Abstract
From July 2022, cases of imported diphtheria with toxigenic Corynebacterium diphtheriae remarkably increased among migrants arriving in Germany. Up to 30 September 2022, 44 cases have been reported to the national public health institute, all laboratory-confirmed, male, and mainly coming from Syria (n = 21) and Afghanistan (n = 17). Phylogeny and available journey information indicate that most cases (n = 19) were infected along the Balkan route. Active case finding, increased laboratory preparedness and epicentre localisation in countries along this route are important.
Keywords: cgMLST, Corynebacterium diphtheriae, diphtheria, infectious disease outbreaks, migration and health, Next Generation Sequencing, outbreak investigation, population surveillance, skin lesions, vaccine-preventable disease
Since the end of July 2022, a remarkable increase of cases of imported, mainly cutaneous diphtheria caused by toxigenic Corynebacterium diphtheriae has been observed among migrants who recently arrived in Germany [1]. Other countries in Europe have reported similar observations, including Austria, Belgium, France, Norway, Switzerland, and the United Kingdom [2]. Here we describe the first epidemiological data for 44 cases officially reported in Germany between 1 January and 30 September 2022, present phylogenetic analyses of 42 isolates respectively obtained from distinct cases and share suggestions for outbreak investigations and response that might be considered in other countries affected by this event.
Descriptive analysis of the outbreak
In Germany, diphtheria is generally laboratory-confirmed when the responsible pathogen such as C. diphtheriae or C. ulcerans is isolated in culture and either the tox gene coding for diphtheria toxin (DT) is detected by nucleic acid amplification (e.g. PCR) or evidence of production of DT is found by Elek–Ouchterlony immunoprecipitation (Elek) test (tox+ or toxigenic C. spp., respectively) [3]. This means that cases with proof of tox+ C. spp. are also considered as confirmed cases despite a negative, pending, or not performed Elek test.
Outbreak case definition
An outbreak investigation was initiated in August 2022 using the following working case definition. A confirmed case is any person who is either: (i) a migrant who has arrived in Germany and received laboratory confirmation of tox+ C. diphtheriae on or after 1 January 2022, regardless of the manifestation, i.e. presenting with respiratory or cutaneous diphtheria or being asymptomatic or with unknown clinical data, or (ii) a close contact (as considered by the local health authority according to national recommendations [4]) of a confirmed case with laboratory confirmation of tox+ C. diphtheriae, regardless of the country of origin and clinical presentation.
Reported outbreak cases and their characteristics
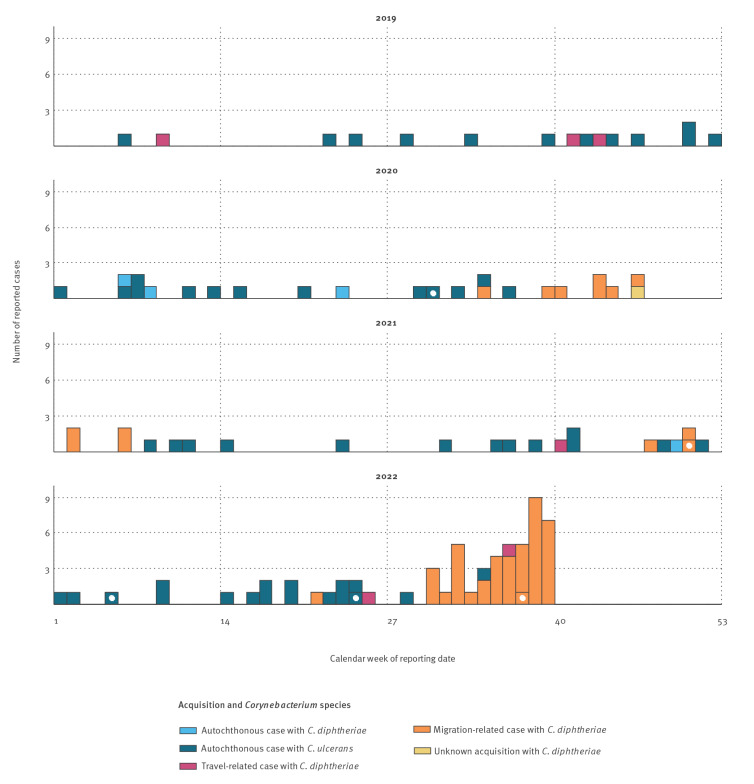
Between 1 January and 30 September 2022, the Robert Koch Institute (RKI), Germany’s national public health institute, received reports of 44 diphtheria cases who met the working outbreak case definition of a confirmed case (data up to 22 October 2022 to allow for delayed reporting and laboratory follow-up). Nineteen local health districts in seven of 16 federal states were affected. One case was reported in May 2022 (calendar week (CW) 21 2022), the other 43 starting from late July 2022 (CW 30–39 2022) (Figure 1). Of the 44 cases, 42 presented with cutaneous diphtheria, one with respiratory diphtheria and one was asymptomatic yet laboratory-confirmed.
