Significance
In this paper, we describe an additional function for KIF2A in the postnatal brain and provide evidence that KIF2A-related pathologies result from defects in neuronal connectivity and early-onset neurodegeneration rather than from impaired neurogenesis as commonly assumed. We conditionally deleted KIF2A from progenitors, nascent and mature cortical neurons and showed that this protein is key for maturation and maintenance of neurons. KIF2A deficiency altered microtubule dynamics and intracellular transport and compromised neuronal connectivity and survival. Our results shed light on the mechanisms by which KIF2A mutations cause brain diseases.
Keywords: neuronal polarity, brain wiring, neurodevelopment, neurodegeneration
Abstract
KIF2A is an atypical kinesin that has the capacity to depolymerize microtubules. Patients carrying mutations in KIF2A suffer from progressive microcephaly and mental disabilities. While the role of this protein is well documented in neuronal migration, the relationship between its dysfunction and the pathobiology of brain disorders is unclear. Here, we report that KIF2A is dispensable for embryogenic neurogenesis but critical in postnatal stages for maturation, connectivity, and maintenance of neurons. We used a conditional approach to inactivate KIF2A in cortical progenitors, nascent postmitotic neurons, and mature neurons in mice. We show that the lack of KIF2A alters microtubule dynamics and disrupts several microtubule-dependent processes, including neuronal polarity, neuritogenesis, synaptogenesis, and axonal transport. KIF2A-deficient neurons exhibit aberrant electrophysiological characteristics, neuronal connectivity, and function, leading to their loss. The role of KIF2A is not limited to development, as fully mature neurons require KIF2A for survival. Our results emphasize an additional function of KIF2A and help explain how its mutations lead to brain disorders.
Microtubules (MTs) are important components of the cytoskeleton. They play crucial roles in cell polarity, adhesion, proliferation, migration, and differentiation (1). MTs undergo cycles of growth (polymerization) and shrinkage (depolymerization) through addition or loss of tubulin-dimer subunits. This dynamic is mediated by severing and depolymerizing enzymes and regulated by MT-binding proteins such as microtubule-associated proteins (MAPs) and plus-end–tracking proteins (+TIPs), as well as by tubulin posttranslational modifications (PTMs). Defective MT dynamics are often associated with neurodevelopmental and neurodegenerative disorders (2, 3).
KIF2A belongs to the kinesin-13 subfamily endowed with an MT depolymerizing activity (4). In mitotic cells, KIF2A is localized at the kinetochore and centrosome, where it depolymerizes the MT’s minus-end and regulates mitotic spindle and cilia assembly/disassembly (5–7). Several studies have investigated the role of KIF2A in cortical neurogenesis. However, the published results are contradictory. In utero electroporation of KIF2A short hairpin RNA (shRNA) decreases the proliferation of neural progenitors and promotes differentiation of neurons (8). In contrast, electroporation of two KIF2A variants (i.e., c.961C > G, p.His321Asp and c.950G > A, p.Ser317Asn; Gene ID: 3796) causes the opposite effect, increasing proliferation of progenitors and decreasing the production of neurons (9). Conditional knock-in (KI) mice expressing one copy of the human KIF2A variant c.961C > G, p.His321Asp have no change in proliferation but display an increase in early embryonic cell death. Consequently, the mutant brain is smaller at birth (10). In human brain organoids, KIF2A, which is expressed in the mother centriole, interacts with CEP170 downstream of WDR62 to promote disassembly of the primary cilium. Disruption of WDR62-CEP170-KIF2A signaling depletes the number of progenitor cells, leading to smaller brains (11). KIF2A is enriched in postmitotic neurons where it forms homodimers that bind to MTs and move toward the plus end, wherein they remove tubulin subunits in an adenosine 5′-triphosphate–dependent manner (4, 12, 13). Functional analysis using Kif2a KO mice revealed that the protein is necessary for neuronal migration, elongation of axon collaterals, and axonal pruning (14–16). KIF2A accumulates in the growth cone where it regulates MT length (14, 17). Early postnatal deletion of KIF2A does not disrupt proliferation or migration of granular cells in the dentate gyrus, but results in mossy fiber sprouting and an epileptic hippocampus (18). KIF2A has several splice isoforms whose expression is developmentally regulated (19, 20), and its activity is regulated by phosphorylation by different kinases (21). These data emphasize the complexity of KIF2A function and suggest that it could have different roles depending on isoform, expression, activity level, and interaction with different partners.
In humans, de novo mutations in KIF2A have been linked to a wide variety of clinical manifestations depending on the affected domain of the protein. Mutations in the nucleotide-binding domain were associated with malformations of cortical development, including microcephaly, lissencephaly, and partial agenesis of the corpus callosum (CC; 22–25). Variants in the motor domain were associated with epilepsy, and variants in other domains were associated with autism spectrum disorder (25–27). Finally, overexpression and one rare variant were associated with Alzheimer’s disease (AD) (28, 29).
While the role of KIF2A in neuronal migration is well established (14, 15, 20), its roles in embryonic neurogenesis and in the postnatal brain remain unclear. We conditionally ablated the gene from cortical progenitors, nascent neurons, and fully mature neurons. We report that KIF2A is dispensable for embryonic neurogenesis but crucial for neuronal maturation, connectivity, and survival.
Results
The Lack of KIF2A Triggers Premature Loss of Neurons.
The role of KIF2A in cortical neurogenesis and its relationship with microcephaly have been controversial (8–11). To explore this, we first examined KIF2A expression in the developing and adult cerebral cortex using in situ hybridization and immunofluorescence in mice. KIF2A is expressed in postmitotic neurons throughout life and to a lesser extent in neural progenitors (SI Appendix, Fig. S1 A–I). We crossed Kif2aF/F mice with Emx1-Cre to inactivate KIF2A in cortical progenitors starting from embryonic day (E) 9.5 (15, 30) and generated cortex-specific conditional knockout mice (hereafter referred to as Emx1-cKO). In these mice, KIF2A expression persisted in sparse neurons, presumably cortical interneurons, which do not express Emx1-Cre (SI Appendix, Fig. S1 D and I). To assess the impact of KIF2A deletion on proliferation of cortical progenitors, we evaluated the number of Ki67-positive cells in the ventricular-subventricular zones, the number of 5-bromo-2′-deoxyuridine (BrdU)–positive cells, 2 h after intraperitoneal injection of BrdU in pregnant females at E14.5, and the ratio between Ki67-positive and BrdU-positive cells. We did not detect any difference between Emx1-cKO embryos and littermate controls (SI Appendix, Fig. S2 A–C). Furthermore, there was no difference in number of apical radial glia (Pax6+), progenitors undergoing mitosis (PHH3+), intermediate progenitors (Tbr2+), early postmitotic neurons (Tbr1+), or apoptotic cells (cleaved Caspase 3 [cCasp3-positive]) during embryonic stages (SI Appendix, Fig. S2 D–I). Accordingly, the number of neurons and brain size were normal at birth, indicating that the loss KIF2A had no effect on neuronal differentiation (SI Appendix, Fig. S2 J–M). However, the distribution of neurons was altered in Emx1-cKO mice due to defective radial migration (SI Appendix, Fig. S2J), which is in line with published data (14). We analyzed cortical layering at P21 and found that upper layer neurons, labeled with Cux1, were the most affected (SI Appendix, Fig. S3 A–D). To test whether this effect was cell autonomous or due to abnormalities in radial glia, we deleted KIF2A only in postmitotic neurons using Nex-Cre mice (31) (hereafter referred to as Nex-cKO) (SI Appendix, Fig. S1 E and I). Like Emx1-cKO, Nex-cKO mice displayed a widespread distribution of Cux1+ neurons, extending from layer I to layer VI (SI Appendix, Fig. S3 A–D).
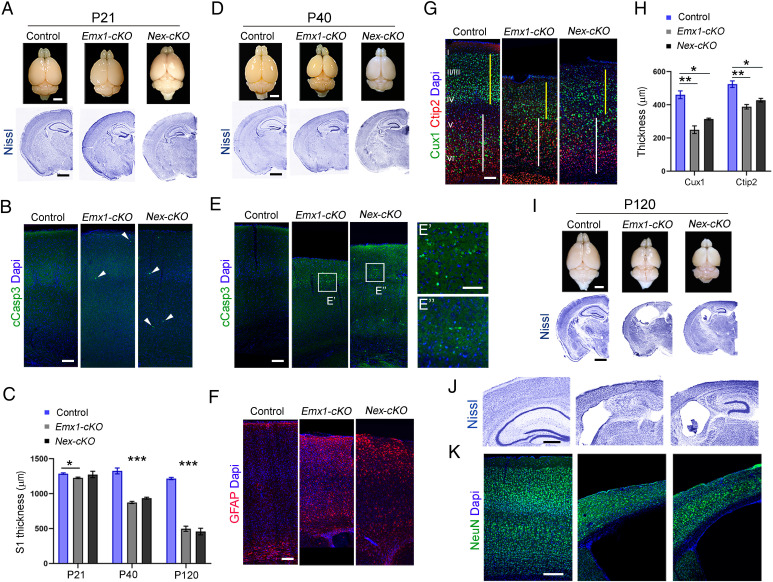
At postnatal day (P) 21, brain size was roughly similar between control and mutant mice (Fig. 1A). However, closer scrutiny revealed the presence of few apoptotic cells (cCasp3-positive; Fig. 1B) along with shrinkage of the somatosensory cortex in Emx1-cKO mice (Emx1-cKO: −4.71 ± 1.22%, P = 0.0026; Nex-cKO: −0.95 ± 3.12%, P = 0.259; Fig. 1C). At P40, the reduction of cortical thickness was exacerbated (Emx1-cKO mice: −32.07 ± 3.9%, P = 0.0003; Nex-cKO: −29.37 ± 3.35%, P = 0.0001) (Fig. 1C). The mutant brains were markedly smaller (Fig. 1D), and many cCasp3-positive cells and reactive astrocytes were detected (Fig. 1 E and F). The upper cortical layers were relatively more affected than the deep layers (Cux1+ layer thickness: Emx1-cKO: −45.8 ± 7.1%, P = 0.003; Nex-cKO: −32.6 ± 10.4%, P = 0.035; Ctip2+ layers thickness: Emx1-cKO: −25.9 ± 4.6%, P = 0.005; Nex-cKO: −18.4 ± 4.4%, P = 0.014) (Fig. 1 G and H). At P120, the reduction reached −59.16 ± 3.30% (P < 0.0001) and −62.50 ± 4.14% (P = 0.0001) in Emx1-cKO and Nex-cKO mice, respectively (Fig. 1 C, I–K). Given that Nex-Cre recombines only in neuronal and not in glial cells (31), the neurodegenerative effect was cell autonomous. Other cortical areas such as the motor and visual cortices as well as hippocampal formation were also reduced in Emx1-cKO and Nex-cKO mice (SI Appendix, Fig. S4).
