Significance
Impairment of the endolymphatic system is a common cause of deafness. Our study of a mouse model of campomelic dysplasia, a syndrome characterized in part by deafness of unknown etiology, reveals the roles of SOX9 and its relative SOX10 in maintaining a functioning endolymphatic system. Early in its development, these SOXE factors are critical for regulating the differentiation of progenitors and the expression of ion transporters and channels essential for ionic homeostasis. Later, they control the expression of Aqp3, a water/glycerol channel. The data provide a rich resource for the discovery of deafness genes. Since SoxE genes have many roles in development and in stem cells, the principles we have found could be applicable to tissue-specific effects in syndromes.
Keywords: SOXE transcription factors, inner ear endolymphatic system, haploinsufficiency, dominant negative, developmental syndrome
Abstract
The in vivo mechanisms underlying dominant syndromes caused by mutations in SRY-Box Transcription Factor 9 (SOX9) and SOX10 (SOXE) transcription factors, when they either are expressed alone or are coexpressed, are ill-defined. We created a mouse model for the campomelic dysplasia SOX9Y440X mutation, which truncates the transactivation domain but leaves DNA binding and dimerization intact. Here, we find that SOX9Y440X causes deafness via distinct mechanisms in the endolymphatic sac (ES)/duct and cochlea. By contrast, conditional heterozygous Sox9-null mice are normal. During the ES development of Sox9Y440X/+ heterozygotes, Sox10 and genes important for ionic homeostasis are down-regulated, and there is developmental persistence of progenitors, resulting in fewer mature cells. Sox10 heterozygous null mutants also display persistence of ES/duct progenitors. By contrast, SOX10 retains its expression in the early Sox9Y440X/+ mutant cochlea. Later, in the postnatal stria vascularis, dominant interference by SOX9Y440X is implicated in impairing the normal cooperation of SOX9 and SOX10 in repressing the expression of the water channel Aquaporin 3, thereby contributing to endolymphatic hydrops. Our study shows that for a functioning endolymphatic system in the inner ear, SOX9 regulates Sox10, and depending on the cell type and target gene, it works either independently of or cooperatively with SOX10. SOX9Y440X can interfere with the activity of both SOXE factors, exerting effects that can be classified as haploinsufficient/hypomorphic or dominant negative depending on the cell/gene context. This model of disruption of transcription factor partnerships may be applicable to congenital deafness, which affects ∼0.3% of newborns, and other syndromic disorders.
The SOXE (Sex-determining region Y [SRY]–related HMG-box, group E) transcription factors SOX9 and SOX10 are essential for the specification and differentiation of many progenitor cell (ProgC) types and for the development of several organs and tissues (1, 2). The expression patterns of the two genes can be distinct (e.g., cartilage) or overlapping (e.g., neural crest, nervous system, inner ear). Misexpression studies in mice suggest that SOXE genes can be partially redundant (3–5). SOX9 and SOX10 have redundant functions in oligodendrocyte precursors (5). SOX8, the third member of the SOXE group in the SRY-related transcription factor (SOX) family, is widely expressed, and although it is not essential for normal embryonic development, it has a role in adult Sertoli cell function (2, 6). SOX8 and SOX10 have also been shown to be partially redundant in oligodendrocyte development (2, 3, 7).
Mutations in SOX9 and SOX10 are dominant, causing several syndromes affecting different organ systems. In humans, heterozygous mutations in SOX10 are associated with sensorineural hearing loss in Waardenburg–Shah syndrome (types II and IV; Online Mendelian Inheritance in Man [OMIM] 611584 and 613266), Kallmann syndrome (8), peripheral demyelinating neuropathy, and central dysmyelinating leukodystrophy (PCWH; OMIM 609136) (9, 10). Heterozygous mutations in SOX9 result in campomelic dysplasia (CD), which is a syndrome characterized by skeletal dysmorphology, XY female sex reversal (OMIM 114290), abnormal development of the kidneys, central nervous system, and sensorineural deafness of unknown etiology (11–13).
Hearing and balance require a functioning sensory and endolymphatic system in the inner ear. The cochlea detects sound, and the vestibular organs detect linear acceleration and gravity. The different sensory organs and the endolymphatic sac (ES) of the endolymphatic system are specified at the otic vesicle stage (14, 15). During the development of the otic vesicle, the elaborate membranous labyrinthine system becomes filled with endolymph, a specialized extracellular fluid essential for sensory function. Dysfunction of this fluid pathway results in endolymphatic hydrops that causes hearing loss, vertigo, tinnitus, aural fullness, and pain. The ES is the first structure of the inner ear to develop and control endolymph volume and pressure (16). The stria vascularis on the lateral wall of the cochlear duct develops later, matures postnatally (postnatal day 3), generates the endocochlear potential between the endolymph and the perilymph, and regulates the ionic content of the endolymph. In a fully developed inner ear, endolymph secreted from the cochlear duct drains into the saccule and enters the endolymphatic duct (ED) and ES, where it is reabsorbed by the vascular system (17).
The roles of SOXE factors in the development of the endolymphatic system are not well defined. SOX9 and SOX10 are both expressed in the developing inner ear (18–20). Sox9 is required for specification of otic placode (21) and essential for maintaining otic progenitors in Xenopus (22). It is widely expressed in the mouse inner ear during development (19, 22) and is required for otic invagination (18). Loss of Sox9 expression in the periotic mesenchyme affects both otic capsule differentiation and cochlear coiling (23). Complete loss of Sox10 in mice results in shortened cochlea ducts in both heterozygotes and homozygotes, which are attributed to the loss of cochlea progenitors, although those remaining are correctly differentiated (20). Although Sox8 acts redundantly with Sox9 in testes development, this may not be the case in the ear. In chicks, Sox8 regulates Sox10 in the otic vesicle, but this is not conserved in mice (24, 25). Sox8 is expressed in the supporting cells in the sensory epithelium of the cochlea, but Sox8-null mice do not have inner ear defects (20, 26–28), suggesting a nonessential role in hearing.
CD is attributed to haploinsufficiency for SOX9 because there is no obvious correlation between the type or position of mutation and disease severity, a concept supported by the recapitulation of the cartilage defects in Sox9 heterozygous null mutant mice (29). However, not all SOX9 mutations result in the absence of protein. For example, some mutations in SOX9 result in a truncated transactivation domain but intact DNA binding domain. Mechanisms, such as dominant negative effects, cannot be ruled out. There is no systematic or definitive comparison between probands affected by different SOX9 mutations in terms of survival (30, 31). CD is usually described as “perinatal lethal,” with some probands surviving to 2 y. However, some probands with the SOX9 Y440X mutation, which truncates the C-terminal transactivation domain, survive for up to 4 to 9 y (32–34). Intriguingly, residual transactivation activity of SOX9Y440X was demonstrated in vitro (13, 32–34), raising the possibility of a hypomorphic mechanism that allows for longer survival. By contrast, in vitro tests of other SOX9 mutants with truncated C termini suggest a dominant negative effect (35).
In vitro assays show that SOX9 and SOX10 interact with each other and form homodimers as well as heterodimers (2, 36). SOXE factors also work with partner factors to regulate gene expression (37, 38). Important questions regarding disease mechanisms underlying heterozygosity for SOXE factors are how the mutation in one allele impacts the function of the normal allele or other coexpressed partners, leading to disease phenotypes. How heterozygous mutations in SOX9 cause sensorineural deafness in CD and whether these impact the function of SOX10, its SOXE partner, are unknown.
To address these questions, we focused on delineating the molecular mechanism(s) underlying the least understood clinical aspect of CD caused by the SOX9Y440X mutation: sensorineural deafness. By comparing the impact on inner ear development of constitutive and otic-specific expression of the SOX9Y440X mutation in heterozygotes and heterozygous otic-specific Sox9-null and heterozygous Sox10-null mutants, we implicate essential roles for both SOXE factors in hearing and vestibular function. Early in inner ear development, both SOX9 and SOX10 control ProgC differentiation and ionic homeostasis for a functioning endolymphatic system of the inner ear. The SOXE factors also act cooperatively later in inner ear development, contributing to the functional properties of the stria vascularis in the cochlea, for example, by controlling the expression of a water/glycerol channel aquaporin 3 (AQP3). Overall, the developmental stage and inner ear cell-type context-dependent impact of the SOX9Y440X mutation combined with the disruption of the SOXE gene regulatory network leads to auditory and vestibular dysfunction.
Results
Sox9Y440X Causes Vestibular Dysfunction and Lack of Endocochlear Potential.
To investigate the underlying basis for deafness in CD, we generated mice carrying a conditional targeted allele (Sox9Y440X-IRES-EGFP; abbreviated Sox9Y440X) comprising Sox9 exon 3 flanked by loxP sites and a mutated exon 3 containing Sox9Y440X linked to an internal ribosome entry site–enhanced green fluorescent protein (IRES-EGFP) reporter, inserted downstream (Fig. 1A and SI Appendix, Fig. S1 A–G). Mice with the conditional allele were phenotypically and behaviorally normal and fertile. Sox9 is expressed in the developing otic epithelium from embryonic day 8.5 (E8.5), the surrounding mesenchyme from E10.5, and the otic capsule at later stages (19, 23). Expression of the mutant allele, reported by GFP expression, was confirmed in the heterozygous embryos carrying an ubiquitously expressed β-actin Cre recombinase (cre)–directed knock-in allele of Sox9Y440X. GFP expression recapitulates the endogenous pattern of Sox9 in the otic vesicle at E10.5 and ES/ED and cochlea at E14.5 (SI Appendix, Fig. S1 C–G). We confirmed the specific and stable expression of truncated SOX9Y440X protein in the developing mutant ES/ED and cochlea using an antibody that recognizes the mutant protein specifically (Fig. 1 B and C). Approximately 10% of ubiquitous cre-induced constitutive Sox9Y440X/+ mice survived, from which a heterozygous mutant line was established. No Sox9Y440X/Y440X homozygotes survived to birth. In addition to the skeletal phenotype typical of CD in newborns (SI Appendix, Fig. S1B), the surviving heterozygous mutant mice had kinked tails, showed circling behavior, and failed swimming tests, typical of vestibular disorders (Movies S1 and S2).
