Abstract
The identification of polysaccharide structures in biological samples remains a unique challenge complicated by the lack of specific tools for polymeric mixtures. In this work, we present a method that dissociates plant polysaccharides to generate diagnostic oligosaccharide markers that are then analyzed by high performance liquid chromatography-quadrupole time-of-flight mass spectrometry (HPLC-QTOF MS). Rapid identification of food polysaccharides was performed by aligning the identified oligosaccharides with a fingerprint library of oligosaccharide markers generated from standard polysaccharides. Measurements of standard and food polysaccharides were performed to obtain the contributions of the identified polysaccharides using percent peak coverage and angle cosine methods. The method was validated using a synthetic mixture of standard polysaccharides including xyloglucan, amylopectin, and arabinoxylan. Reproducibility of the oxidative dissociation method was confirmed with experimental triplicates of butternut squash (Cucurbita moschata) samples, where standard deviation was less than 3% for the relative abundance of oligosaccharides. The method was further employed to examine various foods including coconut flesh (Cocos nucifera), yellow corn meal (Zea mays), jackfruit flesh (Artocarpus heterophyllus), guava flesh (Psidium guajava), yam leaves (Dioscorea sp.), bok choy leaves (Brassica rapa), wheat grass (Triticum sp.), whole grain oat cereal (Avena sativa), horseradish root (Armoracia rusticana), and spent coffee grounds (Coffea arabica).
INTRODUCTION
Plant polysaccharides are the most abundant biomacromolecules found in nature, and they are central to a wide range of biological, agricultural, and industrial applications. Polysaccharides often have bioactive properties when consumed. For example, mushroom polysaccharides, which include chitin, α- and β-glucans, mannans, xylans, and galactans, are found to have antitumor and immunomodulating activities.1 Beyond food, polysaccharides are also components in therapeutics and nutraceutical products.1,2 Polysaccharides can also be used in monitoring agricultural products. Different stages in plant maturity has been associated with changes in polysaccharide compositions.3–5 Apples, for example, are considered ripe when they reach a low glucan and high polyuronide polysaccharide composition.6 Commercialization of polysaccharide products also employ polysaccharide composition analysis primarily for batch-to-batch product validation.7,8 For example, the polysaccharide components in tea-based Chinese herbal medicines are monitored across batches to ensure all products contain the same composition in order to facilitate similar health benefits.9 In such practices, the methods for polysaccharide analysis often requires tedious sample preparation and several instrument platforms, rendering them unsuitable for broad characterization of the different structural classes of polysaccharides.
There remains a clear and considerable need for rapid polysaccharide identification.10 Plant polysaccharides have wide structural diversity as they can be composed of a variety of monosaccharides, linkage types, and degree of branching. As a result, there is no single method that can fully characterize polysaccharide compositions in complex matrices such as food. A traditional method for structural elucidation of plant polysaccharides involves nuclear magnetic resonance spectroscopy.11 However, this technique can only be performed on a highly concentrated and pure polysaccharide sample.12,13 Plant polysaccharide composition analysis has also been performed by Fourier-transform infrared spectroscopy and applying extensive chemometrics.14,15 In this analysis, the majority of the diagnostic peaks are located in the fingerprint region of the spectrum. Even with the use of chemometrics, significant overlap in the fingerprint region causes the analysis to increase in complexity as the number of polysaccharides in the mixture increases.
Mass spectrometry (MS) has been employed for the characterization of plant polysaccharides due to its high sensitivity and the ability to discriminate by mass-to-charge ratios. However, to be amenable to MS, polysaccharides must be dissociated into oligosaccharides through either enzymatic or chemical processes. For example, enzymatically released plant oligosaccharides were used as diagnostic fingerprints to identify polysaccharides. This method used matrix-assisted laser desorption ionization-time of flight MS to characterize the oligosaccharides.16,17 However, the method lacks isomer separation of oligosaccharides which renders branching and linear structures indistinguishable. Additionally, a major drawback for enzymatic hydrolysis is that it requires the use of specific enzymes for each type of polysaccharide present. There is no single enzyme capable of universally cleaving all polysaccharides. Acid hydrolysis with gas-chromatography MS (GC-MS) of monosaccharides has also been used to predict the parent polysaccharide structures.18–21 However, the monosaccharide arrangement information is lost during acid hydrolysis which renders the technique less suitable for overall polysaccharide identification.
In this research, we employed a recently developed chemical method for the dissociation of plant polysaccharides into oligosaccharides.22 The method was optimized to be universal among plant polysaccharides. The resulting oligosaccharides were analyzed using liquid chromatography-mass spectrometry. Generated oligosaccharides were matched against a library of oligosaccharide fingerprints created from standard polysaccharides to determine the polysaccharide composition.
METHODS
Samples and Materials.
Sodium borohydride (NaBH4), sodium hydroxide (NaOH), sodium acetate, glacial acetic acid, trifluoroacetic acid (TFA), hydrogen peroxide (H2O2), and iron(III) sulfate pentahydrate (Fe2(SO4)3·5H2O) were purchased from Sigma-Aldrich (St. Louis, MO). Galactan, amylose, β-glucan, arabinan, xyloglucan, curdlan, arabinoxylan, lichenan, glucomannan, mannan, galactomannan, arabinogalactan and xylan were purchased from Megazyme (Bray, Ireland). Microcrystalline cellulose was purchased from ACROS Organics. Yellow corn meal (Zea mays), wheat grass (Triticum sp.), whole grain oat cereal (Avena sativa), horseradish root (Armoracia rusticana), and coffee grounds (Coffea arabica) were purchased from the Davis Co-op (Davis, CA). Coconut (Cocos nucifera), jackfruit (Artocarpus heterophyllus), guava (Psidium guajava), yam leaves (Dioscorea sp.), bok choy leaves (Brassica rapa) were purchased from 99 Ranch Market (Sacramento, CA). Acetonitrile (ACN, HPLC grade) was purchased from Honeywell (Muskegon, MI). Formic Acid (FA) was purchased from Fisher Scientific (Belgium, UK). Porous graphitized carbon (PGC) solid phase extraction (SPE) plates were purchased from Glygen (Columbia, MD). Nanopure water (18.2 MΩ-cm) was used for all experiments.
Generation of Representative Oligosaccharides.
Polysaccharides were dissociated using an oxidative method optimized toward several polysaccharides.22 The treatment was performed on standard polysaccharides and food samples. Briefly, a reaction solution was prepared using 95% (v/v) 40 mM sodium acetate buffer adjusted to pH 5 with glacial acetic acid, 0.29 M hydrogen peroxide, and 65 μM Fe2(SO4)3. The reaction mixture was vortexed and added to polysaccharide standards and food samples resulting in a final concentration of 1 mg/mL. To initiate the reaction, samples were incubated at 100 °C for 20 mins with a follow-up treatment with equal volume of 2 M NaOH to quench the reaction. Neutralization was performed by the addition of glacial acetic acid.
Reduction of Oligosaccharides.
Oligosaccharides were reduced by treatment with 1 M NaBH4 for 1 hour of incubation at 65 ºC. Purification of oligosaccharides was performed using PGC cartridges. Cartridges were conditioned with 80% ACN and 0.1% (v/v) TFA. After loading the reaction mixture, samples were washed with five column volumes of water. Elution of oligosaccharides was performed using 40% ACN with 0.05% (v/v) TFA. Evaporative centrifugation was used to dry the samples to completion.
High performance liquid chromatography-quadrupole time of flight mass spectrometry (HPLC-QTOF MS) analysis.
