Abstract
The Apocynaceae tree Voacanga thouarsii, native to southern Africa and Madagascar, produces monoterpene indole alkaloids (MIA), which are specialized metabolites with a wide range of bioactive properties. Voacanga species mainly accumulates tabersonine in seeds making these species valuable medicinal plants currently used for industrial MIA production. Despite their importance, the MIA biosynthesis in Voacanga species remains poorly studied. Here, we report the first genome assembly and annotation of a Voacanga species. The combined assembly of Oxford Nanopore Technologies long-reads and Illumina short-reads resulted in 3,406 scaffolds with a total length of 1,354.26 Mb and an N50 of 3.04 Mb. A total of 33,300 protein-coding genes were predicted and functionally annotated. These genes were then used to establish gene families and to investigate gene family expansion and contraction across the phylogenetic tree. A transposable element (TE) analysis showed the highest proportion of TE in Voacanga thouarsii compared with all other MIA-producing plants. In a nutshell, this first reference genome of V. thouarsii will thus contribute to strengthen future comparative and evolutionary studies in MIA-producing plants leading to a better understanding of MIA pathway evolution. This will also allow the potential identification of new MIA biosynthetic genes for metabolic engineering purposes.
Keywords: monoterpene indole alkaloids, tabersonine, wild frangipani
Significance.
Voacanga species are major industrial resources of tabersonine, an important intermediate in the synthesis of aspidosperma-type monoterpene indole alkaloids (MIA), that are of high pharmaceuticals importance. Despite their significant role in the pharmaceutical industry, no previous study reported genomic analysis of MIA metabolism in Voacanga species. Here, we provide the first annotated reference genome of a Voacanga species that, together with the previously published MIA-producing plant genomes, will help understand evolution and diversification of MIA in plants as well as identifying MIA biosynthetic genes to enrich the molecular MIA toolbox used for production of MIA in heterologous hosts.
Introduction
The wild frangipani, Voacanga thouarsii, is a small Apocynaceae tree native to southern Africa and Madagascar. Apocynaceae species are known to accumulate a broad spectrum of specialized metabolites including monoterpene indole alkaloids (MIA; Leeuwenberg, 1980). These compounds are part of the plant defense mechanisms to face both biotic and abiotic pressures (Dugé de Bernonville et al. 2015). Due to the high diversity of their bioactive properties, MIAs are active substances of many drugs such as antihypertensive and anticancer ones (O’Connor and Marseh, 2006; Macabeo et al. 2009).
MIAs originate from the condensation of secologanin and tryptamine yielding strictosidine followed by its subsequent decorations and/or cyclisation (De Luca et al. 1987; Maresh et al. 2007). Their biosynthetic pathways have extensively been studied over the last three decades mainly in Catharanthus roseus (as reviewed by Pan et al. 2016 and Kulagina et al. 2022). More than 100 MIAs have been reported in Voacanga species (see Hussain et al. 2012 for extensive review) including the valuable voacamine, resulting from the dimerization of vobasine and ibogaine (fig. 1A.iii). Voacamine is used in several African countries to fight malaria and also displays strong antimicrobial and cardiotonic properties (Diavara et al. 1984; Ramanitrahasimbola et al. 2001). Voacanga thouarsii and V. africana also stands out for a high accumulation level of the aspidosperma-type MIA tabersonine (fig. 1A.ii) especially in seeds (Dzoyem et al. 2013; Kunesch et al. 1977; Rolland et al. 1975; Goldblatt et al. 1970). Tabersonine is a key intermediate in the synthesis of many important medicinal MIA (e.g., vindoline, pachysiphine). Even though these two Voacanga species are major industrial sources of tabersonine, the biosynthetic routes of their MIAs have not been studied to date. Here, we report the first Voacanga genome assembly. Together with the eight previously published MIA-producing plant genome (Stander et al. 2022), this new genome resource will increase our understanding of MIA diversification as well as the evolution of their biosynthetic pathways.
Fig. 1.
