disease states in the human or animal host. One form of candidiasis is a disseminated disease that primarily affects immunocompromised individuals (15). Much work has been done to define the mechanisms that C. albicans uses to cause disease. Among the attributes of C. albicans that have been positively correlated with pathogenic potential is the adhesive capacity of the organism (1). The ability to adhere to host surfaces contributes to the process of colonization and, therefore, may facilitate tissue invasion. The study of C. albicans adherence to host tissue has identified several adhesion proteins in the organism (2, 4, 6).
Genes of the C. albicans ALS (agglutinin-like sequence) family encode proteins with features of cell surface adhesion glycoproteins (8–10). Cell surface localization of Als proteins on C. albicans grown in vitro was demonstrated by indirect immunofluorescence by using a rabbit polyclonal anti-Als serum (9). The adhesive function of Als proteins has been suggested by two studies in which C. albicans ALS genes expressed in Saccharomyces cerevisiae conferred an adhesive phenotype on the normally nonadherent S. cerevisiae (3, 5). Molecular characterization of the ALS genes demonstrated that they constitute a large family, with several genes remaining to be characterized (8–10). Based on the known ALS gene sequences, the Als protein profile on the surface of any given C. albicans strain is predicted to be highly variable since ALS genes vary in size and number among different C. albicans isolates (8–10). Much of the variability in the predicted Als protein profile is attributable to differences in the overall length of the tandem repeat domain between the same gene in different strains and, in many cases, between alleles of a gene in the same strain (8–10). The N-terminal domain of Als proteins, which may include the adhesive portion of the molecule, is conserved between proteins in the family; however, it is unclear whether all Als proteins share the same function (9).
Molecular analysis of ALS gene expression demonstrated that ALS genes are differentially regulated by physiologically relevant mechanisms, such as growth stage and morphological form (8–10). In addition, mechanisms of ALS gene expression were noted to differ between C. albicans strains. One notable example is lack of expression of ALS1 in the strain WO-1 under growth conditions sufficient for its expression in several other strains (8). Although study of ALS gene expression in vitro has revealed several characteristics of the gene family, documentation of Als protein production in vivo is essential to support the idea that Als proteins are cell surface adhesion glycoproteins with the potential to promote C. albicans pathogenesis. The main goals of this work are to determine whether Als proteins are produced in a murine model of disseminated candidiasis and to examine whether Als protein production varies with host site. An additional goal is to determine if Als protein production varies by strain in vivo, especially in strains such as WO-1, which differs with respect to in vitro ALS1 expression.
Three different C. albicans strains were used in this study: SC5314 (a gift from W. A. Fonzi), B311 (American Type Culture Collection), and WO-1 (a gift from D. R. Soll). For each C. albicans strain, a single colony from a yeast extract-peptone-dextrose agar plate was inoculated into 50 ml of yeast extract-peptone-dextrose medium and grown at 30°C with shaking (200 rpm). The colony selected for strain WO-1 was in the white phase of growth. After overnight incubation, the cultures had a cell density of approximately 6 × 108 cells/ml. Microscopic examination of cells from each culture indicated that all strains grew in the yeast morphology. Ten milliliters of each culture was removed and washed twice in sterile phosphate-buffered saline (PBS). Cells were then resuspended in 10 ml of PBS, placed in a 50-ml flask, and incubated at 37°C for 30 min with shaking. Cells were harvested and resuspended in fresh PBS at a density of 106 cells per 0.5 ml. Cells were kept on ice and then warmed to 37°C immediately prior to injection into mice.
Seven-week-old, female BALB/cByJ mice weighing between 20 and 24 g (Jackson Labs) were inoculated by tail vein injection with 0.5 ml of the yeast suspension; control mice were inoculated with the vehicle alone. Seven mice were inoculated with a given C. albicans strain, and a total of three control mice were used. Because the goal of the experiment was to follow mice over the course of infection and compare what was observed for the three C. albicans strains, mice were killed at various time points by cervical dislocation (Table 1). Animals that died from the C. albicans infection were also evaluated. Mice in moribund condition were killed irrespective of the planned schedule (Table 1).
TABLE 1.