Figure 1.
Weekly distributions of diphtheria cases fulfilling the German reference case definitiona reported to the national public health institute, Germany, 2019–2022b (n = 123)
C.: Corynebacterium.
a Case definitions in Germany are described in [3].
b Data up to 22 October 2022.
Cases reported in 2022 only relate to the time period of 1 January to 30 September 2022. Cases marked with a white dot represent respiratory diphtheria (n = 5).
All outbreak-related cases were male with a median age of 18 years (interquartile range (IQR): 16–22) (Table 1). Date of symptom onset was available for 13 cases. For these cases, the respective dates of symptom onset ranged from 26 June to 23 September 2022. Although the date of symptom onset was only known in the minority of cases, anecdotal evidence indicated that in the case of wound infections these often had occurred weeks to months before arrival at reception centres in Germany (Christiane Wagner-Wiening, personal communication, October 2022). Several cutaneous lesions were co-infected with meticillin-resistant Staphylococcus aureus (MRSA) or Streptococcus pyogenes. The respiratory case was treated with diphtheria antitoxin (DAT).
Table 1. Demographic, clinical, and microbiological characteristics of outbreak-related diphtheria cases reported to the national public health institute, Germany, 1 January–30 September 2022a (n = 44).
| Case classification according to working outbreak case definition | Number of cases |
|---|---|
| Confirmed case | 44 |
| Manifestation and pathogen | |
| Respiratory diphtheria with C. diphtheriae | 1 |
| Cutaneous diphtheria with C. diphtheriae | 42 |
| No clinical confirmation or asymptomatic with C. diphtheriae | 1 |
| Age group (years) | |
| 0–4 | 0 |
| 5–14 | 2 |
| 15–24 | 34 |
| 25–44 | 8 |
| ≥ 45 | 0 |
| Gender | |
| Female | 0 |
| Male | 44 |
| Other/unknown | 0 |
| Country of origin | |
| Afghanistan | 17 |
| Syria | 21 |
| Tunisia | 1 |
| Yemen | 1 |
| Unknown | 4 |
| Laboratory confirmation by National Consiliary Laboratory on Diphtheria | |
| PCR+ and Elek+ | 44 |
C. diphtheriae: Corynebacterium diphtheriae.
a Data up to 22 October 2022.
Most cases were detected by medical examinations in reception centres as it is mandatory for people living in a shared accommodation for migrants and asylum seekers to accept a clinical examination on communicable diseases. Vaccination status generally could not be established due to missing vaccination cards. Among the 44 cases, no human-to-human transmission within Germany could be confirmed. The asymptomatic case had been diagnosed during contact person management of the respiratory case and isolates from the two cases belonged to the same phylogenetic cluster (as shown further in the laboratory analysis and phylogeny section; isolates KL2203 and KL2189 respectively); nevertheless, it could not be determined whether this asymptomatic case was a secondary case infected by the respiratory case, whether they had the same source of infection or whether they got infected independently.
Diphtheria cases 2019–2022
For comparison to the current situation in Germany with regard to diphtheria, Figure 1 depicts the diphtheria cases meeting the German reference case definition (see Supplement S1: Routine and intensified diphtheria surveillance in Germany) [3]. Between 2019 and 2021, cases of diphtheria in Germany were evenly distributed over the years, and average annual numbers of reported diphtheria cases were comparable to those reported in the yet incomplete year of 2022 for the local population and travellers (13.8 vs 18 for autochthonous transmission; 2.6 vs 2 for travel-related transmission), but not for migrants (2.8 vs 43 for migration-related transmission, excluding the asymptomatic case). Notably, all travel- and migration-related, i.e. imported, cases were caused by C. diphtheriae, while the majority of autochthonous cases were caused by C. ulcerans. In 2019, the first human-to-human transmission of cutaneous diphtheria caused by a toxigenic C. diphtheriae strain in almost 40 years within Germany was detected among siblings [5].
