Significance
Causal research on maternal-fetal epigenetic programming in humans is rare and has been hampered by a lack of data that connects early-life maternal insults to offspring health across the life course. This study examines whether early-life exposure to adverse economic conditions during the Great Depression—the worst economic downturn in US history—impacted how fast individuals aged biologically decades later according to their epigenetic aging profiles. Using a quasi-experimental strategy, results show that faster epigenetic aging later in life is associated with worse economic conditions during the prenatal period specifically, suggesting it may be a sensitive window for the development of later-life disparities in aging. As a result, early-life investments may help postpone age-related morbidity and mortality and extend healthy life span.
Keywords: epigenetic aging, fetal programming, Great Depression, aging
Abstract
Research on maternal-fetal epigenetic programming argues that adverse exposures to the intrauterine environment can have long-term effects on adult morbidity and mortality. However, causal research on epigenetic programming in humans at a population level is rare and is often unable to separate intrauterine effects from conditions in the postnatal period that may continue to impact child development. In this study, we used a quasi-natural experiment that leverages state-year variation in economic shocks during the Great Depression to examine the causal effect of environmental exposures in early life on late-life accelerated epigenetic aging for 832 participants in the US Health and Retirement Study (HRS). HRS is the first population-representative study to collect epigenome-wide DNA methylation data that has the sample size and geographic variation necessary to exploit quasi-random variation in state environments, which expands possibilities for causal research in epigenetics. Our findings suggest that exposure to changing economic conditions in the 1930s had lasting impacts on next-generation epigenetic aging signatures that were developed to predict mortality risk (GrimAge) and physiological decline (DunedinPoAm). We show that these effects are localized to the in utero period specifically as opposed to the preconception, postnatal, childhood, or early adolescent periods. After evaluating endogenous shifts in mortality and fertility related to Depression-era birth cohorts, we conclude that these effects likely represent lower bound estimates of the true impacts of the economic shock on long-term epigenetic aging.
Aging is characterized by the gradual accumulation of cellular damage, leading to physiological deterioration, loss of function, and increased vulnerability to death (1). A growing body of research suggests that the beginnings of human aging occur during the initial phases of embryogenesis, a paradigm shift in aging research that anchors the onset of age-related damage accumulation to the prenatal period as opposed to later in the life course after the completion of development and the onset of reproductive age (2, 3). The idea that aging later in life could be linked to in utero programming has important implications for the development of early-life interventions that could postpone age-related morbidity and mortality and significantly extend healthy life span (2).
Early-life programming of aging and longevity has its roots in the developmental origins of health and disease (DOHaD) work of David Barker (4, 5), and in the field of environmental epigenetics, which linked the molecular basis for this mode of inheritance to epigenetic mechanisms in animal models (6). While age-related deterioration and damage is reflected in nearly every biological process at the molecular and cellular level, the dysregulation of these processes is a function of upstream epigenomic changes that control transcriptional and chromatin networks. In humans, the most well-researched type of epigenetic modification is DNA methylation (DNAm), which refers to the addition of a methyl group to a cytosine nucleotide at a cytosine-phosphate-guanine (CpG) site. DNAm regulates transcription without changing the DNA sequence by, for example, recruiting proteins involved in gene repression or by inhibiting the binding of transcriptional machinery to the DNA.
The epigenome plays a critical role during embryogenesis, when the proliferation of cellular diversity and transcriptional networks are calibrated by epigenetic processes that are reproduced in subsequent DNA replication and cell division cycles (7). Because the rapid pace of fetal development exceeds the rate of development during any other stage of the life course, developmental plasticity and malleability are high, and experiences during this period may exert lasting effects on gene expression and subsequent tissue development, known as fetal programming (8). Furthermore, because tissue development occurs in a specific sequence from conception to maturity, adverse exposures during gestation are more likely to disrupt the timing of organogenesis in a manner that has long-lasting consequences for the health of the developing fetus (8). For example, maternal malnutrition, inflammation, and other sources of prenatal stress may contribute to fetal growth restriction and preterm birth, with damage to peripheral organs occurring to protect the developing brain (8, 9). Consequently, the timing of environmental exposures in early life may play a crucial role in the biological embedding of adverse experiences (10, 11).
Recent quantitative tests linking early-life epigenetic programming with trajectories of aging (12, 13) have been bolstered by discoveries in epigenetic profiling and machine learning technologies that facilitated the development of epigenetic aging measures or epigenetic clocks that can accurately track biological aging in utero and across the life course (14–26). Epigenetic clocks are calculated by taking the genome-wide weighted average of DNAm levels at CpG sites that are highly associated with either chronological age (first generation clocks) or phenotypic hallmarks of aging (second generation clocks). Epigenetic clocks accurately predict chronological age (14, 22–24, 27, 28), and numerous studies have linked deviations between DNAm age and chronological age, i.e., epigenetic age acceleration (EAA), with age-related diseases and mortality (29–40), suggesting EAA measures may serve as molecular biomarkers of aging that reflect both resilience and vulnerability during the aging process. More recently, pace of aging measures have been developed using the change in 18 age-associated biomarkers in the same cohort of individuals over time, as opposed to cross-sectional measurements in different individuals (21, 41). Second generation clocks and pace of aging measures have shown more consistent associations with socioeconomic disparities across the life course and are more predictive of morbidity and mortality than first generation clocks that were developed to predict chronological age (42–45). Epigenetic aging measures tend to outperform other biomarkers of aging in predicting lifespans (19, 20, 46), and their correlations with age-related conditions makes them useful in a variety of contexts, including anti-aging interventions (18).
However, despite these advances in molecular aging research, existing causal evidence on the long-term impacts of early-life epigenetic programming in humans at a population level is rare. In addition, few studies have looked at the impact of environmental exposures at different time points in childhood to identify sensitive periods in development in which environmental insults could have their greatest impact on long-term epigenetic signatures. Causal research to date on fetal epigenetic programming is limited to a small handful of natural experiments, including the Dutch Hunger Winter Families Study (47–49), Project Ice Storm (50–53), and research on Holocaust survivors (54), that examined the impact of maternal nutrition or stress on DNAm changes in preselected CpG regions that regulate specific genes. These findings suggested that adverse maternal environments early in human gestation could result in persistent changes in epigenetic information in adulthood, particularly with respect to the regulation of metabolic, immune, or neurological pathways.
