Summary
Efficient pathogen diagnostics and genotyping methods enable effective disease management and breeding, improve crop productivity and ensure food security. However, current germplasm selection and pathogen detection techniques are laborious, time‐consuming, expensive and not easy to mass‐scale application in the field. Here, we optimized a field‐deployable lateral flow assay, Bio‐SCAN, as a highly sensitive tool to precisely identify elite germplasm and detect mutations, transgenes and phytopathogens in <1 h, starting from sample isolation to result output using lateral flow strips. As a proof of concept, we genotyped various wheat germplasms for the Lr34 and Lr67 alleles conferring broad‐spectrum resistance to stripe rust, confirmed the presence of synthetically produced herbicide‐resistant alleles in the rice genome and screened for the presence of transgenic elements in the genome of transgenic tobacco and rice plants with 100% specificity. We also successfully applied this new assay to the detection of phytopathogens, including viruses and bacterial pathogens in Nicotiana benthamiana, and two destructive fungal pathogens (Puccinia striiformis f. sp. tritici and Magnaporthe oryzae Triticum) in wheat. Our results illustrate the power of Bio‐SCAN in crop breeding, genetic engineering and pathogen diagnostics to enhance food security. The high sensitivity, simplicity, versatility and in‐field deployability make the Bio‐SCAN as an attractive molecular diagnostic tool for diverse applications in agriculture.
Keywords: germplasm, phytopathogens, lateral flow assay, CRISPR/dCas9 and dCas9, nucleic acid detection, biotin
Bio‐SCAN‐based point of care system for genotyping of plant genome for pathogens and herbicide resistance alleles and detection of transgenes and phytopathogens on lateral flow strips.
![]()
Introduction
Modern agriculture relies on the identification of gene variants (alleles) that confer better growth and greater stress tolerance and on the production of genetically engineered crops to meet the increasing food demands for the growing human population (Ul Haq and Ijaz, 2020). Wheat (Triticum aestivum L.), for instance, is a widely grown crop that feeds around 35% of people worldwide (Deng et al., 2019; Grote et al., 2021). It has been intensively bred to accumulate desirable traits for making elite cultivars with high productivity (Baenziger, 2016; Bedő and Láng, 2015) and resistance to various disease pathogens, including stem rust (Ai et al., 2015; Aman et al., 2020c; Ciuca et al., 2015; Dakouri et al., 2013). Two natural alleles, Lr34 (encoding an ABC transporter) and Lr67 (encoding a hexose‐proton symporter), are widely used in breeding programs to provide broad‐spectrum resistance against rust (Krattinger et al., 2009; Milne et al., 2018; Moore et al., 2015; Spielmeyer et al., 2013a,b). The resistant alleles of Lr34 and Lr67 differ from the susceptible alleles by only single nucleotide polymorphisms (SNPs; Krattinger et al., 2009; Moore et al., 2015).
In addition to the selection of natural alleles, different methods have been developed to incorporate desired crop traits. Synthetic directed evolution is a recent approach to generate crops resistant to environmental stresses and with desired agronomic traits by inducing changes in localized genes. Other groups and we have recently engineered resistance against herbicides in rice (Oryza sativa) via directed evolution of ALS (ACETOLACTATE SYNTHASE, resistant to herbicide bispyribac sodium) and SF3B1 (SPLICEOSOME FACTOR 3B1, resistant to herboxidiene GEX1A, a naturally occurring splicing inhibitor used as a herbicide; Butt et al., 2019; Rao et al., 2021). The resulting evolved alleles of ALS and SF3B1 differ from the wild‐type alleles by a few changes in the DNA sequence (Butt et al., 2019, 2020).
The evaluation of resistance alleles, either natural or engineered, is a lengthy and tedious procedure that includes field trials for germplasm–pathogen interaction, the selection of resistant cultivars for breeding programs and genotyping for the identification of a specific combination of alleles, and subsequent testing in the progeny (Lorenz et al., 2011). Therefore, a simple detection method to evaluate the mutant lines or to confirm the presence of these alleles in each breeding line and generation would assist the directed evolution and breeding methodologies.
Furthermore, the applications of testing are not limited to the detection of resistant alleles, they expand to benefit food security. For example, through the detection of pythopathogens and molecular markers of GMOs. It is estimated that pythopathogens cause 22%–30% of global yield losses of the three vital food crops viz. wheat, rice and maize (Zea mays; Islam et al., 2016; Nazarov et al., 2020; Peng et al., 2021; Powderly, 2019; Ristaino et al., 2021). The worst offenders are fungi causing losses equivalent to 200 billion dollars (Villamizar‐Gallardo et al., 2019), followed by viruses (over 30 billion dollars) and bacteria (over 1 billion dollars) worldwide every year (Jones and Naidu, 2019; Martins et al., 2018; Peng et al., 2021). Additionally, since several countries have a legislation for crop trading that requires strict testing processes for GMOs, screening for this purpose is equally important (Salisu et al., 2017).
Traditional methods, such as visual observation, microscopy, pure cultures isolation, immunological tests, polymerase chain reaction (PCR), DNA microarrays, matrix‐assisted laser desorption ionization‐time of flight mass spectrometry (MALDI‐TOF MS; Khakimov et al., 2022), restriction fragment length polymorphism (RFLP) and DNA hybridization, are widely used for plant genotyping, detection of plant pathogens and GMO identification in crops (Batley, 2015; Henry, 2014). However, these methods are complex, time consuming, require special laboratory setup and thus not compatible with point‐of‐care diagnostics. Therefore, simple, precise and field‐deployable point‐of‐care rapid detection modules are highly demanded to bypass lab‐based testing and alleviate laborious phenotyping and genotyping of thousands of samples in an extended process.
For the molecular detection of alleles, pathogens and biological contaminants, nucleic acids are the preferred biomarkers due to their stability, reproducible amplification and easy coupling with different reporter systems (Zheng et al., 2021). Even when PCR is the current gold standard for nucleic acid detection, it suffers from several major drawbacks, as it is time‐consuming, relies on sophisticated laboratory facilities and specialized equipment and requires highly trained operators (Zheng et al., 2021; Zhu et al., 2020). By contrast, methods such as isothermal amplification that can be performed easily in one step and are suitable for point‐of‐care testing (POCT) and point‐of‐need testing (PONT) applications provide an attractive alternative to PCR for field detection. Among these methodologies, recombinase polymerase amplification (RPA) and loop‐mediated isothermal amplification (LAMP) show great potential in POCT. In particular, coupling isothermal amplification with CRISPR/CRISPR‐associated nuclease (Cas) systems offers specificity, with the possibility of a visual readout on lateral flow assay (LFA) strips or colorimetric assays. These diagnostic tools can be easily adopted under resource‐limited conditions for low‐cost bulk screening with high sensitivity and easy‐to‐use POCT operations (Aman et al., 2020a; Kaminski et al., 2021; Zheng et al., 2021; Zou et al., 2020).
We and others have previously developed CRISPR‐based biosensing technologies such as DETECTR, a system that couples RT‐LAMP and Cas12 with the use of a reporter to obtain an LFA readout (Broughton et al., 2020); AIOD‐CRISPR, a one‐pot system based on Cas12a for visual fluorescent detections (Ding et al., 2020); iSCAN, a two‐pot system coupling RT‐LAMP and CRISPR‐Cas12a (Ali et al., 2020); iSCAN‐V2, a one‐pot system that combines RT‐RPA and CRISPR/Cas12b (Aman et al., 2022) for the detection of SARS‐CoV‐2; Vigilant, a platform that relies on using a chimeric fusion between the nuclease‐dead Cas9 (dCas9) and VirD2 coupled with an ssDNA reporter as a detection complex (Marsic et al., 2021) and Bio‐SCAN (biotin‐coupled specific CRISPR‐based assay for nucleic acid detection; Ali et al., 2022).
Bio‐SCAN was recently developed as a highly sensitive, cost‐effective and easy‐to‐use platform that requires only a single guide RNA (sgRNA) and recombinant biotin‐labelled nuclease‐dead version of Cas9 (bio‐dCas9) to detect one‐step FAM‐labelled amplicon produced by RT‐RPA for a target sequence with commercially available streptavidin‐biotin‐based LFA strips (Ali et al., 2022). Bio‐SCAN previously demonstrated the ability to detect the human pathogen SARS‐CoV‐2 (Ali et al., 2022). Taking advantage of its simplicity, specificity and sensitivity, we wished to optimize the robust Bio‐SCAN system (Graphical Abstract) as a POCT platform for agriculture to facilitate crop breeding, plant gene editing, transgene detection and the early molecular diagnosis of phytopathogens to enhance crop production and food security.
