Abstract
A sensible control of hormone secretion from pancreatic islets requires concerted inter-cellular communications, but a comprehensive picture of the whole islet interactome is presently missing. Single-cell transcriptomics allows to overcome this and we used here a single-cell dataset from type 2 diabetic (T2D) and non-diabetic (ND) donors to leverage islet interaction networks. The single-cell dataset contains 3046 cells classified in 7 cell types. The interactions across cell types in T2D and ND were obtained and resulting networks analysed to identify high-centrality genes and altered interactions in T2D. The T2D interactome displayed a higher number of interactions (10 787) than ND (9707); 1289 interactions involved beta cells (1147 in ND). High-centrality genes included EGFR, FGFR1 and FGFR2, important for cell survival and proliferation. In conclusion, this analysis represents the first in silico model of the human islet interactome, enabling the identification of signatures potentially relevant for T2D pathophysiology.
INTRODUCTION
Islets of Langerhans are cell clusters located in the pancreas that play a key role in balancing body intermediate metabolism. In human adults the number of islets ranges 0.5–4 million, and islet size may vary from 30 to 40 up to 400–500 μm in diameter (1,2). The endocrine cells (mostly beta, alpha and delta cells, but also PP and ghrelin-producing cells) account for the vast majority of the islet cells (3,4), but these highly heterogeneous micro-organs also contain non-endocrine cells. Altogether, these cellular ‘ecosystems’ engage in complex communication networks able to affect the function of the endocrine cells (5–7). Thus, beta cell function is modulated not only by nutrient concentrations, but also by the action of other islet hormones, in particular glucagon and somatostatin (8).
The effects of such intercellular communications highlight the importance that the whole islet ‘system’ has on the properties of its individual components. However, comprehensive descriptions of the intra-islet cross-talk, including, in particular, ligand–receptor interactions, and how these may be perturbed in type 2 diabetes (T2D) are still missing. This can also be due to the heterogeneous nature of pancreatic islets, which complicates the characterization of individual cell types by high-throughput methods such as transcriptomics and proteomics.
The advancements of single-cell technology filled this gap, allowing to define the transcriptomic features of the human islet cell types and how these are affected by T2D (9–11). CellPhoneDB (12), a recent inclusion among the resources available to analyse such data, is a public repository of intercellular interactions, involving either ligand–receptor pairs or more complex interactions i.e. those in which both partners are receptors (such as interactions between membrane proteins) or involving complexes. For the sake of simplicity, we will refer to these different interaction classes with the term ligand–receptor interactions. The database is provided with scripts allowing to map single-cell expression data, enabling to derive the complete set of interactions occurring between all pairwise combinations of cell types, which will be referred to as the interactome. The methods implemented in CellPhoneDB include a statistical analysis method based on empirical shuffling to compute which ligand–receptor interactions display significant cell-type specificity. Such an approach has been previously used to generate an atlas of interactions occurring at the maternal–fetal interface during early pregnancy (13), to characterize interactions between cancer and immune cells (14), and to investigate the immune mechanisms associated with severe COVID-19 infections (15).
In this work, we applied the CellPhoneDB framework to analyse a single-cell dataset integrating different studies previously published (16). By separately considering cells from non-diabetic (ND) and T2D donors, we assembled two different interaction networks (interactomes) in order to describe the interactions occurring in health (ND) and disease (T2D). We then described these perturbations, focusing on cell types and genes of pathophysiological relevance.
MATERIALS AND METHODS
Single-cell data analysis
A previously published dataset (16) that integrates three single-cell transcriptomic studies (9–11) was loaded in the Scanpy framework (17) as a data frame (AnnData). This data structure stores the expression value of each gene in the single cells, as well as the metadata annotation of cells and genes. The cells have been divided in two groups according to the ‘Disease’ variable, which indicates if the corresponding donor was diabetic or not. For each of the two groups, the read counts and the metadata reporting the cell types have been exported as tab-separated files and analysed with CellPhoneDB 2.1.1 (12) in a Unix environment with the command line: ‘cellphonedb method statistical_analysis meta.tsv counts.tsv’, where ‘meta.tsv’ and ‘counts.tsv’ correspond to the metadata and the read counts files, respectively.
Data from HPAP (18) was used to perform an orthogonal validation of the ND interactome. Briefly, transcriptomic data from two donors (HPAP-054 and HPAP-059) that we previously studied (19), was analysed with CellPhoneDB, separately for each donor, using the same command line and combined in a single interactome (HPAP-ND interactome).
Interactome analysis
The CellPhoneDB output files ‘significant_means.txt’ were analysed in a python environment using the Pandas library (https://pandas.pydata.org). A detailed description of the code used to generate the obtained results is available in a Github repository (https://github.com/EBosi/IsletInteractome). The resulting data frames, reporting the interaction pairs between all cell types pairwise combinations, were reshaped from a wide to a long format, adding two columns indicating the cell source (as-source column) and the cell target (as-target column). Based on the CellPhoneDB annotation, each interaction was defined as ‘directed’ if one of the partners was a ligand and the other was a receptor, and ‘undirected’ otherwise. As an example, the interaction between INS and INSR is directed, whereas the interaction between ADGRL1 and NRG1 is not. The steps taken were as follows: first, by using the as-source and as-target columns from the data frames described above we obtained pivot tables reporting the number of interactions between as-source (columns) and as-target (rows) cell types. With these tables we computed Spearman's Rank Correlation column-wise and row-wise, corresponding to the correlation between as-source and as-target cell types pairs, respectively. Second, we used each data frame to obtain as-source and as-target interaction profiles for each cell type. This was done adding two new columns, obtained by merging the as-source (or as-target) cell types with the interaction id, e.g. merging the as-source cell type ‘ductal cell’ and the interaction id ‘CPI-SC06873D10F’ would result in the string ‘ductal cell CPI-SC06873D10F’. These columns were used to obtain two matrices encoding the presence/absence of the as-source (or as-target) interactions in the different cell types with 1/0 values. These matrices have been used to compute the Spearman's Rank Correlation between the cell types.
The sets of interactions were encoded as multi-partite directed graphs. Such a class of graph is defined as a 3-tuple G = (C,M,E), with the sets C, M, and E being respectively the nodes corresponding to cell types, to molecules (ligands/receptors), and the set of directed edges connecting the various elements of the graph. The graphs have been initialized as empty objects, then populated by iterating on the rows of the interactome data frames and adding the corresponding interactions as paths in the graph. A path is a sequence of edges connecting a source cell to a ligand, a ligand to a receptor, and a receptor molecule to a target cell. As an example, let's consider the directed interaction between the cell type A, expressing the ligand X, and the cell type B, expressing the receptor Y, the corresponding directed path (the order in which the nodes are visited) would be then A→X→Y→B. Undirected interactions are included in the graph by adding two paths with both of the possible directions, e.g. the undirected interaction between the cell type A, expressing the molecule X which interact with the molecule Y expressed in the cell B (A,X,Y,B) corresponds to the paths A→X→Y→B and B→Y→X→A. The graphs have been iteratively constructed starting from the interactome dataframes, iterating on the rows and adding a path for each row. For each edge, a ‘weight’ W was defined to encode the total number of occurrences of the corresponding interactions in the interactome. The two paths of an undirected interaction are counted only once for the purpose of computing W. The networks were assembled using the NetworkX library (20), and analysed with the graph algorithms implemented in the library. The computation of centrality measures was performed as following: the degree, in-degree and out-degree centrality values were computed with the functions ‘degree_centrality’, ‘in_degree_centrality’ and ‘out_degree_centrality’; the ‘betweenness_centrality’ function was used to compute betweenness centrality, using the inverse of the edge weights as a distance measure; the ‘katz_centrality_numpy’ function was used to compute the katz centrality, with the edge weights as the weight measure, and the attenuation factor α as the reciprocal of the absolute value of the largest eigenvalue of the network adjacency matrix.