In this study, we use a quasi-natural experiment that leverages state-year variation in economic conditions during the Great Depression to examine the causal effect of environmental exposures during the prenatal period on late-life epigenetic aging signatures for participants in the US Health and Retirement Study (HRS). This approach compares individuals whose in utero development overlapped relatively worse economic conditions with peers born in the same year and state whose in utero development overlapped relatively better economic conditions because they were born at different times of the year. For example, an individual born in February of 1933 would have been more exposed prenatally to economic conditions in 1932 compared to an individual born in November of 1933 in the same state. Additionally, by linking state-year macroeconomic data on wages, employment, and consumption to the first 16 years of HRS participants’ lives, we were able to condition on economic exposures during the preconception, postnatal, and childhood periods to assess the relative impact of exposure timing at different developmental stages on accelerated aging. HRS is the first population-representative study in the United States to collect CpG-level data that has the sample size and geographic variation necessary to exploit quasi-random variation in economic conditions across states and over time, which expands possibilities for causal research in epigenetics.
We focus on the Great Depression for several reasons. First and foremost is the magnitude of the exposure. The Great Depression was the most devastating macroeconomic recession in US history: from 1929 to 1933, real output contracted by more than 25%, prices fell by 33%, and the unemployment rate increased from 3.2 to 25%, reaching the highest levels ever documented in the United States (55). The extreme nature of the economic shock was a unique failure of the industrial economy that had devastating effects on individuals’ financial and overall well-being (55, 56). Second, at the time, there were few social welfare programs to ameliorate the widespread economic devastation families experienced. The science of prenatal care was in its infancy, and women lacked access to prenatal vitamins or other nutritional supplements that are now considered vital for fetal development, further exacerbating nutritional deprivation and stressful living conditions for expectant mothers and their children. Third, with respect to study design, because the HRS initially surveyed over 12,000 individuals who were born between 1931 and 1941 when the study began in 1992, we can examine epigenetic aging patterns in a relatively large, population-representative sample of surviving cohort members who had their blood drawn in 2016 (n = 832).
Our findings suggest that exposure to economic conditions during the Great Depression had lasting impacts on epigenetic aging signatures, and that these effects were salient in both magnitude and statistical significance for in utero exposures only, strongly suggesting the existence of sensitive periods of development. Results are specific to next-generation epigenetic aging measures, including GrimAge EAA and the DunedinPoAm (Dunedin(P)ace(o)f(A)ging(m)ethylation) measure, both of which incorporate more complex phenotypes into DNAm algorithms that are trained on clinical outcomes and mortality risk (GrimAge) or on the rate of change in system-integrity biomarkers (DunedinPoAm). A one SD (SD) decrease in wages during the 1930s, which is equal to roughly half of the overall decline in wages experienced during the Great Depression, resulted in ∼0.4 SD increase in accelerated epigenetic aging when participants were between the ages of 75 and 84. Results are consistent but attenuated after adjusting estimates for mortality selection using an inverse probability weighted (IPW) estimator (57), suggesting individuals that survived to older ages appear to be positively selected. Finally, we show that compared to other aging and health phenotypes, accelerated GrimAge and DunedinPoAm signatures are more sensitive indicators of both early-life exposure to economic conditions and subsequent mortality, which suggests epigenetic aging measures may contain additional valuable information that could further our understanding of the causes of social disparities in aging and health span.
Results
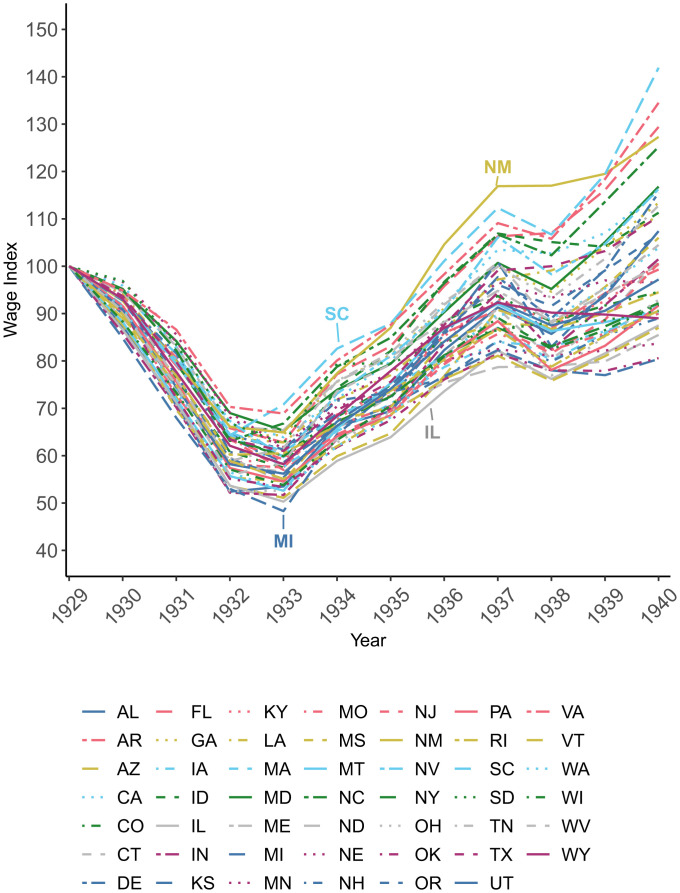
To conduct our analyses, we linked individual-level epigenetic aging measures from the HRS that were profiled in 2016 with macroeconomic data at the state- and year-of-birth level (SI Appendix, Section 1 and Table S1provides more details on the epigenetic aging measures used in this study). Annual state-level data that document the dynamics of the macroeconomy in the 1920s and 1930s are rare. Our preferred exposure measure is a wage index from the Bureau of Economic Analysis (BEA) because it includes both farm and nonfarm wages, which better approximates the economic conditions of families with young children in urban and rural areas (58). In addition, the data are available from 1929 to 1956, which allows us to test the impact of wage fluctuations prenatally through adolescence for individuals born in the 1930s. Fig. 1 documents the variation in the wage index across states relative to 1929 that we exploit in our analysis. Relevant summary statistics for the sample are reported in SI Appendix, Table S2.
Fig. 1.
Variation in the wage index across states, 1929 to 1940. The figure shows fluctuations in nominal, unadjusted farm and nonfarm wages and salaries relative to 1929. Data were obtained from the Bureau of Economic Analysis (BEA) in index form (SAINC7H Wages and Salaries by Industry [Historical] 1929 to 1957).
