Significance
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer lacking targeted therapy. Since the transcript variation in TNBC remains unexplored, we performed systematic analysis of RNA splicing. We identified an oncogenic transcript, macrophage receptor with collagenous structure–tumor-specific transcript (MARCO-TST), which is specifically expressed in TNBC tumor cells and activates the hypoxia pathway. We also illustrated that BRD4-bound superenhancer drives the transcription of MARCO-TST. BET inhibitors result in sustained repression of MARCO-TST function and transcriptional activity. Our study provides a potential therapeutic strategy of TNBC to restrain MARCO-TST activity.
Keywords: triple-negative breast cancer, MARCO, RNA splicing, hypoxia, BET inhibitor
Abstract
The transcription variation, leading to various forms of transcripts and protein diversity, remains largely unexplored in triple-negative breast cancers (TNBCs). Here, we presented a comprehensive analysis of RNA splicing in breast cancer to illustrate the biological function and clinical implications of tumor-specific transcripts (TSTs) arising from these splicing junctions. Aberrant RNA splicing or TSTs were frequently harbored in TNBC and were correlated with a poor outcome. We discovered a tumor-specific splicing variant of macrophage receptor with collagenous structure–TST (MARCO-TST), which was distinguished from myeloid cell-specific wild-type MARCO. MARCO-TST expression was associated with poor outcomes in TNBC patients and could promote tumor progression in vitro and in vivo. Mechanically, MARCO-TST interacted with PLOD2 and enhanced the stability of HIF-1α, which resulted in the metabolic dysregulation of TNBC to form a hypoxic tumor microenvironment. MARCO-TST was initiated from a de novo alternative transcription initiation site that was activated by a superenhancer. Tumors with MARCO-TST expression conferred greater sensitivity to bromodomain and extraterminal protein inhibitors. This treatment strategy was further validated in patient-derived organoids. In conclusion, our results revealed the transcription variation landscape of TNBC, highlighting MARCO-TST as a crucial oncogenic transcript and therapeutic target.
Triple-negative breast cancer (TNBC) is a subtype of breast cancer lacking estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) expression that accounts for ∼15 to 20% of all breast cancers (1). Compared to hormone receptor-positive or HER2-positive breast cancer, TNBC has a higher relapse rate at an early time and fewer treatment options (2). The development of targeted therapies for TNBC is challenging because of its molecular heterogeneity and lack of prevalent therapeutic targets. Therefore, there is an urgent need to understand the heterogeneity within TNBC and identify more actionable targets from multidimensional data.
To date, genomic studies have demonstrated the driving role of genomic alterations in TNBC, such as germline BRCA1/2 mutation, somatic TP53 mutation, and MYC amplification (3–5). However, breast cancer was found to have a relatively low mutation frequency among multiple cancer types (6). Therefore, other driver alterations, such as transcriptome variations and epigenetic modifications, need to be studied (7). RNA splicing is an important mechanism leading to transcription variation and protein diversity. In tumorigenic states, the RNA splicing machinery is often dysregulated and therefore leads to the biogenesis of tumor-specific splice events (8, 9). In addition, targeting the spliceosome is selectively effective in MYC-driven cancers, including TNBC (10, 11). Alternative transcription initiation (ATI) or alternative promoter usage is also a main source of transcription variation. Genome-wide studies have revealed that transcription start sites are frequently differentially used in cancer (12, 13). Recently, a pan-cancer transcriptome analysis revealed that promoter usage was dominated by cancer types and was associated with patient survival (14). A novel isoform of the anaplastic lymphoma kinase (ALK) has been reported to be a variant derived from tumor-specific ATI (ALKATI) that promotes tumorigenesis and conferred ALK inhibitor sensitivity (15). Thus, specific alterations in transcription induce functional impacts that drive tumor progression and confer therapeutic vulnerability.
In this study, we aimed to comprehensively analyze transcriptome variations in TNBC and screen tumor-specific alterations for potential treatment targets. We identified 227 tumor-specific transcripts (TSTs), including a variant of macrophage receptor with collagenous structure (MARCO), which promote tumor progression and imply therapeutic vulnerability in TNBC.
Results
The Landscape of RNA Splicing in TNBC.
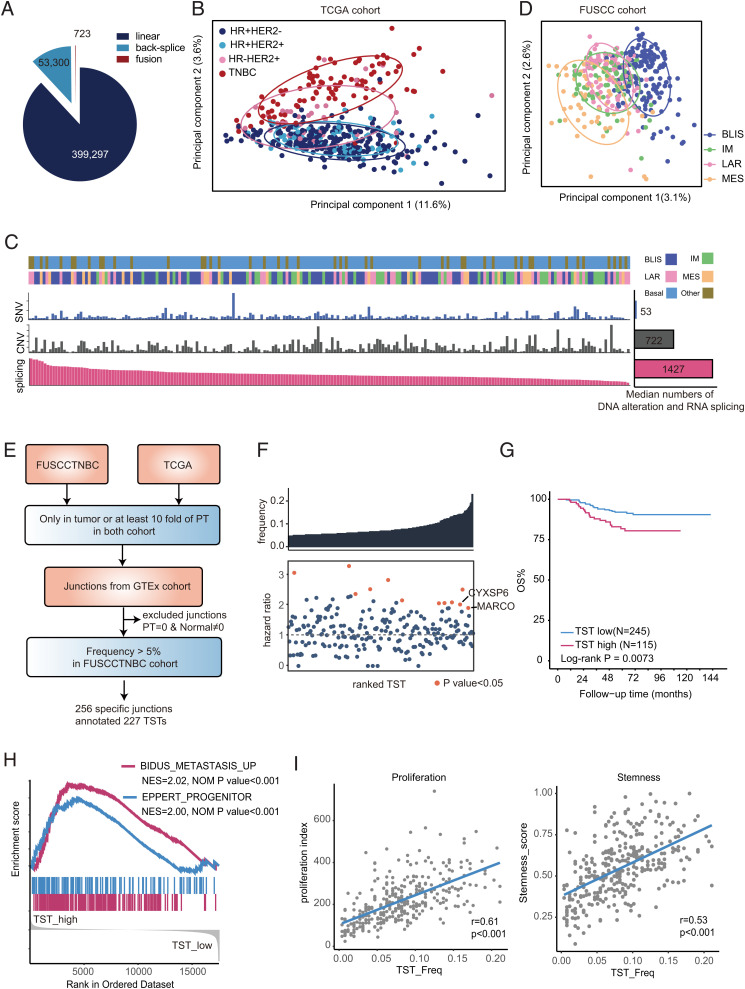
To assess the landscape of RNA splicing in TNBCs, we collected RNA-sequencing (RNA-seq) data of 360 TNBC patients (360 tumors and 88 paired adjacent nontumor tissues) from the Fudan University Shanghai Cancer Center TNBC (FUSCCTNBC) cohort (SI Appendix, Fig. S1A). RNA splicing junctions were identified using Assembling Splice Junctions Analysis (ASJA) software. This software could identify and characterize all splice junctions (including linear junctions, fusion junctions, and back-splice junctions) from high-throughput RNA-seq data (16). Junctions with low confidence (maximum expression value [CPT] of <0.8 or expression frequency of <1%) were filtered. Here, we identified 453,320 splice junctions from the FUSCCTNBC cohort, including 399,297 linear junctions, 723 fusion junctions, and 53,330 back-splice junctions (Fig. 1A), and 16.6% of the junctions were nonannotated junctions (SI Appendix, Fig. S1B). We observed that 80.68% of the junctions overlapped with known genes, and the remaining junctions originated from intergenic regions (SI Appendix, Fig. S1C).
Fig. 1.
The landscape of RNA splicing in TNBC. (A) The fraction and number of linear, fusion, and back-splice junctions in the FUSCCTNBC cohort. (B) PCA of splicing junctions of breast cancer tissues in the TCGA cohort (n = 548) using the CPT of splicing junctions. Ellipses are drawn for the 68% confidence zone. HR, hormone receptor. (C) The number of DNA or RNA alterations across TNBCs in the FUSCCTNBC cohort. The samples were ordered by the number of alternative splicing events per sample from high to low. The bar plot shows the median number of DNA or RNA alterations. SNV, single nucleotide variants; CNV, copy number variation. (D) PCA of splicing junctions of TNBC in the FUSCCTNBC cohort (n = 360) using the CPT of splicing junctions. Ellipses are drawn for the 68% confidence zone. (E) The workflow for the identification and validation of TSTs in TNBC. (F) Detection frequency and hazard ratio of each TST. The Top Box indicates the detection frequency. The Bottom Box shows the hazard ratio of DMFS, and orange points represent survival-associated TSTs. The P values were calculated using the Cox regression model. (G) Kaplan-Meier plots for the overall survival (OS) of patients with high TST burden (>22 transcripts) and low TST burden (≤22 transcripts). The P values were calculated using the log-rank test. (H) GSEA plots showing the enrichment of metastasis- and progenitor-related gene sets in high-TST-burden patients compared to low patients. (I) Correlation of TST burden with proliferation and stemness scores in the FUSCCTNBC cohort. Pearson’s correlation coefficient was used to determine the correlation (SI Appendix, Figs. S1 and S2 and Datasets S1 and S2).