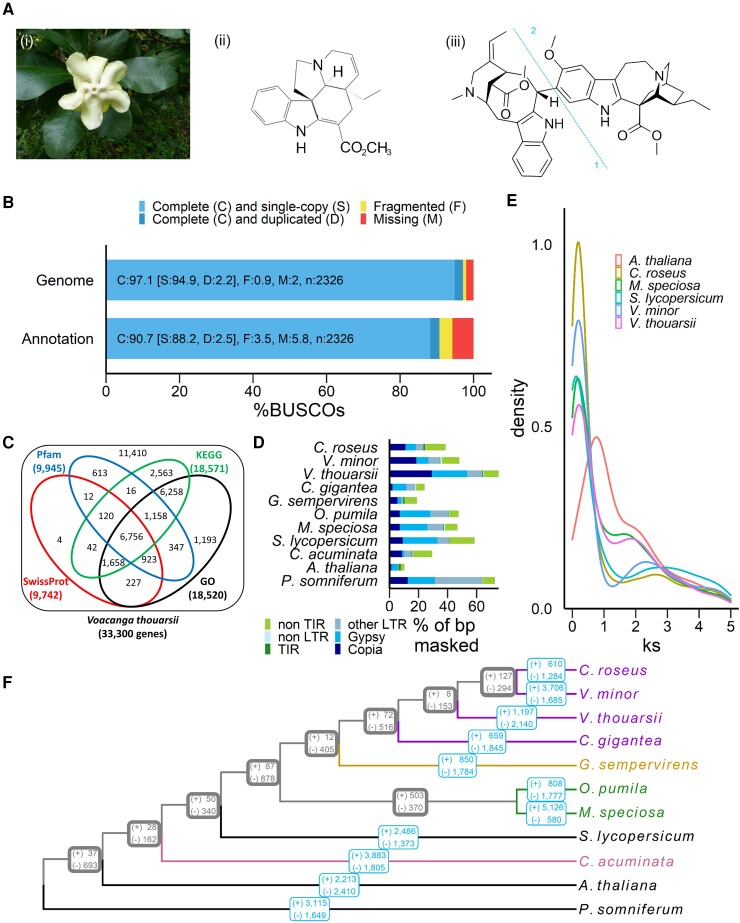
The annotated Voacanga thouarsii genome. (A) Voacanga thoaursii flowers (i) and the molecular structure of two main MIA: tabersonine (ii) and voacamine (iii) which results in the combination of a vobasine (1) and an ibogain (2). (B) BUSCO scores of genome and annotated genes. (C) Functional annotation of genes using SwissProt, Pfam, KEGG, and GO databases. (D) Transposable element proportion and classification. TIR: terminal inverted repeat, LTR: long terminal repeat, other LTR: LTR containing retrotransposons except for Gypsy and Copia, non-LTR: retrotransposons without LTR sequence, non TIR: DNA transposons without TIR sequence. (E) Synonymous substitution (Ks) rate distribution plot for V. thouarsii orthologs compared with other eudicots. (F) Phylogenetic tree of V. thouarsii and 10 other species including three Apocynaceae (purple: C. roseus, V. minor, C. gigantea), one Gelsemiaceae (yellow: G. sempervirens), two Rubiaceae (green: O. pumila, M. speciosa) and one Cornales (pink: C. acuminata). Gene family expansion (+) and contraction (−) were calculated using Cafe5 in each lineage (light bordered blue boxes) and in internal nodes of ancestral population for each taxon (thick bordered grey boxes).
Results and Discussion
Genome Assembly and Annotation
Voacanga thouarsii was assembled into 3,451 contigs with an N50 of 2.91 Mb. The pilon-polished assembly consisted in 1,341.26-Mb distributed across 3,406 scaffolds with an N50 of 3.04 Mb (table 1) and a GC content of 34.31%. Currently reported Apocynaceae assemblies (Calotropis gigantea, Catharanthus roseus, Vinca minor), ranging from 157.28 to 679.10 Mb, are smaller than the one reported here (table 1). The base-level QV of 36.8732, corresponding to more than 99.999% accuracy, and the k-mer completeness of 93.9624% are good indicators of the high quality of the assembled genome.
Table 1.
