Brassica oleracea is an important plant species that includes many globally cultivated vegetable crops (cole crops), such as cabbage, broccoli, cauliflower, kale and Brussels sprouts. These plants provide human beings with not only plentiful nutrients such as carotenoids, minerals, vitamins A and C, dietary fibre but also unique health‐promoting compounds like glucosinolates (Xu et al., 2014).
Heterosis utilization in crops, including cole vegetables, requires the development of homozygous lines usually generated by multiple rounds of selfing or backcrossing (Zhong et al., 2019). Doubled haploid (DH) technology enables the generation of complete homozygous lines within two generations, dramatically accelerating the breeding progress (Zhong et al., 2020). However, traditional haploid induction (HI) in Brassica oleracea depends on an in vitro anther/microspore cultivation approach, which is not only complicated but also highly limited by plant genotype. In recent years, MTL/NLD/ZmPLA1, ZmDMP and ZmPOD65 were found to be responsible for inducing in vivo maternal haploid embryos in maize (Jiang et al., 2022 and references therein). Although orthologues of MTL/NLD/ZmPLA1 have not been found in dicots, ZmDMP‐like genes are present in dicots and have been demonstrated to trigger in vivo maternal HI in Arabidopsis, Medicago truncatula, tomato, rapeseed and tobacco (Li et al., 2022; Wang et al., 2022; Zhong et al., 2020, 2022a,b). However, it is still not known whether this approach can be applied to cole crops.
We found fifteen putative DMP‐like proteins in the Brassica oleracea genome. Among the proteins identified above, BoC04.DMP9 and BoC03.DMP9 were highly similar to ZmDMP, with 61% and 60% sequence identity, respectively, and they were assigned to a subclade together with AtDMP9 and AtDMP8 (Figure 1a). qRT–PCR analysis indicated that both BoC03.DMP9 and BoC04.DMP9 are highly expressed in pollen and flower buds, with BoC03.DMP9 being more highly expressed (Figure 1b). We cloned BoC03.DMP9 and BoC04.DMP9 from multiple cabbage inbred lines. Intriguingly, BoC04.DMP9 was lost in these cabbage lines due to a 1‐bp deletion in exon. We further investigated DMPs in the Brassica genus, which showed that DMP8 was completely lost, and DMP9 experienced duplication and then lost. Both A and B genomes retained two normal DMP9 genes, whereas in the C genome, differential DMP9 orthologues were lost, including BoC04.DMP9 in B. oleracea and BnaC03g03890D in B. napus, indicating that the loss of DMP9 is a recent event after the formation of B. napus.
Figure 1.
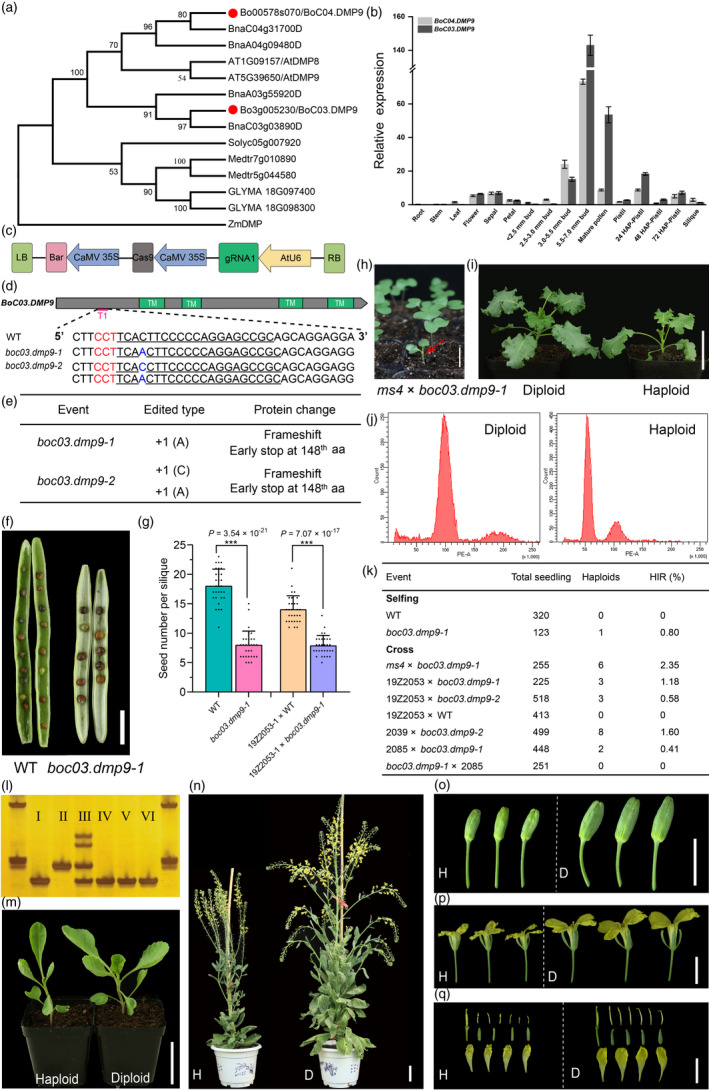
