Abstract
Abnormal gene expression level or expression of genes containing deleterious mutations are two of the main determinants which lead to genetic disease. To obtain a therapeutic effect and thus to cure genetic diseases, it is crucial to regulate the host’s gene expression and restore it to physiological conditions. With this purpose, several molecular tools have been developed and are currently tested in clinical trials. Genome editing nucleases are a class of molecular tools routinely used in laboratories to rewire host’s gene expression. Genome editing nucleases include different categories of enzymes: meganucleses (MNs), zinc finger nucleases (ZFNs), clustered regularly interspaced short palindromic repeats (CRISPR)- CRISPR associated protein (Cas) and transcription activator-like effector nuclease (TALENs). Transposable elements are also a category of molecular tools which includes different members, for example Sleeping Beauty (SB), PiggyBac (PB), Tol2 and TcBuster. Transposons have been used for genetic studies and can serve as gene delivery tools. Molecular tools to rewire host’s gene expression also include episomes, which are divided into different categories depending on their molecular structure. Finally, RNA interference is commonly used to regulate gene expression through the administration of small interfering RNA (siRNA), short hairpin RNA (shRNA) and bi-functional shRNA molecules. In this review, we will describe the different molecular tools that can be used to regulate gene expression and discuss their potential for clinical applications. These molecular tools are delivered into the host's cells in the form of DNA, RNA or protein using vectors that can be grouped into physical or biochemical categories. In this review we will also illustrate the different types of payloads that can be used, and we will discuss recent developments in viral and non-viral vector technology.
Keywords: Genome-editing nuclease, Transposons, Episomes, siRNA, shRNA, Viral vectors, Non-viral vectors
Gene expression regulation as a strategy to treat genetic diseases
A variety of human diseases are defined by underlying genetic determinants, which may be represented by modifications in gene expression patterns, such as upregulation, downregulation or ectopic expression of wild-type or mutant genes. The physiological expression of genes harboring deleterious mutations may also cause loss-of-function genetic disease. For example, mutations in the SMN1 gene lead to spinal muscular atrophy type I (SMA 1), the most common genetic cause of infant mortality [1, 2]. Gain-of-function mutation is, instead, a type of mutation which confers new or enhanced activity on a gene product, such as for PIK3CA E545K. Such mutation results in loss of regulation and constitutive PI3Kα activity, which can lead to oncogenesis [3]. Finally, in some instances, dominant negative mutations can occur, creating a protein which adversely affects the wild-type gene product, within the same cell. A classic example is Huntington's disease caused by the expansion of a CAG trinucleotide repeat stretch in the coding sequence of the HTT gene [4].
To cure genetic diseases, it is crucial to rewire aberrant host’s gene expression back to physiological conditions. With this purpose, researchers developed molecular tools including genome editing nucleases, transposons, episomes and siRNA/shRNA. Such tools are delivered into the host's cells in the form of DNA, RNA or protein using physical or biochemical methods.
In this review, we describe the different molecular tools that can be used to regulate gene expression and discuss their potential for clinical applications. Moreover, we address the advantages and disadvantages of payload types packed by both viral and non-viral vectors. We focus on recent developments in vector technology and outline the requirements for vectors to succeed in cell and gene therapy.
Molecular tools to modify gene expression
A variety of molecular tools are available to regulate gene expression. Some of them have been profusely investigated and adapted to clinical use, while others are still being tested at the preclinical stage.
In this section, we discuss different methods to modify host gene expression, through the delivery of genetic payloads. Such strategies include the delivery of genome editing nucleases, transposons, episomes, siRNA and shRNA. Each category presents advantages and disadvantages summarized in Table 1.
Table 1.
Different systems used to modify gene expression and their main advantages and disadvantages. This is the summary of paragraphs that include references from [1] to [145]
| Options | Key feature | Advantages | Disadvantages | |
|---|---|---|---|---|
| Genome Editing Nucleases |
• MNs • ZFNs • TALENs • CRISPR/Cas |
• Locus-specific |
• One-time treatment • Versatility and multiplex ability |
• Off-target effects leading to mutagenesis and translocations |
| Transposons |
• SB • PB • TcBuster • Tol2 |
• Non locus-specific |
• High cloning capacity • Low toxicity • Biosafety |
• Transposition efficiency • Insertional mutagenesis |
| Episomes |
• MAC, HAC • Putative ORI • S/MAR family |
• Transient (potentially longer) |
• No insertional mutagenesis • Remain extrachromosomal • Low toxicity • High cloning capacity |
• Gene silencing due to host gene regulation mechanisms • Transfection efficiency • Mitotic instability |
| Small RNA molecules |
• siRNA • shRNA • bifunctional shRNA |
• Transient |
• No insertional mutagenesis • High efficiency • Chemical modifications to reduce off-target effects |
• Transient effect |
Genome editing nucleases
Different types of genome-editing nucleases: advantages and disadvantages
Gene editing is a type of genetic engineering that allows the introduction of permanent and locus-specific DNA modifications in the genome. Four types of gene-editing nucleases have been used so far in research: meganucleases (MNs), zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)-associated endonucleases (Cas) [5, 6].
To guide the nuclease to the target site, MNs, ZFNs, and TALENs use a protein-DNA interaction while CRISPR-Cas systems are guided by RNA–DNA interactions. MNs are highly specific endonucleases, recognizing target sequences of about 14–40 base pairs (bp) [7]. A drawback in using MNs is the limited number of target sites that they recognize and therefore the extreme difficulty to use these endonucleases in clinical settings, where a higher level of flexibility is desirable [8]. The creation of new MNs is a laborious process requiring complex protein engineering procedures because the DNA binding and cleavage domains are difficult to separate [7, 9]. Such laborious engineering required for MNs has constrained their widescale use [10].
The main characteristics of genome-editing nucleases are listed in Table 2. Both ZFNs and TALENs utilize the nuclease FokI as a cleavage domain. Each ZFN module is composed of about 18–36 amino acids and recognizes a specific 3 bp sequence [11, 12]. Therefore, several zinc finger modules need to be engineered to recognize the target sequence: each module will bind 3 bp in the target sequence and the FokI nuclease will be coupled to the DNA-binding modules [13–15]. TALENs follow the same principle, but the DNA-binding module recognizes one single nucleotide, instead of 3 [16]. TALENs modules are composed of 30–40 amino acids, resulting in a protein with higher specificity but larger than ZFNs [17]. Additionally, for both ZFNs and TALENs, it is necessary to engineer two different enzymes for each target, one upstream and one downstream of the cut site. This is necessary because FokI dimerization is required for completion of the double-stranded break. Overall, the engineering of ZFNs and TALENs are technically challenging, and time consuming compared to the engineering of CRISPR-Cas nucleases [8, 18]. Furthermore, the target sequence requirements for ZFNs render the selection of an appropriate and specific target difficult [6, 19, 20].
Table 2.
Main features of the different genome-editing nucleases
| ZFNs | TALENs | CRISPR/Cas | |
|---|---|---|---|
| Phylogenetic origin | artificial restriction enzyme [252, 253] | Xanthomonas bacteria [254] | Streptococcus pyogenes [255] |
| DNA binding domain | zinc finger protein [253, 256] | TALE protein [16, 257, 258] | guide RNA [259–261] |
| DNA cleavage | FokI [262] | FokI [257, 258, 262] | Cas9 [259, 260] |
| DNA recognition range | 18–36 bp (3 bp per module) [253] | 30–40 bp (1 bp per module) [257, 258, 263] | 22 bp (DNA-RNA base pairing) [261] |
| DNA cut | dsDNA as a dimer [264] | dsDNA as a dimer [265] | dsDNA complex protein-gRNA [259] |
| Recognition sequence | 5'-GNNGNNGNN-3’ [256] | sequence with 5'-T and A-3' [16, 254, 263] | sequence immediately followed by 5'-NGG-3' [259, 266] |
| Advantages | Small protein size (< 1 Kb), sequence-based module engineering [267] | High specificity, easy selection of target region [268] | Easy to multiplex, simple synthesis of gRNA, easy selection of target region [269] |
| Disadvantages | Difficult sequence selection and protein engineering,, expensive and time consuming [267] | Large protein size (> 3 Kb), expensive and time-consuming [269, 270] | Large protein size (> 4 Kb) [269] |
| Safety concerns | off-target effects: genome mutagenesis and GCRs [271] | off-target effects: genome mutagenesis and GCRs [270] | off-target effects: genome mutagenesis and GCRs [271] |
CRISPR-Cas technology
CRISPR-Cas nucleases have crucial advantages compared with ZFNs and TALENs, including the simplicity of the guide RNA (gRNA) design (Table 2). Such nucleases use a 22 bp gRNA to bind a complementary target sequence, which is subsequently cut by the Cas itself [8, 21, 22]. By designing specific gRNAs, CRISPR-Cas systems could theoretically target any sequence in the genome. Indeed, the ease of target selection and the possibility of multiplexing the gRNAs while maintaining high specificity and efficiency led to the rapid development of CRISPR-Cas methods for clinical purposes [18]. CRISPR-Cas systems allow to rapidly screen a large number of gRNAs and the scalability of this platform permits an accurate optimization of the study system [23–25].
Stable integration of CRISPR-Cas is not necessary to provide a therapeutic effect and long-term expression is usually considered a disadvantage, as it can lead to off-target cleavage. However, the persistence of CRISPR-Cas in the cell must be sufficient to perform the editing function [26].
The delivery of a native Cas protein in complex with a gRNA bypasses the requirement for transcription and translation. It introduces genome editing approximately 3 h after delivery and is degraded after 24–48 h [26]. Circumventing transcription and translation is useful in post-mitotic or hard-to-transfect cells. Such transient functionality allows for rapid editing and reduced off-target effects. However, obtaining pure active protein is a difficult process, and the risk of endotoxin contamination remains of concern [27]. The delivery of Cas proteins offers an improved dose-control compared to mRNA and DNA but, in order to produce a therapeutic effect, a significant amount of protein must be successfully delivered. This is due to the lack of amplification signal which normally occurs with mRNA and DNA. Moreover, the CRISPR-Cas protein is large, which may present a challenge for intracellular delivery.
Another strategy to minimize off-target editing events is through the delivery of Cas mRNA. This process results in rapid genome editing (5–7 h after transfection) and avoids the step of nuclear entry [28]. The mRNA that codes for the Cas protein is immediately translated in the cytosol and the complex Cas-gRNA subsequently enters the nucleus. The transient expression of Cas proteins reduces off-target effects and risk of integration, but the dose and timing of mRNA delivery have to be carefully titrated [29, 30].
Finally, the delivery of genome editing nucleases through plasmids is an easy procedure. Due to the necessity of nuclear entry, subsequent transcription, and translation into protein, the genome editing efficacy is significantly delayed. Plasmids are very stable molecules, therefore Cas protein expression may last several days, leading to a high risk of off-target effects and safety concerns [31, 32]. Plasmid delivery may also trigger cytosolic DNA toxicity [33]. Despite the fact that each of the above-mentioned nucleases present specific advantages and disadvantages, CRISPR-Cas technology has been widely adopted and improved in the last few years and it remains a promising route for preclinical and clinical investigations [8]. Depending on the cell type and experimental conditions, the knockout efficiency for CRISPR-Cas9 varies between 40% (induced pluripotent stem cells, iPSCs) and 99.4% (cortex, hippocampus and spinal cord) [34–37].
CRISPR-Cas9 can also be used for gene or targeted nucleotide knock-in experiments. Such manipulations are usually more challenging to perform and require accurate optimization, including the addition of an extra component in the form of a DNA donor template [38]. Different studies reported a wide range of knock-in efficiencies depending on the method and the cell type used. Liu et al. [39] compared the efficiencies of CRISPR-Cas9 versus ZFN and TALEN, performing knock-ins in fetal fibroblasts [39]. They found that CRISPR-Cas9-mediated gene knock-in (70–80% efficiency) was 5.6 times more efficient than ZFN and around 3 times more efficient than TALEN. Nevertheless, other studies found lower knock-in efficiencies for CRISPR-Cas9, for example ∼20% in human primary T cells [40]. In conclusion, the overall efficiency of knock-in seems to remain lower than the knockout efficiency, using CRISPR-Cas9. Moreover, delivery of CRISPR-Cas9 through viral vectors requires in vitro T cell activation and culture [41, 42]. However, the use of electroporation (EP) methods to deliver Cas9-gRNA protein complex in knockout studies demonstrated the potential to overcome this issue and to achieve gene editing without in vitro T cell activation [42, 43]. Human primary T cells are difficult to manipulate and chemically modified gRNAs were also tested to enhance genome editing efficiency [44].
In the last decade many tools have been developed and optimized to investigate genome functional complexity based on Cas proteins. Among those, a Cas-based tool for epigenome editing (non-gene editing) called “nuclease dead Cas” (dCas) was developed by creating a mutant form of Cas which lacks endonuclease activity. This enzyme still retains the capability to bind the gRNA and it can target Cas-coupled effector proteins to a specific locus of the genome [45]. Coupling dCas with VPR activator (CRISPRa) or KRAB repressor (CRISPRi) of transcription creates a powerful tool for precise epigenetic editing. For example, Schmidt et al. [46] developed a CRISPRa and CRISPRi platform to perform genome-wide screens for functional regulators of cytokine production in response to T cell stimulation [46]. Yang et al. [47] developed a CAR-T cell product called RB-340–1, which was engineered through a CRISPRi circuit to prevent Programmed cell Death protein 1 (PD-1) expression upon antigen-encounter [47]. RB-340–1 is the first application of CRISPRi toward a clinically relevant product and allows the conditional and reversible suppression of PD-1. The reversible nature of this editing also allows fine tuning of the degree of PD-1 expression. RB-340–1 demonstrated resilience to checkpoint inhibition and increased persistence and effectiveness against HER2-expressing cancer xenografts [47].
Safety of genome editing-based techniques
A drawback that should be considered when performing genome editing is the immunogenicity of the nucleases – ZFNs, TALENs and CRISPR-Cas are exogenous proteins and may trigger an immune response in the patient. Regarding SpCas9, in vivo delivery has been found to elicit both antibody and T cell responses in immunocompetent mice [48–51]. Cell therapy which employs products transiently edited ex vivo through plasmids, mRNA or protein, is expected to be safe as the Cas9 is diluted during cell proliferation [52]. Early reports from clinical trials revealed persistence of T cells edited ex vivo through SpCas9, in cancer patients [53–55]. However, in all these reports Cas9-directed immune responses were not directly evaluated [56] and the available safety data derived from patients that had a compromised immune system. Despite the encouraging results, thorough investigations with ex vivo engineered T cell products may be needed to assess humoral and cellular immune response, after infusion.
Off-target genotoxicity together with the risk of creating translocations when multiplex genome editing is performed are a major drawback of genome editing nucleases [57]. Indeed, chromosomal translocations are natural byproducts of inducing simultaneous genomic breaks [58, 59]. Different nuclease combinations or the presence of a homologous single-stranded donor have been suggested as approaches to reduce chromosomal translocations in multiplex editing [58]. For example, Bothmer et al. [58], performed knockout at the TRAC and B2M loci in human T cells, including a single-stranded repair template in the reaction. The repair template presented 70 bp of homology on either side of the double-strand break (DSB) with a 10 bp stop cassette, to achieve functional knockout. With this strategy, the DSB repair mechanism was shifted from Non-Homologous End Joining (NHEJ) which can cause translocations, to single-stranded template repair (SSTR) [58].