Laboratory analysis and phylogeny
The German National Consiliary Laboratory on Diphtheria (NCLD) conducted C. diphtheriae strain identification, antimicrobial susceptibility testing, verified toxigenicity by real-time-PCR and a modified Elek test, and performed whole genome sequencing (WGS, see Supplement S2: Laboratory methods).
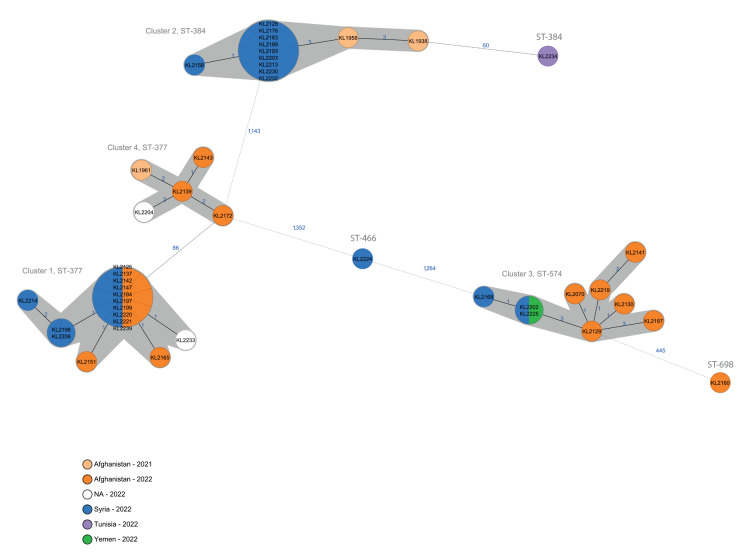
WGS data were analysed by multilocus sequence typing (MLST), using seven target loci, and also by core genome MLST (cgMLST), based on allele typing of 1,553 target loci. Figure 2 shows a minimum spanning tree (MST) based on cgMLST, including the genetic relationships as single-linkage (SL) allelic distances (AD), which display the shortest pairwise AD, rather than every absolute bilateral AD. The cgMLST analysis revealed six sequence types (ST) appearing on the phylogenetic tree as three singletons, and four genetically closely related distinct clusters, hereinafter numbered according to the number of isolates they comprised.
Figure 2.
Minimum spanning tree of the cgMLST allelic profiles of C. diphtheriae isolates detected among migrants, Germany, 2021–2022a (n = 45)
C. diphtheriae: Corynebacterium diphtheriae; cgMLST: core genome multi locus sequence typing; NA: not available (no information on country of origin); SL-AD: single-linkage allelic distance.
a An ad hoc C. diphtheriae cgMLST scheme of 1,553 target loci was used for the cgMLST analysis.
Samples are colour-coded according to the case’s country of origin and collection year if information was available. The blue font is used for SL-ADs. Clusters with a SL-AD of ≤ 5 are shaded in grey.
Cluster 1 consisted of 16 isolates with ST-377 and was characterised by SL-ADs of 0–2 (maximum bilateral AD = 4). Cluster 2 comprised 12 isolates with ST-384, including two isolates from 2021, and had SL-ADs of 0–3 (maximum bilateral AD = 5). Cluster 3 comprised nine isolates with ST-574 and had SL-ADs of 0–3 (maximum bilateral AD = 7). Cluster 4 consisted of five isolates of ST-377 including one isolate from 2021 and had SL-ADs of 1–2 (maximum bilateral AD = 3); this cluster was separated by ≥ 86 SL-AD from Cluster 1, which was characterised by the same ST. The three singletons of ST-698, ST-466 and ST-384 showed an SL-AD of 445, 1,264 and 60 to the nearest cluster, respectively.
Clusters 1, 2, and 3 coincided with representative genome sequences of isolates from Austria and Norway, which were shared in the course of the investigation and in the European Centre for Disease Prevention and Control (ECDC) Rapid Risk Assessment [2] (data not shown). All isolates in Cluster 4 were phenotypically resistant to erythromycin and clindamycin. In line with the phenotypic results and recently published data from a similar cluster with ST-377 among migrants in Switzerland [6], our search for acquired resistance genes in Cluster 4 isolates found, among others, genes for resistance against macrolides/lincosamides and beta-lactams (ermX, OXA-2) (data not shown).
