Abstract
Thousands of human gene regulatory enhancers are composed of sequences with multiple evolutionary origins. These evolutionarily “complex” enhancers consist of older “core” sequences and younger “derived” sequences. However, the functional relationship between the sequences of different evolutionary origins within complex enhancers is poorly understood. We evaluated the function, selective pressures, and sequence variation across core and derived components of human complex enhancers. We find that both components are older than expected from the genomic background, and complex enhancers are enriched for core and derived sequences of similar evolutionary ages. Both components show strong evidence of biochemical activity in massively parallel report assays. However, core and derived sequences have distinct transcription factor (TF)-binding preferences that are largely similar across evolutionary origins. As expected, given these signatures of function, both core and derived sequences have substantial evidence of purifying selection. Nonetheless, derived sequences exhibit weaker purifying selection than adjacent cores. Derived sequences also tolerate more common genetic variation and are enriched compared with cores for expression quantitative trait loci associated with gene expression variability in human populations. In conclusion, both core and derived sequences have strong evidence of gene regulatory function, but derived sequences have distinct constraint profiles, TF-binding preferences, and tolerance to variation compared with cores. We propose that the step-wise integration of younger derived with older core sequences has generated regulatory substrates with robust activity and the potential for functional variation. Our analyses demonstrate that synthesizing study of enhancer evolution and function can aid interpretation of regulatory sequence activity and functional variation across human populations.
Keywords: functional genomics, gene regulation, evolution, human enhancers, genetic variation, sequence age
Significance.
Thousands of human gene regulatory enhancers are mosaics of sequences from multiple evolutionary origins, yet how these different segments combine to contribute to enhancer function is poorly understood. By dissecting their regulatory functions, transcription factor binding, constraint, and human genetic variation, we show that both older “core” and younger “derived” sequences in complex enhancers have strong evidence of gene regulatory function, but derived sequences are more likely to harbor genetic variants that influence function. Together, our results support a model in which the integration of sequences of different origins generates regulatory substrates with robust activity and the potential for functional variation.
Introduction
Enhancers are distal gene regulatory DNA sequences that modulate target gene expression in cell-type and spatiotemporal-specific contexts (Shlyueva et al. 2014). Enhancer function is mediated by the binding of transcription factors (TFs) that recognize DNA sequence motifs and interact with promoters. Changes in enhancer function are major drivers of species divergence and variation within species (Wray 2007; Sholtis and Noonan 2010; Wittkopp and Kalay 2012; Franchini and Pollard 2015; Rebeiz and Tsiantis 2017), yet the evolutionary events underlying the creation and functional evolution of sequences with enhancer activity are less understood.
Studying enhancer sequence evolution poses several challenges. First, enhancer activity turns over rapidly between mammalian species, but most sequences with current enhancer activity have ancient origins (Villar et al. 2015). Furthermore, the conservation of enhancer activity can be maintained without detectable sequence conservation (Wong et al. 2020), as has been proposed in the developmental systems drift hypothesis (True and Haag 2001). Nonetheless, several connections have been discovered between the evolutionary sequence origins and current gene regulatory functions. The age of a regulatory sequence is predictive of the genes that it likely targets, and different periods of regulatory sequence innovation have contributed to vertebrate evolution (Lowe et al. 2011). Moreover, younger mammalian neocortical enhancers are more weakly constrained, and many neocortical enhancers consist of sequences of multiple evolutionary origins (Emera et al. 2016). Underscoring the functional relevance of these evolutionary events, older sequences with gene regulatory activity are more enriched for heritability in a range of human complex traits than younger sequences with regulatory activity (Hujoel et al. 2019). These waves of regulatory change have been driven in large part by the integration of transposable elements (TEs) carrying different TF-binding sites (TFBSs) into the genome at different times (Marnetto et al. 2018).
Mammalian enhancer sequences are often composed of functional units, or modules, that bind different combinations of transcription factors (Long et al. 2016; Jindal and Farley 2021). Recent work has begun to reveal the nature of the modular organization of enhancer functions (Gotea et al. 2010; Farley et al. 2015; Long et al. 2020; Tippens et al. 2020; Wong et al. 2020). Enhancer sequences often result from the integration of different combinations of sequence over time (Emera et al. 2016; Fong and Capra 2021). However, models that synthesize the evolutionary origins of enhancer sequences with an understanding of functional modules are needed.
The potential value of integrating evolution and function to human enhancer sequences is illustrated by the utility of models of protein-coding sequence evolution. Over evolutionary time, protein-coding sequences often generate novel protein functions by integrating functional modules in different combinations. Knowledge of the evolutionary origins of different proteins and domains provides valuable context for interpreting the evolution and function of protein families (Capra et al. 2013). As a result, many statistical frameworks exist for modeling protein domain and family evolution (Stolzer et al. 2015; Forslund et al. 2019). While enhancer functional domains evolve via mechanisms distinct from those of protein domains, we anticipate that expanding knowledge of the relationship between enhancer sequence evolution and function will improve our ability to determine whether changes to specific gene regulatory sequence features produce changes in regulatory function. Thus, deeper understanding of enhancer sequence evolution will contribute valuable context for resolving gene regulatory functions of candidate disease variants of unknown significance, understanding the molecular basis for differences between species, and developing synthetic gene regulatory elements.
We recently explored how the evolutionary origins of an enhancer sequence are reflected in its functional and regulatory features, such as pleiotropy and robustness to perturbation of its biochemical activity by genetic variants (Fong and Capra 2021). We discovered that a significant fraction of enhancer sequences in diverse tissues consist of DNA from multiple evolutionary origins. These “complex” enhancers are the result of genomic integration and rearrangement events over evolutionary time. Complex enhancers are more likely to be active across multiple tissues than their more tissue-specific evolutionarily simpler counterparts. Yet, we emphasize that the term complex only refers to the evolutionary origins of the enhancer and not necessarily its function or architecture. Indeed, the relationship between the sequences of different evolutionary origins in these enhancers and the gene regulatory functions they produce is poorly understood. For example, whether the sequences from different evolutionary periods have independent gene regulatory functions is unclear in most complex enhancers.
Here, we address this gap by contrasting the evolutionary origins, functional characteristics, TF binding, selection pressures, and human genetic diversity of the oldest “core” regions and younger “derived” regions of complex enhancer sequences. We find that both core and derived regions have strong evidence of gene regulatory function, but derived regions have distinct properties in terms of their constraint profiles, TF-binding preferences, and tolerance to variation compared with cores. In addition, complex enhancers show a strong enrichment for sequences of similar evolutionary ages. Overall, our results illustrate that the combination of core and derived regions in enhancer sequences often promotes robust gene regulatory activity while providing a substrate for functional variation in humans.
Results
Enhancers are Commonly Composed of Older Core and Younger Derived Sequences
Thousands of human gene regulatory enhancers are composed of sequences with multiple evolutionary origins. Previous work classified the components of these complex enhancers into two classes—core and derived sequences (fig. 1A; Emera et al. 2016; Fong and Capra 2021). The core sequence(s) are the oldest sequences in an enhancer, and the younger sequence regions are derived. Our goal is to evaluate the function, selective pressures on, and sequence variation across these components of complex human gene regulatory enhancers genome wide (fig. 1A).
Fig. 1.