Clinical trials
Over the past few years, genome editing nucleases made their appearance in clinical trials although so far, no U.S. Food and Drug Administration (FDA) approved treatment based on this technology has been commercialized and no late-stage clinical trial has been approved.
In gene therapy, ZFN and CRISPR-Cas9 are currently being investigated in clinical trials to treat genetic diseases such as Mucopolysaccharidosis (NCT03041324, NCT04628871, NCT02702115), Hemophilia B (NCT02695160), β-Thalassemia (NCT03432364, NCT03728322, NCT03655678), Neurofibromatosis type 1 (NCT03332030), Sickle cell disease (NCT03745287) and LCA10 (NCT03872479). All these clinical trials are at an early stage: phase I or I/II and no clinical trials have been approved for TALEN so far.
The situation is similar for cell therapy: early-stage clinical trials are currently evaluating products to treat both hematological malignancies and solid tumors. Genome editing nucleases are employed to knock out target genes such as IL13Ralpha2, PD-1, CISH, TRAC and B2M. For example, TALEN is currently being tested in a clinical trial to knockout TRAC and CD52 in allogeneic CAR-T cells (NCT02808442). This is to create an off the shelf CAR product for a specific patient population [60]. ZFN has been used to permanently disrupt the glucocorticoid receptor GRm13Z40-2 in anti-IL13Ralpha2 allogeneic CD8 + T cells, used to treat patients with recurrent/refractory malignant glioma (NCT01082926). Products engineered through CRISPR-Cas9 technology have been more frequently adopted in clinical trials. For solid tumors, PDC1 and TRAC knockouts have been tested (NCT02793856, NCT03081715, NCT03044743, NCT03545815, NCT03747965) aiming to decrease CAR-T cells exhaustion. For hematological malignancies CRISPR-Cas9 target genes include TRAC, B2M, CD7, CD28, CD19, CD20 and CD22 (NCT03190278, NCT03166878, NCT03398967, NCT03690011), depending on the cancer characteristics.
Transposons
Transposable elements (TEs), also known as transposons, are sequences of DNA that move from one location to another in the genome. TEs have been identified in all organisms and can comprise a large proportion of a species’ genome [61, 62]. Class I retrotransposon replicates by a copy-and-paste mechanism, producing intermediate mRNA copies that are reverse transcribed [63]. Class I retrotransposons make up 40–50% of the human genome and include long and short interspersed repeats (LINEs and SINEs) and long terminal repeat elements (LTR) [61]. Class II DNA transposons which are not active in humans, utilize a cut-and-paste mechanism to move DNA elements from one location to another [64, 65]. In some cases, such as Helitrons, a peel-and-paste mechanism, involving a circular DNA intermediate, is used to move and replicate DNA elements in the genome [66].
DNA transposons consist of a transposase gene that is flanked at both ends by terminal inverted repeats (TIRs) [67]. The expressed transposase recognizes both ends of TIRs and deploys cut-and-paste mechanisms to move the entire transposon to another location of the genome [68]. Taking advantage of TIR sequences and transposase activity, different types of transposons have been used for genetic studies and serve as gene delivery tools (Table 1). In the design of delivery constructs, the gene of interest (expression cassette) is flanked by TIRs on both ends, and the transposase gene can be delivered separately via mRNA, protein, or DNA.
Different types of transposons
Different types of TEs have been studied, the most common being used as genetic tools in mammalian cells, including Sleeping Beauty (SB), PiggyBac (PB), TcBuster, and Tol2 [67]. All are mobilized through a cut-and-paste mechanism [69–71]. TEs are active in various species from protozoa to vertebrates, including mice, rats, and humans. Several studies demonstrated that in mammalian cells, PB and SB have high transposition activity, with PB having stronger activity [72–74].
Most natural DNA transposons are inactive during evolution for genome stability [72]. Hyperactive transposases have been designed through rational amino acid substitution and codon optimization to increase the transposition efficiency. For example, the hyperactive SB100X and SB150X TEs possess respectively 100-fold and 130-fold higher transposition activity compared to the original SB10 [75–77]. Hyperactive SBs have been used in germline transgenesis in rodents, rabbits, and pigs, and to reprogram mouse embryonic and human fibroblasts into iPSCs [78–85]. The wild-type PB (PBase) was codon-optimized for mammalians (mPBase) demonstrating a 20-fold increase in transposition efficiency, compared to PBase [86]. Subsequently, seven amino acid substitutions were combined, leading to the hyperactive PB called hyPBase [87]. hyPBase demonstrated a 17-fold increase in excision and a ninefold increase in integration, compared to mPBase. Recently, a new hyperactive variant of TcBuster has been commercialized and has been used in a variety of studies [88]. A final example is the 6X-His-tagged variant of Tol2 transposase, which maintains high transposition activity in vitro and in vivo. Such a tag-targeted modification of Tol2 allowed the identification and purification of the Tol2 transposase from cells, a process which is useful for biochemical studies [89]. This is an important result because targeted modification of transposase enzymes often results in reduced enzymatic activity, as was the case for SB [74, 90].
During the past few decades, variants of TIRs such as truncations and changes in their configuration were also generated to improve transposition efficiency [91]. The truncation of PB’s TIR led to the development of IRmicro, which increased the transgene expression in vivo by 1.5-fold [92]. Over the years different miniTol2 were created as well. However, in these studies, the minimal TIR sequence did not improve transposition activity in vivo and in vitro compared to the original TIR [89, 93, 94].
Regarding SB, each TIR unit on both ends is 200–250 bp in length. The left TIR also contains an extra half direct repeat (DR) to enhance element transposition. Modification of the DRs and the space sequence between the DRs lead to the development of pT2, which displayed a fourfold increase in transposition efficiency compared to the first-generation transposon [95]. In the sandwich transposon (SA), the TIR elements consist of two complete transposon elements in a head-to-head configuration, which flank a DNA expression cassette. The SA displayed a 3.7-fold increase in transposition compared to SB10 [96]. The pT3 vector contains a duplication of the left TIR, which acts as a transposition enhancer. Moreover, it has an extra TA dinucleotide flanking the transgene, to promote increased excision. pT3 showed a threefold increase in efficacy in vivo [97]. pT3 showed a threefold increase in efficacy in vivo. The DNA-recognition domain of the transposase contains two subdomains: PAI and RED. The pT4 vector was created by introducing mutations in the PAI interaction motifs of the inner DRs. The new construct showed a twofold increase in activity compared to pT2 [98].
Safety of transposon-based techniques
The members of the Tc1/mariner transposon family, such as SB, present ‘overproduction inhibition’, which refers to loss of transpositional activity in the presence of increasing concentrations of transposase [99]. This negative dosage effect was observed in vitro and in vivo [100] and may be beneficial in clinical settings in terms of biosafety. Data regarding PiggyBac is contradictory, as different studies have inconsistently detected overproduction inhibition [74, 101, 102]. Tol2 and TcBuster transposition is directly proportional to the level of transposase, and these systems lack overproduction inhibition [102, 103].
Since TEs are also randomly integrated in the host genome, like retroviral integration, there is potential insertional mutagenesis risk. Transposon-associated CRISPR-Cas delivery systems have also been developed, using RNA-guided DNA transposition to target the integration of transgenes into specific sites, which would provide further biosafety advantages [104–106].
So far, no evidence has been provided of transposon-mediated transgene integration inducing gross chromosomal rearrangements (GCRs). However, studies have reported cases of rearrangements when a pair of transposition-competent elements is present at the same locus [107, 108]. Remobilization of transposons in the genome may also result in GCRs [109]. More investigations need to be carried to understand the risk.
Transposons that utilize the cut-and-paste mechanism exhibit an effect called ‘local hopping’ when mobilized from a chromosome. Local hopping refers to the preference for cis-integration into sites that are in proximity of the donor locus [110]. Therefore, local hopping confines the target region which is available to a transposon moving from a donor locus. The extent of this phenomenon varies among different TE types, species, and the donor locus itself. SB and Tol2 exhibits local hopping [111, 112]. PB, despite presenting a certain degree of local hopping, has a wider integration range [73, 113, 114]. Local hopping may be a double-edged sword, depending on where the TE is located. If such a region is rich in coding sequences, local hopping might increase the risk of gene mutagenesis.
In conclusion, transposons may be a potential technology to achieve long-term expression of transgenes and may be used in clinical settings. A robust study and construct design are still needed to further validate and avoid undesirable effects in patients.
Clinical trials
One Phase I/II clinical trial (NCT04284254) has been approved, testing the use of SB to treat patients with Hurler syndrome [115]. No clinical trials for PB have been approved in gene therapy. Regarding adoptive cell therapy, the landscape is more dynamic with a variety of active clinical trials for both SB and PB. Currently, SB and PB are being investigated to deliver CD19, BCMA, SLAMF7, CD33 or CD116 CAR for immunotherapy of hematological malignancies (NCT03389035, NCT04499339, NCT03927261, NCT00968760, NCT01497184, NCT03288493, NCT04960579, jRCT2033210029, ACTRN1261700157938, ChiCTR1800018111). All these trials are in Phase I/II.
Episomes
Episomes are defined as closed circular DNA molecules which are autonomously replicating. Episomal vectors may be an interesting clinical strategy due to their long-term expression and intrinsic characteristic of extrachromosomal maintenance, which prevents insertional mutagenesis (Table 1). With proper design and modification, episomes also provide large payload capacity and reduced toxicity [116, 117]. This applies in particular to episomal minicircles (MCs), which harbor reduced amounts of bacterial sequences. However, episomal vectors present some disadvantages, including the possibility of transgene silencing due to epigenetic modifications of the vector [118]. Indeed, episomal genes remain subject to host gene control mechanisms and may undergo modifications such as methylation or deacetylation. Therefore, reducing bacterial sequence and CpG contents may improve transgene expression. Another drawback is their mitotic instability, referred to as episome loss after multiple cycles of cellular division. Proper designs of episomal vectors can be carried to improve mitotic stability, such as pEPI-1 with an S/MAR sequence [119].
Different types of episomes
Episomal vectors are divided into three main categories: (A) mammalian or human artificial chromosomes (MAC, HAC), (B) episomes including putative origins of replication (ORI) and (C) episomes including chromosomal scaffold/matrix attachment region sequences (S/MAR) [116]. MAC or HAC possess the highest cloning capacity and harbor telomeric and centromeric sequences to ensure mitotic stability.
Several efforts have been made to create episomes including putative ORI, for use in mammalian cells and only a small number of attempts have been successful because mammalian ORI do not share sequence homologies [120, 121].
S/MAR are genomic DNA sequences that mediate structural organization of the chromatin, anchoring the chromatin to the nuclear matrix protein SAF-A during interphase and thus facilitating DNA segregation into dividing cells [122, 123]. Therefore, the S/MAR family of episomes presents increased mitotic stability compared to conventional plasmids. A variety of S/MAR episomes have been developed over the years, including pEPI-1, PEPito, pEPI-cHS4, pEPI-UCOE, pEPI-TetON, pEdER-IR and pEP-IR [122, 124–128]. To improve vector efficacy, S/MAR MCs were created lacking the bacterial backbone, antibiotic resistance sequences, and bacterial origins of replication [116, 129, 130]. Minicircles have been successfully used in preclinical studies in numerous cell types including CAR-T cells [131–134]. Recently, a new system called nano-S/MAR (nanovector) has been developed [135]. Nanovectors are easier to produce compared to MCs, with a final product of higher purity. Nano-S/MAR vectors have shown improved efficiency of establishment, transgene expression, and minimal cytotoxicity in preclinical studies [135, 136]. Despite the successful application of S/MAR vectors in vitro and in vivo, no clinical trial involving their use has been approved so far.
siRNA and shRNA
RNA interference (RNAi) is also known as RNA silencing or post-transcriptional gene silencing (PTGS) and it is mediated by three classes of molecules: siRNA, shRNA and bifunctional shRNA (Table 1). RNAi is a strategy to transiently regulate gene expression in a wide range of eukaryotes, without permanently modifying nuclear DNA [137].
SiRNA is a class of double-stranded RNA molecules of typically 20–24 base pairs in length. After being delivered to the cell, the siRNA molecule is incorporated into the RNA-Induced Silencing Complex (RISC) where it is unwounded to a single strand RNA. The less thermodynamically stable RNA strand is then used by RISC to probe and anneal with target complementary mRNA. The target mRNA is subsequently cleaved. Alternatively, the siRNA molecule can be incorporated into an RNA-induced transcriptional silencing (RITS) complex, which triggers heterochromatin formation at the locus where the siRNA binds its complementary sequence of DNA. Allele-specific siRNAs (ASP-siRNA) have also been developed to minimize off-target toxicity and selectively inhibit the mutant allele of a gene [138]. This is possible thanks to their ability to distinguish the target from the wild-type sequence, with single-nucleotide specificity. ASP-siRNAs have the potential to be employed in vivo for gene therapy, to silence the dominant mutant allele and to treat a variety of genetic disorders [138, 139].
SiRNA molecules produce a transient effect, especially in rapidly proliferating cells and this characteristic may be considered as a double-edged sword. In the situation where a more durable effect is desired, the siRNA sequence is modified into a shRNA. ShRNAs contain a tight hairpin turn and are encoded in a DNA vector to be delivered into cells. A shRNA vector can provide a more long-lasting effect than just treating cells with siRNA, as it allows for a durable expression of shRNA. Expression of shRNA may also be controlled through inducible or tissue-specific promoters. ShRNAs are transcribed in the nucleus and then processed by Drosha/DGCR8 complex and Dicer/TRBP/PACT into mature shRNA. Mature shRNAs are loaded on Argonaute protein containing RISC complex and thus provide RNAi activity. Finally, bi-functional shRNAs have been developed to obtain a more rapid and efficient RNAi function [137]. These RNAs can be associated with cleavage-dependent and –independent RISCs, simultaneously inducing target mRNA degradation and inhibiting translation through mRNA sequestration.
Clinical trials
In 2018 the first U.S. siRNA product was approved by the FDA. Onpattro® (patisiran) is a siRNA product aiming to treat the rare hereditary disease transthyretin-mediated amyloidosis, in adult patients. The majority of siRNA products are intended to treat cancer and orphan/rare genetic diseases. Other Alnylam’s commercially available products are Givlaari® (givosiran) for acute hepatic porphyrias and Oxlumo® (lumasiran) as the first treatment for primary hyperoxaluria type 1. Leqvio® (inclisiran) was FDA-approved in December 2021 and is a treatment to lower cholesterol for people with atherosclerotic cardiovascular disease. The most recently approved siRNA is Amvuttra™ (vutisiran) for treatment of the polyneuropathy of hereditary transthyretin-mediated amyloidosis (June 2022).
A product recently tested in Phase 3 clinical trials is Fitusiran (NCT03417245). Data from Phase 3 demonstrated that Fitusiran significantly inhibits bleeding in patients with hemophilia A or B [140]. Another advanced clinical trial is currently testing Teprasiran (Phase 3—NCT03510897), a siRNA which inhibits p53-mediated cell death that underlies acute kidney injury (AKI) in high-risk patients undergoing cardiac surgery. The incidence, severity and duration of AKI were significantly reduced after Teprasiran administration [141]. Cosdosiran (Phase 2/3—NCT02341560) is being evaluated to improve visual function in subjects with recent-onset acute nonarteritic anterior ischemic optic neuropathy (NAION). Lastly, Nedosiran (Phase 2—NCT03847909) is an siRNA that inhibits hepatic lactate dehydrogenase in patients with primary hyperoxaluria and Tivanisiran (Phase 3—NCT04819269) is a therapy for dry eye disease due to Sjögren's Syndrome [142].