Since linear junctions are the main component of RNA splice junctions, we further focused on the characteristics of linear junctions (Fig. 1A). We first visualized the splicing pattern across different breast cancer molecular subtypes in the Cancer Genome Atlas (TCGA) cohort by using principal component analysis (PCA), highlighting the molecular subtype differences in RNA splicing (Fig. 1B). TNBC had distinctive RNA splicing patterns from hormone receptor-positive tumors, whereas hormone receptor-negative HER2-positive tumors had mixed alternative splicing patterns. This difference could also be observed in terms of exon skipping patterns (SI Appendix, Fig. S1D). Next, we sought to compare these transcriptome variations to genomic alterations by analyzing the frequency of RNA splicing, nonsynonymous mutations, and copy-number alteration (CNA) at the gene level in the FUSCCTNBC cohort (17). We observed that the median number of RNA splicing events was higher than that of nonsynonymous mutations and CNAs (Fig. 1C). In addition, no correlation was found between RNA splicing and nonsynonymous mutations, homologous recombination deficiency score, or CNA (SI Appendix, Fig. S1E), suggesting that transcription variations are not due to genomic alterations and may have important oncogenic functions in TNBCs.
According to previous research, TNBCs are highly heterogeneous and can be classified into the following four messenger RNA (mRNA) subtypes: luminal androgen receptor (LAR), immunomodulatory (IM), basal-like immune-suppressed (BLIS), and mesenchymal-like (MES) (18). We further evaluated the heterogeneity of RNA splicing with TNBC mRNA subtypes. PCA illustrated that the BLIS subtype was clearly separated from the other subtypes (Fig. 1D and SI Appendix, Fig. S1F). Subsequently, pathway analysis indicated that genes with RNA splicing contributing to principle component 1 were particularly enriched in DNA damage and repair pathways (SI Appendix, Fig. S1G). Therefore, the mean difference in RNA splicing genes between BLIS and non-BLIS tumors was associated with DNA damage and repair functions, which is consistent with the biological features of BLIS tumors (18). Collectively, our results provide a general picture of transcription variation in breast cancer and suggest that RNA splicing is pronounced in TNBC and the BLIS subtype has a distinct splicing pattern.
TSTs Are Strongly Associated with a Poor Prognosis in TNBC.
To identify transcriptome variations that predominantly occur in tumor samples, we developed a computational pipeline for the identification of tumor-specific junctions (TSJs), which was defined as follows: 1) not detected in normal and paired adjacent nontumor tissues or 2) expression fold change of >10 between tumor and paired adjacent nontumor tissues (Fig. 1E and SI Appendix, Fig. S2A). In the FUSCCTNBC cohort, we identified 3,893 potential TSJs, and 3,521 of those TSJs could be validated in the TCGA cohort. Next, we excluded junctions not expressed in adjacent nontumor tissues but in normal tissues from the Genotype-Tissue Expression (GTEx) database (Fig. 1E). Finally, we included 256 TSJs with an expression frequency of >5% in the FUSCCTNBC cohort for further study (Dataset S1). Notably, most of the TSJs were unannotated (92%) in GENCODE and derived from the intragenic region (64%) (SI Appendix, Fig. S2B). We obtained 227 TSTs from these TSJs, including 85 noncoding RNAs and 142 protein-coding RNAs (Dataset S2). TSTs were frequently expressed in TNBC, and BLIS tumors had the highest TST expression frequency (SI Appendix, Fig. S2C). However, a low association of genes with somatic mutations or TSTs was observed (SI Appendix, Fig. S2D). Known somatic mutations in TNBC, such as TP53 and PIK3CA, displayed no TSTs, while top-ranked TSTs did not display mutations. Similarly, the number of genes with mutations and TSTs showed a low correlation (SI Appendix, Fig. S2E). This pattern supported that factors other than genomic alterations may be major determinants of TST expression in cancer.
To investigate the clinical significance of TSTs, we examined the association of TSTs with distant metastasis-free survival (DMFS) in the FUSCCTNBC cohort. We identified 12 TSTs that showed significant associations with worse DMFS (hazard ratio of >1) (Fig. 1F). Moreover, we found that patients with a high TST burden (TST-high group, TST detection of >22 transcripts per patient) had worse survival than patients with a low TST burden (TST-low group, TST detection of ≤22 transcripts per patient) (Fig. 1G and SI Appendix, Fig. S2F). Multivariate Cox proportional hazard models also revealed that TST burden independently predicted worse DMFS after adjusting for age at diagnosis, tumor size, node status, Ki-67 value, tumor grade, and TNBC mRNA subtype (SI Appendix, Fig. S2G). Meanwhile, the nonsynonymous mutation load was not associated with survival (SI Appendix, Fig. S2H), indicating a more important prognostic role of TSTs in TNBC.
We further investigated the transcriptome features of patients with different TST burdens. Gene Set Enrichment Analysis (GSEA) showed that metastasis- and stemness-related genes were enriched in the TST-high group (Fig. 1H). Moreover, we calculated the correlation of TST burden with different molecular features and observed a positive correlation with the proliferation index and stemness score (Fig. 1I). Meanwhile, the TST-high group was mainly composed of BLIS subtype (SI Appendix, Fig. S2I). In addition to TNBC mRNA subtypes, the TST burden could partly explain the splicing diversity among TNBCs (SI Appendix, Fig. S2J). Collectively, these results illustrate that TST-high patients are an aggressive subgroup with poor outcomes.
A MARCO Splicing Variant, MARCO-TST, Formed by ATI, Is Frequently Expressed in TNBC.
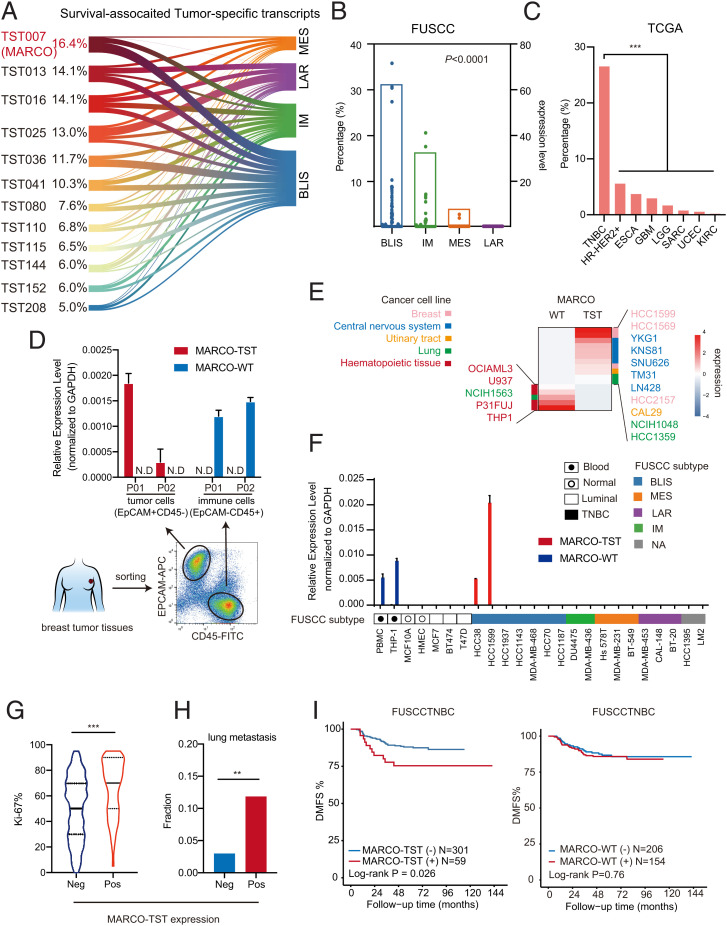
We further screened the survival-associated TSTs in TNBC to identify TSTs with high frequency that may have crucial biological function in TNBC (Fig. 1F and Dataset S2). First, we observed that survival-associated TSTs were most frequently detected in the BLIS subtype (Fig. 2A). Of these TSTs, we indicated that macrophage receptor with collagenous structure-TST (MARCO-TST) showed the highest frequency (16.4%) and was significantly enriched in BLIS (30.9%) tumors (Fig. 2 A and B). Notably, with external validation in the pan-cancer analysis of the TCGA cohort, we noticed that TNBC had the highest frequency of MARCO-TST expression among multiple cancer types (Fig. 2C). MARCO-TST is a splicing variant of MARCO that contains three extra exons at the 5′ end, and the exon 4 is identical to exon 2 of wild-type MARCO (MARCO-WT), which suggested an ATI site upstream of MARCO-WT (SI Appendix, Fig. S3A). We acquired the full-length sequence through short-read and long-read RNA-seq data from HCC1599 cells (SI Appendix, Fig. S3B). Then, PCR and Sanger sequencing were performed to confirm the full-length sequence of MARCO-TST with specific primers (SI Appendix, Fig. S3C). Likewise, qRT-PCR could detect MARCO-TST expression in 15% of TNBCs but not in paired adjacent nontumor tissues or non-TNBC tissues (SI Appendix, Fig. S3D). Therefore, we discovered a variant of MARCO that is selectively expressed in TNBC; thus, we need to further study MARCO-TST expression, regulation, and function in TNBC.
