Abstract
The production of exfoliative toxin B (ET-B), but not ET-A, was shown to be specifically associated with production of a highly conserved two-component lantibiotic peptide system in phage group II Staphylococcus aureus. Two previously studied but incompletely characterized S. aureus bacteriocins, staphylococcins C55 and BacR1, were found to be members of this lantibiotic system, and considerable homology was also found with the two-component Lactococcus lactis bacteriocin, lacticin 3147. sacαA and sacβA, the structural genes of the lantibiotics staphylococcins C55α and C55β and two putative lantibiotic processing genes, sacM1 and sacT, were localized together with the ET-B structural gene to a single 32-kb plasmid in strain C55. Irreversible loss of both ET-B and two-component lantibiotic production occurs during laboratory passage of ET-B-positive S. aureus strains, particularly at elevated temperatures.
Associations between the expression of virulence factors and bacteriocins have been demonstrated in several bacterial pathogens, and in some instances, their genetic determinants have been localized to the same plasmid. For instance, it was shown that both botulinum toxin type G and bacteriocin production could be eliminated from certain Clostridium botulinum strains in association with the loss of an 81-MDa plasmid when cultures were grown at 44°C (6). Hemocin production in Haemophilus influenzae is found in 98% of strains producing type b capsule and not in any nontypeable strains (14). It was suggested that hemocin may play a role in nasopharyngeal colonization by assisting competition against commensal Haemophilus spp. In another study, pathogenic human Enterococcus faecalis strains were demonstrated to frequently produce both hemolysin and bacteriocin activities and the determinants were shown to be encoded by a transmissible plasmid (13). A unique feature of this two-component peptide system is its cytolytic activity against both prokaryotic and eukaryotic cells (2).
The simultaneous elimination of bacteriocin (BacR1) and exfoliative toxin B (ET-B) production from the phage group II strain Staphylococcus aureus U0007 was demonstrated by Warren and associates (26) by either incubation of the bacteria at elevated temperatures or treatment with ethidium bromide. They concluded that both products are encoded by a 37-kb plasmid. The same group of researchers then demonstrated in vitro transduction of the plasmid encoding ET-B into other S. aureus strains and suggested the possibility that a similar transfer occurs within the mixed microflora of the skin (18). They were later able to clone and sequence the gene responsible for the production of ET-B (9, 12). However, the determinant for BacR1 production was not identified.
Production of bacteriocin-like inhibitory activity by S. aureus has been reported on many occasions (21, 24), but primary-structure details are only available for staphylococcins C55α and C55β, isolated from S. aureus C55 (16). The production of antibacterial activity by strain C55 was first reported by Dajani’s group in 1970 (3). Those researchers partially purified an inhibitory agent from strain C55 and described it as a nondialyzable proteinaceous substance. We have recently reported that the majority of the inhibitory activity of strain C55 is due to the synergistic activity of the lantibiotics staphylococcins C55α and C55β (16). In the same communication, we demonstrated that the production of staphylococcins C55α and C55β is dependent on the presence of a 32-kb plasmid. Staphylococcins C55α and C55β have molecular masses of 3,339 and 2,993 Da, respectively. Amino acid composition analyses confirmed the presence of lanthionine and/or β-methyllanthionine in both peptides, but the specific location and orientation of these unusual amino acids in lantibiotic molecules cannot be determined by conventional N-terminal amino acid sequencing.
In the present report, we establish that the bacteriocins produced by Dajani’s strain C55 and Rogolsky’s strain U0007 are identical and that this type of bacteriocin is widely distributed in phage group II S. aureus. Moreover, we demonstrate that the bacteriocin structural genes are closely associated with the ET-B determinant and are located on the same plasmid.
Cloning of genes encoding strain C55 lantibiotic production.
