Abstract
Tumors exploit numerous immune checkpoints, including those deployed by myeloid cells to curtail antitumor immunity. Here, we show that the C-type lectin receptor CLEC-1 expressed by myeloid cells senses dead cells killed by programmed necrosis. Moreover, we identified Tripartite Motif Containing 21 (TRIM21) as an endogenous ligand overexpressed in various cancers. We observed that the combination of CLEC-1 blockade with chemotherapy prolonged mouse survival in tumor models. Loss of CLEC-1 reduced the accumulation of immunosuppressive myeloid cells in tumors and invigorated the activation state of dendritic cells (DCs), thereby increasing T cell responses. Mechanistically, we found that the absence of CLEC-1 increased the cross-presentation of dead cell–associated antigens by conventional type-1 DCs. We identified antihuman CLEC-1 antagonist antibodies able to enhance antitumor immunity in CLEC-1 humanized mice. Together, our results demonstrate that CLEC-1 acts as an immune checkpoint in myeloid cells and support CLEC-1 as a novel target for cancer immunotherapy.
CLEC-1 acts as an immune checkpoint in myeloid cells and represents a target for cancer immunotherapy.
INTRODUCTION
Tumors evolve to escape antitumor immunity by promoting an immunosuppressive microenvironment that exploits numerous inhibitory checkpoints. These immune checkpoints are crucial to maintain self-tolerance and modulate the duration and magnitude of effector immune responses in peripheral tissues to minimize collateral tissue damage. Their critical roles in obstructing antitumor immunity have been illustrated by the remarkable success of the monoclonal antibodies (mAbs) against cytotoxic T lymphocyte–associated protein 4 or programmed cell death protein 1/programmed death-ligand 1 (PD-1/PD-L1) that were proven to be clinically effective in a variety of cancers (1). Most immune checkpoint blockade therapies currently used in the clinic work through targeting T cells to unleash the antitumor potential of CD8+ T cells. However, current strategies have limited success in some patients with cancer, indicating that additional inhibitory pathways are required to complement existing therapeutics.
As the major tumor-infiltrating immune cell population, myeloid cells, which encompass a wide range of cells [monocytes, dendritic cells (DCs), macrophages, and neutrophils], are commonly educated by tumors to favor the establishment of an immunosuppressive tumor microenvironment (TME), limiting effective tumor immunity (2). Tumor-driven distortion of myelopoiesis results in severely altered phenotypes with the skewing of myeloid progenitor differentiation into immunosuppressive cells such as tumor-associated macrophages and so-called myeloid-derived suppressive cells (MDSCs) rather than into mature DCs (2). In addition, compelling evidence indicates that DCs, especially conventional type-1 DCs (cDC1s) that normally phagocytose dead cells and cross-present tumor antigens to CD8+ T cells, are largely defective in their functional activity and contribute to immune suppression in cancer (3). Such myeloid deviation and accumulation of immunosuppressive myeloid cells are strongly associated with tumor progression and unfavorable prognosis across multiple cancer types (2). Therefore, there is an ongoing effort to find immunotherapeutic targets to switch the function of myeloid cells toward an immunostimulatory state, by either overcoming their suppression or enhancing their effector functions of phagocytosis and antigen presentation.
Recent literature demonstrated that pattern recognition receptors represent attractive immune targets to modulate the properties of myeloid cells and enhance cancer immunotherapy (4). Among them, C-type lectin receptors (CLRs) were reported to be hijacked by tumors to suppress myeloid cell activation and promote tumor immune evasion (5). CLRs were primarily described as sensors of pathogen-associated molecular patterns that couple innate and adaptive immunity for host defense. However, these receptors have also recently emerged as orchestrators of sterile inflammation by scanning danger signals such as carbohydrates, proteins, or lipids that are exposed only by stressed, infected, malignant, or dead cells (6, 7). CLRs regulate numerous properties of myeloid cells such as antigen uptake and degradation, maturation, and secretion of cytokines and chemokines, to finely polarize subsequent T cell responses (6, 7).
Among CLRs, we previously characterized CLEC-1 and showed its expression by myeloid cells such as DCs, macrophages, monocytes, and neutrophils and by endothelial cells in both rodents and humans (8, 9). Moreover, we demonstrated that CLEC-1 is enhanced in DCs by the immunosuppressive cytokine transforming growth factor–β (TGFβ) and suppresses immune response during sterile inflammation (8, 9). CLEC-1 encoded by the Clec1a gene belongs to the “Dectin” cluster of genes encoding Mincle and Dectin-1 and, although identified long ago, is the least characterized of this cluster (10, 11). CLEC-1 presents an undefined signaling sequence in its intracellular tail, and functional work has been limited because of the lack of known ligands. However, CLEC-1 might be an important protein judging from its high evolutionary conservation between species (70% homology human/mouse) (12).
The findings presented here reveal a previously unrecognized function for CLEC-1 in myeloid cells as a specific sensor of nonhomeostatic cell death and identified the protein TRIM21 as an endogenous ligand for CLEC-1. We showed that the absence of CLEC-1 suppresses tumor growth and hinders immunosuppressive TME. In addition, we uncovered an important role for CLEC-1 in the negative regulation of antigen cross-presentation by cDC1s to CD8+ T cells. Last, we provide evidence to support the preclinical evaluation of antihuman CLEC-1 antagonist mAbs to enhance antitumor immunity.
RESULTS
CLEC-1 is a sensor of necrotic cells
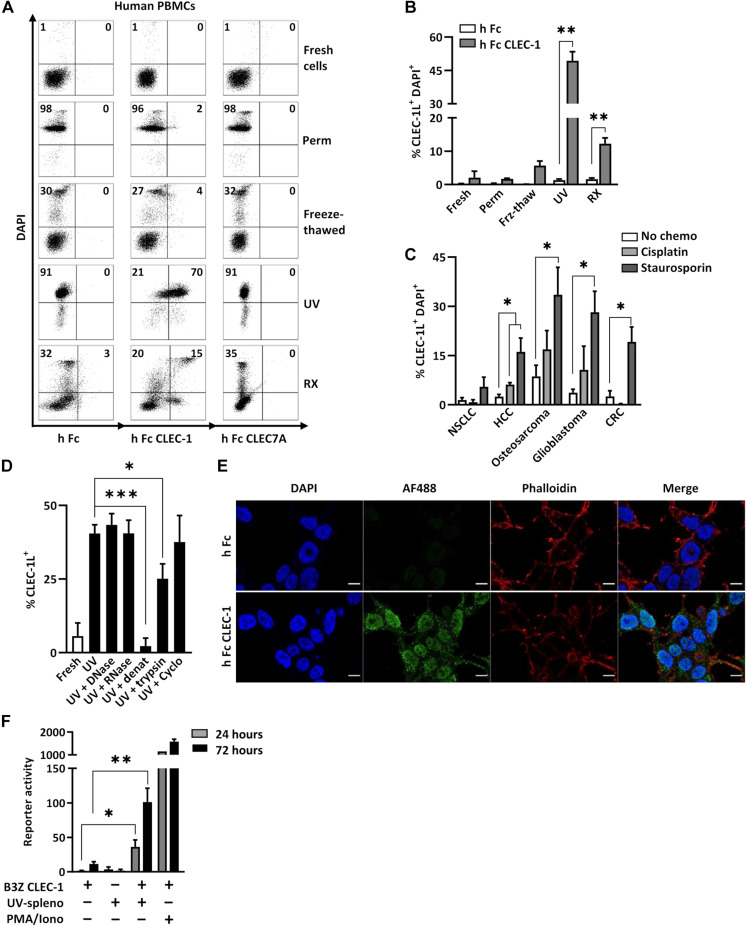
To investigate whether CLEC-1 acts as a receptor sensing cell death similar to other CLRs (7), we generated Fc CLEC-1 fusion proteins and tested their binding to fresh, stressed, or dead cells killed by accidental or programmed necrosis. We observed that human Fc CLEC-1 was able to bind necrotic peripheral blood mononuclear cells (PBMCs) previously treated by ultraviolet (UV) or x-ray radiation (18 hours earlier) that had lost their membrane integrity (Fig. 1, A and B). As controls, the Fc portion alone and the irrelevant CLR Fc CLEC7A, did not bind to dead PBMCs. Human Fc CLEC-1 neither bound fresh permeabilized cells nor dead cells killed by accidental necrosis (freeze-thaw cycles). Similar results were obtained with mouse Fc CLEC-1 on necrotic mouse splenocytes (fig. S1A). Moreover, we noticed that human and mouse Fc CLEC-1 did not bind to the cell surface of early apoptotic or early necrotic cells (6 hours after UV radiation), suggesting that the ligands appeared at a late stage of programmed necrosis (fig. S1B). We also found that the ligands were not restricted to a specific cell type, as human and mouse Fc CLEC-1 recognized secondary necrotic T, B, natural killer, and myeloid cells (fig. S2A). Furthermore, ligands appear conserved between species as human Fc CLEC-1 was able to bind dead murine splenocytes and vice versa (fig. S2B). We found that treatment with chemotherapies such as cisplatin or staurosporine induced strong expression of the ligands in various human tumor cell lines (Fig. 1C and fig. S2C). We observed that binding of human Fc CLEC-1 decreased when dead cells were heat-denaturated or treated with trypsin to digest protein (Fig. 1D). However, binding was still present when cells were treated with cycloheximide, an inhibitor of protein synthesis or with deoxyribonuclease (DNase) or ribonuclease (RNase) to degrade nucleic acids. These results suggest that CLEC-1 ligand(s) correspond(s) to protein(s) that are not de novo synthesized but require a particular conformation to be able to bind CLEC-1. Similar results were obtained with mouse Fc CLEC-1 on mouse splenocytes (fig. S2D). Furthermore, we observed by confocal microscopy that CLEC-1 ligands localized in the cytoplasm and the nucleus of dying cells (Fig. 1E). Last, we showed that the binding of necrotic cells to CLEC-1 was able to trigger a signal in a mouse CLEC-1 reporter cell line expressing the extracellular and transmembrane domains of murine CLEC-1 (Fig. 1F). Overall, these experiments demonstrate that CLEC-1 recognizes endogenous ligands that are neither species- nor cell type–restricted and that are exposed following programmed necrosis.
Fig. 1. CLEC-1 recognizes intracellular ligands exposed upon programmed necrosis.
(A and B) Flow cytometry analysis of human PBMCs either fresh, permeabilized, freeze-thawed, or previously treated (18 hours earlier) by UV or x-ray radiation (RX) and stained with 4′,6-diamidino-2-phenylindole (DAPI) and indicated Fc-AF488 fusion proteins or Fc-AF488 alone as control. Representative dot plots (A) and histograms (B) indicate the percentage of CLEC-1 ligand–positive cells in DAPI+ cells (n = 5 to 10, means ± SEM, paired t test, **P < 0.01). (C) Flow cytometry analysis of human non–small cell lung carcinoma (NSCLC), hepatocarcinoma (HCC), osteosarcoma, glioblastoma, and colon carcinoma (CRC) cell lines treated 18 hours with the cytotoxic chemotherapies cisplatin (20 μM) and staurosporin (1 μM) and stained with DAPI and Fc CLEC-1–AF488 fusion protein or Fc-AF488 alone as control. Histograms indicate the percentage of CLEC-1 ligand–positive cells in DAPI+ cells (Fc Ctrl subtracted) (n = 5, means ± SEM, paired t test, *P < 0.05). (D) Flow cytometry analysis of fresh or UV-treated human PBMCs treated with DNase A (10 μg/ml), RNase I (50 U/ml), heat-denaturated (20 min at 65°C) or treated with trypsin (100 μg/ml) (30 min at 37°C) or cycloheximide (100 μg/ml), and then stained with viability dye and Fc CLEC-1–AF488 fusion protein or Fc-AF488 alone as control. Histograms indicate mean fluorescence intensity (MFI) staining of CLEC-1 ligand–positive cells in dead cells (Fc Ctrl subtracted) (n = 5 to 13, means ± SEM, paired t test, *P < 0.05 and ***P < 0.001). (E) Confocal microscopy analysis and representative images of UV-treated human embryonic kidney (HEK) 293T cells, fixed, permeabilized, and stained with DAPI, phalloidin-TexasRed, and human Fc CLEC-1–AF488 fusion protein or Fc-AF488 alone as control. Scale bars, 10 μm. (F) Interleukin-2 (IL-2) production by CLEC-1 reporter mouse cell line following incubation or not with UV-treated splenocytes (ratio, 1:10) for 24 (gray bar) or 72 hours (black bar) and evaluated by enzyme-linked immunosorbent assay (ELISA). Phorbol 12-myristate 13-acetate (PMA)/ionomycin stimulation was used as a positive control (n = 7, means ± SEM of three independent experiments, paired t test, *P < 0.05 and **P < 0.01).