Table 1 presents results from our baseline specification, which showcases the impact of state-level wages in utero on six epigenetic aging measures constructed from DNAm data profiled when HRS participants born in the 1930s were between the ages of 75 and 84. Comparison across multiple epigenetic aging measures is important for the interpretation of our results because each algorithm was developed using different assumptions that capture different aspects of the biological aging process. As evidence of this, corresponding age-adjusted EAA measures are not highly correlated in the HRS sample (r = 0.069 to 0.605), although we do see stronger correlations among first generation clocks (r = 0.435 to 0.605) and next generation GrimAge and DunedinPoAm measures (r = 0.551) (SI Appendix, Fig. S1) (59). This correlative structure is reflected in Table 1, which indicates that declines in wages during the Great Depression had long-term impacts on GrimAge and DunedinPoAm epigenetic aging signatures specifically as opposed to first generation measures. A 1 SD decline in the wage index increased GrimAge EAA by 0.380 SD and decreased the pace or rate of aging as measured by DunedinPoAm by 0.449 SD (Bonferroni corrected P value < 0.05). To put the magnitude of these effects into perspective, a 1 SD decline in wages is equivalent to approximately half of the overall decline in wages that occurred between 1929 and 1933. The magnitude and significance of these results are robust across empirical specifications (SI Appendix, Table S3).
Table 1.
Effect of wage index declines in utero on EAA and pace of aging measures
| Horvath EAA | SkinBlood EAA | Hannum EAA | PhenoAge EAA | GrimAge EAA | Dunedin- PoAm | |
|---|---|---|---|---|---|---|
| Wage index declines in utero | −0.0868 | 0.0635 | 0.0617 | 0.0248 | 0.3804† | 0.4489† |
| (0.1607) | (0.2112) | (0.1960) | (0.1593) | (0.1056) | (0.1428) | |
| Observations | 832 | 832 | 832 | 832 | 832 | 832 |
| R2 | 0.152 | 0.125 | 0.167 | 0.156 | 0.250 | 0.119 |
Note: The table reports effect sizes from analyses of the association between the wage index and six measures of epigenetic aging. Results are reported in SD units of the aging measure per SD unit of the wage index in utero, interpretable as Pearson's r. The signs on the effect sizes have been flipped so that values correspond to a SD decline in the wage index. The wage index was transformed to real wages using the consumer price index. Robust standard errors clustered at the state of birth level are in parentheses. A cross (†) indicates a significant P value after Bonferroni correction for 6 independent tests at a family-wise error rate of 0.05 (P < 0.0083). All models control for sex, race, maternal education, year-of-birth (YOB) fixed effects (FE), state-of-birth FE, region of birth-specific linear time trends (LTT), and state-level controls interacted with YOB LTT, including the 1928 infant mortality rate, the 1929 maternal mortality rate, and whether a state's share of farmland was in the 75th percentile nationally in 1930. The model also includes YOB FE interacted with an indicator for whether state employment in manufacturing was in the 75th percentile nationally in 1929. Models were estimated using linear regression with weights provided by the HRS for the Venous Blood Study (VBS) sample.
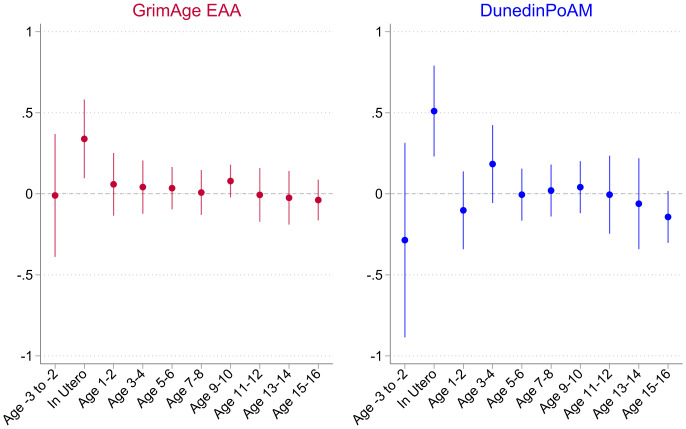
To determine the extent to which these effects were driven by in utero exposures, we estimated a model that conditioned on exposures from the preconception period through age 16. Fig. 2 plots the age-specific exposure coefficients from this specification for GrimAge EAA and DunedinPoAm. Coefficients are significant for the in utero period only and are similar in magnitude and significance to our baseline results. Importantly, we do not see any evidence of differential trends prior to conception, which is given by the null effect of the wage index 2 to 3 years prior to birth, providing support for the identification strategy.
Fig. 2.
Effect of wage index declines during the preconception, in utero, childhood, and adolescent periods on GrimAge epigenetic age acceleration (EAA) and DunedinPoAm. Note: The figure reports estimated coefficients from a model that conditions on exposures to the wage index in the preconception, prenatal, childhood, and early adolescent periods for GrimAge EAA and DunedinPoAm as described in Eq. 3 (n = 832). Results are reported in SD units of the aging measure per SD unit of the wage index in utero, interpretable as Pearson's r. The signs on the effect sizes have been flipped so that values correspond to a one SD decline in the wage index. The wage index was transformed to real wages using the consumer price index. All models control for sex, race, maternal education, year-of-birth (YOB) fixed effects (FE), state-of-birth FE, region of birth-specific linear time trends (LTT), and state-level controls interacted with YOB LTT, including the 1928 infant mortality rate, the 1929 maternal mortality rate, and whether a state's share of farmland was in the 75th percentile nationally in 1930. The model also includes YOB FE interacted with an indicator for whether state employment in manufacturing was in the 75th percentile nationally in 1929. Models were estimated using linear regression with weights provided by the HRS for the Venous Blood Study (VBS) sample. Robust 95% confidence intervals. Standard errors are clustered at the state of birth level.
The salience of exposures during the in utero period is also evident when we use other available state-level data on macroeconomic conditions, including employment and car sales (SI Appendix, Tables S4 and S5). Employment data reflect labor market fluctuations in manufacturing and nonmanufacturing industries (60) and data on car sales proxy household consumption (61). SI Appendix, Figs. S2 and S3 depict state-year variation relative to 1929 for the employment and car sales indices. Since these data were not available after 1940, the models are estimated in a subset of individuals with three years of exposure data before birth and two years after birth (n = 588). Findings are comparable to results with the wage index. Thus, our findings are not specific to wages but appear to reflect a more consistent pattern between adverse economic conditions in utero and accelerated biological aging.
Sensitivity Analysis.