Samples were reconstituted in nanopure water before HPLC Q-TOF MS analysis. Analytical separation was carried out using an Agilent 1260 Infinity II HPLC coupled to an Agilent 6530 Accurate-Mass Q-TOF mass spectrometer operated in the positive mode. Chromatographic separation was performed on an analytical PGC column (Hypercarb, 5 μm, 1 x 150 mm, Thermo Scientific). A binary gradient was employed and consisted of solvent A (3% (v/v) ACN/H2O with 0.1% FA) and solvent B (90% ACN/H2O with 0.1% FA). A 45-min gradient with a flow rate of 0.150 mL/min was used for chromatographic separation: 3-25% B, 0-15 min; 25-25% B, 15-18 min; 25-99% B, 18-30 min; 99-99% B, 30-32 min; 99-3% B, 32-34 min; 3-3% B, 34-45 min. Internal calibrant ions ranged from m/z 121.051 to m/z 2421.914. Drying gas temperature and flow rate were set to 150 ºC and 11 L/min, respectively. Operation voltages for the fragment, skimmer, and octupole 1 RF were 175, 60, and 750 V, respectively. The acquisition rate was set to 0.63 spectra/second. For fragmentation, the linear function, Collision Energy = 1.45*(m/z)-3.5, was employed. Data obtained from the HPLC-QTOF MS was collected using Agilent MassHunter Workstation Data Acquisition version B.06.01. The acquired data was analyzed using Agilent MassHunter Quantitative Analysis version B.06.00. Oligosaccharides were manually identified using tandem MS, where neutral mass losses of the monosaccharides were monitored. The LC-MS profiles were annotated with the number of monosaccharides per monosaccharide class (Hexose or Hex, Pentose or Pnt, 4-O-Methylated Glucuronic Acid or GcaOMe) involved in the makeup of the identified oligosaccharide. The number of monosaccharides was represented as a subscript. For example, oligosaccharides containing a mixture of monosaccharide classes were labeled as HexnPntm, where n represents the number of hexoses and m represents the number of pentoses. Thus, a monosaccharide composition of Hex3Pnt indicated the presence of an oligosaccharide composed of 3 hexoses and 1 pentose resulting in an overall degree of polymerization (DP) of 4.
Measurement of similarity between chromatographic profiles.
Two methods were employed to examine the similarity between the chromatographic profiles of standard and food polysaccharide samples. Peak coverage determines the percentage of oligosaccharide peaks observed in the food sample relative to the polysaccharide standard. Therefore, the higher the number of matched oligosaccharide peaks between the food and standard polysaccharide LC-MS profiles, the higher the percent peak coverage value for that polysaccharide.
A second approach involves a chemometric technique using the angle cosine method. In this method, the two chromatograms under investigation are treated as vectors. The number of oligosaccharide peaks along with the corresponding area are included in the similarity computation. The angle cosine method was applied to measure the similarity between the two vectors which was calculated using Equation (1):
where xi and yi refer to the chromatographic peak areas of oligosaccharide i between the two samples, respectively and n is the number of chromatographic peaks. In this manner, a rcos value of 0 indicates that there is no similarity between the two chromatograms while a value of 1 signifies that the two chromatograms are the same.
RESULTS AND DISCUSSION
The method employs oxidative dissociation of polysaccharides followed by reduction of product oligosaccharides and purification before HPLC-QTOF MS analysis (Figure 1). Dissociation of polysaccharides to oligosaccharides was performed using a metal catalyst, Fe2(SO4)3, and an oxidizing agent, H2O2, to produce hydroxyl radicals. Oligosaccharides were released and neutralized using NaOH and glacial acetic acid, respectively. The generated oligosaccharides were reduced using NaBH4 to prevent chromatographic anomer separation during analysis. A final cleanup procedure employing solid phase extraction was performed to purify the oligosaccharide fraction. Reduced oligosaccharides were then analyzed by HPLC-QTOF MS.
Figure 1.
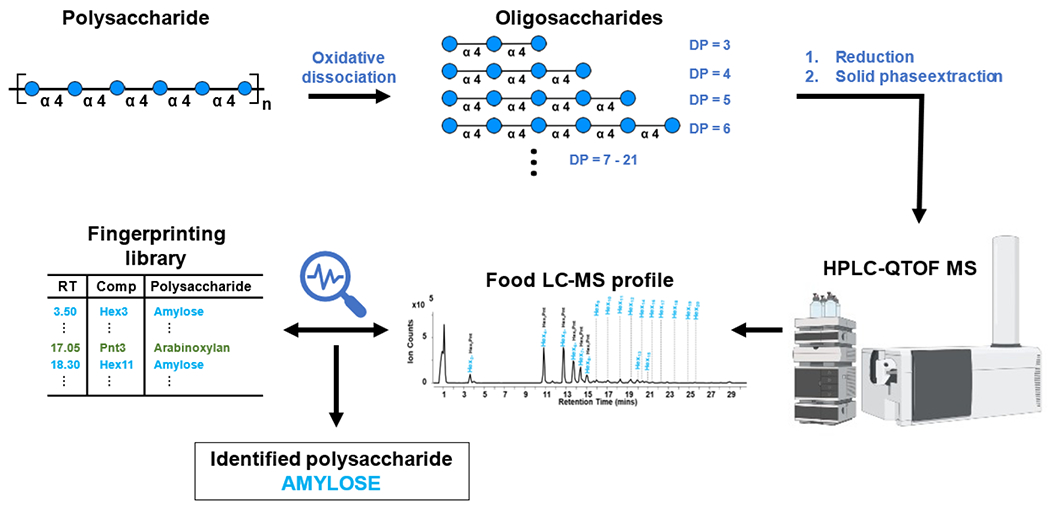
Oxidative dissociation of polysaccharides generates representative oligosaccharides which were reduced and purified for HPLC-QTOF MS analysis. Oligosaccharides identified in the LC-MS profile were aligned with the oligosaccharide fingerprint library to identify the corresponding parent polysaccharide.
Optimization of Reaction Conditions
To achieve the optimal reaction conditions, several parameters were optimized including the concentration of Fe2(SO4)3 and H2O2, pH, reaction time and temperature. A substrate with a diverse monosaccharide and glycosidic linkage composition and a high degree of branching was chosen for the optimization of the reaction conditions. Xyloglucan, a heteropolysaccharide known to contain a β(1→4) glucose backbone with occasional α(1→6) xylose side-chains capped with galactose residues23 was used for the optimization of the concentrations of Fe2(SO4)3 and H2O2, pH, reaction time and temperature. The efficacy of the reaction was monitored by examining the total peak area and average DP of the generated xyloglucan oligosaccharides in the chromatogram.
Concentrations of Fe2(SO4)3 and H2O2 were found to have significant roles on the overall efficiency of the oxidation reaction.24,25 The optimal Fe2(SO4)3 concentration was determined by comparing the total peak areas of the generated xyloglucan oligosaccharides with concentrations of 0.0065, 0.065, 0.65, 1.95, 6.5, and 65 μM. Maximum yield of oligosaccharides was observed at 65 μM of Fe2(SO4)3 (Figure 2A). The optimized Fe2(SO4)3 concentration was then used to optimize for the concentration of H2O2. The H2O2 concentration was varied at concentrations of 0.06, 0.29, 0.58, and 1.16 M. Oligosaccharide yield was highest at 0.29 M of H2O2 (Figure 2B). Correspondingly, the highest average DP of the generated product oligosaccharides was also observed at 0.29 M H2O2.
Figure 2.