Complex enhancers consist of older core and younger derived sequences. (A) Illustration of the approach for mapping enhancer sequence ages and architectures. We quantify the age of a sequence with human enhancer activity based on the oldest MRCA in overlapping syntenic blocks from the MultiZ multiple sequence alignments of 46 vertebrates. Enhancer age is assigned as the oldest, overlapping syntenic block age. Estimates of divergence time in Ma from TimeTree (Hedges et al., 2015) are annotated in the key. (B) Autosomal transcribed enhancers from the FANTOM5 consortium (N = 30,434) were classified as having complex (multi-age) or simple (single-age) architectures. Complex enhancers were further dissected into the oldest core and younger derived sequence regions. (C) A complex developing limb bud enhancer (NC 000007.13) of SSH is located ∼1 Mb away in an intron of LMBR1 and has multiple evolutionary origins. Among 11 variants in OMIM that cause preaxial polydactyly 2 (PPD2), eight variants are in the Vertebrate core region, and three are in the Tetrapod derived region. Common variants (minor allele frequency >1% in 1,000 Genomes Project phase 3) from dbSNP (version 153) are observed only in derived regions. H3K27ac ChIP-seq peaks in H1-ESC and DNase I hypersensitive clusters from 125 cell lines in ENCODE3 are shown for context.
To illustrate the components of a complex enhancer, we dissected evolutionary origins of the zone of polarizing activity regulatory sequence (ZRS), a long-range enhancer of SHH involved in developmental limb bud formation (Lettice et al. 2017). The ZRS sequence achieves its regulatory function via multiple distinct regulatory domains (Lettice 2003; Lettice et al. 2012; Long et al. 2016). The core sequence has origins before the last common ancestor of all vertebrates, and it is flanked on both sides by multiple derived regions with origins in the ancestors of tetrapods, amniotes, and mammals. This enhancer sequence is both strongly conserved and involved in evolutionary variation in limb morphology. Loss of function variants at this locus contributed to limbless evolution in snakes (Kvon et al. 2016), whereas variants in vertebrate and tetrapod sequences are associated with preaxial polydactyly 2 (PPD2; Hill and Lettice 2013; Ushiki et al. 2021). In humans, 8 of the 11 PPD2-causing variants annotated in the Online Mendelian Inheritance in Man (OMIM) catalog are located in the Vertebrate core of the ZRS enhancer sequence, whereas three are located in Tetrapod derived regions (fig. 1C). Common variants (minor allele frequency >1% in 1,000 Genomes Projects from dbSNPv153) are observed in the younger derived amniote and mammal sequences, but not in older tetrapod and vertebrate sequences. This example illustrates that variants in both older core sequences and younger derived regions can cause human disease.
Derived Regions Constitute a Substantial Fraction of Complex Enhancer Sequences
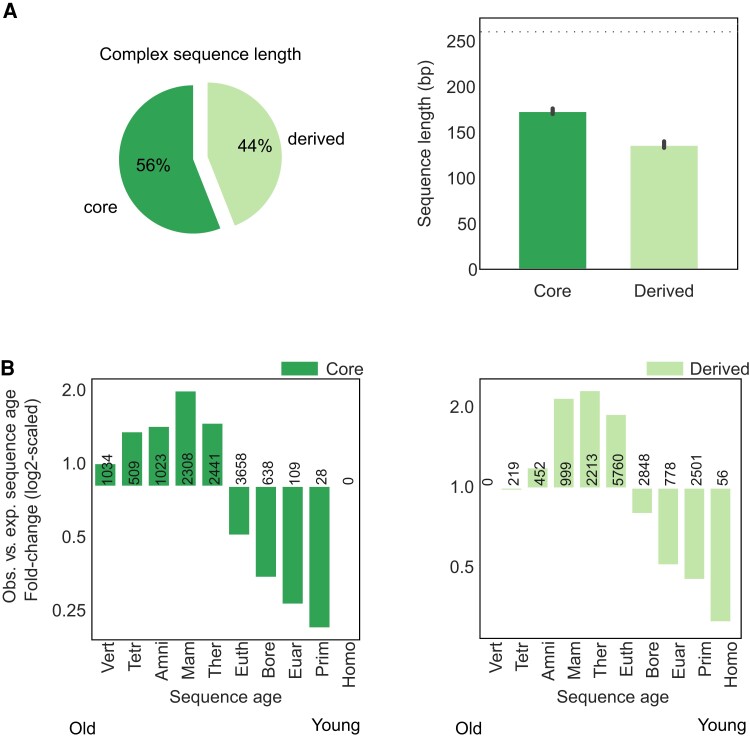
We first evaluated basic features of core and derived sequences in non-coding autosomal transcribed enhancers from 112 diverse tissues and cell samples from the FANTOM5 consortium (N = 10,686; fig. 1B). Derived regions represent 46% of the base pairs (bp) in a typical complex enhancer sequence (fig. 2A, left; median total length of 310 bp), and complex enhancers have a median of one derived region per core region (supplementary fig. S1, Supplementary Material online). However, derived regions are shorter than core regions (fig. 2A, right; median bp 136 derived vs. 174 core). To evaluate whether these patterns are specific to complex enhancer sequences or are generally true for adjacent sequences of different ages, we generated 100 non-coding region sets matched to the length and chromosome distributions of observed enhancers (Materials and Methods). We identified core and derived segments of these regions and used them to establish null distributions for comparison with the observed enhancers’ attributes. We will refer to these as “null,” “background,” or “expected” distributions.
Fig. 2.
Derived sequences are shorter than cores and older than expected from the non-coding genome. (A) Derived regions constitute 44% of complex enhancer sequences (left), but are shorter than core regions (right, median 136 bp derived vs. 174 bp core, MWU P = 4.9e-57). Both core and derived regions are shorter than simple enhancers (dashed line, median 260 bp simple, P< 2.2e-308). (B) Both core and derived sequences are enriched for older sequence ages and depleted of younger sequence ages. Per age, the log2 of the fold change of the observed core (left) and derived (right) sequence ages versus the expected proportion estimated from 100 × sets of length-, chromosome-, and architecture-matched non-coding sequences. Sample size is annotated per bar.
Derived enhancer RNA (eRNA) sequences are shorter than expected from background regions with multiple sequence ages [supplementary fig. S2, Supplementary Material online; median bp 136 observed vs. 157 expected; Mann–Whitney U test (MWU) P = 1.4e-46]. Conversely, core regions are longer than expected (median bp 174 observed vs. 143 expected; MWU P = 2.4e-73; supplementary fig. S2, Supplementary Material online). Stratifying enhancers and background regions by their core ages and repeating these comparisons yielded similar results (supplementary fig. S3, Supplementary Material online). Thus, derived sequences make up less of enhancer sequence than expected, but still contribute a substantial fraction of complex enhancer sequence and are sufficiently long to bind multiple TFs.
Both Derived and Core Regions are Older Than Expected From Matched Background Regions
Enhancer sequences are generally older than expected from the non-coding genomic background, suggesting that many have been maintained due to their function (Lowe et al. 2011; Villar et al. 2015; Emera et al. 2016; Marnetto et al. 2018; The ENCODE Project Consortium et al. 2020; Fong and Capra 2021). We expanded previous analyses of enhancer ages to consider the multiple evolutionary origins of complex enhancers. We compared the distributions of core and derived sequence ages to background regions. Core sequences are enriched for older ages (Therian ancestor and older) compared with expected core sequence ages (fig. 2B left; median age 0.30 observed vs. 0.175 expected; MWU P< 2.2e-238). Derived sequences are also enriched for older ages compared with derived regions of background sequences with matched core ages. The enrichment extends through sequences with Eutherian origins (fig. 2B right; median derived sequence age 0.175 observed vs. 0.152 expected; MWU P< 2.2e-238). These results indicate that both core and derived sequences are older than expected and suggest that both components often have constrained regulatory function.