Fig. 2.
Identifying a TST, MARCO-TST, in TNBC. (A) Expression frequency of survival-associated TSTs across TNBC mRNA subtypes. (B) MARCO-TST expression frequency (bar plot) and level (dot plot) across TNBC mRNA subtypes. (C) Expression frequency of MARCO-TST in the TCGA cohort (n = 9,581, including 33 cancer types). Cancer types without MARCO-TST expression were eliminated. (D) qRT–PCR analysis of MARCO variant expression in EpCAM+CD45− tumor cells and EpCAM−CD45+ immune cells sorted from TNBC tumor tissues (P01 and P02). The expression value was normalized to GAPDH. Data are represented as the mean ± SD. Samples with cycle threshold values over 40 were defined as not detected (N.D.). (E) Heatmap of MARCO-TST and MARCO-WT transcript expression in the CCLE database (n = 933). Cell lines with no expression of either the MARCO-TST or MARCO-WT transcript were eliminated. (F) qRT-PCR analysis of MARCO variant expression in multiple cell lines. Samples with cycle threshold values over 40 were defined as N.D. (G and H) The Ki-67 value (G) and lung metastasis frequency (H) of MARCO-TST-expressing (n = 59) and MARCO-TST-deficient patients (n = 301) in the FUSCCTNBC cohort. (I) Kaplan–Meier plot of the DMFS of TNBC patients with MARCO-TST or MARCO-WT expression in FUSCCTNBC cohort. The P values were calculated using the log-rank test. **P < 0.01; ***P < 0.001. In B, C, and H, the P values were calculated using the two-tailed χ2 test. In G, the P values were calculated using the Wilcoxon rank-sum test. See also SI Appendix, Fig. S3 and Table S1.
MARCO-TST Distinguished from MARCO-WT Is Expressed in Tumor Cells and Is Correlated with Decreased Survival in TNBC Patients.
Previous studies have shown that MARCO is a marker of M2 macrophages (19). To further investigate the expression pattern of MARCO-TST and -WT in different cell populations, we sorted tumor cells and immune cells from TNBC tumor tissues. We observed that MARCO-TST was expressed in EPCAM+CD45− tumor cells, while MARCO-WT was expressed in EPCAM−CD45+ immune cells (Fig. 2D). In addition, RNA splicing junction data of cell lines from the Cancer Cell Line Encyclopedia (CCLE) database showed that MARCO-TST was mainly expressed in solid tumor cell lines, including breast and brain tumors, while MARCO-WT was specifically expressed in hematopoietic cells, including a macrophage cell line (THP-1) (Fig. 2E). Moreover, the qRT-PCR assay showed that two TNBC cell lines (HCC38 and HCC1599) had MARCO-TST expression, which was not detected in normal or luminal breast cancer cell lines (Fig. 2F). On the other hand, MARCO-WT could be detected in peripheral blood mononuclear cells and THP-1 cells but not in breast normal cells or tumor cells, including HCC38 and HCC1599. Collectively, these results suggest that MARCO-TST is a tumor-cell-specific expressed transcript, while MARCO-WT is selectively expressed in immune cells.
We further explored the association of MARCO-TST and MARCO-WT with tumor progression in TNBC. Within the FUSCCTNBC cohort, MARCO-TST expression was associated with prognostic clinical factors, including tumor grade and Ki-67 value of TNBCs but not with tumor size or node status (Fig. 2G and SI Appendix, Table S1). Moreover, MARCO-TST-positive patients had a higher probability of lung metastasis and shorter survival than MARCO-TST-negative patients (Fig. 2 H and I). A similar trend was also observed in the TCGA cohort but with no significant difference due to the small sample size (SI Appendix, Fig. S3E). However, MARCO-WT showed no association with patient outcomes in TNBCs (Fig. 2I and SI Appendix, Fig. S3E). These results suggested that MARCO-TST was distinguished from MARCO-WT as a tumor-specific splicing variant as well as a potential oncogenic transcript in TNBC.
MARCO-TST Promotes TNBC Tumor Growth and Metastasis.
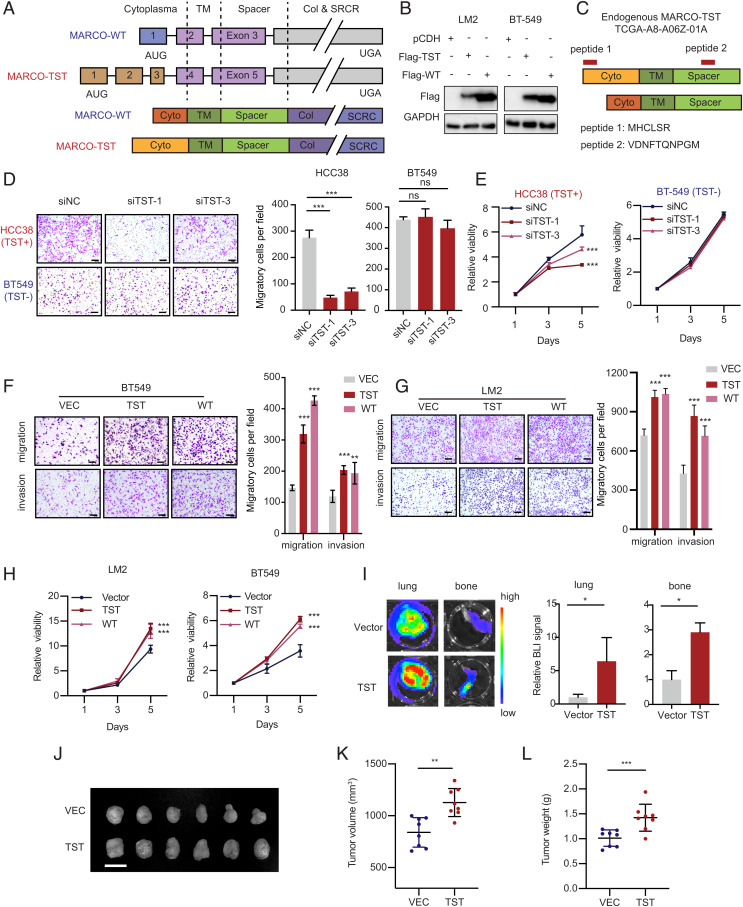
Evaluation of the sequence of MARCO-TST revealed a start codon AUG in frame with the UGA stop codon of MARCO-WT (Fig. 3 A, Upper Panel). Translation from AUG to UGA is predicted to encode a larger form of MARCO of 535 amino acids (SI Appendix, Fig. S4A). The two proteins shared the same transmembrane, spacer, collagen-like, and scavenger receptor cysteine-rich domains but contained different cytoplasmic domains (Fig. 3 A, Lower Panel). Western blotting of LM2 and BT549 cells ectopically expressing full-length MARCO-TST and MARCO-WT showed bands at 70 kDa and 68 kDa, respectively (Fig. 3B). Furthermore, the unique cytoplasmic domain resulting from MARCO-TST was confirmed by searching mass spectrometry data from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) project (Fig. 3C). To explore the oncogenic function of MARCO-TST, we designed three small interfering RNA (siRNAs) targeting the unique sequence of MARCO-TST and screened siRNAs that can readily knockdown MARCO-TST expression (SI Appendix, Fig. S4 B and C). MARCO-TST knockdown resulted in decreased cell migration and proliferation capacity in MARCO-TST-expressing HCC38 cells (Fig. 3 D and E). Moreover, these siRNAs had no effect in BT549 cells lacking MARCO-TST. Next, we examined whether the function of MARCO-TST depends on the genetic context of MARCO-TST-positive cells. We found that ectopic expression of MARCO-TST could also enhance the migration, invasion, and proliferation capacity of LM2 and BT549 cells (Fig. 3 F–H). These effects could be abrogated with MARCO-TST depletion in MARCO-TST-overexpressing HCC38 or LM2 cells (SI Appendix, Fig. S4 D–G). To validate our findings in vivo, an orthotopic injection of LM2 cells stably ectopically expressing MARCO-TST or an empty vector into female NOD/SCID mice was established. We observed that MARCO-TST enhanced tumor growth and lung and bone metastasis in vivo (Fig. 3 I–L). Collectively, these results indicate that the MARCO-TST protein promotes TNBC tumor growth and distant metastasis.