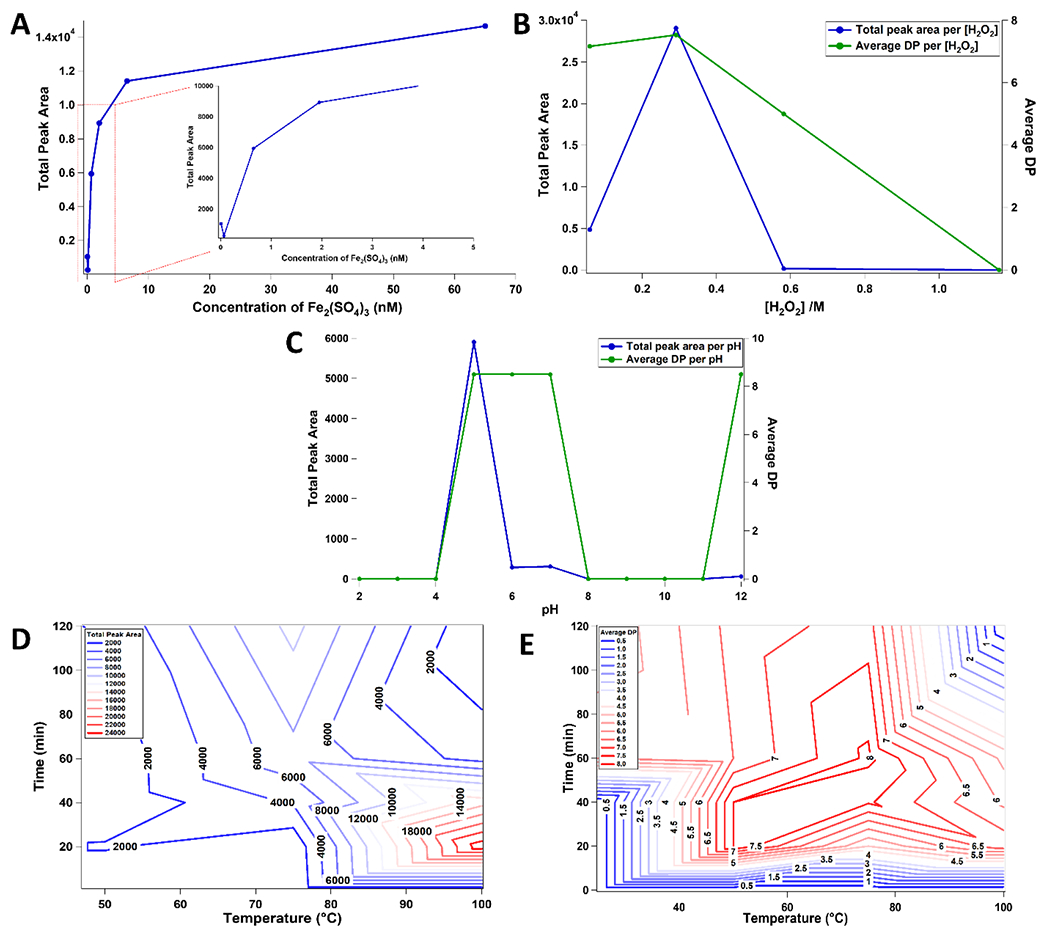
Effective polysaccharide dissociation was performed by optimizing several reaction parameters including the concentration of Fe2(SO4)3 (A) and H2O2 (B), and pH (C). Time and temperature combination were also optimized to ensure high total peak area (D) and average DP (E) of the generated oligosaccharides.
To optimize the buffer pH, a pH range between 2.0 to 12.0 was evaluated with the optimized concentrations of Fe2(SO4)3 and H2O2. Previous studies on similar reactions revealed a strong pH dependency.26,27 The optimal pH for efficient oligosaccharide production was determined to be approximately 5.0 as shown in Figure 2C. At pH 12.0, the average DP increased further. However, the total peak area of oligosaccharides was substantially lower. Under alkaline conditions, the decline in the progress of the oxidation reaction was attributed to the precipitation of Fe(OH)3.24
Reaction time and temperature were also determined to be important parameters in the efficiency of the oxidation reaction. The temperature was varied between 25 °C and 100 °C in increments of 25 °C while time was varied between 0 min and 120 min in increments of 20 min. Optimal yield was obtained at a temperature and time of 100 °C and 20 min (Figure 2D). The highest average DP of product oligosaccharides in the chromatogram was observed at 75 °C after 60 min (Figure 2E). However, the total yield at these conditions was substantially lower. Thus, a temperature of 100 °C and time of 20 min was chosen as the optimal condition for effective polysaccharide dissociation.
Method Validation
The optimized reaction conditions were used to generate a series of oligosaccharides that would fingerprint the corresponding plant polysaccharides. These oligosaccharides generated from commercially available polysaccharide standards were tabulated in Supplementary Information 1, which includes their retention time, monoisotopic mass, oligosaccharide composition, and parent polysaccharide of each identified oligosaccharide.
A validation step was performed to confirm the concept of fingerprinting plant polysaccharides using diagnostic oligosaccharides. The method was validated using a synthetic mixture of polysaccharide standards including arabinoxylan, xyloglucan, and amylopectin. The polysaccharides, mixed at equivalent ratios by mass, were reacted using the optimized conditions for oxidative dissociation to produce representative oligosaccharides. The LC-MS oligosaccharide profile of a mixture of the three polysaccharides is shown in Figure 3D. Peak annotation and matching were performed using the individual LC-MS profiles for arabinoxylan, xyloglucan, and amylopectin as shown in Figure 3A–C, respectively.
Figure 3.
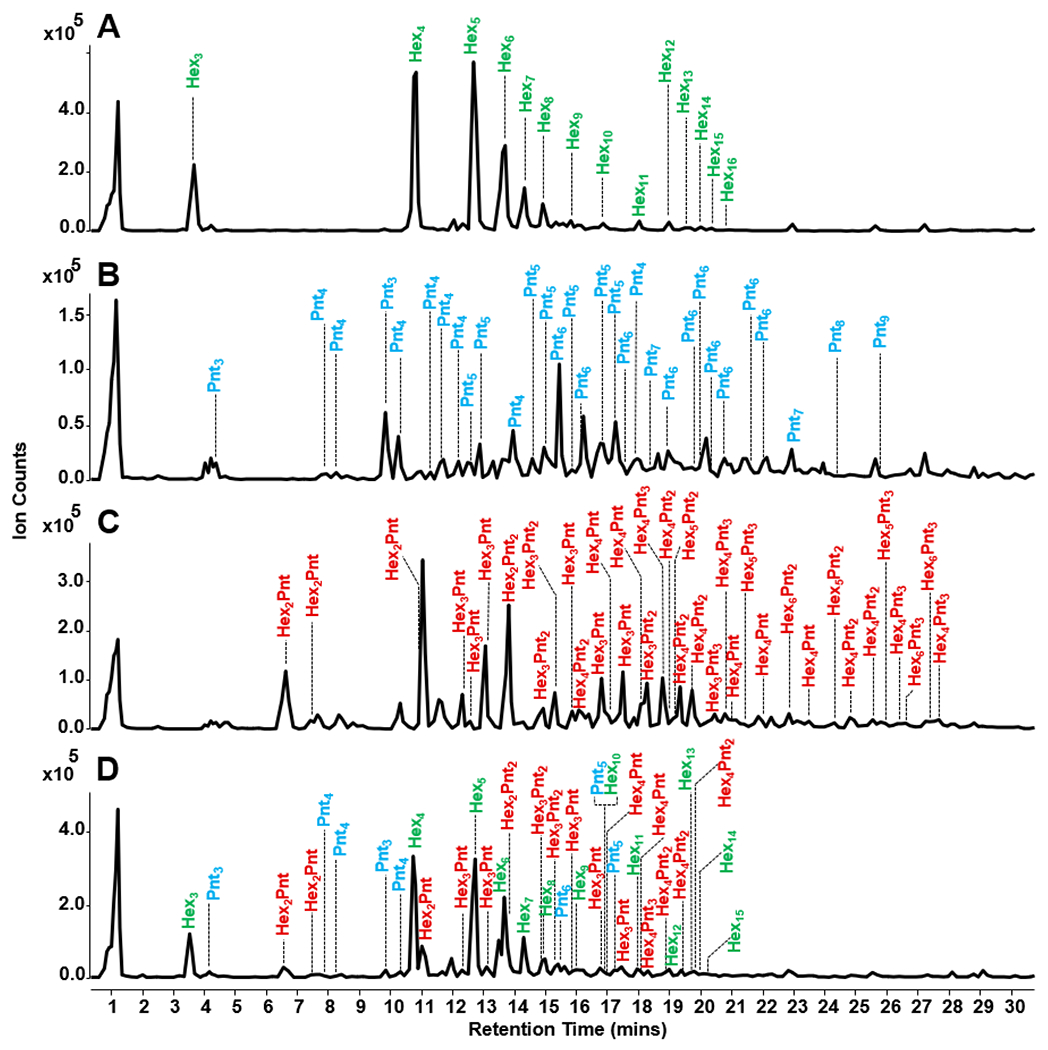
Polysaccharide fingerprinting method validation was performed using a mixture of three polysaccharide standards. Annotated base peak chromatograms of the oligosaccharide profiles of amylopectin (A), arabinoxylan (B), xyloglucan (C), and the mixture (D) are illustrated. Using oligosaccharide fingerprints, all three polysaccharides were successfully identified in the mixture.