Complex Enhancers are Enriched for Core and Derived Sequences from Consecutive Phylogenetic Branches
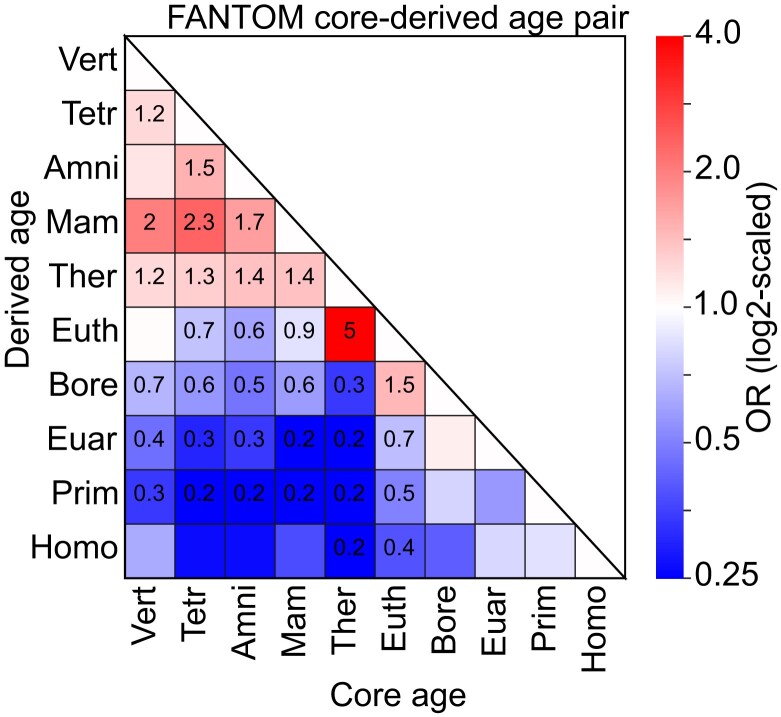
To explore whether core and derived sequences in the same complex enhancer have temporal relationships, we evaluated enrichment for sequence age combinations among observed derived and core sequence pairs. We hypothesized that derived sequence origins would likely occur soon after the origins of the corresponding core sequences.
Overall, enhancers are enriched for core and derived sequences from the consecutive phylogenetic branches compared with background complex regions (fig. 3). This suggests a preference for integration of derived sequences into older core enhancer sequences on contiguous branches, and that integration of much younger derived sequences was less tolerated by old cores. In addition, Mammalian core sequences and older are enriched for Therian derived sequences and older, but depleted of derived sequences from younger ages. The oldest complex enhancers (from the Mammalian ancestor and earlier) are enriched for derived sequences of several ancient origins (from the Therian ancestor and earlier), likely due to their very old ages. Core and derived segments of each age have sequence identities to their most distant homologs similar to background regions of the same age; this suggests that differences in sequence divergence across enhancers are unlikely to systematically bias the assignment of ages or produce these phylogenetic patterns (supplementary fig. S5, Supplementary Material online).
Fig. 3.
Complex enhancers are enriched for core and derived sequences from consecutive phylogenetic branches. For each enhancer core age, the enrichment for derived sequences of each age was measured against the expectation from core-age-matched shuffled sequences using Fisher's exact test. The boxes are colored according to the log2 of the corresponding ORs. Text in a box indicates significant enrichment (OR>1) or depletion (OR<1) after controlling the FDR at 0.05 with the Benjamini–Hochberg procedure. The oldest complex enhancers (pre-placental mammals) are enriched for older derived sequences. Outside of the oldest enhancers, there is consistent significant depletion for complex enhancers with core and derived segments with origins on non-consecutive phylogenetic branches.
These results indicate that the pairing of core and derived sequences within complex enhancers is not random with respect to their origins and that evolution favors the step-wise addition of derived sequences that are near in age to the core sequence.
Derived Sequences have Higher TFBS Density than Cores
Transcription factor binding at enhancer sequences is required for gene regulation, but the relative contributions of core and derived sequences to TF recruitment in complex enhancer sequences is not known. Some derived regions may be non-functional sequences flanking functional enhancer cores that are identified due to the limited resolution of enhancer assays. Alternatively, derived sequences could bind TFs essential for the proper function of the enhancer in specific contexts.
To evaluate the role of derived sequences in binding TFs, we leveraged the ENCODE project's deep characterization of TFBSs and enhancers in HepG2 and K562 cells: 119 and 249 TF chromatin immuno-precipitation sequencing (ChIP-seq) assays and previously identified candidate cis-regulatory elements (cCREs) with enhancer-like signatures based on DNase I hypersensitivity, CCCTC-binding factor (CTCF), and histone mark ChIP-seq assays (The ENCODE Project Consortium et al. 2020). We first confirmed that our findings on complex HepG2 and K562 enhancer architectures are consistent with those in FANTOM5 (supplementary figs. S6 and S9, Supplementary Material online).
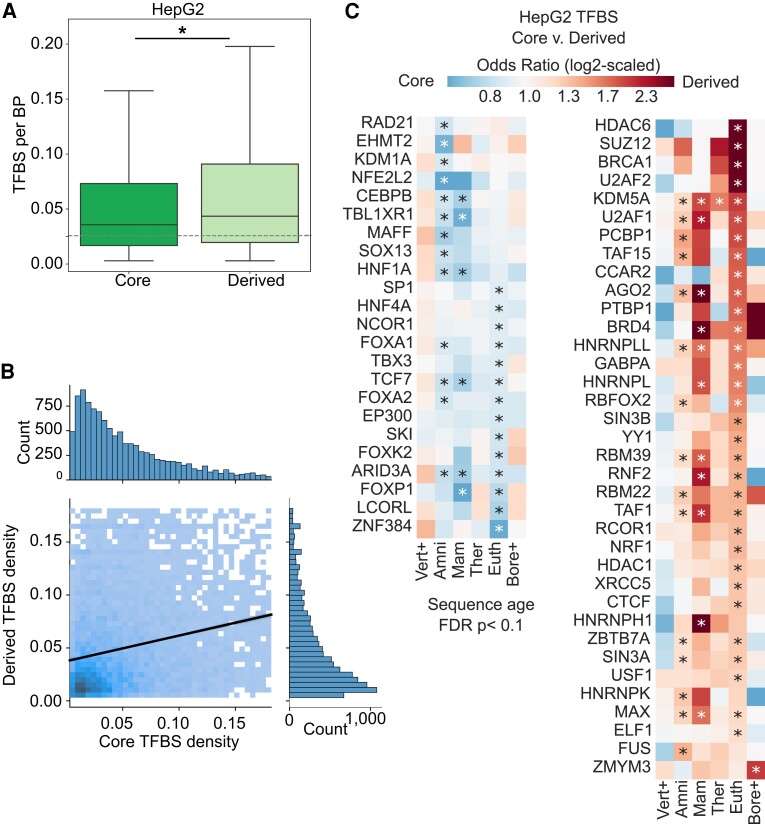
We then quantified TFBS density and enrichment patterns in core and derived regions of these enhancers. In complex HepG2 enhancers, we observe that 46% of derived regions bind TFs compared with 67% of core regions and 87% of simple HepG2 enhancers (supplementary fig. S20, Supplementary Material online). A similar trend was observed in K562 complex enhancers, where 59% of derived, 79% of core, and 93% of simple regions bind TFs. We note that we have better power to detect TFBS in K562 cells because more ChIP-seq assays have been performed in that cell model (249 K562 vs. 119 HepG2 ChIP-seq assays). Complex enhancer regions with no evidence of TF binding occur at similar frequencies across ages for both HepG2 and K562 cells, suggesting that TF-binding evidence is independent of enhancer sequence age (supplementary fig. S7, Supplementary Material online).
In complex HepG2 enhancers with bound TFs, derived regions have higher TFBS densities compared with core regions and simple enhancers (fig. 4A; median 4.3 binding sites/100 bp in derived regions vs. 3.6 binding sites/100 bp in core regions, MWU P = 1.1e-68). We observed a similar trend in complex K562 enhancers (supplementary fig. S10A, Supplementary Material online; median 7.4 binding sites/100 bp in derived regions vs. 6.4 binding sites/100 bp in core regions, MWU P = 3.5e-52). This trend of higher derived region TFBS density is consistent across enhancers of different ages (supplementary fig. S8, Supplementary Material online), suggesting that derived sequences bind TFs and have higher TFBS densities than core sequences across evolutionary ages. Thus, derived sequences have a higher density of assayed TFBSs when a binding site is present, but they are less likely to be bound by a TF than core segments overall.
