Significance
The Wnt pathway is critical for regeneration of tissues such as lung, skin, and intestine. Using a cystine-knot peptide (CKP) drug-discovery platform, we identified a class of peptide agonists that bind to the Wnt receptor LRP6 and stimulate growth of stem cell–derived lung and human intestinal organoids. CKP agonists exhibit a unique mechanism of action because they enhance signaling only in the presence of a specific subset of Wnt proteins. As such, CKPs are differentiated from previously described antibody-based synthetic Wnt agonists and could provide a path for activating tissue-specific Wnt ligands, informing future therapeutic strategies, and possibly improving safety profiles in disease. Our findings advance the important frontier for designing next-generation agonists to modulate cellular signaling pathways.
Keywords: directed evolution, phage display, peptide, agonist, Wnt, stem cells, regeneration
Abstract
Developing peptide-based tools to fine-tune growth signaling pathways, in particular molecules with exquisite selectivity and high affinities, opens up opportunities for cellular reprogramming in tissue regeneration. Here, we present a library based on cystine-knot peptides (CKPs) that incorporate multiple loops for randomization and selection via directed evolution. Resulting binders could be assembled into multimeric structures to fine-tune cellular signaling. An example is presented for the Wnt pathway, which plays a key role in the homeostasis and regeneration of tissues such as lung, skin, and intestine. We discovered picomolar affinity CKP agonists of the human LPR6 receptor by exploring the limits of the topological manipulation of LRP6 dimerization. Structural analyses revealed that the agonists bind at the first β-propeller domain of LRP6, mimicking the natural Wnt inhibitors DKK1 and SOST. However, the CKP agonists exhibit a different mode of action as they amplify the signaling of natural Wnt ligands but do not activate the pathway by themselves. In an alveolosphere organoid model, the CKP agonists induced alveolar stem cell activity. They also stimulated growth in primary human intestinal organoids. The approach described here advances the important frontier of next-generation agonist design and could be applied to other signaling pathways to discover tunable agonist ligands.
Nature relies on a large number of highly specific interactions between ligands and cell surface receptors to transduce extracellular information, switch on intracellular signaling pathways, and control cellular functions. However, natural ligands (e.g., peptide hormones, neurotransmitters) are typically not suitable for use as tools or therapeutics because they are short-lived, and the scarcity of structural information limits an understanding of how receptor conformational states influence ligand function. Today, most of the approved peptide therapeutics are derived from natural peptide hormones (1–3), but they have been extensively optimized by medicinal chemistry approaches to improve their pharmaceutical properties.
While there has been tremendous progress in identifying bioactive peptides by phage display technologies, these ligands typically exhibit low affinities toward their intended targets, in part due to the predominantly linear nature of the peptide libraries available and thus lack of defined 3D conformations. Recent efforts to constrain the peptide backbone to improve affinity seem promising, but require the introduction of chemical modification steps both during and after the phage selection process (4–7). Moreover, the suitability of these scaffolds for drug development, in particular target affinity, stability, and solubility, remains unclear and often could require substantial optimization by medicinal chemistry to improve their target affinities.
Our strategy leverages cystine knot peptides (CKPs) as a starting molecular framework for ligand discovery because they provide many of the desired drug-like characteristics, including a three-dimensional (3D) conformation, exceptional stability, high solubility, and are obtained by both chemical synthesis and recombinant methods (8, 9). CKPs are short peptides, typically approximately 30 residues, with a characteristic motif of three “knotted” disulfide bridges. The first two disulfides form a ring and the third threads through it. The CKP framework offers five loops, which are attractive regions for randomization and selection by directed evolution methods. We postulated that the 3D conformation of CKPs would also enable high-affinity binding and exquisite selectivity, like antibody frameworks, due to the relatively large binding region they could occupy on the target protein. Recent discovery efforts have leveraged library-based approaches for the design or selection of cysteine-stabilized peptides. Examples include (1) computational methods to design peptides with defined structures (10, 11), (2) in silico modeling to evaluate the natural cysteine-rich scaffold repertoire followed by library construction and screening (12–14), or (3) selections from structure-guided random cysteine-rich libraries or chemically modified libraries (15, 16). Such approaches have successfully demonstrated that binders with high specificity and affinity could be identified from libraries enriched with a high diversity of starting scaffolds.
Here, we developed a CKP-based phage display library to discover small, targeted, high-affinity peptides based on the Ecballium elaterium trypsin inhibitor II (EETI-II) scaffold. Our library design was based on a single scaffold because it would provide high diversity and fast turnaround time for library construction (17, 18). We selected EETI-II as the scaffold of choice because it is small in size, with a molecular weight of less than 3.5 kDa; however, it exhibits high stability, solubility, and can be produced either recombinantly or by peptide synthesis (19–21). We reasoned that these characteristics would facilitate the isolation of well-behaved binders with drug-like properties that offer flexibility for manufacturing and further engineering for the development of therapeutics.
We applied the EETI-II-based phage display library to discover high-affinity surrogate antagonists and agonists of the cell surface Wnt co-receptor, low-density lipoprotein receptor–related protein 6 (LRP6). Wnt/β-catenin signaling is initiated when secreted extracellular Wnt proteins form a ternary complex with their cell surface receptors Frizzled (FZD) and LRP5/6, triggering an intracellular response that leads to the accumulation of β-catenin and subsequent transcription of Wnt target genes (22–26). In addition, several inhibitors such as Dickkopf-1 (DKK1) and sclerostin (SOST) regulate Wnt signaling by direct binding to LRP5/6 receptors (27–30). R-spondins (RSPO1 to 4) potentiate Wnt signaling by sequestering the cell surface ubiquitin ligases RNF43 and ZNRF3, two negative regulators of FZD receptors (31–33).
Wnt signaling is central to tissue homeostasis and regeneration in multiple organs (34, 35) and, as such, activation of Wnt signaling is an attractive therapeutic concept for regenerative medicine. Due to unfavorable pharmacologic properties, including poor stability, difficult production, and lack of isoform specificity, natural regulators of Wnt signaling do not lend themselves as therapeutic modalities (36). Artificial activators of Wnt signaling have been developed based on antibodies and other binding domains (37–40). These surrogates function by simultaneously binding to FZD and LRP receptors, thereby mimicking the function of natural Wnts. However, they do not synergize but rather compete with endogenous Wnt ligands to elicit a broad level of signal activation that is not Wnt isoform dependent. Therefore, it is critical to develop high-affinity synthetic agonists with exquisite selectivity and differentiated mechanisms of potentiation to regulate only specific branches of Wnt signaling. Activating tissue-specific single Wnt ligands or particular Wnt subclasses with synthetic ligands could be an effective therapeutic approach that may improve safety outcomes but also enable investigation into the specific roles of the individual Wnts and at the same time inform therapeutic strategies in disease.
The classical Wnt receptor LRP6 contains two distinct Wnt interaction epitopes that are located on the first two (E1-E2) and last two (E3-E4) β-propeller domains of the extracellular domain (41). This architecture offers the possibility of specifically modulating a subset of Wnt ligands by designing binders to one of these interaction epitopes. It also offers the opportunity to explore the limits of topological manipulation of LRP6 dimerization. We identified a high-affinity CKP that binds to the E1 domain of LRP6. This CKP acts as a WNT1 antagonist, while sparing the WNT3a binding site on LRP6; it also mimics DKK1 and SOST, with a slightly distinct mechanism of binding. To drive receptor clustering and activate Wnt signaling pathways, we engineered bivalent CKPs to dimerize LRP6 receptors and fine-tune their signaling output. This afforded a class of tunable agonist ligands that offer clear advantages over natural ligands, including synthetic accessibility, modularity, solubility, and high affinity. The dimeric CKPs potentiate WNT3a signaling, synergize with RSPO, induce stem cell activity in lung alveolospheres, and stimulate growth in primary human intestinal organoids. The overall concepts and technologies explored in this study provide valuable general insights to advance the important frontier of next-generation agonist design.
Results
Design and Construction of CKP Libraries for Targeted Peptide Discovery.
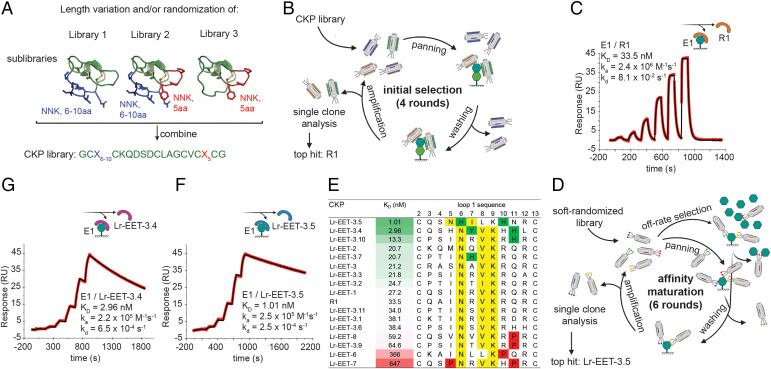
We aimed to display CKP frameworks on the surface of M13 bacteriophages and then use directed evolution to identify CKPs with biological properties that are different from those of the parent framework. The multiple exposed surface loops constitute amino acid stretches that are ideal for randomization to create specific binding interfaces to targets. We selected the EETI-II, a 28-amino acid CKP (20, 42), as the parental framework for building the phage library. EETI-II is accessible by both chemical synthesis and recombinant methods. Moreover, the loop 1 in EETI-II has been shown to be tolerant to changes in length without compromising the 3D conformation of the framework (17). We demonstrated that EETI-II could be displayed on the surface of M13 bacteriophages and that it retained its binding to trypsin (SI Appendix, Fig. S1A), indicative of a properly folded cystine-knot framework. Our library design strategy comprised the randomization of either loop 1, loop 5, or both loops simultaneously. We varied the length of loop 1 from 6 to 10 residues, whereas the native loop 5 length (5 residues) was maintained (Fig. 1A). The three constructed libraries were then displayed on gene 8 of M13 bacteriophages, achieving a total diversity of 3 × 1010 (see Materials and Methods).