Fig. 1.
Severe premature neurodegeneration in Kif2a cKO mice. (A) Representative images of whole brains (Top) and Nissl-stained coronal sections (Bottom) of the indicated genotypes at P21. (B) Coronal sections from P21 brains stained for cCasp3. White arrowheads point to cCasp3-positive cells. (C) Cortical thickness at the level of the somatosensory area (S1) at P21 (control = 1,287 ± 11 µm, n = 7; Emx1-cKO = 1,226 ± 11 µm, n = 6; Nex-cKO = 1,275 ± 46 µm, n = 5); P40 (control = 1,325 ± 42 µm, n = 4; Emx1-cKO = 874 ± 16 µm, n = 3; Nex-cKO = 936 ± 14 µm, n = 4); and P120 (control = 1,217 ± 16 µm, n = 3; Emx1-cKO = 497 ± 37 µm, n = 3; Nex-cKO = 457 ± 48 µm, n = 3). (D) Representative images of whole brains (Top) and Nissl-stained coronal sections (Bottom) at P40. (E) Coronal sections from P40 brains stained for cCasp3. (E’ and E’’) Magnifications of the boxed areas in E from Emx1-cKO and Nex-cKO, respectively, showing cCasp3-positive cells. (F and G) Coronal sections from P40 brains stained for GFAP (F) and Cux1 and Ctip2 (G). The yellow lines in G outline the thickness of upper cortical layers (II to IV), whereas the white lines delineate the thickness of deep cortical layers (V and VI). (H) Quantification of thickness of Cux+/upper layers (control = 460.3 ± 23.4 µm; Emx1-cKO = 249.4 ± 23 µm; Nex-cKO = 310 ± 41.8 µm) and Ctip2+/deep layers (control = 524 ± 20.2 µm; Emx1-cKO = 388.1 ± 13.5 µm; Nex-cKO = 427 ± 11.1 µm); n = 3 animals for each genotype. (I) Representative images of whole brains (Top) and Nissl-stained coronal sections (Bottom) of the indicated genotypes at P120. (J and K) Higher magnification of brain coronal sections stained with Nissl (J) or anti–NeuN antibodies (K) from control (Left), Emx1-cKO (Middle), and Nex-cKO (Right) mice. Scale bars: (A, D, and I) 2 mm (Top), 1 mm (Bottom); (B and E–G) 100 µm; (E’) 50 µm; (J) 500 µm; and (K) 200 µm. Data are represented as mean ± SEM. Values were obtained by unpaired. Student’s t-test; *P < 0.05, **P < 0.01, and ***P < 0.001. Dapi, 4′,6-diamidino-2-phenylindole; GFAP, Glial fibrillary acidic protein.
KIF2A Is Essential for Neuronal Function.
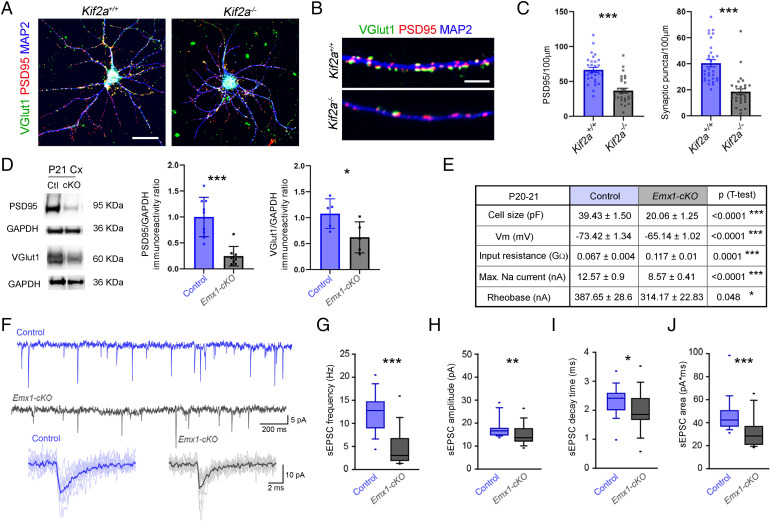
To function properly, neurons must form synapses and be integrated into neuronal networks. To test whether the lack of KIF2A could affect synapse formation, we cultured hippocampal neurons from control and Kif2a−/− embryos at E18.5 and used immunostaining for pre- and postsynaptic markers VGlut1 and PSD95 to identify glutamatergic synapses after 15 d in vitro (15 DIV) (Fig. 2A). Kif2a−/− neurons exhibited lower density of PSD95 terminals (−44.76 ± 7.37%, P < 0.0001) and diminished colocalization of PSD95 and Vglut1 in dendrites compared with control neurons (synaptic puncta: −54.27 ± 8.78%, P < 0.0001; Fig. 2 B and C). In line with this, Western blot analysis of cortical extracts showed a reduction in PSD95 (−75.45 ± 14.8%, P < 0.0001) and Vglut1 (−45.71 ± 18.49%, P = 0.039) in Emx1-cKO mice at P21, suggesting that the loss of KIF2A disturbed the formation of glutamatergic synapses (Fig. 2D). We next analyzed the intrinsic electrophysiological features of neurons by whole-cell recording of pyramidal neurons in acute brain slices. At P21, neurons were smaller and failed to maintain the membrane potential (Vm; Fig. 2E). The input resistance was higher and the maximum Na-current was lower in KIF2A-depleted neurons compared with controls (Fig. 2E), potentially due to the decrease in cell size and/or a change in membrane channel insertion in mutant neurons. Higher input resistance implies lower membrane conductance so that any current entering the neuron could have a larger or faster impact on membrane potential. Indeed, the rheobase of mutant neurons was reduced, and less current was required to induce action potentials (Fig. 2E and SI Appendix, Fig. S5 A–C).
Fig. 2.
KIF2A is essential for glutamatergic synapse maintenance and neuronal function. (A) Hippocampal neurons from Kif2a+/+ and Kif2a−/− embryos at 15 DIV, immunostained with anti-Vglut1, -PSD95, and -MAP2 to visualize glutamatergic synapses. (B) Representative images of a dendrite from Kif2a+/+ (Top) and Kif2a−/− (Bottom) pyramidal neurons showing presynaptic boutons (Vglut1, green) and postsynaptic terminals (PSD95, red). (C) Quantification of PSD95 and synaptic puncta per 100 µm (PSD95: Kif2a+/+ = 66.62 ± 3.56, Kif2a−/− = 36.80 ± 3.39; Puncta: Kif2a+/+ = 40.41 ± 2.79, Kif2a−/− = 18.48 ± 2.2; n = 30 neurons for each genotype). (D) Western blot analysis and quantification of the relative amount of PSD95 and Vglut1 from P21 control (Ctl) and Emx1-cKO cortex extracts. (E) Means ± SEM and P values of intrinsic physiological characteristics of cortical somatosensory L2/3 pyramidal neurons recorded in P21 brain slices. (F) Top: Example traces of sEPSCs in control (blue) and Emx1-cKO (gray) recordings. Bottom: 10 Superimposed individual (shaded) sEPSCs and the averaged (bold) sEPSC in control (blue) and Emx1-cKO (gray) neurons. (G) sEPSC frequency (control = 12.27 ± 0.9 Hz; Emx1-cKO = 5.91 ± 1.3 Hz). (H) sEPSC amplitude (control = 17.46 ± 0.9 pA; Emx1-cKO = 14.94 ± 0.9 pA). (I) sEPSC decay time (control = 2.31 ± 0.1 ms; Emx1-cKO = 1.98 ± 0.1 ms). (J) sEPSC area (control = 45.89 ± 3.3 fC; Emx1-cKO = 31.48 ± 2.5 fC). Sample size for G–J, control n = 21 and Emx1-cKO n = 28 neurons from four different animals. Scale bars: (A) 25 µm; (B) 5 µm. Data are represented as mean ± SEM (C and D) or as tukey plot (G–J). Values were obtained by unpaired Student’s t-test; *P < 0.05, **P < 0.01, and ***P < 0.001. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Max., maximum; Vm, membrane potential.