CLEC-1 interacts with TRIM21
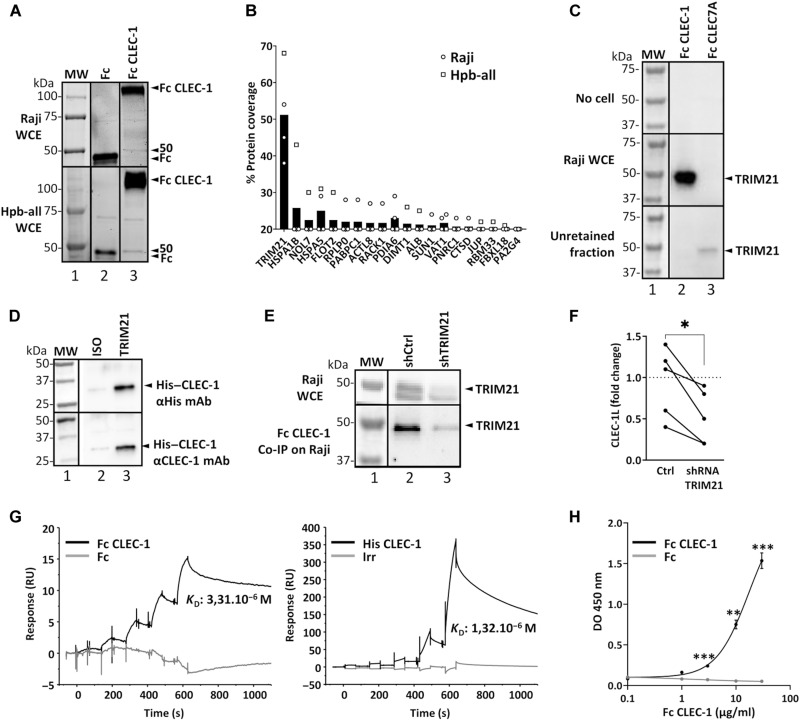
To identify CLEC-1 endogenous ligand(s) from necrotic cells, we performed several affinity purifications coupled with liquid chromatography–mass spectrometry (LC-MS). Eluted proteins from the co–immunoprecipitation (IP) of human Fc CLEC-1 with protein extracts from necrotic human cells revealed one specific band around 50 kDa (Fig. 2A). LC-MS identification performed on this band produced a short list of 20 possible candidates, with among them, TRIM21, an E3 ubiquitin ligase, harboring the highest mean coverage (50%), and whose molecular weight of 52 kDa corresponds to the main band observed (Fig. 2B). We further confirmed the specificity of the interaction of TRIM21 with Fc–CLEC-1 by Western blot, and as an irrelevant control, human Fc CLEC7A did not capture any TRIM21 from the extract (Fig. 2C). As TRIM21 is also a cytosolic Fc receptor (13), we excluded the possibility that the interaction of the Fc CLEC-1 to TRIM21 was due to binding of the Fc fragment by first coimmunoprecipitating human necrotic cells with anti-TRIM21 mAbs and then incubating the retained fraction with human recombinant His-tagged CLEC-1 instead of the Fc CLEC-1 fusion protein. We then revealed the binding of His–CLEC-1 to TRIM21 by Western blot with anti-His or anti–CLEC-1 antibodies, and co-IP with isotype mAbs was used as control (Fig. 2D). Interaction of TRIM21 with CLEC-1 was further confirmed by knocking down TRIM21 via short hairpin–mediated RNA (shRNA) in necrotic cells, leading to a subsequent lower enrichment of TRIM21 following the co-IP of Fc CLEC-1 and to a decrease in the Fc CLEC-1 binding by flow cytometry (Fig. 2, E and F). In addition, we validated the direct interaction of human recombinant TRIM21 with human CLEC-1 by Biacore and enzyme-linked immunosorbent assay (ELISA; Fig. 2, G and H). We observed strong expression of TRIM21 in various human tumor cell lines treated with chemotherapies such as cisplatin or staurosporine, which was consistent with the high level of Fc CLEC-1 binding to these cells (fig. S2E). Collectively, these data demonstrated that TRIM21 expressed by necrotic cells corresponds to a ligand for CLEC-1.
Fig. 2. Identification of TRIM21 as a ligand of CLEC-1.
(A) Representative affinity chromatography elution products (SDS–polyacrylamide gel electrophoresis) after co-IP of UV-treated Raji or Hpb-all whole-cell extract (WCE) with protein G beads coupled to human Fc CLEC-1 or Fc alone (control) (three independent experiments). Arrowheads at 50 kDa indicate a band only present under the Fc CLEC-1 condition. MW, molecular weight. (B) LC-MS analysis (of the 50-kDa gel band) depicting the top hits of Fc CLEC-1 over the Fc alone control condition (n = 4, mean). (C) Representative Western blot of human TRIM21 after co-IP of UV-treated Raji WCE (or without cells as control) with protein G beads coupled to human Fc CLEC-1 or irrelevant Fc CLEC7A (three independent experiments). Arrowheads correspond to TRIM21 detected in the co-IP in the retained and unretained fractions. (D) Representative Western blot of human CLEC-1 in elution product after co-IP of UV-treated Raji WCE and His–CLEC-1 with protein G beads coupled to antihuman TRIM21 mAb and revealed with either anti-His or anti–CLEC-1 mAb (three independent experiments). (E) Representative Western blot of human TRIM21 after co-IP of UV-treated Raji transduced with lentiviral TRIM21 (sh+) or control scrambled shRNA with protein G beads coupled to human Fc CLEC-1 (three independent experiments). (F) Flow cytometry analysis of human Fc CLEC-1 binding on UV-treated Raji transduced with lentiviral TRIM21 or control scrambled shRNA (Ctrl) (data expressed as fold change of binding of Fc CLEC-1 compared to binding of Fc alone) (n = 5, paired t test, *P < 0.05). (G) Sensograms generated by Biacore-based affinity measurement representing interaction of chip-immobilized recombinant human TRIM21 with five increasing concentrations of human Fc CLEC-1 (Fc alone as control) or His–CLEC-1 [irrelevant (irr) protein as control]. (H) Representative ELISA [density of optical (DO)] of recombinant human TRIM21 on coated human Fc CLEC-1 or Fc alone (n = 4, means ± SEM, unpaired t test, **P < 0.01 and ***P < 0.001).
CLEC-1 limits antitumor immunity in immunocompetent mice
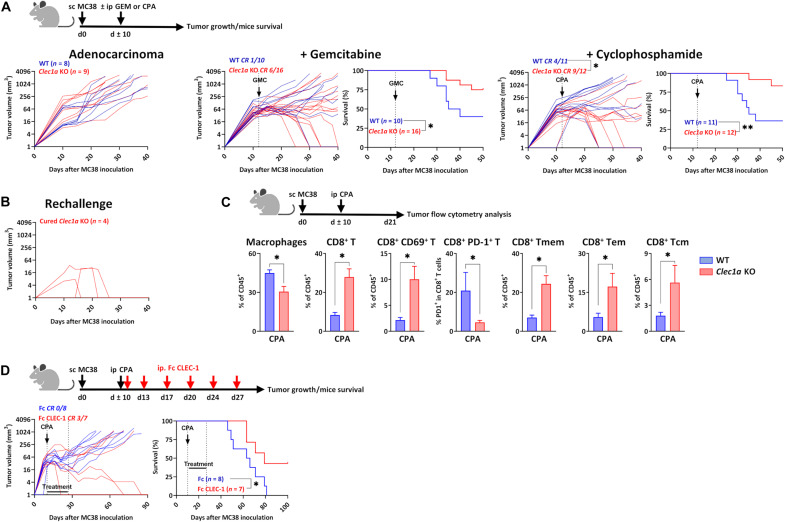
We then raised the hypothesis that CLEC-1 and the release of its ligand(s), due to the abundant necrosis occurring within tumors that can be induced by chemotherapy, could be exploited by cancer cells to suppress the immune response and promote immune evasion. Therefore, we evaluated the role of CLEC-1 in different syngeneic tumor models in mice using Clec1a knockout (KO) mice. These mice exhibited a normal immune cell composition and no obvious developmental abnormalities or signs of pathology at steady state (table S1). We observed in the subcutaneous MC38 colon adenocarcinoma model that the absence of CLEC-1 in combination with chemotherapies such as gemcitabine (GEM) or cyclophosphamide (CPA), reduced tumor growth, and induced complete tumor elimination in 37 and 75% of Clec1a KO mice, respectively [versus 10 and 36% in wild-type (WT) mice], thereby significantly prolonging survival (Fig. 3A). CPA-treated cured Clec1a KO mice that had eradicated their tumors, when re-engrafted with MC38 cells in the opposite flank, spontaneously rejected the new tumors, suggesting that these mice acquired durable antitumor immunity preventing tumor recurrence (Fig. 3B). To understand the cellular mechanisms involved in tumor growth reduction, we examined by flow cytometry the TME immune cell composition of CPA-treated Clec1a KO mice. We noticed a significant decrease in the proportion of macrophages in the TME of Clec1a KO mice compared to WT mice (Fig. 3C). In contrast, we observed more CD8+ T cells exhibiting an effector or memory phenotype and expressing less of the exhaustion marker PD-1. The blockade of CLEC-1 signaling with mouse Fc CLEC-1 fusion protein recapitulated the prolongation of survival observed in Clec1a KO mice in combination with CPA and induced a complete response in 42% of mice (versus 0% in mice treated with control Fc) (Fig. 3D).
Fig. 3. CLEC-1 blockade combines with chemotherapy in mouse model of MC38 adenocarcinoma.
(A) Tumor growth and/or survival curves of WT and Clec1a KO mice subcutaneously (sc) injected with adenocarcinoma MC38 cells and treated or not with GEM (50 mg/kg) or CPA monohydrate (CPA) (150 mg/kg) when tumor diameter reached 65 mm3 (at 7 to 12 days) (n = 8 to 16 of three independent experiments, unpaired t test, *P < 0.05 for tumor growth, and log-rank test, *P < 0.05 and **P < 0.01 for survival). ip, intraperitoneal. (B) Tumor growth of MC38 cell–inoculated cured Clec1a KO mice subcutaneously rechallenged with MC38 cells (90 days after first inoculation) (n = 4). (C) Percentage of macrophages (CD11b+F4/80+Ly6C+), CD8+ T (CD3+CD8+), CD8+ CD69+ T, CD8+ PD-1+ T, CD8+ T memory (Tmem) (CD44+), T effector memory (Tem) (CD44+CD62L−), and T central memory (Tcm) (CD44+CD62L+) evaluated by flow cytometry in the MC38 tumors from WT and Clec1a KO mice at day 20 after tumor inoculation [data are expressed in the percentage of CD45+ (n = 7 to 9, means ± SEM of two independent experiments, unpaired t test, *P < 0.05)]. (D) Tumor growth and survival curves of naïve mice subcutaneously injected with adenocarcinoma MC38 cells and treated with either mouse Fc or mouse Fc CLEC-1 fusion proteins (5 mg/kg; twice a week for 3 weeks) in combination with CPA (150 mg/kg) (n = 7 to 8, log-rank test, *P < 0.05 for survival).