Fig. 1.
CKP library design and phage selection strategy for discovery of high affinity binders to LRP6 E1. (A) Design of CKP library. The randomized regions are highlighted in blue and red for loop 1 and 5, respectively, on the native EETI structure (Protein Data Bank [PDB]: 1W7Z). NNK: randomization by NNK-codon incorporation; aa: amino acid. (B) Schematic of phage display workflow during initial selections. (C, F, and G) Affinity determination of CKP hits by SPR (measured by single-cycle kinetics). E1 was immobilized on sensor chips, and different CKP concentrations were injected. Sensograms of the the parental CKP R1 (C), and the two affinity-matured CKPs Lr-EET-3.5 (F) and Lr-EET-3.4 (G). Fit to a 1:1 model is shown in black. (D) Schematic workflow of phage display during affinity maturation. (E) Comparison of affinity and sequence of affinity matured CKPs. CKPs identified by the affinity maturation and the parental CKP R1 are sorted by affinity (green-white-red gradient), with their loop 1 sequences shown. Nx(I/V)K motifs are highlighted in yellow, and residues detrimental or beneficial to binding are highlighted in red and green, respectively.
Identification of LRP6 E1-E2-Binding CKPs by Directed Evolution.
We selected the E1-E2 domains of LRP6 as a model system to discover small targeted peptides from the CKP libraries that could act as antagonists of WNT1 signaling. Previous efforts to screen linear or cyclic peptide phage libraries against the E1-E2 domains of LRP6 failed to produce any high-affinity binders (43). We speculated that the constrained nature of the peptide loops in CKPs, together with the potential additional interactions from multiple loops within the scaffold, would allow for the generation of high-affinity ligands. The CKP library was screened twice against E1-E2 in two separate phage display selection campaigns (Fig. 1B) (44). In the first campaign, 96 random clones were selected from the enriched phage pool, and they exhibited specific binding to E1-E2 in phage enzyme-linked immunosorbent assays (ELISAs). All of the binders comprised an Nx(I/V)(K/R) motif or slight variations thereof in loop 1 (SI Appendix, Fig. S1 B and C). Remarkably, this motif is similar to that found in the natural Wnt antagonists DKK1 and SOST (43). The loop 5 sequence observed in the isolated binders was either wild type or did not contain a consensus motif. In the second campaign, the selection converged to a single sequence with an NxV motif in loop 1 (SI Appendix, Fig. S1B).
De Novo Discovery of WNT1 Blocking CKPs.
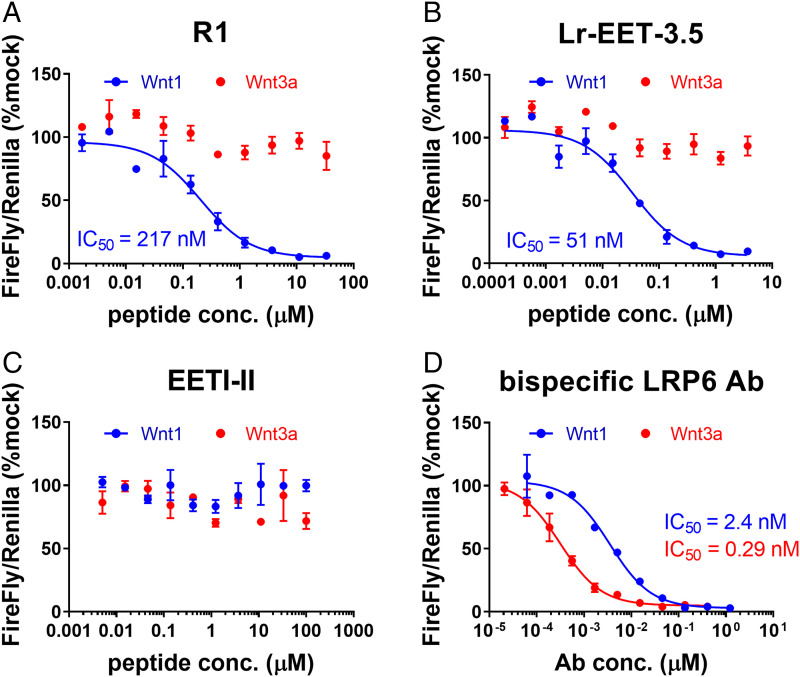
Three CKPs (R1, R19, and R77) from both campaigns were selected for further characterization (SI Appendix, Fig. S1B). The crude linear peptides were obtained by solid-phase peptide synthesis and then refolded in 0.1 M ammonium bicarbonate (pH 9), 1 mM reduced glutathione, 50% dimethyl sulfoxide, resulting in almost complete conversion of the linear peptide to a major species containing three disulfides as observed by liquid chromatography-mass spectrometry after 48 h (see Materials and Methods and extended data file). The folded peptide species were purified by reverse phase-high-performance liquid chromatography to greater than 95% purity. To characterize their bioactivity, the LRP6 E1-E2-binding CKPs were tested in TOPbrite (TB) dual-luciferase reporter assays. HEK293-TB cells were transfected with cDNA encoding either WNT3a or WNT1 and treated with varying concentrations of the CKP hits. WNT1 and WNT3a were used as representatives of the different Wnt classes that bind to E1-E2 and E3-E4 domains, respectively (41, 45). CKPs showed a concentration-dependent specific inhibition of WNT1- but not WNT3a-mediated signaling, as expected of an E1 binder (SI Appendix, Fig. S1B). CKP R1 was the most potent inhibitor of WNT1-mediated signaling, with a half-maximal inhibitory concentration (IC50) of 217 nM (SI Appendix, Fig. S1B); it also displayed a binding constant of 34 nM (for LRP6 E1), as measured by surface plasmon resonance (SPR) (Fig. 1C). The binding epitope of R1 CKP was mapped exclusively to the E1 domain since there was no detectable binding to the E2 domain as measured by SPR (SI Appendix, Fig. S2 A–C). The parent EETI-II CKP did not show any detectable binding to LRP6 E1 or inhibition of WNT1 signaling (SI Appendix, Figs. S1B and S2D). Altogether, these results demonstrate the power of our CKP display platform to evolve the EETI-II framework from a trypsin inhibitory molecule to a targeted, high-affinity CKP antagonist of WNT1 signaling. These CKPs are no longer expected to bind or inhibit trypsin given the extensive changes introduced in loop 1, which is the trypsin-binding region in the native EETI-II framework.
Affinity Maturation Yields Single-Digit Nanomolar Binders to LRP6 E1-E2.
To further improve the affinity of R1, we constructed a “soft-randomized” library focused on loop 1 residues in R1 CKP (see Materials and Methods). The library was displayed as gene 8 or gene 3 fusions (g8 and g3 library, respectively) on M13 bacteriophages. Both libraries were cycled through four rounds of selection with increasing stringency (Fig. 1D). Rounds 3 and 4 consistently showed a strong enrichment of recovered phages from wells immobilized with LRP6 E1 domain compared to blank wells. To further increase the stringency, we designed an off-rate selection round in which the enriched g3 phage pool (obtained after round 3) was incubated with an excess of nonbiotinylated E1 for 1 h before subsequent pull down and intensive washing (Fig. 1D). Limited enrichment of positive selection was observed after this round, and therefore we conducted one additional round with less stringent conditions (“recovery round”). In total, 95 random clones were picked from rounds 3 and 4 of the g8 library and rounds 3 to 6 of the g3 library, and they were subsequently analyzed by phage ELISA. Sequencing revealed a very strong enrichment of the Nx(V/I)K motif (SI Appendix, Figs. S3–S8). However, there was no obvious enrichment of specific residues at positions outside the core motif, with the exception of a slight enrichment observed for aromatic residues at position 7 (x in Nx(V/I)K) and a histidine at position 11 in the clones after the recovery round (SI Appendix, Fig. S8). We selected the top 15 clones that displayed the highest signal over noise in phage ELISA from all of the rounds for synthesis and further characterization (SI Appendix, Figs. S3–S8). We also designed two additional sequences (Lr-EET-3.10 and Lr-EET-3.11) that combined the most enriched residues at specific positions throughout the selection campaign (Fig. 1E).
The binding of all of the CKP hits to LRP6 E1 was first measured by SPR (Fig. 1 E–G). Of the 17 CKPs, 12 exhibited affinities close to R1 CKP (13 to 65 nM), whereas two CKPs showed markedly reduced affinities (366 to 647 nM). We identified two CKPs, Lr-EET-3.4 and Lr-EET-3.5, with markedly improved binding affinities (KD values of 3 and 1 nM, respectively) (Fig. 1 F and G). Notably these were the two CKPs that were identified in the recovery round. The observed gain in affinity is due to a substantial decrease in the dissociation rate (∼120- to 600-fold) and a less pronounced reduction in the association rate (∼15-fold) compared to the parent (SI Appendix, Table S1). This observation is consistent with the off-rate selection strategy we used. Comparison of the loop 1 sequences and the affinities of the 17 synthetic CKPs showed that Lr-EET-3.4 and Lr-EET-3.5 were the only molecules displaying an aromatic residue at position 6/7 within the NxI motif and a histidine residue at position 10/11 downstream of the NxI motif (Fig. 1E), indicating their important contributions to the observed tight binding. However, the presence of proline residues close to the core motif seemed to be detrimental for binding. Interestingly, the Nx(I/V) motif of the tightest binder Lr-EET-3.5 is shifted by one residue toward the N terminus, and the extended motif is NxIL, while all of the other CKPs carry an Nx(I/V)K motif (Fig. 1E).
LRP6 CKPs Block WNT1 but Not WNT3a-Mediated Signaling.