To understand how KIF2A mutant neurons were integrated in the cortical network, we examined spontaneous excitatory and inhibitory postsynaptic currents (sEPSCs and sIPSCs, respectively) (Fig. 2 F–J and SI Appendix, Fig. S5 D–H). KIF2A mutant neurons had significantly reduced sEPSC frequency (−51.83%, P ≤ 0.0001) and amplitude (−14.43%, P = 0.008) (Fig. 2 F–H). The decay time and area were reduced (decay time: −14.47%, P = 0.016; area: −31.40%, P ≤ 0.0001) (Fig. 2 F, I, and J), indicating that glutamatergic synapses were smaller, receptor subunit composition was different, and/or the postsynaptic channel density was reduced. We also analyzed the sIPSCs in mutant P21 cortex. Pyramidal neurons receive inhibitory inputs from cortical interneurons, which are not mutated in Emx1-cKO mice. The frequency and amplitude of sIPSCs were similar in control and mutant neurons (frequency: +11.1%, P = 0.725; amplitude: +1.64%, P = 0.516) (SI Appendix, Fig. S5 D–F). However, the decay time (−28.75%, P ≤ 0.0001) and the total current (area: −26.98%, P = 0.001) were smaller, possibly due to differences in the composition of postsynaptic channels (SI Appendix, Fig. S5 D, G, and H).
KIF2A cKO Mice Exhibit Wiring Abnormalities.
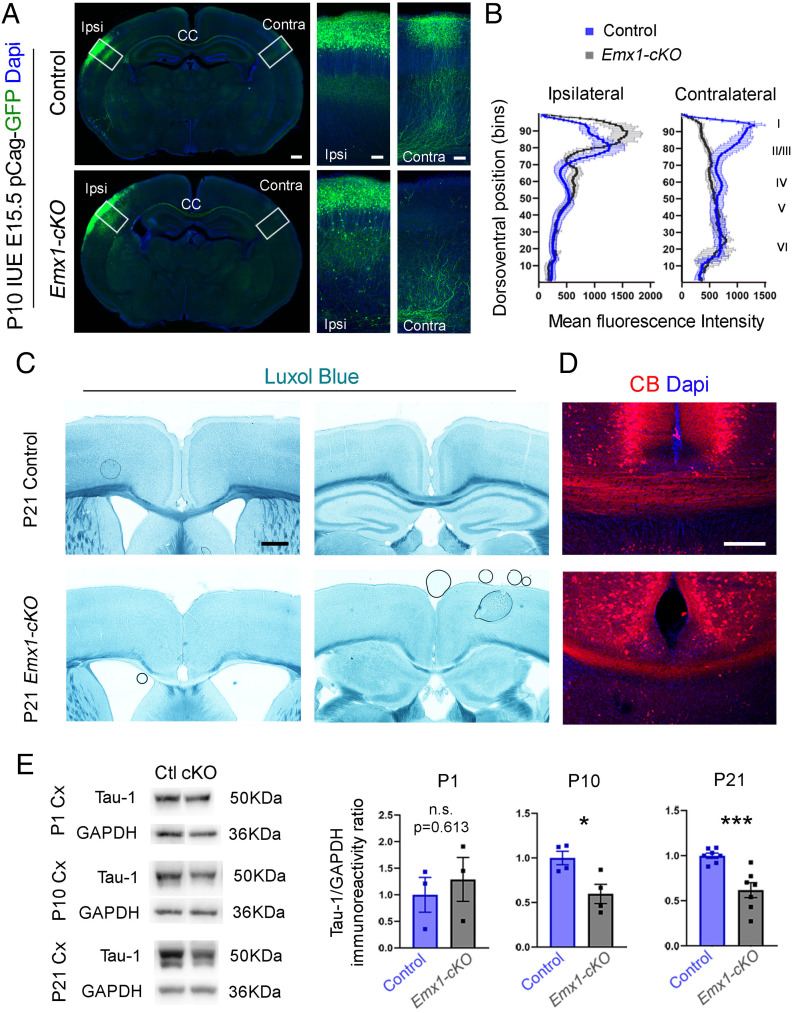
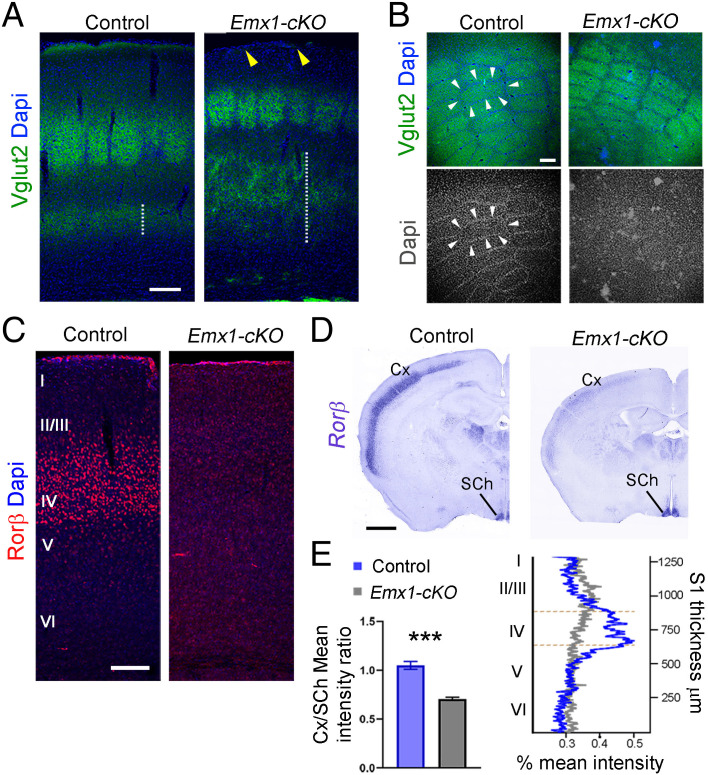
KIF2A is implicated in axonal pruning during development and affects the length of axon collaterals (14, 16). Yet, whether this has an impact on axon targeting and neuronal connectivity is unknown. To address this, we analyzed axon pathfinding and formation of the main tracts. Cortical neurons form corticocortical and cortico-subcortical connections. To evaluate corticocortical projections, we used in utero electroporation at E15.5 to label prospective neurons in layers II and III and analyzed callosal projections at P10. In control animals, axons crossed the CC and ran radially through the contralateral hemisphere, reaching the most upper cortical layers (Fig. 3A). In sharp contrast, mutant callosal axons, although they were able to cross the midline, ended their journey in the white matter or deep cortical layers (Fig. 3A). We evaluated the distribution of the mean fluorescence intensity and found that it was greatly reduced in upper layers of the contralateral side (Fig. 3B). At P21, the CC was thinner in Emx1-cKO mice compared with control mice (Fig. 3 C and D). To assess cortico-subcortical projections, we used the Thy1-YFP transgene to identify pyramidal neurons of layer V and examine their projections (32). Compared with control mice, the number of axons in the corticospinal tract (CST) was significantly reduced in the brain stem, and the pyramidal decussation was almost absent in Emx1-cKO and Nex-cKO mice (SI Appendix, Fig. S6 A–C). CST axons were not detected in the spinal cord in conditional mutant mice (SI Appendix, Fig. S6 A’–C’). The loss of axons in the mutant cortex was confirmed by a gradual decrease in Tau-1 protein levels from P1 to P21 (Fig. 3E). Finally, even though we did not delete KIF2A in the thalamus, we analyzed thalamocortical projections at P21 to assess how wild-type thalamic inputs are integrated in the mutant cortex. In contrast to control animals, more thalamic axon terminals (labeled with Vglut2) were found in deep cortical layers (Fig. 4A, dashed line), and none reached layer I (Fig. 4A, arrowheads). A reduced number of terminals were observed in layer IV, where they shaped lessened barrel fields (Fig. 4A), but stellate cells were not correctly organized along the walls of barrel fields (Fig. 4B). We tested whether layer IV neurons were nevertheless activated by thalamic afferents using expression of retinoic acid–related orphan receptor Beta (Rorβ), a thalamic activity-dependent transcription factor (33, 34). Rorβ immunoreactivity was considerably decreased in mutant layer IV compared with control littermates (Fig. 4C), and this was further confirmed by in situ hybridization that emphasized a robust reduction of Rorβ messenger RNA (mRNA) (−32.40 ± 4.95%, P = 0.0002) (Fig. 4 D and E).
Fig. 3.
Defective connectivity in Kif2a cKO mice. (A) Left: P10 coronal sections from control and Emx1-cKO mice that were electroporated with a GFP-expressing plasmid at E15.5. (Right) Higher magnifications of the boxed areas emphasizing the electroporated side (Ipsi) and contralateral somatosensory cortex (Contra). (B) Mean fluorescence intensity per bin in control and Emx1-cKO mice in the electroporated (Ipsilateral, Left) and (contralateral, Right) sides. (C) Luxol fast blue (myelin) staining of brain coronal sections at P21 underscoring the absence of callosal commissure in Emx1-cKO mice. (D) Immunofluorescence for calbindin (CB) on coronal sections from control and Emx1-cKO mice at the level of the CC at P21. (E) Western blot analysis and quantification of Tau-1 levels at P1 (+29 ± 52%, P = 0.613), P10 (−40 ± 13%, P = 0.022), and P21 (−38.2 ± 9%, P = 0.001) in cortical extracts from control and Emx1-cKO mice. Scale bars: (A, Left and C) 500 µm; (A, Right and D) 100 µm. Data are represented as mean ± SEM. Values were obtained by unpaired Student’s t-test; n.s., not significant, *P < 0.05, **P < 0.01, and ***P < 0.001. IUE, In utero electroporation; Ctl, control; Dapi, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Fig. 4.