Genome assembly metrics
| Species | Family | Assembly size (Mb) | No. of scaff.a | N50 (Mb) | Protein -coding genes | BUSCO C [S; D]; F; Mb | Ref. |
|---|---|---|---|---|---|---|---|
| A. thaliana | Bras. | 119.67 | 7 | 23.46 | 27,564 | 99.6 [98.8; 0.8]; 0.1; 0.3 | [1] |
| C. gigantea | Apo. | 157.28 | 1,536 | 0.81 | 18,197 | 93.0 [91.6; 1.4]; 1.7; 5.3 | [2] |
| C. acuminata | Corn. | 414.95 | 775 | 18.28 | 27,940 | 98.2 [90.5; 2.2]; 0.5; 1.3 | [3] |
| C. roseus | Apo. | 541.13 | 2,090 | 2.58 | 34,363 | 97.0 [95.5; 1.5]; 1.3; 1.7 | [4] |
| G. sempervirens | Gel. | 244.39 | 3,352 | 0.41 | 22,617 | 96.5 [95.1; 1.4]; 0.9; 2.6 | [4] |
| M. speciosa | Rub. | 1,122.52 | 40,370 | 1.02 | 55,746 | 91.2 [37.7; 53.5]; 5.0; 3.8 | [5] |
| O. pumila | Rub. | 440.32 | 13 | 40.57 | 91,162 | 96.9 [93.7; 3.2]; 0.9; 2.2 | [6] |
| P. somniferum | Pap. | 2,715.53 | 34,381 | 204.47 | 62,934 | 94.8 [9.2; 85.6]; 1.2; 4.0 | [7] |
| S. lycopersicum | Solan. | 782.52 | 13 | 65.27 | 34,075 | 98.5 [97.6; 0.9]; 0.6; 0.9 | [8] |
| V. minor | Apo. | 679.10 | 296 | 5.97 | 29,624 | 96.9 [60.3; 36.6]; 1.1; 2.0 | [9] |
| V. thouarsii | Apo. | 1,351.26 | 3,406 | 3.04 | 33,300 | 97.1 [94.9; 2.2]; 0.9; 2.0 | This study |
number of scaffolds; bBUSCO scores (genome mode) % Complete [% Complete and single-copy; % Complete and Duplicated]; % Fragmented; % Missing (n = 2,326).
A. thaliana: Arabidopsis thaliana, C. gigantea: Calotropis gigantea, C. acuminata: Camptotheca acuminata, C. roseus: Catharanthus roseus, G. sempervirens: Gelsemium sempervirens, M. speciosa: Mytragyna speciosa, O. pumila: Ophiorrhiza pumila, P. somniferum: Papaver somniferum, S. lycopersicum: Solanum lycopersicum, V. minor: Vinca minor, V. thouarsii: Voacanga thoaursii.
Bras.: Brassicaceae, Apo: Apocynaceae, Corn.: Cornales, Gel.: Gelsemiaceae, Rub.: Rubiaceae Pap.: Papaveraceae, Solan.: Solanaceae.
[1] Lamesh et al. 2012; [2] Hoopes et al. 2018; [3] Kang et al. 2021; [4] Franke et al. 2019; [5] Brose et al. 2021; [6] Rai et al. 2021; [7] Guo et al. 2018; [8] Hosmani et al. 2019; [9] Stander et al. 2022.
Based on the identification of core Eudicotyledons Benchmarking Universal Single-Copy Orthologs (BUSCO), the assembled genome is 97.1% complete (fig. 1B). Gene prediction with MAKER2 (Holt and Yandell, 2011) annotation tool identified 33,300 protein-coding genes in the assembled genome which is comparable to the previously published Apocynaceae species (table 1). Based on Eudicotyledons BUSCO, this predicted set of genes is 90.7% complete with a very low duplication rate (2.5%, fig. 1B).
The combination of BLASTP and BLASTX against UniProt database and hmmscan against the PFAM database led to the functional annotation of 65.7% of the predicted genes (21,890 of the 33,300 genes, fig. 1C, supplementary table S1, Supplementary Material online). To identify putative orthologs of MIA biosynthetic genes, we used functionally validated MIA pathway genes from Catharanthus roseus, V. minor, Tabernanthe iboga, Gelsemium sempervirens and Rauwolfia species to conduct BLAST searches considering hits of at least 90% coverage and 40% identity (supplementary tables S2-S3, Supplementary Material online). The most probable orthologues were then selected based on best hits and phylogeny analysis (supplementary table S4, Supplementary Material online). Based on this approach, we were able to identify putative orthologs with high confidences (76–94% protein identity) for almost 90% of terpenoids, iridoids, and MIA biosynthetic genes up to tabersonine. Interestingly, putative orthologs of genes from the terpenoids and early iridoid pathway tend to be more expressed in leaves while orthologs of genes from the late iridoid, indole, and central MIA pathways tend to be more expressed in roots, thus suggestion a specialization of MIA synthesis (supplementary table S4, Supplementary Material online). Very poor confidences were obtained for putative orthologs of the MIA pathways genes downstream of tabersonine (<64% protein identity). As an example, identity between orthologs is so weak that we were not able to discriminate putative orthologs of T16H and TEX. This thus suggests two possible evolution scenarios of the genes encoding tabersonine-modifying enzymes relying either on a wide diversification in plants accumulating several tabersonine-derived MIAs such as in Catharanthus roseus or to their non-functionalization in V. thourasii leading to their loss and the high accumulation of tabersonine. Indeed, such an evolutionary process has already been described in benzoxazinoid biosynthesis (Frey et al. 2009). By contrast, confident gene orthologs of the voacamine biosynthesis branch were found in agreement with its accumulation in Voacanga species (Hussain et al. 2012).