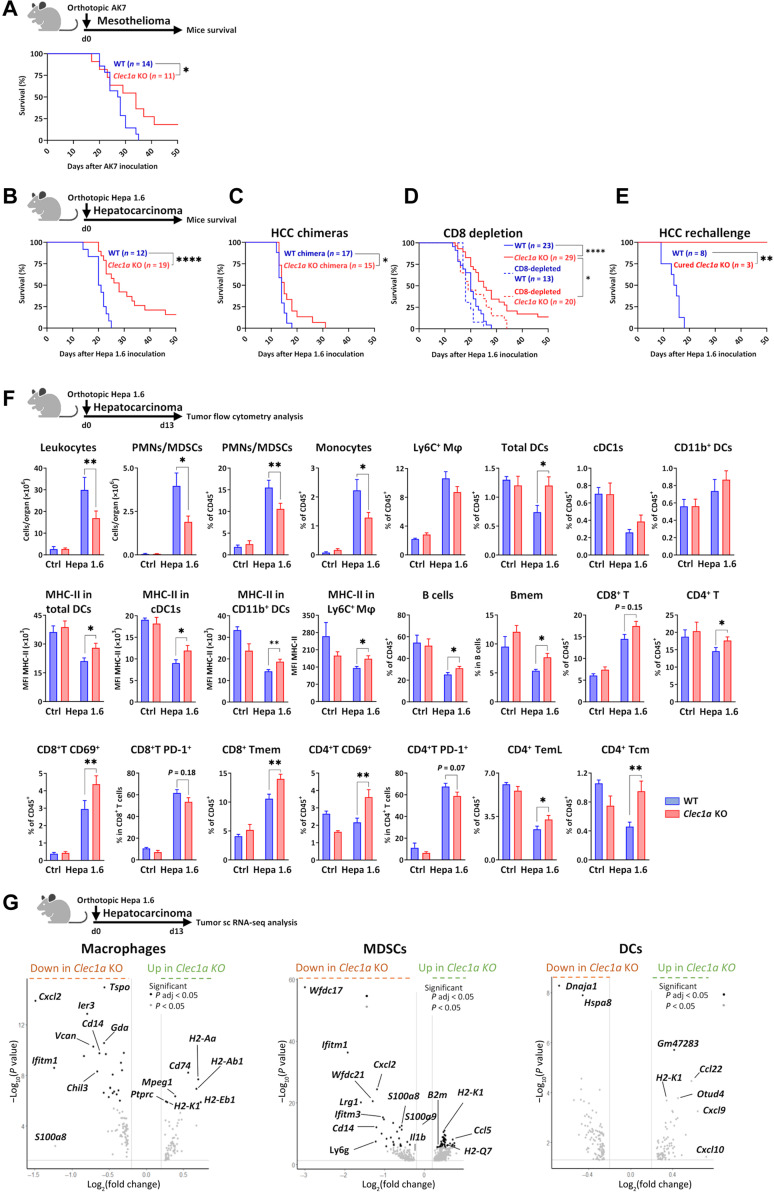
Furthermore, in the orthotopic mesothelioma (AK7) and hepatocarcinoma (HCC) (Hepa 1.6) models, we observed a significant prolongation of survival of Clec1a KO compared to WT mice with 18 and 16% of the mice respectively exhibiting long-term survival (Fig. 4, A and B). As CLEC-1 is expressed not only by myeloid cells but also by endothelial cells (8, 9), we confirmed in the HCC model that the effect on mice survival was due to the absence of CLEC-1 in myeloid cells by generating bone marrow chimeras with the absence of CLEC-1 only in the hematopoietic compartment. We observed prolongation of survival in bone marrow chimeric mice, supporting the role of CLEC-1 in myeloid cells (Fig. 4C). Furthermore, to determine whether the prolongation of survival in this model was CD8+ T cell–mediated, we depleted Clec1a-deficient mice for CD8. We observed that prolongation of survival in Clec1a KO mice was partially abrogated by CD8 depletion, suggesting that CLEC-1 plays an indirect role by enhancing antitumor CD8+ T cell response but that additional mechanisms are also involved (Fig. 4D). Cured Clec1a KO mice resisted a new challenge by splenic Hepa 1.6 inoculation, demonstrating the development of a memory immune response (Fig. 4E). Flow cytometry revealed that tumor-burdened livers from Clec1a KO mice exhibited a substantial reduction in their infiltration by polymorphonuclear neutrophils (PMNs) presumably MDSCs and by monocytes compared to those from WT mice (Fig. 4F). In contrast, we noticed a higher proportion of DCs, regardless of their cDC1 or CD11b+ DC subtypes. These both DC subtypes and the infiltrating macrophages overexpressed major histocompatibility complex class II (MHC-II), suggesting their higher activation state in the absence of CLEC-1. In addition, we found a significant increase in effector and/or memory CD8+, CD4+ T and B cells in tumor-burdened livers of Clec1a KO compared to WT mice. Increased antitumor immunity was not specific to the tumor stroma as we also found fewer PMNs/MDSCs and monocytes, more activated DCs, and more effector or memory CD8+ and CD4+ T cells exhibiting less of the exhaustion marker PD-1 in the spleen (fig. S3A). Overall, these data suggested that the absence of CLEC-1 prevented the tumor-driven distortion of myelopoiesis and led to a profound shift in myeloid cell composition toward fewer immunosuppressive myeloid cells such as MDSCs and macrophages and more mature DCs, thereby promoting infiltration by effector and memory T and B cells.
Fig. 4. Absence of Clec1a enhances antitumor immunity in mouse orthotopic models.
(A) Survival curve of WT and Clec1a KO mice with pleural mesothelioma (AK7) (n = 11 to 14, log-rank test, *P < 0.05). (B) Survival curve of WT and Clec1a KO mice with Hepa 1.6 orthotopic HCC (n = 12 to 19 of three independent experiments, log-rank test, ****P < 0.0001). (C) Survival curve of chimeric mice with the absence of CLEC-1 only in the hematopoietic compartment (Clec1a KO chimeras) or WT chimeras with HCC (n = 15 to 17 of two independent experiments, log-rank test, *P < 0.05). (D) Survival curve of WT, Clec1a KO, CD8-depleted WT, and CD8-depleted Clec1a KO mice injected with HCC (n = 13 to 29 of two independent experiments, log-rank test, *P < 0.05 and ****P < 0.0001). (E) Survival curve of HCC-cured Clec1a KO rechallenged in the spleen with HCC at day 90 after first inoculation (WT naïve mice were used as controls) (n = 3 to 8, log-rank test, **P < 0.01). (F) Total number and/or percentage of leukocytes, PMNs/MDSCs (CD11b+Ly6G+MHC-II−), monocytes (CD11b+Ly6C+MHC-II−), macrophages (Mφ) (CD11b+F4/80+Ly6C+), total DCs (CD11c+MHC-II+), cDC1 (CD11c+MHC-II+CD11b−XCR1+), DCs CD11b+ (CD11c+MHC-II+CD11b+), B memory (Bmem) (CD19+CD45R+CD24+CD38−), CD8+ or CD4+ T (CD3+CD4/8+), CD69+, PD-1+, T memory (Tmem) (CD44+), T effector-memory Late (TemL) (CD44+CD62L−CD27−), and T central memory (Tcm) (CD44+CD62L+) CD8+ or CD4+ T cells evaluated by flow cytometry in the tumor-burdened livers of WT and Clec1a KO mice at day 13 after HCC [data are expressed in cell count, in the percentage of CD45+, in the percentage in particular cell subset or in MFI of MHC-II expression] (n = 5 for controls and n = 15 for Hepa 1.6–treated mice, means ± SEM of three independent experiments, unpaired t test, *P < 0.05 and **P < 0.01). (G) Volcano plot representations of differential gene expression analysis by single-cell RNA sequencing (scRNA-seq) of macrophage, MDSC, and DC clusters from WT and Clec1a KO mice at day 13 after HCC. Data represent the −log10 (P values) versus the log2 (fold change), and genes are colored on the basis of their significance.
CLEC-1 promotes an immunosuppressive TME
To understand further the molecular mechanisms by which CLEC-1 deficiency enhances the antitumor response, we investigated in detail the transcriptional change of distinct immune cell populations infiltrating HCC tumors by single-cell RNA sequencing (scRNA-seq). Unsupervised clustering analysis and subsequent cell type annotation identified 11 clusters: seven lymphoid (γδ T, CD4 T, CD8 T, natural killer T, regulatory T, natural killer, and B cells) and four myeloid (cDCs, macrophages, MDSCs, and neutrophils) (fig. S4A). Resembling the previous flow cytometry results, we observed that the most substantial difference in the cell proportion was the reduction in macrophages and MDSCs in tumor-burden livers of Clec1a KO compared to WT mice (fig. S4B). Evaluation of the distinct transcriptomic profiles further revealed a profound remodeling of gene expression, essentially in the myeloid cell compartment. Notably, we noticed that genes involved in immunosuppressive myeloid cell expansion, recruitment (S100a8/a9, Cxcl2, and Il1b) and function (Wfdc17, Cd14, Ifitm1, Ier3, and Tspo) (14, 15), were significantly down-regulated in infiltrating macrophages or MDSCs from tumor-burden livers of Clec1a KO compared to WT mice (Fig. 4G and data file S1). In contrast, genes involved in immunogenic antigen presentation such as MHC-I– or MHC-II–related genes (H2-Ab1, H2-Aa, H2-Eb1, H2-k1, H2-q6, B2m, and Cd74) or in DC1 and T cell tumor infiltration (Ccl5) (16) were up-regulated (Fig. 5D). “Tumor necrosis factor–α signaling via nuclear factor κB (NF-κB)” and “interleukin-6 (IL-6)/Janus kinase/signal transducers and activators of transcription 3” pathways, both known to promote MDSC expansion and function (17), were down-regulated in tumoral macrophages and MDSCs in the absence of CLEC-1, whereas pathways associated with type I or II interferon (IFN) responses and allograft rejection were up-regulated (fig. S4, C and D). Gene ontology analysis identified the biological processes of “lymphocyte differentiation” and “T cell activation” enriched in macrophages and “regulation of immune effector process,” “adaptive immune response,” and “antigen processing and presentation” enriched in MDSCs (fig. S4, E and F). In the cluster of cDCs, we detected an increased expression of Cxcl9, Cxcl10, and Ccl22 that correspond to T cell chemoattractants (16), in the tumor-burden livers of Clec1a KO compared to WT mice (Fig. 5D). We also noticed up-regulation of Otud4 coding for an enzyme sustaining the production of type I IFN and proinflammatory cytokines in DCs (18). In contrast, Hspa8 and its cochaperone Dnaja1, which regulate the process of antigen trafficking and presentation in cDCs by orienting proteins toward the proteasome or lysosomes for degradation (19), were down-regulated. Collectively, our results suggest that the lack of CLEC-1 hinders immunosuppressive TME by limiting the recruitment and function of suppressive myeloid cells such as MDSCs and macrophages while invigorating the activation state of DCs to express proinflammatory cytokines and chemokines.