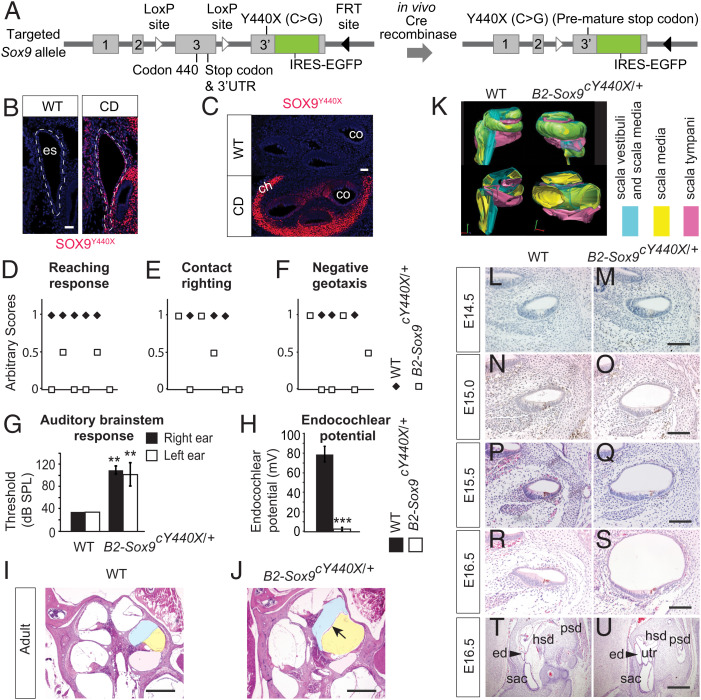
Fig. 1.
Impaired vestibular and auditory function in B2-Sox9cY440X/+ mutants. (A) Schematic diagram of the Sox9cY440X/+ targeted allele comprising exon 3 flanked by LoxP sites followed by exon 3′ carrying the Y440X mutation and IRES-EGFP sequences. FRT, flippase recognition target. UTR, untranslated region. (B and C) Immunostaining of SOX9Y440X in WT and Sox9Y440X/+ (CD) ES (B) and cochlea (C) at E14.5 (n = 2). (D–F) B2-Sox9cY440X/+ mutants displayed impaired vestibular function. (D) Reaching response: forelimbs stretched scored 1, curled ventrally scored 0, and curled slightly scored 0.5. (E) Contact righting: flipped back to upright position within 5 s scored 1, crawled upside down scored 0, and crawled sideways scored 0.5. (F) Negative geotaxis: turned and moved up the slope surface scored 1, failed to stay on the slope surface scored 0, and failed to turn and stayed scored 0.5 (WT n = 5; B2-Sox9cY440X n = 6). (G) Thresholds for auditory brain stem response (sound pressure level [SPL]); 120-dB SPL was the highest level measured (n = 5). (H) Endocochlear potential in 4- to 7-mo-old WT and B2-Sox9cY440X/+ mice (n = 4). (I and J) Cochlear ducts from WT (I) and B2-Sox9cY440X/+ (J) adult animals at 6 mo. (K) Three-dimensional reconstruction of WT and B2-Sox9cY440X/+ adult cochlea. Scala vestibule/scala media, scala media, and scala tympani are in blue, yellow, and pink, respectively. (L–S) Cochlear ducts from WT and B2-Sox9cY440X/+ embryos at E14.5 (L and M), E15.0 (N and O), E15.5 (P and Q), and E16.5 (R and S; n = 2 to 4). (T and U) Transverse sections of WT and B2-Sox9cY440X/+ ED (ed; arrowhead) at E16.5 (n = 2). Data are presented as means ± SEM. Results for the CD mutants are from the constitutively active Sox9Y440X/+ samples unless labeled as conditional. ch, chondrocyte; co, cochlea; es, ES; hsd, horizontal semicircular duct; psd, posterior semicircular duct; sac, saccule; utr, utricle. (Scale bars: 40 µm in B and C; 100 µm in M, O, Q, and S; 400 µm in I, J, and U.) **P < 0.01; ***P < 0.001.
We generated conditional mutants (B2-Sox9cY440X/+) using the HoxB2(r4)-Cre (B2-r4-cre) mouse that expresses CRE in the otic placode from E8.5 (39) to assess the effect of Sox9Y440X on inner ear development. Otic vesicles of E9.5 B2-Sox9cY440X/+ embryos were morphologically normal. However, before weaning, mutants developed vestibular and auditory phenotypes, such as hyperactivity, head tossing and circling behavior (Movie S3), and head tilting (SI Appendix, Fig. S1H). They failed tests for reaching response (Fig. 1D and SI Appendix, Fig. S1I), contact righting, and negative geotaxis (Fig. 1 E and F and Movies S4 and S5). Their auditory brain stem responses showed a hearing deficit of 40 to 80 dB (Fig. 1G and SI Appendix, Fig. S1J), and the endocochlear potential was reduced to 2 mV in mature mutants (Fig. 1H).
Otic Expression of Sox9Y440X Causes Endolymphatic Hydrops.
Endolymphatic hydrops is caused by disruption of endolymph homeostasis. Histology revealed endolymphatic hydrops in B2-Sox9cY440X/+ mice (Fig. 1 I, J, and L–U). In adult mutants, the scala media containing the endolymph were expanded, with a distended Reissner's membrane (Fig. 1 I and J). Reconstruction of the cochlea structure in three dimensions (Fig. 1K and SI Appendix, Fig. S1K) showed that of the three sound-conducting chambers, the scala media in the mutant were twice the volume of that in the wild type (WT), while the scala tympani was one-third smaller; the scala vestibuli were of the same size. Consistent with volume changes in the developing ear (40), expansion of the cochlear duct was not detectable at E14.5 (Fig. 1 L and M), but it was apparent by E15.0 (Fig. 1 N and O) and became more prominent and obvious at E16.5 (Fig. 1 R and S) and in adults (Fig. 1 I and J). At P9, we found otoconia deficiency in the mutant utricle and saccule, an indication of defects in the endolymphatic system (SI Appendix, Fig. S1L). Scanning electron microcopy of the inner ears of B2-Sox9cY440X/+ mice at P4 showed normal arrangement of hair cells and stereocilia in the organ of Corti, suggesting that the hearing deficit is not caused by abnormal prosensory development (SI Appendix, Fig. S1M). Assessment of the relative numbers of bromodeoxyuridine (BrdU)–labeled proliferating cells (ProlCs) and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)–positive apoptotic cells among the total number of cochlea epithelial cells at E14.5 showed no significant differences between WT and mutants (SI Appendix, Fig. S1 N–S), suggesting that cell proliferation and survival were not affected.
Fewer Slc26a4-Expressing Cells in the Sox9Y440X/+ Mutant ES.
The mouse Lmx1a functional null mutant dreher (drJ/drJ) (41–43) lacks an ED, while patients with LMX1A mutations have impaired hearing (44, 45). Normal Lmx1a expression in the prospective ES of the Sox9Y440X/+ mutants at E10.5 indicates proper specification of the rudiment (SI Appendix, Fig. S1T). The ED was enlarged in B2-Sox9cY440X/+ mutants at E16.5 compared with WT (Fig. 1 T and U), reminiscent of hydropic ears (46). In humans, enlarged ES and cochlea are characteristic of Pendred syndrome caused by homozygous mutations in SLC26A4 (Pendrin) encoding an anion exchanger (47, 48); heterozygous mutations in the transcription factor FOXI1 (OMIM 601093), which activates SLC26A4 transcription (49); or compound heterozygous mutations for both SLC26A4 and FOXI1. In mice, disruption of NaCl resorption mediated by FOXI1 and its target Slc26a4 causes enlargement of the endolymphatic compartment, endolymph imbalance, and deafness (50). Mice lacking SLC26A4 in the ES develop endolymphatic hydrops (51). We examined if Sox9Y440X/+ impacts on Slc26a4 expression. The signal intensity of in situ hybridization for Slc26a4 in the mutant ES appeared unchanged, but the preponderance of Slc26a4-expressing cells was significantly reduced by 40% (Fig. 2 A and B), which may contribute to the deafness phenotype.
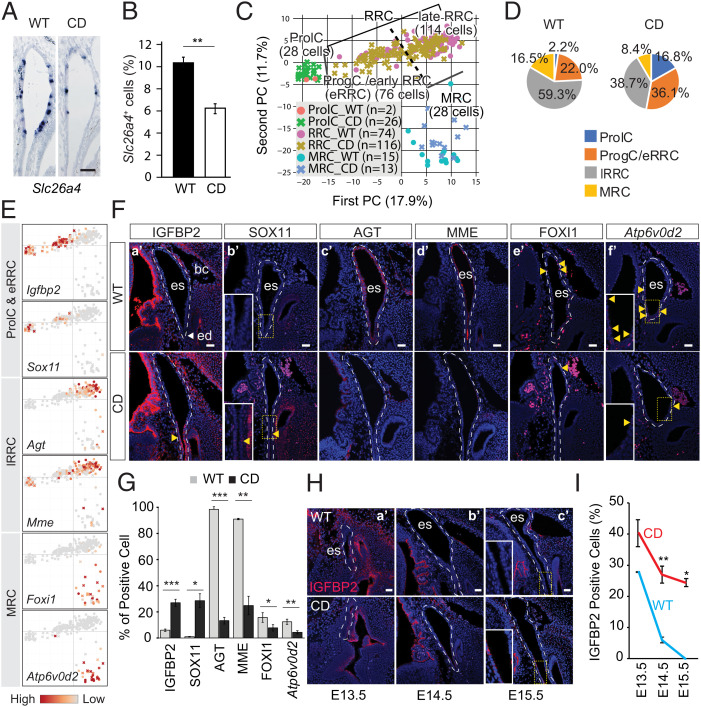
Fig. 2.
Impaired differentiation in the Sox9Y440X/+ (CD) ES. (A) Slc26a4 expression in the ES of WT and CD mutant at E14.5 (n = 3). (B) Percentage of Slc26a4-expressing cells (blue in A) among the total number of epithelial cells (n = 3). (C) Principal component (PC) analysis of 246 cells identifies three distinctive cell populations, whereby RRCs are further divided into early (eRRCs) and late (lRRCs) ones. (D) Pie charts showing the proportion of cell populations per genotype. (E) PCA plots showing marker gene expression for each population identified. (F) Immunostaining or hybridization chain reaction (HCR) of population markers in the WT and CD ES at E14.5: (F, a′) IGFBP2 (n = 3), (F, b′) SOX11 (n = 2), (F, c′) AGT (n = 2), (F, d′) MME (n = 2), (F, e′) FOXI1 (n = 3), and (F, f′) Atp6v0d2 (n = 3). (G) Proportion of cells (percentage) expressing markers in F. (H) Immunostaining of IGFBP2 in E13.5 to E15.5 WT and CD ES: (H, a′) E13.5 (n = 2), (H, b′) E14.5 (n = 3), and (H, c′) E15.5 (n = 2). (I) Quantification of cells (percentage) expressing IGFBP2 in E13.5 to E15.5 WT and CD ES. Data are presented as means ± SEM. Highlighted signals are indicated with yellow arrowheads. bc, blood cell; ed, ED; es, ES. (Scale bars: 50 µm in A; 40 µm in F and H.) *P < 0.05; **P < 0.01; ***P < 0.001.
Immature and Mature Cell Populations Are Present in the Sox9Y440X/+ ES.