Some early-stage clinical trials adopt a combo approach using siRNA and immune cells. In order to knock down the CD3ζ component of the TCR, shRNA was tested in a new allogeneic product (CYAD-211) that co-express an anti-BCMA CAR (NCT04613557). The product was well-tolerated and two partial responses were observed among five evaluable patients [143]. Finally, in the clinical trial NCT03208556, iPD1 CD19 eCAR T cells were tested in relapsed or refractory B-cell lymphoma. A PD1 shRNA-expressing cassette was incorporated in the CAR lentivector with the objective to improve anti-tumor activity by inhibiting PD1 induction.
Type of payloads to deliver molecular tools for gene expression regulation
The molecular tools described above can be delivered into cells using various forms of genetic materials (such as DNA, RNA) and protein, based on the intended purpose (e.g., transient or long-term expression), the different genetic tool design and the delivery methods.
DNA-based payload
Plasmid DNA (pDNA) is the most widely used payload in gene and cell therapy. pDNA is usually composed of a supercoiled double-stranded DNA of variable size (< 1 kb to 200 kb). Typically, pDNA contains an antibiotic resistance gene, a prokaryotic origin of replication for plasmid propagation, and an expression cassette [144, 145]. The cassette is usually composed of a gene of interest, a promoter for its transcription, and a polyadenylation signal for mRNA export. However, the large backbone and antibiotic resistance gene limit their use in gene and cell therapy due to reduced biocompatibility and safety [146]. The unmethylated CpG motif contained in a variety of plasmids has been shown to induce an immune response via TLR9 signaling and may trigger inflammation and tissue damage when administered in vivo [147]. In addition, bacterial sequences in the plasmid can contribute to transgene silencing in host cells. Moreover, the size of the plasmid can significantly impact transfection efficiency. Efforts have been made to downsize the bacterial fragments and antibiotic resistance genes to improve biocompatibility, durability, and safety [148].
Other forms of DNA payload have also been explored for cell therapy. Linear DNA synthesized through PCR has entered the cell therapy arena to challenge the traditional use of bacterial plasmid DNA, however linear DNA also possesses the risk of foreign DNA and endotoxin contamination. Linear DNA can be used in combination with other payloads in EP. Roth et al. [149] demonstrated that electroporation of primary T cells with CRISPR-Cas9 RNPs and linear double-stranded DNA achieved a transfection efficiency of ~ 50% as shown by the GFP reporter signal [149]. As mentioned before, minicircles are supercoiled DNA molecules with very few bacterial sequences [130]. Improvements in transfection efficiency and prolonged expression have been reported for MCs and they are considered having a favorable safety profile. Other DNA forms, such as DNA ministrings [150] and nano/mini-plasmids, have also shown improvements in transfection efficiency by reducing the unnecessary bacterial sequences.
Ministring DNA is a plasmid-derived DNA delivery system which possess linear covalently closed (LCC) ends, DNA targeting sequences at both ends and transgene expression cassette devoid of prokaryotic sequences [150]. Mini and nanoplasmids are small circular DNA constructs and do not possess antibiotic resistance genes [151].
Gene delivery systems should consider the toxicity associated with DNA delivery, which includes the sensing of cytosolic DNA by host cells [152, 153]. Cellular DNA sensors detect cytosolic genetic material, usually due to viral infections or self-DNA leaking from different cellular compartments [154–157]. The detection of cytosolic nucleic acids triggers the host immune response and is pivotal for mammalian organisms to control malignant transformation and to mediate cell-intrinsic onco-suppression [158]. The accumulation of cytoplasmic DNA can lead to cellular senescence or regulated cell death through multiple pathways, involving proteins including STING, ZBP1, AIM2 and IFI16 and therefore creating vector dependent DNA-associated cytotoxicity [159–163].
RNA-based payload
Small mRNA payloads are preferred to larger pDNA regarding post-transfection cell viability. However, there are innate limitations for mRNA-based gene editing. For example, in vitro-transcribed (IVT) mRNA is less stable than DNA. This problem could be mitigated by introducing chemical modifications in the synthetic RNA, such as adding a 5’-methylated cap and poly(A) tail (reviewed in[164]). Similar to DNA, mRNA also elicits innate immune response. The strategy consists of employing modified nucleosides, including pseudouridine, N1-methylpseudouridine, 2-thiouridine, 5-methylcytidine, or N6-methyladenosine to dampen the immunogenicity of the mRNA [165, 166]. Thanks to these improvements, IVT mRNA has been used in CAR-T cell studies on hematological and solid tumors, some of which have advanced to clinical trials (reviewed in [167]).
Another concern is durability: since mRNA does not integrate into the genome, the transgene expression is usually transient and may require multiple doses for long-term expression. Multiple studies showed that EP of mRNA results in CAR expression that lasts for around seven days [168–170], potentially reducing on-target-off-tumor toxicity [171]. It may also be useful to target antigens with specific spatial and temporal patterns, as in the recent example where mRNA engineered CAR-T cells were used to treat cardiac injury [172]. Finally, small regulatory RNAs (microRNA, shRNA, siRNA, etc.) are used in gene and cell therapy to transiently silence a target gene, as discussed above. This is a targeted approach that has been investigated for the treatment of cardiovascular diseases, viral infections, and cancer. Researchers are currently attempting to address the drawbacks of small regulatory RNAs such as stability, extracellular and intracellular barriers, and innate immune stimulation (reviewed in [173–177]).
Protein-based payload
Most macromolecules such as proteins do not enter cells by passive diffusion because of their limited intrinsic capability to internalize. Membrane disruption and cell-penetrating peptides have been used for cross-membrane transport of large molecules. Payload proteins include antibodies, transcription factors, and genome editing nucleases [178]. Most notably, proteins used in cell therapies are usually components of gene-editing nucleases and transposons. The advantage of using protein payload is to precisely control the dose and expression time frame of interest genes. Proteins could also be delivered by nanoscale injection or localized EP devices, leading to minimum cellular damage [179].
Techniques and materials to deliver genetic payloads
As previously mentioned, genetic payloads are delivered into cells through vectors. Gene delivery systems are grouped into two categories: viral and non-viral vectors. In this section we will discuss advantages and disadvantages for both types of vectors, and their current and potential applications in the gene and cell therapy fields.
Viral vectors
Viral vectors are delivery vehicles used to transduce human cells. In 1968, Rogers and Pfuderer were the first to perform proof of concept studies for virus mediated gene transfer using lysates of tobacco leaves infected with tobacco mosaic virus [180] (Table 3). The first instance of viral gene therapy performed on T cells ex vivo was conducted in 1990, on a four-year-old girl suffering from Severe Combined Immunodeficiency Defect (SCID). This treatment provided an encouraging signal to the field, even though the effects were temporary [181]. Despite the original success, a major setback occurred in 1999 when a patient died after receiving in vivo gene therapy [182]. The patient developed an intense inflammatory response against the adenovirus (AV) used as a vector, with blood-clotting followed by kidney, liver, and lung failure. Moreover, in another instance, 4 out of 10 patients with X-linked SCID treated with cell therapy developed vector-related T cell leukemia [183]. The transduction was performed ex vivo into autologous CD34+ hematopoietic cells, using a gammaretroviral vector. It’s worth noting that retroviral and lentiviral vectors have been used and monitored extensively in cell and gene therapy trials, and replication-competent retrovirus/lentivirus (RCR/L) and insertional oncogenesis related risks are considered unlikely to happen in T cell products [184] when FDA guideline criteria [185] are met.
Table 3.
Milestones of gene and cell therapies, as well as notable technologies. Oligonucleotides therapies are not included in this table
| Year | Notable technology | Gene therapy | Cell therapy |
|---|---|---|---|
| 1962 | Szybalski coined the term “gene therapy” [272–275] | ||
| 1968 | Rogers and Pfuderer demonstrated virus mediated gene transfer [180] | ||
| 1972 | Literature outlined the potential and concerns of gene therapy [276] | ||
| 1976 | T cell growth factor interleukin 2 discovered [277] | ||
| 1982 | EP system first described [204, 278] | ||
| 1983 | Identification of human T cell antigen receptor [279–281] | ||
| 1984 | RVV transduced mammalian cells in vitro [282] | ||
| 1989 | AV approved for clinical trial [283] | Concept of CAR-T appeared [284] | |
| 1990 | First successful ex vivo gene therapy performed on an ADA-SCID patient [181] | ||
| 1993 | 1st gen CAR-T cells developed [285] | ||
| 1996 | 1st gen LVVs created [286], ZFN gene editing system available [253] | ||
| 1997 | SB transposon system designed for human cells [287] | FDA approved 1st anti-cancer monoclonal antibody Rituxan for NHL [288] | |
| 1999 | A patient died due to immune response triggered by in vivo AV-based therapy [289] | ||
| 2000 | 4-component LNP reported [235] | Some SCID-X1 patients received ex vivo, RVV-based gene therapy [290] | |
| 2003 | First approved LVV use in a phase 1 trial (ex vivo, for HIV control) [291] | Reports of RVV related tumorigenesis in two patients after gene therapy for SCID-X1 [292] | |
| 2005 | New RNA modification to reduce immunogenicity developed [166], PB transposon system applied to mammalian cells [114] | ||
| 2006 | TCR therapy applied to melanoma [293] | ||
| 2007 | First IND for CD19 CAR-T [294] | ||
| 2009 | Reports of eye disease treated in vivo with AAV-based vector [295] | ||
| 2010 | TALEN gene editing system available [268] | CD19 CAR-T in NHL case reports [296, 297] | |
| 2011 | CD19 CAR-T in CLL & ALL case reports [298–300] | ||
| 2012 | Crispr/Cas gene editing system available[259] [301] | EMA approved Glybera (withdrawn in 2017 for market reason) [302] | |
| 2015 | First BCMA CAR-T clinical trial [303] | ||
| 2016 | CRISPR/Cas9 applied to T cell engineering [304] | EMA approved Strimvelis [305] | |
| 2017 | FDA approved Luxturna [306] | FDA approved first cell therapies Yescarta [307]and Kymriah [308] | |
| 2018 | Multiple CRISPR clinical trials combined CAR-T and PD-1 KO for cancer immunotherapies [304] | ||
| 2019 | FDA approved Zolgensma [309] EMA approves Zynteglo [310] | ||
| 2020 | FDA approved Tecartus [311] | ||
| 2021 | FDA approved Breyanzi [312] and Abecma [313] | ||
| 2022 | FDA approved Carvykti [314] |
These severe adverse events represented a turning point toward the development of alternatives to viral vectors. However, over the years, research on viral vectors also led to significant improvements in their safety. More recently, CAR-T and gene therapy treatments based on viral vectors have been approved and commercialized (Table 4). A variety of viral vectors are currently available for clinical practice and several comprehensive reviews have been written about this topic [186–189].
Table 4.
Details of the approved therapies based on viral vector technology
| Trade name | Agency | First approval | First approved indication | Cargo | Delivery method | Admin | Pivotal clinical trial | Actual completion date if applicable |
|---|---|---|---|---|---|---|---|---|
| Glybera® | EMA | 2012/10 (withdrawn 2017/10) | LPLD | LPL | AAV | in vivo | NCT00891306 | 2009/02—2011/04 |
| Strimvelis® | EMA | 2016/04 | ADA-SCID | ADA | RVV | ex vivo | NCT00598481 | 2008/01—2019/06 |
| Kymriah® | FDA | 2017/08 | r/r B-cell ALL | CD19 CAR | LVV | ex vivo | NCT02435849 | 2015/04—2020/01 |
| Yescarta® | FDA | 2017/10 | r/r LBCL | CD19 CAR | RVV | ex vivo | NCT02348216 | 2015/04—2020/09 |
| Luxturna® | FDA | 2017/12 | biallelic RPE65 mutation-associated IRD | RPE65 | AAV | in vivo | NCT00999609 | 2012/10—2015/07 |
| Zolgensma ® | FDA | 2019/05 | SMA (type I) | SMN | AAV | in vivo | NCT03306277 | 2017/10—2019/11 |
| Zynteglo™ | EMA | 2019/05 (withdrawn 2022/03) | TDT | HBB | LVV | ex vivo | NCT01745120 | 2013/08—2018/02 |
| Tecartus™ | FDA | 2020/07 | r/r MCL | CD19 CAR | RVV | ex vivo | NCT02601313 | 2015/11—2019/07 |
| Breyanzi® | FDA | 2021/02 | r/r LBCL | CD19 CAR | LVV | ex vivo | NCT02631044 | 2016/01—2022/12 |
| Abecma® | FDA | 2021/03 | r/r MM | BCMA CAR | LVV | ex vivo | NCT03361748 | 2017/12—2024/11 |
| Carvykti ™ | FDA | 2022/02 | r/r MM | BCMA CAR | LVV | ex vivo | NCT03548207 | 2018/06—2022/08 |
Despite successful developments, the main drawback for viral vectors remains their immunogenicity, especially in relation to inflammation [190, 191]. For example, there are significant challenges associated with using vectors based on adenovirus serotype 5 and adeno-associated virus type 2 in clinical settings. These viruses are so widespread that a significant number of people bear pre-existing immunity against them [192, 193]. Even repeated administration of viral vectors with low seroprevalence to patients may present serious challenges, due to risks in developing immunity against the vector and degeneration of the transduced tissue. Moreover, ectopic integration of viral DNA can also be responsible for insertional mutagenesis [194, 195]. This process can lead to disruption of tumor suppressor genes or activation of oncogenes, triggering neoplastic transformation of the host cells. Other disadvantages associated with viral vectors include their limited transgenic capacity, manufacturing challenges, and efforts related to scaling up the production process [196, 197]. Viral vector manufacturing presents technical barriers which may create a supply chain shortage and may slow the expansion of cell and gene therapy [198, 199]. Viral vectors also have a lengthy production process in terms of generating and testing the master and working cell banks [199, 200]. Continuous efforts are currently carried out to mitigate this issue. Due to these drawbacks, the development of non-viral vectors and delivery methods with lower immunogenicity remains a viable option.
Non-viral vectors
Since the early 2000s, the use of non-viral vectors in clinical trials has increased steadily. This may be due to improvements in their efficiency, safety, stability of gene expression and specificity. Non-viral vectors have a lower testing burden and smaller storage footprint compared to viral vectors [201, 202]. Moreover, they are amenable for use in personalized therapies that target patients’ personal mutanome [202, 203]. These characteristics are important when deciding on a gene correction platform for treating one or a small number of patients.