LC-MS profiles of the individual standard polysaccharides were compared with the LC-MS profile of the mixture using the peak coverage method. The peak coverage value of a polysaccharide represents the percentage of oligosaccharide peaks observed from a mixture in relation to the oligosaccharide peaks observed from a pure polysaccharide solution. When compared to the pure sample, oligosaccharides in the mixture from amylopectin had a peak coverage value of 93%, while xyloglucan and arabinoxylan had 45% and 25%, respectively. The discrepancy in the peak coverage values was ascribed to the wide distribution of molecular weights of the manufactured pure polysaccharides. There was an inherent difference in the molar ratio of the polysaccharides introduced into the oxidative dissociation reaction. However, the three polysaccharides in the mixture were successfully identified using oligosaccharide fingerprint information, which included monosaccharide composition, retention time, and monoisotopic mass. Thus, employing the oligosaccharide fingerprint library from pure polysaccharides for the identification of unknown polysaccharide compositions in a mixture was successfully validated.
Polysaccharide fingerprinting
The capabilities of the polysaccharide fingerprinting method were probed by analyzing unknown polysaccharide compositions of various foods. The optimized oxidative dissociation method for polysaccharides was applied to food samples to generate representative oligosaccharides. Oligosaccharide fingerprints from food LC-MS profiles were matched with the library of oligosaccharide fingerprints generated from standard polysaccharides to confirm the corresponding polysaccharide composition. For example, if a Hex6 at a retention time of 14.36 min was observed in the food LC-MS profile, it was inferred using the fingerprinting library that the Hex6 was generated from amylose. Retention time shifts were corrected during peak alignment and library matching.
The reproducibility of the oxidative dissociation reaction was determined using a food sample. Experimental triplicates of butternut squash were analyzed to ensure that the method generates reproducible LC-MS profiles and polysaccharide compositions. For each identified oligosaccharide, the retention time, oligosaccharide composition, and polysaccharide of origin are shown in Table 1. Average relative abundances and their corresponding standard deviations were also tabulated. Overall, standard deviation values were less than 3% for relative abundances greater than 1%, demonstrating reproducibility of the method for oxidative dissociation of polysaccharides. Moreover, the polysaccharide composition of butternut squash correctly identified amylose and cellulose, which were in good agreement with literature.28 Oligosaccharide peaks that were not identified using the library were binned until future assignments.
Table 1.
Oxidative dissociation method reproducibility was performed with experimental replicates (n = 3) of whole butternut squash sample. For relative abundances greater than 1%, standard deviations were calculated to be less than 3%, signifying the method reproducibility for polysaccharide dissociation.
| RT (min) | monosaccharide composition | relative abundance (%) | polysaccharide of origin |
|---|---|---|---|
| 10.92 | Hex4 | 17 ± 2 | amylose |
| 12.88 | Hex5 | 14 ± 1 | amylose |
| 13.87 | Hex6 | 9 ± 0 | amylose |
| 14.50 | Hex7 | 7 ± 3 | amylose |
| 3.70 | Hex3 | 5 ± 1 | amylose |
| 10.92 | Hex3Pnt | 4 ± 1 | * |
| 12.88 | Hex4Pnt | 4 ± 0 | * |
| 15.16 | Hex8 | 4 ± 1 | amylose |
| 13.87 | Hex5Pnt | 3 ± 0 | * |
| 3.70 | Hex2Pnt | 2 ± 0 | * |
| 25.50 | Hex5 | 2 ± 3 | cellulose |
| 19.74 | Hex4 | 2 ± 3 | cellulose |
| 14.50 | Hex6Pnt | 2 ± 1 | * |
| 15.16 | Hex7Pnt | 2 ± 2 | * |
| 20.05 | Hex13 | 1 ± 1 | amylose |
| 18.41 | Hex11 | 1 ± 0 | amylose |
| 11.93 | Hex3 # | 1 ± 1 | * |
| 16.16 | Hex9 | 1 ± 0 | amylose |
| 19.50 | Hex12 | 1 ± 0 | amylose |
| 20.55 | Hex14 | 1 ± 1 | amylose |
| 21.64 | Hex16 | 1 ± 1 | amylose |
Not present in the oligosaccharide fingerprinting library
Non-reducing oligosaccharide
In addition to identification of the polysaccharides, an estimation of relative abundances was performed using similarity calculations between the LC-MS profiles of food and standard polysaccharides, specifically, the peak coverage and angle cosine methods. Identified polysaccharides that have high peak coverage values encompassed a greater number of oligosaccharide matches to the corresponding LC-MS profiles of standard polysaccharides. While peak coverage was an adequate method for running a quick measure of similarity by peak count, it did not consider peak areas. Thus, a chemometrics approach, employing the angle cosine method, was additionally used to measure similarity by treating the chromatograms from food and standard polysaccharides as vectors. The angle cosine method calculates values (similarity indices) using Equation 1, where a value of 1 indicates high similarity and a value of 0 indicates low similarity between the two chromatograms. The calculated similarity indices along with the percent peak coverage values between the LC-MS profiles of food and standard polysaccharides are summarized in Table 2.
Table 2.
Similarity indices between the LC-MS profiles of food and standard polysaccharides were examined for coconut flesh, yellow corn meal, jackfruit flesh, guava flesh, yam leaves, bok choy leaves, wheat grass, whole grain oatmeal cereal, horseradish root, and spent coffee grounds. Non-percentage values are similarity indices from the angle cosine method. Percentage values are peak coverage calculations.
| Food Samples |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Standard Polysaccharides | Coconut flesh | Yellow corn meal | Jackfruit flesh | Guava flesh | Yam leaves | Bok choy leaves | Wheat grass | Whole grain oat | Horseradish root | Spent coffee grounds |
| Amylose | * | 0.947, 95% | 0.976, 30% | 0.823, 63% | 0.978, 79% | * | 0.893, 47% | 0.863, 63% | 0.947, 32% | 0.995, 79% |
| Mannan | 0.494, 14% | * | * | * | * | 0.472, 14% | * | 0.349, 7% | * | * |
| Cellulose | 0.011, 33% | * | 0.705, 67% | 0.567, 100% | 0.239, 67% | 0.564, 67% | 0.593, 100% | * | 0.431, 100% | 0.011, 33% |
| Curdlan | * | * | * | * | 0.568, 33% | * | * | * | 0.568, 33% | * |
| Lichenan | 0.014, 3% | * | * | * | * | * | * | * | * | * |
| β-Glucan | 0.004, 4% | * | 0.178, 13% | * | 0.435, 22% | * | * | * | 0.555, 9% | 0.281, 22% |
| Xylan | * | * | * | 0.500, 4% | * | * | 0.500, 4% | * | 0.950, 17% | * |
| Arabinoxylan | * | * | 0.032, 2% | 0.485, 4% | * | * | * | * | * | * |
| Galactomannan | 0.586, 33% | * | 0.381, 7% | * | 0.202, 7% | 0.188, 15% | * | * | * | 0.233, 7% |
| Glucomannan | 0.068, 2% | * | 0.165, 2% | 0.165, 2% | * | 0.103, 3% | * | * | * | * |
| Arabinogalactan | 0.108, 5% | * | * | * | * | * | * | * | * | 0.319, 2% |
| Arabinan | 0.006, 4% | * | * | * | 0.006, 4% | * | 0.006, 4% | 0.006, 4% | 0.245, 13% | * |
| Xyloglucan | * | * | * | 0.658, 2% | * | * | * | * | * | * |
Polysaccharide not present.