Fig. 3.
MARCO-TST drives TNBC tumor growth and metastasis. (A) Illustration of MARCO transcript variants (Top) and corresponding protein isoforms (Bottom). TM, transmembrane domain; Col, collagen-like domain; SRCR, scavenger receptor cysteine-rich domain. (B) Western blots of Flag in LM2 and BT549 cells transfected with Flag-tagged MARCO-TST or -WT. (C) The N-terminal domain of MARCO-TST was detected using mass spectrometry data from the CPTAC database. Peptides detected are depicted in red at their mapping position onto the protein sequence. (D and E) Migration (D) and proliferation (E) assays of HCC38 and BT549 cells transfected with MARCO-TST siRNAs. Scale bar, 100 µm. (F and G) Migration and invasion assays of BT549 (F) and LM2 (G) cells stably expressing control, MARCO-TST, or MARCO-WT plasmids. (H) Proliferation assay of LM2 and BT549 cells stably expressing control, MARCO-TST, or MARCO-WT plasmids. (I) Representative lung and bone images (Left) and BLI quantitative data (Right) of mice 8 wk after tumor cell injection. n = 8 lungs and n = 16 bones per group. (J–L) Images of tumors (J), tumor volume (K), and tumor weight (L) after dissection of MARCO-TST-overexpressing LM2 cells injected orthotopically into the mammary fat pad of NOD/SCID mice (n = 8). Scale bar, 2 cm. Data are represented as the mean ± SD; *P < 0.05; **P < 0.01; ***P < 0.001. The P values were calculated using two-tailed Student’s t test (SI Appendix, Fig. S4).
MARCO-TST Enhances HIF-1α Protein Stability.
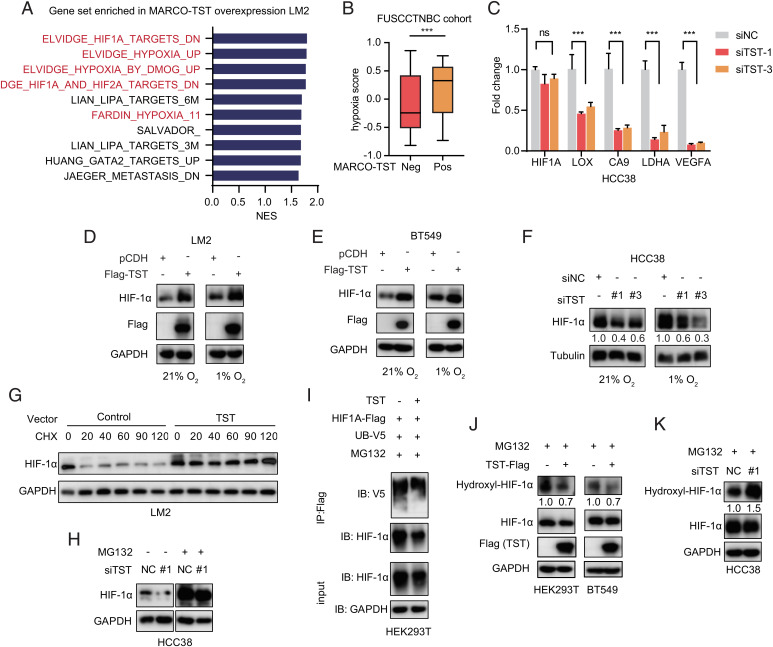
To elucidate the molecular mechanisms by which MARCO-TST promotes cancer progression, we performed RNA-seq gene expression profiling of MARCO-TST-expressing or control LM2 cells. GSEA indicated that the top 4 gene sets enriched in MARCO-TST-expressing cells were all related to the hypoxia pathway (Fig. 4A). Similar results were obtained in MARCO-TST-positive TNBC patients (SI Appendix, Fig. S5 A and B). To further confirm this result, we evaluated the hypoxia status of tumor samples in the FUSCCTNBC cohort using a 15-gene expression signature (20, 21). Indeed, TNBC tumors with MARCO-TST expression showed significantly higher hypoxia scores than those without MARCO-TST expression (Fig. 4B). Collectively, these results suggested that MARCO-TST might contribute to the hypoxic response in tumor.
Fig. 4.
MARCO-TST enhances HIF-1α protein stability. (A) GSEA of pathways significantly enriched (P < 0.05) in MARCO-TST-overexpressing LM2 cells, showing the top ten enriched pathways. NES, normalized enrichment score. (B) The hypoxia score of TNBC tumors with or without MARCO-TST expression in the FUSCCTNBC cohort. Neg, negative; pos, positive. (C) qRT-PCR analysis of HIF1A and its target genes in MARCO-TST-silenced HCC38 cells. siNC, scrambled siRNA; siTST, MARCO-TST siRNA. Data are represented as the mean ± SD; ***P < 0.001. The P values were calculated using two-tailed Student’s t test. (D and E) Immunoblot analysis of HIF-1α protein levels in MARCO-TST-expressing LM2 (D) and BT549 cells (E) under normoxic and hypoxic conditions. (F) HIF-1α protein levels in MARCO-TST-silenced HCC38 cells under normoxic and hypoxic conditions. (G) Immunoblot analysis of HIF-1α protein levels in control and MARCO-TST-expressing LM2 cells treated with CHX (50 µg/mL) for different times. (H) HIF-1α protein levels in MARCO-TST-silenced HCC38 cells with or without proteasome inhibitor (MG132) treatment. NC, scrambled siRNA. (I) Immunoprecipitation and Western blot analysis of HIF-1α ubiquitination in control and MARCO-TST-expressing HEK293T cells treated with MG132 for 8 h. (J and K) Analysis of the hydroxylation level of HIF-1α in MARCO-TST-expressing HEK293T and BT549 cells (J) or MARCO-TST-silenced HCC38 cells (K) (SI Appendix, Fig. S5).
Hypoxia-inducible factor 1α (HIF-1α) is a major mediator of the hypoxic response and regulates the expression of genes involved in controlling O2 homeostasis (22). Therefore, we assessed whether MARCO-TST expression could induce HIF-1α expression. qRT-PCR assays showed that knockdown of MARCO-TST had little effect on HIF1A mRNA levels (Fig. 4C). However, HIF-1α target genes, including LOX, CA9, LDHA, and VEGFA, were reduced in HCC38 cells with MARCO-TST knockdown (Fig. 4C). MARCO-TST expression increased HIF-1α protein levels under both normoxic and hypoxic conditions (Fig. 4 D and E). Similarly, MARCO-TST knockdown decreased the protein expression level of HIF-1α in the HCC38 cell line (Fig. 4F). In addition, ectopic expression of MARCO-WT in tumor cells also increased the HIF-1α protein (SI Appendix, Fig. S5C). Therefore, these results indicated that MARCO-TST positively regulates HIF-1α at the protein level, resulting in hypoxia pathway activation.
Since MARCO-TST had little effect on HIF1A mRNA levels, we hypothesized that MARCO-TST could enhance HIF-1α protein stability. To determine the half-life of HIF-1α, LM2 cells with MARCO-TST expression and control cells were treated with the protein synthesis inhibitor cycloheximide (CHX) for different time periods. The half-life of the HIF-1α protein was significantly increased with MARCO-TST overexpression (Fig. 4G). Importantly, treatment with the proteasome inhibitor MG132 abolished the effect of MARCO-TST on HIF-1α protein levels in MARCO-TST-silenced and MARCO-TST-overexpressing cells (Fig. 4H and SI Appendix, Fig. S5D), suggesting that HIF-1α is subjected to ubiquitination-mediated proteasomal degradation and its protein stability is regulated by MARCO-TST. Consistently, the HIF-1α ubiquitination level was also decreased in MARCO-TST-expressing cells (Fig. 4I). Previous studies have shown that the ubiquitination-dependent degradation of HIF-1α is mediated by its proline hydroxylation status at P402 and P564 (23–25). We used an antibody against hydroxylated HIF-1α (P564), and we found that MARCO-TST overexpression decreased HIF-1α hydroxylation in HEK293T and BT549 cells, while knockdown of MARCO-TST increased proline hydroxylation in HCC38 cells (Fig. 4 J and K). Then, we transfected the HIF1A-P402/P564 mutant vector into HEK293T MARCO-TST-expressing cells and HCC38 MARCO-TST-silenced cells. The HIF1A-P402/P564 mutant abrogated MARCO-TST-mediated HIF-1α activation (SI Appendix, Fig. S5 E and F). Together, these results supported our hypothesis that MARCO-TST enhances HIF-1α protein stability by reducing proline hydroxylation.
MARCO-TST Enhances HIF-1α Stability by Interacting with PLOD2.