Comparison with sequences from diphtheria cases of migrants who had arrived in Germany in previous years revealed that two isolates matched with Cluster 2 and one isolate with Cluster 4 (pale orange colour in Figure 2).
Analysis of migration routes
Countries of origin when known, were mostly Syria (n = 21) and Afghanistan (n = 17), however, there were also cases from Tunisia and Yemen (i.e. one from each country). Information on transit countries on route to Germany was available for 23 of the 44 cases as well as for the three cases from 2021 who were infected by strains belonging to ST clusters identified in the current outbreak (Table 2). Of the resulting 26 individuals, 22 mentioned having transited through one or more countries along the Balkan route, i.e. Albania, Bosnia and Herzegovina, Bulgaria, Croatia, North Macedonia, Romania, Serbia, and Slovenia. Two persons mentioned transiting through Belarus and Poland, i.e. the Belarus route; one of them initially also transited along the Balkan route. Cases mentioning transit countries along the Balkan route, including the three cases from 2021, are mainly distributed among Clusters 1, 2, and 4. In contrast, Cluster 3 contains only one case who transited through Balkan countries. Although another case in this cluster travelled via the Belarus route, it should be noted that most cases (7 of 9) in Cluster 3 had little details available about their journey to Germany. Investigations by local health authorities revealed that migrants were en route often for months, living under very poor hygienic conditions. The majority of them stayed in migration camps or in forests in the Balkans. Injuries acquired while fleeing were not treated due to lack of access to healthcare services.
Table 2. Migration routes, including transit countries, named by diphtheria cases among migrants in Germany, 2021–2022a (n = 47).
| Identifier at NCLD | Year of reporting | Country of origin | ST | Cluster | Migration route to Germany | Balkan route |
|---|---|---|---|---|---|---|
| KL2125 | 2022 | Afghanistan | 377 | 1 | Afghanistan | Unknown |
| KL2142 | 2022 | Afghanistan | 377 | 1 | Afghanistan | Unknown |
| KL2147 | 2022 | Afghanistan | 377 | 1 | Afghanistan | Unknown |
| KL2196 | 2022 | Syria | 377 | 1 | Syria | Unknown |
| KL2220 | 2022 | Syria | 377 | 1 | Syria | Unknown |
| KL2137 | 2022 | Afghanistan | 377 | 1 | Afghanistan, via Pakistan, Iran, Türkiye, Greece, Serbia, Hungary, Austria | Yes |
| KL2151 | 2022 | Afghanistan | 377 | 1 | Afghanistan, via Bulgaria | Yes |
| KL2165 | 2022 | Afghanistan | 377 | 1 | Afghanistan, via Bosnia and Herzegovina, Serbia | Yes |
| KL2197 | 2022 | Syria | 377 | 1 | Syria, via Serbia | Yes |
| KL2199 | 2022 | Syria | 377 | 1 | Syria, via Iraq, Türkiye, Greece, Albania, Serbia | Yes |
| KL2206 | 2022 | Syria | 377 | 1 | Syria, via Bosnia and Herzegovina, Serbia | Yes |
| KL2184 | 2022 | Syria | 377 | 1 | Syria, via Türkiye, North Macedonia, Serbia, Czechia | Yes |
| KL2214 | 2022 | Syria | 377 | 1 | Syria, via Türkiye, Bulgaria, North Macedonia, Serbia, Slovenia | Yes |
| KL2233 | 2022 | Unknown | 377 | 1 | via Syria, Türkiye, Greece, North Macedonia, Serbia, Hungary, Czechia | Yes |
| KL2221 | 2022 | Afghanistan | 377 | 1 | Afghanistan | Unknown |
| KL2239 | 2022 | Syria | 377 | 1 | Syria, via Türkiye, Bulgaria, Serbia, Hungary, Austria | Yes |
| KL1938 | 2021 | Afghanistan | 384 | 2 | Afghanistan, via Iran, Türkiye, Greece, Romania, Serbia, Belarus, Poland | Yes |