Experimental design and survivability
| Mouse no. | C. albicans strain | Days postinfection/dispositiona |
|---|---|---|
| 1 | None | 1/S |
| 2 | None | 1/S |
| 3 | WO-1 | 1/S |
| 4 | WO-1 | 1/S |
| 5 | B311 | 1/S |
| 6 | B311 | 1/S |
| 7 | SC5314 | 1/S |
| 8 | SC5314 | 1/S |
| 9 | SC5314 | 2/D |
| 10 | SC5314 | 2/D |
| 11 | B311 | 2/S |
| 12 | WO-1 | 2/S |
| 13 | SC5314 | 3/D |
| 14 | B311 | 3/S |
| 15 | B311 | 3/D |
| 16 | B311 | 3/S |
| 17 | WO-1 | 3/S |
| 18 | WO-1 | 3/S |
| 19 | SC5314 | 3/D |
| 20 | SC5314 | 3/D |
| 21 | WO-1 | 5/M |
| 22 | B311 | 6/M |
| 23 | None | 7/S |
| 24 | WO-1 | 7/S |
Mice were either sacrificed as scheduled (S), died from the C. albicans infection (D), or were sacrificed off schedule due to moribund condition (M).
The kidneys, spleen, heart, liver, and lungs were removed from each animal, and a small portion of each organ was weighed and homogenized in sterile PBS. The homogenate was plated onto Sabouraud dextrose agar (11) to verify the presence of viable C. albicans in each organ. Rigorous quantitation of CFUs was not undertaken, because the goals of this study were qualitative. Samples from each organ were fixed in 10% neutral-buffered formalin (pH 7.2) for approximately 16 h at 25°C, embedded in paraffin, sliced into 3-μm sections, and mounted on treated glass slides for histopathological and immunohistochemical analyses. Sections were stained with hematoxylin and eosin for routine evaluation, with periodic acid-Schiff stain, which reacts with carbohydrate in the fungal cell wall, and with Gomori methenamine silver (GMS), a silver-based stain that impregnates the fungal cell wall polysaccharides, staining them black (13).
Histopathology analysis was performed to compare the pathogenic processes of strains SC5314, B311, and WO-1 in BALB/cByJ mice. Scores ranging from zero to ++++ were assigned to describe the relative numbers of yeast and hyphae in each tissue and to evaluate features of the pathogenic process including necrosis, presence of inflammatory cells, and suppuration. Analysis of each organ from the 24 mice was conducted, and the results were tabulated by days of infection within each strain. Trends for each strain were compiled from these tables, and an overall summary statement was written for each organ. In general, the progression of experimental disseminated infection was consistent with observations by other researchers (reviewed in reference 15). In order of decreasing severity, lesions were found in the kidneys, spleen, heart, liver, and lungs. For all three strains, lesions in the right and left kidneys were similar. General trends for the three C. albicans strains and some notable observations are summarized here to serve as the basis for interpretation of immunohistochemical staining experiments with the anti-Als serum (see below).
Among the C. albicans strains used in this study, SC5314 was the most pathogenic, causing, within 72 h, the death of all remaining mice injected with this strain (Table 1). The greater severity of SC5314 infection relative to the other two strains was also evidenced by the greater number of C. albicans cells present in tissue sections and by increased inflammation and necrosis. SC5314 formed more hyphae in vivo than the other two strains. Lesions from mice infected with C. albicans B311 were less severe than SC5314 lesions; WO-1 caused the least damage and, quite notably, formed fewer hyphae in vivo than either of the other strains. Differences in the extent of angioinvasion were noted between the three strains, with SC5314 being the most angioinvasive and WO-1 being the least angioinvasive. In mice surviving 5 to 7 days after infection, the overall numbers of C. albicans colonies and cells declined significantly, and most lesions, with the exception of those in the kidneys, were at least partially resolved.
In kidney lesions caused by all three strains, C. albicans colonies first appeared in the cortex, where they extended from the interstitium into the tubules, accompanied by large multifocal (WO-1 and B311) to coalescing (SC5314) areas of liquefactive necrosis with numerous neutrophils. By 5 to 7 days postinoculation, the microabscesses in the cortex and outer medulla contained fewer C. albicans cells, and these were surrounded by macrophages and fibroblasts. In contrast, a florid pyelonephritis with multifocal microabscesses centered around abundant C. albicans colonies scattered in the renal papilla attested to the spread of C. albicans along the nephron. The pelvic urothelium of one B311-infected mouse (number 22) was ulcerated by arborescing fungal hyphae. Prominent vasculitis and thrombosis were observed in mice infected with SC5314.