Fig. 4.
Derived regions have high TFBS densities and bind different transcription factors compared with core regions. (A) Derived regions in HepG2 complex enhancers have higher TFBS densities (defined from ENCODE ChIP-seq data) than core regions (mean 0.043 derived vs. 0.036 core TFBS/bp; MWU P = 1.1e-68). However, derived regions are more likely to have no TFs bound than core regions (supplementary fig. S20, Supplementary Material online). Core and derived regions both have higher TFBS density than simple enhancers (dashed line; 0.026 TFBS/bp). This analysis includes complex enhancers with evidence of TF binding in either core or derived regions (n = 20,263 total, n = 20,210 derived, and n = 19,957 core). Asterisks represent p-value < 0.05. (B) TFBS density is positively correlated between core-derived sequence pairs within complex enhancers with evidence of TF binding in both regions (N = 11,899). Color intensity represents the density of core-derived pairs, and the black line is a linear regression fit (slope = 0.23, intercept = 0.04, r = 0.24, P = 5.1e-140); outliers (>95th percentile) are not plotted for ease of visualization. (C) Derived and core regions of the same age are enriched for binding of different TFs. Enrichment patterns for TFs are generally consistent across ages. TFBS enrichment for each age was tested using Fisher's exact test; only TFs with at least one significant enrichment (FDR < 0.1) are shown. Vertebrate, Sarcopterygii, and Tetrapod enhancer ancestors were grouped into “Vert+.” Boreotherian, Euarchontoglires, and Primate enhancer ancestors were grouped into “Bore+.” Asterisks represent significance at a FDR < 0.1.
Next, we quantified the relationship of TFBS density within core and derived segments of the same complex enhancer. Among HepG2 enhancer sequences with bound TFs in both core and derived sequences (N = 11,899), TFBS density is positively correlated between the core and derived regions (fig. 4B; linear regression slope = 0.23, intercept = 0.04, r = 0.24, P = 5.1e-140). We observed a similar positive correlation in K562 cells (supplementary fig. S10, Supplementary Material online; linear regression slope = 0.39, intercept = 0.056, r = 0.39, P = 0.0, stderr = 0.008). Relaxing our criteria to include core and derived sequences with no evidence of TF binding, we still observe that core and derived density within a single enhancer sequence is positively correlated (supplementary fig. S11, Supplementary Material online). These results show that TFBS density is overall positively correlated in adjacent core and derived regions, and that when bound, derived sequences have a higher TFBS density.
Core and Derived Sequences are Enriched for Distinct TFBS across Ages
Given the differences in TF-binding probability and density between core and derived regions, we hypothesized that regions might also exhibit different TF preferences. Indeed, we found that derived and core HepG2 enhancer regions are enriched for binding of distinct TFs (fig. 4C). Core regions are enriched for the binding of 23 different TFs in at least one age, and derived regions are enriched for the binding of 36 TFs in at least one age. Furthermore, many these TFs are consistently enriched in derived or core regions across multiple sequence ages, suggesting that specific TFs prefer binding core or derived sequence contexts.
We tested these conclusions in another deeply characterized ENCODE cell line, K562, and found similar patterns (supplementary fig. S9, Supplementary Material online), including higher TFBS density in derived sequences and TF-DNA binding biases in core and derived sequences (supplementary fig. S10, Supplementary Material online). TFs specific to core and derived sequences were unique among HepG2 and K562 enhancers, suggesting that core and derived sequence evolution is cell-type specific. Overall, these results indicate that many derived regions have distinct TF-binding partners from their associated cores.
Gene ontology (GO) annotation enrichment analyses did not identify strong specific functional enrichment among TFs with binding preferences for core or derived regions. No GO annotations were enriched among TFs with a preference for binding derived sequences at any age. However, core sequence TFs with preferences for the Amniote and Eutherian ancestors are enriched for “regulation of transcription by RNA polymerase II” [GO:0006357, derived vs. core odds ratio (OR) = 0.13, P = 0.03 for Eutherian and OR = 0.08 P = 0.04 for Amniote sequences, false discovery rate (FDR) < 10%]. This suggests that core TFs are enriched for factors that recruit the RNA polymerase II machinery needed to initiate transcription, whereas derived TFs are depleted and may instead diversify transcriptional activity.
TFBSs vary in their sequence specificity and robustness to mutation (Payne and Wagner, 2014). Thus, we explored whether differences in the TFs enriched in core versus derived regions could lead to differences in constraint. We compared the sequence specificity of each TF's motif (as measured by the relative entropy from the genomic background) between those with enrichment for core versus derived segments. Binding motifs for TFs significantly enriched in derived sequences have higher sequence specificity than TFs enriched in cores in both HepG2 and K562 cell lines (supplementary fig. S13, Supplementary Material online). Thus, differences in the sequence preferences of specific TFs are unlikely to produce substantial differences in constraint on core versus derived sequences.
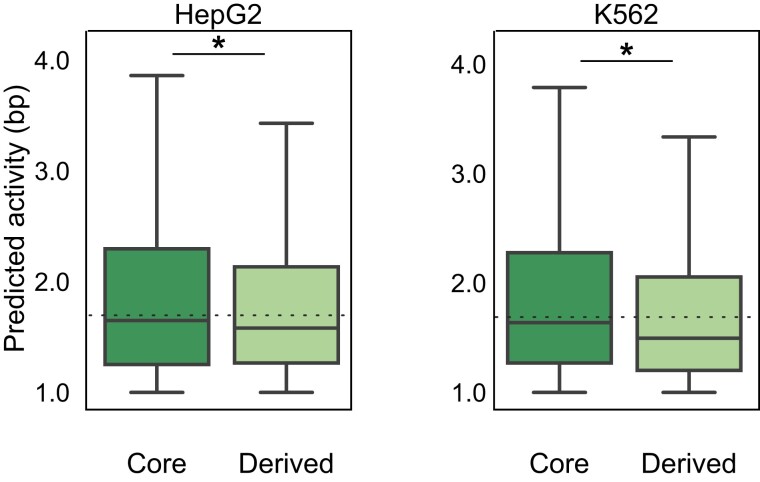
Core and Derived Regions have Similar Activity in Massively Parallel Reporter Assays
Given the TF-binding patterns in derived sequences, we hypothesized that these regions often have functional gene regulatory activity. To evaluate this, we compared the estimated activity of core and derived enhancer sequences from previously published Systematic High-resolution Activation and Repression Profiling with Reporter-tiling (SHARPR) massively parallel reporter assays (MPRAs; Ernst et al. 2016). Briefly, SHARPR uses probabilistic graphical models to estimate bp-level biochemical activity from the levels of transcribed mRNA and corresponding episomal DNA plasmids for 4,000 HepG2 and K562 enhancers. We assigned ages and architectures to the sequences with per bp regulatory activity in SHARPR-MPRA assays (>1:1 ratio of mRNA transcripts to DNA plasmids). Among active bases, derived and core sequences have similar activity per bp in both K562 and HepG2 cells, though core regions are slightly higher (fig. 5; HepG2: median per bp activity 1.58 derived vs. 1.65 core, MWU P = 2.0e-6; K562: 1.50 derived vs. 1.63 core, P = 6.6e-32). Stratified by age, we do not observe any consistent trends in core versus derived activity across evolutionary periods in HepG2 or K562 cells (supplementary fig. S14, Supplementary Material online). Simple enhancers (i.e., enhancers of a single age) show slightly higher activity per bp (median 1.69) than both core and derived segments of complex enhancers. Nonetheless, these data suggest that many derived sequences are biochemically active, have similar levels of activity compared with their adjacent cores, and contribute to gene regulatory function.