Bio‐SCAN successfully detected the resistant alleles of Lr34 and Lr67 in wheat cultivars; the synthetic mutants SGR3, SGR5 and OsmALS in rice; the transgenic promoters Ubiquitin and cauliflower mosaic virus (CaMV) 35S; the plant pathogenic viruses tomato yellow leaf curl virus (TYLCV), tobacco mosaic virus (TMV) and potato virus Y (PVY) in Nicotiana benthamiana; the fungi Puccinia striiformis f. sp. tritici and Magnaporthe oryzae Triticum (MoT) in wheat and the bacterial pathogens Pseudomonas and Agrobacterium tumefaciens in N. benthamiana. This work illustrates the efficacy of Bio‐SCAN as a convenient detection platform that can assist screening during crop breeding, plant transgenesis, plant synthetic evolution and rapid detection of phytopathogens.
Results
Bio‐SCAN‐mediated precise detection of Lr34 alleles in wheat germplasm
To optimize Bio‐SCAN for agricultural purposes, we challenged our platform with the detection of a resistance allele in the wheat genome, one of the most complex plant genomes on Earth. The resistance allele of the Lr34 locus is widely used in breeding programs to confer resistance against common wheat diseases caused by the pathogens Puccinia triticina, P. striiformis and Blumeria graminis. The resistance Lr34 protein is characterized by two amino acid changes caused by a 3‐bp deletion (codon ‘TTC’ in exon 11), resulting in the absence of a Phe residue, as well as a SNP in exon 12 (replacing a Tyr residue with His; Figure 1a), which are sufficient to provide broad‐spectrum resistance (Krattinger et al., 2009).
Figure 1.
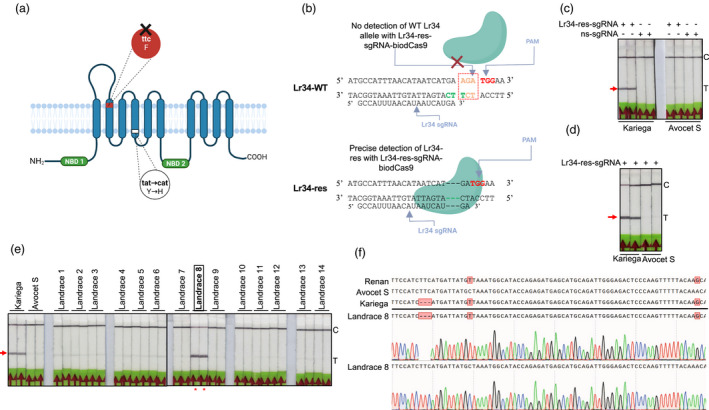
Screening for fungus‐resistant alleles in wheat. (a) Schematic diagram of the ABC transporter Lr34‐res. The resistant allele carries two changes affecting the transmembrane region. The region highlighted in red is the targeted ‘TTC’ deletion present in resistant cultivars. F stands for phenylalanine and is coded by the ‘ttc’ codon, Y represents tyrosine and it is coded by the ‘tat’ codon and H stands for histidine and is coded by the ‘cat’ codon. (b) Detection principle of the Lr34 resistant allele with the Bio‐SCAN module. The Lr34‐res‐sgRNA–biodCas9 ribonucleoprotein (RNP) complex specifically binds to and detects the resistant Lr34 allele, whereas the wild‐type Lr34 allele is not recognized by the Lr34‐res‐sgRNA–biodCas9. The Lr34‐res‐sgRNA and PAM sequences are indicated by arrows. Three dashes represent the deletion of three nucleotides present in the resistant allele. (c) Bio‐SCAN detects the Lr34 resistant allele (with the 3‐bp deletion). FAM‐labelled PCR amplicons from the genomic DNA of the Kariega (harbouring the Lr34 resistant allele) and Avocet S (with the wild‐type Lr34 allele) cultivars were incubated with the Lr34‐res‐sgRNA–biodCas9 RNP complex. Nonspecific sgRNA (ns‐sgRNA) was used as negative control. (d) RPA‐coupled Bio‐SCAN‐based detection of the Lr34 resistant allele. FAM‐labelled RPA amplicons from the genomic DNA of the Kariega and Avocet S cultivars were incubated with the Lr34 sgRNA–biodCas9 RNP complex. (e) Screening of the Lr34 resistant allele in 14 local cultivars through Bio‐SCAN‐based LFA. The varieties Kariega and Avocet S were used as positive and negative controls, respectively. BioSCAN detected the same Lr34‐resistant allele in cultivar Kariega and in landrace 8. C, control line; T, test line. Red arrow, position of the Lr34 allele detection. (f) Confirmation of the presence of the Lr34 resistant allele by Sanger sequencing in landrace 8. In addition to the 3‐bp deletion, Kariega and landrace 8 carry two extra SNPs (red boxes) that are absent in the susceptible cultivar Avocet S. However, the Renan cultivar, reported in the literature as susceptible, also harbours these two SNPs, but not the 3‐bp deletion. The electropherograms demonstrate the presence of both Lr34res and WT sequence.
Following the stringent rule of the binding of the Cas9‐sgRNA RNP complex to its target, sgRNA specific to the Lr34 resistance allele were designed by incorporating the 3‐bp deletion into the target seed region, which was very close to a PAM (Figure 1b). Next, a pair of primers flanking the target region in the Lr34 locus were designed to generate FAM‐labelled amplicons compatible with PCR and RPA reactions. As a proof‐of‐concept, the Lr34 target sequence was amplified by PCR, using genomic DNA extracted from a susceptible (Avocet S) and a resistant (Kariega) wheat variety as template and analysed the resulting PCR amplicons with the Bio‐SCAN platform to assess the sensitivity and specificity of detection. Bio‐SCAN precisely detected the presence of the resistance Lr34 allele, as evidenced by the presence of a band on the LFS at the test line (T line) for the Kariega cultivar that is absent in samples from the Avocet S cultivar (Figure 1c).
Sustainable agriculture requires point‐of‐care testing (POCT) methodologies to identify elite germplasm in the field, seed collections and distribution centers. Therefore, Bio‐SCAN platform was coupled with RPA‐based rapid isothermal amplification to detect the presence of Lr34 allele in the genomic DNA from the susceptible cultivar Avocet S and the resistant cultivar Kariega. As shown in Figure 1d, Bio‐SCAN successfully detected the resistance Lr34 allele in the Kariega genome, but not in the Avocet S genome.
To simplify the lengthy process of allele's selection, Bio‐SCAN offers an alternative method, for the detection of a known allele in wheat germplasm. Accordingly, the RPA‐based Bio‐SCAN platform was applied to detect the resistance Lr34 allele in 14 locally grown wheat varieties, using Avocet S and Kariega as negative and positive controls, respectively. Notably, Bio‐SCAN detected a sequence related to the resistance Lr34 allele at the T line in one local cultivar (Figure 1e). To validate the presence of the resistance Lr34 allele, the region harbouring the 3‐bp deletion was amplified by PCR, cloned the amplicons in a sequencing vector and subjected individual clones to Sanger sequencing, using Kariega and Avocet S as a negative and positive control, respectively. Bio‐SCAN indeed determined that the local cultivar with a positive result for the resistance Lr34 allele harbours the same resistance allele as Kariega (Figure 1f). Next, Sanger sequencing results, confirmed the resistant Lr34 allele (as defined by the 3‐bp deletion) with a frequency of 30% in landrace 8, compared with a frequency of 33% in the resistant control Kariega, which appeared to carry both alleles as well (Figure S1). By contrast, the Avocet S variety contained no resistance allele sequences with a ‘TTC’ deletion, in agreement with the negative result by Bio‐SCAN (Figure 1f).
To confirm and expand the efficacy of the Bio‐SCAN platform to detect elite alleles in crop germplasm, we used the resistance allele of the Lr67 locus, which confers resistance to fungal pathogens. The resistant Lr67 resistant allele has two amino acid substitutions in the encoded protein: a glycine‐to‐arginine (G144R) and a valine‐to‐leucine (V387L; Moore et al., 2015).
For detection confirmation, Lr67‐res allele‐like sequences and its specific sgRNAs were custom synthesized to detect each SNP. RPA‐based Bio‐SCAN efficiently and precisely detected both SNPs for the resistance Lr67 allele with two different sgRNAs for each target in synthetic Lr67 samples (Figure S2). While, Lr67‐res specific sgRNA did not detect a nonspecific FAM‐labelled RPA product. Also, Bio‐SCAN with sgRNA specific to Lr67 resistance allele did not detect the presence of Lr67‐res allele in the available 14 landraces and cultivars Kariega and Avocet S (Figure S3).
Our established Bio‐SCAN assay thus demonstrated its sensitivity to precisely detect changes as small as 1 and 3 bp in the hexaploid wheat genome and that it could provide a simple and low‐cost platform to screen potential candidates in germplasm collections and to include elite alleles into breeding programs.