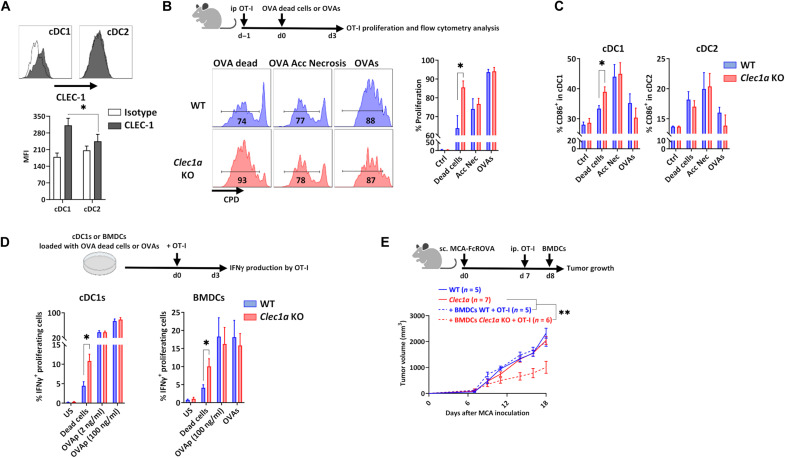
Fig. 5. Loss of CLEC-1 signaling enhances cross-presentation of dead cell–associated antigens by DCs.
(A) Representative overlay and histogram of CLEC-1 expression on human blood cDC1 (CD45+LIN−CD11c+HLA-DRhighBDCA3+) and cDC2 (CD45+LIN−CD11c+HLA-DRhighCD1c+) evaluated by flow cytometry (MFI staining) (n = 12, means ± SEM of three independent experiments, unpaired t test, *P < 0.05). (B) Representative profile and histograms of the in vivo proliferation evaluated by flow cytometry [Cell Proliferation Dye (CPD) dilution] of splenic OVA-specific CD8+ T cells (OT-I) injected in WT and Clec1a KO mice with UV-treated (OVA dead cells) or freeze-thaw cycles treated (accidental necrosis) MCA cells expressing mOVA or following injection with soluble OVA protein (OVAs). Control represents naïve mice without OVA injection (n = 5 to 8, means ± SEM of three independent experiments, unpaired t test, *P < 0.05). (C) Percentage of MHC-II+CD86+ cells in splenic cDC1 (CD45+CD11c+MHC-II+CD11b−XCR1+) and cDC2 (CD45+CD11c+MHC-II+CD11b+XCR1−) evaluated by flow cytometry following injection of WT and Clec1a KO mice with OT-I cells and UV-treated (OVA dead cells) or freeze-thaw cycle–treated (accidental necrosis) MCA cells expressing mOVA (FcROVA) or following injection with soluble OVA protein (OVAs) (n = 5 to 7, means ± SEM of two independent experiments, unpaired t test, *P < 0.05). (D) Percentage of IFNγ+ proliferating OVA-specific CD8+ T (OT-I) cells in response to WT and Clec1a KO cDC1s or BMDCs loaded with UV-treated MCA cells expressing mOVA (dead cells), OVA peptide SIINFEKL (2 and 100 ng/ml; OVAp), or soluble OVA (OVAs) evaluated by flow cytometry (n = 6 to 9, means ± SEM of three independent experiments, unpaired t test, *P < 0.05). (E) Tumor growth of WT and Clec1a KO mice subcutaneously injected with MCA tumor cells expressing mOVA and injected or not with WT or Clec1a KO BMDCs loaded with dead cell–associated OVA antigens (UV-treated MCA FcROVA) and with preactivated OT-I cells (n = 5 to 7, means ± SEM, unpaired t test, **P < 0.01).
CLEC-1 reduces cDC1 cross-presentation of dead cell–associated antigens to CD8+ T cells
We previously described that CLEC-1 is expressed by different types of myeloid cells such as monocytes, macrophages, neutrophils, and DCs (8, 9). We noticed in mouse and human public datasets (ImmGen MNP Open-source) that Clec1a is expressed under steady-state conditions by several subtypes of DCs and that among DCs, the highest expression is by the subset of conventional cDC1 (XCR1+ and CD141/BDCA-3+ in mouse and human, respectively), which are especially efficient at processing material from dead cells (fig. S5A) (20, 21). We confirmed higher cell-surface expression of CLEC-1 at the protein level on cDC1s compared to cDC2s from the blood of healthy individuals (Fig. 5A).
We therefore evaluated whether CLEC-1 in DCs might play a role in the cross-presentation of dead cell–associated antigens to CD8+ T cells. For that, we coinjected WT or Clec1a KO mice with necrotic methylcholanthrene-induced fibrosarcoma (MCA) cells expressing membrane-bound ovalbumin (mOVA), together with OVA-specific OT-I T cell receptor (TCR)–transgenic CD8+ T cells. In this in vivo model, CD8+ T cell activation has been shown to rely primarily on basic leucine zipper ATF-like transcription factor 3 (Batf3)–dependent cDC1s (20). We observed an increased proliferation of the OT-I CD8+ T cells injected in Clec1a KO mice compared to those injected in WT mice (Fig. 5B). Moreover, the cDC1s from the Clec1a KO mice further up-regulated the costimulation marker CD86, suggesting their increased activation state and a positive feedback loop between cDC1s and CD8+ T cells (Fig. 5C). In contrast, when we administrated OVA protein in its soluble form or mOVA from dead cells induced by primary accidental necrosis, we saw no difference in OT-I CD8+ T cell proliferation nor in cDC1 maturation between WT and Clec1a KO mice (Fig. 5, B and C). Furthermore, no difference was observed in the proliferation of OVA-specific TCR–transgenic OT-II CD4+ T cells in response to soluble OVA (fig. S5B). Moreover, as control of cross-presentation of necrotic cells by DC1, no proliferation of OT-II CD4+ T cells was retrieved in response to OVA associated with necrotic cells necrotic form of OVA. These data suggested that cDC1 from Clec1a KO mice induced better cross-presentation of dead cell–associated antigens in vivo only when dead cells were induced by programmed necrosis. To support this hypothesis, we performed similar experiments in vitro using cDC1s purified from the spleen of WT or Clec1a KO mice and evaluated the IFNγ production by cocultured OT-I CD8+ T cells. We observed that cDC1s from Clec1a KO loaded with necrotic dead cell–associated mOVA antigen induced in vitro more IFNγ production by the cocultured OT-I CD8+ T cells compared to cDC1s from WT mice (Fig. 5D). In contrast, IFNγ production by OT-I cells, in response to cDC1 loaded with the synthetic MHC-I–restricted OVA peptide (SIINFEKL), was similar between both genotypes. We obtained similar results with bone marrow–derived DCs (BMDCs) (Fig. 5D) that are known to be able to cross-present antigens and, as in rat (8, 9), down-regulate Clec1a transcripts by proinflammatory stimuli, and up-regulate them by TGFβ (fig. S5C). Moreover, proliferation of OT-I or OT-II cells in response to BMDCs loaded with soluble OVA or specific OVA peptides was similar between WT and Clec1a KO mice (Fig. 5D and fig. S5D). Moreover, to evaluate the cytotoxic potential of the OT-I cells stimulated by Clec1a-deficient BMDCs, we coinjected in vivo preactivated OT-I cells with Clec1a KO BMDCs loaded with necrotic dead cell–associated mOVA antigen in mice previously injected with MCA tumors expressing mOVA. We observed that coinjection with the loaded Clec1a KO BMDCs delayed the growth of MCA tumors expressing mOVA, whereas coinjection of the loaded WT BMDCs had no effect. These data suggested that DCs deficient in Clec1a induced a better cytotoxic potential of OT-I cells, thereby leading to more effective eradication of the tumor (Fig. 5E).
We then investigated whether this better activation of CD8+ T cells observed in the absence of CLEC-1 in DCs was due to an enhanced uptake of dead cells, to a higher activation state driven by necrotic cells, or to an increased processing of dead cell–associated antigens after their uptake. We found no difference in vitro in the engulfment of dead cells over time by Clec1a KO BMDCs compared to WT BMDCs (fig. S5E). Moreover, no difference was observed in the expression of MHC-I or MHC-II or in the expression of costimulatory molecules (CD86 andCD40) in response to stimulation by Toll-like receptor 3 ligand (TLR3-L), by necrotic cells, or by both together (fig. S5F). However, we detected increased expression of some proinflammatory genes in Clec1a KO BMDCs such as Ccl2 and Isg15 in response to TLR3-L stimulation (fig. S5G). These data suggest that CLEC-1 in DCs may act as a negative regulator of the expression of proinflammatory cytokines and chemokines in response to acute inflammation, thereby tempering subsequent CD8+ T cell activation.
Last, as cross-presentation is associated with not only cross-priming to foreign antigen but also with cross-tolerance to self, we explored whether the deficiency of CLEC-1 in cDC1 could break self-tolerance to homeostatic cell death and trigger autoimmunity. Using the RIP mOVA mouse system (22), we observed that the lack of CLEC-1 in cDC1s did not prevent the cross-tolerization of the injected OT-I T cells nor aggravate incidence of diabetes induced by the coadministration with anti-OVA immunoglobulin Gs (fig. S6).
Together, our results demonstrate that the absence of CLEC-1 enhances cross-presentation of dead cell–associated antigens to CD8+ T cells by DC1s during nonhomeostatic cell death but does not break DC1-mediated self-tolerance.
Treatment with antihuman CLEC-1 antagonist mAbs enhances antitumor immunity in preclinical models
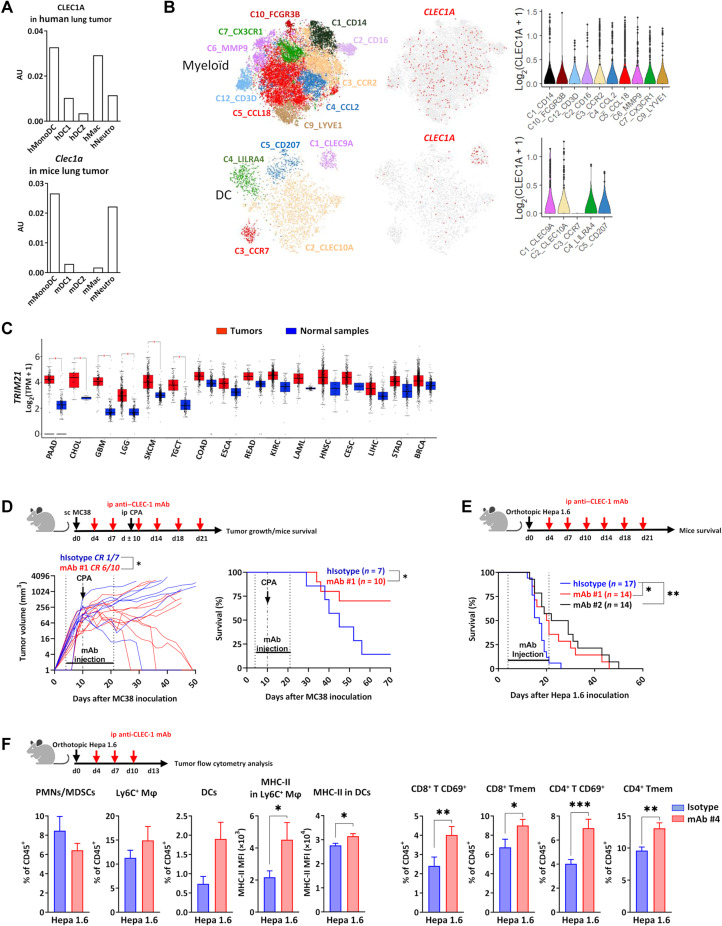
To evaluate the clinical relevance of CLEC-1 in patients with cancer, we analyzed scRNA-seq datasets of myeloid cells from human lung TME (23). We observed the highest expression of CLEC1A by a particular subset of monocytes sharing characteristics of immature DC, by DC1, and by subsets of macrophages and neutrophils displaying protumoral features (Fig. 6A). This expression profile was similar in mice. In addition, pan-cancer analysis (breast, colorectal, lung, and ovarian) identified CLEC1A expression in various subsets of myeloid cells from human TME, such as in CD14+ (C1) and CD16+ (C2) clusters of monocytes and in different subtypes of macrophages expressing antitumoral M1 (C3 and C4) or protumoral M2 (C5 to C7) markers (Fig. 6B) (24). Among DCs, CLEC1A expression was retrieved mostly in the C1 cluster corresponding to cDC1 cells. Analysis of datasets from The Cancer Genome Atlas (TCGA) with GEPIA tool (25) indicated that a broad spectrum of human cancers such as pancreatic adenocarcinoma or glioblastoma overexpress TRIM21 compared to normal tissues (Fig. 6C). These data suggest that tumors may exploit their overexpression of TRIM21 to suppress the function of myeloid cells via CLEC-1.