The N-terminal sequence of the C55α peptide has previously been shown to be XXDhbNXFDhaLXDYWGNKGNWCTA, where X represents an unidentified amino acid residue and Dhb and Dha represent dehydrobutyrine and dehydroalanine, respectively (16). From this sequence, amino acids 10 to 15 (i.e., DYWGNK) were used to design a wobbled 17-mer oligonucleotide probe [5′-GA(CT) TA(CT) TGG GG(AGTC) AA(CT) AA-3′]. The probe was labelled with [γ32P]ATP by using T4 polynucleotide kinase as described by Sambrook et al. (20). Strain C55 was found to contain two plasmids with sizes of approximately 3.5 and 32 kb. A 6.5-kb PstI fragment of the 32-kb plasmid hybridized with the staphylococcin C55α probe by Southern hybridization. This fragment was cloned in pUC19 with Escherichia coli Dh5α′ as the host, and both strands of the plasmid insert were sequenced by the dideoxy-chain termination method.
Nucleotide sequence of strain C55 lantibiotic genes.
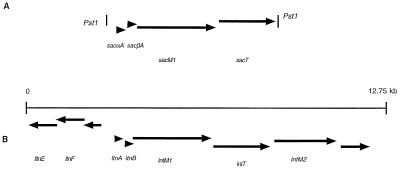
The cloned fragment comprised 6,276 bp, and computer analysis revealed four open reading frames (ORFs) in the same orientation designated sacαA, sacβA, sacM1, and sacT (Fig. 1). Putative ribosomal binding sites were identified in front of sacαA, sacβA, and sacM1, and only a single base was found between sacM1 and sacT (Fig. 2).
FIG. 1.
Comparison of the organization of staphylococcin C55 ORFs with that of the putative gene cluster of lacticin 3147. (A) Organization of the ORFs designated sacαA, sacβA, sacM1, and sacT in a 6,276-bp PstI fragment. (B) Putative lacticin 3147 gene cluster (5).
FIG. 2.
Nucleotide sequence of the structural genes of staphylococcins C55α and C55β. The deduced amino acid sequences of the ORFs are shown below the nucleotide sequence. Vertical arrows indicate the cleavage sites of propeptides. The termination codons are indicated by asterisks. Primers used for amplifications are underlined.
Characterization of sacαA and sacβA.
The previously determined C55α peptide sequence was consistent with the deduced amino acid sequence of the sacαA ORF (Fig. 3). The C55α propeptide starts at the first Cys residue in the predicted prepeptide. The presence of a Thr codon corresponding to the third amino acid residue in the propeptide and a Ser codon at the seventh amino acid residue agrees with the locations of Dhb and Dha residues on N-terminal sequencing of the mercaptoethanol-modified C55α peptide (16). The blank cycles (denoted by the letter X) for amino acid residues 1, 2, 5, and 9 probably represent components of lanthionine and β-methyllanthionine residues, since they correlate with the presence of Thr, Ser, or Cys codons in the nucleotide sequence. The presence of a Cys component of lanthionine or β-methyllanthionine at the N terminus of C55α differs from the arrangement of these modified amino acids in all other known class I lantibiotics. In all other cases, the Cys component is toward the C terminus. The calculated mass of C55α, based on the predicted amino acids and the presence in the peptide of four lanthionine and/or β-methyllanthionine amino acids and three dehydro amino acids, is 3,336 Da, which agrees closely with the actual mass of 3,339 Da determined by mass spectrometry.
FIG. 3.
Elucidation of the primary structure of staphylococcins C55α and C55β. The first and third lines show the sequences derived by N-terminal amino acid sequencing (16). Dhb and Dha represent dehydrobutyrine and dehydroalanine. X represents unidentified cycles during Edman degradation. The second and fourth lines represent the amino acid sequences deduced from the DNA sequences. The arrows indicate cleavage sites.