Sensory input processing in Kif2a cKO mice. (A) Immunofluorescence for Vglut2 on coronal sections from control and Emx1-cKO mice at P21. White dashed lines delineate lower cortical layers innervated by thalamocortical inputs, whereas yellow arrowheads point to the lack of thalamocortical innervation of layer I in Emx1-cKO mice. (B) Immunofluorescence for Vglut2 in transversal sections of P21 control and Emx1-cKO mice emphasizing barrel fields. White arrowheads depict the walls of barrel fields formed by stellate cells. (C) Rorβ immunostaining of P21 coronal sections from control and Emx1-cKO mice. (D) Coronal sections of P21 brains from control and Emx1-cKO mice hybridized with a Rorβ riboprobe showing mRNA expression in the cortex (Cx) and suprachiasmatic nucleus (SCh). (E) Mean intensity of the signal in Cx relative to SCh (Control: 1.05 ± 0.04; Emx1-cKO: 0.71 ± 0.02; n = 3 for each genotype) and distribution of Rorβ-mRNA signal across cortical layers of control (blue) and Emx1-cKO (gray) mice at P21. Orange dashed lines delimit layer IV of the cortex. Scale bars: (A–C) 100 µm; (D) 1 mm. Data is represented as mean ± SEM. Values were obtained by unpaired Student’s t-test; *P < 0.05, **P < 0.01, and ***P < 0.001. Dapi, 4′,6-diamidino-2-phenylindole.
KIF2A Regulates Neuritogenesis.
While analyzing cortical lamination and connectivity, we noticed a substantial reduction of layer I in Emx1-cKO and Nex-cKO mice (SI Appendix, Fig. S3 C and D, orange lines). This layer comprises apical dendrites of pyramidal neurons that receive long-range inputs and few inhibitory interneurons. We stained cortical sections with the dendritic marker MAP2 and observed an abnormal distribution. While MAP2 delineated radially oriented apical dendrites in control mice, it accumulated in cell bodies in Emx1-cKO and Nex-cKO mice (Fig. 5 A and B). To investigate this further, we electroporated a green fluorescent protein (GFP)–expressing plasmid at E15.5 and analyzed neuronal morphology at postnatal stages. At P10, KIF2A-deficient neurons exhibited smaller dendritic trees compared with control neurons (SI Appendix, Fig. S7A). At P21, mutant dendrites frequently displayed swellings and dendritic spines could barely be recognized (Fig. 5C). These abnormalities were not secondary to defects in neuronal migration since they were not restricted to mislocalized neurons. To test if the effect of KIF2A was cell autonomous, we electroporated Cre- and floxed GFP-expressing plasmids in cortices of Kif2aF/F embryos to delete KIF2A only from sparse cortical neurons, identified by Cre-dependent GFP expression. Mutant neurons in control background displayed defects in neuronal shape, suggesting that the function of KIF2A is cell autonomous (SI Appendix, Fig. S7B). Layer V pyramidal neurons labeled with Thy1-YFP also exhibited aberrant morphology (SI Appendix, Fig. S6 D and E).
Fig. 5.
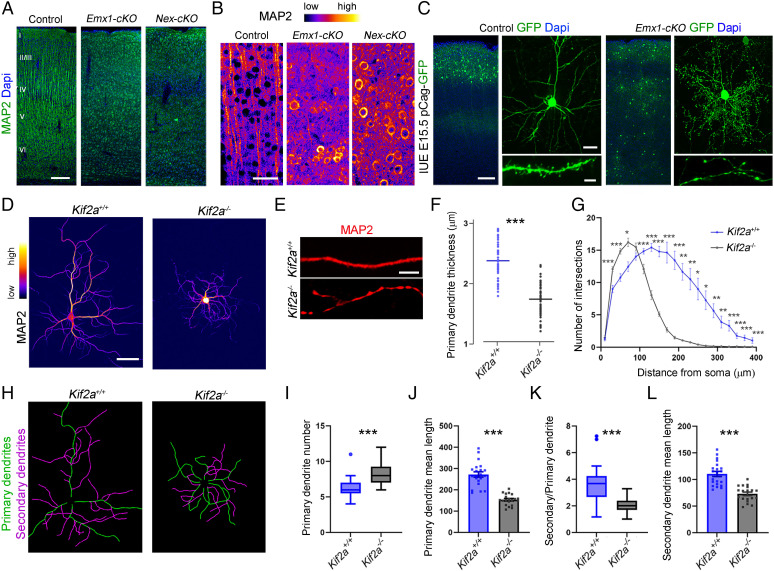
KIF2A shapes neuronal morphology. (A) Immunofluorescence for MAP2 on P21 coronal sections from control, Emx1-cKO, and Nex-cKO mice. (B) Fire look-up table (LUT) representation of MAP2 staining in the cortex underscoring the distribution, intensity, and shape of the signal. (C) Coronal sections of P21 brains from control and Emx1-cKO mice electroporated at E15.5 with a GFP-expressing plasmid. Pyramidal neurons are distributed in the upper layers in control and scattered in all layers in Emx1-cKO. In addition, the dendritic arborization is altered with the presence of swellings and absence of spines. (D) Hippocampal primary culture from E18.5 Kif2a+/+ and Kif2a−/− embryos immunolabeled with anti-MAP2 antibody at 15 DIV and represented in fire LUT to highlight the difference in fluorescence intensity and distribution. (E) Representative images of dendrites from Kif2a+/+ and Kif2a−/− at 15 DIV. (F) Quantification of dendritic thickness (Kif2a+/+ = 2.38 ± 0.05 µm, n = 43 dendrites; Kif2a−/− = 1.74 ± 0.04 µm, n = 50 dendrites). (G) Sholl analysis showing the number of intersections every 20 µm from the cell body until 380 µm (n = 32 and 47 neurons for Kif2a+/+ and Kif2a−/−, respectively). (H) Illustration of primary (green) and secondary (magenta) dendrites in control and mutant neurons. (I) Average number of primary dendrites per neuron (Kif2a+/+ = 6.1 ± 0.3, n = 21 neurons; Kif2a−/− = 8.46 ± 0.3, n = 26 neurons). (J) Mean length of primary dendrites (Kif2a+/+ = 272.3 ± 12.8 µm, n = 20 neurons; Kif2a−/− = 153.4 ± 6 µm, n = 19 neurons). (K) Number of secondary dendrites per primary dendrite per neuron (Kif2a−/− = 3.6 ± 0.3, n = 21 neurons; Kif2a+/+ = 2.1 ± 0.1, n = 26 neurons). (L) Mean length of secondary dendrites (Kif2a−/− = 110.4 ± 5, n = 21 neurons; Kif2a+/+ = 72.9 ± 3.5, n = 19 neurons). All the values are represented as mean ± SEM except for graphs I and K, represented as a Tukey plot. Scale bars: (A) 100 µm; (B) 50 µm; (C, Left) 100 µm; (C, Right Top) 50 µm; (C, Right Bottom) 5 µm; (D) 50 µm; and (E) 5 µm. Data are represented as mean ± SEM (F, G, J and L) or as tukey plot (I and K). Values were obtained by unpaired Student’s t-test; *P < 0.05, **P < 0.01, and ***P < 0.001. Dapi, 4′,6-diamidino-2-phenylindole; IUE, In utero electroporation.
To explore further the function of KIF2A in dendrite formation, we used primary cultures of hippocampal neurons. At 15 DIV, MAP2 was distributed in dendrites in control but not in mutant neurons (Fig. 5D). Some portions were swollen and contained dense signal, whereas others were devoid of MAP2 signal (Fig. 5E). Primary dendrites were significantly thinner in KIF2A-depleted neurons (−26.89 ± 2.48%, P < 0.0001) (Fig. 5F). We conducted a Sholl analysis and found that Kif2a−/− neurons formed more proximal (near the soma) and less distal intersections compared with control neurons (Fig. 5G), indicating a defect in dendritic branching. Mutant neurons formed more but shorter primary dendrites compared to wild-type neurons (number: +37.78 ± 7.8%, P < 0.0001; length: −43.7 ± 5.14%, P < 0.0001) (Fig. 5 H–J). Mutant neurons had significantly fewer and shorter secondary dendrites than controls (number of secondary dendrites per primary dendrite: −43.25 ± 9.1%, P < 0.0001; secondary dendrite length: −33.92 ± 5.46%, P < 0.0001; Fig. 5 H, K, and L).