An estimation of the abundance of the identified polysaccharides was performed using output values from the angle cosine method.29,30 Similarity index were used to estimate which polysaccharides have the highest contribution to the polysaccharide composition of different food samples. The LC-MS profiles of identified polysaccharides that have high similarity indices resembled the profiles from standard polysaccharides. Therefore, such polysaccharides could have higher abundances in the sample relative to others. Validation of the concept was performed by evaluating of how well similarity indices conformed with the abundances of polysaccharides reported in literature as discussed below.
Food polysaccharide composition analysis
Oligosaccharide profiles of food samples including coconut flesh, yellow corn meal, jackfruit flesh, guava flesh, yam leaves, bok choy leaves, wheat grass, whole grain oatmeal cereal, horseradish root, and spent coffee grounds are shown in Figure 4A–J. The results of the identified polysaccharides from food samples were compared to the profiles found in literature. However, literature values typically did not provide deep coverage in the polysaccharide analysis of most foods. In such cases, the reported monosaccharide composition analyses were compared with our findings from the polysaccharide fingerprinting method.
Figure 4.
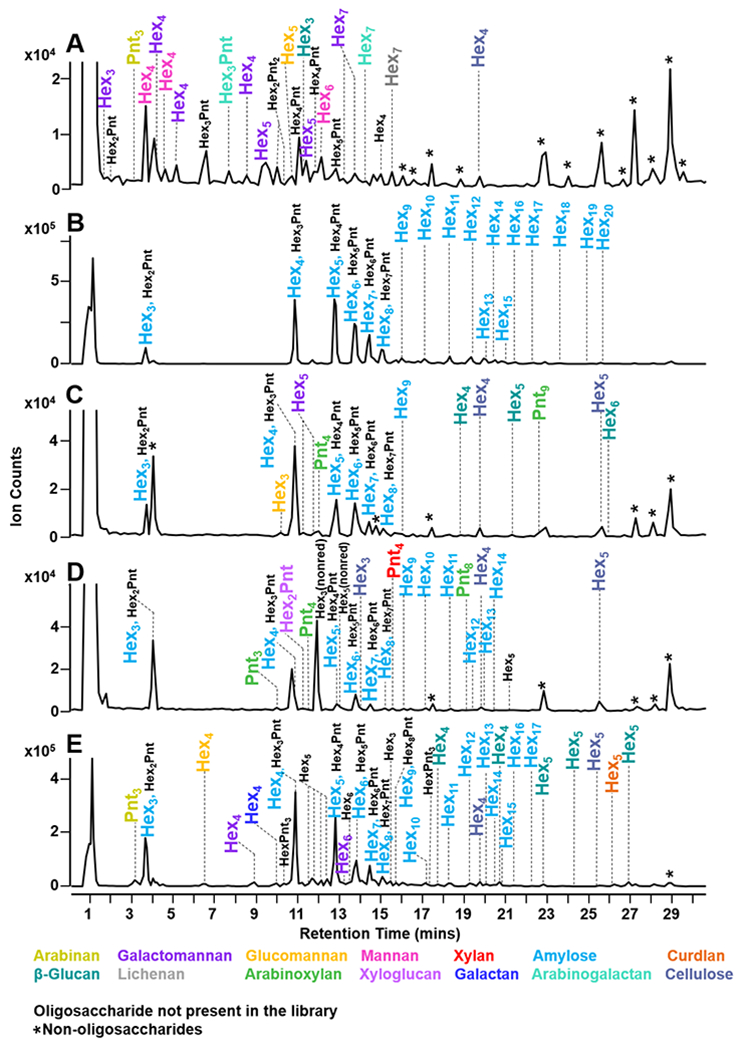
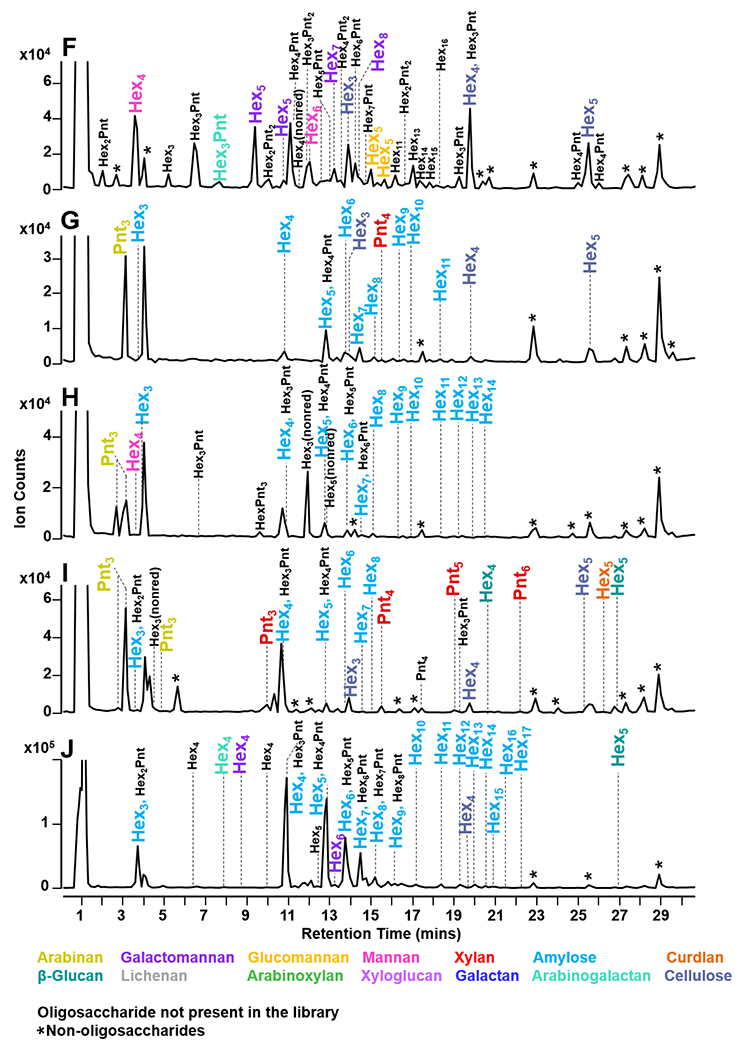
Food polysaccharide fingerprinting was performed with coconut flesh (A), yellow corn meal (B), yam leaves (C), bok choy leaves (D), guava flesh (E), jackfruit flesh (F),wheat grass (G), whole grain oat cereal (H), horseradish roots (I), and spent coffee grounds (J). Corresponding polysaccharides are represented in a color-coded legend. Annotations of co-eluting peaks are separated by a comma. Oligosaccharides that are not present in the library were binned until future assignments. Non-oligosaccharide peaks were denoted with (*).