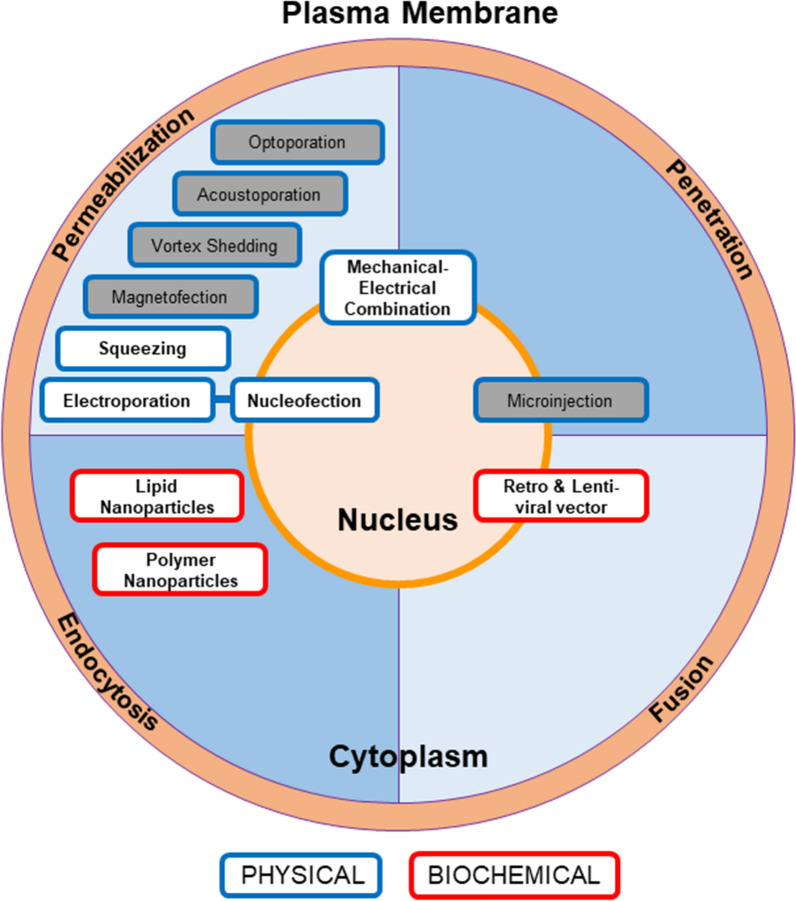
Nevertheless, the optimal non-viral vector and delivery system still need to be tailored to the type of target cells and to the characteristics of the transgene, requiring a customized system. Commonly used non-viral vectors include transposons and episomes, both have been discussed above. Depending on its nature (DNA, mRNA, protein), the payload needs to pass through one or two barriers before reaching the genome: the cellular plasma membrane (the first barrier) and the nuclear membrane (the second barrier). Four different methods have been exploited so far to deliver the payload into the cells: fusion, penetration, permeabilization and endocytosis (Fig. 1). As mentioned above, viral transduction, due to its natural cell entry mechanism through fusion, has been utilized in commercialized cell and gene therapy products. In this section, we will focus on non-viral gene delivery, using physical or biochemical-based techniques. Physical methods utilize permeabilization and penetration to deliver a genetic payload into the cells, while biochemical approaches rely on endocytosis and fusion.
Fig. 1.
Common delivery methods are shown here with color coded boxes. The blue and red borders represent the physical or biochemical nature of the methods. Different delivery methods are positioned in color-coded sections to represent the mechanism of entry. The method with multiple mechanisms is positioned on the border of the mechanism involved. The methods not covered in detail in this review are grayed out. Specific references for each method can be found in the relative sections of the manuscript
Physical delivery methods
Commonly used physical delivery methods include electroporation (EP) and microfluidic-based mechanical methods. These approaches create pores on the cellular membrane for gene entry via electric shock or mechanical stress. The payload is usually prepared in a specific buffer and does not require laborious preparation (e.g., envelope the payload in a viral capsid or coat with nanoparticles), significantly reducing the effort associated with this type of methodology. Physical methods do not have stringent limitations on payload size; however, as payload size increases, the need to obtain larger and more persistent pores may compromise cell viability. In bulk EP transfection, cells are mixed with the payload in a conductive buffer that is connected to two electrodes, then exposed to a brief electrical pulse for a few milliseconds [204, 205]. By combining different electrical voltages, pulse durations, and buffer chemistry, EP can be optimized to maximize transduction efficiency for different types of payloads and minimize cellular damage, even in hard-to-transduce cells, such as stem cells and T cells [206, 207]. Although the delivery efficiency is dependent on payload size, Zhao et al. [170] and Birkholz et al. [208], demonstrated that the efficiency of EP transfection can be up to 80% using pDNA, and higher than 90% in stimulated T cells transduced with mRNA [170, 208]. In another study, Distler et al. [209] designed an advanced EP technique called nucleofection which improves the transfection efficiency in unstimulated T cells, by using a unique combination of conductive buffer and electrical pulses [209].
Proteins can also be successfully delivered through EP. Due to its versatility in payload delivery, EP may be paired with new gene-editing techniques that require multiple components. In the first human clinical trial to assess the safety and feasibility of CRISPR-Cas9 gene editing of human T cells, the researchers used EP to deliver ~ 160 kDa ribonucleoproteins, targeting three genes in primary human T cells with the frequency of editing up to 45% in this trial [53]. KO efficiency could reach 85% to 98% in activated human T cells [42] and > 80% in other leukocytes [210]. CRISPR/cas9 system has been delivered with viral vectors such as lentiviral [211] and adenoviral vectors [212] and exhibited varied KO efficiencies, but few studies have directly compared efficiency of viral and non-viral methods with the same target and same CRISPR system. Multiple studies have also shown that EP is effective in delivering a transposon system into immune cells, with up to 65% transfection efficiency [213, 214]. EP-based transfection technologies have been developed to enable scalability, using different microfluidic approaches. For example, a semi-continuous flow and micro-fluidic EP devices have been developed to process large quantities of cells with high transfection efficiency for clinical applications [189].
EP transfection is also associated with several drawbacks. It requires special equipment and optimization for each cell type [215, 216] since fine tuning of EP conditions to achieve good viability and transfection efficiency is critical. EP conditions have been particularly difficult to optimize for certain cell types. To increase efficiency, high DNA concentration is generally used but can result in DNA toxicity in host cells. For bulk EP, non-uniform electric field distribution may cause Joule heating and bubble formation that could severely affect transfection efficiency and cell viability (reviewed in [217–219]). Recent developments could enhance the local electric field, thus lowering the operating voltage and preventing the formation of bubbles, therefore increasing cell viability up to 90% [220].
In addition to cell viability, multiple reports expressed concerns regarding the impact of EP on immune cells, as it may lead to alteration in gene expression, reduced expansion capacity and cytotoxicity [221–223]. To minimize the impact on primary immune cells, other physical methods have been explored, such as the squeezing method. By passing cells through a microfluidic device with constriction 30–80% smaller than the cell diameter, it is possible to create transient holes in the cell membrane. This method has successfully delivered protein, RNA, and DNA to multiple cell types, including embryonic stem cells and immune cells [224, 225]. It has been shown that the squeezing method has a minimal effect on gene expression and does not interfere with T cell activity, as observed with EP [224].
However, the application of squeezing technologies to human primary T cells still needs further study. Despite these recent advances, squeezing may not be suitable for in vivo gene therapy because target cells need to be isolated and processed ex vivo using specific equipment [226].
Recently, mechanical–electrical combination technology has been developed, combining nanostraw [227] or cell squeezing [228] and electric-field-driven transport. For the latter, cells are passed through microfluidic constrictions to disrupt the plasma membrane, then shocked with an electronic pulse to permeabilize the nuclear envelope. Nuclear delivery of pDNA was detected within 1-h post-treatment, and the integrity of the nuclear envelope was recovered within 15 min post-treatment. Cell viability is similar to the cells exposed to standard EP at the 24 h post-treatment time point [228].
Other mechanical methods commonly used in preclinical studies are fluid shear, vortex shedding, microinjections, acoustoporation, laser optoporation, and magnetofection (reviewed in [217, 229]).
Biochemical delivery methods
Nanoparticle chemistry has application to gene and drug delivery. Nanoparticle-based gene delivery, either lipid-based nanoparticles (LNPs) or polymeric nanoparticles (PNPs), are technologies centered on the encapsulation of the payload [230]. The cellular plasma membrane is negatively charged, which makes it difficult for negatively charged pDNA and mRNA to enter the cells by diffusion. Cationic nanoparticles bind to pDNA and mRNA to form lipoplexes and polyplexes that harbor a net positive charge and can enter the cell through endocytosis [231].
Lipid molecules have been used to transport genetic material into cells for a long time [232, 233]. Modern LNP systems appeared around the year 2000 to deliver DNA. They consist of four components: phospholipid, lipid-anchored polyethylene glycol (PEG), cholesterol, and ionizable lipid [234, 235]. The recent COVID-19 mRNA vaccines utilized this four-component LNP system demonstrating the efficiency and safety of delivering genetic material [236–238].
Their main drawback is the limited capacity to undergo endosomal escape, which affects the amount of genetic material that may reach the cytoplasm [239]. By incorporating the component that could target specific molecules, LNP delivery systems have the potential for precise in vivo delivery. This aspect was highlighted in a recent study using T cell-targeting LNPs to deliver mRNA encoding the CAR in vivo [172]. Overall, LNPs have low toxicity due to the natural and biological origin of the components and, under proper storage conditions, they may be stable for up to 150 days [240].
PNPs are another type of vehicle used for drug delivery, composed of natural carbohydrate polymers or synthetic polymers. Some PNP-based systems may exhibit higher stability and mechanical resistance compared to LNP-based systems [241–243]. The natural polymer chitosan (CS) is a cationic polysaccharide obtained from the exoskeleton of crustaceans such as crabs and shrimps. Chitosan nanoparticles can form electrostatic complexes with DNA, making it an attractive carrier for non-viral application [244]. It has been tested as a carrier for gene therapy in brain tumors [245], but it is also known to trigger an IL-1β response in a variety of cell types [246], which may be a concern for in vivo therapies.
Synthetic polymers present limited batch-to-batch variation. One example is polycationic polyethyleneimine (PEI) which is commonly used for gene delivery, thanks to its high transfection efficiency and high buffer capacity. Such a property is attributed to the proton sponge effect from the partially protonated amines on PEI chains (reviewed in [247]). PEI nanoparticles have been used to transfer large (12–14 kb) payloads, such as self-amplifying replicon RNAs (RepRNA) [248]. Recently, Olden et al. [249] developed an architecture of pDMAEMA polymers with 25% transfection efficiency for mRNA and 18% for pDNA in CD4+ and CD8+ primary T cells [249]. Another study achieved 12% transfection efficiency of pDNA in primary T cells [250].
Degradable synthetic polymers such as polylactide (PLA) and Poly (β-amino ester) (PBAE) have been developed to address concerns regarding the long-term toxicity of non-biodegradable polymers. PBAE exhibited robust transfection capabilities and efficient endosomal escape properties. It also showed promising results in cytosolic protein delivery in vitro and efficient CRISPR-Cas9 delivery in several cell types [251]. PNPs could also be manufactured in combination with ligands for in vivo tissue and cell targeting. A recent study indeed showed that PBAE nanocarriers successfully delivered CAR or TCR-encoding mRNA to circulating human primary T cells. Engineered T cells achieved tumor regression in xenograft mouse models [168].
Conclusion
Multiple options are currently being explored to treat genetic disease using molecular tools, to restore gene expression to physiological conditions. As alternatives to viral vectors, non-viral vectors represent a potentially promising strategy due to comparable efficacy to viral vectors, for clinical applications.
In this review, we summarized the landscape of molecular tools, type of payloads, material and methods used as a strategy to regulate gene expression for gene and cell therapy. Knowledge about non-viral vectors is expanding exponentially and will likely prompt an acceleration in the application of new compounds in different medical specialties.
Acknowledgements
Not applicable.
Abbreviations
- ADA-SCID
adenosine deaminase severe combined immunodeficiency
- ALL
acute lymphocytic leukemia
- ASP-siRNA
allele-specific siRNAs
- AV
adenovirus
- AAV
adeno-associated virus
- bp
base pair
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cas
CRISPR associated protein
- CLL
chronic lymphocytic leukemia
- DSB
double-strand break
- EP
electroporation
- EMA
European Medicines Agency
- GCRs
gross chromosomal rearrangements
- gRNA
guide RNA
- IVT
in vitro-transcribed
- LBCL
large B-cell lymphoma
- LCC
linear covalently closed
- LPLD
lipoprotein lipase deficiency
- LNPs
lipid-based nanoparticles
- LTR
long terminal repeat elements
- LVV
lenti-viral vector
- MAC, HAC
mammalian or human artificial chromosomes
- MCL
mantle cell lymphoma
- MNs
meganucleses
- MM
multiple myeloma
- NHL
non-Hodgkin's lymphoma
- NHEJ
Non-Homologous End Joining
- dCas
nuclease dead Cas
- PB
PiggyBac
- PBAE
poly β-amino ester
- PEI
polycationic polyethyleneimine
- PEG
polyethylene glycol
- PLA
polylactide
- PNPs
polymeric nanoparticles
- PTGS
post-transcriptional gene silencing
- PD-1
programmed cell Death protein 1
- RepRNA
replicon RNAs
- RNAi
RNA interference
- RISC
RNA-Induced Silencing Complex
- RITS
RNA-induced transcriptional silencing
- r/r
relapsed/refractory
- RVV
retro-viral vector
- SA
sandwich transposon
- S/MAR
scaffold/matrix attachment region sequences
- SCID
Severe Combined Immunodeficiency Defect
- shRNA
short hairpin RNA
- SSTR
single-stranded template repair
- SB
Sleeping Beauty
- siRNA
small interfering RNA
- SMA 1
spinal muscular atrophy type I
- TDT
transfusion-dependent beta-thalassemia
- TIRs
terminal inverted repeats
- TALENs
transcription activator-like effector nuclease
- TEs
transposable elements
- FDA
U.S. Food and Drug Administration
- ZFNs
zinc finger nucleases
Author contributions
HT, VP and MP wrote and edited the manuscript, prepared the figures and incorporated the references. HT and VP coordinated writing tasks. SW wrote a part of the manuscript and prepared figures. LZ prepared a table. QC and FM reviewed and commented on the content of the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors consent to the publication of this manuscript.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hsing-Chuan Tsai, Violena Pietrobon and Maoyu Peng contributed equally to this work
References
- 1.Ségalat L. Loss-of-function genetic diseases and the concept of pharmaceutical targets. Orphanet J Rare Dis. 2007;2(1):30. doi: 10.1186/1750-1172-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keinath MC, Prior DE, Prior TW. Spinal muscular atrophy: mutations, testing, and clinical relevance. Appl Clin Genetics. 2021;14:11–25. doi: 10.2147/TACG.S239603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leontiadou H, Galdadas I, Athanasiou C, Cournia Z. Insights into the mechanism of the PIK3CA E545K activating mutation using MD simulations. Sci Rep-uk. 2018;8(1):15544. doi: 10.1038/s41598-018-27044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubinsztein DC. How does the Huntington’s Disease mutation damage cells? Sci Aging Knowl Environ. 2003;2003(37):PE26. doi: 10.1126/sageke.2003.37.pe26. [DOI] [PubMed] [Google Scholar]
