Abstract
Alzheimer’s disease (AD) is a heterogeneous disorder with abnormalities in multiple biological domains. In an advanced machine learning analysis of postmortem brain and in vivo blood multi-omics molecular data (N = 1863), we integrated epigenomic, transcriptomic, proteomic, and metabolomic profiles into a multilevel biological AD taxonomy. We obtained a personalized multilevel molecular index of AD dementia progression that predicts severity of neuropathologies, and identified three robust molecular-based subtypes that explain much of the pathologic and clinical heterogeneity of AD. These subtypes present distinct patterns of alteration in DNA methylation, RNA, proteins, and metabolites, identifiable in the brain and subsequently in blood. In addition, the genetic variations that predispose to the various AD subtypes in brain predict distinct spatial patterns of alteration in cell types, suggesting a unique influence of each putative AD variant on neuropathological mechanisms. These observations support that an individually tailored multi-omics molecular taxonomy of AD may represent distinct targets for preventive or treatment interventions.
Integration of multilayer molecular data provides a robust assessment and classification of the Alzheimer’s disease spectrum.
INTRODUCTION
The development of increasingly sophisticated high-throughput tools to examine genome-wide multilevel omics has led to new approaches to define the molecular taxonomy of diseases (1, 2). This trend has been most fruitful in cancer, which long ago witnessed the dawn of precision medicine (2, 3). In recent years, several large efforts have been funded to generate open science, multi-omic, postmortem, and in vivo brain and matched blood datasets. These include the Accelerating Medicines Partnership–Alzheimer’s Disease (AMP-AD) (4, 5) and the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (6, 7). These large-scale initiatives are shifting the traditional paradigm for development of new biologically informed diagnostic tools and therapies for Alzheimer’s disease (AD) (4, 5, 8).
A challenging aspect of working with postmortem brain relates to the metric of time. While tumors can be biopsied repeatedly over time, including before and after administration of a potential therapeutic agent, this is not possible with brain and other central nervous system tissues. Instead, brains are typically aligned using actual time, e.g., age at death, biologic age such as epigenomic clock, extent of neuropathology, or clinical disease severity. Each of these measures has its advantages and disadvantages. The concept of disease pseudo-time is an alternative approach that infers sequence from cross-sectional data (9–11). An early example is the Guttman scale (12), which orders cross-sectional data. Pseudo-time has recently been applied to omic data from cancer and brain (9–11). This approach has recently been explored using RNA sequencing (RNAseq) in several AMP-AD projects including the Religious Orders Study (ROS) and the Rush Memory and Aging Project (MAP) (13, 14).
A further challenge is posed by the interindividual heterogeneity of AD with the implied existence of several possible disease subtypes. This heterogeneity suggests that the study and treatment of AD should consider not only the timeline of its long-term progression but also the possibility of distinct variants. Recent postmortem neuropathological and in vivo neuroimaging studies have identified several such subtypes associated with differences in clinical presentation (15–17). Thus, previous work in ROSMAP used cognitive profile mixture models to identify latent classes of clinical disease progression, which, as expected, are related to extent of neuropathology, as well as latent classes of residual cognitive decline after regressing out the effects of common neuropathologies (18, 19). At a molecular level, network analysis of transcriptome data also revealed distinct RNA-based AD subgroups (20), suggesting different combinations of dysregulated pathways and subtype-specific genetic drivers that result in a similar progressive loss of cognition. Similarly, independent methylome-wide association analysis has uncovered both brain- and blood-based genes with AD subgroup–specific associations (21). Nevertheless, to date, molecular omics analysis of AD variability has typically been limited to the study of individual omic layers without multi-omics integration.
We recently used ROSMAP and ADNI data to infer a series of sequential alterations in gene expression over years of AD progression (13). This allowed us to align individual brains according to an RNA disease index associated with the development of dementia and the severity of AD pathology. However, the diversity of molecular alterations underlying AD progression and heterogeneity goes far beyond RNA modifications alone. Here, we extend previous molecular characterizations of the AD spectrum in three fundamental ways. First, we used a novel machine learning (ML) framework to integrate several layers of molecular omics data from brain epigenomic, transcriptomic, proteomic, and metabolomic databases. Second, relying on these aggregated data, we developed and cross-validated an algorithm that assigns each participant to both a multilevel molecular estimate of the progression of AD and a putative AD subtype. Each of such subtype is associated with a distinct pathologic disease pseudo-trajectory, defined as a concatenated subset of cognitively impaired and/or AD subjects following a unique pattern in multi-omics molecular data integrated space. Third, to examine the generalizability and utility of our results, we translated this brain-based multi-omics algorithm to peripheral blood data. We assessed whether the identified multilevel molecular estimate of the AD progression and subtypes aligned with postmortem neuropathological data and in vivo positron emission tomography/magnetic resonance imaging (PET/MRI) AD biomarkers. All resulting analytic tools are shared with the scientific and clinical community via a user-friendly software for further translation and validation of our findings.
RESULTS
Multimodal data origin and unification approach
We obtained DNA methylation (DNAm), RNAseq, proteomic, and/or metabolomic data; neuropathologic or biomarker data; and clinical data from 1863 with a wide range of cognition in two independent large-scale studies (see Fig. 1 and the “Dataset 1” and “Dataset 2” sections). In dataset 1 (ROSMAP; N = 822), multi-omics molecular evaluations were performed on the material from the dorsolateral prefrontal cortex (DLPFC) of autopsied brains, with a subset (N = 168) also having blood monocyte RNA quantification. Assignment of no cognitive impairment (NCI), mild cognitive impairment (MCI), or AD dementia categories corresponded with most likely clinical diagnosis at the time of death. Dataset 2 (ADNI; N = 1041) included in vivo blood samples for multi-omics molecular characterization, with a subset of 610 also having one or more types of brain imaging evaluations including amyloid PET, tau PET, and/or structural MRI (table S1). Participants were clinically diagnosed at baseline as NCI, early MCI (EMCI), late MCI (LMCI), or probable AD patient.
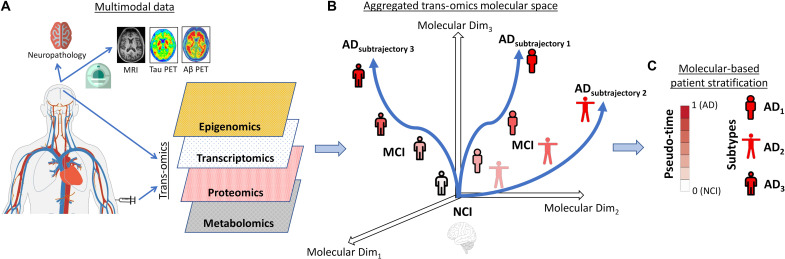
Fig. 1. Schematic approach for multi-omics molecular integration and patient stratification in the late-onset AD dementia spectrum.
(A) Postmortem brain (N = 822) and in vivo blood (N = 1041) tissues were obtained from previous studies (see the “Data”), including DNAm (from 420,132 to 865,918 CpG islands), RNAseq (about 48,000 transcripts), proteins (from 149 to 186 proteins), and metabolite concentrations (from 205 to 430). (B) Integrated multimodal molecular disease space, where subjects are stratified by the algorithm (in terms of advance on the path to develop AD dementia and subtrajectories) according to their position (see the “mcTI definition” and “Detailed mcTI algorithm” sections). (C) Two trans-omics molecular-based estimates are obtained for each participant, a pseudo-time or disease advance score (i.e., multi-omics mDPS) and the most likely molecular AD subtype (corresponding to a distinctive disease subtrajectory in the aggregated molecular space). The validity of this approach in providing a useful molecularly defined assessment and classification of the AD spectrum is subsequently explored in terms of capacity to reflect severity of neuropathologies and cell type alterations, as well as generalizability across different tissue samples (brain and blood).
To identify the multimodal molecular reconfigurations underlying neurodegenerative advance and heterogeneity in late-onset AD, we unified, reordered, and stratified the multi-omics patterns as shown in Fig. 1 (A and B). To do so, we implemented a novel cross-validated ML algorithm for detection of disease-associated multimodal data patterns anchored in those with NCI relative to MCI and AD dementia (see the “mcTI definition” section; Fig. 1A). Each molecular feature/marker was first adjusted for potentially confounding covariates (e.g., age, sex, education, and postmortem time interval). The generic algorithm then aggregated the different molecular data types while accounting for relevant AD patterns (2). We hypothesized that the position of each subject in this aggregated multi-omics space would predict individual severity of AD neuropathology and degree of relatedness to distinctive emergent disease pseudo-subtrajectories. Accordingly, a multi-omics molecular disease progression score (multi-omics mDPS) was calculated for each subject, which ranged from 0 (which should be NCI) to 1 (which should be late AD dementia) (see the “mcTI definition” section). Relatively low or high values indicate lesser or greater distance on the path to develop AD dementia (Fig. 1, B and C). In addition, each subject was molecularly subtyped according to the maximum probability of “belonging” to an identified disease pseudo-subtrajectory in the multimodal molecular space (see Fig. 1B). Thus, for each participant, a separate molecular disease advance score (i.e., multi-omics mDPS) and the most likely molecular AD subtype were obtained (Fig. 1, A to C). The multi-omics mDPS proved to be valid across the entire AD clinical spectrum, without need for subtype-specific variation in its calculation.
Capturing multimodal pathomolecular progression in the AD brain
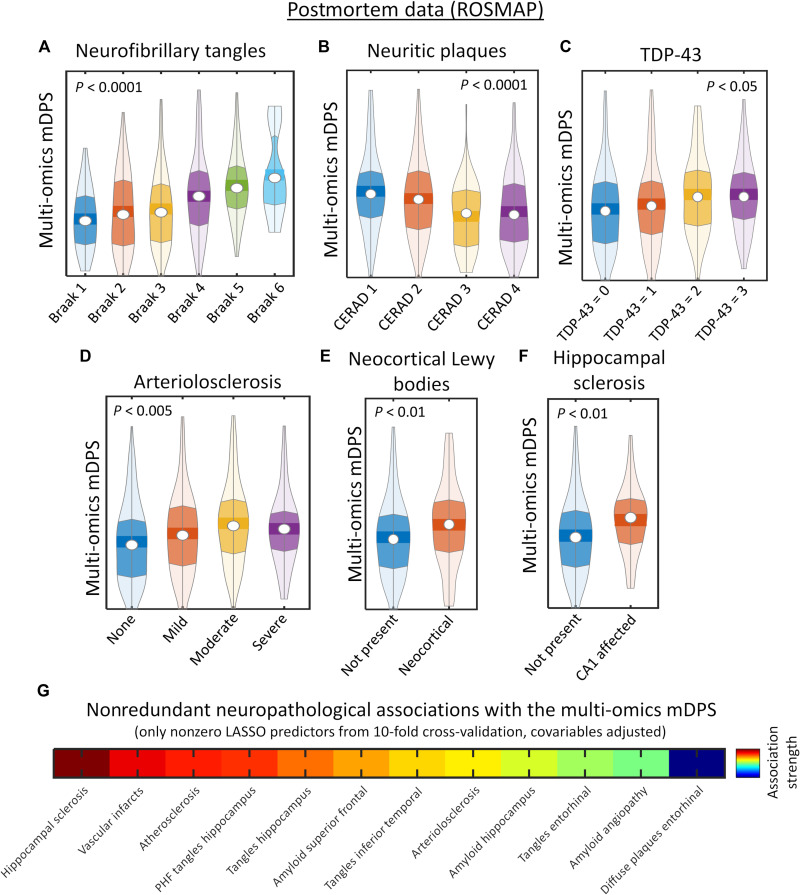
A useful molecularly defined stratification system should predict disease severity at the point of data evaluation. First, we investigated whether the postmortem brain-based multi-omics mDPS could serve as a marker of individual severity of AD-related neuropathologies. The ROSMAP-based results (Fig. 2, A to F) show clear statistical associations between the multimodal molecular disease score and postmortem neurofibrillary tangles (NFTs), β-amyloid neuritic plaques (NPs), TDP-43, arteriolosclerosis, neocortical Lewy bodies, and hippocampal sclerosis, with higher scores in each case indicating more advanced neuropathology. Specifically, differences in molecular disease scores across the contiguous neuropathological stages (for NFT, NP, TDP-43, arteriolosclerosis, neocortical Lewy bodies, and hippocampal sclerosis) were statistically tested via Kruskal-Wallis tests with permutations, previously adjusting for age, sex, and education (see the “Statistical analyses” section). We found statistically significant associations with Braak stages (χ2 = 47.05, P < 0.0001, Family Wise Error corrected [FWE-corrected]; Fig. 2A), CERAD stages (χ2 = 32.01, P < 0.0001, FWE-corrected; Fig. 2B), TDP-43 cytoplasmatic inclusions in neurons and glia (χ2 = 10.27, P < 0.05, FWE-corrected; Fig. 2C), arteriolosclerosis (χ2 = 14.50, P < 0.005, FWE-corrected; Fig. 2D), presence of neocortical Lewy bodies (χ2 = 7.59, P < 0.01, FWE-corrected; Fig. 2E), and hippocampal sclerosis in region CA1 (χ2 = 7.22, P < 0.01, FWE-corrected; Fig. 2F).