The reduced proportion of Slc26a4-expressing cells in the ES could arise from an effect of Sox9Y440X on the activation of gene expression and/or cell identity. To examine both possibilities, we sequenced the transcriptomes of single cells isolated from WT (91 cells, n = 3) and Sox9Y440X/+ (155 cells, n = 2) ESs at E14.5 before an apparent phenotype. Using principal component analysis (52), we identified three cell populations and defined cluster-specific signatures by determining the differentially expressed genes (DEGs) of each cluster vs. all other clusters (Dataset S1) and by reference to single-cell transcriptomes for E12.5 and E16.5 ES (53) (SI Appendix, Fig. S2 A–C). The clusters were ProlCs (marked by Mki67 and Birc5), ribosome-rich cells (RRCs; marked by Dach2, Oc90, Agt, and Mme), and mitochondrial-rich cells (MRCs; marked by Atp6v0d2, Foxi1, and Slc26a4). Functional annotation by gene ontology (GO) analyses was consistent with current knowledge; ProlCs were enriched in cell cycle and mitotic activities, and MRCs and RRCs were enriched in epithelial cell properties. GO for MRCs reflected the enrichment for mitochondrial activity and expression of membrane transporters and ion channels that maintain fluid homeostasis in the inner ear. For RRCs, GO reflected enrichment for cell–cell communication and the extracellular matrix (matrisome) (54) (SI Appendix, Fig. S2D). All three populations expressed genes encoding ionic transporters or membrane channels (GO: 0015075, total of 851 genes), especially RRCs and MRCs. MRCs expressed more active transporters and/or membrane channels than the other two clusters: ProlCs and RRCs (SI Appendix, Fig. S2E).
Density contour plots identified two subpopulations within the RRC cluster (SI Appendix, Fig. S2 F and G). One subpopulation was similar to reported immature cell types, early ribosome-rich cells (eRRCs; odds ratio = 31.4), and ProgCs (odds ratio = 44.1), which we annotated as ProgCs/eRRCs (SI Appendix, Fig. S2H). The other subpopulation was similar (odds ratio = 21.7) to reported more mature ribosome-rich cells (late RRCs, or lRRCs) (53). Genes marking the ProgCs/eRRCs include Igfbp2, Aldh1a3, and Fam132a, and those marking lRRCs include Mme and Agt (Dataset S1 and SI Appendix, Fig. S2I). Pseudotime trajectory plots and empirical entropy estimation predicted lineage progression from ProgCs/eRRC to RRC and then, MRC (SI Appendix, Fig. S2 J–M), consistent with the suggested lineage origin of MRC from RRCs (53).
Increased Proportion of Immature Cells in the Sox9Y440X/+ ES.
To examine if the Sox9Y440X mutation affects RRCs, MRC fate, and/or differentiation progression, we compared the proportion of different cell clusters. We observed a lower proportion of MRCs (mature cells) in the mutant (8.4%) than in WT (16.5%). The proportion of lRRCs (mature cells) was also reduced by 20.6 percentage points in the mutant (Pearson χ2 P = 0.003) compared with WT. By contrast, the proportion of immature cells increased (ProlCs [14.6 percentage points] and ProgCs/eRRCs [14.1 percentage points]) in the mutant (Pearson χ2 P = 0.001 and P = 0.029, respectively) (Fig. 2 C and D), resulting in a deficiency in mature cells.
To assess whether this deficiency was associated with differentiation states, we assessed the sequence of differentiation in the ES. IGFBP2 and SOX11 mark ProlCs/eRRCs at the proximal end of the ES. AGT and MME mark lRRCs, while FOXI1 and Atp6v0d2 mark MRCs. Both cell types are evenly distributed with more lRRCs in the sac (Fig. 2 E and F). Quantification of these different cell types confirmed the reduced proportions of mature cells (lRRCs and MRCs) vs. immature cells in the mutant compared with WT (Fig. 2G and SI Appendix, Table S1). FOXI1 directly transactivates Slc26a4 (49). In line with the reduced proportion of Slc26a4+ cells (Fig. 2 A and B), there were fewer Foxi1-expressing cells (8% in mutants vs. 16% in WT) (SI Appendix, Table S1). However, transcript levels for both genes were comparable in the MRCs of both genotypes (Fig. 2A and SI Appendix, Fig. S2N), suggesting that the reduced proportion of Slc26a4+ cells was not due to the down-regulation of FOXI1.
Analyses of our data and published single cell RNA sequencing (scRNA-seq) data (53) suggest that lRRCs and MRCs originate from ProgCs/eRRCs (SI Appendix, Fig. S2 J and K), which are Igfbp2+. We assessed the number of IGFBP2+ cells from E13.5 to E15.5 in the WT and mutant ED. While the proportion of WT IGFBP2+ between E13.5 and E15.5 reduced from 27.8 to 0%, in mutants the frequency of these cells was significantly higher at all stages (E13.5 [40.3%]) and at E14.5 and E15.5, remained at 27.0 and 24.4%, respectively (Fig. 2 H and I), suggesting that differentiation was delayed. Sox11 is essential for inner ear development (55) and is expressed by E12.5 ES progenitors (53). Consistent with our hypothesis of delayed differentiation, we found that Sox11 expression in the mutant ProlCs/ProgCs/eRRCs persisted in the mutant ES/ED at E14.5 (Fig. 2 E and F).
Dysregulated Genes Regulating Fluid Regulation, Cell–Cell Interaction, and Wnt Signaling in Sox9Y440X/+ RRCs.
We next identified DEGs in RRCs caused by Sox9Y440X. Over a hundred DEGs (103 genes) were found between WT and mutants (Fig. 3A, Dataset S1, and SI Appendix, Fig. S3A), of which 58 genes were also DEGs in RRCs against all cell clusters (odds ratio = 94.6, Pearson χ2 P < 2.2 × 10−16). Functional annotation (GO) of genes down-regulated in mutant RRCs identified processes related to extracellular structure organization, sensory organ and ear/otolith development, and neurogenesis, while the up-regulated genes were associated with cell junctions and cell–cell adhesion (SI Appendix, Fig. S3B). Importantly, several genes involved in fluid regulation were down-regulated in the Sox9Y440X/+ mutant ES, such as genes encoding ion transporters (e.g., Slc24a4 and Slc15a1), channels (Ttyh1), gap junction protein (Gjb2), and members of the renin-angiotensin system (e.g., Mme, Agt) (Fig. 3A, Dataset S1, and SI Appendix, Fig. S3 A, C, and D). These changes are consistent with the observed ear hydrops. Genes associated with the Wingless-related integration site (Wnt) pathway were also affected. Dkk3, a Wnt antagonist, was down-regulated, while Wnt targets and ligands Ccnd2 and Wnt6/10a were up-regulated in the mutant (Fig. 3A, Dataset S1, and SI Appendix, Fig. S3A). Coexpression analyses confirmed that Ccnd2, Wnt6, and Igfbp2 were negatively correlated with Dkk3, indicating a coordinated dysregulation and a net up-regulation of the pathway (SI Appendix, Fig. S3E).
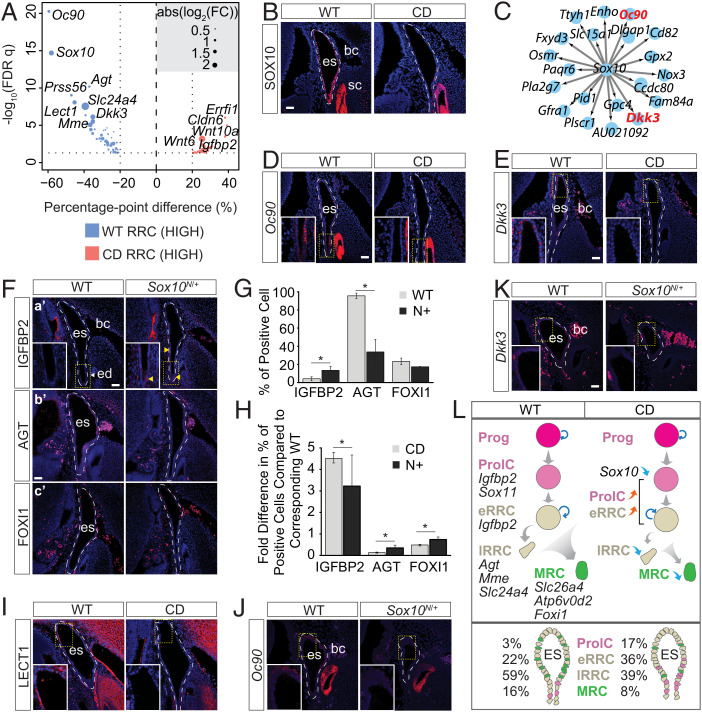
Fig. 3.
Impaired Sox10 regulon disrupts ES maturation. (A) DEGs between the two genotypes in RRCs. FC, fold change. (B) SOX10 immunostaining in WT and CD E14.5 ES (n = 3). (C) SOX10 target genes down-regulated in CD cells. (D and E) Sox10 regulon expression (D) Oc90 (n = 2) and (E) Dkk3 (n = 2) in CD ES at E14.5. (F) Population markers expressed in Sox10N/+ (N+) E14.5 ES. (F, a′) IGFBP2 (n = 3). (F, b′) AGT (n = 2). (F, c′) FOXI1 (n = 2). (G) Proportion of cells (percentage) expressing population markers in WT and N+ ES at E14.5. (H) Fold difference in the percentage of expressing cell (population markers) compared with the corresponding wild-type control for CD and N+ E14.5 ES. (I) Immunostaining of LECT1 in E14.5 WT and CD ES (n = 2). (J and K) Sox10 regulon (J) Oc90 (n = 2) and (K) Dkk3 (n = 2) in WT and N+ E14.5 ES. (L) Schematic diagram showing the regulatory and cellular dynamics of normal and defective differentiation of ES cells in WT and CD. Data are presented as means ± SEM. Highlighted signals are indicated with yellow arrowhead. bc, blood cell; ed, ED; es, ES; sc, semicircular canal; FDR, false discovery rate. (Scale bars: 40 µm.) *P < 0.05.
Sox9Y440X Impacts Sox10 Expression and Its Regulon.
Sox10 expression is initiated later than Sox9 expression in the mouse otic placode and is absent in conditional Sox9-null otocysts (18). Sox10 is coexpressed with Sox9 in the otic epithelium (19, 56). Strikingly, expression of Sox10 was highly affected by the Sox9Y440X mutation in the RRCs (Fig. 3 A and B). Almost all ES/ED cells express SOX10 from E13.5 to E15.5. However, the proportion of SOX10+ cells in the mutant was low and decreased from 27.2% at E13.5 to less than 20% at E15.5 (SI Appendix, Fig. S3F). Single-Cell Regulatory Network Inference and Clustering (SCENIC) regulon analysis also identified reduced SOX10 activity in the mutant RRCs (SI Appendix, Fig. S3G). Transcriptional targets (Oc90, Dkk3, and Nox3) of the SOX10 regulon, known to be critical for ES/inner ear function, were affected (Fig. 3 C–E).
