Abstract
While brains of patients with Alzheimer’s disease and related tauopathies have evidence of altered RNA processing, we lack mechanistic understanding of how altered RNA processing arises in these disorders and if such changes are causally linked to neurodegeneration. Using Drosophila melanogaster models of tauopathy, we find that overall activity of nonsense-mediated mRNA decay (NMD), a key RNA quality control mechanism, is reduced. Genetic manipulation of NMD machinery significantly modifies tau-induced neurotoxicity, suggesting that deficits in NMD are causally linked to neurodegeneration. Mechanistically, we find that deficits in NMD are a consequence of aberrant RNA export and RNA accumulation within nuclear envelope invaginations in tauopathy. We identify a pharmacological activator of NMD that suppresses neurodegeneration in tau transgenic Drosophila, indicating that tau-induced deficits in RNA quality control are druggable. Our studies suggest that NMD activators should be explored for their potential therapeutic value to patients with tauopathies.
Keywords: Tauopathy, Alzheimer’s disease, nonsense-mediated RNA decay, neurodegeneration, Drosophila, nucleus
1 |. NARRATIVE
1.1 |. Contextual Background
There are over 20 neurodegenerative disorders known collectively as “tauopathies,” which are pathologically defined by characteristic inclusions of the microtubule-associated protein tau (MAPT) in brains of affected individuals. Mutations in the MAPT gene cause autosomal dominant forms of frontotemporal dementia, collectively known as frontotemporal lobar dementia (FTLD)-tau with MAPT mutation1–4. While most tauopathies, including Alzheimer’s disease, feature deposition of wild-type tau protein, this genetic link between MAPT mutation and neurodegeneration clearly demonstrate that tau dysfunction is sufficient to drive neurodegeneration. Despite lack of MAPT mutation in sporadic tauopathies, animal models featuring transgenic expression of human tau harboring disease-associated mutations have been quite valuable for identifying disease mechanisms that are conserved in Alzheimer’s disease. In Drosophila, for example, transgenic expression of human wild-type tau or MAPT mutations drive neurodegeneration through conserved mechanisms including but not limited to actin-overstabilization5,6, heterochromatin decondensation7, disruption in nuclear architecture8, and deficits in autophagy6. Importantly, these features of Drosophila models of tauopathy are also present in brains of patients with Alzheimer’s disease7–11. Importantly, animal models of tauopathy allow us to determine whether such observations in human brain are causally linked to neurodegeneration. Identifying novel cellular mediators of tau-induced neurodegeneration is essential for the development of disease-modifying, mechanism-based therapies that will improve outcomes for patients who, to date, are treated symptomatically with inconsistent outcomes.
The cellular nucleus is a membrane-bound organelle that encapsulates the genome and houses the transcriptional machinery that activates or silences gene expression. When a gene is activated, transcriptional machinery home to the gene and generate a pre-messenger RNA transcript. Within the nucleus, pre-messenger RNAs are processed by A) Splicing, a process whereby introns are removed and exons are retained within the transcript, B) Polyadenylation, in which a string of adenosines are added to the 3’ end of the transcript, and C) 5’ capping of the transcript with a guanine. After processing, mature messenger RNA (mRNA) transcripts are exported from the cell nucleus through nuclear pores into the cytoplasm where they are translated by ribosomes into protein.
While the above described “DNA makes RNA makes protein” is the central dogma of biology, not all protein-coding RNA transcripts are translated into functional proteins. Errors in transcription and RNA processing are common in healthy cells12,13. Cells have multiple strategies that dictate which RNAs are translated into protein, including quality control mechanisms that surveil the transcriptome and degrade or stop the translation of error-containing, “faulty” RNAs. While expression of a single faulty RNA is most often inconsequential, failure to clear faulty RNAs generated from error-prone transcription of thousands of genes can have catastrophic consequences for organismal fitness. For example, RNA transcripts harboring nonsense errors that generate a premature termination codon (PTC) contribute to about one third of inherited genetic disorders14. NMD is an evolutionarily conserved, translation-dependent RNA surveillance mechanism that recognizes and degrades PTC- and other error-containing transcripts as well as many naturally-occurring, error-free transcripts. The ability of NMD to also target ‘normal’ mRNA transcripts is important for a wide range of physiological pathways including differentiation and proliferation15,16, development17–20, viral defense21,22, stress response23–25, and neuronal activity (e.g. synaptic plasticity and axonal guidance)26–28. NMD machinery consists of several core factors that were first discovered in yeast as the up-frameshift-1 (Upf1), Upf2, and Upf3 genes29. Four additional conserved NMD factors were subsequently discovered in C. elegans: suppressor of morphological defects on genitalia 1 (Smg1), Smg5, Smg6, and Smg730. Knockdown of NMD core factors Upf119, Upf231, Smg132,33 or Smg634 are lethal in various model systems, indicating that proper function of NMD factors is essential for survival.
Studies in C. elegans report that neurons, unlike other cell types, retain NMD activity throughout aging35. Mutations in genes encoding NMD core factors Upf3B36–38, Upf3A39, Upf240 and Smg639 are associated with a wide clinical spectrum of intellectual disability in humans39, suggesting that proper clearance of RNAs by NMD is critical for brain health. Why might neurons require more persistent NMD throughout aging than other cell types? Unlike skin cells that can be replaced if lost or damaged, mature neurons are postmitotic, meaning they do not divide and thereby cannot ‘dilute’ toxic RNA during mitosis. In addition, neurons are morphologically complex and have a higher level of alternative splicing than other cell types in the body, leading to more diversity within the transcriptome and proteome41. Given that approximately one third of alternative transcripts harbor PTCs42, a significant portion of alternatively spliced transcripts are cleared via NMD.
Clearance of RNAs via NMD occurs at the cytoplasmic face of nuclear pores during or soon after mRNA export43– 45, suggesting that there is a spatially-defined checkpoint beyond which RNAs could evade NMD. The speed at which an RNA is exported from the nucleus is largely dependent on its proximity to nuclear pores that are embedded within the nuclear envelope46. Unlike the generally spherical shape of nuclei in healthy cells, nuclei of aged cells47, laminopathies48– 50, various cancers51–53, and neurons from patients with Alzheimer’s disease54 have morphological changes including lobulations and invaginations of the nuclear membrane into the nuclear interior. Analyses of post-mortem human Alzheimer’s disease brain tissue reveal that nuclear envelope invaginations are lined with nuclear pore complexes54, indicating that both the inner and outer nuclear membranes invaginate and that nuclear envelope invaginations contain a core of cytoplasm. Nuclear envelope invaginations are also present in Drosophila54 and mouse55 models of tauopathy as well as in induced pluripotent stem cell-derived neurons from patients carrying MAPT mutations56, suggesting that nuclear envelope invaginations observed in human Alzheimer’s disease brain are a consequence of pathological forms of tau. Genetic approaches to repair nuclear architecture significantly suppress neurodegeneration in brains of tau transgenic Drosophila, indicating that tau-induced disruption of nuclear morphology is a causal factor driving neuronal death54,57. Polyadenylated (polyA) RNA accumulates within nuclear envelope invaginations in neurons of tau transgenic Drosophila, and genetic or pharmacologic reduction of RNA export reduces the burden of RNA within nuclear envelope invaginations and suppresses tau-induced neurodegeneration58. In the current study, we utilize tau transgenic Drosophila to test the overall hypothesis that accumulation of RNA within nuclear envelope invaginations limits the extent to which faulty RNAs are cleared from the cell.
1.2 |. Study design and main results
Using Drosophila models of tauopathy, we find that transgenic expression of disease-associated mutant human tau (tauR406W) and wild-type human tau (tauWT)59 cause an overall deficit in NMD, and that NMD-sensitive transcripts that evade clearance can be translated into protein. We find that NMD targets accumulate within tau-induced nuclear envelope invaginations in brains of tau transgenic Drosophila. Experimentally activating NMD decreases accumulation of RNAs within nuclear envelope invaginations and suppresses neurotoxicity in tau transgenic Drosophila, suggesting that tau-induced deficits in NMD are causally associated with neurodegeneration. Mechanistically, we find that genetically decreasing RNA export reduces levels of transcripts that are normally cleared by NMD, raising the possibility that tau-induced increases in RNA export overwhelm the NMD machinery.
Recent work reports that tranilast (N-[3’,4’-dimethoxycinnamoyl]-anthranilic acid), an analog of a tryptophan metabolite approved for the treatment of bronchial asthma in Japan and South Korea60, is an effective activator of NMD in Drosophila61. We find that a ten-day treatment of tau transgenic Drosophila with tranilast activates NMD and suppresses tau-induced neurodegeneration and locomotor dysfunction, indicating that tau-induced deficits in NMD are druggable. Overall, our data provide new insights into how tau drives neuronal death and identify RNA quality control as a target for further investigation in the context of Alzheimer’s disease and related tauopathies.
1.3 |. Study conclusions, translational relevance, and therapeutic opportunities
Cellular homeostasis relies on genes being expressed at the right time, in the right place, at the right amount. Clearance of error-containing or faulty transcripts by NMD is integral to ensuring faithful expression of the genome at the protein level. Alterations in RNA processing and transport can lead to an abundance of error-containing transcripts that are likely to be degraded by NMD62,63. Elevation of such error-containing transcripts are observed in tauopathy64 and as a consequence of physiological aging in C. elegans35, suggesting that faulty RNA is either produced in higher quantities and/or that faulty RNA is not properly cleared from the cell in the context of tauopathy and aging. In tau transgenic mice and post-mortem human Alzheimer’s disease brain tissue, pathogenic forms of tau are reported to disrupt proper nucleocytoplasmic transport of RNA and protein55,57. Studies in a Drosophila model of tauopathy indicate that tau-induced increase in RNA export is a causal factor driving neurodegeneration and that RNAs accumulate within tau-induced invaginations of the nuclear envelope58. In the current study, we identify deficits in RNA quality control as a novel mechanism connecting aberrant RNA export to nuclear envelope invagination and neurodegeneration in tau transgenic Drosophila.
Overall, we find that mutant and wild-type forms of human tau limit NMD in the Drosophila brain, that genetic manipulation of NMD modifies tau-induced neuronal death, and that tau-induced deficits in NMD are amenable to pharmacological intervention. Our finding that overexpression of the NMD core factor Upf1 is neuroprotective in tauopathy is similar to reports in yeast65, rodent66,67, fly and human61,68 models of ALS-FTD, suggesting an overlapping pathophysiology of tauopathies and ALS, a motor neuron disease involving RNA dysregulation. Unlike our study, however, recent work in human cells harboring C9orf72 expansion68, ALS-causing mutations in FUS69, and C9orf72 motor neurons cultured from ALS patients report an increase in NMD activity in the disease context. Our studies point toward an alternative mechanism in tauopathy in which RNAs escape clearance via NMD as a consequence of increased RNA export and accumulation within nuclear envelope invaginations.
We58 and others68,69 detect RNA and NMD machinery within nuclear envelope invaginations. While these data suggest that RNA surveillance can occur within nuclear invaginations, we find that most RNA-enriched invaginations lack Upf1 in brains of tau transgenic Drosophila. As Upf1 is required for NMD in Drosophila18,70, these data suggest that a large majority of RNAs within invaginations are not subject to RNA surveillance via NMD in the context of tauopathy. We further find that genetic reduction of RNA export significantly reduces transcript levels of NMD targets in tau transgenic Drosophila. We speculate that existing NMD machinery is saturated in tauopathy due to increased RNA export, and that genetically elevating Upf1 suppresses tau neurotoxicity by increasing the pool of Upf1 that is available to clear RNA. Based on our studies to date, we favor a simple model in which RNA is exported through nuclear pores directly into tau-induced nuclear envelope invaginations, where it is shielded from cellular RNA surveillance mechanisms and is translated into faulty protein. To directly test this model, live cell imaging will be necessary to clarify the export path of RNAs, to quantify the longevity of NMD-sensitive RNAs that are exported into nuclear invaginations in tauopathy, and to monitor the location and degree to which faulty RNA transcripts are translated into protein.