Fig. 2. Integrated multi-omics molecular predictions of progressive cognitive and neuropathological deterioration.
(A to F) Postmortem brain-based molecular disease progression predictions (ROSMAP data) of tau neurofibrillary tangles (A), neuritic plaques (B), TDP-43 cytoplasmatic inclusions in neurons and glia (C), arteriolosclerosis stages (D), presence of neocortical Lewy bodies (E), and hippocampal sclerosis (F). All P values are FWE-corrected via randomized permutation tests. (G) Nonredundant associations with neuropathological phenotypes (i.e., LASSO regression results, with 10-fold cross-validation, and adjusted by age, sex, and educational level).
In addition, the brain-based multi-omics mDPS correlated with greater neuropathology in multiple brain areas [q < 0.05, false discovery rate (FDR)–corrected, covariables adjusted]. These findings included positive associations with the accumulation of NFT, β-amyloid, and/or paired helical filament (PHF) tangles in temporal, hippocampal, cingulate, angular, and entorhinal cortices. However, regional neuropathological measurements are usually highly intercorrelated (22), suggesting the need for further assessing their nonoverlapping relationship with the multi-omics mDPS. Thus, we proceeded to identify nonredundant neuropathological associations with mDPS via LASSO regression, a regularization technique that focuses on decoding dependencies among the predictors to enhance model prediction and interpretability (23). In the presence of collinear regressors, LASSO iteratively detects the least contributing (or redundant) markers, which are assigned a zero or near-zero coefficient. Notably, while an association of these regressors with the dependent variable cannot be categorically rejected, their individual contributions are outperformed by other predictors, which define a more parsimonious model without compromising prediction accuracy. Each regional neuropathological measurement was considered a competing regressor of the multi-omics mDPS in a 10-fold cross-validation LASSO analysis, adding age, sex, and educational level as covariables. From the 32 initially considered neuropathological measurements (text S1), 12 (i.e., 37%) showed nonredundant significant association with the multimodal molecular disease score (Fig. 2G). The strongest nonredundant significant predictors included the individual levels of hippocampal sclerosis, β-amyloid in superior frontal, PHF tangles in hippocampus, NFT in entorhinal cortex and inferior temporal, vascular infarcts, atherosclerosis, and cerebral amyloid angiopathy. These results suggest that, rather than aligning with the advance in a unique neuropathological process (e.g., β-amyloid burden), the estimated molecular-based disease progression score (and its related “timeline”) reflects the aggregated effect of multiple brain alterations, notably including hippocampus sclerosis, NFT, NP, and PHF accumulation levels in commonly affected AD areas (entorhinal, hippocampus, inferior temporal, and superior frontal) and cerebrovascular abnormalities.
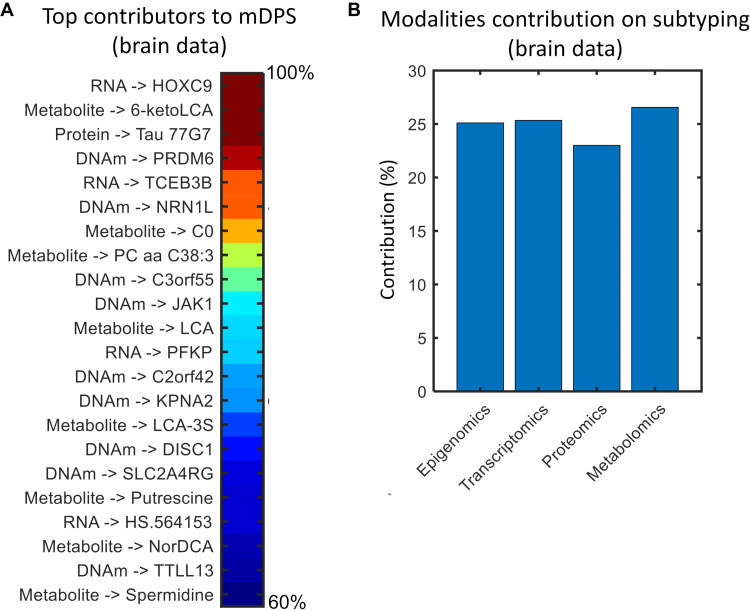
Next, we identified the top molecular markers contributing to the prediction of AD progression. Notably, the multi-omics mDPS can provide quantitative mapping of the most influential CpGs, genes, proteins, and metabolites during the process of AD trajectory inference. Specifically, the method’s internal loadings (or weights) reflect how much each specific biological marker, in the original high-dimensional multi-omics space, contributed to the reduced low-dimensional space from which the trajectories were obtained (see the “Assessing marker contributions on mDPS” section). We found (Fig. 3A and table S2) a varied list of predictors distributed across the four omics layers, with most previously associated with AD. For instance, at the RNA level, gene HOXC9 is known as a key regulator of endothelial cell quiescence and vascular morphogenesis (24) and has been identified earlier in AD genome-wide association studies (25). Gene PFKP is related with impaired neuronal glucose metabolism observed in AD (26). With an abnormal methylation level here, gene DISC1 is commonly associated with schizophrenia, but its ectopic (out of place) expression has been suggested to delay the progression of AD by protecting synaptic plasticity and down-regulating BACE1 (27). Protein 77G7 is a monoclonal antibody with the capacity to recognize tau filament cores (28). At the metabolomic level, plasma concentration of lithocholic acid is significantly associated with clinical deterioration across MCI and AD individuals (29). Therapeutic administration of spermidine, with hypothesized cardioprotective and neuroprotective effects (30), is suggested to improve cognitive performance in subjects with mild and moderate dementia (31). These results suggest that the multi-omics mDPS comprises complementary information, which coexists across multiple biological scales and is traditionally analyzed separately.
Fig. 3. Molecular omics contributions to AD stratification in postmortem brain tissue.
(A) Top influential epigenetic, transcriptomic, proteomic, and metabolomic markers during the process of AD trajectory inference. Values are percentages, normalized with regard to the maximum (only markers over the 99 percentile are shown; for an extended list, see table S2). (B) Modality-specific contributions (in percentages) to the identified AD subtypes. For technical details, see the “Assessing markers contributions on mDPS” and “Assessing omics contributions on subtyping” sections.
Last, the observed strong associations with neuropathologic levels (Fig. 2) lead us to question whether the multi-omics mDPS could reflect AD dementia beyond the overall neuropathologic burden. To further investigate this, we adjusted the individual mDPS by several neuropathologic traits (NFT, NP, TDP-43, arteriolosclerosis, Lewy bodies, hippocampal sclerosis, amyloid angiopathy, atherosclerosis, vascular infarcts, and chronic microinfarcts) and demographics (age, sex, and education). Next, we compared the residual multi-omics mDPS values among NCI and AD subjects via Kruskal-Wallis tests with permutations, still observing a very strong clinical association with the residual mDPS (χ2 = 89.78, P < 0.0001, FWE-corrected). In addition, we verified that all together (global and regional neuropathological traits, plus demographics) can only explain about 11% of mDPS’s population variance (i.e., R2 = 0.11 in the additive regression analysis). Both observations support the multi-omics mDPS’s capacity to capture deep pathomolecular processes in AD beyond the severity of neuropathologic burden, as also reflected by the top contributing markers (Fig. 3).
Brain multimodal molecular information reveals distinctive AD subtypes
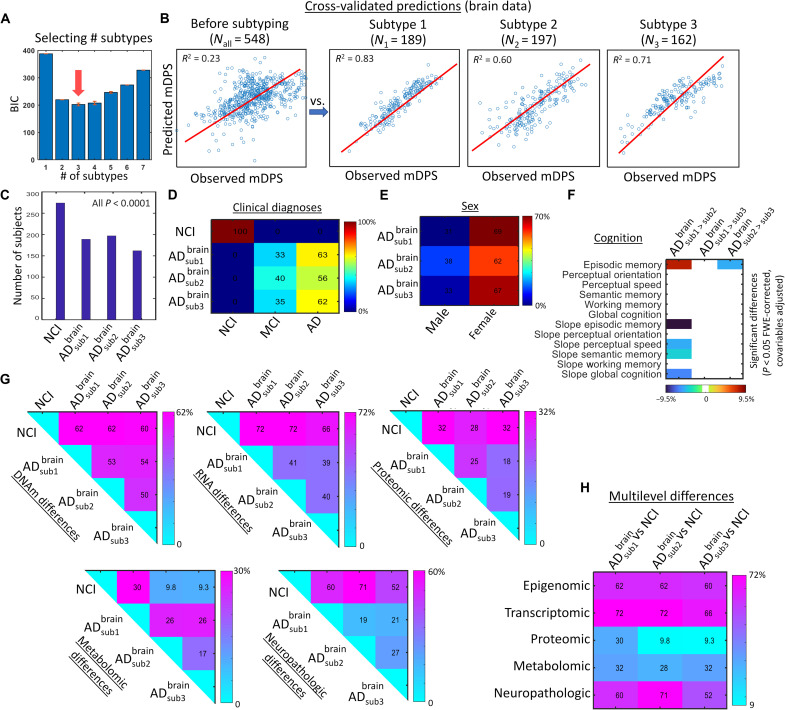
Another desired attribute of a clinically useful biologically defined stratification system is an ability to detect distinctive disease subtrajectories. We inferred putative AD subtypes by identifying distinctive multimodal molecular pseudo-subtrajectories in the aggregated multi-omics disease space. A cross-validated expectation-maximization (EM) algorithm was used to detect subgroups of subjects consistently aligned to a single disease timeline (see the “mcTI definition” section). The Bayesian information criterion (BIC) (32), which allows optimizing the trade-off between data explanation and model complexity, was used to identify the optimum number of putative subtypes (from a minimum of 1 up to a maximum of 7; see Fig. 4A). Here, we used an internal leave-one-out prediction analysis. The algorithm identified each individual’s most likely subtype configuration in terms of predictability of clinical progression and model simplicity. The stability and significance of each subtype configuration was tested via a permutation procedure (20). Specifically, subtype stability was defined as the rate at which sample pairs group together into the same subtypes upon repeated clustering on random subsets of the input data (20).
Fig. 4. Three distinctive AD subtypes identified with multi-omics molecular data from the brain (ROSMAP).
(A) BIC values obtained for each considered number of putative subtypes. In statistics, the model with lowest BIC is preferred (here, three subtypes). For analyzing the stability of this selection, the subtyping was repeated 500 times via bootstrapping with replacement (presented BIC values correspond to the bootstrap average). Note that, across all bootstrap repetitions, the method always selected three as the optimum number of subtypes. (B) Cross-validation analysis for identification of the most compelling classification structure in terms of predictability of pathological advance (as quantified by the multi-omics mDPS). Note that the deconstruction of the whole population into smaller/stable subtypes brought a significant improvement in internal data homogeneity and multi-omics mDPS predictability. (C) Number of samples per subtype and corresponding significance obtained with randomization testing (all P < 0.001, few-corrected). (D and E) Subtype-specific clinical diagnoses and sex proportions, respectively. (F) Inter-subtype differences in cognitive domains and their rates of change over time [all P < 0.05, FWE-corrected, based on analysis of variance (ANOVA) tests with permutations]. Only significant differences are shown, with values corresponding to explained variance and signs reflecting direction (for slopes, a positive value would indicate stronger cognitive decline for the first specified subtype, while a negative value would indicate the contrary). (G and H) Total inter-subgroup molecular and neuropathological differences. For each omics data type and comparison with the control population, each matrix element corresponds to the percent of significantly different features (all q < 0.05, FDR-corrected, based on ANOVA tests with subtype as grouping variable; see the “Statistical analyses” section). For each AD subtype-subtype matrix element, the reported value represents the percent of data features that are abnormal for one subtype but not for the other (mismatch level).