Fig. 6. Treatment with antihuman CLEC-1 antagonist mAb enhances antitumor immunity in CLEC-1 humanized mice.
(A) CLEC1A or Clec1a mRNA expression in different subsets of myeloid cells from human or mouse lung tumors from Open-source data of scRNA-seq. AU, arbitrary units. (B) Two-dimensional Uniform Manifold Approximation and Projection and violin plot representations of CLEC1A expression (red scale projection) among clusters of myeloid cells and DCs from scRNA-seq dataset of pan-cancer (breast, colorectal, lung, and ovarian) TME. (C) Box plots representation of log2 [transcript count per million (TMP)+1] values for TRIM21 expression in different cancer types from TCGA analyzed by GEPIA tool. BRCA, breast-invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower-grade glioma; LIHC, liver hepatocellular carcinoma; PAAD, pancreatic adenocarcinoma; READ, rectum adenocarcinoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors. Unpaired t test, *P < 0.05. (D and E) Tumor growth or survival curve of CLEC-1 humanized mice with subcutaneous MC38 adenocarcinoma in combination with CPA (150 mg/kg) (D) or with Hepa 1.6 LIHC (E) and treated with irrelevant isotype control or antihuman CLEC-1 antagonist mAbs #1 or #2 (100 μg per mouse) twice a week from days 4 to 21 (n = 14 to 17 of two independent experiments, unpaired t test, *P < 0.05 for tumor growth, and log-rank test, *P < 0.05 and **P < 0.01 for survival). (F) Percentage of PMNs/MDSCs (CD11b+Ly6G+MHC-II−), macrophages (CD11b+F4/80+Ly6C+), DCs (CD11c+MHC-II+), CD69+or Tmem (CD44+) CD8+, or CD4+ (CD3+CD4/8+) T cells evaluated by flow cytometry at day 13 after LIHC in the tumor-burdened livers of CLEC-1 humanized mice treated with isotype control or antihuman CLEC-1 antagonist mAbs #4 (100 μg per mouse) (data are expressed in the percentage of CD45+ or in MFI of MHC-II expression) (n = 8 to 13, means ± SEM of three independent experiments, unpaired t test *P < 0.05, **P < 0.01, and ***P < 0.001).
Therefore, to evaluate the translational potential of CLEC-1 as an immunotherapeutic target, we generated several antihuman CLEC-1 mAbs able to block the interaction of human CLEC-1 with its putative ligand(s) in human necrotic cells (fig. S7, A and C). Knowing the cross-reactivity of the ligands, we tested the therapeutic potential of these antihuman CLEC-1 antagonist mAbs in the context of a fully functional immune system using a newly generated CLEC-1 humanized mouse model. These CLEC-1 knock-in (KI) mice have the extracellular domain of mouse CLEC-1 replaced by its human counterpart, while the transmembrane and intracellular domains remain intact. We confirmed that these humanized mice exhibited no difference in immune cell composition compared to regular naïve mice (table S2). Moreover, we confirmed with our mAbs, the expression of the chimeric CLEC-1 protein in myeloid cells such as macrophages and cDCs from lung or spleen, with cDC1s showing the highest expression of CLEC-1 among cDCs (fig. S7D). We observed that treatment with an antihuman CLEC-1 antagonist mAb was able to reduce tumor growth in the subcutaneous model of MC38 colon adenocarcinoma in combination with CPA, with a complete response in 60% of the mice (versus 15% in mice treated with isotype control), thereby significantly prolonging survival (Fig. 6D). Similar results were observed in the orthotopic model of HCC, with a significant enhancement of the survival of CLEC-1 humanized mice treated with antihuman CLEC-1 antagonist mAb compared to treatment with isotype control (Fig. 6E). We confirmed this prolongation of mice survival with a second antihuman CLEC-1 antagonist mAb. Flow cytometry analysis of the TME revealed the presence of more mature macrophages and DCs (MHC-II expression) and of more effector and memory CD4+ and CD8+ T cells in the tumor-burdened livers of mice treated with antihuman CLEC-1 antagonist mAbs compared to mice treated with isotype control (Fig. 6F). These results resembled what we obtained with Clec1a KO mice and validated the translational potential of antihuman CLEC-1 antagonist mAbs to enhance antitumor immunity. Collectively, these data represent a proof of principle of the therapeutic efficiency of antihuman CLEC-1 antagonist mAbs to restore properties of myeloid cells and enhance antitumor immunity.
DISCUSSION
Tumors hijack physiological mechanisms that normally restrain tissue damage to suppress myeloid cell function and promote tumor immune evasion. Given the role of myeloid cells in T cell activation, targeting checkpoint molecules expressed in this population represents a promising therapeutic strategy to enhance antitumor immunity. In this study, we revealed that the C-type lectin CLEC-1 expressed by myeloid cells acts as a sensor of death and may contribute to tumor progression by promoting an immunosuppressive TME and by hampering the cross-presentation of dead cell–associated antigens by DCs to CD8+ T cells. Moreover, we provided a rationale for the clinical development of anti–CLEC-1 mAbs to improve antitumor immunity.
Our results showed that CLEC-1 binds conserved protein structures exposed upon programmed cell death. Other CLRs such as CLEC12A and DNGR-1 have been described as sensors of death able to inhibit myeloid cell activation triggered by heterologous receptors following tissue injury (26, 27). However, in contrast to DNGR-1 that recognizes F-actin in its ubiquitous form (28), CLEC-1 does not seem to recognize endogenous molecules concealed intracellularly under normal conditions. This suggests that ubiquitous preformed ligand(s) of CLEC-1 may exist and be sequestered in healthy cells and subjected to conformational or enzymatic posttranslational modifications following stress and/or upon loss of membrane integrity. Similarly, CLEC12A was shown to bind monosodium urate crystals formed by crystallization of soluble uric acid following contact with extracellular sodium ions, once the cells are dying (26). We observed that CLEC-1 ligand(s) are strongly expressed by necrotic tumor cells induced by radiation or chemotherapy and identified TRIM21, as an endogenous ligand for CLEC-1. TRIM21 is a highly conserved IFN-inducible protein ubiquitously expressed in hematopoietic, endothelial, and epithelial cells and localized in the nucleus and cytoplasm along the microtubule network (13). TRIM21 was shown to be up-regulated and subjected to posttranslational and conformational changes following IFNβ treatment or stress such as radiation or chemotherapy. TRIM21 induces necroptosis and is associated with autoimmune diseases by acting as an autoantigen released by necrotic cells (13, 29–31). Furthermore, TRIM21 is an E3 ubiquitin ligase catalyzing the ubiquitination of proteins such as IFN regulatory factors and plays a role in the negative regulation of innate and adaptive signaling. As a result, Trim21 KO mice develop an excessive inflammatory response and systemic autoimmunity following tissue damage (13). Moreover, DCs and macrophages from Trim21 KO mice have been shown to up-regulate IFN signaling and NF-κB–mediated proinflammatory cytokines and chemokines (32, 33). We observed an up-regulation of TRIM21 expression in numerous human cancers, and TRIM21 has been associated with carcinogenesis, tumor aggressiveness, and therapy resistance in various cancers such as HCC or glioma (34, 35). Therefore, we hypothesize that TRIM21 released by necrotic cells, for example, in the context of chemotherapy may correspond to a damage-associated molecular pattern that suppresses the cellular response of myeloid cells via CLEC-1, thereby restraining antitumor immunity. However, we do not rule out the possibility that TRIM21 could act as an intracellular partner of CLEC-1 following dead cell internalization and phagosomal rupture in the cytosol. It has recently been shown that the E3 ubiquitin ligase Ring Finger Protein 41 acts as a negative regulator of cross-presentation of dead cell–derived antigens by directly binding and ubiquitinating the extracellular domains of DNGR-1 within the cDC1s (36). Moreover, CLEC-1, similar to other CLRs, could recognize several endogenous ligands (6, 7). Recently, studies reported that histidine-rich glycoprotein suppresses inflammatory signaling pathways and regulates phagocytosis of necrotic cells via CLEC-1 (37, 38).
Our data indicated that abrogation of CLEC-1 signaling by genetic ablation or antibody blockade enhanced survival and antitumor immunity in immunocompetent mice. We showed that this phenotype was due to the presence of CLEC-1 in hematopoietic cells presumably myeloid cells and was in part due to a better CD8+ T cell response. This suggests that CLEC-1, in addition to its role in DCs in the cross-presentation of dead cell–associated antigens to CD8+ T cells, may play other roles in myeloid cells and contribute to additional mechanisms leading to this phenotype. Consistent with this hypothesis, we observed that CLEC-1 is expressed by several other subsets of myeloid cells from the TME such as monocytes, neutrophils, and macrophages. Moreover, we found that remodeling of the myeloid TME is a key mechanism underlying CLEC-1 blockade resulting in fewer immature myeloid cells such as PMNs/MDSCs and macrophages and more activated DCs. CLEC-1 blockade may therefore prevent the tumor-driven distortion of myelopoiesis and avoid the skewing of myeloid progenitor cell differentiation into MDSCs rather than into mature DCs (2). We also found that DCs overexpressed the Cxcl9, Cxcl10, and Ccl22 chemokines. CXCL9 and CXCL10 are expressed mostly by cDC1s following exposure to IFNγ produced by T cells to amplify recruitment of additional CXCR3+ effector CD8+ T cells in the TME (3, 16). Moreover, we noticed in myeloid cells an increased expression of Ccl5, a chemokine promoting cDC1 and effector T cell tumor infiltration (39). Expression of CXCL9, CXCL10, CCL22, and CCL5 was shown to be associated with potent antitumoral cDC1s and improved clinical outcome and responses to chemotherapy in patients (16, 40). cDC1s, by their superior ability to cross-present antigens, play a crucial role in antitumor immunity (3). Their tumor infiltration is strongly correlated with improved overall survival of patients in different types of cancer (21). Therefore, CLEC-1 in DC1s may suppress some key steps of cross-presentation and the secretion of cytokines and chemokines that are important for DC-T cross-talk (3). Several lectins have been shown to regulate the cross-presentation of antigens by DCs. DNGR-1, which is specific to cDC1, has been shown to favor phagosome rupture and the escape of phagosomal contents into the cytosol for efficient cross-presentation (41). In contrast, SIGLEC-10, whose expression is enhanced by IL-10 and TGFβ, was shown in cDC1 to restrain cross-presentation of antigen by triggering the SHP-1 inhibitory axis (42, 43). CLEC-1 does not contain a classical immunoreceptor tyrosine–based inhibition motif in its cytoplasmic tail but rather a tyrosine residue in a noncharacterized signaling sequence [YSST], which could potentially be used for downstream signaling (44). However, CLEC-1 contains also a tri-acidic motif [DDD]. This motif, also carried by other CLRs (DEC205, DC-SIGN, BDCA2, and DECTIN-1) promotes endosomal acidification and premature antigen degradation that could prevent efficient cross-presentation (45). Therefore, further studies are warranted to elucidate whether CLEC-1 in cDC1s could induce excessive antigen degradation of cell corpses as a mechanism to restrict cross-presentation to CD8+ T cells. In particular, a conditional KO in cDC1 would help to define the role of CLEC-1 in antigen cross-presentation and to assign the increased antitumor immunity phenotype directly to these cells.