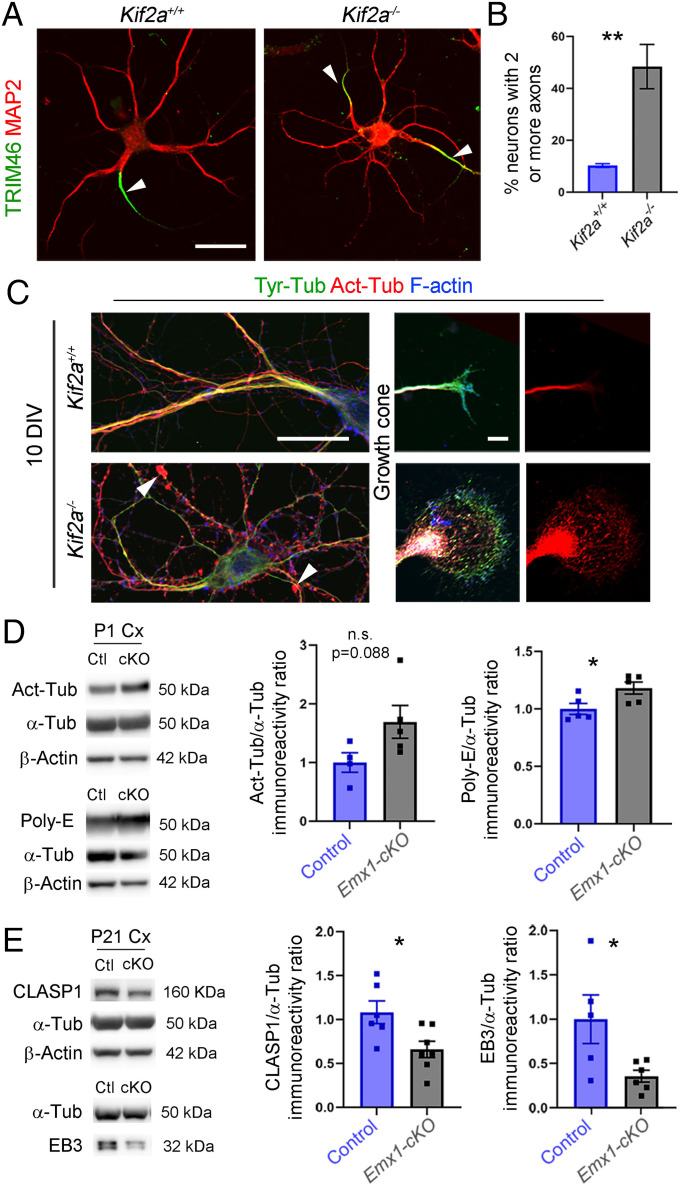
The presence of longer axon collaterals in Kif2a KO mice (14) prompted us to assess axonogenesis in KIF2A-deficient neurons. We used TRIM46 antibodies that label the proximal axon and found that 48.44 ± 8.52% of mutant neurons versus 10.27 ± 0.71% of control neurons had two or more axons (Fig. 6 A and B; P = 0.0043). A key determinant of axon versus dendrite formation is the stabilization of MTs (35). During the first steps of neuronal polarization, the neurite which accumulates more stable MTs, forms the axon (35). Since KIF2A regulates MT dynamics (13, 15, 16), we analyzed the PTMs of tubulin using immunostaining for acetylated tubulin (long-lived/stable MT) and tyrosinated tubulin (freshly polymerized/dynamic MT). At 10 DIV, Kif2a−/− hippocampal neurons accumulated more acetylated tubulin and less tyrosinated tubulin than control neurons (Fig. 6C). Acetylated tubulin was not evenly distributed along dendrites in the mutant neurons. Instead, it clustered locally, forming a pearl necklace-like structure (Fig. 6C, arrowheads). The axon’s growth cone was considerably larger and contained more stable MTs (Fig. 6C). Western blot analysis of cortical extracts from control and Emx1-cKO mice at P1 confirmed an increase in stable MTs in the mutant (acetylated tubulin: +69.4 ± 35%, P = 0.088; polyglutamylated tubulin: +18.1 ± 7%, P = 0.035) (Fig. 6D). We assessed the level of CLASP1 and EB3, two +TIP proteins important for MT dynamics (36). Both were significantly reduced in the mutant cortex (CLASP1: −42.4 ± 15%, P = 0.02; and EB3: −64 ± 26%, P = 0.034) (Fig. 6E). These results emphasize the importance of KIF2A for MT dynamics, neuronal polarization, and neurite morphogenesis.
Fig. 6.
Polarization and MT content of KIF2A-deficient neurons. (A) Hippocampal neurons from Kif2a+/+ and Kif2a−/− embryos at 10 DIV, immunolabeled with anti-TRIM46 and anti-MAP2 antibodies to highlight the proximal axon (white arrowheads) and dendrites, respectively. (B) Percentage of neurons with two or more axons (Kif2a+/+ = 10.27 ± 0.7%, n = 79 neurons; Kif2a−/− = 48.44 ± 8.52%, n = 94 neurons from four different experiments). (C) Hippocampal neurons from Kif2a+/+ and Kif2a−/− embryos at 10 DIV, immunolabeled with anti-tyrosinated tubulin (Tyr-Tub), anti-acetylated tubulin (Act-Tub), and anti–F-Actin antibodies. White arrowheads indicate accumulation of acetylated-tubulin in dendrites of Kif2a−/− neurons. (D) Western blot analysis and quantification of the relative amount of Act-Tub and polyglutamylated tubulin (Poly-E) in P1 cortical extracts from control (Ctl) and Emx1-cKO. (E) Western blot analysis and quantification of the relative amount of CLASP1 and EB3 from P21 control and Emx1-cKO cortical extracts. Scale bars: (A) 25 µm; (C, Left) 25 µm; and (C, Right) 5 µm. Data are represented as mean ± SEM. Values were obtained by unpaired Student’s t-test; n.s., not significant, *P < 0.05, **P < 0.01, and ***P < 0.001.
KIF2A Is Essential for Transport of Lysosomes.
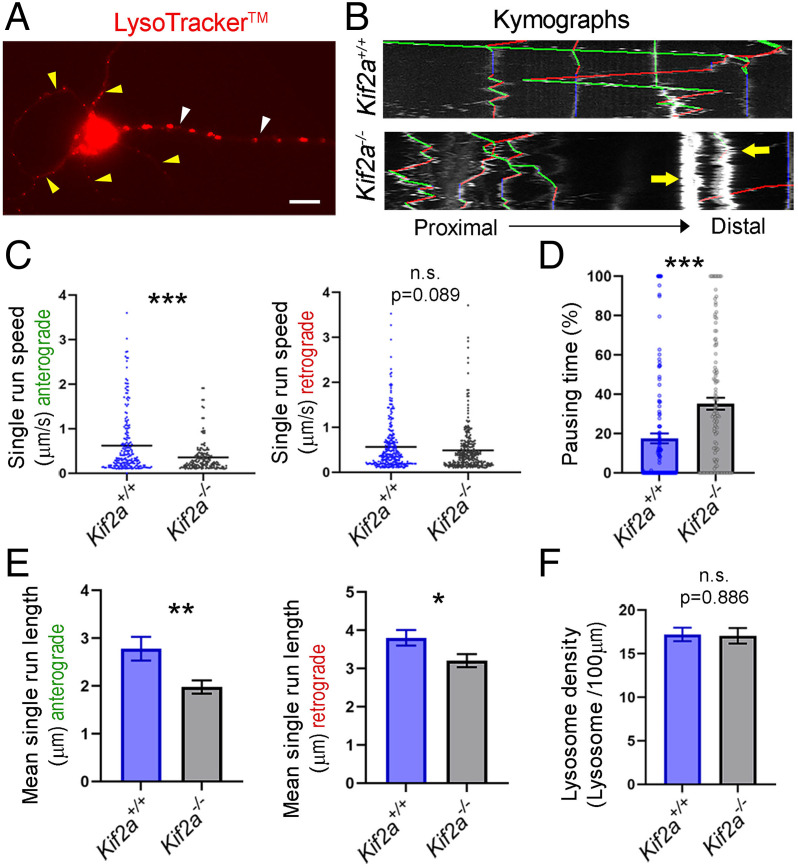
MTs serve as a railway for organelle transport. Thus, any change in MT composition or dynamics can affect intracellular transport. To test whether transport is affected in the absence of KIF2A, we labeled lysosomes in primary cultures of hippocampal neurons with Lysotracker DND-99 and evaluated their movement along a segment of the axon (Fig. 7A, white arrowheads) from time-lapse videos. Particle displacements were converted to kymographs, and different transport parameters were analyzed (Fig. 7B). The mean speed in single anterograde runs was significantly lower in mutant neurons compared with controls (−42.6 ± 9.3%, P < 0.0001) (Fig. 7C). The speed of retrograde movements was also reduced, but this was not statistically significant (−13.21 ± 7.7%, P = 0.089) (Fig. 7C). The pausing time (% of the time the lysosomes were static) was higher in mutant neurons than in controls (+50.14 ± 11.14%, P < 0.0001) (Fig. 7D). In line with reduced velocity and increased pausing time, KIF2A mutant neurons showed a significant reduction in single run length (anterograde movement: −28.79 ± 10.8%, P = 0.008; retrograde movement: −15.79 ± 7.1%, P = 0.025) (Fig. 7E). Many lysosomes were trapped in axonal beadings (yellow arrows in Fig. 7B), but the overall density of lysosomIs in the axon was similar between control and mutant neurons (Fig. 7F). These results show that the velocity and traveled distance were reduced in mutant neurons and that more lysosomes were static, suggesting that intracellular transport is affected by the loss of KIF2A.
Fig. 7.
Disrupted lysosomal transport in Kif2a−/− neurons. (A) Hippocampal neuron at 5 DIV transfected with LysoTracker (red). White and yellow arrowheads point to the axon and dendrites, respectively. (B) Kymograph generated from 1.5-min live imaging of lysosomes moving anterogradely (green lines), moving retrogradely (red lines), or static (blue lines). Yellow arrows depict trapped lysosomes. (C) Distribution and mean of the speed of antegrade and retrograde movements (anterograde: Kif2a+/+ = 0.621 ± 0.05 µm/s, n = 221 runs; Kif2a−/− = 0.357 ± 0.03 µm/s, n = 160 runs; retrograde: Kif2a+/+ = 0.564 ± 0.03 µm/s, n = 293 runs; Kif2a−/− = 0.490 ± 0.03 µm/s, n = 296 runs). (D) Pausing time of lysosomes (Kif2a+/+ = 17.54 ± 2.5%, n = 131 runs; Kif2a−/− = 35.18 ± 3.1%, n = 113 runs). (E) Mean of length of anterograde and retrograde movements of single runs (anterograde: Kif2a+/+ = 2.78 ± 0.25 µm, n = 264 runs; Kif2a−/− = 1.98 ± 0.14 µm, n = 215 runs; retrograde: Kif2a+/+ = 3.8 ± 0.2 µm, n = 499 runs; Kif2a−/− = 3.2 ± 0.17 µm, n = 517 runs). (F) Lysosome density in the axon (Kif2a+/+ = 17.21 ± 0.8, n = 20 axons; Kif2a−/− = 17.04 ± 0.9, n = 15 axons). Scale bar: (A) 10 µm. Data are represented as mean ± SEM. Values were obtained by unpaired Student’s t-test; n.s., not significant, *P < 0.05, **P < 0.01, and ***P < 0.001.