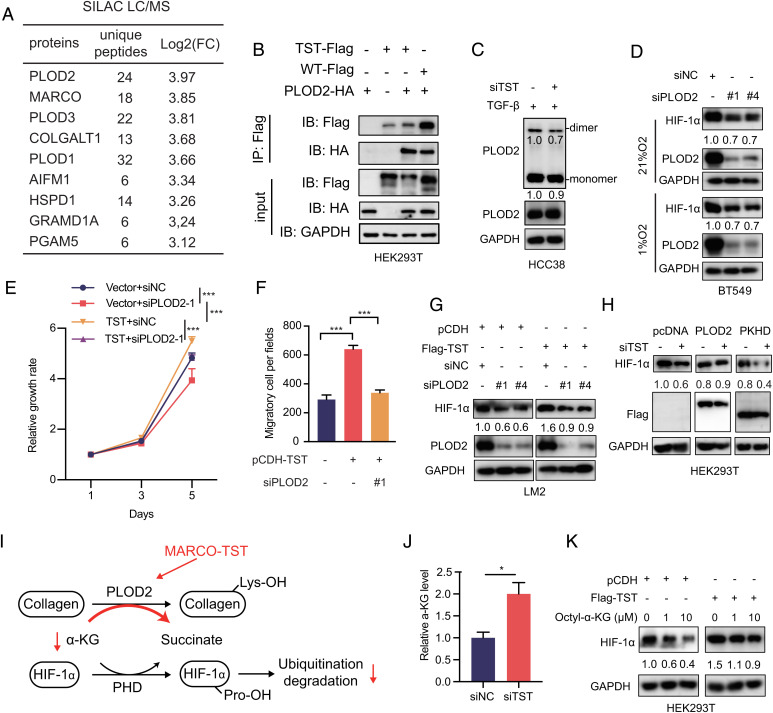
To investigate the mechanism by which MARCO-TST enhances HIF-1α stability, we performed coimmunoprecipitation (Co-IP) assays combined with stable isotope labeling with amino acids in cell culture (SILAC)-based quantitative proteomics to identify proteins associated with MARCO-TST. Stable isotopes of Lys8 and Arg10 (heavy) and Lys0 and Arg0–labeled (light) HEK293T cells were transfected with Flag-tagged MARCO-TST plasmid and empty vector, respectively. We identified 9 proteins that may interact with MARCO-TST (Fig. 5A), and the procollagen-lysine, 2-oxoglutarate 5-dioxygenase (PLOD) family had the highest binding ratio (heavy/light). The PLOD family is a collagen-modifying enzyme and plays an important role in cancer progression (26–29). Co-IP assays further confirmed the binding of PLOD2 and PLOD3 to MARCO-TST (Fig. 5B and SI Appendix, Fig. S6 A and B). However, silencing PLOD2 but not PLOD3 repressed cell migration in TNBC cell lines (SI Appendix, Fig. S5C). Therefore, we inferred that MARCO-TST could mediate tumor progression by interacting with PLOD2. In addition, PLOD2 could also bind to MARCO-WT in HEK293T cells with ectopic expression of MARCO-WT (SI Appendix, Fig. S6 A and B). However, PLOD2 and MARCO variants showed different expression patterns between different cell types (SI Appendix, Fig. S6D). PLOD2 is selectively coexpressed with MARCO-TST in tumor cells instead of MARCO-WT. Therefore, MARCO-WT does not facilitate its function by interacting with PLOD2. To determine the binding region of MARCO-TST necessary for its association with PLOD2, we obtained a series of MARCO-TST truncation mutants (SI Appendix, Fig. S6 E, Upper Panel) according to the domains of MARCO (30). We found that the transmembrane domain of MARCO-TST was the major region binding to PLOD2 (SI Appendix, Fig. S6 E, Lower Panel). Furthermore, confocal microscopy assays showed colocalization of MARCO-TST and PLOD2 in the cytoplasm (SI Appendix, Fig. S6F). Together, these results indicated that the transmembrane domain of MARCO-TST was responsible for binding with PLOD2.
Fig. 5.
MARCO-TST and PLOD2 cooperatively regulate HIF-1α stability. (A) Summary table of binding proteins of MARCO-TST detected by SILAC subjected to mass spectrometry (MS). FC, fold change. (B) HEK293T cells expressing Flag-tagged MARCO-TST or MARCO-WT with HA-tagged PLOD2 were harvested for immunoprecipitation with anti-Flag beads, and proteins were assessed by immunoblotting (IB) as indicated. (C) Immunoblot detection of PLOD2 monomers/dimers by native PAGE with nondenatured lysates (Upper) and SDS/PAGE with denatured lysates (Lower) in HCC38 cells transfected with MARCO-TST siRNA and treated with TGFβ1 for 48 h. (D) HIF-1α protein levels in PLOD2-silenced BT549 cells under normoxic and hypoxic conditions. (E and F) Growth rate (E) and migration assay (F) of MARCO-TST-overexpressing LM2 cells cotransfected with PLOD2 siRNA. (G) Immunoblot analysis of HIF-1α protein levels in LM2 cells expressing MARCO-TST with PLOD2 depletion. (H) HIF-1α protein levels in MARCO-TST knockdown HCC38 cells transfected with full-length PLOD2 or mutants lacking the hydroxylase domain (ΔPKHD). (I) Schematic of the mechanism by which MARCO-TST regulates HIF-1α stability via PLOD2 and α-KG. Lys, lysine. (J) Cytoplasmic α-KG levels were measured in control and MARCO-TST-silenced HCC38 cells. (K) HIF-1α protein levels were assessed in control and MARCO-TST-expressing LM2 cells in the presence or absence of octyl-α-KG. Data are represented as the mean ± SD; *P < 0.05; ***P < 0.001. The P values were calculated using two-tailed Student’s t test (SI Appendix, Figs. S6 and S7).
With the observation that MARCO-TST interacts with PLOD2, we further explored the function of this interaction. We first found that MARCO-TST had no effect on PLOD2 protein stability (SI Appendix, Fig. S7A). PLOD2 is an intracellular enzyme that catalyzes procollagen lysyl hydroxylation, which is a critical step of collagen biosynthesis (31, 32). Previous reports showed that PLOD2 activity relied on homodimerization formation (33–35). Therefore, we speculated that MARCO-TST might bind with PLOD2 and affect its dimerization. Nonreduced immunoblot analysis of endogenous PLOD2 showed that dimerization was decreased with the depletion of MARCO-TST in HCC38 cells (Fig. 5C). Since the dimerization of PLOD2 was increased by MARCO-TST, we hypothesized that the modulation of procollagen might be promoted. Indeed, we found that the secretion of type IV collagen was increased by the overexpression of MARCO-TST (SI Appendix, Fig. S7B), and collagen deposition was also increased in MARCO-TST-expressing tumor tissues (SI Appendix, Fig. S7C). These results indicated that MARCO-TST could interact with PLOD2 and enhance its biological function.
Similar to MARCO-TST, PLOD2 knockdown decreased HIF-1α protein levels regardless of oxygen conditions, while PLOD3 depletion showed no impact on HIF-1α (Fig. 5D and SI Appendix, Fig. S7D). The hydroxylation of HIF-1α was reduced in PLOD2-expressing HEK293T cells, and hydroxylation-deficient mutants restored this effect (SI Appendix, Fig. S7 E and F). These results suggested that PLOD2 could modulate HIF-1α protein stability. Silencing PLOD2 completely abolished the induction of the proliferation and migration elicited by MARCO-TST overexpression (Fig. 5 E and F), indicating that PLOD2 is a critical mediator of this phenotype. On the other hand, repressing MARCO-TST partly abolished the phenotype induced by PLOD2 overexpression (SI Appendix, Fig. S7 G and H). Silencing PLOD2 abrogated the HIF-1α protein increase induced by MARCO-TST overexpression (Fig. 5G). However, the PLOD2 mutant lacking the hydroxylase domain (ΔPKHD) failed to restore the HIF-1α protein in MARCO-TST-depleted HCC38 cells (Fig. 5H). Therefore, MARCO-TST increased HIF-1α protein levels by relying on PLOD2 enzyme activity. Consistently, the assessment of HEK293T cells with MARCO-TST truncated mutants showed that truncated mutants Δ2 and Δ3, lacking PLOD2 binding sites, failed to increase HIF-1α (SI Appendix, Fig. S7I). Collectively, these results illustrated that the association with PLOD2 is required for MARCO-TST to promote tumor progression and enhance HIF-1α stability.
HIF-1α is hydroxylated by an α-ketoglutarate (α-KG)-dependent prolyl hydroxylase domain (PHD) (36–38), and α-KG can increase PHD activity, leading to increased degradation of HIF-1α (39, 40). P4HA1, an α-KG-dependent dioxygenase that catalyzes the 4-hydroxylation of procollagen, reduces PHD activity to enhance the stability of HIF-1α by modulating α-KG levels (41). We therefore inferred that MARCO-TST might reduce PHD activity by reducing the concentration of α-KG via PLOD2 (Fig. 5I). The α-KG quantification assay showed that depletion of MARCO-TST increased the α-KG levels (Fig. 5J). Notably, HIF-1α activation was abrogated in MARCO-TST-overexpressing HEK293T cells with the addition of octyl-α-KG. (Fig. 5K). Collectively, MARCO-TST increases HIF-1α protein levels by modulating α-KG levels.