SOX factors interact with a range of partners, allowing for individual factors to regulate different genes and alternative developmental programs (2, 37, 57, 58). β-catenin activates gene expression via interaction with T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors. SOX9, through its C-terminal transactivation domain, interacts physically with β-catenin, stimulates its degradation, and inhibits TCF/LEF gene activation (59, 60). The truncation of SOX9 at Y440 deletes most of the interacting domain (amino acids 382 to 509) and likely compromises the inhibitory activity of SOX9 on β-catenin. The increase in LEF regulon activity observed in lRRCs and MRCs supports the notion that SOX9 and TCF/LEF compete for binding to β-catenin (59). Differential gene expression analyses and marker analyses indicate dysregulation of Wnt signaling in the Sox9Y440X/+ mutant ES as reflected by changes in pathway-associated genes, such as Gjb2, Wnt6, Wnt10a, Ccnd2, and Dkk3. Interestingly, there are two conserved enhancers in the Sox10 locus that are active in the chick otic vesicle, one of which bears SOXE binding sites with overlapping TCF/LEF sites (24, 25), consistent with the possibility of impaired WNT signaling control of Sox10. In skeletogenic cells, SOXC proteins, such as SOX11, contribute to the synergistic stabilization of β-catenin and negative regulation of Sox9 (61), which raises the question about the contribution of Sox11 to persisting progenitors in the Sox9Y440X/+ ES/ED and to the dysregulation of canonical Wnt signaling.
SOXE genes act dosage dependently (62). We asked to what degree the downstream gene expression changes were attributable to down-regulation of Sox10 and/or impaired SOX9 function. As in Sox9Y440X/+ mutants but to a lesser degree, more IGFBP2+ cells (∼10%) were present in the E14.5 ES/ED of Sox10N/+ (Sox10-null) mutants than in WT, suggesting that reduced SOX10 levels contribute to the persistence of IGFBP2+ progenitors in Sox9Y440X/+ mutants. There was also a corresponding decrease in the proportion of mature RRCs and MRCs identified by their expression of Agt and Foxi1, respectively (Fig. 3 F–H and SI Appendix, Table S2). Chondromodulin (Lect1), a known downstream target of SOX9 in chondrocytes (63, 64), was strongly down-regulated in the Sox9Y440X/+ mutant RRCs/ES (Fig. 3 A and I) but unaffected in Sox10N/+ mutant ES/ED cells (SI Appendix, Fig. S3H). We hypothesize that SOX9Y440X impacts the ES/ED cells by specific effects on SOX9 direct targets (e.g., on Lect1 and Sox10) combined with downstream changes caused by down-regulation of SOX10. For example, Oc90 and Dkk3 are down-regulated not only in Sox9Y440X/+ but also, in Sox10N/+ mutants (Fig. 3 J and K) and may be common targets. By contrast, expression of the SOX9 target Lect1 was unaffected in Sox10N/+ ES/ED, suggesting that it is not regulated by SOX10. The more severe impact of the SOX9Y440X mutation could be caused by the combined failure of SOX9 to activate its own specific target genes and impaired maintenance of Sox10 expression. Together, these changes culminate to disrupt the differentiation of mature cells (RRCs and MRCs) for proper ES/ED function (Fig. 3L).
Sox9 and Sox10 Interact Genetically, with Sox9Y440X Exerting the Most Severe Effects.
SOX proteins work dose dependently (37). To gain insight into the functional relationship between Sox9, Sox10, and gene dosage effects, we studied the genetic interaction between Sox9 and Sox10 in single and compound mutants: conditional heterozygous Sox9-null mutant (B2-Sox9c/+), conditional heterozygous Sox9Y440X mutant (B2-Sox9cY440X/+), hetero- and homozygous Sox10-null mutants Sox10N/+ and Sox10N/N, compound heterozygous conditional Sox9-null and Sox10-null mutants (B2-Sox9c/+;Sox10N/+), and compound heterozygous Sox9Y440X and Sox10-null mutants (B2-Sox9cY440X/+;Sox10N/+). Using the area of the basal cochlea lumen as the readout revealed significant enlargement in some of the mutants (Fig. 4 A–G). At E15.5, the area of the basal cochlea lumen was normal in the B2-Sox9c/+ mutant (Fig. 4B), suggesting that haploinsufficiency for SOX9 was not responsible for the enlargement. However, the lumen was larger in both Sox10N/+ (P < 0.05) (Fig. 4C) and B2-Sox9c/+;Sox10N/+ mutants (P < 0.05) (Fig. 4 D and H), but there was no statistical difference in the basal lumen area between Sox10N/+ and B2-Sox9c/+;Sox10N/+ mutants suggesting haploinsufficiency for SOX10 as the cause of enlargement. However, the basal lumen area was increased 3.2 times (P < 0.01) for B2-Sox9cY440X/+, 2.0 times (P < 0.01) for Sox10N/N, and 2.7 times (P < 0.01) for B2-Sox9Y440X/+;Sox10N/+ compared with WT (Fig. 4 E–G). The Sox10N/+ allele in combination with Sox9cY440X/+ did not significantly exacerbate the lumen enlargement (P = 0.4), whereas the impact of Sox9cY440X/+ was significantly greater than for Sox10N/N (P = 0.01). Overall, Sox9Y440X alone resulted in the most severe hydrops, raising the possibility that it may interfere in a dominant manner with normal SOX9 and SOX10 function. Consistent with this notion is the lack of auditory and vestibular deficits in the otic conditional Sox9-null mutant (B2-Sox9c/+) (SI Appendix, Fig. S4 H–K) in contrast to B2-Sox9cY440X/+ mutants.
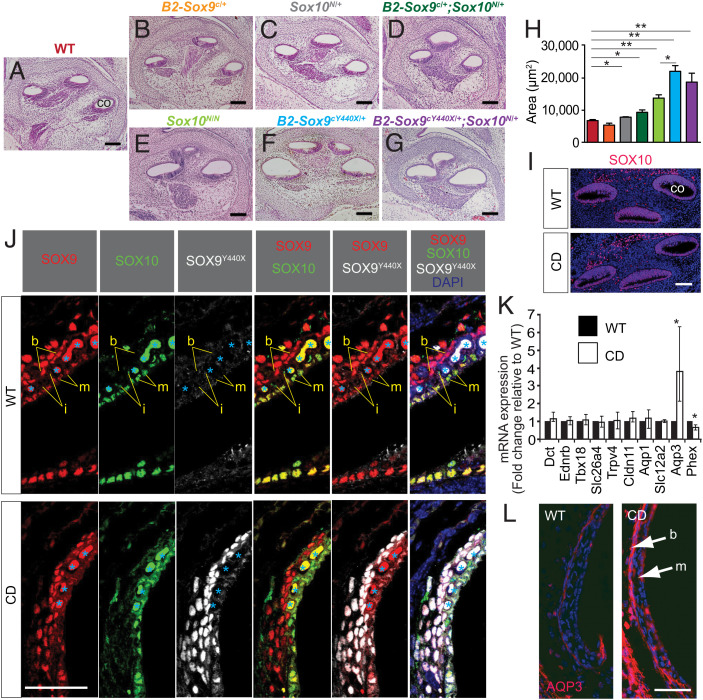
Fig. 4.
Prenatal hydrops in SoxE mutant cochleae and increased Aqp3 expression in the Sox9Y440X/+ (CD) mutant. (A–G) Histology of WT and SoxE mutant cochleae at E15.5 (n = 4 to 5). (H) Basal cochlear luminal area measured for cochleae in A–G (n = 4 to 5). (I) Immunostaining of SOX10 in WT and CD cochlea at E14.5 (n = 3). (J) Immunostaining of SOX9, SOX10, and SOX9Y440X in WT and CD stria vascularis at P5 (n = 2). Blood cells are indicated by light blue asterisks. DAPI, 4′,6-diamidino-2-phenylindole. (K) Relative messenger RNA (mRNA) expression (normalized to Hprt) of candidate SOX9 targets in P3 cochleae by qRT-PCR (n = 3). (L) Increased expression of AQP3 in the marginal and basal layers of stria vascularis of CD mutant cochlea at P3 shown by immunostaining (n = 2). Data are presented as means ± SEM. b, basal cell; co, cochlea; i, intermediate cell; m, marginal cell. (Scale bars: 100 µm in A–G; 40 µm in I, J, and L.) *P < 0.05; **P < 0.01.
AQP3 Is Derepressed in the Postnatal Sox9Y440X/+ Stria Vascularis.
We asked whether Sox10 was also down-regulated in the E14.5 cochlea. Unexpectedly, despite the presence of SOX9Y440X protein in the cochlea (Fig. 1C), SOX10, LECT1, and MME expressions were unaffected at E14.5 (Fig. 4I and SI Appendix, Fig. S5 A and B), even though the genes encoding these proteins were down-regulated in the ES. Since the ES/ED is continuous with the cochlea, the enlarged cochlea lumen may be a secondary consequence of dysregulated ionic/fluid balance in the ES/ED starting from E15. The reason for the divergent impact of SOX9Y440X on Sox10 in the ES and E14.5 cochlea is unclear. Expression of the mutant allele and SOX9Y440X protein in the ES/ED appeared higher than in the cochlea. The contrasting effect on the cochlea could be due to the lower SOX9Y440X protein expression in the E14.5 cochlea epithelium compared with the cartilaginous capsule, where high levels of SOX9Y440X were detected (Fig. 1C) and LECT1 expression was down-regulated (SI Appendix, Fig. S5A).
The stria vascularis is essential for generating endocochlear potential, develops later in inner ear development, and matures at postnatal stages around P3. There are three distinct cell layers of the stria vascularis: the marginal layer derived from the otic epithelium, the intermediate layer composed of melanocytes, and the basal layer derived from the otic mesenchyme. Basal cells express both SOX9 and SOX9Y440X proteins. SOX9/SOX9Y440X are coexpressed with SOX10 in the marginal cells, while the intermediate cells express only SOX10 (Fig. 4J and SI Appendix, Fig. S5F). Given the marked swelling of the scala media in adults (Fig. 1 I and J) and the expression of SOX9 and SOX10, it was possible that these factors have a later function in addition to the ES/ED, and molecular changes could occur as the stria vascularis matures that could impact endocochlear function. All three layers developed in Sox9Y440X/+ mutants as identified by corresponding marker proteins (SI Appendix, Fig. S5 C–E). We examined genes for the following: ion channels; cotransporters, and water channels (Slc12a2, Trpv4, Aqp1, Aqp3); genes expressed by melanocytes (Ednrb, Dct); factors important for fibrocyte development (Tbx18); tight junction components (Cldn11); and genes in which mutations cause hydropic ears (Slc26a4, Phex) by qRT-PCR. Among these, Phex, a reported SOX9 target (65), was down-regulated, while Aqp3 was found to be significantly up-regulated, with a four-time increase in the Sox9Y440X/+ mutant cochlea as compared with the WT (Fig. 4K). AQPs transport water molecules across the plasma membrane and are essential for the establishment and maintenance of the inner ear fluid. Their dysregulation has been implicated in the development of endolymphatic hydrops in mice (66) and humans, and they are potential drug targets for the condition (67, 68). Immunostaining confirmed stronger signals for AQP3 in both marginal and basal cell layers in the mutant compared with WT (Fig. 4L), which could contribute to unbalanced fluid transport to the ductal lumen and fluid accumulation. These data also raise the possibility that SOX9Y440X may dominantly interfere with SOX9 and SOX10 cooperation in the marginal and basal layers.