Deletion of KIF2A in Mature Neurons Is Sufficient to Cause Their Loss.
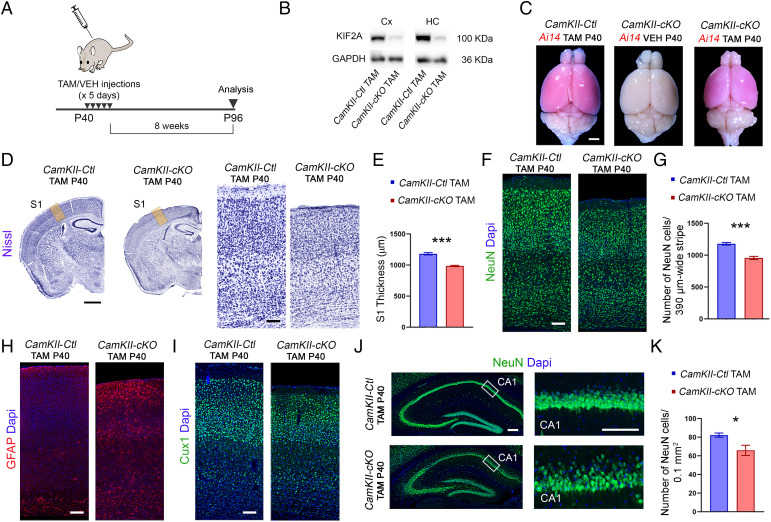
The expression of KIF2A is maintained in the adult brain (SI Appendix, Fig. S1 H and I; Allen brain atlas: https://mouse.brain-map.org/). To assess the protein’s function in mature neurons and circumvent the developmental effect, we ablated it from glutamatergic neurons of the cerebral cortex and hippocampus using CamKIIa-CreERT2, while expressing Tomato (Ai14) in Cre-expressing neurons (37). We injected CamKII-cKO (Kif2aF/F; tdTomato; CamKII-CreERT2) and control (Kif2a+/+; tdTomato; CamKIIa-CreERT2) mice with tamoxifen for 5 consecutive d starting from P40 and analyzed their brains 8 wk after the last injection (Fig. 8 A and B). CamKII-cKO brains (post-tamoxifen injections) were markedly smaller, and the neocortex was thinner compared with control brains (−16.7 ± 2%, P < 0.0001; Fig. 8 C–E). The number of NeuN-positive cells in the somatosensory cortex was significantly reduced in mutant compared with control mice (−18.9 ± 2.7%, P = 0.0005; Fig. 8 F and G). Moreover, we observed reactive astrocytes in mutant cortex, suggesting an increase of astrogliosis, likely secondary to cell death (Fig. 8H). As expected, neuronal migration was not affected (Fig. 8 I and J). However, the mutant hippocampus had a lower neuronal density especially in the CA1 (−20 ± 6.7%, P = 0.02; Fig. 8 J and K). These results indicate that KIF2A, independent of its developmental function, is necessary for neuronal survival.
Fig. 8.
Postnatal deletion of KIF2A causes neuronal loss. (A) Schematic diagram for experimental procedure used to induce KIF2A deletion. Tamoxifen (TAM) or vehicle (VEH) was injected for 5 consecutive d starting from P40, and brains were analyzed 8 wk after the last injection (P96). (B) Western blot analysis of KIF2A from cortical (Cx) and hippocampal (HC) protein extracts in control and mutant mice. (C) Representative images of whole brains of the indicated genotypes. (D) Nissl-stained coronal sections in control and mutant mice. Right panels are higher magnification of the yellow areas in left panels of the primary somatosensory cortex (S1) for each genotype. (E) Quantification of the S1 thickness (CamKII-Ctl TAM P40 = 1,148 ± 19.9 µm, n = 5; CamKII-cKO TAM P40 = 968.3 ± 10.3 µm, n = 4). (F) NeuN staining in the S1 of control and mutant mice. (G) Number of NeuN-positive cells per 390-µm-wide stripes (CamKII-Ctl TAM P40 = 1,179 ± 20.30 cells, n = 4; CamKII-cKO TAM P40 = 956 ± 25.23 cells, n = 4). (H) GFAP staining in coronal sections showing reactive astrocytes in mutant cortex. (I) Cux1 immunostaining in coronal sections. (J) Left: NeuN immunostaining in the hippocampus. Right: Magnification of the areas boxed in Left panels showing NeuN–positive cells in the pyramidal layer of the CA1 area. (K) Quantification of NeuN-positive cell density in the CA1 area (CamKII-Ctl TAM P40 = 82.31 ± 2.2 cells per 0.1 mm2, n = 5; CamKII-cKO TAM P40 = 65.83 ± 5.6 per 0.1 mm2, n = 4). Scale bars: (C) 1.7 mm, (D, Left) 1 mm; (D, Right); 100 µm, (F, H, and I) 100 µm; (J, Left) 200 µm; and (J, Right) 100 µm. Data are represented as mean ± SEM. Values were obtained by unpaired Student’s t-test; *P < 0.05, **P < 0.01, and ***P < 0.001. Dapi, 4′,6-diamidino-2-phenylindole; GFAP, Glial fibrillary acidic protein.
Discussion
KIF2A Regulates MT Dynamics and Intracellular Transport.
KIF2A catalyzes tubulin disassembly, and its loss not only reduces depolymerization of the MT but also disrupts its function, since KIF2A-depleted neuroblasts display slower displacement of EB3 comets (15). Down-regulation of KLP10A, the Drosophila ortholog of KIF2A, reduced MT dynamics and increased MT pausing time in Drosophila S2 cells (38). Abnormalities in MT dynamics correlate with enhanced PTM of tubulin, such as acetylation and polyglutamylation. +TIP proteins interact with the labile domain of the MT (39). Therefore, increased acetylation and polyglutamylation along with decreased levels of +TIP proteins CLASP1 and EB3 indicate that the lack of KIF2A triggers changes in the composition of the MT, enhancing stable MTs at the expense of dynamic MTs. CLASP1 modulates MT dynamics and promotes neurite extension (40, 41). EB proteins bind the MT plus-end, mediate MT–protein interactions, and increase growth velocity (42–44). In KIF2A mutant neurons, MAP2 accumulates in the cell body instead of dendrites. MAP2 is important for MT assembly, and its loss results in shorter dendrites (45, 46), a phenotype found in Kif2a−/− neurons. Higher levels of stable MTs and lower levels of MAP2, CLASP1, and EB3 could account for the aberrant polarization, supernumerary axons (35), and dendritic abnormalities. The delicate balance between tubulin PTM and MAPs is important for intracellular transport (47–50). For instance, an increase in tubulin polyglutamylation leads to a reduction in axonal transport and neurodegeneration (51–53). Mutant neurons for spastin, an MT-severing protein, have increased levels in tubulin polyglutamylation that, in turn, reduce KIF5 binding to MTs (54). Given that the lysosomes are transported by KIF5, the aberrant lysosome transport observed in KIF2A-depleted neurons could be due to increased polyglutamylation. Therefore, KIF2A is critical to maintain MT dynamics, intracellular transport, and neuronal survival.
Loss of KIF2A Triggers Neurodegeneration.
Our results show that KIF2A is important during development for polarization, maturation, and survival of neurons. Conditional ablation of KIF2A from progenitor or early postmitotic neurons triggers neurodegeneration, leading to severe shrinkage of the cerebral cortex (less than half the volume of a normal cortex by the age of 4 mo) and cortical dysfunction. Deletion of KIF2A in the mature brain also triggers neurodegeneration, although to a lesser extent, indicating that KIF2A is not only necessary during developmental but also in the mature brain. Neurodegenerative diseases have in common defective MTs, loss of functional synapses, axon degeneration, and disintegration of neuronal networks. All these features are found in KIF2A conditional knockouts. Deletion of KIF2A affects the intracellular transport of lysosomes, and defective lysosome transport was associated with autophagic stress and neurodegeneration (55, 56). KIF2A was recently linked to AD (28, 29), and its activity depends on phosphorylation by different kinases such as CDK5, PKA, and TTBK2 (21, 57). Importantly, all these kinases have been implicated in Tau phosphorylation and neurodegenerative diseases.
KIF2A Links Neurodevelopment to Neurodegeneration.
While the function of KIF2A in neuronal migration is well established (14, 58), its role in neurogenesis remains controversial. In utero electroporation of KIF2A shRNA decreases the proliferation of progenitors and promotes neuronal differentiation (8). In contrast electroporation of KIF2A variants c.961C > G/p.His321Asp and c.950G > A/p.Ser317Asn causes the opposite effect, enhancing the proliferation of progenitor cells and decreasing the production of neurons (9). KI mice expressing one copy of the first variant (c.961C > G, p.His321Asp; Gene ID: 3796) have no change in proliferation but an enhanced embryonic cell death. Consequently, the mutant brain is smaller at birth (10). In the current work, we deleted KIF2A specifically in cortical progenitors and did not observe any change in either proliferation or survival of these cells. Mutant mice were born with normal cortical size. Hence, human variants could have a dominant negative activity and/or could behave as a gain-of-function mutation during embryonic neurogenesis, since the mutant protein interacts with different partners compared with the wild type (20). Adult (4-mo-old) mice carrying the human variant c.961C > G, p.His321Asp exhibited a slight reduction of cortex and hippocampal area that was ascribed to embryonic cell death. Kif2a cKO mice (Emx1-cKO and Nex-cKO), by contrast, displayed a severe neurodegeneration at the same age (4 mo). The somatosensory cortex was reduced by 60%, whereas the motor and visual cortex and hippocampal area were reduced by 75%. Postnatal deletion of KIF2A confirmed that KIF2A, independent of its role during development, is essential for neuronal survival. It is worth mentioning that microcephaly due to KIF2A mutations in patients worsens postnatally (25), and in some cases, patients are diagnosed with secondary microcephaly (DECIPHER patient 280143, c.1762G > T, p.Val588Phe), suggesting that these mutations affect postnatal processes such as neuronal maturation or survival and not only embryonic development.