MARCO-TST Is Activated by Its Upstream Superenhancer.
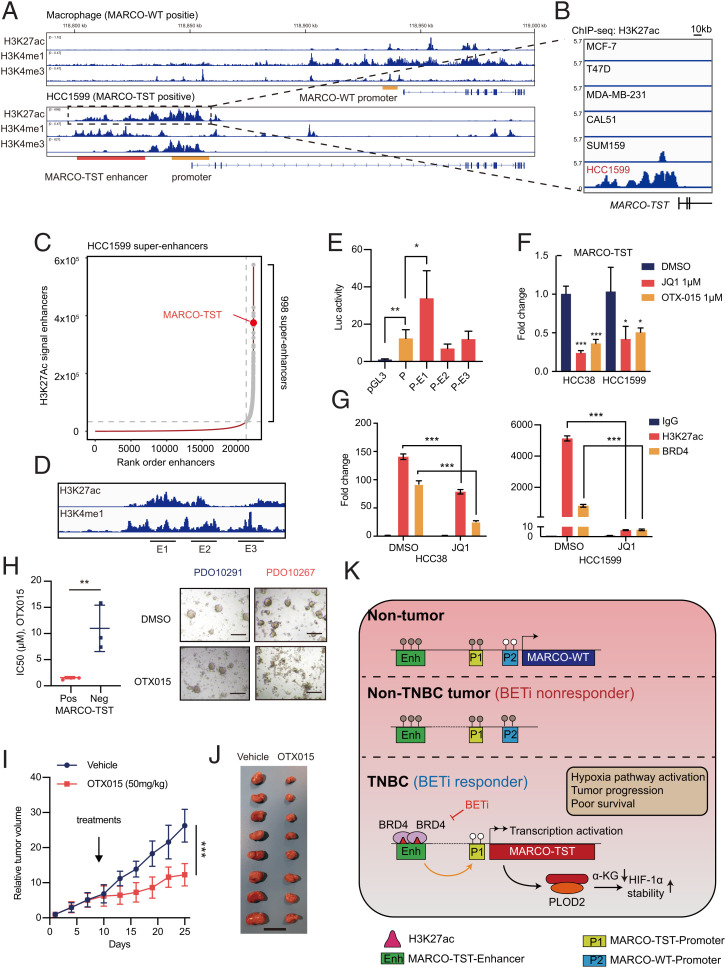
To characterize the transcriptional regulation of MARCO-TST, we examined chromatin immunoprecipitation sequencing (ChIP-seq) profiles of H3K27ac, H3K4me1, and H3K4me3 in macrophages (MARCO-WT-expressing) and HCC1599 (MARCO-TST-expressing) cell lines. ChIP-seq and ChIP-qCPR data revealed that only MARCO-TST-expressing cells had a significant enrichment of markers of active promoters (H3K27ac and H3K4me3) at the promoter of MARCO-TST but not MARCO-WT (Fig. 6A and SI Appendix, Fig. S8 A and B). These data indicated that MARCO-TST originates from a newly established transcription initial site. Notably, ChIP-seq data on H3K27ac and K3K4me1 indicated that a putative enhancer (occupied by H3K27ac and H3K4me1) exists in the upstream region of MARCO-TST in HCC1599 cells (Fig. 6A and SI Appendix, Fig. S8C). This enhancer existed only in MARCO-TST-positive cells but not in MARCO-TST-negative or non-TNBC cells (Fig. 6B). The landscape of enhancers in HCC1599 cells showed that this enhancer was a superenhancer and ranked as one of the most H3K27ac-enriched superenhancers (Fig. 6C). Enhancer RNAs (eRNAs) are products of active enhancers and are associated with enhancer activity (42, 43). To validate the activity of the MATCO-TST superenhancer in patient samples, we extracted the eRNA expression profile of the TCGA cohort from the eRic database (http://hanlab.uth.edu/eRic) (44). We identified a cluster of eRNAs located at the MARCO-TST superenhancer that showed a positive correlation with MARCO-TST mRNA expression (SI Appendix, Fig. S9 A and B). Indeed, MARCO-TST-associated eRNAs exhibited much higher expression levels in MARCO-TST-positive tumors than in MARCO-TST-negative tumors (SI Appendix, Fig. S9C). High-throughput chromosome conformation capture (Hi-C) data showed a significant increase (3.68-fold) in MARCO-TST promoter-enhancer contact in HCC1599 cells compared to human mammary epithelial cells (SI Appendix, Fig. S9D). The luciferase reporter assay confirmed higher transcription-enhancing activity in HCC38 cells transfected with the MARCO-TST promoter-enhancer vector than in those transfected with the MARCO-TST promoter vector (Fig. 6 D and E). To verify the role of superenhancers in regulating MARCO-TST transcription, we quantified the mRNA level of MARCO-TST with bromodomain (BRD) and extraterminal (BET) inhibitor treatment (JQ1 and OTX015), a competitive inhibitor that blocks BRD4 from binding to acetylated lysine residues at superenhancers and specifically diminishes the expression of superenhancer-driven genes (45, 46). Indeed, the mRNA level of MARCO-TST was repressed by JQ1 and OTX015 treatment (Fig. 6F). Similarly, the expression of MARCO-TST-associated eRNA could be depleted by BET inhibitors (SI Appendix, Fig. S9E). Moreover, ChIP-qPCR assays showed that JQ1 decreased the occupancy of H3K27ac and BRD4 at the MARCO-TST superenhancer site in HCC38 and HCC1599 cells (Fig. 6G). Together, our data indicated that the superenhancer-driven expression of MARCO-TST and BET inhibitors could abolish this mechanism.
Fig. 6.
MARCO-TST expression is regulated by its proximal upstream superenhancer and conferred BET inhibitor sensitivity. (A) ChIP-seq tracks showing the H3K27ac, H3K4me1, and H3K4me3 binding patterns of the MARCO-TST and MARCO-WT loci in macrophages and HCC1599 cells. The promoter and enhancer regions were shadowed in yellow and red, respectively. (B) ChIP-seq tracks showing the H3K27ac binding pattern of the MARCO-TST enhancer region in MCF-7, T47D, MDA-MB-231, CAL51, SUM159, and HCC1599 cells. (C) Rank order of the H3K27ac signal at enhancer loci in HCC1599 cells. Superenhancers are shown in gray points, and the SE at the proximal upstream of MARCO-TST is labeled. (D) Schematic of MARCO-TST superenhancers showing E1 to E3 DNA segments. (E) Luciferase activity of MARCO-TST superenhancers in HCC38 cells. (F) qRT-PCR analysis of MARCO-TST mRNA and eRNA levels in HCC38 or HCC1599 cells treated with 1 μM JQ1 or 1 μM OTX015 for 24 h. (G) ChIP-qPCR analysis of H3K27ac and BRD4 occupancy on the enhancer region of MARCO-TST in HCC38 cells treated with 1 μM JQ1 for 24 h. Primers (E4) used in this assay are shown in SI Appendix, Fig. S8A. (H) Viability assays in six TNBC patient-derived organoid models. The individual IC50 values and representative bright-field images of organoids treated with OTX015 are shown. Scale bars, 200 μm. PDO, patient-derived organoid. (I and J) Tumor growth (I) and image of tumors (J) of HCC1599 cell-implanted NOD/SCID mice (n = 8) treated with vehicle (n = 8) or OTX015 (n = 8). Scale bar, 2 cm. (K) Model of transcriptional regulation of MARCO-TST in TNBC and targeting MARCO-TST-positive TNBC tumors with BET inhibitors. BETi, bromodomain and extraterminal inhibitor. Data are presented as the mean ± SD; *P < 0.05; **P < 0.01; ***P < 0.001. The P values were calculated using two-tailed Student’s t test (SI Appendix, Figs. S8–S11).
Previous studies illustrated that DNA methylation is an important mechanism activating ATI (15, 47). We retrieved methylation data from the TCGA cohort. We found a significantly higher methylation level at the promoter regions in MARCO-TST-negative tumor samples than in MARCO-TST-positive samples (SI Appendix, Fig. S10 A and B). To test whether demethylation was sufficient to induce MARCO-TST expression, we treated MDA-MB-231 cells with the DNA methyltransferase inhibitor 5-aza-deoxycytidine (5-AZA). qRT-PCR assays showed that MARCO-TST transcription was increased under 5-AZA treatment in both MARCO-TST-positive and MARCO-TST-negative cells (SI Appendix, Fig. S10C).