- 5.Zhang HX, Zhang Y, Yin H. Genome editing with mRNA encoding ZFN, TALEN, and Cas9. Mol Ther J Am Soc Gene Ther. 2019;27(4):735–746. doi: 10.1016/j.ymthe.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P, et al. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther. 2011;11(1):11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. 2020;5(1):1. doi: 10.1038/s41392-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belfort M, Bonocora RP. Homing endonucleases: from genetic anomalies to programmable genomic clippers. Methods Mol Biology Clifton N J. 2014;1123:1–26. doi: 10.1007/978-1-62703-968-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mittal RD. Gene editing in clinical practice. Ind J Clin Biochem. 2018;33(1):1–4. doi: 10.1007/s12291-017-0720-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27(9):851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5(1):97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 14.Kim JS, Lee HJ, Carroll D. Genome editing with modularly assembled zinc-finger nucleases. Nat Methods. 2010;7(2):91–91. doi: 10.1038/nmeth0210-91b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300(5620):764–764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 16.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, et al. Breaking the code of DNA binding specificity of TAL-Type III effectors. Science. 2009;326(5959):1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 17.Feng Y, Zhang S, Huang X. A robust TALENs system for highly efficient mammalian genome editing. Sci Rep-uk. 2014;4(1):3632. doi: 10.1038/srep03632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan SH. Genome-editing technologies: concept, pros, and cons of various genome-editing techniques and bioethical concerns for clinical application. Mol Ther Nucleic Acids. 2019;16:326–334. doi: 10.1016/j.omtn.2019.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc National Acad Sci. 2001;98(13):7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grover A, Pande A, Choudhary K, Gupta K, Sundar D. Re-programming DNA-binding specificity in zinc finger proteins for targeting unique address in a genome. Syst Synth Biol. 2010;4(4):323–329. doi: 10.1007/s11693-011-9077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25(1):1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azangou-Khyavy M, Ghasemi M, Khanali J, Boroomand-Saboor M, Jamalkhah M, Soleimani M, et al. CRISPR/Cas: from tumor gene editing to T cell-based immunotherapy of cancer. Front Immunol. 2020;11:2062. doi: 10.3389/fimmu.2020.02062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang T, Lander ES, Sabatini DM. Large-scale single guide RNA library construction and use for CRISPR–Cas9-based genetic screens. Cold Spring Harb Protoc. 2016;2016(3):pdb.top086892. doi: 10.1101/pdb.top086892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Covarrubias S, Vollmers AC, Capili A, Boettcher M, Shulkin A, Correa MR, et al. High-throughput CRISPR screening identifies genes involved in macrophage viability and inflammatory pathways. Cell Rep. 2020;33(13):108541. doi: 10.1016/j.celrep.2020.108541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spangler JR, Leski TA, Schultzhaus Z, Wang Z, Stenger DA. Large scale screening of CRISPR guide RNAs using an optimized high throughput robotics system. Sci Rep-uk. 2022;12(1):13953. doi: 10.1038/s41598-022-17474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeWitt MA, Corn JE, Carroll D. Genome editing via delivery of Cas9 ribonucleoprotein. Methods. 2017;121:9–15. doi: 10.1016/j.ymeth.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarz H, Schmittner M, Duschl A, Horejs-Hoeck J. Residual endotoxin contaminations in recombinant proteins are sufficient to activate human CD1c+ Dendritic cells. PLoS ONE. 2014;9(12):e113840. doi: 10.1371/journal.pone.0113840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecker JG. Non-Viral, lipid-mediated DNA and mRNA gene therapy of the central nervous system (CNS): chemical-based transfection. Methods Mol Biology Clifton N J. 2016;1382:307–324. doi: 10.1007/978-1-4939-3271-9_23. [DOI] [PubMed] [Google Scholar]
- 29.Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, et al. Non-Viral CRISPR/Cas Gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angewandte Chemie Int Ed. 2017;56(4):1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA, Siegwart DJ. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat Nanotechnol. 2020;15(4):313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merkle FT, Neuhausser WM, Santos D, Valen E, Gagnon JA, Maas K, et al. Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep. 2015;11(6):875–883. doi: 10.1016/j.celrep.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han HA, Pang JKS, Soh BS. Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing. J Mol Med. 2020;98(5):615–632. doi: 10.1007/s00109-020-01893-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Znidar K, Bosnjak M, Semenova N, Pakhomova O, Heller L, Cemazar M. Tumor cell death after electrotransfer of plasmid DNA is associated with cytosolic DNA sensor upregulation. Oncotarget. 2018;9(27):18665–18681. doi: 10.18632/oncotarget.24816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro-Guerrero E, Tay C, Whalley JP, Cowley SA, Davies B, Knight JC, et al. Genome-wide CRISPR/Cas9-knockout in human induced Pluripotent Stem Cell (iPSC)-derived macrophages. Sci Rep-uk. 2021;11(1):4245. doi: 10.1038/s41598-021-82137-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hana S, Peterson M, McLaughlin H, Marshall E, Fabian AJ, McKissick O, et al. Highly efficient neuronal gene knockout in vivo by CRISPR-Cas9 via neonatal intracerebroventricular injection of AAV in mice. Gene Ther. 2021;28(10–11):646–658. doi: 10.1038/s41434-021-00224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li XL, Li GH, Fu J, Fu YW, Zhang L, Chen W, et al. Highly efficient genome editing via CRISPR–Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Res. 2018;46(19):gky04. doi: 10.1093/nar/gky804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen W, Cheng X, Fu Y, Meng F, Zhang JP, Zhang L, et al. High-Level Precise Knockin of iPSCs by simultaneous reprogramming and genome editing of human peripheral blood mononuclear cells. Stem Cell Rep. 2018;10(6):1821–1834. doi: 10.1016/j.stemcr.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang JP, Li XL, Li GH, Chen W, Arakaki C, Botimer GD, et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017;18(1):35. doi: 10.1186/s13059-017-1164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Liu C, Zhao Y, Han X, Zhou Z, Wang C, et al. Comparing successful gene knock-in efficiencies of CRISPR/Cas9 with ZFNs and TALENs gene editing systems in bovine and dairy goat fetal fibroblasts. J Integr Agr. 2018;17(2):406–14. [Google Scholar]
- 40.Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. P Natl Acad Sci Usa. 2015;112(33):10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rezalotfi A, Fritz L, Förster R, Bošnjak B. Challenges of CRISPR-Based gene editing in primary T cells. Int J Mol Sci. 2022;23(3):1689. doi: 10.3390/ijms23031689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seki A, Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J Exp Med. 2018;215(3):985–997. doi: 10.1084/jem.20171626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nüssing S, House IG, Kearney CJ, Chen AXY, Vervoort SJ, Beavis PA, et al. Efficient CRISPR/Cas9 gene editing in uncultured naive mouse T cells for in vivo studies. J Immunol. 2020;204(8):2308–2315. doi: 10.4049/jimmunol.1901396. [DOI] [PubMed] [Google Scholar]
- 44.Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33(9):985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt R, Steinhart Z, Layeghi M, Freimer JW, Bueno R, Nguyen VQ, et al. CRISPR activation and interference screens decode stimulation responses in primary human T cells. Sci New York N Y. 2022;375(6580):4008. doi: 10.1126/science.abj4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z, Li L, Turkoz A, Chen P, Harari-Steinfeld R, Bobbin M, et al. Contextual reprogramming of CAR-T cells for treatment of HER2+ cancers. J Transl Med. 2021;19(1):459. doi: 10.1186/s12967-021-03132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li A, Tanner MR, Lee CM, Hurley AE, Giorgi MD, Jarrett KE, et al. AAV-CRISPR gene editing is negated by pre-existing immunity to Cas9. Mol Ther. 2020;28(6):1432–1441. doi: 10.1016/j.ymthe.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D, Mou H, Li S, Li Y, Hough S, Tran K, et al. Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum Gene Ther. 2015;26(7):432–442. doi: 10.1089/hum.2015.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ajina R, Zamalin D, Zuo A, Moussa M, Catalfamo M, Jablonski SA, et al. SpCas9-expression by tumor cells can cause T cell-dependent tumor rejection in immunocompetent mice. Oncoimmunology. 2019;8(5):1–11. doi: 10.1080/2162402X.2019.1577127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chew WL, Tabebordbar M, Cheng JKW, Mali P, Wu EY, Ng AHM, et al. A multifunctional AAV–CRISPR–Cas9 and its host response. Nat Methods. 2016;13(10):868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amini L, Wagner DL, Rössler U, Zarrinrad G, Wagner LF, Vollmer T, et al. CRISPR-Cas9-Edited tacrolimus-resistant antiviral T Cells for advanced adoptive immunotherapy in transplant recipients. Mol Ther. 2021;29(1):32–46. doi: 10.1016/j.ymthe.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E, et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367(6481):eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu L, Wang J, Liu Y, Xie L, Su B, Mou D, et al. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. New Engl J Med. 2019;381(13):1240–1247. doi: 10.1056/NEJMoa1817426. [DOI] [PubMed] [Google Scholar]
- 55.Lu Y, Xue J, Deng T, Zhou X, Yu K, Deng L, et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat Med. 2020;26(5):732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 56.Wagner DL, Peter L, Schmueck-Henneresse M. Cas9-directed immune tolerance in humans—a model to evaluate regulatory T cells in gene therapy? Gene Ther. 2021;28(9):549–559. doi: 10.1038/s41434-021-00232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blattner G, Cavazza A, Thrasher AJ, Turchiano G. Gene editing and genotoxicity: targeting the off-targets. Frontiers Genome Ed. 2020;2:613252. doi: 10.3389/fgeed.2020.613252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bothmer A, Gareau KW, Abdulkerim HS, Buquicchio F, Cohen L, Viswanathan R, et al. Detection and modulation of DNA translocations during multi-gene genome editing in T cells. Crispr J. 2020;3(3):177–187. doi: 10.1089/crispr.2019.0074. [DOI] [PubMed] [Google Scholar]
- 59.Brunet E, Jasin M. Induction of chromosomal translocations with CRISPR-Cas9 and other nucleases: understanding the repair mechanisms that give rise to translocations. Adv Exp Med Biol. 2018;1044:15–25. doi: 10.1007/978-981-13-0593-1_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qasim W, Ciocarlie O, Adams S, Inglott S, Murphy C, Rivat C, et al. Preliminary results of UCART19, an allogeneic Anti-CD19 CAR T-Cell Product in a First-in-Human Trial (PALL) in pediatric patients with CD19+ relapsed/refractory B-cell acute lymphoblastic leukemia. Blood. 2017;130:887. [Google Scholar]
- 61.SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274(5288):765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 62.Mobile DNA III . Editor-in-Chief: Nancy L. Craig; Editors:Michael Chandler, Martin Gellert, Alan M. Lambowitz, Phoebe A. Rice, and Suzanne B. Sandmeyer. Washington (DC): ASM Press. $160.00. xxiv + 1321 p.; ill.; index. ISBN: 978–1–55581–920–0. 2015. Q Rev Biology. 2017;92(2):203–203.
- 63.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 64.Greenblatt IM, Brink RA. Transpositions of modulator in maize into divided and undivided chromosome segments. Nature. 1963;197(4865):412–413. [Google Scholar]
- 65.Rubin GM, Kidwell MG, Bingham PM. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982;29(3):987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- 66.Grabundzija I, Messing SA, Thomas J, Cosby RL, Bilic I, Miskey C, et al. A Helitron transposon reconstructed from bats reveals a novel mechanism of genome shuffling in eukaryotes. Nat Commun. 2016;7(1):10716. doi: 10.1038/ncomms10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muñoz-López M, García-Pérez JL. DNA transposons: nature and applications in genomics. Curr Genomics. 2010;11(2):115–128. doi: 10.2174/138920210790886871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, et al. Ten things you should know about transposable elements. Genome Biol. 2018;19(1):199. doi: 10.1186/s13059-018-1577-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ochmann MT, Ivics Z. Jumping ahead with sleeping beauty: mechanistic insights into cut-and-paste transposition. Viruses. 2021;13(1):76. doi: 10.3390/v13010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawakami K, Koga A, Hori H, Shima A. Excision of the Tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish. Danio rerio Gene. 1998;225(1–2):17–22. doi: 10.1016/s0378-1119(98)00537-x. [DOI] [PubMed] [Google Scholar]
- 71.Woodard LE, Li X, Malani N, Kaja A, Hice RH, Atkinson PW, et al. Comparative analysis of the recently discovered hAT transposon TcBuster in human cells. PLoS ONE. 2012;7(11):e42666. doi: 10.1371/journal.pone.0042666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshida J, Akagi K, Misawa R, Kokubu C, Takeda J, Horie K. Chromatin states shape insertion profiles of the piggyBac, Tol2 and Sleeping Beauty transposons and murine leukemia virus. Sci Rep-uk. 2017;7(1):43613. doi: 10.1038/srep43613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. P Natl Acad Sci Usa. 2008;105(27):9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu SCY, Meir YJJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, et al. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. P Natl Acad Sci Usa. 2006;103(41):15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mátés L, Chuah MKL, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41(6):753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 76.Kowarz E, Löscher D, Marschalek R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol J. 2015;10(4):647–653. doi: 10.1002/biot.201400821. [DOI] [PubMed] [Google Scholar]
- 77.Voigt K, Gogol-Döring A, Miskey C, Chen W, Cathomen T, Izsvák Z, et al. Retargeting sleeping beauty transposon insertions by engineered zinc finger DNA-binding domains. Mol Ther. 2012;20(10):1852–1862. doi: 10.1038/mt.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ivics Z, Hiripi L, Hoffmann OI, Mátés L, Yau TY, Bashir S, et al. Germline transgenesis in rabbits by pronuclear microinjection of Sleeping Beauty transposons. Nat Protoc. 2014;9(4):794–809. doi: 10.1038/nprot.2014.009. [DOI] [PubMed] [Google Scholar]
- 79.Ivics Z, Mátés L, Yau TY, Landa V, Zidek V, Bashir S, et al. Germline transgenesis in rodents by pronuclear microinjection of Sleeping Beauty transposons. Nat Protoc. 2014;9(4):773–793. doi: 10.1038/nprot.2014.008. [DOI] [PubMed] [Google Scholar]