Bio‐SCAN precisely detects synthetically evolved herbicide‐resistant alleles
Bio‐SCAN can precisely detect naturally occurring alleles (Ali et al., 2022). We wished to optimize Bio‐SCAN to detect ‘synthetically evolved’ herbicide‐resistant alleles. To this end, three rice lines SGR3 (SF3B1‐GEX1A‐Resistant 3), SGR5 (SF3B1‐GEX1A‐Resistant 5) and OsmALS (bispyribac‐resistant) previously generated by our group (Butt et al., 2019, 2020) were selected. Line SGR3 harbours a deletion of a Lys residue at position 1050 (K1050–) that evolved from the wild type (WT) via a deletion of the corresponding ‘AAG’ codon, while line SGR5 differs from the WT by a H1048Q substitution and a K1049 deletion (Figure 2a). The SGR3‐sgRNA and SGR5‐sgRNA were designed and produced. The target sequences were amplified with RPA and subjected to Bio‐SCAN detection with their respective sgRNAs, using WT genomic DNA as a negative control. As shown in Figure 2b, Bio‐SCAN detected the SGR3‐ and SGR5‐specific alleles, but returned a negative result with WT DNA.
Figure 2.
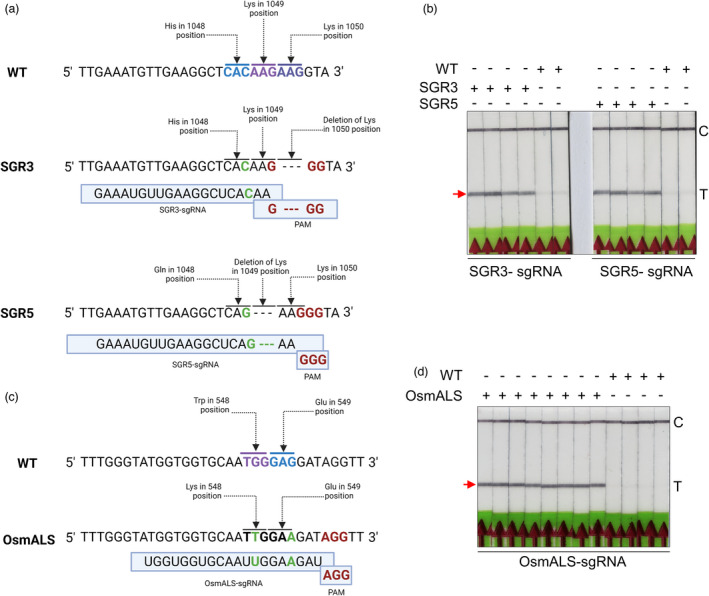
Detection of synthetically produced herbicide resistance mutations in rice. (a) Partial sequence alignment of the evolved gene SF3B1 in rice lines SGR3 and SGR5 and in the wild type (WT). SGR3 harbours a deletion of residue K1050. SGR5 differs from the WT by a H1048Q substitution and a deletion of residue K1049. (b) Bio‐SCAN specifically detects both synthetically evolved sequences in the SGR3 and SGR5 lines with their respective sgRNAs. Neither SGR3‐sgRNA nor SGR5‐sgRNA detected the WT sequence for the SF3B1 gene. (c) Partial sequence alignment of ALS in the mutant and the wild type. mALS differs from the WT protein by a W548L substitution and a silent G‐to‐A substitution that does not affect the E549 residue. (d) Bio‐SCAN specifically detects the mALS sequence. C, control line; T, test line. The arrowhead indicates the location of the expected band at the T line.
To determine whether the Bio‐SCAN assay can be applied to other induced mutant sequences, a new sgRNA that recognizes the synthetically evolved OsALS mutant (OsmALS; Figure 2c) was designed. As with SGR3 and SGR5 above, Bio‐SCAN efficiently detected the OsmALS allele sequence only in the genome of evolved herbicide‐resistant rice plants, but not in the WT rice genome (Figure 2d).
These results established that Bio‐SCAN can specifically recognize evolved sequences with a single nucleotide change and can provide a simple tool to confirm specific alleles created through directed evolution methodologies.
Bio‐SCAN detects most of GMOs
Public perception and governmental regulations regarding GMOs for human consumption are very extensive. In some countries, food, seeds and plant‐based material trading are regularly tested for the presence of GMOs and genome‐edited crops. (Turnbull et al., 2021). Such tests mainly rely on the detection in laboratory settings of transgenic DNA or immunological methods, such as ELISA, to detect proteins encoded by the transgenes (Salisu et al., 2017). As an alternative, we propose Bio‐SCAN as a quick protocol to detect the commonly used cauliflower mosaic virus (CaMV) 35S promoter, used to express transgenes in dicot plants and the maize Ubiquitin promoter (Ubipro), which is primarily used in monocots for the same purpose.
To test the efficacy of Bio‐SCAN for the detection of commonly used transgenic sequences, total genomic DNA was extracted from transgenic Nicotiana benthamiana plants harbouring a T‐DNA with both CaMV 35S and Ubipro (Figure 3a). The target sequences was amplified via RPA, followed by a Bio‐SCAN assay on the resulting amplicons. sgRNAs complementary to the CaMV 35S and Ubi promoters specifically detected the transgenic sequences, as shown by the appearance of a test band on LFS. Separate Bio‐SCAN assays with both sgRNAs failed to produce a positive test band in samples amplified from genomic DNA extracted from nontransgenic N. benthamiana plants (Figure 3b).
Figure 3.
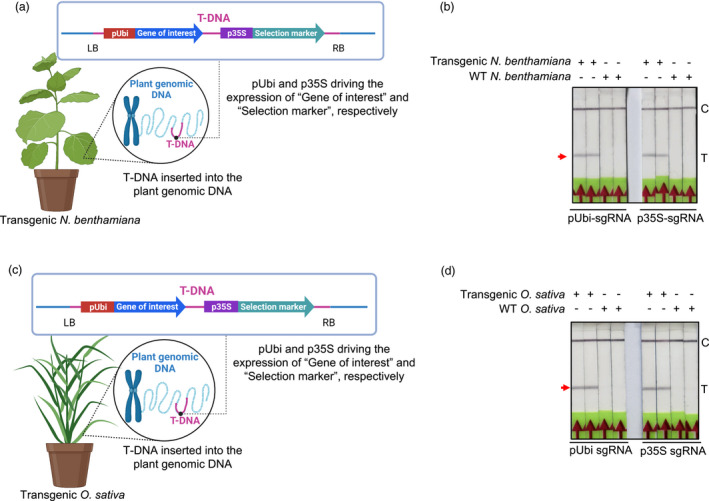
BioSCAN efficiently detects transgenic sequences in plants. (a and c) Schematic diagram of the T‐DNA harbouring the CaMV 35S and Ubiquitin promoters. Target sequences were amplified via RPA using FAM‐labelled primers. (b) Bio‐SCAN detects the Ubiquitin and CaMV 35S promoters in genomic DNA isolated from transgenic Nicotiana benthamiana plants. Nontransgenic N. benthamiana plants were used as control. (d) Detection of the transgene in rice plants. Bio‐SCAN detects the Ubiquitin and CaMV 35S promoters in transgenic rice, but not in nontransgenic plants. C, control line; T, test line.
The same Bio‐SCAN assay was performed on total genomic DNA extracted from a transgenic rice line with a T‐DNA harbouring both the Ubi and CaMV 35S promoters (Figure 3c). Each target sequence was amplified with their respective set of RPA primers and subjected to their individual sgRNA‐coupled Bio‐SCAN assay, using genomic DNA extracted from nontransgenic rice plants as control. As shown in Figure 3d, Bio‐SCAN successfully detected both promoter sequences from the genomic DNA of stably transformed transgenic rice plants, but not in nontransgenic plants.
These results confirm that the Bio‐SCAN platform can be used as a cost‐effective field‐deployable easy detection system that can fulfil the requirements of quick screening of GMOs for public concerns and food security established by governments worldwide (Bak and Emerson, 2020; Christensen and Quail, 1996).
Bio‐SCAN is a convenient tool for the rapid detection of phytopathogens
Phytopathogens are one of the main constraints in crop production, whose effects are exacerbated by climate change (Bebber et al., 2013). According to the Food and Agriculture Organization (FAO), plant diseases cost the global economy of around 220 billion dollars each year (Editorial, 2021). Therefore, early, fast, accurate and low‐cost diagnostics methods are required to support disease management through pathogen identification, treatment and monitoring (Buja et al., 2021).
Detection of plant viruses
We selected the DNA virus TYLCV and the RNA viruses TMV and PVY for detection with the Bio‐SCAN platform in N. benthamiana plants. Individual sgRNAs were targeting the sequence encoding the coat protein (CP) of each virus were designed. As a proof‐of‐concept for the functionality of our platform in detecting phytopathogenic viral nucleic acid sequences, Bio‐SCAN platform was tested for the detection of TYLCV, TMV and PVY using PCR amplicons obtained using plasmids carrying each virus genome as a template. Bio‐SCAN specifically detects each amplicon with its respective sgRNA (Figure 4a). Next, N. benthamiana plants were infected with infectious clones for each virus via Agrobacterium (Agrobacterium tumefaciens)‐mediated infiltration. Seven days after infiltration, total DNA (for TYLCV) or total RNA (for TMV and PVY) was extracted from infected plants. To detect TYLCV genomic DNA, we applied an RPA‐based Bio‐SCAN assay with an sgRNA targeting the sequence encoding the TYLCV coat protein. As controls, wild‐type plants and plants infected with the TMV were used as control. The RPA‐based Bio‐SCAN module detected the TYLCV genome with 100% specificity and did not return a positive T band when tested with genomic DNA from WT plants or with plants infected with another virus (TMV; Figure 4b).