RESULTS
Defining the human islet interactomes
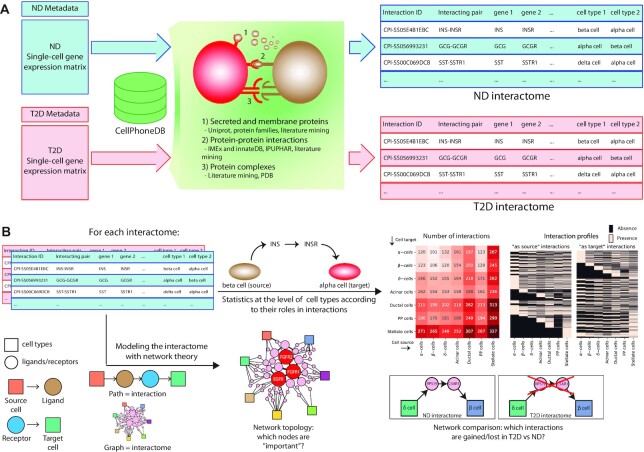
The human islet single-cell dataset used in this work features a total of 3046 cells, expressing 27 931 genes (16). The cells, obtained and sequenced in previous studies (9–11), have been obtained from 52 donors, 19 of which were T2D and 33 ND. The metadata annotation reports a total of 7 cell types (alpha, beta, delta, PP, acinar, ductal and stellate cells), 1611 cells from T2D donors and 1435 cells from ND. The ND and T2D cells were separately analysed with CellPhoneDB, resulting in two sets of interactions between the seven cell types (Interactomes), represented as multi-partite directed graphs (Figure 1).
Figure 1.
A general description for the workflow of the interactome analysis. (A) the single-cell datasets of cells from ND and T2D donors are separately analysed with CellPhoneDB, leading to two separate tables defining the comprehensive sets of interactions in ND and T2D (the interactomes). (B) The interactomes have been analysed with two separate approaches. In the first approach (top-right of the panel) the tables are used to perform statistical analyses at the level of cell types, considering also their role in the interactions, i.e. considering as-source and as-target cells separately. The second approach (bottom-right) models the interactomes as networks, with each interaction representing a path in the network (bottom-left), to identify relevant network topology patterns.
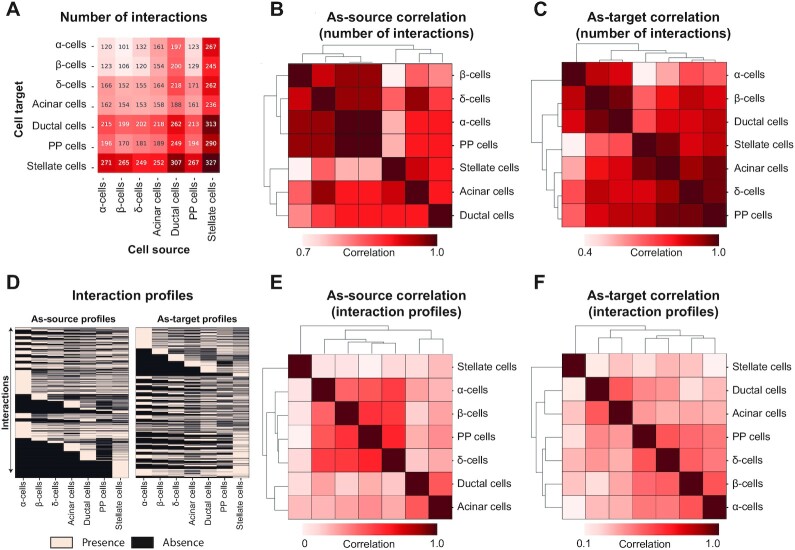
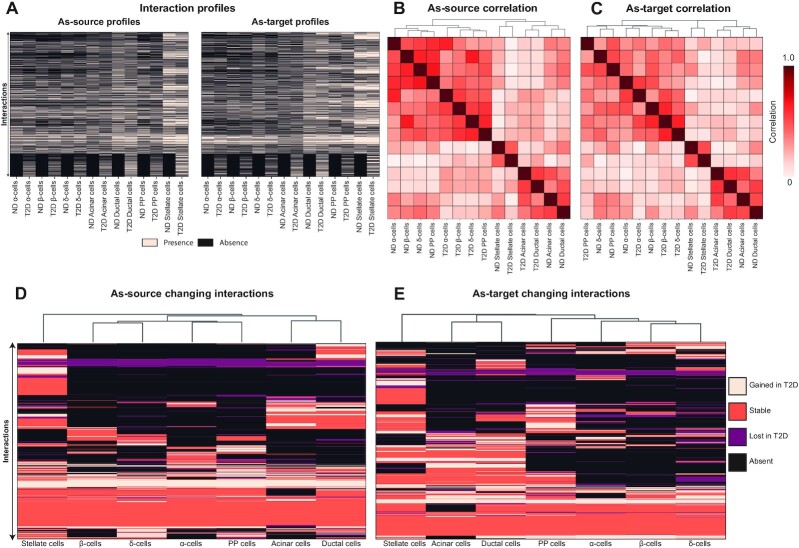
The ND interactome (Supplementary Table S1) embeds the human islet inter-cellular interactions likely occurring under non-pathological conditions: there were a total of 9707 interactions occurring between all the pairwise combinations of the cell types (n = 49) (Figure 2A), involving a total number of 355 genes. Of these, 101 and 155 genes encode ligands and receptors, respectively. A total of 109 genes are associated with more complex, undirected interactions involving two receptors (e.g. interactions between two membrane proteins) or multimeric complexes. A correlation analysis between the cell types performed considering the number of interactions in which they are sources (as-source, Figure 2B) revealed that endocrine cell types (alpha, beta, delta and PP cells) have similar interaction profiles as compared with non-endocrine cells, whereas these latter show more variegated interaction profiles (Supplementary Table S2): ductal and acinar cells are more similar each other, while the stellate cells have their own distinct profile. The same analysis performed on the interactions in which the cell types are targets (as-target, Figure 2C) delivered completely different results, as the distinction between endocrine and non-endocrine cells is lost: beta and ductal cells display similar as-target interaction profiles, acinar cells have a profile comparable to that of alpha and PP cells, and stellate cells feature more similarity with acinar cells (Supplementary Table S2).
Figure 2.
The ND interactome. (A) the heatmap reports the number of interactions between cell types, as cell sources and targets. (B, C) the correlation matrices report the similarity between cell types, in terms of spearman correlation between the number of interactions in which the cell types act as cell sources (B) or as cell targets (C). The matrices are symmetrical and rows/columns have been clustered according to their similarities. The clustering results are indicated as dendrograms on the matrix margins. (D) The presence/absence matrices report the presence and absence of the interactions (rows) in each cell type (columns). (E, F) the correlation matrices report the similarity between cell types, in terms of spearman correlation between the interaction profiles, as-source (E) or as-target (F). The matrices are symmetrical and rows/columns have been clustered according to their similarities. The clustering results are indicated as dendrograms on the matrix margins.