Fig. 5.
Both core and derived regions have regulatory activity in massively parallel reporter assays. Derived sequences had enhancer activity in a previous HepG2 MPRA analysis (activity score ≥1 for entire enhancer; N = 2,000 enhancers for HepG2 and N = 2,000 for K562; Ernst et al. 2016). However, derived activity was modestly, but significantly lower than core sequences (median 1.58 derived vs. 1.65 core activity per bp; N = 9,076 bp tested; MWU P = 2.0e-6) Patterns were similar in K562 cells (mean 1.50 derived vs. 1.64 core; P = 6.6e-32). Both core and derived segments of complex enhancers had lower activity per bp than simple enhancers (dashed lines, median 1.69). For HepG2, N = 6,498 derived and N = 9,076 core active bp were tested, whereas for K562, N = 7,568 derived and N = 9,846 core bp were tested. Asterisks represent p-value < 0.05.
Derived Sequences are Less Evolutionarily Constrained than Core Sequences
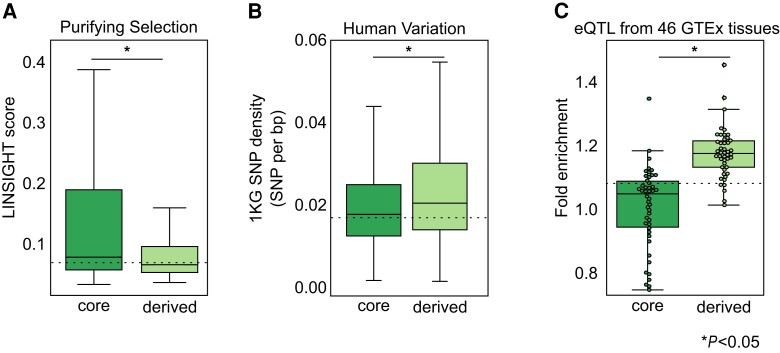
We next evaluated evolutionary constraints on core and derived sequences. To do this, we compared LINSIGHT per bp estimates of purifying selection (Huang et al. 2017) for derived sequences and associated cores in the FANTOM data set. Overall, derived sequences have slightly, but significantly lower LINSIGHT scores than adjacent cores (fig 6A; median 0.07 derived vs. 0.08 core LINSIGHT score; derived vs. core MWU P< 2.2e-238), suggesting that derived regions experience weaker purifying selection than adjacent enhancer cores. This pattern also holds when stratifying complex enhancers by sequence age (supplementary fig. S15, Supplementary Material online). As older enhancer sequences are generally under stronger evolutionary constraint, we also compared core and derived sequences of the same age and found that derived regions also have consistently lower LINSIGHT scores than age-matched core sequences (supplementary fig. S16, Supplementary Material online).
Fig. 6.
Derived regions experience weaker purifying selection, have more genetic variation, and are enriched for eQTL compared with adjacent core sequences. (A) Derived regions have significantly lower LINSIGHT purifying selection scores than adjacent core regions (median 0.08 core vs. 0.07 derived per bp LINSIGHT score; n = 2,271,279 and 2,021,098 bp, respectively; MWU P< 2.2e-308). The dashed line represents median simple enhancer LINSIGHT score (0.07, n = 5,398,405 bp). (B) Derived regions have higher genetic variant densities than associated core regions (median 0.020 derived vs. 0.018 core variants per bp; n = 26,451 and 27,691 variants, respectively; MWU P = 1.4e-202). Variant densities were calculated as the number of variants from the 1,000 Genomes Project in each enhancer region divided by its length. The dashed line represents median density in simple enhancers (n = 71,415 variants). (C) Derived regions are significantly more enriched for eQTL than core regions (Kruskal–Wallis P = 9.4e-12). eQTL from the GTEx consortium v6 from 46 tissues were intersected with enhancers. Enrichment for eQTL from each tissue in core and derived components was estimated from 1,000 length-matched, chromosome-matched permutations, and confidence intervals were estimated from 10,000 bootstraps. Each dot represents enrichment for eQTL of a tissue in core or derived enhancer regions. Derived, core, and simple enhancers have significantly different eQTL enrichments (MWU P = 1.3e-11). The dashed line represents the median simple enhancer eQTL enrichment across 46 tissues. Asterisks represent p-value < 0.05..
To evaluate the strength of sequence constraint across enhancer sequences, we binned each enhancer sequence into 10 equal-size bins (median 37 bp per bin) and computed the LINSIGHT scores in each bin. Sequence constraint is significantly lower in the six bins on the edges compared with the central four bins for complex enhancer sequences (supplementary fig. S18, Supplementary Material online; median weighted LINSIGHT score of 0.80 for outer vs. 0.86, Welch's P = 3.4e-24). However, these patterns were similar in simple enhancers (0.081 vs. 0.89; Welch's P = 3.4e-24) suggesting that they do not drive the distinction between these regions.
Together, these results indicate that derived sequences are under slightly weaker purifying selection than neighboring core regions in the same complex enhancer and than core regions of the same age.
Derived Enhancer Regions have More Genetic Variation than Core Regions
Given the modest differences in purifying selection between core and derived sequences, we compared their variant densities using genetic variants segregating in diverse human populations from the 1,000 Genomes Project. As expected, derived sequences have modestly higher variant densities than complex core regions (fig. 6B; median 0.020 vs. 0.018 variants per bp; MWU P = 1.4e-202, supplementary fig. S19, Supplementary Material online) and than simple enhancers (median 0.017 variants per bp). Consistent with this, global minor allele frequencies are also slightly higher in derived sequences compared with core and simple sequences (supplementary fig. S17, Supplementary Material online). This implies that derived sequences accumulate more genetic variants than core sequences, consistent with our observation that derived regions are under weaker purifying selection than adjacent cores.
Derived Enhancer Regions are Enriched for Expression Quantitative Trait Loci
To explore whether variation in derived regions is associated with changes in their effects on gene regulation, we quantified enrichment of expression quantitative trait loci (eQTL) in derived and core regions using eQTL from GTEx for 46 tissues (GTEx Consortium 2017). As expected, all enhancer architecture components are enriched for eQTL compared with the genomic background (fig. 6C; median OR 1.20 derived, 1.05 core, 1.10 simple; MWU core vs. derived, P = 1.3e-11). However, derived regions have the strongest enrichment. This is consistent with the higher minor allele frequencies (supplementary fig. S17, Supplementary Material online) and lower purifying selection pressure (fig. 6A) in derived regions. Nonetheless, eQTL enrichment in derived sequences indicates that variation in these regions of complex enhancers contributes to gene expression variability in human populations.
Discussion
Our analyses of human transcribed enhancers reveal that a substantial fraction (∼35%) is composed of sequences that originate from multiple evolutionary periods. We demonstrate that both the older core and younger derived sequences in these evolutionarily complex enhancers often show evidence of biochemical function and evolutionary constraint. Complex enhancers are enriched for core and derived sequences of similar ages. This suggests that the evolution of complex enhancer sequences proceeded in a step-wise and temporally constrained manner. However, we observe important differences in core versus derived regions, including the density and identity of TFs that bind, evolutionary constraint, and genetic variation. We confirm previous results from neocortical enhancers that derived regions are generally under less constraint (Emera et al. 2016). We also find that they are more likely to harbor genetic variation in human populations and variants that are associated with gene expression levels. Thus, both core and derived sequences appear to often be functional, but they also exhibit different evolutionary and functional attributes.