The LC-MS profile of the copra of coconut (Cocos nucifera) was determined to be composed of galactomannan, cellulose, mannan, arabinogalactan, β-glucan, arabinan, lichenan, and glucomannan (Figure 4A). In literature, it was reported that the coconut flesh was composed of mannan-based polysaccharides, which were the highest in abundance.31 From our results, similarity indices from the angle cosine method for galactomannan and mannan were 0.586 and 0.494, respectively. Thus, the similarity indices were most likely directly proportional to the true abundance of the observed polysaccharides. Additionally, our method was capable of differentially identifying mannan, glucomannan, and galactomannan polysaccharides. Based on the results, it was evident that coconut flesh was mainly composed of mannan and galactomannan polysaccharides. Yellow corn meal (Zea mays), which is largely composed of corn starch, was expected to contain amylose polysaccharides.32 Similarly, the results indicated that yellow corn meal was mainly composed of amylose as shown in Figure 4B. The peak coverage was 95% with a similarity index of 0.947, indicating a high abundance of amylose. Additionally, several HexnPentm peaks were observed that did not correspond to any reference polysaccharides. These peaks were binned for future assignments of currently unknown polysaccharides.
Jackfruit flesh (Artocarpus heterophyllus) polysaccharides were determined to be composed of amylose, cellulose, β-glucan, galactomannan, arabinoxylan, and glucomannan (Figure 4C). Based on literature, jackfruit flesh was mainly composed of glucose, arabinose, and galactose monosaccharide components.33,34 While the literature results did not include polysaccharide compositions, the overall polysaccharide composition from the fingerprinting method matched with the reported monosaccharide compositions. Both amylose and cellulose were comparable to the standard profiles as represented by similarity indices of 0.976 and 0.705, respectively. Previous reports indicated a high abundance of glucose,33,34 which could be attributed to amylose and cellulose. Moreover, several HexnPentm oligosaccharides were also identified. Based on the reported monosaccharide composition,33,34 arabinose could be one of the potential sources of the HexnPentm oligosaccharides.
Guava flesh (Psidium guajava) was composed of amylose, cellulose, arabinoxylan, xyloglucan, xylan, and glucomannan. A previous report found that guava flesh was primarily composed of glucose, xylose, and arabinose constituents,35 confirming the results from the fingerprinting method (Figure 4D). Here, cellulose and amylose were the large contributors of the glucose content with similarity indices of 0.823 and 0.567, respectively. Additionally, one of the polysaccharides isolated from guava flesh in a previous study was characterized to contain a combination of 3-linked arabinose, 5-linked arabinose, 2,3,5-linked arabinose backbone with occasional glucose branching.36 Thus, the binned HexnPentm peaks could potentially correspond to the presence of a ‘glucoarabinan’ type polysaccharide.
Polysaccharides from yam leaves (Dioscorea sp.) are composed of β-glucan, cellulose, curdlan, galactomannan, amylose, and arabinan, as shown in Figure 4E. There have been no report to our best knowledge of polysaccharides in yam leaves, however several studies have reported the presence of mannan in yam.37,38 Using our method, galactomannan was found with a peak coverage of 7% and a similarity index of 0.202. Based on this observation, galactomannan was present at a lower abundance in yam leaves. Moreover, polysaccharide analysis of other types of yam have found β(1→3) linked glucose residues,39 which was consistent with the presence of β-glucan and curdlan polysaccharides with similarity indices of 0.435 and 0.568, respectively.
The diverse composition of bok choy leaves (Brassica rapa) included cellulose, mannan, galactomannan, and glucomannan (Figure 4F). Previous neutral monosaccharide analysis indicated a high abundance of glucose, galactose, and mannose residues.40 Therefore, it was consistent with the presence of cellulose, mannan, glucomannan, and galactomannan. The similarity indices for cellulose, mannan, glucomannan, and galactomannan were 0.564, 0.472, 0.103, and 0.188, respectively. This indicated that cellulose and mannan contributed to the high concentration of glucose and mannose. A significant amount of arabinose was previously reported,40 which could be a possible component of the observed HexnPentm oligosaccharides.
The polysaccharide composition of wheat grass (Triticum sp.) included cellulose, xylan, arabinan, and amylose (Figure 4G). Previous studies reported xylan, arabinan, and β-glucan as significant polysaccharide components of wheat grass.41,42 Literature from hydrolysis experiments determined high abundance of glucose, xylose, and arabinose residues.43 The similarity indices of amylose, cellulose, xylan, and arabinan were 0.893, 0.593, 0.500, and 0.006, respectively. Thus, the largest contribution to the overall polysaccharide concentration originated from amylose, cellulose, and xylan. The results were similarly consistent with the previously reported compositions.
Whole grain oat (Avena sativa) cereal was composed of amylose, mannan, and arabinan as shown in Figure 4H. Several reports indicated the presence of amylose,44 arabinan,45 and β-glucans.46,47 Using the fingerprinting method, amylose and mannan had the highest similarity indices, with 0.863 and 0.349, respectively. The results indicate that whole grain oat is largely abundant in amylose. Polysaccharides from horseradish (Armoracia rusticana) root included arabinan, β-glucan, curdlan, cellulose, xylan, and amylose (Figure 4I). In literature, cellulose and starch were the most notable components of horseradish roots.48 Results from the polysaccharide fingerprinting method indicated a predominant presence of amylose and cellulose with similarity indices of 0.947 and 0.431, respectively. The percent peak coverage values were 32% for amylose and 100% for cellulose.
The polysaccharide fingerprinting method was also applied to spent coffee grounds (Coffea arabica) (Figure 4J). Spent coffee grounds were composed of amylose, cellulose, β-glucan, galactomannan, and arabinogalactan. Monosaccharide composition analysis from literature indicated that glucose, galactose, and mannose were major residues present.49 These results were in agreement with the corresponding polysaccharides from the fingerprinting method. The angle cosine similarity values for amylose, β-glucan, and arabinogalactan were 0.995, 0.568, and 0.319, respectively. Here, amylose and β-glucan are likely to be the more abundant polysaccharide structures which is comparable to the reported monosaccharide analysis.49 The percent peak coverage values for amylose, β-glucan, and arabinogalactan were 79%, 33%, and 2%, respectively.
CONCLUSIONS
We have developed a method for determining the polysaccharide composition of food based on polysaccharide dissociation and oligosaccharide fingerprints generated from polysaccharide standards. This method represented a substantial improvement to the slow and stepwise methods for polysaccharide analysis. The oligosaccharide fingerprinting method was validated using a synthetic mixture of standard polysaccharides comprising of xyloglucan, amylose, and arabinoxylan. Method reproducibility was confirmed with experimental triplicates of butternut squash samples, where the overall standard deviation values were calculated to be less than 3% for oligosaccharides with relative abundances greater than 1%. Successful polysaccharide composition identification was performed for ten various food samples. The identified polysaccharide list was validated by comparison with the known compositions in literature. Similarity index from angle cosine method was proven to be consistent with previously reported polysaccharide compositions of food samples and demonstrated to be an effective measure of similarity between pure standard and food polysaccharide LC-MS profiles.
Conventional methods for polysaccharide analysis mostly rely on monosaccharide and linkage information to predict the polysaccharide structures in the sample, which often results in several predicted candidates of the parent polysaccharide structures.50 In contrast to previous techniques, the presented method is capable of differentially identifying polysaccharides in food matrices such as glucose polysaccharides including amylose, cellulose, curdlan, lichenan, and β-glucan, which would otherwise be rendered indistinguishable from monosaccharide composition data.
Extending the current polysaccharide fingerprinting method for quantitation of polysaccharides would require an orthogonal tool to measure the concentration of identified polysaccharides. In combination with the polysaccharide fingerprinting method, quantitation of polysaccharides using an LC-MS platform is currently being developed and will be the topic of future reports. The presented optimized oxidative method for polysaccharide dissociation and the comprehensive library of oligosaccharide fingerprints will allow for a more targeted and rapid workflow for profiling polysaccharides.
Supplementary Material
ACKNOWLEDGEMENTS
Funding provided by the Mars, Incorporated is gratefully acknowledged.