Many cellular phenotypes of Drosophila tauopathy models are conserved in human Alzheimer’s disease, including but not limited to oxidative stress71, DNA damage72, actin overstabilization5, nuclear pleomorphisms54, epigenetic changes7, transposable element activation73,74, aberrant activation of the cell cycle in postmitotic neurons75, and progressive neurodegeneration. There are clear limitations, however, of using Drosophila to model human neurological disorders. Drosophila neurons are smaller and have less processes extending from the cell body than their vertebrate counterparts76,77, lack axon-specific neurofilaments78, and represent the majority of cells in the central nervous system79. Glia are ten times more abundant than neurons in the human brain, suggesting that an increased glia:neuron ratio represents greater brain complexity. Vertebrate-specific neuronal features may explain why large aggregates of misfolded tau protein form neurofibrillary tangles in brains of Alzheimer’s disease patients but not in Drosophila models of tauopathy80. Abnormally phosphorylated tau and disease-associated conformations of tau do, however, accumulate with age in brains of tau transgenic Drosophila similar to early stages of human Alzheimer’s disease80,81.
Based on our reductionist approach that leverages simple Drosophila models of tauopathy in which transgenic human tau is expressed in all neurons of the adult fly brain, we speculate that cells harboring pathogenic forms of tau in Alzheimer’s disease and related tauopathies would exhibit a deficit in NMD. Ideally, one would sort pathogenic tau-containing neurons from patient brain samples and perform long-read RNA sequencing to quantify the burden of NMD targets in pathogenic tau-containing neurons versus those that lack pathogenic forms of tau. Similarly, one could analyze NMD targets in specific circuits of the human brain that are particularly affected in tauopathies. These experiments will, however, be costly and will require large amounts of brain tissue. In the absence of such studies, two recent analyses provide some clue that NMD may be functioning at a deficit in human Alzheimer’s disease. Both studies report increased intron retention in transcriptomes from patients with Alzheimer’s disease82,83. Drosophila models of tauopathy also exhibit intron retention82, suggesting that human Alzheimer’s disease-associated intron retention is a consequence of pathogenic forms of tau. Intron retention is known to trigger NMD, as introns often introduce a PTC into a mature mRNA84,85. While no analyses of human Alzheimer’s disease have specifically focused on NMD, an increase in transcripts with intron retention in brains of patients with human Alzheimer’s disease brains is consistent with deficits in NMD.
Aged Drosophila have significantly altered splicing patterns, including increased intron retention of genes, some of which are associated with Alzheimer’s disease82. Since intron retention may trigger NMD, these data suggest a potential link between aberrant splicing, NMD, and age-related diseases like Alzheimer’s disease. Analysis of gene expression profiles in cells lacking core NMD factors indicate that NMD targets about 10% of the transcriptome in Drosophila18 and humans86, suggesting that while the select targets may differ, the biological impact of deficits in NMD is shared between the two species. The steady-state level of RNA depends on the ratio between synthesis and degradation, making the use of steady-state RNA levels to infer NMD activity intrinsically limited. These limitations can be overcome by methods more amenable to cell culture that compare a PTC-containing, presumably NMD-targeted transcript relative to the errorless, NMD-insensitive transcript87,88. Our studies in Drosophila models of tauopathy should thus be validated in other model systems and critically and cautiously applied to human Alzheimer’s disease and related tauopathies.
Overall, our data identifies tau-induced deficits in NMD as a pharmacologically targetable driver of neurodegeneration that occurs early in the disease process. We find that tranilast, a well-tolerated and low toxicity drug that penetrates the blood brain barrier60, activates NMD and suppresses tau-induced neurotoxicity in Drosophila. The mechanism of tranilast-induced NMD activation is unclear and additional research on its action in the brain is required to assess the potential of tranilast as a tau therapy. Based on our findings that deficits in NMD activity occur prior to neurodegeneration in tau transgenic Drosophila, it is possible that NMD activators have disease-modifying potential during the early asymptomatic stages of Alzheimer’s disease and related tauopathies. Our experiments are thus designed to identify and target early events, and to focus on mechanisms in which genetic manipulation modifies tau-induced neuronal death. While compounds that activate NMD would also be attractive for some cancer types89–92, there are no publicly available NMD-activating compounds. A previous screen for NMD inhibitors happened to also identify 14 candidate compounds that enhance NMD activity in human cells87, among which four were further studied in an ALS-FTD model. Only tranilast was validated as an NMD activator in vivo61. To our knowledge, there are no other studies testing NMD activators in the context of neurodegeneration.
A major challenge to the development of NMD activators is knowing which NMD machinery to target. The mechanisms by which Upf1 is phosphorylated93,94, PTCs are recognized95,96, and mRNA is degraded97,98 vary across species. In fact, the efficiency of NMD is tissue-specific35,99 and highly variable from cell-to-cell100 and between individuals38,101. The search for small molecules that increase NMD activity will require more rigorous testing of the current model in which aberrant translation termination is thought to be the primary activation signal for NMD. Genetic modifier screening and small molecule screening using cell-based reporters of NMD activity may identify additional targets and approaches for further therapeutic development. The data described herein suggest that small molecules that specifically activate NMD in the brain, or gene therapy designed to modulate expression of NMD core factors in neurons are plausible NMD activation strategies for tauopathies. While our findings require further testing in vertebrate models, our study suggest that patients with Alzheimer’s disease and related tauopathies could potentially benefit from therapy based on NMD activation.
2 |. CONSOLIDATED RESULTS AND STUDY DESIGN
We detect deficits in NMD in brains of adult tau transgenic Drosophila using two complementary approaches. We find that panneuronal expression of human tauR406W and tauWT in the Drosophila brain disrupts the ability of NMD to clear a NMD-sensitive fluorescent reporter102 and RNA transcripts of endogenous NMD targets Gadd4561,103,104 and Arc161. As phenotypes of tauWT transgenic Drosophila are delayed compared to tauR406W transgenic Drosophila, and tauR406W transgenic Drosophila feature a moderate degree of neurodegeneration at 10 days of age that is convenient for genetic analyses59, we used this MAPT-mutation based model for subsequent mechanistic inquiry into the cause and consequence of blunted NMD in tauopathy. We find that NMD activity is at a deficit in tauR406W transgenic Drosophila compared to age-matched controls prior to overt neurodegeneration.
To determine if NMD deficits causally mediate tau-induced neurodegeneration in Drosophila, we next asked if genetic manipulation of NMD core factors alters the degree of neurotoxicity in brains of tauR406W transgenic Drosophila. We quantified tauR406W-induced neurodegeneration in response to genetic manipulation of NMD core factors Upf1, Upf2, Upf3 and Smg5 using terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), which detects DNA fragmentation associated with apoptosis7,75,80. Panneuronal RNAi-mediated depletion of Upf1, Upf3 and Smg5 and expression of a dominant-negative form of Upf2 significantly enhance tauR406W-induced neurodegeneration in Drosophila, while panneuronal overexpression of wild-type Upf1 and Upf2 significantly suppress tauR406W-induced neurodegeneration.
Having previously reported that polyA RNA accumulates within nuclear envelope invaginations in tauopathy58, we next asked if faulty RNAs also accumulate within invaginations. We detect protein encoded by a reporter transcript that is normally cleared by NMD within nuclear invaginations of tauR406W transgenic Drosophila. This finding suggests that NMD-sensitive targets are present within nuclear envelope invaginations in brains of tauR406W transgenic Drosophila, and that such NMD-sensitive transcripts are effectively translated into protein within invaginations. We hypothesized that RNA within nuclear invaginations may not have access to NMD machinery. We visualized polyA RNA, Upf1, and the lamin nucleoskeleton using fluorescence in situ hybridization combined with immunofluorescence (FISH-IF). A quantitative evaluation revealed that Upf1 is largely excluded from RNA-enriched nuclear envelope invaginations in brains of tauR406W transgenic Drosophila, suggesting that Upf1 is physically restricted from RNA within these invaginations, that faulty RNA is in excess to endogenous Upf1, or that Upf1 is recruited to some but not all RNAs within invaginations.
To determine if overexpression of Upf1 prevents neurodegeneration by enhancing the ability of NMD to clear RNA transcripts, in particular RNA within nuclear invaginations, we quantified the fraction of nuclear envelope invaginations that harbor RNA in brains of tauR406W transgenic Drosophila with and without panneuronal overexpression of Upf1. We find that genetic activation of NMD significantly decreases the incidence of RNA-containing invaginations, as predicted. These results suggest that RNAs within nuclear invaginations are subject to degradation by NMD, and that activation of NMD may be an effective strategy to decrease neuronal loss in tauopathy.
Given that many of the clinical trial failures for Alzheimer’s disease and related tauopathies stem from an inadequate understanding of the mechanisms driving neurodegeneration, we took additional steps to examine a potential mechanism underlying tau-induced deficits in NMD. Having previously reported that reducing RNA export clears RNA from nuclear envelope invaginations and suppresses tau-induced neurotoxicity58, we hypothesized that tau-mediated increase in RNA export overwhelms the ability of NMD machinery to clear target RNA transcripts. We find that panneuronal RNAi-mediated reduction of nuclear export factors sbr and Nxt1, Drosophila homologues of human nuclear transport factor 2 like export factor 1 (NXT1) and nuclear RNA export factor 1 (NXF1), significantly reduces protein levels of the NMD-sensitive fluorescent reporter in Drosophila and reduces transcript levels of Gadd45 and Arc1 back to control levels in tauR406W transgenic Drosophila. These findings suggest that an increase in nuclear RNA export limits NMD in tauopathy.
Equally important for future drug development efforts is to determine if tau-induced deficits in NMD are pharmaceutically targetable. Tranilast is an anti-inflammatory drug and NMD activator in Drosophila61. We find that tauR406W transgenic Drosophila fed tranilast have greater NMD activity and significantly reduced neurodegeneration and locomotor deficits. Taken together, our findings reveal a new, non-canonical function of tau and highlight tau-induced deficits in NMD as a pharmacologically targetable, mechanistic driver of neurodegeneration with therapeutic potential for Alzheimer’s disease and related tauopathies.
3 |. DETAILED METHODS AND RESULTS
3.1.1 |. Drosophila genetics
Drosophila melanogaster crosses and aging were performed at 25°C with a 12 hour light/dark cycle on standard fly food (Bloomington formulation) or fly food containing drug (as described below). Aging flies were transferred to fresh food every other day. In all experiments, expression of transgenes and RNAi-mediated knockdown in Drosophila was achieved using the panneuronal elav promoter and the GAL4-UAS system. Drosophila lines were obtained from Bloomington Drosophila Stock Center. Drosophila RNAi Screening Center (DRSC) - Transgenic RNAi Project (TRiP)105 and Vienna Drosophila Resource Center (VDRC, www.vdrc.at)106 supplied transgenic RNAi fly lines (Supplemental Table 1). UAS-tauR406W and UAS-tauWT flies were provided by Dr. Mel Feany54 and Drosophila expressing the NMD fluorescent reporter were provided by Dr. Mark Metzstein102. All experiments used an equal number of male and female flies unless otherwise noted.
3.1.2 |. Fluorescence in situ hybridization with immunofluorescence
For fluorescence in situ hybridization combined with immunofluorescence, Drosophila brains were dissected in 1X PBS and fixed in 4% PFA for 30 minutes. After washing once in PBS plus 0.3% Triton, samples were incubated with 1 ng/μL 5’3’-DIG labelled probe in pre-heated hybridization buffer for at least three hours at 37°C. Samples were washed in 2X SSC and blocked in PBS plus 2% milk and 0.3% Triton for at least 30 mins. Primary antibodies (Supplemental Table 2) were added to samples for overnight incubation at room temperature. After washing, secondary antibodies donkey anti-mouse Alexa Fluor 555, anti-rabbit Alexa Fluor 488, or anti-goat Alexa Fluor 647 (1:200, Thermo Fisher Scientific) were incubated at room temperature for two hours before mounting samples with Fluoromount G with DAPI (Southern Biotech). The number of nuclear invaginations containing polyA RNA were scored as previously reported54,58. RNA was observed as a focal increase contained completely within a nuclear envelope invagination as imaged with lamin staining. All comparisons were made between samples processed at the same time.
For Upf1 and lamin immunofluorescence, we used Drosophila heads fixed in formalin, embedded in paraffin and serially sectioned at 4 μm through the entire brain. Slides were subjected to antigen retrieval in sodium citrate for 20 minutes. After washing, brains were blocked with 2% milk in 0.3% Triton in PBS and incubated with a primary antibody overnight in a humidifier chamber at room temperature. Slides were washed again prior to incubation with Alexa Fluor-conjugated secondary antibodies for two hours at room temperature. Drosophila brains were visualized on a Zeiss confocal microscope using Airyscanning technology to image nuclei in a single slice with high resolution.