SOX9 and SOX10 Bind Cooperatively to an Aqp3 Cis-Regulatory Element.
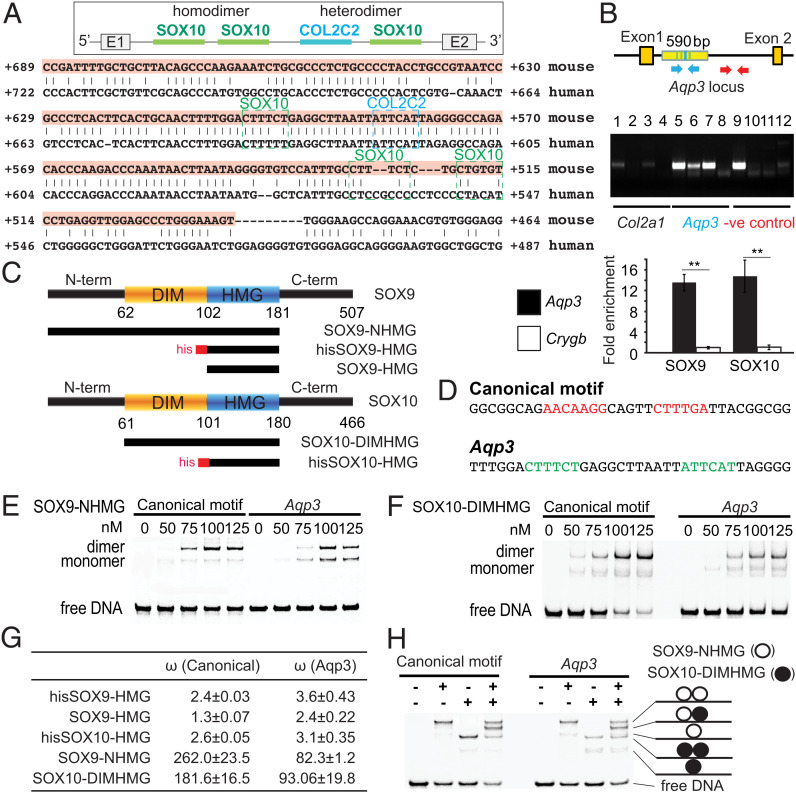
The genetic interaction between Sox9 and Sox10 mutants (Fig. 4) raised the possibility that the two factors cooperate molecularly in regulating transcription, which leads us to hypothesize that this cooperation would be relevant for those targets, such as Aqp3, that harbor regulatory elements that can bind both SOXE factors. We asked whether SOX9 and SOX10 could both regulate Aqp3 directly. We identified a functional in vivo SOX9 binding site [COL2C2 (69)] in a conserved region in intron 1 of Aqp3 (70) (SI Appendix, Fig. S6 A and B). Three SOX10 binding sites were predicted in the region using motifs of the JASPAR database (71, 72) (Fig. 5A and SI Appendix, Fig. S6 A and B) and overlap with a The Encyclopedia of DNA Elements (ENCODE) candidate cis-regulatory element. By chromatin immunoprecipitation (ChIP) qPCR assays, we found that SOX9 and SOX10 bound DNA fragments containing the SOX9/SOX10 binding sites in the Aqp3 locus (Fig. 5B) but not unrelated sequences (negative control, Crygb), suggesting that SOX9 and SOX10 directly regulate Aqp3.
Fig. 5.
Aqp3 is potentially regulated by SOXE dimers with strong positive cooperativity. (A) Conserved SOX9 and SOX10 binding sites predicted by bsConserve and Jasper over a 205-bp conserved noncoding region by Vista in intron 1 of Aqp3. A COL2C2 (ATTCAT) SOX9 binding site (69) is detected in intron 1. COL2C2 and SOX10 binding sites are boxed in blue and green, respectively. (B) ChIP assays for SOX9 and SOX10 on the WT inner ear at E14.5 (n = 2). Col2a1 (positive control) is the known SOX9 binding region. Arrows in blue indicate primer sites flanking the Aqp3 intron 1 region with predicted SOX9 and SOX10 binding sites. Arrows in red indicate primer sites flanking 1 kb downstream of the predicted SOX9 or SOX10 binding site. Real-time PCR-amplified DNA fragments were immunoprecipitated by SOX9 or SOX10 antibody, the Aqp3 intron1 region (blue arrows), or the Crygb (gamma-b-crystallin) promoter (negative control; n = 3). Lanes 1, 5, and 9: genomic DNA (gDNA); lanes 2, 6, and 10: α-SOX10 antibody (Ab); lanes 3, 7, and 11: α-SOX9 Ab; lanes 4, 8, and 12: control serum. (C) Domain composition of SOX9 and SOX10 proteins and (D) DNA sequences of oligos used in EMSAs. (E) The SOX9-NHMG and (F) SOX10-DIMHMG constructs containing the DIM domain dimerized with strong positive cooperativity (n = 4). (G) The homodimer cooperativity factors were estimated as described (98, 99). (H) SOX9 and SOX10 proteins heterodimerized effectively on the Aqp3 sequences (n = 4). -, not added. +, added. Data are presented as means ± SEM. **P < 0.01.
To test whether SOX9 and SOX10 are physically associated, we tested for positive cooperativity for interactions by electrophoretic mobility shift assays (EMSAs) with protein constructs purified to homogeneity (36) (Fig. 5C). We selected a sequence with a palindromic (reverse–forward) configuration of canonical SOX half-sites (termed the “CD-Rap” motif) (73) (Fig. 5D). We also tested probes encoding the Aqp3 SOX9–SOX10 motif harboring a COL2C2-like SOX9 site juxtaposed to a degenerate SOX10 binding site, which resembles a hitherto unknown “forward–forward” configuration with an 11–base pair spacer. High mobility group (HMG) box constructs devoid of the dimerization domain migrated predominantly as monomers on both the canonical CD-Rap and the Aqp3 element (SI Appendix, Fig. S6 C and D). By contrast, the SOX9-NHMG (N terminus to HMG) and SOX10-DIM (dimerization) constructs, including the dimerization domains, migrated predominantly as dimers, indicating highly cooperative homodimerization that depends on the presence of the DIM domain (Fig. 5 E–G).
We next asked whether SOX9 and SOX10 could form heterodimers on the noncanonical Aqp3 element. We incubated canonical or Aqp3 EMSA probes simultaneously with SOX9 and SOX10 protein constructs, both of which contain the DIM domain but that are of different lengths, such that the various protein/DNA complexes are discernible on EMSA gels (Fig. 5 C and H). On both motifs, we observed prominent SOX9/SOX10 complexes as well as homodimeric SOX9/SOX9 or SOX10/SOX10 complexes (Fig. 5H). These biophysical/biochemical interaction assays suggest that SOX9 and SOX10 can form highly cooperative homodimers as well as heterodimers on the Aqp3 enhancer DNA. This suggests that both proteins work together to suppress Aqp3 expression. Whether possible bindings of SOX9 homodimers, SOX10 homodimers, and SOX9–SOX10 heterodimers are interchangeable is an important question to address in the future.
SOX9Y440X Compromises Cooperative Repression of Aqp3 by SOX9 and SOX10.
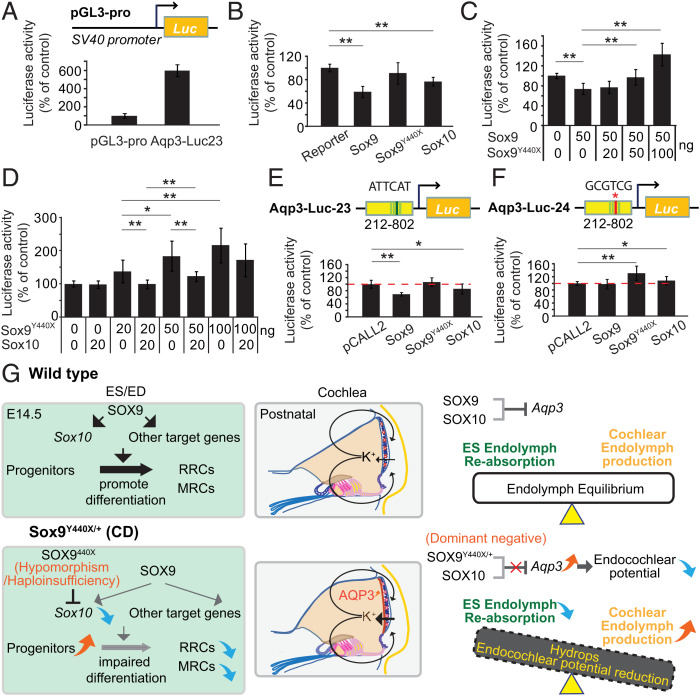
The inappropriate expression of AQP3 in the stria vascularis led us to hypothesize that SOX9 and SOX10 can repress Aqp3 expression in part via the first intron element. We, therefore, investigated the effect of SOX9, SOX9Y440X, and SOX10 on regulation of the Aqp3 gene. We constructed a 590-base pair (bp) reporter vector from intron 1 of Aqp3 (Aqp3-Luc-23) containing a 205-bp sequence harboring the heterodimer site with an 11-bp spacing and one SOX10 homodimer site with 3-bp spacing. Aqp3-Luc-23 was transfected into mouse inner medullary collecting duct (mIMCD) cells, which express high levels of Aqp3 but little Sox9 and Sox10 and are, therefore, suitable for testing the inhibitory effect of SOXE factors. Higher luciferase expression was observed in cells transfected with the reporter construct compared with cells transfected with the empty luciferase plasmid (Fig. 6A). Hence, the fragment acted like an enhancer for Aqp3 in cells expressing the gene. However, when Aqp3-Luc-23 was transfected together with an SOX9 or SOX10 expression construct, luciferase reporter activity was inhibited, with SOX9 exerting a stronger negative effect. Transfection of SOX9Y440X with Aqp3-Luc-23 had no effect (Fig. 6B). Cotransfection of expression constructs for SOX9Y440X and SOX9 reversed the inhibition of Aqp3 expression and even increased it above control levels when the SOX9Y440X concentration was twice the amount of SOX9 (Fig. 6C). This result suggests that the competition of SOX9 and SOX9Y440X proteins in binding to the Aqp3 regulatory element allows some transactivation activity. The transactivation effect of SOX9Y440X on reporter expression was inhibited by SOX10 dose dependently (Fig. 6D). Collectively, our results suggest that SOX9Y440X and WT SOX9/10 factors compete with one another to regulate Aqp3.