| KL1958 | 2021 | Afghanistan | 384 | 2 | Afghanistan, via Syria, Türkiye, Greece | Yes |
| KL2128 | 2022 | Syria | 384 | 2 | Syria | Unknown |
| KL2158 | 2022 | Syria | 384 | 2 | Syria | Unknown |
| KL2176 | 2022 | Syria | 384 | 2 | Syria | Unknown |
| KL2189 | 2022 | Syria | 384 | 2 | Syria | Unknown |
| KL2203 | 2022 | Syria | 384 | 2 | Unknown | Unknown |
| KL2183 | 2022 | Syria | 384 | 2 | Syria, via Türkiye, Bulgaria, Serbia, Hungary, Slovakia, Czechia | Yes |
| KL2193 | 2022 | Syria | 384 | 2 | Syria, via Türkiye, Serbia, Croatia | Yes |
| KL2213 | 2022 | Syria | 384 | 2 | Syria, via Türkiye, Bulgaria, North Macedonia, Serbia, Slovenia, Czechia | Yes |
| KL2230 | 2022 | Syria | 384 | 2 | Unknown | Unknown |
| KL2232 | 2022 | Syria | 384 | 2 | Syria, via Türkiye, Bulgaria, Serbia, Hungary, Austria | Yes |
| KL2070 | 2022 | Afghanistan | 574 | 3 | Afghanistan | Unknown |
| KL2129 | 2022 | Afghanistan | 574 | 3 | Afghanistan | Unknown |
| KL2130 | 2022 | Afghanistan | 574 | 3 | Afghanistan | Unknown |
| KL2141 | 2022 | Afghanistan | 574 | 3 | Afghanistan, via Bulgaria | Yes |
| KL2168 | 2022 | Syria | 574 | 3 | Syria | Unknown |
| KL2187 | 2022 | Afghanistan | 574 | 3 | Afghanistan, via Türkiye, Belarus, Poland | No |
| KL2202 | 2022 | Yemen | 574 | 3 | Yemen, via Syria | Unknown |
| KL2219 | 2022 | Afghanistan | 574 | 3 | Afghanistan, via Türkiye | Unknown |
| KL2225 | 2022 | Syria | 574 | 3 | Unknown | Unknown |
| KL1961 | 2021 | Afghanistan | 377 | 4 | Afghanistan, via Pakistan, Iran, Türkiye, Bulgaria, Serbia, Hungary, Austria | Yes |
| KL2139 | 2022 | Afghanistan | 377 | 4 | Afghanistan, via Bulgaria | Yes |
| KL2143 | 2022 | Afghanistan | 377 | 4 | Afghanistan | Unknown |
| KL2172 | 2022 | Afghanistan | 377 | 4 | Afghanistan, via Bulgaria | Yes |
| KL2204 | 2022 | Unknown | 377 | 4 | Unknown | Unknown |
| KL2160 | 2022 | Afghanistan | 698 | None | Afghanistan, via Serbia, Hungary, Austria | Yes |
| KL2224 | 2022 | Syria | 466 | None | Syria, via Balkan route | Yes |
| KL2234 | 2022 | Tunisia | 384 | None | Tunisia, via Italy, Austria | No |
| KL2249 | 2022 | Unknown | Pending | Pending | Unknown | Unknown |
| KL2257 | 2022 | Unknown | Pending | Pending | Unknown | Unknown |
NCLD: German National Consiliary Laboratory on Diphtheria; ST: sequence type.
a Data up to 22 October 2022.
This table comprises all cases of diphtheria reported to Germany’s national public health institute, from 1 January to 30 September 2022, that fulfil the working case definition of this outbreak investigation (n = 44) as well as additional cases from 2021 whose isolates belong to the same STs or clusters relevant in this outbreak investigation (n = 3). The migration route starts with the country of departure and continues via transit countries if such countries were reported.
Discussion
Diphtheria is a vaccine-preventable disease caused by DT producing, i.e. toxigenic, C. spp. Diphtheria predominantly presents as a respiratory or a cutaneous manifestation. In Germany, the majority of cases notified in recent years have been cutaneous diphtheria caused by zoonotic, toxigenic C. ulcerans [7]. This is likely due to incidental findings following improvements in the microbiological diagnostic methods [8] and its wider use. However, since July 2022, we observed an unprecedented increase of diphtheria cases caused by toxigenic C. diphtheriae among migrants who recently arrived in Germany.