To quantify TUNEL-positive neurons in Drosophila brains, we used a commercially available kit for TUNEL staining (Calbiochem, TdT FragEL) on formalin-fixed, paraffin embedded Drosophila heads sectioned at 4 μm. Following the kit protocol, DAB was used for secondary detection of biotin-labelled deoxynucleotides at exposed ends of DNA fragments. Brightfield microscopy was used to quantify TUNEL-positive neurons throughout the entire fly brain.
3.1.3 |. Immunoblotting
One male and one female Drosophila head were homogenized in 2X Laemmelli buffer (Invitrogen) and boiled for 10 minutes. Samples were analyzed by 10% SDS-PAGE and transferred to nitrocellulose membranes in transfer buffer with methanol. Ponceau S was added to membranes to assess equal loading before blocking membranes for 10–30 mins with 2% milk in PBS plus 0.05% Tween. Membranes were incubated overnight at 4°C with primary antibody. After washing with 1X PBS plus 0.05% Tween, membranes were incubated with HRP-conjugated secondary antibodies for two hours at room temperature and developed with an enhanced chemiluminescent substrate. Blots were visualized with a FluorChem HD2 imager (ProteinSimple) and the relative density of each band was quantified in ImageJ.
3.1.4 |. Drug treatments
Tranilast (Sigma-Aldrich, T0318) was dissolved in DMSO and 1 μL was added to 1 mL fly food to a final concentration of 10μM. Drosophila were transferred to vials containing Tranilast- or DMSO-treated food at day one of adulthood and flipped into fresh drug or vehicle treated food every other day. At day ten of adulthood, flies were frozen or fixed for subsequent analyses.
3.1.5 |. Locomotor Assay
Drosophila fed tranilast or vehicle (DMSO) for ten or twenty days were individually transferred to drug-treated food at least 24 hours prior to measuring walking activity, as described previously7.
3.1.6 |. NMD reporter assays
Drosophila harboring UAS-nlsDsRed2::SV40 3ʹUTR on the X chromosome (NMD reporter) were balanced with FM7. Virgins of genotype UAS-nlsDsRed2::SV40 3ʹUTR/FM7 were mated to elav-Gal4/+ or elav-Gal4;+/+;TauR406W/+ males. Female progeny of genotype elav-Gal4/UAS-nlsDsRed2::SV40 3ʹUTR;+/+;TauR406W/+, elav-Gal4/UAS-nlsDsRed2::SV40 3ʹUTR;+/+;TauWT/+ and elav-Gal4/UAS-nlsDsRed2::SV40 3ʹUTR;+/+;+/+ were aged to ten or twenty days post-eclosion and frozen at −80°C prior to digital PCR (dPCR) or ELISA assays.
Since experiments utilizing the NMD reporter require two genetic elements (NMD reporter and elav-Gal4) on the X chromosome, only female flies were used for analysis. For each biological sample, five frozen Drosophila heads were homogenized in 15 μL RIPA buffer with protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, 78440), diluted in 185 μL Assay Diluent, and 200 μL of each sample added to the anti-RFP antibody-coated plate in duplicate for quantification of RFP according to the RFP ELISA protocol (Cell BioLabs, AKR-122). Absorbance for each well was measured 5 mins after stopping the enzyme reaction on a GloMax Discover Plate Reader (Promega) using 450 nm as the primary wavelength.
3.1.7 |. Digital PCR
Absolute quantification of Gadd45, Arc1, and NMD factors (Upf1, Upf2, Upf3, and Smg5) was performed using the Applied Biosystems QuantStudio 3D Digital PCR system. To quantify RNA expression, six Drosophila heads were homogenized in RIPA buffer plus protease and phosphatase inhibitor cocktail and RNA extracted using TRIzol (Invitrogen, 15596026) according to the manufacturer’s protocol. RNA concentrations were measured using Nanodrop 8000 spectrophotometer (Thermo Scientific) and equal amounts of RNA added to a reverse transcription reaction (cDNA Reverse Transcription Kit, Applied Biosystems). cDNA was loaded into a PCR chip, sealed, and amplified (up to 24 chips at one time) before analysis to determine the absolute concentration in copies/μL. The primers and probe for each Drosophila mRNA target were predesigned TaqMan gene expression (Supplemental Table 3).
3.1.8 |. Statistical analyses
For all experiments, we performed either G*Power analysis107 or used previously reported sample sizes7,54,58,73 to determine sample size for each experiment. All reported n’s represent biological replicates. Statistical analyses were performed using Prism 7 (GraphPad Software Inc., La Jolla, CA). Unless otherwise specified, results were considered significant if P<0.05 by ANOVA and Tukey’s test for multiple comparisons, and student’s t-test for single comparisons. To limit bias, samples were randomized and investigators were blinded to genotype in all immunofluorescence, immunohistochemistry, and locomotor assays when possible.
3.2 |. RESULTS
3.2.1 |. Deficits in NMD occur prior to neurodegeneration in tauR406W transgenic Drosophila
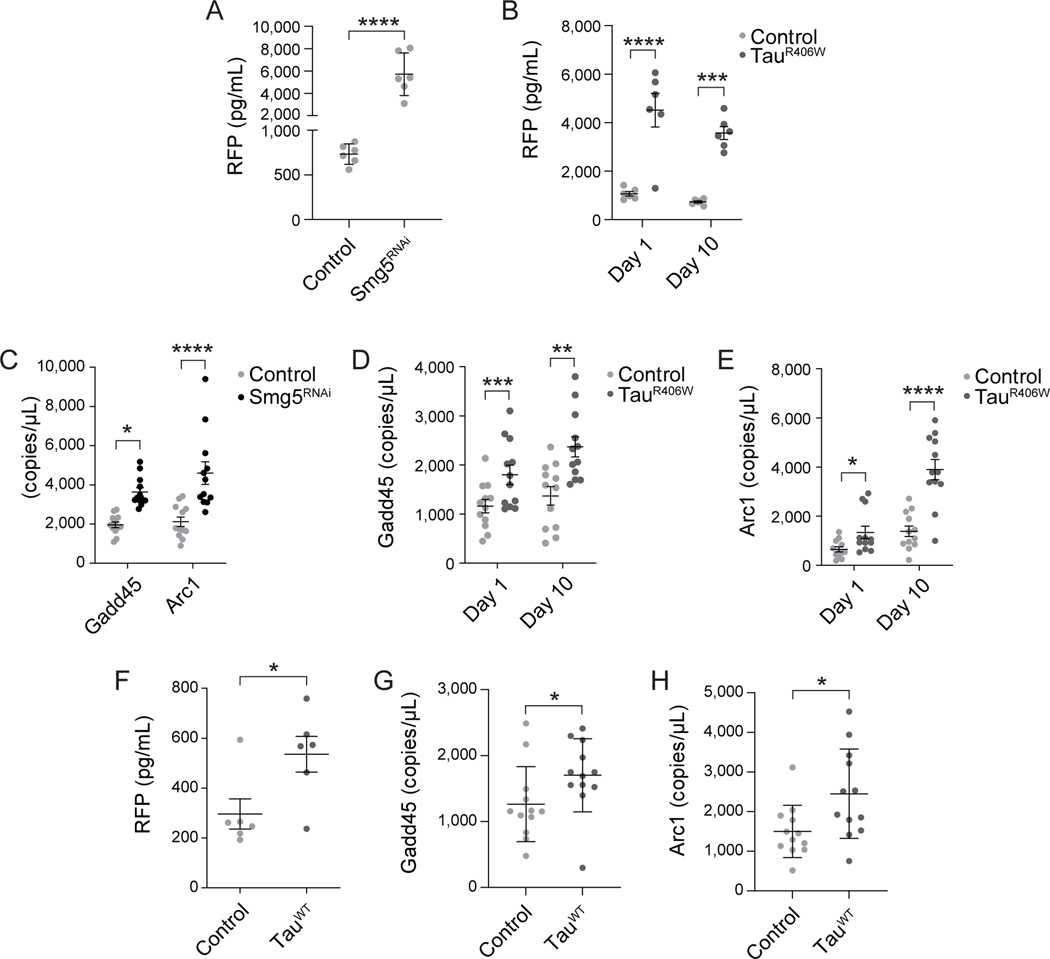
To evaluate overall activity of NMD in tauopathy, we quantified NMD at days one and ten of adulthood in brains of control and tauR406W transgenic Drosophila using two complementary approaches. We first utilized a fluorescent reporter previously developed to monitor NMD activity in Drosophila102. The NMD reporter utilizes the SV40 3’ small T antigen intron and a polyA signal in the 3’ UTR to trigger NMD-mediated degradation of a red fluorescent protein (RFP) transcript. High RFP levels indicate that the transcript has evaded clearance by NMD and has been translated into protein. We first validated that the NMD reporter is activated as expected when NMD is dysfunctional in the adult Drosophila brain. We find that panneuronal knockdown of an NMD core factor, Smg5, effectively activates the NMD-sensitive reporter based on RFP ELISA, thus validating the utility of the reporter in the adult Drosophila brain (Fig. 1A). Similarly, we find robust activation of the NMD reporter in brains of tauR406W transgenic Drosophila compared to control, indicating that tauR406W transgenic flies fail to clear the “faulty” RNA encoded by the reporter and that the faulty RNA is effectively translated into protein. We find that tauR406W-induced deficits in NMD occur early in the disease process, as the NMD reporter is activated at day one of adulthood, which is prior to neurodegeneration in this model, and at day 10 of adulthood, which is approximately 33% of the maximum lifespan of tauR406W transgenic Drosophila80 (Fig. 1B).
Figure 1 |. Reduced clearance of aberrant transcripts by NMD in tau transgenic Drosophila.
A) Panneuronal RNAi-mediated reduction of Smg5 limits NMD in Drosophila based on activation of an NMD sensitive reporter. n=6 biological replicates per genotype, per age. B) Panneuronal expression of transgenic human tauR406W limits NMD in Drosophila at day one and day ten of adulthood based on activation of an NMD sensitive reporter. n=6 biological replicates per genotype, per age. Transcript levels of Gadd45 and Arc1 are significantly elevated in Drosophila with panneuronal RNAi-mediated reduction of Smg5 (C) and in tauR406W transgenic Drosophila at day one and day ten of adulthood (D, E). F) Panneuronal expression of transgenic human tauWT limits NMD in Drosophila at day twenty of adulthood based on activation of an NMD sensitive reporter. n=6 biological replicates per genotype. Panneuronal expression of tauWT elevates Gadd45 (G) and Arc1 (H) transcript levels compared to controls. n=12 biological replicates per genotype. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, unpaired t-test and multiple unpaired t-test. Error bars=SEM. Full genotypes are included in Supplemental Table 1.
As a second approach to quantify NMD activity, we measured RNA levels of Gadd45 and Arc1, which are well-established normal, non-error containing transcripts that are known to be cleared by NMD61,103,104. As expected, we find that panneuronal knockdown of Smg5 increases transcript levels of Gadd45 and Arc1 in the adult Drosophila brain, thus validating that these RNAs are indeed NMD targets (Fig. 1C). Consistent with our findings using the NMD-sensitive reporter, we find that Gadd45 (Fig. 1D) and Arc1 (Fig. 1E) transcript levels are elevated at both day one and day ten of adulthood in brains of tauR406W transgenic Drosophila. Taken together, these data suggest that tauR406W transgenic Drosophila fail to clear faulty and normal transcripts that are typically degraded via NMD.
While Drosophila featuring panneuronal expression of wild-type tau, which models sporadic tauopathies, feature delayed phenotypes and a milder degree of neurotoxicity compared to tauR406W Drosophila, previous studies in Drosophila report that transgenic expression of human wild-type tau or MAPT mutations drive neurodegeneration through conserved mechanisms5–8. To determine if deficits in NMD occur in a Drosophila model of sporadic tauopathy, we analyzed NMD in flies overexpressing wild-type human tau108. We analyzed NMD at day twenty of adulthood due to the milder phenotypes of this model. We find that panneuronal transgenic expression of tauWT significantly increases fluorescence of the NMD-sensitive reporter at twenty days of adulthood (Fig. 1F), indicating defective NMD. In addition, we find elevated levels of Gadd45 and Arc1 transcripts in head of twenty-day-old tauWT flies (Fig. 1G, H), further suggesting blunted NMD in this model. These findings suggest that the NMD deficits we observe in tauR406W transgenic Drosophila are also relevant to sporadic forms of tauopathy.