We tested the top three CKPs with the highest affinities against LRP6-E1 (Fig. 1E; Lr-EET-3.4, -3.5, and -3.10) for their ability to specifically inhibit Wnt-mediated signaling in HEK293-TB cells that were transfected with either WNT3a or WNT1 plasmids. All of the LRP6 E1-binding CKPs showed selective inhibition of WNT1- but not WNT3a-mediated signaling (SI Appendix, Table S1). The potency of cellular inhibition was roughly correlated with the observed in vitro affinity of the different CKPs as determined by SPR, with the tightest binders displaying the highest cellular potencies. The IC50 of WNT1 signaling inhibition of the best binder Lr-EET-3.5 was ∼50 nM (KD = 1 nM), while the parental molecule R1 had IC50 and KD of ∼220 and 34 nM, respectively (Figs. 1 C, 1 F, and 2). The parent EETI-II CKP showed no effect on either WNT1- or WNT3a-mediated signaling as anticipated (Fig. 2C). The WNT1-specific antagonism of the CKPs distinguishes them from previously reported antagonists, such as a bispecific LRP6 antibody, which binds to both E1 and E3-E4 domains of LRP6 (46), as well as a FZD7-binding peptide (dFz7-21) (47). Both controls showed inhibition of WNT1- and WNT3a-mediated signaling in luciferase reporter assays (Fig. 2D and SI Appendix, Table S1). The Lr-EET-3.5 CKP also blocked paracrine signaling in cells that were stimulated with the recombinant WNT1/sFRP-1 complex (see below) but not exogenous recombinant WNT3a (SI Appendix, Fig. S9 and Table S1). To further validate the mechanism of action of Lr-EET-3.5 CKP, we took advantage of murine small intestinal organoids, which rely on the Paneth cell endogenous secretion of Wnt3a. As expected, the treatment of these cells with the pan-Wnt inhibitor LGK974 or bispecific LRP6 antibody had a profound effect on organoid growth and gene expression, with an apparent decrease in various stem cell and Wnt-related target genes (SI Appendix, Fig. S10). Treatment with antigen-binding fragment antibodies (Fabs) that bind to E3-E4 (FabYW211) (45) also decreased the transcriptional activity of Wnt target gene and stem cell markers. However, no transcriptional changes were detected when the organoids were treated with Lr-EET-3.5 or a control Fab that binds to E1-E2 (FabYW210) (45) (SI Appendix, Fig. S10). These observations support the notion that WNT3a is the major mediator of stem cell activity in mouse intestinal organoids and further validate the specificity of the CKP, as exemplified by the apparent lack of Wnt3a-blocking activity. To assess the isoform selectivity of Lr-EET-3.5, we determined its binding to LRP5, a highly conserved isoform of LRP6 (73.6% sequence identity within the four β-propeller domains). SPR measurements demonstrated that Lr-EET-3.5 bound to the E1-E4 domain of LRP5 with affinities in the range of 0.7 to 1.0 nM (SI Appendix, Fig. S11), indicating that it binds equally well to both LRP5 and LRP6 receptors.
Fig. 2.
LRP6 E-1 targeting CKPs block Wnt1- but not Wnt3a-mediated signaling. Representative examples of Wnt reporter assays are shown. HEK293 TB cells were transfected with either Wnt1 (blue) or WNT3a (red) and treated with R1 (A), Lr-EET-3.5 (B), EETI-II (C), or bispecific α-LRP6 antibody (D). Calculated IC50 values are derived from three to five independent experiments, each run with five technical replicates. Error bars represent SDs.
CKPs Compete with WNT1 for Binding to LRP6.
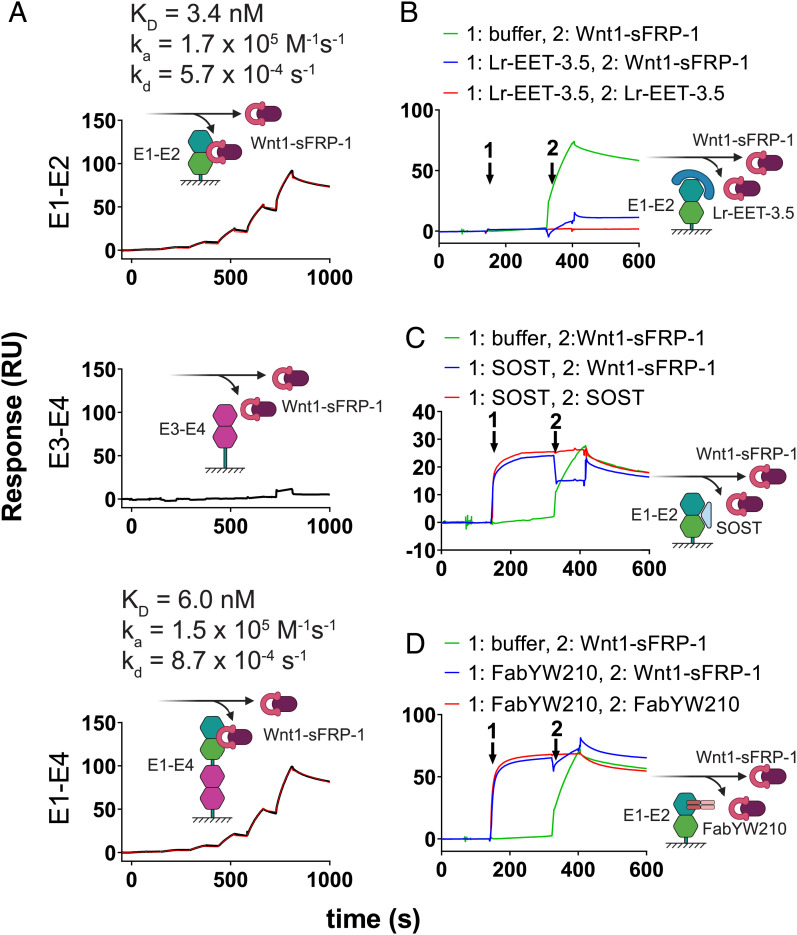
To determine the mechanism of inhibition of WNT1 signaling, we analyzed the effect of Lr-EET-3.5 CKP on the binding of WNT1 to LRP6. Due to the lack of availability of a fully functional recombinant WNT1 protein, we used instead a purified recombinant WNT1/secreted frizzled related protein 1 (sFRP-1) complex (48). sFRP1 is a mimic of the FZD cysteine-rich domain and helps to stabilize WNT1 in solution but is not expected to interfere with WNT1 binding to LRP6. We validated the interaction of WNT1/sFRP-1 with LRP6. The observed affinities to the E1-E2 and E1-E4 domains were very similar, exhibiting KD values of 3.4 and 6.0 nM, respectively. However, there was no interaction detected between WNT1/sFRP-1 and E3-E4 (Fig. 3A), consistent with the notion that WNT1 binds to the E1-E2 domain of LRP6 (41, 43). Competition assays revealed that Lr-EET-3.5 CKP blocked the binding of WNT1/sFRP-1 to LRP6 (Fig. 3B), suggesting an overlapping epitope with WNT1. This observation is also consistent with the antagonistic effect of SOST and a Fab (FabYW210), both of which bind to the E1 domain (Fig. 3 C and D). These results support the notion that the observed cellular activity of the CKP is mediated by its direct competition with WNT1 for binding to LRP6 E1-E2, and potentially LRP5.
Fig. 3.
Wnt1/sFRP-1 binding to LRP6 E1 domain is blocked by CKP. (A) Binding of Wnt1/sFRP-1 to different domains of LRP6. Fit to a 1:1 binding model is indicated (red). Sensogram plots for competition experiments between Wnt1/sFRP-1 and Lr-EET-3.5 (B) SOST (C) and FabYW210 (D) are shown. The surface was coated with LRP6-E1-E2. The surface was then saturated with competitor or treated with buffer only (first arrow), and then immediately afterward either Wnt1/sFRP-1 (green), competitor (red), or a mixture of competitor and Wnt1/sFRP-1 (blue) was injected. The resulting binding signal of Wnt1/sFRP-1 in the presence or absence of competitors was inspected manually. Note that the binding signal of Lr-EET-3.5 alone is not visible due to very low molecular weight compared to other reagents.
X-Ray Crystal Structure of Lr-EET-3.5 CKP in Complex with E1 LRP6.