Fig. 6.
SOXE regulation of Aqp3 expression and development of the endolymphatic system. (A–F) Impact of SOXE and SOX9Y440X on Aqp3 regulatory element–driven luciferase reporter transactivation. (A) Reporter activity of the expression vector comprising the fragment of intron 1 of Aqp3 (+212 to +802) with SOX9 and SOX10 binding sites, driving expression of a luciferase reporter pGL3-pro (Aqp3-Luc-23), in mIMCD3 cells (n = 3). (B) Impact of SOX9, SOX9Y440X, and SOX10 on Aqp3-Luc-23 activity. Luciferase activity was normalized to activity with the reporter only. SOX9 but not SOX9Y440X dampened reporter activity by 40% (n = 3). (C) The effect of increasing amounts of Sox9Y440X expression vector (0 to 100 ng) on Sox9 expression vector (50-ng) inhibition of Aqp3-Luc23 activity. SOX9 inhibition of reporter was reversed by increasing concentrations of transfected Sox9Y440X (n = 3). (D) Effect of increasing amounts of Sox9Y440X expression vector (0 to 100 ng) on Sox10 expression vector (20-ng) inhibition of Aqp3-luc23. SOX9Y440X overcame SOX10 inhibition of Aqp3-luc23 (n = 3). (E and F) SOX9 and SOX10 inhibition of Aqp3-luc23 activity was abolished when the SOX9 binding site COL2C2 was mutated (from ATTCAT to GCGTCG [red asterisk]; Aqp3-luc24; n = 3). (G) Model of SOXE control of endolymphatic development and the impact of SOX9Y440X mutation on inner ear fluid homeostasis via two independent mechanisms in the cochlea and ES. Data are presented as means ± SEM. *P < 0.05; **P < 0.01.
Neither SOX9 nor SOX10 were able to suppress the reporter activity in mIMCD cells when the SOX9 in vivo binding consensus (COL2C2-like sequence site) (69) or the SOX10 binding sites were mutated (Fig. 6 E and F and SI Appendix, Fig. S6 E and F). Interestingly, when the COL2C2 site was mutated, expression of SOX9Y440X resulted in transactivation of the reporter. Similarly, transactivation of the reporter was increased when the SOX10 site was mutated, consistent with the requirement for both SOXEs for inhibition. These results may reflect partial recovery of the enhancer activity due to the reduced binding of SOX9 and altered interactions with partner factors via the truncated transactivation domain. Overall, the data suggest that the specific binding sequences are critical for the inhibition of Aqp3 expression by SOX9 and SOX10, which is mediated in part by the activity of heterodimers in concert with appropriate partner factors (Fig. 6G).
Discussion
Defining in vivo cellular and molecular mechanisms underlying dominant syndromic disorders is complicated by unclear genotype–phenotype correlations and compensatory or dominant negative effects in multiple affected tissues. This is particularly relevant where heterozygosity involves a mutation that could lead to both hypomorphic and dominant effects in a developmentally important transcription factor, such as SOX9. While SOX9 and SOX10 are coexpressed in the inner ear and other cell types, such as the neural crest, SOX9, but not SOX10, is expressed in chondrocytes. Heterozygosity for SOX9 causes endochondral bone defects (29), which are not seen in the Sox10Dom mutants (74, 75), consistent with independent roles for each SOXE factor. In mouse, Xenopus, and zebrafish, complete loss of either Sox9 or Sox10 impacts otic development (18, 20, 76). SOX9 and SOX10 have both transactivator and repressor activity depending on the target gene and cell type (1, 77, 78), although such activity has not been demonstrated in the inner ear. Another level of complexity is that SOXE works in various dimer configurations and partners with other factors to regulate gene expression (37, 38). To gain insights into mechanisms, we have addressed the impact of heterozygosity for SOXE mutations, singly and in combination, when either or both of them are mutated by studying three types of mutations: heterozygous null mutations in both Sox9 and Sox10 and the Y440X truncating mutation in Sox9, which gives a CD phenotype different from that of the null.
A key finding is the lack of inner ear phenotype in heterozygous conditional Sox9+/−-null mutants. This suggests that haploinsufficiency for SOX9 is not the underlying cause of the inner ear defects and implicates instead a dominant negative or neomorphic mode of action for SOX9Y440X. The impact of Sox9Y440X on enlargement of the cochlear lumen was more marked than for Sox10N/+ (which must be haploinsufficient), consistent with a dominant negative impact of SOX9Y440X on Sox10 itself, on SOX10 target genes, and also, on other targets regulated by SOX9 independent of SOX10. The differing impact of the SOX9Y440X mutation in two critical structures of the inner ear, the ES and the stria vascularis, could also implicate hypomorphic effects in the former and dominant negative effects in the latter, which combine to affect the development of the endolymphatic system and the later maintenance of fluid homeostasis/endocochlear potential and thereby, hearing.
We have provided in vivo genetic evidence for the essential roles of cooperation of SOX9 and SOX10 in controlling the development of the endolymphatic system and the ionic composition of the endolymph. We found that these SOXE factors function early in the developing ES/ED. SOX9 has a critical role in maintaining Sox10 expression and the timely differentiation of progenitors as well as controlling the expression of genes important for ionic homeostasis. The major impact of heterozygosity or homozygosity for Sox10 deficiency in the mouse inner ear is on the survival of progenitors in the cochlea, but no gross defects were observed in the ES of these mutants (20). Notably, different hypomorphic mutations in Sox10 that impair the transactivation domain or DNA-dependent dimerization impact distinctive aspects of satellite glia and Schwann cells development (79). Our study shows that, compared with Sox9Y440X, a similar, albeit more mild differentiation delay was seen in the heterozygous Sox10-null ES/ED, suggesting that hypomorphism/haploinsufficiency for SOX10 contributes to the developmental defect but is not solely responsible for those seen in the Sox9Y440X/+ inner ear. The different impact of SOX9Y440X vs. deficiency for SOX10 (in Sox10N/+ mutants) on the expression of Lect1 (a known direct target of SOX9 in chondrocytes) in the ES/ED (64) highlights independent targets for each SOXE factor. The lack of impact of SOX9Y440X on Lect1 expression in the cochlea correlated with lower levels of protein expression as compared with the ES/ED and chondrocytes, suggesting that the impact may depend on the level of expression of the mutant protein. The reduction of Sox10 expression may also be exacerbated by decreased levels of SOX10 protein since its transcription is autoregulated, at least in some cell types (80).
The differential impact on Sox10 expression level in the Sox9Y440X ES/ED vs. the cochlea may be related to developmental stage/cell-type differences in Sox10 enhancer utilization (25). The level and cell type–restricted expression of Sox9 are regulated by many enhancers distributed over 2 Mb of the genome (81). These enhancers control the exquisite sensitivity of the phenotype to levels of Sox9 expression. The divergent levels of expression of Sox9Y440X seen in the ES/ED, cochlear, and cartilage may be controlled by separable enhancers. The different phenotypic impact may be attributable to differing sensitivity to perturbation in SOX9 dosage and activity similar to the neural crest (81).
Our molecular data and analyses on the postnatal stria vascularis indicate that SOX9 and SOX10 cooperate and may function as heterodimers as well as homodimers in vivo. Therefore, multiple modes of regulation by SOX9 and SOX10 may operate to maintain appropriate gene expression in the inner ear. Both transactivation and repression activity for SOXE factors have been reported in cells and in vivo models (77, 78, 82–86), although such activity has not been demonstrated in the inner ear. Here, we provide evidence that for a target gene that harbors binding regions for both SOX factors, SOX9 and SOX10 cooperate to repress Aqp3 expression in postnatal stria vascularis; this is another example of context-dependent transactivation and repression activity of SOXE (1, 77, 78). Canonical SOXE dimers predominantly activate genes (57). Yet, in the context of the noncanonical Aqp3 element, we observe transcriptional repression. Such regulation in vivo may involve possible interchangeable binding of SOX9 homodimers, SOX10 homodimers, and SOX9–SOX10 heterodimers in repressing Aqp3 expression. Dimerization of SOXE is mediated by reciprocal interactions between DIM and HMG box domains (36). Because the truncated C terminus is far away from the dimerization domain, it is unlikely that Y440X changes whether SOX9Y440X directly interferes with SOX9:SOX10 heterodimerization or SOX9 homodimerization in vivo. Rather, SOX9Y440X likely impacts interactions with appropriate partner factors.
Where Sox9 is expressed in the stria vascularis basal layer but not Sox10 (87), SOX9 may act alone in these cells to inhibit Aqp3 expression, but this inhibition is impaired in Sox9Y440X/+ either by interference with cofactor/partner interaction or by haploinsufficiency/hypomorphism. In WT, Sox9 and Sox10 are both expressed in the marginal cells (87), and Aqp3 expression is completely absent; however, its expression is derepressed when SOX9Y440X is present. Here, SOX9Y440X may also interfere with the cooperation of SOX9 and SOX10 in directly repressing transcription of Aqp3. Whether such cooperation operates in vivo via combinations of heterodimers and homodimers, and the degree with which these are additive and/or redundant is unclear. Whether a target gene is regulated by SOXE dimers and monomers in vivo is also probably dependent on the appropriate binding regions for these factors in the target gene. Questions for the future are to what extent the SOX9Y440X mutation affects the common SOXE bipartite transactivation mechanism (57), in which a transactivation domain in the middle of the protein synergizes with a C-terminal region, thereby contributing to hypomorphism/haploinsufficiency for SOX9 and/or dominant negative effects.
A similar mechanism may underlie the endolymphatic hydrops in pig SOX10 R109W mutants with phenotypic similarities to human Mondini dysplasia (88). It is notable that a dominant negative mechanism has been proposed for the mouse Sox10Dom mutation that truncates SOX10 but spares the DNA binding domain (89), raising the question of to what extent the SOX10Dom mutation interferes with SOX9–SOX10 cooperation and impacts inner ear development.
Our findings have implications for understanding disorders affecting the ionic composition, which are the most common causes of deafness and are often associated with an enlarged vestibular aqueduct (EVA; OMIM 600791). To date, dysfunction in fluid homeostasis leading to EVA (or Pendred syndrome) has been mostly attributed to mutations in FOXI1 (OMIM 601093) and its target gene SLC26A4. Our work extends the range of candidate causative mutations for common causes of dysregulated fluid homeostasis in childhood congenital deafness and also identifies disease genes regulated by the SOXE factors, which could be a guide for the diagnosis of deafness and vestibular problems (Dataset S1). Mutations of GJB2 that encodes connexin 26, a component of the gap junction, result in deafness due to fluid imbalance (90). Our study identifies SOXE as a regulator of Gjb2, which is down-regulated in Sox9Y440X/+ RRCs. We also identify potential candidate EVA genes downstream of SOX9, such as genes encoding ion transporters (e.g., Slc24a4 and Slc15a1), channels (Ttyh1), and components of the renin-angiotensin system, involved in ion transport and water regulation represented by Agt and Mme. The Sox9Y440X model demonstrates that having appropriate numbers of mature cells, including the Slc26a4+ ion transport cells, is key to ionic homeostasis and a functioning endolymphatic system.