To conclude, our study identifies CLEC-1 as a novel immune suppressor of myeloid cells and provides support for a translational potential of CLEC-1 as a pharmaceutical target for cancer immunotherapy.
MATERIALS AND METHODS
Experimental design
In this study, we designed Fc CLEC-1 fusion proteins to identify ligands of CLEC-1 by flow cytometry and by co-IP followed by mass spectrometry. We studied the function of CLEC-1 in vitro and in vivo on immune responses and antitumor immunity using Clec1a KO mice. Last, we generated antihuman CLEC-1 antagonist mAbs and evaluated their therapeutic potential on antitumor immunity in fully immunocompetent CLEC-1 humanized mice. For animal studies, sample size was based on prior knowledge of the intragroup variation of tumor growth or mouse survival and according to experimental procedures carried out in strict accordance with protocols approved by the Committee on the Ethics of Animal Experiments of Pays de la Loire and authorized by the French Government’s Ministry of Higher Education and Research (Apafis no. 12578, 27773v3). Mice were bred in Centre de Distribution, Typage et Archivage animal (Orléans, France) under specific pathogen–free conditions and used at 8 to 12 weeks old (20 to 25 g), and littermates were used for experiments. Animals were anesthetized with a mixture of 5% isoflurane and oxygen at 1 liter/min and euthanized by CO2 intoxication and/or cervical dislocation after anesthesia. Blinding was done for measurement of tumor growth. For all experiments, number of samples, experiment replicates, and statistical test used are reported in the figure legends.
Animals and genotyping
C57Bl/6 Clec1a KO mice (by insertion of eGFP-CreERT2 into Clec1a to generate null alleles) were rederived from sperm (provided by D. J. Rossi, Harvard, USA) (46). CLEC-1 humanized KI C57Bl/6 mice were generated by Centre d’immuno-phénomique (Marseille, France). These mice express a chimeric CLEC-1 consisted of human extracellular and murine transmembrane and intracellular domains of CLEC-1. Briefly, a hybrid CLEC1A cDNA (human extracellular domain appended to mouse sequences coding for the transmembrane and cytosolic domains) was introduced in the first coding exon of the mouse Clec1a locus (Clec1a-201 ENSMUST00000037481.9 transcript) between bases Chr6:129451711-129451713 and Chr6:129451715-129451717. Clec1a KO and CLEC-1 KI mice were genotyped by standard polymerase chain reaction (PCR) with specific primers designed to discriminate between the mutant and WT alleles (CRE and Neo cassette, respectively) (table S3). OT-I.Ly5.1 homozygous mice were obtained by intercrossing OVA-specific TCR-transgenic OT-I mice [C57BL/6-Tg (TcraTcrb) 1100Mjb/Crl] (Charles River) with Ly5.1 mice (B6.SJL-PtprcaPepcb/BoyCrl) (Charles River) and genotyped by CD45.1 staining by flow cytometry. To generate chimeric mice, 5 × 106 bone marrow cells collected from femurs and tibiae of WT or Clec1a KO mice were intravenously injected into lethally whole-body x-irradiated (Faxitron CP 160, Faxitron X-Ray Corp., Wheeling) [day −1, 11 grays (Gy)] recipient C57Bl/6 naïve mice. Reconstituted mice were subcutaneously injected with terramycin (200 mg/kg; Sigma-Aldrich), and drinking water was supplemented with neomycin trisulfate (2 mg/ml; Coophavet) for 2 weeks. Chimeras were used for tumor inoculation 8 weeks after reconstitution.
Cell lines and stimulation
Human non–small cell lung carcinoma (NSCLC), HepG2, U2OS, U373, DLD-1, human embryonic kidney (HEK) 293T, Hpb-all, Raji, and C57Bl/6 mouse AK7, Hepa 1.6, MC38 (purchased from American Type Culture Collection or Leibniz Institute DSMZ), and MCA101-FcROVA mouse cells expressing mOVA (gifted by C. Thery, Curie, Paris, France) (47) were cultured according to the supplier’s instructions in Dulbecco’s modified Eagle’s medium or RPMI 1640 medium (Life Technologies), supplemented with [10% endotoxin-free fetal bovine serum (Thermo Fisher Scientific), 2 mM l-glutamine (Sigma-Aldrich), penicillicin (100 U/ml), and streptamycin (100 μg/ml; Life Tecnologies)] at 37°C and 5% CO2. CLEC-1 reporter B3Z cells (provided by D. Sancho, Madrid, Spain) expressing an NFAT-LacZ construct with the extracellular and transmembrane domains of the murine CLEC-1 and green fluorescent protein (GFP) and the cytoplasmic tail of the CD3ζ chain were cultured in complete RPMI 1640 medium. Reporter cells were untreated or stimulated with necrotic UV-treated splenocytes (1:10 ratio) or phorbol 12-myristate 13-acetate (PMA; 100 ng/ml) and ionomycin (2 μg/ml) (BD Biosciences). IL-2 was assessed in supernatants by commercial IL-2 ELISA kit (#M200, Bio-Techne).
Cell death induction and treatments
Cells were treated with UV-C light (150 mJ/cm2; CL-1000 UV Crosslinker, Analytik Jena), x-ray Faxitron CP 160 (10 Gy; Faxitron X-Ray Corp., Wheeling), or chemotherapies [staurosporine (1 μM; Sigma-Aldrich) and cisplatin (20 μM; Merck)] and subsequently incubated under culture conditions for 18 hours at 37°C or were killed by three freeze-thaw cycles (dry ice/37°C). UV-treated cells were incubated with RNase A (10 μg/ml; Sigma-Aldrich) or DNase I (50 U/ml; QIAGEN) to degrade nucleic acids or with trypsin (100 μg/ml; Gibco) to degrade proteins (30 min at 37°C). For heat denaturation, dead cells were fixed with paraformaldehyde (PFA) (2% VWR Chemicals for 20 min) and heat-treated (65°C for 20 min). To prevent new protein synthesis, cells were UV-treated and immediately treated with cycloheximide during culture (100 μg/ml; Sigma-Aldrich).
Flow cytometry analysis and sorting
Antibodies and panels used for fluorescence-activated cell sorting (FACS) analysis and cell sorting are listed in table S4 (5 μg/ml for 30 min). Cells were treated with Fc receptor–blocking reagents (CD16/32 mAbs; BD Biosciences), and dead cells were excluded using Fixable Viability Dye (eBioscience) or 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific). Human PBMCs, tumor cells, and mouse splenocytes were stained with Fc-AF488 recombinant proteins (5 μg/ml). Generation of human and mouse Fc CLEC-1 fusion proteins is described in Supplementary Methods. Cells were sorted by ARIA II and FACS analysis was performed using BD FACSCanto or BD FACSymphony (BD Biosciences) and analyzed with FlowJo v.10 software (Tree Star).
Immunofluorescence
UV-C light (150 mJ/cm2) treated HEK293T cells (after 18 h of culture) in eight-well μ-Slides (Ibidi) were fixed (4% PFA) (Electron Microscopy Science), permeabilized (0.1% Triton X-100) (Sigma-Aldrich), and stained with human Fc CLEC-1 or Fc ctrl (5 μg/ml), in phosphate-buffered saline (PBS), 1% fetal bovine serum, and 1% bovine serum albumin (Sigma-Aldrich). Cells were stained with DAPI, fixed in 1% PFA, and observed (Prolong Gold antifade, Thermo Fisher Scientific) by confocal fluorescence microscopy (Nikon A1-RSi) at the MicroPICell platform in Nantes. Images were obtained (360 Plan Apo numerical aperture: 1.4, zoom 2) with sequential mode and analyzed with ImageJ software.
RNA extraction and quantitative reverse transcription PCR
Total RNA was isolated using QIAGEN RNeasy mini or micro kit (QIAGEN, Hilden) and subjected to reverse transcription [Moloney Murine Leukemia Virus Reverse Transcriptase and pdT24 (Thermo Fisher Scientific)]. Reverse transcription PCR was performed using ViiA 7 Real-Time PCR System and SYBR Green PCR Master Mix (Applied Biosystems, Thermo Fisher Scientific) with primers described in table S5. Hprt was used as an endogenous control gene for normalization. Relative expression was calculated using the 2−ΔΔCt method and expressed in arbitrary units.
Reverse co-IP and analysis of TRIM21/CLEC-1 interaction
Identification of CLEC-1 ligand by co-IP of Fc CLEC-1 with necrotic cells followed by mass spectrometry is described in Supplementary Methods, and final shortlist of LC-MS was filtered for candidates displaying over 20% sequence coverage and at least seven specific peptides. Reverse co-IP was performed as co-IP, except that beads were coated with anti-TRIM21 antibody (R&D Systems) or control isotype (BD Pharm) and coincubated with His CLEC-1 protein (Bio-Techne). Western blot was performed with antihuman TRIM21 (AB01/1G5, Bio-Rad), anti-His (291-3, MBL International), or antihuman CLEC-1 (MAB1704, Bio-Techne) mAbs followed by anti-mouse horseradish peroxidase conjugate (the Jackson Laboratory) and revealed by chemiluminescence (Clarity ECL substrate, Bio-Rad). TRIM21 knockdown was obtained by lentiviral transduction of Raji cells with pGFP-shLenti–based constructs (Origen 29-mer HUSH) (CTAGGATTCACGCAGAGTTTGTGCAGCAA) and scrambled sequence (GCACTACCAGAGCTAACTCAGATAGTACT) used as control. ELISA was performed by standard protocol by coating recombinant human TRIM21 (SinoBiological) on 96-well Polysorp plates [at 2 μg/ml in carbonate buffer (pH 9.2)] and then saturated with superblock buffer (Thermo Fisher Scientific). Biotinylated human CLEC-1 [Fc CLEC-1 or His CLEC-1 (Bio-Techne)] was added and revealed with streptavidin-peroxidase (Jackson ImmunoResearch) by conventional methods. Biacore-based affinity measurement was performed on T200 (Cytiva, France) by Platform PP2I (Montpellier, France). Briefly, recombinant human TRIM21 SinoBiological, USA) was immobilized on a CM5 sensor chip (Cytiva) at 20 to 40 μg/ml in 10 mM acetate (pH 5) buffer. Five increasing concentrations (74 to 600 nM) of human Fc CLEC-1 or Fc alone were injected by single-cycle kinetic titration at 100 μl/min with injection times of 60 s, followed by a final dissociation step of 400 s in running buffer (PBS + 0.05% P20 surfactant). Sensograms were evaluated by a bivalent model (dimerization of Fc part) after subtracting the low signal from the control reference surface and buffer blank injections.