A more granular analysis of the similarities between cell type connections was performed considering, for each cell type, the combination of all 9707 possible interactions with all cell types, both as-source and as-target. Such profiles, encoding presence/absence values (Figure 2D), were used to compute how similar the cell types are to each other (Supplementary Table S2, Figure 2E and F): the cell types clustered in three groups which are (i) endocrine cells, (ii) acinar and ductal cells and (iii) stellate cells. Notably, the clusters were consistent for both as-source and as-target profiles. A closer look at the relationships between endocrine cell types revealed beta cells to be most similar to delta cells, as indicated by the as-source profiles, with a Jaccard Similarity (JS) of 0.62, followed by PP (JS = 0.59) and alpha cells (JS = 0.47). In other words, since the as-source interactions of a cell reflect the interactions in which the cell participates as the signal sender, the beta and delta cells use similar methods to interact with related targets. Considering the similarities of as-target profiles instead, the cells that are more similar to beta cells are alpha cells (JS = 0.47), followed by PP (JS = 0.43) and delta cells (JS = 0.40). Together with the points discussed above, this suggests that beta and alpha cells are receiving similar signals from islet, stellate, ductal, and acinar cells.
Network analysis
By representing the ND interactome as a multi-partite graph, we were able to apply algorithms from graph theory to gain insights into the importance of genes. Such a network presents 301 nodes and 2272 edges (983 between cell types and ligands, 289 between ligands and receptors, and 1000 between receptors and cell types). Each edge is associated with a weight, quantifying how many times the corresponding connection was found in the cell interactome. As an example, the ligand insulin (INS), produced by beta cells, significantly interacts with the insulin receptor (INSR) of all seven cell types. Therefore, the edge connecting beta cells with INS will have a weight 7 (as there are seven connections), as well as the edge connecting INS and INSR. The edges connecting INSR with cell targets will have weight 1. The collections of nodes, edges, and the relative attributes (i.e. weights, identification codes, descriptions) are provided as GEXF files for further investigations (Supplementary File 1–2). The relative importance of ligands and receptors is reflected by different centrality measures: computation of various centrality measures allows to infer the relative importance of ligands and receptors, and of corresponding genes encoding these molecules.
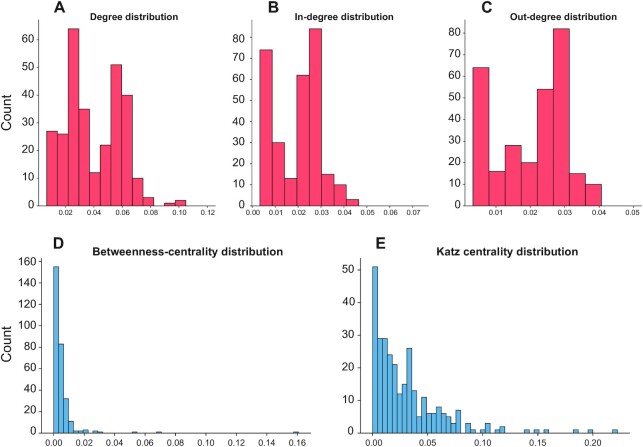
The degree centrality (K) measures the number of connections associated to a ligand/receptor (Supplementary Table S3): higher K values indicate molecules that are involved in many interactions within the islet, and thus likely to hold a relative importance for the communication network. The K distribution (Figure 3A), computed for ligands and receptors, displays bimodality and an average K of 0.040, with a standard deviation of 0.019. Decomposing the degree into out- and in-degree (K-out and K-in, respectively) allowed to track which interacting molecules were more important in the different aspects of inter-cellular communications, i.e. which are the ‘forward’ communication of a signal, or its reception. The K-in and K-out distributions (Figure 3B and C) display bimodality and an increased skewness (from 1.16 to 3.10 and 4.28, respectively) to the left side. The ligands/receptors with extreme K values are likely to represent important actors of the islet communication network. The genes with the highest K (top 5% quantile) were 17, whereas the top genes according to K-in and K-out were 16 and 15, respectively (Supplementary Table S3). These three distributions yielded mostly consistent results and included genes of proved relevance in the context of islet function and diabetes. In particular, the top four genes of K and K-in encode three receptors associated with islet cell survival and proliferation (FGFR2, FGFR1 and EGFR) and one ligand (FAM3C or ILEI); the top 4 genes of K-out instead were WNT5A, EGFR, FAM3C and FGFR2. All these genes have been previously demonstrated to play relevant roles in beta cell pathophysiology (21–24).
Figure 3.
Node centrality measures of the ND interaction network. (A) The histogram reports the distribution of the degree values of the nodes. (B) The histogram reports the distribution of the in-degree values of the nodes. (C) The histogram reports the distribution of the out-degree values of the nodes. (D) The histogram reports the distribution of the betweenness-centrality values of the nodes. (E) The histogram reports the distribution of the Katz centrality values of the nodes.
Although degree centrality can provide meaningful insights regarding the importance of ligands and receptors, some crucial features cannot be captured with this index alone. For instance, ligands and receptors with low degree can still be important for the overall network structure if they connect regions that would be disjointed otherwise. Therefore, different centrality measures, that are betweenness-centrality (BC) and Katz centrality (KC), were used to identify central players of the interactome that could have been overlooked by the K analysis. Considering the distributions of these parameters (Figure 3D and E), the obtained values fit the so-called ‘power-law’, that is, most of the ligand/receptors have a low betweenness, whereas few of them exhibit large values. Such property is important, as it has been repeatedly found in association with complex biological networks, and it has been linked to the emergence of robustness (25). Considering the genes that were associated with extreme BC values (Supplementary Table S4), there were 15 genes, with FGFR1 as the one with the highest BC, followed by FGFR2, FAM3C and WNT5A, thus overlapping with extreme genes found with the K metrics. Regarding the genes with the highest KC, we found slight differences with the other distributions, such as TNF being the gene with the second highest centrality value (Supplementary Table S5).
To draw more general conclusions from the comparison of these metrics, we reported the genes in the top 5 percentile in all the distributions, along with those high-scoring only in specific distributions (Supplementary Table S6). The union of all the distributions' right tails encompassed 32 extreme-value genes, of which 5 were shared between all distributions (EGFR, FAM3C, FGFR1, FGFR2 and PVRL3) and 14 were unique to some distributions (TNFRSF14, MDK, JAG2, EPHA4, FLT4, NOTCH3, CEACAM1, GCG, DSG2, COPA, CEACAM6, MIF, FGF9, GHRL). A considerable proportion of these genes are associated with cellular proliferation and survival.
Orthogonal validation of the ND interactome
To assess the extent to which the results obtained for the interactome represent a generalizable system, we produced another version of the ND interactome with an orthogonal dataset, using single-cell transcriptomics data of human islets from the HPAP database (18). With this dataset, embedding a total of 1525 cells, we produced an interactome (ND-HPAP) which was used to test whether the general features of the ND interactome could be retrieved.
First, we considered if the cell types with a higher number of interactions were consistent between the ND and ND-HPAP interactomes (Supplementary Figure S1). Non-endocrine cell types displayed similar behaviour in both datasets, with stellate and ductal cells having the highest number of interactions, followed by acinar cells and endocrine cell types. Interestingly, PP cells showed a different trend in the two datasets, i.e. in the ND interactome they had a higher number of interactions (comparable to those of acinar cells), whereas in ND-HPAP they had fewer interactions (similarly to endocrine cells). This difference could be due to the fact that PP cells are less represented in the HPAP samples (17 PP cells, accounting for 1.1% of the total) (19), therefore some features of this cell type might have been lost.