Because DNAm was profiled in whole blood, DNAm measures of aging may reflect differences in the white blood cell (WBC) composition of samples from which the DNA were extracted. Since the relative composition of WBCs changes with age, we tested the sensitivity of our analysis to this variation by adjusting for the percentage of WBCs present and their interactions with year of birth fixed effects (SI Appendix, Table S6). Adjusting for WBC composition reduced, but did not fully mediate, the magnitude and significance of our results, suggesting our findings may be driven more by extrinsic epigenetic age acceleration (EEAA) as opposed to intrinsic epigenetic age acceleration (IEAA). The EEAA terminology has been used to refer to the observation that clocks trained in whole blood may be more reflective of immune system aging or age-related changes in leukocyte composition that have been more closely linked to metabolic health and environmental stressors, whereas multitissue clocks like the Horvath clock that adjust for cell composition are more reflective of cell-intrinsic aging (62, 63).
Additionally, results do not appear to be driven by outliers, or individuals in the top and bottom 1% of the GrimAge EAA and DunedinPoAm distributions (SI Appendix, Table S7). Finally, because DNAm is in part regulated by genetic polymorphisms, we confirmed that our results are robust to confounding from population stratification in a subsample of European ancestry individuals by adjusting for the first 10 principal components of the genetic data and their interaction with the treatment (SI Appendix, Table S8).
Impacts of Other Co-Occurring Historical Events.
The 1930s and 1940s were a historically rich period characterized by several coinciding events. We analyzed our results in the context of the Dust Bowl, New Deal relief spending, the spread of rural electrification, World War II mobilization rates, and variation in extreme weather conditions to determine the degree to which they may be influencing our estimates (SI Appendix, Tables S9–S13). Overall, our results do not appear to be driven by these co-occurring events, suggesting that economic fluctuations from the Great Depression had an independent effect on biological aging.
Early-Life Exposure to Economic Shocks and Old-Age Mortality.
Since DNAm was profiled in our sample at older ages, we conducted additional analyses to understand how mortality selection may be biasing our estimates. While studies have shown that economic conditions in early life can affect mortality (64, 65), evidence on the long-term impacts of the Great Depression on mortality has been mixed (66–69). We re-examined these patterns in the HRS by regressing age-specific survival probabilities on wage index declines for all respondents born between 1929 and 1940 (n = 7,898). Results reveal a negative and significant association between worsening economic conditions at birth and the probability of survival from age 75 onwards (SI Appendix, Table S14), which overlaps with the age range of our sample in 2016 when epigenetic profiling was conducted. Survival probabilities are also positively linked to higher maternal education (a key proxy of family resources in childhood) (SI Appendix, Table S14). Regarding the cause of death, earlier mortality appears to be driven primarily by metabolic disorders, which have been linked to intrauterine growth disruptions (SI Appendix, Table S15) (5, 47–49). Taken together, these results suggest that our sample is positively selected for survival at older ages.
To investigate the extent that mortality is biasing our estimates, we used fitted values from regression models of survival as inverse probability weights to adjust our estimates so they are more reflective of the HRS sample just prior to mortality selection (SI Appendix, section 9 provides more details) (57, 70). Survival was modeled using a probit specification under two scenarios: 1) survival until age 75 (the age that we first observe mortality selection in our sample), and 2) survival until 2016 (the year epigenetics were profiled in the HRS). For both scenarios, we present inverse probability weighted (IPW) estimates that use weights constructed with and without adjustments for maternal education in the survival model (Table 2). After adjusting for mortality selection using the IPW estimator, results are attenuated by ∼9% to 38%, which indicates that conditional on survival into the HRS, mortality selection appears to be biasing our estimates downward.
Table 2.
Effect of wage index declines in utero on GrimAge EAA and DunedinPoAm, IPW estimates
| Outcome: GrimAge EAA | ||||
|---|---|---|---|---|
| Survival to 2016 | Survival to age 75 | |||
| IPW, no maternal education | IPW, with maternal education | IPW, no maternal education | IPW, with maternal education | |
| Wage index declines in utero | 0.2657* | 0.2602* | 0.3167** | 0.3131** |
| (0.1000) | (0.0996) | (0.0969) | (0.0965) | |
| Outcome: DunedinPoAm | ||||
| Survival to 2016 | Survival to age 75 | |||
| IPW, no maternal education | IPW, with maternal education | IPW, no maternal education | IPW, with maternal education | |
| Wage index declines in utero | 0.3877** | 0.3877** | 0.4081** | 0.4081** |
| (0.1224) | (0.1224) | (0.1428) | (0.1428) | |
Note: The table reports effect sizes from analyses of the association between the wage index and GrimAge EAA and DunedinPoAm using inverse probability weights (n = 832). Estimates are from separate linear regressions. Results are reported in SD units of the aging measure per SD unit of the wage index in utero, interpretable as Pearson's r. The signs on the effect sizes have been flipped so that values correspond to a 1 SD decline in the wage index. The wage index was transformed to real wages using the consumer price index. Weights were calculated by taking the inverse of fitted values from probit models that adjusted for the wage index in utero, year- of-birth fixed effects (FE), state-of-birth FE, sex, and race. Weights applied in Columns 2 and 4 were derived from probit models that also accounted for maternal education and its interaction with the wage index, where maternal education was a dichotomous variable equal to one if the respondent’s mother had no degree or maternal education was missing and zero otherwise. The first two columns report estimates that use IPW weights to adjust for survival until 2016, and the second two columns use IPW weights to adjust for survival until age 75. Results are from the fully specified model (see Table 1 footnote for model details). Robust standard errors clustered at the state of birth level are in parentheses. *P < 0.05, **P < 0.01.
Economic Shocks and Changes in Fertility and Mortality at Birth.
Business cycles can affect fertility due to changes in income and/or the opportunity cost of time (71, 72). If fertility responses vary across groups, e.g., if more educated women had more children because of the shock, this may bias our estimates downward as children of more advantaged mothers may experience less extreme nutritional deprivation or stress-related hardships while pregnant. Using data from the 1% representative sample of the 1940 Census, we examined whether declines in the wage index are associated with the number of household births in the 1930s and/or whether this association varies by social class or other demographic characteristics. Results suggest that as wages declined, women without a degree or at most a high school degree had fewer children than college educated women, suggesting a small but positive selection on fertility (SI Appendix, Table S16). Similarly, worsening economic conditions may have affected the probability that a pregnancy was carried to term, particularly for male fetuses, who are more susceptible to disease or death than females (73, 74). Data from the 1940 Census indicate that a 1 SD decline in the wage index reduced both cohort size and the male-to-female sex ratio of births by 7% and 12%, respectively (SI Appendix, Table S17), suggesting antenatal selection may be another key channel through which our estimates are downwardly biased.
Effects on Other Aging Outcomes and Longevity.