Tumor models and treatments
MC38 colon adenocarcinoma (1 × 106 per mouse) or mOVA-expressing MCA fibrosarcoma (5 × 105 per mouse) was subcutaneously injected in the left flank of WT, Clec1a KO, and CLEC-1 humanized mice. For secondary tumor challenge of cured mice, a second dose of MC38 cells (1 × 106) was subcutaneously injected in the right flank 90 days after first tumor inoculation. Mice were injected intraperitoneally with CPA (150 mg/kg; Sigma-Aldrich) or GEM (50 mg/kg; Sigma-Aldrich) when tumor diameter reached 65mm3 (at 7 to 12 days) and coinjected intraperitoneally with mouse Fc control or Fc CLEC-1 fusion proteins (5 mg/kg; twice a week for 3 weeks). Tumor growth was measured with calipers and expressed as the volume based on two perpendicular diameters [(length × width)1.5] × π/6. Mice were euthanized when tumor size reached the maximum permitted size. For mesothelioma, AK7 cells (3 × 106 per mouse) were injected into the pleural cavities of female WT and Clec1a KO mice. For HCC, Hepa 1.6 (2.5 × 106 per mouse) were injected through the portal vein of male WT, Clec1a KO, bone marrow chimeric mice with the absence of Clec1a only in the hematopoietic compartment and CLEC-1 humanized mice as previously described (48). Tumor rechallenge of cured mice was performed by injecting 2.5 × 106 Hepa 1.6 cells into the spleen. CD8 depletion was performed by intraperitoneal injection of 200 μg of anti-CD8 antibody (BE0061, Bio X Cell) on day −1 and 1 and then twice a week for 3 weeks.
CLEC-1 humanized KI C57Bl/6 mice were inoculated with tumors and treated with antihuman CLEC-1 antagonist mAbs #1 or #2 that we generated as described in Supplementary Methods or with isotype control mAb (MOTA clone, Evitria, Zürich, Switzerland) (100 μg per mouse) intraperitoneally twice a week from days 4 to 21. For subsequent analysis, tumors were digested with collagenase IV (Sigma-Aldrich), and leukocytes were enriched by Percoll (GE Healthcare) density gradient centrifugation.
Single-cell RNA sequencing
CD45.2+ cells from tumor-burdened livers of two WT and two Clec1a KO mice (day 13 after tumor inoculation) were sorted. Cells were encapsulated in droplets using the Chromium Controller (10X Genomics, San Francisco, CA) (load of 20,000 cells) and marked with anti-mouse TotalSeq hashtag antibodies (BioLegend). scRNA-seq libraries were prepared with the Chromium Single Cell 3′ Library and Gel Bead Kit v2 (PN-120267) and Chromium Next GEM Single Cell 3′ Library and Gel Bead Kit v3.1 (PN-1000121) (10X Genomics). Library was prepared by sequencing in a NovaSeq 6000 (Illumina, SFR, Nantes). The Cell Ranger Suite (version 6.0.0; 10X Genomics) was used to perform sample demultiplexing, barcode processing, and single-cell gene unique molecular identifier counting. The gene cell barcode matrices were merged into one matrix (20,473 genes × 31,976 cells) and then loaded into the R package Seurat, version 4.0.1 (https://satijalab.org/seurat) as described previously (49). scRNA-seq data were obtained for a total of 31,976 sorted individual cells (5536 from WT and 6065 from Clec1a KO tumor–burdened livers), at an average of 11,748 uniquely mapped read pairs per cell.
In vivo and in vitro cross-presentation assays
In vivo, mice were intravenously injected with purified OVA-specific CD8+ T cells from OT-I.Ly5.1 mice (CD8+ T cell isolation Kit II, Miltenyi Biotec) (1 × 106 cells per mouse) labeled with Cell Proliferation Dye (CPD) eFluor670 (eBioscience). Mice were then injected intraperitoneally 1 day later with 100 μg of EndoFit OVA (InvivoGen) or with 1 × 107 MCA101-FcROVA treated by UV-C light (150 mJ/cm2) or killed by three freeze-thaw cycles. Proliferation of splenic CD8+ T cells was assessed 3 days later by flow cytometry.
In vitro, cDC1 were enriched from the spleen using XCR1 microbeads (Miltenyi Biotec) according to the manufacturer’s instructions. BMDCs were generated from the bone marrow (5 × 105 cells/ml), cultured for 8 days in complete RPMI 1640 medium supplemented with mouse granulocyte-macrophage colony-stimulating factor (20 ng/ml; Miltenyi Biotec). cDC1 and nonadherent BMDCs were incubated 5 hours in 96-well plates (1.25 × 104 cells per well) with EndoFit OVA (500 μg/ml), OVA257–264 (2 or 100 ng/ml; SIINFEKL) peptides (GenScript), or UV-treated (150 mJ/cm2) MCA101-FcROVA (ratio, 1:1). Pulsed cDC1 or BMDCs were washed (three times) and coincubated for 3 days with purified CPD-labeled CD8+ T cells (ratio, 1:2). Intracellular IFNγ production was evaluated in fix/perm (BD Biosciences) OT-I cells by flow cytometry following PMA (50 ng/ml) and ionomycin (1 μg/ml) restimulation for 4 hours in the presence of Golgi stop (BD Biosciences).
For the in vivo effect on tumor growth, BMDCs (2 × 106 cells per mouse) were injected intravenously, together with preactivated OT-1 [activation with 1 nM OVA257–264 (SIINFEKL) peptide (GenScript) and recombinant IL-2 (100 U/ml; Proleukine) for 3 days] in mice previously subcutaneously inoculated (day 7) in the flank with MCA101-FcROVA (5 × 105 cells per mouse).
Statistical analysis
Statistical analyses were performed using GraphPad Prism software (La Jolla, CA, USA) using the Mann-Whitney, Wilcoxon t test, Kruskal-Wallis test, or log-rank (Mantel-Cox) test (for survival rates). P < 0.05 was considered significant.
Acknowledgments
We thank D. McIlroy for language editing. We acknowledge the IBISA MicroPICell facility (Biogenouest), member of the national infrastructure France-Bioimaging supported by the French national research agency (ANR-10-INBS-04), the Genomics Core Facility GenoA, member of Biogenouest and France Genomique and the Bioinformatics Core Facility BiRD, member of Biogenouest and Institut Français de Bioinformatique (IFB) (ANR-11-INBS-0013) for the use of their resources and their technical support.
Funding: This work was supported by LIGUE 44, 56, and 29 (2018) and LIGUE 44 (2021) (to E.C.); LabEX IGO programs (ANR-11-LABX-0016-01) from National Research Agency (ANR) for French government (to E.C.); and ANR generique PRCE 2018 from National Research Agency (ANR) for French government (to E.C. and N.P.).
Author contributions: Conceptualization: E.C. and N.P. Formal analysis: M.D., J.S., V.G., B.E., G.T., S.P., C.M., A.D., V.T., E.W., E.M., C.Li, I.G,. M.-D.L., D.S., J.P., and E.C. Funding acquisition: E.C. and N.P. Investigation: M.D., J.S., V.G., B.E., G.T., S.P., C.M., A.D., V.T., E.W., E.M., C.Li, M.-D.L., C.F., N.P., and E.C. Methodology: M.D., J.S, V.G., B.E., G.T., S.P., C.M., A.D., V.T., E.W., E.M., C.Li, M.-D.L., C.F., I.B, A.M., C.Lo, R.J., J.P., N.P., and E.C. Supervision: E.C. and N.P. Visualization: M.D., J.S., V.G., N.P., and E.C. Writing–original draft: E.C. and M.D. Writing–review and editing: E.C.
Competing interests: M.D., E.C., N.P., V.G., G.T., C.M., S.P., A.D., V.T., and E.W are authors of patents related to CLEC-1 inhibition: “Methods and pharmaceutical compositions for promoting T cells response,” granted patent. Organizations: OSE Immunotherapeutics, Université de Nantes, Institut national de la santé et de la recherche médicale. Authors on this paper who are also authors on the patent: E.C., N.P., A.D., V.T., C.M., V.G., and S.P. Filed 21 October 2016, EP16306381, WO/2018/073440. “Anti–CLEC-1 antibodies and antigen-binding fragment thereof,” published patent. Organizations: OSE Immunotherapeutics, Université de Nantes, Institut national de la santé et de la recherche médicale. Authors on this paper who are also authors on the patent: E.C., N.P., A.D., V.T., C.M., V.G., and S.P. Filed 5 December 2019, EP 19306583.6, WO/2021/110990A1. “Humanized anti–CLEC-1A antibodies and antigen-binding fragments thereof and mimetics thereof,” filed patent. Organizations: OSE Immunotherapeutics, Université de Nantes, Institut national de la santé et de la recherche médicale. Authors on this paper who are also authors on the patent: E.C., N.P., M.D., C.M., V.G., and S.P. Filed 8 June 2021, EP 21305777.1. “Identification of CLEC-1 ligand and uses thereof”, filed patent. Organizations: OSE Immunotherapeutics, Université de Nantes, Institut national de la santé et de la recherche médicale. Authors on this paper who are also authors on the patent: E.C., N.P., E.W., G.T., C.M., and V.G. Filed 9 November 2021, EP 21306571.7. N.P., V.G., G.T., S.P., C.M., A.D., V.T., E.W., I.G., and I.B are employee and shareholders of OSE Immunotherapeutics, a company involved in immunotherapy development in immuno-oncology. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. scRNA-seq data of HCC in WT and Clec1a KO mice are available at Zenodo DOI: 10.5281/zenodo.7030954. Fc CLEC-1 fusion proteins and anti–CLEC-1 mAbs could be provided by OSE Immunotherapeutics pending scientific review (depending of the research subject, university, potential of valorization, etc.) and a completed material transfer agreement. Requests for the Fc CLEC-1 fusion proteins and anti–CLEC-1 mAbs should be submitted to: nicolas.poirier@ose-immuno.com.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S7
Tables S1 to S5
References
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Couzin-Frankel J., Cancer immunotherapy. Science 342, 1432–1433 (2013). [DOI] [PubMed] [Google Scholar]
- 2.De Vlaeminck Y., González-Rascón A., Goyvaerts C., Breckpot K., Cancer-associated myeloid regulatory cells. Front. Immunol. 7, 113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böttcher J. P., Reis e Sousa C., The role of type 1 conventional dendritic cells in cancer immunity. Trends in Cancer 4, 784–792 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shekarian T., Valsesia-Wittmann S., Brody J., Michallet M. C., Depil S., Caux C., Marabelle A., Pattern recognition receptors: Immune targets to enhance cancer immunotherapy. Ann. Oncol. 28, 1756–1766 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Yan H., Kamiya T., Suabjakyong P., Tsuji N. M., Targeting C-type lectin receptors for cancer immunity. Front. Immunol. 6, 408 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiffoleau E., C-type lectin-like receptors as emerging orchestrators of sterile inflammation represent potential therapeutic targets. Front. Immunol. 9, 227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drouin M., Saenz J., Chiffoleau E., C-type lectin-like receptors: Head or tail in cell death immunity. Front. Immunol. 11, 251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thebault P., Lhermite N., Tilly G., Le Texier L., Quillard T., Heslan M., Anegon I., Soulillou J.-P., Brouard S., Charreau B., Cuturi M.-C., Chiffoleau E., The C-type lectin-like receptor CLEC-1, expressed by myeloid cells and endothelial cells, is up-regulated by immunoregulatory mediators and moderates T cell activation. J. Immunol. 183, 3099–3108 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Robles M. D. L., Pallier A., Huchet V., Le Texier L., Remy S., Braudeau C., Delbos L., Moreau A., Louvet C., Brosseau C., Royer P. J., Magnan A., Halary F., Josien R., Cuturi M. C., Anegon I., Chiffoleau E., Cell-surface C-type lectin-like receptor CLEC-1 dampens dendritic cell activation and downstream Th17 responses. Blood Adv. 1, 557–568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobanov Y., Bernreiter A., Derdak S., Mechtcheriakova D., Schweighofer B., Düchler M., Kalthoff F., Hofer E., A novel cluster of lectin-like receptor genes expressed in monocytic, dendritic and endothelial cells maps close to the NK receptor genes in the human NK gene complex. Eur. J. Immunol. 