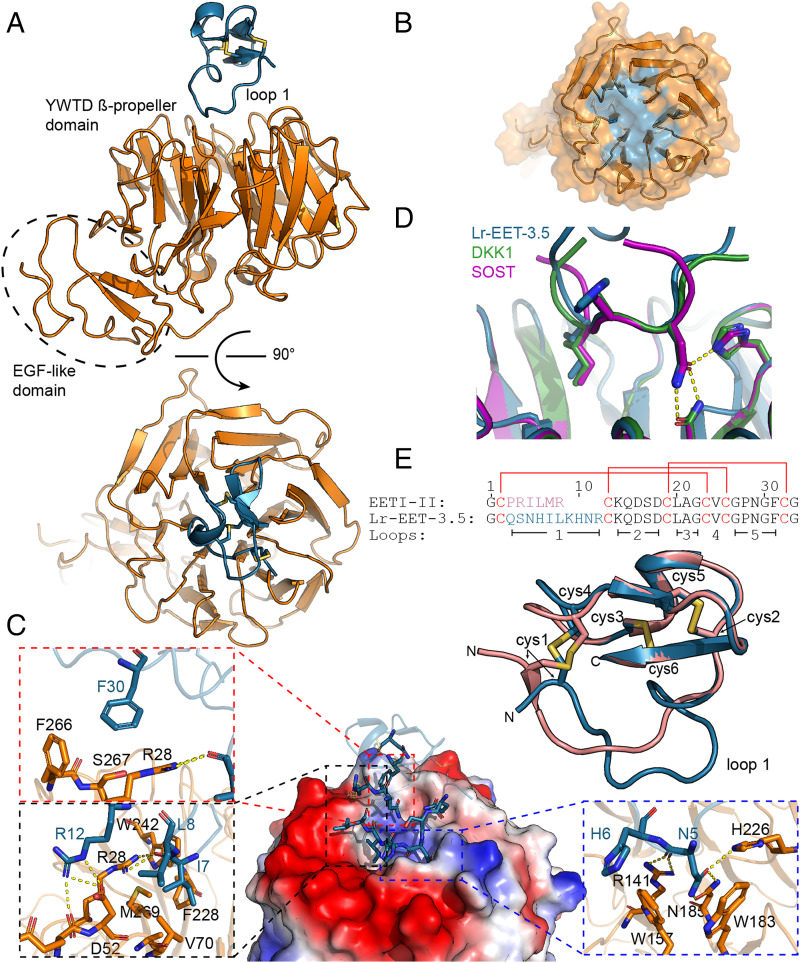
To further understand the inhibitory mechanism of the CKP, we determined the X-ray cocrystal structure of Lr-EET-3.5 CKP in complex with the E1 domain of LRP6 to 1.6 Å resolution (Fig. 4A; data collection and refinement statistics are given in SI Appendix, Table S2). Clear electron density was observed for both CKP and the E1 domain (SI Appendix, Fig. S12A), revealing a 1:1 stoichiometry. The E1 domain comprised the YWTD β-propeller and epidermal growth factor–like domains. The Lr-EET-3.5 CKP was positioned at the top surface of the six-bladed β-propeller domain, making electrostatic and hydrophobic contacts primarily through the loop 1 residues (Fig. 4A). Loop 1 protruded into the putative WNT1 binding site on E1 and buried 570 Å2 of surface area (Fig. 4 B and C). The binding hotspots clustered around the Asn-His-Ile motif. Asn5 formed a series of hydrogen bonds to residues H226, N185, and R141 on the E1 domain (Fig. 4C). Moreover, His6 residue was involved in a π–π interaction with W157, whereas Ile7 was placed in a hydrophobic pocket lined by V70, F228, W242, and M269 (Fig. 4C). Superposition of the NxI motif in Lr-EET-3.5 with that found in the peptides derived from DKK1 and SOST (43) revealed virtually identical binding modes (Fig. 4D), indicating that the Lr-EET-3.5 CKP recapitulates the binding mechanism of DKK1 and SOST at this epitope. However, there were additional interactions formed by residues outside the NxI motif in loop 1. For instance, Leu8 packed against the hydrophobic portions of Asp52 and Val70 side chains, and Arg12 formed hydrogen bonds with Asp52 (Fig. 4C). Remarkably, we observed interactions for the native residues outside loop 1. For instance, residue Phe30 in loop 5 exhibited hydrophobic packing with residues Arg28, Phe266, and Ser267 on E1 (Fig. 4C). Substitutions introduced at key positions involved in the interaction such as Asn5, Ile7, Leu8, or Phe30 led to a complete loss in the binding of Lr-EET-3.5 CKP to LRP6 E1 as determined by SPR measurements, validating the structural observations (SI Appendix, Table S1). We also pursued a structure-based design approach to improve the affinity of the CKP. Replacement of the Arg12 residue with l-methylarginine (Lr-EET-3.5.R12MeR) led to 1.5-fold improvement in affinity (KD = 645 pM) compared to the parental molecule Lr-EET-3.5 (SI Appendix, Table S1), and therefore we used Lr-EET-3.5.R12MeR CKP in subsequent functional studies.
Fig. 4.
Structure of the complex between LRP6 E1 and Lr-EET-3.5. (A) Structure overview from two perpendicular angles. E1 is shown in orange and Lr-EET-3.5 in blue. (B) Interaction surface of Lr-EET-3.5 (blue) projected on the surface of E1 (orange). (C) Interaction of E1 and Lr-EET-3.5. Overview of the interaction is shown in the center. Surface rendering of E1 with electrostatic coloring (blue, positive; red, negative) is shown, with Lr-EET-3.5 (ribbon) in blue, with interacting residues highlighted as stick. Insets: Interaction details of hotspot residues. Hydrogen bonds are shown as yellow dashes. (D) Superposition of the NHI motif of Lr-EET-3.5 (blue) with NxI motifs derived from DKK1 and SOST. (E) Superposition of Lr-EET-3.5 (blue) with the parental scaffold EETI (green). Cysteines (cys) are shown as sticks and numbered. Sequence alignment is given on the bottom. Disulfide connectivity is indicated by red lines and loops by black bars.
It is noteworthy that Lr-EET-3.5 CKP exhibited a disulfide bond connectivity that is identical to that found in the parental EETI-II framework (Fig. 4E) (root mean square excluding the randomized loop 1 is 0.57 Å; alignment of 19 Cα atoms), despite having a longer loop 1 length compared to the native loop (10 versus 6 residues). All of the intramolecular hydrogen bonds outside of loop 1 were conserved between EETI-II and Lr-EET-3.5 (SI Appendix, Fig. S12B). The main difference between EETI-II and Lr-EET-3.5 structures was observed in the variable loop 1 region, which formed an extensive network of 9 intramolecular hydrogen bonds in Lr-EET-3.5 but displayed only 2 hydrogen bonds in the parental EETI-II framework (SI Appendix, Fig. S12B). Altogether, these findings demonstrate the utility of the CKP platform to identify small, targeted, highly potent, and selective CKPs with biological properties that are distinct from EETI-II, but they still maintain the native cystine-knot fold. Based on the structural findings, most of the residues in the CKP binding epitope are conserved between LRP6 and LRP5 (SI Appendix, Fig. S12 C and D), consistent with the above findings demonstrating similar affinities of Lr-EET-3.5 CKP to LRP6 and LRP5. Moreover, it is expected that the CKP no longer binds to or inhibits trypsin, given the substantial structural changes observed in loop 1 compared to native EETI-II.
LRP6 CKP Mimics Natural Wnt Inhibitors but Displays a Distinct Mechanism of Binding.
We used the CKP as a specific tool to further investigate outstanding questions regarding DKK1 and SOST interaction with LRP6. We compared the binding mode of LRP6 CKP to the natural Wnt inhibitors DKK1 and SOST, given the similarities observed above in their NxI motifs. DKK1 and SOST have been implicated in several diseases (49, 50), and hence their molecular binding mode is a subject of great interest. It has been established that DKK1 binds to two distinct epitopes on LRP6. The E1 domain is recognized by an NAI peptide motif in the N-terminal region of DKK1, while the C-terminal domain interacts with LRP6 E3-E4 (43, 51, 52). SOST also contains an NAI motif that has been shown to interact with LRP6 E1 (43). However, recent structural investigations revealed that SOST additionally interacts via its C-terminal tail with the LRP6 E2 domain and that this interaction appears to be stronger than the binding to the E1 domain (53). By using SPR, we characterized the binding of each of these ligands to immobilized E1-E2, E3-E4, and the full-length extracellular domain of LRP6 (E1-E4) in the presence and absence of Lr-EET-3.5 CKP.
We observed that SOST bound to E1-E2 (KD = 3.3 nM) and E1-E4 (KD = 3.8 nM) with comparable affinities, but there was no interaction detected with E3-E4 (SI Appendix, Fig. S13), in agreement with previous reports (43, 54). We also observed direct binding of SOST to E1 alone (KD = 46 nM), and this interaction was completely blocked by the addition of Lr-EET-3.5. However, the binding of SOST to immobilized E1-E2 or E1-E4 could still be detected in the presence of Lr-EET-3.5, albeit with reduced affinity (KD values of 47 and 41 nM, respectively), indicating that SOST also recognizes an additional binding site on the E2 domain of LRP6. The use of the CKP to block the E1 binding site revealed the contribution of E2 binding to SOST, and the competition result with CKP indicated roughly equal contributions of E1 and E2 to SOST binding (SI Appendix, Fig. S13). Previously, the binding of SOST to E2 alone was reported to have a KD of 1,300 nM (43). The difference between these values and our measured affinities could be due to differences in the quality of E2 protein used or its behavior during the studies. Regardless, our data demonstrate that SOST uses both E1 and E2 to bind to LRP6 and are consistent with the notion that SOST has two distinct binding sites on LRP6 E1 and E2 (53). They also highlight the overlapping yet distinct mode of inhibition of the CKP; it binds exclusively to E1, yet it fully blocks WNT1 binding to LRP6 and exhibits superior binding affinity to LRP6 compared to SOST.
DKK1 displayed high-affinity binding to E1-E4 (KD = 3.6 nM), but weaker binding to either E1-E2 (KD = 135 nM) or E3-E4 (KD = 43 nM) (SI Appendix, Fig. S14), consistent with prior findings (41, 43, 51, 52). Also, there was no detectable binding of DKK1 to E1, likely due to the low DKK1 concentrations used in the measurements (<100 nM). However, blocking the E1 binding site via the addition of LRP6 CKP Lr-EET-3.5 ablated the binding of DKK1 to E1-E2, indicating weak contributions from both E1 and E2 that otherwise could not be detected individually. These findings are corroborated by the observation that DKK1 binds to E1-E4 with an affinity of 10 nM in the presence of the E1-specific binder Lr-EET-3.5 CKP, which likely represents the contributions of domains E2-E4 (SI Appendix, Fig. S14). It is noteworthy that this affinity lies between the high affinity observed in E3-E4 (25 to 43 nM) and to E1-E4 (3.6 nM). Overall, the binding of DKK1 to LRP6 appears to be mainly mediated by E3-E4, with slight contributions from both E1 and E2 (SI Appendix, Fig. S14). Altogether, using the CKP as a tool, we characterized the binding specificity of DKK1 and SOST to individual domains of LRP6 and also revealed the involvement of the E2 domain in both interactions. Moreover, even though the synthetic CKP does not fully recapitulate the binding mechanism of DKK1 and SOST, it offers potential advantages over these natural inhibitors. Unlike DKK1, which blocks both WNT1 and WNT3a signaling, the LRP6 CKP exclusively blocks WNT1 signaling and displays an affinity that is ∼8-fold tighter than DKK1 to LRP6.
LRP6 CKP Inhibits Differentiation of Osteoblasts.