Figure 4.
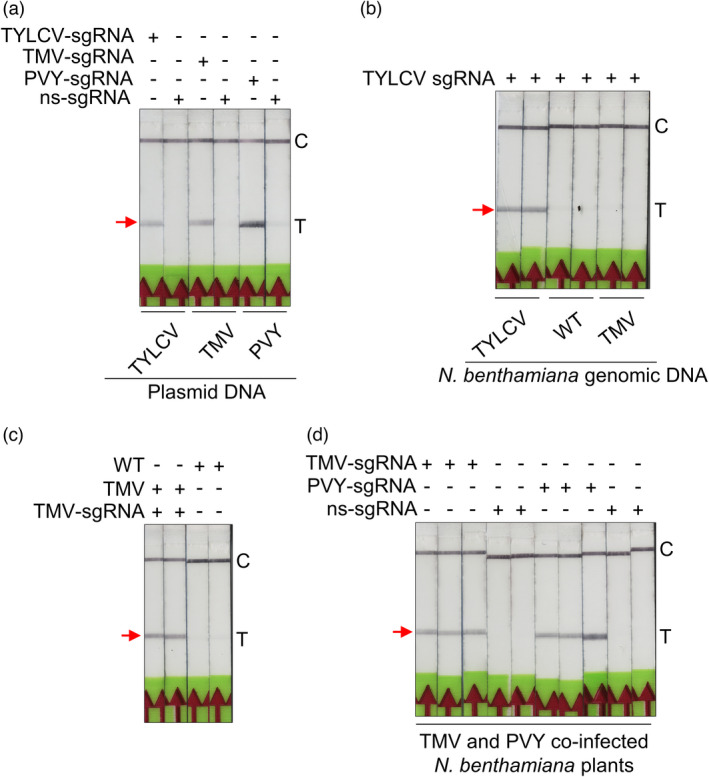
Efficient BioSCAN‐based detection of phytopathogens. (a) Bio‐SCAN specifically detects the sequences from the viral genomes of TYLCV, TMV and PVY when using plasmid DNA. The FAM‐labelled amplicons were generated through PCR. BioSCAN specifically detected TYLCV, TMV and PVY with their respective sgRNAs. (b) Bio‐SCAN specifically detects the TYLCV virus genome in samples isolated from TYLCV‐infected Nicotiana benthamiana plants and amplified with RPA. (c) Bio‐SCAN specifically detects the TMV virus RNA genome in RT‐RPA amplicons from N. benthamiana samples infected with TMV infectious clones. (d) Bio‐SCAN detects both the TMV and PVY RNA genomes in samples isolated from N. benthamiana plants co‐infected with TMV and PVY. Noninfected N. benthamiana plants were used as a control. C, control line; T, test line. Arrowheads indicate the location of the expected band at the T line on the LFA strip.
Encouraged by this result, we attempted to test whether Bio‐SCAN might exhibit sensitivity and specificity to detect the TMV genomic RNA. Accordingly, total RNA extracted from plants infected with TMV and PVY and from uninfected plants was subjected to RT‐RPA‐based Bio‐SCAN assay with a TMV‐specific sgRNA to detect the TMV genomic RNA on plants only infected with TMV. Bio‐SCAN specifically detected the TMV RNA with the TMV‐specific sgRNA but did not observe the formation of a positive T band when using a PVY‐specific sgRNA, indicating the specificity of the assay (Figure 4c). To ascertain the applicability of Bio‐SCAN to detect multiple viruses, N. benthamiana plants were co‐infected with TMV and PVX, using nonspecific sgRNA as a negative control. Bio‐SCAN successfully detected both viruses in total RNA extracted from the same plant using the respective primer sets and sgRNAs for TMV or PYX, but not with nonspecific sgRNA (Figure 4d).
Detection of fungal pathogens
For this purpose, we selected the two most devastating fungal pathogens [Puccinia striiformis f. sp. tritici and Magnaporthe oryzae Triticum (MoT)] infecting wheat. For the detection of P. striiformis f. sp. tritici, specific sequences were selected to avoid cross‐contamination from the wheat genome or from other fungal species. Next, two sets of FAM‐labelled RPA primers and two sgRNAs targeting the fungus‐specific internal transcribed spacer (ITS) region (Zhao et al., 2007) and the fungal β‐tubulin gene (Lihua et al., 2008) were designed for Bio‐SCAN assay with synthetic DNA to test the activity and specificity of sgRNAs. The ITS sgRNA against the ITS and the β‐tubulin FAM‐labelled amplicons, showed 100% specificity for the ITS DNA fragment (Figure 5a). We conducted the same experiment to test the fungal β‐tubulin sgRNA, which detected the β‐tubulin fragment, but not the ITS fragment, confirming high specificity (Figure 5a). To make sure that Bio‐SCAN can detect P. striiformis f. sp. tritici DNA in total genomic DNA extracted from potentially infected wheat plants, synthetic DNA was spiked in wheat genomic DNA and repeated the Bio‐SCAN assay. As shown in Figure 5b, Bio‐SCAN specifically detected P. striiformis tritici DNA in wheat genomic DNA spiked with synthetic P. striiformis f. sp. tritici DNA, but did not return a positive result on wild‐type wheat genomic DNA alone.
Figure 5.
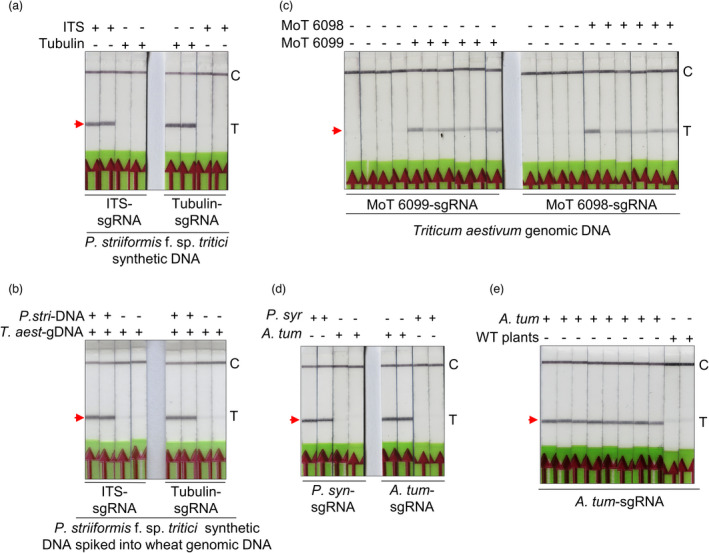
Bio‐SCAN efficiently detects pathogenic fungi of wheat and bacterial pathogens in Nicotiana benthamiana. (a) Bio‐SCAN detects specific sequences (ITS and β‐tubulin) of Puccinia striiformis f. sp. tritici in synthetic DNA. The pathogen‐specific sequence was detected on LFA strips via RPA‐based Bio‐SCAN assay. A nonspecific sgRNA was used as a control. (b) BioSCAN‐based detection of Puccinia striiformis f. sp. tritici in wheat genomic DNA. Synthetic Puccinia striiformis f. sp. tritici DNA was spiked into wheat genomic DNA and detected via RPA‐Bio‐SCAN. P. stri (Puccinia striiformis f. sp. tritici): T. aest (Triticum aestivum). (c) Detection of Magnaporthe oryzae Triticum in wheat samples. Wheat tissues infected with M. oryzae triticum were used for total genomic DNA isolation. The extracted genomic DNA was subjected to an RPA‐based Bio‐SCAN assay. Bio‐SCAN specifically detected both conserved sequences (MoT 6099 and MoT 6098) of M. oryzae Triticum in samples isolated from infected wheat plants. Noninfected wheat tissues were used as a control. (d) Bio‐SCAN detects specific sequences, AvrE of Pseudomonas syringae and ChvA of Agrobacterium tumefaciens in synthetic DNA. P. syn (Pseudomonas syringae); A. tum (Agrobacterium tumefaciens). The pathogen‐specific sequence was detected on LFA strips via RPA‐based Bio‐SCAN assay. A nonspecific sgRNA was used as a control. (e) BioSCAN‐based detection of Agrobacterium tumefaciens in the genomic DNA of N. benthamiana plants. Total DNA is isolated from agroinfiltrated N. benthamiana plants and detected via RPA‐Bio‐SCAN. Noninfiltrated N. benthamiana tissues were used as a control. C, control line; T, test line. The arrowheads indicate the location of the expected band at the T line on the LFA strip. [Correction added on 27 October 2022, after first online publication: Figure 5 has been corrected in this version.]