Then, the centrality of genes has been evaluated to verify if the same high-centrality genes are consistently found in the two interactomes. Considering all different centrality metrics, there were a total of 26 central genes, 6 less than those identified in the ND interactome (Supplementary Table S6). Of these, 15 are shared among the two interactomes, including all the highest centrality genes previously found (EGFR, FAM3C, FGFR1, FGFR2 and PVRL3) which are top-ranked also in the ND-HPAP interactome.
The interactome perturbations in T2D
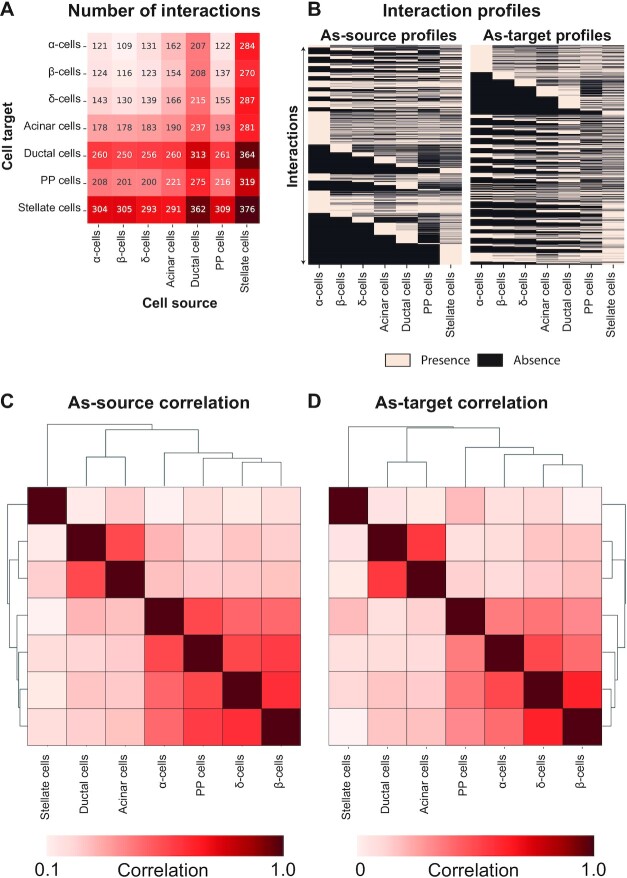
The network analysed so far (ND interactome) has been assembled with the interactions in cells from ND individuals, thus representing cross-talk normally occurring in the islet. To understand the extent of the perturbations of such network in T2D, we used cells from T2D donors to compile the corresponding interaction network (T2D interactome). The system we obtained comprises an 11% increased number of interactions with respect to ND, for a total of 10 787 interactions (Supplementary Table S7, Figure 4A). The number of genes involved in such connections also increased with a similar proportion (9%), with a total of 386 genes present in the T2D interactome. To understand if the T2D interaction patterns were similar to that of the ND interactome, we performed correlation analyses between the cell types (Supplementary Table S8). This enabled to test whether the interaction patterns between cell types were affected, and in particular if the distinction previously observed between endocrine, non-endocrine and stellate cells was disrupted. The analysis was performed considering all possible interactions between all cell types, encoded in a large presence/absence matrix to compute similarities between cell types (Figure 4B, Supplementary Table S8). The correlation analysis identified three clusters for both as-source and as-target interaction profiles, corresponding to endocrine, non-endocrine and stellate cells (Figure 4C and D). The cluster composition is highly similar in both series, within a slight difference in the endocrine cluster.
Figure 4.
The T2D interactome. (A) The heatmap reports the number of interactions between cell types, as cell sources and targets. (B) The presence/absence matrices report the presence and absence of the interactions (rows) in each cell type (columns). (C, D) The correlation matrices report the similarity between cell types, in terms of spearman correlation between the interaction profiles, as-source (E) or as-target (F). The matrices are symmetrical and rows/columns have been clustered according to their similarities. The clustering results are indicated as dendrograms on the matrix margins.
We then compared the interaction profiles, both as-source and as-target, of the ND and T2D interactomes (Figure 5A), to test if cells clustered together according to their cell type composition or to their interactome source (ND versus T2D). This analysis resulted in different patterns for the as-source and as-target profiles (Figure 5B and C): in the former, we found two main clusters, corresponding to endocrine vs non-endocrine cell types. Within each cluster, the cells are divided first according to the disease (T2D versus ND), then according to cell types. For the as-target profiles, we found a first separation between endocrine and non-endocrine cell types. While the non-endocrine cluster is the same as that found for the as-source profiles, endocrine cells cluster differently, in that the cells do not clearly separate according to the tested variables (disease, cell types): alpha cells from ND and T2D donors cluster together, interaction profiles of T2D beta and delta cells, close to each other, are similar to that of ND beta cells.
Figure 5.
Differences between ND and T2D interactome. (A) The presence/absence matrices report the presence and absence of the interactions (rows) in each cell type (columns) of the ND and T2D interactome. (B, C) The correlation matrices report the similarity between cell types of the ND and T2D interactome, in terms of spearman correlation between the interaction profiles, as-source (B) or as-target (C). The matrices are symmetrical and rows/columns have been clustered according to their similarities. The clustering results are indicated as dendrograms on the matrix margins. (D, E) The heatmaps report the changing interactions (rows) in each cell type (columns) in terms of spearman correlation between the interaction profiles, as-source (D) or as-target (E). The changing interaction profiles have been clustered according to their similarities, as indicated by the dendrograms on the top margins.
Contrasting the ND and T2D interactomes we were able to classify each interaction of cell types in four categories, namely absent, stable, lost, and gained. An interaction is absent in a cell type when it is neither present in T2D nor in ND, whereas the stable interactions are present in both series. The lost and gained interactions are present, respectively, in ND or in T2D interactomes only. In general, all cell types displayed a considerable proportion of interactions that were changing from ND to T2D (Table 1). Comparing the profiles of changing reactions (Supplementary Table S9) between cell types revealed consistent results for as-source and as-target interactions (Figure 5D and E): endocrine cells cluster together, with beta and delta cells displaying most similarities; the other two groups are formed by acinar and ductal cells, and stellate cells.
Table 1.
number of changing interactions. The table reports, for each cell type (rows), the number of interactions classified in each category, separately for as-source and as-target interactions
| As-source interactions | As-target interactions | |||||||
|---|---|---|---|---|---|---|---|---|
| Absent | Gained | Lost | Stable | Absent | Gained | Lost | Stable | |
| α-cells | 1821 | 319 | 234 | 1019 | 1847 | 331 | 296 | 805 |
| β-cells | 1924 | 322 | 180 | 967 | 1908 | 294 | 239 | 838 |
| δ-cells | 1763 | 438 | 305 | 887 | 1594 | 397 | 450 | 838 |
| Acinar cells | 1633 | 464 | 316 | 980 | 1601 | 466 | 238 | 974 |
| Ductal cells | 1267 | 505 | 309 | 1312 | 1115 | 542 | 200 | 1422 |
| PP cells | 1672 | 463 | 328 | 930 | 1319 | 491 | 320 | 1149 |
| Stellate cells | 920 | 533 | 292 | 1648 | 818 | 523 | 221 | 1717 |
Focus on the beta cell interactome
Considering the centrality of beta cells for the development and progression of T2D, we focused on the subset of interactions in which beta cells are involved (beta cell interactome) to gain insights into the direct perturbations of beta cell interactome.