A second lantibiotic structural gene, sacβA, was found immediately downstream of sacαA, and its deduced amino acid sequence (Fig. 2) agrees with the C55β sequence obtained by N-terminal sequencing (Fig. 3). As was found for C55α, the presence of Thr in residues 2 and 12 correlates with the position of Dhb in the N-terminal sequence of the mercaptoethanol-modified C55β peptide. The presence of Ser and Cys in positions equivalent to propeptide amino acids 5, 19, and 23 correlates with blank cycles on N-terminal sequencing of C55β. The calculated mass, based on the predicted amino acids and the presence of three lanthionine and/or β-methyllanthionine amino acids and two dehydro amino acids, was 2,993 Da, which agrees with the mass of 2,993 Da determined by mass spectrometry.
Comparison of sacαA and sacβA with the structural genes of other lantibiotics.
Marked differences were observed when the structural genes sacαA and sacβA were compared with those of other known staphylococcal lantibiotics. All of the well-studied staphylococcal lantibiotics (epidermin, gallidermin, epilancin, Pep5, and epicidin) (8, 19) can be classified as class AI (4) or type FNLD (19) lantibiotics and neither staphylococcin C55α nor C55β is related to any of the lantibiotics in this group. However, the presence of Gly and Ala in the −2 and −1 positions of the C55α prepeptide is consistent with the cleavage sites of class AII or double-Gly-type lantibiotics (4, 19). The presence of Ala in positions −1 and −2 at the cleavage site of the C55β prepeptide has not been described before in double-Gly-type lantibiotics but does occur in mersacidin, a type B lantibiotic (1). A computer-aided homology search indicated that these two peptides have no homologies with other lantibiotics listed in the data banks, but both have very high homology with two putative lantibiotic peptides encoded by Lactococcus lactis DPC3147 plasmid pMRC01 (5, 15). Comparison of the deduced amino acid sequences of the sacαA and ltnA ORFs gave 65.5% identity and 77.6% similarity. Also, the sacβB- and ltnB-encoded peptides showed 44.6% identity and 63.1% similarity. These results indicate that strains C55 and DPC 3147 produce closely related two-component lantibiotic systems.
ORFs downstream of sacαA and sacβA.
An ORF encoding a putative protein comprising 965 amino acids was found in the same orientation and 18 bp 3′ to sacβA (Fig. 2). This ORF, named sacM1, has some homology (20% identity and 38% similarity) with the lctM gene, located downstream of lctA, the structural gene for lactococcin DR (17) (now called lacticin 481). lctM encodes the protein that modifies the lacticin 481 propeptide. Based on the comparison of lantibiotic M genes done by Siezen et al. (22), some residues within the amino acid sequence of C55M1 correlate with conserved amino acids and segments found in other lantibiotic M gene products. A further ORF, sacT, identified downstream of sacM1 encodes a protein of 720 amino acids. This has strong homology with the genes for several transporters, including lctT, which have been shown to be involved in both the transport and the processing of this type of lantibiotic (7, 22).
Just as there was found to be similarity between the staphylococcin C55 and lacticin 3147 structural genes, sacM1 has 45% identity and 63% similarity to ltnM1 and sacT has 49.3% identity and 69.2% similarity to ltnT. The staphylococcin C55 ORFs identified in this study are arranged identically to the corresponding ORFs thought to encode lacticin 3147 (Fig. 1). It might be anticipated that the remainder of the staphylococcin C55 gene arrangement is also closely similar to that found for lacticin 3147. Interestingly, both the lacticin 3147 plasmid (5) and the staphylococcin C55 ORFs have a G+C content of 28% and, moreover, this value is closer to the range associated with DNA from S. aureus (32 to 38%) (23) than that from L. lactis (36 to 38%) (11).
Screening for etb and for bacteriocin production similar to that of strain C55.