MARCO-TST Expression Confers Sensitivity to BET Inhibitors in TNBC Tumors.
BET inhibitors are a class of compounds targeting the BRD family (including BRD2, BRD3, and BRD4) and have entered several clinical trials for solid tumors and hematological malignancies (48). Preclinical studies have demonstrated the preferential sensitivity of selective TNBC cells to BET inhibitors (49, 50). The selectivity of BET inhibition arises from the localization of BRD proteins to superenhancers that regulate oncogenic transcriptional programs (51, 52). Since BET inhibitors could regulate MARCO-TST transcription, we first investigated whether MARCO-TST-expressing cells were sensitive to BET inhibitors. We observed that TNBC cell lines with MARCO-TST expression (HCC1599 and HCC38) were more sensitive to BET inhibitors (OTX015) (SI Appendix, Fig. S11A). Validating these findings, we observed a similar association in six patient-derived organoids (Fig. 6H and SI Appendix, Fig. S11B), which exhibited differential sensitivity to OTX015. To determine the antitumor efficacy of OTX015 in vivo, we transplanted HCC1599 cells into the mammary fat pads of female NOD/SCID mice. Treatment with OTX015 significantly induced tumor regression (Fig. 6 I and J and SI Appendix, Fig. S11C). Further immunohistochemical analysis showed that OTX015 significantly reduced cell growth and HIF-1α protein levels in tumor tissues (SI Appendix, Fig. S11 D and E). Overall, these data support that BET inhibitors are a promising treatment strategy for TNBCs with MARCO-TST expression.
Discussion
Transcription variation has been shown to have an important role in TNBC and multicancer development and progression (53–55), but a complete picture of transcription variation in TNBC is still missing. In this study, we focused on the transcription variation in TNBC using the largest multiomics TNBC cohort (to date). We found that TSTs are commonly expressed in TNBC and have important prognostic value. Then, we identified a high-frequency oncogenic TST, MARCO-TST, derived from an alternation transcription initiation site for MARCO and sporadically expressed in other cancer types. We found that MARCO-TST cooperates with PLOD2 to promote tumor growth and metastasis by activating the hypoxia pathway. Furthermore, we discovered that MARCO-TST is driven by a superenhancer on which MARCO-TST-positive tumors depend the most. Importantly, preclinical models illustrated that MARCO-TST could reflect the response to BET inhibitors in TNBC patients (Fig. 6K).
ATI led to the expression of MARCO-TST. Similarly, ALK has been reported to be a variant derived from tumor-specific ATI (15). In our study, MARCO-TST, but not MARCO-WT, was frequently expressed in TNBC and was identified as a predictor of worse survival. However, according to our data, the function of MARCO-TST was similar to that of ectopically expressed MARCO-WT in tumor cells, as it promoted cell proliferation, migration, and invasion. Since studies of MARCO have mainly been conducted in macrophages (56, 57), we hypothesize that the difference in prognostic value between MARCO-TST and MARCO-WT is due to the mutually exclusive expression pattern. We found that MARCO-TST and MARCO-WT were predominantly expressed in tumor cells and macrophages, respectively. Consistent with our study, ALK variants (ALKATI, ALKF1174L, EML4–ALK, and wild-type ALK) have similar functional effects in terms of activating the ALK pathway and tumorigenesis (15).
In contrast to other alternative splicing events, such as exon skipping, which regulate gene isoform expression posttranscriptionally by splicing factors, alternative transcription initially provides a way to regulate gene isoform expression pretranscriptionally. Therefore, changes in genomic and epigenomic landscapes affect the initial transcription event (58–60). As expected, we found an H3K27ac signal at the promoter site of MARCO-TST, and this signal was only enriched in MARCO-TST-expressing cells. Surprisingly, the H3K27Ac signal spanning as much as 40 kb on the chromosome suggested an enhancer site upstream of MARCO-TST. The luciferase reporter assay further confirmed that the core enhancer region could enhance the activity of the MARCO-TST promoter. A previous study demonstrated that CpG hypomethylation at the promoter site was the mechanism of ATI activation (15, 47, 61). This mechanism was also validated at the promoter region of MARCO-TST. In addition, demethylation treatment (5-AZA) not only induced the expression of MARCO-TST but also increased enhancer activity according to the eRNA level.
Since BET BRDs are transcriptional regulators of MARCO-TST, we found that BET inhibitors are also preferentially effective at inhibiting MARCO-TST-positive tumor cell and organoid growth. BET inhibitors are currently under phase I/II clinical trials for patients with solid tumors and hematologic malignancies (62). Preclinical studies have demonstrated that TNBC has preferential sensitivity to BET inhibitors compared to other breast cancer subtypes (49, 50), but biomarkers of BET inhibitors are still lacking. Considering that the mechanism underlying the antitumor activity of BET inhibitors is attributed to selective disruption of superenhancer-associated proteins (63, 64), we speculate that BET inhibitors have selective antitumor activity in MARCO-TST-expressing TNBC. Upon BET inhibitor treatment, BET was displaced from the MARCO-TST enhancer and repressed tumor growth. Our study shows that MARCO-TST, as a target gene of BET, predicts the sensitivity of tumors to BET inhibitors.
Several limitations of this study should be considered. We have revealed the dysregulation of transcription among breast cancer, but the mechanism driving this phenomenon was undetermined. However, we proposed a correlation analysis between genetic alterations and transcription variation, which showed a limited positive relationship. Although we used patient-derived organoids to partly mimic the treatment effect on patients, the clinical effect of BET inhibitors in MARCO-TST-positive TNBC patients should be further investigated. An important extension of the present work is to evaluate the potential of TST-generating neoantigens. We have been performing the proteome analysis of this TNBC cohort, which can provide opportunities to assess the immunogenicity of splicing-derived neoantigens and the significance of TST neoantigen burden as a biomarker of immunotherapy in TNBC.
In summary, in this study, we assessed the alterations in the transcriptome of TNBC and identified MARCO-TST, which is required for TNBC cell proliferation and metastasis induced by activating the hypoxia pathway. On the basis of our preclinical results, we suggest that BET inhibitors could repress the growth of MARCO-TST-expressing TNBC tumors and that MARCO-TST could be a potential biomarker for patient treatment selection. As transcriptome alterations in TNBC have not been described before, our study opens opportunities to explore the contribution of transcription variation to tumor progression, diagnosis, and treatment.
Materials and Methods
To systematically analyze RNA splicing in TNBC, we analyzed the RNA-seq data for 360 TNBC tumors and 88 paired nontumor tissues (Sequence Read Archive [SRA]: SRP157974) by using ASJA software. To detect MARCO-TST and -WT transcripts, we designed specific primers for exons 1 and 2 of MARCO-TST and exon 1 of MARCO-WT. The primer sequences are listed in SI Appendix, Table S2. The full-length complementary DNAs of MARCO-TST and -WT were amplified from HCC1599 cells and cloned into a pcDNA3.1 vector. Transwell migration and Matrigel invasion assays were performed to assess cell invasiveness, and CCK-8 assays were performed to assess cell viability. Transcriptome sequencing of the LM2 cells was performed on the NovaSeq 6000 sequencing system. For xenograft models, tumor cells were orthotopically injected directly into the inguinal mammary fat pads of NOD/SCID mice. To assess the in vivo drug sensitivity, OTX015 (50 mg/kg) was given orally daily. More detailed methods are provided in the SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Key Research and Development Project of China (2020YFA0112304, 2021YFA1300500), the National Natural Science Foundation of China (81922048, 81874113, 82072922, 81902684, 91959207, and 92159301), the Program of Shanghai Academic/Technology Research Leader (20XD1421100), the Natural Science Foundation of Shanghai (22ZR1479200), the Shanghai Key Laboratory of Breast Cancer (12DZ2260100), the Clinical Research Plan of Shanghai Hospital Development Center (SHDC2020CR4002, SHDC2020CR5005), and the SHDC Municipal Project for Developing Emerging and Frontier Technology in Shanghai Hospitals (SHDC12021103). We also thank Dr. Gonghong Wei and Dr. Xianghuo He for editing the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. C.-M.C. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120393119/-/DCSupplemental.
Data, Materials, and Software Availability
The microarray data and sequence data for the FUSCCTNBC cohort have been deposited in the NCBI Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo; OncoScan array; GEO: GSE118527) and SRA (https://www.ncbi.nlm.nih.gov/sra; whole exome sequencing and RNA-seq; SRA: SRP157974). The RNA-seq data for the TCGA cohort, CCLE, and GTEx project are available in a public repository from the https://gdc.cancer.gov/, https://sites.broadinstitute.org/ccle, and https://www.gtexportal.org/home/ websites. The ChIP-seq data used in this research could be downloaded from the NCBI GEO (https://www.ncbi.nlm.nih.gov/geo; GSE214133, GSE69112, GSE116871, and GSE109440). Other data needed to evaluate the conclusions of this paper are presented in the paper and the SI Appendix.