Materials and Methods
Animals.
All animal procedures were carried out in accordance with European guidelines (2010/63/UE) and were approved by the animal ethics committee of the Université catholique de Louvain under agreement 2019/UCL/MD/006. Mice were housed in a standard 12-h dark/12-h light cycle. The temperature was between 20 and 24 °C and humidity between 40% and 60%. We used the following mouse lines: Emx1-Cre (30), Nex-Cre (31), CamKIIa-CreERT2 (37) Kif2aF/F (15), Thy1-YFP (32), Ai14 (59), and Kif2a−/−. To generate the Kif2a+/− mouse line, we crossed Kif2aF/+ males with PGK-Cre females (60) and intercrossed Kif2a+/− females and males to obtain Kif2a−/− and Kif2a+/+ embryos. To produce Kif2aFlF;Emx1-Cre (Emx1-cKO) and Kif2aFlF;Nex-Cre (Nex-cKO) mice, we crossed Emx1-Cre;Kif2aF/+ and Nex-Cre;Kif2aF/+ males with Kif2aF/F females. Kif2aF/F littermates (without Cre) were considered as control mice. To produce Kif2aFlF;CamKII-CreERT2 (CamKII-cKO) mice, we crossed CamKII-CreERT2;Kif2aF/+ males with Kif2aF/+ females. CamKII-CreERT2 Kif2a+/+ littermates were considered control mice. The day of the vaginal plug was considered E0.5. Tamoxifen (Sigma-Aldrich, T5648) was diluted in 10% ethanol in sunflower seed oil (Sigma-Aldrich, S5007) (30 mg/mL) and intraperitoneally injected at 180 mg/kg/d for 5 consecutive d.
Brain Tissue Preparation and Sectioning.
Embryos were fixed in 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB), pH 7.4, at room temperature (RT) for 2 h. Mice were perfused transcardially with PFA 4% in 0.1 phosphate-buffered saline (PBS), pH 7.4. Brains were harvested and postfixed in the same fixative for 2 h at RT for immunohistochemistry and overnight (ON) at 4 °C for In situ hybridization (ISH), Nissl, and myelin staining. Embryonic and postanal brains were washed in PBS after fixation, embedded in 4% agarose diluted in PBS, and sectioned with a Leica VT1000S vibratome (80 μm and 40 μm for embryonic and postnatal brains, respectively). For barrel field visualization, P21 cerebral cortices were separated from basal ganglia and other brain tissue, flattened between two slides, postfixed ON at 4 °C, washed with PBS, and sectioned at 80 μm using a vibratome.
Immunofluorescence.
Immunohistochemical staining was performed as previously described (61). Briefly, vibratome floating sections were blocked in 4% bovine serum albumin (Amresco, 9048–46-8), 3% goat serum (Sigma-Aldrich, G9023), and 0.2% Triton X-100 (Sigma-Aldrich, T8787) in PBS at RT for 1 h and incubated with the primary antibodies diluted in blocking solution at 4 °C ON and with the secondary antibodies for 2 h.
BrdU Injection and Immunolabeling.
Pregnant females were injected intraperitoneally at E14.5 with BrdU (50 mg/kg body weight; Sigma-Aldrich, B5002). Embryos were collected after 2 h, fixed in PFA 4% in PB 0.1 M. Vibratome sections were pretreated with HCl 2 N for 30 min, followed by a 10-min incubation with borate buffer and immunodetection using Rat anti-BrdU (Serotec, MCA2060GA; 1:200).
In utero Electroporation.
Plasmids used in this study were as follows: pCag-DsRed (Addgene, #11151; 2 µg/µL), pCag-GFP (Addgene, #11150; 1 µg/µL), pCALNL-GFP (Addgene, #13770; 1 µg/µL), and pCag-Cre (Addgene, #13775; 4 ng/µL) (62, 63). Plasmids were purified with a Maxiprep Endofree Kit (Macherey-Nagel) and diluted in 1x PBS. pCAG-DsRed was added to the mix of electroporation as a control of efficiency. The DNA solution (with 0.05% Fast-green) was injected into the lateral ventricle of E15.5 embryos using pulled glass pipettes. The embryos were electroporated using tweezer-type electrodes (CUY650-5). Five square electric pulses were passed (40V, 50-ms interval cycle length, 950-ms interval pause). Electroporated animals, expressing DsRed, were selected using a fluorescent flashlight lamp (Nightsea, DFP-1).
Quantification of Callosal Axons.
Slices from controls and Emx1-cKO electroporated brains (n = 3 mice for each genotype) were analyzed with a laser scanning confocal microscope (Olympus, Fluoview FV1000). To quantify fluorescence, we analyzed an area of 350 µm width per 1,150 µm height in Fiji (ImageJ). This frame was subdivided into 100 equal bins of 350 µm width per 11.5 µm height, and fluorescence intensity in each bin was quantified. The mean fluorescence intensity for each bin was calculated using three animals and represented as mean ± SEM.
Axonal Transport of Lysosomes.
Intracellular transport was assessed using an adapted protocol from ref. 64. Hippocampal neurons from Kif2a+/+ and Kif2a−/− littermate embryos were cultured in a glass-bottomed cell culture dish 35/10 mm (627860, CELLview, Greiner bio-one) coated with laminin (5 μg/mL; Sigma-Aldrich, L2020) and poly-lysine (100 μg/mL; Sigma-Aldrich, P2636). At 5 DIV, the medium was replaced by warm Neurobasal (NB) containing 50 nM LysoTracker (LysoTracker Red DND-99; Thermo Fisher Scientific, L7528). Neurons were incubated for 20 min at 37 °C with LysoTracker, washed with 1 mL warmed NB medium, and supplemented with 1 mL NB medium. They were imaged using an inverted Zeiss Axio Observer microscope equipped with an environmental chamber (37 °C, 5% CO2; Pecon) and Zeiss Axiocam 503 mono camera and 63x (numerical aperture = 1.2) water immersion objective (Zeiss). Time-lapse series were captured every 1 s for 1.5 min from LysoTracker–positive neurons. Neurons with a pyramidal shape were considered. Images were analyzed using Fiji software (ImageJ). First, the axon of the positive neuron was straightened from the cell body to the distal axon using the “segmented line” tool of Fiji. We used the KymoToolBox plugin (65) to analyze the movement of lysosomes. Stationary vesicles (moving <0.1 µm/s) and run length of less than 0.3 µm (the size of a lysosome) were not considered for analysis. We assessed the mean speed and length of individual runs and percentage of time pause of each vesicle. Eleven neurons with pyramidal shape from each genotype were analyzed from three independent experiments.
Imaging, Quantification, and Statistical Analyses.
Images were captured with a digital camera coupled with an inverted Zeiss Axio Observer microscope or in a Laser Scanning confocal microscope (Olympus, Fluoview FV1000). Figures were prepared using Adobe Photoshop and Adobe Illustrator CC 2019, and two-dimensional mosaic reconstructions were produced when needed using the Photomerge tool of Photoshop software package. Cell counting was conducted manually using Fiji software (ImageJ). For NeuN counting in the cortex, images were pretreated in Fiji with gray scale attribute filtering (MorpholibJ plugin) and watershed process before automatic counting. Cells were counted in a 180-µm-wide stripe (E14.5), 340-µm-wide stripes (P1), and 390-µm-wide stripes (P21 and mature brains). A minimum of three animals and three slices of each animal were used for all analyses and quantifications. The exact sample size is specified in the figure legends. Prism 9 (GraphPad) software was used for statistical analysis. We described the percentage of difference between control and mutant conditions in the Results. The mean value and the SEM for each quantification are indicated in the figure legends. All the quantifications are represented as mean ± SEM except for graphs in Figs. 2 G–J and 5 I and K and SI Appendix, Fig. S5 E–H (Tukey plot). Values were obtained by unpaired two-tailed t test, two-tailed Mann-Whitney rank sum tests for sEPSC and sIPSC frequency, amplitude, and area, and unpaired two-tailed t test for sEPSC and sIPSC decay time. ns, not significant; * P < 0.05; ** P < 0.01; and *** P < 0.001. The P values are reported in the Results. Observations without quantification were validated and successfully reproduced in a minimum of three different animals.
Supplementary Material
Acknowledgments
We thank Isabelle Lambermont and Clarisse Fouss for technical assistance. This work was supported by the following grants: Belgian Fund for Scientific Research (FNRS) PDR T00075.15, FNRS PDR T0236.20, FNRS-FWO EOS 30913351, Fondation Médicale Reine Elisabeth, and Fondation Jacques Et Danièle Espinasse (JED)-Belgique. G.C. and N.R.-R are Research Fellow and Postdoctoral Researcher, respectively, at the Belgian FNRS. F.T. is an Honorary Research Director at FNRS.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120393119/-/DCSupplemental.
Data, Materials, and Software Availability
Mouse lines and other resources used in the study are available through material transfer agreements. All data generated during this study have been included in the manuscript and SI Appendix.