Next, to expand the utility of Bio‐SCAN to fungal pathogens in infected wheat plants, we selected another most economically important fungal pathogen, M. oryzae Triticum (MoT) of wheat (Islam et al., 2016). The MoT‐6098 and MoT‐6099 sequences were recently proposed as MoT pathogen‐specific biomarkers (Kang et al., 2021). Thus two sgRNAs targeting the conserved MoT‐6098 and MoT‐6099 sequences, were, respectively. Total DNA from MoT‐infected wheat plants was subjected to an RPA‐based Bio‐SCAN assay. DNA isolated from noninfected wheat plants was used as a negative control. Both sgRNAs specific for M. oryzae Triticum detected MoT infection in wheat tissues (Figure 5c).
Detection of pathogenic bacteria
Lastly, we tested whether Bio‐SCAN could detect the bacteria Pseudomonas syringae and A. tumefaciens in N. benthamiana plants in a sensitive and specific manner. To confirm, synthetic DNA fragments of P. syringae and A. tumefaciens were subjected to RPA‐based isothermal amplification with FAM‐labelled primers specific for each bacterial species. Bio‐SCAN assay with related sgRNA was applied to detect the species‐specific FAM‐labelled amplicons. Our results confirm successful and precise detection of both, P. syringae and A. tumefaciens sequences (Figure 5d). To demonstrate that our optimized system can detect bacterial pathogens in samples from infected plants, total genomic DNA isolated from A. tumefaciens infiltrated N. benthamiana plants was subjected to RPA‐based Bio‐SCAN assay. As shown in Figure 5e, Bio‐SCAN assay successfully detected Agrobacterium‐specific sequence on LFA from the DNA samples isolated from N. benthamiana plants.
Collectively, these results demonstrate that Bio‐SCAN is a powerful and specific method for detecting pathogens independently of their nature: viruses, bacteria or fungi. The wide range of applications for Bio‐SCAN makes it an essential low‐cost tool that can ease the complexity of screening protocols in crop breeding and crop genetic engineering and show that it can assist in food security regulations for GMOs and enhance crop diagnostics practices in the field.
Discussion
The incorporation of economically significant agronomic traits/alleles and the management of diseases caused by phytopathogens are two strategies for achieving sustainable agriculture and food security. In this study, we demonstrated that Bio‐SCAN is a new, convenient, rapid and robust field deployable detection method which is significantly advantageous over other currently available tools to detect elite alleles and phytopathogens. Bio‐SCAN can be efficiently applied in fundamental plant biology research and disease prevention in crop plants. It is rapid, sensitive, specific, low‐cost and easily adaptable. Multiple phytopathogens detection platforms have recently been developed, such as for direct detection with protein‐based serological and nucleic acid‐based isothermal amplification point‐of‐care tests; however, the complexity of the reactions, their low sensitivity, relative nonspecificity and the time commitment associated are limiting factors restricting the broad use of these methods in agriculture (Fujita et al., 2021).
Importantly, recent advances in CRISPR/Cas‐based systems for diagnostics can now expedite the development and deployment of tests in the field (Henry, 2014). However, despite the development of various CRISPR‐based detection platforms for phytopathogens, no such platform is currently available for the detection and selection of germplasm or elite varieties. A recent study demonstrated the successful use of Bio‐SCAN diagnostics for the sensitive and specific detection of variants in the genomes of RNA viruses with a sensitivity down to a single nucleotide (Ali et al., 2022), confirming the potential use of Bio‐SCAN for the detection of point mutations for known elite alleles in crop plants.
Elite germplasm and disease prevention constitute the core of modern agriculture. Over time, researchers have developed genetic collections of unique mutations, alleles, genes and chromosomal rearrangements, translocations and inversions (Ul Haq and Ijaz, 2020). The search for an elite allele that brings a specific agronomic trait for food, feed, fibre, forages and industrial use from global landraces or wild species is laborious and time‐consuming. The recent developments in sequencing technologies have helped tremendously in identifying the specific genetic factors present in a particular landrace that underlie these agronomic traits. However, identifying one or several traits by sequencing thousands of individuals in a vast germplasm collection and breeding programs is not efficient for agricultural purposes. Alternatively, allele/germplasm‐specific molecular markers or germline‐specific random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP) and protein markers can be used to identify elite germplasm, but they are laborious, time‐consuming and suffer from low throughput (Gao et al., 2010; Souframanien and Gopalakrishna, 2004).
Nevertheless, there is no molecular tool to detect point mutations in the plant genome, specifically in a point‐of‐care setting. Bio‐SCAN can easily detect known alleles in a set of landraces or in a wild relative of a particular crop. As a proof of concept, we used Bio‐SCAN to successfully detect the naturally occurring resistant alleles for the Lr34 and Lr67 loci in wheat, as well as the synthetically evolved SGR3, SGR5 and OsmALS herbicide‐resistant alleles in rice plants.
Multiple CRISPR/Cas systems have been harnessed to develop point‐of‐care nucleic acid diagnostics systems. Most of these CRISPR‐Dx systems rely on the collateral activity of type‐V and ‐VI Cas nucleases triggered by target recognition. Taking advantage of the simplicity and ease of adaptability of CRISPR‐Dx systems, Cas12 and Cas13 variants have been used to detect transgenes and phytopathogens (fungi and viruses; Abudayyeh et al., 2019; Aman et al., 2020b; Mahas et al., 2021). In contrast to the collateral activity of Cas12 and Cas13 for nucleic acid detection, Cas9 and its catalytically inactive variant dCas9 were shown to detect pathogen sequences directly or when coupled to an LFA (Liu et al., 2022; Wang et al., 2020). Although significant progress has been made in the use of Cas9, Cas12 and Cas13 in detection, these systems are still complicated, require additional steps and require sensitive and unstable reagents. These concerns make most of these systems impractical for their large‐scale field application in agriculture.
A recent study (Kang et al., 2021) has reported a CRISPR‐Cas12‐based method to detect the wheat pathogen M. oryzae Triticum. The developed detection method is based on two independent RPA reactions for target amplification that requires multiple sample handling steps and three probes: ssDNA, FAM labelled left primer and biotin‐labelled right primer. Furthermore, Kang et al. (2021) method is Cas12a dependent that relies on nonspecific digestion of an ssDNA probe used in second round RPA. Thus, contamination of any nuclease can lead to a false positive result. Two‐step RPA, multi‐step processing and additional oligonucleotides/probes add extra complexity to the diagnostic setup and increase the cost of the detection method.
In contrast, Bio‐SCAN requires only one RPA reaction. No probe is required (bio‐dCas9 is already biotinylated). Also, only one FAM‐labelled primer (either left or right) is enough for FAM labeling via RPA and LFA‐based detection. Moreover, the results on the lateral flow strip obtained in Bio‐SCAN platform are direct and coherent with commercially available lateral flow devices: a control line and a test line, directly confirming the presence or absence of the target nucleic acid. In contrast, in the method by Kang et al. (2021), the presence of pathogen (Cas12a‐based digestion of ssDNA) did not develop any band on the test line because of the lack of biotin‐ and FAM‐ labelled fragments, while the absence of pathogen will give band at the test line. Furthermore, Bio‐SCAN is very stable, user‐friendly, easy to interpret and can be adopted for field‐based applications. Importantly, Bio‐SCAN can detect any nucleic acid sequence in <1 h from sample collection to result output and require no equipment and laboratory settings (Ali et al., 2022).
The successful detection of natural and synthetically evolved alleles, transgenic DNA and phytopathogens, demonstrate the potential and versatile use of Bio‐SCAN module in the field of agriculture. However, target selection, primers design for RPA‐based amplification, sgRNAs synthesis and LFA strips must be optimized to reduce false detection, minimize the background and enhance specificity in field experiments.
In conclusion, our developed Bio‐SCAN toolkit fulfils all the basic features of a field‐deployable point‐of‐care detection system. Bio‐SCAN will facilitate the rapid, sensitive and specific mass screening of transgenes, synthetic modifications and early detection of pathogens for better crop management and sustainable agriculture.
Materials and methods
Plant materials
Nicotiana benthamiana, rice (Oryza sativa cultivar Nipponbare) and wheat (Triticum aestivum cultivars Avocet S and Kariega and local landraces) plants were grown on soil in the greenhouse at 28 °C and under an 11‐h‐light/13‐h‐dark photoperiod. The seeds were germinated on a 3 : 1 Stender® soil substrate : rocks mix.
Leaves were collected from 5‐week‐old wild‐type (WT) and transgenic N. benthamiana and O. sativa plants for extraction of genomic DNA for the detection of the Ubi and 35S CaMV promoters. Leaves from O. sativa WT and O. sativa plants containing the synthetically evolved sequences SGR3, SGR5 and OsmALs were collected for extraction of genomic DNA for their detection.