3.2.2 |. Genetic manipulation of NMD modifies tauR406W-induced neurodegeneration
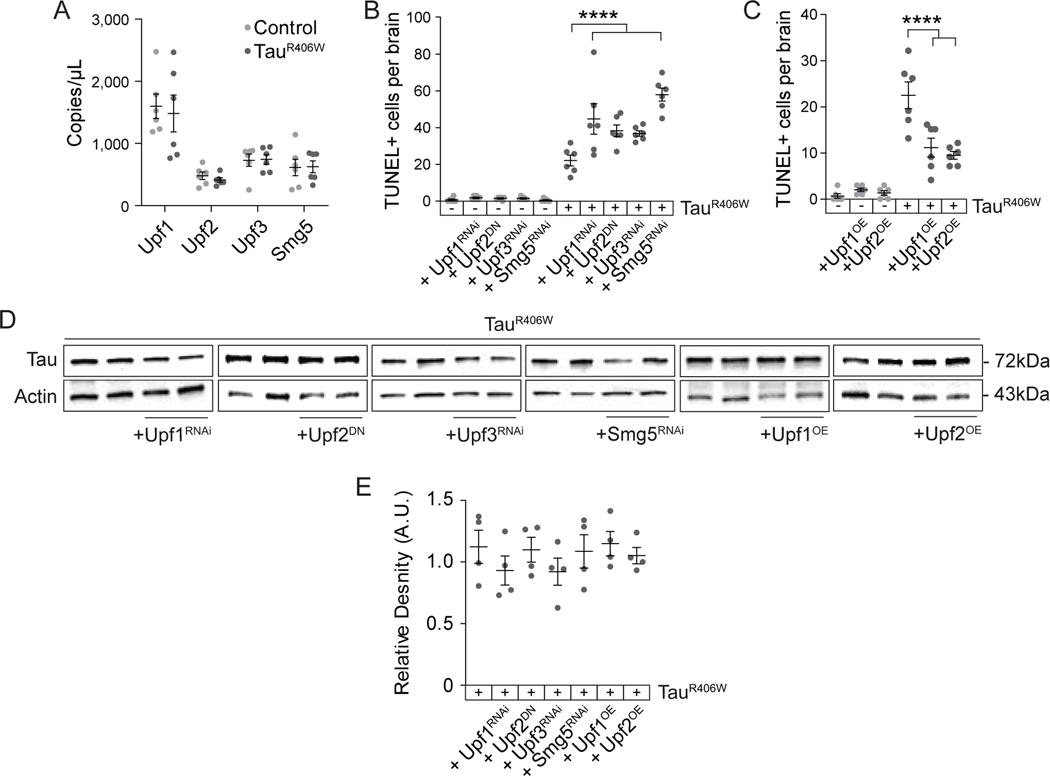
We next determined if tau-induced deficits in NMD are causally associated with neurodegeneration. For this analysis and subsequent experiments, we utilized the tauR406W transgenic flies, as this model features a moderate level of degeneration at day ten of adulthood that is convenient for genetic analyses and precedes an exponential decline in survivorship80,109. We first quantified overall transcript levels of NMD core factors Upf1, Upf2, Upf3 and Smg5 using dPCR, and find that these transcripts are unchanged in brains of control versus tauR406W transgenic Drosophila (Fig. 2A). We genetically manipulated NMD core factors Upf1, Upf2, Upf3 and Smg5 in tauR406W transgenic Drosophila and quantified tauR406W-induced neurotoxicity using TUNEL. We find that panneuronal expression of a dominant-negative form of Upf2 (Upf2E801R, henceforth referred to as “Upf2DN”)17 and panneuronal RNAi-mediated reduction of Upf1, Upf3 or Smg5 significantly enhance tauR406W-induced neurodegeneration (Fig. 2B). Conversely, we find that panneuronal overexpression of wild-type Upf1 or Upf2 significantly suppress tauR406W-induced neurodegeneration (Fig. 2C). Overall protein levels of transgenic tauR406W are unchanged as a consequence of genetic manipulation of NMD core factors (Fig. 2D, E), excluding the possibility that modification of tauR406W neurotoxicity is a result of differential transgenic tauR406W expression. These findings indicate that tau-induced deficits in NMD causally mediate neurodegeneration.
Figure 2 |. Genetic manipulation of NMD modifies tauR406W-induced neurodegeneration in Drosophila.
A) Transcripts levels of Upf1, Upf2, Upf3, and Smg5 are unchanged in control versus tauR406W Drosophila head lysates based on dPCR. B) RNAi-mediated reduction of Upf1, Upf3, and Smg5, and overexpression of Upf2 harboring a dominant-negative mutation significantly enhance tauR406W-induced neuronal death based on TUNEL staining in Drosophila brains. C) Panneuronal overexpression of wild-type Upf1 or Upf2 significantly suppress tauR406W-induced neuronal death based on TUNEL staining in Drosophila brains. D) Genetic manipulation of NMD core factors does not change total protein levels of transgenic tauR406W based on western blotting. E) Quantification of (D), total levels of tau protein normalized to actin. n=4–6 biological replicates per genotype. All assays were performed at day ten of adulthood. ****p<0.0001, one-way ANOVA. Error bars=SEM. Full genotypes are included in Supplemental Table 1.
3.2.3 |. Tau-induced nuclear envelope invaginations lack NMD machinery and accumulate protein encoded by faulty RNA in the Drosophila brain.
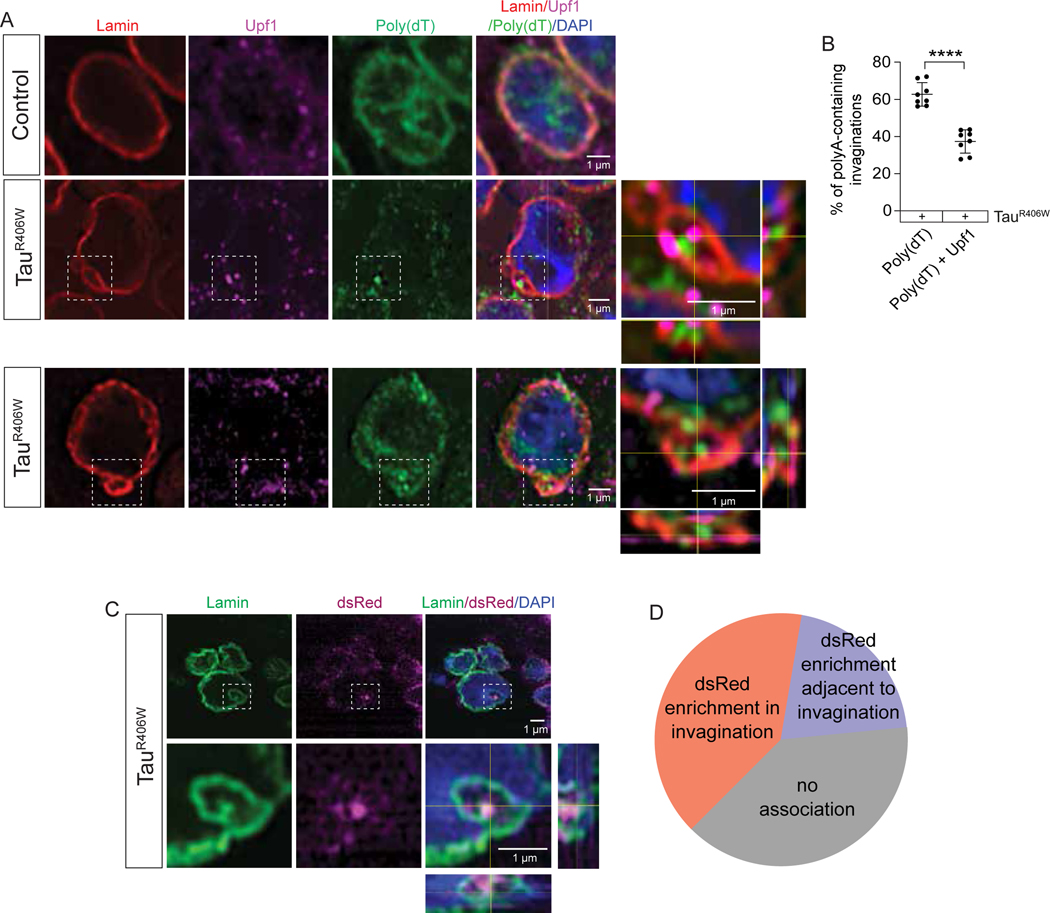
We became interested in a potential link between failed NMD and tau-induced nuclear envelope invaginations based on previous studies from our lab and others reporting that pathogenic forms of tau cause nuclear envelope invaginations56,109 and that RNA accumulates within and adjacent to tau-induced nuclear envelope invaginations58. We hypothesized that RNA may accumulate within such invaginations because it is failed to be cleared by NMD. We first determined if NMD machinery is present alongside the RNA that accumulates within tauR406W-induced nuclear invaginations. We visualized RNA, Upf1, and their relationship with nuclear envelope invaginations by combining poly(dT) FISH with immunofluorescence-based detection of the lamin nucleoskeleton, which lines nuclear invaginations44, and Upf1 (Fig. 3A). In line with previous studies, we find that Upf1 and polyA RNA are enriched at the nuclear envelope in cells of the Drosophila brain and are also present within the cytoplasm and nucleus (Supplemental Fig. 1). While we detect both Upf1 and RNA within about 38% of invaginations in brains of tauR406W transgenic Drosophila, we find that the majority (∼62%) of RNA-enriched invaginations lack Upf1 (Fig. 3B). As Upf1 is required for NMD in Drosophila18,70, these data indicate that most RNA-containing nuclear envelope invaginations are not actively clearing faulty RNA via NMD.
Figure 3 |. TauR406W-induced nuclear envelope invaginations lack NMD machinery and accumulate protein encoded by faulty RNA in the Drosophila brain.
A) FISH/IF based detection of Upf1, Lamin and polyA RNA in brains of tauR406W transgenic Drosophila shows polyA RNA-enriched nuclear envelope invaginations containing Upf1 and lacking Upf1. Orthogonal views of the zoom region depicted show polyA RNA and/or Upf1 staining within lamin invaginations. B) Quantification of (A), percentage of polyA RNA-positive nuclear envelope invaginations containing versus lacking Upf1. C) Co-staining brains of tauR406W transgenic Drosophila harboring the NMD-sensitive reporter with antibodies detecting lamin and dsRed indicate that reporter expression is present within nuclear envelope invaginations. D) Quantification of (C). All assays were performed at day ten of adulthood. n=6–8 biological replicates per genotype. p****<0.0001, unpaired t-test. Error bars=SEM. Full genotypes are included in Supplemental Table 1.
We next asked if NMD targets accumulate within nuclear envelope invaginations of tauR406W transgenic Drosophila. Using the NMD-sensitive reporter as measure of general NMD, we stained brains of tauR406W transgenic Drosophila with antibodies detecting dsRed, which detects protein expressed by the NMD reporter when its transcript fails to be cleared by NMD, and lamin. We observe dsRed puncta within ∼40% of nuclear invaginations of tauR406W Drosophila (Fig. 3C, D), suggesting that NMD targets are not only present within nuclear invaginations, but that they are actively translated into protein.