Three putative AD subtypes (each corresponding to a distinct subtrajectory or concatenation of subjects in the integrated molecular space) were identified, showing high internal data homogeneity compared to the whole population and significant subtype stability (P < 0.001, FWE-corrected; Fig. 4, B and C). Because the subtypes were defined in terms of “continuous” disease pseudo-subtrajectories in the integrated molecular space (i.e., each being a concatenation of individuals following a coherent data pattern; Materials and Methods), they covered the spectrum of the disease from normality to AD dementia (Fig. 4D). However, substantial subtype-subtype differences were observed for omic layers and neuropathologic traits. Specifically, we calculated the proportion of features that differed between any pair of subtypes for each biological data modality, i.e., epigenomic, transcriptomic, proteomic, metabolomic, or neuropathology. As is shown in Fig. 4 (G and H), percent levels of subtype-subtype differences are notable across the four studied molecular omic levels and the neuropathological features, with difference percentages ranging from 9.3 to 72%. In addition, the specific multimodal molecular and neuropathological features differentiating the subtypes can be seen in Fig. 5. The subtypes did not significantly differ among themselves by clinical diagnosis (MCI and AD proportions; Fig. 4D), sex (female and male proportions; Fig. 4E), and global cognition (Fig. 4F). However, they did significantly differ in various cognitive domains (all P < 0.05, FWE-corrected; Fig. 4F). AD subtype 1 was significantly more affected than subtype 2 in episodic memory and decline in perceptual speed over time, episodic memory, semantic memory, and global cognition. AD subtype 3 was also more significantly affected than subtype 2 in episodic memory.
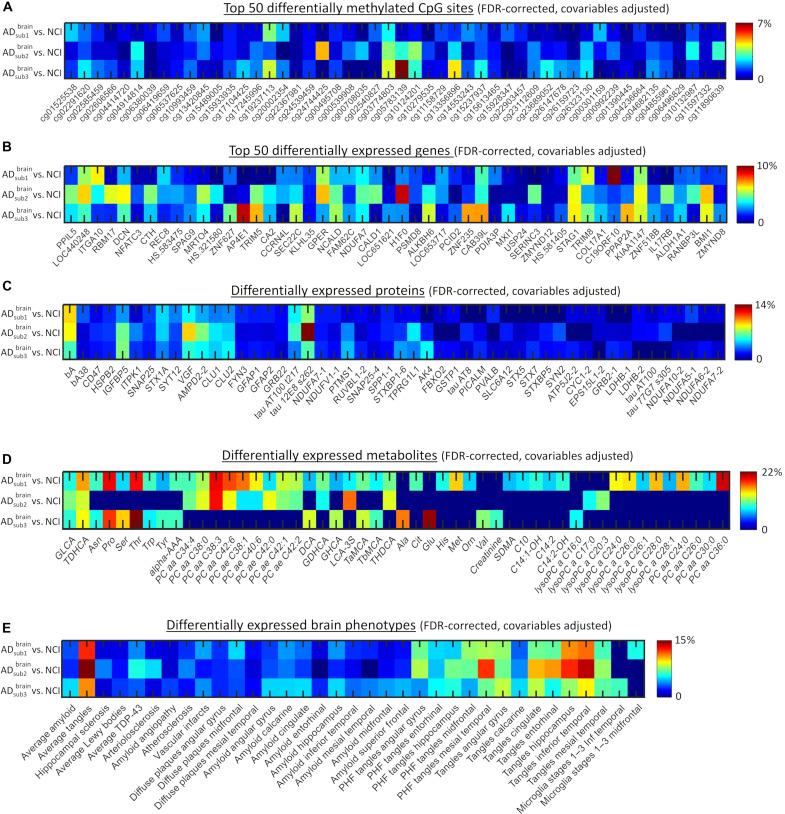
Fig. 5. Differentially expressed CpG sites, genes, proteins, metabolites, and brain phenotypes in brain-based AD subtypes.
(A to E) Only significantly expressed features are presented (q < 0.05, FDR-corrected). ANOVA tests with subtype as grouping variable were used (data were previously adjusted by age, sex, educational level, and experimental confounders; see the “mcTI definition” section). Color scale corresponds to the explained variance. For DNAm and RNA, only the top 50 most significant CpG sites and genes are presented (see table S3 for a complete list).
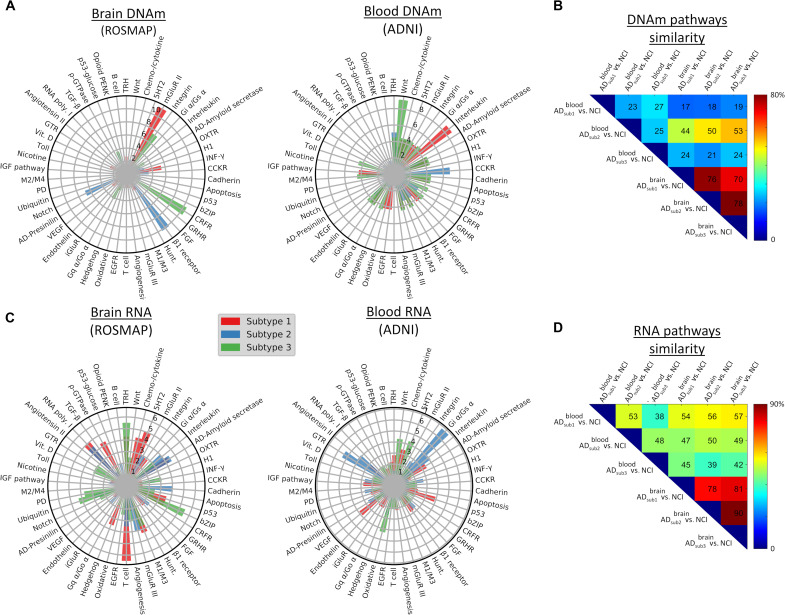
Furthermore, we performed large-scale gene functional analyses with the Protein Annotation Through Evolutionary Relationship (PANTHER) classification system (33) for each subtype and its differentially expressed DNAm CpGs or RNA genes. The three AD subtypes shared 70 to 78% of epigenetic and 78 to 90% of transcriptomic molecular pathways affected, respectively (Fig. 6, B to D). Across subtypes, prominent transcriptomic pathways associated with generalized cellular and molecular processes, such as general transcription regulation (GTR), ubiquitination, transcription regulation by bZIP, RNA synthesis by RNA polymerase, cytoskeletal regulation by ρ-GTPase (guanosine triphosphatase), oxidative stress response, and cellular signaling via the Gs α subunit. Several specialized AD-relevant pathways included amyloid secretase, cholinergic M1/M3 receptor signaling, and metabolic glutamatergic receptors (mGluR III). Epigenetic modifications also involved specialized pathways, with prominent examples including neurotransmitter receptors (metabolic glutamate receptors in subtype 1 and the adrenergic β1 receptor in subtype 2), and the corticotropin-releasing factor receptor (CRFR) pathway (affecting behavioral, autonomic, endocrinic, and immunologic responses in AD subtype 3).
Fig. 6. Top subtype-specific molecular pathways associated with differentially methylated or expressed genes.
(A and C) Radial plots for methylation- or RNA-based pathways, respectively. The radio represents the presence level (as a percentage) of pathways in each tissue-specific AD subtype. The full molecular pathway names are listed in table S4. (B and D) Molecular-based similarity (overlapping) across brain-based (ROSMAP) and blood-based (ADNI) AD subtypes. Each pairwise value was calculated as the percentage of common number of differentially expressed pathways relative to the total.
In addition, the three AD subtypes shared similar alterations in protein concentrations (Fig. 5C), including tau 12E8 phosphorylated at S262, β-amyloid, and nerve growth factor. Nevertheless, each subtype presented an additional set of uniquely altered proteins. AD subtype 1’s distinctively expressed proteins associated with electron transport chain in mitochondria and response to stressful conditions (e.g., heat shock, cold, and ultraviolet light), including ubiquinone oxidoreductase subunit A10 (Ndufa10) and heat shock family A member 8 (Hspa8) proteins, respectively. AD subtypes 2 and 3 included greater protein alterations in cardiovascular development and lactate-pyruvate catalysis, such as bridging integrator-1 (BIN1) and lactate dehydrogenase A (LDHA), respectively. Notably, AD subtype 1 presented a larger number of altered metabolites (Fig. 5D), involving phosphatidylcholines such as ae-C38:4, aa-C36:0, and ae-C24:0, and basic amino acids such as methionine, histidine, ornithine, and citrulline. Although less predominantly, subtypes 2 and 3 also presented several altered metabolites (Fig. 5D), including glutamate (Glu), proline (Pro), threonine (Thr), and valine (Val). These particular findings suggest that previously reported metabolomic alterations in AD (34, 35), and the proposed need to account for them in therapeutic interventions, may depend on the specific AD variant(s) under study. Furthermore, the three subtypes shared widespread neuropathologic alterations (Fig. 5E). However, contrary to AD subtypes 1 and 3, subtype 2 did not present significantly abnormal levels of microglia activation across the evaluated brain regions, suggesting that it may be associated with distinct immune and neuroinflammatory pathomechanisms.
Together, the multilevel (molecular and macroscopic) differential analysis of the identified AD subtypes (Figs. 4, G and H, 5, and 6) revealed that AD subtype 1 is mainly characterized by widespread metabolic alterations (about 41% of its molecular alterations). Contrary, subtypes 2 and 3 are distinctively associated with extensive RNA and epigenetic alterations, covering about 50 and 39% of their multilayer molecular alterations, respectively. The three AD subtypes presented similar widespread proportions of proteomic and neuropathological alterations.
Last, we aimed to investigate the extent to which the observed subtype-specific multi-omics alterations would survive statistical adjustment for individual neuropathologies. For each molecular marker, analysis of variance (ANOVA) tests with subtype as the grouping variable were used while adjusting by several AD-associated neuropathologic traits (NFT, NP, TDP-43, arteriolosclerosis, Lewy bodies, hippocampal sclerosis, amyloid angiopathy, atherosclerosis, vascular infarcts, and chronic microinfarcts; molecular data were previously adjusted by age, sex, educational level, and experimental confounders; Materials and Methods). Again, we found a large set of significant epigenetic (748 CpGs), transcriptomic (417 genes), proteomic (19 proteins), and metabolomic (28 metabolites) alterations spread across the three AD subtypes (all q < 0.05, FDR-corrected). Note, however, that this analysis is, to some extent, statistically circular and is only performed here for exploratory purposes. Neuropathology levels partly correlate with AD dementia, and statistically removing them can mask (or attenuate) relevant clinical components that biologically defined AD subtypes should be reflecting. Although often used in the literature, current “statistical adjustment” techniques do not account for causal mechanisms, and “controlling” effects from specific factors is not entirely accurate and requires deeper causal analyses (36, 37). Furthermore, neuropathological alterations may be collinear with many molecular changes, without necessarily causing them (and vice versa), an effect difficult to account by using traditional statistical perspectives.