Different wheat landraces were germinated, and the leaves were harvested 5 weeks later for extraction of genomic DNA to identify the Lr34‐res allele.
For virus detection, 5‐week‐old N. benthamiana plants were inoculated with infectious virus clones via agroinfiltration. The plant samples were collected after 1 week of agroinfiltration with the virus. Total genomic DNA was isolated using our lab‐established protocol (Ali et al., 2015).
Protein purification and in vitro sgRNA transcription
The plasmid pET28a‐dCas9‐AviTag‐BirA was used to produce the biotinylated dCas9‐AviTag protein (Ali et al., 2022). Since the binding of Cas9‐sgRNA RNP complex to the target DNA relies on the presence of a protospacer adjacent motif (PAM, NGG) sequence and does not tolerate mismatches in the seed region next to PAM (Sternberg et al., 2014), the sgRNAs for the detection were designed targeting the respective regions with specific focus on PAM or the seed region of the target DNA sequence.
Regarding the detection of resistant alleles in wheat germplasm, the Lr34‐res sgRNA was designed by incorporating the 3 bp deletion into the target seed region in close proximity to PAM (Figure 1b). Similarly, the Lr67‐res sgRNAs were intended to detect one single nucleotide conversion in two different regions: of G to C (target in the 144 position) and G to T (target in the position 387) in the seed region. For the detection of synthetically evolved herbicide‐resistant alleles, the SGR3‐sgRNA (5′ GAAAUGUUGAAGGCUCACAA) was designed by incorporating the 3 bp deletion (AAG) that generates a new PAM different from the WT sequence, the SGR5‐sgRNA (5′ GAAAUGUUGAAGGCUCAGAA) was designed by incorporating the 3 bp deletion (AAG) in the seed region (Figure 2b), the OsmALS‐sgRNA (5′ UGGUGGUGCAAUUGGAAGAU 3′) incorporates two nucleotide differences with the wild‐type sequence in the seed region (Figure 2c). The sgRNAS for the detection of GMOs and pathogens were designed to detect DNA sequences that are exogenous to the plant and that correspond to particular elements of the transgene (such as the CaMV 35 S or Ubiquitin promoter) or the pathogen.
For the production of sgRNAs, DNA fragments were designed and synthesized as g‐blocks containing the target recognition, the T7 promoter at the 5′ end and the Cas9 binding scaffold at the 3′ end. The g‐blocks were PCR amplified and purified using the QIAquick PCR Purification Kit (QIAGEN). DNA concentration and quality were determined with a NanoDrop UV/Vis spectrophotometer and electrophoresis on a 1% (w/v) agarose gel. To obtain sgRNAs, 1 μg of purified DNA was transcribed in vitro at 37 °C for 8 h using a HiScribe T7 Quick, high‐yield RNA synthesis kit (E2050S; New England Biolabs, Inc., Ipswich, MA, USA) according to the manufacturer's instructions. The resulting sgRNA was purified with the Direct‐zol RNA MiniPrep Kit (Zymo Research, Irvine, CA, USA). The stability of sgRNAs was confirmed on a 2% (w/v) agarose gel run in Tris borate EDTA (TBE) buffer before sgRNA concentration was determined with a NanoDrop UV/Vis spectrophotometer. The sgRNAs were diluted into a working stock of 5 μm, flash‐frozen and stored at −80 °C. The primer sequences used for PCR amplification are provided in Table S1, and the sequences of g‐blocks are given in Table S2.
Detection of PCR‐amplified products with the Bio‐SCAN platform
Target DNA was amplified by PCR using the FAM‐labelled primers listed in Table S1. For the Bio‐SCAN assay, a 50‐μL reaction was set up in a PCR tube containing 100 ng of the FAM‐labelled amplicon, 8 μL of 0.15 μm dCas9‐bio (250 nm), 2.5 μL of 5 μm sgRNA and 5 μL of 10× NEB Buffer 3.1, with the final volume adjusted to 50 μL with ultrapure nuclease‐free water. The reaction mixture was incubated for 5 min followed by 2 min at 60 °C. Briefly, 50 μL of dipstick assay buffer (HybriDetect Milenia biotec, Milenia Biotec GmbH, Gießen, Germany) was added to each reaction and gently mixed. HybriDetect Dipsticks were placed in the reaction tube with the Gold‐NP anti‐FAM antibody region immersed in the mixture, which starts flowing by capillarity toward the absorption pad. The dipsticks were removed after the control line appeared, and the result was interpreted and recorded immediately. For a negative result, only the control band, which is closer to the absorption pad, will appear; for a positive result, two bands will appear, the control band and the test band, which is closer to the Gold‐NP anti‐FAM antibody region.
Detection of RPA and RT‐RPA‐amplified product with the Bio‐SCAN platform
The RPA and RT‐RPA reactions were performed with the Twist‐Amp basic kit (Twist‐Dx Limited, Maidenhead, UK) according to the manufacturer's instructions. For the RPA reactions, the lyophilized RPA reagents in one tube were resuspended in 29.5 μL of rehydration buffer, to which 1 μL of 25 μm FAM‐labelled forward primer, 1 μL of 25 μm unlabeled reverse primer and 6.5 μL of ultra‐pure nuclease‐free water was added. For the RT‐RPA reactions, one tube of lyophilized RPA reagent was resuspended in 29.5 μL of rehydration buffer, after which 0.5 μL of SuperScript IV reverse transcriptase, 0.5 μL of RNase H, 0.5 μL of RNase Out, 5 μL of nuclease‐free water, 1 μL of 25 μm FAM‐labelled forward and 1 μL of 25 μM unlabeled reverse primer was added. The mixture was homogenized and divided in two tubes, and 2 μL of magnesium acetate (280 mm) was added and mixed by vortexing. Then, 4 μL of the sample of interest (for RT‐RPA: total gRNA for the TMV, TuMV, PVX and Nicotiana benthamiana WT; for RPA: total gDNA samples) was added. The mixture was mixed thoroughly and briefly collected by centrifugation before incubation at 42 °C for 15 min. The RT‐RPA DNA products were quality‐assessed on a 1.5% (w/v) agarose gel. A 50‐μL reaction was set up with 2 μL of RT‐RPA product, 250 nm (8 μL of 0.15 μm stock) of dCas9‐bio, 2.5 μL of 5 μm sgRNA and 5 μL 10× NEB Buffer 3.1, before the final volume was adjusted to 50 μL with ultrapure nuclease‐free water. The mixture was mixed thoroughly and briefly collected by centrifugation before incubation at 37 °C for 5 min and then at 60 °C for 2 min. Then, 50 μL of dipstick assay buffer (HybriDetect Milenia biotec) was added. The mixture was mixed and centrifuged briefly, the lateral flow assay was performed and the results were interpreted as described earlier.
Total DNA extraction
Total DNA from plant tissue was extracted using our established protocol (Ali et al., 2015). Briefly, frozen plant tissue was ground and collected in an Eppendorf tube. Extraction buffer (0.1 m Tris–HCl pH 8.0, 1 mm EDTA, 0.1 m NaCl, 0.1 m LiCl, 0.1 m β‐mercaptoethanol; 0.4% [w/v] RNase I) was added in a proportion of 500 μL per 300 μL of ground tissue. Samples were vortexed for 1 min and incubated at 65 °C for 10 min. One volume of phenol:chloroform:isoamyl alcohol (25 : 24 : 1) was added to each sample and vortexed for 1 min. The samples were centrifuged at 13500 g at room temperature for 10 min. The supernatant was transferred to an Eppendorf tube containing 500 μL of chloroform, and the samples were vortexed and centrifuged for 10 min at 13500 g at 4 °C. The supernatant was transferred to an Eppendorf tube containing two volumes of pre‐chilled 99% ethanol and 0.1 volume of sodium acetate before gentle mixing by inversion. For ethanol precipitation, the tubes were placed at −20 °C overnight. The samples were then centrifuged at 13500 g at 4 °C for 30 min. The pellets were washed twice with 1 mL of 70% (v/v) ethanol as follows: vortex for 1 min, centrifugation for 30 min at 13500 g and 4 °C, removal of the supernatant and air‐dry the pellet for 20 min. The gDNA samples were assessed for concentration and quality using a NanoDrop UV/Vis spectrophotometer and 1% (w/v) agarose gels.
Total RNA extraction
Total RNA was extracted from plant tissue samples using the Direct‐zol™ RNA Miniprep Plus system (Zymo Research) according to the manufacturer's recommendations: 800 μL of TRIzol® was added to 300 μL of tissue, mixed until homogenized and centrifuged for 30 s at 13500 g and room temperature. The supernatant was transferred to a new tube, to which an equal volume of 99% ethanol was added and mixed. The mixture was transferred onto a Zymo‐Spin™ IIICG Column in a collection tube and centrifuged at 13500 g for 30 s. Each column was pre‐washed twice with 400 μL Direct‐zol™ RNA Pre wash buffer and centrifuged for 30 s at 13500 g. Each column was then washed with 700 μL of wash buffer and centrifuged for 2 min. The column was transferred to a new tube, and the RNA was eluted with DNase/RNase‐Free Water by centrifugation. The samples were assessed for concentration and quality using a NanoDrop UV/Vis spectrophotometer and 2% (w/v) agarose gel.