- 80.Garrels W, Mátés L, Holler S, Dalda A, Taylor U, Petersen B, et al. Germline transgenic pigs by sleeping beauty transposition in porcine zygotes and targeted integration in the pig genome. PLoS ONE. 2011;6(8):e23573. doi: 10.1371/journal.pone.0023573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prommersberger S, Reiser M, Beckmann J, Danhof S, Amberger M, Quade-Lyssy P, et al. CARAMBA: a first-in-human clinical trial with SLAMF7 CAR-T cells prepared by virus-free Sleeping Beauty gene transfer to treat multiple myeloma. Gene Ther. 2021;28(9):560–571. doi: 10.1038/s41434-021-00254-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh H, Moyes JSE, Huls MH, Cooper LJN. Manufacture of T cells using the Sleeping Beauty system to enforce expression of a CD19-specific chimeric antigen receptor. Cancer Gene Ther. 2015;22(2):95–100. doi: 10.1038/cgt.2014.69. [DOI] [PubMed] [Google Scholar]
- 83.Magnani CF, Gaipa G, Lussana F, Belotti D, Gritti G, Napolitano S, et al. Sleeping Beauty–engineered CAR T cells achieve antileukemic activity without severe toxicities. J Clin Invest. 2020;130(11):6021–6033. doi: 10.1172/JCI138473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sebe A, Ivics Z. Reprogramming of human fibroblasts to induced pluripotent stem cells with sleeping beauty transposon-based stable gene delivery. Methods Mol Biology Clifton N J. 2016;1400:419–427. doi: 10.1007/978-1-4939-3372-3_26. [DOI] [PubMed] [Google Scholar]
- 85.Grabundzija I, Wang J, Sebe A, Erdei Z, Kajdi R, Devaraj A, et al. Sleeping Beauty transposon-based system for cellular reprogramming and targeted gene insertion in induced pluripotent stem cells. Nucleic Acids Res. 2013;41(3):1829–1847. doi: 10.1093/nar/gks1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cadiñanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35(12):e87–e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc National Acad Sci. 2011;108(4):1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pomeroy EJ, Lahr WS, Chang JW, Krueger J, Wick BJ, Slipek NJ, et al. Non-viral engineering of CAR-NK and CAR-T cells using the Tc Buster transposon systemTM. Biorxiv. 2021 doi: 10.1101/2021.08.02.454772. [DOI] [Google Scholar]
- 89.Ni J, Wangensteen KJ, Nelsen D, Balciunas D, Skuster KJ, Urban MD, et al. Active recombinant Tol2 transposase for gene transfer and gene discovery applications. Mobile Dna-uk. 2016;7(1):6. doi: 10.1186/s13100-016-0062-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yant SR, Huang Y, Akache B, Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35(7):e50–e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sandoval-Villegas N, Nurieva W, Amberger M, Ivics Z. Contemporary transposon tools: a review and guide through mechanisms and applications of sleeping beauty, piggybac and tol2 for genome engineering. Int J Mol Sci. 2021;22(10):5084. doi: 10.3390/ijms22105084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matteo MD, Samara-Kuko E, Ward NJ, Waddington SN, Waddingon SN, McVey JH, et al. Hyperactive PiggyBac transposons for sustained and robust liver-targeted gene therapy. Mol Ther. 2014;22(9):1614–1624. doi: 10.1038/mt.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Urasaki A, Morvan G, Kawakami K. Functional Dissection of the Tol2 transposable element identified the minimal cis -sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174(2):639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts A, Sivasubbu S, et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. Plos Genet. 2006;2(11):e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cui Z, Geurts AM, Liu G, Kaufman CD, Hackett PB. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol. 2002;318(5):1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 96.Zayed H, Izsvák Z, Walisko O, Ivics Z. Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther. 2004;9(2):292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 97.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational Analysis of the N-Terminal DNA-Binding domain of sleeping beauty transposase: critical Residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol. 2004;24(20):9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y, Pryputniewicz-Dobrinska D, Nagy EÉ, Kaufman CD, Singh M, Yant S, et al. Regulated complex assembly safeguards the fidelity of Sleeping Beauty transposition. Nucleic Acids Res. 2017;45(1):311–326. doi: 10.1093/nar/gkw1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bire S, Casteret S, Arnaoty A, Piégu B, Lecomte T, Bigot Y. Transposase concentration controls transposition activity: myth or reality? Gene. 2013;530(2):165–171. doi: 10.1016/j.gene.2013.08.039. [DOI] [PubMed] [Google Scholar]
- 100.Yant SR, Meuse L, Park J, Kay MA. 1014. The Sleeping Beauty transposase is regulated by overproduction inhibition in vitro and in vivo. Mol Ther. 2002;5(5):329–330. [Google Scholar]
- 101.Wilson MH, Coates CJ, George AL. PiggyBac Transposon-mediated gene transfer in human cells. Mol Ther. 2007;15(1):139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 102.Li X, Ewis H, Hice RH, Malani N, Parker N, Zhou L, et al. A resurrected mammalian hAT transposable element and a closely related insect element are highly active in human cell culture. P Natl Acad Sci Usa. 2012;110(6):E478–E487. doi: 10.1073/pnas.1121543109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kawakami K, Noda T. Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics. 2004;166(2):895–899. doi: 10.1534/genetics.166.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.He YZ, Yan JR, He B, Ren H, Kuang X, Long TF, et al. A Transposon-Associated CRISPR/Cas9 system specifically eliminates both chromosomal and plasmid-borne mcr-1 in Escherichia coli. Antimicrob Agents Ch. 2021;65(10):e01054–e1121. doi: 10.1128/AAC.01054-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Karvelis T, Druteika G, Bigelyte G, Budre K, Zedaveinyte R, Silanskas A, et al. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature. 2021;599(7886):692–696. doi: 10.1038/s41586-021-04058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peters JE, Makarova KS, Shmakov S, Koonin EV. Recruitment of CRISPR-Cas systems by Tn7-like transposons. P Natl Acad Sci USA. 2017;114(35):E7358–E7366. doi: 10.1073/pnas.1709035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang J, Yu C, Pulletikurti V, Lamb J, Danilova T, Weber DF, et al. Alternative Ac/Ds transposition induces major chromosomal rearrangements in maize. Gene Dev. 2009;23(6):755–765. doi: 10.1101/gad.1776909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Geurts AM, Collier LS, Geurts JL, Oseth LL, Bell ML, Mu D, et al. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. Plos Genet. 2006;2(9):e156. doi: 10.1371/journal.pgen.0020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Keng VW, Yae K, Hayakawa T, Mizuno S, Uno Y, Yusa K, et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat Methods. 2005;2(10):763–769. doi: 10.1038/nmeth795. [DOI] [PubMed] [Google Scholar]
- 110.Ivics Z, Izsvák Z. The expanding universe of transposon technologies for gene and cell engineering. Mobile Dna-uk. 2010;1(1):25. doi: 10.1186/1759-8753-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Keng VW, Ryan BJ, Wangensteen KJ, Balciunas D, Schmedt C, Ekker SC, et al. Efficient transposition of Tol2 in the mouse germline. Genetics. 2009;183(4):1565–1573. doi: 10.1534/genetics.109.100768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Carlson CM, Dupuy AJ, Fritz S, Roberg-Perez KJ, Fletcher CF, Largaespada DA. Transposon mutagenesis of the mouse germline. Genetics. 2003;165(1):243–256. doi: 10.1093/genetics/165.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liang Q, Kong J, Stalker J, Bradley A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis. 2009;47(6):404–408. doi: 10.1002/dvg.20508. [DOI] [PubMed] [Google Scholar]
- 114.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient Transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122(3):473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 115.Amberger M, Ivics Z. Latest advances for the sleeping beauty transposon system: 23 years of insomnia but prettier than ever. BioEssays. 2020;42(11):2000136. doi: 10.1002/bies.202000136. [DOI] [PubMed] [Google Scholar]
- 116.Mulia GE, Picanço-Castro V, Stavrou EF, Athanassiadou A, Figueiredo ML. Advances in the development and the applications of nonviral, episomal vectors for gene therapy. Hum Gene Ther. 2021;32(19–20):1076–1095. doi: 10.1089/hum.2020.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Conese M, Auriche C, Ascenzioni F. Gene therapy progress and prospects: episomally maintained self-replicating systems. Gene Ther. 2004;11(24):1735–1741. doi: 10.1038/sj.gt.3302362. [DOI] [PubMed] [Google Scholar]
- 118.Riu E, Chen ZY, Xu H, He CY, Kay MA. Histone modifications are associated with the persistence or silencing of vector-mediated transgene expression in vivo. Mol Ther. 2007;15(7):1348–1355. doi: 10.1038/sj.mt.6300177. [DOI] [PubMed] [Google Scholar]
- 119.Piechaczek C, Fetzer C, Baiker A, Bode J, Lipps HJ. A vector based on the SV40 origin of replication and chromosomal S/MARs replicates episomally in CHO cells. Nucleic Acids Res. 1999;27(2):426–428. doi: 10.1093/nar/27.2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jackson DA, Juranek S, Lipps HJ. Designing nonviral vectors for efficient gene transfer and long-term gene expression. Mol Ther. 2006;14(5):613–626. doi: 10.1016/j.ymthe.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 121.Ehrhardt A, Haase R, Schepers A, Deutsch M, Lipps H, Baiker A. Episomal vectors for gene therapy. Curr Gene Ther. 2008;8(3):147–161. doi: 10.2174/156652308784746440. [DOI] [PubMed] [Google Scholar]
- 122.Baiker A, Maercker C, Piechaczek C, Schmidt SBA, Bode J, Benham C, et al. Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix. Nat Cell Biol. 2000;2(3):182–184. doi: 10.1038/35004061. [DOI] [PubMed] [Google Scholar]
- 123.Verghese SC, Goloviznina NA, Skinner AM, Lipps HJ, Kurre P. S/MAR sequence confers long-term mitotic stability on non-integrating lentiviral vector episomes without selection. Nucleic Acids Res. 2014;42(7):e53–e53. doi: 10.1093/nar/gku082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stavrou EF, Giannakopoulos A, Spyridonidis A, Athanassiadou A. A bona fide mammalian replicator enhances all aspects of episomal gene transfer into human hematopoietic progenitor cells. Mol Ther. 2015;23:S97. [Google Scholar]
- 125.Rupprecht S, Hagedorn C, Seruggia D, Magnusson T, Wagner E, Ogris M, et al. Controlled removal of a nonviral episomal vector from transfected cells. Gene. 2010;466(1–2):36–42. doi: 10.1016/j.gene.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 126.Hagedorn C, Antoniou MN, Lipps HJ. Genomic cis-acting sequences improve expression and establishment of a nonviral vector. Mol Ther - Nucleic Acids. 2013;2(8):e118. doi: 10.1038/mtna.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Haase R, Argyros O, Wong SP, Harbottle RP, Lipps HJ, Ogris M, et al. pEPito: a significantly improved non-viral episomal expression vector for mammalian cells. Bmc Biotechnol. 2010;10(1):20. doi: 10.1186/1472-6750-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Giannakopoulos A, Stavrou EF, Zarkadis I, Zoumbos N, Thrasher AJ, Athanassiadou A. The Functional Role of S/MARs in episomal vectors as defined by the stress-induced destabilization profile of the vector sequences. J Mol Biol. 2009;387(5):1239–1249. doi: 10.1016/j.jmb.2009.02.043. [DOI] [PubMed] [Google Scholar]
- 129.Chen ZY, He CY, Ehrhardt A, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther. 2003;8(3):495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 130.Darquet AM, Rangara R, Kreiss P, Schwartz B, Naimi S, Delaère P, et al. Minicircle: an improved DNA molecule for in vitro and in vivo gene transfer. Gene Ther. 1999;6(2):209–218. doi: 10.1038/sj.gt.3300816. [DOI] [PubMed] [Google Scholar]
- 131.Han J, Gao F, Geng S, Ye X, Wang T, Du P, et al. Minicircle DNA-Engineered CAR T cells suppressed tumor growth in mice. Mol Cancer Ther. 2020;19(1):178–186. doi: 10.1158/1535-7163.MCT-19-0204. [DOI] [PubMed] [Google Scholar]
- 132.Wang H, Ye X, Ju Y, Cai Z, Wang X, Du P, et al. Minicircle DNA-Mediated CAR T Cells Targeting CD44 suppressed hepatocellular carcinoma both in vitro and in vivo. Oncotargets Ther. 2020;13:3703–3716. doi: 10.2147/OTT.S247836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hudecek M, Gogishvili T, Monjezi R, Wegner J, Shankar R, Kruesemann C, et al. Minicircle-based engineering of chimeric antigen receptor (CAR) T cells. Recent Results Cancer Res Fortschritte Der Krebsforschung Progres Dans Les Recherches Sur Le Cancer. 2016;209:37–50. doi: 10.1007/978-3-319-42934-2_3. [DOI] [PubMed] [Google Scholar]
- 134.Monjezi R, Miskey C, Gogishvili T, Schleef M, Schmeer M, Einsele H, et al. Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia. 2017;31(1):186–194. doi: 10.1038/leu.2016.180. [DOI] [PubMed] [Google Scholar]
- 135.Bozza M, Green EW, Espinet E, Roia AD, Klein C, Vogel V, et al. Novel Non-integrating DNA Nano-S/MAR vectors restore gene function in isogenic patient-derived pancreatic tumor models. Mol Ther - Methods Clin Dev. 2020;17:957–968. doi: 10.1016/j.omtm.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bozza M, Roia AD, Correia MP, Berger A, Tuch A, Schmidt A, et al. A nonviral, nonintegrating DNA nanovector platform for the safe, rapid, and persistent manufacture of recombinant T cells. Sci Adv. 2021;7(16):eabf1333. doi: 10.1126/sciadv.abf1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs. shRNA: similarities and differences. Adv Drug Deliver Rev. 2009;61(9):746–59. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 138.Monga I, Qureshi A, Thakur N, Gupta AK, Kumar M. ASPsiRNA A Resource of ASP-siRNAs having therapeutic potential for human genetic disorders and algorithm for prediction of their inhibitory efficacy. G3 Genes Genomes Genetics. 2017;7(9):2931–43. doi: 10.1534/g3.117.044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rodriguez-Lebron E, Paulson HL. Allele-specific RNA interference for neurological disease. Gene Ther. 2006;13(6):576–581. doi: 10.1038/sj.gt.3302702. [DOI] [PubMed] [Google Scholar]
- 140.Sanofi - Data from two Phase 3 studies demonstrating fitusiran significantly reduced bleeds in people with hemophilia A or B, with or without inhibitors, were featured at ASH’s plenary and late-breaking sessions [Internet]. [cited 2022 Aug 12]. Available from: https://www.sanofi.com/en/media-room/press-releases/2021/2021-12-14-14-00-00-2351761.
- 141.Thielmann M, Corteville D, Szabo G, Swaminathan M, Lamy A, Lehner LJ, et al. Teprasiran, A Small Interfering RNA, for the prevention of acute kidney injury in high-risk patients undergoing cardiac surgery: a randomized clinical study. Circulation. 2021;144(14):1133–1144. doi: 10.1161/CIRCULATIONAHA.120.053029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang MM, Bahal R, Rasmussen TP, Manautou JE, Zhong X. The growth of siRNA-based therapeutics: updated clinical studies. Biochem Pharmacol. 2021;189:114432. doi: 10.1016/j.bcp.2021.114432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Celyad Oncology - Celyad Oncology presents preliminary data from phase 1 IMMUNICY-1 trial of shRNA-based allogeneic CAR T candidate CYAD-211 in relapsed/refractory multiple myeloma at the European Hematology Association virtual congress [Internet]. [cited 2022 Aug 12]. Available from: https://celyad.com/2021/06/11/celyad-oncology-presents-preliminary-data-from-phase-1-immunicy-1-trial-of-shrna-based-allogeneic-car-t-candidate-cyad-211-in-relapsed-refractory-multiple-myeloma-at-the-european-hematology-associatio/.