AD subtypes associated with distinct cell type patterns
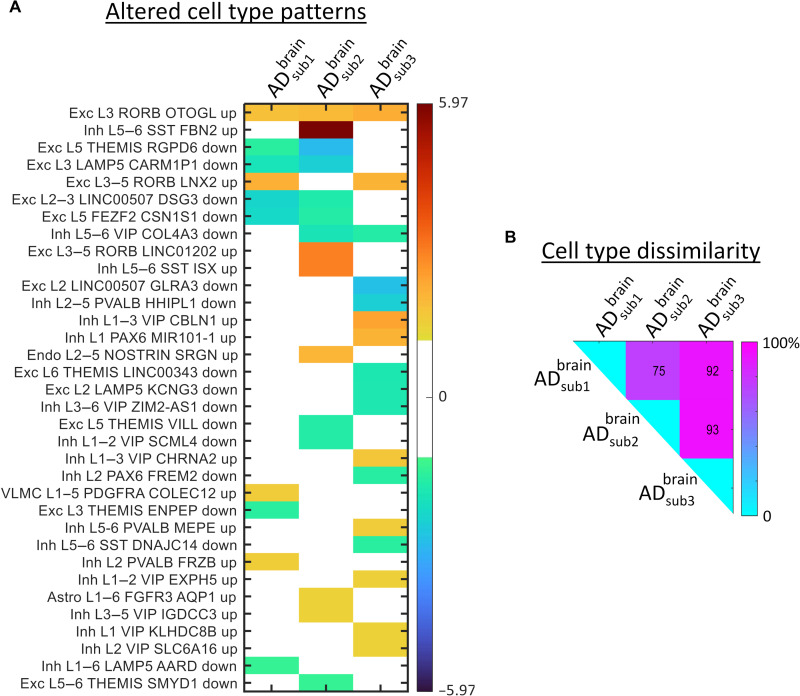
Next, we hypothesized that AD subtypes corresponding to different disease subtrajectories would be associated with distinct cell type differences. Therefore, we performed cell type deconvolution with Enrichr software and the Allen Brain Atlas 10x scRNA 2021 on the subtype-specific differentially expressed genes (38). For each AD subtype, a ranked list of about 200 potentially down- or up-regulated brain cells was obtained, with a z score value per cell indicating the statistical likelihood of being enriched compared to a random background (see the “Brain cell type analysis” section). A subsequent multiple-comparison analysis of these z scores revealed 34 different cell types potentially altered (q < 0.05, FDR-corrected) for the three subtypes (Fig. 7A). All AD subtypes presented a varied pattern of down- and up-regulated genes at excitatory and inhibitory neurons in all cortical layers. Subtype 2 also presented up-regulated genes at astrocytes and endothelial cells, while subtype 3 presented up-regulated alterations in vascular and leptomeningeal cells. Next, we quantified the subtypes’ dissimilarity regarding their cellular patterns. As shown in Fig. 7B, each pair of subtypes had a high level (75 to 93%) of mismatch in terms of unique down-/up-regulated cell types. These results suggest a distinct pattern of cellular vulnerability across the AD subtrajectories, which may be associated with different underlying pathological mechanisms, and to some extent may explain the observed multilevel (molecular and macroscopic) differences (Figs. 4, G and H,5, and 6).
Fig. 7. Altered cell type patterns in brain-based multimodal molecular AD subtypes.
(A) Significantly enriched cell types (q < 0.05, FDR-corrected) across the three brain-based AD subtypes (values correspond to enrichment z scores). Included cell types are excitatory neuron (Exc), inhibitory neuron (Inh), endothelial cell (Endo), vascular and leptomeningeal cell (VLMC), and astrocyte cell (Astro). Each cell entry includes the affected cortical layers (L), a broad marker gene, and a subclass-specific marker gene. (B) Subtype-subtype mismatch matrix (element i,j represents the percent of uncommon altered cell types among subtypes i and j).
Postmortem brain-based AD stratification is translatable to in vivo peripheral data
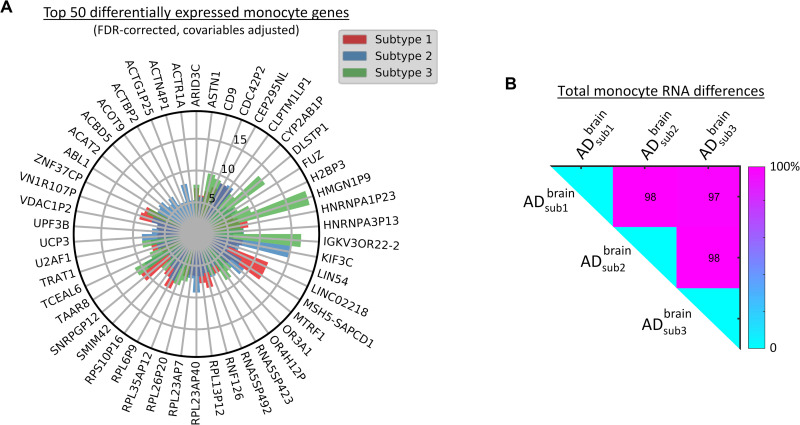
We next aimed to examine the generalizability and potential clinical utility in living persons of the pseudo-time subtrajectories using data from in vivo blood samples from ROSMAP and ADNI data. First, we explored whether the brain-based classification could yield characteristic molecular patterns in the periphery of the same studied subjects. A subset (N = 168) of the ROSMAP subjects had undergone blood monocyte RNA quantification. By comparing these persons’ identified AD subtypes, we confirmed that they also presented distinct patterns of monocyte RNA alteration (all q < 0.05, FDR-corrected; Fig. 8). Together, the subtypes accounted for 1110 differentially expressed monocyte transcripts of 28,923 (3.83%; Fig. 8A and table S5). Furthermore, each AD subtype had a high proportion of uniquely affected monocyte features, with all three possible pairs of subtypes sharing fewer than 3% of all their corresponding differentially expressed genes (Fig. 8B). The identification of strong subtype-specific monocyte signal alterations supports the notion that a biologically defined classification of the AD spectrum may be feasible based solely on peripheral multimodal molecular data.
Fig. 8. Distinctively expressed genes in blood monocyte cells across putative AD subtypes (ROSMAP).
(A) Top 50 differentially expressed genes (all q < 0.05, FDR-corrected; for the complete list, see table S5). Results are based in ANOVA tests with subtype as grouping variable, adjusting by age, sex, and educational level. The radio represents the explained variance (as a percentage, covering the scale [0, 14%]). (B) Subtype-subtype mismatch matrix (element i,j represents the percent of uncommon altered monocyte genes among subtypes i and j).
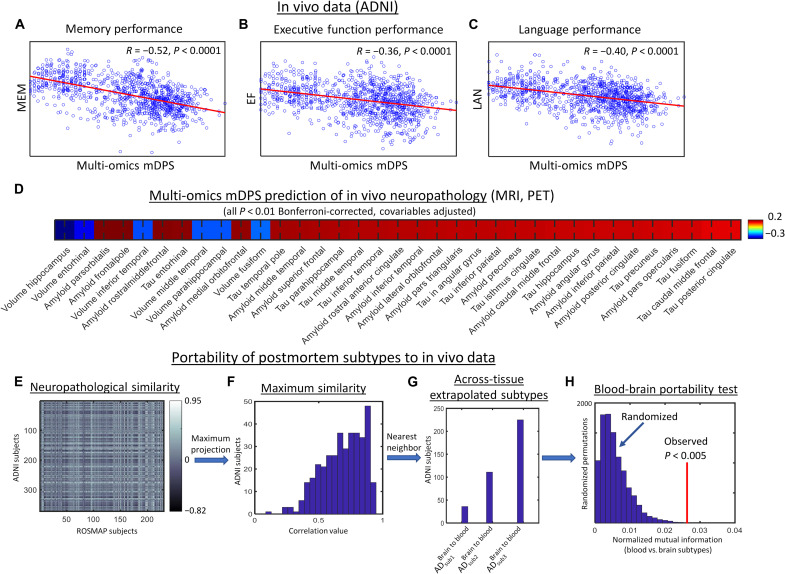
We subsequently applied the multi-omics contrastive trajectory inference (mcTI) algorithm to whole-blood multi-omics molecular data from the ADNI cohort (N = 1041). The results (Fig. 9, A to C) confirmed that the in vivo multimodal molecular disease score significantly predicts memory, executive function, and language performance (all P < 0.0001). In addition, we observed (Fig. 9D) strong correlations (all P < 0.01, Bonferroni-corrected, adjusted by age, sex, and educational level) with MRI, amyloid PET, and tau PET. In line with our postmortem brain data, the blood-based multi-omics mDPS associated negatively with the volume of hippocampus, entorhinal, inferior temporal, middle temporal, fusiform, and parahippocampal cortices, while it associated positively with amyloid and tau in several brain areas, including hippocampus, entorhinal, frontal, temporal, and cingulate cortices, among others.
Fig. 9. Generalizability analysis of brain-based stratification to in vivo blood samples.
(A to D) Integrated blood-based multi-omics molecular predictions of cognitive performance (A to C) and neuropathological biomarkers (D) in ADNI subjects (N = 1041). (E) Neuropathological similarity between ROSMAP and ADNI subjects (based on 16 common brain regional amyloid and tau deposition measurements). (F) Maximum similarity projection for ADNI subjects (note that, on average, ADNI subjects strongly correlate with at least one ROSMAP subject). (G) Distribution of extrapolated brain-based AD subtypes from the postmortem population to the alive participants (i.e., from ROSMAP to ADNI). (H) Portability test comparing mutual information between the two independently obtained AD stratifications.
Also aligned with results for the postmortem brain data, the top molecular features contributing to the blood-derived multi-omics DPS covered the four omics layers (fig. S1A and table S2), with most previously associated with AD. Top transcriptomic, proteomic, and metabolomic contributing factors related to metabolism (proteins insulin and leptin and gene HMG20A), immune cell infiltration (protein MCP-2), immune response and inflammation (protein PARC), antioxidant signaling (gene KIAA0319 and metabolite α-tocopherol), and protein cleavage (metabolite asparagine). Several epigenetic and transcriptomic factors representing generalized cellular and molecular processes were also identified as relevant mDPS contributors, including cell growth and proliferation (OAZ3), posttranscriptional regulation of gene expression (MIR548H4), signal transduction and protein phosphorylation (HUNK), Wnt signaling (RSPO2), gene expression regulation and cytoskeletal structural organization (LMO3), and cellular signal transduction (TENM1).
In addition, on the basis of our mcTI’s cross-validated EM algorithm, three statistically consistent AD subtypes were identified in the ADNI population (fig. S2). The BIC (32) was used to select the number of putative subtypes (up to a maximum of seven), and the statistical stability and significance of the subtypes were confirmed via permutation tests (all P < 0.001, FWE-corrected; see the “mcTI definition” section). Similar to the postmortem brain data, the in vivo blood-based subtypes differed at the multimodal molecular and neuropathological level, each showing a distinctive pattern of blood DNAm, RNA, proteomic, metabolomic, and molecular pathway alterations, along with unique brain phenotypic (PET/MRI) changes (see Fig. 6 and figs. S2, F and G, and S3, A to E). Like brain-derived subtypes, the blood subtypes did not significantly differ by proportions of clinical diagnosis (MCI and AD; fig. S2C) and sex (fig. S2D), while they did significantly differ in various cognitive domains (all P < 0.05, FWE-corrected; fig. S2F). AD subtype 2 was significantly more affected than subtype 1 in memory performance and overall cognitive function. AD subtype 3 was more significantly affected than subtype 1 in long-term decline in memory performance.
Ideally, AD subtypes should be comparable when obtained from different tissues and cohorts. Hence, we tested the portability of the identified putative AD subtypes across the two studied populations. For this, we used 16 neuropathologic traits available in both ROSMAP and ADNI to (i) extrapolate the postmortem brain-based subtypes to the in vivo population and (ii) evaluate the mutual information among the two independently obtained stratifications. On the basis of autopsied neuropathological evaluations or PET/MRI acquisitions, both ROSMAP and ADNI datasets included amyloid in the angular gyrus and calcarine, cingulate, entorhinal, hippocampal, inferior temporal, mesial temporal, midfrontal and superior frontal cortices, and tangles in the angular gyrus, calcarine, cingulate, entorhinal, hippocampal, inferior temporal, and mesial temporal cortices. The person-specific similarity of ADNI to ROSMAP was calculated as the Pearson correlation with the 16 corresponding values (Fig. 9, E and F). Next, each ADNI participant was assigned the brain-based AD subtype of the most neuropathologically correlated ROSMAP subject (Fig. 9G). This provided a second stratification/subtype for each ADNI participant.
We then determined whether the two independently obtained stratifications in ADNI would share common or complementary information. For this, we compared both classifications via the normalized mutual information (nMI; a measure of mutual dependence between the two classification configurations) and the variation of information (VI; amount of information lost and gained) (39). The significances of the nMI and VI values were tested via a randomized permutation procedure (i.e., comparing to the null distributions defined by randomly permuting the individual subtypes, with 10,000 repetitions). We observed (Fig. 9H) nonrandom convergent information (P = 0.0015, FWE-corrected) from the stratification of the two samples, confirming the portability of our multimodal molecular-defined AD classification system not only across different tissue samples (brain and blood) but also in independent cohorts.