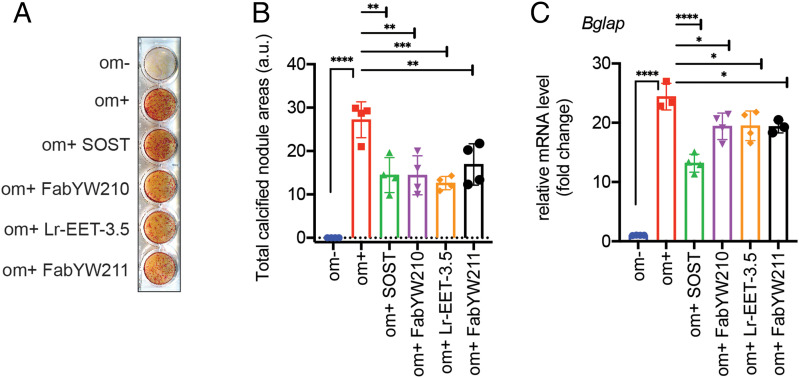
The Wnt pathway is a key regulator of bone metabolism. Mutations in WNT1 have been observed in patients with skeletal disorders, although the potential involvement of other Wnt proteins in osteoblast activity remains elusive (55–57). To evaluate the contribution of the E1 domain of LRP6 to osteogenic differentiation, we used LRP6 CKP as a pharmacological tool, given its exquisite selectivity to the E1 domain and its specificity in disrupting WNT1 class interaction with LRP6/5. Differentiation of MC3T3-E1, a murine-derived osteoblast cell line, was induced with ascorbic acid and inorganic phosphate (58). LRP6 CKP inhibited matrix mineralization by ∼50%, as determined by the observed reduction in alizarin red-S staining levels (Fig. 5 A and B). The effect was comparable to the effect of SOST and a Fab binding to E1-E2 (FabYW210) (Fig. 5 A and B). It is noteworthy that a E3-E4-binding Fab (FabYW211) also showed comparable inhibitory activity, suggesting that WNT3 class proteins are involved in this osteogenic model. The CKP-mediated inhibition of osteoblast differentiation was accompanied by the down-regulation of the osteogenic marker osteocalcin (Bglap) (Fig. 5C). The observed 50% maximal inhibition of osteoblast differentiation by these reagents is likely due to this process being driven by multiple Wnt ligands as well as other LRP6-independent pathways such as platelet-derived growth factor receptor signaling (59–61). Altogether, our pharmacological data highlight the contribution of the E1 domain of LRP6 to Wnt-mediated osteoblast differentiation.
Fig. 5.
Inhibition of osteoblast differentiation. (A) MC3T3-E1 cells were treated with LRP6-binding reagents (1 µg SOST, 1 µM FabYW210 and FabYW211, 5 µM Lr-EET-3.5) for 21 d, and mineralized nodule formation was assessed by alizarin red S staining. FabYW210 binds to the E1-E2 domain of LRP6. The differentiation medium (om+) includes 50 µg/mL ascorbate-2-phosphate and 10 mM β-glycerophosphate (see Materials and Methods). Om- refers to the control medium without the added differentiation factors. (B) Quantification of calcified nodule area. The total calcified nodule area above a threshold defined by the background from om- wells was measured. a.u.: arbitrary unit. (C) Effect on the mRNA expression level of bone marker, osteocalcin (Bglap). The cells were treated for 1 wk before qRT-PCR. Bars represent the means of four independent assays. Error bars represent SDs (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA).
Engineered Bivalent LRP6 CKP Agonist Potentiates WNT3a/β-Catenin Signaling.
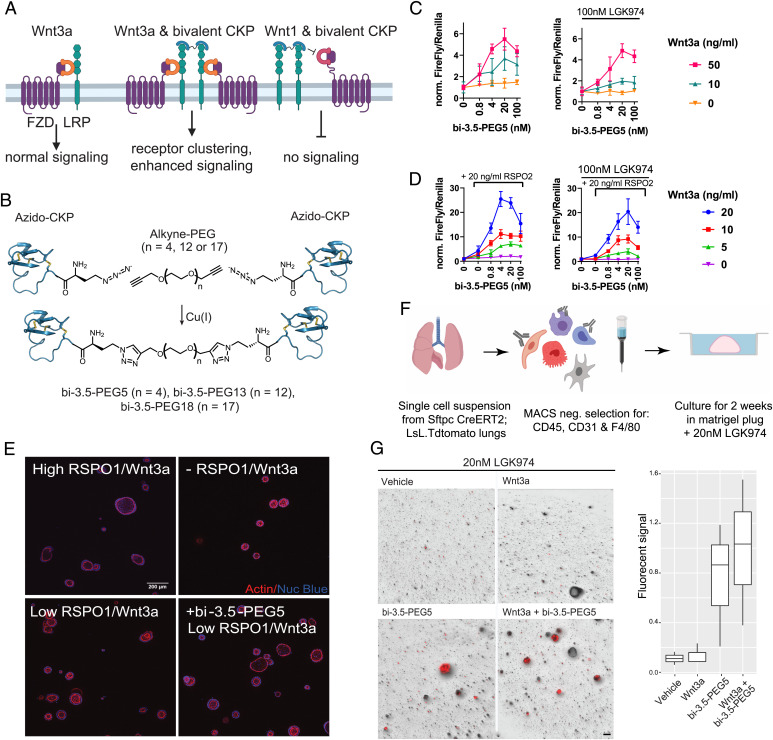
The observation that the LRP6 E1-binding CKP did not influence WNT3a signaling prompted us to explore whether a bivalent CKP format could enhance WNT3a-mediated signaling by recruiting additional LRP6 receptors to signaling complexes (Fig. 6A). To activate Wnt signaling, we developed a modular strategy that enables the robust generation of bivalent CKPs with different linker lengths. We designed and synthesized an analog of Lr-EET-3.5 R12MeR incorporating an N-terminal l-azidohomoalanine residue (Lr-EET-3.5 NAha.R12MeR). This unnatural amino acid could be subsequently conjugated via click chemistry to different polyethylene glycol (PEG)–based linkers that are functionalized with alkyne groups (Fig. 6B). Three bivalent CKPs with different linker lengths (PEG5, PEG13, and PEG18) were synthesized and purified to greater than 95% purity (bi-3.5-PEG5, bi-3.5-PEG13, and bi-3.5-PEG18, respectively). All of the CKP dimers were observed to potentiate Wnt/β-catenin signaling to varying levels in HEK293-TB cells in the presence of exogenous recombinant human Wnt3a, with bi-3.5-PEG5 showing the maximal signaling activation (greater than fivefold increase in Wnt3a-mediated signaling at 4 to 20 nM concentration of bi-3.5-PEG5) (Fig. 6C and SI Appendix, Fig. S15). Modest activation was also observed by bi-3.5-PEG5 in the absence of exogenous Wnt3a, and this was fully blocked by the overnight pretreatment of cells with LGK974, which inhibits endogenous Wnt activity (Fig. 6C). Although weaker, the PEG13 and PEG18 linkers still achieved up to twofold cellular activation (SI Appendix, Fig. S15). In all of the cases, the degree of activation was also dependent on the amount of WNT3a present. The extent of Wnt/β-catenin activation was reduced when cells were treated with high concentrations of the CKP dimer (Fig. 6C). This hook effect is likely due to the monovalent behavior of the ligand observed at high CKP concentrations, leading to the saturation of the LRP6 receptors and thereby ablating receptor clustering. It is noteworthy that the monovalent LRP6 CKP (Lr-EET-3.5) did not show any potentiation or inhibition of Wnt3a-mediated signaling (Fig. 2B), consistent with its binding mechanism. However, the bivalent LRP6 CKP (bi-3.5-PEG5) still inhibited WNT1-mediated signaling with similar potency as the monovalent CKPs (SI Appendix, Fig. S16).
Fig. 6.
Activation of Wnt signaling by bivalent LRP6 CKPs. (A) Proposed mechanism of WNT3a signaling activation by bivalent CKPs. A bivalent CKP brings together two WNT3/FZD/LRP6 signaling complexes by binding to the E1 domains of and dimerizing two LRP receptors. The same bivalent CKP competes with Wnt1 class ligands to block Wnt1 signaling. (B) Synthesis scheme of bivalent CKPs. (C) Activation of Wnt signaling by bivalent CKPs in the absence and presence of LGK974 (100 nM). HEK293-TB cells were preincubated overnight in the absence or presence of LGK974 and then treated with CKP and WNT3a at the indicated concentration for 6 h before assay readout. A representative example from three biological replicates with five technical replicates is shown. Error bars represent SDs. All of the data were normalized to the control without CKP treatment in each group. (D) Activation of Wnt signaling by bi-3.5-PEG5 in the presence of varying concentrations of WNT3a and RSPO2 and with or without LGK974 (100 nM) treatment. HEK293-TB cells were preincubated overnight in the absence or presence of LGK974 and then treated with CKP, RSPO2, and WNT3a at the indicated concentration for 6 h before assay readout. A representative example from three biological replicates with five technical replicates is shown. Error bars represent SDs. All of the data were normalized to the control with no added CKP and RSPO2 in each group. (E) Representative images of human duodenal organoid cultures taken 5 d after treatment with 500 nM of Wnt-C59 in the absence or presence of WNT3a conditioned media, RSPO1, and bi-3.5-PEG5 [4 nM] as indicated. High RSPO1/WNT3a = 250 ng/mL/50% WNT3a conditioned medium. Low RSPO1/WNT3a = 2.5 ng/mL/25% WNT3a conditioned medium. Staining for nuclei and actin cytoskeleton are depicted in blue and red, respectively. (Scale bar, 200 μm.) (F) Workflow of alveolosphere experiment. Fibroblasts and AT2 cells were isolated from adult (2 mo) Sftpc.Cre.ERT2;LSL;Tdtomato mice and then cultured in Matrigel plugs. (G) Representative images of alveolosphere cultures taken 2 wk after treatment with LGK974, LGK974+WNT3a, LGK974+CKP, and LGK974+CKP+WNT3a. WNT3a, 50 ng/mL bi-3.5-PEG5, 48 nM final concentration. Quantification of AT2 growth as measured by total Tdtomato fluorescent signal (three biological replicates) across the different treatment conditions. (Scale bar, 100 μm.)