Transposable Element Annotation
Transposable elements (TE) have well-known roles in genome evolution, genetic instability and gene expression regulation (Sahebi et al. 2018), prompting us to analyze TE composition in V. thouarsii. This analysis showed that 75.16% of the genome consists of TE (fig. 1D), which mainly corresponds to long terminal repeat retrotransposons (63.9% of total TE) with a similar proportion of Copia and Gypsy elements (29.1% and 24.4%, respectively, fig. 1D). Interestingly, V. thouarsii genome displayed the highest proportion of TE compared with all other MIA-producing plants studied that could be a reason for the low scaffolding levels. Indeed, V. thouarsii genome is composed of 3,406 scaffolds when all other Apocynaceae genomes we studied ranged between 296 and 2,090 scaffolds (table 1). Moreover, a similar phenomenon is observed with Papaver somniferum genome, which has a similar TE content to V. thoaursii (72.58%), a genome size that is almost twice as large and a high number of scaffolds (table 1, fig. 1D).
Whole-Genome Duplication Analysis
We then searched for whole-genome duplication (WGD) events by calculating synonymous substitution per synonymous sites (Ks) for paralogous gene pairs across different plant species (fig. 1E). Here, we detected the conserved γ whole-genome triplication (Jiao et al. 2012) common to all eudicots at a Ks of around 2 in all studied species. No other secondary peak could be observed indicating that V. thouarsii did not go through any additional WGD.
Comparison of Orthologous Genes
A maximum-likelihood phylogenetic tree of the 11 studied species was constructed from 680 single-copy orthogroups obtained from OrthoFinder. Lineage-specific (fig. 1F, blue) and ancestral (fig. 1F, grey) gene family evolution was determined using Cafe5. Even though a similar number of genes was annotated in V. Thouarsii genome compared with other Apocynaceae, V. thouarsii showed the highest decrease in orthogroups (2,140) among all investigated MIA-producing plants. Such a difference may result from putative variations in the copy-number of several genes For instance, 404 V. thouarsii genes were annotated as putative cytochrome P450 while 225 cytochrome P450 are annotated in Catharanthus roseus genome (Franke et al. 2019). Among the 2,140 decreased orthogroups, 1,608 orthogroups completely disappeared in V. thouarsii including 952 existing in the three studied Apocynaceae (supplementary table S5, Supplementary Material online). These losses could be linked to the high proportion of TE in V. thouarsii compared with other Apocynaceae, in agreement with their key roles in genome evolution (Catlin and Josephs, 2022). Indeed, several studies conducted in plants (Tang et al. 2012; Bariah et al. 2020; Boatwright et al. 2021) and animals (Pantzartzi et al. 2018; Bourgeois et al. 2020) have highlighted the impact of TE on the loss of functional genes. Such dynamics could also explain the differences in MIAs from Voacanga species compared with other MIA-producing plants.
Conclusions
Here, we described the genome of the wild frangipani, Voacanga thouarsii, which will be a valuable resource for future evolutionary and functional studies. Our genomic analysis showed that despite some similarities (e.g., similar gene content, absence of post-γ whole-genome duplication), V. thouarsii genome displays specific genomic features such as a higher TE content and a bigger size compared with other Apocynaceae. This new Apocynaceae genome thereby paves the way for a better understanding of MIA biosynthesis as well as the identification of new and/or more efficient MIA biosynthetic enzymes that can be used in the developing yeast cell factories producing MIAs (Guirimand et al. 2021; Kulagina et al. 2021; Zhang et al. 2022).
Materials and Methods
Sample Collection, DNA Extraction and Sequencing
Voacanga thouarsii seeds were obtained from Boutique Végétale (https://boutique-vegetale.com/). Seeds were soaked for 16 h before sowing. Plant were greenhouse-grown for three months before sampling. DNA was extracted from V. thouarsii leaves using Qiagen Plant DNeasy kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Illumina sequencing library were built using the Nextera Flex kit (Illumina, San Diego, USA) by Future Genomics Technologies (Leiden, The Netherlands) and subsequently sequenced in paired-end mode (2 × 150 bp) using Illumina NovaSeq 6,000 technology. Future Genomics Technologies (Leiden, The Netherland) constructed ONT library using ONT 1D ligation sequencing kit (Oxford Nanopore Technologies Ltd, Oxford, United-Kingdom) subsequently sequenced on Nanopore PromethION flowcell (Oxford Nanopore Technologies Ltd, Oxford, United-Kingdom) with the guppy version 4.0.11 high-accuracy basecaller. A total of 281,539,400 reads were obtained from the Illumina NovaSeq 6,000 sequencing and 11,390,893 from the ONT PromethION sequencing.