In summary, our study reveals that depending on the target gene context, SOX9 and SOX10 act both independently and co-operatively to ensure proper development of the endolymphatic system and ionic homeostasis (Fig. 6G). These functions are important both early and later in the development of the endolymphatic system and also suggest a cause of the sensorineural deafness pathology in CD. Since SoxE genes have many roles in embryogenesis and in adult tissue-specific stem cells, the principles we have uncovered are relevant to the effects of heterozygous dominant mutations on other cell types where SOX9 and SOX10 are coexpressed. The lessons learned from the pathogenic phenotypes that arise from disrupting transcription factors important for development are also relevant to understanding the impact of mutations in developmental transcription factors on different cells and/or tissues.
Materials and Methods
Animals.
Animal care and experiments were in accordance with the protocols approved by the Committee on the Use of Live Animals in Teaching and Research of the University of Hong Kong. Sox9Y440X/+ mutants were generated from a variety of crosses: floxed mutant (Sox9floxedY440X) with the β-actin Cre driver (91) (provided by G. Martin, University of California, San Francisco, CA) to generate constitutive mutants. While most of these mutants died at birth, about 10% of the constitutive mutants survived. Surviving mutants were crossed to WT (C57/BL6) animals to obtain Sox9Y440X/+ heterozygotes for analysis at postnatal stages. In addition, we also generated constitutive β-actin Cre;Sox9Y440X/+ heterozygote fetuses for molecular marker analyses independently of the surviving line. Otic-specific mutant was generated by crossing the transgenic B2-r4-Cre mouse line (39) to Sox9fY440X/+ mice to generate B2-Sox9cY440X/+ and control littermates. Sox9c/+ (92) mice were a gift of Andreas Schedl, Institut de Biologie Valrose, Nice, France, and Richard Behringer, MD Anderson Cancer Center, Houston, TX. SoxE compound mutants were generated by intercrossing, Sox9cY440X/+, Sox9c/+, and Sox10N/+ (93) mice. In all cases, we compared WT and mutant littermates. All mice analyzed were of mixed genetic background. Sox9Y440X/+, Sox9c/+, and Sox10N/+ mice were of mixed genetic background 129Sv/Ev; C57/BL6. The genetic background of beta-actin Cre and B2-r4-Cre mouse lines was Institute of Cancer Research and C57/BL6;CBA respectively (39).
SOX9Y440X Antibody.
The rabbit polyclonal SOX9Y440X antibody was generated against the 14-aa peptide sequence before the Y440X mutation from 426 to 439, YSPSYPPITRSQYD, which was predicted to generate a neoepitope, synthesized and raised by Covalab UK Ltd. (https://www.covalab.com). Specificity of the antibody was confirmed by immunostaining on WT and Sox9Y440X/+ mutant tissues (Fig. 1 B, C, and J).
A detailed description of other methods can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Gigi Wong, Becky C. M. Cheng, and Nelson W. F. Dung for technical support; Simon Chan for performing hearing and endocochlear potential tests; J.-N. Lu and Nelson W. F. Dung for assistance with single cell RNA sequencing; Calista Ng for EMSA reagents; and Reinhard Faessler, Patrick Tam, Doris Wu, Kevin Yip, Pak Sham, and Michael Zhang for helpful discussion. R.J. was supported by National Natural Science Foundation of China Grant 31771454. B.F. is supported by NIH Grant R01 AG060504. This work was supported by Research Grants Council, Hong Kong Grants HKU7222/97M (to K.S.E.C.), HKU2/02C (to K.S.E.C.), HKU4/05C (to K.S.E.C.), AoE/M-04/04 (to K.S.E.C.), and T12-708/12N (to K.S.E.C.) and the Jimmy and Emily Tang Professorship (K.S.E.C.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. S.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120393119/-/DCSupplemental.
Data, Materials, and Software Availability
Sequencing data have been deposited on NCBI GEO (accession nos. GSE131196 and GSE139587) (94, 95), with the processed data interactively hosted at https://www.sbms.hku.hk/kclab/sox9-ear/ (96). Some of the bioinformatics scripts were deposited in GitHub at https://github.com/hkukclab/sox9-ear (97). All other data are included in the article and/or supporting information.
References
- 1.Lefebvre V., Roles and regulation of SOX transcription factors in skeletogenesis. Curr. Top. Dev. Biol. 133, 171–193 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weider M., Wegner M., SoxE factors: Transcriptional regulators of neural differentiation and nervous system development. Semin. Cell Dev. Biol. 63, 35–42 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Kellerer S., et al. , Replacement of the Sox10 transcription factor by Sox8 reveals incomplete functional equivalence. Development 133, 2875–2886 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Polanco J. C., Wilhelm D., Davidson T. L., Knight D., Koopman P., Sox10 gain-of-function causes XX sex reversal in mice: Implications for human 22q-linked disorders of sex development. Hum. Mol. Genet. 19, 506–516 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Finzsch M., Stolt C. C., Lommes P., Wegner M., Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor alpha expression. Development 135, 637–646 (2008). [DOI] [PubMed] [Google Scholar]
- 6.She Z. Y., Yang W. X., Sry and SoxE genes: How they participate in mammalian sex determination and gonadal development? Semin. Cell Dev. Biol. 63, 13–22 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Stolt C. C., Lommes P., Friedrich R. P., Wegner M., Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development 131, 2349–2358 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Suzuki E., et al. , Loss-of-function SOX10 mutation in a patient with Kallmann syndrome, hearing loss, and iris hypopigmentation. Horm. Res. Paediatr. 84, 212–216 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Pingault V., et al. , Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 31, 391–406 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Song J., et al. , Hearing loss in Waardenburg syndrome: A systematic review. Clin. Genet. 89, 416–425 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Unger S., Scherer G., Superti-Furga A., “Campomelic dysplasia” in GeneReviews, Adam M. P., et al., Eds. (University of Washington, Seattle, WA, : 1993), pp. 1–16. [PubMed] [Google Scholar]
- 12.Foster J. W., et al. , Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372, 525–530 (1994). [DOI] [PubMed] [Google Scholar]
- 13.Wagner T., et al. , Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79, 1111–1120 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Fekete D. M., Wu D. K., Revisiting cell fate specification in the inner ear. Curr. Opin. Neurobiol. 12, 35–42 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Raft S., et al. , Ephrin-B2 governs morphogenesis of endolymphatic sac and duct epithelia in the mouse inner ear. Dev. Biol. 390, 51–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lundquist P. G., Aspects on endolymphatic sac morphology and function. Arch. Otorhinolaryngol. 212, 231–240 (1976). [DOI] [PubMed] [Google Scholar]
- 17.Salt A. N., Regulation of endolymphatic fluid volume. Ann. N. Y. Acad. Sci. 942, 306–312 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Barrionuevo F., et al. , Sox9 is required for invagination of the otic placode in mice. Dev. Biol. 317, 213–224 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Mak A. C., Szeto I. Y., Fritzsch B., Cheah K. S., Differential and overlapping expression pattern of SOX2 and SOX9 in inner ear development. Gene Expr. Patterns 9, 444–453 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breuskin I., et al. , Sox10 promotes the survival of cochlear progenitors during the establishment of the organ of Corti. Dev. Biol. 335, 327–339 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Saint-Germain N., Lee Y. H., Zhang Y., Sargent T. D., Saint-Jeannet J. P., Specification of the otic placode depends on Sox9 function in Xenopus. Development 131, 1755–1763 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Park B. Y., Saint-Jeannet J. P., Long-term consequences of Sox9 depletion on inner ear development. Dev. Dyn. 239, 1102–1112 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trowe M. O., et al. , Loss of Sox9 in the periotic mesenchyme affects mesenchymal expansion and differentiation, and epithelial morphogenesis during cochlea development in the mouse. Dev. Biol. 342, 51–62 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Betancur P., Bronner-Fraser M., Sauka-Spengler T., Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc. Natl. Acad. Sci. U.S.A. 107, 3570–3575 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betancur P., Sauka-Spengler T., Bronner M., A Sox10 enhancer element common to the otic placode and neural crest is activated by tissue-specific paralogs. Development 138, 3689–3698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolla L., et al. , Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat. Commun. 11, 2389 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheffer D. I., Shen J., Corey D. P., Chen Z. Y., Gene expression by mouse inner ear hair cells during development. J. Neurosci. 35, 6366–6380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S., Lee M. P., Jones S., Liu J., Waldhaus J., Mapping the regulatory landscape of auditory hair cells from single-cell multi-omics data. Genome Res. 31, 1885–1899 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bi W., et al. , Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc. Natl. Acad. Sci. U.S.A. 98, 6698–6703 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherer G., Zabel B., Nishimura G., Clinical utility gene card for: Campomelic dysplasia. Eur. J. Hum. Genet., 10.1038/ejhg.2012.228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unger S., Scherer G., Superti-Furga A., “Campomelic dysplasia” in GeneReviews, Pagon R. A., Adam M. P., Ardinger H. H., Eds. (University of Washington, Seattle, WA: 2008), pp. 1–16. [PubMed] [Google Scholar]
- 32.Hageman R. M., Cameron F. J., Sinclair A. H., Mutation analysis of the SOX9 gene in a patient with campomelic dysplasia. Hum. Mutat. 1 (suppl. 1), S112–S113 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Meyer J., et al. , Mutational analysis of the SOX9 gene in campomelic dysplasia and autosomal sex reversal: Lack of genotype/phenotype correlations. Hum. Mol. Genet. 6, 91–98 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Pop R., Zaragoza M. V., Gaudette M., Dohrmann U., Scherer G., A homozygous nonsense mutation in SOX9 in the dominant disorder campomelic dysplasia: A case of mitotic gene conversion. Hum. Genet. 117, 43–53 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Csukasi F., et al. , Dominant-negative SOX9 mutations in campomelic dysplasia. Hum. Mutat. 40, 2344–2352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y. H., Jankowski A., Cheah K. S., Prabhakar S., Jauch R., SOXE transcription factors form selective dimers on non-compact DNA motifs through multifaceted interactions between dimerization and high-mobility group domains. Sci. Rep. 5, 10398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamachi Y., Kondoh H., Sox proteins: Regulators of cell fate specification and differentiation. Development 140, 4129–4144 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Sugahara S., Fujimoto T., Kondoh H., Uchikawa M., Nasal and otic placode specific regulation of Sox2 involves both activation by Sox-Sall4 synergism and multiple repression mechanisms. Dev. Biol. 433, 61–74 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Szeto I. Y., Leung K. K., Sham M. H., Cheah K. S., Utility of HoxB2 enhancer-mediated Cre activity for functional studies in the developing inner ear. Genesis 47, 361–365 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Kopecky B. J., Jahan I., Fritzsch B., Correct timing of proliferation and differentiation is necessary for normal inner ear development and auditory hair cell viability. Dev. Dyn. 242, 132–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chizhikov V. V., Iskusnykh I. Y., Fattakhov N., Fritzsch B., Lmx1a and Lmx1b are redundantly required for the development of multiple components of the mammalian auditory system. Neuroscience 452, 247–264 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koo S. K., et al. , Lmx1a maintains proper neurogenic, sensory, and non-sensory domains in the mammalian inner ear. Dev. Biol. 333, 14–25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nichols D. H., et al. , Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell Tissue Res. 334, 339–358 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schrauwen I., et al. , A variant in LMX1A causes autosomal recessive severe-to-profound hearing impairment. Hum. Genet. 