Finally, we tested whether in utero exposure to wages is predictive of other self-reported or doctor diagnosed measures of aging in our sample, including frailty, metabolic syndrome, self-reported health status (SRHS), and the number of chronic disease conditions (SI Appendix, Table S18). These measures are less precise and, apart from the number of chronic disease conditions, the magnitude of their effect sizes (per 1 SD decline in the wage index) are ∼40% to 50% lower relative to GrimAge EAA and DunedinPoAm effect sizes. We then examined the degree to which these measures are associated with the probability of dying in the following HRS wave (∼7% of our sample died between 2016 and 2018). GrimAge EAA displays the strongest association in terms of both magnitude and significance ( = 0.173 SD increase in the probability of death per 1 SD increase in GrimAge; P < 0.001), followed by SRHS (=0.154 SD; P < 0.001), DunedinPoAm (=0.101 SD; P < 0.05), and the number of chronic disease conditions (=0.077 SD; P < 0.05) (SI Appendix, Table S19). Thus, GrimAge and DunedinPoAm appear to be more sensitive indicators of both in utero exposures to the wage index and subsequent longevity than other commonly used measures of aging and health, suggesting they contain additional information on the connection between social disparities in early-life environments and mortality.
Discussion
In a population-representative sample of over 800 individuals born in the 1930s, we find a significant association between early-life exposure to economic conditions during the Great Depression and late-life epigenetic age acceleration as captured by the GrimAge and DunedinPoAm algorithms. These findings are robust across empirical specifications that account for additional state-level controls and region-specific linear time trends, supporting a causal interpretation. Using the few available sources of state-level variation in macroeconomic conditions from the 1930s, we show that these results are not sensitive to how the economic shock was measured but rather reflect a consistent pattern across changes in wages, consumption, and employment. After evaluating endogenous shifts in mortality and fertility related to Depression-era birth cohorts, we conclude that these effects likely represent lower bound estimates of the true impacts. Finally, we shed light on the existence of sensitive periods, which is increasingly acknowledged but often difficult to test empirically (10, 11), by demonstrating that these effects were isolated to the in utero period specifically.
We did not identify any effects for the Horvath, SkinBlood, Hannum, or PhenoAge clocks, which is consistent with prior research that found stronger associations between socioeconomic disadvantage and GrimAge EAA and DunedinPoAm (42, 45, 75). Because epigenetic aging measures are composite indicators that are comprised of many different DNAm patterns, a major drawback of their application is a lack of mechanistic understanding of what they are capturing, both in terms of how environmental processes may be initiating these changes as well as their connection to disease etiology (76, 77). Of note, a recent study deconstructed over 5,000 clock CpGs into twelve distinct submodules that display different biological underpinnings and vary considerably in their proportion across clocks (76). GrimAge and DunedinPoAm, which were trained in whole blood to predict mortality or physiological changes with aging, share a very similar composition of submodules that are stronger predictors of mortality and cardiovascular related outcomes. Likewise, while the Horvath, PhenoAge, SkinBlood, and Hannum clocks, which were all trained in some manner on chronological age, are also comprised of mortality-associated modules, they contain additional submodules that have weak or inverse associations with mortality (76). This suggests a connection between our findings and mortality or cardiovascular risk as opposed to tumorigenesis or other age-related cellular processes that are captured more strongly by other clocks.
Overall, it is difficult to disentangle whether the connection between in utero exposures and accelerated biological aging later in life that we observe is operating from epigenetic alterations induced during early development that result in consistently higher incidence of damage throughout life (i.e., fetal programming) (4, 5) or epigenetic signs of aging processes that are accelerated by insults in utero but that continue to develop across the life course (i.e., the idea of high initial damage load or the HIDL hypothesis) (2). In both cases, any downstream consequences of early-life insults will not be readily apparent until we can observe aging at a phenotypic level when progressive accumulation of damage and loss of physiological integrity begin to take hold later in life. Thus, although we cannot use the clocks to disentangle specific mechanistic pathways, we show that they may be particularly sensitive indicators of early-life programming and subsequent mortality risk at later ages. Moreover, this appears to be the case in a subsample of relatively healthier, surviving cohort members that outlived their counterparts. More research is needed, but these results suggest that composite measures of epigenetic aging may be especially useful for the detection of disparities in aging prior to the emergence of disease or death.
Along these lines, our findings diverge from prior work on the long-term health effects of the Dust Bowl and the Great Depression that were not able to detect a relationship between in utero exposures and an array of health outcomes in the 1992 to 2004 HRS waves (69). In part, we hypothesize that differences in the authors’ empirical strategy, which relied on exploiting variation in economic conditions at the region- and year-of-birth level, may have masked substantial heterogeneity in economic activity at the state level that affected the precision of their results. However, effects may also be biased downward due to the use of self-reported measures that either suffered from measurement error or were not able to detect more subtle differences in aging that are connected to in utero insults.
Some limitations of our study are worth mentioning. First, because monthly macroeconomic data at the state level were not collected in the 1930s, we were not able to identify specific time windows or trimesters during pregnancy that may have been especially vulnerable to economic shocks. Second, we cannot identify why and how aggregate economic exposures were affecting the fetal environment, or if our results were driven more by nutritional deprivation, maternal stress, a depletion of economic resources, or a combination of these factors. Future research will be better poised to address these limitations as the cost of epigenetic profiling continues to fall, enabling longitudinal collection in larger population-representative studies and field experiments.
Materials and Methods
Data.
The HRS.
The HRS is a nationally representative, biannual, longitudinal panel study of individuals over the age of 50 and their spouses that began in 1992. The study is sponsored by the National Institute on Aging (NIA U01AG009740) and is conducted by the University of Michigan (78). Comprehensive information about participants’ socioeconomic background, income, assets, and employment is collected from the time of respondent entry until death. The HRS introduces a new cohort of participants every 6 years and interviews around 20,000 participants every 2 years.
DNA methylation data were collected as part of the 2016 HRS Venous Blood Study (VBS). The DNAm sample is racially and socioeconomically diverse and representative of the full HRS sample (79). In sensitivity analyses, we adjusted for cell-type proportions using results from a WBC differential assay (80). Demographic and socioeconomic data were taken from the RAND HRS Longitudinal File 2018 (V1) (81). Information on cause of death was taken from HRS exit interview files (82).
State-level measures of economic conditions.
The following available measures were linked to HRS participants at the year-of-birth and state-of-birth levels: 1) Wage Index (1929-1956): farm and nonfarm wages and salaries from the Bureau of Economic Analysis (BEA) (58); 2) Employment Index (1929-1940): employment in manufacturing and nonmanufacturing sectors (60); 3) Car Sales Index (1929-1940): total number of car sales from the annual statistical issues of the industry trade publication, Automotive Industries (61). Measures were converted into indices by dividing the variable by its 1929 level and multiplying by 100 so each state has a value of 100 in 1929. For all analyses, we transformed the wage index to real wages using the consumer price index (base year = 2011).