Last, a comparison of altered molecular pathways in different subtypes indicated a considerable across-tissue overlapping (Fig. 6, B and D). Blood-derived AD subtype 2 showed the most consistent multi-omics similarity with brain-derived subtypes (with 44 to 53% epigenetic and 47 to 49% transcriptomic shared pathways). In addition, blood-based AD subtype 1 and brain-based subtype 3 shared over 57% of their altered RNA molecular pathways. In both tissues, the immunological interleukin signaling, the gastric cholecystokinin receptor (CCKR), and the Wnt signal transduction pathways were epigenetically altered in the identified AD subtypes (Fig. 6A). Common to brain-derived subtypes, prominent transcriptomic pathways in blood (Fig. 6C) reflected generalized cellular and molecular processes, such as GTR, ubiquitination, transcription regulation by bZIP, RNA synthesis by RNA polymerase, cytoskeletal regulation by ρ-GTPase, oxidative stress response, and cellular signaling via the Gs α subunit. The finding of these common pathways (see table S4 for a complete list across subtypes) evidences the direct relationship between the central nervous system and the body, and further supports the portability of our AD classification system.
DISCUSSION
We proposed a novel multimodal molecular taxonomy for the assessment and classification of AD progression. Efforts to develop a comprehensive multi-omics characterization of AD’s substantial heterogeneity are in their infancy. We started from a multilayer characterization of heterogeneity in the postmortem brain with NCI as the biological reference, and we extended our analyses to independent peripheral samples from living participants. We found that our approach (i) predicted the person-specific severity of AD pathology as quantified by the molecular pseudo-time score; (ii) detected distinct, biologically differentiable, and statistically stable AD subtrajectories/subtypes, each associated with a unique pattern of multilevel molecular, neuropathological, cell type, and cognitive alterations; and for potential clinical utility (iii) was applicable to peripheral blood samples from living patients from a different cohort.
Our results are in accord with a previously reported RNA-based AD classification (20). Expectedly, given the notable complexity of AD dementia, we observed that AD heterogeneity cannot be entirely explained by neuropathological patterns, clinical severity, or differences in age or sex. RNA provides additional information, but we show here that consideration of other molecular information including epigenomics, proteomics, and metabolomics can offer substantial complementary information over RNA alone. Consistent results support that our identified multi-omics molecular AD progression index, subtypes, and associated multi-omics differences cannot be assumed to be a mere reflection of neuropathology severity. This should not be surprising. Although consideration of neuropathologies is crucial for understanding and potentially preventing and treating AD, recent studies have evidenced their limited capacity to explain the clinical decline observed in neurodegeneration (22). In a complementary analysis, we also verified that our multi-omics molecular AD subtypes cannot be replaced (and vice versa) by the neuropathology-derived Murray-Dickson AD subtypes based on brain NFT distributions (15, 40), with no significant convergent information among both classifications (see fig. S4; see the “Comparative analysis with neuropathological subtypes” section). This result strengthens the importance of further considering complementary disease processes across different biological scales. Even with the remarkable recent advances in AD marker detection using biofluids (41, 42), a fast and accurate multilevel classification of the entire AD spectrum should offer substantial advances in AD research and therapeutics such as facilitating the identification of subtype-specific therapeutic targets when population-based targets do not generalize well from subtype to subtype. Our cross-validated extension from brain to blood multi-omics data and identification of robust subtypes are a promising step toward minimally invasive multilevel patient profiling in the clinical setting and in clinical trials.
In addition to simultaneously uncovering disease dynamics and heterogeneity, the mcTI approach overcomes many of the traditional limitations of ML to enable the identification of the most informative molecular substrates including CpGs, genes, proteins, and metabolites (Fig. 3A and fig. S4A). This approach deals with high-dimensional data by including an intrinsic contrastive dimensionality reduction technique (43). This technique allows detection of disease-associated patterns in populations of interest while it adjusts for confounding components in the background control population (e.g., concurrent aging or experimental effects). We have previously observed (13) that, in the context of disease trajectory inference, this technique [contrastive principal components analysis (cPCA) (43)] is more sensitive for detection of pathological progression than other popular methods of dimensionality reduction [e.g., PCA and Uniform Manifold Approximation and Projection (UMAP)]. mcTI can be applied to any sort of neuroscience data including molecular, histopathological, neuroimaging, electrophysiological, and clinical data (37). Furthermore, while this method predicts neuropathology severity, no model training is performed to fit the neuropathological data. Contrary, other recently proposed ML techniques (44) focus on the supervised identification of molecular predictors of AD neuropathology. A comparison of both families of methods (without and with model training) is not straightforward and depends on the related scientific question or target application. However, mcTI and other unsupervised approaches for studying both disease progression and heterogeneity (14, 37) allow the molecular-based discovery of putative disease subtypes, which are typically not uncovered by the supervised models.
Our study also has a number of limitations. We used the first generation of large-scale multi-omics data from autopsied AD brains (ROSMAP) (45). However, these data come from a unique brain region (DLPFC). This choice of regional source is logical given that it is a neocortical hub of cognitive circuitry, with a central function of controlling executive functions (e.g., working memory and cognitive flexibility) (46). Furthermore, a growing body of evidence supports a key role of the DLPFC in AD progression, as it can present several molecular and phenotypic alterations related to clinical deterioration (45). Our findings from postmortem data are limited to the present investigation of ROSMAP data. To date, no other study in AMP-AD or similar data repository presents all four types of molecular information used in this study, hindering replication in independent postmortem samples. However, given the significant mutual sharing of our observations with the blood-based subtypes (ADNI data), the DLPFC may suffice to represent brain-body heterogeneity in AD. We urge caution, nonetheless, because of the regional restriction of postmortem multi-omics data currently available and because our method of testing the validation of extrapolation across cohorts relied on information from only 16 regional values for amyloid and tau. Furthermore, while our combined use of (epi)genetic, proteomic, and metabolomic data for subject profiling represents a significant extension of earlier work, we have used only a limited portion of data potentially available from the last two modalities. Specifically, the remaining technical limitations in screening have also resulted in significantly less information for metabolites/proteins (149 to 430 analytes) than for (epi)genetic features (48,000 to 865,918 potentially usable). However, for both the brain and the blood multi-omics data, a quantitative analysis of each modality’s contribution to the obtained AD subtypes revealed similar across-omics influence (Fig. 3B and fig. S4B; see the “Assessing omics contributions on subtyping” section). Notably, data from the proteome and metabolome implicate several proteins linking AD with metabolic disorders and immune response. These results suggest that all included molecular layers may be containing relevant and complementary biological information, without any specific modality dominating the multi-omics AD stratification. However, we expect that our disproportionate reliance on (epi)genetic markers will be successively improved in the near future with the accessibility to new proteomic and metabolomic quantification techniques/data (47). Similarly, although the study of single-cell molecular data in AD is still at the small-scale population level (48, 49), the increasing collection and accessibility to such data modalities in the near future should facilitate a deeper multilevel characterization of the disease.
Our study also had many strengths. Both ROS and MAP have extraordinarily high follow-up and autopsy rates, ensuring excellent internal validity. We leveraged multilevel omic data from hundreds of cases with each layer, far more than all other studies of which we are aware. The studies are unbiased by selection of cases and controls or pathology, allowing inferences to older persons in general. We also obtained the same omics modalities from blood samples (ADNI data) and corresponding multimodal brain imaging evaluations (molecular PET and MRI) for cross-validation with in vivo data. Application of a novel ML method to these independent datasets (N = 1863) allowed us to examine the disease’s marked multilevel molecular complexity and heterogeneity. Furthermore, we are encouraged by recent developments in cancer research that have been characterized by the successful integration of multiple omics technologies and their subsequent application in precision medicine (2, 3, 50). As similar advances have improved the detection and treatment of cancer (2), we hope that our fused molecular information can lead to a refined representation and understanding of neurodegeneration, thereby facilitating the identification of individual disease mechanisms and therapeutic requirements. We note that the analytic tools described here have been made freely available as part of the user-friendly cross-platform Neuroinformatics for Personalized Medicine software [NeuroPM-box (37); neuropm-lab.com/neuropm-box.html]. This multi-tool computational application allows for advanced analytical modeling for molecular, histopathological, brain imaging, and/or clinical evaluations, allowing the characterization of multiscale and multifactorial neuropathological mechanisms. We would be gratified if the molecularly informed AD stratification framework described here paves the way for deeper multidimensional profiling of patients in clinical research, and still more pleased if our methods can someday be used as a clinical tool. In this last context, we note that these methods, developed for research into AD, should also be readily adaptable to the study of many other neurological and neuropsychiatric conditions.
MATERIALS AND METHODS
Data
Ethics statement
ROSMAP was approved by an Institutional Review Board (IRB) of the Rush University Medical Center. All participants signed an informed consent and Anatomical Gift Act; in addition, they signed a repository consent allowing their data to be shared. Data documentation and sharing documents can be obtained at www.radc.rush.edu. The study was conducted according to Good Clinical Practice guidelines, the Declaration of Helsinki, and IRBs (adni.loni.usc.edu). Study subjects (table S1) and/or authorized representatives gave written informed consent at the time of enrollment for sample collection and completed questionnaires approved by each participating site IRB. The authors obtained approval from the ADNI Data Sharing and Publications Committee for data use and publication; see documents http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Data_Use_Agreement.pdf and http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Manuscript_Citations.pdf, respectively.
This study used multi-omics molecular data (Ntotal = 1863) from two large-scale databases (see table S1 for demographic characteristics). Each dataset was processed and analyzed independently.
Dataset 1
Dataset 1 includes multimodal molecular, neuropathological, and/or clinical data from a total of 822 participants (table S1) enrolled in ROS (51) or MAP (52). The multi-omics data contain RNA expression, DNAm, proteins, and metabolomics concentration from DLPFC of subsets of 489, 708, 111, and 1225 autopsied subjects, respectively. All these data were generated in previous studies as described in (35, 45, 53–56) and downloaded from the AMP-AD knowledge portal (www.synapse.org), using the following Synapse IDs: syn3157275 (for epigenomic data), syn3800853 (transcriptomic), syn10468856 (proteomic), and syn10235595 and syn10235594 (metabolomic). Only participants with at least two molecular data modalities were considered. The metabolomic data were generated by the Alzheimer’s Disease Metabolomics Consortium (ADMC; see also Acknowledgments). Blood monocyte RNA data for a subset of 615 subjects were also included here (syn22024496). Annual administration of cognitive tests was incorporated into summary measures of five domains of cognitive function [episodic memory, visuospatial ability, perceptual speed, semantic memory, and working memory (57)] and a global cognition measure computed by averaging the five summary scores. For each cognitive measure, the person-specific random slope was estimated as the rate of change in the variable over time. It comes a from linear mixed-effects model with annual variable as the longitudinal outcome. The model controls for age at baseline, sex, and years of education (58). Assignment of NCI, MCI, or AD dementia categories was performed in correspondence with most likely clinical diagnosis at the time of death. All available clinical data were reviewed by a neurologist with expertise in dementia, and a summary diagnostic opinion was rendered regarding the most likely clinical diagnosis. Case conferences including one or more neurologists and a neuropsychologist were used for consensus (59). All subjects underwent postmortem neuropathologic evaluations, including uniform structured assessment of AD pathology, cerebral infarcts, Lewy body disease, TDP-43 cytoplasmatic inclusions in neurons and glia, and other pathologies common in aging and dementia. Brain regional and average data can be requested at www.radc.rush.edu/requests.htm. The pathologic diagnosis of AD uses NIA-Reagan and modified CERAD criteria, and the staging of neurofibrillary pathology uses Braak staging (60).