In the spleen, remarkably few C. albicans cells were detected at 24 h postinoculation. Hyphal colonies were more prominent in mice infected with strain SC5314 than in other strains. In mouse 10, SC5314 cells were associated with areas of necrosis and angioinvasion. By 5 to 7 days postinfection, fungal hyphae were no longer apparent in the spleen of mice inoculated with B311 or WO-1, and the yeast numbers had declined considerably in the splenic sinusoids.
Cardiac lesions, characterized by small multifocal areas of cardiomyocyte necrosis and aggregates of neutrophils, were obvious within 24 h postinfection for all three strains. Lesions were directly caused by C. albicans, as evidenced by the presence of yeasts and/or germ tubes in the fragmented cardiomyocytes. One B311-infected mouse (number 14) exhibited atrial thrombosis associated with C. albicans colonies. After 7 days, WO-1- and B311-induced lesions had mainly progressed to multifocal fibrosis with few or no detectable organisms.
In the liver, a few C. albicans cells from all three strains were present at 24, 48, and 72 h postinfection in the sinusoids, predominantly in the Kupffer cells. They elicited only minimal tissue reaction, being occasionally associated with minute aggregates of neutrophils. By 6 to 7 days, the rare remaining yeasts were detected almost exclusively in Kupffer cells. Only SC5314, at 48 h postinoculation, formed small colonies which arose multifocally from Kupffer cells, hepatocytes, or sinusoids.
Pulmonary tissue was the least affected by the experimental C. albicans infection. For all three C. albicans strains, rare yeasts and germ tubes were present multifocally in the pulmonary parenchyma, mostly in alveolar macrophages. Only rarely was an inflammatory response (accumulation of neutrophils) noted; this response was observed in SC5314-infected mouse 13 and WO-1-infected mice 17 and 18. In only one mouse (number 13) did colony formation occur in which Candida developed within a thrombus in a branch of the bronchial artery and spread to the adjacent pulmonary parenchyma. By 6 to 7 days, C. albicans cells were no longer detectable in the lungs of WO-1- and B311-infected mice.
Immunohistochemical analysis was performed on serial sections of all tissues by using a polyclonal rabbit anti-Als serum raised against four keyhole limpet hemocyanin (KLH)-linked 10-mer peptides from the N-terminal domain of Als1p (9). Serial sections were also stained with negative-control sera, including the matched preimmune serum from the rabbit in which the anti-Als serum was raised and a commercially purchased anti-KLH serum (ICN). Preimmune and anti-KLH sera were included to detect nonspecific staining and staining due to the anti-KLH portion of the anti-Als serum, respectively. A fourth serial section was GMS-stained to visualize all fungi that were present and also to highlight the cell wall for comparisons to anti-Als staining.
Immunohistochemical staining followed the protocol outlined in the Vectastain ABC Kit (Vector Labs). Briefly, paraffin-embedded tissue sections were deparaffinized in a series of xylene baths and rehydrated in a series of ethanol baths of decreasing concentration. Endogenous peroxidase activity was quenched with 0.5% hydrogen peroxide in methanol. Nonspecific antibody binding was blocked with normal goat serum in PBS as specified by the manufacturer. Tissues were incubated overnight at 4°C with anti-Als antiserum diluted 1:500 in PBS. Control preimmune and anti-KLH (2 mg/ml) sera were also used at 1:500 dilutions. Biotinylated goat anti-rabbit serum was diluted 1:2,000 and incubated for 1 h at room temperature. Slides were treated with the Vectastain ABC reagent according to the manufacturer’s instructions. 3,3′-Diaminobenzidine (DAB) color substrate (Kirkegaard and Perry) was added to visualize the antigen by peroxidase reaction in the presence of H2O2. Slides from a given animal were processed as a group, since the goal of these experiments was to detect differences in staining between the anti-Als-treated and control (preimmune and anti-KLH sera) slides. The color reaction on the anti-Als slide from each animal was developed first, and an identical time of exposure was used to develop control slides from the same animal. Slides were counterstained for 10 s in Gill’s hematoxylin (Sigma), dehydrated in an ethanol series, passed through three xylene baths, and coverslipped with Permount (Sigma). Immunohistochemical staining with the DAB substrate and Gill’s hematoxylin counterstain yields brown signals on a purple background.