Fifty strains previously reported to be ET producers and belonging to phage group II (and including Rogolsky’s strain U0007) and 15 phage group II strains negative for ET production were tested for bacteriocin production and for the presence of cross-immunity to the bacteriocin produced by strain C55 (Table 1). For specific amplification of sacαA and sacβA, primers AGC GTG GTG ATT CTT ATG and TCT GAT TTA TTT AGT TCT GGA T were designed by using the sequence given in Fig. 2. DNA extraction for PCR was done by the method of Unal et al. (25). The PCR amplification was performed in a total volume of 100 μl with each deoxyribonucleotide triphosphate at 200 μM and each primer at 1 μM in 1× reaction buffer. Each reaction mixture was heated to 72°C for 5 min before the addition of 2.5 U of Taq polymerase. A total of 30 cycles (1 cycle being 30 s at 94°C, 30 s at 48°C, and 1 min at 72°C) and a 5-min final extension at 72°C were performed on a DNA thermal cycler. The PCR products (499 bp) were analyzed by electrophoresis using a 2% agarose gel in Tris-acetate electrophoresis buffer and then stained with ethidium bromide. The ET-A and ET-B genes, eta and etb, were amplified by use of the primers described by Johnson et al. (10).
TABLE 1.
Production of inhibitory activity similar to that of strain C55 and presence of eta and etb in S. aureus strains originally found to produce ET-A and/or ET-Ba
| Original ET-A and ET-B production status of strains | No. of strains | No. with inhibitor productionb | No. with immunity to C55 | Detection by PCR of:
|
||
|---|---|---|---|---|---|---|
| eta | etb | sacαA and sacβA | ||||
| ET-A | 8 | 0 | 0 | 8 | 0 | 0 |
| ET-B | 9 | 4 | 4 | 0 | 4 | 4 |
| ET-A + ET-B | 33 | 16 | 16 | 33 | 16 | 16 |
| ET-A and ET-B negative | 15 | 0 | 0 | 0 | 0 | 0 |
S. aureus strains were supplied by P. M. Schlievert, S. Poston, and the Institute of Environmental Science and Research Ltd., Porirua, New Zealand.
Inhibitor production was tested by the simultaneous-antagonism test using M. luteus as the indicator. An inhibitory zone of greater than 7 mm was taken as a positive result.
Two procedures commonly used to detect bacteriocin-producing strains are the simultaneous-antagonism test and the deferred-antagonism test (16). In the present study, Columbia agar base (GIBCO, Ltd., Paisley, United Kingdom) was used as the growth medium. Briefly, simultaneous-antagonism testing involves stab inoculation of the strains being evaluated for bacteriocin production into a freshly seeded lawn of Micrococcus luteus and then, following incubation for 18 h, examination of the plate for zones of inhibited lawn growth surrounding individual stab cultures. For deferred-antagonism testing, the test strain is grown as a 1-cm diametric streak culture and then, following removal of the cells and sterilization of the agar surface with chloroform vapor, a series of strains being tested for bacteriocin sensitivity are inoculated across the line of the original test strain growth. Following incubation, the range and extent of inhibition of the indicator strains by the test strain can be assessed. The results of the initial screening for bacteriocin production by the simultaneous-antagonism test and of the deferred-antagonism cross-testing for immunity to C55 are given together with the PCR results in Table 1. All 20 strains (and only those strains) producing an inhibitory zone of greater than 7 mm in the simultaneous-antagonism test and also showing cross-immunity to strain C55 in the deferred-antagonism test (16) were confirmed to be positive for sacαA and sacβA by PCR. Sequencing of the PCR products established that there were no variations in the nucleotide sequence of either sacαA or sacβA in any of the 20 strains found to contain both of these genes. Our results thus demonstrate that this lantibiotic system is not unique to strain C55 but that it is also present in strain U0007 and various other phage group II S. aureus strains. Several strains with zones of inhibition of less than 4 mm were found to be negative for sacαA and sacβA, suggesting that they produce different types of inhibitory agents.