To test whether activation of Wnt signaling by the natural Wnt agonist RSPO1 to 4 could be further potentiated by bi-3.5-PEG5, we treated HEK293-TB cells with different concentrations of Wnt3a, RSPO2, and bi-3.5-PEG5. RSPO2 potentiates Wnt signaling by sequestering the cell surface ubiquitin ligases RNF43 and ZNRF3, leading to higher cell surface levels of the LRP6 and FZD receptors (31–33). As expected, the addition of RSPO2 on its own led to a concentration-dependent increase (∼1.5- to 4-fold) in WNT3a signaling (Fig. 6D). The treatment of cells with both RSPO2 and bi-3.5-PEG5 led to ∼26-fold signaling activation compared to WNT3a alone, and this effect was observed both in the absence and presence of LGK974 treatment (Fig. 6D). The optimal potency of bi-3.5-PEG5 was ∼4 to 20 nM in the presence of RSPO2, with higher concentrations resulting in relatively reduced levels of activation. Modest potentiation was also observed by RSPO2 and bi-3.5-PEG5 even in the absence of exogenous WNT3a. This effect was abolished by the overnight pretreatment of cells with LGK974 before the RSPO2 and bi-3.5-PEG5 treatment (Fig. 6D), indicating that the synergistic effect in this case is dependent on the presence of very low levels of endogenous WNT3/WNT3a secreted by HEK293 cells. Altogether, our findings demonstrate that monovalent LRP6 CKPs could be turned into Wnt pathway agonists through dimerization, and that we could fine tune their activity by varying the linker length. Moreover, the observed synergistic rather than additive effects could be explained by the CKP-mediated clustering of LRP6 receptors, and potentially Wnt signaling complexes, combined with the increase in LRP6 cell surface receptor levels due to RSPO2 treatment.
Bivalent LRP6 CKP Agonist Stimulates Growth in Primary Human Intestinal Organoids and Stem Cell Activity in Alveolospheres.
We sought to test whether the LRP6 CKP dimer could affect physiological processes that depend on Wnt signaling. We took advantage of human primary duodenal organoids, an established culture system known to rely on elevated Wnt signaling for culture expansion. In the presence of a high concentration of RSPO1 and Wnt3a, organoids presented with cystic growth and a well-delineated lumen (Fig. 6E). However, both features were lost in the absence of or under low (suboptimal) concentrations of these growth factors but were restored when a low concentration of bi-3.5-PEG5 was added to the culture media (Fig. 6E). The biological impact of bi-3.5-PEG5 was assessed quantitatively through high content imaging. The addition of bi-3.5-PEG5 (4 nM) in the presence of low concentrations of RSPO1 and WNT3a resulted in organoid growth that was equivalent to cultures treated with high concentrations of RSPO1 and WNT3a (SI Appendix, Fig. S17). Consistent with the HEK293-TB assay, higher concentrations of bi-3.5-PEG5 resulted in reduced levels of growth stimulation (SI Appendix, Fig. S17). To further validate the activity of bi-3.5-PEG5 CKP in a separate biological system, we focused on the regeneration of the lung alveolar epithelium, where alveolar epithelial type 2 cells (AT2) require Wnt signaling to act as stem cells and respond to mitogenic signals (62, 63). We modeled alveolar regeneration using the alveolosphere model, in which AT2 cells were cocultured with stromal cells that provided paracrine signals activating AT2 stem cell activity (64) (Fig. 6F). Consistent with previous mouse studies showing that Wnt is required for AT2 stem cell activity, the inhibition of Wnt secretion with LGK974 abrogated AT2 proliferation and the formation of alveolospheres (Fig. 6G). The addition of WNT3a at 50 ng/mL did not rescue this phenotype, suggesting that recombinant Wnt may not be sufficient to induce signaling in this setting. However, the addition of bi-3.5-PEG5 to cultures that contained LGK974 rescued this phenotype and restored proliferation and alveolosphere formation, highlighting the ability of this reagent to stimulate alveolar stem cell activity in alveolospheres (Fig. 6G).
The ability of bivalent CKP to rescue LGK974 inhibition is surprising, since reporter cell line experiments suggested that it could potentiate but not phenocopy Wnt ligands. One potential reason for the observed differences may be the cellular source of Wnt ligands in the two assays. In the alveolosphere assay, lung fibroblasts that produce Wnt ligands have the ability to decrease intracellular small-molecule concentrations through efflux pumps (65), potentially leading to an incomplete inhibition of Wnt secretion that is sufficient to inhibit AT2 stem cell activity. This small amount of Wnt ligand remaining is sufficient, however, for the CKP dimerizer to restore Wnt signaling levels in AT2 cells and drive stem cell activity.
Discussion
Here, we show that CKPs could be formatted into activators of Wnt signaling by facile multimerization. Our strategy to drive receptor dimerization rested on varying the distance between the monovalent CKPs to fine-tune the signaling output of LRP6. The bivalent molecules demonstrated strong agonistic activity in potentiating cellular WNT3a signaling and promoting the growth of alveolospheres and human intestinal organoids. The mechanism of this class of activators is fundamentally different from previously described surrogate Wnt agonists that consist of multimeric antibody binders to both FZD and LRP receptors (37–40). While on the one hand, such agonists activate Wnt signaling independent of the natural Wnt ligands, the bivalent CKPs on the other hand specifically enhance Wnt signaling in the presence of a subset of noncompeting endogenous Wnt ligands (i.e., WNT3a or WNT3). Such an approach may enable more precise signaling activation for therapeutic intervention in a tissue-specific manner. The substantially smaller molecular size of the CKPs could enable better tissue penetration compared to their larger antibody counterparts, in addition to the synthetic accessibility of CKPs and flexibility to modulate their molecular properties as needed.
The isoform-selective potentiation of Wnt signaling by E1-binding reagents was observed previously with LRP6-binding immunoglobulin Gs (IgGs) (45). An E1-binding monoclonal antibody (mAb) demonstrated 3.8-fold potentiation of WNT3a signaling in HEK293-TB cells that were transfected with a WNT3a-expressing plasmid. Our findings indicate that bi-3.5-PEG5 CKP dimer seems to exhibit superior cellular potency compared to the E1-binding mAb, likely due to a more optimal linker spacing and geometry leading to more productive clustering of LRP6 receptors at the cell surface. It is noteworthy that in our assays bi-3.5-PEG5 was tested in the presence of exogenously added recombinant WNT3a protein at concentrations that were lower than what is typically expected from transfection methods. Given that the potentiation observed with CKP agonists is dependent on the levels of Wnt protein, it is conceivable that the CKP agonists could show an even higher degree of potentiation under conditions in which cells are transfected with Wnt-expressing plasmids. Nonetheless, the modular CKP agonist design strategy described here provides an advantage over antibody formats as it enables the rapid assembly of multiple molecules with varying architectures to optimize cellular potency. The high affinity and selectivity observed for the CKPs are comparable to those typically obtained for antibody or other protein frameworks (66). However, the ease of manufacturing and feasibility to incorporate synthetic unnatural amino acids at specific sites within the CKP framework could provide an advantage during drug development campaigns.
Notably, the directed evolution of CKP libraries led to the enrichment of an NxI motif that is found in the natural Wnt inhibitors. Structural studies revealed that the binding mode of the NxI motif in the identified CKPs was identical to DKK1 and SOST. Our structural findings are consistent with the notion that the constrained nature of the cystine-knot scaffold enables the display of recognition motifs on surface exposed loops. The loop 1 region, displaying the NxI motif, is stabilized by several hydrogen bonds and by interactions with other loops within the cystine-knot framework, thereby minimizing the entropic penalty upon binding. Remarkably, there are additional residues outside of loop 1 that are also involved in the peptide–protein interaction, including the native F30 residue in loop 5, which is optimally positioned for binding. Several residue substitutions confirmed the involvement of these hot spot residues in the binding to LRP6. Altogether, these molecular observations help explain the superior affinity obtained for the isolated CKPs against the E1 β-propeller domain of LRP6/5. Moreover, they act as synthetic surrogate SOST mimetics, but offer advantages over the natural Wnt inhibitors because they exhibit good solubility and can be generated with high yields. The utilization of CKPs as tools helped to shine insights onto the recognition of LRP6 E2 domain by DKK1 and SOST, in addition to establishing a role for the E1 domain in osteogenic differentiation.
Recently, several library-based approaches have been developed for the design or selection of cysteine-stabilized peptides. For instance, cystine-dense libraries have been created using mammalian surface display to isolate binders to TEAD proteins (13). Phage-displayed libraries based on disulfide-rich peptides with engineered cysteine-proline-proline-cysteine motifs have been developed and selected to identify binders against CD28 (15). In both of these examples, the generated libraries did not contain the native cystine-knot peptide scaffolds but instead used artificial peptide frameworks that were identified by either Rosetta or structure-guided protein design. Truncated forms of CKPs have also been used to generate leads that bind to alpha(v)beta (3) integrins with high affinity and specificity by using yeast-surface display (18). In this case, the hits were expressed recombinantly in Pichia pastoris to generate peptides for functional screening. Moreover, large combinatorial libraries of bicyclic peptides displayed on rigid small molecule scaffolds were screened to identify binders to tumor necrosis factor-α (16). However, this approach required extensive chemical synthesis of the library, not to mention the limited diversity of the library compared to phage-displayed libraries.
Other types of cysteine-free miniprotein scaffolds have also been leveraged for peptide engineering and as display scaffolds (14, 67, 68). Examples include centyrins based on fibronectin type 3 domains (69), fynomers based on the SH3 domain of Fyn kinase (70), and three-helix domain affibodies (71), to name a few. These frameworks readily fold in bacteria and, unlike CKPs, do not need oxidative folding methods. However, they are typically in the 7 to 10 kDa size range and therefore are more topologically similar to antibody fragments, comprising variable loops for target recognition that are displayed on rigid protein structures. Importantly, these frameworks do not often exhibit drug-like properties and hence may be challenging to optimize and develop into therapeutics. For instance, avimers require the coordination of metal ions, which may not be compatible with downstream therapeutic development efforts. In this study, we decided to focus on small frameworks with a molecular weight of less than 4 kDa because they could provide flexibility during drug discovery and development campaigns, including medicinal chemistry efforts and optimization of their pharmaceutical properties. Our studies demonstrated that the phage display system is effective at allowing the CKP scaffold to fold properly, similar to other display systems such as mammalian and bacterial display, as mentioned above. Our phage display methods also enabled the generation of CKPs with diverse 3D structures, potentially yielding leads for biological applications. In our experience, we did not need to construct libraries comprising many distinct CKP scaffolds as has been reported earlier because the EETI-II library on its own was sufficient to yield high-affinity binders to LRP6.