These results motivate further investigation of how the evolutionary origins of enhancer sequences relate to their functions and suggest that, as for proteins, sequences of independent origins are often juxtaposed in functional enhancers. However, many fundamental questions remain to be resolved about the modularity of enhancer evolution and function.
What is the Functional Importance of Derived Enhancer Sequences to Their Core Regions?
Our results suggest that core and derived sequences often both have gene regulatory functions. However, we do not know how often core and derived sequences alone are sufficient for stand-alone regulatory activity. Previous work has proposed that promoters and enhancers have many similar features, including transcription start sites, bidirectional transcription, and GC-rich sequences (Andersson and Sandelin 2020), even though promoters require enhancer sequences to increase gene expression. Derived regions have slightly higher GC content than cores (supplementary fig. S21, Supplementary Material online), have higher activity, and are less evolutionarily conserved than core sequences. Thus, it is possible that derived regions may function to enhance the promoter-like activity of core enhancer regions. In other words, derived sequences may enhance core enhancer activity.
We previously observed that human liver enhancers with multi-aged sequences are more often active in other placental mammal livers than simple enhancer sequences (Fong and Capra 2021), suggesting that younger derived sequences can be found at loci with conserved gene regulatory activity. In these cases, derived sequences may serve to reinforce or modulate existing gene regulatory function over evolutionary time, rather produce species-specific activity. We also observe sequence conservation in older, derived sequences (supplementary fig. S15, Supplementary Material online, suggesting derived sequences may drift for only relatively short periods before becoming conserved. Future work is needed to determine when derived sequences reinforce or diversify gene regulatory function across species.
Future studies should assess how often core enhancer sequences are sufficient for gene regulatory activity without flanking derived regions, and when core and derived regions cooperate to specify regulatory function. We anticipate that both scenarios may be common among complex enhancers. Further, the molecular mechanisms by which the core and derived regions contribute to regulatory function (e.g., changing chromatin accessibility, binding different TFs) must be determined. Many of these questions can be answered with evolution-aware reporter assays and gene editing strategies that disrupt core or derived sequences while preserving other sequence properties.
Are Evolutionary Modules Functional Modules?
Functional dissection of enhancer sequences suggests the modular organization of many enhancers (Long et al. 2016; Dukler et al. 2017; Sabarıs et al. 2019). Previous work has focused on this modularity in the context of TFs and other functional genomic markers. These have revealed the importance of transcriptional units (Tippens et al. 2020), the organization of its TFBS into clusters (Gotea et al. 2010), and the spatial distribution between TFBS (Farley et al. 2015; Grossman et al. 2018) to enhancer sequence modularity. Taking an evolutionary perspective, we demonstrate that many enhancers consist of distinct evolutionary modules. Yet, how these evolutionary modules relate to functional modules must be further clarified. For example, different evolutionary modules could have distinct modular regulatory functions that are combined. The independent biochemical activity for many derived enhancer sequences suggests that this scenario occurs. Further, core and derived sequences may develop synergistic regulatory functions. A recent analysis of SOX9 gene regulation has demonstrated that two sub-regions of the EC1.45 enhancer (from Therian and Vertebrate common ancestors, respectively) synergistically activate human SOX9 expression (Long et al. 2020). The extent to which synergy is observed between core and derived regions of complex enhancer sequences should be explored further. We speculate that the combination of sequences from different evolutionary origins often enables gene regulatory innovation while conserving core regulatory functions. As suggested in the previous section, future work should combine evolutionary analysis with high-resolution assays of regulatory function to assess the relationship between evolutionary sequence modules and function.
Can Considering Enhancer Evolutionary Architecture Aid Interpretation of Rare and Common Genetic Non-coding Variation?
Our work suggests that considering the evolutionary history of core and derived regions may provide valuable context for interpreting the function and disease relevance of human variation. The SHH enhancer (Lettice et al. 2017) provides an example where rare variants causing PPD2 are more prevalent in the core region and common variants are only present in the derived segments. Whether deleterious rare variation is generally concentrated in enhancer cores must be explored further. However, the small number of known non-coding Mendelian variants makes enrichment analyses challenging. Regarding common variation and associations with complex traits, we observed that eQTL are enriched in derived sequences. Derived regions also have higher variant density and slightly higher minor allele frequency than core regions; thus, we have greater power to detect effects on gene expression. Given the presence of linkage disequilibrium, whether variants in derived sequences directly affect gene expression variation must be tested to estimate their true contribution. Recent work has reported that the heritability of common variants is overrepresented in older gene regulatory elements (Hujoel et al. 2019), but whether this signal is due to variation in older complex enhancers and more specifically in cores, derived regions, or both remains to be explored. In general, more work is needed to understand the implications of common and rare variation in enhancer cores, derived regions, and their association with human traits.
Limitations
Our work has several limitations. The available sequence, TF, and functional data limit the scope and resolution of some analyses. First, the sampling of species with available genome sequences, the depth of sequencing, and the quality of available genome assemblies all influence estimates of sequence age (Margulies and Birney 2008; Sholtis and Noonan 2010). It is also possible that some enhancers classified as simple contain components that arose at different times along the same branch, especially for long branches. Moreover, varying levels of constraint over time also influence sequence age estimates. It is also possible that very different rates of evolution within the same enhancer could produce differences in alignability that appear to indicate different ages. However, we show that there are not systematic differences in the sequence divergence levels in core and derived segments compared with the expectation for regions of similar age (supplementary fig. S4, Supplementary Material online). Nonetheless, the age estimates should be considered a lower bound. Second, we emphasize that the estimated age of sequences with human enhancer activity is not necessarily the age when the sequence first gained enhancer activity. It is also possible that some enhancers have maintained conserved activity without detectable sequence similarity as in the developmental drift model (True and Haag 2001). Third, we leveraged previously published MPRA data; however, these only covered a few thousand enhancer regions in two cellular contexts. Without further biochemical assays, we cannot test whether most core and derived sequences have regulatory activity when separated. This is an important avenue for future work to determine whether derived sequences enhance pre-existing enhancer activity or if they work with core sequences to nucleate enhancer activity. Fourth, due to the challenges of linking regulatory elements to genes, we do not evaluate the gene targets associated with complex enhancers. Given their age and persistence over long evolutionary time, we speculate that complex enhancers often regulate genes involved in essential processes (Berthelot et al. 2018). Finally, in the TFBS analyses, we are limited to TFs with binding data in the relevant contexts. Some enhancers lacking TFBS in core or derived regions may be misclassified simple enhancers, but given that many TFs do not have available binding data, we anticipate that most such enhancers bind TFs, or spatial combinations of TFs, that have not been characterized. Given that we focus on comparisons of TFs with binding data between core and derived regions, we do not anticipate that this should influence our main conclusions.
Conclusion
Variation in gene regulatory sequences underlies much of the phenotypic variation between individuals and species. However, unlike protein sequences, we do not understand how enhancer sequence origin and evolution relate to functional activity. Here, we show that enhancers commonly consist of sequences from multiple evolutionary epochs and that both core and derived segments exhibit hallmarks of gene regulatory function. Thus, our results support and extend previous models of modular enhancer evolution by sequence accretion (Emera et al. 2016; Fong and Capra 2021) and suggest that enhancers composed of sequences of distinct evolutionary origins may promote gene regulatory function and variability in gene expression. Our work motivates the further study of the evolution of gene regulatory elements and the functional interaction of sequences of different origins over evolutionary time.