Footnotes
COMPETING INTERESTS
The authors declare no competing financial interest.
REFERENCES
- (1).Singdevsachan SK; Auroshree P; Mishra J; Baliyarsingh B; Tayung K; Thatoi H Mushroom Polysaccharides as Potential Prebiotics with Their Antitumor and Immunomodulating Properties: A Review. Bioact. Carbohydrates Diet. Fibre 2016, 7 (1), 1–14. 10.1016/j.bcdf.2015.11.001. [DOI] [Google Scholar]
- (2).Kothari D; Patel S; Kim SK Anticancer and Other Therapeutic Relevance of Mushroom Polysaccharides: A Holistic Appraisal. Biomed. Pharmacother 2018, 105 (May), 377–394. 10.1016/j.biopha.2018.05.138. [DOI] [PubMed] [Google Scholar]
- (3).Gross KC; Wallner SJ Degradation of Cell Wall Polysaccharides during Tomato Fruit Ripening’. 1979, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Yashoda HM; Prabha TN; Tharanathan RN Mango Ripening - Chemical and Structural Characterization of Pectic and Hemicellulosic Polysaccharides. Carbohydr. Res 2005, 340 (7), 1335–1342. 10.1016/j.carres.2005.03.004. [DOI] [PubMed] [Google Scholar]
- (5).Redgwell RJ; Melton LD; Brasch DJ Changes to Pectic and Hemicellulosic Polysaccharides of Kiwifruit During Ripening. In II International Symposium on Kiwifruit; International Society for Horticultural Science: Palmerston North, 1991; pp 627–634. [Google Scholar]
- (6).Knee M Polysaccharide Changes in Cell Walls of Ripening Apples. Phytochemistry 1973, 12 (7), 1543–1549. 10.1016/0031-9422(73)80365-6. [DOI] [Google Scholar]
- (7).Guan J; Li SP Discrimination of Polysaccharides from Traditional Chinese Medicines Using Saccharide Mapping-Enzymatic Digestion Followed by Chromatographic Analysis. J. Pharm. Biomed. Anal 2010, 51 (3), 590–598. 10.1016/j.jpba.2009.09.026. [DOI] [PubMed] [Google Scholar]
- (8).Jing P; Zhao SJ; Lu MM; Cai Z; Pang J; Song LH Multiple-Fingerprint Analysis for Investigating Quality Control of Flammulina Velutipes Fruiting Body Polysaccharides. J. Agric. Food Chem 2014, 62 (50), 12128–12133. 10.1021/jf504349r. [DOI] [PubMed] [Google Scholar]
- (9).Wang Y; Xian J; Xi X; Wei X Multi-Fingerprint and Quality Control Analysis of Tea Polysaccharides. Carbohydr. Polym 2013, 92 (1), 583–590. 10.1016/j.carbpol.2012.09.004. [DOI] [PubMed] [Google Scholar]
- (10).Amicucci MJ; Nandita E; Lebrilla CB Function without Structures: The Need for In-Depth Analysis of Dietary Carbohydrates. J. Agric. Food Chem 2019, 67, 4418–4424. 10.1021/acs.jafc.9b00720. [DOI] [PubMed] [Google Scholar]
- (11).Nie SP; Wang C; Cui SW; Wang Q; Xie MY; Phillips GO A Further Amendment to the Classical Core Structure of Gum Arabic (Acacia Senegal). Food Hydrocoll. 2013, 31 (1), 42–48. 10.1016/j.foodhyd.2012.09.014. [DOI] [Google Scholar]
- (12).Dourado F; Cardoso SM; Silva AMS; Gama FM; Coimbra MA NMR Structural Elucidation of the Arabinan from Prunus Dulcis Immunobiological Active Pectic Polysaccharides. Carbohydr. Polym 2006, 66 (1), 27–33. 10.1016/j.carbpol.2006.02.020. [DOI] [Google Scholar]
- (13).Merkx DWH; Westphal Y; van Velzen EJJ; Thakoer KV; de Roo N; van Duynhoven JPM Quantification of Food Polysaccharide Mixtures by 1H NMR. Carbohydr. Polym 2018, 179, 379–385. 10.1016/j.carbpol.2017.09.074. [DOI] [PubMed] [Google Scholar]
- (14).Rocha M; Delgadillo I; Marcela C; Jana C Use of FT-IR Spectroscopy as a Tool for the Analysis of Polysaccharide Food Additives. 2003, 51, 383–389. [Google Scholar]
- (15).Coimbra MA; Gonçalves F; Barros AS; Delgadillo I Fourier Transform Infrared Spectroscopy and Chemometric Analysis of White Wine Polysaccharide Extracts. J. Agric. Food Chem 2002, 50 (12), 3405–3411. 10.1021/jf020074p. [DOI] [PubMed] [Google Scholar]
- (16).Lerouxel O; Choo TS; Se M; Lerouge P; Pauly M Rapid Structural Phenotyping of Plant Cell Wall Mutants by Enzymatic Oligosaccharide Fingerprinting. Plant Physiol. 2002, 130, 1754–1763. 10.1104/pp.011965.Progress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Obel N; Erben V; Schwarz T; Kühnel S; Fodor A; Pauly M Microanalysis of Plant Cell Wall Polysaccharides. Mol. Plant 2009, 2 (5), 922–932. 10.1093/mp/ssp046. [DOI] [PubMed] [Google Scholar]
- (18).Doco T; O’Neill MA; Pellerin P Determination of the Neutral and Acidic Glycosyl-Residue Compositions of Plant Polysaccharides by GC-EI-MS Analysis of the Trimethylsilyl Methyl Glycoside Derivatives. Carbohydr. Polym 2001, 46 (3), 249–259. 10.1016/S0144-8617(00)00328-3. [DOI] [Google Scholar]
- (19).Guadalupe Z; Martínez-Pinilla O; Garrido Á; Carrillo JD; Ayestarán B Quantitative Determination of Wine Polysaccharides by Gas Chromatography-Mass Spectrometry (GC-MS) and Size Exclusion Chromatography (SEC). Food Chem. 2012, 131 (1), 367–374. 10.1016/j.foodchem.2011.08.049. [DOI] [Google Scholar]
- (20).Xia YG; Wang TL; Sun HM; Liang J; Kuang HX Gas Chromatography–Mass Spectrometry-Based Trimethylsilyl-Alditol Derivatives for Quantitation and Fingerprint Analysis of Anemarrhena Asphodeloides Bunge Polysaccharides. Carbohydr. Polym 2018, 198 (February), 155–163. 10.1016/j.carbpol.2018.06.066. [DOI] [PubMed] [Google Scholar]
- (21).Amicucci MJ; Galermo AG; Nandita E; Vo TTT; Liu Y; Lee M; Xu G; Lebrilla CB A Rapid-Throughput Adaptable Method for Determining the Monosaccharide Composition of Polysaccharides. Int. J. Mass Spectrom 2019, 438, 22–28. 10.1016/j.ijms.2018.12.009. [DOI] [Google Scholar]
- (22).Amicucci MJ; Nandita E; Galermo AG; Castillo JJ; Chen S; Park D; Smilowitz JT; German JB; Mills DA; Lebrilla CB A Nonenzymatic Method for Cleaving Polysaccharides to Yield Oligosaccharides for Structural Analysis. Nat. Commun 2020, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Nishinari K; Takemasa M; Zhang H; Takahashi R Storage Plant Polysaccharides: Xyloglucans, Galactomannans, Glucomannans. In Comprehensive glycoscience: from chemistry to systems biology; Kamerling H, Ed.; Elsevier, 2007; pp 613–652. [Google Scholar]
- (24).Fischbacher A; von Sonntag C; Schmidt TC Hydroxyl Radical Yields in the Fenton Process under Various PH, Ligand Concentrations and Hydrogen Peroxide/Fe(II) Ratios. Chemosphere 2017, 182, 738–744. 10.1016/j.chemosphere.2017.05.039. [DOI] [PubMed] [Google Scholar]