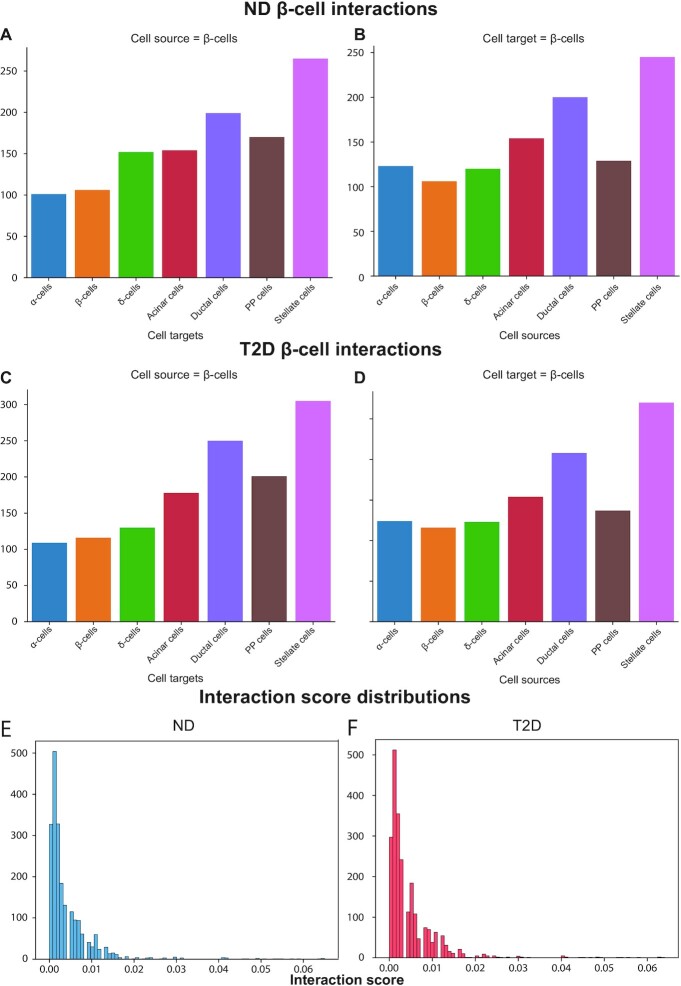
Looking at the number of interactions, the beta cells display: 1147 as-source and 1077 as-target interactions in ND; 1289 as-source and 1132 as-target interactions in T2D. A breakdown of which cell types engage in these interactions with beta cells, both as-source and as-target, showed that the higher number of interactions occur with non-endocrine cells, with stellate and ductal cells displaying the highest number of interactions (Figure 6A and B). This pattern is also observed in the T2D interactome (Figure 6C and D). These interactions have been ranked on the basis of the Interaction Score (IS), a metric reported by CellPhoneDB that quantifies the average expression of ligands and receptors. Looking at the distribution of such scores for the beta cell interactions revealed that both in ND and T2D interactomes most of these connections had a low score (Figure 6E and F). The same behaviour is observed when looking at the complete interactome (that is, not limited to the subset of beta cell interactions).
Figure 6.
Beta cell interactions in ND and T2D. (A, B) The barplots report the number of interactions in the ND interactome between the beta cells, either as source (A) or as target (B), and all the cell types. (C, D) The barplots report the number of interactions in the T2D interactome between the beta cells, either as source (C) or as target (D), and all the cell types. (E, F) The histograms report the distribution of the interaction scores in the ND (E) and T2D (F).
The beta cell interactions with high score values (HSI, percentile ≥ 99), 23 and 27 in the ND and T2D beta cell interactome, respectively, reflect the ‘important’ interactions of beta cells within the islet: among the top interactions in the ND interactome we found INS-INSR, GCG-GCGR and SST-SSTR. Interestingly, the interactions between Insulin Degrading Enzyme (IDE), a zinc metalloprotease that degrades different bioactive peptides, and insulin, amylin and glucagon are all extremely high-scoring. The HSIs were compared between T2D and ND: considering the beta cell as-target HSIs, there were 8 interactions in ND, all also found in T2D, whereas one autocrine interaction between beta cells and beta cells, involving IAPP and IDE, was present only in T2D. Considering the beta cell as-source HSIs, there were 17 interactions shared between ND and T2D, whereas T2D presented four interactions between IAPP and IDE with alpha, beta, delta and PP cells. The observed differences were due to the percentile threshold used to define HSI: comparing the IAPP-IDE interaction scores between ND and T2D, there was no significant difference.
Although overall beta cells display more interactions with non-endocrine than endocrine cell types, the beta cell high-scoring as-target interactions displayed a significant enrichment of endocrine cells compared with the other interactions, whereas the as-source interactions did not display any significant enrichment for both ND and T2D beta cell interactomes.
Finally, we analysed the interactions changing (from ND to T2D) in beta cells to find genes with functional implications. There were 566 gained interactions (Supplementary Table S10), 297 of which as-source and 269 as-target. The lost interactions were 389, with 165 as-source and 224 as-target interactions. The interaction score was used to rank interactions, allowing to focus the search of interactions related to beta cell function in the top ranks. Consistent with previous results, the score distribution of these interactions had a power-law behavior, i.e. there was a low number of changing interactions with a high score (Supplementary Figure S2). Looking at the interactions with extremely high scores (95th percentile and above) there were 59 high-score changing interactions, 32 of which were gained (18 as-source, 14 as-target) and 27 lost (15 as-source and 12 as-target). Interestingly, the gained and lost interactions exhibited an opposing trend regarding cell type partners: of the 32 gained interactions, the majority (19) involved non-endocrine cell types, whereas most (17) of the 27 lost interactions involved endocrine cells, the majority of which (12) were delta cells. Finally, we found highly ranked interactions that were previously associated with beta cell function: the highest-score lost interaction involved the ligand RPS19, produced by delta cells, and the beta cell receptor of the C5 protein, C5aR1, which was demonstrated to enhance glucose-stimulated insulin secretion and protect from apoptosis (26). Most of the lost as-source interactions occurred between beta and delta cells (9), 8 of which involved FGFR1 and 1 EGFR, probably with implications for delta cell survival.
DISCUSSION
The present study is the first attempt to chart the global landscape of islet intercellular interactions, with a focus on alterations in T2D. Importantly, the presently used methods can be applied to investigate other conditions, such as T1D or islets exposed to novel therapeutic agents. The approach can also be expanded by the inclusion of methods already used to gain insight into inter-cellular interactions (27,28) to obtain a more accurate description of the interactome and to facilitate the validation of results obtained with single-cell transcriptomics. The provided results represent a novel platform to generate hypotheses and to drive the design of future experiments focusing on interactions of interest for both islet function and dysfunction. Additional information regarding a specific interaction can be obtained from the CellPhoneDB database (12), which provides cross-links with a number of databases to facilitate biological insights at the molecular and cellular levels.
Analysing the patterns of cell type connectivity, we found that the endocrine/non-endocrine classification is recapitulated by interaction patterns, as beta cells display most similarities with delta cells when acting as signal senders (as-source) and with alpha cells when acting as signal recipients (as-target). On the other side of the spectrum, alpha cells showed a unique pattern of interactions compared with other endocrine and non-endocrine cell types and, interestingly, appear as the cell type with the highest number of interactions. Indeed, it was described that pancreatic islets can contain specific sub-populations of non-endocrine cells, including the so-called islet stellate cells (24), that can interact with endocrine cells and affect their function. Increasing evidence associates stellate cells with islet fibrosis (29–32), pancreatitis and T2D progression (33). The intercellular interactions that were the aim of our study are relevant in this context, as the mechanisms linking stellate cells to amyloid deposits actually encompass intercellular signaling via the renin-angiotensin system (31,34). The interactome can therefore be useful to focus on specific cell types aiming at validation or discovery of interactions to be associated with pathologies.