References
- 1.Hakanen J., Ruiz-Reig N., Tissir F., Linking cell polarity to cortical development and malformations. Front. Cell. Neurosci. 13, 244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parrini E., Conti V., Dobyns W. B., Guerrini R., Genetic basis of brain malformations. Mol. Syndromol. 7, 220–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubey J., Ratnakaran N., Koushika S. P., Neurodegeneration and microtubule dynamics: Death by a thousand cuts. Front. Cell. Neurosci. 9, 343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai A., Verma S., Mitchison T. J., Walczak C. E., Kin I kinesins are microtubule-destabilizing enzymes. Cell 96, 69–78 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Ganem N. J., Compton D. A., The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 166, 473–478 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uehara R., et al. , Aurora B and Kif2A control microtubule length for assembly of a functional central spindle during anaphase. J. Cell Biol. 202, 623–636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyamoto T., et al. , The microtubule-depolymerizing activity of a mitotic kinesin protein KIF2A drives primary cilia disassembly coupled with cell proliferation. Cell Rep. 10, 664–673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun D., et al. , Regulation of neural stem cell proliferation and differentiation by Kinesin family member 2a. PLoS One 12, e0179047 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broix L., et al. , Ciliogenesis and cell cycle alterations contribute to KIF2A-related malformations of cortical development. Hum. Mol. Genet. 27, 224–238 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Gilet J. G., et al. , Conditional switching of KIF2A mutation provides new insights into cortical malformation pathogeny. Hum. Mol. Genet. 29, 766–784 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W., et al. , Modeling microcephaly with cerebral organoids reveals a WDR62-CEP170-KIF2A pathway promoting cilium disassembly in neural progenitors. Nat. Commun. 10, 2612 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walczak C. E., Gayek S., Ohi R., Microtubule-depolymerizing kinesins. Annu. Rev. Cell Dev. Biol. 29, 417–441 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Trofimova D., et al. , Ternary complex of Kif2A-bound tandem tubulin heterodimers represents a kinesin-13-mediated microtubule depolymerization reaction intermediate. Nat. Commun. 9, 2628 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homma N., et al. , Kinesin superfamily protein 2A (KIF2A) functions in suppression of collateral branch extension. Cell 114, 229–239 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Hakanen J., et al. , The Celsr3-Kif2a axis directs neuronal migration in the postnatal brain. Prog. Neurobiol. 208, 102177 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Maor-Nof M., et al. , Axonal pruning is actively regulated by the microtubule-destabilizing protein kinesin superfamily protein 2A. Cell Rep. 3, 971–977 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Noda Y., et al. , Phosphatidylinositol 4-phosphate 5-kinase alpha (PIPKα) regulates neuronal microtubule depolymerase kinesin, KIF2A and suppresses elongation of axon branches. Proc. Natl. Acad. Sci. U.S.A. 109, 1725–1730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Homma N., et al. , KIF2A regulates the development of dentate granule cells and postnatal hippocampal wiring. eLife 7, e30935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ince-Dunn G., et al. , Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron 75, 1067–1080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akkaya C., et al. , Roles of developmentally regulated KIF2A alternative isoforms in cortical neuron migration and differentiation. Development 148, dev192674 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Ogawa T., Hirokawa N., Microtubule destabilizer KIF2A undergoes distinct site-specific phosphorylation cascades that differentially affect neuronal morphogenesis. Cell Rep. 12, 1774–1788 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Poirier K., et al. , Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet. 45, 639–647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian G., et al. , A patient with lissencephaly, developmental delay, and infantile spasms, due to de novo heterozygous mutation of KIF2A. Mol. Genet. Genomic Med. 4, 599–603 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavallin M., et al. , Recurrent KIF2A mutations are responsible for classic lissencephaly. Neurogenetics 18, 73–79 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Hatano M., et al. , Variants in KIF2A cause broad clinical presentation; the computational structural analysis of a novel variant in a patient with a cortical dysplasia, complex, with other brain malformations 3. Am. J. Med. Genet. A. 185, 1113–1119 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Yuen R. K., et al. , Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med. 21, 185–191 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Costain G., Cordeiro D., Matviychuk D., Mercimek-Andrews S., Clinical application of targeted next-generation sequencing panels and whole exome sequencing in childhood epilepsy. Neuroscience 418, 291–310 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Prokopenko D., et al. ; Alzheimer’s Disease Neuroimaging Initiative (ADNI), Whole-genome sequencing reveals new Alzheimer’s disease-associated rare variants in loci related to synaptic function and neuronal development. Alzheimers Dement. 17, 1509–1527 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cáceres A., González J. R., Female-specific risk of Alzheimer’s disease is associated with tau phosphorylation processes: A transcriptome-wide interaction analysis. Neurobiol. Aging 96, 104–108 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Gorski J. A., et al. , Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goebbels S., et al. , Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis 44, 611–621 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Feng G., et al. , Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Jabaudon D., Shnider S. J., Tischfield D. J., Galazo M. J., Macklis J. D., RORβ induces barrel-like neuronal clusters in the developing neocortex. Cereb. Cortex 22, 996–1006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark E. A., et al. , Cortical RORβ is required for layer 4 transcriptional identity and barrel integrity. eLife 9, e52370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Witte H., Neukirchen D., Bradke F., Microtubule stabilization specifies initial neuronal polarization. J. Cell Biol. 180, 619–632 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van de Willige D., Hoogenraad C. C., Akhmanova A., Microtubule plus-end tracking proteins in neuronal development. Cell. Mol. Life Sci. 73, 2053–2077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erdmann G., Schütz G., Berger S., Inducible gene inactivation in neurons of the adult mouse forebrain. BMC Neurosci. 8, 63 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mennella V., et al. , Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. 7, 235–245 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Peris L., et al. , Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J. Cell Biol. 174, 839–849 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mimori-Kiyosue Y., et al. , CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168, 141–153 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marx A., et al. , Xenopus cytoplasmic linker-associated protein 1 (XCLASP1) promotes axon elongation and advance of pioneer microtubules. Mol. Biol. Cell 24, 1544–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maurer S. P., et al. , EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr. Biol. 24, 372–384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaworski J., et al. , Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron 61, 85–100 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Straube A., Merdes A., EB3 regulates microtubule dynamics at the cell cortex and is required for myoblast elongation and fusion. Curr. Biol. 17, 1318–1325 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caceres A., Mautino J., Kosik K. S., Suppression of MAP2 in cultured cerebellar macroneurons inhibits minor neurite formation. Neuron 9, 607–618 (1992). [DOI] [PubMed] [Google Scholar]
- 46.Harada A., Teng J., Takei Y., Oguchi K., Hirokawa N., MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J. Cell Biol. 158, 541–549 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janke C., Bulinski J. C., Post-translational regulation of the microtubule cytoskeleton: Mechanisms and functions. Nat. Rev. Mol. Cell Biol. 12, 773–786 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Janke C., Magiera M. M., The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 21, 307–326 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Moutin M. J., Bosc C., Peris L., Andrieux A., Tubulin post-translational modifications control neuronal development and functions. Dev. Neurobiol. 81, 253–272 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bora G., et al. , Microtubule-associated protein 1B dysregulates microtubule dynamics and neuronal mitochondrial transport in spinal muscular atrophy. Hum. Mol. Genet. 29, 3935–3944 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Magiera M. M., et al. , Excessive tubulin polyglutamylation causes neurodegeneration and perturbs neuronal transport. EMBO J. 37, e100440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mullen R. J., Eicher E. M., Sidman R. L., Purkinje cell degeneration, a new neurological mutation in the mouse. Proc. Natl. Acad. Sci. U.S.A. 73, 208–212 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greer C. A., Shepherd G. M., Mitral cell degeneration and sensory function in the neurological mutant mouse Purkinje cell degeneration (PCD). Brain Res. 235, 156–161 (1982). [DOI] [PubMed] [Google Scholar]
- 54.Lopes A. T., et al. , Spastin depletion increases tubulin polyglutamylation and impairs kinesin-mediated neuronal transport, leading to working and associative memory deficits. PLoS Biol. 18, e3000820 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lie P. P. Y., Nixon R. A., Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol. Dis. 122, 94–105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roney J. C., et al. , Lipid-mediated impairment of axonal lysosome transport contributing to autophagic stress. Autophagy 17, 1796–1798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe T., et al. , TTBK2 with EB1/3 regulates microtubule dynamics in migrating cells through KIF2A phosphorylation. J. Cell Biol. 210, 737–751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hakanen J., et al. , The Celsr3-Kif2a axis directs neuronal migration in the postnatal brain. Prog. Neurobiol. 208, 102177 (2022). [DOI] [PubMed] [Google Scholar]
- 59.Madisen L., et al. , A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lallemand Y., Luria V., Haffner-Krausz R., Lonai P., Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 7, 105–112 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Touzot A., Ruiz-Reig N., Vitalis T., Studer M., Molecular control of two novel migratory paths for CGE-derived interneurons in the developing mouse brain. Development 143, 1753–1765 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Matsuda T., Cepko C. L., Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. U.S.A. 101, 16–22 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsuda T., Cepko C. L., Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. U.S.A. 104, 1027–1032 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bodakuntla S., Magiera M. M., Janke C., Measuring the impact of tubulin posttranslational modifications on axonal transport. Methods Mol. Biol. 2101, 353–370 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Zala D., et al. , Vesicular glycolysis provides on-board energy for fast axonal transport. Cell 152, 479–491 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mouse lines and other resources used in the study are available through material transfer agreements. All data generated during this study have been included in the manuscript and SI Appendix.