Upon investigating this event, we considered among others the following four aspects. Firstly, a change in the incidence of diphtheria in the countries of origin. For 2021, Afghanistan reported 61 cases to the World Health Organization (WHO), after several years with zero or single-digit case counts, or no reporting [9]. Whether the increase in 2021 is due to an increased circulation of C. spp. remains unknown. We also lack information on the epidemiology of cutaneous diphtheria in Afghanistan and Syria in 2022. Coverage of a diphtheria-containing vaccine represented by the third dose of diphtheria, tetanus and pertussis vaccines (DTP3) was 66% in Afghanistan, 78% in Iraq, and 48% in Syria, as WHO and United Nations Children's fund (UNICEF) estimated for 2021 [10].
Secondly, an increase in the number of migrants arriving in Germany. In 2022, more migrants came to Germany than in 2021, but not in the order of magnitude as imported diphtheria cases increased [11-13]. A proxy for the number of arriving migrants is the number of first asylum applications. In the third quarter (July–September) of 2022, approximately 4% more Afghans and 20% more Syrians applied for asylum in Germany for the first time, compared with the third quarter of 2021. Interestingly, no diphtheria case in this outbreak has been reported among migrants from Iraq as country of origin although Iraqis are another main group seeking asylum in Germany [13]. Migrants from Iraq to Europe more commonly arrive via the Belarus route which passes via Moldova, Ukraine, Belarus, and Poland [14].
Thirdly, a possible detection bias due to increased diagnostics of skin lesions in the wake of the monkeypox (MPX) outbreak [15]. This outbreak and increased awareness to detect and diagnose skin lesions especially among young males since May 2022 might have helped to detect the diphtheria upsurge early. Local health authorities reported that several diphtheria cases were diagnosed after initial suspicion of MPX which was ruled out.
Fourthly we considered, human-to-human transmission in reception centres in Germany. Germany’s national public health institute continuously raises awareness and advises local and federal health authorities to follow the general recommendations for the management of diphtheria cases and contact persons as well as the particular recommendation for outbreaks of infectious diseases in shared accommodations [4,16]. Among the 44 cases reported by 19 different local health authorities until 30 September 2022, no secondary case could be confirmed. Nevertheless, in October 2022, after the data cut-off time for this report, two local health authorities reported one secondary case each deriving from a respiratory diphtheria case.
All the listed aspects might have contributed to the remarkable upsurge of migration-related diphtheria cases but do not fully explain it. Our sequencing and epidemiological data as well as information regarding the migration routes confirm that the majority of cases most likely acquired C. diphtheriae during migration, particularly along the Balkan route. Nevertheless, individual persons could also have already been colonised with C. diphtheriae in their country of origin, been injured en route, and only then developed cutaneous diphtheria.
That cases are imported rather than occurring as secondary cases within Germany is supported by further evidence. For one, in each cluster, individuals came from different countries of origin and then resided in different cities in Germany. Also, isolates from Austria matched with Clusters 1, 2, and 3, and an isolate from Norway matched with Cluster 1. Additionally, the molecular clock also indicates an origin abroad; in a recent publication of a similar outbreak situation in Switzerland, a molecular clock model of 1.67 × 10 − 6 substitutions per site and year [17] was used in a phylogeny based on single nucleotide polymorphism (SNP) to date back the occurrence of 1 SNP between two isolates to ≥ 6 weeks, 2 SNPs to 5 months and 5 SNPs to 1 year [6]. In our study, the phylogenetic relationship was analysed by cgMLST, based on AD, not SNPs, which have been shown to be within a similar range as the used cgMLST scheme [18], although they generally tend to be slightly smaller in numbers than corresponding SNP distances. Therefore, the model proposed in the Swiss study [6] can be used as an approximation. Maximum AD in Clusters 1, 2, 3, and 4 were 4, 5, 7, and 3, respectively, suggesting transmission events within the last year.
Potential limitations of this study are, besides those mentioned earlier, that migration route is not known for all cases, and that time between arrival in reception centres and medical examination is not reported and therefore human-to-human transmission in the accommodation could remain unidentified.
Conclusions
In 2022 three times more diphtheria cases were reported in a 10-week period (CW 30 to 39) than in the 3 previous years altogether. This event, as per the ECDC definition [19], constitutes an outbreak. Based on our data, it seems likely that this outbreak originated abroad, not in Germany, and that there are multiple sources of infection, mainly along the Balkan route. Nevertheless, no epicentres have been identified yet. Considering that 19 of 23 cases from 2022 with known migration route came along the Balkan route and since similar cases occurred in other European countries, this outbreak likely involves multiple countries.