RNA Sequencing and Assembly
RNA was extracted from liquid nitrogen flash-frozen roots, young and old leaves using NucleoSpin RNA Plant and Fungi mini kit (Macherey-Nagel, Düren, Germany) and purified using RNase-free TURBO DNase set (Thermo Fisher Scientific, Illkirch-Graffenstaden, France), both according to the suppliers’ instructions. RNA library construction and sequencing was performed at FGTech using Illumina NovaSeq 6,000 technology. Raw RNA-seq data have been deposited under the SRA accession numbers SRR19972991, SRR19972992, and SRR19972993. Transcriptome was assembled using CLC assembler (v.4.4.1) with a word size of 60 and a bubble size of 250.
De Novo Genome Assembly, Gene Model Prediction and Gene Functional Annotation
The V. thouarsii genome assembly and gene model prediction were performed by Future Genomics Technologies (Leiden, The Netherlands). Adapters were removed using porechop (Wick et al. 2017). ONT reads were first assembled into contig using Flye assembler (v.2.8.2, Kolmogorov et al. 2019) with the following options: –min-overlap 10,000 -i 2. Redundant contigs were removed using purge_haplotigs (v.1.1.0) followed by two rounds of polishing with Illumina paired-end reads using pilon (v.1.23, Walker et al. 2014). Gene modeling was performed using MAKER2 pipeline (v.3.01.02, Holt and Yandell, 2011) using CLC assembled transcriptome as evidence. Putative function for each gene model was then assigned via a combination of similarity search (BLASTX of predicted transcript and BLASTP of TransDecoder (v.5.5.0, Haas et al. 2013) predicted ORFs against the UniProt database) and hmmscan (v.3.1b2, Finn et al. 2011) against the PFAM database (https://pfam.xfam.org/).
Assembly Completeness Assessment
Assembly quality was assessed using the stat program from BBMap tool (v.38.94, Bushnell, 2014). Complementary quality metrics were obtained from merqury (v. 1.3, Rhie et al. 2020). Briefly, 20-mers database was constructed from Illumina short-reads using count function from meryl (v.1.3, Koren et al. 2017). K-mer survival rate was then used to estimate base-level consensus quality score (QV). K-mer completeness was evaluated considering the fraction of reliable k-mers in read database also found in the assembly. Genome and gene models completeness were assessed by applying Benchmarking Universal Single-Copy Orthologs (BUSCO v.5.2.2, Simão et al. 2015) with default settings using a plant-specific database of 2,326 single-copy orthologs (eudicots_odb10). Gene models statistics were obtained using agat_sp_statistics from the AGAT package (v.0.8.0, Dainat, 2022).
Transposable Elements Prediction and Annotation
Extensive de novo TE annotator (EDTA v.1.9.5, Ou et al. 2019) was used to identify and annotate transposable element (TE). Sensitive option using RepeatModeler (v.2.0.1, Smit and Hubley, 2015) was used to identify remaining TEs. Classification consistency was evaluated using evaluate option. alluniRefprexp082813 curated database was used to perform TE annotation.
Whole-Genome Duplication Analysis
To infer whole-genome duplication (WGD) events, transcript sequences of V. thouarsii, V. minor (Stander et al. 2022), Arabidopsis thaliana (Lamesh et al. 2012), Catharanthus roseus (Franke et al. 2019), Mytragyna speciosa (Brose et al. 2021), Solanum lycopersicum (Hosmani et al. 2019), Camptotheca acuminata (Kang et al. 2021), Calotropis gigantea (Hoopes et al. 2018), G. sempervirens (Franke et al. 2019), Ophiorrhiza pumila (Rai et al. 2021), and P. somniferum (Guo et al. 2018) were input to the DupPipe pipeline (Barker et al. 2010). For each dataset, discontiguous MegaBLAST (Ma et al. 2002, Zhang et al. 2004) was used to identify duplicated gene pairs (40% sequence similarity over 300 bp). For each gene pair, the open reading frame was infer from the NCBI's plant RefSeq protein database (May 21, 2021) using BLASTx (v.2.6.0-1, Camacho et al. 2009). Only the best hit sequence was retained (sequence similarity threshold: 30% over 150 aa). DNA sequence alignment against its best hit homologous protein sequence and its translation was performed using GeneWise (Birney et al. 2004). Resulting amino acid sequences for each gene pair were aligned using MUSCLE (v.3.6, Edgar, 2004). This alignment further guided nucleic acid alignment using RevTrans (v.1.4, Wernersson and Perdersen, 2003). Codeml's F3 × 4 model from PAML package (v.4.9, Yang, 1997) was used to calculate substitutions per synonymous site (Ks) and thus determine divergence times between gene pairs.