Conflict of interest
The authors declare no competing financial interests.
Authors contributions
MM and ZA conceived the research. MM, ZA and ES designed the research. ES, TI and ZA built and conducted the research. MM, ZA, TI and ES wrote and edited the article with input from all authors.
Funding
This work was supported, in part, by BAS/1/1035‐01‐01 grant from the KAUST to MM. TI received partial funding through the OFANS project of the Krishi Gobeshona Foundation of Bangladesh.
Supporting information
Figure S1 Alignment of Lr34 Sanger sequencing.
Figure S2 Detection of Lr67 fungus‐resistant allele in wheat.
Figure S3 Bio‐SCAN‐based Lr67‐res detection in wheat.
Table S1 Primers used in this study.
Table S2 The g‐blocks used in this study.
Acknowledgements
We would like to thank professor Simon Krattinger and his team at the Biological and Environmental Science and Engineering Division, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia, for providing the Lr34 referenced wheat germplasm. We would like to thank Nur Uddin Mahmud of the Institute of Biotechnology and Genetic Engineering (IBGE), Bangabandhu Sheikh Mujibur Rahman Agricultural University, Gazipur, Bangladesh, for assistance in extracting DNA from wheat blast fungus, Magnaporthe oryzae Triticum isolates. We would like to thank Dr. Haroon Butt at the Biological and Environmental Science and Engineering Division, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia, for providing the SGR3, SGR5 and OsmALS rice lines. We would like to thank Dr Nasir Saeed, Wheat Biotechnology Lab, NIBGE, Pakistan for providing wheat landraces and genomic DNA samples. We also thank members of the genome engineering and synthetic biology laboratory for insightful discussions and technical support.
References
- Abudayyeh, O.O. , Gootenberg, J.S. , Kellner, M.J. and Zhang, F. (2019) Nucleic acid detection of plant genes using CRISPR‐Cas13. CRISPR J. 2, 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai, F. , Am, E.‐S. and Wm, E.‐O. (2015) Leaf rust resistance and molecular identification of Lr 34 gene in Egyptian wheat. J. Microbial Biochem. Technol. 07, 338–343. [Google Scholar]
- Ali, Z. , Abul‐Faraj, A. , Li, L. , Ghosh, N. , Piatek, M. , Mahjoub, A. , Aouida, M. et al. (2015) Efficient virus‐mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant, 8, 1288–1291. [DOI] [PubMed] [Google Scholar]
- Ali, Z. , Aman, R. , Mahas, A. , Rao, G.S. , Tehseen, M. , Marsic, T. , Salunke, R. et al. (2020) iscan: An Rt‐lamp‐coupled CRISPR‐Cas12 module for rapid, sensitive detection of Sars‐CoV‐2. Virus Res. 288, 198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, Z. , Sánchez, E. , Tehseen, M. , Mahas, A. , Marsic, T. , Aman, R. , Sivakrishna Rao, G. et al. (2022) Bio‐Scan: a CRISPR/dCas9‐based lateral flow assay for rapid, specific, and sensitive detection of Sars‐CoV‐2. ACS Synth. Biol. 11, 406–419. [DOI] [PubMed] [Google Scholar]
- Aman, R. , Mahas, A. and Mahfouz, M. (2020a) Nucleic acid detection using CRISPR/Cas biosensing technologies. ACS Synth. Biol. 9, 1226–1233. [DOI] [PubMed] [Google Scholar]
- Aman, R. , Mahas, A. , Marsic, T. , Hassan, N. and Mahfouz, M.M. (2020b) Efficient, rapid, and sensitive detection of plant rna viruses with one‐pot Rt‐Rpa–CRISPR/Cas12a assay. Front. Microbiol. 11, 610872. 10.3389/fmicb.2020.610872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman, Y. , Khalid, F. , Shaukat, M. , Mahmood, T. , Hasan, S.W. and Mirza, J.I. (2020c) Characterization of seedling and adult plant resistance to stripe rust in recombinant inbred lines derived from wheat landrace Pi388222 × Avocet cross. Plant Genet. Resour. 18, 11–18. [Google Scholar]
- Aman, R. , Marsic, T. , Sivakrishna Rao, G. , Mahas, A. , Ali, Z. , Alsanea, M. , Al‐Qahtani, A. et al. (2022) iscan‐V2: a one‐pot Rt‐Rpa–CRISPR/Cas12b assay for point‐of‐care Sars‐CoV‐2 detection. Front. Bioeng. Biotechnol. 9, 800104. 10.3389/fbioe.2021.800104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baenziger, P.S. (2016) Wheat Breeding and Genetics. Reference Module in Food Science. Lincoln, NE: Elsevier. [Google Scholar]
- Bak, A. and Emerson, J.B. (2020) Cauliflower mosaic virus (CaMV) biology, management, and relevance to GM plant detection for sustainable organic agriculture. Front. Sustain. Food Syst. 4, 00021. 10.3389/fsufs.2020.00021 [DOI] [Google Scholar]
- Batley, J. (2015) Plant Genotyping: Methods and Protocols. Crawley, Australia: Springer. [Google Scholar]
- Bebber, D.P. , Ramotowski, M.A.T. and Gurr, S.J. (2013) Crop pests and pathogens move polewards in a warming world. Nat. Clim. Change, 3, 985–988. [Google Scholar]
- Bedő, Z. and Láng, L. (2015) Wheat Breeding: Current Status and Bottlenecks. In Alien Introgression in Wheat: Cytogenetics, Molecular Biology, and Genomics ( Molnár‐Láng, M. , Ceoloni, C. and DoleŽel, J. , eds), pp. 77–101. Cham: Springer International Publishing. [Google Scholar]
- Broughton, J.P. , Deng, X. , Yu, G. , Fasching, C.L. , Servellita, V. , Singh, J. , Miao, X. et al. (2020) CRISPR–Cas12‐based detection of Sars‐CoV‐2. Nat. Biotechnol. 38, 870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buja, I. , Sabella, E. , Monteduro, A.G. , Chiriacò, M.S. , De Bellis, L. , Luvisi, A. and Maruccio, G. (2021) Advances in plant disease detection and monitoring: from traditional assays to in‐field diagnostics. Sensors, 21, 2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt, H. , Eid, A. , Momin, A.A. , Bazin, J. , Crespi, M. , Arold, S.T. and Mahfouz, M.M. (2019) CRISPR directed evolution of the spliceosome for resistance to splicing inhibitors. Genome Biol. 20, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt, H. , Rao, G.S. , Sedeek, K. , Aman, R. , Kamel, R. and Mahfouz, M. (2020) Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J. 18, 2370–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, A.H. and Quail, P.H. (1996) Ubiquitin promoter‐based vectors for high‐level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5, 213–218. [DOI] [PubMed] [Google Scholar]
- Ciuca, M. , Cristina, D. , Turcu, A.G. , Contescu, E.L. , Ionescu, V. and Saulescu, N.N. (2015) Molecular detection of the adult plant leaf rust resistance GeneLr34in romanian winter wheat germplasm. Cereal Res. Commun. 43, 249–259. [Google Scholar]
- Dakouri, A. , Mccallum, B.D. , Radovanovic, N. and Cloutier, S. (2013) Molecular and phenotypic characterization of seedling and adult plant leaf rust resistance in a world wheat collection. Mol. Breed. 32, 663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, G. , Zou, Q. , Chen, Y. , Wang, L. , Huang, G. , Cui, Y. , Ding, M. et al. (2019) The complete mitochondrial genome of Cochliobolus miyabeanus (Dothideomycetes, Pleosporaceae) causing brown spot disease of rice. Mitochondrial DNA B Resour. 4, 2832–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, X. , Yin, K. , Li, Z. , Lalla, R.V. , Ballesteros, E. , Sfeir, M.M. and Liu, C. (2020) Ultrasensitive and visual detection of Sars‐CoV‐2 using all‐in‐one dual CRISPR‐Cas12a assay. Nat. Commun. 11, 4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editorial, R. (2021) Pathogens, precipitation and produce prices. Nat. Clim. Change, 11, 635. [Google Scholar]
- Fujita, T. , Nagata, S. and Fujii, H. (2021) Protein or ribonucleoprotein‐mediated blocking of recombinase polymerase amplification enables the discrimination of nucleotide and epigenetic differences between cell populations. Commun. Biol. 4, 988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, L. , Ma, W. , Chen, J. , Wang, K. , Li, J. , Wang, S. , Bekes, F. et al. (2010) Characterization and comparative analysis of wheat high molecular weight glutenin subunits by SDS‐PAGE, RP‐HPLC, HPCE, and MALDI‐TOF‐MS. J. Agric. Food Chem. 58, 2777–2786. [DOI] [PubMed] [Google Scholar]