137, 471–478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wesdorp M., et al. ; DOOFNL Consortium, Heterozygous missense variants of LMX1A lead to nonsyndromic hearing impairment and vestibular dysfunction. Hum. Genet. 137, 389–400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim H. M., Wangemann P., Failure of fluid absorption in the endolymphatic sac initiates cochlear enlargement that leads to deafness in mice lacking pendrin expression. PLoS One 5, e14041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albert S., et al. , SLC26A4 gene is frequently involved in nonsyndromic hearing impairment with enlarged vestibular aqueduct in Caucasian populations. Eur. J. Hum. Genet. 14, 773–779 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Khan M. R., Bashir R., Naz S., SLC26A4 mutations in patients with moderate to severe hearing loss. Biochem. Genet. 51, 514–523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acampora D., et al. , Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development 126, 3795–3809 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Hulander M., et al. , Lack of pendrin expression leads to deafness and expansion of the endolymphatic compartment in inner ears of Foxi1 null mutant mice. Development 130, 2013–2025 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Griffith A. J., Wangemann P., Hearing loss associated with enlargement of the vestibular aqueduct: Mechanistic insights from clinical phenotypes, genotypes, and mouse models. Hear. Res. 281, 11–17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobak D., Berens P., The art of using t-SNE for single-cell transcriptomics. Nat. Commun. 10, 5416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Honda K., et al. , Molecular architecture underlying fluid absorption by the developing inner ear. eLife 6, e26851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peters T. A., Tonnaer E. L., Kuijpers W., Curfs J. H., Changes in ultrastructural characteristics of endolymphatic sac ribosome-rich cells of the rat during development. Hear. Res. 176, 94–104 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Gnedeva K., Hudspeth A. J., SoxC transcription factors are essential for the development of the inner ear. Proc. Natl. Acad. Sci. U.S.A. 112, 14066–14071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mao Y., Reiprich S., Wegner M., Fritzsch B., Targeted deletion of Sox10 by Wnt1-cre defects neuronal migration and projection in the mouse inner ear. PLoS One 9, e94580 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haseeb A., Lefebvre V., The SOXE transcription factors-SOX8, SOX9 and SOX10-share a bi-partite transactivation mechanism. Nucleic Acids Res. 47, 6917–6931 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hou L., Srivastava Y., Jauch R., Molecular basis for the genome engagement by Sox proteins. Semin. Cell Dev. Biol. 63, 2–12 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Akiyama H., et al. , Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 18, 1072–1087 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bastide P., et al. , Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J. Cell Biol. 178, 635–648 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhattaram P., et al. , SOXC proteins amplify canonical WNT signaling to secure nonchondrocytic fates in skeletogenesis. J. Cell Biol. 207, 657–671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Symon A., Harley V., SOX9: A genomic view of tissue specific expression and action. Int. J. Biochem. Cell Biol. 87, 18–22 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Liu C. F., Angelozzi M., Haseeb A., Lefebvre V., SOX9 is dispensable for the initiation of epigenetic remodeling and the activation of marker genes at the onset of chondrogenesis. Development 145, dev164459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ohba S., He X., Hojo H., McMahon A. P., Distinct transcriptional programs underlie Sox9 regulation of the mammalian chondrocyte. Cell Rep. 12, 229–243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu S., Guo R., Quarles L. D., Cloning and characterization of the proximal murine Phex promoter. Endocrinology 142, 3987–3995 (2001). [DOI] [PubMed] [Google Scholar]
- 66.Cowan C. A., Yokoyama N., Bianchi L. M., Henkemeyer M., Fritzsch B., EphB2 guides axons at the midline and is necessary for normal vestibular function. Neuron 26, 417–430 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Eckhard A., et al. , Co-localisation of K(ir)4.1 and AQP4 in rat and human cochleae reveals a gap in water channel expression at the transduction sites of endocochlear K(+) recycling routes. Cell Tissue Res. 350, 27–43 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Takeda T., Taguchi D., Aquaporins as potential drug targets for Meniere’s disease and its related diseases. Handb. Exp. Pharmacol. 190, 171–184 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Bell D. M., et al. , SOX9 directly regulates the type-II collagen gene. Nat. Genet. 16, 174–178 (1997). [DOI] [PubMed] [Google Scholar]
- 70.Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I., VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32, W273-9 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mathelier A., et al. , JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 42, D142–D147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sandelin A., Alkema W., Engström P., Wasserman W. W., Lenhard B., JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 32, D91–D94 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sock E., et al. , Loss of DNA-dependent dimerization of the transcription factor SOX9 as a cause for campomelic dysplasia. Hum. Mol. Genet. 12, 1439–1447 (2003). [DOI] [PubMed] [Google Scholar]
- 74.Herbarth B., et al. , Mutation of the Sry-related Sox10 gene in Dominant megacolon, a mouse model for human Hirschsprung disease. Proc. Natl. Acad. Sci. U.S.A. 95, 5161–5165 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Southard-Smith E. M., Kos L., Pavan W. J., Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat. Genet. 18, 60–64 (1998). [DOI] [PubMed] [Google Scholar]
- 76.Dutton K., et al. , A zebrafish model for Waardenburg syndrome type IV reveals diverse roles for Sox10 in the otic vesicle. Dis. Model. Mech. 2, 68–83 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cruz-Solis I., et al. , Glutamate-dependent transcriptional control in Bergmann glia: Sox10 as a repressor. J. Neurochem. 109, 899–910 (2009). [DOI] [PubMed] [Google Scholar]
- 78.Leung V. Y., et al. , SOX9 governs differentiation stage-specific gene expression in growth plate chondrocytes via direct concomitant transactivation and repression. PLoS Genet. 7, e1002356 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schreiner S., et al. , Hypomorphic Sox10 alleles reveal novel protein functions and unravel developmental differences in glial lineages. Development 134, 3271–3281 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Wahlbuhl M., Reiprich S., Vogl M. R., Bösl M. R., Wegner M., Transcription factor Sox10 orchestrates activity of a neural crest-specific enhancer in the vicinity of its gene. Nucleic Acids Res. 40, 88–101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Long H. K., et al. , Loss of extreme long-range enhancers in human neural crest drives a craniofacial disorder. Cell Stem Cell 27, 765–783.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee P. C., et al. , SUMOylated SoxE factors recruit Grg4 and function as transcriptional repressors in the neural crest. J. Cell Biol. 198, 799–813 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schock E. N., LaBonne C., Sorting Sox: Diverse roles for Sox transcription factors during neural crest and craniofacial development. Front. Physiol. 11, 606889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taylor K. M., Labonne C., SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev. Cell 9, 593–603 (2005). [DOI] [PubMed] [Google Scholar]
- 85.Hernández-Hernández J. M., et al. , Sox9 represses alpha-sarcoglycan gene expression in early myogenic differentiation. J. Mol. Biol. 394, 1–14 (2009). [DOI] [PubMed] [Google Scholar]
- 86.Pozniak C. D., et al. , Sox10 directs neural stem cells toward the oligodendrocyte lineage by decreasing Suppressor of Fused expression. Proc. Natl. Acad. Sci. U.S.A. 107, 21795–21800 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Korrapati S., et al. , Single cell and single nucleus RNA-Seq reveal cellular heterogeneity and homeostatic regulatory networks in adult mouse stria vascularis. Front. Mol. Neurosci. 12, 316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hai T., et al. , Pilot study of large-scale production of mutant pigs by ENU mutagenesis. eLife 6, e26248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cantrell V. A., et al. , Interactions between Sox10 and EdnrB modulate penetrance and severity of aganglionosis in the Sox10Dom mouse model of Hirschsprung disease. Hum. Mol. Genet. 13, 2289–2301 (2004). [DOI] [PubMed] [Google Scholar]
- 90.Hochman J. B., et al. , Prevalence of Connexin 26 (GJB2) and Pendred (SLC26A4) mutations in a population of adult cochlear implant candidates. Otol. Neurotol. 31, 919–922 (2010). [DOI] [PubMed] [Google Scholar]
- 91.Lewandoski M., Meyers E. N., Martin G. R., Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 62, 159–168 (1997). [PubMed] [Google Scholar]
- 92.Akiyama H., Chaboissier M. C., Martin J. F., Schedl A., de Crombrugghe B., The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16, 2813–2828 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bondurand N., Sham M. H., The role of SOX10 during enteric nervous system development. Dev. Biol. 382, 330–343 (2013). [DOI] [PubMed] [Google Scholar]
- 94.Szeto I. Y. Y., et al. , SOX9 and SOX10 control fluid homeostasis in the inner ear for hearing through independent and cooperative mechanisms (scRNA-seq: batch2). NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139587. Deposited 10 October 2022. [DOI] [PMC free article] [PubMed]
- 95.Szeto I. Y. Y., et al. , SOX9 and SOX10 control fluid homeostasis in the inner ear for hearing through independent and cooperative mechanisms. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131196. Deposited 10 October 2022. [DOI] [PMC free article] [PubMed]
- 96.Szeto I. Y. Y., et al. , SOX9 and SOX10 control fluid homeostasis in the inner ear for hearing through independent and cooperative mechanisms. KC Lab Data Portal. https://www.sbms.hku.hk/kclab/sox9-ear/. Deposited 14 September 2022. [DOI] [PMC free article] [PubMed]
- 97.Szeto I. Y. Y., et al. , Source codes for SOX9 and SOX10 control fluid homeostasis in the inner ear for hearing through independent and cooperative mechanisms. GitHub. https://github.com/hkukclab/sox9-ear. Deposited 11 October 2022. [DOI] [PMC free article] [PubMed]
- 98.BabuRajendran N., et al. , Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-beta effectors. Nucleic Acids Res. 38, 3477–3488 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jerabek S., et al. , Changing POU dimerization preferences converts Oct6 into a pluripotency inducer. EMBO Rep. 18, 319–333 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited on NCBI GEO (accession nos. GSE131196 and GSE139587) (94, 95), with the processed data interactively hosted at https://www.sbms.hku.hk/kclab/sox9-ear/ (96). Some of the bioinformatics scripts were deposited in GitHub at https://github.com/hkukclab/sox9-ear (97). All other data are included in the article and/or supporting information.