We also modeled interactions using network-based methods to derive insights into emerging properties of ligands and receptors, aiming in particular at identifying which molecular actors were most important for overall network connectivity. Considering the genes with the highest centrality values (percentile > = 95%), 5 were found across all metrics tested: EGFR, FAM3C, FGFR1, FGFR2 and PVRL3. Of these, EGFR, FGFR1 and FGFR2 encode growth factor receptors that were previously associated with islet development and survival (35,36). Accordingly, inactivation of these genes in animal models resulted in decreased beta cell mass and diabetes (22,37,38). Although the other two genes were not previously described in islet biology, they may have pathophysiological relevance: the protein encoded by FAM3C, also known as ILEI, has a protective role in Alzheimer's disease by destabilizing the amyloid-β precursor (39), facilitates cancer progression by favoring epithelial-mesenchymal transition (40), and is involved in glucose and lipid homeostasis in liver (41). Regarding PVRL3, also known as NECTIN3, there is little information: this gene encodes a ligand that interacts with TIGIT to suppress T-cell activity (42), but we found no obvious connection with T2D.
Among the other identified genes, namely CEACAM6, CEACAM1, SORT1, JAG2, GDNF, GRN, DSG2, WNT5A, ERBB4, CD209 and HBEGF, some were already described in relation with islet physiology. HBEGF enables glucose-stimulated beta cell proliferation in rat models (43), while signaling by WNT5A stimulates insulin secretion (44); SORT1 encodes the receptor sortilin that binds with high affinity to neurotensin (45), a regulator involved with insulin secretion and beta cell survival (46,47). Besides playing a role during islet development (36), the ERBB receptor kinases mediate the action of betacellulin to promote neogenesis of endocrine cells (48); the GDNF ligand promotes islet survival in vitro (49). Finally, some of these genes (FGFR2, CEACAM6, GRN, DSG2) have been implicated in cancer development (50–53). This latter finding probably reflects the general importance of these communication mechanisms for tissue organization as they all stand at the crossroad between cell proliferation and death. As these genes are potentially relevant for islet homeostasis, they could play a role in T2D; for instance indirect associations have been reported between GRN and T2D (54–56).
Finally, by comparing the ND and T2D beta cell interactomes we highlighted the impact of disease on interaction patterns. The clustering analysis of interaction profiles highlighted how, for the as-source interactions, cells clustered first according to endocrine/non-endocrine cell types, then according to the disease variable, and then according to cell types, indicating that T2D affected considerably the as-source interactions of all cell types. The as-target interactome was diversely affected, with some cell types (alpha cells) having similar interactions in both conditions, whereas for others (delta cells) interaction profiles were remarkably different. These findings suggest that T2D diversely affects signal reception (as-target interactions) in endocrine cells with some, e.g. alpha cells, being less altered than others, e.g. delta cells.
Regarding the changing interactions in beta cells, we found the score provided by CellPhoneDB to be a good proxy for assessing the importance of interactions since the interactions with the highest ranks recapitulate the major mechanisms described for islet cells (57). Therefore, we ranked changing interactions in T2D using this metric to highlight changes more likely to have strong implications for pathophysiology. Among these, the lost interaction with the highest score, the C5AR1-RPS19 interaction between delta and beta cells, has been linked with increased insulin secretion and apoptosis protection in human islets (26). Other high-ranked interactions not previously associated with islet function might represent good candidates to be evaluated in future studies.
In conclusion, this work represents the first attempt to comprehensively define intercellular interactions within the human islet. Taking a top-down approach, our study provides the basis to generate hypotheses to be tested in future studies, including the discovery of ligands and receptors that are likely to have important implications for T2D pathophysiology.
Limitations of the study
Despite its novelty, this study has a number of shortcomings and thus should be seen as a first step towards an increased understanding of the complex network regulating the islet system. A first limitation, that is intrinsically linked to systems-wide studies, lies in the fact that our results provide a comprehensive description of the whole system features rather than a detailed understanding of the biological relevance of the individual components of the system. In this sense, future bottom-up studies, including specific perturbations in the system (e.g. deletion or inactivation of specific interactions) will prove crucial to validate our results and assess the roles of the individual interactions for the islet function. Secondly, although CellPhoneDB represents one of the most comprehensive databases of intercellular interactions, it was not specifically designed for pancreatic islets and it is thus quite possible that some of the specific interactions occurring in the islet are absent in CellPhoneDB. Therefore, it will be important to replicate this work as soon as the knowledge of the islet interactomes expands. Finally, single-cell technologies are continually improving and it is very likely that future characterizations of the human islet transcriptome, detecting a larger number of genes will deliver a more accurate picture of the interactome.
DATA AVAILABILITY
The single-cell dataset used in this study, as a H5AD file, is available on the Github repository scPanBetaT2D (https://github.com/EBosi/scPanBetaT2D). The files produced in this work are available as supplementary files or tables. The code used to generate the results as well as the intermediate files to replicate the analyses are available on the Github repository IsletInteractome (https://github.com/EBosi/IsletInteractome).
Supplementary Material
Contributor Information
Emanuele Bosi, Department of Experimental and Clinical Medicine, Pancreatic islets laboratory, University of Pisa, Pisa, Italy; Department of Earth, Environmental and Life Sciences (DISTAV), University of Genoa, Genoa, Italy.
Lorella Marselli, Department of Experimental and Clinical Medicine, Pancreatic islets laboratory, University of Pisa, Pisa, Italy.
Mara Suleiman, Department of Experimental and Clinical Medicine, Pancreatic islets laboratory, University of Pisa, Pisa, Italy.
Marta Tesi, Department of Experimental and Clinical Medicine, Pancreatic islets laboratory, University of Pisa, Pisa, Italy.
Carmela De Luca, Department of Experimental and Clinical Medicine, Pancreatic islets laboratory, University of Pisa, Pisa, Italy.
Silvia Del Guerra, Department of Experimental and Clinical Medicine, Pancreatic islets laboratory, University of Pisa, Pisa, Italy.
Miriam Cnop, ULB Center for Diabetes Research, Université Libre de Bruxelles, Brussels, Belgium; Division of Endocrinology, Erasmus Hospital, Université Libre de Bruxelles, Brussels, Belgium.
Decio L Eizirik, ULB Center for Diabetes Research, Université Libre de Bruxelles, Brussels, Belgium.
Piero Marchetti, Department of Experimental and Clinical Medicine, Pancreatic islets laboratory, University of Pisa, Pisa, Italy.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
M.C. is funded by Fonds National de la Recherche Scientifique; Walloon Region SPW-EER Win2Wal project BetaSource, Belgium; Francophone Foundation for Diabetes Research (FFRD, that is sponsored by the French Diabetes Federation, Abbott, Eli Lilly, Merck Sharp & Dohme and Novo Nordisk), France; D.L.E. acknowledges the support of a grant from the Welbio-FNRS (Fonds National de la Recherche Scientifique), Belgium; Dutch Diabetes Fonds (DDFR), Holland; D.L.E. and M.C. are supported by the Brussels Capital Region Innoviris project DiaType; D.L.E., M.C., P.M. and L.M. receive support from the Innovative Medicines Initiative 2 Joint Undertaking [115,797 (INNODIA) and 945,268 (INNODIA HARVEST)]; these Joint Undertakings receive support from the Union's Horizon 2020 research and innovation program and ‘EFPIA’, ‘JDRF’ and ‘The Leona M. and Harry B. Helmsley Charitable Trust’; E.B., P.M. and L.M. are supported by the Italian Ministry of University and Research, PRIN 2017 [2017KAM2R5_005].