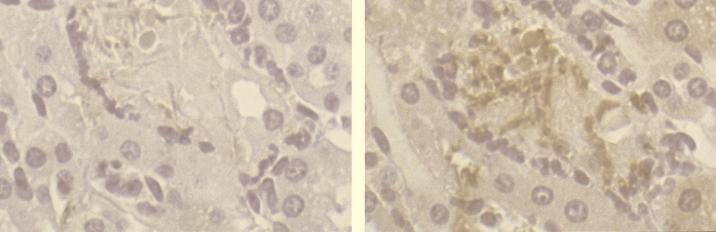
The specificity of anti-Als staining of C. albicans cells was demonstrated with the anti-KLH and preimmune control sera. Staining of murine tissues and C. albicans cells with the anti-KLH serum was negative for all mice in the project. The preimmune serum showed light, nonspecific background staining of murine tissue and very light background staining of C. albicans cells. Nonspecific background staining could be readily distinguished from staining with the hyperimmune anti-Als serum, which produced a signal far above background levels (Fig. 1). The specificity of anti-Als staining was further demonstrated in an additional experiment which showed that preadsorption of the anti-Als serum with a soluble N-terminal fragment of Als1p (9) eliminated staining of C. albicans cells in murine tissue. Preadsorption of the anti-Als serum with KLH did not affect the anti-Als staining.
FIG. 1.
Light micrographs of serial kidney sections from mouse 7 stained with preimmune serum (left) or anti-Als serum (right). C. albicans hyphal cells that failed to stain with the preimmune serum appear light purple, while cells staining with the DAB substrate are brown.
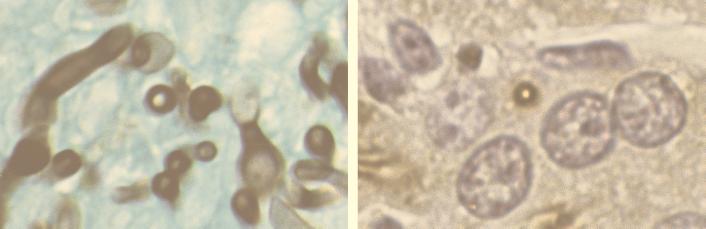
Evaluation of immunohistochemical staining used a scale ranging from zero to +++. Scores were assigned based on the intensity of staining with the preimmune, anti-Als, and anti-KLH sera. Results were compiled by organ for each mouse in the study. All fungi that stained with GMS also stained with the anti-Als serum, suggesting that there were no Als-negative fungi present in the mice. Staining with the anti-Als serum was observed across the cell wall of both the yeast and hyphal forms of all three C. albicans strains, as judged by comparison to the GMS-stained cells (Fig. 2). Direct comparisons between staining of yeast and hyphal forms, observed in tissue sections where the two forms were found in close proximity to each other, showed that yeast forms consistently stained more intensely than hyphal forms. Staining of the C. albicans cell wall was consistent over time, suggesting that no detectable changes occurred as infection progressed. These observations further strengthen our conclusion of cell wall localization for Als proteins (9) and suggest that Als proteins are continuously present on the surface of C. albicans cells found in the organs examined by using the mouse model of disseminated disease.
FIG. 2.
Light micrographs of serial kidney sections from mouse 7 stained with the GMS method (left) or with the anti-Als serum (right). The full thickness of the fungal cell wall is stained by each method, supporting the conclusion that Als proteins are localized in the C. albicans cell wall.