Phylogenetic Tree Reconstruction
Gene families were constructed by comparing the protein sequences of V. thouarsii with ten other plant species: V. minor (Stander et al. 2022), A. thaliana (Lamesh et al. 2012), Catharanthus roseus (Franker et al. 2019), M. speciosa (Brose et al. 2021), S. lycopersicum (Hosmani et al. 2019), C. acuminata (Kang et al. 2021), Calotropis gigantea (Hoopes et al. 2018), G. sempervirens (Franke et al. 2019), O. pumila (Rai et al. 2021) and P. somniferum (Guo et al. 2018). Protein sequences of less than 30 amino acids were removed. For each species, the longest representative protein was selected in each CD-HIT (v.4.7; Fu et al. 2012) cluster. These sequences were then used as input for OrthoFinder (v.2.5.4; Emms and Kelly, 2019) using the following parameters: -S diamond -M msa -A muscle -T raxml-ng. 680 single-copy orthogroups were used to build a maximum-likelihood phylogenetic tree. Orthogroup gain and expansion were determined across the phylogenetic tree using Cafe5 (v.4.2.1, Mendes et al. 2021).
Transcript Abundance Estimation
Reads were pseudo-aligned onto the predicted transcripts and counted using Salmon (v.0.6.0; Patro et al. 2017) using -biasCorrect and -vbo options. Abundance estimates were established as transcripts per million (TPM) and are presented in supplementary table S6, Supplementary Material online.
Supplementary Material
Acknowledgements
We thank access and support to the CCSC computing resources (Cascimodot Federation, CNRS, Orléans). We acknowledge funding from the EU Horizon 2020 research and innovation program (MIAMi project-Grant agreement N°814645), ARD-CVL Biopharmaceutical program of the Région Centre Val de Loire (ETOPOCentre project), and ANR (project MIACYC—ANR-20-CE43-0010).
The authors benefitted from the use of the cluster at the Centre de Calcul Scientifique en région Centre-Val de Loire.
Contributor Information
Clément Cuello, Biomolécules et Biotechnologies Végétales, EA2106, Université de Tours, 37200 Tours, France.
Emily Amor Stander, Biomolécules et Biotechnologies Végétales, EA2106, Université de Tours, 37200 Tours, France.
Hans J Jansen, Future Genomics Technologies, 2333 BE Leiden, The Netherlands.
Thomas Dugé de Bernonville, Biomolécules et Biotechnologies Végétales, EA2106, Université de Tours, 37200 Tours, France.
Arnaud Lanoue, Biomolécules et Biotechnologies Végétales, EA2106, Université de Tours, 37200 Tours, France.
Nathalie Giglioli-Guivarc'h, Biomolécules et Biotechnologies Végétales, EA2106, Université de Tours, 37200 Tours, France.
Nicolas Papon, Univ Angers, Univ Brest, IRF, SFR ICAT, F-49000 Angers, France.
Ron P Dirks, Future Genomics Technologies, 2333 BE Leiden, The Netherlands.
Michael Krogh Jensen, Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, Kgs Lyngby, Denmark.
Sarah Ellen O'Connor, Department of Natural Product Biosynthesis, Max Planck Institute for Chemical Ecology, Jena 07745, Germany.
Sébastien Besseau, Biomolécules et Biotechnologies Végétales, EA2106, Université de Tours, 37200 Tours, France.
Vincent Courdavault, Biomolécules et Biotechnologies Végétales, EA2106, Université de Tours, 37200 Tours, France.
Supplementary Material
Supplementary data are available online at Genome Biology and Evolution online.
Author Contributions
S.E.O., M.K.J., T.D.D.B., S.B., V.C. designed the research. E.A.S., R.P.D. and H.J.J. acquired the data. C.C., E.A.S., H.J.J., A.L., N.G.G., N.P. and R.P.D. analyzed the data. C.C., S.B. and V.C. wrote the article. All authors read and approved the final manuscript.
Data Availability
The annotated genome has been deposited in the NCBI database under the Bioproject accession number PRJNA860765. RNAseq raw reads have been deposited in the NCBI database under the SRA accession numbers SRR19972991, SRR19972992, and SRR19972993. Genome annotation predicted transcripts and proteins, and transcript expression abundances are available on figshare: https://doi.org/10.6084/m9.figshare.20223093.v1
Funding Information
This work was supported by Horizon 2020 research and innovation program [MIAMi project-Grant agreement N 814645]; ARD CVL Biopharmaceutical program of the Région Centre-Val de Loire [ETOPOCentre project]; and ANR (project MIACYC—ANR-20-CE43-0010).