Based on our findings, it seems crucial to identify the source(s) of this outbreak. Outbreak investigations in further countries could provide necessary data on the epidemiology, phylogeny and detailed migration routes. Analyses from pooled molecular data from different countries may also help narrow down where infections might have been acquired. In this respect, a study on diphtheria outbreaks in a Swiss asylum centre has just been published [6], with identification of a number of identical STs to our study. Moreover, active case finding is necessary, not only in Germany and other destination countries, but also in the main countries of origin and countries along the Balkan route. In contrast to severe respiratory diphtheria, cutaneous diphtheria may have unspecific symptoms and be masked by co-infections but is a potential source for secondary cases. Case and contact management should be in accordance with respective national guidelines. Laboratory preparedness should also be increased in possibly affected countries, including personnel and reagents. Laboratory confirmation by PCR and Elek test is important to reveal the true magnitude of the ongoing outbreak, and sequencing of isolates from both migration-related cases as well as autochthonous cases is necessary for further in-depth analysis. Altogether, this could contribute to effectively respond to this multi-country diphtheria outbreak and to implement infection prevention and control measures such as targeted and efficient vaccination campaigns.
Ethical statement
Ethical approval was not necessary for this study as it was performed as part of routine surveillance activities, intensified diphtheria surveillance activities and diphtheria diagnostics in Germany.
Funding statement
Henrieke Prins is a fellow of the ECDC Fellowship Programme, supported financially by the European Centre for Disease Prevention and Control. The views and opinions expressed herein do not state or reflect those of ECDC. ECDC is not responsible for the data and information collation and analysis and cannot be held liable for conclusions or opinions drawn.
Data availability statement
Whole genome sequencing (WGS) data are available under BioProject ID PRJNA898270 at http://www.ncbi.nlm.nih.gov/bioproject/898270 . Detailed accession numbers are given in Supplementary Table S3.
Acknowledgements
We would like to acknowledge all involved local and federal health authorities in Germany for case finding and data collection, in particular the local health authorities of Baden-Wuerttemberg, Germany. We would also like to acknowledge the LGL teams of bacteriology and NGS Core Unit for their laboratory work and sample management. We would like to thank Jan Walter for his supervising role. Furthermore, we would like to thank Christa Bedwin for editing.
Supplementary Data
Conflict of interest: None declared.
Authors’ contributions: Conceptualisation: FB, AB, ADo, CS, HP, WKS, OW, ASi. Methodology: FB, AB, ASi. Line list: CS, HP, AB, FB. Epidemiological data analysis and visualization: FB, ADo. Migration route analysis: CWW, HP, CS, FB. Laboratory diagnostics: AB, BH. Sequencing and phylogenetic analysis: AB, ADa, ASp. Writing the draft: FB, AB, HP, CWW. Commenting and editing: all authors.
References
- 1. Robert Koch Institute (RKI) . Häufung von Fällen mit Hautdiphtherie in Deutschland und Europa. [Upsurge of cases of cutaneous diphtheria in Germany and Europe]. Epidemiol Bull. 2022; (36):27-8. Available from: https://www.rki.de/DE/Content/Infekt/EpidBull/Archiv/2022/Ausgaben/36_22.pdf [Google Scholar]
- 2.European Centre for Disease Prevention and Control (ECDC). Increase of reported diphtheria cases among migrants in Europe due to Corynebacterium diphtheriae, 2022 – 06 October 2022. Stockholm: ECDC; 2022. Available from: https://www.ecdc.europa.eu/en/publications-data/increase-reported-diphtheria-cases-among-migrants-europe-due-corynebacterium
- 3.Robert Koch Institute (RKI). Falldefinitionen des Robert Koch-Instituts zur Übermittlung von Erkrankungs- oder Todesfällen und Nachweisen von Krankheitserregern. [Case definitions of RKI for reporting of cases of disease or death and confirmation of pathogens]. Berlin: RKI; 2019. Available from: www.rki.de/falldefinitionen