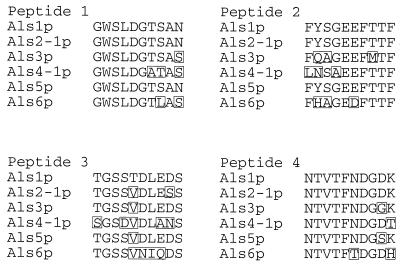
The anti-Als serum used for detection of Als proteins in this experiment was raised against four KLH-linked, 10-mer peptide sequences from the N-terminal domain of Als1p as previously described (9). Characterization of additional ALS genes demonstrated that their predicted proteins are very similar to Als1p within the N-terminal domain (9), and amino acid sequences of the immunogenic 10-mer peptides are often highly conserved between proteins in the family (Fig. 3). The 10-mer sequences of Als2p and Als5p are so highly conserved with those of Als1p that it is highly likely that these proteins will be recognized by the anti-Als serum. Specificity of the antiserum was tested against proteins with less-conserved 10-mer sequences by producing N-terminal protein fragments and Western blotting these fragments with the anti-Als serum.
FIG. 3.
Amino acid sequences of the four Als1p 10-mer peptides used to raise anti-Als serum aligned with the corresponding sequences predicted from other characterized ALS genes (7–10). Amino acids that differ from the Als1p sequence are boxed. Coordinates of peptides selected from Als1p are Peptide 1 (amino acids 53 to 62), Peptide 2 (amino acids 98 to 107), Peptide 3 (amino acids 139 to 148), and Peptide 4 (amino acids 156 to 165). The Als5p sequence shown was originally designated Ala1p (5).
The method for producing a secreted soluble form of the N-terminal domain of Als1p by using the S. cerevisiae expression vector p138NB was described previously (9). The same method was used to produce the N-terminal 433 amino acids of Als3p, Als4p, and Als6p. Briefly, PCR fragments produced with Pfu polymerase (Stratagene) were cloned into p138NB, which contains the copper-sulfate-inducible CUP1 promoter (12, 14). Plasmids free of PCR-incorporated DNA sequence errors were transformed into S. cerevisiae YPH274 (16), and transformants were selected as previously described (9). Growth of the transformants, induction of protein expression, and processing of the secreted protein from culture supernatant also followed the published methods (9). One microgram of total protein from each clone was run in duplicate sets of lanes on a sodium dodecyl sulfate–17.5% polyacrylamide gel; one-half of the gel was silver stained (Bio-Rad), and the other half was Western blotted with a 1:5,000 dilution of the anti-Als serum and the ECL Western blotting kit (Amersham).
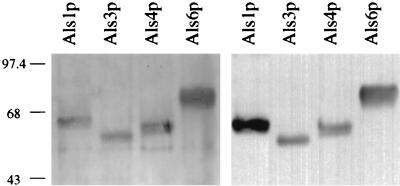
Comparison of the resulting silver-stained acrylamide gel lanes with those from the Western blot indicate that the anti-Als serum preferentially recognizes Als1p, but strongly recognizes the other Als proteins tested (Fig. 4). The specificity of Als protein recognition by the anti-Als serum is supported by noting that long exposures of the Western blot failed to show signals from other proteins found in the S. cerevisiae culture supernatants. The anti-Als serum also failed to recognize any proteins in the concentrated culture supernatant of a strain containing the blank p138NB vector (data not shown). Because the anti-Als serum recognizes multiple Als proteins, the immunohistochemical staining observed in tissue sections is likely to arise from recognition of multiple Als proteins. Development of monospecific anti-Als sera to study the relationship between production of individual Als proteins and particular host contexts will require a complete characterization of the ALS gene family and knowledge of which ALS genes are found in the C. albicans strain studied. As this information becomes available, a more thorough understanding of individual Als proteins and their interactions with host tissue will result. The data presented here describe production of Als proteins in vivo, in tissue samples from mice with experimentally induced disseminated disease. Despite the potential for in vitro gene expression differences between C. albicans strains, a similar pattern of Als protein production was observed between strains in vivo. The widespread distribution of Als-expressing C. albicans cells in vivo suggests that Als proteins are well positioned to play a role in the host-fungus interaction and in C. albicans pathogenesis.
FIG. 4.
Analysis of the specificity of the anti-Als serum. An N-terminal, 433-amino-acid fragment of each Als protein was heterologously produced in S. cerevisiae and run in duplicate sets of lanes on a sodium dodecyl sulfate-polyacrylamide gel. One-half of the gel was silver stained (left panel) and the other half Western blotted with the anti-Als serum (right panel). The variability in migration of each protein is attributable to differential glycosylation of the Als proteins (7). Molecular size (in kilodaltons) is shown at the left of the figure.