References
- 1.Garrido-Castro A. C., Lin N. U., Polyak K., Insights into molecular classifications of triple-negative breast cancer: Improving patient selection for treatment. Cancer Discov. 9, 176–198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foulkes W. D., Smith I. E., Reis-Filho J. S., Triple-negative breast cancer. N. Engl. J. Med. 363, 1938–1948 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network, Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wooster R., et al. , Identification of the breast cancer susceptibility gene BRCA2. Nature 378, 789–792 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Miki Y., et al. , A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266, 66–71 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Kandoth C., et al. , Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., et al. , CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 163, 174–186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frampton G. M., et al. , Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 5, 850–859 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Awad M. M., et al. , MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-met overexpression. J. Clin. Oncol. 34, 721–730 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Hsu T. Y.-T., et al. , The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 525, 384–388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowling E. A., et al. , Spliceosome-targeted therapies trigger an antiviral immune response in triple-negative breast cancer. Cell 184, 384–403.e21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashimoto K., et al. , CAGE profiling of ncRNAs in hepatocellular carcinoma reveals widespread activation of retroviral LTR promoters in virus-induced tumors. Genome Res. 25, 1812–1824 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muratani M., et al. , Nanoscale chromatin profiling of gastric adenocarcinoma reveals cancer-associated cryptic promoters and somatically acquired regulatory elements. Nat. Commun. 5, 4361 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Demircioğlu D., et al. , A pan-cancer transcriptome analysis reveals pervasive regulation through alternative promoters. Cell 178, 1465–1477.e17 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Wiesner T., et al. , Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature 526, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J., et al. , ASJA: A program for assembling splice junctions analysis. Comput. Struct. Biotechnol. J. 17, 1143–1150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calabrese C., et al. ; PCAWG Transcriptome Core Group; PCAWG Transcriptome Working Group; PCAWG Consortium, Genomic basis for RNA alterations in cancer. Nature 578, 129–136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y.-Z., et al. , Genomic and transcriptomic landscape of triple-negative breast cancers: Subtypes and treatment strategies. Cancer Cell 35, 428–440.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Georgoudaki A.-M., et al. , Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis. Cell Rep. 15, 2000–2011 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Buffa F. M., Harris A. L., West C. M., Miller C. J., Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br. J. Cancer 102, 428–435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye Y., et al. , Characterization of hypoxia-associated molecular features to aid hypoxia-targeted therapy. Nat. Metab. 1, 431–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Heer E. C., Jalving M., Harris A. L., HIFs, angiogenesis, and metabolism: Elusive enemies in breast cancer. J. Clin. Invest. 130, 5074–5087 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxwell P. H., Eckardt K.-U., HIF prolyl hydroxylase inhibitors for the treatment of renal anaemia and beyond. Nat. Rev. Nephrol. 12, 157–168 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Epstein A. C., et al. , C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Bruick R. K., McKnight S. L., A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Chen Y., et al. , Lysyl hydroxylase 2 induces a collagen cross-link switch in tumor stroma. J. Clin. Invest. 125, 1147–1162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisinger-Mathason T. S. K., et al. , Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer Discov. 3, 1190–1205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du H., et al. , PLOD2 regulated by transcription factor FOXA1 promotes metastasis in NSCLC. Cell Death Dis. 8, e3143 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baek J.-H., et al. , PLOD3 suppression exerts an anti-tumor effect on human lung cancer cells by modulating the PKC-delta signaling pathway. Cell Death Dis. 10, 156 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsay H.-J., et al. , Identifying N-linked glycan moiety and motifs in the cysteine-rich domain critical for N-glycosylation and intracellular trafficking of SR-AI and MARCO. J. Biomed. Sci. 23, 27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi Y., Xu R., Roles of PLODs in collagen synthesis and cancer progression. Front. Cell Dev. Biol. 6, 66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scietti L., et al. , Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3. Nat. Commun. 9, 3163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scietti L., Campioni M., Forneris F., SiMPLOD, a structure-integrated database of collagen lysyl hydroxylase (LH/PLOD) enzyme variants. J. Bone Miner. Res. 34, 1376–1382 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Gjaltema R. A. F., van der Stoel M. M., Boersema M., Bank R. A., Disentangling mechanisms involved in collagen pyridinoline cross-linking: The immunophilin FKBP65 is critical for dimerization of lysyl hydroxylase 2. Proc. Natl. Acad. Sci. U.S.A. 113, 7142–7147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo H.-F., et al. , Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding. Nat. Commun. 9, 512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srinivas V., Zhang L. P., Zhu X. H., Caro J., Characterization of an oxygen/redox-dependent degradation domain of hypoxia-inducible factor alpha (HIF-alpha) proteins. Biochem. Biophys. Res. Commun. 260, 557–561 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Masson N., Willam C., Maxwell P. H., Pugh C. W., Ratcliffe P. J., Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 20, 5197–5206 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bargiela D., Burr S. P., Chinnery P. F., Mitochondria and hypoxia: Metabolic crosstalk in cell-fate decisions. Trends Endocrinol. Metab. 29, 249–259 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Xu W., et al. , Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao S., et al. , Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science 324, 261–265 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong G., et al. , Collagen prolyl 4-hydroxylase 1 is essential for HIF-1α stabilization and TNBC chemoresistance. Nat. Commun. 9, 4456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sartorelli V., Lauberth S. M., Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 27, 521–528 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikhaylichenko O., et al. , The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 32, 42–57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z., et al. , Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat. Commun. 10, 4562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L., et al. , Repression of BET activity sensitizes homologous recombination-proficient cancers to PARP inhibition. Sci. Transl. Med. 9, eaal1645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wimalasena V. K., Wang T., Sigua L. H., Durbin A. D., Qi J., Using chemical epigenetics to target cancer. Mol. Cells 78, 1086–1095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo W., et al. , A LIN28B tumor-specific transcript in cancer. Cell Rep. 22, 2016–2025 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Bechter O., Schöffski P., Make your best BET: The emerging role of BET inhibitor treatment in malignant tumors. Pharmacol. Ther. 208, 107479 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Shu S., et al. , Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529, 413–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shu S., et al. , Synthetic lethal and resistance interactions with BET bromodomain inhibitors in triple-negative breast cancer. Mol. Cell 78, 1096–1113.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delmore J. E., et al. , BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Filippakopoulos P., et al. , Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng Y.-Z., et al. , PHF5A epigenetically inhibits apoptosis to promote breast cancer progression. Cancer Res. 78, 3190–3206 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Yang L.-F., et al. , Discrete functional and mechanistic roles of chromodomain Y-like 2 (CDYL2) transcript variants in breast cancer growth and metastasis. Theranostics 10, 5242–5258 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonnal S. C., López-Oreja I., Valcárcel J., Roles and mechanisms of alternative splicing in cancer - implications for care. Nat. Rev. Clin. Oncol. 17, 457–474 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Brunner J. S., et al. , The PI3K pathway preserves metabolic health through MARCO-dependent lipid uptake by adipose tissue macrophages. Nat. Metab. 2, 1427–1442 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Pinheiro da Silva F., et al. , CD16 promotes Escherichia coli sepsis through an FcR γ inhibitory pathway that prevents phagocytosis and facilitates inflammation. Nat. Med. 13, 1368–1374 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Pal S., et al. , Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 21, 1260–1272 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun H., et al. , Genome-wide mapping of RNA Pol-II promoter usage in mouse tissues by ChIP-seq. Nucleic Acids Res. 39, 190–201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pal S., Gupta R., Davuluri R. V., Alternative transcription and alternative splicing in cancer. Pharmacol. Ther. 136, 283–294 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Loriot A., et al. , A novel cancer-germline transcript carrying pro-metastatic miR-105 and TET-targeting miR-767 induced by DNA hypomethylation in tumors. Epigenetics 9, 1163–1171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stathis A., Bertoni F., BET proteins as targets for anticancer treatment. Cancer Discov. 8, 24–36 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Chapuy B., et al. , Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24, 777–790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lovén J., et al. , Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data and sequence data for the FUSCCTNBC cohort have been deposited in the NCBI Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo; OncoScan array; GEO: GSE118527) and SRA (https://www.ncbi.nlm.nih.gov/sra; whole exome sequencing and RNA-seq; SRA: SRP157974). The RNA-seq data for the TCGA cohort, CCLE, and GTEx project are available in a public repository from the https://gdc.cancer.gov/, https://sites.broadinstitute.org/ccle, and https://www.gtexportal.org/home/ websites. The ChIP-seq data used in this research could be downloaded from the NCBI GEO (https://www.ncbi.nlm.nih.gov/geo; GSE214133, GSE69112, GSE116871, and GSE109440). Other data needed to evaluate the conclusions of this paper are presented in the paper and the SI Appendix.