Due to their favorable physical properties, the CKP agonists discovered in this study could serve as pharmacological tools to further investigate the molecular basis for how Wnt signaling regulates stem cell growth and maintenance. Future efforts will be aimed at further elucidating the genetic circuits that regulate stem cell growth in various tissue systems. Moreover, since WNT1 plays a critical role in bone homeostasis (72, 73), engineered CKP agonists that recognize different domains on LRP6 could serve as WNT1 surrogates for increasing bone mineral density, an area of future investigation. Collectively, our studies establish a class of receptor dimerizers that synergizes with a subclass of endogenous WNT3 ligands and pave the way for next-generation agonist design.
One limitation of CKP-based therapeutics is their expected short systemic half-life. This can be overcome by (1) adding a binding domain to serum albumin or IgGs or (2) chemical modifications such as lipidation or polymer conjugation (74, 75). The short serum half-life could limit exposure in off-target tissues if CKPs could be delivered specifically to certain target organs. For instance, formulation approaches to deliver CKPs to the lungs via inhalation or to the intestine via oral delivery could enable their application in lung and intestinal regeneration for chronic disease. Because the dimeric CKP agonists lose activity at high concentrations, likely due to the single occupancy of LRP6, their in vivo dosing will need to be finely adjusted for optimal activity. In general, it is conceivable that the pharmacodynamic effects of agonist ligands would be longer lasting compared to antagonists. Triggering receptor activation at the cell surface will lead to a cascade of downstream signaling events and transcription of Wnt target genes, which amplify the pathway and turn on cellular differentiation mechanisms. Unlike antagonist ligands in which maintaining high trough concentrations of the ligand at all times is required for neutralizing the target protein, the agonist mechanism is expected to have a different profile in which pharmacokinetics is decoupled from pharmacodynamics. In other words, sufficient stimulation of the pathway, even for short periods of time, is expected to lead to durable outcomes manifested in cellular differentiation, which typically takes days to weeks.
In summary, we explored strategies in this study to activate cell surface receptors by exploiting natural receptor activation mechanisms and ligand engineering. We also leveraged organoid models to ascertain the physiological relevance of receptor agonism. The strategy described here for the generation of CKP binders and their dimerization could have wide applications beyond Wnt pathway modulation such as other transmembrane signaling events involving cytokine receptors and growth factors, to name a few. The versatility of CKP-based agonists could also be beneficial in cases in which antibody discovery campaigns have historically not yielded functional molecules due to limitations with antibody formats and the specific mechanism of action of receptors of interest. Methods incorporating protein modeling software for the structural prediction of CKP frameworks combined with in silico screening could help accelerate the pace of discovery of CKP binders against targets of interest (12). This approach could be powerful, especially when paired with experimental display screening methods, such as mammalian or phage display, to identify binders that exhibit good folding properties and stable structural features. The development of robust methods to screen for drug-like properties of CKPs, along with in-depth understanding of the biodistribution and target exposure profiles of diverse CKP scaffolds, could enable the therapeutic development and clinical translation of these scaffolds.
Materials and Methods
Protein Expression and Purification.
LRP6 constructs were expressed and purified as described previously (41). Detailed methods are described in the SI Appendix.
Peptide Synthesis.
CKPs were synthesized using standard 9-fluorenylmethoxycarbonyl protocols as described earlier (76). Detailed methods are described in the SI Appendix.
Bivalent CKP Synthesis.
Bivalent CKPs were synthesized by Cu(I)-catalyzed azide-alkyne cycloaddition reaction as described in the SI Appendix.
Phage Display.
Display of EETI-II on M13 Phage.
EETI-II was displayed on the surface of theM13 bacteriophage by modifying a previously described phagemid pS2202b (77), and the propagated phage was purified according to standard protocols (44). Detailed methods are described in the SI Appendix.
Library Construction and Sorting.
The EETI-II libraries were constructed following the Kunkel mutagenesis method (78). Details of the libraries and their construction are described in the SI Appendix.
Initial Selections.
The initial EETI library had the sequence GC(X5–10)CKQDSDCLAGCVCX5CG, where X denotes residues in loops 1 and 5 that were randomized by incorporating NNK codons at these positions. M13 phages displaying this library as fusions to g8 were applied to rounds of binding selections with LRP6-E1-E2 that was directly immobilized on plates following standard protocols (44). After four rounds of selection, single-phage clones were amplified and analyzed in a phage ELISA against wells with immobilized targets and blank wells missing the target (n). Only hits with Abs. 450 nm ratios (s/n) > 5 and absolute Abs. 450 nm >0.3 in target-positive wells were considered positive hits.
Affinity Maturation.
A library based on the most potent hit from the initial selections R1 was created by soft randomization of loop 1. Detailed methods are described in the SI Appendix.
SPR.
A Biacore S200 instrument was used for all of the measurements in 50 mM Tris pH 7.5, 300 mM NaCl, 0.05% Triton X, 5% glycerol. Detailed methods are described in the SI Appendix.
X-Ray Crystallography.
Purified protein was concentrated to 10 mg/mL and mixed with fivefold molar excess of peptides in in 10 mM 2-(N-morpholino)ethanesulfonic acid pH 5.5, 300 mM NaCl. Detailed methods for crystallization screens are described in the SI Appendix. The data collection and refinements statistics are listed in the SI Appendix, Table S2.
Wnt Reporter Assays.
Wnt signaling assays were performed in HEK293 cells with stably integrated firefly-luciferase-based Wnt reporter (TB) (77) and pRL-SV40 Renilla luciferase (Promega) as described in the SI Appendix.
Intestinal Organoids.
Mouse organoids were established from isolated crypts collected from the entire length of the small intestine and maintained as previously described (79). Detailed methods are described in the SI Appendix.
RNA Sequencing (RNA-Seq).
RNA-seq libraries were prepared using TruSeq Stranded Total RNA Library prep kit (Illumina). Detailed methods are described in the SI Appendix.
Osteogenic Assays.
Detailed methods for culturing MC3T3-E1 cells and treatments with reagents are described in the SI Appendix.
qRT-PCR Analysis.
MC3T3-E1 cells were treated as described above and RNAs were isolated using the RNeasy kit (Qiagen). Detailed methods are described in the SI Appendix.
Human Duodenal Organoids.
Human primary organoids experiments were performed at Ocello, B.V., as described in detail in the SI Appendix.
Isolating Lung Cells.
Lung cells were isolated as described in the SI Appendix.
Lung Alveolospheres.
Lung cells were grown in Matrigel in the presence of growth factors and treated with CKP agonist for 2 wk as described in the SI Appendix.
Supplementary Material
Footnotes
Competing interest statement: All of the authors were employees of Genentech, a member of the Roche group, at the time the research was performed.
This article is a PNAS Direct Submission. C.P.H. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120393119/-/DCSupplemental.
Data, Materials, and Software Availability
Diffraction data and coordinates have been deposited at the PDB using accession code 7NAM (80).
All of the study data are included in the article and/or supporting information.