31, 3493–3503 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Colonna M., Samaridis J., Angman L., Molecular characterization of two novel C-type lectin-like receptors, one of which is selectively expressed in human dendritic cells. Eur. J. Immunol. 30, 697–704 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Sattler S., Reiche D., Sturtzel C., Karas I., Richter S., Kalb M. L., Gregor W., Hofer E., The human C-type lectin-like receptor CLEC-1 is upregulated by TGF-β and primarily localized in the endoplasmic membrane compartment. Scand. J. Immunol. 75, 282–292 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Yoshimi R., Ishigatsubo Y., Ozato K., Autoantigen TRIM21/Ro52 as a possible target for treatment of systemic lupus erythematosus. Int. J. Rheumatol. 2012, 718237 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davidov V., Jensen G., Mai S., Chen S. H., Pan P. Y., Analyzing one cell at a TIME: Analysis of myeloid cell contributions in the tumor immune microenvironment. Front. Immunol. 11, 1842 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller S., Kohanbash G., Liu S. J., Alvarado B., Carrera D., Bhaduri A., Watchmaker P. B., Yagnik G., Di Lullo E., Malatesta M., Amankulor N. M., Kriegstein A. R., Lim D. A., Aghi M., Okada H., Diaz A., Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 18, 234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghislat G., Cheema A. S., Baudoin E., Verthuy C., Ballester P. J., Crozat K., Attaf N., Dong C., Milpied P., Malissen B., Auphan-Anezin N., Manh T. P. V., Dalod M., Lawrence T., NF-κB–dependent IRF1 activation programs cDC1 dendritic cells to drive antitumor immunity. Sci. Immunol. 6, 3570 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Veglia F., Sanseviero E., Gabrilovich D. I., Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 21, 485–498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liuyu T., Yu K., Ye L., Zhang Z., Zhang M., Ren Y., Cai Z., Zhu Q., Lin D., Zhong B., Induction of OTUD4 by viral infection promotes antiviral responses through deubiquitinating and stabilizing MAVS. Cell Res. 29, 67–79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stricher F., Macri C., Ruff M., Muller S., HSPA8/HSC70 chaperone protein: Structure, function, and chemical targeting. Autophagy 9, 1937–1954 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Hildner K., Edelson B. T., Purtha W. E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B. U., Unanue E. R., Diamond M. S., Schreiber R. D., Murphy T. L., Murphy K. M., Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broz M. L., Binnewies M., Boldajipour B., Nelson A. E., Pollack J. L., Erle D. J., Barczak A., Rosenblum M. D., Daud A., Barber D. L., Amigorena S., Van’tVeer L. J., Sperling A. I., Wolf D. M., Krummel M. F., Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell 26, 638–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harbers S. O., Crocker A., Catalano G., D’Agati V., Jung S., Desai D. D., Clynes R., Antibody-enhanced cross-presentation of self antigen breaks T cell tolerance. J. Clin. Invest. 117, 1361–1369 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zilionis R., Engblom C., Pfirschke C., Savova V., Zemmour D., Saatcioglu H. D., Krishnan I., Maroni G., Meyerovitz C. V., Kerwin C. M., Choi S., Richards W. G., De Rienzo A., Tenen D. G., Bueno R., Levantini E., Pittet M. J., Klein A. M., Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 50, 1317–1334.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian J., Olbrecht S., Boeckx B., Vos H., Laoui D., Etlioglu E., Wauters E., Pomella V., Verbandt S., Busschaert P., Bassez A., Franken A., Vanden Bempt M., Xiong J., Weynand B., van Herck Y., Antoranz A., Bosisio F. M., Thienpont B., Floris G., Vergote I., Smeets A., Tejpar S., Lambrechts D., A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 30, 745–762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z., GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann K., Castiñ Eiras-Vilariñ M., Hö Ckendorf U., Hannesschlä Ger N., Lemeer S., Kupka D., Meyermann S., Lech M., Anders H.-J., Kuster B., Busch D. H., Gewies A., Naumann R., Groß O., Ruland J., Clec12a is an inhibitory receptor for uric acid crystals that regulates inflammation in response to cell death. Immunity 40, 389–399 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Del Fresno C., Saz-Leal P., Enamorado M., Wculek S. K., Martínez-Cano S., Blanco-Menéndez N., Schulz O., Gallizioli M., Miró-Mur F., Cano E., Planas A., Sancho D., DNGR-1 in dendritic cells limits tissue damage by dampening neutrophil recruitment. Science 362, 351–356 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Ahrens S., Zelenay S., Sancho D., Hanč P., Kjær S., Feest C., Fletcher G., Durkin C., Postigo A., Skehel M., Batista F., Thompson B., Way M., Reis e Sousa C., Schulz O., F-actin is an evolutionarily conserved damage-associated molecular pattern recognized by DNGR-1, a receptor for dead cells. Immunity 36, 635–645 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Li L., Wei J., Mallampalli R. K., Zhao Y., Zhao J., TRIM21 mitigates human lung microvascular endothelial cells’ inflammatory responses to LPS. Am. J. Respir. Cell Mol. Biol. 61, 776–785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espinosa A., Dardalhon V., Brauner S., Ambrosi A., Higgs R., Quintana F. J., Sjöstrand M., Eloranta M. L., Gabhann J. N., Winqvist O., Sundelin B., Jefferies C. A., Rozell B., Kuchroo V. K., Wahren-Herlenius M., Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J. Exp. Med. 206, 1661–1671 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simoes Eugénio M., Faurez F., Kara-Ali G. H., Lagarrigue M., Uhart P., Bonnet M. C., Gallais I., Com E., Pineau C., Samson M., Le Seyec J., Dimanche-Boitrel M. T., TRIM21, a new component of the TRAIL-induced endogenous necrosome complex. Front. Mol. Biosci. 8, 139 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshimi R., Chang T.-H., Wang H., Atsumi T., Morse H. C., Ozato K., Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-κB-dependent cytokine expression in fibroblasts. J. Immunol. 182, 7527–7538 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z., Bao M., Lu N., Weng L., Yuan B., Liu Y. J., The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat. Immunol. 14, 172–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang F., Zhang Y., Shen J., Yang B., Dai W., Yan J., Maimouni S., Daguplo H. Q., Coppola S., Gao Y., Wang Y., Du Z., Peng K., Liu H., Zhang Q., Tang F., Wang P., Gao S., Wang Y., Ding W. X., Guo G., Wang F., Zong W. X., The ubiquitin E3 ligase TRIM21 promotes hepatocarcinogenesis by suppressing the p62-Keap1-Nrf2 antioxidant pathway. Cell Mol. Gastroenterol. Hepatol. 11, 1369–1385 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Z., Wang Y., Yun D., Huang Q., Meng D., Li Q., Zhang P., Wang C., Chen H., Lu D., TRIM21 overexpression promotes tumor progression by regulating cell proliferation, cell migration and cell senescence in human glioma. Am. J. Cancer Res. 10, 114–130 (2020). [PMC free article] [PubMed] [Google Scholar]
- 36.Tullett K. M., Tan P. S., Park H. Y., Schittenhelm R. B., Michael N., Li R., Policheni A. N., Gruber E., Huang C., Fulcher A. J., Danne J. C., Czabotar P. E., Wakim L. M., Mintern J. D., Ramm G., Radford K. J., Caminschi I., O’Keeffe M., Villadangos J. A., Wright M. D., Blewitt M. E., Heath W. R., Shortman K., Purcel A. W., Nicola N. A., Zhang J. G., Lahoud M. H., RNF41 regulates the damage recognition receptor Clec9a and antigen cross-presentation in mouse dendritic cells. eLife 9, e63452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao S., Wake H., Sakaguchi M., Wang D., Takahashi Y., Teshigawara K., Zhong H., Mori S., Liu K., Takahashi H., Nishibori M., Histidine-rich glycoprotein inhibits high-mobility group box-1-mediated pathways in vascular endothelial cells through CLEC-1A. iScience 23, 101180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi Y., Wake H., Sakaguchi M., Yoshii Y., Teshigawara K., Wang D., Nishibori M., Histidine-rich glycoprotein stimulates human neutrophil phagocytosis and prolongs survival through CLEC1A. J. Immunol. 206, 737–750 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huffman A. P., Lin J. H., Kim S. I., Byrne K. T., Vonderheide R. H., CCL5 mediates CD40-driven CD4+ T cell tumor infiltration and immunity. JCI Insight 5, e137263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denkert C., Loibl S., Noske A., Roller M., Müller B. M., Komor M., Budczies J., Darb-Esfahani S., Kronenwett R., Hanusch C., Von Törne C., Weichert W., Engels K., Solbach C., Schrader I., Dietel M., Von Minckwitz G., Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 28, 105–113 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Canton J., Blees H., Henry C. M., Buck M. D., Schulz O., Rogers N. C., Childs E., Zelenay S., Rhys H., Domart M., Collinson L., Alloatti A., Ellison C. J., Amigorena S., Papayannopoulos V., Thomas D. C., Randow F., Reis C., The receptor DNGR-1 signals for phagosomal rupture to promote cross-presentation of dead-cell-associated antigens. Nat. Immunol. 22, 140–153 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barkal A. A., Brewer R. E., Markovic M., Kowarsky M., Barkal S. A., Zaro B. W., Krishnan V., Hatakeyama J., Dorigo O., Barkal L. J., Weissman I. L., CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 572, 392–396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding Y., Guo Z., Liu Y., Li X., Zhang Q., Xu X., Gu Y., Zhang Y., Zhao D., Cao X., The lectin Siglec-G inhibits dendritic cell cross-presentation by impairing MHC class I-peptide complex formation. Nat. Immunol. 17, 1167–1175 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Plato A., Willment J. A., Brown G. D., C-type lectin-like receptors of the dectin-1 cluster: Ligands and signaling pathways. Int. Rev. Immunol. 32, 134–156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Embgenbroich M., Burgdorf S., Current concepts of antigen cross-presentation. Front. Immunol. 9, 1643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gazit R., Mandal P. K., Ebina W., Ben-Zvi A., Nombela-Arrieta C., Silberstein L. E., Rossi D. J., Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J. Exp. Med. 211, 1315–1331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeelenberg I. S., van Maren W. W. C., Boissonnas A., Van Hout-Kuijer M. A., Den Brok M. H. M. G. M., Wagenaars J. A. L., van der Schaaf A., Jansen E. J. R., Amigorena S., Théry C., Figdor C. G., Adema G. J., Antigen localization controls T cell-mediated tumor immunity. J. Immunol. 187, 1281–1288 (2011). [DOI] [PubMed] [Google Scholar]
- 48.McIlroy D., Barteau B., Cany J., Richard P., Gourden C., Conchon S., Pitard B., DNA/amphiphilic block copolymer nanospheres promote low-dose DNA vaccination. Mol. Ther. 17, 1473–1481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurts C., Heath W. R., Carbone F. R., Allison J., Miller J. F. A. F., Kosaka H., Constitutive class I-restricted exogenous presentation of self antigens in vivo. J. Exp. Med. 184, 923–930 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Z., Bailey A., Kuleshov M. V., Clarke D. J. B., Evangelista J. E., Jenkins S. L., Lachmann A., Wojciechowicz M. L., Kropiwnicki E., Jagodnik K. M., Jeon M., Ma’ayan A., Gene set knowledge discovery with enrichr. Curr. Protoc. 1, e90 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y., Wang J., Jaehnig E. J., Shi Z., Zhang B., WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S7
Tables S1 to S5
References
Data S1