Conflict of Interest
Ron Dirks and Hans Jensen are CEO and CTO of Future Genomics Technologies, respectively.
Literature Cited
- Bariah I, Keidar-Friedman D, Kashkush K. 2020. Where the wild things are: transposable elements as drivers of structural and functional variations in the wheat genome. Front Plant Sci. 11:585515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, et al. 2010. Evopipes.net: bioinformatic tools for ecological and evolutionary genomics. Evol Bioinforma Online 6:143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. 2004. Genewise and genomewise. Genome Res. 14:988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatwright JL, et al. 2021. Trajectories of homoeolog-specific expression in allotetraploid Tragopogon castellanus populations of independent origins. Front Plant Sci. 12:679047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois Y, Ruggiero RP, Hariyani I, Boissinot S. 2020. Disentangling the determinants of transposable elements dynamics in vertebrate genomes using empirical evidences and simulations. PLoS Genet. 16:e1009082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose J, et al. 2021. The Mitragyna speciosa (kratom) genome: a resource for data-mining potent pharmaceuticals that impact human health. G3 (Bethesda) 11:jkab058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B (2014). BBMap: a fast, accurate, splice-aware aligner. (No. LBNL-7065E). Berkeley (CA): Lawrence Berkeley National Lab. (LBNL). [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinf. 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi L, et al. 2018. Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Science 360:1235–1239. [DOI] [PubMed] [Google Scholar]
- Catlin NS, Josephs EB. 2022. The important contribution of transposable elements to phenotypic variation and evolution. Curr Opin Plant Biol. 65:102140. [DOI] [PubMed] [Google Scholar]
- Dainat J, Hereñú D, LucileSol, pascal-git. 2022. NBISweden/AGAT: AGAT-v0.8.1. Zenodo. https://zenodo.org/record/5834795#.Y2jXNNLMKV4. [Google Scholar]
- De Luca V, Balsevich J, Tyler RT, Kurz WGW. 1987. Characterization of a novel N-methyltransferase (NMT) from Catharanthus roseus plants. Plant Cell Rep. 6:458–461. [DOI] [PubMed] [Google Scholar]
- Diavara D, Pyuskyulev B, Kuzmanov B. 1984. Alkaloid-bearing plants in the flora of Guinea. Alkaloids from Voacanga africana stapf. Izvestiya po Khimiya 17:364–371. [Google Scholar]
- Dugé de Bernonville T, et al. 2015. Phytochemical genomics of the Madagascar periwinkle: unravelling the last twists of the alkaloid engine. Phytochemistry 113:9–23. [DOI] [PubMed] [Google Scholar]
- Dzoyem JP, Tshikalange E, Kuete V. 2013. Medicinal plants market and industry in Africa. In: Kuete V, editors. Medicinal plant research in Africa—pharmacology and chemistry: Elsevier. p. 859–890. [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. Orthofinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. 2011. HMMER Web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke J, et al. 2019. Gene discovery in Gelsemium highlights conserved gene clusters in monoterpene indole alkaloid biosynthesis. ChemBioChem. 20:83–87. [DOI] [PubMed] [Google Scholar]
- Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A. 2009. Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70:1645–1651. [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldblatt A, Hootele C, Pecher J. 1970. Indole alkaloids. XXIII. Alkaloids of Voacanga thouarsii. Phytochemistry 9:1293–1298. [Google Scholar]
- Guirimand G, Julagina N, Papon N, Hasunuma T, Courdavault V. 2021. T innovative tools and strategies for optimizing yeast cell factories. Trends Biotechnol 39:488–504. [DOI] [PubMed] [Google Scholar]
- Guo L, et al. 2018. The opium poppy genome and morphinan production. Science 362:343–347. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq: reference generation and analysis with TRinity. Nat Protoc. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinf. 12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes GM, et al. 2018. Genome assembly and annotation of the medicinal plant Calotropis gigantea, a producer of anticancer and antimalarial cardenolides. G3 (Bethesda) 8:385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmani PS, et al. 2019. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, hi-C proximity ligation and optical maps. [Google Scholar]
- Hussain H, Hussain J, Al-Harrasi A, Green IR. 2012. Chemistry and biology of the genus Voacanga. Pharm Biol. 50:1183–1193. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. 2012. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 13:R3–R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, et al. 2021. A chromosome-level Camptotheca acuminata genome assembly provides insights into the evolutionary origin of camptothecin biosynthesis. Nat Commun. 121:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 375:540–546. [DOI] [PubMed] [Google Scholar]