- 4.Robert Koch Institute (RKI). RKI-Ratgeber Diphtherie. [RKI recommendations on diphtheria]. Berlin: RKI; 2018. [Accessed 10 Nov 2022]. Available from: https://www.rki.de/DE/Content/Infekt/EpidBull/Merkblaetter/Ratgeber_Diphtherie.html
- 5. Berger A, Dangel A, Schober T, Schmidbauer B, Konrad R, Marosevic D, et al. Whole genome sequencing suggests transmission of Corynebacterium diphtheriae-caused cutaneous diphtheria in two siblings, Germany, 2018. Euro Surveill. 2019;24(2):1-4. 10.2807/1560-7917.ES.2019.24.2.1800683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kofler J, Ramette A, Iseli P, Stauber L, Fichtner J, Droz S, et al. Ongoing toxin-positive diphtheria outbreaks in a federal asylum centre in Switzerland, analysis July to September 2022. Euro Surveill. 2022;27(44):1-5. 10.2807/1560-7917.ES.2022.27.44.2200811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robert Koch Institute (RKI). Infektionsepidemiologisches Jahrbuch meldepflichtiger Krankheiten für 2020. [Yearbook of notifiable diseases 2020]. Berlin: RKI; 2021. Available from: www.rki.de/jahrbuch
- 8. Konrad R, Berger A, Huber I, Boschert V, Hörmansdorfer S, Busch U, et al. Matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry as a tool for rapid diagnosis of potentially toxigenic Corynebacterium species in the laboratory management of diphtheria-associated bacteria. Euro Surveill. 2010;15(43):1-5. 10.2807/ese.15.43.19699-en [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization (WHO). Diphtheria reported cases and incidence. Geneva: WHO. [Accessed 9 Oct 2022]. Available from: https://immunizationdata.who.int/pages/incidence/diphtheria.html
- 10.World Health Organization (WHO). Immunization dashboard. Most recent update: 15 July 2022. Geneva: WHO; 2022. [Accessed 9 Nov 2022]. Available from: https://immunizationdata.who.int/pages/coverage/dtp.html
- 11.Bundesamt für Migration und Flüchtlinge (BAMF) [Federal Office for Migration and Refugees]. Aktuelle Zahlen. Ausgabe: Juli 2022. [Update asylum seekers, July 2022]. Nürnberg: BAMF; 2022. Available from: https://www.bamf.de/SharedDocs/Anlagen/DE/Statistik/AsylinZahlen/aktuelle-zahlen-juli-2022.pdf
- 12.Bundesamt für Migration und Flüchtlinge (BAMF) [Federal Office for Migration and Refugees]. Aktuelle Zahlen. Ausgabe: August 2022. [Update asylum seekers, August 2022]. Nürnberg: BAMF; 2022. Available from: https://www.bamf.de/SharedDocs/Anlagen/DE/Statistik/AsylinZahlen/aktuelle-zahlen-august-2022.pdf
- 13.Bundesamt für Migration und Flüchtlinge (BAMF) [Federal Office for Migration and Refugees]. Aktuelle Zahlen. Ausgabe: September 2022. [Update asylum seekers, September 2022]. Nürnberg: BAMF; 2022.
- 14.European Union Agency for Asylum (EUAA). Asylum Report 2022. Luxembourg: EUAA; 2022. Available from: https://euaa.europa.eu/publications/asylum-report-2022
- 15.World Health Organization (WHO). Monkeypox outbreak 2022 - Global. Geneva: WHO; 2022. [Accessed 9 Nov 2022]. Available from: https://www.who.int/emergencies/situations/monkeypox-oubreak-2022
- 16.Robert Koch Institute (RKI). Management von Ausbrüchen in Gemeinschaftsunterkünften für Geflüchtete. [Management of outbreaks in shared accommodations for migrants]. Berlin: RKI; 2022. Available from: https://www.rki.de/DE/Content/GesundAZ/F/Flucht/Management_Ausbruch.pdf
- 17. Badell E, Alharazi A, Criscuolo A, Almoayed KAA, Lefrancq N, Bouchez V, et al. NCPHL diphtheria outbreak working group . Ongoing diphtheria outbreak in Yemen: a cross-sectional and genomic epidemiology study. Lancet Microbe. 2021;2(8):e386-96. 10.1016/S2666-5247(21)00094-X [DOI] [PubMed] [Google Scholar]
- 18. Dangel A, Berger A, Konrad R, Bischoff H, Sing A. Geographically diverse clusters of nontoxigenic corynebacterium diphtheriae infection, Germany, 2016-2017. Emerg Infect Dis. 2018;24(7):1239-45. 10.3201/eid2407.172026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.European Centre for Disease Prevention and Control (ECDC). Outbreak Investigations. Stockholm: ECDC. [Accessed 9 Oct 2022]. Available from: https://wiki.ecdc.europa.eu/fem/Pages/Outbreak%20Investigations.aspx
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.