Acknowledgments
We thank George Livi and Megan McLaughlin of SmithKline Beecham Pharmaceuticals for their gift of the anti-Als antiserum, the histology section of the Laboratory of Veterinary Diagnostic Medicine for preparation of histology slides, and Paul Vancutsem for advice on immunohistochemistry techniques.
This work was supported by Public Health Service grant AI39441 (to L.L.H.); by Cooperative State Research, Education and Extension, U.S. Department of Agriculture, under Project No. ILLU-70-0305 (to L.L.H.); and by the University of Illinois Campus Research Board (to L.L.H.).
REFERENCES
- 1.Calderone R A, Braun P C. Adherence and receptor relationships of Candida albicans. Microbiol Rev. 1991;55:1–20. doi: 10.1128/mr.55.1.1-20.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaffin W L, Lopez-Ribot J L, Casanova M, Gozalbo D, Martinez J P. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol Mol Biol Rev. 1998;62:130–180. doi: 10.1128/mmbr.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu Y, Rieg G, Fonzi W A, Belanger P H, Edwards J E, Jr, Filler S G. Expression of the Candida albicans gene ALS1 in Saccharomyces cerevisiae induces adherence to endothelial and epithelial cells. Infect Immun. 1998;66:1783–1786. doi: 10.1128/iai.66.4.1783-1786.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukazawa Y, Kagaya K. Molecular bases of adhesion of Candida albicans. J Med Vet Mycol. 1997;35:87–99. doi: 10.1080/02681219780000971. [DOI] [PubMed] [Google Scholar]
- 5.Gaur N K, Klotz S A. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect Immun. 1997;65:5289–5294. doi: 10.1128/iai.65.12.5289-5294.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hostetter M K. Adhesins and ligands involved in the interaction of Candida spp. with epithelial and endothelial surfaces. Clin Microbiol Rev. 1994;7:29–42. doi: 10.1128/cmr.7.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoyer, L. L., and J. E. Hecht. 1999. Unpublished data.
- 8.Hoyer L L, Payne T L, Bell M, Myers A M, Scherer S. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr Genet. 1998;33:451–459. doi: 10.1007/s002940050359. [DOI] [PubMed] [Google Scholar]
- 9.Hoyer L L, Payne T L, Hecht J E. The ALS2 and ALS4 genes of Candida albicans and localization of Als proteins to the fungal cell surface. J Bacteriol. 1998;180:5334–5343. doi: 10.1128/jb.180.20.5334-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoyer L L, Scherer S, Shatzman A R, Livi G P. Candida albicans ALS1: domains related to a Saccharomyces cerevisiae sexual agglutinin separated by a repeating motif. Mol Microbiol. 1995;15:39–54. doi: 10.1111/j.1365-2958.1995.tb02219.x. [DOI] [PubMed] [Google Scholar]
- 11.Larone D H. Medically important fungi. 3rd ed. Washington, D.C: American Society for Microbiology; 1995. [Google Scholar]
- 12.Livi G P, Kmetz P, McHale M M, Cieslinski L B, Sathe G M, Taylor D P, Davis R L, Torphy T J, Balcarek J M. Cloning and expression of cDNA for a human low-Km, rolipram-sensitive cyclic AMP phosphodiesterase. Mol Cell Biol. 1990;10:2678–2686. doi: 10.1128/mcb.10.6.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luna L, editor. Manual of histologic staining methods of the Armed Forces Institute of Pathology. 3rd ed. New York, N.Y: McGraw-Hill; 1968. [Google Scholar]
- 14.McHale M M, Cieslinski L B, Eng W-K, Johnson R K, Torphy T J, Livi G P. Expression of human recombinant cAMP phosphodiesterase isozyme IV reverses growth arrest phenotypes in phosphodiesterase-deficient yeast. Mol Pharmacol. 1991;39:109–113. [PubMed] [Google Scholar]
- 15.Odds F C. Candida and candidosis. 2nd ed. London, England: Balliere Tindall; 1988. [Google Scholar]
- 16.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]