3.2.4 |. Experimental activation of NMD decreases RNA accumulation in nuclear invaginations in brains of tauR406W transgenic Drosophila
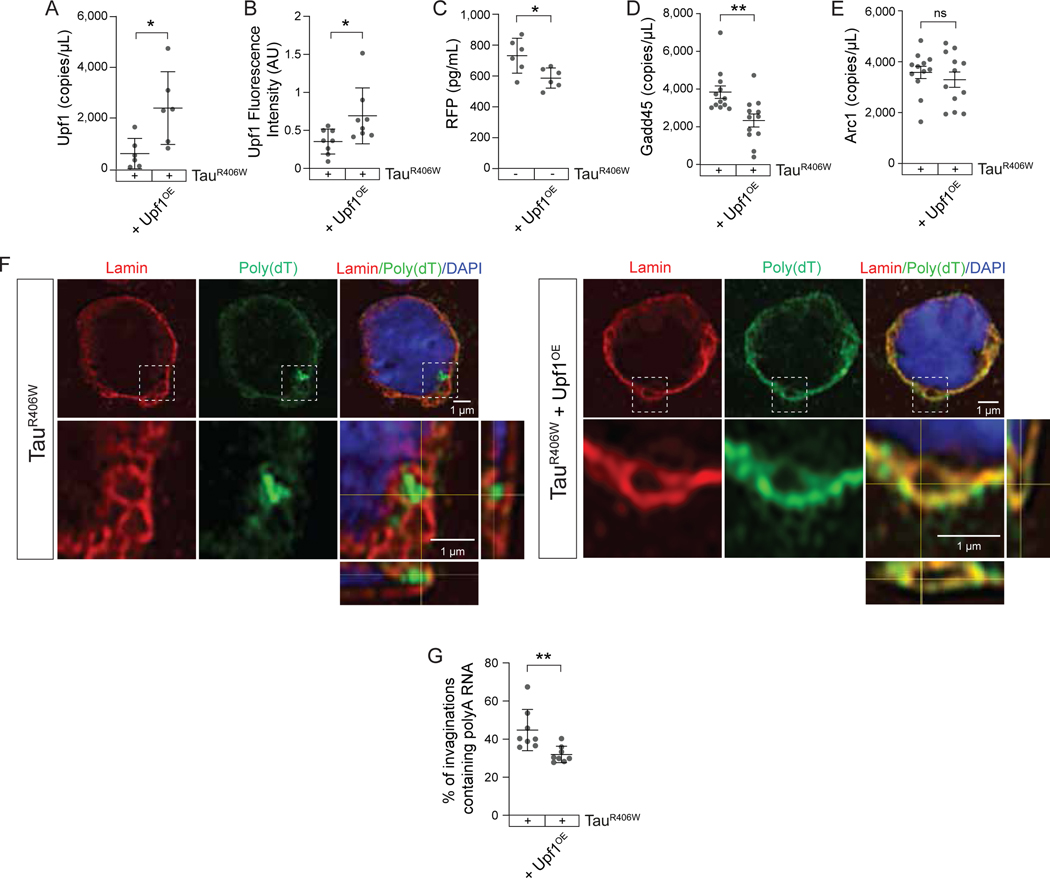
Having established that genetically increasing NMD suppresses tauR406W-induced neurotoxicity, that tauR406W-induced nuclear envelope invaginations generally lack Upf1, and that protein encoded by faulty RNA is present within tauR406W-induced nuclear envelope invaginations, we next asked if RNA can be cleared from nuclear envelope invaginations by experimentally increasing NMD via Upf1 overexpression. We first quantified the degree of Upf1 overexpression and NMD activity in tau transgenic Drosophila with panneuronal overexpression of Upf1. We detect a four-fold increase in Upf1 RNA levels in heads of Upf1OE tauR406W transgenic Drosophila (Fig. 4A) alongside a two-fold increase in Upf1 protein levels (Fig. 4B). To determine the extent to which Upf1 overexpression generally functions to clear NMD targets, we determined the ability of Upf1 overexpression to reduce activation of the NMD-sensitive reporter. This experiment was performed in the absence of the human tauR406W transgene due to the number of transgenes that would be required for this analysis in tauR406W transgenic Drosophila. We find that panneuronal overexpression of Upf1 significantly reduces activation of the NMD sensitive reporter (Fig. 4C), suggesting that Upf1 overexpression is sufficient to activate NMD in the adult Drosophila brain. We further find that panneuronal Upf1 overexpression is sufficient to significantly reduce Gadd45 (Fig. 4D), but not Arc1 (Fig. 4E) transcript levels in brains of tauR406W transgenic Drosophila, suggesting that Upf1 overexpression effectively clears some, but not all, endogenous targets of NMD. Having found that panneuronal Upf1 overexpression effectively increases Upf1 protein levels and activates NMD, we next asked if Upf1 overexpression affects the burden of RNAs that accumulate within tauR406W-induced nuclear envelope invaginations. While we frequently detect enrichment of RNA within nuclear invaginations based on poly(dT)/lamin FISH-IF in brains of tauR406W transgenic Drosophila, we find significantly less polyA RNA enrichment within nuclear envelope invaginations in brains of tauR406W transgenic Drosophila with Upf1 overexpression (Fig. 4F, G), providing additional evidence that RNAs that accumulate in nuclear envelope invaginations are targets of NMD.
Figure 4 |. Experimentally elevating NMD decreases RNA accumulation in nuclear envelope invaginations in brains of tauR406W transgenic Drosophila.
A) Upf1 RNA levels in tauR406W Drosophila with and without Upf1 overexpression. B) Immunofluorescence-based quantification of Upf1 protein levels in tauR406W Drosophila with and without Upf1 overexpression. C) ELISA-based NMD-sensitive reporter activity in control Drosophila with and without Upf1 overexpression. Gadd45 (D) and Arc1 (E) transcript levels based on dPCR in tauR406W Drosophila with and without Upf1 overexpression. F) FISH/IF-based detection of polyA RNA and lamin in tauR406W Drosophila with and without Upf1 overexpression. Representative images include an orthogonal view of zoom region depicted showing polyA RNA-positive and -negative invaginations. G) Quantification of (F), percentage of nuclear envelope invaginations containing polyA RNA in flies of the indicated genotype. All assays were performed at day ten of adulthood. n=6–12 biological replicates per genotype. p*<0.05, one-way ANOVA. Error bars=SEM. Full genotypes are included in Supplemental Table 1.
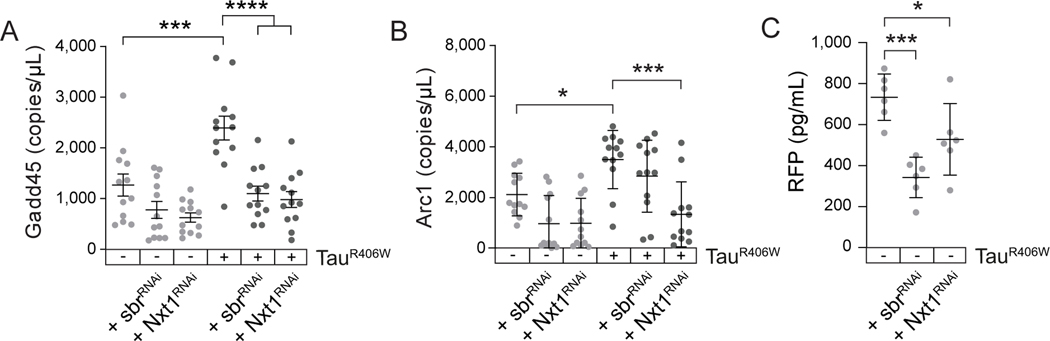
3.2.5 |. Genetic reduction of RNA export suppresses NMD deficits in tauR406W transgenic Drosophila
We have previously reported that experimentally reducing RNA export factors sbr and Nxt1 significantly diminishes deposition of RNA within tau-induced nuclear envelope invaginations and suppresses tau neurotoxicity in Drosophila58, suggesting that RNAs are actively transported into invaginations. We hypothesized that the tau-induced increase in RNA export could perhaps overwhelm NMD machinery and thus reduce the ability of NMD to effectively degrade its targets. To test this hypothesis, we analyzed Gadd45 and Arc1 transcript levels in tauR406W transgenic Drosophila in response to panneuronal RNAi-mediated knockdown of sbr or Nxt1. Indeed, we find that reduction of sbr or Nxt1 significantly reduces RNA levels of Gadd45 in tauR406W transgenic Drosophila (Fig. 5A), and that Nxt1RNAi effectively reduces RNA levels of Arc1 in tauR406W transgenic Drosophila (Fig. 5B), suggesting that genetic manipulation of RNA export mediates NMD in tauR406W transgenic Drosophila. While experimentally reducing RNA export is not sufficient to significantly reduce Gadd45 or Arc1 RNA levels in flies lacking transgenic tau, we find that that experimentally reducing RNA export is sufficient to reduce protein encoded by the NMD reporter in healthy controls (Fig. 5C). While this finding raises the possibility that experimentally decreasing RNA export may allow NMD to more effectively clear some, but not all, faulty RNAs in healthy fly brains, it is also possible that experimentally reducing RNA export may simply result in less reporter RNA that is available for translation. Overall, these data suggest that reduction and/or slowing of RNA export in tauR406W transgenic Drosophila decreases levels of RNAs that are usually cleared by NMD, and mechanistically links tau-induced increase in nuclear RNA export to a functional reduction in NMD.
Figure 5 |. Genetic reduction of RNA export significantly increases NMD in brains of tauR406W transgenic Drosophila.
RNAi-mediated reduction of sbr or Nxt1 stabilizes expression of NMD-sensitive Gadd45 (A) and Arc1 (B) in tauR406W transgenic Drosophila. n=12 biological replicates per genotype. C) Panneuronal RNAi-mediated reduction of sbr or Nxt1 increase NMD in control flies based on the NMD-sensitive reporter. n=6 biological replicates per genotype. All assays were performed at day ten of adulthood. *p<0.05, ***p<0.001, ****p<0.0001, one-way ANOVA. Error bars=SEM. Full genotypes are included in Supplemental Table 1.
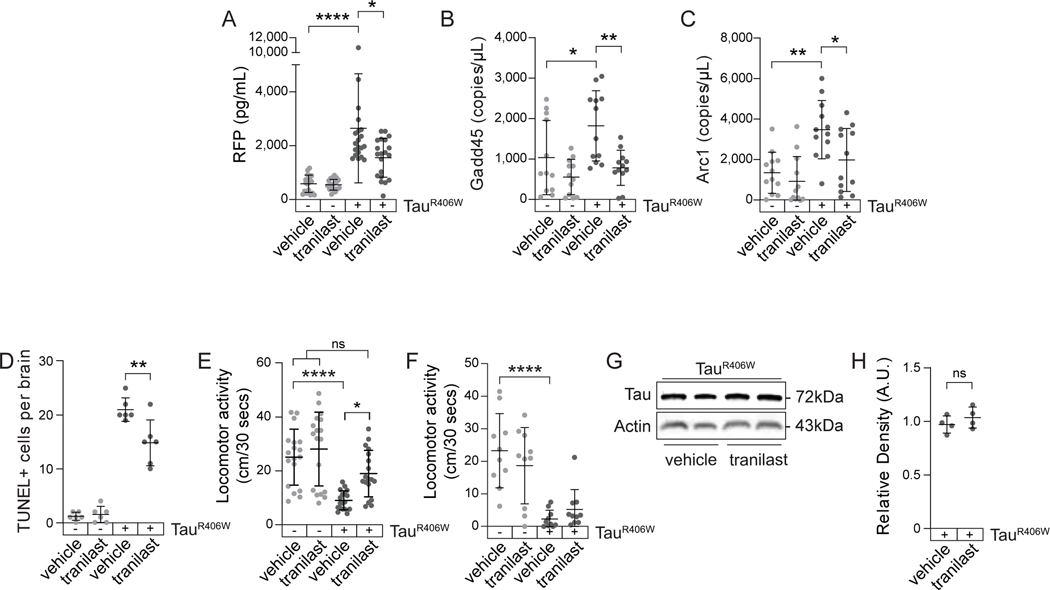
3.2.6 |. Pharmacological activation of NMD is neuroprotective in tauR406W transgenic Drosophila
We next asked if tau-induced deficits in NMD are pharmacologically targetable. While an increasing number of preclinical drugs that inhibit NMD are in development for the treatment of cancer and human genetic disorders caused by a single-base pair mutation110–113, NMD-activating drugs are less common. We first determined if tranilast, a previously identified NMD activator in Drosophila61, effectively activates NMD in tauopathy. We treated control and tauR406W transgenic Drosophila with vehicle (dimethyl sulfoxide, DMSO) or 10 μM tranilast in food from day one to ten of adulthood. We find that tranilast significantly activates general NMD in brains of tauR406W transgenic Drosophila based on decreased expression of the NMD-sensitive reporter (Fig. 6A). In addition, tranilast treatment significantly decreases transcript levels of Gadd45 and Arc1 in tauR406W transgenic Drosophila but does not alter their levels in controls (Fig. 6B, C). These results confirm the NMD-activating effects of tranilast and suggest that tranilast-mediated activation of NMD is efficacious only in an NMD-deficient background. Having established that tranilast activates NMD in tauR406W transgenic Drosophila, we next asked if pharmacological activation of NMD protects against tau-induced toxicity. We find that ten days of tranilast treatment significantly ameliorates tau-induced neurodegeneration based on TUNEL staining (Fig. 6D). As an overall measure of organismal fitness, we analyzed the effects of tranilast on locomotor activity. We find that ten days of tranilast treatment significantly suppresses tau-induced locomotor deficits (Fig. 6E), but that this effect is lost after 20 days of treatment, an age at which tauR406W transgenic Drosophila are significantly impaired (Fig. 6F). Total levels of transgenic tauR406W protein are not changed by tranilast treatment (Fig. 6G, H). Together, these results suggest that NMD is a pharmacologically targetable pathway with therapeutic potential for tauopathy.