Dataset 2
Blood-based multi-omics molecular screening, multimodal brain imaging, and/or clinical data from 1041 alive participants (table S1) were obtained from ADNI (adni.loni.usc.edu). Molecular data included blood RNA expression, DNAm, proteins, and metabolomics concentration of subsets of 658, 595, 635, and 551 subjects, respectively. Only participants with at least two molecular data modalities were included. ADNI was launched in 2003 as a public-private partnership, led by principal investigator M. W. Weiner. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of CI and AD. All the participants were also characterized cognitively, including the mini-mental state examination (MMSE) and composite scores of executive function (EF) and memory integrity (MEM) (61). In addition, they were clinically diagnosed at baseline as healthy control (NCI), EMCI, LMCI, or probable AD patient. Gene expression profiling used the Affymetrix Human Genome U219 Array (www.affymetrix.com), with quality-controlled gene expression data including activity levels for 49,293 transcripts. The Illumina Infinium HumanMethylationEPIC BeadChip Array (www.illumina.com) was used for methylation profiling, covering about 866,000 CpGs. DNAm IDAT files were read into R v3.5.1 (R Core Team, 2018) using the minfi package and annotated with the Infinium MethylationEPIC v1.0 B5 Manifest File (https://support.illumina.com/downloads.html). Normalized DNAm was measured using a bivariate gamma distribution method (62), which has been shown to outperform the traditional beta and M value regression algorithm. Bile acids, lipidomic, and/or purine metabolites data were generated by ADMC (see Acknowledgments), as extensively described in (35, 55, 56). Different sets of previously quantified proteins were combined, including the 190-analyte multiplex immunoassay panel developed on the Luminex xMAP platform (ADNI data file “Biomarkers Consortium Plasma Proteomics Data Primer 02Aug2013 FINAL”), the tau protein phosphorylated at threonine-181 (P-tau181; ADNI data file “University of Gothenburg Longitudinal plasma P-tau181”), the axonal protein tau (ADNI data file “Blennow Lab – ADNI-1 – Plasma tau”), and the plasma neurofilament light [NFL; ADNI data file “Blennow Lab ADNI1-2 Plasma neurofilament light (NFL) longitudinal”]. Last, molecular PET and MRI images quantifying three different biological properties were mapped in vivo using the following techniques: structural MRI (for structural tissular properties), florbetapir PET (for Aβ deposition), and 18F-AV-1451 PET (for tau deposition). For both Aβ and tau, corresponding mean standardized uptake value ratio values were extracted for 34 gray matter regions of interest defined by Freesurfer v7.1.1 (see ADNI data files “UC Berkeley - AV45 Analysis [ADNI1,G–,2,3]” and “UC Berkeley - AV1451 Analysis [ADNI1,GO,2,3],” respectively). MRI-based regional volumes were also quantified with Freesurfer. See table S1 for the corresponding demographic and data characteristics.
Methods
Data preprocessing
Before applying the mcTI approach, each molecular feature’s quality-controlled values (gene abundance, CpG site methylation level, and metabolite or protein concentration) was adjusted for relevant covariates using robust additive linear models. Covariables included age, sex, and educational level for both in vivo and postmortem data, plus postmortem interval in hours, sample pH, RNA integrity number, and batch number when applicable.
mcTI definition
Given a set of multiple “omic” data types, this method provides two estimations for each participant (see detailed algorithm below): (i) a personalized multi-omics mDPS, reflecting how close (in terms of multilevel molecular alterations) each participant is to developing AD dementia, and (ii) a putative disease subtype, corresponding to a distinctive disease trajectory that the participant may be developing. Initially, each data modality’s high number of features/biomarkers is reduced by identifying the low-dimensional pattern enriched in subjects diagnosed with AD dementia relative to subjects with NCI. For this, cPCA (43) is performed, reducing each data modality to a few components capturing the AD-associated patterns (13). Next, all the molecular data modalities’ disease-enriched components are aggregated by similarity network fusion (SNF) (2), an ML algorithm that combines diverse types of measurements, here, DNAm, RNA expression, metabolites, and protein concentrations. SNF constructs a robust fused subject-subject similarity network for different data scales, collection bias, and measurement error. This network constitutes the integrated data space where the population stratification is performed for disease progression and subtrajectories. Essentially, each subject’s pseudo-time value represents a personalized multi-omics mDPS calculated as the distance over the network to the NCI subjects (i.e., for each subject, adding the network links through any intermediary AD or MCI participant until reaching the NCI subgroup). A continuous standardized value between 0 and 1 is obtained for each participant, reflecting the proximity to the cognitively healthy state. Next, participants are assigned to distinct molecular subtypes/subtrajectories, which are identified via an EM algorithm that maximizes the alignment of the subjects within each specific subtype based on their molecular data [i.e., each subtype corresponds to a subtrajectory defined as a concatenated subset of cognitively affected subjects (MCI and AD) following a similar pattern in the multi-omics data’s integrated space; see Fig. 1B]. Last, a statistical subtypes’ stability and significance analysis is performed via randomized permutations. Each disease subtype’s stability (20) is calculated as the rate at which pairs of subjects group together into the same cluster upon repeated clustering on random subsets of the input data. The comparison of each subtype’s intrinsic stability with a generated null distribution allows testing its significance.
Detailed mcTI algorithm
The inference of multi-omics contrastive pseudo-temporal subtrajectories/subtypes consisted of six main steps (see simplified definition above):
1) Optional initial selection of features most likely to be involved in a trajectory across the entire diseased population. We only applied this step for the transcriptomic and epigenomic data (with considerably higher dimensionality than the population’s sample size and the metabolomic/proteomic data). For each of these two data types, only the top 1000 differentially expressed features (transcripts or CpGs) were selected for the subsequent analyses [for this, a T score value per feature was preliminarily calculated, reflecting its likelihood to be altered in the cognitively affected subpopulation (MCI or AD) in comparison with the subjects with NCI].
2) For each molecular data type, data exploration and visualization were performed via cPCA (43). This technique identified low-dimensional patterns that are enriched in the target dataset (here, the subjects with AD dementia) relative to a comparison background dataset (the NCI). By controlling the effects of characteristic patterns in the background, cPCA allows the visualization of specific data structures missed by standard data exploration and visualization methods (e.g., traditional PCA and Kernel PCA). Specifically, if Ctarget and Cbackground are the covariance matrices of the target and background data, the directions returned by cPCA are the singular vectors of the weighted difference of the covariance matrices: Ctarget – α·Cbackground. The contrast parameter α represents the trade-off between having high target variance and low background variance. Multiple values of α are used (i.e., 100 logarithmically equally spaced points between 10−2 and 102). Instead of choosing a single α, the resulting subspaces for all the – values are clustered [based on their proximity in terms of the principal angle and spectral clustering (63)] in a few subspaces. The data are then projected onto each of these few subspaces, revealing different trends within the target data. While the original cPCA algorithm (43) selects the final subspace via visual examination, we automatically select the subspace that maximizes the clustering tendency in the projected target data, relative to the clustering tendency in the background population.
3) Aggregation of the different dimensionally reduced molecular data modalities via SNF (2). The well-known SNF algorithm, originally proposed in the context of cancer research (2), allows us to combine diverse types of measurements (here, DNAm, RNA expression, and metabolite and protein concentrations) for a given population. It first creates a sample similarity network for each of the data types (here, each of the four dimensionally reduced omics data) and then iteratively integrates these networks into a fused subject-subject similarity network (FN). SNF is robust to different data scales, collection bias, and noise in different measurement types. Here, this nonlinear data fusion algorithm allowed the utilization of common and complementary information in the different molecular data types.
4) Individual multi-omics mDPS calculation according to the distance over the network (FN) to the background NCI subpopulation [i.e., for each cognitively affected subject (MCI or AD), adding the network links through any intermediary participant until reaching the NCI subgroup]. For this, we first calculated the minimum spanning tree of the FN (FN-MST). The FN-MST is then used to calculate the shortest path from any participant to the background subjects. Each shortest path is defined as the concatenation of relatively similar subjects in the integrated multimodal molecular space that minimizes the distance to the background. The position of each subject in her/his corresponding shortest path reflects the individual distance to the NCI subpopulation and, if analyzed in the inverse direction, to advanced disease state (AD dementia). Thus, to quantify the distance to these two extremes (NCI or AD dementia), the individual multi-omics mDPS is calculated as the shortest distance value to the background’s centroid, relative to the maximum population value (i.e., values are standardized between 0 and 1). Relatively low or high values indicate greater or lesser distance on the path to develop AD dementia (Fig. 1, B and C).
5) Subtyping via EM. This step focuses on detecting subgroups of cognitively affected subjects aligned to distinctive molecular subtrajectories in the integrated multi-omics space. Preliminarily, spectral clustering (63) is performed over the FN and provided as initial solution to the EM subtyping. In addition, using multidimensional scaling [MDS; Matlab function mdscale (64)], the multimodal molecular information contained in the FN is translated to an abstract Cartesian space (here, on FN-MDS) where each subject is represented by a set of coordinate values. Note that the FN-MDS space preserves the distances/similarities between all the subjects in the FN, providing a numerical representation of the integrated molecular information at the whole population level. The FN-MDS’s optimum number of dimensions was automatically determined using BIC (32). Next, the EM is applied by repeating two steps: (i) leave-one-out predictions of each subject’s multi-omics mDPS (obtained in step 4). Training independent subtype-specific models with the individual FN-MDS coordinates as predicting variables, each subject receives as many mDPS predictions as the number of available subtypes, always keeping the subject outside of the training step. For each subject i and subtype j, this provides a prediction error, reflecting how well the subject i aligns with the subtype j’s internal multimodal molecular data. (ii) The subjects’ subtypes are updated according to the obtained prediction errors (i.e., each subject is reassigned now to the subtype that better predicted its molecular disease progression index). Steps (i) and (ii) are repeated until the subtypes’ configurations reach a small level of variation, providing potentially stable subtypes.
6) Last, subtype stability and significance evaluation via randomized permutations. Following the method proposed in (20), subtype stability was defined as the rate at which pairs of subjects group together into the same subtypes upon repeated clustering on random subsets of the input data. Extended to the multilevel molecular information, the reasoning behind this method (20) is that if true subtypes are reflected in the data, then a robust subtyping method should provide the same set of clustered samples on repeated reevaluation using fewer samples or molecular features. Contrarily, if no distinctive signature is reflected in the data and/or the method is not robust, then different sets of subtyped samples will result from repeated reclustering. In practice, we calculated the rate at which each pair of subjects shared the same subtype across all 50 bootstrapping and the resulting average pairwise sample reclustering rate for all pairs of samples within the sample subtypes. Similarly, a null distribution of average pairwise sample reclustering rates per subtype was calculated across 5000 randomized permutations. We then calculated the empirical likelihood that the subtype stability rate and the null stability rate are the same (for each subtype, defining a significance P value as one minus the proportion of cases in which the observed stability rate was higher than the null distribution values).
Assessing marker contributions on mDPS
For each dataset and molecular omics modality, the total contribution Ci of each modality-specific marker i to the obtained reduced representation space (and the multi-omics mDPS) was quantified as (13)
| (1) |
where is the normalized eigenvalue of the contrasted principal component j, min_λ is the minimum obtained eigenvalue, Ntotal is the original number of contrasted principal components, NcPC is the number of contrasted principal components with over a predefined cutoff value (i.e., 0.025), ωi, j is the loading/weight of the marker i on the component j, and Nfeatures is the total number of modality-specific markers considered in the dimensionality reduction analysis (step 2 in mcTI algorithm). For comparison across datasets/modalities, obtained values were normalized by the maximum and expressed as percentages (Fig. 3A and fig. S4A).
Assessing omics contributions on subtyping
For each molecular omics modality i (epigenome, transcriptome, proteome, or metabolome), its contribution to the obtained subtypes was calculated as:
1) i was removed from the mcTI algorithm’s input data, and a new AD classification was obtained (i.e., without any information from i).
2) An index reflecting the level of dependence in modality i for obtaining the original AD classification was calculated as: 1 − nMI, where nMI is the normalized mutual information between the two obtained AD classification configurations (i.e., with and without considering modality i). Note that a value of 1 would imply a high level of dependency in modality i, and 0 otherwise.
The indexes were normalized to express percentages across the four molecular modalities (Fig. 3B and fig. S4B).
Brain cell type analysis
Brain-based subtype-specific differentially expressed genes (from the original no-preselected transcripts, dataset 1) were used to perform a cell type identification analysis with the Enrichr software (38) and the Allen Brain Atlas 10x scRNA 2021. For each putative subtype, a ranked list of about 200 potentially down- or up-regulated brain cells was obtained, with a z score value per cell indicating the statistical likelihood to be enriched in comparison with a random background. An FDR multiple-comparison analysis of these z scores was performed to identify highly likely altered cell types. Subtype-subtype dissimilarity regarding their significantly altered cells was calculated as the relative mismatch (in percent) regarding unique down-/up-regulated cell types.