boc03.dmp9 mutants trigger maternal haploid induction. (a) Phylogenetic analysis of DMP homologues in Brassica oleracea (Bo), Arabidopsis (At), Brassica napus (Bna), soybean (Glyma), Medicago truncatula (Medtr) and Solanum lycopersicum (Soly). BoC03.DMP9 and BoC04.DMP9 are indicated with red dots. A neighbour‐joining phylogenetic tree (1000 bootstrap replications) was constructed using MEGA7 software. (b) Relative expression levels of BoC03.DMP9 and BoC04.DMP9 in cabbage tissues. Three independent biological replicates were performed. Error bars represent mean ± SD. HAP, hours after pollination. (c) Schematic diagram of the CRISPR/Cas9 construct targeting BoC03.DMP9. Bar, bialaphos resistance gene; AtU6, Arabidopsis U6‐26 promoter. (d) Schematic of BoC03.DMP9. Grey blocks, gene coding region; green blocks, predicted transmembrane domains (TMs); red lines, the region (T1) targeted by sgRNA. Sequences from the wild‐type (WT) and mutants are shown below the overview. The target sequences are underlined. Insertions are highlighted in blue, and PAM sequences are highlighted in red. (e) Two representative plants edited at the target region of BoC03.DMP9. (f) Representative siliques from selfed MW (WT) and boc03.dmp9 mutants. (g) Seed number per silique of selfed WT and boc03.dmp9 mutants. Bars represent mean ± SD (n = 30); asterisks indicate significant differences (***P < 0.001, Student's t‐test). (h) Six‐day old seedlings from ms4 × boc03.dmp9‐1. Purple plants are typical F1 hybrids. Red arrow indicates a haploid with non‐purple phenotype similar to ms4. (i) Diploid and haploid from the tester line ms4. (j) Flow cytometry analysis verification of putative haploids. (k) HIR of boc03.dmp9 determined by selfing or crossing. (l) Putative haploids were genotyped with molecular markers. Left and right lanes, DNA marker; I‐III, PCR bands of the 19Z2053, boc03.dmp9 mutant, a F1 hybrid from 19Z2053 × boc03.dmp9‐1, representatively; IV‐VI, PCR bands of three haploids from 19Z2053. (m–q) Phenotypes of haploid and diploid 19Z2053. H, haploid; D, diploid. Scale bars: 1 cm (f, h, o, p and q), 5 cm (i, m and n).
We employed the CRISPR/Cas9 approach to knock out BoC03.DMP9 in the cabbage ‘MW’ background. A CRISPR/Cas9 construct with a specific guide RNA sequence targeting the exon of BoC03.DMP9 was generated and introduced into cabbage by Agrobacterium‐mediated transformation (Figure 1c). We obtained 8 lines with mutations in the target region, among which two homozygous or biallelic boc03.dmp9 mutants with deletions/insertions that led to frameshift and premature termination were selected for further studies (Figure 1d,e). Upon selfing or serving as pollen donors for crossing, the boc03.dmp9 mutants showed significantly reduced seed sets (Figure 1f,g).
To test whether boc03.dmp9 mutants could induce the production of haploids, we crossed boc03.dmp9 mutants (as male parents) with the tester line ms4, a curly kale male‐sterile line. ms4 is an ideal HI testing material owing to its two characteristics: (i) completely green, a natural phenotype resulting from the abolishment of anthocyanin accumulation (Figure 1h), and (ii) male sterility, which prevents the occurrence of selfing. We found that six out of 255 progenies exhibited a completely green phenotype (Figure 1i). Flow cytometry analysis revealed that all six non‐purple plants were true haploids, which corresponded to a haploid induction rate (HIR) of 2.35% (Figure 1j,k).