All of the producers of staphylococcins C55α and C55β included in Table 1 were confirmed to be positive for etb by PCR amplification. None of the other S. aureus strains were positive for either etb, sacαA, or sacβA. Thus, a simple screen for ET-B-positive S. aureus is to detect strains that show specific immunity when tested for sensitivity to S. aureus C55 or any other ET-B producer strain in a deferred-antagonism test. We found by PCR that 22 (52%) of 42 strains originally thought to produce ET-B had, in fact, lost the gene and that all of these were also negative for bacteriocin production and did not contain sacαA or sacβA. This was thought to be due to loss of the bacteriocin–ET-B plasmid during storage or subculture. By contrast, there was no evidence of spontaneous loss of ET-A production by any of the 41 ET-A-positive strains tested in this study, and this is consistent with the known chromosomal location of eta (12). Previously, we have demonstrated 100% curing of bacteriocin production on incubation of S. aureus C55 at 42°C (16). As a result of our studies, we suggest that avoidance of the incubation of suspected ET-B producers at elevated temperatures will aid in the maintenance of the toxin-encoding plasmid.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in the GenBank nucleic acid sequence database under accession no. AF147744.
Acknowledgments
This work was supported by a grant from the Health Research Council of New Zealand and a travel grant from the German Ministry for Research and Technology (BMBF) through DLR.
Thanks are due to John Sullivan and Clive Ronson for advice and to Michaela Yorsten and Armgard Viebahn for technical assistance.
REFERENCES
- 1.Bierbaum G, Brotz H, Koller K P, Sahl H-G. Cloning, sequencing and production of the lantibiotic mersacidin. FEMS Microbiol Lett. 1995;127:121–126. doi: 10.1111/j.1574-6968.1995.tb07460.x. [DOI] [PubMed] [Google Scholar]
- 2.Booth M C, Bogie C P, Sahl H-G, Siezen R J, Hatter K L, Gilmore M S. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol Microbiol. 1996;21:1175–1184. doi: 10.1046/j.1365-2958.1996.831449.x. [DOI] [PubMed] [Google Scholar]
- 3.Dajani A S, Wannamaker L W. Demonstration of a bacteriocidal substance against β-hemolytic streptococci in supernatant fluids of staphylococcal cultures. J Bacteriol. 1969;97:985–991. doi: 10.1128/jb.97.3.985-991.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Vos W M, Kuipers O P, van der Meer J R, Siezen R J. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by gram-positive bacteria. Mol Microbiol. 1995;17:427–437. doi: 10.1111/j.1365-2958.1995.mmi_17030427.x. [DOI] [PubMed] [Google Scholar]
- 5.Dougherty B A, Hill C, Weidman J F, Richardson D R, Venter J C, Ross R P. Sequence and analysis of the 60kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol Microbiol. 1998;29:1029–1038. doi: 10.1046/j.1365-2958.1998.00988.x. [DOI] [PubMed] [Google Scholar]
- 6.Eklund M W, Poysky F T, Mseitif L M, Strom M S. Evidence for plasmid-mediated toxin and bacteriocin production in Clostridium botulinum type G. Appl Environ Microbiol. 1988;54:1405–1408. doi: 10.1128/aem.54.6.1405-1408.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havarstein L S, Diep D B, Nes I F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 8.Heidrich C, Pag U, Josten M, Metzger J, Jack R W, Bierbaum G, Jung G, Sahl H-G. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl Environ Microbiol. 1998;64:3140–3146. doi: 10.1128/aem.64.9.3140-3146.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson M P, Iandolo J J. Cloning and expression of the exfoliative toxin B gene from Staphylococcus aureus. J Bacteriol. 1986;166:574–580. doi: 10.1128/jb.166.2.574-580.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson W M, Tyler S D, Ewan E P, Ashton F E, Pollard D R, Rozee K R. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J Clin Microbiol. 1991;29:426–430. doi: 10.1128/jcm.29.3.426-430.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilpper-Balz R, Fischer G, Schleifer K H. Nucleic acid hybridisation of group N and group D streptococci. Curr Microbiol. 1982;7:245–250. [Google Scholar]