In utero exposure measure.
Calendar year does not always correspond well with the prenatal period, however monthly state-level macroeconomic data were not available in the 1930s. To generate a more precise measure, we constructed a weighted average of in utero exposure as follows:
| [1] |
Where and reflect the approximate number of months individual spent in utero in and relative to their month of birth, and values of the wage index are assigned according to ’s state () and year of birth (). For example, an individual born in March received an in utero wage index equal to . Since we do not know the exact number of months spent in utero, our exposure variable is still subject to measurement error; however, our results are larger in magnitude and more precise than in utero measures that correspond to quarter of birth (SI Appendix, Table S20).
Epigenetic aging measures.
We used six different epigenetic clocks and pace of aging measures that were constructed by the HRS from individual CpG-level data and are publicly available (79). Epigenetic age acceleration (EAA) was computed by using the residuals from regressions of each clock on chronological age. Residualization was not applied to DunedinPoAm since it already quantifies deviations in chronological age from the expected sample norm. SI Appendix, Table S1 provides more details on the methods and number of CpG sites that were used to compute epigenetic aging measures according to author-specific algorithms.
Empirical Framework.
Our baseline specification is as follows:
| [2] |
where is the epigenetic age acceleration outcome in 2016 for individual i, born in state s in year c. Wages represents the aggregate wage index at the state and year levels for the in utero period as defined in Eq. 1. The matrix contains individual characteristics at baseline including sex, race, and dichotomous indicators for maternal education (no degree and high school degree, omitted category is college degree). To avoid attrition bias from listwise deletion we also include a dichotomous indicator for missing maternal education. is a vector of state-level characteristics around 1930 interacted with year of birth, including the maternal mortality rate in 1929 (83), the infant mortality rate in 1928 (84), and whether the percent of farmland in a state was above the 75th percentile nationally in 1930 (85). The term represents a state’s share of wage earners in manufacturing in 1929 (86, 87) interacted with year of birth fixed effects. We include these controls because the severity of cyclical fluctuations across states was driven in part by state differences in the proportion of manufacturing and agricultural industries (88). The terms θs and ηc are state- and year-of-birth fixed effects, respectively. The geographic fixed effects help absorb time-invariant differences at the state level, while the time fixed effects absorb factors that vary over time but are invariant across states. To control for changes in regional conditions throughout the 1930s we include region-specific linear time trends, or the term . Robust standard errors are clustered at the state-of-birth level. All models were estimated using HRS sample weights for the 2016 VBS sample to adjust for sample composition. In tables and figures, the coefficient on the wage index was flipped so that higher values correspond to wage declines. Results are reported in SD units of the aging measure per SD unit of the wage index in utero, interpretable as Pearson's r.
Coefficients reported in Fig. 2 are from our second specification:
| [3] |
This model is identical to the baseline specification in Eq. 2 except in addition to the in utero term, the model also conditions on a pretrend for average wages in years and and eight additional two-year averaged terms for state-level wage exposures when individuals were between the ages of one and sixteen.
Supplementary Material
Acknowledgments
The authors thank Jason Fletcher, Dan Belsky, Dalton Conley, and Josh Hausman for their valuable feedback, Liz Duffie and Vikas Gawai for their research assistance, HRS investigators and staff for their contributions, and HRS study participants for sharing their lives and making this research possible.
Footnotes
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120393119/-/DCSupplemental.
Data, Materials, and Software Availability
This study used restricted individual level information from the HRS and our contractual agreement does not permit public dissemination of the data. Details on how to access restricted HRS data can be found at https://hrs.isr.umich.edu/data-products/restricted-data (89). All code and publicly available, historical state-level data used in this study are posted on github: https://github.com/laurenschmitz/great-depression-epigenetic-aging (90).
Funding
Funding for this project was generously provided by the National Institute on Aging (K99 AG056599, R00 AG056599, P30 AG012846, P30 AG017265, and P30 AG017266), the Center for Retirement Research Steven H. Sandell Grant Program pursuant to a grant from the U.S. Social Security Administration (BC20-S2), and the University of Michigan Marshall Weinberg Endowment (G002832). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Social Security Administration or the National Institute on Aging.
References
- 1.López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavrilov L. A., Gavrilova N. S., Early-life programming of aging and longevity: The idea of high initial damage load (the HIDL hypothesis). Ann. N. Y. Acad. Sci. 1019, 496–501 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Gladyshev V. N., The ground zero of organismal life and aging. Trends Mol. Med. 27, 11–19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker D. J., The fetal and infant origins of adult disease. BMJ 301, 1111 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker D. J., Mothers, Babies and Health in Later Life (Churchill Livingstone, ed. 2, 1998). [Google Scholar]
- 6.Perera B. P. U., Faulk C., Svoboda L. K., Goodrich J. M., Dolinoy D. C., The role of environmental exposures and the epigenome in health and disease. Environ. Mol. Mutagen. 61, 176–192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyce W. T., Kobor M. S., Development and the epigenome: The ‘synapse’ of gene-environment interplay. Dev. Sci. 18, 1–23 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Vohr B. R., Poggi Davis E., Wanke C. A., Krebs N. F., Neurodevelopment: The impact of nutrition and inflammation during preconception and pregnancy in low-resource settings. Pediatrics 139 (suppl. 1), S38–S49 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Sandman C. A., Glynn L. M., Davis E. P., “Neurobehavioral consequences of fetal exposure to gestational stress” in Fetal Development, Reissland N., Kisilevsky B. S., Eds. (Springer International, 2016), pp. 229–265. [Google Scholar]
- 10.Boyce W. T., Levitt P., Martinez F. D., McEwen B. S., Shonkoff J. P., genes, environments, and time: The biology of adversity and resilience. Pediatrics 147, e20201654 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Boyce W. T., Sokolowski M. B., Robinson G. E., Genes and environments, development and time. Proc. Natl. Acad. Sci. U.S.A. 117, 23235–23241 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerepesi C., Zhang B., Lee S.-G., Trapp A., Gladyshev V. N., Epigenetic clocks reveal a rejuvenation event during embryogenesis followed by aging. Sci. Adv. 7, eabg6082 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinzina E. D., Podolskiy D. I., Dmitriev S. E., Gladyshev V. N., Patterns of aging biomarkers, mortality, and damaging mutations illuminate the beginning of aging and causes of early-life mortality. Cell Rep. 29, 4276–4284.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Bocklandt S., et al. , Epigenetic predictor of age. PLoS One 6, e14821 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garagnani P., et al. , Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell 11, 1132–1134 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Yang Z., et al. , Correlation of an epigenetic mitotic clock with cancer risk. Genome Biol. 17, 205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., et al. , DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat. Commun. 