- Grote, U. , Fasse, A. , Nguyen, T.T. and Erenstein, O. (2021) Food security and the dynamics of wheat and maize value chains in Africa and Asia. Front. Sustain. Food Syst. 4, 617009. 10.3389/fsufs.2020.617009 [DOI] [Google Scholar]
- Henry, R. (2014) Genomics strategies for germplasm characterization and the development of climate resilient crops. Front. Plant Sci. 5, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M.T. , Croll, D. , Gladieux, P. , Soanes, D.M. , Persoons, A. , Bhattacharjee, P. , Hossain, M.S. et al. (2016) Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae . BMC Biol. 14, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R.A.C. and Naidu, R.A. (2019) Global dimensions of plant virus diseases: current status and future perspectives. Ann. Rev. Virol. 6, 387–409. [DOI] [PubMed] [Google Scholar]
- Kaminski, M.M. , Abudayyeh, O.O. , Gootenberg, J.S. , Zhang, F. and Collins, J.J. (2021) CRISPR‐based diagnostics. Nat. Biomed. Eng. 5, 643–656. [DOI] [PubMed] [Google Scholar]
- Kang, H. , Peng, Y. , Hua, K. , Deng, Y. , Bellizzi, M. , Gupta, D.R. , Mahmud, N.U. et al. (2021) Rapid detection of wheat blast pathogen Magnaporthe oryzae Triticum pathotype using genome‐specific primers and Cas12a‐mediated technology. Engineering, 7, 1326–1335. [Google Scholar]
- Khakimov, A. , Salakhutdinov, I. , Omolikov, A. and Utaganov, S. (2022) Traditional and current‐prospective methods of agricultural plant diseases detection: a review. IOP Conf. Series Earth Environ. Sci. 951, 012002. [Google Scholar]
- Krattinger, S.G. , Lagudah, E.S. , Spielmeyer, W. , Singh, R.P. , Huerta‐Espino, J. , Mcfadden, H. , Bossolini, E. et al. (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science, 323, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Lihua, C. , Shichang, X. , Ruiming, L. , Taiguo, L. and Wanquan, C. (2008) Early molecular diagnosis and detection of Puccinia striiformis f. sp. tritici in China. Lett. Appl. Microbiol. 46, 501–506. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Hussain, M. , Dai, J. , Li, Y. , Zhang, L. , Yang, J. , Ali, Z. et al. (2022) Programmable biosensors based on RNA‐guided CRISPR/Cas endonuclease. Biol. Proced. Online, 24, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, A.J. , Chao, S. , Asoro, F.G. , Heffner, E.L. , Hayashi, T. , Iwata, H. , Smith, K.P. et al. (2011) Chapter two – genomic selection in plant breeding: knowledge and prospects. In Advances in Agronomy ( Sparks, D.L. , ed). Delaware: Academic Press. [Google Scholar]
- Mahas, A. , Hassan, N. , Aman, R. , Marsic, T. , Wang, Q. , Ali, Z. and Mahfouz, M.M. (2021) Lamp‐coupled CRISPR–Cas12a module for rapid and sensitive detection of plant DNA viruses. Viruses, 13, 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsic, T. , Ali, Z. , Tehseen, M. , Mahas, A. , Hamdan, S. and Mahfouz, M. (2021) Vigilant: an engineered VirD2‐Cas9 complex for lateral flow assay‐based detection of SARS‐CoV2. Nano Lett. 21, 3596–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins, P.M.M. , Merfa, M.V. , Takita, M.A. and De Souza, A.A. (2018) Persistence in phytopathogenic bacteria: do we know enough? Front. Microbiol. 9, 1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne, R.J. , Dibley, K.E. , Schnippenkoetter, W. , Mascher, M. , Lui, A.C.W. , Wang, L. , Lo, C. et al. (2018) The wheat Lr67 gene from the sugar transport protein 13 family confers multipathogen resistance in Barley. Plant Physiol. 179, 1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J.W. , Herrera‐Foessel, S. , Lan, C. , Schnippenkoetter, W. , Ayliffe, M. , Huerta‐Espino, J. , Lillemo, M. et al. (2015) A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. [DOI] [PubMed] [Google Scholar]
- Nazarov, P.A. , Baleev, D.N. , Ivanova, M.I. , Sokolova, L.M. and Karakozova, M.V. (2020) Infectious plant diseases: etiology, current status, problems and prospects in plant protection. Acta Nat. 12, 46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. , Li, S.J. , Yan, J. , Tang, Y. , Cheng, J.P. , Gao, A.J. , Yao, X. et al. (2021) Research progress on phytopathogenic fungi and their role as biocontrol agents. Front. Microbiol. 12, 670135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powderly, W.G. (2019) How infection shaped history: lessons from the Irish famine. Trans. Am. Clin. Climatol. Assoc. 130, 127–135. [PMC free article] [PubMed] [Google Scholar]
- Rao, G.S. , Jiang, W. and Mahfouz, M. (2021) Synthetic directed evolution in plants: unlocking trait engineering and improvement. Synth. Biol. 6, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristaino, J.B. , Anderson, P.K. , Bebber, D.P. , Brauman, K.A. , Cunniffe, N.J. , Fedoroff, N.V. , Finegold, C. et al. (2021) The persistent threat of emerging plant disease pandemics to global food security. Proc. Natl. Acad. Sci. USA, 118, e2022239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisu, I.B. , Shahid, A.A. , Yaqoob, A. , Ali, Q. , Bajwa, K.S. , Rao, A.Q. and Husnain, T. (2017) Molecular approaches for high throughput detection and quantification of genetically modified crops: a review. Front. Plant Sci. 8, 1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souframanien, J. and Gopalakrishna, T. (2004) A comparative analysis of genetic diversity in blackgram genotypes using RAPD and ISSR markers. Theor. Appl. Genet. 109, 1687–1693. [DOI] [PubMed] [Google Scholar]
- Spielmeyer, W. , Mago, R. , Wellings, C. and Ayliffe, M. (2013a) Lr67 and Lr34 rust resistance genes have much in common – they confer broad spectrum resistance to multiple pathogens in wheat. BMC Plant Biol. 13, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmeyer, W. , Mago, R. , Wellings, C. and Ayliffe, M. (2013b) Lr67 and Lr34rust resistance genes have much in common – they confer broad spectrum resistance to multiple pathogens in wheat. BMC Plant Biol. 13, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg, S.H. , Redding, S. , Jinek, M. , Greene, E.C. and Doudna, J.A. (2014) DNA interrogation by the CRISPR RNA‐guided endonuclease Cas9. Nature, 507, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull, C. , Lillemo, M. and Hvoslef‐Eide, T.A.K. (2021) Global regulation of genetically modified crops amid the gene edited crop boom – a review. Front. Plant Sci. 12, 630396. 10.3389/fpls.2021.630396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ul Haq, I. and Ijaz, S. (2020) History and Recent Trends in Plant Disease Control: An Overview. Faisalabad, Pakistan: Springer International Publishing. [Google Scholar]
- Villamizar‐Gallardo, R. , Osma, J.F. and Ortíz‐Rodriguez, O.O. (2019) Regional evaluation of fungal pathogen incidence in Colombian cocoa crops. Agriculture, 9, 44. [Google Scholar]
- Wang, M. , Zhang, R. and Li, J. (2020) CRISPR/cas systems redefine nucleic acid detection: principles and methods. Biosens. Bioelectron. 165, 112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Wang, X.J. , Chen, C.Q. , Huang, L.L. and Kang, Z.S. (2007) A PCR‐based assay for detection of Puccinia striiformis f. sp. tritici in wheat. Plant Dis. 91, 1669–1674. [DOI] [PubMed] [Google Scholar]
- Zheng, C. , Wang, K. , Zheng, W. , Cheng, Y. , Li, T. , Cao, B. , Jin, Q. et al. (2021) Rapid developments in lateral flow immunoassay for nucleic acid detection. Analyst, 146, 1514–1528. [DOI] [PubMed] [Google Scholar]
- Zhu, H. , Zhang, H. , Xu, Y. , Laššáková, S. , Korabečná, M. and NeuŽil, P. (2020) PCR past, present and future. Biotechniques, 69, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Y. , Mason, M.G. and Botella, J.R. (2020) Evaluation and improvement of isothermal amplification methods for point‐of‐need plant disease diagnostics. PLoS One, 15, e0235216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Alignment of Lr34 Sanger sequencing.
Figure S2 Detection of Lr67 fungus‐resistant allele in wheat.
Figure S3 Bio‐SCAN‐based Lr67‐res detection in wheat.
Table S1 Primers used in this study.
Table S2 The g‐blocks used in this study.