- 12.Lee C Y, Schmidt J J, Wineger A D J-, Spero L, Iandolo J. Sequence determination and comparison of the exfoliative toxin A and toxin B genes from Staphylococcus aureus. J Bacteriol. 1987;169:3904–3909. doi: 10.1128/jb.169.9.3904-3909.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Libertin C R, Dumitru R, Stein D S. The hemolysin/bacteriocin produced by enterococci is a marker of pathogenicity. Diagn Microbiol Infect Dis. 1992;15:115–120. doi: 10.1016/0732-8893(92)90033-p. [DOI] [PubMed] [Google Scholar]
- 14.LiPuma J J, Richman H, Stull T L. Haemocin, the bacteriocin produced by Haemophilus influenzae: species distribution and role in colonization. Infect Immun. 1990;58:1600–1605. doi: 10.1128/iai.58.6.1600-1605.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McAuliffe O, Ryan M P, Ross R P, Hill C, Breeuwer P, Abee T. Lacticin 3147, a broad-spectrum bacteriocin which selectively dissipates the membrane potential. Appl Environ Microbiol. 1998;64:439–445. doi: 10.1128/aem.64.2.439-445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navaratna, M. A. D. B., H.-G. Sahl, and J. R. Tagg. Two-component anti-Staphylococcus aureus lantibiotic activity produced by Staphylococcus aureus C55. Appl. Environ. Microbiol. 64:4803–4808. [DOI] [PMC free article] [PubMed]
- 17.Rince A, Dufour A, Le Pogam S, Thuault D, Bourgeois C M, Le Pennec J-P. Cloning, expression, and nucleotide sequence of genes involved in production of lactococcin DR, a bacteriocin from Lactococcus lactis. subsp. lactis. Appl Environ Microbiol. 1994;60:1652–1657. doi: 10.1128/aem.60.5.1652-1657.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogolsky M, Beall B W, Wiley B B. Transfer of the plasmid for exfoliative toxin B synthesis in mixed cultures on nitrocellulose membranes. Infect Immun. 1986;54:265–268. doi: 10.1128/iai.54.1.265-268.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahl H-G, Bierbaum G. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from Gram-positive bacteria. Annu Rev Microbiol. 1998;52:41–79. doi: 10.1146/annurev.micro.52.1.41. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Scott J C, Sahl H-G, Carne A, Tagg J R. Lantibiotic-mediated anti-lactobacillus activity of a vaginal Staphylococcus aureus isolate. FEMS Microbiol Lett. 1992;72:97–102. doi: 10.1016/0378-1097(92)90496-b. [DOI] [PubMed] [Google Scholar]
- 22.Siezen J R, Kuipers O P, de Vos W M. Comparison of the lantibiotic gene clusters and encoded proteins. Antonie Leeuwenhoek. 1996;69:171–184. doi: 10.1007/BF00399422. [DOI] [PubMed] [Google Scholar]
- 23.Sneath, P., P. H., N. S. Mair, M. E. Sharpe, and J. G. Holt. Bergey’s manual of systemic bacteriology, vol. 2, p. 1212. Williams & Wilkins Co., Baltimore, Md.
- 24.Tagg J R, Dajani A S, Wannamaker L W. Bacteriocins of gram-positive bacteria. Bacteriol Rev. 1976;40:722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Unal S, Hoskins J, Flokowitsch J E, Wu C Y, Preston D A, Skatrud P L. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J Clin Microbiol. 1992;30:1685–1691. doi: 10.1128/jcm.30.7.1685-1691.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warren R, Rogolsky M, Wiley B B, Glasgow L A. Isolation of extrachromosomal deoxyribonucleic acid for exfoliative toxin production from phage group II Staphylococcus aureus. J Bacteriol. 1975;122:99–105. doi: 10.1128/jb.122.1.99-105.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]