Materials and Methods
Assigning Ages to Sequences based on Alignment Syntenic Blocks
The genome-wide hg19 46-way and hg38 100-way vertebrate MultiZ multiple species alignment was downloaded from the UCSC genome browser. Each syntenic block was assigned an age based on the most recent common ancestor (MRCA) of the species present in the alignment block in the UCSC all species tree model (fig. 1A). For most analyses, we focus on the MRCA-based age, but when a continuous estimate is needed, we use evolutionary distances from humans to the MRCA node in the fixed 46-way or 100-way neutral species phylogenetic tree. Estimates of the divergence times of species pairs in Ma were downloaded from TimeTree (Hedges et al. 2015). Sequence age provides a lower bound on the evolutionary age of the sequence block. Sequence ages could be estimated for 93% of the autosomal bp in the hg19 human genome and 94% of the autosomal bp in the hg38 human genome.
eRNA Enhancer Data, Age Assignment, and Architecture Mapping
We considered eRNAs identified across 112 tissues and cell lines by high-resolution cap analysis of gene expression sequencing carried out by the FANTOM5 consortium (Andersson et al. 2014). This yielded a single set of 30,439 autosomal enhancer coordinates. We assigned ages to enhancer sequences by intersecting their genomic coordinates with aged syntenic blocks using Bedtools v2.27.1 (Quinlan and Hall 2010). Syntenic blocks that overlapped at least 6 bp of an enhancer sequence reflecting the minimum size of a TFBS (Lambert et al. 2018) were considered when assigning the enhancer's age and architecture. We considered enhancers with one age observed across its syntenic block(s) as “simple” enhancer architectures and enhancers overlapping syntenic blocks with different ages as complex enhancer architectures. We assigned complex enhancers ages according to the oldest block. Sequences without an assigned age were excluded from this analysis.
cCRE Enhancer Data, Age Assignment, and Architecture Mapping
We considered HepG2 and K562 ENCODE3 candidate cCRE enhancer loci annotated with proximal or distal enhancer-like signatures (pELS or dELS, with and without CTCF binding; The ENCODE Project Consortium et al. 2020). This yielded 53,864 HepG2 and 46,188 K562 cCREs coordinates. As for eRNA, we assigned ages and architectures to enhancer sequences by intersecting their locations with hg38 syntenic blocks and evaluating the diversity of syntenic ages. Syntenic blocks that overlapped at least 6 bp of an enhancer sequence were considered when assigning the enhancer's age and architecture. Complex enhancer architectures were defined as sequences with more than one age.
MPRA Activity Data
MPRA activity data and tile coordinates as assayed by the SHARPR-MPRA approach (Ernst et al. 2016) were downloaded and filtered for “Enh,” “EnhF,” “EnhW,” and “EnhWF” ChromHMM annotations. All tiles were 295 bp in length. We intersected autosomal MPRA tile coordinates with syntenic blocks and assigned ages and architectures as described above for other enhancers.
Genome-wide Shuffles to Determine Expected Background Distributions
To generate null distributions for expected properties of FANTOM and cCRE complex enhancers, we shuffled each set 100 × in the background non-coding genome (hg19 or hg38, respectively) using Bedtools. These shuffled sets were matched to the chromosome and length distribution of the observed regions in each data set. Coding sequences and ENCODE blacklist regions were excluded (Amemiya et al. 2019, https://www.encodeproject.org/annotations/ENCSR636HFF/). Each set of shuffled non-coding background genomic regions was then assigned ages and architectures with the same strategy used for the observed enhancers.
For example, applying this procedure to the FANTOM data set, we assigned ages to 2,567,773 shuffled regions from the genomic background (across all 100 matched sets). We identified 1,129,917 multi-aged, shuffled regions, and further classified their components as core and derived. These shuffled complex (i.e., multi-aged) sequences provided context for inferring whether the attributes of complex enhancer sequences differ from multi-aged sequences in the non-coding genomic background. When noted, we matched the ages of the core or derived background regions to those of the enhancers analyzed.
TFBS Density and Enrichment
Coordinates for ENCODE3 ChIP-seq peaks for 119 and 249 transcription factors assayed in HepG2 and K562, respectively, were downloaded from the ENCODE project's SCREEN interface (https://screen.encodeproject.org, last downloaded February 14, 2021). To assign TFBS to enhancer components, we intersected the 30 bp around the peak midpoint with simple and complex enhancer coordinates from the matching cell line. ChIP-seq peaks overlapping enhancers by ≥6 bp were counted as overlapping and peak overlap counts were normalized by syntenic length to estimate the density of TFBS per bp for each enhancer component.
For TFBS density and binding site enrichment, we only considered complex enhancers where TFBS overlapped enhancers. To correlate core and derived TFBS density, some complex enhancers have multiple derived sequences, which complicates the comparison of core and derived TFBS density. Thus, for this analysis, we calculated TFBS density as the sum of TFBS sites divided by the sum of the length of derived or core regions. We observed similar result when considering pair-wise syntenic TFBS densities and summed core-derived TFBS densities (supplementary figs. S11 and S12, Supplementary Material online). For TFBS enrichment, we used regions matched on core and derived sequence ages to compare TFBS enrichment among sequences that emerged in the same evolutionary period. Per age TFBS enrichment in derived versus core regions was calculated as the number of TFBS peaks that bind these regulatory regions versus all other TFBS loci that bind regulatory regions in that evolutionary period. Fisher's exact test was used to compute P-values for the observed ORs, and the P-values were corrected for multiple hypothesis testing to control the FDR at 5% using the Benjamini–Hochberg procedure.
1,000 Genome Variant Density and Minor Allele Frequency Analyses
Genetic variants from 2,504 diverse humans were downloaded from the 1,000 Genomes Project phase 3 (shapeit2 mvncall integrated v5a release 20130502). We intersected all variants with FANTOM enhancers and stratified by core and derived regions. Variant density was estimated as the number of SNPs overlapping a syntenic block divided by the length of the syntenic block. Singletons, that is alleles observed only once in a single individual, were removed from this analysis.
LINSIGHT Purifying Selection Estimates
Pre-computed LINSIGHT scores were downloaded from http://compgen.cshl.edu/LINSIGHT/. LINSIGHT provides per bp estimates of the probability of negative selection (Huang et al. 2017). We intersected FANTOM enhancers with LINSIGHT bp scores to determine the levels of constraint on bases within core and derived sequences.
TFBS Motif Sequence Specificity
We evaluated the sequence specificity of JASPAR core vertebrate non-redundant sequences with significant ChIP-seq TFBS enrichment in core or derived HepG2 or K562 enhancers. Specifically, we calculate the Kullback–Leibler divergence of the motif from genomic background nucleotide frequencies for A/T (0.3) and GC (0.2), similar to the previously described procedure (Li and Wunderlich 2017). For all ChIP-seq TFBS motifs (regardless of significant enrichment), we assigned these motifs to core or derived regions if they were more often enriched in core over derived sequences and vice versa.
eQTL Enrichment
The enrichment for GTEx eQTL from 46 tissues (last downloaded July 23, 2019) in core and derived enhancer sequences was tested against matched background sets. In this analysis, we considered 500 matched sets. Median fold change was calculated as the number of eQTLs overlapping enhancer sequence components (i.e., core or derived) compared with the appropriate random sets. Confidence intervals (95%) were generated by 10,000 bootstraps. P-values were corrected for multiple hypothesis testing by controlling the FDR at 5% using the Benjamini–Hochberg procedure.
Supplementary Material
Contributor Information
Sarah L Fong, Vanderbilt Genetics Institute, Vanderbilt University, Nashville, Tennessee.
John A Capra, Department of Biological Sciences, Vanderbilt University, Nashville, Tennessee; Bakar Computational Health Sciences Institute and Department of Epidemiology and Biostatistics, University of California, San Francisco.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Funding
This work was supported by the National Institutes of Health (grants R35GM127087 to J.A.C. and T32GM080178 to S.F.).
Data Availability
Sequence age Data Sets
Hg19 syntenic age data (including aged FANTOM eRNAs) underlying this article are available in Zenodo, at https://dx.doi.org/10.5281/zenodo.4618495.
Hg38 syntenic age data underlying this article are available in Zenodo, at https://doi.org/10.5281/zenodo.5809634.