- 144.Helinski DR. A Brief History of Plasmids. Ecosal Plus. 2022;eESP-0028–2021. [DOI] [PMC free article] [PubMed]
- 145.Volkert FC. Plasmids of eukaryotes. Fundamentals and applications. Q Rev Biology. 1988;63(1):76–76. [Google Scholar]
- 146.Hodges BL, Taylor KM, Joseph MF, Bourgeois SA, Scheule RK. Long-term transgene expression from plasmid DNA Gene therapy vectors is negatively affected by CPg dinucleotides. Mol Ther. 2004;10(2):269–278. doi: 10.1016/j.ymthe.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 147.Luo Z, Shi H, Zhang H, Li M, Zhao Y, Zhang J, et al. Plasmid DNA containing multiple CpG motifs triggers a strong immune response to hepatitis B surface antigen when combined with incomplete Freund’s adjuvant but not aluminum hydroxide. Mol Med Rep. 2012;6(6):1309–1314. doi: 10.3892/mmr.2012.1079. [DOI] [PubMed] [Google Scholar]
- 148.Hardee CL, Arévalo-Soliz LM, Hornstein BD, Zechiedrich L. Advances in non-viral DNA vectors for gene therapy. Genes-basel. 2017;8(2):65. doi: 10.3390/genes8020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Roth TL, Puig-Saus C, Yu R, Shifrut E, Carnevale J, Li PJ, et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559(7714):405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nafissi N, Alqawlaq S, Lee EA, Foldvari M, Spagnuolo PA, Slavcev RA. DNA Ministrings: highly safe and effective gene delivery vectors. Mol Ther Nucleic Acids. 2014;3(6):e165. doi: 10.1038/mtna.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mitdank H, Tröger M, Sonntag A, Shirazi NA, Woith E, Fuchs H, et al. Suicide nanoplasmids coding for ribosome-inactivating proteins. Eur J Pharm Sci. 2022;170:106107. doi: 10.1016/j.ejps.2021.106107. [DOI] [PubMed] [Google Scholar]
- 152.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38(5):870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Abe T, Marutani Y, Shoji I. Cytosolic DNA-sensing immune response and viral infection. Microbiol Immunol. 2019;63(2):51–64. doi: 10.1111/1348-0421.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, et al. A Toll-like receptor–independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7(1):40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 155.Stetson DB, Medzhitov R. Recognition of Cytosolic DNA Activates an IRF3-dependent innate immune response. Immunity. 2006;24(1):93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 156.Amadio R, Piperno GM, Benvenuti F. Self-DNA Sensing by cGAS-STING and TLR9 in autoimmunity: is the cytoskeleton in control? Front Immunol. 2021;12:657344. doi: 10.3389/fimmu.2021.657344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Okabe Y, Kawane K, Akira S, Taniguchi T, Nagata S. Toll-like receptor–independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J Exp Medicine. 2005;202(10):1333–1339. doi: 10.1084/jem.20051654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vanpouille-Box C, Demaria S, Formenti SC, Galluzzi L. Cytosolic DNA Sensing in organismal tumor control. Cancer Cell. 2018;34(3):361–378. doi: 10.1016/j.ccell.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 159.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1 activating inflammasome with ASC. Nature. 2009;458(7237):514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yu L, Liu P. Cytosolic DNA sensing by cGAS: regulation, function, and human diseases. Signal Transduct Target Ther. 2021;6(1):170. doi: 10.1038/s41392-021-00554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 162.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17(10):1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 164.Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics — developing a new class of drugs. Nat Rev Drug Discov. 2014;13(10):759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 165.Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, et al. Incorporation of Pseudouridine Into mRNA Yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16(11):1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA Recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23(2):165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 167.Foster JB, Barrett DM, Karikó K. The emerging role of in vitro-transcribed mRNA in adoptive T cell immunotherapy. Mol Ther. 2019;27(4):747–756. doi: 10.1016/j.ymthe.2019.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Parayath NN, Stephan SB, Koehne AL, Nelson PS, Stephan MT. In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat Commun. 2020;11(1):6080. doi: 10.1038/s41467-020-19486-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Foster JB, Choudhari N, Perazzelli J, Storm J, Hofmann TJ, Jain P, et al. Purification of mRNA encoding chimeric antigen receptor is critical for generation of a robust T-cell response. Hum Gene Ther. 2019;30(2):168–178. doi: 10.1089/hum.2018.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhao Y, Zheng Z, Cohen CJ, Gattinoni L, Palmer DC, Restifo NP, et al. High-Efficiency transfection of primary human and mouse T lymphocytes using RNA electroporation. Mol Ther. 2006;13(1):151–159. doi: 10.1016/j.ymthe.2005.07.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Beatty GL, Haas AR, Maus MV, Torigian DA, Soulen MC, Plesa G, et al. Mesothelin-specific chimeric antigen receptor MRNA-engineered T cells induce antitumor activity in solid malignancies. Cancer Immunol Res. 2014;2(2):112–120. doi: 10.1158/2326-6066.CIR-13-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rurik JG, Tombácz I, Yadegari A, Fernández POM, Shewale SV, Li L, et al. CAR T cells produced in vivo to treat cardiac injury. Science. 2022;375(6576):91–96. doi: 10.1126/science.abm0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen X, Mangala LS, Rodriguez-Aguayo C, Kong X, Lopez-Berestein G, Sood AK. RNA interference-based therapy and its delivery systems. Cancer Metast Rev. 2018;37(1):107–124. doi: 10.1007/s10555-017-9717-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mainini F, Eccles MR. Lipid and polymer-based nanoparticle siRNA delivery systems for cancer therapy. Molecules. 2020;25(11):2692. doi: 10.3390/molecules25112692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wahane A, Waghmode A, Kapphahn A, Dhuri K, Gupta A, Bahal R. Role of lipid-based and polymer-based non-viral vectors in nucleic acid delivery for next-generation gene therapy. Molecules. 2020;25(12):2866. doi: 10.3390/molecules25122866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, et al. Therapeutic siRNA: state of the art. Signal Transduct Target Ther. 2020;5(1):101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Saw PE, Song EW. siRNA therapeutics: a clinical reality. Sci China Life Sci. 2020;63(4):485–500. doi: 10.1007/s11427-018-9438-y. [DOI] [PubMed] [Google Scholar]
- 178.Slastnikova TA, Ulasov AV, Rosenkranz AA, Sobolev AS. Targeted intracellular delivery of antibodies: the state of the art. Front Pharmacol. 2018;9:1208. doi: 10.3389/fphar.2018.01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Moncalvo F, Espinoza MIM, Cellesi F. nanosized delivery systems for therapeutic proteins: clinically validated technologies and advanced development strategies. Front Bioeng Biotechnol. 2020;8:89. doi: 10.3389/fbioe.2020.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rogers S, Pfuderer P. Use of viruses as carriers of added genetic information. Nature. 1968;219(5155):749–751. doi: 10.1038/219749a0. [DOI] [PubMed] [Google Scholar]
- 181.Blaese RM, Culver KW, Miller AD, Carter CS, Fleisher T, Clerici M, et al. T Lymphocyte-directed gene therapy for ADA− SCID: initial trial results after 4 years. Science. 1995;270(5235):475–480. doi: 10.1126/science.270.5235.475. [DOI] [PubMed] [Google Scholar]
- 182.Sibbald B. Death but one unintended consequence of gene-therapy trial. Cmaj Can Medical Assoc J J De L’association Medicale Can. 2001;164(11):1612. [PMC free article] [PubMed] [Google Scholar]
- 183.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Marcucci KT, Jadlowsky JK, Hwang WT, Suhoski-Davis M, Gonzalez VE, Kulikovskaya I, et al. Retroviral and lentiviral safety analysis of gene-modified T Cell products and infused HIV and oncology patients. Mol Ther. 2018;26(1):269–279. doi: 10.1016/j.ymthe.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.(CBER) G for I US Department of Health and Human Services, Food and Drug Administration, Center for Biologics Evaluation and Research. Supplemental Guidance on Testing for Replication-Competent Retrovirus in Retroviral Vector-Based Gene Therapy Products and During Follow-up of Patients in Clinical Trials Using Retroviral Vectors. Hum Gene Ther. 2001;12(3):315–20. [DOI] [PubMed]
- 186.Zhao Z, Anselmo AC, Mitragotri S. Viral vector-based gene therapies in the clinic. Bioeng Transl Medicine. 2022;7(1):e10258. doi: 10.1002/btm2.10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bulcha JT, Wang Y, Ma H, Tai PWL, Gao G. Viral vector platforms within the gene therapy landscape. Signal Transduct Target Ther. 2021;6(1):53. doi: 10.1038/s41392-021-00487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ghosh S, Brown AM, Jenkins C, Campbell K. Viral Vector systems for gene therapy: a comprehensive literature review of progress and biosafety challenges. Appl Biosaf. 2020;25(1):7–18. doi: 10.1177/1535676019899502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Goswami R, Subramanian G, Silayeva L, Newkirk I, Doctor D, Chawla K, et al. Gene therapy leaves a vicious cycle. Frontiers Oncol. 2019;9:297. doi: 10.3389/fonc.2019.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bessis N, GarciaCozar FJ, Boissier MC. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 2004;11(Suppl 1):S10–S17. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- 191.Shirley JL, de Jong YP, Terhorst C, Herzog RW. Immune responses to viral gene therapy vectors. Mol Ther. 2020;28(3):709–722. doi: 10.1016/j.ymthe.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chirmule N, Propert KJ, Magosin SA, Qian Y, Qian R, Wilson JM. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 1999;6(9):1574–1583. doi: 10.1038/sj.gt.3300994. [DOI] [PubMed] [Google Scholar]
- 193.Fausther-Bovendo H, Kobinger GP. Pre-existing immunity against Ad vectors: humoral, cellular and innate response, what’s important? Hum Vacc Immunother. 2014;10(9):2875. doi: 10.4161/hv.29594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Uren AG, Kool J, Berns A, van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24(52):7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 195.David RM, Doherty AT. Viral Vectors: The road to reducing genotoxicity. Toxicol Sci. 2017;155(2):315–325. doi: 10.1093/toxsci/kfw220. [DOI] [PubMed] [Google Scholar]
- 196.Ali M, Lemoine NR, Ring CJ. The use of DNA viruses as vectors for gene therapy. Gene Ther. 1994;1(6):367–384. [PubMed] [Google Scholar]
- 197.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4(5):346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 198.2021 C. Strategies to address the viral vector manufacturing shortage [Internet]. [cited 2021]. https://www.pharmaceutical-technology.com/sponsored/addressing-viral-vector-manufacturing-shortage/
- 199.McKinsey & Company - Viral-vector therapies at scale: Today’s challenges and future opportunities | McKinsey [Internet]. [cited 2022 Aug 12]. https://www.mckinsey.com/industries/life-sciences/our-insights/viral-vector-therapies-at-scale-todays-challenges-and-future-opportunities.
- 200.van der Loo JCM, Wright JF. Progress and challenges in viral vector manufacturing. Hum Mol Genet. 2016;25(R1):R42–52. doi: 10.1093/hmg/ddv451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ramamoorth M, Narvekar A. Non viral vectors in gene therapy- an overview. J Clin Diagnostic Res. 2015;9(1):01–6. doi: 10.7860/JCDR/2015/10443.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kanvinde S, Kulkarni T, Deodhar S, Bhattacharya D, Dasgupta A. Non-viral vectors for delivery of nucleic acid therapies for cancer. Biotech. 2022;11(1):6. doi: 10.3390/biotech11010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Löwer M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547(7662):222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 204.Neumann E, Schaefer-Ridder M, Wang Y, Hofschneider PH. Gene transfer into mouse lyoma cells by electroporation in high electric fields. Embo J. 1982;1(7):841–845. doi: 10.1002/j.1460-2075.1982.tb01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chang DC. Encyclopedia of molecular cell biology and molecular medicine. 2006. 2nd Edition by Robert A. Meyers (Editor). Publisher Wiley-Blackwell. ISBN-13: 978-3527305476 ISBN-10: 3527305475 Chapter author: Chang DC.
- 206.Smits E, Ponsaerts P, Lenjou M, Nijs G, Bockstaele DRV, Berneman ZN, et al. RNA-based gene transfer for adult stem cells and T cells. Leukemia. 2004;18(11):1898–1902. doi: 10.1038/sj.leu.2403463. [DOI] [PubMed] [Google Scholar]
- 207.Barrett DM, Zhao Y, Liu X, Jiang S, Carpenito C, Kalos M, et al. Treatment of advanced leukemia in mice with mRNA Engineered T Cells. Hum Gene Ther. 2011;22(12):1575–1586. doi: 10.1089/hum.2011.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Birkholz K, Hombach A, Krug C, Reuter S, Kershaw M, Kämpgen E, et al. Transfer of mRNA encoding recombinant immunoreceptors reprograms CD4+ and CD8+ T cells for use in the adoptive immunotherapy of cancer. Gene Ther. 2009;16(5):596–604. doi: 10.1038/gt.2008.189. [DOI] [PubMed] [Google Scholar]
- 209.Distler JHW, Jüngel A, Kurowska-Stolarska M, Michel BA, Gay RE, Gay S, et al. Nucleofection: a new, highly efficient transfection method for primary human keratinocytes*. Exp Dermatol. 2005;14(4):315–320. doi: 10.1111/j.0906-6705.2005.00276.x. [DOI] [PubMed] [Google Scholar]
- 210.Freund EC, Lock JY, Oh J, Maculins T, Delamarre L, Bohlen CJ, et al. Efficient gene knockout in primary human and murine myeloid cells by non-viral delivery of CRISPR-Cas9. J Exp Med. 2020;217(7):e20191692. doi: 10.1084/jem.20191692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS ONE. 2014;9(12):e115987. doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Li C, Guan X, Du T, Jin W, Wu B, Liu Y, et al. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. J Gen Virol. 2015;96(8):2381–2393. doi: 10.1099/vir.0.000139. [DOI] [PubMed] [Google Scholar]
- 213.Singh H, Huls H, Kebriaei P, Cooper LJN. A new approach to gene therapy using Sleeping Beauty to genetically modify clinical-grade T cells to target CD19. Immunol Rev. 2014;257(1):181–190. doi: 10.1111/imr.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Maiti SN, Huls H, Singh H, Dawson M, Figliola M, Olivares S, et al. Sleeping Beauty System to Redirect T-cell specificity for human applications. J Immunother. 2013;36(2):112–123. doi: 10.1097/CJI.0b013e3182811ce9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Harris E, Elmer JJ. Optimization of electroporation and other non-viral gene delivery strategies for T cells. Biotechnol Progr. 2021;37(1):e3066. doi: 10.1002/btpr.3066. [DOI] [PubMed] [Google Scholar]
- 216.Zhang Z, Qiu S, Zhang X, Chen W. Optimized DNA electroporation for primary human T cell engineering. Bmc Biotechnol. 2018;18(1):4. doi: 10.1186/s12896-018-0419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Stewart MP, Langer R, Jensen KF. Intracellular delivery by membrane disruption: mechanisms, strategies, and concepts. Chem Rev. 2018;118(16):7409–7531. doi: 10.1021/acs.chemrev.7b00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Napotnik T, Polajžer T, Miklavčič D. Cell death due to electroporation—a review. Bioelectrochemistry. 2021;141:107871. doi: 10.1016/j.bioelechem.2021.107871. [DOI] [PubMed] [Google Scholar]
- 219.Weaver JC, Chizmadzhev Yu. Theory of electroporation: a review. Bioelectroch Bioener. 1996;41(2):135–60. [Google Scholar]
- 220.Cao Y, Ma E, Cestellos-Blanco S, Zhang B, Qiu R, Su Y, et al. Nontoxic nanopore electroporation for effective intracellular delivery of biological macromolecules. Proc National Acad Sci. 2019;116(16):201818553. doi: 10.1073/pnas.1818553116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.DiTommaso T, Cole JM, Cassereau L, Buggé JA, Hanson JLS, Bridgen DT, et al. Cell engineering with microfluidic squeezing preserves functionality of primary immune cells in vivo. P Natl Acad Sci Usa. 2018;115(46):E10907–E10914. doi: 10.1073/pnas.1809671115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhang M, Ma Z, Selliah N, Weiss G, Genin A, Finkel TH, et al. The impact of Nucleofection® on the activation state of primary human CD4 T cells. J Immunol Methods. 2014;408:123–131. doi: 10.1016/j.jim.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Beane JD, Lee G, Zheng Z, Mendel M, Abate-Daga D, Bharathan M, et al. Clinical Scale zinc finger nuclease-mediated gene editing of PD-1 in tumor infiltrating lymphocytes for the treatment of metastatic melanoma. Mol Ther. 2015;23(8):1380–1390. doi: 10.1038/mt.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sharei A, Zoldan J, Adamo A, Sim WY, Cho N, Jackson E, et al. A vector-free microfluidic platform for intracellular delivery. Proc National Acad Sci. 2013;110(6):2082–2087. doi: 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sharei A, Cho N, Mao S, Jackson E, Poceviciute R, Adamo A, et al. Cell squeezing as a robust, microfluidic intracellular delivery platform. J Vis Exp. 2013;81:e50980. doi: 10.3791/50980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.SQZ Biotech - SQZ Biotechnologies Announces FDA Clearance of Investigational New Drug (IND) Application for SQZ-eAPC-HPV, a Novel mRNA-based Cell Therapy for the Treatment of HPV16 Positive Solid Tumors. https://investors.sqzbiotech.com/news/news-details/2022/SQZ-Biotechnologies-Announces-FDA-Clearance-of-Investigational-New-Drug-IND-Application-for-SQZ-eAPC-HPV-a-Novel-mRNA-based-Cell-Therapy-for-the-Treatment-of-HPV16-Positive-Solid-Tumors/default.aspx. Accessed 25 Mar 2022.