8, 14617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath S., et al. , Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging (Albany NY) 10, 1758–1775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine M. E., et al. , An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 10, 573–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu A. T., et al. , DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 11, 303–327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belsky D. W., et al. , Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. eLife 9, e58470 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvath S., DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannum G., et al. , Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weidner C. I., et al. , Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 15, R24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Q., et al. , DNA methylation levels at individual age-associated CpG sites can be indicative for life expectancy. Aging (Albany NY) 8, 394–401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vidal-Bralo L., Lopez-Golan Y., Gonzalez A., Simplified assay for epigenetic age estimation in whole blood of adults. Front. Genet. 7, 126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horvath S., et al. , Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 13, R97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Florath I., Butterbach K., Müller H., Bewerunge-Hudler M., Brenner H., Cross-sectional and longitudinal changes in DNA methylation with age: An epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum. Mol. Genet. 23, 1186–1201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marioni R. E., et al. , DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 16, 25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christiansen L., et al. , DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell 15, 149–154 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horvath S., Levine A. J., HIV-1 infection accelerates age according to the epigenetic clock. J. Infect. Dis. 212, 1563–1573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horvath S., Ritz B. R., Increased epigenetic age and granulocyte counts in the blood of Parkinson’s disease patients. Aging (Albany NY) 7, 1130–1142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine M. E., Lu A. T., Bennett D. A., Horvath S., Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging (Albany NY) 7, 1198–1211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine M. E., et al. , DNA methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging (Albany NY) 7, 690–700 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perna L., et al. , Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin. Epigenetics 8, 64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horvath S., et al. , Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging (Albany NY) 7, 1159–1170 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breitling L. P., et al. , Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin. Epigenetics 8, 21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marioni R. E., et al. , The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int. J. Epidemiol. 44, 1388–1396 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horvath S., et al. , Obesity accelerates epigenetic aging of human liver. Proc. Natl. Acad. Sci. U.S.A. 111, 15538–15543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horvath S., et al. , Accelerated epigenetic aging in Down syndrome. Aging Cell 14, 491–495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belsky D. W., et al. , DunedinPACE, a DNA methylation biomarker of the pace of aging. eLife 11, e73420 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitz L. L., et al. , The socioeconomic gradient in epigenetic ageing clocks: Evidence from The Multi-Ethnic Study of Atherosclerosis and The Health and Retirement Study. Epigenetics 17, 1–23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raffington L., et al. , Socioeconomic disadvantage and the pace of biological aging in children. Pediatrics 147, e2020024406 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graf G. H., et al. , Testing black-white disparities in biological aging among older adults in the United States: Analysis of DNA-methylation and blood-chemistry methods. Am. J. Epidemiol. 191, 613–625 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crimmins E. M., Thyagarajan B., Levine M. E., Weir D. R., Faul J., Associations of age, sex, race/ethnicity, and education with 13 epigenetic clocks in a nationally representative U.S. sample: The Health and Retirement Study. J. Gerontol. A Biol. Sci. Med. Sci. 76, 1117–1123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verschoor C. P., et al. , Comparing biological age estimates using domain-specific measures from the Canadian longitudinal study on aging. J. Gerontol. A Biol. Sci. Med. Sci. 76, 187–194 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heijmans B. T., et al. , Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. U.S.A. 105, 17046–17049 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tobi E. W., et al. , Early gestation as the critical time-window for changes in the prenatal environment to affect the adult human blood methylome. Int. J. Epidemiol. 44, 1211–1223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tobi E. W., et al. ; Biobank-based Integrative Omics Studies Consortium, DNA methylation as a mediator of the association between prenatal adversity and risk factors for metabolic disease in adulthood. Sci. Adv. 4, eaao4364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao-Lei L., et al. , DNA methylation signatures triggered by prenatal maternal stress exposure to a natural disaster: Project Ice Storm. PLoS One 9, e107653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao-Lei L., et al. , Pregnant women’s cognitive appraisal of a natural disaster affects DNA methylation in their children 13 years later: Project Ice Storm. Transl. Psychiatry 5, e515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao-Lei L., et al. , DNA methylation mediates the impact of exposure to prenatal maternal stress on BMI and central adiposity in children at age 13½ years: Project Ice Storm. Epigenetics 10, 749–761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao-Lei L., et al. , DNA methylation mediates the effect of exposure to prenatal maternal stress on cytokine production in children at age 13½ years: Project Ice Storm. Clin. Epigenetics 8, 54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yehuda R., et al. , Holocaust exposure induced intergenerational effects on FKBP5 methylation. Biol. Psychiatry 80, 372–380 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Temin P., “The Great Depression” in The Cambridge Economic History of the United States, Engerman S., Gallman R., Eds. (Cambridge University Press, 1996), pp. 301–328. [Google Scholar]
- 56.Terkel S., Hard Times, An Illustrated Oral History of the Great Depression (The New Press, 2012). [Google Scholar]
- 57.Solon G., Haider S. J., Wooldridge J. M., What are we weighting for? J. Hum. Resour. 50, 301–316 (2015). [Google Scholar]
- 58.U.S. Bureau of Economic Analysis, SAINC7H Wages and Salaries by Industry (Historical) 1929-1957. https://www.bea.gov/itable/regional-gdp-and-personal-income. Accessed 20 October 2022.
- 59.Belsky D. W., et al. , Eleven telomere, epigenetic clock, and biomarker-composite quantifications of biological aging: Do they measure the same thing? Am. J. Epidemiol. 187, 1220–1230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallis J. J., Employment in the Great Depression: New data and hypotheses. Explor. Econ. Hist. 26, 45–72 (1989). [Google Scholar]
- 61.Hausman J. K., Fiscal policy and economic recovery: The case of the 1936 Veterans’ Bonus. Am. Econ. Rev. 106, 1100–1143 (2016). [Google Scholar]
- 62.Horvath S., Raj K., DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371–384 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Quach A., et al. , Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 9, 419–446 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van den Berg G. J., Lindeboom M., Portrait F., Economic conditions early in life and individual mortality. Am. Econ. Rev. 96, 290–302 (2006). [DOI] [PubMed] [Google Scholar]
- 65.D. M. Cutler, W. Huang, A. Lleras-Muney, Economic conditions and mortality: Evidence from 200 years of data. National Bureau of Economic Research, No. w22690, (2016).