We further carried out a set of tests using boc03.dmp9 mutants to cross cabbage materials, including two inbred lines (19Z2053 and 2039) and one hybrid (2085). Molecular markers showing InDel polymorphism between boc03.dmp9 and the female parents were developed to screen all the progenies. Haploid would show genotype identical to the corresponding female (Figure 1l). Potential haploids identified by molecular markers were further confirmed by cytometry analysis and plant phenotyping. The HIRs ranged from 0.41% to 1.60% (Figure 1k). Haploids derived from the 19Z2053 × boc03.dmp9 and 2039 × boc03.dmp9 crosses were morphologically similar to the corresponding female parent (Figure 1m,n) but had smaller organs and were sterile (Figure 1o–q). We also tested the HI ability by the use of the boc03.dmp9 as a female, but no haploids were identified after crossing (Figure 1k).
In summary, we demonstrated that boc03.dmp9 mutants could induce in vivo maternal haploids in cole crops. The reported DMP‐based in vivo HI system offers a novel, simple and cost‐effective DH technology without genotype recalcitrance. Importantly, this system is applicable to one‐step creation of homozygous male‐sterile lines for hybrid seed production. In summary, this HI system could accelerate cultivar improvement and genetic studies of these important vegetable crops and provides reference information for extending this system to other dicotyledonous crop species.
Conflicts of interest
The authors declare no conflict of interest.
Author contributions
F.H. and H.L. conceived and designed the work. X.Z., K.Y., Y.L. and N.Z. performed the experiments. F.H. and X.Z. wrote and revised the manuscript. L.Y., Y.Z., Y.W., J.J. and Z.F. analysed the data and revised the manuscript. All authors have read and approved the final manuscript.
Acknowledgements
This work was supported by grants from the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS‐ASTIP‐IVFCAAS) and China Agriculture Research System of MOF and MARA (CARS−23).
Contributor Information
Fengqing Han, Email: hanfengqing@caas.cn.
Honghao Lv, Email: lvhonghao@caas.cn.
References
- Jiang, C. , Sun, J. , Li, R. , Yan, S. , Chen, W. , Guo, L. , Qin, G. et al. (2022) A reactive oxygen species burst causes haploid induction in maize. Mol. Plant, 15, 943–955. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Li, D. , Xiao, Q. , Wang, H. , Wen, J. , Tu, J. , Shen, J. et al. (2022) An in planta haploid induction system in Brassica napus . J. Integr. Plant Biol. 64, 1140–1144. [DOI] [PubMed] [Google Scholar]
- Wang, N. , Xia, X. , Jiang, T. , Li, L. , Zhang, P. , Niu, L. , Cheng, H. et al. (2022) In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula . Plant Biotech. J. 20, 22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F. , Zheng, Y. , Yang, Z. , Cao, S. , Shao, X. and Wang, H. (2014) Domestic cooking methods affect the nutritional quality of red cabbage. Food Chem. 161, 162–167. [DOI] [PubMed] [Google Scholar]
- Zhong, Y. , Liu, C. , Qi, X. , Jiao, Y. , Wang, D. , Wang, Y. , Liu, Z. et al. (2019) Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants, 5, 575–580. [DOI] [PubMed] [Google Scholar]
- Zhong, Y. , Chen, B. , Li, M. , Wang, D. , Jiao, Y. , Qi, X. , Wang, M. et al. (2020) A DMP‐triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis . Nat. Plants, 6, 466–472. [DOI] [PubMed] [Google Scholar]
- Zhong, Y. , Chen, B. , Wang, D. , Zhu, X. , Li, M. , Zhang, J. , Chen, M. et al. (2022a) In vivo maternal haploid induction in tomato. Plant Biotech. J. 20, 250–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Y. , Wang, Y. , Chen, B. , Liu, J. , Wang, D. , Li, M. , Qi, X. et al. (2022b) Establishment of a dmp based maternal haploid induction system for polyploid Brassica napus and Nicotiana tabacum . J. Integr. Plant Biol. 64, 1281–1294. [DOI] [PubMed] [Google Scholar]