- 227.Xie X, Xu AM, Leal-Ortiz S, Cao Y, Garner CC, Melosh NA. Nanostraw-electroporation system for highly efficient intracellular delivery and transfection. ACS Nano. 2013;7(5):4351–4358. doi: 10.1021/nn400874a. [DOI] [PubMed] [Google Scholar]
- 228.Ding X, Stewart MP, Sharei A, Weaver JC, Langer RS, Jensen KF. High-throughput nuclear delivery and rapid expression of DNA via mechanical and electrical cell-membrane disruption. Nat Biomed Eng. 2017;1(3):0039. doi: 10.1038/s41551-017-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Fajrial AK, He QQ, Wirusanti NI, Slansky JE, Ding X. A review of emerging physical transfection methods for CRISPR/Cas9-mediated gene editing. Theranostics. 2020;10(12):5532–5549. doi: 10.7150/thno.43465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Cullis PR, Hope MJ. Lipid nanoparticle systems for enabling gene therapies. Mol Ther. 2017;25(7):1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Fröhlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int J Nanomed. 2012;7:5577–5591. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc National Acad Sci. 1987;84(21):7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Tenchov R, Bird R, Curtze AE, Zhou Q. Lipid Nanoparticles from liposomes to mRNA vaccine delivery, a landscape of research diversity and advancement. ACS Nano. 2021;15(11):16982–17015. doi: 10.1021/acsnano.1c04996. [DOI] [PubMed] [Google Scholar]
- 234.Kulkarni JA, Cullis PR, van der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. 2018;28(3):146–157. doi: 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
- 235.Dolgin E. The tangled history of mRNA vaccines. Nature. 2021;597(7876):318–324. doi: 10.1038/d41586-021-02483-w. [DOI] [PubMed] [Google Scholar]
- 236.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines—a new era in vaccinology. Nat Rev Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Pilkington EH, Suys EJA, Trevaskis NL, Wheatley AK, Zukancic D, Algarni A, et al. From influenza to COVID-19: Lipid nanoparticle mRNA vaccines at the frontiers of infectious diseases. Acta Biomater. 2021;131:16–40. doi: 10.1016/j.actbio.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Schoenmaker L, Witzigmann D, Kulkarni JA, Verbeke R, Kersten G, Jiskoot W, et al. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int J Pharmaceut. 2021;601:120586. doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Heyes J, Palmer L, Bremner K, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Control Release. 2005;107(2):276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 240.Ball RL, Bajaj P, Whitehead KA. Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization. Int J Nanomed. 2016;12:305–315. doi: 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Discher BM, Won YY, Ege DS, Lee JCM, Bates FS, Discher DE, et al. Polymersomes: tough vesicles made from diblock copolymers. Science. 1999;284(5417):1143–1146. doi: 10.1126/science.284.5417.1143. [DOI] [PubMed] [Google Scholar]
- 242.Meins JFL, Sandre O, Lecommandoux S. Recent trends in the tuning of polymersomes’ membrane properties. European Phys J E. 2011;34(2):14. doi: 10.1140/epje/i2011-11014-y. [DOI] [PubMed] [Google Scholar]
- 243.Martinelli C, Pucci C, Ciofani G. Nanostructured carriers as innovative tools for cancer diagnosis and therapy. Apl Bioeng. 2019;3(1):011502. doi: 10.1063/1.5079943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Baghdan E, Pinnapireddy SR, Strehlow B, Engelhardt KH, Schäfer J, Bakowsky U. Lipid coated chitosan-DNA nanoparticles for enhanced gene delivery. Int J Pharmaceut. 2018;535(1–2):473–479. doi: 10.1016/j.ijpharm.2017.11.045. [DOI] [PubMed] [Google Scholar]
- 245.Lara-Velazquez M, Alkharboosh R, Norton ES, Ramirez-Loera C, Freeman WD, Guerrero-Cazares H, et al. Chitosan-based non-viral gene and drug delivery systems for brain cancer. Front Neurol. 2020;11:740. doi: 10.3389/fneur.2020.00740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Bueter CL, Lee CK, Wang JP, Ostroff GR, Specht CA, Levitz SM. Spectrum and mechanisms of inflammasome activation by Chitosan. J Immunol. 2014;192(12):5943–5951. doi: 10.4049/jimmunol.1301695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Gallops C, Ziebarth J, Wang Y. Polymers in Therapeutic Delivery. Acs Sym Ser. 2020;1–12.
- 248.Démoulins T, Milona P, Englezou PC, Ebensen T, Schulze K, Suter R, et al. Polyethylenimine-based polyplex delivery of self-replicating RNA vaccines. Nanomed Nanotechnol Biol Med. 2016;12(3):711–722. doi: 10.1016/j.nano.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 249.Olden BR, Cheng Y, Yu JL, Pun SH. Cationic polymers for non-viral gene delivery to human T cells. J Control Release. 2018;282:140–147. doi: 10.1016/j.jconrel.2018.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Raup A, Stahlschmidt U, Jérôme V, Synatschke CV, Müller AHE, Freitag R. Influence of Polyplex formation on the performance of star-shaped polycationic transfection agents for mammalian cells. Polymers-basel. 2016;8(6):224. doi: 10.3390/polym8060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Rui Y, Wilson DR, Choi J, Varanasi M, Sanders K, Karlsson J, et al. Carboxylated branched poly(β-amino ester) nanoparticles enable robust cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci Adv. 2019;5(12):eaay3255. doi: 10.1126/sciadv.aay3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. Embo J. 1985;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc National Acad Sci. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Boch J. TALEs of genome targeting. Nat Biotechnol. 2011;29(2):135–136. doi: 10.1038/nbt.1767. [DOI] [PubMed] [Google Scholar]
- 255.Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42(10):6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Kim YG, Chandrasegaran S. Chimeric restriction endonuclease. Proc National Acad Sci. 1994;91(3):883–887. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, et al. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39(1):359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39(14):6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. P Natl Acad Sci Usa. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. P Natl Acad Sci USA. 1998;95(18):10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Morbitzer R, Römer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc National Acad Sci. 2010;107(50):21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28(17):3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 267.Genome CD, Nucleases E-F. genome engineering with zinc-finger nucleases. Genetics. 2011;188(4):773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Becker S, Boch J. TALE and TALEN genome editing technologies. Gene Genome Ed. 2021;2:100007. [Google Scholar]
- 269.Shim G, Kim D, Park GT, Jin H, Suh SK, Oh YK. Therapeutic gene editing: delivery and regulatory perspectives. Acta Pharmacol Sin. 2017;38(6):738–753. doi: 10.1038/aps.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Nemudryi AA, Valetdinova KR, Medvedev SP, Zakian SM. TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Naturae. 2014;6(3):19–40. [PMC free article] [PubMed] [Google Scholar]
- 271.Yee J. Off-target effects of engineered nucleases. Febs J. 2016;283(17):3239–3248. doi: 10.1111/febs.13760. [DOI] [PubMed] [Google Scholar]
- 272.Roots SW. Use of the HPRT gene and the HAT selection technique in DNA-mediated transformation of mammalian cells: first steps toward developing hybridoma techniques and gene therapy. BioEssays. 1992;14(7):495–500. doi: 10.1002/bies.950140712. [DOI] [PubMed] [Google Scholar]
- 273.Szybalska EH, Szybalski W. Genetics of human cell lines, iv. Dna-mediated heritable transformation of a biochemical trait. Proc national Acad Sci. 1962;48(12):2026–34. doi: 10.1073/pnas.48.12.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Dulak J. Gene therapy. The legacy of Wacław Szybalski. Acta Biochim Pol. 2021;68(3):359–75. doi: 10.18388/abp.2020_5805. [DOI] [PubMed] [Google Scholar]
- 275.Bigda JJ, Koszałka P. Wacław Szybalski’s contribution to immunotherapy: HGPRT mutation & HAT selection as first steps to gene therapy and hybrid techniques in mammalian cells. Gene. 2013;525(2):158–161. doi: 10.1016/j.gene.2013.03.099. [DOI] [PubMed] [Google Scholar]
- 276.Friedmann T, Roblin R. Gene therapy for human genetic disease? Science. 1972;175(4025):949–955. doi: 10.1126/science.175.4025.949. [DOI] [PubMed] [Google Scholar]
- 277.Morgan DA, Ruscetti FW, Gallo R. Selective in vitro growth of t lymphocytes from normal human bone marrows. Science. 1976;193(4257):1007–1008. doi: 10.1126/science.181845. [DOI] [PubMed] [Google Scholar]
- 278.Meijerink MR, Scheffer HJ, Narayanan G (eds.) Irreversible electroporation in clinical practice. 2018. 285 p.
- 279.Meuer SC, Fitzgerald KA, Hussey RE, Hodgdon JC, Schlossman SF, Reinherz EL. 1983. Clonotypic structures involved in antigen-specific human T cell function. Relationship to the T3 molecular complex. J Exp Medicine. 157(2):705–19. [DOI] [PMC free article] [PubMed]
- 280.Reinherz EL, Meuer SC, Fitzgerald KA, Hussey RE, Hodgdon JC, Acuto O, et al. Comparison of T3-associated 49- and 43-kilodalton cell surface molecules on individual human T-cell clones: evidence for peptide variability in T-cell receptor structures. Proc National Acad Sci. 1983;80(13):4104–4108. doi: 10.1073/pnas.80.13.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Meuer SC, Acuto O, Hussey RE, Hodgdon JC, Fitzgerald KA, Schlossman SF, et al. Evidence for the T3-associated 90K heterodimer as the T-cell antigen receptor. Nature. 1983;303(5920):808–810. doi: 10.1038/303808a0. [DOI] [PubMed] [Google Scholar]
- 282.Cone RD, Mulligan RC. High-efficiency gene transfer into mammalian cells: generation of helper-free recombinant retrovirus with broad mammalian host range. Proc National Acad Sci. 1984;81(20):6349–6353. doi: 10.1073/pnas.81.20.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.NCT00004498. Phase I Study of Adenoviral Vector Mediated Gene Transfer for Ornithine Transcarbamylase in Adults With Partial Ornithine Transcarbamylase Deficiency [Internet]. https://clinicaltrials.gov/ct2/show/NCT00004498.
- 284.Gross G, Waks T, Eshhar Z. Expression of immunoglobulin-T-cell receptor chimeric molecules as functional receptors with antibody-type specificity. Proc National Acad Sci. 1989;86(24):10024–10028. doi: 10.1073/pnas.86.24.10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Eshhar Z, Waks T, Gross G, Schindler DG. Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc National Acad Sci. 1993;90(2):720–724. doi: 10.1073/pnas.90.2.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 287.Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. Molecular reconstruction of sleeping beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91(4):501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 288.Drugs@FDA: FDA-Approved Drug. https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&ApplNo=103705. Accessed 12 Aug 2022
- 289.Raper SE, Chirmule N, Lee FS, Wivel NA, Bagg A, Gao G, et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab. 2003;80(1–2):148–58. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 290.Cavazzana-Calvo M, Hacein-Bey S, de Basile GS, Gross F, Yvon E, Nusbaum P, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288(5466):669–72. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 291.McGarrity GJ, Hoyah G, Winemiller A, Andre K, Stein D, Blick G, et al. Patient monitoring and follow-up in lentiviral clinical trials. J Gene Medicine. 2013;15(2):78–82. doi: 10.1002/jgm.2691. [DOI] [PubMed] [Google Scholar]
- 292.Hacein-Bey-Abina S, Kalle CV, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal t cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 293.NCT00393029. Phase II Study of Metastatic Cancer That Overexpresses P53 Using Lymphodepleting Conditioning Followed by Infusion of Anti-P53 TCR-Gene Engineered Lymphocytes [Internet]. https://clinicaltrials.gov/ct2/show/NCT00393029 T Cells with Chimeric Antigen Receptors Have Potent Antitumor Effects and Can Establish Memory in Patients with Advanced Leukemia
- 294.Sadelain M. CD19 CAR T Cells. Cell. 2017;171(7):1471. doi: 10.1016/j.cell.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 295.Cideciyan AV, Hauswirth WW, Aleman TS, Kaushal S, Schwartz SB, Boye SL, et al. Human RPE65 gene therapy for leber congenital amaurosis: persistence of early visual improvements and safety at 1 Year. Hum Gene Ther. 2009;20(9):999–1004. doi: 10.1089/hum.2009.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Jensen MC, Popplewell L, Cooper LJ, DiGiusto D, Kalos M, Ostberg JR, et al. Antitransgene rejection responses contribute to attenuated persistence of adoptively transferred CD20/CD19-specific chimeric antigen receptor redirected t cells in humans. Biol Blood Marrow Tr. 2010;16(9):1245–1256. doi: 10.1016/j.bbmt.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Kochenderfer JN, Wilson WH, Janik JE, Dudley ME, Stetler-Stevenson M, Feldman SA, et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood. 2010;116(20):4099–4102. doi: 10.1182/blood-2010-04-281931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, et al. T Cells with Chimeric Antigen Receptors Have Potent Antitumor Effects and Can Establish Memory in Patients with Advanced Leukemia. Sci Transl Med. 2011;3(95):9573. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified t cells in chronic lymphoid leukemia. New Engl J Medicine. 2011;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Brentjens RJ, Rivière I, Park JH, Davila ML, Wang X, Stefanski J, et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood. 2011;118(18):4817–4828. doi: 10.1182/blood-2011-04-348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc National Acad Sci. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Glybera - European Medicines Agency. https://www.ema.europa.eu/en/medicines/human/EPAR/glybera. Accessed 12 Aug 2022.
- 303.NCT02546167. CART-BCMA Cells for Multiple Myeloma [Internet]. 2020. [cited 2022 Aug 12]. Available from: https://clinicaltrials.gov/ct2/show/NCT02546167. Accessed 12 Aug 2022.
- 304.McGowan E, Lin Q, Ma G, Yin H, Chen S, Lin Y. PD-1 disrupted CAR-T cells in the treatment of solid tumors: promises and challenges. Biomed Pharmacother. 2020;121:109625. doi: 10.1016/j.biopha.2019.109625. [DOI] [PubMed] [Google Scholar]
- 305.Strimvelis -European Medicine Agency. https://www.ema.europa.eu/en/medicines/human/EPAR/strimvelis. Accessed 12 Aug 2022.
- 306.Approval letter Luxturna. https://www.fda.gov/media/109487/download. Accessed 12 Aug 2022.
- 307.Approval letter Yescarta. https://www.fda.gov/media/108458/download. Accessed 12 Aug 2022.
- 308.Approval letter Kymriah. https://www.fda.gov/media/106989/download. Accessed 12 Aug 2022.
- 309.Approval letter Zolgensma. https://www.fda.gov/media/126130/download. Accessed 12 Aug 2022.
- 310.Zynteglo European Medicines Agency. https://www.ema.europa.eu/en/medicines/human/EPAR/zynteglo. Accessed 12 Aug 2022.
- 311.Approval letter Tecartus. https://www.fda.gov/media/152696/download. Accessed 12 Aug 2022.
- 312.Approval letter Breyanzi. https://www.fda.gov/media/159473/download. Accessed 12 Aug 2022.
- 313.Approval letter ABECMA. https://www.fda.gov/media/147062/download. Accessed 12 Aug 2022.
- 314.Approval letter Carvykti. https://www.fda.gov/media/156572/download. Accessed 12 Aug 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.