- Koren S, et al. 2018. De novo assembly of haplotype-resolved genomes with trio binning. Nat Biotechnol. 36(12):1174–1182. doi: 10.1038/nbt.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulagina N, et al. 2021. Enhanced bioproduction of anticancer precursor vindoline by yeast cell factories. Microb Biotechnol 14:2693–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulagina N, Méteignier LV, Papon N, O'Connor SE, Courdavault V. 2022. More than a Catharanthus plant: a multicellular and pluri-organelle alkaloid-producing factory. Curr Opin Plant Biol. 67:102200. [DOI] [PubMed] [Google Scholar]
- Kunesch N, et al. 1977. Alkaloids of Voacanga. XVIII. Alkaloids of Callichilia subsessilis. IV. Organic natural products. 165. The structure of bisindoline alkaloids of a novel type. Helv Chim Acta 60:2854–2859. [Google Scholar]
- Lamesch P, et al. 2012. The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40:D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuwenberg AJM. 1980. The taxonomic position of some genera in the Loganiaceae, Apocynaceae, and Rubiaceae, related families which contain indole alkaloids. In: Phillipson JD and Zenk MH, editors. Indole and biogenetically related alkaloids: Academic Press. p. 1–10. [Google Scholar]
- Ma B, Tromp J, Li M. 2002. Patternhunter: faster and more sensitive homology search. Bioinformatics 18:440–445. [DOI] [PubMed] [Google Scholar]
- Macabeo AP, Alejandro GJ, Hallare AV, Vidar WS, Villaflores OB. 2009. Phytochemical survey and pharmacological activities of the indole alkaloids in the genus Voacanga thouars (Apocynaceae)—an update. Pharmacogn Rev. 3:132–142. [Google Scholar]
- Maresh JJ, et al. 2007. Strictosidine synthase: mechanism of a pictet-spengler catalyzing enzyme. J Am Chem Soc. 130:710–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes FK, Vanderpool D, Fulton B, Hahn MW. 2021. CAFE 5 Models variation in evolutionary rates among gene families. Bioinformatics 36:5516–5518. [DOI] [PubMed] [Google Scholar]
- Mistry V, Darji S, Tiwari P, Sharma A. 2022. Engineering Catharanthus roseus monoterpenoid indole alkaloid pathway in yeast. Appl Microbiol Biotechnol 106:2337–2347. [DOI] [PubMed] [Google Scholar]
- O’Connor SE, Maresh JJ. 2006. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep. 23:532–547. [DOI] [PubMed] [Google Scholar]
- Ou S, et al. 2019. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Mustafa NR, Tang K, Choi YH, Verpoorte R. 2016. Monoterpenoid indole alkaloids biosynthesis and its regulation in Catharanthus roseus: a literature review from genes to metabolites. Phytochem Rev 15:221–250. [Google Scholar]
- Pantzartzi C, Pergner J, Kozmik Z. 2018. The role of transposable elements in functional evolution of amphioxus genome: the case of opsin gene family. Sci Rep. 8:2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A, et al. 2021. Chromosome-level genome assembly of Ophiorrhiza pumila reveals the evolution of camptothecin biosynthesis. Nat Commun. 121:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanitrahasimbola D, et al. 2001. Biological activities of the plant-derived bisindole voacamine with reference to malaria. Phytother Res. 15:30–33. [DOI] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM. 2020. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21:245. doi: 10.1186/s13059-020-02134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland Y, Kunesch N, Libot F, Poisson J, Budzikiewicz H. 1975. Alkaloids of Voacanga. XV. Structure of new alkaloids from the leaves of Voacanga thouarsii. Bull Soc Chim Fr. 11:2503–2506. [Google Scholar]
- Sahebi M, et al. 2018. Contribution of transposable elements in the plant's Genome. Gene 665:155–166. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R. 2015. RepeatModeler Open-1.0. http://www.repeatmasker.org.
- Stander EA, et al. 2022. The Vinca minor genome highlights conserved evolutionary traits in monoterpene indole alkaloid synthesis. G3 (Bethesda):jkac268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, et al. 2012. Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics 190:1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. 2003. Revtrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 31:3537–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genom 3:e000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Bioinformatics 13:555–556. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. 2022. A microbial supply chain for production of the anti-cancer drug vinblastine. Nature 609:341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. 2004. A greedy algorithm for aligning DNA sequences. J Comput Biol. 7:203–214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The annotated genome has been deposited in the NCBI database under the Bioproject accession number PRJNA860765. RNAseq raw reads have been deposited in the NCBI database under the SRA accession numbers SRR19972991, SRR19972992, and SRR19972993. Genome annotation predicted transcripts and proteins, and transcript expression abundances are available on figshare: https://doi.org/10.6084/m9.figshare.20223093.v1