Conflict of interest statement. None declared.
REFERENCES
- 1. Walker J.T., Saunders D.C., Brissova M., Powers A.C.. The human islet: mini-organ with mega-impact. Endocr. Rev. 2021; 42:605–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marchetti P., Suleiman M., De Luca C., Baronti W., Bosi E., Tesi M., Marselli L.. A direct look at the dysfunction and pathology of the β cells in human type 2 diabetes. Semin. Cell Dev. Biol. 2020; 103:83–93. [DOI] [PubMed] [Google Scholar]
- 3. Cabrera O., Berman D.M., Kenyon N.S., Ricordi C., Berggren P.-O., Caicedo A.. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:2334–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brissova M., Fowler M.J., Nicholson W.E., Chu A., Hirshberg B., Harlan D.M., Powers A.C.. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J. Histochem. Cytochem. 2005; 53:1087–1097. [DOI] [PubMed] [Google Scholar]
- 5. Barreto S.G., Carati C.J., Toouli J., Saccone G.T.P.. The islet-acinar axis of the pancreas: more than just insulin. Am. J. Physiol. Gastrointest. Liver Physiol. 2010; 299:G10–G22. [DOI] [PubMed] [Google Scholar]
- 6. Zang G., Sandberg M., Carlsson P.-O., Welsh N., Jansson L., Barbu A.. Activated pancreatic stellate cells can impair pancreatic islet function in mice. Ups. J. Med. Sci. 2015; 120:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Narayanan S., Loganathan G., Dhanasekaran M., Tucker W., Patel A., Subhashree V., Mokshagundam S., Hughes M.G., Williams S.K., Balamurugan A.N.. Intra-islet endothelial cell and β-cell crosstalk: implication for islet cell transplantation. World J. Transplant. 2017; 7:117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Noguchi G.M., Huising M.O.. Integrating the inputs that shape pancreatic islet hormone release. Nat. Metab. 2019; 1:1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xin Y., Kim J., Okamoto H., Ni M., Wei Y., Adler C., Murphy A.J., Yancopoulos G.D., Lin C., Gromada J.. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metab. 2016; 24:608–615. [DOI] [PubMed] [Google Scholar]
- 10. Segerstolpe Å., Palasantza A., Eliasson P., Andersson E.-M., Andréasson A.-C., Sun X., Picelli S., Sabirsh A., Clausen M., Bjursell M.K.et al.. Single-Cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016; 24:593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lawlor N., George J., Bolisetty M., Kursawe R., Sun L., Sivakamasundari V., Kycia I., Robson P., Stitzel M.L.. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res. 2017; 27:208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R.. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020; 15:1484–1506. [DOI] [PubMed] [Google Scholar]
- 13. Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.-E., Stephenson E., Polański K., Goncalves A.et al.. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature. 2018; 563:347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumar M.P., Du J., Lagoudas G., Jiao Y., Sawyer A., Drummond D.C., Lauffenburger D.A., Raue A.. Analysis of single-cell RNA-Seq identifies cell-cell communication associated with tumor characteristics. Cell Rep. 2018; 25:1458–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T.et al.. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020; 38:970–979. [DOI] [PubMed] [Google Scholar]
- 16. Bosi E., Marselli L., De Luca C., Suleiman M., Tesi M., Ibberson M., Eizirik D.L., Cnop M., Marchetti P.. Integration of single-cell datasets reveals novel transcriptomic signatures of β-cells in human type 2 diabetes. NAR Genom Bioinform. 2020; 2:lqaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolf F.A., Angerer P., Theis F.J.. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018; 19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaestner K.H., Powers A.C., Naji A., Consortium HPAP, Atkinson M.A. NIH initiative to improve understanding of the pancreas, islet, and autoimmunity in type 1 diabetes: the human pancreas analysis program (HPAP). Diabetes. 2019; 68:1394–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bosi E., Marchetti P., Rutter G.A., Eizirik D.L.. Human alpha cell transcriptomic signatures of types 1 and 2 diabetes highlight disease-specific dysfunction pathways. Iscience. 2022; 25:105056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hagberg A., Swart P., Chult S.D.. Exploring network structure, dynamics, and function using networkx los alamos national lab (LANL). 2008; Los Alamos, NM (United States).
- 21. Miettinen P., Ormio P., Hakonen E., Banerjee M., Otonkoski T.. EGF receptor in pancreatic beta-cell mass regulation. Biochem. Soc. Trans. 2008; 36:280–285. [DOI] [PubMed] [Google Scholar]
- 22. Hart A.W., Baeza N., Apelqvist A., Edlund H.. Attenuation of FGF signalling in mouse beta-cells leads to diabetes. Nature. 2000; 408:864–868. [DOI] [PubMed] [Google Scholar]
- 23. Zhang X., Yang W., Wang J., Meng Y., Guan Y., Yang J.. FAM3 gene family: a promising therapeutical target for NAFLD and type 2 diabetes. Metabolism. 2018; 81:71–82. [DOI] [PubMed] [Google Scholar]
- 24. Xu W., Jones P.M., Geng H., Li R., Liu X., Li Y., Lv Q., Liu Y., Wang J., Wang X.et al.. Islet stellate cells regulate insulin secretion via wnt5a in min6 cells. Int. J. Endocrinol. 2020; 2020:4708132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barabási A.-L. Network science. Philos. Trans. A Math. Phys. Eng. Sci. 2013; 371:20120375. [DOI] [PubMed] [Google Scholar]
- 26. Atanes P., Ruz-Maldonado I., Pingitore A., Hawkes R., Liu B., Zhao M., Huang G.C., Persaud S.J., Amisten S.. C3aR and C5aR1 act as key regulators of human and mouse β-cell function. Cell. Mol. Life Sci. 2018; 75:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nishida-Aoki N., Gujral T.S.. Emerging approaches to study cell-cell interactions in tumor microenvironment. Oncotarget. 2019; 10:785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bechtel T.J., Reyes-Robles T., Fadeyi O.O., Oslund R.C.. Strategies for monitoring cell-cell interactions. Nat. Chem. Biol. 2021; 17:641–652. [DOI] [PubMed] [Google Scholar]
- 29. Omary M.B., Lugea A., Lowe A.W., Pandol S.J.. The pancreatic stellate cell: a star on the rise in pancreatic diseases. J. Clin. Invest. 2007; 117:50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Apte M.V., Haber P.S., Darby S.J., Rodgers S.C., McCaughan G.W., Korsten M.A., Pirola R.C., Wilson J.S.. Pancreatic stellate cells are activated by proinflammatory cytokines: implications for pancreatic fibrogenesis. Gut. 1999; 44:534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ko S.-H., Hong O.-K., Kim J.-W., Ahn Y.-B., Song K.-H., Cha B.-Y., Son H.-Y., Kim M.-J., Jeong I.-K., Yoon K.-H.. High glucose increases extracellular matrix production in pancreatic stellate cells by activating the renin-angiotensin system. J. Cell. Biochem. 2006; 98:343–355. [DOI] [PubMed] [Google Scholar]