Figure 6 |. 10 μM tranilast activates NMD and suppresses tauR406W-induced neurodegeneration in Drosophila.
A) Tranilast alleviates NMD deficits in tauR406W transgenic Drosophila based on reduced expression of the NMD reporter. n=12 biological replicates per genotype, per treatment. RNA levels of the NMD-sensitive transcript Gadd45 (B) and Arc1 (C) are reduced by tranilast treatment of tauR406W transgenic Drosophila. n=12 biological replicates per genotype, per treatment. D) Pharmacological activation of NMD by tranilast significantly suppresses neuronal death in tauR406W transgenic Drosophila based on TUNEL staining. n=6 biological replicates per genotype, per treatment. E) Ten days of tranilast treatment significantly suppresses tauR406W-induced locomotor deficits. n=18 biological replicates per genotype, per treatment. F) Twenty days of tranilast treatment does not suppress tauR406W -induced locomotor deficits. n=10 biological replicates per genotype, per treatment. G) Transgenic tau protein levels in DMSO- and tranilast-treated tauR406W flies based on western blotting. H) Quantification of (G). n=4 biological replicates per treatment. *p<0.05, **p<0.01, unpaired t-test and one-way ANOVA. Error bars=SEM. Full genotypes are included in Supplemental Table 1.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic review: Pathological forms of tau drive neuronal death in Alzheimer’s disease and related tauopathies. Accumulating evidence indicate that tau pathology impairs RNA processing in the brain and may contribute to downstream neuronal loss. We previously reported that increased RNA export contributes to tau-induced neurodegeneration through an unknown mechanism.
Interpretation: We have discovered that tau-induced increase in RNA export leads to a deficit in NMD, a key RNA surveillance mechanism that clears faulty transcripts. We leverage genetic and pharmacological approaches in Drosophila to demonstrate that deficits in NMD causally mediate tau-induced neurotoxicity.
Future direction: Our study provides rationale for determining if NMD is at a deficit in human tauopathies. Given that activation of NMD is neuroprotective in Drosophila, future studies include 1) screening for and identifying compounds that activate NMD, and 2) investigating the therapeutic effect of highly selective NMD activators in a vertebrate model of tauopathy.
ACKNOWLEDGEMENTS
This project was supported by R01 AG057896 (BF) and T32 AG021890, T32 GM113896, TL1 TR002647, and T32 NS082145 (GZ). We thank the Bloomington Drosophila Stock Center, VDRC and DRSC-TriP for providing transgenic fly strains. The rabbit LamDmo antibody was a kind gift from Dr. Paul Fisher. Transgenic human tau fly lines were provided by Dr. Mel Feany, and flies expressing the NMD fluorescent reporter were provided by Dr. Mark Metzstein.
HUMAN SUBJECTS
Human subjects were not involved in this study and consent was not necessary.
DISCLOSURES
Frost Disclosures
Support received for the submitted work
Federal (paid to institution)
Period: 02/15/19 – 11/30/23
Project #: 1 R01 AG057896
Funding Agency: NIA/NINDS
Title: Mechanisms of tau- and aging-induced neurological dysfunction: Focus on the nucleus
Status: Active
Role: Principal Investigator
Support received outside the submitted work
Grants and contracts over the past 36 months, all paid to institution:
Federal
Period: 02/15/19 – 11/30/23
Project #: 1 R01 AG057896
Funding Agency: NIA/NINDS
Title: Mechanisms of tau- and aging-induced neurological dysfunction: Focus on the nucleus
Status: Active
Role: Principal Investigator
Period: 09/01/19 – 05/31/24
Project #: 1 R01 AG062475-01A1
Funding Agency: NIA/NINDS
Title: Regulation of neuronal clearance pathways via nuclear calcium signaling in Alzheimer’s disease
Status: Active
Role: Co-Investigator, PI: Radek Dobrowolski
Period: 07/01/19 – 06/30/24
Project #: 1 RF1 NS112391-01
Funding Agency: NIA/NINDS
Title: Investigating the role of transposable element dysregulation as a driver of neurotoxicity in tauopathy
Status: Active
Role: Principal Investigator
Period: 03/01/19 – 11/30/23
Project #: 1 R01 AG058778-01
Funding Agency: NIH/NINDS
Title: Regulation of ER-stress Activated UPR kinase PERK in Neurodegenerative Diseases
Status: Active
Role: Co-Investigator, PI: James Lechleiter
Total Direct Costs to Frost: $325,050
Grant Detail: The major goal of this grant is to understand how cytosolic and luminal regulation of PERK activity contribute to neurodegenerative disorders
Period: 08/01/14 – 11/30/19
Project #: 1 K99/R00 NS 088429
Funding Agency: NIH/NINDS
Title: Mechanisms and Consequences of Heterochromatin Loss in Tauopathies
Status: Complete
Role: Principal Investigator
Institutional
Period: 09/01/19 – 08/31/21
Project #: N/A
Funding Agency: UTHSCSA Pepper Center
Title: Evaluating the extent of transposable element activation in brain and fluid from patients with Alzheimer’s disease
Status: Active
Role: Principal Investigator
Period: 01/01/19 – 12/31/19
Project #: N/A
Funding Agency: Center for Biomedical Neurosciences Pilot Grant
Title: Testing 3TC as a novel therapeutic for transposable element activation and consequent neurodegenerationin a mouse model of tauopathy
Status: Complete
Role: Principal Investigator
Private
Period: 08/01/19 – 07/31/21
Project #: N/A
Funding Agency: Rainwater Foundation/Tau Consortium
Title: Development of a Drosophila model of prion-like tau propagation for future genetic modifier and drug screening Status: Active
Role: Principal Investigator
Period: 06/01/19 – 05/31/21
Project #: N/A
Funding Agency: William and Ella Owens Medical Research Foundation
Title: Dysregulation of Transposable Element Sequences as a Novel Disease Mechanism in Alzheimer’s Diseaseand Related Tauopathies
Status: Active
Role: Principal Investigator
Period: 08/01/18 – 08/31/20
Project #: N/A
Funding Agency: Rainwater Foundation/Tau Consortium
Title: Investigating transposable element activation as a causal mediator of neuronal death in tauopathy
Status: Active
Role: Principal Investigator
Sponsored Research Agreements
Period: 01/01/21 – 12/31/21
Project #: N/A
Funding Agency: MD Anderson Neurodegeneration Consortium
Title: Nuclear architecture and transposable element activation as a therapeutic target in human Alzheimer’s disease and associated tauopathies
Status: Active
Role: Principal Investigator
Period: 01/01/20 – 12/31/20
Project #: N/A
Funding Agency: MD Anderson Neurodegeneration Consortium
Title: Transposable element activation as a therapeutic target in human Alzheimer’s disease and associated tauopathies
Status: Complete
Role: Principal Investigator
Period: 06/01/20 – 05/31/21
Project #: N/A
Entity: Transposon Therapeutics
Title: Testing reverse transcriptase inhibitors in Drosophila models of tauopathy
Status: Complete
Role: Principal Investigator
Period: 05/01/19 – 04/31/20
Project #: N/A
Funding Agency: MD Anderson Neurodegeneration Consortium
Title: Reverse transcriptase inhibitors for Alzheimer’s disease
Status: Complete
Role: Principal Investigator
Consulting over the past 36 months, paid to institution:
2019: $50,000.00
2021: $50,000.00
Source: MD Anderson Neurodegeneration Consortium
Paid honoraria over the past 36 months, paid to individual:
2020: University of Texas Austin, Department of Neuroscience Seminar Series, invited speaker
2019: University of Texas Southwestern, Symposium on Neurodegenerative Diseases, invited speaker
Paid travel over the past 36 months, paid to individual:
2020: Tau2020 and Tau Consortium Investigator’s Meeting, Washington, DC, Paid travel from Rainwater Foundation, invited speaker.
2019: Gerontological Society of America, Austin, TX. Paid travel from GSA, invited speaker.
2019: Alzheimer’s Association International Conference, Los Angeles, CA. Paid travel from Alzheimer’s Association, invited speaker.
2019: Tau Consortium Meeting, Dallas, TX. Paid travel from Rainwater Foundation.
2018: American Federation for Aging Research New Investigator in Alzheimer’s Disease Meeting, Santa Barbara, CA. Paid travel from AFAR, invited speaker.
2018: PSP & CBD International Research Symposium, London, United Kingdom. Paid travel from CurePSP, keynote speaker.
2018: Keystone Meeting, Advances in Neurodegenerative Disease Research, Keystone, CO (Invited Speaker), Paid travel from Keystone, invited speaker.
Leadership roles on boards over the past 36 months (unpaid)
2021: Scientific Advisory Board for CurePSP
Zuniga Disclosures
Support received for the submitted work
Federal (paid to institution)
Period: 07/01/18 – 06/30/23
Project #: T32 GM113896
Funding Agency: NIGMS
Title: South Texas Medical Scientist Training Program (STX-MSTP)
Status: Active
Role: Trainee
Principal Investigator: José Cavazos, M.D., Ph.D.
Period: 05/24/18– 04/30/23
Project #: TL1 TR002647
Funding Agency: NCATS
Title: NRSA Training Core (TL1)
Status: Complete
Role: Trainee
Principal Investigator: Linda McManus, Ph.D. and Christopher Frei, Ph.D.
Period: 07/01/13 – 06/30/18
Project #: T32 NS082145
Funding Agency: NINDS
Title: Integrated Graduate Training Program in Neuroscience, UTHS
Status: Complete
Role: Trainee
Principal Investigator: David Morilak, Ph.D.
Support received outside the submitted work
Grants and contracts over the past 36 months, all paid to institution:
Federal
Period: 07/01/21 – 06/30/22
Project #: T32 AG021890
Funding Agency: NIA
Title: Training Grant on Biology of Aging
Status: Active
Role: Trainee
Principal Investigator: Nicolas Musi, M.D.
Paid travel over the past 36 months, paid to individual:
2020: Tau2020 and Tau Consortium Investigator’s Meeting, Washington, DC, Paid travel from Rainwater Foundation, invited fellow.
2019: Alzheimer’s Drug Discovery Foundation Young Investigators Scholarship, 13th Annual Drug Discovery for Neurodegeneration Conference, An Educational Course on Translating Research into Drugs. Long Beach, CA, Paid travel from ADDF, invited fellow.
Levy Disclosures
Paid travel over the past 36 months, paid to individual:
2019: Tau Consortium Investigators’ Meeting, San Diego, CA, Paid travel from Rainwater Foundation, invited fellow.
2019: Keystone Symposium on Neurodegenerative Disorders, Keystone, CO, Paid travel from UT Health San Antonio Graduate School of Biomedical Sciences Summer Travel Award
Ramirez Disclosures
Support received outside the submitted work
Grants and contracts over the past 36 months, all paid to institution:
Federal
Period: 08/01/19 – 08/31/21
Project #: R25 GM095480-10
Funding Agency: NIGMS
Title: Recruiting and Retaining Underrepresented Students
Status: Active
Role: Trainee
Principal Investigator: Nicquet Blake, PhD
Footnotes
De Mange Disclosures
No disclosures to report.
Gonzalez Disclosures
No disclosures to report.
Gamez Disclosures
No disclosures to report.