References
- 1.Lau J. L., Dunn M. K., Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 26, 2700–2707 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Muttenthaler M., King G. F., Adams D. J., Alewood P. F., Trends in peptide drug discovery. Nat. Rev. Drug Discov. 20, 309–325 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Fosgerau K., Hoffmann T., Peptide therapeutics: Current status and future directions. Drug Discov. Today 20, 122–128 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Deyle K., Kong X. D., Heinis C., Phage selection of cyclic peptides for application in research and drug development. Acc. Chem. Res. 50, 1866–1874 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Derda R., Ng S., Genetically encoded fragment-based discovery. Curr. Opin. Chem. Biol. 50, 128–137 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Chen S., et al. , Identification of highly selective covalent inhibitors by phage display. Nat. Biotechnol. 39, 490–498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ekanayake A. I., et al. , Genetically encoded fragment-based discovery from phage-displayed macrocyclic libraries with genetically encoded unnatural pharmacophores. J. Am. Chem. Soc. 143, 5497–5507 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Craik D. J., Daly N. L., Waine C., The cystine knot motif in toxins and implications for drug design. Toxicon 39, 43–60 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Craik D. J., Du J., Cyclotides as drug design scaffolds. Curr. Opin. Chem. Biol. 38, 8–16 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Bhardwaj G., et al. , Accurate de novo design of hyperstable constrained peptides. Nature 538, 329–335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchko G. W., et al. , Cytosolic expression, solution structures, and molecular dynamics simulation of genetically encodable disulfide-rich de novo designed peptides. Protein Sci. 27, 1611–1623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crook Z. R., et al. , Ex silico engineering of cystine-dense peptides yielding a potent bispecific T cell engager. Sci. Transl. Med. 14, eabn0402 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crook Z. R., et al. , Mammalian display screening of diverse cystine-dense peptides for difficult to drug targets. Nat. Commun. 8, 2244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crook Z. R., Sevilla G. P., Mhyre A. J., Olson J. M., Mammalian surface display screening of diverse cystine-dense peptide libraries for difficult-to-drug targets. Methods Mol. Biol. 2070, 363–396 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Wu Y., et al. , Structure-guided design of CPPC-paired disulfide-rich peptide libraries for ligand and drug discovery. Chem. Sci. (Camb.) 13, 7780–7789 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lian W., Upadhyaya P., Rhodes C. A., Liu Y., Pei D., Screening bicyclic peptide libraries for protein-protein interaction inhibitors: Discovery of a tumor necrosis factor-α antagonist. J. Am. Chem. Soc. 135, 11990–11995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura R. H., Levin A. M., Cochran F. V., Cochran J. R., Engineered cystine knot peptides that bind alphavbeta3, alphavbeta5, and alpha5beta1 integrins with low-nanomolar affinity. Proteins 77, 359–369 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silverman A. P., Levin A. M., Lahti J. L., Cochran J. R., Engineered cystine-knot peptides that bind alpha(v)beta(3) integrin with antibody-like affinities. J. Mol. Biol. 385, 1064–1075 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiche L., et al. , Use of restrained molecular dynamics in water to determine three-dimensional protein structure: Prediction of the three-dimensional structure of Ecballium elaterium trypsin inhibitor II. Proteins 6, 405–417 (1989). [DOI] [PubMed] [Google Scholar]
- 20.Heitz A., Chiche L., Le-Nguyen D., Castro B., 1H 2D NMR and distance geometry study of the folding of Ecballium elaterium trypsin inhibitor, a member of the squash inhibitors family. Biochemistry 28, 2392–2398 (1989). [DOI] [PubMed] [Google Scholar]
- 21.Kojima S., Miyoshi K., Miura K., Synthesis of a squash-type protease inhibitor by gene engineering and effects of replacements of conserved hydrophobic amino acid residues on its inhibitory activity. Protein Eng. 9, 1241–1246 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Bhanot P., et al. , A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature 382, 225–230 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Pinson K. I., Brennan J., Monkley S., Avery B. J., Skarnes W. C., An LDL-receptor-related protein mediates Wnt signalling in mice. Nature 407, 535–538 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Tamai K., et al. , LDL-receptor-related proteins in Wnt signal transduction. Nature 407, 530–535 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Clevers H., Wnt/beta-catenin signaling in development and disease. Cell 127, 469–480 (2006). [DOI] [PubMed] [Google Scholar]
- 26.van Amerongen R., Mikels A., Nusse R., Alternative wnt signaling is initiated by distinct receptors. Sci. Signal. 1, re9 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Bafico A., Liu G., Yaniv A., Gazit A., Aaronson S. A., Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat. Cell Biol. 3, 683–686 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Mao B., et al. , LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature 411, 321–325 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Li X., et al. , Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J. Biol. Chem. 280, 19883–19887 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Semenov M. V., He X., LRP5 mutations linked to high bone mass diseases cause reduced LRP5 binding and inhibition by SOST. J. Biol. Chem. 281, 38276–38284 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Hao H. X., et al. , ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200 (2012). [DOI] [PubMed] [Google Scholar]
- 32.de Lau W., Peng W. C., Gros P., Clevers H., The R-spondin/Lgr5/Rnf43 module: Regulator of Wnt signal strength. Genes Dev. 28, 305–316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koo B. K., et al. , Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Clevers H., Loh K. M., Nusse R., An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 54 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Whyte J. L., Smith A. A., Helms J. A., Wnt signaling and injury repair. Cold Spring Harb. Perspect. Biol. 4, a008078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nusse R., Clevers H., Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Tao Y., et al. , Tailored tetravalent antibodies potently and specifically activate Wnt/Frizzled pathways in cells, organoids and mice. eLife 8, e24903 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H., et al. , Development of potent, selective surrogate WNT molecules and their application in defining frizzled requirements. Cell Chem. Biol. 27, 598–609.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Janda C. Y., et al. , Surrogate Wnt agonists that phenocopy canonical Wnt and β-catenin signalling. Nature 545, 234–237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao Y., et al. , Next-generation surrogate Wnts support organoid growth and deconvolute frizzled pleiotropy in vivo. Cell Stem Cell 27, 840–851.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bourhis E., et al. , Reconstitution of a frizzled8.Wnt3a.LRP6 signaling complex reveals multiple Wnt and Dkk1 binding sites on LRP6. J. Biol. Chem. 285, 9172–9179 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krätzner R., et al. , Structure of Ecballium elaterium trypsin inhibitor II (EETI-II): A rigid molecular scaffold. Acta Crystallogr. D Biol. Crystallogr. 61, 1255–1262 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Bourhis E., et al. , Wnt antagonists bind through a short peptide to the first β-propeller domain of LRP5/6. Structure 19, 1433–1442 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Tonikian R., Zhang Y., Boone C., Sidhu S. S., Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat. Protoc. 2, 1368–1386 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Gong Y., et al. , Wnt isoform-specific interactions with coreceptor specify inhibition or potentiation of signaling by LRP6 antibodies. PLoS One 5, e12682 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian H., et al. , Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 11, 33–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nile A. H., et al. , A selective peptide inhibitor of Frizzled 7 receptors disrupts intestinal stem cells. Nat. Chem. Biol. 14, 582–590 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Xiong L., et al. , The first bioactive Wnt1 protein: Purification of Wnt1/sFRP complexes. https://resources.rndsystems.com/images/site/bio-techne-bioactive-wnt1-wp.pdf.
- 49.Evenepoel P., D’Haese P., Brandenburg V., Sclerostin and DKK1: New players in renal bone and vascular disease. Kidney Int. 88, 235–240 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Mason J. J., Williams B. O., SOST and DKK: Antagonists of LRP family signaling as targets for treating bone disease. J. Osteoporos. 2010, 460120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahn V. E., et al. , Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. Dev. Cell 21, 862–873 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng Z., et al. , Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nat. Struct. Mol. Biol. 18, 1204–1210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J., et al. , Sclerostin inhibits Wnt signaling through tandem interaction with two LRP6 ectodomains. Nat. Commun. 11, 5357 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holdsworth G., et al. , Characterization of the interaction of sclerostin with the low density lipoprotein receptor-related protein (LRP) family of Wnt co-receptors. J. Biol. Chem. 287, 26464–26477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fahiminiya S., et al. , Mutations in WNT1 are a cause of osteogenesis imperfecta. J. Med. Genet. 50, 345–348 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Laine C. M., et al. , WNT1 mutations in early-onset osteoporosis and osteogenesis imperfecta. N. Engl. J. Med. 368, 1809–1816 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pyott S. M., et al. , WNT1 mutations in families affected by moderately severe and progressive recessive osteogenesis imperfecta. Am. J. Hum. Genet. 92, 590–597 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang D., et al. , Isolation and characterization of MC3T3-E1 preosteoblast subclones with distinct in vitro and in vivo differentiation/mineralization potential. J. Bone Miner. Res. 14, 893–903 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Thouverey C., Caverzasio J., Sclerostin inhibits osteoblast differentiation without affecting BMP2/SMAD1/5 or Wnt3a/β-catenin signaling but through activation of platelet-derived growth factor receptor signaling in vitro. Bonekey Rep. 4, 757 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kedlaya R., et al. , Sclerostin inhibition reverses skeletal fragility in an Lrp5-deficient mouse model of OPPG syndrome. Sci. Transl. Med. 5, 211ra158 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang M. K., et al. , Reversing LRP5-dependent osteoporosis and SOST deficiency-induced sclerosing bone disorders by altering WNT signaling activity. J. Bone Miner. Res. 29, 29–42 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Nabhan A. N., Brownfield D. G., Harbury P. B., Krasnow M. A., Desai T. J., Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zacharias W. J., et al. , Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barkauskas C. E., et al. , Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chee Y. C., et al. , Intrinsic xenobiotic resistance of the intestinal stem cell niche. Dev. Cell 46, 681 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Binz H. K., Amstutz P., Plückthun A., Engineering novel binding proteins from nonimmunoglobulin domains. Nat. Biotechnol. 23, 1257–1268 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Ciesiołkiewicz A., Lizandra Perez J., Berlicki Ł., Miniproteins in medicinal chemistry. Bioorg. Med. Chem. Lett. 71, 128806 (2022). [DOI] [PubMed] [Google Scholar]
- 68.Baker E. G., Bartlett G. J., Porter Goff K. L., Woolfson D. N., Miniprotein design: Past, present, and prospects. Acc. Chem. Res. 50, 2085–2092 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Sachdev E., Gong J., Rimel B., Mita M., Adnectin-targeted inhibitors: Rationale and results. Curr. Oncol. Rep. 17, 35 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Schlatter D., et al. , Generation, characterization and structural data of chymase binding proteins based on the human Fyn kinase SH3 domain. MAbs 4, 497–508 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frejd F. Y., Kim K. T., Affibody molecules as engineered protein drugs. Exp. Mol. Med. 49, e306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joeng K. S., et al. , Osteocyte-specific WNT1 regulates osteoblast function during bone homeostasis. J. Clin. Invest. 127, 2678–2688 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang F., Rummukainen P., Heino T. J., Kiviranta R., Osteoblastic Wnt1 regulates periosteal bone formation in adult mice. Bone 143, 115754 (2021). [DOI] [PubMed] [Google Scholar]
- 74.Bech E. M., Pedersen S. L., Jensen K. J., Chemical strategies for half-life extension of biopharmaceuticals: Lipidation and its alternatives. ACS Med. Chem. Lett. 9, 577–580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zaman R., et al. , Current strategies in extending half-lives of therapeutic proteins. J. Control. Release 301, 176–189 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Zhang Y., et al. , Inhibition of Wnt signaling by Dishevelled PDZ peptides. Nat. Chem. Biol. 5, 217–219 (2009). [DOI] [PubMed] [Google Scholar]
- 77.Skelton N. J., et al. , Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain. J. Biol. Chem. 278, 7645–7654 (2003). [DOI] [PubMed] [Google Scholar]
- 78.Kunkel T. A., Roberts J. D., Zakour R. A., Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154, 367–382 (1987). [DOI] [PubMed] [Google Scholar]
- 79.Sato T., et al. , Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 80.Hansen S., et al. , Directed evolution identifies high-affinity cystine-knot peptide agonists and antagonists of Wnt/β-catenin signaling. PDB. https://www.rcsb.org/structure/7NAM. Deposited 21 June 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Diffraction data and coordinates have been deposited at the PDB using accession code 7NAM (80).
All of the study data are included in the article and/or supporting information.