- 32. Yang Y., Kim J.-W., Park H.-S., Lee E.-Y., Yoon K.-H.. Pancreatic stellate cells in the islets as a novel target to preserve the pancreatic β-cell mass and function. J. Diabetes Investig. 2020; 11:268–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jurgens C.A., Toukatly M.N., Fligner C.L., Udayasankar J., Subramanian S.L., Zraika S., Aston-Mourney K., Carr D.B., Westermark P., Westermark G.T.et al.. β-cell loss and β-cell apoptosis in human type 2 diabetes are related to islet amyloid deposition. Am. J. Pathol. 2011; 178:2632–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ko S.-H., Kwon H.-S., Kim S.-R., Moon S.-D., Ahn Y.-B., Song K.-H., Son H.-S., Cha B.-Y., Lee K.-W., Son H.-Y.et al.. Ramipril treatment suppresses islet fibrosis in otsuka long–evans tokushima fatty rats. Biochem. Biophys. Res. Commun. 2004; 316:114–122. [DOI] [PubMed] [Google Scholar]
- 35. Arany E., Hill D.J.. Ontogeny of fibroblast growth factors in the early development of the rat endocrine pancreas. Pediatr. Res. 2000; 48:389–403. [DOI] [PubMed] [Google Scholar]
- 36. Miettinen P.J., Huotari M., Koivisto T., Ustinov J., Palgi J., Rasilainen S., Lehtonen E., Keski-Oja J., Otonkoski T.. Impaired migration and delayed differentiation of pancreatic islet cells in mice lacking EGF-receptors. Development. 2000; 127:2617–2627. [DOI] [PubMed] [Google Scholar]
- 37. Miettinen P.J., Ustinov J., Ormio P., Gao R., Palgi J., Hakonen E., Juntti-Berggren L., Berggren P.-O., Otonkoski T.. Downregulation of EGF receptor signaling in pancreatic islets causes diabetes due to impaired postnatal beta-cell growth. Diabetes. 2006; 55:3299–3308. [DOI] [PubMed] [Google Scholar]
- 38. Hakonen E., Ustinov J., Eizirik D.L., Sariola H., Miettinen P.J., Otonkoski T.. In vivo activation of the PI3K-Akt pathway in mouse beta cells by the EGFR mutation L858R protects against diabetes. Diabetologia. 2014; 57:970–979. [DOI] [PubMed] [Google Scholar]
- 39. Hasegawa H., Liu L., Tooyama I., Murayama S., Nishimura M.. The FAM3 superfamily member ILEI ameliorates alzheimer's disease-like pathology by destabilizing the penultimate amyloid-β precursor. Nat. Commun. 2014; 5:3917. [DOI] [PubMed] [Google Scholar]
- 40. Waerner T., Alacakaptan M., Tamir I., Oberauer R., Gal A., Brabletz T., Schreiber M., Jechlinger M., Beug H.. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell. 2006; 10:227–239. [DOI] [PubMed] [Google Scholar]
- 41. Chen Z., Ding L., Yang W., Wang J., Chen L., Chang Y., Geng B., Cui Q., Guan Y., Yang J.. Hepatic activation of the FAM3C-HSF1-CaM pathway attenuates hyperglycemia of obese diabetic mice. Diabetes. 2017; 66:1185–1197. [DOI] [PubMed] [Google Scholar]
- 42. Yu X., Harden K., Gonzalez L.C., Francesco M., Chiang E., Irving B., Tom I., Ivelja S., Refino C.J., Clark H.et al.. The surface protein TIGIT suppresses t cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 2009; 10:48–57. [DOI] [PubMed] [Google Scholar]
- 43. Maachi H., Fergusson G., Ethier M., Brill G.N., Katz L.S., Honig L.B., Metukuri M.R., Scott D.K., Ghislain J., Poitout V.. HB-EGF signaling is required for glucose-induced pancreatic β-Cell proliferation in rats. Diabetes. 2020; 69:369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu W., Liang J., Geng H.F., Lu J., Li R., Wang X.L., Lv Q., Liu Y., Wang J., Liu X.K.et al.. Wingless-Type MMTV integration site family member 5a is a key secreted islet stellate cell-derived product that regulates islet function. Int. J. Endocrinol. 2019; 2019:7870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blondeau N., Béraud-Dufour S., Lebrun P., Hivelin C., Coppola T.. Sortilin in glucose homeostasis: from accessory protein to key player?. Front. Pharmacol. 2018; 9:1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Béraud-Dufour S., Abderrahmani A., Noel J., Brau F., Waeber G., Mazella J., Coppola T.. Neurotensin is a regulator of insulin secretion in pancreatic beta-cells. Int. J. Biochem. Cell Biol. 2010; 42:1681–1688. [DOI] [PubMed] [Google Scholar]
- 47. Béraud-Dufour S., Coppola T., Massa F., Mazella J.. Neurotensin receptor-2 and -3 are crucial for the anti-apoptotic effect of neurotensin on pancreatic beta-TC3 cells. Int. J. Biochem. Cell Biol. 2009; 41:2398–2402. [DOI] [PubMed] [Google Scholar]
- 48. Lee Y.-S., Song G.J., Jun H.-S.. Betacellulin-Induced α-Cell proliferation is mediated by erbb3 and erbb4, and may contribute to β-Cell regeneration. Front. Cell Dev. Biol. 2020; 8:605110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mwangi S.M., Usta Y., Shahnavaz N., Joseph I., Avila J., Cano J., Chetty V.K., Larsen C.P., Sitaraman S.V., Srinivasan S.. Glial cell line-derived neurotrophic factor enhances human islet posttransplantation survival. Transplantation. 2011; 92:745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Szybowska P., Kostas M., Wesche J., Wiedlocha A., Haugsten E.M.. Cancer mutations in FGFR2 prevent a negative feedback loop mediated by the ERK1/2 pathway. Cells. 2019; 8:518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pandey R., Zhou M., Islam S., Chen B., Barker N.K., Langlais P., Srivastava A., Luo M., Cooke L.S., Weterings E.et al.. Carcinoembryonic antigen cell adhesion molecule 6 (CEACAM6) in pancreatic ductal adenocarcinoma (PDA): an integrative analysis of a novel therapeutic target. Sci. Rep. 2019; 9:18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bateman A., Bennett H.P.J.. The granulin gene family: from cancer to dementia. Bioessays. 2009; 31:1245–1254. [DOI] [PubMed] [Google Scholar]
- 53. Zhou G., Yang L., Gray A., Srivastava A.K., Li C., Zhang G., Cui T.. The role of desmosomes in carcinogenesis. Onco. Targets. Ther. 2017; 10:4059–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fradin D., Heath S., Lathrop M., Bougnères P.. Quantitative trait loci for fasting glucose in young europeans replicate previous findings for type 2 diabetes in 2q23-24 and other locations. Diabetes. 2007; 56:1742–1745. [DOI] [PubMed] [Google Scholar]
- 55. Qu H., Deng H., Hu Z.. Plasma progranulin concentrations are increased in patients with type 2 diabetes and obesity and correlated with insulin resistance. Mediators Inflamm. 2013; 2013:360190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tönjes A., Fasshauer M., Kratzsch J., Stumvoll M., Blüher M.. Adipokine pattern in subjects with impaired fasting glucose and impaired glucose tolerance in comparison to normal glucose tolerance and diabetes. PLoS One. 2010; 5:e13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huising M.O. Paracrine regulation of insulin secretion. Diabetologia. 2020; 63:2057–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single-cell dataset used in this study, as a H5AD file, is available on the Github repository scPanBetaT2D (https://github.com/EBosi/scPanBetaT2D). The files produced in this work are available as supplementary files or tables. The code used to generate the results as well as the intermediate files to replicate the analyses are available on the Github repository IsletInteractome (https://github.com/EBosi/IsletInteractome).