HepG2 and K562 aged cCRE sequences underlying this article are available in Zenodo, at https://doi.org/10.5281/zenodo.5809629.
Data Sets Derived From Sources in the Public Domain
FANTOM5 eRNAs (Andersson et al. 2014): http://slidebase.binf.ku.dk/human_enhancers/.
ENCODE cCREs and TFBS ChIP-seq (The ENCODE Project Consortium et al. 2020): https://screen.encodeproject.org.
HepG2 and K562 MPRAs (Ernst et al. 2016): GSE71279.
Hg19 46-way vertebrate species multiz alignment: https://hgdownload.soe.ucsc.edu/gbdb/hg19/multiz46way/.
Hg38 100-way vertebrate species multiz alignment: https://hgdownload.soe.ucsc.edu/gbdb/hg38/multiz100way/.
LINSIGHT (Huang et al. 2017): http://compgen.cshl.edu/LINSIGHT/LINSIGHT.bw
Source code is freely available at: https://github.com/slifong08/enh_ages.
References
- GTEx Consortium . 2017. Genetic effects on gene expression across human tissues. Nature 550(7675):204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, et al . 2020. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583(7818):699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya HM, et al. 2019. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep. 9(1):9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507(7493):455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Sandelin A. 2020. Determinants of enhancer and promoter activities of regulatory elements. Nat Rev Genetics. 21(2):71–87. [DOI] [PubMed] [Google Scholar]
- Berthelot C, et al. 2018. Complexity and conservation of regulatory landscapes underlie evolutionary resilience of mammalian gene expression. Nat Ecol Evol. 2(1):152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, et al. 2013. How old is my gene? Trends Genet. 29(11):659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukler N, et al. 2017. Is a super-enhancer greater than the sum of its parts? Nat Genet. 49(1):2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emera D, et al. 2016. Origin and evolution of developmental enhancers in the mammalian neocortex. Proc Natl Acad Sci U S A. 113(19):E2617–E2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, et al. 2016. Genome-scale high-resolution mapping of activating and repressive nucleotides in regulatory regions. Nat Biotechnol. 34(11):1180–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley EK, et al. 2015. Suboptimization of developmental enhancers. Science 350(6258):325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong SL, Capra JA. 2021. Modeling the evolutionary architectures of transcribed human enhancer sequences reveals distinct origins, functions, and associations with human-trait variation. Mol Biol Evol. 38(9):3681–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund SK, et al. 2019. Evolution of protein domain architectures. In: Anisimova M, editor. Evolutionary genomics. Vol. 1910.New York, NY: Springer. p.469–504. [Google Scholar]
- Franchini LF, Pollard KS. 2015. Genomic approaches to studying human-specific developmental traits. Development 142(18):3100–3112. [DOI] [PubMed] [Google Scholar]
- Gotea V, et al. 2010. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 20(5):565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, et al. 2018. Positional specificity of different transcription factor classes within enhancers. Proc Natl Acad SciU S A. 115(30):E7222–E7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, et al. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 32(4):835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RE,, Lettice LA. 2013. Alterations to the remote control of Shh gene expression cause congenital abnormalities. Philos Trans R Soc Lond B Biol Sci 368(1620):20120357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-F, et al. 2017. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat Genet. 49(4):618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hujoel MLA, et al. 2019. Disease heritability enrichment of regulatory elements is concentrated in elements with ancient sequence age and conserved function across species. Am J Hum Genet. 104(4):611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal GA, Farley EK. 2021. Enhancer grammar in development, evolution, and disease: dependencies and interplay. Dev Cell. 56(5):575–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, et al. 2016. Progressive loss of function in a limb enhancer during snake evolution. Cell 167(3):633–642.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SA, et al. 2018. The human transcription factors. Cell 172(4):650–665. [DOI] [PubMed] [Google Scholar]
- Lettice LA. 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 12(14):1725–1735. [DOI] [PubMed] [Google Scholar]
- Lettice LA, et al. 2012. Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly. Dev Cell. 22(2):459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, et al. 2017. The conserved sonic hedgehog limb enhancer consists of discrete functional elements that regulate precise spatial expression. Cell Rep. 20(6):1396–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Lily, Wunderlich Z. 2017. An enhancer's Length and composition are shaped by its regulatory task. Front Genet. 8:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, et al. 2016. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167(5):1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, et al. 2020. Loss of extreme long-range enhancers in human neural crest drives a craniofacial disorder. Cell Stem Cell. 27(5):765–783.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, et al. 2011. Three periods of regulatory innovation during vertebrate evolution. Science 333(6045):1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies EH, Birney E. 2008. Approaches to comparative sequence analysis: towards a functional view of vertebrate genomes. Nat Rev Genet. 9(4):303–313. [DOI] [PubMed] [Google Scholar]
- Marnetto D, et al. 2018. Evolutionary rewiring of human regulatory networks by waves of genome expansion. Am J Hum Genet. 102(2):207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne JL, Wagner A. 2014. The robustness and evolvability of transcription factor binding sites. Science 343(6173):875–877. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Tsiantis M. 2017. Enhancer evolution and the origins of morphological novelty. Curr Opin Genet Dev. 45:115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabarıs G, et al. 2019. Actors with multiple roles: pleiotropic enhancers and the paradigm of enhancer modularity. Trends Genet. 35(6):423–433. [DOI] [PubMed] [Google Scholar]
- Shlyueva D, et al. 2014. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15(4):272–286. [DOI] [PubMed] [Google Scholar]
- Sholtis SJ, Noonan JP. 2010. Gene regulation and the origins of human biological uniqueness. Trends Genet. 26(3):110–118. [DOI] [PubMed] [Google Scholar]
- Stolzer M, et al. 2015. Event inference in multidomain families with phylogenetic reconciliation. BMC Bioinformatics 16(S14):S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippens ND, et al. 2020. Transcription imparts architecture, function and logic to enhancer units. Nat Genet. 52(10):1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Haag ES. 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 3(2):109–119. [DOI] [PubMed] [Google Scholar]
- Ushiki A, et al. 2021. Deletion of CTCF sites in the SHH locus alters enhancer-promoter interactions and leads to acheiropodia. Nat Commun. 12(1):2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160(3):554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G. 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 13(1):59–69. [DOI] [PubMed] [Google Scholar]
- Wong ES, et al. 2020. Deep conservation of the enhancer regulatory code in animals. Science 370(6517):eaax8137. [DOI] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Gen. 8(3):206–216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence age Data Sets
Hg19 syntenic age data (including aged FANTOM eRNAs) underlying this article are available in Zenodo, at https://dx.doi.org/10.5281/zenodo.4618495.
Hg38 syntenic age data underlying this article are available in Zenodo, at https://doi.org/10.5281/zenodo.5809634.
HepG2 and K562 aged cCRE sequences underlying this article are available in Zenodo, at https://doi.org/10.5281/zenodo.5809629.
Data Sets Derived From Sources in the Public Domain
FANTOM5 eRNAs (Andersson et al. 2014): http://slidebase.binf.ku.dk/human_enhancers/.
ENCODE cCREs and TFBS ChIP-seq (The ENCODE Project Consortium et al. 2020): https://screen.encodeproject.org.
HepG2 and K562 MPRAs (Ernst et al. 2016): GSE71279.
Hg19 46-way vertebrate species multiz alignment: https://hgdownload.soe.ucsc.edu/gbdb/hg19/multiz46way/.
Hg38 100-way vertebrate species multiz alignment: https://hgdownload.soe.ucsc.edu/gbdb/hg38/multiz100way/.
LINSIGHT (Huang et al. 2017): http://compgen.cshl.edu/LINSIGHT/LINSIGHT.bw
Source code is freely available at: https://github.com/slifong08/enh_ages.