Comparative analysis with neuropathological subtypes
Similar to (40), criteria for three neuropathological AD subtypes were adapted from Murray et al. (15). On the basis of the cortical and hippocampal density and distribution of NFT in postmortem brains, the Murray-Dickson AD classification consists of three subtypes: typical AD (tAD), limbic predominant (LP), and hippocampal sparing (HpSp). In dataset 1, a subset of participants (N = 131) satisfied inclusion criteria (i.e., AD confirmed, Braak stage V or VI, and no hippocampal sclerosis). NFT counts for the subiculum and CA1 regions of the hippocampus were considered together as a single region rather than separated and averaged (40). Four cortical regions with NFT were considered (inferior temporal, angular, calcarine, and cingulate cortices). From all participants, 98 (74.81%) met the three required criteria for tAD: (i) the ratio of the hippocampal NFT counts to the average cortical NFT to be less than the 25th percentile of all AD cases, (ii) the hippocampal NFT counts to be less than the population’s median value, and (iii) at least three of the cortical NFT count values to be greater than or equal to the median values. Nineteen subjects (14.50%) met the three criteria for LP: (i) the ratio of the hippocampal NFT counts to the average cortical NFT to be greater than the 75th percentile of all AD cases, (ii) the hippocampal NFT counts to be greater than the population’s median value, and (iii) at least three of the cortical NFT count values to be less than or equal to the median values. Fourteen subjects (10.69%) satisfied the criteria for HpSp: AD participants not classified as tAD or LP. Last, these Murray-Dickson AD subtypes were quantitatively compared with our multi-omics molecular AD subtypes via the nMI (a measure of mutual dependence between the two classification configurations) and the VI (amount of information lost and gained) (39). The significances of the obtained nMI and VI values were tested via a randomized permutation procedure (i.e., comparing to the randomized subtype distributions, with 10,000 repetitions; fig. S4).
Statistical analyses
Multi-omics mDPS associations with selected neuropathological staging variables (e.g., Braak, CERAD, and TDP-43) were tested via Kruskal-Wallis tests, previously adjusting the pseudo-time for age, sex, and educational level. For each neuropathological staging variable, a randomized permutation procedure (5000 repetitions) was used to construct a robust null distribution and obtain associated FWE-corrected Kruskal-Wallis significance values. In addition, all statistical subtype-subtype comparisons were performed via ANOVA tests, with subtype as the grouping variable (age, sex, and educational level were included as covariables when the data were not already adjusted for these confounding factors, e.g., for neuropathological features). The resulting ANOVA P values underwent FDR correction (with significance cutoff q < 0.05) (65). Analyses were performed in MATLAB version R2019b.
Acknowledgments
We thank the two anonymous reviewers for many insightful comments and suggestions.
Funding: This project was undertaken thanks, in part, to the following funding awards to Y.I.-M.: the Canada Research Chair tier-2, the CIHR Project Grant 2020, the Weston Family Foundation’s Transformational Research in AD 2020, and the New Investigator start-up grant from McGill University’s Healthy Brains for Healthy Lives Initiative (Canada First Research Excellence Fund). In addition, we used the computational infrastructure of the McConnell Brain Imaging Center at the Montreal Neurological Institute, supported, in part, by the Brain Canada Foundation, through the Canada Brain Research Fund, with the financial support of Health Canada and sponsors. In addition, the RNAseq, epigenetic, proteomic, and neuropathology data for dataset 1 (ROSMAP) were provided by the Rush Alzheimer’s Disease Center (Rush University Medical Center, Chicago) and generated by grants to D.A.B. at the Rush University Medical Center, P.L.D.J. at the Columbia University Medical Center, and V.A.P. at Pacific Northwest National Laboratories. The metabolomic data for both dataset 1 (ROSMAP) and dataset 2 (ADNI) were generated by ADMC (ADMC members list: https://sites.duke.edu/adnimetab/team/), led by R. Kaddurah-Daouk at Duke University, using biospecimens provided by the Rush Alzheimer’s Disease Center and ADNI, respectively. Support for the biospecimen processing and metabolomic data generation conducted by ADMC was provided by the following: National Institute on Aging grants R01AG046171, RF1AG051550, RF1AG057452, RF1AG059093, RF1AG058942, and U01AG061359 and Foundation for the NIH (FNIH) grant DAOU16AMPA. In addition, dataset 2 (ADNI) collection and sharing for this project was funded by ADNI (NIH grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging and the National Institute of Biomedical Imaging and Bioengineering and through contributions from the following: AbbVie, Alzheimer’s Association, Alzheimer’s Drug Discovery Foundation, Araclon Biotech, BioClinica Inc., Biogen, Bristol-Myers Squibb Company, CereSpir Inc., Eisai Inc., Elan Pharmaceuticals Inc., Eli Lilly and Company, EuroImmun, F. Hoffmann–La Roche Ltd. and its affiliated company Genentech Inc., Fujirebio, GE Healthcare, IXICO Ltd., Janssen Alzheimer Immunotherapy Research & Development LLC, Johnson & Johnson Pharmaceutical Research & Development LLC, Lumosity, Lundbeck, Merck & Co. Inc., Meso Scale Diagnostics LLC, NeuroRx Research, Neurotrack Technologies, Novartis Pharmaceuticals Corporation, Pfizer Inc., Piramal Imaging, Servier, Takeda Pharmaceutical Company, and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by FNIH (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for NeuroImaging at the University of Southern California.
Author contributions: Y.I.-M. conceived the study, implemented the analytical source codes (for mcTI), preprocessed and analyzed the presented data, and wrote the first manuscript draft. D.A.B. participated in study conception. D.A.B. and J.B. actively contributed to manuscript writing and revision. Q.A. performed the RNA cell type deconvolution analysis, preprocessed the blood monocyte RNA data, and revised the manuscript. A.F.K. contributed to the figure and table preparation and result interpretation. All authors contributed to constructive discussions regarding study design, results, and manuscript preparation.
Competing interests: S.D. has participated in sponsored research in the field of Alzheimer’s disease (Biogen, Ionis Pharmaceuticals, Novo Nordisk, and Janssen) in addition to advisory boards (Biogen and Eisai) and paid consultancy/speaking honorarium (Eisai and QuRALIS). The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All ROSMAP molecular data are available for general research at the AMP-AD knowledge portal (Synapse IDs syn3800853, syn3157275, syn10235595, syn10235594, syn10468856, and syn22024496; www.synapse.org/) according to the following requirements for data access and attribution (https://adknowledgeportal.synapse.org/DataAccess/Instructions). Detailed ROSMAP neuropathological and clinical data are available at the RADC Research Resource Sharing Hub (radc.rush.edu), pending scientific review and a completed material transfer agreement (see radc.rush.edu/requests.htm). All the ADNI data are available at the web portal (adni.loni.usc.edu). The used multi-omics stratification method (mcTI) will be freely available with article publication as part of the NeuroPM-box software (neuropm-lab.com/neuropm-box.html) (37). User-friendly standalone applications for Linux, macOS, and Windows systems are provided (programming expertise is not required). A detailed easy-to-follow tutorial is also provided.
Supplementary Materials
This PDF file includes:
Supplementary Text S1
Figs. S1 to S4
Tables S1 to S5
REFERENCES AND NOTES
- 1.Park J., Hescott B. J., Slonim D. K., Towards a more molecular taxonomy of disease. J. Biomed. Semantics 8, 25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang B., Mezlini A. M., Demir F., Fiume M., Tu Z., Brudno M., Haibe-Kains B., Goldenberg A., Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 11, 333–337 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Collisson E. A., Bailey P., Chang D. K., Biankin A. V., Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 16, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Hodes R. J., Buckholtz N., Accelerating medicines partnership: Alzheimer’s disease (AMP-AD) knowledge portal aids Alzheimer’s drug discovery through open data sharing. Expert Opin. Ther. Targets 20, 389–391 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Greenwood A. K., Montgomery K. S., Kauer N., Woo K. H., Leanza Z. J., Poehlman W. L., Gockley J., Sieberts S. K., Bradic L., Logsdon B. A., Peters M. A., Omberg L., Mangravite L. M., The AD knowledge portal: A repository for multi-omic data on Alzheimer’s disease and aging. Curr. Protoc. Hum. Genet. 108, e105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jagust W., Bandy D., Chen K., Foster N., Landau S., Mathis C. A., Price J. C., Reiman E. M., Skovronsky D., Koeppe R. A.; Alzheimer’s Disease Neuroimaging Initiative , The Alzheimer’s disease neuroimaging initiative positron emission tomography core. Alzheimers Dement. 6, 221–229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trojanowski J. Q., Vandeerstichele H., Korecka M., Clark C. M., Aisen P. S., Petersen R. C., Blennow K., Soares H., Simon A., Lewczuk P., Dean R., Siemers E., Potter W. Z., Weiner M. W., Jack C. R., Jagust W., Toga A. W., Lee V. M.-Y., Shaw L. M.; Alzheimer’s Disease Neuroimaging Initiative , Update on the biomarker core of the Alzheimer’s disease neuroimaging initiative subjects. Alzheimers Dement. 6, 230–238 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee S., Perumal T. M., Daily K., Sieberts S. K., Omberg L., Preuss C., Carter G. W., Mangravite L. M., Logsdon B. A., Identifying and ranking potential driver genes of Alzheimer’s disease using multiview evidence aggregation. Bioinformatics 35, i568–i576 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannoodt R., Saelens W., Saeys Y., Computational methods for trajectory inference from single-cell transcriptomics. Eur. J. Immunol. 46, 2496–2506 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Magwene P. M., Lizardi P., Kim J., Reconstructing the temporal ordering of biological samples using microarray data. Bioinformatics 19, 842–850 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Gupta A., Bar-Joseph Z., Extracting dynamics from static cancer expression data. IEEE/ACM Trans. Comput. Biol. Bioinform. 5, 172–182 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Guttman L., A basis for scaling qualitative data. Am. Sociol. Rev. 9, 139–150 (1944). [Google Scholar]
- 13.Iturria-Medina Y., Khan A. F., Adewale Q., Shirazi A. H.; Alzheimer’s Disease Neuroimaging Initiative , Blood and brain gene expression trajectories mirror neuropathology and clinical deterioration in neurodegeneration. Brain 143, 661–673 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherjee S., Heath L., Preuss C., Jayadev S., Garden G. A., Greenwood A. K., Sieberts S. K., De Jager P. L., Ertekin-Taner N., Carter G. W., Mangravite L. M., Logsdon B. A., Molecular estimation of neurodegeneration pseudotime in older brains. Nat. Commun. 11, 5781 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray M. E., Graff N. R., Ross O. A., Petersen R. C., Duara R., Dickson D. W., Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: A retrospective study. Lancet Neurol. 10, 785–796 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charil A., Shcherbinin S., Southekal S., Devous M. D., Mintun M., Murray M. E., Miller B. B., Schwarz A. J., Tau subtypes of Alzheimer’s disease determined in vivo using flortaucipir PET imaging. J. Alzheimers Dis. 71, 1037–1048 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Vogel J. W., Young A. L., Oxtoby N. P., Smith R., Ossenkoppele R., Strandberg O. T., La Joie R., Aksman L. M., Grothe M. J., Iturria-Medina Y.; Alzheimer’s Disease Neuroimaging Initiative, Pontecorvo M. J., Devous M. D., Rabinovici G. D., Alexander D. C., Lyoo C. H., Evans A. C., Hansson O., Four distinct trajectories of tau deposition identified in Alzheimer’s disease. Nat. Med. 27, 871–881 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayden K. M., Reed B. R., Manly J. J., Tommet D., Pietrzak R. H., Chelune G. J., Yang F. M., Revell A. J., Bennett D. A., Jones R. N., Cognitive decline in the elderly: An analysis of population heterogeneity. Age Ageing 40, 684–689 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu L., Boyle P. A., Segawa E., Leurgans S., Schneider J. A., Wilson R. S., Bennett D. A., Residual decline in cognition after adjustment for common neuropathologic conditions. Neuropsychology 29, 335–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neff R. A., Wang M., Vatansever S., Guo L., Ming C., Wang Q., Wang E., Horgusluoglu-Moloch E., Song W. M., Li A., Castranio E. L., Julia T. C. W., Ho L., Goate A., Fossati V., Noggle S., Gandy S., Ehrlich M. E., Katsel P., Schadt E., Cai D., Brennand K. J., Haroutunian V., Zhang B., Molecular subtyping of Alzheimer’s disease using RNA sequencing data reveals novel mechanisms and targets. Sci. Adv. 7, eabb5398 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nazarian A., Yashin A. I., Kulminski A. M., Summary-based methylome-wide association analyses suggest potential genetically driven epigenetic heterogeneity of Alzheimer’s disease. J. Clin. Med. 9, 1489 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyle P. A., Yu L., Wilson R. S., Leurgans S. E., Schneider J. A., Bennett D. A., Person-specific contribution of neuropathologies to cognitive loss in old age. Ann. Neurol. 83, 74–83 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tibshirani R., Regression selection and shrinkage via the Lasso. J. R. Stat. Soc. B 58, 267–288 (1996). [Google Scholar]