- (25).Chan KH; Chu W The Dose and Ratio Effects of Fe(II) and H2O2 in Fenton’s Process on the Removal of Atrazine. Environ. Technol 2003, 24 (6), 703–710. 10.1080/09593330309385606. [DOI] [PubMed] [Google Scholar]
- (26).Barb WG; Baxendale JH; George P; Hargrave KR Reactions of Ferrous and Ferric Ions with Hydrogen Peroxide Part I: The Ferrous Ion Reaction. Trans. Faraday Soc 1951, 47, 462–500. [Google Scholar]
- (27).Weiss J; Humphrey CW Reaction between Hydrogen Peroxide and Iron Salts. Nature 1949, 163 (4148), 691. [Google Scholar]
- (28).Phillips TG Changes in the Composition of Squash During Storage. Plant Physiol. 1946, 21 (4), 533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Xu S; Yang L; Tian R; Wang Z; Liu Z; Xie P; Feng Q Species Differentiation and Quality Assessment of Radix Paeoniae Rubra (Chi-Shao) by Means of High-Performance Liquid Chromatographic Fingerprint. J. Chromatogr. A 2009, 1216 (11), 2163–2168. 10.1016/j.chroma.2008.04.064. [DOI] [PubMed] [Google Scholar]
- (30).Sun X; Wang H; Han X; Chen S; Zhu S; Dai J Fingerprint Analysis of Polysaccharides from Different Ganoderma by HPLC Combined with Chemometrics Methods. Carbohydr. Polym 2014, 114, 432–439. 10.1016/j.carbpol.2014.08.048. [DOI] [PubMed] [Google Scholar]
- (31).Khuwijitjaru P; Pokpong A; Klinchongkon K; Adachi S Production of Oligosaccharides from Coconut Meal by Subcritical Water Treatment. Int. J. Food Sci. Technol 2014, 49 (8), 1946–1952. 10.1111/ijfs.12524. [DOI] [Google Scholar]
- (32).Gwirtz JA; Garcia-Casal MN Processing Maize Flour and Corn Meal Food Products. Ann. N. Y. Acad. Sci 2014, 1312 (1), 66–75. 10.1111/nyas.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhu K; Zhang Y; Nie S; Xu F; He S; Gong D; Wu G; Tan L Physicochemical Properties and in Vitro Antioxidant Activities of Polysaccharide from Artocarpus Heterophyllus Lam. Pulp. Carbohydr. Polym 2017, 155, 354–361. 10.1016/j.carbpol.2016.08.074. [DOI] [PubMed] [Google Scholar]
- (34).Tan YF; Li HL; Lai WY; Zhang JQ Crude Dietary Polysaccharide Fraction Isolated from Jackfruit Enhances Immune System Activity in Mice. J. Med. Food 2013, 16 (7), 663–668. 10.1089/jmf.2012.2565. [DOI] [PubMed] [Google Scholar]
- (35).Jiménez-Escrig A; Rincón M; Pulido R; Saura-Calixto F Guava Fruit (Psidium Guajava L.) as a New Source of Antioxidant Dietary Fiber. J. Agric. Food Chem 2001, 49 (11), 5489–5493. 10.1021/jf010147p. [DOI] [PubMed] [Google Scholar]
- (36).Zhang Z; Kong F; Ni H; Mo Z; Wan JB; Hua D; Yan C Structural Characterization, α-Glucosidase Inhibitory and DPPH Scavenging Activities of Polysaccharides from Guava. Carbohydr. Polym 2016, 144, 106–114. 10.1016/j.carbpol.2016.02.030. [DOI] [PubMed] [Google Scholar]
- (37).Myoda T; Matsuda Y; Suzuki T; Nakagawa T; Nagai T; Nagashima T Identification of Soluble Proteins and Interaction with Mannan in Mucilage of Dioscorea Opposita Thunb. (Chinese Yam Tuber). Food Sci. Technol. Res 2006, 12 (4), 299–302. 10.3136/fstr.12.299. [DOI] [Google Scholar]
- (38).Fu YC; Chen S; Lai YJ Centrifugation and Foam Fractionation Effect on Mucilage Recovery from Dioscorea (Yam) Tuber. J. Food Sci 2004, 69 (9), 471–477. [Google Scholar]
- (39).Yang W; Wang Y; Li X; Yu P Purification and Structural Characterization of Chinese Yam Polysaccharide and Its Activities. Carbohydr. Polym 2015, 117, 1021–1027. 10.1016/j.carbpol.2014.09.082. [DOI] [PubMed] [Google Scholar]
- (40).Vollendorf NW; Marlett JA Comparison of Two Methods of Fiber Analysis of 58 Foods. Journal of Food Composition and Analysis. 1993, pp 203–214. 10.1006/jfca.1993.1023. [DOI] [Google Scholar]
- (41).Monono EM; Haagenson DM; Pryor SW Developing and Evaluating NIR Calibration Models for Multi-Species Herbaceous Perennials. Ind. Biotechnol 2012, 8 (5), 285–292. 10.1089/ind.2012.0018. [DOI] [Google Scholar]
- (42).Monono EM; Nyren PE; Berti MT; Pryor SW Variability in Biomass Yield, Chemical Composition, and Ethanol Potential of Individual and Mixed Herbaceous Biomass Species Grown in North Dakota. Ind. Crops Prod 2013, 41 (1), 331–339. 10.1016/j.indcrop.2012.04.051. [DOI] [Google Scholar]
- (43).Zheng Y; Pan Z; Zhang R; Wang D; Labavitch J; Jenkins BM Dilute Acid Pretreatment and Enzymatic Hydrolysis of Saline Biomass for Sugar Production. In 2006 ASABE Annual International Meeting; American Society of Agricultural and Biological Engineers: Portland, OR, 2006. [Google Scholar]
- (44).Wang LZ; White PJ Structure and Properties of Amylose, Amylopectin, and Intermediate Materials of Oat Starches. Cereal Chemistry. 1994, pp 263–268. [Google Scholar]
- (45).Pronyk C; Mazza G Fractionation of Triticale, Wheat, Barley, Oats, Canola, and Mustard Straws for the Production of Carbohydrates and Lignins. Bioresour. Technol 2012, 106, 117–124. 10.1016/j.biortech.2011.11.071. [DOI] [PubMed] [Google Scholar]
- (46).Wood PJ Cereal β-Glucans in Diet and Health. J. Cereal Sci 2007, 46 (3), 230–238. 10.1016/j.jcs.2007.06.012. [DOI] [Google Scholar]
- (47).Butt MS; Tahir-Nadeem M; Khan MKI; Shabir R; Butt MS Oat: Unique among the Cereals. Eur. J. Nutr 2008, 47 (2), 68–79. 10.1007/s00394-008-0698-7. [DOI] [PubMed] [Google Scholar]
- (48).Varo P; Laine R; Veijalainen K; Espo A; Wetterhoff A; Koivistoinen P Dietary Fibre and Avaialable Carbohydrates in Finnish Vegetables and Fruits. J. Agric. Sci. Finl 1984, 56, 49–59. [Google Scholar]
- (49).Ballesteros LF; Cerqueira MA; Teixeira JA; Mussatto SI Characterization of Polysaccharides Extracted from Spent Coffee Grounds by Alkali Pretreatment. Carbohydr. Polym 2015, 127, 347–354. 10.1016/j.carbpol.2015.03.047. [DOI] [PubMed] [Google Scholar]
- (50).Pettolino FA; Walsh C; Fincher GB; Bacic A Determining the Polysaccharide Composition of Plant Cell Walls. Nat. Protoc 2012, 7 (9), 1590–1607. 10.1038/nprot.2012.081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