REFERENCES
- 1.Hutton M, Lendon CL, Rizzu P, et al. Association of missense and 5ʹ-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393(6686):702–705. doi: 10.1038/31508 [DOI] [PubMed] [Google Scholar]
- 2.Spillantini MG, Crowther RA, Kamphorst W, Heutink P, Van Swieten JC. Tau pathology in two Dutch families with mutations in the microtubule- binding region of tau. Am J Pathol. 1998. doi: 10.1016/S0002-9440(10)65721-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poorkaj P, Bird TD, Wijsman E, et al. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol. 1998;43(6):815–825. doi: 10.1002/ana.410430617 [DOI] [PubMed] [Google Scholar]
- 4.Forrest SL, Kril JJ, Stevens CH, et al. Retiring the term FTDP-17 as MAPT mutations are genetic forms of sporadic frontotemporal tauopathies. Brain. 2018. doi: 10.1093/brain/awx328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fulga TA, Elson-Schwab I, Khurana V, et al. Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat Cell Biol 2006 92. 2006;9(2):139–148. doi: 10.1038/ncb1528 [DOI] [PubMed] [Google Scholar]
- 6.Bardai FH, Wang L, Mutreja Y, Yenjerla M, Gamblin TC, Feany MB. A Conserved Cytoskeletal Signaling Cascade Mediates Neurotoxicity of FTDP-17 Tau Mutations In Vivo. J Neurosci. 2018;38(1):108–119. doi: 10.1523/JNEUROSCI.1550-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frost B, Hemberg M, Lewis J, Feany MB. Tau promotes neurodegeneration through global chromatin relaxation. Nat Neurosci. 2014;17(3):357–366. doi: 10.1038/nn.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frost B, Bardai FH, Feany MB. Lamin Dysfunction Mediates Neurodegeneration in Tauopathies. Curr Biol. 2016;26(1). doi: 10.1016/j.cub.2015.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano A. Hirano bodies and related neuronal inclusions. Neuropathol Appl Neurobiol. 1994. doi: 10.1111/j.1365-2990.1994.tb00951.x [DOI] [PubMed] [Google Scholar]
- 10.El Hajjar J, Chatoo W, Hanna R, et al. Heterochromatic genome instability and neurodegeneration sharing similarities with Alzheimer’s disease in old Bmi1+/− mice. Sci Rep. 2019;9(1):594. doi: 10.1038/s41598-018-37444-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nixon RA, Wegiel J, Kumar A, et al. Extensive involvement of autophagy in Alzheimer disease: an immune-electron microscopy study. J Neuropathol Exp Neurol. 2005;64(2):113–122. doi: 10.1093/jnen/64.2.113 [DOI] [PubMed] [Google Scholar]
- 12.De Rosa M, Morelli G, Cesaro E, et al. Alternative splicing and nonsense-mediated mRNA decay in the regulation of a new adenomatous polyposis coli transcript. Gene. 2007;395(1–2):8–14. doi: 10.1016/j.gene.2006.10.027 [DOI] [PubMed] [Google Scholar]
- 13.Gout J-F, Li W, Fritsch C, et al. The Landscape of Transcription Errors in Eukaryotic Cells; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frischmeyer PA, Dietz HC. Nonsense-Mediated mRNA Decay in Health and Disease. Hum Mol Genet. 1999;8(10):1893–1900. doi: 10.1093/HMG/8.10.1893 [DOI] [PubMed] [Google Scholar]
- 15.Lou CH, Shao A, Shum EY, et al. Posttranscriptional Control of the Stem Cell and Neurogenic Programs by the Nonsense-Mediated RNA Decay Pathway. Cell Rep. 2014;6(4):748. doi: 10.1016/J.CELREP.2014.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruno IG, Karam R, Huang L, et al. Identification of a MicroRNA that Activates Gene Expression by Repressing Nonsense-Mediated RNA Decay. Mol Cell. 2011;42(4):500. doi: 10.1016/J.MOLCEL.2011.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avery P, Vicente-Crespo M, Francis D, Nashchekina O, Alonso CR, Palacios IM. Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. RNA. 2011;17(4):624–638. doi: 10.1261/rna.2404211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11(10):1530–1544. doi: 10.1261/rna.2160905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a Trans-Effector of Nonsense-Mediated MRNA Decay, Is Essential for Mammalian Embryonic Viability. Vol 10.; 2001. [DOI] [PubMed] [Google Scholar]
- 20.Wittkopp N, Huntzinger E, Weiler C, et al. Nonsense-Mediated mRNA Decay Effectors Are Essential for Zebrafish Embryonic Development and Survival. Mol Cell Biol. 2009;29(13):3517. doi: 10.1128/MCB.00177-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balistreri G, Horvath P, Schweingruber C, et al. The Host Nonsense-Mediated mRNA Decay Pathway Restricts Mammalian RNA Virus Replication. Cell Host Microbe. 2014;16(3):403–411. doi: 10.1016/J.CHOM.2014.08.007 [DOI] [PubMed] [Google Scholar]
- 22.Ramage HR, Kumar GR, Verschueren E, et al. A Combined Proteomics/Genomics Approach Links Hepatitis C Virus Infection with Nonsense-Mediated mRNA Decay. Mol Cell. 2015;57(2):329. doi: 10.1016/J.MOLCEL.2014.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner LB. Hypoxic Inhibition of Nonsense-Mediated RNA Decay Regulates Gene Expression and the Integrated Stress Response. Mol Cell Biol. 2008;28(11):3729. doi: 10.1128/MCB.02284-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Usuki F, Yamashita A, Fujimura M. Environmental stresses suppress nonsense-mediated mRNA decay (NMD) and affect cells by stabilizing NMD-targeted gene expression. Sci Reports 2019 91. 2019;9(1):1–15. doi: 10.1038/s41598-018-38015-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 2004 3610. 2004;36(10):1073–1078. doi: 10.1038/NG1429 [DOI] [PubMed] [Google Scholar]
- 26.Johnson JL, Stoica L, Liu Y, et al. Inhibition of Upf2-Dependent Nonsense-Mediated Decay Leads to Behavioral and Neurophysiological Abnormalities by Activating the Immune Response. Neuron. 2019;104(4):665–679.e8. doi: 10.1016/j.neuron.2019.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Notaras M, Allen M, Longo F, et al. UPF2 leads to degradation of dendritically-targeted mRNAs to regulate synaptic plasticity and cognitive function. Mol Psychiatry. 2020;25(12):3360. doi: 10.1038/S41380-019-0547-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colak D, Ji S-J, Porse BT, Jaffrey SR. Regulation of Axon Guidance by Compartmentalized Nonsense-Mediated mRNA Decay. Cell. 2013;153(6):1252. doi: 10.1016/J.CELL.2013.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12(5):2165–2177. http://www.ncbi.nlm.nih.gov/pubmed/1569946. Accessed March 14, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LA T GA N, J W, et al. UPF2 is a critical regulator of liver development, function and regeneration. PLoS One. 2010;5(7). doi: 10.1371/JOURNAL.PONE.0011650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.TL R UH, J L, et al. Smg1 haploinsufficiency predisposes to tumor formation and inflammation. Proc Natl Acad Sci U S A. 2013;110(4). doi: 10.1073/PNAS.1215696110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mcilwain DR, Pan Q, Reilly PT, et al. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. doi: 10.1073/pnas.1007336107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.T L Y S, P W, et al. Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J. 2015;34(12):1630–1647. doi: 10.15252/EMBJ.201489947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Son HG, Seo M, Ham S, et al. RNA surveillance via nonsense-mediated mRNA decay is crucial for longevity in daf-2/insulin/IGF-1 mutant C. Elegans. Nat Commun. 2017;8(1):1–11. doi: 10.1038/ncomms14749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X, Zhang L, Tong P, et al. Exome sequencing identifies UPF3B as the causative gene for a Chinese non-syndrome mental retardation pedigree. Clin Genet. 2013;83(6):560–564. doi: 10.1111/CGE.12014 [DOI] [PubMed] [Google Scholar]
- 37.Tarpey PS, Raymond FL, Nguyen LS, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39(9):1127. doi: 10.1038/NG2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen LS, Jolly L, Shoubridge C, et al. Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol Psychiatry. 2012;17(11):1103–1115. doi: 10.1038/mp.2011.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen LS, Kim H-G, Rosenfeld JA, et al. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet. 2013;22(9):1816–1825. doi: 10.1093/hmg/ddt035 [DOI] [PubMed] [Google Scholar]
- 40.Gulsuner S, Walsh T, Watts AC, et al. Spatial and Temporal Mapping of De novo Mutations in Schizophrenia To a Fetal Prefrontal Cortical Network. Cell. 2013;154(3):518. doi: 10.1016/J.CELL.2013.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol 2004 510. 2004;5(10):1–15. doi: 10.1186/GB-2004-5-10-R74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100(1):189. doi: 10.1073/PNAS.0136770100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trcek T, Sato H, Singer RH, Maquat LE. Temporal and spatial characterization of nonsense-mediated mRNA decay. 2013:541–551. doi: 10.1101/gad.209635.112.NMD [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh AK, Choudhury SR, De S, et al. The RNA helicase UPF1 associates with mRNAs co-transcriptionally and is required for the release of mRNAs from gene loci. Elife. 2019;8. doi: 10.7554/eLife.41444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Jubran K, Wen J, Abdullahi A, et al. Visualization of the joining of ribosomal subunits reveals the presence of 80S ribosomes in the nucleus. RNA. 2013;19(12):1669–1683. doi: 10.1261/RNA.038356.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mor A, Shav-Tal Y. Dynamics and kinetics of nucleo-cytoplasmic mRNA export. Wiley Interdiscip Rev RNA. 2010;1(3):388–401. doi: 10.1002/wrna.41 [DOI] [PubMed] [Google Scholar]
- 47.Whitton H, Singh LN, Patrick MA, et al. Changes at the nuclear lamina alter binding of pioneer factor Foxa2 in aged liver. Aging Cell. 2018;17(3):e12742. doi: 10.1111/ACEL.12742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldman RD, Shumaker DK, Erdos MR, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2004;101(24):8963. doi: 10.1073/PNAS.0402943101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fong LG, Ng JK, Meta M, et al. Heterozygosity for Lmna deficiency eliminates the progeria-like phenotypes in Zmpste24-deficient mice. Proc Natl Acad Sci U S A. 2004;101(52):18111. doi: 10.1073/PNAS.0408558102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shumaker DK, Dechat T, Kohlmaier A, et al. From the Cover: Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A. 2006;103(23):8703. doi: 10.1073/PNAS.0602569103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broers JL, Raymond Y, Rot MK, Kuijpers H, Wagenaar SS, Ramaekers FC. Nuclear A-type lamins are differentially expressed in human lung cancer subtypes. Am J Pathol. 1993;143(1):211. /pmc/articles/PMC1886958/?report=abstract. Accessed August 6, 2021. [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer AH, Bond JA, Taysavang P, Battles OE, Wynford-Thomas D. Papillary Thyroid Carcinoma Oncogene (RET/PTC) Alters the Nuclear Envelope and Chromatin Structure. Am J Pathol. 1998;153(5):1443. doi: 10.1016/S0002-9440(10)65731-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwertheim S, Theurer S, Jastrow H, et al. New insights into intranuclear inclusions in thyroid carcinoma: Association with autophagy and with BRAFV600E mutation. PLoS One. 2019;14(12):e0226199. doi: 10.1371/JOURNAL.PONE.0226199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frost B, Bardai FH, Feany MB. Lamin Dysfunction Mediates Neurodegeneration in Tauopathies. Curr Biol. 2016;26(1):129–136. doi: 10.1016/j.cub.2015.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eftekharzadeh B, Daigle JG, Kapinos LE, et al. Tau protein disrupts nucleocytoplasmic transport in Alzheimer’s disease HHS Public Access. Neuron. 2018;99(5):925–940. doi: 10.1016/j.neuron.2018.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paonessa F, Evans LD, Solanki R, Hardy J, Jackson SP, Livesey Correspondence FJ. Microtubules Deform the Nuclear Membrane and Disrupt Nucleocytoplasmic Transport in Tau-Mediated Frontotemporal Dementia. 2019. doi: 10.1016/j.celrep.2018.12.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paonessa F, Evans LD, Solanki R, Hardy J, Jackson SP, Livesey Correspondence FJ. Microtubules Deform the Nuclear Membrane and Disrupt Nucleocytoplasmic Transport in Tau-Mediated Frontotemporal Dementia. CellReports. 2019;26:582–593.e5. doi: 10.1016/j.celrep.2018.12.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cornelison GL, Levy SA, Jenson T, Frost B. Tau-induced nuclear envelope invagination causes a toxic accumulation of mRNA in Drosophila. Aging Cell. 2019;18(1):e12847. doi: 10.1111/acel.12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wittmann CW. Tauopathy in Drosophila: Neurodegeneration Without Neurofibrillary Tangles. Science (80- ). 2001;293(5530):711–714. doi: 10.1126/science.1062382 [DOI] [PubMed] [Google Scholar]