- 24.Stoll S. J., Kroll J., HOXC9: A key regulator of endothelial cell quiescence and vascular morphogenesis. Trends Cardiovasc. Med. 22, 7–11 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Karch C. M., Ezerskiy L. A., Bertelsen S.; Alzheimer’s Disease Genetics Consortium (ADGC), Goate A. M., Alzheimer’s disease risk polymorphisms regulate gene expression in the ZCWPW1 and the CELF1 loci. PLOS ONE 11, e0148717 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito E. R., Miller J. B., Harari O., Cruchaga C., Mihindukulasuriya K. A., Kauwe J. S. K., Bikman B. T., Alzheimer’s disease alters oligodendrocytic glycolytic and ketolytic gene expression. Alzheimers Dement. 17, 1474–1486 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J., Huang R., Peng Y., Wang H., Feng Z., Fan Y., Zeng Z., Wang Y., Wei J., Wang Z., Effects of DISC1 on Alzheimer’s disease cell models assessed by iTRAQ proteomics analysis. Biosci. Rep. 42, BSR20211150 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlomagno Y., Manne S., DeTure M., Prudencio M., Zhang Y. J., Hanna Al-Shaikh R., Dunmore J. A., Daughrity L. M., Song Y., Castanedes-Casey M., Lewis-Tuffin L. J., Nicholson K. A., Wszolek Z. K., Dickson D. W., Fitzpatrick A. W. P., Petrucelli L., Cook C. N., The AD tau core spontaneously self-assembles and recruits full-length tau to filaments. Cell Rep. 34, 108843 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marksteiner J., Blasko I., Kemmler G., Koal T., Humpel C., Bile acid quantification of 20 plasma metabolites identifies lithocholic acid as a putative biomarker in Alzheimer’s disease. Metabolomics 14, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madeo F., Eisenberg T., Pietrocola F., Kroemer G., Spermidine in health and disease. Science 359, eaan2788 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Pekar T., Wendzel A., Flak W., Kremer A., Pauschenwein-Frantsich S., Gschaider A., Wantke F., Jarisch R., Spermidine in dementia: Relation to age and memory performance. Wien. Klin. Wochenschr. 132, 42–46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gideon S., Estimating the dimension of a model. Ann. Stat. 6, 461–464 (1978). [Google Scholar]
- 33.Mi H., Muruganujan A., Casagrande J. T., Thomas P. D., Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varma V. R., Oommen A. M., Varma S., Casanova R., An Y., Andrews R. M., O’Brien R., Pletnikova O., Troncoso J. C., Toledo J., Baillie R., Arnold M., Kastenmueller G., Nho K., Doraiswamy P. M., Saykin A. J., Kaddurah-Daouk R., Legido-Quigley C., Thambisetty M., Brain and blood metabolite signatures of pathology and progression in Alzheimer disease: A targeted metabolomics study. PLOS Med. 15, e1002482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.John-Williams L. S., Mahmoudiandehkordi S., Arnold M., Massaro T., Blach C., Kastenmüller G., Louie G., Kueider-Paisley A., Han X., Baillie R., Motsinger-Reif A. A., Rotroff D., Nho K., Saykin A. J., Risacher S. L., Koal T., Moseley M. A., Tenenbaum J. D., Thompson J. W., Kaddurah-Daouk R.; Alzheimer’s Disease Neuroimaging Initiative; Alzheimer’s Disease Metabolomics Consortium , Bile acids targeted metabolomics and medication classification data in the ADNI1 and ADNIGO/2 cohorts. Sci. Data 6, 212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan A. F., Adewale Q., Baumeister T. R., Carbonell F., Zilles K., Palomero-Gallagher N., Iturria-Medina Y.; for the Alzheimer’s Disease Neuroimaging Initiative , Personalized brain models identify neurotransmitter receptor changes in Alzheimer’s disease. Brain 145, 1785–1804 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iturria-medina Y., Carbonell F., Assadi A., Adewale Q., Khan A. F., Baumeister T. R., Sanchez-Rodriguez L., NeuroPM toolbox: Integrating molecular, neuroimaging and clinical data for characterizing neuropathological progression and individual. Commun. Biol. 4, 614 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R., Ma’ayan A., Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubinov M., Sporns O., Complex network measures of brain connectivity: Uses and interpretations. Neuroimage 52, 1059–1069 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Uretsky M., Gibbons L. E., Mukherjee S., Trittschuh E. H., Fardo D. W., Boyle P. A., Keene C. D., Saykin A. J., Crane P. K., Schneider J. A., Mez J., Longitudinal cognitive performance of Alzheimer’s disease neuropathological subtypes. Alzheimer’s Dement. 7, e12201 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson E. C. B., Dammer E. B., Duong D. M., Ping L., Zhou M., Yin L., Higginbotham L. A., Guajardo A., White B., Troncoso J. C., Thambisetty M., Montine T. J., Lee E. B., Trojanowski J. Q., Beach T. G., Reiman E. M., Haroutunian V., Wang M., Schadt E., Zhang B., Dickson D. W., Ertekin-Taner N., Golde T. E., Petyuk V. A., De Jager P. L., Bennett D. A., Wingo T. S., Rangaraju S., Hajjar I., Shulman J. M., Lah J. J., Levey A. I., Seyfried N. T., Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med. 26, 769–780 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashton N. J., Janelidze S., Al Khleifat A., Leuzy A., van der Ende E. L., Karikari T. K., Benedet A. L., Pascoal T. A., Lleó A., Parnetti L., Galimberti D., Bonanni L., Pilotto A., Padovani A., Lycke J., Novakova L., Axelsson M., Velayudhan L., Rabinovici G. D., Miller B., Pariante C., Nikkheslat N., Resnick S. M., Thambisetty M., Schöll M., Fernández-Eulate G., Gil-Bea F. J., de Munain A. L., Al-Chalabi A., Rosa-Neto P., Strydom A., Svenningsson P., Stomrud E., Santillo A., Aarsland D., van Swieten J. C., Palmqvist S., Zetterberg H., Blennow K., Hye A., Hansson O., A multicentre validation study of the diagnostic value of plasma neurofilament light. Nat. Commun. 12, 3400 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abid A., Zhang M. J., Bagaria V. K., Zou J., Exploring patterns enriched in a dataset with contrastive principal component analysis. Nat. Commun. 9, 2134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beebe-Wang N., Celik S., Weinberger E., Sturmfels P., De Jager P. L., Mostafavi S., Lee S. I., Unified AI framework to uncover deep interrelationships between gene expression and Alzheimer’s disease neuropathologies. Nat. Commun. 12, 5369 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Jager P. L., Ma Y., McCabe C., Xu J., Vardarajan B. N., Felsky D., Klein H. U., White C. C., Peters M. A., Lodgson B., Nejad P., Tang A., Mangravite L. M., Yu L., Gaiteri C., Mostafavi S., Schneider J. A., Bennett D. A., Data descriptor: A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci. Data 5, 180142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaplan J. T., Gimbel S. I., Harris S., Neural correlates of maintaining one’s political beliefs in the face of counterevidence. Sci. Rep. 6, 39589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., Bridgland A., Cowie A., Meyer C., Laydon A., Velankar S., Kleywegt G. J., Bateman A., Evans R., Pritzel A., Figurnov M., Ronneberger O., Bates R., Kohl S. A. A., Potapenko A., Ballard A. J., Romera-Paredes B., Nikolov S., Jain R., Clancy E., Reiman D., Petersen S., Senior A. W., Kavukcuoglu K., Birney E., Kohli P., Jumper J., Hassabis D., Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grubman A., Chew G., Ouyang J. F., Sun G., Choo X. Y., McLean C., Simmons R. K., Buckberry S., Vargas-Landin D. B., Poppe D., Pflueger J., Lister R., Rackham O. J. L., Petretto E., Polo J. M., A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 22, 2087–2097 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Mathys H., Davila-Velderrain J., Peng Z., Gao F., Mohammadi S., Young J. Z., Menon M., He L., Abdurrob F., Jiang X., Martorell A. J., Ransohoff R. M., Hafler B. P., Bennett D. A., Kellis M., Tsai L. H., Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.T. Ma, A. Zhang, Integrate multi-omic data using affinity network fusion (ANF) for cancer patient clustering, in Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM 2017) (IEEE, 2017), pp. 398–403.
- 51.Bennett D., Schneider J., Arvanitakis Z., Wilson R., Overview and findings from the religious orders study. Curr. Alzheimer Res. 9, 628–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bennett D., Schneider J., Buchman A., Barnes L., Boyle P., Wilson R., Overview and findings from the rush memory and aging project. Curr. Alzheimer Res. 9, 646–663 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett D. A., Yu L., De Jager P. L., Building a pipeline to discover and validate novel therapeutic targets and lead compounds for Alzheimer’s disease. Biochem. Pharmacol. 88, 617–630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Jager P. L., Srivastava G., Lunnon K., Burgess J., Schalkwyk L. C., Yu L., Eaton M. L., Keenan B. T., Ernst J., McCabe C., Tang A., Raj T., Replogle J., Brodeur W., Gabriel S., Chai H. S., Younkin C., Younkin S. G., Zou F., Szyf M., Epstein C. B., Schneider J. A., Bernstein B. E., Meissner A., Ertekin-Taner N., Chibnik L. B., Kellis M., Mill J., Bennett D. A., Alzheimer’s disease: Early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci. 17, 1156–1163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.St John-Williams L., Blach C., Toledo J. B., Rotroff D. M., Kim S., Klavins K., Baillie R., Han X., Mahmoudiandehkordi S., Jack J., Massaro T. J., Lucas J. E., Louie G., Motsinger-Reif A. A., Risacher S. L., Saykin A. J., Kastenmüller G., Arnold M., Koal T., Moseley M. A., Mangravite L. M., Peters M. A., Tenenbaum J. D., Thompson J. W., Kaddurah-Daouk R., Targeted metabolomics and medication classification data from participants in the ADNI1 cohort. Sci. Data 4, 170140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barupal D. K., Fan S., Wancewicz B., Cajka T., Sa M., Showalter M. R., Baillie R., Tenenbaum J. D., Louie G., Kaddurah-Daouk R., Fiehn O., Generation and quality control of lipidomics data for the Alzheimer’s disease neuroimaging initiative cohort. Sci. Data 5, 180263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bennett D. A., Schneider J. A., Buchman A. S., Mendes de Leon C., Bienias J. L., Wilson R. S., The rush memory and aging project: Study design and baseline characteristics of the study cohort. Neuroepidemiology 25, 163–175 (2005). [DOI] [PubMed] [Google Scholar]
- 58.De Jager P. L., Shulman J. M., Chibnik L. B., Keenan B. T., Raj T., Wilson R. S., Yu L., Leurgans S. E., Tran D., Aubin C., Anderson C. D., Biffi A., Corneveaux J. J., Huentelman M. J., Rosand J., Daly M. J., Myers A. J., Reiman E. M., Bennett D. A., Evans D. A., A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiol. Aging 33, 1017.e1–1017.e15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schneider J. A., Arvanitakis Z., Bennett D. A., Mixed brain pathologies account for most dementia cases in community- dwelling older persons. Neurology 69, 2197–2204 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Braak H., Braak E., Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991). [DOI] [PubMed] [Google Scholar]
- 61.Gibbons L. E., Carle A. C., Mackin R. S., Harvey D., Mukherjee S., Insel P., Curtis S. M. K., Mungas D., Crane P. K.; Alzheimer’s Disease Neuroimaging Initiative , A composite score for executive functioning, validated in Alzheimer’s Disease Neuroimaging Initiative (ADNI) participants with baseline mild cognitive impairment. Brain Imaging Behav. 6, 517–527 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weinhold L., Wahl S., Pechlivanis S., Hoffmann P., Schmid M., A statistical model for the analysis of beta values in DNA methylation studies. BMC Bioinformatics 17, 480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.P. Hespanha, An efficient MATLAB algorithm for graph partitioning technical report graph partitioning (2004); https://web.ece.ucsb.edu/~hespanha/published/tr-ell-gp.pdf.
- 64.Mead A., Review of the development of multidimensional scaling methods. J. R. Stat. Soc. 41, 27 (1992). [Google Scholar]
- 65.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text S1
Figs. S1 to S4
Tables S1 to S5