- 66.Tapia Granados J. A., Diez Roux A. V., Life and death during the Great Depression. Proc. Natl. Acad. Sci. U.S.A. 106, 17290–17295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stuckler D., Meissner C., Fishback P., Basu S., McKee M., Banking crises and mortality during the Great Depression: Evidence from US urban populations, 1929-1937. J. Epidemiol. Community Health 66, 410–419 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Fishback P. V., Haines M. R., Kantor S., Births, deaths, and New Deal relief during the Great Depression. Rev. Econ. Stat. 89, 1–14 (2007). [Google Scholar]
- 69.Cutler D. M., Miller G., Norton D. M., Evidence on early-life income and late-life health from America’s Dust Bowl era. Proc. Natl. Acad. Sci. U.S.A. 104, 13244–13249 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Domingue B. W., et al. , Mortality selection in a genetic sample and implications for association studies. Int. J. Epidemiol. 46, 1285–1294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Currie J., Schwandt H., Short- and long-term effects of unemployment on fertility. Proc. Natl. Acad. Sci. U.S.A. 111, 14734–14739 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sobotka T., Skirbekk V., Philipov D., Economic recession and fertility in the developed world. Popul. Dev. Rev. 37, 267–306 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Pongou R., Why is infant mortality higher in boys than in girls? A new hypothesis based on preconception environment and evidence from a large sample of twins. Demography 50, 421–444 (2013). [DOI] [PubMed] [Google Scholar]
- 74.I. Waldron, "Sex differences in infant and early childhood mortality: Major causes of death and possible biological causes" in Too Young to Die: Genes or Gender? (United Nations, 1998), pp. 64–83.
- 75.Raffington L., Belsky D. W., Integrating DNA methylation measures of biological aging into social determinants of health research. Curr. Environ. Heal. Rep. 9, 196–210 (2022). [DOI] [PubMed] [Google Scholar]
- 76.Levine M. E., Higgins-Chen A., Thrush K., Minteer C., Niimi P., Clock work: Deconstructing the epigenetic clock signals in aging, disease, and reprogramming. bioRxiv [Preprint] (2022). https://www.biorxiv.org/content/10.1101/2022.02.13.480245v1 (Accessed 12 August 2022).
- 77.Liu Z., et al. , Underlying features of epigenetic aging clocks in vivo and in vitro. Aging Cell 19, e13229 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sonnega A., et al. , Cohort profile: The health and retirement study (HRS). Int. J. Epidemiol. 43, 576–585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.E. Crimmins, J. K. Kim, J. Fisher, J. D. Faul, HRS Epigenetic Clocks – Release 1. Ann Arbor, MI: Survey Research Center, Institute for Social Research, University of Michigan, 2020.
- 80.E. Crimmins, J. D. Faul, B. Thyagarajan, D. R. Weir, Venous blood collection and assay protocol in the 2016 Health and Retirement Study. Ann Arbor, MI: Survey Research Center, Institute for Social Research, University of Michigan, 2017.
- 81.RAND HRS Longitudinal File 2018 (V1), produced by the RAND Center for the Study of Aging, with funding from the National Institute on Aging and the Social Security Administration. Santa Monica, CA (February 2021).
- 82.Health and Retirement Study (1996-2018 HRS Exit) public use dataset, produced and distributed by the University of Michigan with funding from the National Institute on Aging (grant number NIA U01AG009740). Ann Arbor, MI (2021).
- 83.Jayachandran S., Lleras-Muney A., Smith K. V., Replication data for: Modern medicine and the twentieth century decline in mortality: Evidence on the impact of sulfa drugs. Nashville, TN: Am. Econ. Assoc. [publisher] (2010). Ann Arbor, MI: Inter-university Consort. for Pol. Soc. Rese [distributor], 2019. 10.3886/E113743V1. [DOI]
- 84.D. Norton, "Data on infant mortality and births, 1920–1945". Available at https://www.nber.org/research/data/vital-statistics-births-and-infant-mortality-1920-1945. Deposited 22 January 2007. [Google Scholar]
- 85.U.S. Census Bureau, 1930 Census: Agriculture Volume 2. Reports by States, with Statistics for Counties and a Summary for the United States (1932). https://www.census.gov/library/publications/1932/dec/1930d-vol-02-agriculture.html. Accessed 20 October 2022.
- 86.U.S. Census Bureau, 1930 Census: Manufacturers, 1929 Volume 3. Reports by States. Statistics for Industrial Areas, Counties, and Cities (1933). https://www.census.gov/library/publications/1933/dec/1930f-vol-03-manufactures.html. Accessed 20 October 2022. [Google Scholar]
- 87.U.S. Census Bureau, 1930 Census: Volume 4. Occupations, by States. Reports by States, Giving Statistics for Cities of 25,000 or More. United States Summary. (1933). https://www.census.gov/library/publications/1933/dec/1930a-vol-04-occupations.html. Accessed 20 October 2022.
- 88.Borts G. H., Regional cycles of manufacturing employment in the United States, 1914–1953. J. Am. Stat. Assoc. 55, 151–211 (1960). [Google Scholar]
- 89.Health and Retirement Study, Cross-Wave Geographic Information (State) [1992-2018] restricted dataset. Produced and distributed by the University of Michigan with funding from the National Institute on Aging (grant number NIA U01AG009740). Ann Arbor, MI, (2022). [Google Scholar]
- 90.L. L. Schmitz, V. Duque, In Utero Exposure to the Great Depression is Reflected in Late-Life Epigenetic Aging Signatures. GitHub. https://github.com/laurenschmitz/great-depression-epigenetic-aging. Deposited 14 August 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study used restricted individual level information from the HRS and our contractual agreement does not permit public dissemination of the data. Details on how to access restricted HRS data can be found at https://hrs.isr.umich.edu/data-products/restricted-data (89). All code and publicly available, historical state-level data used in this study are posted on github: https://github.com/laurenschmitz/great-depression-epigenetic-aging (90).
Funding for this project was generously provided by the National Institute on Aging (K99 AG056599, R00 AG056599, P30 AG012846, P30 AG017265, and P30 AG017266), the Center for Retirement Research Steven H. Sandell Grant Program pursuant to a grant from the U.S. Social Security Administration (BC20-S2), and the University of Michigan Marshall Weinberg Endowment (G002832). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Social Security Administration or the National Institute on Aging.