- 60.Darakhshan S, Pour AB. Tranilast: A review of its therapeutic applications. Pharmacol Res. 2015;91:15–28. doi: 10.1016/j.phrs.2014.10.009 [DOI] [PubMed] [Google Scholar]
- 61.Reactivation of nonsense-mediated mRNA decay protects against C9orf72 dipeptide-repeat neurotoxicity. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6487333/. Accessed March 27, 2020. [DOI] [PMC free article] [PubMed]
- 62.Baek D, Green P. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc Natl Acad Sci U S A. 2005;102(36):12813–12818. doi: 10.1073/pnas.0506139102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boeckmann B, Bairoch A, Apweiler R, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31(1):365–370. doi: 10.1093/nar/gkg095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsieh YC, Guo C, Yalamanchili HK, et al. Tau-Mediated Disruption of the Spliceosome Triggers Cryptic RNA Splicing and Neurodegeneration in Alzheimer’s Disease. Cell Rep. 2019;29(2):301–316.e10. doi: 10.1016/j.celrep.2019.08.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ju S, Tardiff DF, Han H, et al. A Yeast Model of FUS/TLS-Dependent Cytotoxicity. Weissman JS, ed. PLoS Biol. 2011;9(4):e1001052. doi: 10.1371/journal.pbio.1001052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barmada SJ, Ju S, Arjun A, et al. Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proc Natl Acad Sci U S A. 2015;112(25):7821–7826. doi: 10.1073/pnas.1509744112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jackson KL, Dayton RD, Orchard EA, et al. Preservation of forelimb function by UPF1 gene therapy in a rat model of TDP-43-induced motor paralysis. Gene Ther. 2015;22(1):20–28. doi: 10.1038/gt.2014.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ortega JA, Daley EL, Kour S, et al. Nucleocytoplasmic Proteomic Analysis Uncovers eRF1 and Nonsense-Mediated Decay as Modifiers of ALS/FTD C9orf72 Toxicity. Neuron. 2020;106(1):90–107.e13. doi: 10.1016/j.neuron.2020.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kamelgarn M, Chen J, Kuang L, Jin H, Kasarskis EJ, Zhu H. ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay. Proc Natl Acad Sci U S A. 2018;115(51):E11904–E11913. doi: 10.1073/pnas.1810413115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frizzell KA, Rynearson SG, Metzstein MM. Drosophila mutants show NMD pathway activity is reduced, but not eliminated, in the absence of Smg6. Rna. 2012;18(8):1475–1486. doi: 10.1261/rna.032821.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dias-Santagata D, Fulga TA, Duttaroy A, Feany MB. Oxidative stress mediates tau-induced neurodegeneration in Drosophila. J Clin Invest. 2007;117(1):236–245. doi: 10.1172/JCI28769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khurana V, Merlo P, Duboff B, et al. A neuroprotective role for the DNA damage checkpoint in tauopathy. Aging Cell. 2012;11(2):360–362. doi: 10.1111/j.1474-9726.2011.00778.x [DOI] [PubMed] [Google Scholar]
- 73.Sun W, Samimi H, Gamez M, Zare H, Frost B. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat Neurosci. 2018;21(8):1038–1048. doi: 10.1038/s41593-018-0194-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo C, Jeong HH, Hsieh YC, et al. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep. 2018;23(10):2874–2880. doi: 10.1016/j.celrep.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khurana V, Lu Y, Steinhilb ML, Oldham S, Shulman JM, Feany MB. TOR-Mediated Cell-Cycle Activation Causes Neurodegeneration in a Drosophila Tauopathy Model. Curr Biol. 2006;16(3):230–241. doi: 10.1016/J.CUB.2005.12.042 [DOI] [PubMed] [Google Scholar]
- 76.Tuthill JC. Lessons from a Compartmental Model of a Drosophila Neuron. J Neurosci. 2009;29(39):12033–12034. doi: 10.1523/JNEUROSCI.3348-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rolls MM, Jegla TJ. Neuronal polarity: an evolutionary perspective. J Exp Biol. 2015;218(4):572. doi: 10.1242/JEB.112359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hummel T, Krukkert K, Roos J, Davis G, Klämbt C. Drosophila Futsch/22C10 Is a MAP1B-like Protein Required for Dendritic and Axonal Development. Neuron. 2000;26(2):357–370. doi: 10.1016/S0896-6273(00)81169-1 [DOI] [PubMed] [Google Scholar]
- 79.Ou J, Gao Z, Song L, Ho MS. Analysis of Glial Distribution in Drosophila Adult Brains. Neurosci Bull. 2016;32(2):162. doi: 10.1007/S12264-016-0014-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wittmann CW, Wszolek MF, Shulman JM, et al. Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science. 2001;293(5530):711–714. doi: 10.1126/science.1062382 [DOI] [PubMed] [Google Scholar]
- 81.Thangavel R, Van Hoesen GW, Zaheer A. The Abnormally Phosphorylated Tau Lesion of Early Alzheimer’s Disease. Neurochem Res 2008 341. 2008;34(1):118–123. doi: 10.1007/S11064-008-9701-1 [DOI] [PubMed] [Google Scholar]
- 82.Adusumalli S, Ngian ZK, Lin WQ, Benoukraf T, Ong CT. Increased intron retention is a post-transcriptional signature associated with progressive aging and Alzheimer’s disease. Aging Cell. 2019;18(3). doi: 10.1111/ACEL.12928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li H-D, Funk CC, McFarland K, et al. Integrative functional genomic analysis of intron retention in human and mouse brain with Alzheimer’s disease. Alzheimers Dement. 2021;17(6):984–1004. doi: 10.1002/alz.12254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Braunschweig U, Barbosa-Morais NL, Pan Q, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24(11):1774–1786. doi: 10.1101/gr.177790.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ge Y, Porse BT. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. Bioessays. 2014;36(3):236–243. doi: 10.1002/bies.201300156 [DOI] [PubMed] [Google Scholar]
- 86.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36(10):1073–1078. doi: 10.1038/ng1429 [DOI] [PubMed] [Google Scholar]
- 87.Nickless A, Jackson E, Marasa J, et al. Intracellular Calcium Regulates Nonsense-Mediated mRNA Decay. Nat Med. 2014;20(8):961. doi: 10.1038/NM.3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paillusson A, Hirschi N, Vallan C, Azzalin CM, Mühlemann O. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res. 2005;33(6):e54–e54. doi: 10.1093/NAR/GNI052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu J, Plank T-D, Su F, et al. The nonsense-mediated RNA decay pathway is disrupted in inflammatory myofibroblastic tumors. J Clin Invest. 2016;126(8):3058. doi: 10.1172/JCI86508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet. 2002;11(23):2805–2814. doi: 10.1093/HMG/11.23.2805 [DOI] [PubMed] [Google Scholar]
- 91.Wang D, Zavadil J, Martin L, et al. Inhibition of Nonsense-Mediated RNA Decay by the Tumor Microenvironment Promotes Tumorigenesis. Mol Cell Biol. 2011;31(17):3670. doi: 10.1128/MCB.05704-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cao L, Qi L, Zhang L, et al. Human nonsense-mediated RNA decay regulates EMT by targeting the TGF-ß signaling pathway in lung adenocarcinoma. Cancer Lett. 2017;403:246–259. doi: 10.1016/J.CANLET.2017.06.021 [DOI] [PubMed] [Google Scholar]
- 93.Grimson A, O’Connor S, Newman CL, Anderson P. SMG-1 Is a Phosphatidylinositol Kinase-Related Protein Kinase Required for Nonsense-Mediated mRNA Decay in Caenorhabditis elegans . Mol Cell Biol. 2004;24(17):7483–7490. doi: 10.1128/MCB.24.17.7483-7490.2004/ASSET/D9423F9B-EBC5-4040-81CA-D03FE37495CB/ASSETS/GRAPHIC/ZMB0170443240006.JPEG [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yamashita A. Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay. Genes to Cells. 2013;18(3):161–175. doi: 10.1111/GTC.12033 [DOI] [PubMed] [Google Scholar]
- 95.Kebaara BW, Atkin AL. Long 3ʹ-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37(9):2771–2778. doi: 10.1093/nar/gkp146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3ʹ-UTR promotes aberrant termination and triggers nonsense- mediated mRNA decay. Nat 2004 4327013. 2004;432(7013):112–118. doi: 10.1038/nature03060 [DOI] [PubMed] [Google Scholar]
- 97.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila:at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22(15):3960–3970. doi: 10.1093/emboj/cdg371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen C- YA, Shyu A-B. Rapid Deadenylation Triggered by a Nonsense Codon Precedes Decay of the RNA Body in a Mammalian Cytoplasmic Nonsense-Mediated Decay Pathway. Mol Cell Biol. 2003;23(14):4805. doi: 10.1128/MCB.23.14.4805-4813.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weischenfeldt J, Waage J, Tian G, et al. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 2012;13(5):R35. doi: 10.1186/GB-2012-13-5-R35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sato H, Singer RH. Cellular variability of nonsense-mediated mRNA decay. Nat Commun 2021 121. 2021;12(1):1–12. doi: 10.1038/s41467-021-27423-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Resta N, Susca FC, Di Giacomo MC, et al. A homozygous frameshift mutation in the ESCO2 gene: Evidence of intertissue and interindividual variation in Nmd efficiency. J Cell Physiol. 2006;209(1):67–73. doi: 10.1002/JCP.20708 [DOI] [PubMed] [Google Scholar]
- 102.Metzstein MM, Krasnow MA. Functions of the Nonsense-Mediated mRNA Decay Pathway in Drosophila Development. PLoS Genet. 2006;2(12):e180. doi: 10.1371/journal.pgen.0020180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nelson JO, Moore KA, Chapin A, Hollien J, Metzstein MM. Degradation of Gadd45 mRNA by nonsense-mediated decay is essential for viability. Elife. 2016;5. doi: 10.7554/eLife.12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nickless A, Bailis JM, You Z. Control of gene expression through the nonsense-mediated RNA decay pathway. Cell Biosci. 2017;7(1):26. doi: 10.1186/s13578-017-0153-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hu Y, Comjean A, Roesel C, et al. FlyRNAi.org-the database of the Drosophila RNAi screening center and transgenic RNAi project: 2017 update. Nucleic Acids Res. 2016;(1). doi: 10.1093/nar/gkw977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dietzl G, Chen D, Schnorrer F, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–156. doi: 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- 107.Erdfelder E, FAul F, Buchner A, Lang AG. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav Res Methods. 2009;41(4):1149–1160. doi: 10.3758/BRM.41.4.1149 [DOI] [PubMed] [Google Scholar]
- 108.Dujardin S, Lécolle K, Caillierez R, et al. Neuron-to-neuron wild-type Tau protein transfer through a trans-synaptic mechanism: Relevance to sporadic tauopathies. Acta Neuropathol Commun. 2014;2(1):1–14. doi: 10.1186/2051-5960-2-14/FIGURES/7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Frost B, Bardai FH, Feany MB. Lamin Dysfunction Mediates Neurodegeneration in Tauopathies. Curr Biol. 2016;26(1):129–136. doi: 10.1016/j.cub.2015.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat. 2008;29(8):1037–1047. doi: 10.1002/humu.20763 [DOI] [PubMed] [Google Scholar]
- 111.Bokhari A, Jonchere V, Lagrange A, et al. Targeting nonsense-mediated mRNA decay in colorectal cancers with microsatellite instability. Oncogenesis. 2018;7(9):70. doi: 10.1038/s41389-018-0079-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gudikote J, Cascone T, Poteete A, et al. Inhibition of nonsense-mediated decay rescues functional p53β/γ isoforms in MDM2-amplified cancers. bioRxiv. April 2020:2020.04.06.026955. doi: 10.1101/2020.04.06.026955 [DOI] [Google Scholar]
- 113.Erwood S, Laselva O, Bily TMI, et al. Allele-Specific Prevention of Nonsense-Mediated Decay in Cystic Fibrosis Using Homology-Independent Genome Editing. Mol Ther - Methods Clin Dev. 2020;17:1118–1128. doi: 10.1016/j.omtm.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.